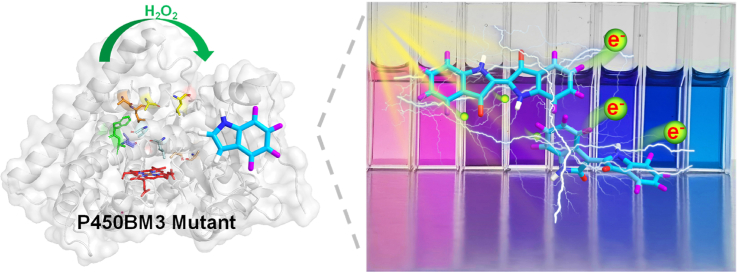
Abstract
Indigoids, a class of bis-indoles, have long been applied in dyeing, food, and pharmaceutical industries. Recently, interest in these ‘old’ molecules has been renewed in the field of organic semiconductors as functional building blocks for organic electronics due to their excellent chemical and physical properties. However, these indigo derivatives are difficult to access through chemical synthesis. In this study, we engineer cytochrome P450 BM3 from an NADPH-dependent monooxygenase to peroxygenases through directed evolution. A select number of P450 BM3 variants are used for the selective oxidation of indole derivatives to form different indigoid pigments with a spectrum of colors. Among the prepared indigoid organic photocatalysts, a majority of indigoids demonstrate a reduced band gap than indigo due to the increased light capture and improved charge separation, making them promising candidates for the development of new organic electronic devices. Thus, we present a useful enzymatic approach with broad substrate scope and cost-effectiveness by using low-cost H2O2 as a cofactor for the preparation of diversified indigoids, offering versatility in designing and manufacturing new dyestuff and electronic/sensor components.
Keywords: Cytochrome P450 enzymes, P450 BM3 peroxygenase, Indigoids, Dyestuff, Organic electronics
Graphical abstract
1. Introduction
The natural pigment indigo is an old and widely used vat dyestuff due to its safety and extraordinary spectral properties [1]. The intermolecular N–H‧‧‧O C hydrogen bonds play an essential role in molecular packing [2]. Indigo is also an ambipolar organic semiconductor with well-balanced electron and hole mobilities in transistors [3,4]. The natural semiconductor appears to be a promising option for fabrication of fully biodegradable and biocompatible organic electronics. Indigo used to be made from the indigo and woad plants. Today, natural indigo is almost entirely replaced by synthetic indigo.
Substituted indigo derivatives are termed as indigoids, which are potential candidate compounds in dyeing, pharmaceutical, food, and semiconductor industries [5]. For example, the indigo derivatives 6,6′-difluoroindogo, 6,6′-dichloroindigo, 5,5′-dibromoindigo, 6,6′-dibromoindigo (Tyrian purple), 6,6′-diiodoindigo, and 5,5′-diiodoindigo have shown prospective applications in organic electronics [[6], [7], [8]]. The introduction of substituents could induce varied optical and electronic properities such as absorption, charge carrier transport properties, transistor characteristics, LUMO energy levels, as well as the electrical performance in organic field-effect transistors (OFETs) [7].
Indigo has extremely low solubility in water and most of organic solvents, and is only soluble in a small number of polar protic solvents such as DMSO and DMF. It has a high melting point >300 °C, because of stabilization resulted from inter- and intramolecular hydrogen bonding and π–π stacking [9]. These properties largely limit its use and further derivatization. Indigo is chemically synthesized from 2-nitrobenzaldehyde and acetone in alkaline media [10]. Indigo carmine is made from indigo and sulfuric acid [11]. Multiple dihalogenoindigos, such as 5,5’/6,6′-dibromoindigo, 5,5’/6,6′-dichloroindigo, and 5,5’/6,6′-difluoroindigo, have also been synthesized via different approaches [12,13]. However, these classical synthetic approaches for indigo and indigoids would cause irreversible environmental pollution, some of which start from expensive raw materials, thus being unsustainable. By contrast, the enzymatic synthesis of indigo/indigoids with controllable regioselectivity under mild conditions is environmentally friendly and economically attractive.
Many oxygenases have been reported to catalyze the oxidative coupling of indoles to form indigo [[14], [15], [16], [17]], such as naphthalene dioxygenase [18], phenol hydroxylases [18], xylene oxygenase [19], toluene-2/4-monooxygenase [20,21], peroxygenases [18], flavin-containing monooxygenases [22], and cytochrome P450 monooxygenases (P450s) [23]. Among these oxidative enzymes, P450s are very attractive targets due to their broad substrate spectra, catalytic versatility, and function fabricability by multiple engineering strategies [18,[24], [25], [26]]. Thus, a growing number of applications of P450s in biocatalysis and biotransformation, as well as in green synthetic chemistry have been emerging [27]. In particular, P450 BM3 (CYP102A1) originated from Bacillus megaterium is a preferred candidate for P450-based biocatalysis because of its superb catalytic efficiency (turnover number: ∼200 sec−1 towards some fatty acid substrates) and expandable substrate scope [28]. This bacterial soluble enzyme consists of a P450 domain naturally fused to a diflavin NADPH reductase domain, thus circumventing the requirement of separate redox partner proteins by most of native P450s. Moreover, it has been reported that some P450 BM3 mutants could use H2O2 as an alternative cofactor to costly NAD(P)H [[29], [30], [31]], thereby further lowering the cost of P450-mediated biosyntheses of indigo/indigoids and other compounds.
In our previous study, we designed and validated a novel “multi-enzymes-for-multi-substrates” (MEMS) directed evolution model, by which a number of P450 BM3 mutants were identified to be capable of efficiently converting indole into indigo by using NADPH as electron donor [32]. In this work, we adopted colorimetric method to detect the oxidative products of indole and its derivatives by using the alternative cofactor H2O2. As a result, we successfully developed several productive P450 BM3 peroxygenases for enzymatic synthesis of diversified indigoids. The different electrical substituents (electron-withdrawing and electron-donating groups) and positions of these functional groups (substituents at 4-, 5-, 6-, and 7-positions of the phenyl ring) changed the intramolecular proton transfer, thus resulting in various photophysical properties of indigoids.
2. Materials and methods
2.1. Materials
Antibiotics and chemicals were purchased from SolarBio (Beijing, China) and Sigma Aldrich (St. Louis, MO, USA). KOD-Plus Neo DNA Polymerase was obtained from TOYOBO (Osaka, Japan). The kits for plasmid extraction and DNA purification were purchased from OMEGA Bio-Tek (Norcross, GA, USA) and Promega (Madison, WI, USA). His-tagged protein purification used Qiagen Ni-NTA resin (Valencia, CA, USA), Millipore Amicon Ultra centrifugal filters (Billerica, MA, USA), and PD-10 desalting columns from GE Healthcare (Piscataway, NJ, USA). Oligonucleotides synthesis and DNA sequencing were conducted by Sangon Biotech (Shanghai, China).
2.2. General experimental procedures
High resolution Q-TOF mass spectrometry data were acquired on a maXis ultrahigh-resolution TOF system (Bruker Daltonik, Germany). Masses of negatively charged ions were calibrated using aqueous sodium formate as an internal standard. The reaction samples were analyzed by HPLC/HRMS with a Triart C18 column (5 μm, 4.6 mm × 250 mm, YMC Co., Ltd., Japan). Two solvent reservoirs, containing solvent A, water +0.1% (v/v) TFA and solvent B, methanol + 0.1% (v/v) TFA, were used to separate the indigoid components, under gradient elution. The gradient elution profile was as follow: 0–8 min, 60% solvent B; 8–23 min, 60–100% solvent B; 23–25 min, 100% solvent B; 25–26 min, 100-60% solvent B; 26–30 min, 60% solvent B. The analytes were quantified at 254, 550 and 600 nm. Separation of products was performed with a flow rate of 1 mL min−1. Nuclear magnetic resonance (NMR) spectra were acquired on a Bruker 600 MHz spectrometer (Bruker BioSpin GmbH Co., Rheinstetten, Germany). NMR data were processed using MestReNova software.
