Abstract
Background and Objective:
The endoscopic diagnosis of pathological changes in the gastroesophageal junction including esophagitis and Barrett's mucosa is based on the visual detection of two boundaries: mucosal color change between esophagus and stomach, and top endpoint of gastric folds. The presence and pattern of mucosal breaks in the gastroesophageal mucosal junction (Z line) classify esophagitis in patients and the distance between the two boundaries points to the possible columnar lined epithelium. Since visual detection may suffer from intra- and interobserver variability, our objective was to define the boundaries automatically based on image processing algorithms, which may enable us to measure the detentions of changes in future studies.
Methods:
To demarcate the Z-line, first the artifacts of endoscopy images are eliminated. In the second step, using SUSAN edge detector, Mahalanobis distance criteria, and Gabor filter bank, an initial contour is estimated for the Z-line. Using region-based active contours, this initial contour converges to the Z-line. Finally, by applying morphological operators and Gabor Filter Bank to the region inside of the Z-line, gastric folds are segmented.
Results:
To evaluate the results, a database consisting of 50 images and their ground truths were collected. The average dice coefficient and mean square error of Z-line segmentation were 0.93 and 3.3, respectively. Furthermore, the average boundary distance criteria are 12.3 pixels. In addition, two other criteria that compare the segmentation of folds with several ground truths, i.e., Sweet-Spot Coverage and Jaccard Index for Golden Standard, are 0.90 and 0.84, respectively.
Conclusions:
Considering the results, automatic segmentation of Z-line and gastric folds are matched to the ground truths with appropriate accuracy.
Keywords: Adenocarcinoma, Barrett's esophagus, demarcating Z-line and gastric folds boundary, segmentation of lower esophageal sphincter endoscopy images
Introduction
Today with the tendency of societies toward new diets, the prevalence of gastroesophageal reflux disease as a common digestive disorder has been on rise.[1] Fortunately, the development of various endoscopic imaging systems allows the diagnosis of these diseases. The most common methods used for the diagnosis and categorization of these diseases are based on the physician's estimation from a suspected tissue length during the endoscopy process.[2]
As shown in Figure 1, the anatomical line indicates the gastroesophageal junction and the histological Z-line reveals the gastrointestinal junction tissue. In normal cases, the transition region (i.e., where the stomach tissue ends and the esophagus tissue begins) is located right at the end of the esophagus. In the abnormal case, as shown in Figure 1, these two lines are separated from each other. Therefore, it is highly important to identify two regions in lower esophagus sphincter (LES):
Figure 1.

