Abstract
Deuterodon is a genus of the subfamily Stethaprioninae, a group of Neotropical fish known as tetras. Deuterodon hastatus represents a species complex, which is supported by cytogenetic and molecular data. In this study, we show the results of comparative evolutionary analyses of the ATP synthase subunit 6 gene in four Deuterodon species, in addition to ribosomal markers (18S rDNA and 5S rDNA), of a new population of the D. hastatus species complex from the Angra dos Reis/RJ region. The study population comprised a new cytotype, which we refer to as cytotype D, in D. hastatus, with 2n = 50 = 6M+8SM+8ST+28A. We obtained three different clades of D. hastatus in our phylogeny, one of them composed only by specimens of cytotype D. By using molecular clock dating, we observed that the radiation of Deuterodon from southeastern Brazil seemed to be associated with neotectonic events that occurred during the Miocene-Pliocene and Pliocene-Pleistocene transitions, marked by the capture of headwater streams and marine transgressions. The results obtained reinforce the idea that D. hastatus is a species complex, and at least three evolutionary significant units were identified in this group.
Keywords: 5S rDNA, 18S rDNA, chromosomal polymorphisms, molecular clock, phylogeny
Introduction
Deuterodon is a genus of small fish, known as tetras, described by Eigenmann et al. (1907). Of complex taxonomy, for a long time, some species belonging to the genus were considered incertae sedis in the family Characidae (Lima, 2003). Morphological features that are diagnostic indicators of the genus are also found in some species historically attributed to the genus Astyanax, and some studies over the years have shown that these species are phylogenetically closer to Deuterodon species than to other species of Astyanax, like Astyanax giton, Astyanax hastatus, Astyanax intermedius, Astyanax ribeirae, Astyanax taeniatus among others (Lucena and Lucena, 1992; Silva et al., 2017; Terán et al., 2020).
After an extensive revision, conducted by Terán et al. (2020), several coastal species of Astyanax, including the species mentioned in the paragraph above, as well as Hyphessobrycon luetkenii, were transferred to the genus Deuterodon. In addition, the authors also proposed that Myxiopis and Probolodus were synonyms of Deuterodon. Currently, the genus includes 24 valid species, whereas Deuterodon potaroensis remains as incertae sedis (Fricke et al., 2022). Of these, nine have described karyotypes (Table 1) and 20 have sequences deposited in GenBank, four of which are of the ATP synthase subunit 6 gene (Benson et al., 2015). Among Deuterodon, Deuterodon hastatus constitutes a species complex (Kavalco et al., 2009), which was identified based on karyotype differences between populations, biological limits between specimens, and the absence of hybridism (Kavalco et al., 2009; Pazza et al., 2018).
Table 1 -. Cytogenetic characteristics of nine Deuterodon species.
| Species | Sample procedence | 2n | Karyotypic formulae | FN | C-banding | Ag-NORs | GC-rich sites | 18S | 5S | As-51 | References |
|---|---|---|---|---|---|---|---|---|---|---|---|
| D. giton a | Jacuí stream/SP - Paraíba do Sul river basin | 50 | 6M+8SM+8ST+28A | 72 | Pericentromeric and interstitial. Many chromosomes. | 6 | absent | 10 | 10 | absent | Kavalco and Moreira-Filho (2003) Kavalco et al. (2004) Kavalco et al. (2007) |
| Latão Creek/MG Doce river basin | 50 | 6M+8SM+24ST+12A | 88 | Multiple markes many chromosomes. | 2 | 2 | 10 | 2 | - | Aguiar et al. (2011) | |
| D. intermedius a | Jacuí stream/SP - Paraíba do Sul river basin | 50 | 6M+8SM+4ST+32A | 68 | Pericentromeric and interstitial. Many chromosomes. | 6 | absent | 12 | 10 | absent | Kavalco and Moreira-Filho (2003) Kavalco et al. (2004) Kavalco et al. (2007) |
| D. hastatus a | Ypiranga Community/RJ Guapimirim-Macacu river basin | 50 | 4M+8SM+10ST+28A (Cytotype A) | 72 | Pericentromeric and interstitial. Many chromosomes. | 3 | - | 6 | - | absent | Kavalco et al. (2009) |
| Santana de Japuíba County/RJ Guapimirim-Macacu river basin | 50 | 8M+10SM+14ST+18A (Cytotype B) | 82 | Pericentromeric and interstitial. Many chromosomes. | 3 | - | 8 | - | absent | Kavalco et al. (2009) | |
| Macacu River Guapimirim-Macacu river basin | 50 | 6M+8SM+4ST+32A (Cytotype C) | 68 | Pericentromeric and interstitial. Many chromosomes. | 2 | - | 6 | - | absent | Kavalco et al. (2009) | |
| Town of Chachoeiras de Macacu/RJ Guapimirim-Macacu river basin | 50 | 6M+8SM+4ST+32A (Cytotype C) | 68 | Pericentromeric and interstitial. Many chromosomes. | 2 | - | 6 | - | absent | Kavalco et al. (2009) | |
| Angra dos Reis/RJ Ariró river basin | 50 | 6M+8SM+8ST+28A (Cytotype D) | 72 | - | - | - | 6 | 7 | - | present work | |
| D. ribeirae a | Poço Grande Community, Iporanga City/SP Ribeira de Iguape river basin | 50 | 4M+10SM+6ST+30A | 70 | Pericentromeric. Some chromosomes. | 2 | - | 4 | 6 | absent | Kavalco et al. (2010) |
| Town of Registro/SP Ribeira de Iguape river basin | 50 | 4M+10SM+6ST+30A | 70 | Pericentromeric. Some chromosomes. | 2 | - | 4 | 6 | absent | Kavalco et al. (2010) | |
| Town of Sete Barras/SP Ribeira de Iguape river basin | 50 | 4M+10SM+6ST+30A | 70 | Pericentromeric. Some chromosomes. | - | 6 | 4 | 6 | absent | Kavalco et al. (2010) | |
| D. taeniatus a | Hydroelectric Risoleta Neves Reservoir/MG Doce river basin | 50 | 14M+12SM+16ST+8A | 92 | Centromeric and pericentromeric. Most SM and ST. | 4-8 | - | 10 | 8 | - | Da Cunha et al. (2016) |
| 50 | 10M+14SM+18ST+8A | 92 | Centromeric and pericentromeric. Most SM and ST. | 4-8 | - | 8 | 8 | - | Da Cunha et al. (2016) | ||
| D. pedri | Sant’Anna dos Ferros/MG Santo Antônio river basin | 50 | 12M+12SM+20ST+6A | 94 | Centromeric many chromosomes. Instersticial some chromosomes. | 2-4 | - | 10 | 2 | - | Coutinho-Sanches and Dergam (2015) |
| D. iguape b | Ipiranga River/SP Ribeira do Iguape river basin | 50 | 14M/SM+36ST/A | - | - | - | - | - | - | - | Portela et al. (1988) |
| D. stigmaturus | Maquiné River/RS Tramandaí river basin | 50 | 8M+6SM+2ST+34A | 66 | Pericentromeric. All chromossomes. | 4-7 | Many A short arms. | 8 | - | - | Mendes et al. (2011) |
| D. janeiroensis a | Açungui River/PR Ribeira do Igapé river basin | 50 | 6M+14SM+14SM+16A | 84 | Centromeric and telomeric. Most chromosomes | 3-7 | 16 | 22 | 2 | 14 | Carvalho et al. (2002) Vicari et al. (2008) |
| Sacovão River/PR Ribeira do Igapé river basin | 50 | 6M+14SM+14SM+16A | 84 | Centromeric and telomeric. Most chromosomes | 3-7 | 16 | 22 | 2 | 14 | Vicari et al. (2008) |
In the reference, named as Astyanax. b In the reference, it is named as Deuterodon pedri.