2.3. Construction of expression vectors
In our previous study [13], we constructed a P450 BM3*(i.e., P450 BM3 F87A) mutants library and identified several mutant enzymes capable of oxidizing indole through high-throughput screening. In these reactions, the catalytically required electrons are provided by NADPH and transferred to the heme-iron reactive center via the fused reductase domain of P450 BM3*. To use H2O2 as an alternative cofactor to costly NADPH, in this study we also initiated these enzymatic reactions with H2O2. To further investigate the potential use of H2O2 as a cofactor in place of a redox partner system, we constructed a truncated version of P450 BM3* with the 1–455 amino acid residues (covering the whole heme domain, while losing the reductase domain), referred to as P450 BM3T, by amplifying the full-length DNA sequence of the heme domain using the primers 5′-ATGACAATTAAAGAAATGCC-3′ and 5′-AAGCGGAATTTTTTTCGATTTTG-3’.
2.4. Reaction screening in 96-well plates
Single colonies were randomly picked and inoculated into 300 μL of LB medium containing 50 μg/mL kanamycin (LBkan) in sterilized 96-deepwell plates. The cultures were shaking incubated at 37 °C, 220 rpm overnight. The overnight culture (40 μL) in each well was transferred into a new sterilized 96-deepwell plate pre-containing 400 μL of TBkan media (supplemented with 50 μg/mL kanamycin, 1 mM thiamine, and the rare salt solution: 25 μM FeCl3·6H2O, 4 μM ZnCl2, 2 μM CoCl2·6H2O, 2 μM Na2MoO4·2H2O, 2 μM CaCl2, 3 μM CuSO4 and 2 μM H3BO3) [33] and shaked for 3 h at 37 °C, 220 rpm. Then, the P450 enzyme expression was induced by addition of 0.2 mM (final concentration) isopropyl-β-d-thiogalactopyranoside (IPTG) and 0.5 mM (final concentration) 5-aminolevulinic acid into each individual wells and cultivated at 20 °C, 220 rpm for 20 h. The cells were pelleted by centrifugation at 3700 g for 10 min and stored at −80 °C for later use. The freeze-thaw cell pellets were resuspended in 50 mM potassium phosphate buffer (pH 7.4) (200 μL/well) containing 100 mg/L lysozyme, 300 U/mL DNase I, and 10% Triton X-100. The 96-well plates were centrifuged and the supernatants were transferred to a new microtiter 96-well plate, to which 0.5 mM of indole and 20 mM H2O2 were added. The plates were incubated at 30 °C for 1 h before reactions measurement.
2.5. Protein expression and purification
Escherichia coli BL21(DE3) cells carrying a plasmid encoding a P450 BM3T variant were grown overnight in LBkan media (37 °C, 220 rpm). The overnight seed culture was used for 1:100 (v/v) inoculation of 0.5 L TBkan medium containing 1 mM thiamin and the rare salt solution. The E. coli cells were grown at 37 °C for additional 2–3 h until OD600 reached 0.4–0.6, at which IPTG was added to a final concentration of 0.2 mM to induce gene expression, and 0.5 mM 5-aminolevulinic acid was supplemented as the heme synthetic precursor. Expression was conducted at 20 °C, 150 rpm, for 20 h. Cultures were then centrifuged at 6000 g for 10 min to pellet cells. The protein purification was carried out by following the previously developed procedure [34]. Purified proteins were flash-frozen by liquid nitrogen and stored at −80 °C for later use.
2.6. P450 enzyme concentration determination
P450 BM3T enzymes were purified according to the procedures described previously [35,36]. The functional P450 concentrations were determined from the CO-reduced difference spectra [37] using the extinction coefficient of ϵ450–490 = 91,000 M−1 cm−1.
2.7. In vitro reactions
In vitro reactions of P450 BM3T mutants were performed according to the previous report [32] using H2O2 as cofactor. The standard assay contained 1 μM P450, 1 mM substrate, and 60 mM H2O2 in 100 mM potassium phosphate buffer (pH 8.0). The reactions for turnover number (TON) determination contained 0.1 μM P450 BM3T enzyme, 1 mM indole, and 60 mM H2O2. The reaction was stopped after incubation of the reaction mixture at 30 °C for 30 min or 1 h by adding 100 μL methanol. The supernatants were used for calculation of substrate consumption.
2.8. Indigoids extraction and UV/Vis spectroscopy
The engineered P450 BM3T mutant was used to synthesize indigoids. However, all resulting indigoids had poor solubility in water and common organic solvents. The indigoid-containing precipitates were thoroughly washed three times with water to remove ions and denatured proteins. Purified precipitates were then dissolved in DMSO for UV/Vis spectroscopic analysis. The absorption spectra of samples were recorded in the 250–800 nm range using an Agilent 1220 HPLC system with integrated UV–Vis detector. The isomer ratios were calculated based on their maximum absorption peaks.
2.9. Structural determination of products
Indigoid products were isolated from the in vitro enzymatic reactions. The products 6,6-dibromoindigo and 6,6-dibromoindirubin were characterized by 1H NMR on a Bruker AVANCE NEO 600 MHz spectrometer with a 5 mm TCI cryo-probe. For 6,6-dibromoindigo, the reduced soluble leuco forms were synthesized for 1H NMR analysis due to the low solubility. About 2 mg of 6,6-dibromoindigo was reacted in a solution of 20 mg NaOH, 20 mg Na2S2O4 and 500 μL D2O at 80 °C [38]. All the obtained indigo derivatives exhibited an extremely low solubility in common organic solvents, making their NMR characterization quite challenging. Thus, most of these compounds were monitored by UV–Vis detector and confirmed by high-resolution mass spectroscopy.
2.10. Electronic properties of products
Photocurrent-time curves were tested using a potentiostat PGSTAT302 N Metrohm Autolab electrochemical station (Utrecht, Netherlands). The standard three-electrode configuration was used in electrochemical test with the photocatalyst-coated FTO as the working electrode, a Pt plate as the counter electrode and a saturated calomel electrode as the reference electrode. A 0.1 M NaOH solution was used as the electrolyte. FTO glasses were coated via the following steps: 4 mg of sample was added to 1 mL of alcohol and 10 μL of Nafion mixture, and 200 μL suspension was dropped onto FTO glass and dried in a vacuum oven. Photocurrent values were expressed according to the following equation.
Absorption spectra were calculated from the acquired UV/Vis data via the Kubelka–Munk transformation [39]:
where h is the Planck constant, A is the characteristic constant, Eg is the optical band gap energy, and v is the frequency.