Shematic Ilustration of abnormal LES image, adapted from Figure 1 of[3]
The junction of gastroesophageal constructions, which marks the end of stomach folds.
The junction of gastrointestinal tissues, which is marked with the red-pink line (Z-line).
The distance between these two regions is so crucial for a clinician's diagnosis. However, since physicians tend to estimate the tissue length without any benchmark, in many cases, the precise location of these two regions is determined erroneously.[2] On the other hand, common methods developed to diagnose and categorize these diseases are based on measuring the length of these regions. Both Los Angeles (LA) classification for esophagitis[4] and Prague criteria for Barrett's esophagus (BE)[5] are based on quantitative criteria to measure distances. Therefore, it is clear that increasing the accuracy of measurements improves the diagnosis quality of these two methods. For this purpose and after consultation with gastrointestinal clinicians, we introduced an image processing method to segment Z-line and gastric folds to reinforce accurate diagnosis. In the following, we will first review the relevant literature.
Several methods have been proposed to help physicians diagnose esophagitis. A systematic review of recent studies is presented in.[6] It focuses on the automatic detection of the neoplastic region for classification purposes under different endoscopy modalities. Another study that discusses advances in esophagitis computer-aided diagnosis systems is presented.[7] It starts with a brief introduction to endoscopy modalities used for esophageal examination and deals with the detection methods recently developed for BE detection. One of the first studies that directly employed image processing techniques for BE detection was proposed by Kim et al. in 1995.[8] The goal of this paper was to prepare a two-dimensional map of the esophagus to demarcate Barrett's metaplasia. The authors examined their method both in a cylindrical model of the esophagus and also in human samples. Seo et al.[9] classified esophagitis in four grades based on the LA classification criteria. Their algorithm separated these four grades with a success ratio of 96% using a neural network and texture analysis. Santosh et al.[10] attempted to classify the esophagitis by LA classification using low-level image features and a neural network classifier. According to their results, as grades approach to level D, the texture changes grew more intense and therefore the features could be distinguished with greater accuracy. Hence, in grades like level A where there are small changes in the texture, the results are inaccurate. Thus, as the authors suggested, accurate segmentation of the Z-line and stomach would improve the distinguishing ability of the LA classification. In a study by Serpa-Andrade et al.,[11] an approach based on the analysis of esophageal irregularities (the Z-line) was proposed. To analyze these irregularities, they applied the Fourier transform to the shape signature of the Z-line and classified images into the healthy and the diseased categories. Yousefi et al.[12] proposed a hybrid algorithm that uses spatial fuzzy c-mean and level set methods for BE segmentation. They concluded that this approach was suitable for the automatic segmentation of esophagus metaplasia. However, their method was only tested on four LES images and might not provide accurate results for different patients with almost different LES images. In a study by Van Der Sommen et al.,[13] a supportive automatic system was proposed to annotate early esophageal cancerous tissue in high-definition endoscopic images. The proposed algorithm computes local color and texture features both in the original image and also in the Gabor-filtered image. Then, by exploring the spectral characteristics of the image and by applying a trained support vector machine (SVM), the suspected tissue was segmented. Souza et al.[14] proposed a method to examine the feasibility of adenocarcinoma classification in endoscopic images. The algorithm starts with the extraction of speed-up robust features (SURF), which are employed with SVM for training and testing purposes. Souza et al.[15] investigated the use of the optimum-path forest (OPF) classifier for the automatic identification of BE. They consider describing endoscopic images using feature extractors based on key information, such as the SURF and scale-invariant feature transform. They further designed a bag of visual words that is used to feed both OPF and SVM classifiers. The best results were obtained using the OPF classifier for both feature extraction methods. Mendel et al.[16] investigated the application of deep learning in specialist-annotated images containing adenocarcinoma and BE's disease. The convolutional neural network was applied to a set of images by adopting a transfer learning approach in a leave-one-patient-out cross-validation. The study demonstrated the generalizability of results to the BE's segmentation domain.
Following these studies, our contributions are as follows:
First, since there was no dataset for this study, we collected a dataset for this purpose. Therefore, we gathered and annotated LES images for both gastric folds and Z-line by three physicians. Second, as we noticed there is a huge disagreement between physicians themselves for determining the annotation of gastric folds. Therefore, we collected three Ground Truths for each image and compared our algorithms with the intersection of them to handle inter-observer variability. Third, we evaluate the feasibility of developing machine vision algorithms to assist physicians for a reliable segmentation the Z-line and Gastric folds in disease diagnosis. For this purpose, a classic image processing algorithm is developed and its results are compared with U-Net,[17] which is a popular State-of-the-Art convolutional neural network (CNN) architecture in semantic segmentation models.
Obviously, these automatic segmentations do not resolve all the problems, as we cannot decide whether mucus is cancerous or not. We only segment Z-line and gastric folds and these segmentations indirectly lead to enhanced diagnosis of esophagitis. In Section 2, our suggested method for Z-line and gastric folds segmentation is presented. In Section 3.1, we will explain the collected database. Finally, in Section 3, the results of our algorithm are compared to those presented by other gastroenterologists.
Methods
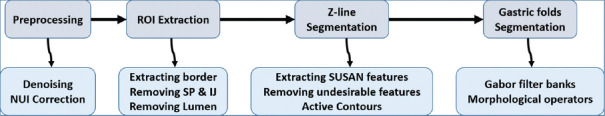
Figure 2 shows our proposed method consists of four major parts including preprocessing, extraction of the region of interest (ROI), Z-line, and gastric folds segmentation. In the preprocessing step, noise, and nonuniform illumination (NUI) are removed using nonlinear diffusion (NLD) filtering and local normalization. In the next step, regions with no useful information (e.g., regions with an intense lightening), are omitted. In the last two steps, Z-line and gastric folds are segmented using SUSAN edge detector, active contours, and Gabor filters. It should be noted that as gastric folds are usually inside the Z-line, we first offer a rough estimation of Z-line and then gastric folds are searched inside the Z-line region.
Figure 2.
Proposed method
Preprocessing
One major source of noise in the endoscopic images is noise induced by cables and imaging devices. To eliminate the noise while preserving the edges, the NLD filter was used. NLD which was proposed by Perona and Malik,[18] removes noise by diffusing the image gradient in the neighboring pixels. This diffusion could be suppressed at the boundaries by a proper selection of filter parameters. In each dimension, this process could be formulated mathematically as follows:

