Abstract
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) is a potent lung carcinogen present in tobacco products and exposure to it is likely one of the factors contributing to the development of lung cancer in cigarette smokers. To exert its carcinogenic effects, NNK must be metabolically activated into highly reactive species generating a wide spectrum of DNA damage. We have identified a new class of DNA adducts, DNA-RNA crosslinks found for the first time in NNK-treated mice lung DNA using our improved high-resolution accurate mass segmented full scan data-dependent neutral loss MS3 screening strategy. The levels of these DNA-RNA crosslinks were found to be significantly higher in NNK-treated mice compared to the corresponding controls, which is consistent with higher levels of formaldehyde due to NNK metabolism as compared to endogenous levels. We hypothesize that this DNA-RNA crosslinking occurs through reaction with NNK-generated formaldehyde, and speculate that this phenomenon has broad implications for NNK-induced carcinogenesis. The structures of these crosslinks were characterized using high-resolution LC/MS2 and -MS3 accurate mass spectra analysis and comparison to a newly synthetized standard. Taken together, our data demonstrate a previously unknown link between DNA-RNA crosslink adducts and NNK and provide a unique opportunity to further investigate how these novel NNK-derived DNA-RNA crosslinks contribute to carcinogenesis in the future.
Keywords: DNA-RNA crosslinks, DNA:RNA hybrids, R loops, genome instability, DNA damage, DNA adducts, mass spectrometry, DNA adductomics, tobacco-induced carcinogenesis, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), lung cancer, metabolic activation, formaldehyde
Graphical Abstract

Introduction
The tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), is a potent lung carcinogen present at high levels in tobacco products.1–3 This compound is classified as a Group 1 human carcinogen by the International Agency for Research on Cancer (IARC) and is believed to play a key role in tobacco-induced lung cancer in humans. To exert its carcinogenic effects, NNK must be metabolically activated into highly reactive species capable of interacting and modifying cellular biomolecules such as DNA, RNA, and proteins.1, 4–7 The modifications occurring in DNA (DNA adducts) can lead to increased genomic instability which, if not repaired can ultimately result into cancer development. Thus, identifying NNK-induced DNA modifications that are critical drivers of malignant transformation in chemical-induced carcinogenesis, enables a better understanding of the mechanisms underlying cancer initiation.
Metabolic activation of NNK leads to DNA damage and impacts important cellular functions. There are two well understood pathways involved in DNA adduct formation in NNK-induced lung carcinogenesis in laboratory animals: the methylating and pyridyloxobutylating pathways. The methylating pathway produces methyl DNA adducts in vivo which includes O6-methyl-deoxyguanosine (O6-methyl-dG), N7-methyl-guanine, and O2-methyl-deoxythymidine (O2-methyl-dT), 8, 9 while the pyridyloxobutylating pathway yields pyridyloxobutylated DNA adducts including O6-POB-dG and O2-POB-dT.10, 11 Recently, methyl phosphate adducts and pyridyloxobutyl phosphate DNA adducts were also identified and quantified in rats and lung DNA of smokers.12, 13
The mechanisms of NNK-induced lung tumorigenesis as demonstrated in various animal models, involves both direct and indirect genotoxic mechanisms.3, 8, 10, 14–21 In A/J mice, a strain susceptible to developing spontaneous lung tumors, this process is enhanced by chemical carcinogens.20 Specifically, the mechanism of NNK-induced lung carcinogenesis is believed to involve a direct genotoxic mechanism via the formation of O6-methyl-dG.14–16, 19–22 This DNA adduct is highly mutagenic causing G to A transitions and G to T transversions in codon 12 of the K-ras oncogene.8, 14, 16, 20, 21 In contrast, in animals that are less susceptible to the formation of spontaneous tumors, for instance in rats, the induction of pulmonary tumors by NNK does not solely appear to involve the ras pathway.19–21 The methylated DNA adducts alone do not account for the high tumorigenicity of NNK in rat lung and nasal mucosa,23 and the numerous additional DNA adducts resulting from the reaction of NNK metabolites with DNA may play a key role in these processes. In addition, NNK can also illicit indirect genotoxic effects such as increased oxidative stress, which lead to lipid peroxidation resulting in the production of reactive species and additional DNA adduct formation.24 This broad spectrum of DNA damage by NNK and its prevalence in tobacco is likely one of the many factors contributing to the development of lung cancer in cigarette smokers.
NNK bioactivation also yields two aldehydes, formaldehyde and 4-oxo-4-(3-pyridyl)-butanal (OPB).2, 25 Formaldehyde-induced DNA adducts have been previously identified and characterized.10, 14, 18, 26–36 Although these aldehydes interact directly to form covalent modifications with DNA bases, it is hypothesized that formaldehyde and OPB also interfere with DNA adduct repair and increase the persistence and genotoxicity of methylating species.37 In addition, these aldehydes induce DNA single strand breaks (SSBs) and the frequency of DNA SSBs induced by NNK, formaldehyde, or OPB has been shown to be higher in GSH-depleted hepatocytes.37 Formaldehyde also generates detectable levels of both DNA SSBs and DNA-protein crosslinks as well as much higher frequency of genetic damage compared to other aldehydes such as acrolein.29, 30, 38–40 Formaldehyde is formed through various cellular processes and ubiquitous in the environment and it has been shown to generate a wide array of DNA adducts including DNA-protein and DNA-DNA crosslinks. 29–36 Using our comprehensive and improved segmented full scan high-resolution accurate mass (HR/AM) data-dependent MS3 neutral loss (NL) screening strategy, we were able to detect and identify, for the first time, a new class of DNA adducts, namely DNA-RNA crosslinks, which were induced by formaldehyde generated in situ upon metabolic activation of NNK. We hypothesize that they are generated through the interaction of formaldehyde with naturally occurring DNA:RNA hybrids formed during replication and/or transcription in cells.41, 42 The covalent and unscheduled formaldehyde-induced crosslinking of DNA and RNA may be a source of genomic instability and could be one of the broad mechanisms of lung carcinogenicity of NNK.
Experimental
Caution: NNK is carcinogenic and should be handled with proper care, personal protective equipment, and ventilation.
Materials.
4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK; 98%) was synthesized as previously described.6 Recombinant deoxyribonuclease I (DNase I) expressed from Pichia pastoris, phosphodiesterase-1 (PDE-1) from Crotalus adamanteus, recombinant alkaline phosphatase from Pichia pastoris, and calf thymus DNA (CT DNA) were purchased from Roche (St. Louis, MO, USA). DNA nucleoside standards (dC, dG, dT, dA) and the RNA nucleoside standard, guanosine, were obtained from Sigma Aldrich (Milwaukee, WI). Formaldehyde (37%) was obtained from Fisher Scientific (Pittsburgh, PA). DMSO-d6 for NMR analysis was purchased from Cambridge Isotopes Inc (Tewksbury, MA). Mouse anti-ds DNA antibody and secondary antibody: Goat Anti-Mouse IgG H&L (HRP) were obtained from Abcam (Cambridge, MA). RNase H was obtained from New England BioLabs (Ipswich, MA). Positively charged nylon transfer membrane (Hybond-N+, Amersham) was purchased from Cytiva (Marlborough, MA). Bio-Rad Clarity™ Western ECL substrate and milk for the membrane blocking step in the immunodetection of DNA:RNA hybrids were obtained from Bio-Rad (Hercules, CA). [15N5] N2-Ethyl-deoxyguanosine ([15N5] N2-ethyl-dG), [15N5] (6R/S)-3-(2′-deoxyribos-1′-yl)-5,6,7,8-tetrahydro-6-pyrimido[1,2-a]-purine-10(3H)one ([15N5] R/S-pro-dG), [D4] O6-[4-(3-pyridyl)-4-oxobut-1-yl]-2′-deoxyguanosine ([D4] O6-POB-dG), O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dT), [D3] O6-methyl-deoxyguanosine ([D3] O6-methyl-dG), O6-methyl-deoxyguanosine (O6-methyl-dG), [D4] N7-PHB-guanine, [15N5]-alpha-hydroxy-Acro-dG, [13C10-15N5] γ-hydroxy-Acro-dG, N6-etheno-dG, 4-HNE-dG, 4-hydroxy-butyl-dG, 8-oxo-dG, O6-ethyl-guanine, [15N5] N6-methyl-deoxyadenosine ([15N5] N6-methyl-dA) and di-(N6-deoxyadenosyl)-methane (dAdo-CH2-dAdo) were synthesized and obtained as previously described.43, 44 All acids and organic solvents were MS grade.
Animal experiment.
Female A/J mice, 6 weeks of age, were purchased from the Jackson Laboratory (Bar Harbor, ME). The mice were housed in pathogen-free animal quarters upon arrival at the Research Animal Resources, University of Minnesota Academic Health Center. The animal experiment was approved by the Institutional Animal Care and Use Committee of the University of Minnesota (Protocol ID: 1602-33469A). Mice were treated with NNK (100 mg/kg) via intraperitoneal injection (IP).45 Control mice were treated with the vehicle, phosphate buffered saline (PBS). The mice (5 mice each time point per group) were euthanized by an overdose of carbon dioxide at different time points (6 h, 1 d, 2 d, 4d, 8 d, and 16 d) after NNK administration (Figure S1). Subsequently, lung tissues were harvested and stored at −80°C until analysis.
DNA isolation.
