ABSTRACT
Background
Accumulating studies demonstrated that resistance of colon cancer (CC) to 5-fluorouracil (5-FU) contributes to adverse prognosis. We investigated how Kruppel-like factor 4 (KLF4) affected 5-FU resistance and autophagy in CC cells.
Methods
KLF4 expression and its downstream target gene RAB26 in CC tissues was analyzed by bioinformatics analysis, and the effect of abnormal KLF4 expression on prognoses of CC patients was predicted. Luciferase reporter assay detected the targeted relationship between KLF4 and RAB26. The viability and apoptosis of CC cells were analyzed by CCK-8 and flow cytometry. The formation of intracellular autophagosomes was detected by confocal laser scanning microscopy and immunofluorescence staining. The mRNA and protein levels were assayed by qRT-PCR and western blot. A xenograft animal model was constructed to verify the function of KLF4. Rescue assay was employed to verify whether KLF4/RAB26 could affect 5-FU resistance in CC cells through autophagy.
Results
KLF4 and RAB26 were lowly expressed in CC. KLF4 correlated with patients’ survival. KLF4 was down-regulated in 5-FU resistant CC cells. KLF4 overexpression suppressed the proliferation and 5-FU resistance of CC cells, and inhibited LC3 II/I expression and autophagosome formation. Autophagy activator Rapamycin or sh-RAB26 treatment reversed the impact of KLF4 overexpression on 5-FU resistance. In vivo assay verified that KLF4 inhibited 5-FU resistance in CC cells. Rescue experiments revealed that KLF4 targeted RAB26 to inhibit CC cell autophagy, resulting in decreasing the resistance to 5-FU.
Conclusion
KLF4 strengthened the sensitivity of CC cells to 5-FU by targeting RAB26 to restrain autophagy pathway.
KEYWORDS: KLF4, RAB26, 5-FU resistance, colon cancer, autophagy
1. Introduction
Colon cancer (CC) is a major cause of cancer deaths worldwide1, and 5-fluorouracil (5-FU) is a common pyrimidine analogue of CC that blocks thymidylate synthase and further inhibits DNA and RNA synthesis2. 5-FU remains one of the widely used first-line treatments for CC3. 5-FU treatment is able to shrink the tumor mass of CC. But 5-FU was also toxic to normal cells, causing serious adverse reactions, which severely limited its clinical application4. Studies have shown that about 50% of CC patients treated with 5-FU develop resistance, which is the leading cause of poor prognosis in colon cancer patients5. Apoptosis of cancer cells in 5-FU resistant CC patients was hindered and cancer cells were viable even with high concentration of 5-FU6. If resistance to 5-FU in cancer patients can be overcome, this finding will provide new insights into the therapeutic strategies of CC7. The molecular mechanism of resistance of CC cells to 5-FU chemotherapy has not been completely elucidated, which needs to be deeply explored.
Kruppel-like factor 4 (KLF4) belongs to the KLF family and is involved in regulating many biological processes8. Accumulating evidence suggests that KLF4 plays a cancer-suppressive role. For example, KLF4 inhibits malignant progression of meningioma by mediating apoptosis, invasion, proliferation, and cell cycle9. While KLF4 overexpression can reduce colorectal cancer cell migration, invasion, and tumorigenicity, thereby inhibiting tumor metastasis10,11. In addition, KLF4 also plays an imperative role in chemoresistance. For example, by altering HMGB1 and hTERT expression, KLF4 strengthens the sensitivity of CC cells HCT-15 to cisplatin12. Deng et al. 13 discovered that KLF4 was down-regulated in oxaliplatin-resistant colorectal cancer cells, and KLF4 negatively regulated PiHL expression, while PiHL induced HMGA2 up-regulation to promote PI3K/Akt phosphorylation, thereby enhancing oxaliplatin resistance in colorectal cancer. Moreover, Irene put forward that KLF4 was up-regulated in carfilzomib resistant myeloma cells, and KLF4 bound to the promoter region of SQSTM1, which increased autophagy level and thereby enhancing the resistance of myeloma cells to carfilzomib14. Taken together, KLF4 played an imperative role in tumorigenesis and chemoresistance, but the molecular mechanism underlying the effect of KLF4 on 5-FU resistance in CC cells was ill-defined.
Autophagy takes an important part in cellular homeostasis maintenance by degrading unwanted cellular components. When cells are exposed to unfavorable environments, like starvation, hypoxia, and metabolic disorders, autophagy will be activated to maintain cell survival15. Inhibition of autophagy has been found to reduce previously activated cellular defense mechanisms, and then increases sensitivity to therapy, whereas autophagy activation leads to cell death via lysosomal overactivation16. Studies have shown that autophagy is vital in tumor chemoresistance. Garcia-Mayea et al. 17 found that acquisition of drug resistance of laryngeal carcinoma was associated with overexpression of autophagy-related proteins (LC3, ATG5), and increased autophagy contributes to drug resistance development. Gao et al. 18 proved that PRKCI-Akt-mTOR pathway was activated by circPARD3 via suppressing miR-145-5p, which in turn inhibited autophagy in laryngeal scaly cell carcinoma, and promoted chemoresistance and malignant progression of this disease. The above studies have suggested that autophagy exerts a key function in tumorigenesis and chemoresistance. This study was aimed at investigating whether autophagy could affect 5-FU resistance in CC cells.
In this work, we revealed that KLF4 was lowly expressed in CC by bioinformatics analysis and correlated with CC patients’ survival. KLF4 was also significantly down-regulated in 5-FU resistant CC cells, and KLF4 suppressed 5-FU resistance by targeting RAB26 to suppress the autophagy pathway. These findings further deepened our understanding of KLF4 and autophagy in the 5-FU resistance and progression of CC, and revealed an important role of KLF4 in regulating autophagy and 5-FU resistance in CC cells.
2. Materials and methods
2.1. Bioinformatics approaches
Expression of KLF4 and predicted target genes in CC patients was analyzed using TCGA-COAD database (T (tumor) = 275; N (normal) = 349) by ‘R’ (version 4.1.1)19 language from TCGA website (https://portal.gdc.cancer.gov/)20. Survival analysis were carried out using the ‘survival’ package21. The target gene predictions on KLF4 were conducted by utilizing DEmRNA-down, ChIPBase (http://deepbase.sysu.edu.cn/chipbase/)22, and hTFtarget (http://bioinfo.life.hust.edu.cn/hTFtarget#!/target)23 databases. Correlation analysis was employed for predicting the targeted genes of KLF4. The predicted target gene was overlapped with KLF4 to obtain a target gene which shared a binding site with KLF4.
2.2. Cell culture and transfection
Human normal colon fibroblast CCD-18Co (BNCC337724) and human CC cell lines SW480 (BNCC100604), SW620 (BNCC337664), and HCT116 (BNCC351970) were offered by BeNa Culture Collection (BNCC, China) and named as 5-FU sensitive CC cell lines. The above cell lines were cultured with RPMI-1640 (with 1% penicillin-streptomycin and 10% fatal bovine serum (FBS)) medium at 37°C with 5% CO2.
Next, 5-FU resistant CC cell lines SW620/FU (MXC858) bought from Shanghai Meixuan Biotechnology Co., Ltd., (China) and HCT116/FU (BNCC342640) purchased from BNCC (China) were incubated in RPMI-1640 (with 10% FBS and 1% penicillin-streptomycin) medium with 5-FU (25 µg/mL).
We also purchased oe-KLF4 overexpression vector and corresponding negative control (oe-NC), sh-RAB26 silencing vector and corresponding negative control (sh-NC) from Ribobio (China). The oe-KLF4 cell lines were constructed by transfecting oe-KLF4 into 5-FU resistant cell lines (SW620/FU and HCT116/FU), respectively, using Lipofectamine 3000 kit (Thermo Fisher Scientific, USA).
SW620/FU and HCT116/FU cell lines were treated with the autophagy activator Rapamycin (100 nM)18 and relevant parameters were measured after 24 h.