In this equation, the diffusion strength is controlled by a constant coefficient.  and ∇I2(x) are image gradients at the right and left side of point x respectively. These gradients are computed and added to the current image intensity in each iteration after passing through the diffusion function fexp. The discussion for the selection of this function is as follows. Indeed, two different functions for f are already proposed:
and ∇I2(x) are image gradients at the right and left side of point x respectively. These gradients are computed and added to the current image intensity in each iteration after passing through the diffusion function fexp. The discussion for the selection of this function is as follows. Indeed, two different functions for f are already proposed:

Figure 3 shows these two functions for k = 5 and α = 1. As illustrated in this figure, for an equal gradient, the exponential function has a greater descent corresponding to the fractional one. Therefore, this function is more useful for preserving edges. In addition, by taking derivative from these two functions, it is obvious that their maximum happens at ∇li (x) = k for ffrac and  for fexp (if α = 1) respectively.
for fexp (if α = 1) respectively.
Figure 3.

Exponential and fractional functions used in non-linear diffusion filters for k = 5 and α = 1
According to this exponential function, we can effectively control the noise diffusion procedure and simultaneously preserve image edges by κ and α parameters. For our dataset, these two parameters are set to 0.125 and 1 respectively. The result of our proposed denoising algorithm in a sample data is shown in Figure 4.
Figure 4.

(a) Original image (b) Denoised image, artifacts and other highpass noises are removed but Z-line edge is preserved
In the next step, the local NUI is corrected. Figure 5a shows a sample image with increased brightness from the left to the right. These NUI can influence the final results of image segmentation. One way to remove these effects is to normalize each pixel's brightness based on its neighbor information. For this purpose, the average and standard deviation for a Gaussian window around each image pixel were calculated. Then, the brightness of this pixel is normalized according to the following equation:
Figure 5.

(a) Original LES image, (b) Output of NUI correction

where mI (x, y) and σI (x, y) are average and standard deviation of pixels in the window, respectively. The window size determines the neighbor information used for correcting the local NUI. As the window size is increased, more far pixels would influence the mean and variance, thereby making the algorithm nonlocal. For our data set, the window size was set to 55 pixel.
Extracting region of interest
In the endoscopy images, there are usually areas that either do not contain any information or have false information, which affects the subsequent processing. As an example, when the lower LES is opened, there is a black region with no information in the image. In the following section, we attempt to identify these areas and then remove them from the ROI.
Extracting image border
In endoscopy images, there is a black border that contains some information about the patient and imaging data. A simple approach to segment this border is thresholding. To this goal, 18,000 samples were selected randomly from different regions of gastrointestinal tissue. The same number of samples were extracted from the border of images. The histograms of these two groups of samples are simultaneously shown in Figure 6a. As illustrated in the figure, there is a clear distinction between these two regions in the R channel of RGB color space. By applying a threshold level of 30 to this channel, the main region of border was segmented. In the next step, the output of this binary image was expanded with the morphology operator dilation. As a result, the text information inside the image border was completely removed.
Figure 6.
(a) Histogram of Red channel, taken from border (mean = 20, variance = 15.48) and from normal tissue (mean = 184.13, variance = 50.37), (b) Histogram of saturation channel, taken from SR and IJ (mean = 7.69, variance = 19.91) and normal tissue (mean = 101.52, variance = 19.68)
Removing specular reflections and intestinal juices
Due to the proximity of the endoscope lamp to the gastrointestinal tissue, SR usually appear in endoscopic images. In addition, there is usually some IJ leaving the stomach during the endoscopy, which impedes the direct vision of the tissue. For extracting these regions, 2500 pixels were extracted from both SR and IJ areas. Furthermore, the same number of pixels were extracted from normal regions of gastric tissue. A comparison of the histograms of these two groups of pixels in different color spaces exhibits a clear discrimination between these two groups of pixels in S channel of HSV color space. As illustrated in Figure 6b, specular reflections (SR) and intestinal juices (IJ) regions belong to the lower bins of histogram; thus, a threshold level of 35 in S channel would be an appropriate choice to segment these regions. In the following, the extracted regions were expanded and denoised using dilation morphological operation.
Extracting lumen
Digestive tract is a dynamic organ that is in interaction with the physician during endoscopy. Moreover, its members are not rigid and exhibit mobility and activities. For example, in most of the LES images, the opened stomach sphincter makes a relatively large area of the image dark. This opened sphincter, which is called lumen, must be segmented and removed from the ROI. As shown in Figure 7, the lumen histogram is mostly in the lower bins of all three components of RGB color space. In this figure, the histogram of 41000 pixels collected from lumen is shown. The same number of samples is gathered from the gastrointestinal tissue. These two regions are more distinct in R channel as the gastrointestinal tissue is mostly red, orange, and pink. Hence, we use the R channel for lumen segmentation. Another challenge in segmenting the lumen is that the lightening conditions of imaging and the opening/closure state of the lumen alter the location and width of the lumen peak in the image histogram. Particularly, in images where the lumen is not entirely open, it would be more difficult to detect this peak. For these reasons, lumen segmentation is not a trivial task. The following method is used to segment the lumen:
Figure 7.
Comparison of red, blue and green histograms for lumen and tissue (a) red channel (b) green channel (c) blue channel
All edges in the image are detected using SUSAN edge detector
The edges located in gastric folds (gastric folds are discussed in Section 2.3.3) are removed
A circle is fitted to the remainder of edges as a rough estimation of lumen. An example of this estimation is shown in Figure 8a
In most cases, the first local maxima in the red channel histogram of this estimated region is a feature for lumen segmentation. More precisely, the histogram is smoothed by an averaging kernel with length 7. Figure 8c shows an image with a lumen peak intensity of 53
The corresponding pixels of the detected peak are fed as an initial seed to a region-growing algorithm. The output of this region growing is the whole lumen.
Figure 8.