Kavalco et al. (2009) described three distinct cytotypes (A, B and C) of D. hastatus in the Guapimirim-Macacu river basin (Table 1). In addition to karyotype formulas, different combinations in the patterns of active nucleolar organizer regions (three or two sites) and 18S rDNA sites (six or eight sites) can differ the cytotypes (Table 1), despite the number of chromosomes (2n = 50) and patterns of constitutive heterochromatin (few positive markings in pericentromeric regions) remaining conserved. Variations in the karyotype formula and different distribution patterns with 18S rDNA were also reported for two sympatric karyomorphs of the species Deuterodon taeniatus (Cunha et al., 2016). In turn, variations in the karyotype formula can be observed among allopatric populations of Deuterodon giton from the Paraíba do Sul River (Kavalco and Moreira-Filho, 2003; Kavalco et al., 2007) and Doce River basins, and for the latter, hybridization with individuals of Oligosarcus argenteus (Aguiar, 2011) was reported.
In the present study, we investigated the phylogenetic relationships between different populations of the D. hastatus species complex and other species distributed on the southeastern Brazilian coast, through an integrated approach comparing cytogenetic and molecular data. Moreover, we present a new cytotype of D. hastatus, named cytotype D. We also estimated the divergence time within the group, using a molecular clock analysis, and discuss the data considering both the karyotypic and molecular evolution of the group.
Material and Methods
We analyzed cell suspensions from eight specimens of D. hastatus that were deposited in the Tissues and Suspension bank of the Federal University of Viçosa Campus Rio Paranaíba. The specimens were collected by Kavalco, K.F. in 2008, in the basin of the Ariró River (East Atlantic watershed) near the municipality of Angra dos Reis/RJ (W 22º54ʹ36.1”/S 44º19’50.3”), and were deposited in the ichthyological collection of the Museum of Zoology of the Federal University of Rio Grande do Sul (Universidade Federal do Rio Grande do Sul - UFRGS) after identification (at the time, designated as Astyanax hastatus), under the code USP 3665-3694.
The metaphase chromosomes were obtained using the protocol of Gold et al. (1990). We then characterized, from a morphological point of view, each chromosome type, according to the arm ratio proposed by Levan (1964). Subsequently, the 18S rDNA (Hatanaka and Galetti, 2004) and 5S rDNA (Martins and Galetti, 1999) probes were mapped to D. hastatus chromosomes via fluorescence in situ hybridization (Pinkel et al., 1986; modified by Pazza et al., 2006). The probes were labeled with biotin-14-dATP via nick translation using the BioNick labeling kit according to the manufacturer’s instructions (Invitrogen LT, Carlsbad, CA, USA).
We randomly chose six individuals from the sample population of D. hastatus for ATP synthase subunit 6 sequencing. DNA from those individuals were isolated from liver tissue using the Purelink Genomic Kit extraction kit (Invitrogen LT), according to the manufacturer’s instructions. After DNA quantification, we diluted the aliquots to a working concentration of 10 ng/μL and amplified the ATPase 6 sequences using the primers ATP 8.2_L8331 (5′-AAAGCRTYRGCCTTTTAAGC) and CO3.2_H9236 (5′-GTTAGTGGTCAKGGGCTTGGRTC) (Sivasundar et al., 2001).
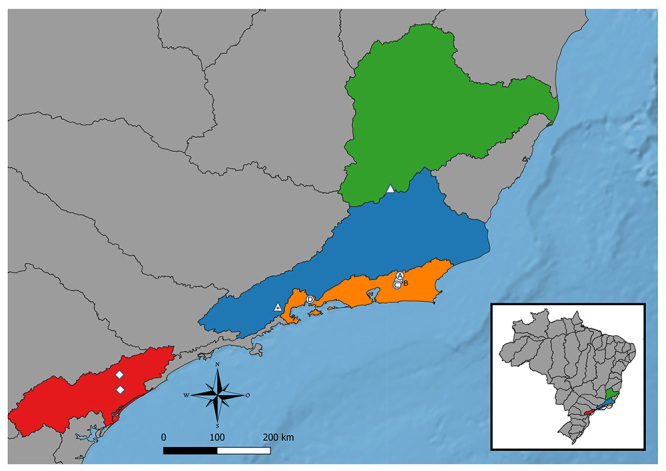
We used the MEGA-X software (Kumar et al., 2018) to perform sequence visualization, editing, and alignment, employing the MUSCLE algorithm (Edgar, 2004), in addition to calculating the interpopulation p distance (Thompson et al., 1994). For phylogenetic analyses, we employed the maximum likelihood estimation method, using IQ-TREE v2.1.2 software with 1000 ultrafast bootstrap replications (Minh et al., 2020). In our dataset, we included sequences of the ATPase 6 gene from four specimens of D. giton (collected from basin streams of the Doce River and the Paraíba do Sul River), three specimens of Deuterodon intermedius (basin of Paraíba do Sul River), seven specimens of Deuterodon ribeirae (two localities in the basin of Ribeira de Iguape River), and 18 specimens of D. hastatus (four localities in the basin of the Guapimirim-Macacu River), in addition to the other 70 sequences from 24 species of the genera Astyanax, Psalidodon, Roeboides, Bryconamericus, Eretmobrycon, and Triportheus (outgroup) present in the NCBI database. The information regarding all sequences used is summarized in supplementary material Table S1. The geographic location of all Deuterodon populations used in our phylogeny can be seen in Figure 1.