2.3. qRT-PCR
Total RNA was extracted by Trizol method, which was reversely transcribed into cDNA with PrimeScript RT reagent Kit (Takara, Japan) and Oligo (dt) 18 primers. The mRNA relative expression of KLF4 and RAB26 were examined by utilizing qRT-PCR kit (Takara, Japan), with β-actin as the internal control and calculated using the 2−ΔΔCt method. The reaction conditions of qRT-PCR were as follows: 94°C, 5 min 1 cycle; 94°C, 10 s, 55°C, 30 s, 94°C, 15 s, 40 cycles. All the primer sequences used in our study are listed in Table 1.
Table 1.
Primers for Qrt-PCR.
| Primer | Forward | Reverse |
|---|---|---|
| KLF4 | 5’-GATTATAAATTAAGGGGAGAGTGGG-3’ | 5’-TCCCTAAAAAATAACCATATACCAAAA-3’ |
| RAB26 | 5′-GTCTGCTGGTGCGATTCAAG-3′ | 5′-GCATGGGTAACACTGCGGA-3′ |
| β-actin | 5’-AGATGTGGATCAGCAAGCAG-3’ | 5’-GCGCAAGTTAGGTTTTGTCA-3’ |
2.4. Western blot
Total proteins were first extracted using RIPA lysate, then total protein concentration was detected with a BCA kit (Thermo Fisher Scientific, USA). Next, proteins were separated by SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked by TBS plus 0.05% Tween 20 (TBST) and 5% dry nonfat dry milk powder in Tris-buffered saline (pH = 7.6) for 1 h and then cultured with primary antibodies anti-rabbit KLF4 (1:1000, ab215036), anti-rabbit RAB26 (1:1000, ab198202), anti-rabbit β-actin (1:1000, ab8227), anti-rabbit p62 (1: 10000, ab109012), and anti-rabbit LC3B (1: 2000, ab192890) at 4°C overnight. After washing three times, Goat anti-rabbit horseradish peroxidase secondary antibody (HRP, 1: 2000, ab6721) was supplemented for 2 h cell culture at room temperature (All antibodies used here were from Abcam (UK)). In the end, we detected the brightness of protein bands24.
2.5. CCK-8 assay
SW620, SW620/FU, HCT116, HCT116/FU cells (1 × 103 cells/well) were seeded in 96-well plates with 100 μL of cell solution per well. For resistant cell lines, 5-FU was added at final concentrations of 0, 50, 100, 200, 400, 600, and 800 μg/mL after overnight culture. For sensitive cell lines, 5-FU was added at final concentrations of 0, 10, 20, 40, 60, 80, and 100 μg/mL. After all cells were cultured for 72 h, 10 μL of CCK-8 solution was supplemented for 2 h of culture. The absorbance value at 450 nm was detected by a microplate reader. We plotted the concentration effect curve with drug concentration and relative cell viability as the horizontal and vertical axes. Then we calculated the half maximal inhibitory concentration (IC50) values.
2.6. Flow cytometry
The transfected SW620/FU and HCT116/FU cells were grown to the logarithmic phase, which were cultured with RPMI-1640 medium (serum-free) with 5-FU (25 μg/mL) for 24 h. Next, we collected these cells, which were washed and diluted to 1 × 106 cells/ml. We then utilized the apoptosis kit (Thermo Fisher Scientific, USA) to detect them. Cells were resuspended with 100 μL labeling buffer, and then 5 μL Annexin V-FITC and 10 μL propidium iodide (PI) were added. Cells were maintained in the dark at room temperature for 15 min, and eventually we accessed the apoptosis rate utilizing flow cytometry.
2.7. Immunofluorescence and immunohistochemical detection
In this study, immunohistochemical13 and immunofluorescence24 assays were performed following the processes described in the former articles. The antibodies of KLF4 (ab215036), LC3B (ab63817), Ki67 (ab15580) were bought from Abcam (UK).
2.8. Construction of the tumor xenograft model
The animal experiment was conducted strictly complying with the Guide for the Care and Use of Laboratory Animals, which had been approved by the Fujian Medical University Laboratory Animal Ethics Committee. A total of 12 athymic BALB/C mice (4-week-old, male) were randomized (n = 3/group). The subcutaneous injection was conducted on the mice with SW620/oe-KLF4 or SW620/oe-NC cells (5 × 106 CC cells were injected into each mouse). When the tumor volume grew to about 0.1 cm3 (recorded as Day 0), the tumorigenicity test of 5-FU treatment was performed. The mice in the above groups were classified into four groups (3 mice/group) for treatment: PBS+oe-NC, PBS+oe-KLF4, 5-FU+oe-NC, 5-FU+oe-KLF4. The mice in 5-FU+oe-NC and 5-FU+oe-KLF4 groups were injected with 5-FU (20 mg/kg/week) intraperitoneally, while the mice in the other two groups were injected with the same volume of PBS intraperitoneally. Tumor weight, tumor length (L) and width (W) were measured every 4 days. The formula for tumor volume was: V = 0.5 × LW2. After 16 days, mice were euthanized. We dissected all the mice for the subcutaneous tumors which were then weighed and we collected the tumor tissues for autophagy and apoptosis analysis25.
2.9. Dual-luciferase reporter gene analysis
The pGL3 Basic-RAB26-WT and pGL3 Basic-RAB26-MUT luciferase reporter vectors (Promega, USA) were constructed first, and then the RAB26-WT/RAB26-MUT, oe-NC/oe-KLF4 and Renilla luciferase vector pRL-TK (Promega, USA) were co-transfected into SW620 cells, mixed with 4 μL lipofectamine 2000 and placed in a 37°C, 5% CO2 environment for an additional culture for 24–48 h. Fluorescence intensity was detected using the Dual-Luciferase Reporter Assay System (Promega, USA).
2.10. Statistical analysis
Data analysis was carried out using GraphPad Prism8.0 software, and data were presented in the form of mean ± standard deviation. Pearson correlation analysis was performed on RAB26 and KLF4. We utilized the Kaplan-Meier method for plotting the survival curve, and compared by the log-rank test using the R (version 4.1.1) package survival. The experimental data were pre-tested using analysis of variance, and then compared between the two groups using Student’s-t test. Each experiment was performed in triplicate. Statistically significant differences were indicated when p < .05.
3. Results
3.1. KLF4 is less expressed in 5-FU resistant CC cells
First, KLF4 expression was analyzed using TCGA-COAD database, and the result exhibited the significant down-regulation of KLF4 in CC tissues (Figure 1a). Next, Kaplan-Meier survival analysis observed that CC patients with low KLF4 expression had relatively shorter overall survival and disease-free survival compared to those with high KLF4 expression, but this difference was not statistically significant (Figure 1b, c). Subsequently, qRT-PCR and western blot were used to analyze mRNA and protein expression of KLF4 in normal colon fibroblast and CC cell lines. It was illustrated that KLF4 was remarkably down-regulated in CC cells with a contrast to the normal colon fibroblast (Figure 1d). In the meantime, we examined the KLF4 expression in 5-FU sensitive and resistant CC cells. It was indicated that KLF4 was notably down-regulated in SW620/FU and HCT116/FU resistant cells with a contrast to SW620 and HCT116 sensitive cells (Figure 1e). In summary, KLF4 was lowly expressed in CC and may be involved in modulating 5-FU resistance in CC cells.
Figure 1.
