SUMMARY
The mechanoelectrical transduction (MET) channel of cochlear hair cells is gated by the tip link, but the mechanisms that establish the exquisite force sensitivity of this MET channel are not known. Here, we show that the tetraspan lipoma HMGIC fusion partner-like 5 (LHFPL5) directly couples the tip link to the MET channel. Disruption of these interactions severely perturbs MET. Notably, the N-terminal cytoplasmic domain of LHFPL5 binds to an amphipathic helix in TMC1, a critical gating domain conserved between different MET channels. Mutations in the amphipathic helix of TMC1 or in the N-terminus of LHFPL5 that perturb interactions of LHFPL5 with the amphipathic helix affect channel responses to mechanical force. We conclude that LHFPL5 couples the tip link to the MET channel and that channel gating depends on a structural element in TMC1 that is evolutionarily conserved between MET channels. Overall, our findings support a tether model for transduction channel gating by the tip link.
In brief
Qiu et al. demonstrate that the tetraspan LHFPL5 tethers the tip link to the MET channel to establish maximal force sensitivity of this ion channel. Channel gating depends on a structural element in TMC1 that is evolutionarily conserved between MET channels.
Graphical abstract

INTRODUCTION
Mechanoelectrical transduction (MET), the conversion of mechanical stimuli into electrical signals, is essential for organisms to perceive a wide range of external and internal stimuli. In recent years, a variety of MET channels have been identified.1 How these channels are gated by mechanical force still needs further study. MET channels in hair cells are localized at the lower end of tip links,2 the extracellular filaments that connect the stereocilia of a hair cell near their tips (Figure 1A). During sound-induced deflection of stereocilia, tip links are thought to transmit mechanical force onto MET channels.3–8 Tip links might couple directly to MET channels, affect membrane curvature, or both.
Figure 1. LHFPL5 is unique among LHFP family members to facilitate hair cell MET.

(A) Diagram of a hair cell and the MET complex.
(B) Phylogenetic tree and membrane topology of LHFP family members.
(C) Examples of OHCs from Lhfpl5 +/− mice at P3 + 1 day in vitro (DIV) after injectoporation of the indicated constructs and stained for phalloidin (red) and MYC or HA (white). Scale bar, 5 μm.
(D) MET currents in OHCs from Lhfpl5−/− ice at P3 + 1 DIV after injectoporation of the indicated constructs. MET currents are in response to 10-ms hair bundle deflections by a stiff glass probe from −400 nm to 1,000 nm.
(E) Current/displacement (I/X) plots from data as in (D). All plots were fitted with a double Boltzmann equation (n = number of cells, mean ± SEM).
(F) Quantification of peak MET currents at 1,000-nm hair bundle deflections obtained from data in (D) and (E) (mean ± SEM; one-way ANOVA with Dunnett’s multiple comparisons test; n.s., not significant; p > 0.05; ***p < 0.001).
See also Figures S1 and S2.
The study of genes linked to deafness has led to identification of molecular components of the MET machinery of hair cells. CDH23 and PCDH15 form the upper and lower parts of tip links, respectively.9–12 TMC1/2 and TMIE localize near the lower end of tip links13,14 and are obligatory subunits of the MET channel.14–19 The tetraspan lipoma HMGIC fusion partner-like 5 (LHFPL5; TMHS) is another component of the MET channel complex. LHFPL5 binds to PCDH15, TMIE, and TMC1.20–23 In Lhfpl5−/− mice, PCDH15 and TMC1 are present in stereocilia at reduced levels, suggesting that LHFPL5 is required for assembly/stability of the MET channel complex.20,23 Electrophysiological recordings from Lhfpl5−/− hair cells have demonstrated altered properties of the remaining MET channels, including changes in resting open probability, rise kinetics, unitary channel conductance, and adaptation.20,23 Based on these findings, it has been proposed that LHFPL5 is a regulatory subunit of the MET channel, similar to the structurally related TARP proteins that regulate the function of glutamate receptors.23
To define the mechanism by which LHFPL5 regulates MET, we took advantage of the fact that LHFPL5 is a member of a small family of six proteins. Among the members of the LHFP gene family, only mutations in the gene for Lhfpl5 have been linked to inherited forms of deafness.24–26 Consistent with this finding, we demonstrate here that LHFPL5 has a unique role in supporting MET by cochlear hair cells that is not shared by other LHFP family members. We then generated a chimera between LHFPL5 and its relative LHFPL3 to map protein domains that are essential for LHFPL5 functions. We demonstrate that the N-terminal part of LHFPL5 is critical for efficient binding of LHFPL5 to the tip link and MET channel as well as for normal MET. To our surprise, four amino acids in the N-terminal cytoplasmic domain of LHFPL5 are essential for establishing optimal force sensitivity of the MET channel. These N-terminal amino acids of LHFPL5 mediate interactions with an amphipathic helix in TMC1, a structural motif that is consistently found in MET channels and required for channel gating.1 We demonstrate that this domain in TMC1 is essential for channel gating by a mechanism that involves interactions with LHFPL5. LHFPL5 thus directly anchors the MET channel to the tip link and gates the channel via an amphipathic helix in TMC1.
RESULTS
Among LHFP proteins, LHFPL5 has a unique role in supporting hair cell MET
LHFPL5 is a member of the LHFP gene family, which is comprised of 6 proteins.25 These proteins share significant sequence homology and contain four predicted transmembrane domains (Figures 1B and S1A). In hair cells from Lhfpl5−/− mice, MET is drastically impaired but not completely eliminated.20,23 We therefore determined whether other LHFP family members can compensate for the loss of LHFPL5. Cochlear hair cells express, at postnatal day 0 (P0) and P1, high amounts of LHFPL5 transcripts and modest amounts of LHFPL2 and LHFPL4. No transcripts for LHFP, LHFPL1, and LHFPL3 were observed (https://umgear.org/).
We first determined whether LHFP family members can localize to stereocilia. We expressed, by injectoporation, LHFP family members tagged with hemagglutinin (HA) or MYC in outer hair cells (OHCs) from Lhfpl5+/− mice at P3. Cochlear explants were stained 1 day later with antibodies to HA or MYC. LHFPL1–LHFPL5, but not LHFP, were detected in stereocilia of OHCs (Figures 1C and S1B).
Next, we analyzed the extent to which LHFP family members can restore MET in OHCs from Lhfpl5−/− hair cells. We focused this analysis on LHFPL2 and LHFPL4, which are expressed at low levels in hair cells (https://umgear.org/), and on LHFPL3, the closest LHFPL5 homolog (Figures 1B and S1A). OHCs were injectoporated at P3 to express LHFP proteins as well as GCaMP3 to identify, by GFP fluorescence, the cells that had taken up the plasmid DNA. We observed that epitope tags slightly affected LHFPL5 function by reducing peak currents and thus carried out rescue experiments with proteins lacking an epitope tag. MET currents were recorded after 1 day in vitro (DIV) by patching on the cell body of OHCs and stepwise deflection of their stereocilia from −400 to 1,000 nm using a stiff glass probe. MET currents were only rescued in OHCs injectoporated to express LHFPL5 (currents at 1 μm deflection: 89.2 ± 15.5 pA for non-transfected cells, 50.7 ± 5.2 pA for LHFPL2, 66.0 ± 16.6 pA for LHFPL3, 71.7 ± 20.8 pA for LHFPL4, and 506.1 ± 41.8 pA for LHFPL5) (Figures 1D–1F).
LHFPL3 is not essential for normal MET
The fact that LHFPL5 has a role in hair cell MET that is not shared by other LHFP family members allowed us to interrogate the unique properties of LHFPL5 by constructing chimeras between LHFPL5 and its closest homolog, LHFPL3. Prior to initiating these experiments, we wanted to ascertain that LHFPL3 is not critical for MET or hearing function. We therefore obtained Lhfpl3 knockout mice generated by replacing the first coding exon of Lhfpl3 with a LacZ cassette.27
Hearing function was normal in Lhfpl3−/− mice, as determined by measurement of the auditory brain stem response (ABR) to click and pure tone stimuli (Figure S2A). MET currents could be evoked in OHCs from Lhfpl3−/− mice at P6 without significant differences compared with Lhfpl3+/− controls (currents at 1 μm deflection: 737.8 ± 55.3 pA for Lhfpl3+/− and 690.9 ± 37.1 pA for Lhfpl3−/−) (Figures S2B–S2D). To further confirm that LHFPL3 does not contribute to MET by hair cells, we recorded MET currents of OHCs from Lhfpl3−/−;Lhfpl5 −/− double mutants. Previous studies have shown that MET currents are drastically reduced in OHCs from Lhfpl5−/− mice.23 There was no further significant current reduction in OHCs from Lhfpl3−/−;Lhfpl5 −/− double mutants (currents at 1 μm deflection: 133.8 ± 15.8 pA for Lhfpl3+/−;Lhfpl5 −/− and 120.9 ± 20.3 pA for Lhfpl3−/−;Lhfpl5 −/−; Figures S2B–S2D). Plots of the open probability of the transducer channel against displacement and fitted with a double Boltzmann equation showed that the response of the MET channel to force and/or the kinetics of channel gating were affected by LHFPL5 but not by LHFPL3 (Figure S2C).
The N-terminal half of LHFPL5 is required for binding to PCDH15, TMIE, and TMC1
LHFPL5 can bind to PCDH15, TMIE, and TMC1.14,20,23,28 In addition, structural data suggest that the N-terminal part of LHFPL5, including the first transmembrane domain and the first extracellular loop of LHFPL5, are in close proximity to the transmembrane and extracellular membrane-proximal domain of PCDH15, respectively.29 We hypothesized that critical interactions with the transduction machinery were not maintained in other LHFP family members because of specific amino acid substitutions. To test this model, we generated chimeric proteins between LHFPL5 and LHFPL3.
We first swapped the N- and C-terminal halves of LHFPL3 and LHFPL5 to generate L5-L3 and L3-L5, respectively (Figure 2A). We also introduced FLAG tags into the C terminus of LHFPL5, LHFPL3, L5-L3, and L3-L5 to facilitate protein detection. Next, we expressed the different proteins alone or together with PCDH15 in HEK293 cells. We used, for all biochemical experiments, the PCDH15-CD2 splice variant that is a component of tip links.30,31 Cell extracts were prepared ~36 h after transfection, and protein complexes were isolated by immunoprecipitation with anti-FLAG antibodies and resolved on SDS-PAGE gels. PCDH15 was detected with an antibody that we have described previously.30 As controls, we analyzed the input amount of the epitope-tagged proteins prior to immunoprecipitation and total amount of co-precipitating proteins (Figure 2B). Quantification of the results revealed that PCDH15 was efficiently co-immunoprecipitated with LHFPL5 and L5-L3 but not with LHFPL3 or L3-L5 (Figure 2B), indicating that the N-terminal half of LHFPL5 is important to mediate strong interactions with PCDH15.
Figure 2. The N-terminal half of LHFPL5 mediates protein-protein interactions and is required for MET.

(A) Schematic depicting LHFPL3 (L3, black) and LHFPL5 (L5, red) chimeras.
(B–D) HEK293 cells were transfected with the constructs indicated at the top of each panel. Immunoprecipitation (IP) was carried out with FLAG-conjugated beads, followed by western blotting with antibodies to PCDH15-CD2 or MYC epitopes (top rows, coIP; center rows, IP; bottom rows, input). Shown is quantification of coIP results from at least 3 independent experiments. Binding of LHFPL proteins and chimeras to PCDH15, TMIE, and TMC1 was normalized to LHFPL5 values (Rel. Int., relative intensity; mean ± SEM; Student’s t test; n.s., p > 0.05; *p < 0.05; **p < 0.01).
(E) Examples of OHCs from Lhfpl5 −/− mice at P3 + 1 DIV after injectoporation of the indicated constructs and immunostained for phalloidin (red) and HA (white). Scale bar, 5 μm.
(F) MET currents in OHCs from Lhfpl5 −/− mice at P3 + 1 DIV after injectoporation of the indicated constructs. Shown are MET currents in response to hair bundle deflections.
(G and H) I/X and plots from data as in (F) and Figure 1D (n = number of cells, mean ± SEM).
(I) Quantification of peak MET currents at 1,000-nm hair bundle deflections obtained from (G).
(J and K) Slopes (Z1 and Z2) (J) and set point (X0) (K) plots obtained from double Boltzmann fitting of the data from (G).
(I–K) Mean ± SEM; one-way ANOVA with Dunnett’s multiple comparisons test; n.s., p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001.
Next, we compared binding of TMIE and TMC1 to LHFPL5, LHFPL3, and the chimeric proteins. We expressed FLAG-tagged LHFPL5, LHFPL3, L5-L3, and L3-L5 together with MYC-tagged TMIE and TMC1 in HEK293 cells and carried out co-immunoprecipitation experiments. LHFPL5 and L5-L3 bound significantly stronger to TMIE compared with LHFPL3 and L3-L5. TMC1 also bound significantly stronger to LHFPL5 compared with LHFPL3, but interactions were reduced with L5-L3 and L3-L5 (Figures 2C and 2D). We conclude that the N-terminal half of LHFPL5 is required to mediate efficient interactions of LHFPL5 with PCDH15 and TMIE, while efficient interactions with TMC1 depend on additional amino acids in the C-terminal LHFPL5 half.