(a) Rough estimation of lumen, (b) detected lumen, (c) Locating the intensity of the initial seed in the histogram of estimated region
Figures 8b and 9 show the extracted lumen and final ROI, respectively.
Figure 9.
Procedure of automatic extraction of ROI, (a) Original image (b) Border extraction, (c) Part b, plus SR and IJ regions excluded, (d) Part c, plus lumen region excluded
Z-line segmentation
After extracting the ROI, we focus on the Z-line and gastric folds segmentation. Our approach involves segmentation of the Z-line and then searching the folds inside its region. We used active contours for Z-line segmentation. Since active contour segmentations are sensitive to their initial contours, we developed the following procedure to find an initial contour that was as close as possible to the Z-line. The general idea behind finding this initial contour is shown in Figure 10. Therefore, Our approach consists of four steps:
Figure 10.

Initial contour extraction
Extracting image edges using SUSAN edge detector
Removing edges that are not located on the Z-line using Mahalanobis distance
Removing gastric folds edges
Interpolating a curve to the remainder of edge pixels and use it as an initial contour
In the following, we will explain these four steps.
Extracting edges using smallest univalue assimilating nucleus edge detector
The basic idea behind the smallest univalue assimilating nucleus (SUSAN) edge detector,[19] is to compare each pixel similarity with its neighboring pixels. For this purpose, SUSAN puts a mask on each image pixel and compares its central pixel (called nucleus) to other pixels inside the mask. An example of SUSAN edge detector output is illustrated in Figure 11a.
Figure 11.
Procedure of removing unwanted edges: (a) SUSAN edge detector output, (b) Removing unwanted edges using Mahalonobis distance, (c) Circle fitting for a rough estimation of gastric folds, (d) Output of Gabor filter bank, (e) First estimation of gastric folds, (f) Final initial contour for Z-line segmentation
Removing edges that do not belong to the Z-line using Mahalanobis distance criteria
A careful analysis of edge pixels located on the Z-line reveals that when we move perpendicular to the edge, we will find two different groups of pixels. The group of pixels inside the Z-line, which are closer to red and orange, and pixels that are outside the Z-line and are closer to white and pink. Considering these color features, Mahalanobis distance criteria are used to
Mahabalonis distance of an arbitrary point P to a cluster C is defined as:

where M and Σ−1 are mean and covariance of cluster C respectively. Using these distance criteria, the following method is used to remove undesirable edges:
Two clusters of 1200 sample pixels (one cluster inside and another outside the Z-line) are randomly collected from 85 LES images. We call them C1 and C2, respectively. To display the distinction between C1 and C2, their green and blue components are shown in Figure 12
Considering each edge pixel direction, two foursome groups of pixels are chosen around each edge pixel using masks, as displayed in Figure 13. For both of these two groups, we first calculate the average intensity and then measure the Mahalanobis distance of these averaged intensities from C1 and C2. We call these two distances MD1 and MD2, respectively. Using these two distances, the procedure of removing or preserving edges was as follows: To preserve an edge pixel, it is checked that if the pixels on one side of the pixel edge belong to C1, the other side pixels must belong to C2. In fact, in this case, pixels of one side of the edge are related to the interior side of Z-line and pixels of other side are related to the exterior side of Z-line. Therefore, we expect that the current edge pixel is located on the Z-line. Figure 11b shows a sample of this subsection output.
Figure 12.

Two clusters formed by pixels inside (Orange) and outside (Pink) of Z-line
Figure 13.
Masks used for choosing pixels around each edge. Yellow square shows the detected edge, orange and pink squares denote chosen pixels around each edge. (a) 0° mask, (b) 45° mask, (c) 90° mask, (d) 135° mask
Removing gastric folds edges
Most of the remained edge pixels belong to the Z-line and gastric folds. Thus, if gastric folds edges are removed, it is possible to fit a curve to the remaining edge pixels and use it as an initial contour to segment Z-line. This could be implemented in four steps:
-
To extract the ROI a circle is fitted to the remaining pixel edges using Pratt method.[20] In this method, a circle is fitted to a collection of 2D data by minimizing:

where B, C, and D are parameters corresponding to the center and radius of this circle and (xi, yi) is the position of any arbitrary data observation. An example of this estimated circle is illustrated in Figure 11c.
Using the bottom hat morphology operator, the contrast between gastric folds and gastrointestinal tissue is increased.
-
Gastric folds are bulges that originate from the stomach and spread in different directions of the esophagus. These folds are reminiscent of a Gabor filter bank with a small modulation frequency and varying widths. Using an appropriate Gabor filter bank, we can detect several folds in different directions. The spatial form of Gabor filter is defined as:

In this equation, σ and β are standard deviation of the elliptic Gaussian function in x and y directions, respectively. In addition, u0 and v0 are Gabor filter frequencies in 2D space, which are defined as:

These two frequencies could be defined in Polar coordinates as:

Therefore, F0 is the amplitude of these frequencies and θ0 is their direction. By changing these two parameters, it is possible to create a filter bank, which could be applied to an image for extracting objects in various directions. An example of this filter bank is illustrated in Figure 14e. To localize gastric folds, the output of these 24 Gabor filters is summed together and then thresholded by applying a global threshold obtained using Otsu method [Figure 11d].
A convex hull polygon is fitted to the output of the previous step, as illustrated in Figure 14e. The edge pixels in this area are considered as folds and are removed from desirable Z-line edge pixels. The remained edge pixels are expected to be mainly located on the Z-line. Thus, as it is shown in Figure 11f, a curve is fitted to the remaining edge pixels and used as our initial contour to segment the Z-line.
Figure 14.
Real parts of Gabor filter for 0° (a) to 157.5° (h) with step of 22.5°, (F0 = 4cycle/pixel, σ= β= 2)
After determining this initial contour, the LES images are then expanded to prevent active contour segmenting border instead of the Z-line. An example of this expansion is illustrated in Figure 15. Furthermore, to increase the segmentation speed, images are down-sampled with a rate of four. Finally, the images are segmented using the obtained initial contour and local active contours method introduced by Lankton et al.[21] Figure 16 shows the results of this automatic annotation and its ground truth for three sample images.
Figure 15.