3. Results
3.1. Screening of P450 BM3* mutants for indigo production
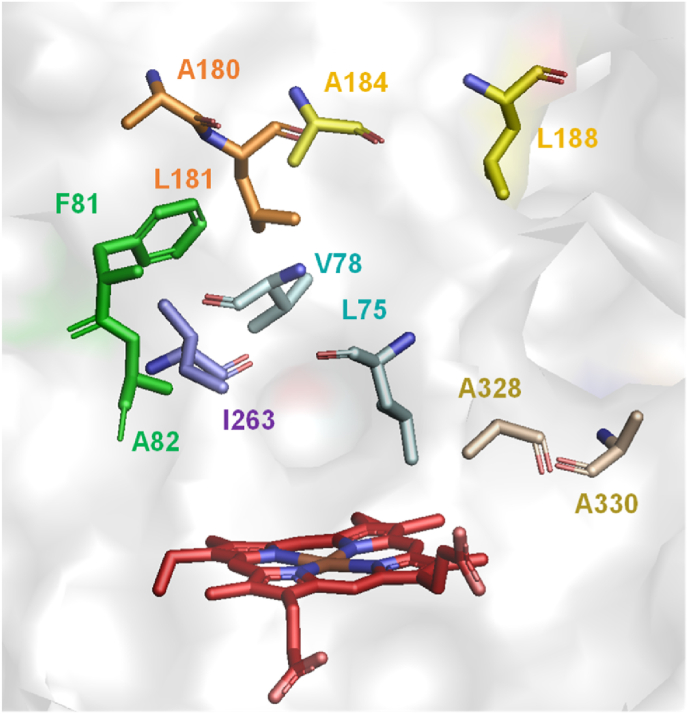
In our previous study, we constructed a P450 BM3* mutant library containing 2016 mutants using the combinatorial active-site saturation test (CAST) strategy by selecting eleven active-site residues. The eleven active-site residues of P450 BM3* were divided into six groups, including L75/V78, F81/A82, A180/L181, A184/L188, A328/A330 and I263 [32] (Fig. 1). This grouping could enhance the cooperative effects with minimal screening efforts. The screening numbers were lowered by adopting the NDT codon degeneracy, which would theoretically attain 100% and 85% coverage for the mutation sites with one and two residues, respectively. The NDT codon degeneracy would generate 12 amino acids (Phe, Leu, Ile, Val, Tyr, His, Asn, Asp, Cys, Arg, Ser, Gly) with diverse side chain properties, while excluding most cases of structurally similar amino acids [40]. The starting enzyme P450 BM3* was termed for convenience as the F87A mutant of P450 BM3 with an enlarged active site to accommodate a broader spectrum of substrates. Notably, P450 BM3* was unable to recognize indole with either NADPH or H2O2 as cofactor (Fig. S1). Of the screened 2016 variants, 625 mutant enzymes from the P450 BM3*mutant library were able to oxidize indole into differently colored products [32]. It is well-known that indoxyl can spontaneously dimerize to form the blue stain indigo and a minority of indirubin as side product (Fig. 2). In this study, by re-screening the 625 P450 BM3* mutants with cheap H2O2 instead of expensive NADPH using a colorimetric method, we identified ten mutant enzymes that exhibited-relatively high activity to hydroxylate indole into indoxyl. Next, we expressed the ten mutant enzymes in the truncated version (referred as to P450 BM3T) without the whole reductase domain (unnecessary when using H2O2 as cofactor). As expected, these ten P450 BM3T mutants were indeed capable of catalyzing the indole dimerization in presence of H2O2 (Table 1). By sequencing the ten expression plasmids of the P450 BM3T-derived indoxyl-producing mutants, we found that the key mutation sites included A82, A180, L181, A184, and L188, which could serve as starting points for further engineering to obtain more active enzymes.
Fig. 1.
The selected mutational sites of P450 BM3* (PDB ID code: 2X80). The key amino acids grouped into six sites are shown as sticks with different colors.
Fig. 2.
Dimerization of various indole derivates by P450 BM3T1. Each product is a mixture of indigo-like and indirubin-like derivatives.
Table 1.
Indole oxidation catalyzed by P450 BM3T mutants in the presence of H2O2.
| Mutant names | Mutations | TON [a] | Indigo : indirubin [b] |
|---|---|---|---|
| P450 BM3T1 | A184N L188S | 1226 ± 63 | 92:8 |
| P450 BM3T2 | A184N L188Y | 1002 ± 46 | 92:8 |
| P450 BM3T3 | A184N L188F | 883 ± 52 | 91:9 |
| P450 BM3T4 | A184N L188H | 810 ± 43 | 90:10 |
| P450 BM3T5 | A180I L181F | 732 ± 52 | 94:6 |
| P450 BM3T6 | A180R L181H | 523 ± 32 | 91:9 |
| P450 BM3T7 | A184N L188N | 407 ± 26 | 93:7 |
| P450 BM3T8 | A180I L181Y | 321 ± 22 | 95:5 |
| P450 BM3T9 | A82F | 303 ± 26 | 94:6 |
| P450 BM3T10 | A180F L181V | 240 ± 18 | 95:5 |
TON: Turnover numbers (TON) were determined based on the decreased amounts of indole from triplicated reactions in 30 min.
Indigo : indirubin: the ratios between indigo and indirubin were quantitatively determined by monitoring their maximum absorption peaks using HPLC. Reaction conditions: 0.1 μM P450 BM3T enzyme, 1 mM indole, and 60 mM H2O2 in 100 mM potassium phosphate buffer (pH 8.0), at 30 °C.
It is noteworthy that a small group of P450s (e.g., CYP152s) adopt the peroxide shunt pathway for catalysis by using H2O2 as cofactor [41]. However, this pathway is inefficient or even inaccessible for most P450s, probably due to their inherent functional and structural limitations. P450 BM3 can be transformed into the peroxygenase versions by introducing an acid–base functional group through site-directed mutagenesis or by engineering the water tunnels to enhance H2O2 access to the heme-iron reactive center [42]. The mutated residues for the ten active mutant enzymes (i.e., P450 BM3T1-T10) involve asparagine, serine, tyrosine, phenylalanine, histidine, isoleucine, arginine, and valine (Table 1). The relatively high H2O2–dependent activities of these mutants might result from the improvement of H2O2 accessibility to the reactive center.
Motivated by these screening results, we overexpressed and purified the ten mutants from Escherichia coli BL21(DE3), all of which showed a signature peak at the wavelength of 450 nm in their CO-reduced difference spectra (Fig. S2). Using 96-well plates for indole reaction screening of the 625 mutant enzymes, we employed 20 mM H2O2 based on a previous report [31]. To optimize the P450 activity, 5, 20, 40, 60, and 70 mM of H2O2 were chosen to drive the indole oxidation reactions mediated by the best P450 BM3T mutant A184N L188S (named as P450 BM3T1). Among the tested concentrations, 60 mM H2O2 exhibited the highest activity (Fig. S3). Of note, when 60 mM H2O2 was fed in batches (20 mM H2O2 was added to the P450 BM3T1 reaction system every 20 min within 1 h), the substrate conversion ratio was determined to be 61.3 ± 3.4%, which was almost the same as that of the reaction (65.3 ± 4.2%) with 60 mM H2O2 added at once in the beginning. Under the optimal H2O2 concentration, all the mutants showed high regioselectivity (>90%). P450 BM3T1 achieved a maximum turnover number (TON) of 1226 (Table 1). The yields and rations of indigo and indirubin were determined by monitoring their maximum absorption peaks at 609 and 541 nm using authentic standards as references (Fig. S4).
3.2. Expanding the substrate scope to yield indigo derivatives
In the past decade, indigoids have been found to exhibit promising semiconductor properties [43]. Indigo, tyrian purple, and several other indigo derivetives have been successfully incorporated as active layers in organic field-effect transistors (OFETs) and shown good performance in inverter circuits [3,43]. In this study, enzymatic syntheses of indigo derivatives substituted with various functional groups were investigated by using the optimal mutant enzyme P450 BM3T1. The indole derivatives with hydrogen atoms at 4-, 5-, 6-, and 7-position of the phenyl ring replaced by different functional groups were selected to form potential organic semiconductor molecules upon the P450-catalyzed dimerization (Fig. 2). The substituent groups included the electron withdrawing groups (–NO2, –CN, –CF3, –COOH, –F/Cl/Br/I, –COOC2H5, and –CHO) and electron donating groups (–OCH3) to achieve different photophysical and electrochemical properties of the resultant indigoids. Due to hyperconjugation, the –OCH3 group acted as electron donors when attached to a π system. We did not test the substituents at some specific positions due to their commercial unavailability.