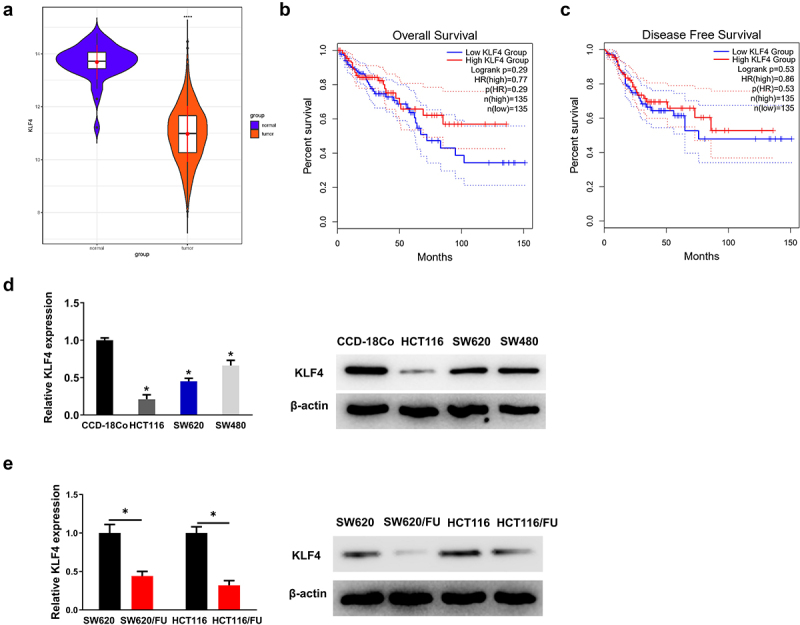
(a) the two violin plots show that KLF4 is significantly downregulated in the colon cancer tissue samples; (b-c) the two Kaplan-Meier survival analysis show that patients with low expression of KLF4 in colon cancer have relatively shorter overall survival and disease-free survival; (d) the expression of KLF4 in three colon cancer cell lines (SW480, HCT116, SW620) were lower than normal colon fibroblast CCD-18Co, detected by qRT-PCR and western blot; (e) the expression of KLF4 in SW620/FU and HCT116/FU were lower than SW620 and HCT116, detected by qRT-PCR and western blot.
3.2. Overexpressed KLF4 promotes apoptosis and suppresses 5-FU resistance in CC cells
To investigate whether there was resistance of SW620/FU and HCT116/FU cells to 5-FU, we calculated IC50 values of 5-FU for resistant cells (SW620/FU and HCT116/FU) and sensitive cells (SW620 and HCT116). It was shown that with a contrast to the sensitive cells, SW620/FU and HCT116/FU cells presented higher IC50 values (Figure 2a), indicating that SW620/FU and HCT116/FU cells had higher resistance to 5-FU relative to their parental cells.
Figure 2.
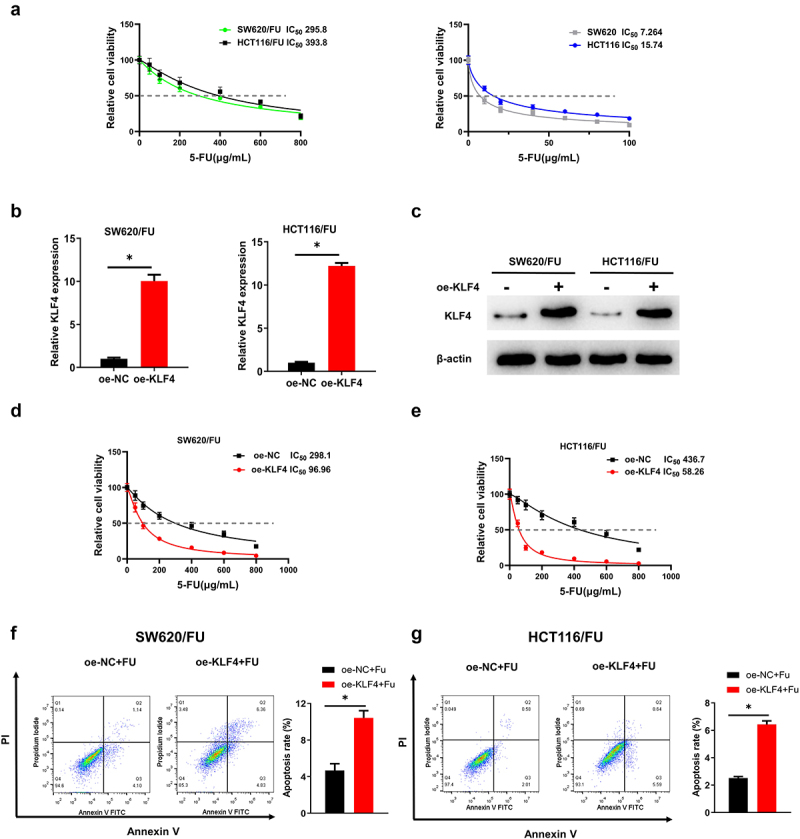
(a) SW620/FU and HCT116/FU cells presented higher IC50 values than SW620 and HCT116; (b-c) After overexpression of KLF4 in SW620/FU and HCT116/FU, its expression levels were significantly increased by qRT-PCR and western blot; (d-e) After overexpression of KLF4 in SW620/FU and HCT116/FU, the IC50 value was significantly reduced by CCK-8; (f-g) After overexpression of KLF4 in SW620/FU and HCT116/FU, the apoptosis was significantly increased by flow cytometry.
To investigate the effect of KLF4 on 5-FU-resistant CC cells, we transfected oe-NC/oe-KLF4 into SW620/FU and HCT116/FU cells, respectively. We then utilized qRT-PCR and western blot to detect the transfection efficiency, the result of which demonstrated that the after treated with oe-KLF4, the mRNA and protein levels of KLF4 were dramatically up-regulated (Figure 2b, c). Then, CCK-8 results showed that the IC50 values of CC cells to 5-FU were significantly reduced in oe-KLF4 group, indicating that KLF4 overexpression inhibited SW620/FU and HCT116/FU resistance to 5-FU (Figure 2d, e). Flow cytometry analysis revealed that KLF4 overexpression promoted apoptosis of SW620/FU and HCT116/FU resistant cells in 5-FU treatment groups (Figure 2f, g). In summary, KLF4 promoted apoptosis and improved 5-FU sensitivity in CC cells.
3.3. KLF4 overexpression inhibits autophagy of CC cells
Previous researches demonstrated that autophagy affected tumor chemoresistance, and inhibition of autophagy could inhibit tumor chemoresistance17,18. Therefore, we speculated that KLF4 inhibited 5-FU resistance in CC cells by repressing autophagy. First, the autophagy protein expression levels of p62 and LC3 in 5-FU sensitive and 5-FU resistant CC cells were analyzed using western blot. The protein expression of LC3II/LC3I was evidently elevated in SW620/FU and HCT116/FU resistant cells with a contrast to the sensitive cells (SW620 and HCT116). However, SW620/FU and HCT116/FU resistant cells had a decreased protein level of p62 (Figure 3a). Besides, KLF4 overexpression significantly suppressed expression of autophagy marker LC3II/LC3I, while significantly promoted expression of autophagy marker p62 in SW620/FU and HCT116/FU cells (Figure 3b). Besides, immunofluorescence results indicated that LC3 accumulation was significantly reduced in SW620/FU and HCT116/FU cells upon overexpression KLF4 (Figure 3c, d). These results confirmed that KLF4 repressed autophagy in CC cells.
Figure 3.