The N-terminal half of LHFPL5 is required for normal MET
To analyze the extent to which the N-terminal half of LHFPL5 is critical for MET by hair cells, we injectoporated L5-L3 and L3-L5 chimeras into OHCs from Lhfpl5−/− mice at P3. For immunolocalization studies, we used proteins containing a C-terminal epitope tag, and for functional studies, we used proteins without an epitope tag but co-expressed GCaMP3 to identify cells that had taken up the plasmids as described previously.23,32 Immunolocalization studies and electrophysiological recordings were carried out after 1 DIV. The chimeric proteins localized to the stereocilia of the injectoporated cells (Figure 2E) and L5-L3, but not L3-L5, significantly rescued MET currents compared with non-injectoporated hair cells (Figures 2F, 2G, and 2I). Statistical analysis showed that peak MET-current amplitude in OHCs injectoporated to express L5-L3 was slightly reduced compared with OHCs injectoporated to express wild-type LHFPL5, while peak current amplitude in OHCs expressing L3-L5 was not significantly different from peak currents in non-transfected OHCs (currents at 1 μm deflection: 89.2 ± 15.5 pA for non-transfected cells, 506.1 ± 41.8 pA for LHFPL5, 368.2 ± 35.6 pA for L5-L3, and 69.0 ± 11.6 pA for L3-L5) (Figures 2G and 2I). The slight reduction of peak currents in hair cells expressing L5-L3 is likely explained by reduced binding to TMC1 (Figure 2D), while the strong reduction in currents observed in L3-L5-expressing hair cells is likely due to perturbed interactions of the chimeric proteins with PCDH15, TMIE, and TMC1.
We also plotted the of the MET channel against displacement and fitted the data with a double Boltzmann equation (Figure 2H). As previously reported,23,33 the curve in non-injectoporated OHC from Lhfpl5−/− mice was significantly shifted to the right and broadened with changes in set point and slope compared with OHCs injectoporated with wild-type LHFPL5 (Figures 2H, 2J, and 2K). The relationship was fully restored in OHCs expressing L5-L3 but not L3-L5 (Figures 2H, 2J, and 2K).
In conclusion, the N-terminal part of LHFPL5 is critical for efficient binding to PCDH15, TMC1, and TMIE, suggesting that MET defects observed in hair cells expressing LHFPL3 and L3-L5 are caused at least in part by defects in coupling of proteins within the MET complex.
Different requirements for the binding of LHFPL5 to PCDH15 and TMIE
We carried out additional experiments to narrow down domains in the N-terminal LHFPL5 half that are required for efficient interactions with its binding partners. Since both the N- and C-terminal part of LHFPL5 are required for efficient interactions with TMC1 (Figure 2D), we focused our experiments on PCDH15 and TMIE. We generated a chimera between LHFPL5 and LHFPL3 containing the N-terminal cytoplasmic domain, transmembrane domain 1 (TM1) and the first extracellular loop of LHFPL5 (L5-NcytoTM1Lp1). A second chimera retained the N-terminal cytoplasmic domain and TM1 of LHFPL5 (L5-Ncy-toTM1) (Figure S3A). L5-NcytoTM1Lp1 and L5-NcytoTM1 no longer efficiently interacted with PCDH15, while both constructs still bound to TMIE, albeit with slightly reduced efficiency compared with wild-type LHFPL5 (Figures S3B and S3C).
Next, we expressed the chimeras by injectoporation in OHCs of Lhfpl5−/− mice at P3. Epitope-tagged versions of the proteins were used for immunolocalization and non-epitope-tagged versions for functional analysis. The two chimeras localized to stereocilia of OHCs (Figure S3D), but only L5-NcytoTM1Lp1 slightly rescued MET currents compared with non-transfected cells, albeit less efficiently compared with wild-type LHFPL5 (currents at 1 μm deflection: 89.2 ± 15.5 pA for non-transfected cells, 506.1 ± 41.8 pA for LHFPL5, 103.2 ± 12.1 pA for L5-NcytoTM1, and 209.7 ± 35.6 pA for L5-NcytoTM1Lp1) (Figures S3E, S3F, and S3H). Plots of the of the MET channel against displacement revealed that both chimeras partially restored the relationship compared with non-transfected OHCs (Figures S3G, S3I, and S3J).
We conclude that the N-terminal cytoplasmic domain of LHFPL5 and/or TM1 are sufficient to promote interactions with TMIE, while interactions with PCDH15 required additional domains of LHFPL5, including extracellular loop 1 and TM2. The findings also suggest that perturbations of interactions of LHFPL5 with PCDH15 alone without significantly affecting binding to TMIE are sufficient to affect MET by hair cells.
The N-terminal cytoplasmic domain of LHFPL5 is critical for normal MET
The N-terminal cytoplasmic domain of LHFPL5 differs from LHFPL3 by an addition of three amino acids in the extreme N terminus of LHFPL5 and substitution of an aliphatic Ile at amino acid 13 with an aliphatic Leu (Figure 3A). To determine the extent to which the N termini of LHFPL5 and LHFPL3 are functionally interchangeable, we swapped the N-terminal cytoplasmic domains of LHFPL5 and LHFPL3 to generate NTL3-L5 and NTL5-L3, respectively (Figure 3A). We expressed FLAG-tagged LHFPL5, LHFPL3, and chimeras in HEK293 cells for co-immunoprecipitation experiments with PCDH15, TMIE-MYC, and TMC1-MYC. NTL3-L5 strongly bound to PCDH15, TMIE, and TMC1 similarly as to LHFPL5 (Figures 3B–3D). In contrast, NTL5-L3 showed weak binding to all three proteins, similar to LHFPL3 (Figures 3B–3D). These findings suggest that the N-terminal cytoplasmic domain of LHFPL5 is not essential to mediate protein-protein interactions of LHFPL5 with PCDH15, TMIE, and TMC1.
Figure 3. The N-terminal cytoplasmic domain of LHFPL5 is critical for MET.
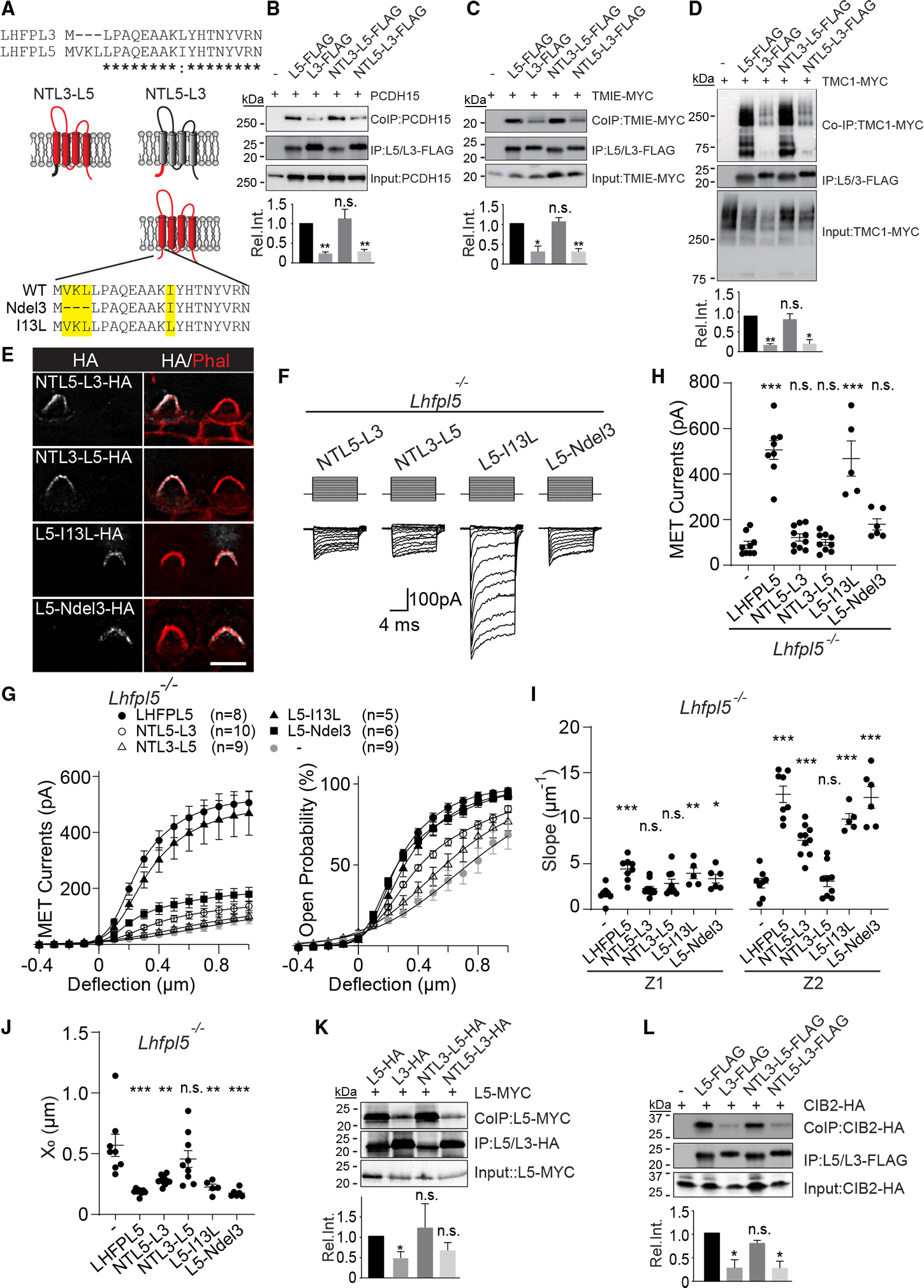
(A) Top: sequence alignment of LHFPL3 and LHFPL5 N-terminal cytoplasmic domains (asterisks, identical amino acids; colon, amino acids with similar properties). Center: LHFPL3 (L3, black) and LHFPL5 (L5, red) chimeras. Bottom: LHFPL5 N-terminal mutants.
(B–D) HEK293 cells were transfected with the constructs indicated at the top of each panel. Experimental details and labeling of panels is as in Figures 2B–2D (mean ± SEM; Student’s t test; n.s., p > 0.05; *p < 0.05; **p < 0.01).
(E) OHCs from Lhfpl5−/− mice at P3 + 1 DIV after injectoporation of the indicated constructs and immunostained for phalloidin (red) and HA (white). Scale bar, 5 μm.
(F) MET currents in OHCs from Lhfpl5−/− mice at P3 + 1 DIV after injectoporation of the indicated constructs.
(G) I/X (left) and (right) plots from data similar as in (F) and Figure 1D (n = number of cells, mean ± SEM).
(H) Quantification of peak MET currents at 1,000-nm hair bundle deflections obtained from (D).
(I and J) Slopes (Z1 and Z2) (I) and set point (X0) (J) plots obtained from double Boltzmann fittings of data from (F) (mean ± SEM; one-way ANOVA with Dunnett’s multiple comparisons test. n.s., p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
(K and L) Analysis of LHFPL5 dimerization (K) and CIB2 binding (L). Experimental conditions, quantification, and panel labeling are as in (B)–(D).
See also Figure S3.
We next expressed the chimeras by injectoporation in OHCs of Lhfpl5−/− mice at P3 and observed protein localization to stereocilia after 1 DIV (Figure 3E). MET recorded from OHCs injectoporated with NTL5-L3 showed a similarly low MET current amplitude as in control Lhfpl5−/− hair cells (currents at 1 μm deflection: 89.2 ± 15.5 pA for non-transfected cells and 121.7 ± 15.8 pA for NTL5-L3) with slightly rescued relationship (Figures 3F–3J), further indicating that the interactions with the tip link and MET channel are required for normal MET. To our surprise, NTL3-L5, which binds to PCDH15, TMIE, and TMC1, failed to rescue MET currents and the relationship (currents at 1 μm deflection: 89.2 ± 15.5 pA for non-transfected cells and 100.1 ± 13.0 pA for NTL3-L5) (Figures 3F–3J), suggesting that MET channels in hair cells expressing NTL3-L5 were less sensitive to hair bundle deflections.
We also analyzed the function of LHFPL5 proteins engineered to only contain the three-amino-acid deletion at the extreme N terminus (L5-Ndel3) or only the Ile13Leu substitution (L5-I13L) (Figure 3A). L5-I13L rescued MET to near-normal levels, while L5-Ndel3 was impaired in its ability to rescue MET in Lhfpl5 −/− hair cells, although not as severely as NTL3-L5, which contained both mutations (currents at 1 μm deflection: 506.1 ± 41.8 pA for LHFPL5, 468.2 ± 77.4 pA for L5-I13L, and 179.3 ± 24.3 pA for L5-Ndel3) (Figures 3F–3H). Failure to rescue MET was not caused by defects in protein trafficking since both mutant proteins were detected in stereocilia of injectoporated hair cells (Figure 3E). We also plotted the and fitted the data with a double Boltzmann equation. Unlike for NTL3-L5, the relationship was restored by L5-I13L and L5-Ndel3 (Figures 3H–3J).
We conclude that the N-terminal cytoplasmic domain of LHFPL5 is critical to regulate the sensitivity of MET channels to hair bundle deflection without noticeably affecting interactions of LHFPL5 with PCDH15, TMIE, and TMC1.
The N-terminal cytoplasmic domain of LHFPL5 is not essential for dimerization or CIB2 binding
Structural data suggest that the lower insertion point of tip links contains two LHFPL5 proteins that interact with two PCDH15 molecules.29 We wondered whether LHFPL5 proteins form dimers and whether defects in homodimerization of NTL3-L5 might explain why this chimera failed to rescue MET in OHCs of Lhfpl5−/− mice. We therefore carried out co-immunoprecipitation experiments with extracts from HEK293 cells transfected to express LHFPL5-MYC with LHFPL5-HA, LHFPL3-HA, and NTL3-L5-HA (Figure 3K). Immunoprecipitation was carried out with antibodies to HA, followed by detection of co-immunoprecipitated proteins with antibodies to MYC. LHFPL5 formed homodimers but bound less efficiently to LHFPL3 (Figure 3K). Homodimer formation was maintained with NTL3-L5-HA, indicating that the N-terminal cytoplasmic domain of LHFPL5 is not critical for homodimerization (Figure 3K).