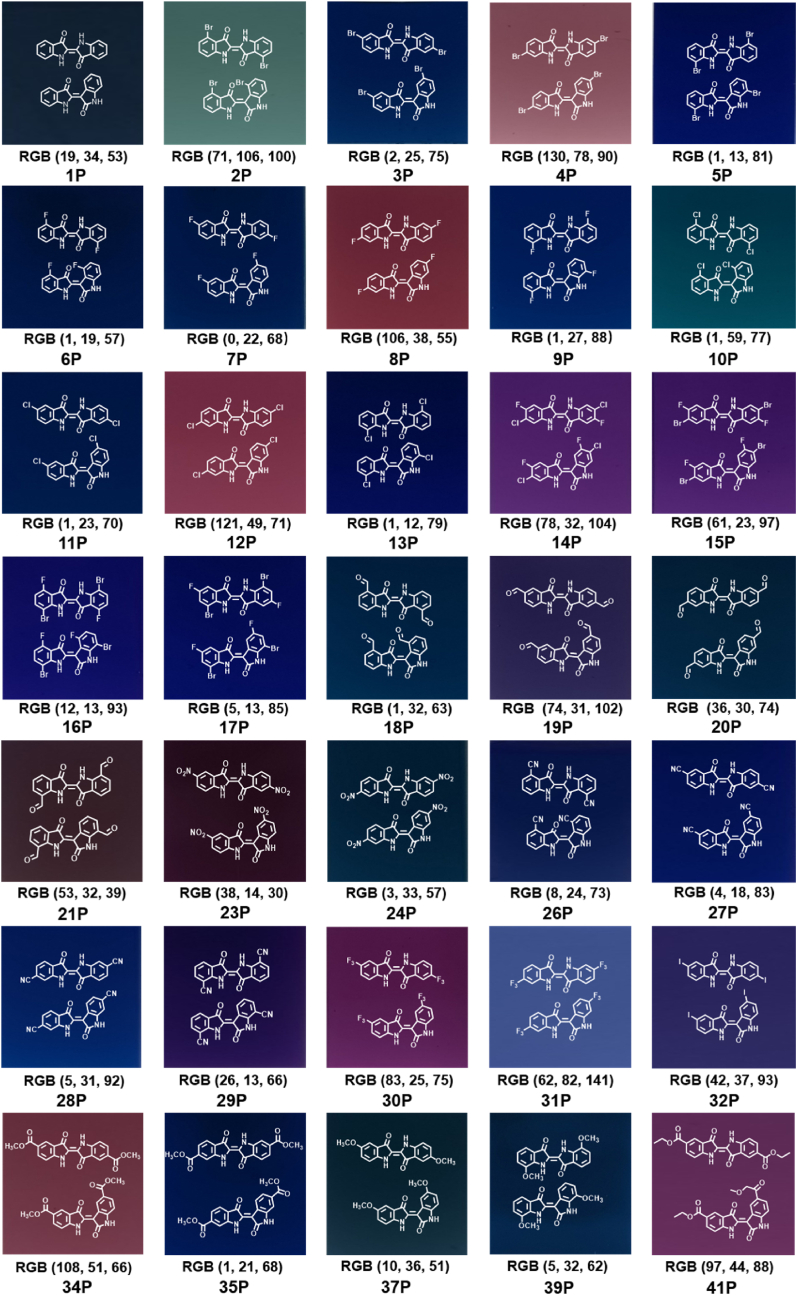
Among the 42 tested indole and derivatives, 35 monomers were dimerized into the corresponding dyes via the oxidative coupling reactions mediated by P450 BM3T1 (Fig. 3). It was shown that the selected mutant could convert almost all indole derivatives (1–21, 26–32) with substituents at 4-, 5-, 6- and 7-position of the phenyl ring.
Fig. 3.
The structures and colors of the indigoids produced by P450 BM3T1. RGB values of the prepared dyes are obtained using the Photoshop's Color Picker.
P450 BM3T1 predominantly produced symmetric indigoids for all reacting substrates, with relatively fewer asymmetric indigoids. Substrates with a Cl substituent at C4 position (10), an I substituent at C5 position (32), and two substituents of F and Br at C5 and C7 positions, respectively (17) exhibited the highest product specificity (symmetric : asymmetric = 99:1). Exceptions included 12 and 34, which had a symmetrical : asymmetrical product ratio of 41:59 and 38:62 (Table 2), respectively. With regard to the activities of P450 BM3T1 towards the reacted indole derivatives, only 37 exhibited a relatively low TON (<1000). For substrates 2–5, the Br substituent at positions 5 and 7 gave a higher TON than the same substitution at positions 4 and 6. The dihalogenoindigoids 2P and 3P with a Br substituent at C4 or C5 positions showed high regioselectivity (>90%) than at C6 or C7 positions (the ratios of symmetrical : asymmetrical derivatives were 59:41 or 68:32, respectively). As for the substrates 6–9 bearing F substituents, the corresponding products 6P–9P were obtained with TONs reaching up to 1500 and a symmetrical : asymmetrical ratio of 98:2, 91:9, 86:14, and 70:30 respectively. For substrates 10–13, the substitution positions gave little effect on the ratio of symmetric and asymmetric indigoid products, with the proportion of symmetric indigoids >89%. Substrates with F or Cl substituents at the C6 position appeared to exhibit lower TTNs compared to those with substituents at the C4, C5, or C7 positions. The transformation was also successful with dihalogen-containing substrates 14–17, trifluoro substrates 30 and 31, and nitrile-containing substrates 26–29, achieving >1200 TONs with a symmetrical : asymmetrical ratio >92%. The substrates with an aldehyde substituent at C4 or C5 positions of the benzene ring (18 and 19) exhibited low selectivity compared with the C6 or C7 positions (20 and 21). However, when the substituent groups were –NO2 (22–25), –COOCH3 (33–35), –OCH3 (36–39), and –COOC2H5 (40–42), the P450 enzyme could only oxidize a part of these substrates with the substituents at some specific positions. For substrates 22–25, the NO2 substituent at the C5 and C6 positions (23 and 24) led to the selectivity of 61:39 and 76:24 respectively. However, when the NO2 group was located at the C4 or C7 position (22 and 25), no reaction occurred. The presence of an electron-donating OCH3 group at the C5 or C7 positions of the benzene ring (37 and 39) resulted in TONs of 965 ± 14 and 2036 ± 24 and the selectivity of 96:4 and 85:15, respectively. However, when the OCH3 group was located at the C4 or C6 position (36 and 38), no product was detected. Of the tested substrates containing ethyl 1H-indole-carboxylate substituents at the C4, C5, or C7 positions (40–42), only those with the C5 substitution yielded the indigoid product (41P). Taken together, these reactivity results suggest that the effects of different substituent groups and their positions on the benzene ring on product formation do not follow an obvious pattern. The mechanisms behind the substrate specificity and oxidative selectivity remain unknown and require further studies.
Table 2.
P450 BM3T1 as the biocatalyst in the oxidative transformation of indole and its derivatives.
| Substrates | TON a | symmetric : asymmetric b |
|---|---|---|
| 1 | 1226 ± 63 | 92:8 |
| 2 | 1206 ± 33 | 98:2 |
| 3 | 2263 ± 29 | 92:8 |
| 4 | 1557 ± 42 | 59:41 |
| 5 | 2142 ± 43 | 68:32 |
| 6 | 1535 ± 22 | 98:2 |
| 7 | 1970 ± 17 | 91:9 |
| 8 | 2014 ± 42 | 86:14 |
| 9 | 2291 ± 55 | 70:30 |
| 10 | 1677 ± 20 | 99:1 |
| 11 | 1495 ± 39 | 90:10 |
| 12 | 1978 ± 29 | 41:59 |
| 13 | 2191 ± 16 | 90:10 |
| 14 | 1761 ± 23 | 89:11 |
| 15 | 1953 ± 19 | 88:12 |
| 16 | 2082 ± 34 | 93:7 |
| 17 | 1760 ± 22 | 99:1 |
| 18 | 1073 ± 16 | 80:20 |
| 19 | 2331 ± 37 | 52:48 |
| 20 | 1433 ± 21 | 92:8 |
| 21 | 1810 ± 63 | 90:10 |
| 23 | 1783 ± 40 | 61:39 |
| 24 | 1697 ± 18 | 76:24 |
| 26 | 2061 ± 22 | 91:9 |
| 27 | 1679 ± 19 | 72:28 |
| 28 | 1767 ± 35 | 70:30 |
| 29 | 1086 ± 10 | 93:7 |
| 30 | 1663 ± 15 | 98:2 |
| 31 | 1292 ± 23 | 92:8 |
| 32 | 1107 ± 15 | 99:1 |
| 34 | 1182 ± 37 | 38:62 |
| 35 | 1713 ± 23 | 87:13 |
| 37 | 965 ± 14 | 96:4 |
| 39 | 2036 ± 24 | 85:15 |
| 41 | 1756 ± 26 | 92:8 |
TONs were calculated by averaging at least three independent experiments. General reaction condition: 0.1 μM P450-BM3T1 enzyme, 1 mM substrate, and 60 mM H2O2 in 100 mM phosphate buffer (pH 8.0), 30 °C. The reactions were performed for 30 min in order to calculate TONs.