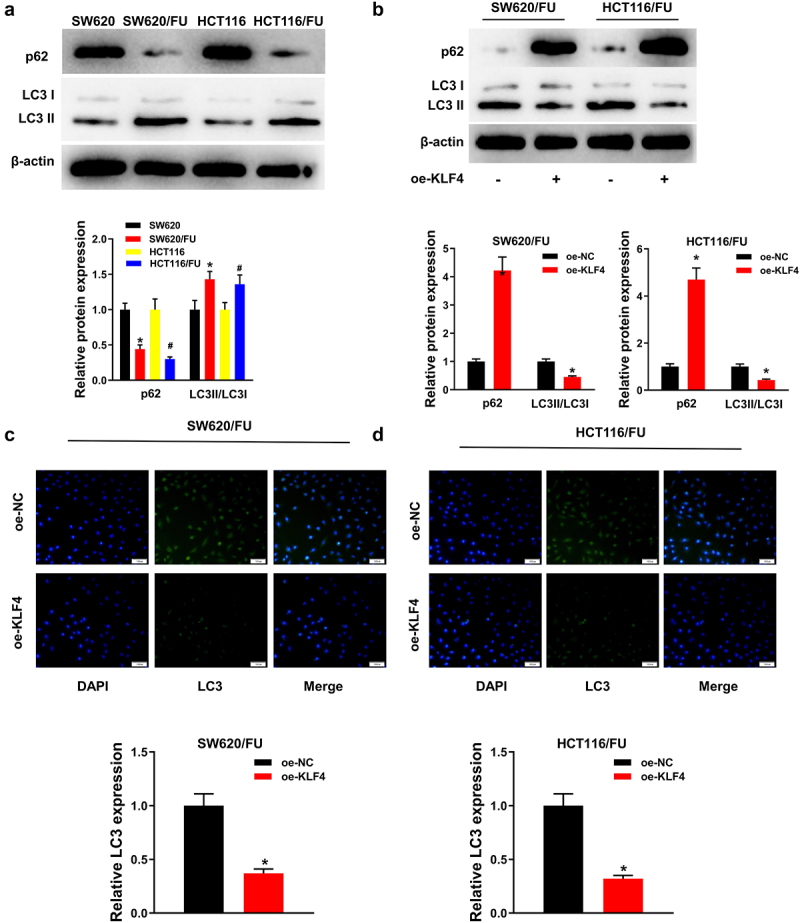
(a) Protein expression of LC3II/LC3I was significantly increased, and the p62 protein level was decreased in SW620/FU and HCT116/FU; (b) Overexpression of KLF4 significantly inhibited the expression of LC3II/LC3I in SW620/FU and HCT116/FU, and promoted the expression of p62; (c-d) Immunofluorescence analysis shows a significant decrease in LC3 after overexpression of KLF4 in SW620/FU and HCT116/FU.
3.4. KLF4 strengthens the cell sensitivity of CC to 5-FU by suppressing autophagy
To further explore the effect of autophagy inhibition by KLF4 on 5-FU resistance in CC cells, we treated SW620/FU resistant cells with the autophagy activator Rapamycin (referred to as Rap) and set the cell groups as follows: oe-NC+PBS, oe-KLF4+PBS, oe-NC+Rap, and oe-KLF4+Rap. First, the p62 and LC3 protein expression levels in SW620/FU cells were examined by western blot (Figure 4a). and we observed that in oe-KLF4 group, the protein level of LC3II/LC3I was notably down-regulated and that of p62 was remarkably up-regulated. Conversely, the LC3II/LC3I protein level was remarkably increased and p62 protein level was notably decreased in Rap treatment group. The protein levels of p62 and LC3 were restored to the level of oe-NC+PBS group after treatment with oe-KLF4+Rap. In addition, immunofluorescence result showed that LC3 aggregation was notably reduced in oe-KLF4 group, but significantly increased in Rap treatment group, and oe-KLF4+Rap treatment could reverse the impact of overexpressed KLF4 on LC3 aggregation (Figure 4b). It could be seen that KLF4 overexpression inhibited autophagy, while which could be reversed by Rap. In addition, the results of CCK-8 and flow cytometry (Figure 4c, d) showed that KLF4 overexpression decreased the IC50 value of CC cells to 5-FU and promoted apoptosis, while oe-KLF4+Rap treatment reversed the influence of overexpressed KLF4 on apoptosis and drug resistance of CC cells. Altogether, KLF4 inhibited resistance of CC cells to 5-FU by suppressing autophagy.
Figure 4.
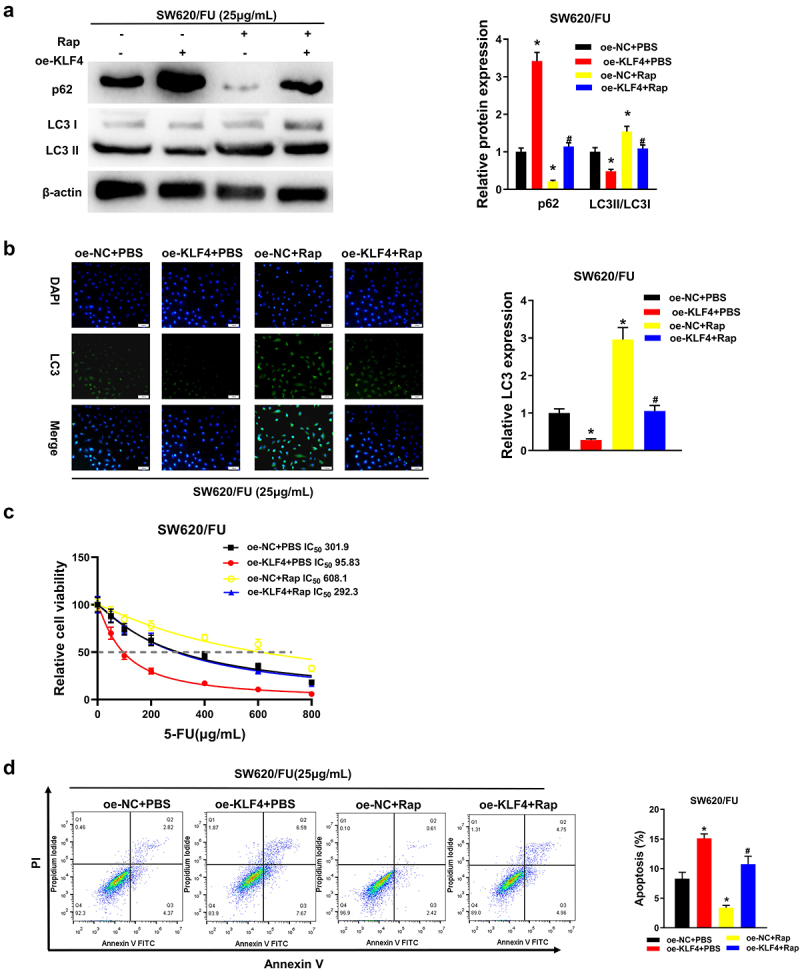
(a) Four groups, treatment with rap restores the effect of KLF4 overexpression on the protein levels of LC3II/LC3I and p62 by western blot; (b) Four groups, immunofluorescence analysis shows treatment with rap restores the effect of KLF4 overexpression on the LC3 fluorescence expression; (c) Four groups, CCK-8 shows treatment with rap restores the effect of KLF4 overexpression on the viability of colon cancer cells; (d) Four groups, treatment with rap restores the effect of KLF4 overexpression on the apoptosis of colon cancer cells detected by flow cytometry.
3.5. KLF4 overexpression inhibits 5-FU resistance in CC cells through in vivo experiment
The above-mentioned experiments demonstrated that KLF4 regulated 5-FU resistance of CC cells in vitro, but it still remained an unanswered question regarding the unction of KLF4 in vivo. To answer this question, we separated mice to four groups with different treatments: (1) PBS+oe-NC, (2) PBS+oe-KLF4, (3) 5-FU+oe-NC, (4) 5-FU+oe-KLF4. In animals without 5-FU treatment, tumor weight and volume were prominently reduced in PBS+oe-KLF4 treatment group compared with PBS+oe-NC treatment group. After 5-FU treatment, tumor weight and volume were dramatically reduced in 5-FU+oe-KLF4 treatment group compared with 5-FU+oe-NC treatment group. More interestingly, tumor weight and volume were significantly reduced with 5-FU+oe-KLF4 treatment compared with the other three groups (Figure 5a–c). What’s more, through qRT-PCR and western blot assays, we observed that KLF4 mRNA and protein expression levels were prominently up-regulated in the oe-KLF4 ×enograft model (Figure 5d, e). Immunohistochemistry displayed that the cell proliferation of Ki67 positive tumor cells was dramatically attenuated by 5-FU+oe-KLF4 treatment compared with the control group (Figure 5f). Additionally, we discovered that LC3 II formation was significantly reduced in 5-FU+oe-KLF4 group compared to 5-FU+oe-NC group (Figure 5f, g). Altogether, KLF4 overexpression inhibited 5-FU resistance in CC cells in vivo.
Figure 5.