We wondered whether the N-terminus of LHFPL5 might bind to CIB2, which interacts with TMC1 and is an accessory protein of the MET channel of cochlear hair cells.34–36 Interactions of CIB2 with LHFPL5 had not been evaluated previously. Co-immunoprecipitation experiments demonstrated that LHFPL5 bound more efficiently to CIB2 compared with LHFPL3 (Figure 3L). This interaction was not affected by NTL3-L5 (Figure 3L), suggesting that the interaction depends on domains of LHFPL5 distinct from the N-terminal cytoplasmic domain.
We conclude that it is unlikely that disruption of the formation of LHFPL5 homodimers or binding of LHFPL5 to CIB2 causes MET defects in OHCs expressing an LHFPL5 protein carrying the Ndel3 and Ile13Leu mutations.
Mutations in the N-terminal cytoplasmic domain of LHFPL5 affect hearing
To investigate the mechanisms by which the N-terminal cytoplasmic domain of LHFPL5 affects MET, we used CRISPR/Cas9 to generate a knockin mouse line that converts the N-terminal cytoplasmic domain of LHFPL5 into LHFPL3 (Lhfpl5Ndel3-I13L) (Figure 4A). Heterozygous Lhfpl5Ndel3-I13L/+ mice had normal hearing (data not shown), while homozygous Lhfpl5Ndel3-I13L/Ndel3-I13L mice were deaf (Figure 4B). Unlike in Lhfpl5−/− mutant mice,23 the morphology of hair cell stereocilia from homozygous Lhfpl5Ndel3-I13L/Ndel3-I13L mice appeared normal at the lightmicroscopy level, at least up to P15 (Figures 4C, 4D–4F, and 4H–4J), the last time point analyzed.
Figure 4. Hearing defects but normal protein localization in Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice.

(A) Schematic depicting the CRISPR/Cas9-generated genomic modifications to alter the N-terminal cytoplasmic domain of LHFPL5.
(B) ABR thresholds in response to click and pure tone stimuli for wild-type (WT) and Lhfpl5 Ndel3-I13L/Ndel3-I13L mice at 4–6 weeks of age (n = number of animals, mean ± SEM).
(C) Cochlear whole mounts from mice of the indicated genetic background at P8 and P15, stained with phalloidin (green) to reveal hair bundles. Scale bar, 5 μm.
(D–F and H–J). Cochlear whole mounts from mice of the indicated genetic background were stained with phalloidin (red) and antibodies to PCDH15-CD2 (D), LHFPL5 (E), HA (F and I), PMCA2 (H), and MYC (J) (white). Mice in (D), (I), and (J) also contained genetic modifications to introduce epitope tags into TMC1 (HA), TMIE (HA), and TMC2 (MYC).15 Scale bars,10 μm.
(G) Quantification of expression levels of PCDH15, LHFPL5, and TMC1-HA in hair bundles (n = number of images quantified from at least 2 mice; mean ± SEM; one-way ANOVA with Sidak’s multiple comparisons test; n.s., p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001).
The localization of PCDH15 and TMC1 to stereocilia is affected in Lhfpl5−/− mice.20,23 To test whether the localization of components of the MET machinery is also affected in Lhfpl5Ndel3-I13L/Ndel3-I13L mice, we stained cochlear whole mounts from Lhfpl5−/− mice and Lhfpl5Ndel3-I13L/Nde3-I13L mice at P6 with antibodies to PCDH15 and LHFPL5. For a quantitative assessment of fluorescence signals, we used Imaris 9.7 to analyze expression in optical sections through the entire hair bundle of OHCs as described previously.34 At P6, PCDH15 was largely reduced, and LHFPL5 was not detectable in hair bundles in Lhfpl5−/− mice compared with controls (Figures 4D, 4E, and 4G). In contrast, amounts of PCDH15 and LHFPL5 in hair bundles of Lhfpl5Ndel3-I13L/Ndel3-I13L mice showed similar expression levels as in controls (Figures 4D, 4E, and 4G).
To analyze effects of Lhfpl5 mutants on TMC1 distribution, we took advantage of Tmc1HA/HA mice, which express TMC1 containing an HA epitope.15 We generated them by crossing Lhfpl5Ndel3-I13L/Ndel3-I13L;Tmc1HA/HA and Lhfpl5−/−;Tmc1HA/HA mice and stained cochlear whole mounts at P4 and P6 with HA antibodies. Consistent with earlier data, TMC1-HA was rarely detected in the stereocilia of hair cells in Lhfpl5−/−; Tmc1HA/HA mice, but it was present in the stereocilia of Lhfpl5Ndel3-I13L/Ndel3-I13L;Tmc1HA/HA mice (Figure 4F). Quantification of TMC1-HA fluorescence in hair bundles of OHCs in the middle to basal part of the cochlea using Imaris 9.7 showed decreased TMC1-HA expression levels in Lhfpl5−/−;Tmc1HA/HA mice starting at P6. Compared with wild-type controls, TMC1-HA levels in Lhfpl5Ndel3-I13L/Ndel3-I13L;Tmc1HA/HA were normal at P4 and slightly reduced at P6 (Figure 4G).
Next, we analyzed expression of TMIE and TMC2 in Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice. As a further control, we evaluated the localization of the plasma membrane calcium ATPase 2 (PMCA2), which is not directly linked to the MET complex. PMCA2 expression was evaluated with antibodies to PMCA2. TMC2 and TMIE expression was evaluated by crossing Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice with Tmc2MYC/MYC mice and TmieHA/HA mice that contain a Tmc2 and Tmie gene modified to encode a MYC tag or HA tag at the C-terminus of TMC2 and TMIE, respectively.15 OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L; Tmc2MYC/MYC mice and Lhfpl5 Ndel3-I13L/Ndel3-I13L;TmieHA/HA mice were analyzed with antibodies to MYC and HA, respectively. We found similar levels of PMCA2, TMIE, and TMC2 in hair cells from Lhfpl5Ndel3-I13L/Ndel3-I13L mice compared with wild-type control mice (Figures 4H–4J).
Thus, unlike in Lhfpl5−/− mice, proteins of the MET channel complex are expressed at normal levels in stereocilia of Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice at early postnatal stages, although TMC1 levels are somewhat decreased by P6.
Mutations in the N-terminal cytoplasmic domain of LHFPL5 affect MET
We recorded MET currents in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice in the mid-apical cochlea at P5–P6 OHCs. Peak MET currents were significantly reduced in the mutant OHCs and similar to those of Lhfpl5−/− mutants (currents at 1 μm deflection: 888.5 ± 50.4 pA for Lhfpl5+/+ and 233.2 ± 24.1 pA for Lhfpl5Ndel3-I13L/Ndel3-I13L) (Figures 5A–5C).
Figure 5. Defects in MET in cochlear hair cells from Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice.

(A) MET currents of P6 OHCs in the mid-apical cochlea of WT and Lhfpl5Ndel3-I13L/Ndel3-I13L mice.
(B) I/X plots from similar data as in (A). Plots fitted with a double Boltzmann equation (n = number of cells, mean ± SEM).
(C) MET current amplitude at 1,000-nm deflection from data as in (B) (mean ± SEM, one-way ANOVA with Sidak’s multiple comparisons test, ***p < 0.001).
(D) MET currents evoked with sinusoidal fluid deflection of hair bundles from OHCs of indicated mice. The recording location along the cochlea is indicated at the top; P4–P6 for apex and middle and P2–P4 for base.
(E) MET current amplitude/relative distance along cochlea plots obtained from data as in (D) (n = number of cells from apex, middle, and base; mean ± SEM).
(F) and G) Single-channel currents of TMC1-containing MET channels from OHCs measured from Tmc2−/−-background mice.
(F) Top: representative single-channel events. Bottom: amplitude histograms generated from the middle representative traces. Gaussian fits of the two peaks in the histograms determine a single-channel current of 7.9 pA and 8.2 pA for the WT and mutant, respectively.
(G) Summary plot of single-channel currents (n = 113 events from 8 cells for WT controls and 82 events from 6 cells for Lhfpl5Nde3-I13L/Ndel3-I13L mutant mice) (mean ± SEM, Student’s two-tailed unpaired t test, **p < 0.01).
The amplitude of macroscopic MET currents in OHCs gradually decreases from the base to the apex of the murine cochlea. This tonotopic gradient is affected in OHCs from Lhfpl5−/− mice.20 To evaluate tonotopic MET current changes in Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice, we stimulated their hair bundles with a fluid jet and recorded maximal MET currents of OHCs from the apical, middle, and basal part of the cochlea (Figure 5D). Current size increased significantly from the apex to base in wild-type but not Lhfpl5−/− mice (Figure 5E). The amplitudes of MET currents in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice were smaller than in wild-type and similar to Lhfpl5−/− mice in the cochlear apex. In the basal cochlea, currents from Lhfpl5Ndel3-I13L/Ndel3-I13L mutants had smaller amplitudes compared with wild-type mice but were significantly larger than the currents in the apical cochlea of Lhfpl5Ndel3-I13L/Ndel3-I13L mutants or in the basal cochlea of Lhfpl5−/− mice (Figures 5D and 5E). We conclude that, compared with wild-type mice, MET currents are reduced in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice at all tonotopic locations but that a tonotopic current gradient is maintained.
Reduced numbers of active MET channels in Lhfpl5Ndel3-I13L/Ndel3-I13L mice
MET currents were reduced in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice without a similar reduction in the amounts of proteins of the MET complex within stereocilia. This could be explained by reductions in single-channel currents. Alternatively, the mechanical stimulus provided by hair bundle deflection might not properly activate the channel. To analyze changes in channel properties, we determined single-channel currents. The experiments were carried out in Lhfpl5Ndel3-I13L/Ndel3-I13L mice on a Tmc2−/− background to measure TMC1-dependent currents that start to predominate by P6.13,16,17,37 Single-channel currents were slightly but significantly lower in Lhfpl5Ndel3-I13L/Ndel3-I13L;Tmc2−/− mice compared with Lhfpl5+/+;Tmc2−/− controls (8.23 ± 1.91 pA for Lhfpl5+/+, Tmc2−/− and 7.42 ± 1.79 pA for Lhfpl5Ndel3-I13L/Ndel3-I13L;Tmc2−/−) (Figures 5F and 5G). The slight change in channel conductance cannot explain the overall reduction in macroscopic MET currents in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice. Instead, the findings suggest that the number of channels in OHCs that can be activated by mechanical force is significantly reduced in mutants.
MET channels in Lhfpl5Ndel3-I13L/Ndel3-I13L mice are less sensitive to mechanical force
Plots of the of the MET channel against displacement and fitted with a double Boltzmann equation revealed that the relationship in Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice was shifted to the right and broadened with changed set points and slopes compared with controls (Figures 6A–6C). Similar to Lhfpl5−/− mutant OHCs,23 we observed an increase in resting in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice compared with controls (3.8% ± 0.4% for Lhfpl5+/+ and 7.2% ± 0.6% for Lhfpl5Ndel3-I13L/Ndel3-I13L) (Figures 6A and 6D). However, it should be noted that current levels in mutants were low, which makes it challenging to determine . To evaluate adaptation, which could affect the set point of the relationship and channel resting , we evoked MET currents by 400-nm probe deflections for 50 ms. The decay from the peak of the inward current to the end of the mechanical stimulus was fitted with a double-exponential equation (Figure 6E). Comparing the parameters from exponential fittings, the fast and slow time constant (Figure 6F), fast and slow component fraction (Figure 6G), and extent of adaptation (Figure 6H) were not significantly different between OHCs from wild-type and Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice. We conclude that MET channels in OHCs of the mutant mice were less sensitive to hair bundle deflection or less well coordinated in their response to force without noticeable effects on channel adaptation.
Figure 6. Altered gating properties of MET channels in Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice.

(A) plots from data as in Figure 5A. Plots were fitted with a double Boltzmann equation (n = number of cells, mean ± SEM).
(B and C) Slope (Z1 and Z2) (B) and set point (X0) (C) plots obtained from double Boltzmann fitting in (A).
(D) Resting plots obtained at zero deflection in (A).
(E) MET currents (top) and normalized currents (bottom) of OHCs from WT and Lhfpl5Ndel3-I13L mice as indicated during 50-ms, 400-nm bundle deflection. Averaged traces are shown in red.
(F and G) Fast and slow component time constants (F) and fractions (G) obtained by a double-exponential fitting of data in (E).
(H) Extent of adaptation calculated as the ratio of reduced current at steady state to the peak current from data in (E).
(I) Rise time of MET currents calculated as the time required for currents rising from 10% to 90% of peak from data in (E).
(J) MET currents of OHCs from WT, Lhfpl5−/−, and Lhfpl5Ndel3-I13L/Ndel3-I13L mice at P5–P6 in the mid-apical cochlear turn. Stiff-probe stimulations were used as a set of 10-ms hair bundle deflections ranging from −400 nm to 1,000 nm with 100-nm steps (black) and 1,200–2,000 nm with 200-nm steps (red).
(K and L) I/X and plots obtained from data as in (J). Plots were fitted with a double Boltzmann equation (n = number of cells).