symmetrical : asymmetric: the ratios of indigo and indirubin derivetives were determined by monitoring their maximum absorption peaks using HPLC.
3.3. Structural identification
The two products of the 1 dimerization reaction catalyzed by P450 BM3T1 were confirmed to be indigo and indirubin (98:2) by retention time comparison with authentic standards. Mechanistically, indigo is produced via indoxyl, which subsequently condenses into indigo, while indirubin is generated by the reaction of indoxyl with isatin (i.e., indole-2,3-dione, which is resulted from 3-hydroxylation and subsequent 2-hydroxylation of indole) (Fig. 2). Thus, we speculated that all the indigoid dyes could be formed in the same dimerization manners. To confirm the hypothesis, the two dimerization products of 4 were purified by semi-preparative HPLC. Their structures were determined to be 6,6′-dibromoindigotin and 6,6′-dibromoindirubin by 1H NMR analysis (Figs. S5 and S6). Due to commercial unavailability of authentic standards, other indigoids were identified based on LC-MS analysis and their UV–Vis spectral properties derived from different dimerization modes (Figs. S7 and S8). Moreover, both indigo and indirubin derivatives are potential organic semiconductor molecules that can produce even richer hues when combined. Consequently, we did not separate the mixture resulting from the enzymatic reaction and directly used it for the following transient photocurrent response tests and quantitative color analysis.
3.4. Photophysical properties
The color and absorption maximum of an indigoid dye depend on the type and position of the substituents on the benzene rings. The spectroscopic properties of the derivatives (Fig. 3 and S7) were found to be similar to those of indigo and indirubin. Prior to characterizing indigo/indirubin derivatives, we examined the optical and electrochemical properties of indigo and indirubin. The absorption maxima of indigo and indirubin were determined to be 609 and 541 nm in 80% methanol, respectively, consistent with the previous report [44]. All the indigoid dyes prepared by P450 BM3T1 were formed through the same dimerization ways. The spectral properties for symmetrical/asymmetric dimerization products with the same substituent(s) could be readily distinguished by their UV–Vis spectroscopy. In this study, all absorption spectra of indigo derivatives were recorded using HPLC UV–Vis spectroscopy in 80% methanol (Fig. S7). The colorful indigo derivatives exhibited maximum absorbance at 570–650 nm, while indirubin derivatives showed an absorption band between 510 and 580 nm, depending on the identity and location of the substituents. These results are well consistent with previously reported spectroscopic parameters [[45], [46], [47]].
Specifically, the absorption spectra of 2P, 3P, 10P, and 11P with their Br or Cl substituents at C4 or C5 positions appeared quite similar to those of indigo and indirubin. The dihalogenoindigos 4P, 5P, 8P, 9P, 12P and 13P with their halogen substituents at C6 or C7 positions showed bathochromic shifts when compared with indigo and indirubin, and the magnitude of these shifts became bigger with more electronegative halogen substituents: F (8P and 9P) > Cl (12P and 13P) ≈ Br (4P and 5P). Other strongly electron deficient substituents such as NO2 (23 and 24), CF3 (30 and 31), and CN (27–28) resulted in comparable absorption band shifts when positioned at the same location. Notably, when these substitutes were located at C5, there was a decrease in the absorption band around 590 and 530 nm for the indigo and indirubin derivatives (23P, 27P, and 30P), while a stronger absorption peak appeared at 643/550 nm (24P) or 625/556 nm (28P) with the C6 substituents. However 6,6′-bis(trifluoromethyl)indigo/indirubin (31P) showed similar absorption spectra at 610/539 nm compared to indigo and indirubin. The absorption spectra of 26P and 29P, with CN substituents at the C4 or C7 positions, exhibited absorptions at 620/515 nm and 593/534 nm, respectively. The tetrafluoroindigo (14–17P) containing F/Cl/Br substituents at positions C5/6, C5/6, C4/7, and C5/7 caused opposite electronic effects. The absorption band of 16P had its maximum at 582/519 nm, while 14P, 15P, and 17P exhibited maximum absorption bands similar to those of indigo and indirubin (1P). For the –COOCH3 group, the absorption spectra of C5-substituted indigoids (34P) had a maximum absorption wavelength at 593/530 nm, while the absorption spectra of C6 substituted indigoids (35P) were similar to those of indigo/indirubin (1P) at 610/551 nm. The ethyl carboxylate group at the C5 position resulted in a products mixture (41P) with absorption maxima at 589/530 nm. The electron-donating –OCH3 group resulted in hypsochromic shifts when located at the C5 or C7 position, with peaks blueshifted to 646/595 nm (37P) and 647/621 nm (39P), respectively. The spectral changes were attributed to both the type(s) and position(s) of substitutes. Thus, the different substituent positions exert significant impacts on the maximum absorption wavelength of indigoids even with the same substituents. For example, the substitution of an electron deficient group –NO2 at the C6 position (24), resulted in an increase in the absorption band to 643/550 nm, while substitution at the C5 position (23) led to a decrease to 596/535 nm. Such a bathochromic/hypsochromic shift could be explained by the extension of the conjugated system during the complexation process.
Both indigo and indirubin derivatives have intriguing colors. When combined in a mixture, they have the ability to modulate and produce even richer colors. The combination of these colors can create amazing effects and add more colors to our lives. Therefore, in this study, we did not separate the mixtures resulting from the enzymatic reactions. Interestingly, the final colors of the enzymatic products were not only determined by the types and positions of substituents, but also related to the ratio between the symmetrical/asymmetrical products. For example, 4P exhibited absorption bands at approximately 595/540 nm in 80% methanol with the ratio of 59:41 (Table 2). A bathochromic shift was observed in the absorption spectrum of a mixture when the enzymatic products were not separated. This also happened when the halogen substitutions (F, Cl, and Br) were located at the C6 position. Taken together, we envision that this enzymatic approach could be applied to generate a variety of dyes in the future.
We utilized a red-green-blue (RGB) color model to quantitatively analyze the enzyme-synthesized dyes. The RGB values were acquired using Photoshop's Color Picker. Indigo is a mesmerizing hue that strikes a balance between violet and blue with the RGB value of (63, 15, 183) (https://www.color-name.com/indigo-blue.color). The RGB value of indirubin is (187, 36, 172) [48]. In this work, the enzymatic reaction mixture containing indigo and small amounts of indirubin (1P) displayed an RGB color code of (19, 34, 53). An impressive triadic color palette was generated by indigoids with varying absorption properties through an enzymatically controlled process. When these mixed colored components reconstructed, the blue hues could be more brighter and the red hues could be more intense. Enzymatically adjusting the concentrations of the blue indigo derivatives and the reddish indirubin derivatives resulted in a visually stunning color spectrum (Fig. 3).