Figure 1 -. Map demonstrating the Deuterodon populations used in the phylogeny. Diamonds indicate populations of D. ribeirae, triangles indicate populations of D. giton (asterisk symbolizing sympatry with D. intermedius) and circles indicate populations of D. hastatus (letters representing the different cytotypes). In red the Ribeira de Iguape river basin, in blue the Paraíba do Sul river basin, in green the Doce river basin and in orange the coastal drainages of the state of Rio de Janeiro.
The divergence times of the group were estimated based on a Bayesian relaxed clock model, using BEAUti v. 2.6.6 and BEAST v. 2.6.6 software (Drummond and Bouckaert, 2015). The relaxed clock model used presented a log-normal distribution (non-correlated). For the “Tree Prior” parameter, we used the macroevolutionary Birth-Death model, and as a nucleotide substitution model, we used HKY+G, estimated using ModelFinder (Kalyaanamoorthy et al., 2017). We used four calibration points, which are as follows: 1) the fossil characid †Paleotetra spp. from the Eocene-Miocene (Weiss et al., 2012), used to limit the minimum age of the clade of all characids included in our analysis by implementing a log-normal prior offset of 33.9 million years ago, with a standard deviation of 1; 2) the fossil Triportheidae †Lignobrycon ligniticus from the late Oligocene (Woodward, 1898), used to limit the clade containing all Triportheus species included in our analysis by implementing a log-normal prior offset of 27.5 million years ago, with a standard deviation of 1; 3) the fossil characid †Megacheirodon unicus from the late Oligocene (Travassos and Santos, 1955), used to limit the clade containing all Stervadiinae (Eretmobrycon spp. + Bryconamericus spp.) in our analysis by implementing a lognormal prior offset of 27.5 million years ago, with a standard deviation of 1; 4) and finally, we used, as a calibration point, the origin of Astyanax species in Central America, dated by Ornelas-García et al. (2008) to 7.8-8.1 million years ago. This last calibration point was used to restrain the minimum age of the clade containing all Astyanax and Psalidodon species in our analysis by implementing a log-normal prior offset of 8 million years ago, with a standard deviation of 0.7. We constructed a haplotype network with the Deuterodon species, using the Haplotype Viewer software (Salzburger et al., 2011).
Results
Cytogenetics
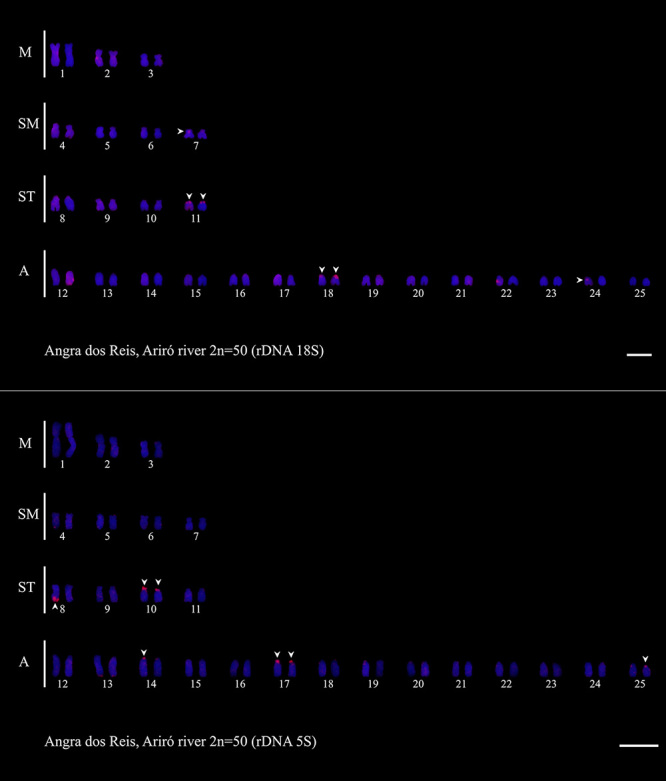
The population analyzed in this study presented 2n = 50 chromosomes, with the karyotype formula 6M+8SM+8ST+28A and NF = 72 (Figure 2). In relation to the 18S rDNA, we observed subtelomeric markers on the short arms of chromosome pairs 11 and 18 and interstitial markers on one of the chromosomes of pairs 7 and 24. We also observed 5S rDNA sites in the subtelomeric region of the short arms of chromosome pairs 10 and 17 and in the terminal region of one of the chromosomes of pairs 8, 14, and 25 (Figure 3).
Figure 2 -. Karyotype of D. hastatus from the Ariró river, Angra dos Reis-RJ. Scale bar: 5 µm.
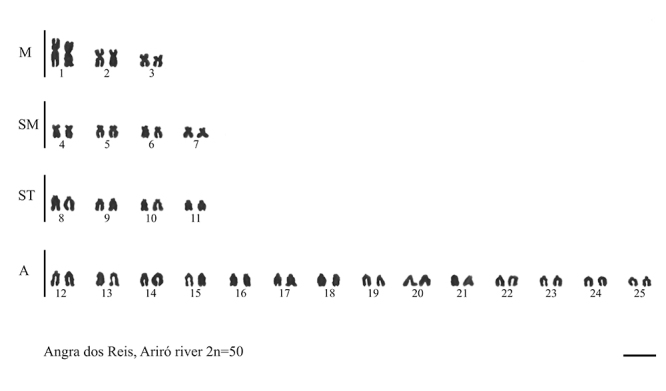
Figure 3 -. Karyotype of D. hastatus after FISH with 18S rDNA (A) and 5S rDNA (B) probes. Scale bar: 5 μm.