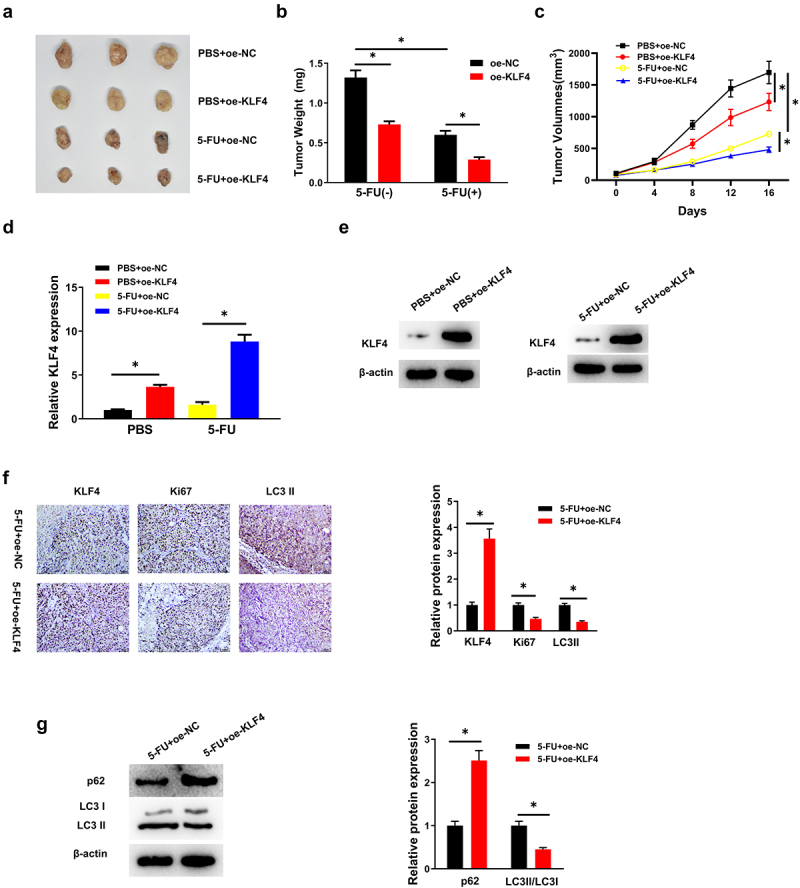
(a-c) Four groups, the tumor weight and volume were dramatically reduced in 5-FU+oe-KLF4 group; (d-e) the expression levels of KLF4 in the 5-FU+oe-KLF4 group were significantly increased tested by Qrt-PCR and western blot; (f) Ki67 positive tumor cells was dramatically attenuated in 5-FU+oe-KLF4 group by Immunohistochemistry; (G) LC3 II formation was significantly reduced in 5-FU+oe-KLF4 group by western blot.
3.6. KLF4 targets RAB26 in CC cells
We predicted target genes downstream of KLF4 in CC by bioinformatics analysis and obtained 134 potential target genes (Figure 6a). It has been mentioned that RAB26 has a close connection with autophagy26, so we chose RAB26 as a downstream gene of KLF4 for subsequent studies. Next, we utilized TCGA-COAD database to perform Pearson correlation analysis between KLF4 and RAB26, the results of which displayed that KLF4 had a markedly positive correlation with RAB16 (correlation coefficient = 0.51) (Figure 6b). The RAB26 expression in CC tissues and adjacent non-cancerous tissues was also analyzed using TCGA database, and it was shown that in CC tumor tissues, RAB26 expression was remarkably down-regulated (Figure 6c). Subsequently, we analyzed the promoter sequence of the RAB26 gene and obtained a binding site and motif between KLF4 and RAB26 using the JASPAR database (Figure 6d). To further validate the targeted relationship between KLF4 and RAB26, the dual-luciferase assay was conducted. It was demonstrated that overexpressed KLF4 substantially enhanced the luciferase activity of WT-RAB26, but having no noticeable effect on the luciferase activity of MUT-RAB26, indicating that KLF4 targeted and bound RAB26 (Figure 6e). We then performed qRT-PCR to detect mRNA expression of RAB26 in normal colon fibroblast and CC cells, which illustrated that RAB26 expression was considerably lower in CC cells with a contrast to that in normal colon fibroblast (Figure 6f). To dive deeper into the effect of KLF4 on RAB26 expression, qRT-PCR and western blot were employed to detect the mRNA and protein expression levels of RAB26 in SW620 and HCT116 cells with oe-KLF4 transfected. It was discovered that RAB26 mRNA and protein levels of RAB26 were remarkably higher in SW620 and HCT116 cells overexpressing KLF4 than in controls, indicating that KLF4 exerted promotive role in RAB26 expression (Figure 6g, h). These results indicated that KLF4 targeted and regulated RAB26 positively.
Figure 6.
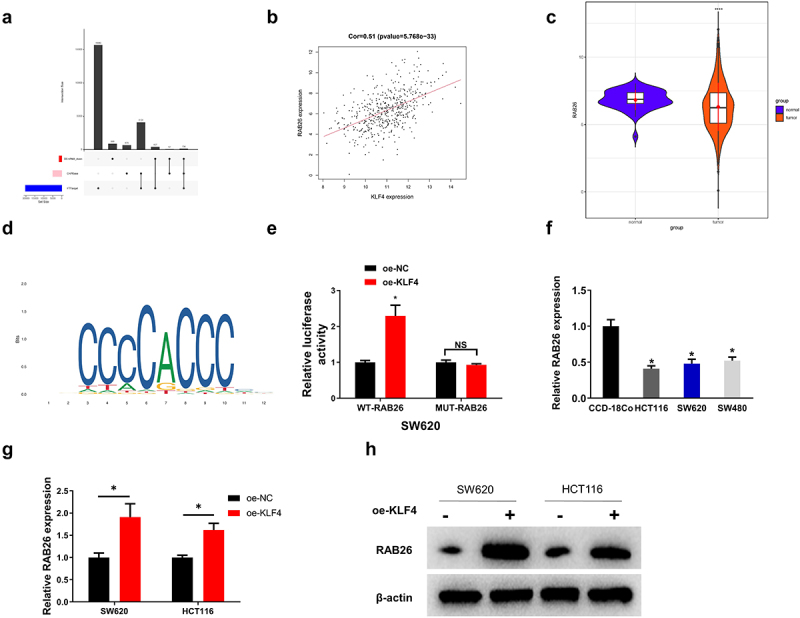
(a) KLF4 in colon cancer has 134 potential target genes by bioinformatics analysis; (b) KLF4 had a markedly positive correlation with RAB26 by Pearson correlation analysis; (c) the two violin plots show that RAB26 is significantly downregulated in the colon cancer tissue samples; (d) Obtained a binding site and motif between KLF4 and RAB26 using the JASPAR database; (e) KLF4 targeted and bound RAB26 by dual-luciferase assay; (f) the expression of RAB26 in three colon cancer cell lines (SW480, HCT116, SW620) were lower than normal colon fibroblast CCD-18Co, detected by qRT-PCR; (G-H) RAB26 levels were remarkably higher in SW620 and HCT116 cells overexpressing KLF4 tested by qRT-PCR and western blot.