(a) Original image, (b) Border expansion
Figure 16.
Four examples of automatic Z-line segmentation. Columns 1 (a and e) and 3 (c and g) are our output and columns 2 (b and f) and 4 (d and h) are clinicians ground truths
Gastric folds segmentation
Since gastric folds are always inside the Z-line, the following steps are used for gastric folds segmentation:
Z-line interior is considered as ROI
Steps two, three, and four are again incorporated as described in Section 2.3.3
In images with a visible lumen, the output of the previous step would be the union of lumen and folds.
Two samples of gastric folds segmentation are shown in Figure 17 along with the ground truth results from three clinicians. As it can be seen, there are significant differences even between clinicians in demarcating the region of gastric folds. This poses a challenge in the evaluation section that we will discuss later.
Figure 17.
Two examples of automatic gastric folds segmentation. Left column (a and e) shows our output and three next columns (b, c, d, f, g, and h) show the ground truth of three different clinicians
Results
Dataset
As there was no standard dataset for our defined problem, LES images were collected under the supervision of gastroenterologists in two independent clinics of digestive diseases. The endoscope device was a Fujinon EG-530FP with a resolution of 320 × 240 pixels per image. The following points were considered during the data collection:
Given the pencil-shaped structure of esophageal, its lower section linked to the stomach is smaller than other sections. As a result, gastric folds may cause some wrinkles in the esophageal tissue. During the endoscopy, the clinician needs to stretch the esophagus tissue to prevent these wrinkles. As a result, gastric folds and their wrinkles are limited to the inside of Z-line.
-
Since LES images have various structures in the pull and push endoscopy, all of the images were collected in the pull stage.
Depending on the patient's request, endoscopy was conducted either with complete or local anesthesia. Finally, 50 images taken from 20 patients were selected as our dataset. The Z-line of all images was demarcated by one clinician and the gastric folds were segmented by three clinicians.
To evaluate the proposed algorithm, the dataset is divided randomly into three sets of train, validation, and test with the ratio of 0.3, 0.3, 0.4, respectively. To compare this method with a modern deep learning neural network, we selected the U-Net architecture, which is a popular CNN model in medical image segmentation tasks. Furthermore, as the deep neural networks are usually data-hungry, and preparing more data could increase the efficiency of the architecture dramatically, a bunch of data augmentations including rotation, vertical and horizontal filliping, zooming, and shearing (all of them with the probability of 0.2) performed. In addition, the dataset is again divided into three sets randomly, but this time with the ratio of 0.5, 0.25, 0.25 respectively. For this method, this random selection is repeated ten times and the final results are the average of these ten experiments. Furthermore, gastric folds training labels are collected randomly from three clinicians' annotations.
Evaluation of Z-line segmentation
As there was no significant difference between clinicians in Z-line demarcating, just one ground truth is considered for each image. Thus, to measure the quality of the segmentation method, we first calculated the dice coefficient and Mean Squared Error (MSE) of the automatically annotated images.
In addition, we computed the average boundary distance (ABD). To do so, we compared the boundaries of our segmentation and the ground truth. That is, for each pixel on the boundary of our segmented image, we identified the nearest pixel in the ground truth and then the Euclidean distance between these two pixels was calculated. This process was repeated for all pixels on the boundary. Finally, the average of these distances indicates ABD. Table 1 shows the average and standard deviation of these three metrics for our dataset. The U-net segmentation architecture is also used as a well-known state-of-the-art alternative to segment Z-line. We used the U-Net architecture as it was proposed.[17] with a single exception that we resized our images to 512 × 512. LES images are fed to the neural network once with preprocessing and once without passing through this step. The results are shown in Table 1 separately. Accordingly, based on these results, the segmented Z-line matches the ground truth with reasonable accuracy in both the classical and the U-Net methods. It is also clear that the preprocessing step greatly increased the segmentation results. Indeed, this step thoroughly increased the performance of the neural network by removing the artifacts, extracting the informative ROI, and also by increasing the contrast between esophageal tissue and the z-line.
Table 1.
Mean and standard deviation of pixel based evaluation metrics
| Mean-SD | |||
|---|---|---|---|
|
| |||
| Classic method | U-Net (with preprocessing) | U-Net (without preprocessing) | |
| Dice coefficient | 0.9310-0.067 | 0.9334-0.053 | 0.87-0.18 |
| ABD | 8.6-5.93 | 16.035-14.5 | 31.1-26.5 |
| MSE | 4.3-3.15 | 2.25-3.5 | 14.1-12.3 |
ABD - Average boundary distance; MSE - Mean square error; SD - Standard deviation
Evaluation of gastric folds segmentation
The evaluation and interpretation of gastric folds segmentation results are not a straightforward issue. That is because the region determined by one clinician could be significantly different from another one. One way to address this issue is to obtain various ground truths from different clinicians and use all of them to evaluate the algorithm results. Columns 2 to 4 in Figure 16 show ground truths for an LES image. As can be seen, clinicians had significantly divergent views about locating the boundary of gastric folds. Hence, two metrics introduced[22] were used to compare our results with ground truths obtained from the three clinicians. The first metric is called Sweet-Spot Coverage (SSC), which is defined as:

In this equation, A is the result of image processing algorithm and M is a set of binary masks, where each binary mask Mi contains ground truth of clinician number i. As it is clear, this metric compares the results of our algorithm with those approved by all clinicians. This metric was computed also between the clinicians themselves. To do so, we put one clinician aside and used the other two clinicians as ground truth to compute SSC. This was repeated three times and each time another clinician was set aside. Finally, the average of these three SSCs was considered as the SSC between clinicians. The average and standard deviation of SSC for both of the classic and the U-Net architecture are shown in Table 2.
Table 2.
Mean and standard deviation of gastric folds segmentation results
| Mean–SD | |||
|---|---|---|---|
|
| |||
| Classic method | U-Net (with preprocessing) | U-Net (without preprocessing) | |
| SSC | 0.90-0.09 | 0.85–0.11 | 0.78–0.19 |
| JIGS | 0.85-0.09 | 0.82–0.12 | 0.76–0.19 |
SCC - Sweet-Spot Coverage; JIGS - Jaccard Index for Golden Standard; SD - Standard deviation
Another metric used to evaluate the results was Jaccard Index for Golden Standard (JIGS), which is defined as:

As it is clear, JIGS would be one when the algorithm segmentation is between the intersection and the union of all three ground truths annotations. This metric was once again computed between the clinicians themselves as described above. The average and standard deviation of JIGS for both of the classic and the U-Net architecture are computed and shown in Table 2. Based on these results, the output of our segmentation algorithms confirms the clinician's annotations.
Discussion
In this research, an automatic method was developed for Z-line and gastric folds segmentation. It was shown that Z-line segmentation is in good agreement with the ground truth. All of the classical pixel-based metrics had averages above 90% with standard deviations less than 1%. Furthermore, ABD had an average of 4.18 pixels with a standard deviation of 2.93. It means that on average, the segmented Z-line had just 4 pixels distance to the ground truth. As there is a disagreement between clinicians for boundary of folds, three different ground truths were gathered for each image of our data set. Then, SSC and JIGS metrics were used to compare our results with the three ground truths. Based on Table 3, the average of SSC was 90% with a standard deviation of just 1%. A comparison of this average with the SSC between clinicians showed that our gastric folds segmentation resembles that of clinicians. This is also true for JIGS metric where our algorithm results are comparable to that of clinicians. To evaluate the performance of deep learning approaches the U-Net architecture is used. The advantage of using these methods is that we get free of choosing a number of parameters that must be determined in classical approaches, but on the other hand, these approaches are usually data-hungry. To overcome this, we applied augmentation methods to increase data samples. Note that this study aims to evaluate the feasibility of machine vision algorithms in segmenting the Z-line and gastric folds. This is clear that for a more robust and reliable algorithm, more amount of LES images collected from a variety of clinics is needed. According to gastroenterologists, results show that it is feasible to accurately segment the Z-line and gastric folds using image processing algorithms and this leads to a more accurate diagnosis of the disease.
Table 3.
Mean and standard deviation of gastric folds segmentation results
| Mean-SD | ||
|---|---|---|
|
| ||
| Between algorithm and clinicians | Between clinicians themselves | |
| SSC | 0.90-0.09 | 0.89-0.05 |
| JIGS | 0.85-0.09 | 0.82-0.07 |
SCC - Sweet-Spot Coverage; JIGS - Jaccard Index for Golden Standard; SD - Standard deviation
Financial support and sponsorship
None.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Rawla P, Barsouk A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz Gastroenterol. 2019;14:26–38. doi: 10.5114/pg.2018.80001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nasseri-Moghaddam S, Razjouyan H, Nouraei M, Alimohammadi M, Mamarabadi M, Va-Hedi H, et al. Inter-and intra-observer variability of the los angeles classification: A reassessment. Arch Iran Med. 2007;10:48–53. [PubMed] [Google Scholar]