3.5. Electrochemical properties
Organic semiconductor photocatalytic materials require high efficiency and a wide spectrum of optical absorption performance. The light absorption performance of an organic semiconductor depends on its high occupied molecular orbital (HOMO)-low occupied molecular orbital (LUMO) energy level (band gap). As the band gap becomes narrow, the molecule becomes easy to be excited, and the wavelength of light absorbed correspondingly becomes longer. Accordingly, the band gap values were determined by Tauc plots calculated from UV–vis spectra of the prepared indigoid organic photocatalysts (Fig. S9). Indigo and indirubin have been reported to have a band gap value of about 2.5 eV and 2.75 eV, respectively [49]. Herein, we report the syntheses of symmetrical and asymmetrical indigoids.
Compared with indigo, the prepared indigoids had narrower band gap values ranging from 1.51 to 1.82 eV, indicating that electrons and holes of indigoid systems were more easily separated under light irradiation. The presence of an electron attractor group NO2 on the indigo molecules 23P and 24P had the lowest bandgap values of 1.51 and 1.56 eV, respectively, which were approximately 1.0 eV lower than those of indigo and indirubin. The narrower band state of indigo derivatives, less radiation electron-hole recombination, and broad light-harvesting capability would promote the photoactivity of the materials. Furthermore, photocurrent response tests were used to study the photocarrier separation efficiency, which is considered a reliable basis for evaluating the photoelectrochemical performance of photocatalysts. Considering that both indigo derivatives and indirubin derivatives are potential organic semiconductor molecules, the resulting mixtures were used for transient photocurrent response tests without further purification. Under cyclic on/off illumination, the transient photocurrent curves of the enzymatically prepared indigoids were determined. The photocurrent values of the prepared indigoids were no less than 0.07 μA cm−2 (Fig. 4, Fig. S10). With values of most indigo derivatives outclassing the indigo, the photocurrent values of most prepared indigoids were greater than 0.12 μA cm−2. Moreover, more than one-third of the indigoids gave the photocurrent values that were more than twice the photocurrent value of indigo. Notably, 4P showed the highest photocurrent value of 0.84 ± 0.0058, which is 6 times higher than that of indigo. This indicates that compared with indigo, 4P has more efficient photocarrier transfer process and higher charge separation efficiency. These results suggest that in addition to altering the energy difference between HOMO and LUMO, the substituted functional groups also play a key role in the charge transport that is essential for the photoelectric properties of the prepared indigo derivatives. Using the tuned catalysts, we expect that these indigo derivatives will provide some applicable electronics and sensors candidates.
Fig. 4.
Photocurrents and bandgap values for indigoids of the Tauc plot.
4. Discussion
Indigoids are a fascinating class of natural products which have attracted interests in several application areas, such as dyeing, pharmaceutical, food and semiconductor industries. Indigo and Tyrian purple are two dyes that have been used worldwide for quite a long time. Indirubin is an active component of Chinese Medicine Danggui Longhui used to treat the symptoms of leukemia, inflammation, psoriasis and skin rashes [50]. 5,5′-Dimethoxyindirubin has been found to be a potent inhibitor of human cyclin-dependent kinases (CDK) and glycogen synthase kinase-3 (GSK-3) [51]. In addition, indigoid dyes have been considered as good active materials for battery applications and organics electronics, due to their good stability and electrical properties [8]. Indigoids with a broad range of biological activities are also high-mobility organic semiconductors and photoresponsive materials [52].
Indigo is a planar molecule with a symmetrical trans structure isomer condensing two indole rings, in which two intramolecular hydrogen bonds are formed between the adjacent carbonyl and imino groups. Indirubin is an asymmetric isomer of indigo via condensation of two indole rings, and there is only one intramolecular hydrogen bond in indirubin. Indigo is a more conjugated planar structure than indirubin, thereby having a lower HOMO-LUMO energy gap possess, thereby having higher absorption wavelength [53]. The insolubility of indigoids warrants them excellent properties of washfastness, but it may also be the reason for why little structural analysis data are available. Nonetheless, in this study we found that the indigoid (indigo/indirubin) derivatives could be conveniently identified by LC-MS and UV–Vis spectroscopy.
Modified indigo derivatives are difficult to synthesize chemically. The improved processability and electron-transport property of indigo derivatives by enzymatic modifications could transform them into novel and inexpensive building blocks for organic electronics. The demand for biocatalysts to prepare indigoids with novel properties is rising.
5. Conclusions
In this study, we reported successful production of a set of indigoids by a simple P450/H2O2 system, providing a biotechnological alternative to chemical synthesis. The enzymatic reactions described herein are efficient under mild conditions without the requirement of expensive redox partner proteins and NADPH [54]. Importantly, the electron density of indigoids could be conveniently altered by introduction of electron-donating or electron-withdrawing groups on the benzene ring by substituting the hydrogen atoms at the 4-, 5-, 6- and/or 7-positions of indole precursors. We found that the introduction of both electron-withdrawing and electron-donating groups can affect HOMO and LUMO energy levels of indigoids. These observations could be helpful in the design of new indigoids with superior charge-transport properties and ambient stability. A series of indigo derivatives with extended π-conjugated system have been synthesized, characterized and investigated as semiconductor materials [8]. The use of the H2O2 as electron acceptor will significantly lower the dye production costs in large-scale industrial process. Altogether, our results suggest that the enzyme engineering of P450 BM3T1 should be further explored for enzymatic synthesis of a larger number of promising indigoids. Also, the revealed structure-properties relationships will be useful for the rational design of a new generation of indigo-based semiconductors with improved electronic properties for sustainable and biocompatible organic electronics.
Funding
This work was supported by the National Key Research and Development Program of China (2019YFA0905100), the National Natural Science Foundation of China (32025001, 32071266 and 32170088), the Shandong Provincial Natural Science Foundation (ZR2019ZD20), and the State Key Laboratory of Microbial Technology Open Projects Fund (Project NO. M2022-01).
CRediT authorship contribution statement
Li Ma: conceived, and designed the experiment, performed the experiment, prepared the manuscript. Tianjian Sun: performed the experiment. Yunjie Liu: performed the experiment. Yue Zhao: performed the experiment. Xiaohui Liu: performed the experiment. Yuxuan Li: analyzed the data. Xinwei Chen: analyzed the data. Lin Cao: measured the electrochemical properties of the prepared indigoids. Qianqian Kang: measured the electrochemical properties of the prepared indigoids. Jiawei Guo: analyzed the data. Lei Du: analyzed the data. Wei Wang: measured the electrochemical properties of the prepared indigoids, prepared the manuscript. Shengying Li: conceived, and designed the experiment, prepared the manuscript, All authors discussed the results and approved the final version of the manuscript.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgements
We thank Jingyao Qu and Guannan Lin from the Core Facilities for Life and Environmental Sciences, State Key Laboratory of Microbial Technology at Shandong University for their help in HRMS testing.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.synbio.2023.06.006.
Contributor Information
Wei Wang, Email: wangwei8038@ouc.edu.cn.