3.7. KLF4 targets RAB26 and mediates autophagy in CC cells, thereby inhibiting 5-FU resistance
To further clarify the ability of KLF4 to attenuate 5-FU resistance in CC cells by inhibiting autophagy through regulation of RAB26, we set the following groups: oe-NC+sh-NC, oe-KLF4+sh-NC, oe-NC+sh-RAB26, and oe-KLF4+sh-RAB26. First, the protein expression levels of autophagy-related genes in the cells of each transfection group were measured by western blot. The results illustrated that in the oe-KLF4- group, LC3II/LC3I was markedly down-regulated and p62 protein level up-regulated. On the contrary, in the sh-RAB26-treat group, LC3II/LC3I was remarkably up-regulated and p62 protein level down-regulated. When oe-KLF4+sh-RAB26 was co-transfected, the expression levels of LC3II/LC3I protein and p62 protein returned to be comparable to those in the oe-NC+sh-NC group (Figure 7a). Immunofluorescence was then used to detect the accumulation of LC3 fluorescence in each transfection group. It was demonstrated that LC3 accumulation was considerably reduced in the oe-KLF4 group and substantially increased in the sh-RAB26 group, and LC3 accumulation returned to the level of the oe-NC+sh-NC group after co-transfection with oe-KLF4 and sh-RAB26 (Figure 7b). Afterwards, CCK-8 assay was utilized to detect the cell viability in different treatment groups, the result of which demonstrated that 5-FU resistance of tumor cells was markedly reduced after oe-KLF4 treatment alone, and greatly enhanced after sh-RAB26 treatment alone. While co-transfection of oe-KLF4 and sh-RAB26 could reverse the weakening effect of oe-KLF4 on 5-FU resistance of tumor cells (Figure 7c). Finally, we examined the apoptosis level of SW620/5-FU cells in each group after transfection, and found that the apoptosis level of SW620/5-FU cells was increased dramatically after oe-KLF4 treatment alone, and decreased remarkably after sh-RAB26 treatment alone. While it was increased substantially after oe-KLF4+sh-RAB26 treatment and recovered to be comparable with the oe-NC+sh-NC group (Figure 7d). Combined with previous results of the autophagy activator Rap, we could draw the corollary that KLF4 inhibited cellular autophagy by targeting RAB26 and then inhibited 5-FU resistance in CC cells.
Figure 7.
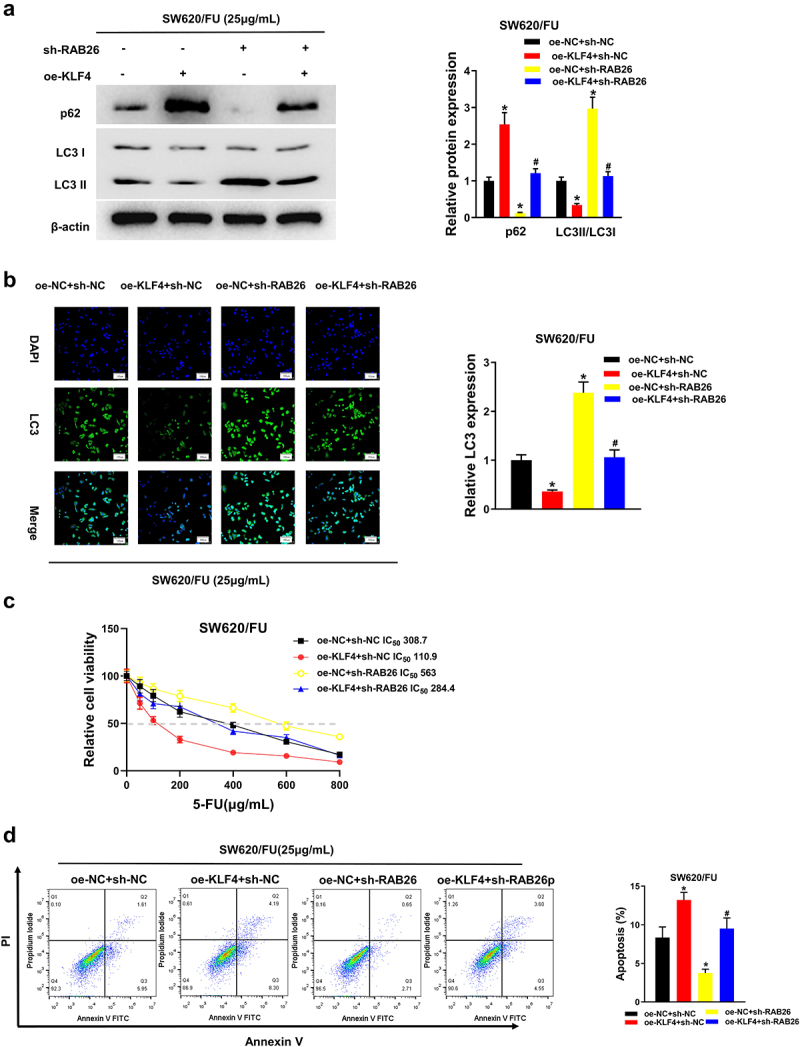
(a) Four groups, treatment with sh-RAB26 restores the effect of KLF4 overexpression on the protein levels of LC3II/LC3I and p62 by western blot; (b) Four groups, immunofluorescence analysis shows treatment with sh-RAB26 restores the effect of KLF4 overexpression on the LC3 fluorescence expression; (c) Four groups, CCK-8 shows treatment with sh-RAB26 restores the effect of KLF4 overexpression on the viability of colon cancer cells; (d) Four groups, treatment with sh-RAB26 restores the effect of KLF4 overexpression on the apoptosis of colon cancer cells detected by flow cytometry.
4. Discussion
Studies have demonstrated that overexpressed KLF4 could hamper invasion and migration of gastric cancer27 and prostate cancer28 cells, while KLF4 overexpression restrain esophageal cancer cell proliferation, promotes apoptosis29, and reduce poor prognosis. Ma et al. 30 discovered a low level of KLF4 in colorectal cancer, which could inhibit cancer cell proliferation via NDRG2. The results of bioinformatics analysis and cellular experimental examination suggested that KLF4 level was significantly reduced in CC tissues and cells, and it had a close connection with patient’s survival. The results of CCK-8 as well as flow cytometry demonstrated that KLF4 overexpression restrained CC cell proliferation and promoted apoptosis, and the same results were obtained in vivo. By comparison, our experimental results were found to be consistent with published relevant studies. 5-FU is the first-line chemotherapeutic agent for CC. Congenital or acquired 5-FU resistance seriously affects the therapeutic efficacy of 5-FU. Therefore, in-depth exploration of the molecular mechanisms of CC cell resistance to 5-FU is required. This study revealed that KLF4 level was significantly reduced in 5-FU resistant cells, while the IC50 values of 5-FU were significantly increased, indicating that 5-FU resistant cells had reduced sensitivity to 5-FU. Moreover, patients with low KLF4 level had shorter survival time and poorer prognosis, indicating that KLF4 may be a 5-FU resistant target. Our study showed that KLF4 overexpression inhibited CC cell proliferation, facilitated apoptosis, and enhanced CC cell sensitivity to 5-FU. Jing-Wen Yang et al. 31 found that inducing cell autophagy could enhance the anti-cancer effect of 5-FU in CC cells. In the meantime, Jia et al. discovered that KLF4 overcame tamoxifen resistance of breast cancer by inhibition of MAPK signaling pathway32. These findings suggested that KLF4 was a key suppressor in tumor and overexpressed KLF4 could suppress tumor drug resistance, which agree with the results of our study.
Autophagy exerts a key regulatory role in invasion, metastasis and chemosensitivity of various tumors33–35. For example, Garcia-Mayea et al. 17 revealed that enhance autophagy could improve tumor drug resistance. Gao et al. 18 revealed that circPARD3 activated the PRKCI-Akt-mTOR signaling pathway could be activated by circPARD3 via suppressing miR-145-5p, and inhibited autophagy in LSCC to enhance cell resistance to cisplatin. It can be seen that the role of autophagy on drug resistance is tumor heterogeneous. While the results of in vitro and in vivo assays demonstrated that KLF4 hindered CC cell resistance to 5-FU by inhibiting autophagy.
RAB26 is a small GTPase in the RAB family that impacts cell survival and stress responses36. It has been shown that RAB26 is differentially expressed in breast cancer cell lines with different degrees of invasion (RAB26 is relatively highly expressed in poorly invasive tumor cells), and inhibit autophagy in breast cancer cells by can phosphorylated Src37. Our results suggested that RAB26 was lowly-expressed in CC cells and that silencing RAB26 attenuated the suppressive effect of overexpression of KLF4 on autophagy. The discovery of this phenomenon may shed light on the mechanism involved in KLF4/RAB26 resistance to 5-FU in CC cells.