Lung DNA samples were isolated from mice treated with NNK (n=6) and controls (n=6). The 6 mice from each group correspond to the different time points after treatment (6 time points; 6 h, 1 d, 2 d, 4 d, 8 d, 16 d). The DNA extraction was performed following a protocol previously described.46 Approximately 100 mg - 1000 mg frozen lung tissue was minced and homogenized in 6 mL lysis buffer using the Tissue-Ruptor (Qiagen, Valencia, CA). After which, 30 μL of Proteinase K was added and incubated overnight at RT in a mechanical shaker. The next day, 30 μL RNase A was added and incubated for 2 h at RT. Two mL protein precipitation solution was added and vortexed vigorously for 20-30 s. The homogenate was allowed to sit for 15 min and centrifuged (highest possible speed) at 4225 x g and 4°C for 20 min. The supernatant was poured into 8 mL cold isopropanol (IPA). The mixture was mixed by inverting gently until the DNA strands were visible. The DNA was fished out from solution using a Pasteur pipet and transferred into clean tubes containing 1 mL Tris-EDTA (10 mM Tris, 1 mM EDTA) solution to dissolve the DNA. One mL of chloroform/isoamyl alcohol (24:1) was added and vortexed for 1 min. The mixture was centrifuged at 12,000 rpm for 15 min to separate the layers. The upper layer (containing the DNA) was carefully pipetted out and transferred to new Eppendorf tubes. One hundred microliters (100 μL) of 5 M NaCl was added and the mixture was vortexed vigorously. The DNA was precipitated by adding 1 mL cold IPA. The DNA was pelleted by centrifugation at 12,000 rpm for 3 min and washed with 70% IPA and finally with 100% IPA. The DNA was air-dried and stored at −80°C until further processing. The DNA concentration and purity was estimated using a nanodrop spectrophotometer. If necessary, additional RNase digestion was performed to eliminate contaminating RNA.
Enzymatic digestion of DNA to nucleosides and offline automated HPLC-fraction collection.
Mice lung DNA (100 μg) dissolved in 400 microliters of 10 mM Tris·HCl, 5 mM MgCl2, pH = 7.5, was hydrolyzed enzymatically to nucleosides using recombinant DNase I (25 U), phosphodiesterase I (0.002 U), and recombinant alkaline phosphatase (20 U) as previously described.46 Labeled synthetic standards were added at 10 fmol μL−1 including [15N5] N2-ethyl-dG, [15N5] R/S-pro-dG, [D4] O6-POB-dG, [D3] O6-methyl-dG, [D4] N7-PHB-guanine, [15N5] α-hydroxy-Acro-dG, [13C10, 15N5] γ-hydroxy-Acro-dG, and [15N5] N6-methyl-dA. After digestion, the samples were filtered to remove the enzymes and 10 μL of the sample was used for dG quantitation using HPLC-UV. The rest of the hydrolysate was enriched using automated offline HPLC fraction collection to remove abundant unmodified deoxyribonucleosides (dC, dG, dT, and dA), which could potentially interfere in the LC-MS analysis. For the HPLC fraction collection, mobile phase A consisted of water and mobile phase B was methanol. Reversed-phase separation of the hydrolysate was performed using a Luna C18 (5 μm, 100 Å, 250 mm x 4.6 mm) (Phenomenex, Torrance, CA) column at a temperature of 45°C and flow rate of 800 μL min−1. Gradient elution was started with 2% B, followed by a linear gradient to 30% B for 30 min, and to 98% B for 15 min. Mobile phase B composition was held constant at 98% B for 10 min and the column was re-equilibrated to 2% B for another 10 min. Total run time was 50 min. Fractions that did not contain the four DNA bases were pooled, dried, and resuspended in 20 μL water for LC-MS analysis.
LC-MS conditions.
All LC and MS analyses of purified samples were performed on a Thermo Orbitrap Fusion mass spectrometer coupled to a Dionex RSLCnano HPLC with a Nanospray Flex ion source (Thermo Scientific, Waltham, MA). A capillary column (200 mm x 75 μm, New Objective, Woburn, MA) was custom-packed with reversed-phase Luna C18 (5 μm, 120 Å, Thermo Scientific, Waltham, MA) and separation was performed at room temperature with a flow rate of 0.3 μL/min using 0.05% formic acid as mobile phase A and acetonitrile as mobile phase B. Chromatographic separation was performed using a stepwise gradient elution starting with 2% B for the first 6 min at a flow rate of 0.9 μL/min. The flow rate was reduced to 0.3 μL/min with the injection valve position switching at 6 min into the run to take the 5 μL injection loop out of the flow path. A linear gradient from 2% to 20% B over 24 min was employed, followed by an increase to 60% B over 10 min. The gradient was then increased to 98% B over 2 min and held constant for 3 min. Finally, the gradient was decreased from 95% to 5% B over 1 min and the flow rate was increased to 0.9 μL/min. The column was re-equilibrated at 5% B with a 0.9 μL/min flow rate for another 5 min before the next injection. The total run time was 55 min. The electrospray voltage was set to 2.2 kV with a source temperature of 350°C. The ion focusing and transfer elements were optimized for maximum signal intensity, which resulted in an S-Lens RF setting of 60%. In addition, the mass accuracy (< 5 ppm) of the signal of an abundant background ion, decamethylcyclopentasiloxane (m/z 371.1012), was evaluated before analysis. Instrument sensitivity and mass accuracy were checked using a mixture of unlabeled and labeled synthetic DNA adduct standards before all LC-MS analyses.
Screening DNA adducts using HR/AM segmented full scan (SFS) data acquisition.
The screening for covalent modifications of DNA (DNA adducts) was performed on a set of samples comprising of the control and NNK-treated mice at different time points (6 h, 1 d, 2 d, 4 d, 8 d, 16 d) after exposure using high-resolution segmented full scan data-dependent MS3 neutral loss screening approach. This method involves monitoring of the neutral loss of deoxyribose (dR) (116.0474 Da) and the four DNA bases (C, 111.0433 Da; G, 151.0494 Da; T, 126.0429 Da; A, 135.0545 Da) in the MS2 spectra with MS3 fragmentation upon observation of one of the neutral losses. This is performed with repeated full scan detection of three narrow and overlapping mass ranges (m/z 150-300, m/z 295-500, m/z 495-750) to increase the DNA adduct detection sensitivity, as compared to a single m/z 150-750 scan range (Figure S2). For the MS1 full scans, the Orbitrap mass analyzer resolution was 120000 (at m/z 240) with automatic gain control (AGC) setting of 1 × 106, and a maximum injection time of 100 ms. MS2 CID fragmentation was performed in the ion trap using the “Top Speed” setting of 2 s. The MS2 fragment ions were then detected at an Orbitrap resolution of 15000 and AGC target of 2 × 105. MS2 parameters include isolation width of 2.0 m/z units, normalized collision energy of 30%, activation Q of 0.25, and activation time of 10 ms. MS2 fragment ions resulting from the characteristic loss of dR or one of the four DNA bases (± 5 ppm) undergo MS3 fragmentation in the ion trap and detection in the Orbitrap at a resolution of 15000. MS3 scan event parameters include isolation width of 3 m/z units, normalized collision energy of 30%, activation Q of 0.25, and activation time of 10 ms. An in-house developed MZmine 2 data analysis module, DFBuilder,47, 48,49 was used for processing mass spectral data from the neutral loss screening method to generate a list of all potential adducts detected in the samples. Briefly, input raw data files were processed to identify all MS2 spectra exhibiting one of the diagnostic neutral losses, and the corresponding precursor ion masses were used to build targeted chromatograms that were deconvoluted into individual compound peaks. A suitable retention time tolerance was used to account for acquired MS2 spectra triggered off the apex of the chromatographic peak to ensure the entirety of the compound peak was retained without capturing neighboring peaks. Additional parameterization was utilized to further curate the search for diagnostic patterns and limit the detection of false positives. An optional exclusion list was included to remove previously defined signals or contaminants detected in blank injections. To mitigate false positive detection, an appropriate signal threshold was used to limit the interference of noise in MS2 screening. Following diagnostic ion screening, the resultant feature list was further processed to remove duplicate hits, group in-source ions and compound complexes, and align peaks between multiple raw data files before data interrogation and interpretation.
Proposed identification of candidate DNA adducts was performed by comparing the high-resolution accurate masses of the MS1 and MSn mass spectral data with internal standards when possible, with an in-house library of endogenous DNA adducts, data available from the literature, and through manual structural evaluation of the MS2 and MS3 mass spectral fragmentation data.
In vitro incubation of DNA and RNA with formaldehyde and characterization of the DNA-RNA crosslink N2-guanosyl–N6-deoxyadenosyl)methane (guanosine-CH2-dA)
To determine whether DNA forms crosslinks with RNA in the presence of formaldehyde in vitro, guanosine (RNA base, 0.5 mM) was incubated with either the single DNA base deoxyadenosine (dA, 0.5 mM) or the mixture of the four DNA bases (dC, dG, dT, dA, 0.5 mM each) with and without formaldehyde (120 mM) (Figure S3). Formaldehyde was added in excess to ensure detection of DNA-RNA crosslinks and the solution was incubated at 37°C for 24 h. The resulting mixture was analyzed by HPLC-UV and targeted LC-MS2 as described below.
The reaction product from the reaction of guanosine and deoxyadenosine in the presence of formaldehyde and attributed to guanosine-CH2-dA, was purified by reverse phase HPLC with a Waters Associates (Milford, MA) system equipped with a Shimadzu SPD-10A 0.2 mm Prep UV-Vis detector set to 268 nm. Separation was performed using a Luna 5 μm C18(2) 100 Å 250 × 10 mm column purchased from Phenomenex (Torrance, CA). A 38 min method was developed, with a constant flow rate of 4 mL/min and a mobile phase gradient starting at 5% MeOH in H2O and ramping up to 30% MeOH in H2O over 18 min. The gradient was then increased to 90% MeOH in H2O over 6 min. Finally, the gradient was increased to 98% MeOH in H2O over 3 min. The instrument was then equilibrated for 8 min at 5% MeOH in H2O before the next injection. The product was collected at a retention time of 23.5 min as a white solid after evaporation of solvents (3.33% yield from 14 mg guanosine).