Shengying Li, Email: lishengying@sdu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Glowacki E.D., Voss G., Sariciftci N.S. 25th anniversary article: progress in chemistry and applications of functional indigos for organic electronics. Adv Mater. 2013;25(47):6783–6799. doi: 10.1002/adma.201302652. [DOI] [PubMed] [Google Scholar]
- 2.Kettner F., Huter L., Schafer J., Roder K., Purgahn U., Krautscheid H. Selective crystallization of indigo B by a modified sublimation method and its redetermined structure Acta. Crystallography: Crystallogr Commun. 2011;67:o2867. doi: 10.1107/S1600536811040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irimia-Vladu M., Glowacki E.D., Troshin P.A., Schwabegger G., Leonat L., Susarova D.K., Krystal O., Ullah M., Kanbur Y., Bodea M.A., Razumov V.F., Sitter H., Bauer S., Sariciftci N.S. Indigo-a natural pigment for high performance ambipolar organic field effect transistors and circuits. Adv Mater. 2012;24(3):375–380. doi: 10.1002/adma.201102619. [DOI] [PubMed] [Google Scholar]
- 4.Bisri S.Z., Piliego C., Gao J., Loi M.A. Outlook and emerging semiconducting materials for ambipolar transistors. Adv Mater. 2014;26(8):1176–1199. doi: 10.1002/adma.201304280. [DOI] [PubMed] [Google Scholar]
- 5.Ma Q., Zhang X.W., Qu Y.Y. Biodegradation and biotransformation of indole: advances and perspectives. Front Microbiol. 2018;9:2625. doi: 10.3389/fmicb.2018.02625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glowacki E.D., Voss G., Leonat L., Irimia-Vladu M., Bauer S., Sariciftci N.S. Indigo and tyrian purple - from ancient natural dyes to modern organic semiconductors. Isr J Chem. 2012;52(6):540–551. doi: 10.1002/ijch.201100130. [DOI] [Google Scholar]
- 7.Klimovich I.V., Leshanskaya L.I., Troyanov S.I., Anokhin D.V., Novikov D.V., Piryazev A.A., Ivanov D.A., Dremova N.N., Troshin P.A. Design of indigo derivatives as environment-friendly organic semiconductors for sustainable organic electronics. J Mater Chem C. 2014;2(36):7621–7631. doi: 10.1039/c4tc00550c. [DOI] [Google Scholar]
- 8.Klimovich I.V., Zhilenkov A.V., Кuznetsova L.I., Frolova L.A., Yamilova O.R., Troyanov S.I., Lyssenko K.A., Troshin P.A. Novel functionalized indigo derivatives for organic electronics. Dyes Pigments. 2021;186 [Google Scholar]
- 9.Glowacki E.D., Romanazzi G., Yumusak C., Coskun H., Monkowius U., Voss G., Burian M., Lechner R.T., Demitri N., Redhammer G.J., Sunger N., Suranna G.P., Sariciftci S. Epindolidiones-versatile and stable hydrogen-bonded pigments for organic field-effect transistors and light-emitting diodes. Adv Funct Mater. 2015;25(5):776–787. doi: 10.1002/adfm.201402539. [DOI] [Google Scholar]
- 10.Li J.J. In: Name reactions: a collection of detailed reaction mechanisms. Li J.J., editor. Springer Berlin Heidelberg; Berlin, Heidelberg: 2002. Baeyer-Drewson indigo synthesis; pp. 14–15. [DOI] [Google Scholar]
- 11.Shadi I.T., Chowdhry B.Z., Snowden M.J., Withnall R. Analysis of the conversion of indigo into indigo carmine dye using. SERRS Chem Commun. 2004;(12):1436–1437. doi: 10.1039/b403601h. [DOI] [PubMed] [Google Scholar]
- 12.Hu Y.Z., Zhang D., Thummel R.P. Friedländer approach for the incorporation of 6-bromoquinoline into novel chelating ligands. Org Lett. 2003;5(13):2251–2253. doi: 10.1021/ol034559q. [DOI] [PubMed] [Google Scholar]
- 13.Tanoue Y., Sakata K., Hashimoto M., Hamada M., Kai N., Nagai T. A facile synthesis of 6,6 '- and 5,5 '-dihalogenoindigos Dyes. Pigm. 2004;62(2):101–105. doi: 10.1016/S0143-7208(03)00150-5. [DOI] [Google Scholar]
- 14.Gillam E.M.J., Aguinaldo A.M.A., Notley L.M., Kim D., Mundkowski R.G., Volkov A.A., Arnold F.H., Soucek P., DeVoss J.J., Guengerich F.P. Formation of indigo by recombinant mammalian cytochrome P450. Biochem Biophys Res Commun. 1999;265(2):469–472. doi: 10.1006/bbrc.1999.1702. [DOI] [PubMed] [Google Scholar]
- 15.Choi H.S., Kim J.K., Cho E.H., Kim Y.C., Kim J.I., Kim S.W. A novel flavin-containing monooxygenase from Methylophaga sp strain SK1 and its indigo synthesis in Escherichia coli. Biochem Biophys Res Commun. 2003;306(4):930–936. doi: 10.1016/S0006-291x(03)01087-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X.W., Qu Y.Y., Ma Q., Zhou H., Li X.L., Kong C.L., Zhou J.T. Cloning and expression of naphthalene dioxygenase genes from Comamonas sp MQ for indigoids production Process. Biochemist. 2013;48(4):581–587. doi: 10.1016/j.procbio.2013.02.008. [DOI] [Google Scholar]
- 17.Ensley B.D., Ratzkin B.J., Osslund T.D., Simon M.J., Wackett L.P., Gibson D.T. Expression of naphthalene oxidation genes in Escherichia Coli results in the biosynthesis of indigo. Science. 1983;222(4620):167–169. doi: 10.1126/science.6353574. [DOI] [PubMed] [Google Scholar]
- 18.Fabara A.N., Fraaije M.W. An overview of microbial indigo-forming enzymes. Appl Microbiol Biotechnol. 2020;104(3):925–933. doi: 10.1007/s00253-019-10292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermod N., Harayama S., Timmis K.N. New route to bacterial production of indigo. Nat Biotechnol. 1986;4:321–324. doi: 10.1038/nbt0486-321. [DOI] [Google Scholar]
- 20.Rui L.Y., Reardon K.F., Wood T.K. Protein engineering of toluene ortho-monooxygenase of Burkholderia cepacia G4 for regiospecific hydroxylation of indole to form various indigoid compounds. Appl Microbiol Biotechnol. 2005;66(4):422–429. doi: 10.1007/s00253-004-1698-z. [DOI] [PubMed] [Google Scholar]
- 21.McClay K., Boss C., Keresztes I., Steffan R.J. Mutations of toluene-4-monooxygenase that alter regiospecificity of indole oxidation and lead to production of novel indigoid pigments. Appl Environ Microbiol. 2005;71(9):5476–5483. doi: 10.1128/AEM.71.9.5476-5483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Berkel W.J.H., Kamerbeek N.M., Fraaije M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol. 2006;124(4):670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 23.Gillam E.M.J., Notley L.M., Cai H.L., De Voss J.J., Guengerich F.P. Oxidation of indole by cytochrome P450 enzymes. Biochemistry. 2000;39(45):13817–13824. doi: 10.1021/bi001229u. [DOI] [PubMed] [Google Scholar]
- 24.Zhang X., Guo J., Cheng F., Li S. Cytochrome P450 enzymes in fungal natural product biosynthesis. Nat Prod Rep. 2021;38(6):1072–1099. doi: 10.1039/d1np00004g. [DOI] [PubMed] [Google Scholar]
- 25.Li Z., Jiang Y., Guengerich F.P., Ma L., Li S., Zhang W. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J Biol Chem. 2020;295(3):833–849. doi: 10.1074/jbc.REV119.008758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Durairaj P., Li S. Functional expression and regulation of eukaryotic cytochrome P450 enzymes in surrogate microbial cell factories. Eng Microbiol. 2022;2(1) [Google Scholar]
- 27.Guengerich F.P. Common and uncommon cytochrome P450 reactions related to metabolism and chemical toxicity. Chem Res Toxicol. 2001;14(6):611–650. doi: 10.1021/tx0002583. [DOI] [PubMed] [Google Scholar]