(M) Quantification of current ratio at 2- to 1-mm deflection, determined from data as in (K).
Data are mean ± SEM; Student’s two-tailed unpaired t test. n.s., p > 0.05; ***p < 0.001).
Since the curve for Lhfpl5Ndel3-I13L/Ndel3-I13L mice indicated that MET currents were not saturated at 1,000-nm deflection as in wild-type mice (Figure 6A), we wondered whether stronger mechanical stimulation of the hair bundle of Lhfpl5Ndel3-I13L/Ndel3-I13L mice might lead to opening of additional MET channels. MET currents in OHCs from wild-type mice saturated at deflections of 1,000 nm or less (Figures 6J, 6K, and 6M). In contrast, MET currents in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice continued to increase with increasing hair bundle deflection of up to 2,000 nm, the most extreme deflection evaluated (ratio of MET current at 2- to 1-μm deflection: 99.8 ± 2.7% for wild-type mice and 154.1 ± 7.9% for Lhfpl5Ndel3-I13L/Ndel3-I13L mice) (Figures 6J, 6K, and 6M). Plots of the of the transducer channel against displacement and fitted with a double Boltzmann equation showed that currents in OHCs from mutants were not even saturated at 2,000-nm deflection (Figure 6L). These findings could be explained in different ways. As one possibility, force might not be efficiently transferred to MET channels because of reduced coupling of channels to tip links or to changes in bundle stiffness. Alternatively, the intrinsic sensitivity of the MET channel to force might be altered.
To gain further insights into the mechanisms by which LHFPL5 regulates the sensitivity of the MET channel to hair bundle deflection, we analyzed the kinetics of channel activation. MET channels in wild-type hair cells open rapidly in response to mechanical stimulation, within a few microseconds.38,39 Because of the limitations in the speed of the stimulation probe, it is difficult to determine the exact rise kinetics of the MET current.40 However, activation of the MET channel is so severely affected in Lhfpl5−/− OHCs that a 3- to 4-fold delay is noticeable in current traces.23 In contrast, within the constraints of our recording equipment, we did not observe a significant increase in rise time in OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice compared with wild-type mice (10%–90% current rise time: 81.4 ± 5.9 ms for wild-type mice and 103.6 ± 9.8 ms for Lhfpl5Ndel3-I13L/Ndel3−113Lmice) (Figure 6I). This finding, together with the fact that the interaction with the MET channel and tip link is not impaired by the mutation, suggests that, within the noticed limitations, it is unlikely that a major defect in force transfer onto the MET channel in OHCs of Lhfpl5Ndel3-I13L/Ndel3-I13L mutant mice causes the observed shift and broadening of the . Instead, we favor the hypothesis that the mutation in LHFPL5 affects the force sensitivity of the channel.
The LHFPL5 N-terminus mediates interactions with an amphipathic helix in TMC1 that is critical for channel function
Common structural elements of all known MET channels are amphipathic helices that are positioned to bind to the cytoplasmic leaflet of the plasma membrane. These amphipathic helices are critical for mechanical gating.1 Structural studies of the C. elegans TMC1 protein complex have identified an amphipathic helix within the N terminus of TMC1 that is conserved in mammalian TMC1 and TMC2. This helix has been named H3 and is preceded by two additional helices, H1 and H2, and is followed by a short helix, H4 (Figure 7A).41 We hypothesized that the amphipathic H3 in TMC1 might be critical for the function of the MET channel. The sequence of H3 is not well conserved in mammalian TMC3 (Figure S4A), and TMC3 cannot rescue MET defects when expressed in hair cells from TMC1/TMC2 double mutants (Figure S4B). We therefore replaced H3 from TMC1 with H3 from TMC3. As controls, we substituted H1 and H2 of TMC1 with helices from TMC3 (Figures 7A and S4A). We named the constructs TMC1-H1-TMC3, TMC1-H2-TMC3, TMC1-H1/2-TMC3, and TMC1-H3-TMC3. MET recorded from OHCs injectoporated to express the chimera demonstrated significant transducer currents in TMC1-H1-TMC3-, TMC1-H2-TMC3-, and TMC1-H1/2-TMC3-expressing hair cells but not in hair cells expressing TMC1-H3-TMC3 (Figures 7B–7E). We conclude that the H3 helix of TMC1 is critical for the function of TMC1 and that H3 of TMC3 cannot substitute for this function.
Figure 7. The N-terminal cytoplasmic domain of LHFPL5 binds to an amphipathic helix in TMC1.

(A) Diagram indicating helix 1 (H1)–H4 in the N-terminal cytoplasmic domain of TMC1 and TMC1/TMC3 chimeras.
(B) MET currents in OHCs from Tmc1dn/dn; Tmc2−/− mice at P3 + 2 DIV after injectoporation of the indicated constructs.
(C and D) I/X and plots from data as in (B).
(E) Quantification of peak MET currents at 1,000-nm hair bundle deflections obtained from (C) (mean ± SEM; one-way ANOVA with Sidak’s multiple comparisons test; n.s., p > 0.05; ***p < 0.001).
(F) Diagram of PAC-HA and PAC-TMC1-HA.
(G and H) CoIP experiments and quantification as in Figures 2B–2D. Quantifications were from at least 3 independent experiments (Rel. Int., relative intensity; mean ± SEM; Student’s t test; n.s., p > 0.05; *p < 0.05; **p < 0.01).
(I) Model for LHFPL5 function. LHFPL5 couples the MET channel to the tip link; the N-terminal cytoplasmic domain of LHFPL5 interacts with the amphipathic H3 in the MET channel near the cytoplasmic leaflet to regulate channel gating. In Lhfpl5−/− mice, coupling between the tip link and MET channel is affected. In Lhfpl5Ndel3-I13L/Ndel3-I13L mice, LHFPL5 still couples the MET channel to the tip link through transmembrane and extracellular domains, but interactions between the amphipathic helix in the TMC1 N terminus and the N-terminal cytoplasmic domain of LHFPL5 are disrupted.
See also Figure S4.
We next wondered whether defects in MET in hair cells expressing LHFPL5-Ndel3-I13L might be caused by defects in the interaction of the cytoplasmic N terminus of LHFPL5 with H3 of TMC1. Defects in these interactions might not be observed in co-immunoprecipitation experiments of full-length LHFPL5-Ndel3-I13L with full-length TMC1(Figure 3D) because interactions between other protein domains, such as the transmembrane domains, may be sufficiently strong to mask defects in the interaction between the N termini of the two proteins. To assay interactions of H3 with LHFPL5 in isolation from other TMC1 domains, we transplanted the N-terminal cytoplasmic domain of TMC1 onto an unrelated transmembrane protein. We reasoned that this would allow for membrane targeting of the TMC1 cytoplasmic domain while maintaining its normal orientation relative to the cell membrane. We substituted the N-terminal cytoplasmic domain of PAC with the N-terminal cytoplasmic domain of TMC1 (Figure 7F). PAC contains two transmembrane domains and forms a proton-activated chloride channel.42 We chose PAC because our co-immunoprecipitation experiments demonstrated that this protein does not bind to LHFPL5 (Figure S4C).
We transfected HEK293 cells to express PAC-TMC1-HA with LHFPL5-FLAG, LHFPL3-FLAG, and NTL3-LHFP5-FLAG. Immunoprecipitation was carried out with antibodies to FLAG, followed by detection of co-immunoprecipitated proteins with antibodies to HA. PAC-TMC1-HA interacted with LHFPL5-FLAG but not with LHFPL3-FLAG or NTL3-LHFP5-FLAG (Figure 7G). Next, we substituted H1, H2, and H3 helices of TMC1 within the PAC-TMC1-HA construct with H1, H2, and H3 from TMC3 to mimic the constructs used for functional studies (Figure 7B). Co-immunoprecipitation with LHFPL5-FLAG demonstrated that H3, but not H1 or H2, of TMC1 was required for interactions of LHFPL5 with PACTMC1-HA.
In summary, the N-terminal cytoplasmic domain of LHFPL5 mediates interactions with the amphipathic H3 in TMC1, which is an essential structural element for TMC1 function. These interactions depend on amino acids in the N-terminal cytoplasmic domain of LHFPL5 that are not conserved in LHFPL3, which also cannot substitute for LHFPL5 in promoting MET by cochlear hair cells. We conclude that interactions between the N terminus of LHFPL5 and H3 of TMC1 are critical for MET by cochlear hair cells.
DISCUSSION
Here, we show that the tetraspan LHFPL5 is required to establish maximal force sensitivity of the MET channel of cochlear hair cells. Our findings support a model where LHFPL5 tethers the MET channel to the tip link to allow efficient force transfer during mechanical stimulation. Amino acids in the N-terminal LHFPL5 cytoplasmic domain are crucial for normal activation of the MET channel by hair bundle deflections and mediate their effect on TMC1 through an evolutionarily conserved structural element of MET channels that is critical for their gating by mechanical force (Figure 7I).1
Previous biochemical data obtained with proteins expressed in heterologous cells have demonstrated that LHFPL5 binds to PCDH15, TMIE, and TMC1.14,20,23,28 We now provide insights into the molecular determinants that are required for binding of LHFPL5 to its partners. Consistent with structural data,29 we demonstrate that the N-terminal LHFPL5 half is required for efficient binding of LHFPL5 to PCDH15. This domain of LHFPL5 also mediates interactions with TMIE, while additional amino acids in the C-terminal half of LHFPL5 contribute to TMC1 binding. LHFPL5/LHFPL3 chimeras with reduced binding affinity for PCDH15, TMC1 and TMIE are functionally impaired, leading to reduced MET current amplitude and a shifted and broadened curve. These phenotypic changes are similar to those in Lhfpl5−/− mice23,33 and are consistent with defects in the coupling of the MET channel to the tip link. The coupling defects could lead to a defect in the stability of the transduction complex and loss of some channels from stereocilia. Coupling defects could also explain the reduction in sensitivity of the MET channel to stereocilia deflection, as reflected by the shifted curves. Notably, our studies do not exclude a role of the membrane in regulating channel function. Recent studies show that the membrane of stereocilia forms a viscoelastic element important for regulating MET.43 Structural studies of the C. elegans TMC1 complex have shown that transmembrane domains of TMIE and TMC1 form a striking intramembranous cavity occupied by lipid molecules near the putative pore-forming TMC-1 helices.41 In addition, a cysteine at the cytosolic boundary of the TMIE transmembrane domain is palmitoylated and close to the TMC1 pore,41 suggesting a possible role of TMIE and lipids in regulating MET channel function. Indeed, the phospholipid binds to TMIE and is required for normal MET.15,44 Our findings show that the cytoplasmic domains of LHFPL5 also interact with several lipids (CC and UM; unpublished data). Further studies are thus necessary to analyze effects of LHFPL5 on the cell membrane.
Our studies demonstrate that the function of LHFPL5 cannot easily be explained by an adaptor function of LHFPL5 alone. Accordingly, mutations in the N-terminal cytoplasmic domain of LHFPL5 in Lhfpl5Ndel3-I13L/Ndel3-I13L mice affect MET without noticeably disrupting interactions with PCDH15, TMIE, TMC1, and CIB2. In OHCs from Lhfpl5Ndel3-I13L/Ndel3-I13L mice at P6, MET current amplitude is reduced, and the relationship is shifted to the right and broadened, similar to what we observed in Lhfpl5−/− mice.23,33 Unlike in Lhfpl5−/− mice, the localization of MET components to stereocilia and single-channel currents of MET channels from Lhfpl5Ndel3-I13L/Ndel3-I13L mice are largely unaffected at P6. This is consistent with the model where not all channels in OHC stereocilia from Lhfpl5Ndel3-I13L/Ndel3-I13L mice are efficiently activated by the mechanical stimulus. Recruitment of additional MET channels is only observed upon deflection of hair bundles beyond 1,000 nm when currents in wild-type bundles are saturated. This could be a consequence of a defect in the transfer of force to the MET channel or in the channel’s ability to respond to force. In support of the latter hypothesis, we did not observe an increase in the rise time of the MET current in Lhfpl5Ndel3-I13L/Ndel3-I13L mice, at least within the limitations of our recording equipment. This differs dramatically from results with OHCs from Lhfpl5−/− mice, where we observed a large delay in rise time.23 In addition, we demonstrate that the N terminus of LHFPL5 binds to an amphipathic helix in TMC1 that is critical for TMC1 function. Such amphipathic helices are structural motives common to all known MET channels, and they are critical for the response of these channels to mechanical force.1 Our findings are thus consistent with a model where LHFPL5 provides a mechanical link between the tip link and the MET channel, where interactions of the N-terminal cytoplasmic domain with the amphipathic helix are crucial for channel gating (Figure 7I).
The structure of a TMC1-containing ion channel complex from C. elegans has been determined,41 consisting of two subunits each of the nematode TMC1, TMIE, and CIB2 proteins. However, orthologs of PCDH15 and CDH23 are not present in the C. elegans genome, and tip links have so far not been observed in worms. In addition, in C. elegans, TMC1 is required for a background leak current, pH sensing, and touch sensitivity.45–47 It is not clear whether the reported structure of the C. elegans ion channel complex reflects TMC1 complexes for all three functions and how it relates to the tip-link-gated ion channel in mammals. Further structural studies with mammalian proteins will be important to determine the spatial relationship of MET channel subunits within the ion channel complex and coupling to the tip link. Our findings suggest a highly interconnected structure where LHFPL5 links several proteins of the ion channel complex into a tight unit.