For additional purification the product was redissolved in H2O, and injected on the HPLC. A 35 min method was developed with a constant flow rate of 4 mL/min starting at 5% MeOH in H2O and increasing to 35% MeOH in H2O over 10 min. The mobile phase was held constant at 35% MeOH in H2O over 15 min, and then returned to 5% MeOH in H2O and allowed to equilibrate before the next injection. The product was collected at a retention time of 16.4 min as a white solid after evaporation of solvents.
Guanosine-CH2-dA was recorded and interpreted on a Bruker 700 MHz spectrometer equipped with a triple resonance cryoprobe dissolving the analyte in DMSO-d6. Chemical shifts are reported as parts per million (ppm). Residual solvent reference peaks were for DMSO-d6 1H NMR (2.50 ppm DMSO-d6, 3.33 ppm residual H2O) and 13C NMR (39.52 ppm). Peak splitting uses the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, ddd = doublet of doublet of doublets), and m = multiplet. 1H NMR (700 MHz, DMSO-d6) δ 10.96 (s, 1H, gua-N1-H), 8.60-8.23 (m, 3H, ade-N6-H, ade-C8-H, ade-C2-H), 7.98 (s, 1H, gua-C8-H), 7.21 (s, 1H, gua-N2-H), 6.38 (s, 1H, dR-1’-H), 5.83 (s, 1H, R-1’-H), 5.46 (d, J = 6.4 Hz, 1H, R-2’-OH), 5.39-5.35 (m, 1H, dR-3’-OH), 5.23 (m, 1H, R-3’-OH), 5.19 (s, 1H, dR-5’-OH), 5.11 (s, 1H, crosslink-CH2), 5.09 (m, 1H, R-5’-OH), 5.01 (s, 1H, crosslink-CH2), 4.53-4.47 (m, 1H, R-2’-H), 4.43 (s, 1H, dR-3’-H), 4.16 (q, J = 4.6 Hz, 1H, R-3’-H), 3.94 (q, J = 3.7 Hz, 1H, R-4’-H), 3.90 (s, 1H, dR-4’-H), 3.66 and 3.58 (m, 2H, R-5’-Ha and R-5’-Hb), 3.63 and 3.53 (m, 2H, dR-5’-Ha and dR-5’-Hb), 2.77-2.69 (m, 1H, dR-2’-Ha), 2.33-2.25 (m, 1H, dR-2’-Hb); 13C NMR (176 MHz, DMSO-d6) δ 157.1 (gua-C=O), 154.5 (ade-C6), 152.6 (ade-C2), 152.5 (gua-C2), 150.9 (gua-C4), 149.2 (ade-C4), 140.7 (ade-C8), 136.8 (gua-C8), 120.1 (ade-C5), 117.7 (gua-C5), 88.4 (dR-C4), 87.4 (R-C1), 85.7 (R-C4), 84.4 (dR-C1), 74.3 (R-C2), 71.3 (dR-C3), 70.8 (R-C3), 62.7 (dR-C5), 61.8 (R-C5), 47.5 (crosslink CH2), 39.7. Structure assignments were confirmed by comparison with COSY, HSQC, HMBC and TOCSY-HSQC experiments, all the NMR spectra can be found in Figure S13–S19. High-resolution mass spectrometry analysis was performed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, Waltham, MA, USA) Guanosine-CH2-dA: [M + H]+ calc’d 547.2008; found 547.2013.
Targeted LC-MS2 of DNA-RNA crosslinks.
To identify and structurally characterize anticipated DNA-RNA crosslinks and other formaldehyde-induced DNA adducts, we performed targeted LC-MS2 of samples from the in vitro incubation of DNA and RNA bases with and without formaldehyde. We then compared the MS2 fragmentation data of the DNA-RNA crosslinks detected in the in vitro incubation with that of the crosslinks identified in mice lung DNA from NNK treated animals (6 h after treatment). A list of target masses and their identities is listed in Table S1. LC conditions and the full scan and MS2 data acquisition parameters were the same as those used for the data-dependent MS3 neutral loss screening method. The targeted LC-MS2 was performed on samples from the control (n=3) and NNK-treated (n=3) mice at 6 h time point.
Immunodetection of DNA:RNA hybrids by dot blot.
For the dot blot analysis, the lung DNA isolated from NNK-treated mice (6 h time point after treatment) and the corresponding mice lung DNA from the control group (6 h time point) was used. DNA:RNA hybrids were detected in these samples using the DNA:RNA hybrid-specific S9.6 antibody. The S9.6 antibody was expressed and purified as previously described.50 The DNA concentration was estimated using a nanodrop spectrophotometer and 1000 ng of DNA was used for the dot blot analysis. Two sets of samples (without and with RNaseH) comprised of lung DNA from NNK-treated mice NNK and control mice were incubated for 1 h at 37°C. The RNaseH treatment was used to confirm the specificity of S9.6 antibody. Samples were blotted in triplicates on two sets of positively charged nylon transfer membranes (Amersham). One set of membranes was used for S9.6 antibody detection and the other set for anti-dsDNA antibody detection. The dot blots were dried and the membranes crosslinked using UV (120 mJ/cm2) at 254 nm for 10 min. The membranes were blocked with 5% milk in TBST buffer (1xTBS with 0.1% Tween-20) for 1 h at RT. The membranes were then incubated with either anti-dsDNA antibody (1:15,000) or anti-S9.6 antibody in blocking solution overnight at 4°C. The membranes were washed with TBST buffer (3-4 times, 10 min washes). After the incubations in dsDNA and S9.6 antibodies, the blots were incubated with HRP (horseradish peroxidase)-conjugated secondary antibody in TBST buffer (1:15,000) for 2 h at RT. Finally, the membranes were incubated with HRP substrate for 5 min at RT. The blots were visualized for dsDNA and S9.6 signals using Li-Cor (LI-COR Biosciences, Lincoln, NE) and images were analyzed using Image One software (LI-COR Biosciences). The amounts of DNA used from different samples for S9.6 dot blots were normalized based on the dsDNA signals.
Statistical analysis.
Comparisons between two groups (NNK and control) in the LC-MS2 and immunodetection experiments were performed using a two-tailed Student’s t-test (Microsoft Excel) with p < 0.05 being considered significant.
Results
Detection and characterization of novel formaldehyde-induced DNA-RNA crosslinks.
Using our comprehensive and improved segmented full scan HR/AM data-dependent MS3 neutral loss screening strategy, we have detected more than six hundred analytes corresponding to potential DNA adducts in mice lung DNA. These putative adducts exhibited in their fragmentation one or more of the diagnostic neutral losses (loss of deoxyribose, loss of ribose, loss of DNA bases such as guanine, adenine, thymine, and cytidine), had a minimum relative abundance of 1.0 x105, and corresponded to at least three independent observations in the experiment.
This list included previously described NNK-derived DNA adducts (methyl- and POB- derived DNA adducts) and a large number of adducts with m/z and fragmentation patterns, consistent with cross-links, due to the presence for example, of the methylene-DNA base (e.g. m/z 164.0567 for methylene-guanine or m/z 148.0618 for methylene-adenine. These typical fragments were identified upon manual scrutiny of the spectra and lead to the identification of a list of putative formaldehyde-induced DNA-RNA crosslinks. Figure 1A shows the extracted ion chromatogram (EIC) of putative DNA-RNA crosslinks at m/z 547.2012. Three chromatographic peaks at tR: 21.2 min, 22.4 min, and 22.8 min were observed upon extraction of the HR/AM (< 5 ppm mass tolerance) of the putative crosslink from the full scan MS data. The first two peaks exhibited MS2 and MS3 mass spectral data suggesting potential isobaric DNA adducts (Figure 1A, I–III).
Fig. 1.
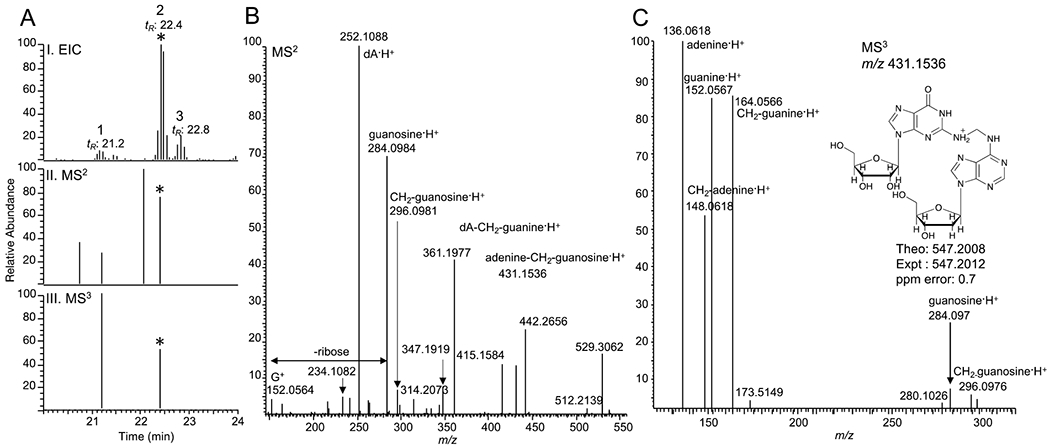
Putative DNA-RNA crosslink detected in NNK-treated mice lung DNA. (A.I) Extracted ion chromatogram, (A.II) MS2, and (A.III) MS3 triggered scan events for m/z 547.2012 at 22.4 min (The corresponding events are marked with the asterisk *). (B) MS2 spectrum of m/z 547.2012 at 22.4 min. and (C) MS3 spectrum of m/z 431.1536 (loss of deoxyribose, dR) from m/z 547.2012 at 22.4 min.