- 28.Narhi L.O., Fulco A.J. Identification and characterization of two functional domains in cytochrome P-450BM-3, a catalytically self-sufficient monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1987;262(14):6683–6690. [PubMed] [Google Scholar]
- 29.Li Q.S., Ogawa J., Schmid R.D., Shimizu S. Indole hydroxylation by bacterial cytochrome P450 BM-3 and modulation of activity by cumene hydroperoxide. Biosci Biotechnol Biochem. 2005;69(2):293–300. doi: 10.1271/bbb.69.293. [DOI] [PubMed] [Google Scholar]
- 30.Li Q.S., Ogawa J Fau - Shimizu S., Shimizu S. Critical role of the residue size at position 87 in H2O2- dependent substrate hydroxylation activity and H2O2 inactivation of cytochrome P450BM-3. Biochem Biophys Res Commun. 2001;280(5):1258–1261. doi: 10.1006/bbrc.2001.4261. [DOI] [PubMed] [Google Scholar]
- 31.Kong F., Chen J., Qin X., Liu C., Jiang Y., Ma L., Xu H., Li S., Cong Z. Evolving a P450BM3 peroxygenase for the production of indigoid dyes from indoles. ChemCatChem. 2022;14(24) doi: 10.1002/cctc.202201151. [DOI] [Google Scholar]
- 32.Ma L., Li F., Zhang X., Chen H., Huang Q., Su J., Liu X., Sun T., Fang B., Liu K., Tang D., Wu D., Zhang W., Du L., Li S. Development of MEMS directed evolution strategy for multiplied throughput and convergent evolution of cytochrome P450 enzymes. Sci China Life Sci. 2022;65(3):550–560. doi: 10.1007/s11427-021-1994-1. [DOI] [PubMed] [Google Scholar]
- 33.Anzai Y., Li S., Chaulagain M.R., Kinoshita K., Kato F., Montgomery J., Sherman D.H. Functional analysis of MycCI and MycG, cytochrome P450 enzymes involved in biosynthesis of mycinamicin macrolide antibiotics. Chem Biol. 2008;15(9):950–959. doi: 10.1016/j.chembiol.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xue Y.Q., Wilson D., Zhao L.S., Liu H.W., Sherman D.H. Hydroxylation of macrolactones YC-17 and narbomycin is mediated by the pikC-encoded cytochrome P450 in Streptomyces venezuelae. Chem Biol. 1998;5(11):661–667. doi: 10.1016/S1074-5521(98)90293-9. [DOI] [PubMed] [Google Scholar]
- 35.Poulos T.L., Finzel B.C., Howard A.J. High-resolution crystal structure of cytochrome P450cam. J Mol Biol. 1987;195(3):687–700. doi: 10.1016/0022-2836(87)90190-2. [DOI] [PubMed] [Google Scholar]
- 36.Boddupalli S.S., Estabrook R.W., Peterson J.A. Fatty acid monooxygenation by cytochrome P-450BM-3. J Biol Chem. 1990;265(8):4233–4239. [PubMed] [Google Scholar]
- 37.Guengerich F.P., Martin M.V., Sohl C.D., Cheng Q. Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nat Protoc. 2009;4(9):1245–1251. doi: 10.1038/nprot.2009.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voss G. Photophysical and spectroscopic studies of indigo derivatives in their keto and leuco forms. J Soc Dye Colour. 2000;116:87–90. doi: 10.1021/jp049076y. [DOI] [Google Scholar]
- 39.Makuła P., Pacia M., Macyk W. How to correctly determine the band gap energy of modified semiconductor photocatalysts based on UV-vis spectra. J Phys Chem Lett. 2018;9(23):6814–6817. doi: 10.1021/acs.jpclett.8b02892. [DOI] [PubMed] [Google Scholar]
- 40.Reetz M.T., Kahakeaw D., Lohmer R. Addressing the numbers problem in directed evolution. Chem BioChem. 2008;9(11):1797–1804. doi: 10.1002/cbic.200800298. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y., Li Z., Wang C., Zhou Y.J., Xu H., Li S. Biochemical characterization of three new alpha-olefin-producing P450 fatty acid decarboxylases with a halophilic property. Biotechnol Biofuels. 2019;12:79. doi: 10.1186/s13068-019-1419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao P., Kong F., Jiang Y., Qin X., Tian X., Cong Z. Enabling peroxygenase activity in cytochrome P450 monooxygenases by engineering hydrogen peroxide tunnels. J Am Chem Soc. 2023;145(9):5506–5511. doi: 10.1021/jacs.3c00195. [DOI] [PubMed] [Google Scholar]
- 43.Anokhin D.V., Leshanskaya L.I., Piryazev A.A., Susarova D.K., Dremova N.N., Shcheglov E.V., Ivanov D.A., Razumov V.F., Troshin P.A. Towards understanding the behavior of indigo thin films in organic field-effect transistors: a template effect of the aliphatic hydrocarbon dielectric on the crystal structure and electrical performance of the semiconductor. Chem Commun. 2014;50(57):7639–7641. doi: 10.1039/c4cc02431a. [DOI] [PubMed] [Google Scholar]
- 44.Yin H., Chen H., Yan M., Li Z., Yang R., Li Y., Wang Y., Guan J., Mao H., Wang Y., Zhang Y. Efficient bioproduction of indigo and indirubin by optimizing a novel terpenoid cyclase XiaI in Escherichia coli. ACS Omega. 2021;6(31):20569–20576. doi: 10.1021/acsomega.1c02679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shriver J.A., Kaller K.S., Kinsey A.L., Wang K.R., Sterrenberg S.R., Van Vors M.K., Cheek J.T., Horner J.S. A tunable synthesis of indigoids: targeting indirubin through temperature. RSC Adv. 2022;12(9):5407–5414. doi: 10.1039/d2ra00400c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sadler P.W. Absorption spectra of indigoid dyes. J Org Chem. 1956;21(3):316–318. doi: 10.1021/jo01109a014. [DOI] [Google Scholar]
- 47.Watanabe M., Uemura N., Ida S., Hagiwara H., Goto K., Ishihara T. 5,5 '-alkylsubsituted indigo for solution-processed optoelectronic devices. Tetrahedron. 2016;72(29):4280–4287. doi: 10.1016/j.tet.2016.05.069. [DOI] [Google Scholar]
- 48.Hartl A., Proaño Gaibor A.N., van Bommel M.R., Hofmann-de Keijzer R. Searching for blue: experiments with woad fermentation vats and an explanation of the colours through dye analysis. J Archaeol Sci. 2015;2:9–39. doi: 10.1016/j.jasrep.2014.12.001. [DOI] [Google Scholar]
- 49.Ju Z., Sun J., Liu Y. Molecular structures and spectral properties of natural indigo and indirubin: experimental and DFT studies. Molecules. 2019;24(21):3831. doi: 10.3390/molecules24213831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoessel R., Leclerc S., Endicott J.A., Nobel M.E.M., Lawrie A., Tunnah P., Leost M., Damiens E., Marie D., Marko D., Niederberger E., Tang W.C., Eisenbrand G., Meijer L. Indirubin, the active constituent of a Chinese antileukaemia medicine, inhibits cyclin-dependent kinases. Nat Cell Biol. 1999;1(1):60–67. doi: 10.1038/9035. [DOI] [PubMed] [Google Scholar]
- 51.Guengerich F.P., Sorrells J.L., Schmitt S., Krauser J.A., Aryal P., Meijer L. Generation of new protein kinase inhibitors utilizing cytochrome P450 mutant enzymes for indigoid synthesis. J Med Chem. 2004;47(12):3236–3241. doi: 10.1021/jm030561b. [DOI] [PubMed] [Google Scholar]
- 52.Choi K.-Y. A review of recent progress in the synthesis of bio-indigoids and their biologically assisted end-use applications Dyes. Pigm. 2020;181 doi: 10.1016/j.dyepig.2020.108570. [DOI] [Google Scholar]
- 53.Jacquemin D., Preat J., Wathelet V., Perpete E.A. Substitution and chemical environment effects on the absorption spectrum of indigo. J Chem Phys. 2006;124(7) doi: 10.1063/1.2166018. [DOI] [PubMed] [Google Scholar]
- 54.Du L., Li S. Compartmentalized biosynthesis of fungal natural products. Curr Opin Biotechnol. 2021;69:128–135. doi: 10.1016/j.copbio.2020.12.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.