Phylogeny
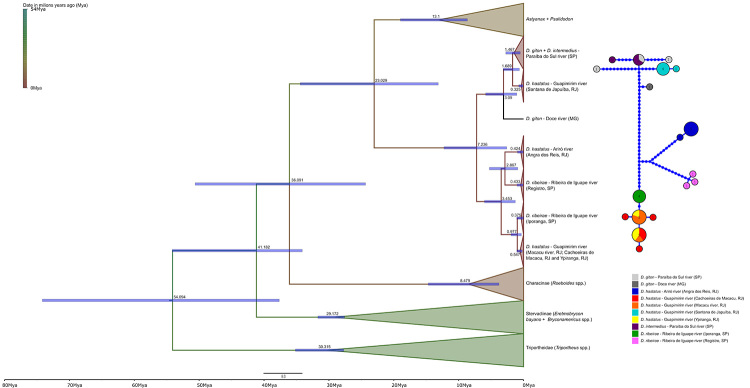
The phylogenetic analysis based on sequences of the ATPase 6 region showed three clusters containing different populations of D. hastatus as follows: (1) one in which individuals from D. hastatus (cytotype D, presented in this study) from the Ariró River population, Angra dos Reis/RJ, were grouped with D. ribeirae (PP) from the Pesqueiro Paraíso, city of Registro/SP (bootstrap = 95 %); (2) one in which individuals of three populations of D. hastatus, two collected from the main channel of the Macacu river, in the municipality of Cachoeiras de Macacu (cytotype C sensu Kavalco et al., 2009) and one in the community Ypiranga (cytotype A sensu Kavalco et al., 2009), from the basin of the Guapimirim-Macacu/RJ River, are grouped with individuals of D. ribeirae (PG) from the community of Poço Grande, Iporanga/SP (bootstrap = 99%); and finally, (3) on in which the sequences from individuals of D. hastatus from Santana de Japuíba population (cytotype B sensu Kavalco et al., 2009) were found to be more closely related to those of D. intermedius and D. giton from the region of Cunha/SP, basin of the Paraíba do Sul River (bootstrap = 99%). In the maximum likelihood phylogram, D. giton from the basin of the Doce River diverged before the clade that harbored the other Deuterodon species used in the analysis (Figure 4). Nevertheless, the same phenomenon was not observed in the Bayesian tree from the molecular clock analysis (Figure 5).
Figure 4 -. Phylogenetic tree of five populations of D. hastatus, two of D. ribeirae, two of D. giton and one of D. intermedius plus 70 sequences from 24 species outside the genus Deuterodon. Bootstrap values are demonstrated on internal nodes. The best evolutionary model, used in the analysis, was the TIM3+F+I+G4 according to ModelFinder.
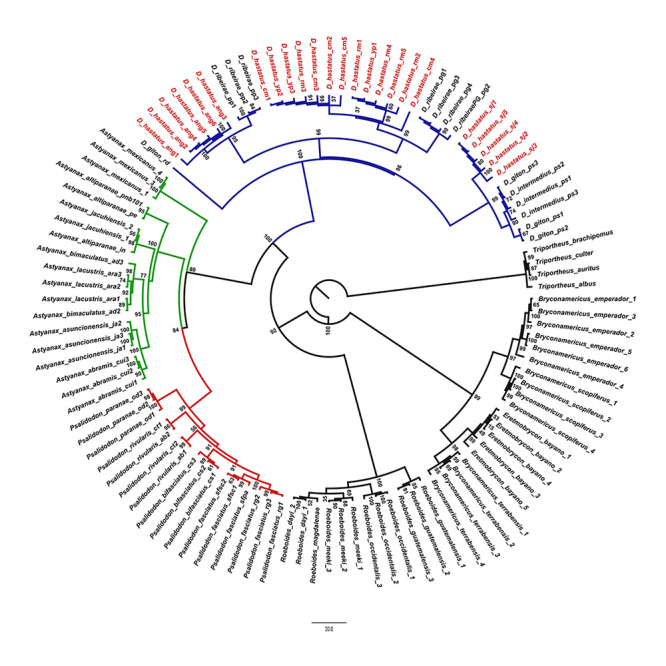
Figure 5 -. On the left, calibrated Bayesian tree, based on the ATP synthase subunit 6 gene, demonstrating the evolutionary relationships between the species of Deuterodon analyzed plus outgroups. Timescale indicated on the x-axis in millions of years. Numbers on inner nodes represent divergence estimates and blue bars represent 95% highest posterior density interval. On the right, haplotype network of the four Deuterodon species analyzed, length of branches proportional to the number of mutations, number of sequences per haplotype demonstrated within each haplotype.
According to the molecular clock analysis, the Deuterodon group diverged from the clade containing Astyanax and Psalidodon at some point in the Oligocene-to-Miocene transition, approximately 23 million years ago (95% HPD, 13.1-34.4 Mya). The first divergence among the analyzed Deuterodon species occurred approximately 7.2 million years ago (95% HPD, 2.6-12.6 Mya), between the Miocene and Pliocene, and it was during this period that the lineage of the population identified as D. hastatus in Santana de Japuíba (cytotype B), together with D. giton and D. intermedius, diverged from the other populations of D. hastatus and D. ribeirae. The separation of D. hastatus of Santana de Japuíba from D. giton and D. intermedius found in the Paraíba do Sul River occurred approximately 1.7 million years ago (95% HPD, 0.6-3.1 Mya), during the Pliocene-to-Pleistocene transition. In relation to the other populations of D. hastatus and D. ribeirae, the first divergence occurred between the Pliocene and Pleistocene, approximately 3.4 million years ago (1.3-6 Mya), and was responsible for separating the populations of D. hastatus from the basin of the Guapimirim-Macacu River (cytotypes A and C) and D. ribeirae of Iporanga (SP) from D. hastatus of the Ariró River (cytotype D) and D. ribeirae of Registro (SP). The final branching between the remaining lineages of D. hastatus of the Guapimirim-Macacu River and D. ribeirae found in Iporanga occurred approximately 1 million years ago (95% HPD, 0.3-1.9 Mya), during the Pleistocene, and that between D. hastatus found in Ariró River and D. ribeirae collected in Registro occurred approximately 2.9 million years ago (95% HPD, 0.9-5.3 Mya) between the Pliocene and Pleistocene.