The MS2 spectrum at tR: 22.4 min exhibited the characteristic fragment ion of m/z 431.1536, which corresponds to the loss of deoxyribose (dR; 116.0474 Da) from the precursor ion, m/z 547.2012 (Figure 1B). The MS2 spectrum further showed characteristic fragment ions corresponding to dA·H+ (m/z 252.1088), guanosine·H+ (m/z 284.0984), and guanine·H+ (m/z 152.0564). In addition, specific fragment ions were observed in the MS2 spectrum, including m/z 415.1584, which corresponds to a loss of ribose (132.0422 Da) from the precursor ion and m/z 296.0981, which corresponds to methylene-guanosine·H+ (CH2-guanosine·H+). Moreover, the presence of m/z 431.1536 (loss of dR) triggered its MS3 fragmentation (Figure 1C). The MS3 spectrum showed characteristic fragment ions including adenine·H+, guanine·H+, and guanosine·H+. The associated methylene counterparts with m/z 148.0618 (CH2-adenine·H+), m/z 164.0566 (CH2-guanine·H+), and m/z 296.0976 (CH2-guanosine·H+) were also observed in the MS3 spectrum. The fragment ions from the MS2 and MS3 strongly support the structure of m/z 547.2012 as deoxyadenosine-CH2-guanosine·H+ (dA-CH2-guanosine·H+). Figure 2 shows the proposed structures of the observed high-resolution accurate masses of the precursor ion and fragment ions in the MS2 and MS3 spectra of dA-CH2-guanosine·H+.
Figure 2.
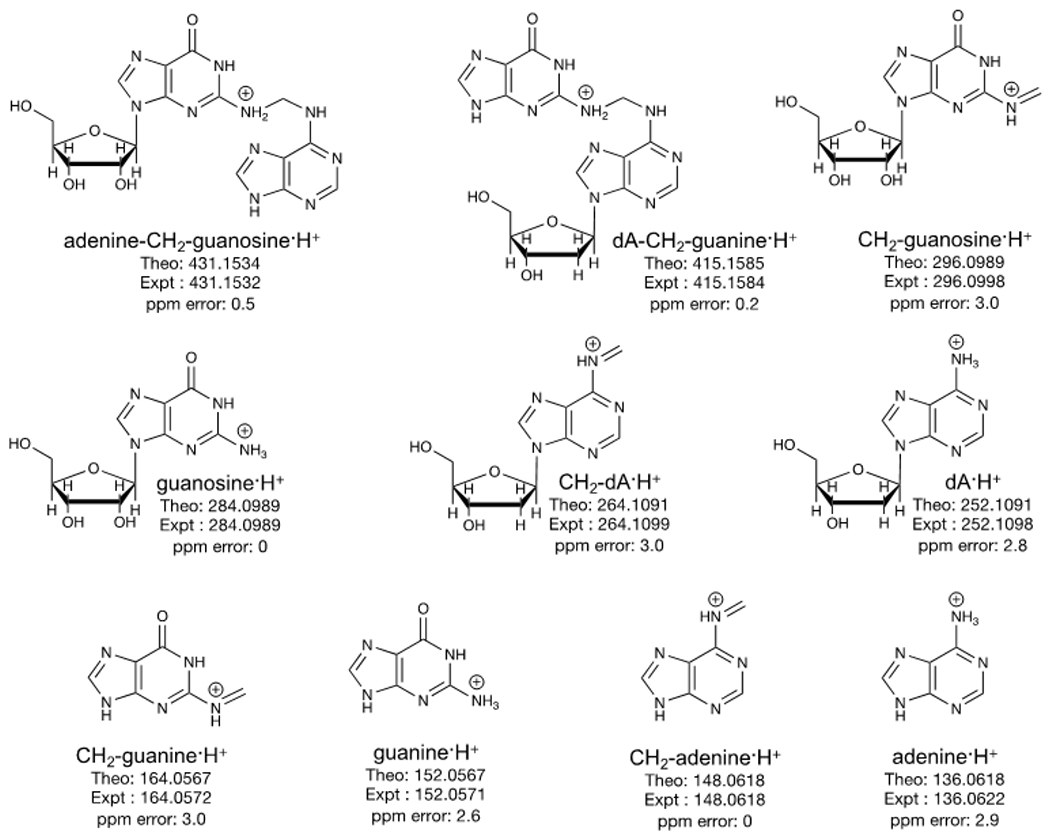
Proposed structures of the MS2 and MS3 fragment ions of the putative formaldehyde-induced DNA-RNA crosslink, guanosine-CH2-dA, detected in A/J mice lung DNA.
Likewise, the first peak at tR: 21.2 min, which is isobaric with dA-CH2-guanosine, exhibited MS2 and MS3 mass spectral data suggesting another DNA adduct. The MS2 spectrum exhibited fragment ions including losses of one dR (m/z 431.1536) and two dRs (m/z 315.1059), dG·H+ (m/z 268.1039), CH2-dG·H+ (m/z 280.1039), CH2-guanine·H+ (m/z 164.0567), and guanine·H+ (m/z 152.0566). In addition, the MS3 spectrum of m/z 431.1536 (loss of dR) showed fragment ions corresponding to guanine·H+ and CH2-guanine·H+ only. No adenine·H+ or CH2-adenine·H+ were observed in the MS3 spectrum (Figure 3C). The fragmentation pattern of this putative formaldehyde-induced DNA crosslink is consistent with the structure of deoxyguanosine-CH2-deoxyguanosine (dG-CH2-dG·H+, m/z 547.2008), which is isomeric with that of dA-CH2-guanosine·H+ (m/z 547.2008). The third peak at tR: 22.8 is an isobaric compound, however, no MS2 and MS3 spectra were observed and therefore no structure characterization is possible.
Fig. 3.
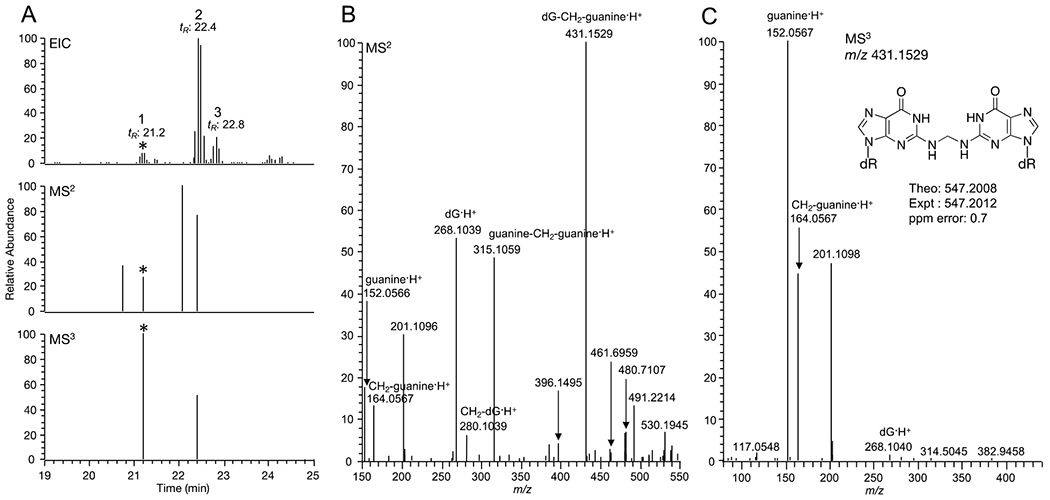
Putative DNA-DNA crosslink detected in NNK-treated mice lung DNA (A) Extracted ion chromatogram, MS2, and MS3 triggered scan events for dG-CH2-dG (The corresponding events are marked with the asterisk *) (B) MS2 spectrum of dG-CH2-dG (C) MS3 spectrum of dG-CH2-dG.
Formaldehyde induces DNA-RNA crosslinks in vitro.
To confirm the identity of the putative formaldehyde-induced DNA-RNA crosslink, dA-CH2-guanosine, we performed in vitro experiments using purified DNA and RNA bases with and without formaldehyde. First, we performed a simple incubation of the DNA base deoxyadenosine (dA) and the RNA base guanosine in equal amounts and added excess formaldehyde. Second, we performed incubation of the four DNA bases (dC, dT, dG, and dA) and guanosine with and without formaldehyde. The reaction mixtures were incubated overnight at 37 °C and the resulting mixtures were analyzed by HPLC-UV and targeted LC-MS2. Figure S4A shows the HPLC-UV chromatograms of dA and guanosine with and without formaldehyde. New chromatographic peaks were observed in the reaction mixture containing formaldehyde, and decreased levels of the corresponding unmodified bases (dA and guanosine) relative to that of the reaction mixture without formaldehyde were observed as well. Furthermore, new chromatographic peaks were also observed in the reaction of the four DNA bases with guanosine in the presence of formaldehyde. These new chromatographic peaks were not observed in the sample without formaldehyde (Figure S4B). To confirm the identities and structurally characterize the resulting new peaks and crosslink products in vitro, we performed LC-MS2 of samples containing dA and guanosine with and without formaldehyde, and targeted expected formaldehyde-induced DNA adducts and crosslinks, including the novel dA-CH2-guanosine crosslink. The target masses and their identities are listed in Table S1. Figure S5 shows extracted MS2 fragment ion chromatograms assigned to the putative DNA-RNA crosslink, dA-CH2-guanosine, with their presence in the sample with formaldehyde and absence in the sample without formaldehyde crosslink, confirming its formation and detection in vitro. In addition, the same profile was observed in the reaction of the four bases and guanosine in the presence of formaldehyde and absent in the sample without formaldehyde (Figure S6). Furthermore, the negative control containing only water with and without formaldehyde did not show any signal corresponding to putative DNA-RNA crosslinks (Figure S7). Figure 4A–B show the comparison of the MS2 fragmentation spectra of dA-CH2-guanosine detected in vitro (A) and that of the crosslink detected in mice lung DNA (B). The MS2 spectrum and extracted ion chromatograms of m/z 547.2012 detected in the in vitro incubation and then purified and thoroughly characterized via NMR (1H, 13C, HSQC, HMBC and TOCSY-HSQC, as shown in Figures S13–S18), matched with that of the m/z 547.2008 detected and characterized in mice lung DNA treated with NNK 6 h after administration, thus confirming the formation and identity of this novel DNA-RNA crosslink (Figures S5, S8 and S19).