Previous studies have shown that the conductance of MET channels in OHCs increases from the apex to the base of the cochlea.48,49 It has subsequently been reported that OHCs at the basal end of the cochlea contain three times as many or more TMC1 molecules per stereocilium compared with OHCs at the apical end.50 These data suggest that the number of MET channels per tip link varies. Simultaneous activation of several ion channel pores by mechanical force would require tight coordination between subunits. How such tight coupling could be achieved is unclear, but LHFPL5 might play a role. Consistent with this model, the gradient in MET channel conductance is dramatically diminished in Lhfpl5−/− mice.20 Perhaps the tips of stereocilia contain a cluster of LHFPL5 molecules. Some of these molecules might not interact with the tip link but might couple several TMC1-containing complexes into a larger unit. However, the gradient in TMC1 molecules along the tonotopic axis was revealed with a transgenic mouse overexpressing a GFP-tagged TMC1 molecule.50 It would be important to confirm these findings with a mouse expressing TMC1 at physiological levels.
Limitations of the study
Mechanisms of MET channel gating have been inferred by characterizing channel properties using electrophysiological measures within OHCs. Ultimately, these studies need to be complemented with an analysis of hair bundle mechanics and structural studies. In addition, reconstitution of the channel complex in heterologous systems, which has so far eluded the field, will be critical to probe channel function and gating mechanisms.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ulrich Mueller (umuelle3@jhmi.edu).
Materials availability
Newly generated materials such as genetically modified mouse lines are available to qualified researchers upon request and after providing a signed MTA agreement.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mouse strains
All animal experiments were approved by the Institutional Animal Care and Use Committee at Johns Hopkins University School of Medicine (#M016M257). All mice were group-housed in pathogen-free facilities with regulated temperature and humidity and given ad libitum access to food and water. Mice were maintained on a 14 h light/10 h dark cycle. Both male and female mice were used for experiments, and no obvious differences were observed between the sexes. All mice used were seemingly free of infection, health abnormalities, or immune system deficiencies. None of the mice used had been used for previous experiments. Mice were used between P0 and adulthood with specific ages for each experiment listed in the results section for each experiment. Littermates of both sexes were randomly assigned to experimental groups. Littermates were used as controls as indicated in the results section of the main text. Mice with loss-of-function alleles of Lhfpl5 has been described as previously (Lhfpl5−/− also referred to as Tmhstm1kjn51 Tmc1-HA, Tmc2-MYC, Tmie-HA mice which introduce epitope tags to the endogenous proteins have been described and tested without affect hair cell function.15,34
Mice with a loss-of function allele of Lhfpl3 were generated with sperm obtained from MMRRC (B6;129S5-Lhfpl3tm1Lex/Mmucd; identification number 032433-UCD). This mouse line was generated by homologous recombination replacing the first coding exon of Lhfpl3 with a LacZ expression cassette.27 Correct gene targeting was confirmed by Southern blot, PCR analysis and DNA sequencing. Loss of expression of the normal L3 transcript was confirmed by RT-PCR analysis.
CRISPR/CAS9 technology was used to generate the Lhfpl5Ndel3-I13L mouse lines. Exons were analyzed for potential sgRNA target sites using the website CRISPOR (crispor.tefor.net). Target sites were chosen based on proximity to desired genomic region and minimal number of predicted off-target sites. Target specific crRNA was ordered from Integrated DNA Technologies (IDT), along with tracrRNA. To generate mutations, ssDNAs containing desired insertions/mutations were designed containing 60 bp homology arms flanking the region of interest. ssDNAs also included silent mutations to PAM sites to prevent excessive cleavage by Cas9 after integration. Pronuclear injection of one-cell C57BL/6J embryos (Jackson Laboratories) was performed by the JHU Transgenic Core using standard microinjection techniques using a mix of Cas9 protein (30ng/ul, PNABio), tracrRNA (0.6μM, Dharmacon), crRNA (0.6μM, IDT) and ssDNA oligo (10ng/ul, IDT) diluted in RNAse free injection buffer (10 mM Tris-HCl, pH 7.4, 0.25 mM EDTA). Injected embryos were transferred into the oviducts of pseudopregnant ICR females (Envigo). Pseudopregnant mice were allowed to give birth, offspring resulting from embryo injections were tail-clipped at P21, and genomic DNA was collected. Genomic DNA was screened using PCR and sequencing to determine presence of deletion and point mutation. Founder mice were bred with C57BL/6J mice (RRID:IMSR JAX:000,664) and offspring were screened to verify germ-line transmission of mutations.
Genetic modification for mutant stain
Lhfpl5Ndel3-I13L:5′-GTGAAGTTGCTGCCAGCCCAGGAGGCCGCCAAGATC-3′ (encoding 2–13 VKLLPAQEAAKI), were mutated to 5′-CTGCCAGCCCAGGAGGCCGCCAAGCTC-3′(encoding 2–10 LPAQEAAKL)
Cell lines
The HEK293 (ATCC# CRL-1573) cell line was used for heterologous expression. Cells were grown at 37°C, 5% CO2 in 1X DMEM+ Glutamax (Gibco) containing 10% Fetal Bovine Serum and 1X Antibiotic-Antimycotic (Gibco). Cells used in experiments were passaged a maximum of twenty times.
METHOD DETAILS
Auditory brainstem response (ABR) measurements
ABR measurements were performed following our published procedures.15,34 Briefly, 4–6 weeks old mice were anesthetized with Ketamine (100 mg/kg) and Xylazine (20 mg/kg) and placed on a heating pad inside of a sound attenuating chamber. Body temperature was monitored throughout the test session with a rectal temperature probe. Subdermal platinum needle electrodes (E2, GrassTechnologies, West Warwick, RI) were placed on the left pinna (inverting), vertex (non-inverting), and on the leg muscle (ground). ABR stimuli were delivered through a free-field speaker (FD28D, Fostex, Tokyo, Japan), placed 10 cm away from the animal’s head. ABR stimuli generation and signal acquisition were controlled by a BiosigRz software interfacing TDT WS4 high performance computer workstation. ABR stimuli consisted of clicks or 5 ms tone pips of varying frequencies (4,8,16, 24, 32kHz) and presented at a rate of 21/s. Stimuli were presented in descending sound levels from maximum speaker output level in 10dB increments. Responses were collected using a low impedance head stage (RA4L1; TDT), pre-amplified and digitized (RA4PA preamp; TDT) and sent to an RZ6 processing module. The signal was filtered (300 −3000 Hz) and averaged over 512 presentations. Threshold was determined statistically based on ABR input/output function and defined as the sound level at which the peak-to-peak ABR signal magnitude was two standard deviations above the average background noise level.
Injectoporations of cochlear hair cells
Injectoporation experiments to express exogenous DNA in cochlear hair cells were carried out following our published procedures.15,32,34 Briefly, the Organ of Corti was dissected from P3 mice and cultured as explants in 1X DMEM/F12 containing 1.5 μg/mL ampicillin. Various plasmid DNA constructs (1 g/mL, described below) were injected in explants using glass pipettes (2 μm diameter) between rows of hair cells. For electrophysiological recording purpose, GCamp3 construct (0.5 μg/mL) was co-transfected as an indicator. Simultaneously with injection, explants were electroporated with four pulses at 60 V (15 msec pulse length, 1 s inter-pulse intervals, ECM 830 square wave electroporator; BTX). After electroporation, half of the media was replaced with DMEM/F12 containing 10% fetal bovine serum. Explants were incubated for 1–2 days at 37°C, 5% CO2 before electrophysiological recording and/or immunostaining (see details below).
Electrophysiology
Mechanotransduction (MET) current recording were performed following our published procedures from P2-P6 old mice.15,34 Apical perfusion was used during recording to perfuse artificial perilymph (in mM): 144 NaCl, 0.7 NaH2PO4, 5.8 KCl, 1.3 CaCl2, 0.9 MgCl2, 5.6 glucose, and 10 H-HEPES, pH 7.4. Borosilicate glass pipettes with filament (Sutter, CA) were pulled with a P-2000 pipette puller (Sutter, CA), and polished with an MF-930 microforge (Narishige, Japan) to resistances of 2–3 MΩ. Mechanical stimulation of hair bundles were applied with a stiff glass probe mounted on a piezoelectric stack actuator (Thorlab, Newton, NJ) or fluid Jet from a glass pipette that was produced by a 27-mm-diameter piezoelectric disc. For stiff glass probe stimuli, the actuator was driven by voltage steps that were low-pass filtered at a frequency of 10 kHz with a 900CT eight-pole Bessel filter (Frequency Devices) to diminish the resonance of the piezo stack. For Fluid Jet, sinusoids (40 Hz) were applied and filtered at 1.0 kHz. Whole-cell recordings were carried out with an EPC 10 USB patch-clamp amplifier (HEKA, Germany) and signals were sampled at 100 kHz. To record macroscopic currents, the patch pipette was filled with intracellular solution (in mM): 140 mM CsCl, 1 mM MgCl2, 1 mM BAPTA, 2 mM Mg-ATP, 0.3 mM Na-GTP and 10 mM H-HEPES, pH 7.2. In rescue experiments, intracellular Ca2+ buffer was used as 0.1mM EGTA. Hair cells were voltage clamped at −84 mV (except for rescue experiments and Lhfpl3 Lhfpl5 double mutations recording that clamped at −74mV). Uncompensated series resistance was less than 5 MΩ (except for rescue experiments). For single-channel recordings, we followed our published procedures.15,34 Hair cells were hold at −80mV whole cell configuration with the intracellular solution (140 mM CsCl, 1 mM MgCl2, 1 mM EGTA, 2 mM Mg-ATP, 0.3 mM Na-GTP and 10 mM H-HEPES, pH 7.2) and briefly treated with Ca2+ free solution (144 mM NaCl, 0.7 mM NaH2PO4, 5.8 mM KCl, 5 mM EGTA, 0.9 mM MgCl2, 5.6 mM glucose, and 10 mM H-HEPES, pH 7.4) by fluid jet. Fluid jet stimuli were applied to hair bundle and single channel events were recorded and offline filtered at 2KHz for further analysis. The events with significant plateau of open status were chosen for amplitude analysis.
Whole-mount and explant immunohistochemistry and imaging
Whole mount cochlea or cultured explants after injectoporation were dissected, fixed, and immunostained.15,34 Briefly, for whole mount cochlea, temporal bones were removed from the skull of P4-P8 mice in 1x HBSS containing 0.01 mM CaCl2. Slightly openings were made in the bony cochlear shell at the apex and through the oval and round windows. Temporal bones were then incubated in 1x HBSS containing 4% PFA and 0.01 mM CaCl2 for 30 min at room temperature (RT) with gentle agitation. For injectoporated explants, dishes containing explants were incubated with 1x HBSS containing 4% PFA and 0.01 mM CaCl2 for 30 min at RT. Fixed tissues were washed in 1x PBS for 3 times, the bony cochlear shell was removed, and the Organ of Corti was separated from the modiolus and collected for immunostaining (for whole mounts). Reissner’s membrane and the Tectorial membrane were dissected away, and the Organ of Corti was permeabilized in PBS containing 0.5% Triton for 30 min at RT with agitation. After permeabilization, the tissue was blocked in PBS containing 10% Goat Serum (GS) for 4–6h at RT. The tissue was then incubated in PBS containing 5% GS and primary antibodies (see below) for 24–48 h at 4°C. The tissue was then washed three times in PBS and incubated for 1 h at RT in PBS containing 5% GS, secondary antibodies (1:5000, Invitrogen, see below), and fluorescently-conjugated Phalloidin to label actin-rich stereocilia (Life Technologies, Phalloidin 488, 555, 1:1000). After secondary antibody incubation, the tissue was washed three times and mounted using ProLong Gold (Invitrogen). For tissues stained only for phalloidin, all steps were the same except for omission of primary antibody step. Tissues were imaged using 100x lenses on a widefield fluorescence deconvolution microscope (Deltavision, GE Life Sciences). Imaris software (Bitplane, Inc) was used to quantify expression levels of PCDH15, LHFPL5 and TMC1-HA in hair bundles as previously described.34 Briefly, all images were acquired with similar exposure parameters for comparison. The outer hair cell bundle was reconstructed in three dimensions based on phalloidin staining. Then PCDH15, LHFPL5 or TMC1-HA puncta within the bundle area defined by this reconstruction were identified with significant high intensity. Within a bundle, each puncta intensity subtracted by mean background of whole bundle and summed to create a measure of expression level for that bundle.
Primary antibodies were as follows: rabbit anti-HA (1:200, Cell signaling, RRID: RRID:AB_1549585), rabbit anti-MYC (1:200, Cell Signaling, RRID:AB_490778), rabbit anti PCDH15-CD2 (1:500, (Webb et al., 2011)), rabbit anti-LHFPL5/TMHS (1:500, (Xiong et al., 2012), and rabbit anti-PMCA2 (1:200, Abcam, RRID:AB_303878).
Secondary antibodies were as follows: Goat anti-rabbit IgG F(ab’)2, Alexa Fluor 488 (1:5000, Invitrogen, Cat#: A11070), Goat anti-Rabbit IgG F(ab’)2, Alexa Fluor 555 (1:5000, Invitrogen, Cat#: A21430).