Clinicians and scientists have tried to explore new therapeutic strategies for combatting cancer via regulating autophagy. For example, Li et al. 38 treated CC cells using autophagy inhibitor 3-MA and found that 3-MA combined with 5-FU prominently increased apoptosis, and proposed that inhibition of autophagy may be a strategy for adjuvant chemotherapy of CC. Tan et al. 39 found that the combination of oxaliplatin and 3-MA inhibits autophagy and increase apoptosis of CC cells. Our research dived into the effect of KLF4 overexpression combined with the autophagy activator Rapamycin on apoptosis and 5-FU resistance of SW620/FU cells. KLF4 overexpression significantly inhibited autophagy level, and KLF4 overexpression suppressed tumor growth and promoted apoptosis in vivo and in vitro. While KLF4 overexpression combined with Rapamycin could reverse the effect of KLF4 overexpression. KLF4 overexpression combined with Rapamycin could significantly activate autophagy and inhibit apoptosis. In addition, combined treatment of KLF4 overexpression and 5-FU chemotherapy remarkably restrained tumor growth and prolonged mouse survival compared with 5-FU chemotherapy treatment alone. In the meantime, KLF4 overexpression and 5-FU combination treatment decreased LC3 II formation in xenograft tumor tissues. Therefore, KLF4 improved the sensitivity of tumor cells to 5-FU by impeding autophagy.
In summary, this work proposed and elucidated the mechanism of KLF4 in CC that KLF4 targeted RAB26 to inhibited autophagy and promoted apoptosis, thus suppressing CC cell resistance to 5-FU (Figure 7). The results may assist the development of biomarkers to refine treatment strategies for CC patients. Admittedly, this study did not further verify the regulation of KLF4/RAB26 axis on 5-FU resistance in CC by in vivo experiments. In the follow-up study, we will carry out more relevant exploration.
Biographies
Yu Zheng, associate chief physician, good at the standardized treatment and comprehensive treatment of gastrointestinal tumors.
Jiansheng Wu, gastrointestinal surgeon, specializing in gastrointestinal diseases, acute appendicitis, intestinal obstruction, perforation of the digestive tract.
Hong Chen, gastrointestinal surgeon, specializes in gastrointestinal disorders.
Dajia Lin, associate chief physician, specializing in gastrointestinal stromal tumor surgery and comprehensive treatment.
Hongyuan Chen, gastrointestinal surgeon, specializing in gastrointestinal surgery-related diseases.
Junyin Zheng, gastrointestinal surgeon, diagnosis and treatment of gastrointestinal surgery related diseases.
Haoyun Xia, the attending physician, specializes in the diagnosis and treatment of gastric ulcer, gastric bleeding, intestinal cancer, gastroenteritis, digestive tract polyp, oblique hernia, direct hernia, appendicitis, pancreatitis, colitis, peritonitis, digestive tract ulcer, intestinal tuberculosis and other diseases.
Liangxiang Huang, director of the department, has been engaged in the clinical work of gastrointestinal surgery for more than 30 years. He is currently the deputy chairman of the Gastrointestinal Tumor Professional Committee of Fujian Anticancer Association, the deputy leader of the Gastrointestinal Group of the Surgical Branch of Fujian Medical Association, and a member of the expert group of Fujian Provincial Cadre Health Committee. She has rich clinical experience in the diagnosis and surgical treatment of various gastrointestinal diseases, especially in-depth research on the diagnosis and treatment of gastrointestinal tumors. She is one of the leaders in minimally invasive laparoscopic surgery for gastroenter-related tumors in Fujian Province.
Changqing Zeng, chief physician, mainly engaged in the comprehensive surgical treatment of gastrointestinal benign and malignant tumors, especially good at gastric cancer, colorectal cancer, gastrointestinal stromal tumor surgical minimally invasive treatment.
Funding Statement
Natural Science Foundation of Fujian Province (2020J011103).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Availability of data and materials
The datasets generated during and analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The animal experiment was performed strictly according to the Guide for the Care and Use of Laboratory Animals and were approved by the Fujian Medical University Laboratory Animal Ethics Committee.
References
- 1.Das V, Kalita J, Pal M.. Predictive and prognostic biomarkers in colorectal cancer: a systematic review of recent advances and challenges. Biomed Pharmacother. 2017;87:8–13. doi: 10.1016/j.biopha.2016.12.064. [DOI] [PubMed] [Google Scholar]
- 2.Joag MG, Sise A, Murillo JC, Sayed-Ahmed I, Wong JR, Mercado C, Galor A, Karp CL. Topical 5-Fluorouracil 1% as primary treatment for ocular surface squamous neoplasia. Ophthalmol. 2016;123(7):1442–1448. doi: 10.1016/j.ophtha.2016.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cho YH, Ro EJ, Yoon JS, Mizutani T, Kang DW, Park JC, Il Kim T, Clevers H, Choi KY. 5-FU promotes stemness of colorectal cancer via p53-mediated WNT/β-catenin pathway activation. Nat Commun. 2020;11(1):5321. doi: 10.1038/s41467-020-19173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hong S, Cai W, Huang Z, Wang Y, Mi X, Huang Y, Lin Z, Chen X. Ginsenoside Rg3 enhances the anticancer effect of 5‑FU in colon cancer cells via the PI3K/AKT pathway. Oncol Rep. 2020;44:1333–1342. doi: 10.3892/or.2020.7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo Z, Chen R, Hu S, Huang X, Huang Z. PVT1 promotes resistance to 5‑FU in colon cancer via the miR‑486‑5p/CDK4 axis. Oncol Lett. 2022;24(2):280. doi: 10.3892/ol.2022.13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao SJ, Ren SN, Liu YT, Yan HW, Chen XB. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330-3p-hexokinase 2 axis. Mol Ther Oncolytics. 2021;23:14–25. doi: 10.1016/j.omto.2021.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Bao Y, Yang GK, Wan J, Du LJ, Ma ZH. MiR-214 sensitizes human colon cancer cells to 5-FU by targeting Hsp27. Cell Mol Biol Lett. 2019;24(1):22. doi: 10.1186/s11658-019-0143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Safaei S, Shanehbandi D, Zafari V, Eghbali E, Sadeghzadeh M, Mohammad Reza Khani H, Sadrazar A, Faghihdinevari M, Shirmohamadi M. Evaluation of miR-107, DAPK1, and KLF4 expression in colorectal tumors and effect of oxaliplatin and 5-FU on their levels in colorectal cancer cell lines. J Middle East J Cancer. 2021;12:190–197. doi: 10.30476/mejc.2020.83091.1131. [DOI] [Google Scholar]
- 9.Tang H, Zhu H, Wang X, Hua L, Li J, Xie Q, Chen X, Zhang T, Gong Y. KLF4 is a tumor suppressor in anaplastic meningioma stem-like cells and human meningiomas. J Mol Cell Biol. 2017;9(4):315–324. doi: 10.1093/jmcb/mjx023. [DOI] [PubMed] [Google Scholar]
- 10.Chen HY, Lin YM, Chung HC, Lang YD, Lin CJ, Huang J, Wang WC, Lin FM, Chen Z, Huang HD, et al. MiR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2012;72(14):3631–3641. doi: 10.1158/0008-5472.CAN-12-0667. [DOI] [PubMed] [Google Scholar]
- 11.Correction: miR-103/107 promote metastasis of colorectal cancer by targeting the metastasis suppressors DAPK and KLF4. Cancer Res. 2017;77(23):6788. doi: 10.1158/0008-5472.CAN-17-3079. [DOI] [PubMed] [Google Scholar]