Fig. 4.
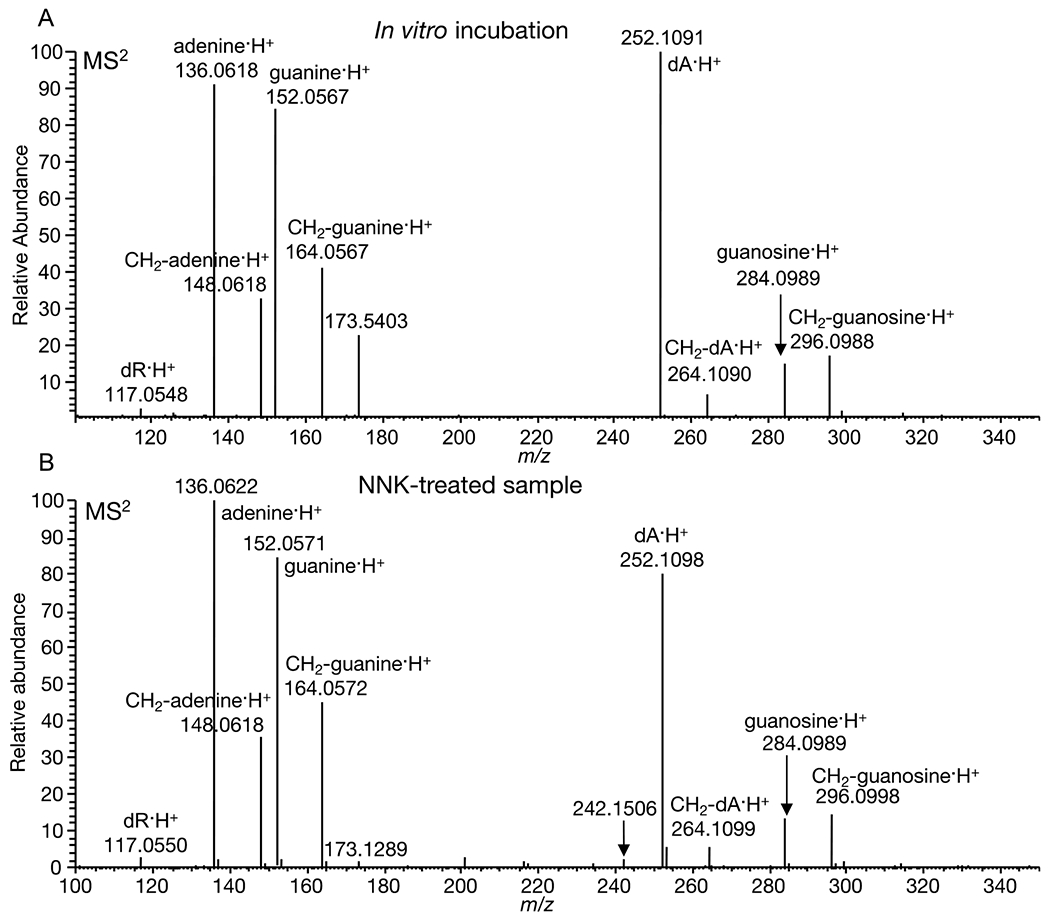
(A) MS2 fragmentation of guanosine-CH2-dA detected in vitro (B) MS2 fragmentation of guanosine-CH2-dA detected in NNK-treated mice lung DNA.
Additional formaldehyde-induced DNA-RNA crosslinks formed in vitro (Table S1) were identified. Most notably, the putative DNA-RNA crosslink with m/z 563.1957 (Figures S9–S10) was observed in mice lung DNA exposed to NNK. Figure 5 shows the MS2 fragment ion chromatograms and MS2 mass spectral data of m/z 563.1957 identified in mice lung DNA after NNK administration. Other crosslinks were observed in vivo and confirmed in vitro including dC-CH2-guanosine and dT-CH2-guanosine, however, the levels of these crosslinks were fairly low in vivo and were not triggered for MS3 during LC/MS analysis (Figures S11–S12). All the DNA adducts and crosslinks identified in vitro and in mice lung DNA, including DNA-DNA and DNA-RNA, are listed in Table S1.
Fig. 5.
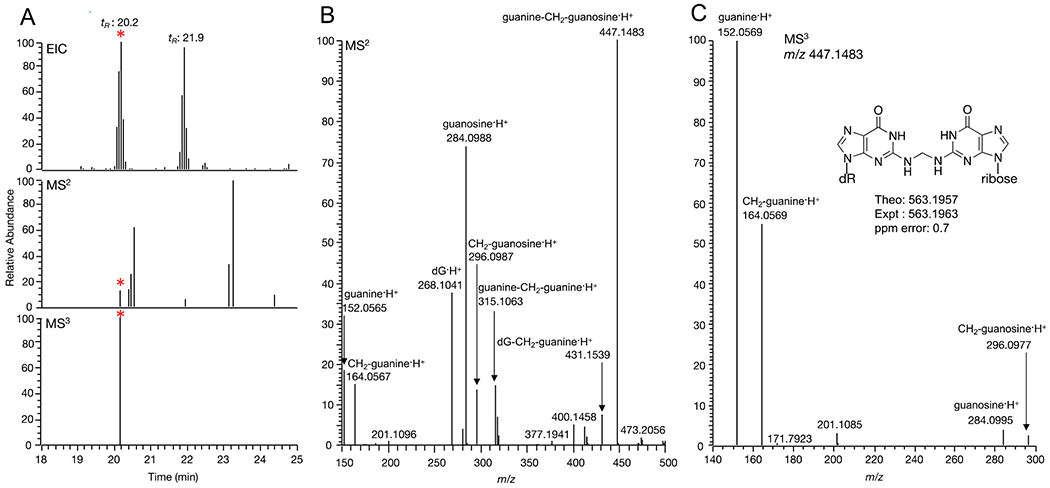
(A) Extracted ion chromatograms of guanosine-CH2-dG (*) in an NNK-treated mouse sample (B) MS2 spectrum of guanosine-CH2-dG (C) MS3 spectrum of guanosine-CH2-dG.
DNA-RNA crosslink adducts elevated in NNK-treated mice.
The levels of the previously unidentified dA-CH2-guanosine and dG-CH2-guanosine crosslinks were found to be elevated in the NNK-treated mice lung DNA as compared to the control group (Figure 6). The relative abundances of DNA-RNA crosslinks detected in vivo are consistent with that seen in vitro, with dA-CH2-guanosine being the most abundant crosslink followed by dG-CH2-guanosine, suggesting that the formation of them in vivo is determined by the relative reactivity of formaldehyde with the individual bases.
Fig. 6.
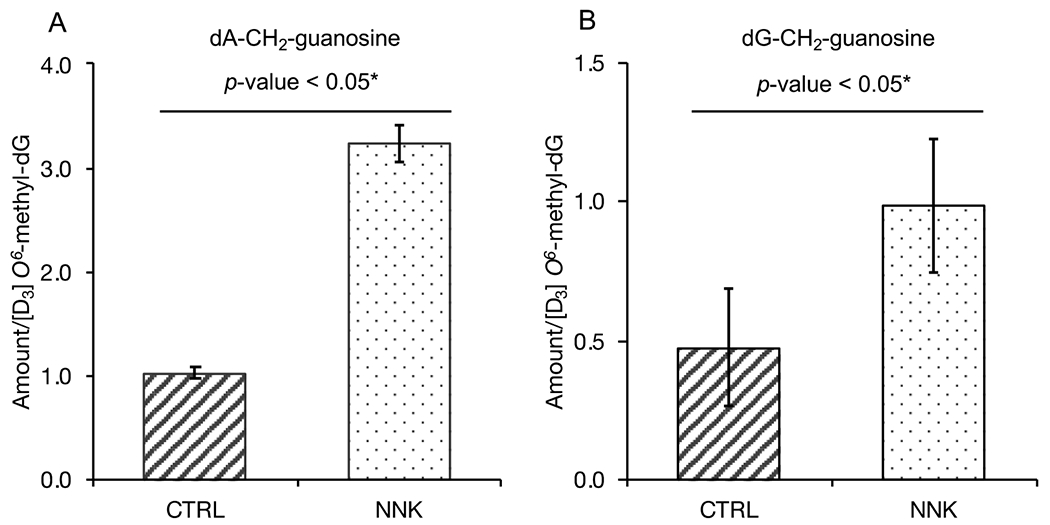
(A) Relative levels of guanosine-CH2-dA - and (B) guanosine-CH2-dG in NNK-treated mouse samples (6 h) compared to the corresponding control. Levels were normalized to [D3] O6-methyl-dG as internal standard. Error bars represent standard deviation (SD) of 3 technical replicate measurements.
DNA:RNA hybrids detected in mice lung DNA.