We obtained 18 haplotypes from the four species of Deuterodon, the polymorphic sites (S) were 83, the nucleotide diversity (Pi) was 0.04476 and the haplotype diversity (Hd) was 0.936. In the haplotype network (Figure 5), it was possible to observe at least four major haplogroups among the Deuterodon lineages. However, these haplogroups did not correspond at all to the taxonomic names of the four studied species. The first haplogroup (dark green, orange, red and yellow) was found to include three populations of D. hastatus (cytotypes A and C) found in the Guapimirim-Macacu River (Macacu River, RJ; Cachoeiras de Macacu, RJ; and Ypiranga community, RJ) and D. ribeirae collected from the Ribeira de Iguape River (Iporanga, SP). Seven haplotypes were found to form this haplogroup, with all being very close to each other. The second haplogroup (pink) was composed of only three haplotypes of D. ribeirae from the Ribeira de Iguape River (Registro, SP). The third haplogroup (dark blue) was composed of two haplotypes of D. hastatus from the Ariró River (cytotype D, Angra dos Reis, RJ). The fourth haplogroup (light gray, dark gray, light blue and purple) was composed of four haplotypes of D. intermedius and D. giton from the Paraíba do Sul River (Cunha, SP) and two haplotypes of D. hastatus from the Guapimirim-Macacu River (cytotype B, Santana de Japuiba, RJ). One of these haplotypes is shared by D. intermedius and D. giton from the rio Paraíba do Sul, with two other haplotypes from each of these species being derived from this one. Another haplotype of D. giton from the Paraíba do Sul river and two other haplotypes of D. hastatus from Santana de Japuíba seem to diverge from this first one from an ancestral haplotype. The genetic distances among all analyzed Deuterodon populations are summarized in Table 2.
Table 2 -. Bottom triangle: genetic distances between analytical populations. Upper triangle: standard deviations.
| 1. | 2. | 3. | 4. | 5. | 6. | 7. | 8. | 9. | 10. | |
|---|---|---|---|---|---|---|---|---|---|---|
| 1. D. giton - Paraíba do Sul river (Cunha, SP) | 0.00584 | 0.01007 | 0.01137 | 0.01135 | 0.00588 | 0.01133 | 0.00364 | 0.01089 | 0.01214 | |
| 2. D. giton - Doce river (Latão Creek, MG) | 0.02511 | 0.01071 | 0.01091 | 0.01088 | 0.00611 | 0.01084 | 0.00514 | 0.01035 | 0.01195 | |
| 3. D. hastatus - Ariró river (Angra dos Reis, RJ) | 0.08014 | 0.06560 | 0.00766 | 0.00750 | 0.01135 | 0.00756 | 0.01075 | 0.00750 | 0.00916 | |
| 4. D. hastatus - Guapimirim river (Cachoeiras de Macacu, RJ) | 0.07721 | 0.06215 | 0.03773 | 0.00125 | 0.01259 | 0.00111 | 0.01187 | 0.00290 | 0.00786 | |
| 5. D. hastatus - Guapimirim river (rio Macacu, RJ) | 0.07684 | 0.06177 | 0.03584 | 0.00218 | 0.01251 | 0.00111 | 0.01185 | 0.00257 | 0.00802 | |
| 6. D. hastatus - Guapimirim river (Santana de Japuiba, RJ) | 0.02109 | 0.02298 | 0.07916 | 0.07382 | 0.07345 | 0.01246 | 0.00503 | 0.01196 | 0.01241 | |
| 7. D. hastatus - Guapimirim river (Ypiranga, RJ) | 0.07596 | 0.06089 | 0.03672 | 0.00201 | 0.00113 | 0.07257 | 0.01176 | 0.00273 | 0.00780 | |
| 8. D. intermedius - Paraíba do Sul river (Cunha, SP) | 0.01381 | 0.01820 | 0.07627 | 0.07282 | 0.07244 | 0.01281 | 0.07156 | 0.01137 | 0.01205 | |
| 9. D. ribeirae - Ribeira de Iguape river (Iporanga, SP) | 0.07345 | 0.05838 | 0.03547 | 0.00603 | 0.00414 | 0.07006 | 0.00502 | 0.06905 | 0.00799 | |
| 10. D. ribeirae - Ribeira de Iguape river (Registro, SP) | 0.08475 | 0.06968 | 0.04488 | 0.04746 | 0.04557 | 0.07947 | 0.04645 | 0.08035 | 0.04143 |
Discussion
In this work, we describe a new cytotype of D. hastatus based on a study of a population from Ariró River, Angra dos Reis/RJ. We referred to it as cytotype D, showing the extensive range of karyotype variation in this group. Herein, we also present the first data of the physical mapping of 5S rDNA in D. hastatus. Our phylogeny also showed paraphyletic groups in at least three species of Deuterodon, namely D. hastatus, D. ribeirae, and D. giton, suggesting the existence of cryptic species in these groups. We also provide the first proposal of a molecular clock for the genus and demonstrate that the evolution of Deuterodon in southeastern Brazil was strongly influenced by the neotectonic events that marked the Pliocene-to-Pleistocene transition.
Despite differences in the karyotype formula (Table 1), cytotype D of D. hastatus has the same number of 18S rDNA sites (six) as the other cytotypes of the species, with the exception of cytotype B from Santana de Japuíba, which has eight sites (Kavalco et al., 2009). In addition, the genetic distance of this cytotype to the others is 3.67% for cytotype A, 7.9% for cytotype B and 3.58-3.77% for cytotype C (Table 2). If we integrate the observed phylogenetic, karyotypic and genetic distances data, we can conclude the existence of at least three different evolutionary significantly units (ESUs) in D. hastatus: one composed of specimens of cytotypes A and C, which have genetic distances of 0.1 and 0.2% between them, another composed by cytotype B, whose genetic distance in relation to the others is between 7.2% and 7.9%, and finally another composed by cytotype D presented here.