- 12.Yadav SS, Kumar M, Varshney A, Yadava PK. KLF4 sensitizes the colon cancer cell HCT-15 to cisplatin by altering the expression of HMGB1 and hTERT. Life Sci. 2019;220:169–176. doi: 10.1016/j.lfs.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Deng X, Kong F, Li S, Jiang H, Dong L, Xu X, Zhang X, Yuan H, Xu Y, Chu Y, et al. A KLF4/PiHL/EZH2/HMGA2 regulatory axis and its function in promoting oxaliplatin-resistance of colorectal cancer. Cell Death Dis. 2021;12(5):485. doi: 10.1038/s41419-021-03753-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riz I, Hawley TS, Hawley RG. KLF4-SQSTM1/p62-associated prosurvival autophagy contributes to carfilzomib resistance in multiple myeloma models. Oncotarget. 2015;6(17):14814–14831. doi: 10.18632/oncotarget.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dalby K, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the pro-death and pro-survival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010;6(3):322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taylor MA, Das BC, Ray SK. Targeting autophagy for combating chemoresistance and radioresistance in glioblastoma. Apoptosis. 2018;23(11–12):563–575. doi: 10.1007/s10495-018-1480-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Mayea Y, Mir C, Muñoz L, Benavente S, Castellvi J, Temprana J, Maggio V, Lorente J, Paciucci R, LLeonart ME, et al. Autophagy inhibition as a promising therapeutic target for laryngeal cancer. Carcinogenesis. 2019;40:1525–1534. doi: 10.1093/carcin/bgz080. [DOI] [PubMed] [Google Scholar]
- 18.Gao W, Guo H, Niu M, Zheng X, Zhang Y, Xue X, Bo Y, Guan X, Li Z, Guo Y, et al. circPARD3 drives malignant progression and chemoresistance of laryngeal squamous cell carcinoma by inhibiting autophagy through the PRKCI-Akt-Mtor pathway. Mol Cancer. 2020;19(1):166. doi: 10.1186/s12943-020-01279-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du J, Gu J, Deng J, Kong L, Guo Y, Jin C, Bao Y, Fu D, Li J. The expression and survival significance of sodium glucose transporters in pancreatic cancer. Bmc Cancer. 2022;22(1):116. doi: 10.1186/s12885-021-09060-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang Z, Cai H, Lin H, Guan B, Wu Y, Zhang Y, Liu X, Zhuang J, Guan G. Development and validation of a robust pyroptosis-related signature for predicting prognosis and immune status in patients with colon cancer. J Oncol. 2021;2021:1–20. doi: 10.1155/2021/5818512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sonabend R, Kiraly FJ, Bender A, Bischl B, Lang M, Wren J. Mlr3proba: an R package for machine learning in survival analysis. Bioinformat. 2021;37(17):2789–2791. doi: 10.1093/bioinformatics/btab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang JH, Li JH, Jiang S, Zhou H, Qu LH. ChIPBase: a database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Nucleic Acids Res. 2013;41(D1):D177–D187. doi: 10.1093/nar/gks1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fu W, Kadeer G, He Y, Feng Y. The regulatory network of potential transcription factors and MiRNAs of mitochondria-related genes for sarcopenia. Front Genet. 2022;13:975886. doi: 10.3389/fgene.2022.975886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang R, Jin Y, Zou C, Chen Y, Wang G, et al. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9(12):1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie P, Yuan FQ, Huang MS, Zhang W, Zhou HH, Li X, Liu ZQ. DCBLD2 affects the development of colorectal cancer via EMT and angiogenesis and modulates 5-FU drug resistance. Front Cell Dev Biol. 2021;9:669285. doi: 10.3389/fcell.2021.669285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Wang Y, Li C, Xu Y, Wang X, Wu D, Gao Z, Qian H, You Z, Zhang Z, et al. Extracellular CIRP-Impaired Rab26 restrains EPOR-mediated macrophage polarization in acute lung injury. Front Immunol. 2021;12:768435. doi: 10.3389/fimmu.2021.768435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kong F, Sun T, Kong X, Xie D, Li Z, Xie K. Krüppel-like factor 4 suppresses serine/threonine kinase 33 activation and metastasis of gastric cancer through reversing epithelial–mesenchymal transition. Clin Cancer Res. 2018;24(10):2440–2451. doi: 10.1158/1078-0432.CCR-17-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Place RF, Huang V, Wang X, Noonan EJ, Magyar CE, Huang J, Li LC. Prognostic value and function of KLF4 in prostate cancer: rNAa and vector-mediated overexpression identify KLF4 as an inhibitor of tumor cell growth and migration. Cancer Res. 2010;70(24):10182–10191. doi: 10.1158/0008-5472.CAN-10-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang Y, Goldstein BG, Chao HH, Katz JP. KLF4 and KLF5 regulate proliferation, apoptosis and invasion in esophageal cancer cells. Cancer Biol Ther. 2005;4(11):1216–1221. doi: 10.4161/cbt.4.11.2090. [DOI] [PubMed] [Google Scholar]
- 30.Ma Y, Wu L, Liu X, Xu Y, Shi W, Liang Y, Yao L, Zheng J, Zhang J. KLF4 inhibits colorectal cancer cell proliferation dependent on NDRG2 signaling. Oncol Rep. 2017;38(2):975–984. doi: 10.3892/or.2017.5736. [DOI] [PubMed] [Google Scholar]
- 31.Yang JW, Zhang QH, Liu T. Autophagy facilitates anticancer effect of 5-fluorouracil in HCT-116 cells. J Cancer Res Ther. 2018;14(12):S1141–S1147. doi: 10.4103/0973-1482.204898. [DOI] [PubMed] [Google Scholar]
- 32.Jia Y, Zhou J, Luo X, Chen M, Chen Y, Wang J, Xiong H, Ying X, Hu W, Zhao W, et al. KLF4 overcomes tamoxifen resistance by suppressing MAPK signaling pathway and predicts good prognosis in breast cancer. Cell Signal. 2018;42:165–175. doi: 10.1016/j.cellsig.2017.09.025. [DOI] [PubMed] [Google Scholar]
- 33.Song P, Li Y, Dong Y, Liang Y, Qu H, Qi D, Lu Y, Jin X, Guo Y, Jia Y, et al. Estrogen receptor β inhibits breast cancer cells migration and invasion through CLDN6-mediated autophagy. J Exp Clin Cancer Res. 2019;38(1):354. doi: 10.1186/s13046-019-1359-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell ES, Coelho PP, Ratcliffe CDH, Rajadurai CV, Peschard P, Vaillancourt R, Zuo D, Park M. Lc3C-mediated autophagy selectively regulates the met RTK and HGF-stimulated migration and invasion. Cell Rep. 2019;29(12):4053–4068 e4056. doi: 10.1016/j.celrep.2019.11.063. [DOI] [PubMed] [Google Scholar]
- 35.Levy JM, Thompson JC, Griesinger AM, Amani V, Donson AM, Birks DK, Morgan MJ, Mirsky DM, Handler MH, Foreman NK, et al. Autophagy inhibition improves chemosensitivity in BRAF(V600E) brain tumors. Cancer Discov. 2014;4(7):773–780. doi: 10.1158/2159-8290.CD-14-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, Yuan M, Huang C, Xu Z, Li M, Zhang C, Gao Z, Zhang M, Xu J, Qian H, et al. Endothelial cell inflammation and barriers are regulated by the Rab26-mediated balance between β 2-AR and TLR4 in pulmonary microvessel endothelial cells. Mediators Inflamm. 2019;2019:1–10. doi: 10.1155/2019/7538071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu H, Zhou Y, Qiu H, Zhuang R, Han Y, Liu X, Qiu X, Wang Z, Xu L, Tan R, et al. Rab26 suppresses migration and invasion of breast cancer cells through mediating autophagic degradation of phosphorylated Src. Cell Death Dis. 2021;12(4):284. doi: 10.1038/s41419-021-03561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li J, Hou N, Faried A, Tsutsumi S, Takeuchi T, Kuwano H. Inhibition of autophagy by 3-MA enhances the effect of 5-FU-induced apoptosis in colon cancer cells. Ann Surg Oncol. 2009;16(3):761–771. doi: 10.1245/s10434-008-0260-0. [DOI] [PubMed] [Google Scholar]
- 39.Tan S, Peng X, Peng W, Zhao Y, Wei Y. Enhancement of oxaliplatin-induced cell apoptosis and tumor suppression by 3-methyladenine in colon cancer. Oncol Lett. 2015;9(5):2056–2062. doi: 10.3892/ol.2015.2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and analyzed during the current study are not publicly available, but are available from the corresponding author on reasonable request.