The presence of DNA-RNA crosslinks in NKK-treated mice raises an interesting question as to where such DNA-RNA crosslinks occur in cells, and one possibility is at the DNA:RNA hybrids of R-loops. An R-loop is a three-stranded nucleic acid structure consisting of an RNA:DNA hybrid and a displaced single-stranded DNA. R-loops are normally formed at transcription regions where the pre-mRNA hybridizes to the template strand DNA, and formaldehyde treatment of cells has been shown to lead to higher levels of R-loops. 51 We hypothesize that naturally occurring or NNK-induced RNA:DNA hybrids in cells are possible sites for formaldehyde DNA-RNA crosslinks. Using immunodetection and dot blot, we evaluated relative levels of RNA:DNA hybrids in NNK-treated mice lung DNA 6 h after treatment compared to the corresponding control (without NNK treatment, 6 h time point). Using the DNA:RNA hybrid-specific S9.6 antibody and RNaseH treatment, we have measured the levels of DNA:RNA hybrids and the confirmed the specificity of the S9.6 antibody, respectively. Figure 7A–C shows the dot blots and the relative levels of DNA:RNA hybrids in NNK-treated sample compared to the control. Treatment of the samples with RNaseH, which degrades the RNA moiety within the DNA:RNA hybrids, completely removed all S9.6 signal, confirming the presence of DNA:RNA hybrids. The difference in hybrid levels of the control and NNK-treated mice was not statistically significant.
Fig. 7.
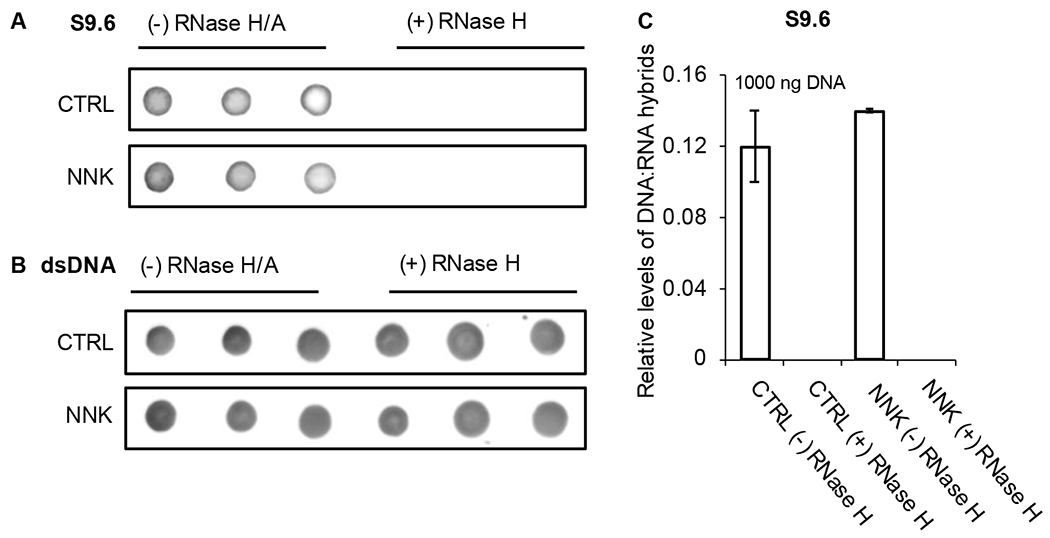
Immunodetection and dot blot of DNA:RNA hybrids in mice lung DNA. (A) DNA:RNA hybrids in NNK-treated mice and control with and without RNase H treatment. (B) Levels of input DNA detected using dsDNA-specific antibody. (C) Relative levels of DNA:RNA hybrids in NNK-treated mice compared to the corresponding control mice. Levels are normalized against dsDNA signals. Error bars represent the standard deviation of the triplicate measurements.
Discussion
Previous studies have extensively characterized the metabolism and DNA adduct formation driven by NNK exposure in animal models of lung carcinogenesis. NNK bioactivation generates reactive species including methylating agents, pyridyloxobutylating species, and reactive aldehydes resulting in a broad spectrum of DNA damage. To date, the well-characterized NNK-induced mono-adducts include the methylated and pyridyloxobutylated DNA adducts. In addition, pyridyloxobutyl dinucleotide phosphate adducts were also identified in vivo 12, 13, 26, 28 and dinucleotide methyl phosphate adducts were detected in lung tissues of smokers and non-smokers.12 Formaldehyde generated in situ upon NNK activation was reported to result in the formation of the abundant DNA adduct N6-hydroxymethyl-dA, which is measured as N6-methyl-dA after reduction to its stable form, by LC-MS2 detection using selective reaction monitoring (SRM).9 Likewise, previous studies have demonstrated that formaldehyde induces DNA-DNA and DNA-protein crosslinks in vitro and in vivo.29–36 However, to the best of our knowledge, a clear characterization of formaldehyde-induced DNA-RNA crosslinks has not been reported. Although it is speculated that DNA-RNA crosslinks could form in the presence of formaldehyde or mitomycin C in cells without a functioning Fanconi anemia (FA) pathway, their detection and characterization remain elusive.52
Using our comprehensive and improved segmented full scan high-resolution accurate mass data-dependent MS3 neutral loss screening strategy, we have detected and identified putative formaldehyde-induced DNA-RNA crosslinks in mice lung DNA. The levels of these crosslinks are significantly higher in NNK-treated mice compared to their corresponding controls when comparing the levels 6 h after NNK exposure. The high-resolution accurate LC-MSn mass spectral data provided robust structural characterization of these novel formaldehyde-induced DNA-RNA crosslinks. These results were further supported by the in vitro experiments using purified DNA bases and the RNA base, guanosine which confirmed the formation and existence of formaldehyde-induced DNA-RNA crosslinks and allowed to produce a synthetic standard for guanosine-CH2-dA . The occurrenceand biological relevance of the novel DNA-RNA crosslinks reported here are largely unknown and their implications in NNK-induced carcinogenesis are poorly understood.
Formaldehyde derived from endogenous and exogenous sources induces DNA adducts, protein modifications, DNA-DNA crosslinks, and DNA-protein crosslinks, which are the basis of its toxicity and carcinogenicity. The reaction of formaldehyde with the free amino group of DNA bases generates hydroxymethyl groups followed by Schiff base formation resulting from partial dehydration of these groups. Depending on the proximity and interaction of other nucleophilic sites with the activated reactive groups, intermolecular or intramolecular crosslinks can occur via formation of a methylene bridge with the exocylic amino groups of nucleic acids , or with the free amino group of amino acid side chains in proteins (e.g. Lys, Arg,).29, 53 Due to its reactivity with biological molecules, formaldehyde has been extensively used in probing DNA-protein interactions and protein-protein interactions.29, 40
DNA-protein crosslinks are the most studied covalent modifications induced by formaldehyde. For example the identities and structures of DNA-protein crosslinks (DPCs) have been previously reported in rats exposed to labeled formaldehyde.30 The measurement of these DPCs was performed using a triple quadrupole instrument and SRM, which used one of the diagnostic MS2 fragment ions corresponding to methylene-guanine·H + (m/z 164.1). This fragment ion has been identified in the MS2 and MS3 mass spectral data of guanine containing DNA-RNA crosslinks identified and reported here and further confirmed by comparison with the fragmentation obtained by analyzing the synthetic standard of the DNA-DNA crosslink analog dA-CH2-dA. Furthermore, the diagnostic ions, m/z 296.0989 for methylene-guanosine·H + and m/z 264.1091 for methylene-dA·H + can be used to confirm the identity of the DNA or RNA nucleoside. For adenine containing crosslinks, the diagnostic ion is m/z 148.0618. The neutral loss of deoxyribose or ribose and the presence of these methylene base moieties can be used as diagnostic ions for the detection and quantitation of either DNA-DNA or DNA-RNA crosslinks induced by formaldehyde. These diagnostic MS2 fragment ions of formaldehyde-induced DNA-RNA crosslinks will enable the comprehensive screening, identification, and structural analysis of these novel crosslinks by MS in various biological matrices from epidemiological studies.
After we have confirmed the formation and structures of these novel crosslinks in vitro we then sought to identify the sources of these DNA-RNA crosslinks. These newly detected crosslinks could represent a much larger family of structures for which the modifications we detected are only what is left after treatment of the DNA with RNAse during the DNA isolation and purification steps. Possible sources of these crosslinks are the naturally occurring DNA:RNA hybrids (R-loops) formed during transcription, the DNA:RNA hybrids formed during replication, and specific DNA-RNA interactions during gene expression. R-loops are transient and naturally occurring DNA:RNA hybrid structures in the genome, which are comprised of a nascent pre-mRNA hybridized with the template DNA and an exposed single-stranded non-template DNA.41, 54 These hybrids are more stable than double-stranded DNA and are abundant at sites with high transcriptional activity, including transcription start sites, repression sites, as well as elongation sites and are associated with various cellular processes such as chromatin remodeling, Ig gene recombination, DNA double strand breaks (DSBs), and genome instability.42, 55 Furthermore, R-loops play critical roles in cancer-associated processes including genome and epigenome maintenance.52 The changes in R-loop frequency, stability, or genomic location/position have been linked to cancer development including activation of oncogenes or loss of tumor suppressor genes in various model systems.52 For instance, previous studies have demonstrated that the FA pathway plays a crucial role in the repair of R-loop-mediated damage or replication fork blockage.56 In cells bearing heterozygous BRCA2 mutations, formaldehyde stalls and destabilizes DNA replication forks, leading to genomic instability,51 while the complete inactivation of BRCA2 increases the levels of R-loops. In cells without a functional FA pathway, formaldehyde treatment results in increased R-loops formation, suggesting a mechanism by which this compound could contribute to genome instability.57 Because formaldehyde reacts with the base moiety of both DNA and RNA, it is possible that both DNA–DNA and DNA–RNA crosslinks are drivers of the observed R-loop induction and may require specific mechanisms to suppress R-loop-associated genomic instability. In the future, identifying specific R-loop response pathways that resolves formaldehyde induced R-loops and/or the potential presence of DNA-RNA crosslinks at R-loops may present opportunities for novel therapeutic strategies that would be relevant in FA-deficient cancers.52 Overall, the results from these studies may illustrate the need to further investigate the roles of tumor suppressors in preventing R loops, which are major sources of replication stress and hence cell death and cancer.58
Although previous studies have shown that NNK exerts its carcinogenic effects via K-ras oncogene activation, only about 30% of human lung adenocarcinomas from smokers have an activated K-ras strongly linking the activation of this oncogene with smoking. It is apparent that the mutational K-ras activation may be a direct consequence of various carcinogens present in tobacco smoke and other mechanisms are involved in tobacco-induced lung carcinogenesis.59 Given the link between activated K-ras (used here in the A/J mouse strain) and unscheduled R-loops as potential oncogenic events in lung adenocarcinomas, we hypothesized that a significant amount of the observed NNK-induced DNA-RNA crosslinking was occurring on the R-loop DNA:RNA hybrids and would result in the accumulation and therefore higher levels of R-loops in the same mice where higher levels of DNA-RNA crosslink adducts were observed.56 Using the DNA:RNA hybrid-specific S9.6 antibody, we were able to detect these DNA:RNA hybrids in mice lung DNA. However, NNK-induced DNA-RNA crosslinks levels as quantified by MS did not correlate with significant increase in global R-loop levels. This may be due to the differential quantitative precision and accuracy of R-loops by dot blot analysis and of DNA-RNA crosslinks by quantitative MS. Alternatively, NNK-generated formaldehyde induced DNA-RNA crosslinks may contribute to carcinogenesis beyond R-loop stabilization; instead, it is still possible that the transient RNA:DNA hybrids at R-loops may become a substrate of DNA-RNA crosslinks downstream of R-loop formation. Future experiments are needed to explore the association of DNA-RNA crosslinks at R-loops by comparing different experimental conditions and higher levels of exposures when measuring the R-loops. Aside from R-loop structures, the formaldehyde-induced DNA-RNA crosslinks could be generated in other contexts such as during DNA replication and specific DNA-RNA interactions during gene expression. The exact contributions of different sources of exposures to the formation of DNA-RNA crosslinks, their potential mutagenicity and their overall biological relevance in lung carcinogenesis remains unknown and warrants further careful investigation in future studies.