As reported for D. giton, D. intermedius, and D. ribeirae (Kavalco and Moreira-Filho, 2003; Kavalco et al., 2007, 2010), D. hastatus lacks the marker chromosome, a metacentric pair carrying the 5S rDNA site in the pericentromeric region, identified by Kavalco et al. (2004, 2010) as a common feature of the other Stethaprioninae genera present in the continent, and first described for some Astyanax species by Almeida-Toledo et al. (2002). Our result corroborates the hypothesis of Pazza et al. (2018) that coastal Stethaprioninae, diverged before the emergence of this site in the interstitial position (i.e., before the fixation of the marker chromosome). The large interspecific variation in 5S rDNA of the Deuterodon species which this marker is disponible is also remarkable. The studied population of D. hastatus possess three more 5S rDNA sites than D. ribeirae (Kavalco et al., 2010), three site less than D. giton found in the Paraíba do Sul River (Kavalco et al., 2004), five more than D. giton from the Doce River (Aguiar, 2011), three less than D. intermedius (Kavalco et al., 2004), and one fewer than D. taeniatus (Da Cunha et al., 2016). The first studies based on this ribosomal DNA sites seemed to indicate a conserved pattern in Characiformes. However, the idea that it is a homogeneous marker in fish was certainly due to the low representativeness in front of the high number of existing species. Even for Deuterodon, it is still necessary to analyze species mainly from the southern Atlantic and northeastern Brazilian coasts and from Guyana, since chromosomal data are concentrated on populations from the southeastern Brazilian coast (Table 1).
According to Pazza et al. (2018), species of the genus Deuterodon (referred to as Astyanax Clade 1) are known to present some symplesiomorphic cytogenetic features, such as the diploid number of 2n = 50, a low FN (FN = 66 - 84), and up to 10 5S rDNA sites, all located in the terminal region of the chromosomes. All of these features can be observed in different Deuterodon species, except for the low FN, since the populations of the genus in Minas Gerais state are likely to have a high FN (FN = 88-94, Table 1) (Aguiar, 2011; Coutinho-Sanches and Dergam, 2015; Da Cunha et al., 2016). Another feature that was mentioned is the absence of a positive hybridization signal for As51 satellite DNA in many species of the genus (Kavalco et al., 2009), except for Deuterodon janeiroensis (Carvalho et al., 2002; Vicari et al., 2008a).
The cytotype B of D. hastatus from the city of Santana de Japuíba had already been suggested by Kavalco et al. (2009) to be a possible cryptic species of D. hastatus, owing to its karyotype differences. As observed by Pazza et al. (2018), this population was closer to D. giton and D. intermedius than to D. ribeirae and the other populations of D. hastatus (Figures 4 and 5). In addition to differences in the karyotype formula, this population was also found to have a higher number of 18S rDNA sites than D. hastatus and D. ribeirae (8 sites vs 6 and 4 respectively) but a lower number than D. giton and D. intermedius (which have 10 and 12 respectively) (Table 1). Thus, our phylogram (Figure 4) can be separated into two subclades, one characterized by Deuterodon populations with a stable number of six 18S rDNA sites (D. hastatus and D. ribeirae) and the other with a variable number of eight or more 18S rDNA sites (D. hastatus SJ, D. giton, and D. intermedius).
Another finding was the absence of monophyly from D. giton haplotypes, as D. giton from the Paraíba do Sul River is paraphyletic in relation to D. intermedius and D. giton from the Doce River does not belong to this group. These specimens were also used in cytogenetic analyses, and in addition to the discrepant karyotype formulas (Table 1), the population from Paraíba do Sul River had more number of active NORs (Kavalco and Moreira-Filho, 2003) and 5S rDNA sites (Kavalco et al., 2004) than the population of the Doce River (Aguiar, 2011) (Table 1). Therefore, we propose that the population of the Doce River, owing to its molecular and cytogenetic features, is a different cryptic species from that found in the Paraíba do Sul River. Thus, it is very likely that the diversity observed in D. hastatus, composed of cryptic species, is similar to that found in D. giton.
The geographic distribution of the D. hastatus populations can be explained by vicariance events related to two hypotheses, which are non-mutually exclusive. One is the capture of headwaters of one river by another that could lead to the fixation of different karyomorphs in nearby locations (Vicari et al., 2008 b ). This hypothesis seems to fit well with the karyotype and molecular differentiation of D. hastatus found in the population of Santana de Japuíba in relation to other populations from the basin of the Guapimirim-Macacu River (Kavalco et al., 2009). The other hypothesis refers to sea level fluctuations that occurred in the Pleistocene, which might have led to the isolation of coastal basins. Radiation chronologically shaped by these fluctuations in the sea level has been proposed for the Odontesthes perugiae complex (Atheriniformes, Atherinopsidae) (Beheregaray et al., 2002). This second hypothesis seems to align more with the situation of D. hastatus from the Ariró River and the other populations of D. hastatus from the Guapimirim-Macacu River (Kavalco et al., 2009), as well as with that of D. giton from the Paraíba do Sul and Doce Rivers (Kavalco and Moreira-Filho, 2003; Kavalco et al., 2007; Aguiar, 2011).
The oldest cladogenic events among Deuterodon species, observed in this study (7.2 Mya), seem to match the oldest cladogenic events among coastal fish of the genus Mimagoniates, estimated at 6.8 Mya (Camelier et al., 2018). These events coincide with strong tectonic activities that occurred in the Neogene, which caused fluvial capture by coastal drainages (Menezes et al., 2008; Ribeiro, 2006). The more recent cladogenic events, observed between D. hastatus from Santana de Japuíba and D. giton/D. intermedius from Paraíba do Sul River, between D. hastatus from Ariró River and D. ribeirae from Iporanga, and finally, between D. hastatus from Macacu/Ypiranga and D. ribeirae from Registro, seem to all date back to the transition epoch between the Pliocene and Pleistocene (1.7, 2.9, and 1 Mya, respectively). This date seems to coincide with the estimated divergence of Astyanax lacustris and Astyanax altiparanae species (Cunha et al., 2019) and with coastal Oligosarcus species in southeastern and southern Brazil (Wendt et al., 2019). This epoch was characterized by intense tectonic activities, which caused several drainage rearrangements through stream capture events and marine transgressions (Ribeiro, 2006).