Co-immunoprecipitation and western blotting
Co-immunoprecipitations were performed following our published procedures.14,15,34,52,53 Briefly, HEK293 cells were transfected with various plasmids using Lipofectamine 3000 (ThermoFisher). After 48 h, cells were lysed using a modified RIPA buffer containing 150 mM NaCl, 50 mM Tris (pH 8), 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% Sodium Dodecyl Sulfate (SDS) and a Roche Complete Protease Inhibitor Tablet. After lysis, lysates were rotated for 30 min at 4°C followed by centrifugation at 20,000 rcf for 15 min at 4°C. At this point, 10% of the lysate was set aside for use as an input control. The rest of the lysate was immunoprecipitated for 1 h at 4°C using EZ View Red HA Affinity Gel (Sigma-Aldrich Cat# E6779, RRID:AB_10109562) or EZ View Red Flag M2 Affinity Gel ((Sigma-Aldrich Cat# F2426, RRID:AB_2616449). After immunoprecipitation, the affinity gel was washed three times with lysis buffer and mixed with 4x Bolt LDS Sample Buffer (Life Technologies) containing 10x Bolt Sample Reducing Agent (Life Technologies) to elute protein complexes. Eluted immunoprecipitated protein was run in parallel with input lysate on 4–12% Bolt Bis-Tris plus gels (Life Technologies) in Bolt MOPS Running Buffer (Life Technologies) and transferred to PVDF membranes for 1 h at 4°C using the Mini Blot Module containing Bolt transfer buffer (Life technologies) with 10% Methanol. Membranes were blocked for 1 h with 2% ECL Prime blocking reagent (GE Life Sciences) in 1X TBST (containing 20 mM Tris-HCl pH 7.5, 150 mM NaCl and 0.1% Tween 20). Membranes were incubated with primary antibodies (see below) in 2% ECL Prime in 1X TBST at 4°C overnight. After primary antibody incubation, membranes were washed three times with 1X TBST, followed by incubation for 1 h at RT in ECL Prime solution containing secondary antibodies (see below) in 1X TBST. Membranes were washed three times in 1X TBST and then imaged with Clarity Substrate (Biorad) on a G-Box ECL imager (Syngene). Quantification of CoIP experiments was done using ImageJ and Microsoft Excel. Western blot band intensity values were obtained using ImageJ for IP bands and whole cell lysate bands for both IP and CoIP proteins. For each experiment, CoIP intensity values were normalized for expression and immunoprecipitation efficacy and divided by controls to generate relative CoIP Intensity values. Mean relative CoIP intensity values were calculated by combining relative values for each construct across independent experiments. For each construct, a mean value was calculated, and statistical significance relative to control constructs were evaluated using a Students’ t-test.
Primary antibodies were as follows: mouse anti-Flag M2 (1:500, Sigma, RRID:AB_262044), rabbit anti-MYC (1:500, Cell Signaling, RRID:AB_490778), and rabbit anti-HA (1:500, Cell signaling, RRID: RRID:AB_1549585).
Secondary antibodies were Veriblot (1:5000, Abcam) and Veriblot anti-mouse (1:5000, Abcam).
DNA constructs and plasmids
DNA constructs were generated as described below. All constructs were sequence-verified.
TMIE, TMC1, CIB2 and PCDH15 constructs for biochemical experiments
Expression vectors for pN3-TMIE-MYC, pCAGEN-TMC1-MYC, pN3-HA-CIB2 and pcDNA-PCDH15-CD2 were described previously.14,15,34,54
LHFP family members, chimeras and LHFPL5 mutant constructs
Expression vectors for pcDNA-LHFPL5 were described previously (Xiong et al., 2012). pCMV-LHFP-MYC, pCMV-LHFPL1-MYC, pCMV-LHFPL2-MYC were purchased from Origene. All constructs were generated by PCR-cloning using primers containing kozak sequence and restriction enzyme sites EcoRI at 5′ and XhoI at 3’. Inserts were subcloned into pcDNA vector with EcoRI and XhoI restriction enzymes. Mutations were introduced with Quik-Change PCR (Stratagene). Specific details for each construct are below.
pcDNA-LHFPL2
pCMV-LHFPL2-MYC-DDK (Origene MR202553) was used as a PCR template to generate LHFPL2. Primers were.
5′- TTGAATTCCCACCATGTGTCATGTCATTGTCACCTG-3′;
5′- TTCTCGAGTTAAAGGAGGCAGACGAGGTTTTTCCCCT-3′
pcDNA-LHFPL3, pcDNA-LHFPL3-HA pcDNA-LHFPL3-MYC, pcDNA-LHFPL3-FLAG
Mouse cochlear cDNA was used as a PCR template to generate LHFPL3 and epitope tagged LHFPL3. Primers were.
5′- TTGGAATTCCCACCATGCTCCCGGCTCAGGA-3′;
5′- GCATGCTCGAGTTAAGCATTTCCATCATCTTTGTTTTC-3′ for LHFPL3; 5′-TTCTCGAGTTAAGCGTAATCNGGAACATCGTATGGGTAAGCATTTCCATCATCTTTGT-3′ for LHFPL3-HA; 5′-TCTCGAGTTACAGATCCTCTTCTGAGATGAGTTTCTGCTCAGCATTTCCATCATCTTTGT-3′ for LHFPL3-MYC; 5′-GCTCGAGTCACTTATCGTCGTCATCCTTGTAATCAGCATTTCCATCATCTTTGTTTTCTGCTTT-3′ for LHFPL3-FLAG
pcDNA-LHFPL4, pcDNA-LHFPL4-MYC
Mouse cochlear cDNA was used as a PCR template to generate LHFPL4 and LHFPL4-MYC. Primers were.
5′- TTGGAATTCCCACCATGCTGCCCTCGCAGGAA-3’
5′- TTCTCGAGTCAGGGTCCCTGTGTGTGAGCAA-3′ for LHFPL4; 5′- TTCTCGAGTCACAGATCCTCTTCTGAGATGAGTTTCTGCTCGGGTCCCTGTGTGTGAGCAA-3′ for LHFPL4-MYC pcDNA-LHFPL5-HA, pcDNA-LHFPL5-MYC, pcDNA-LHFPL5-FLAG
pcDNA-LHFPL5 was used as a PCR template to generate epitope tagged LHFPL5. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3’
-
5′- ATGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTAGACTTCCTCATTCCCATCT-3′ for LHFPL5-HA;
5′- TGCTCGAGTCACAGATCCTCTTCTGAGATGAGTTTCTGCTCGACTTCCTCATTCCCATCT-3′ for LHFPL5-MYC;
5′- TGCTCGAGTCACTTATCGTCGTCATCCTTGTAATCGACTTCCTCATTCCCATCTG-3′ for LHFPL5-FLAG.
pcDNA-L5-L3
L5-L3 contained LHFPL5 1–125 and LHFPL3 123–218. pcDNA-LHFPL3 and pcDNA-LHFPL5 were used as PCR templates. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3′;
5′- GCTTCAGCCTGTTCTTCGTCTGTAATACAGCCACCGTGTACAAGATCTGTGCTT-3′;
5′- AAGCACAGATCTTGTACACGGTGGCTGTATTACAGACGAAGAACAGGCTGAAGC-3′;
5′- GCATGCTCGAGTTAAGCATTTCCATCATCTTTGTTTTC-3′
pcDNA-L3-L5
L3-L5 contained LHFPL3 1–122 and LHFPL5 126–219. pcDNA-LHFPL3 and pcDNA-LHFPL5 were used as PCR templates. Primers were.
5′- TTGGAATTCCCACCATGCTCCCGGCTCAGGA-3′;
5′- TTCTGCAACACAGCCACCGTCTACAAGATTTGCGCCT-3′;
5′- AGGCGCAAATCTTGTAGACGGTGGCTGTGTTGCAGAA-3′;
5′- GCATGCTCGAGTCAGACTTCCTCATTCCCATC-3′.
pcDNA-NTL5-L3
NTL5-L3 contained LHFPL5 1–18 and LHFPL3 16–218. pcDNA-LHFPL3 was used as a PCR template. Primers were.
5′- AAAAGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAGGAGGCCGCCAAGATCTACCACACCAACTATGTGCGGAACTCGCGG-3’
5′- GCATGCTCGAGTTAAGCATTTCCATCATCTTTGTTTTC-3′
pcDNA-NTL3-L5
NTL3-L5 contained LHFPL3 1–15 and LHFPL5 19–219, also named as L5-Ndel3-I13L. pcNDA-LHFPL5 was used as a PCR template. Primers were.
5′- TGGAATTCCCACCATGCTCCCGGCTCAGGAGGCTGCCAAGCTGTACCACACCAACTATGTGCGGAACTCGCGGGCCGTGGGAGTGATGTGGGGCACGCTC −3’
5′- GCATGCTCGAGTCAGACTTCCTCATTCCCATC-3
pcDNA-L5-Ndel3, pcDNA-L5-Ndel3-HA
L5-Ndel3 delete N-terminal 3 amino acid VKL. pcNDA-L5 was used as a PCR template. Primers were.
5′- GGAATTCCCACCATGCTGCCAGCCCAGGAGGCCGCCAAG-3’
5′- GCATGCTCGAGTCAGACTTCCTCATTCCCATC-3′ for L5-Ndel3
5′- ATGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTAGACTTCCTCATTCCCATCT-3′ for L5-Ndel3-HA.
pcDNA-L5-I13L, pcDNA-L5-I13L-HA
pcDNA-LHFPL5 or pcDNA-LHFPL5-HA was used as a PCR template with Quik-Change PCR (Stratagene). Primers were.
5′- AGGAGGCCGCCAAGTTGTACCACACCAACTATGT-3’
5′- ATAGTTGGTGTGGTAGAACTTGGCGGCCTCCTGGGCT-3′
pcDNA-L5NcytoTM1
pcDNA-L5NcytoTM1 contain LHFPL5 1–52 and LHFPL3 50–218. pcDNA-LHFPL3 and pcDNA-LHFPL5 were used as PCR templates. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3’
5′-ATAGGCGACGGCGTGGACACCCCGCAAGCC-3’
5′-TGTCCACGCCGTCGCCTATCCAGTAGGGCTGGA-3’
5′- GCATGCTCGAGTTAAGCATTTCCATCATCTTTGTTTTC-3′
pcDNA-L5NcytoTM1Lp1
pcDNA-L5NcytoTM1Lp1 contain LHFPL5 1–92 and LHFPL3 90–218. pcDNA-LHFPL3 and pcDNA-LHFPL5 were used as PCR templates. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3’
5′-CCAGAGCCTTCAAAGCCGCCTCCTTCTTCAT-3’
5′-GGCTTTGAAGGCTCTGGAGGGGATGGAG-3’
5′- GCATGCTCGAGTTAAGCATTTCCATCATCTTTGTTTTC-3′
pcDNA-L5-L3-HA, pcDNA-NTL5-L3-HA, pcDNA-L5NcytoTM1-HA, pcDNA-L5NcytoTM1Lp1-HA
Non-tagged version of constructs were used as PCR template for pcDNA-L5-L3-HA, pcDNA-NTL5-L3-HA, pcDNA-L5NcytoTM1-HA, pcDNA-L5NcytoTM1Lp1-HA. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3’
5′-TTCTCGAGTTAAGCGTAATCNGGAACATCGTATGGGTAAGCATTTCCATCATCTTTGT-3′
pcDNA-L5-L3-FLAG, pcDNA-NTL5-L3-FLAG, pcDNA-L5NcytoTM1- FLAG, pcDNA-L5NcytoTM1Lp1- FLAG
Non-tagged version of constructs were used as PCR template for pcDNA-L5-L3-FLAG, pcDNA-NTL5-L3- FLAG, pcDNA-L5Ncy-toTM1- FLAG, pcDNA-L5NcytoTM1Lp1- FLAG. Primers were.
5′- GGAATTCCCACCATGGTGAAGTTGCTGCCAGCCCAG-3’
5′-GCTCGAGTCACTTATCGTCGTCATCCTTGTAATCAGCATTTCCATCATCTTTGTTTTCTGCTTT-3′
pcDNA-L3-L5-HA, pcDNA-NTL3-L5-HA
Non-tagged version of constructs were used as PCR template pcDNA-L3-L5-HA, pcDNA-NTL3-L5-HA. Primers were.
5′- TTGGAATTCCCACCATGCTCCCGGCTCAGGA-3′;
5′- ATGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGGTAGACTTCCTCATTCCCATCT-3′
pcDNA-L3-L5-FLAG, pcDNA-NTL3-L5-FLAG
Non-tagged version of constructs were used as PCR template pcDNA-L3-L5-FLAG, pcDNA-NTL3-L5-FLAG. Primers were.
5′- TTGGAATTCCCACCATGCTCCCGGCTCAGGA-3′;
5′- TGCTCGAGTCACTTATCGTCGTCATCCTTGTAATCGACTTCCTCATTCCCATCTG-3′
TMC1, TMC2, TMC3 and chimera constructs for electrophysiological experiments
N-terminal MYC tagged TMC1, TMC2, TMC3 and TMC1/3 chimera were subcloned into pCAGENE-IRES-GFP vector (addgene) with NEBuilder HiFi DNA Assembly (New England Biolabs).
pCAGENE-MYC-TMC1-IRES-GFP, pCAGENE-MYC-TMC2-IRES-GFP, pCAGENE-MYC-TMC3-IRES-GFP
Primers were.
5′-TTTTGGCAAAGAATTCCTCGAGGATCGCCACCATGGAACAAAAACTCATCTCAGAAGAGG-3′;
5′- GGAATTTACGTAGCGGCCGCGATTTACTGGCCACCAGCAGCTGCAGCTGCTC-3′; for TMC1
5′- GGAATTTACGTAGCGGCCGCGATTCAGTTGTGAGGCCTCTGGGTTCTCTTTCCA-3′; for TMC2
5′- GGAATTTACGTAGCGGCCGCGATTTAGACATTTGAACAAATTAGATCATTTAGA-3′; for TMC3
pCAGENE-MYC-TMC1-H1-TMC3-IRES-GFP
Primers were.