Although it is not reported and not within the scope of this work, we have also detected and structurally characterized DNA-DNA and DNA-protein crosslinks induced by NNK-generated and endogenous formaldehyde. Further studies are needed to elucidate the exact roles of these crosslinks in cancer initiation and specifically the contribution, if any, of the novel DNA-RNA crosslinks identified here, in NNK-induced carcinogenesis.
Conclusion
Here, we have identified and structurally characterized, for the first time, a new class of DNA adducts, DNA-RNA crosslinks, using high-resolution accurate mass LC-MSn approaches in mice treated with NNK. The levels of these DNA-RNA crosslinks are significantly higher in NNK treated mice compared to the controls. We have demonstrated that these crosslinks are formed in vitro with purified DNA and RNA bases in the presence of formaldehyde. The covalent and unscheduled formaldehyde-induced crosslinking of DNA and RNA could be one of the broad mechanisms of enhanced lung carcinogenicity of NNK in A/J mouse model system. Understanding the roles and consequences of DNA-RNA crosslinking leading to DNA damage and genome instability are important in elucidating how these structures may interfere and alter genome dynamics and function. In addition, equally important is gaining insights through which these novel DNA-RNA crosslinks are implicated in chemically-induced carcinogenesis. Finally, DNA-RNA crosslinks can be explored and used as biomarkers for assessing sensitivity and efficacy of various chemotherapeutic agents, especially in diseases caused by deficiency in aldehyde metabolizing enzymes and crosslink repair enzymes, such as those relevant to Fanconi Anemia patients..
Supplementary Material
Acknowledgments
This work was supported by the U.S. National Institute of Health and National Cancer Institute [NCI-CA220376]. Mass spectrometry was carried out in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by the U.S. National Institute of Health and National Cancer Institute [Cancer Center Support Grant CA-77598]. Salary support for P.W.V. was provided by the National Cancer Institute (Grant R50-CA211256). H.D.N is a EvansMDS Young Investigator (Edward P. Evans Foundation) and an ASH Scholar (American Society of Hematology). H.D.N is supported by the National Institutes of Health’s National Center for Advancing Translational Sciences, grants KL2TR002492 and UL1TR002494 and National Heart, Lung, and Blood Institute, R01HL163011. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health’s National Center for Advancing Translational Sciences. We thank Prof. Kassie’s group for assistance in the animal experiment and Andrew C. Floeder for the DNA extraction. Finally, we thank Dr Todd Rappe from the Minnesota NMR Center for support with the NMR spectra acquisition.
Footnotes
Supporting Information
The Supporting Information is available free of charge. Figures S1–S11 with the scheme of animal experiment, in vitro incubation of DNA and RNA with formaldehyde and HPLC-UV chromatograms, targeted LC-MS2 data for the DNA-RNA crosslinks, NMR spectra for the synthetized standard and Table S1 with the list of target DNA adducts and crosslinks for LC-MS2 assay.
References
- 1.Hecht SS (1997) Approaches to cancer prevention based on an understanding of N-nitrosamine carcinogenesis. Proc Soc Exp Biol Med 216, 181–191. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS (1998) Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem Res Toxicol 11, 559–603. [DOI] [PubMed] [Google Scholar]
- 3.Hecht SS, Carmella SG, Foiles PG, Murphy SE, and Peterson LA (1993) Tobacco-specific nitrosamine adducts: studies in laboratory animals and humans. Environ Health Perspect 99, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hecht SS, Carmella SG, Foiles PG, and Murphy SE (1994) Biomarkers for human uptake and metabolic activation of tobacco-specific nitrosamines. Cancer Res 54, 1912s–1917s. [PubMed] [Google Scholar]
- 5.Hecht SS (1994) Environmental tobacco smoke and lung cancer: the emerging role of carcinogen biomarkers and molecular epidemiology. J Natl Cancer Inst 86, 1369–1370. [DOI] [PubMed] [Google Scholar]
- 6.Upadhyaya P, Carmella SG, Guengerich FP, and Hecht SS (2000) Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis 21, 1233–1238. [PubMed] [Google Scholar]
- 7.Dator R, von Weymarn LB, Villalta PW, Hooyman CJ, Maertens LA, Upadhyaya P, et al. (2018) In Vivo Stable-Isotope Labeling and Mass-Spectrometry-Based Metabolic Profiling of a Potent Tobacco-Specific Carcinogen in Rats. Anal Chem 90, 11863–11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson LA, and Hecht SS (1991) O6-methylguanine is a critical determinant of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone tumorigenesis in A/J mouse lung. Cancer Res 51, 5557–5564. [PubMed] [Google Scholar]
- 9.Hecht SS (1997) Tobacco and cancer: approaches using carcinogen biomarkers and chemoprevention. Ann N Y Acad Sci 833, 91–111. [DOI] [PubMed] [Google Scholar]
- 10.Peterson LA, Mathew R, B1P6Murphy SE, Trushin N, and Hecht SS (1991) In vivo and in vitro persistence of pyridyloxobutyl DNA adducts from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 12, 2069–2072. [DOI] [PubMed] [Google Scholar]
- 11.Peterson LA, Liu XK, and Hecht SS (1993) Pyridyloxobutyl DNA adducts inhibit the repair of O6-methylguanine. Cancer Res 53, 2780–2785. [PubMed] [Google Scholar]
- 12.Ma B, Villalta PW, Hochalter JB, Stepanov I, and Hecht SS (2019) Methyl DNA phosphate adduct formation in lung tumor tissue and adjacent normal tissue of lung cancer patients. Carcinogenesis 40, 1387–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma B, Zarth AT, Carlson ES, Villalta PW, Upadhyaya P, Stepanov I, and Hecht SS (2018) Methyl DNA Phosphate Adduct Formation in Rats Treated Chronically with 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone and Enantiomers of Its Metabolite 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem Res Toxicol 31, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peterson LA (2017) Context Matters: Contribution of Specific DNA Adducts to the Genotoxic Properties of the Tobacco-Specific Nitrosamine NNK. Chem Res Toxicol 30, 420–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mijal RS, Kanugula S, Vu CC, Fang Q, Pegg AE, and Peterson LA (2006) DNA sequence context affects repair of the tobacco-specific adduct O(6)-[4-Oxo-4-(3-pyridyl)butyl]guanine by human O(6)-alkylguanine-DNA alkyltransferases. Cancer Res 66, 4968–4974. [DOI] [PubMed] [Google Scholar]
- 16.Ronai ZA, Gradia S, Peterson LA, and Hecht SS (1993) G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis 14, 2419–2422. [DOI] [PubMed] [Google Scholar]
- 17.Wichmann AE, Thomson NM, Peterson LA, and Wattenberg EV (2003) Genotoxic methylating agents modulate extracellular signal regulated kinase activity through MEK-dependent, glutathione-, and DNA methylation-independent mechanisms in lung epithelial cells. Chem Res Toxicol 16, 87–94. [DOI] [PubMed] [Google Scholar]
- 18.Thomson NM, Kenney PM, and Peterson LA (2003) The pyridyloxobutyl DNA adduct, O6-[4-oxo-4-(3-pyridyl)butyl]guanine, is detected in tissues from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone-treated A/J mice. Chem Res Toxicol 16, 1–6. [DOI] [PubMed] [Google Scholar]
- 19.Belinsky SA, Foley JF, White CM, Anderson MW, and Maronpot RR (1990) Dose-response relationship between O6-methylguanine formation in Clara cells and induction of pulmonary neoplasia in the rat by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res 50, 3772–3780. [PubMed] [Google Scholar]
- 20.Belinsky SA, Devereux TR, Maronpot RR, Stoner GD, and Anderson MW (1989) Relationship between the formation of promutagenic adducts and the activation of the K-ras protooncogene in lung tumors from A/J mice treated with nitrosamines. Cancer Res 49, 5305–5311. [PubMed] [Google Scholar]
- 21.Belinsky SA, Devereux TR, and Anderson MW (1990) Role of DNA methylation in the activation of proto-oncogenes and the induction of pulmonary neoplasia by nitrosamines. Mutat Res 233, 105–116. [DOI] [PubMed] [Google Scholar]
- 22.Devereux TR, Anderson MW, and Belinsky SA (1991) Role of ras protooncogene activation in the formation of spontaneous and nitrosamine-induced lung tumors in the resistant C3H mouse. Carcinogenesis 12, 299–303. [DOI] [PubMed] [Google Scholar]
- 23.Hecht SS, Trushin N, Castonguay A, and Rivenson A (1986) Comparative tumorigenicity and DNA methylation in F344 rats by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and N-nitrosodimethylamine. Cancer Res 46, 498–502. [PubMed] [Google Scholar]