The successive marine regressions that occurred in the Pliocene-to-Pleistocene transition were associated with frequent fauna exchanges between Brazilian coastal basins (Wendt et al., 2019). Thus, the most recent cladogenic events among Deuterodon from southeastern Brazil can be explained by several exchanges of fauna between the river basins of Ribeira de Iguape, Paraíba do Sul, Doce and coastal rivers of the State of Rio de Janeiro, such as the basin of the Guapimirim-Macacu River (Figure 1). Thus, it is possible that the phylogeographic patterns observed in this study could be explained by events comprising the reciprocal migration of D. ribeirae to the Guapimirim-Macacu basin and D. hastatus to the basin of Ribeira de Iguape. This would explain not only the apparent taxonomic confusion between the two species but also the groups of populations between these two basins, rather than populations within each basin. A similar pattern can be seen in the phylogeny of Mimagoniates macrolepis, where a sample from Itanhaém, SP, is closer to one from the Macacu River, RJ, than to another sample from the Ribeira de Iguape basin, SP (Camelier et al., 2018).
In this work, we were able to observe, based on both cytogenetic and molecular data, the genetic and chromosomal diversity present in some Deuterodon species, even at the intraspecific level. We corroborated our hypothesis referring to the existence of cryptic species complexes in the genus, such as D. hastatus and D. giton. Further studies involving taxonomic and morphological analyses are required to formally classify and describe these units as new species.
Acknowledgments
The authors would like to thank the research funding agencies CAPES and FAPEMIG and the graduate programs of Ecology, UFV; Management and Conservation of Natural and Agricultural Ecosystems, UFV - Forestry; and Zoology, UFMG
Supplementary material
References
- Aguiar HJACD. First report on spontaneous hybridization between Astyanax giton Baird & Girard 1854 and Oligosarcus argenteus Günther 1864 (Pisces: Characidae): Ecological and phylogenetic inferences. Universidade Federal de Viçosa; Viçosa: 2011. 134 M. Sc. Thesis. [Google Scholar]
- Almeida-Toledo LF, Ozouf-Costaz C, Foresti F, Bonillo C, Porto-Foresti F, Daniel-Silva MFZ. Conservation of the 5S-bearing chromosome pair and co-localization with major rDNA clusters in five species of Astyanax (Pisces, Characidae) Cytogenet Genome Res. 2002;97:229–233. doi: 10.1159/000066609. [DOI] [PubMed] [Google Scholar]
- Beheregaray LB, Sunnucks P, Briscoe DA. A rapid fish radiation associated with the last sea-level changes in southern Brazil: The silverside Odontesthes perugiae complex. Proc Biol Sci. 2002;269:65–73. doi: 10.1098/rspb.2001.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2015;43:D30. doi: 10.1093/nar/gku1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camelier P, Menezes NA, Costa-Silva GJ, Oliveira C. Molecular phylogeny and biogeographic history of the Neotropical tribe Glandulocaudini (Characiformes: Characidae: Stevardiinae) Neotrop Ichthyol. 2018;16:e170157 [Google Scholar]
- Carvalho ML, Oliveira C, Foresti F. Cytogenetic analysis of five species of the subfamily Tetragonopterinae (Teleostei, Characiformes, Characidae) Caryologia. 2002;55:181–188. [Google Scholar]
- Coutinho-Sanches N, Dergam JA. Cytogenetic and molecular data suggest Deuterodon pedri Eigenmann, 1907 (Teleostei: Characidae) is a member of an ancient coastal group. Zebrafish. 2015;12:357–365. doi: 10.1089/zeb.2014.1068. [DOI] [PubMed] [Google Scholar]
- Cunha MS, Reis VJC, Dergam JA. Closely related syntopic cytotypes of Astyanax taeniatus (Jenyns, 1842) from the upper Piranga River, upper Doce Basin in southeastern Brazil. Zebrafish. 2016;13:112–117. doi: 10.1089/zeb.2015.1163. [DOI] [PubMed] [Google Scholar]
- Cunha MS, Fregonezi AR, Fava L, Hilsdorf AW, Campos LA, Dergam JA. Phylogeography and historical biogeography of the Astyanax bimaculatus species complex (Teleostei: Characidae) in coastal southeastern South America. Zebrafish. 2019;16:115–127. doi: 10.1089/zeb.2018.1668. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Bouckaert RR. Bayesian Evolutionary Analysis with BEAST. Cambridge University Press; Cambridge: 2015. [Google Scholar]
- Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenmann CH, McAtee WL, Ward PD. On further collections of fishes from Paraguay. American Carnegie Mus. 1907;4:110–157. [Google Scholar]
- Fricke R, Eschmeyer WN, Van der Laan R, editors. Eschmeyer’s Catalog of Fishes: Genera, species, references. California Academy of Sciences. 2022. [23 November 2022]. Fricke R, Eschmeyer WN and Van der Laan R (eds) (2022) Eschmeyer’s Catalog of Fishes: Genera, species, references, California Academy of Sciences, http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp/
- Gold JR, Li YC, Shipley NS, Powers PK. Improved methods for working with fish chromosomes with a review of metaphase chromosome banding. J Fish Biol. 1990;37:563–575. [Google Scholar]
- Hatanaka T, Galetti PM. Mapping of the 18S and 5S ribosomal RNA genes in the fish Prochilodus argenteus Agassiz, 1829 (Characiformes, Prochilodontidae) Genetica. 2004;122:239–244. doi: 10.1007/s10709-004-2039-y. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;4:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalco KF, Moreira- O., Filho Cytogenetical analyses in four species of the genus Astyanax (Pisces, Characidae) from Paraíba do Sul river basin. Caryologia. 2003;56:453–461. [Google Scholar]