5′-TTTTGGCAAAGAATTCCTCGAGGATCGCCACCATGGAACAAAAACTCATCTCAGAAGAGG-3’
5′-GATCACTGCTATCAATTTCTTCTTCTTCTTCCGC-3’
5′-AAGAAGAAATTGATAGCAGTGATCCGGAACAAATC-3’
5′- TCCAAGGTTTACAGCGGATATTTGCCATGAGA-3’
5′- CAAATATCCGCTGTAAACCTTGGAAAATGGAGAAGAA-3’
5′- GGAATTTACGTAGCGGCCGCGATTTACTGGCCACCAGCAGCTGCAGCTGCTC-3′
pCAGENE-MYC-TMC1-H2-TMC3-IRES-GFP
5′-TTTTGGCAAAGAATTCCTCGAGGATCGCCACCATGGAACAAAAACTCATCTCAGAAGAGG-3’
5′- AGTTTCTGCCCCATTTTCCAAGGTTTACATTTGAC-3’
5′-TTGGAAAATGGGGCAGAAACTGCGGGCACT-3’
5′-TCCTTTCCCAAGCCTCCCTTCAAACTTCAGCAC-3’
5′-TTTGAAGGGAGGCTTGGGAAAGGAAAGGGAAAGA-3’
5′- GGAATTTACGTAGCGGCCGCGATTTACTGGCCACCAGCAGCTGCAGCTGCTC-3′
pCAGENE-MYC-TMC3-H3-TMC3-IRES-GFP
5′-TTTTGGCAAAGAATTCCTCGAGGATCGCCACCATGGAACAAAAACTCATCTCAGAAGAGG-3’
5′- CTGCTGCCTGGTATCCCTTTCCTTTCCCAAGAGCG-3’
5′- AGGAAAGGGATACCAGGCAGCAGGAGCAGA-3’
5′- TTCCCATGGGATGAAGATGACCACAAAGTTACA-3’
5′- GTGGTCATCTTCATCCCATGGGAAAACAAAATCAA-3’
5′- GGAATTTACGTAGCGGCCGCGATTTACTGGCCACCAGCAGCTGCAGCTGCTC-3’
pCAGENE-MYC-TMC1-H1/2-TMC3-IRES-GFP
5′-TTTTGGCAAAGAATTCCTCGAGGATCGCCACCATGGAACAAAAACTCATCTCAGAAGAGG-3’
5′- GATCACTGCTATCAATTTCTTCTTCTTCTTCCGC-3’
5′- AAGAAGAAATTGATAGCAGTGATCCGGAACAAATC-3’
5′-TCCTTTCCCAAGCCTCCCTTCAAACTTCAGCAC-3’
5′-TTTGAAGGGAGGCTTGGGAAAGGAAAGGGAAAGA-3’
5′- GGAATTTACGTAGCGGCCGCGATTTACTGGCCACCAGCAGCTGCAGCTGCTC-3′
pN3-PAC-HA, pN3-PAC-TMC1-HA, pN3-PAC-TMC1-H1-TMC3-HA, pN3-PAC-TMC1-H2-TMC3-HA, pN3-PAC-TMC1-H1/2-TMC3-HA, pN3-PAC-TMC1-H3-TMC3-HA were synthesized and cloned by Twist Bioscience (San Francisco, CA). PAC N-terminal cytoplasmic domain amino acids 1–65 were replaced by TMC1 N-terminal cytoplasmic domain amino acids 1–183.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data analysis was performed using Excel (Microsoft) and Igor pro 7 (WaveMetrics, Lake Oswego, OR). Transduction current/displacement curves (I/X) and open probability/displacement curves were fitted with the following double Boltzmann55,56:
where and are the slope factors and represents the set point.
MET currents decay was fitted with a double exponential function:
where and were the decay time constant, and were the respective amplitude. Each fraction of amplitude were and .
Statistical details are described in the results, Figures, and Figure Legends. All data are mean ± SEM. Student’s two-tailed unpaired t test and one-way ANOVA test were used, as indicated in each figure, to determine statistical significance (n.s. p > 0.05, *, p < 0.05, **, p < 0.01, ***, p < 0.001). Exact values of n are reported where appropriate. Depending on the experiment, n represents number of animals, number of cells, number of experiments, or number of single channel current events.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Rabbit anti-PCDH15-CD2 | Webb et al.30 | N/A |
| Rabbit anti-LHFPL5/TMHS | Xiong et al.23 | N/A |
| Rabbit anti-PMCA2 | Thermo Fisher Scientific | Cat# PA1-915, RRID:AB_2243199 |
| Rabbit anti-HA | Cell Signaling | Cat#: 3724; RRID:AB_1549585 |
| Rabbit anti-MYC | Cell Signaling | Cat#: 2278; RRID:AB_490778 |
| Mouse anti-HA | Cell Signaling | Cat#: 2367S; RRID: AB_10691311 |
| Mouse anti-MYC | Cell Signaling | Cat#: 2276S; RRID: AB_331783 |
| Mouse anti-FLAG | Cell Singaling | Cat#: 8146S; RRID: AB_10950495 |
| Rabbit anti-FLAG | Cell Signaling | Cat#: 14793S; RRID: AB_2572291 |
| Goat anti-Rabbit IgG F(ab’)2, Alexa Fluor 488 | Invitrogen | Cat#: A-11070; RRID:AB_2534114 |
| Goat anti-Rabbit IgG F(ab’)2, Alexa Fluor 555 | Invitrogen | Cat#: A-21430; RRID:AB_2535851 |
| Goat anti-Mouse IgG F(ab’)2, Alexa Fluor 647 | Invitrogen | Cat#: A-21237; RRID:AB_2535806 |
| EZ-View Red HA Affinity Gel | Sigma | Cat#: E6779-1ML; RRID:AB_10109562 |
| EZ View Red anti-c-MYC Affinity Gel | Sigma | Cat#: E6654-1ML; RRID:AB_10093201 |
| Veriblot | Abcam | Cat#: ab131366 |
| Veriblot anti-Mouse | Abcam | Cat#: ab131368 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Cas9 Protein | PNA Bio | Cat#: CP01-20 |
| Tris-HCl | Sigma | Cat#: T5941 |
| EDTA | Sigma | Cat#: E5134 |
| DMEM+Glutamax medium | Gibco | Cat#: 10,569–010 |
| Antibiotic-Antimycotic supplement | Gibco | Cat#: 15,240–062 |
| DMEM/F12 medium | Gibco | Cat#: 11,330–032 |
| Fetal Bovine Serum, Heat Inactivated | Sigma | Cat#: F4135 |
| Ampicillin | Sigma | Cat#: A9518 |
| Sodium Chloride | VWR | Cat#: 0241 |
| Sodium phosphate monobasic | Sigma | Cat#: S8282 |
| Potassium Chloride | Fisher Scientific | Cat#: P217 |
| Calcium Chloride | Fisher Scientific | Cat#: M13841 |
| Magnesium Chloride | Sigma | Cat#: M8266 |
| Glucose | Sigma | Cat#: G7021 |
| HEPES | Sigma | Cat#: H4034 |
| BAPTA | Sigma | Cat#: A4926 |
| EGTA | VWR | Cat#: 0732 |
| Magnesium-ATP | Sigma | Cat#: A9187 |
| Sodium-GTP | Sigma | Cat#: G8877 |
| Cesium Chloride | Sigma | Cat#: 203025 |
| DMSO | Sigma | Cat#: D2650 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat#: 15714 |
| 1X HBSS medium | Gibco | Cat#: 14175095 |
| Normal Goat Serum, Heat Inactivated | Gemini Biosciences | Cat#: 100–109 |
| Triton X-100 | Sigma | Cat#: T9284 |
| Alexa Fluor 555 Phalloidin | Life Technologies | Cat#: A34055 |
| Alexa Fluor 488 Phalloidin | Life Technologies | Cat#: A12379 |
| Prolong Gold | ThermoFisher | Cat#: P10144 |
| Lipofectamine 3000 | ThermoFisher | Cat#: L3000015 |
| Tris/Tham | Fisher | Cat#: T370-3 |
| NP-40 | Sigma | Cat#: I3021 |
| Sodium Deoxycholate | Sigma | Cat#: 6750 |
| Sodium Dodecyl Sulfate | Sigma | Cat#: 75746 |
| Complete Mini Protease Inhibitor Tablet, EDTA-free | Roche | Cat#: 11836170001 |
| 4× Laemmli Sample Buffer | Biorad | Cat#: 1610747 |
| 2-Mercaptoethanol | Biorad | Cat#: 1610710 |
| Novex™ WedgeWell™ 4 to 20%, Tris-Glycine, 1.0 mm, Mini Protein Gel | Life Technologies | Cat#: XP04202BOX |
| 10× Tris/Glycine/SDS | Biorad | Cat#: 1610732 |
| 10× Tris/Glycine Buffer for Western Blots and Native Gels | Biorad | Cat#: 1610734 |
| Methanol | VWR | Cat#: BDH1135-4LP |
| ECL Prime Blocking Reagent | GE | Cat#: RPN418 |
| Tween 20 | Sigma | Cat#: P7949 |
| Clarity Western ECL Substrate | Bio-Rad | Cat#: 1705060 |
| BSA, fatty acid free | Sigma | Cat#: A8806 |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| Human: HEK-293 | ATCC | Cat#: CRL-1573 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Lhfpl5 −/− (Tmhs tm1kin) | Longo-Guess et al.25 | MGI:3718680 |
| Lhfpi3−/− mice | Tang et al.27 | RRID:MMRRC_032433-UCD |
| Lhfpl5-Ndel3-I13L mice | This paper | N/A |
| C57BL/6J mice | Jackson Laboratories | Cat#: 000664; RRID:IMSR JAX:000,664 |
| ICR mice | Envigo | Cat#: Hsd:ICR (CD-1) |
|
| ||
| Oligonucleotides | ||
|
| ||
| Target-specific crRNAs | This paper/IDT | N/A |
| tracrRNA | Dharmacon | Cat#: U-002005-05 |
| Target-specific ssDNA | This paper/IDT Ultramer DNA Oligos | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pN3-TMIE-MYC | Zhao et al.14 | N/A |
| pN3-HA-CIB2 | Liang et al.34 | N/A |
| pCAGEN-TMC1-MYC | Cunningham et al.15 | N/A |
| pcDNA-PCDH15-CD2 | Xiong et al.23 | N/A |
| pCMV-LHFP-MYC | Origene | MR202027 |
| pCMV-LHFPL1-MYC | Origene | MR202519 |
| pCMV-LHFPL2-MYC | Origene | MR202553 |
| pcDNA-LHFPL2 | This paper | N/A |
| pcDNA-LHFPL3 | This paper | N/A |
| pcDNA-LHFPL3-HA | This paper | N/A |
| pcDNA-LHFPL3-MYC | This paper | N/A |
| pcDNA-LHFPL3-FLAG | This paper | N/A |
| pcDNA-LHFPL4 | This paper | N/A |
| pcDNA-LHFPL4-MYC | This paper | N/A |
| pcDNA-LHFPL5 | Xiong et al.23 | N/A |
| pcDNA-LHFPL5-HA | This paper | N/A |
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
| pcDNA-LHFPL5-MYC | This paper | N/A |
| pcDNA-LHFPL5-FLAG | This paper | N/A |
| pcDNA-L5-L3 | This paper | N/A |
| pcDNA-L5-L3-HA | This paper | N/A |
| pcDNA-L5-L3-FLAG | This paper | N/A |
| pcDNA-L3-L5 | This paper | N/A |
| pcDNA-L3-L5-HA | This paper | N/A |
| pcDNA-L3-L5-FLAG | This paper | N/A |
| pcDNA-NTL5-L3 | This paper | N/A |
| pcDNA-NTL5-L3-HA | This paper | N/A |
| pcDNA-NTL5-L3-FLAG | This paper | N/A |
| pcDNA-NTL3-L5 | This paper | N/A |
| pcDNA-NTL3-L5-HA | This paper | N/A |
| pcDNA-NTL3-L5-FLAG | This paper | N/A |
| pcDNA-L5-Ndel3 | This paper | N/A |
| pcDNA-L5-Ndel3-HA | This paper | N/A |
| pcDNA-L5-I13L | This paper | N/A |
| pcDNA-L5-I13L-HA | This paper | N/A |
| pcNDA-L5NcytoTM1 | This paper | N/A |
| pcNDA-L5NcytoTM1-HA | This paper | N/A |
| pcNDA-L5NcytoTM1-FLAG | This paper | N/A |
| pcNDA-L5NcytoTM1Lp1 | This paper | N/A |
| pcDNA-L5NcytoTM1Lp1-HA | This paper | N/A |
| pcDNA-L5NcytoTM1Lp1-FLAG | This paper | N/A |
| pCAGEN-IRES-GFP | Addgene | Cat#: Addgene #11159; RRID:Addgene_11159 |
| pCAGEN-MYC-TMC1-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC2-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC3-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC1-H1-TMC3-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC1-H2-TMC3-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC1-H3-TMC3-IRES-GFP | This paper | N/A |
| pCAGEN-MYC-TMC1-H1/2-TMC3-IRES-GFP | This paper | N/A |
| pN3-PAC-HA | This paper | N/A |
| pN3-PAC-TMC1-HA | This paper | N/A |
| pN3-PAC-TMC1-H1 -TMC3-HA | This paper | N/A |
| pN3-PAC-TMC1-H2-TMC3-HA | This paper | N/A |
| pN3-PAC-TMC1-H1/2-TMC3-HA | This paper | N/A |
| pN3-PAC-TMC1-H3-TMC3-HA | This paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| Deltavision Elite Software (SoftWoRx Resolve 3D) | GE | https://www.gelifesciences.com/en/ee/shop/deltavision-elite-high-resolution-microscope-p-04420 |
| Imaris 9.7 | Oxford Instruments | https://imaris.oxinst.com/packages |
| Igor pro 7 | WaveMetrics | https://www.wavemetrics.com/ |
| TDT System 3 | Tucker-Davis Technology | https://www.tdt.com/products/ |
| Patchmaster 2.35 | HEKA | http://www.heka.com/downloads/downloads_main.html |
| Adobe Illustrator 2020 | Adobe | https://www.adobe.com/ |
| Adobe Photoshop 2020 | Adobe | https://www.adobe.com/ |
Highlights.