- 24.Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91, 1194–1210. [DOI] [PubMed] [Google Scholar]
- 25.Demkowicz-Dobrzanski K, and Castonguay A (1992) Modulation by glutathione of DNA strand breaks induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and its aldehyde metabolites in rat hepatocytes. Carcinogenesis 13, 1447–1454. [DOI] [PubMed] [Google Scholar]
- 26.Ma B, Villalta PW, Zarth AT, Kotandeniya D, Upadhyaya P, Stepanov I, and Hecht SS (2015) Comprehensive High-Resolution Mass Spectrometric Analysis of DNA Phosphate Adducts Formed by the Tobacco-Specific Lung Carcinogen 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol 28, 2151–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai D, Kagan SS, Amin S, Carmella SG, and Hecht SS (1993) Identification of 4-(methylnitrosamino)-1-[3-(6-hydroxypyridyl)]-1-butanone as a urinary metabolite of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in rodents. Chem Res Toxicol 6, 794–799. [DOI] [PubMed] [Google Scholar]
- 28.Ma B, Zarth AT, Carlson ES, Villalta PW, Upadhyaya P, Stepanov I, and Hecht SS (2018) Identification of more than 100 structurally unique DNA-phosphate adducts formed during rat lung carcinogenesis by the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 39, 232–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu K, Boysen G, Gao L, Collins LB, and Swenberg JA (2008) Formaldehyde-induced histone modifications in vitro. Chem Res Toxicol 21, 1586–1593. [DOI] [PubMed] [Google Scholar]
- 30.Lai Y, Yu R, Hartwell HJ, Moeller BC, Bodnar WM, and Swenberg JA (2016) Measurement of Endogenous versus Exogenous Formaldehyde-Induced DNA-Protein Crosslinks in Animal Tissues by Stable Isotope Labeling and Ultrasensitive Mass Spectrometry. Cancer Res 76, 2652–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu CW, Chang YJ, Cooke MS, and Chao MR (2019) DNA Crosslinkomics: A Tool for the Comprehensive Assessment of Interstrand Crosslinks Using High Resolution Mass Spectrometry. Anal Chem 91, 15193–15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang H, and Hopkins PB (1993) DNA Interstrand Cross-Linking by Formaldehyde: Nucleotide Sequence Preference and Covalent Structure of the Predominant Cross-Link Formed in Synthetic Oligonucleotides J. Am. Chem. Soc 115, 9402–9408. [Google Scholar]
- 33.Huang H, Solomon MS, and Hopkins PB (1992) Formaldehyde Preferentially Interstrand Cross-Links Duplex DNA through Deoxyadenosine Residues at the Sequence 5’-d(AT) J. Am. Chem. Soc 114, 9240–9241. [Google Scholar]
- 34.Wang M, Cheng G, Villalta PW, and Hecht SS (2007) Development of liquid chromatography electrospray ionization tandem mass spectrometry methods for analysis of DNA adducts of formaldehyde and their application to rats treated with N-nitrosodimethylamine or 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Chem Res Toxicol 20, 1141–1148. [DOI] [PubMed] [Google Scholar]
- 35.Casanova M, Morgan KT, Gross EA, Moss OR, and Heck HA (1994) DNA-protein cross-links and cell replication at specific sites in the nose of F344 rats exposed subchronically to formaldehyde. Fundam Appl Toxicol 23, 525–536. [DOI] [PubMed] [Google Scholar]
- 36.Cheng G, Shi Y, Sturla SJ, Jalas JR, McIntee EJ, Villalta PW, et al. (2003) Reactions of formaldehyde plus acetaldehyde with deoxyguanosine and DNA: formation of cyclic deoxyguanosine adducts and formaldehyde cross-links. Chem Res Toxicol 16, 145–152. [DOI] [PubMed] [Google Scholar]
- 37.Castonguay A, and Rossignol G (1992) Modulation of the activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone by hamster liver microsomes to protein alkylating species. Toxicol In Vitro 6, 397–404. [DOI] [PubMed] [Google Scholar]
- 38.Saladino AJ, Willey JC, Lechner JF, Grafstrom RC, LaVeck M, and Harris CC (1985) Effects of formaldehyde, acetaldehyde, benzoyl peroxide, and hydrogen peroxide on cultured normal human bronchial epithelial cells. Cancer Res 45, 2522–2526. [PubMed] [Google Scholar]
- 39.Grafström RC (1990) In vitro studies of aldehyde effects related to human respiratory carcinogenesis. Mutat Res 238, 175–184. [DOI] [PubMed] [Google Scholar]
- 40.Hoffman EA, Frey BL, Smith LM, and Auble DT (2015) Formaldehyde crosslinking: a tool for the study of chromatin complexes. J Biol Chem 290, 26404–26411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stolz R, Sulthana S, Hartono SR, Malig M, Benham CJ, and Chedin F (2019) Interplay between DNA sequence and negative superhelicity drives R-loop structures. Proc Natl Acad Sci U S A 116, 6260–6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.García-Muse T, and Aguilera A (2019) R Loops: From Physiological to Pathological Roles. Cell 179, 604–618. [DOI] [PubMed] [Google Scholar]
- 43.Balbo S, Hecht SS, Upadhyaya P, and Villalta PW (2014) Application of a high-resolution mass-spectrometry-based DNA adductomics approach for identification of DNA adducts in complex mixtures. Anal Chem 86, 1744–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng G, Wang M, Upadhyaya P, Villalta PW, and Hecht SS (2008) Formation of formaldehyde adducts in the reactions of DNA and deoxyribonucleosides with alpha-acetates of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), and N-nitrosodimethylamine (NDMA). Chem Res Toxicol 21, 746–751. [DOI] [PubMed] [Google Scholar]
- 45.Melkamu T, Qian X, Upadhyaya P, O’Sullivan MG, and Kassie F (2013) Lipopolysaccharide enhances mouse lung tumorigenesis: a model for inflammation-driven lung cancer. Vet Pathol 50, 895–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carrà A, Guidolin V, Dator RP, Upadhyaya P, Kassie F, Villalta PW, and Balbo S (2019) Targeted High Resolution LC/MS. Front Chem 7, 658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walsh JP, Renaud JB, Hoogstra S, McMullin DR, Ibrahim A, Visagie CM, et al. (2019) Diagnostic fragmentation filtering for the discovery of new chaetoglobosins and cytochalasins. Rapid Commun Mass Spectrom 33, 133–139. [DOI] [PubMed] [Google Scholar]
- 48.Pluskal T, Castillo S, Villar-Briones A, and Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murray KJ, Carlson ES, Stornetta A, Balskus EP, Villalta PW, and Balbo S (2021) Extension of Diagnostic Fragmentation Filtering for Automated Discovery in DNA Adductomics. Anal Chem 93, 5754–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, and Zou L (2017) Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Mol Cell 65, 832–847.e834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan SLW, Chadha S, Liu Y, Gabasova E, Perera D, Ahmed K, et al. (2017) A Class of Environmental and Endogenous Toxins Induces BRCA2 Haploinsufficiency and Genome Instability. Cell 169, 1105–1118.e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells JP, White J, and Stirling PC (2019) R Loops and Their Composite Cancer Connections. Trends Cancer 5, 619–631. [DOI] [PubMed] [Google Scholar]
- 53.Lu K, Ye W, Zhou L, Collins LB, Chen X, Gold A, et al. (2010) Structural characterization of formaldehyde-induced cross-links between amino acids and deoxynucleosides and their oligomers. J Am Chem Soc 132, 3388–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skourti-Stathaki K, and Proudfoot NJ (2014) A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 28, 1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crossley MP, Bocek M, and Cimprich KA (2019) R-Loops as Cellular Regulators and Genomic Threats. Mol Cell 73, 398–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.García-Rubio ML, Pérez-Calero C, Barroso SI, Tumini E, Herrera-Moyano E, Rosado IV, and Aguilera A (2015) The Fanconi Anemia Pathway Protects Genome Integrity from R-loops. PLoS Genet 11, e1005674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schwab RA, Nieminuszczy J, Shah F, Langton J, Lopez Martinez D, Liang CC, et al. (2015) The Fanconi Anemia Pathway Maintains Genome Stability by Coordinating Replication and Transcription. Mol Cell 60, 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bhatia V, Barroso SI, García-Rubio ML, Tumini E, Herrera-Moyano E, and Aguilera A (2014) BRCA2 prevents R-loop accumulation and associates with TREX-2 mRNA export factor PCID2. Nature 511, 362–365. [DOI] [PubMed] [Google Scholar]
- 59.Rodenhuis S, Slebos RJ, Boot AJ, Evers SG, Mooi WJ, Wagenaar SS, et al. (1988) Incidence and possible clinical significance of K-ras oncogene activation in adenocarcinoma of the human lung. Cancer Res 48, 5738–5741. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