- Kavalco KF, Pazza R, Bertollo LAC, Moreira- O., Filho Gene mapping of 5S rDNA sites in eight fish species from the Paraíba do Sul river basin, Brazil. Cytogenet Genome Res. 2004;106:107–110. doi: 10.1159/000078567. [DOI] [PubMed] [Google Scholar]
- Kavalco KF, Pazza R, Bertollo LAC, Moreira- O., Filho Satellite DNA sites in four species of the genus Astyanax (Teleostei, Characiformes) Genet Mol Biol. 2007;30:529–535. [Google Scholar]
- Kavalco KF, KdO Brandão, Pazza R, Almeida-Toledo LF. Astyanax hastatus Myers, 1928 (Teleostei, Characidae): A new species complex within the genus Astyanax? Genet Mol Biol. 2009;32:477–483. doi: 10.1590/S1415-47572009005000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavalco KF, Pazza R, de Almeida-Toledo LF. Molecular cytogenetics of Astyanax ribeirae (Teleostei, Characidae), an endemic characin of the Atlantic rainforest. Nucleus (India) 2010;53:51–54. [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levan A. Nomenclature for centromeric position on chromosomes. Hereditas. 1964;52:201–220. [Google Scholar]
- Lima FCT. In: Check List of the Freshwater Fishes of South and Central America. Reis RE, Kullander SO, Ferraris CJ, editors. Edipucrs; Porto Alegre: 2003. Genera incertae sedis in Characidae; pp. 134–141. [Google Scholar]
- Lucena ZD, Lucena CD. Revisão das espécies do gênero Deuterodon Eigenmann, 1907 dos sistemas costeiros do sul do Brasil com a descrição de quatro espécies novas (Ostariophysi, Characiformes, Characidae) Comun Mus Ciênc Tecnol PUCRS, Sér Zool. 1992;5:123–168. [Google Scholar]
- Martins C, Galetti PM. Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes) Chromosome Res. 1999;7:363–367. doi: 10.1023/a:1009216030316. [DOI] [PubMed] [Google Scholar]
- Menezes NA, Ribeiro AC, Weitzman S, Torres RA. Biogeography of Glandulocaudinae (Teleostei: Characiformes: Characidae) revisited: Phylogenetic patterns, historical geology and genetic connectivity. Zootaxa. 2008;1726:33–48. [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornelas-García CP, Domínguez-Domínguez O, Doadrio I. Evolutionary history of the fish genus Astyanax baird & Girard (1854) (Actinopterygii, Characidae) in Mesoamerica reveals multiple morphological homoplasies. BMC Evol Biol. 2008;8:340. doi: 10.1186/1471-2148-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazza R, Kavalco KF, Bertollo LAC. Chromosome polymorphism in Astyanax fasciatus (Teleostei, Characidae). 1. Karyotype analysis, Ag-NORs and mapping of the 18S and 5S ribosomal genes in sympatric karyotypes and their possible hybrid forms. Cytogenet Genome Res. 2006;112:313–319. doi: 10.1159/000089886. [DOI] [PubMed] [Google Scholar]
- Pazza R, Dergam JA, Kavalco KF. Trends in karyotype evolution in Astyanax (Teleostei, Characiformes, Characidae): Insights from molecular data. Front Genet. 2018;9:131. doi: 10.3389/fgene.2018.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AC. Tectonic history and the biogeography of the freshwater fishes from the coastal drainages of eastern Brazil: An example of faunal evolution associated with a divergent continental margin. Neotrop Ichthyol. 2006;4:225–246. [Google Scholar]
- Pinkel D, Straume T, Gray JW. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc Natl Acad Sci U S A. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Ewing GB, von Haeseler A. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Mol Ecol. 2011;20:1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x. [DOI] [PubMed] [Google Scholar]
- Silva PC, Malabarba MC, Malabarba LR. Using ancient DNA to unravel taxonomic puzzles: The identity of Deuterodon pedri (Ostariophysi: Characidae) Neotrop Ichthyol. 2017;15:e160141 [Google Scholar]
- Sivasundar A, Bermingham E, Ortí G. Population structure and biogeography of migratory freshwater fishes (Prochilodus: Characiformes) in major South American rivers. Mol Ecol. 2001;10:407–417. doi: 10.1046/j.1365-294x.2001.01194.x. [DOI] [PubMed] [Google Scholar]
- Terán GE, Benitez MF, Mirande JM. Opening the Trojan horse: Phylogeny of Astyanax, two new genera and resurrection of Psalidodon (Teleostei: Characidae) Zool J Linnean Soc. 2020;190:1217–1234. [Google Scholar]
- Thompson JD, HIiggins DG, Gibson TJ. CLUSTALW: Improving the sensitivity of progressive multiple sequence through weighing, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travassos H, Santos RS. Caracídeos fósseis da Bacia do Paraíba. An Acad Brasil Ciênc. 1955;27:297–322. [Google Scholar]
- Vicari MR, Artoni RF, Moreira O, Filho, Bertollo LAC. Colocalization of repetitive DNAs and silencing of major rRNA genes. A case report of the fish Astyanax janeiroensis. Cytogenet Genome Res. 2008;122:67–72. doi: 10.1159/000151318. [DOI] [PubMed] [Google Scholar]
- Vicari MR, Noleto RB, Artoni RF, Moreira- O, Filho, Bertollo LAC. Comparative cytogenetics among species of the Astyanax scabripinnis complex: Evolutionary and biogeographical inferences. Genet Mol Biol. 2008;31:173–179. [Google Scholar]
- Weiss FE, Malabarba LR, Malabarba MC. Phylogenetic relationships of Paleotetra, a new characiform fish (Ostariophysi) with two new species from the Eocene-Oligocene of south-eastern Brazil. J Syst Palaeotol. 2012;10:73–86. [Google Scholar]
- Wendt EW, Silva PC, Malabarba LR, Carvalho TP. Phylogenetic relationships and historical biogeography of Oligosarcus (Teleostei: Characidae): Examining riverine landscape evolution in southeastern South America. Mol Phylogenet Evol. 2019;140:106604. doi: 10.1016/j.ympev.2019.106604. [DOI] [PubMed] [Google Scholar]
- Woodward AS. Considerações sobre alguns peixes Terciários dos schistos de Taubaté, Estado de São Paulo, Brasil. Rev Mus Paulista. 1898;3:63–70. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.