LHFPL5 tethers tip links and transduction channels
The N-terminal cytoplasmic domain of LHFPL5 is critical for channel gating
TMC1 contains an amphipathic helix critical for channel gating
Channel gating is consistent with a tethered channel model
ACKNOWLEDGMENTS
We thank members of the laboratory for comments; Guihong Peng, Michelle Monroe, and Kaiping Zhang for assistance with mouse colonies; and Chip Hawkins for generating mouse mutants. This work was supported by the NIH, United States (F32DC015724 to C.C.; R21DC019195; and RO1DC005965, RO1DC019514, and RO1DC016960 to U.M.), a Gilliam Graduate Fellowship from the Howard Hughes Medical Institute, United States (to J.P.L.), and the David M. Rubenstein Fund for Hearing Research (to U.M.). U.M. is a Bloomberg Distinguished Professor for Neuroscience and Biology.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Footnotes
DECLARATION OF INTERESTS
U.M. is a co-founder of Decibel Therapeutics.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112245.
REFERENCES
- 1.Kefauver JM, Ward AB, and Patapoutian A (2020). Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beurg M, Fettiplace R, Nam JH, and Ricci AJ (2009). Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat. Neurosci. 12, 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basu A, Lagier S, Vologodskaia M, Fabella BA, and Hudspeth AJ (2016). Direct mechanical stimulation of tip links in hair cells through DNA tethers. Elife 5, e16041. 10.7554/eLife.16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Yamoah EN, and Gillespie PG (1996). Regeneration of broken tip links and restoration of mechanical transduction in hair cells. Proc. Natl. Acad. Sci. USA 93, 15469–15474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Assad JA, Shepherd GM, and Corey DP (1991). Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron 7, 985–994. [DOI] [PubMed] [Google Scholar]
- 6.Pickles JO, Comis SD, and Osborne MP (1984). Cross-links between stereocilia in the Guinea pig organ of Corti, and their possible relation to sensory transduction. Hear. Res. 15, 103–112. [DOI] [PubMed] [Google Scholar]
- 7.Shotwell SL, Jacobs R, and Hudspeth AJ (1981). Directional sensitivity of individual vertebrate hair cells to controlled deflection of their hair bundles. Ann. N. Y. Acad. Sci. 374, 1–10. [DOI] [PubMed] [Google Scholar]
- 8.He DZZ, Jia S, and Dallos P (2004). Mechanoelectrical transduction of adult outer hair cells studied in a gerbil hemicochlea. Nature 429, 766–770. [DOI] [PubMed] [Google Scholar]
- 9.Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Müller U, and Kachar B (2007). Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449, 87–91. [DOI] [PubMed] [Google Scholar]
- 10.Siemens J, Lillo C, Dumont RA, Reynolds A, Williams DS, Gillespie PG, and Müller U (2004). Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature 428, 950–955. [DOI] [PubMed] [Google Scholar]
- 11.Söllner C, Rauch GJ, Siemens J, Geisler R, Schuster SC, Müller, and Nicolson T.; Tübingen 2000 Screen Consortium (2004). Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature 428, 955–959. [DOI] [PubMed] [Google Scholar]
- 12.Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, et al. (2006). The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is protocadherin-15. J. Neurosci. 26, 7022–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurima K, Ebrahim S, Pan B, Sedlacek M, Sengupta P, Millis BA, Cui R, Nakanishi H, Fujikawa T, Kawashima Y, et al. (2015). TMC1 and TMC2 localize at the site of mechanotransduction in mammalian inner ear hair cell stereocilia. Cell Rep. 12, 1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao B, Wu Z, Grillet N, Yan L, Xiong W, Harkins-Perry S, and Müller U (2014). TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron 84, 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham CL, Qiu X, Wu Z, Zhao B, Peng G, Kim YH, Lauer A, and Müller U (2020). TMIE defines pore and gating properties of the mechanotransduction channel of mammalian cochlear hair cells. Neuron 107, 126–143.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawashima Y, Géléoc GSG, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, and Griffith AJ (2011). Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J. Clin. Invest. 121, 4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim KX, and Fettiplace R (2013). Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. J. Gen. Physiol. 141, 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pan B, Akyuz N, Liu XP, Asai Y, Nist-Lund C, Kurima K, Derfler BH, Gyö rgy B., Limapichat W., Walujkar S., et al. (2018). TMC1 forms the pore of mechanosensory transduction channels in vertebrate inner ear hair cells. Neuron 99, 736–753.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan B, Géléoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, and Holt JR (2013). TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79, 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beurg M, Xiong W, Zhao B, Müller U, and Fettiplace R (2015). Subunit determination of the conductance of hair-cell mechanotransducer channels. Proc. Natl. Acad. Sci. USA 112, 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Yu X, Chen X, Liu Z, Wang G, Li C, Wong EYM, Sham MH, Tang J, He J, et al. (2019). Localization of TMC1 and LHFPL5 in auditory hair cells in neonatal and adult mice. Faseb. J. 33, 6838–6851. [DOI] [PubMed] [Google Scholar]
- 22.Mahendrasingam S, Fettiplace R, Alagramam KN, Cross E, and Furness DN (2017). Spatiotemporal changes in the distribution of LHFPL5 in mice cochlear hair bundles during development and in the absence of PCDH15. PLoS One 12, e0185285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiong W, Grillet N, Elledge HM, Wagner TFJ, Zhao B, Johnson KR, Kazmierczak P, and Müller U (2012). TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151, 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dror AA, and Avraham KB (2010). Hearing impairment: a panoply of genes and functions. Neuron 68, 293–308. [DOI] [PubMed] [Google Scholar]
- 25.Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, and Johnson KR (2005). A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc. Natl. Acad. Sci. USA 102, 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shabbir MI, Ahmed ZM, Khan SY, Riazuddin S, Waryah AM, Khan SN, Camps RD, Ghosh M, Kabra M, Belyantseva IA, et al. (2006). Mutations of human TMHS cause recessively inherited non-syndromic hearing loss. J. Med. Genet. 43, 634–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang T, Li L, Tang J, Li Y, Lin WY, Martin F, Grant D, Solloway M, Parker L, Ye W, et al. (2010). A mouse knockout library for secreted and transmembrane proteins. Nat. Biotechnol. 28, 749–755. [DOI] [PubMed] [Google Scholar]
- 28.Yu X, Zhao Q, Li X, Chen Y, Tian Y, Liu S, Xiong W, and Huang P (2020). Deafness mutation D572N of TMC1 destabilizes TMC1 expression by disrupting LHFPL5 binding. Proc. Natl. Acad. Sci. USA 117, 29894–29903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge J, Elferich J, Goehring A, Zhao H, Schuck P, and Gouaux E (2018). Structure of mouse protocadherin 15 of the stereocilia tip link in complex with LHFPL5. Elife 7, e38770. 10.7554/eLife.38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb SW, Grillet N, Andrade LR, Xiong W, Swarthout L, Della Santina CC, Kachar B, and Müller U (2011). Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138, 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pepermans E, Michel V, Goodyear R, Bonnet C, Abdi S, Dupont T, Gherbi S, Holder M, Makrelouf M, Hardelin JP, et al. (2014). The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol. Med. 6, 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong W, Wagner T, Yan L, Grillet N, and Müller U (2014). Using injectoporation to deliver genes to mechanosensory hair cells. Nat. Protoc. 9, 2438–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fettiplace R, Furness DN, and Beurg M (2022). The conductance and organization of the TMC1-containing mechanotransducer channel complex in auditory hair cells. Proc. Natl. Acad. Sci. USA 119, e2210849119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X, Qiu X, Dionne G, Cunningham CL, Pucak ML, Peng G, Kim YH, Lauer A, Shapiro L, and Müller U (2021). CIB2 and CIB3 are auxiliary subunits of the mechanotransduction channel of hair cells. Neuron 109, 2131–2149.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michel V, Booth KT, Patni P, Cortese M, Azaiez H, Bahloul A, Kahrizi K, Labbé M, Emptoz A, Lelli A, et al. (2017). CIB2, defective in isolated deafness, is key for auditory hair cell mechanotransduction and survival. EMBO Mol. Med. 9, 1711–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giese APJ, Tang YQ, Sinha GP, Bowl MR, Goldring AC, Parker A, Freeman MJ, Brown SDM, Riazuddin S, Fettiplace R, et al. (2017). CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat. Commun. 8, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, et al. (2002). Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat. Genet. 30, 277–284. [DOI] [PubMed] [Google Scholar]
- 38.Corey DP, and Hudspeth AJ (1983). Kinetics of the receptor current in bullfrog saccular hair cells. J. Neurosci. 3, 962–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Corey DP, and Hudspeth AJ (1979). Response latency of vertebrate hair cells. Biophys. J. 26, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ricci AJ, Kennedy HJ, Crawford AC, and Fettiplace R (2005). The transduction channel filter in auditory hair cells. J. Neurosci. 25, 7831–7839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong H, Clark S, Goehring A, Dehghani-Ghahnaviyeh S, Rasouli A, Tajkhorshid E, and Gouaux E (2022). Structures of the TMC-1 complex illuminate mechanosensory transduction. Nature 610, 796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Chen J, Del Carmen Vitery M, Osei-Owusu J, Chu J, Yu H, Sun S, and Qiu Z (2019). PAC, an evolutionarily conserved membrane protein, is a proton-activated chloride channel. Science 364, 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.George SS, Steele CR, and Ricci AJ (2020). Rat auditory inner hair cell mechanotransduction and stereociliary membrane diffusivity are similarly modulated by calcium. iScience 23, 101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Effertz T, Becker L, Peng AW, and Ricci AJ (2017). Phosphoinositol-4,5-Bisphosphate regulates auditory hair-cell mechanotransductionchannel pore properties and fast adaptation. J. Neurosci. 37, 11632–11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang YQ, Lee SA, Rahman M, Vanapalli SA, Lu H, and Schafer WR (2020). Ankyrin is an intracellular tether for TMC mechanotransduction channels. Neuron 107, 759–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang X, Li G, Liu J, Liu J, and Xu X (2016). TMC-1 mediates alkaline sensation in C. elegans through nociceptive neurons. Neuron 91, 146–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue X, Zhao J, Li X, Fan Y, Duan D, Zhang X, Zou W, Sheng Y, Zhang T, Yang Q, et al. (2018). TMC proteins modulate egg laying and membrane excitability through a background leak conductance in C. elegans. Neuron 97, 571–585.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricci AJ, Crawford AC, and Fettiplace R (2003). Tonotopic variation in the conductance of the hair cell mechanotransducer channel. Neuron 40, 983–990. [DOI] [PubMed] [Google Scholar]
- 49.Beurg M, Evans MG, Hackney CM, and Fettiplace R (2006). A largeconductance calcium-selective mechanotransducer channel in mammalian cochlear hair cells. J. Neurosci. 26, 10992–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beurg M, Cui R, Goldring AC, Ebrahim S, Fettiplace R, and Kachar B (2018). Variable number of TMC1-dependent mechanotransducer channels underlie tonotopic conductance gradients in the cochlea. Nat. Commun. 9, 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Longo-Guess CM, Gagnon LH, Fritzsch B, and Johnson KR (2007). Targeted knockout and lacZ reporter expression of the mouse Tmhs deafness gene and characterization of the hscy-2J mutation. Mamm. Genome 18, 646–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cunningham CL, Wu Z, Jafari A, Zhao B, Schrode K, Harkins-Perry S, Lauer A, and Müller U (2017). The murine catecholamine methyl-transferase mTOMT is essential for mechanotransduction by cochlear hair cells. Elife 6, e24318. 10.7554/eLife.24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao B, Wu Z, and Müller U (2016). Murine Fam65b forms ring-like structures at the base of stereocilia critical for mechanosensory hair cell function. Elife 5, e14222. 10.7554/eLife.14222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dionne G, Qiu X, Rapp M, Liang X, Zhao B, Peng G, Katsamba PS, Ahlsen G, Rubinstein R, Potter CS, et al. (2018). Mechanotransduction by PCDH15 relies on a novel cis-dimeric architecture. Neuron 99, 480–492.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Peng AW, Gnanasambandam R, Sachs F, and Ricci AJ (2016). Adaptation independent modulation of auditory hair cell mechanotransduction channel open probability implicates a role for the lipid bilayer. J. Neurosci. 36, 2945–2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peng AW, Effertz T, and Ricci AJ (2013). Adaptation of mammalian auditory hair cell mechanotransduction is independent of calcium entry. Neuron 80, 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