- 3.Spechler SJ. Barrett's esophagus. Principles of Deglutition. :723–738. [Google Scholar]
- 4.Armstrong, David, et al. The endoscopic assessment of esophagitis: a progress report on observer agreement. Gastroenterology 111.1. 1996:85–92. doi: 10.1053/gast.1996.v111.pm8698230. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Dent J, Armstrong D, Bergman JJ, Gossner L, Hoshihara Y, et al. The development and validation of an endoscopic grading system for barretts esophagus: The prague C & M criteria. Gastroenterology. 2006;131:1392–9. doi: 10.1053/j.gastro.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 6.de Souza LA, Jr, Palm C, Mendel R, Hook C, Ebigbo A, Probst A, et al. A survey on barrett's esophagus analysis using machine learning. Comput Biol Med. 2018;96:203–13. doi: 10.1016/j.compbiomed.2018.03.014. [DOI] [PubMed] [Google Scholar]
- 7.Ghatwary N, Ahmed A and Ye X. Annual conference on medical image understanding and analysis. Cham: Springer; 2017. Automated detection of Barrett's esophagus using endoscopic images: a survey. [Google Scholar]
- 8.Kim R, Baggott BB, Rose S, Shar AO, Mallory DL, Lasky SS, et al. Quantitative endoscopy: Precise computerized measurement of meta-plastic epithelial surface area in Barrett's esophagus. Gastroenterology. 1995;108:360–6. doi: 10.1016/0016-5085(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 9.Seo KW, Min BR, Kim HT, Lee SY, Lee DW. Classification of esophagitis grade by neural network and texture analysis. J Mech Sci Technol. 2008;22:2475–80. [Google Scholar]
- 10.Saraf SS, Udupi G, Hajare SD. Los angeles classification of esophagitis using image processing techniques. Int J Comput Appl. 2012;42:45–50. [Google Scholar]
- 11.Serpa-Andrade L, et al. An approach based on Fourier descriptors and decision trees to perform presumptive diagnosis of esophagitis for educational purposes. 2015 IEEE International Autumn Meeting on Power, Electronics and Computing (ROPEC). IEEE. 2015 [Google Scholar]
- 12.Yousefi-Banaem H, Rabbani H, Adibi P. Barrett's mucosa segmentation in endoscopic images using a hybrid method: Spatial fuzzy c-mean and level set. J Med Signals Sens. 2016;6:231. [PMC free article] [PubMed] [Google Scholar]
- 13.Van Der Sommen F, Zinger S, Schoon EJ, de With P. Supportive automatic annotation of early esophageal cancer using local Gabor and color features. Neurocomputing. 2014;144:92–106. [Google Scholar]
- 14.Souza L. Bildverarbeitung für die Medizin 2017. Berlin, Heidelberg: Springer Vieweg; 2017. Barrett's esophagus analysis using SURF features; pp. 141–6. [Google Scholar]
- 15.De Souza, Luis Antonio. Barrett's esophagus identification using optimum-path forest. 2017 30th SIBGRAPI conference on graphics, patterns and images (SIBGRAPI) 2017 Ieee. [Google Scholar]
- 16.Mendel, Robert . Bildverarbeitung für die Medizin 2017. Berlin, Heidelberg: Springer Vieweg; 2017. Barrett's esophagus analysis using convolutional neural networks; pp. 80–5. [Google Scholar]
- 17.Ronneberger O, Fischer P, and Brox T. International Conference on Medical image computing and computer-assisted intervention. Cham: Springer; 2015. U-net: Convolutional networks for biomedical image segmentation. [Google Scholar]
- 18.Perona P, Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE Trans Pattern Anal Mach Intell. 1990;12:629–39. [Google Scholar]
- 19.Smith SM, Brady JM. Susana new approach to low level image processing. Int J Comput Vis. 1997;23:45–78. [Google Scholar]
- 20.Pratt V. Direct least-squares fitting of algebraic surfaces. ACM SIGGRAPH computer graphics 21.4. 1987:145–152. [Google Scholar]
- 21.Lankton S, Tannenbaum A. Localizing region-based active contours. IEEE Trans Image Proc. 2008;17:2029–39. doi: 10.1109/TIP.2008.2004611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Sommen F, Zinger S, and Erik J. Sweet-spot training for early esophageal cancer detection. Medical Imaging 2016: Computer-Aided Diagnosis. 2016;9785 SPIE. [Google Scholar]