Summary
As an endosymbiont of the ecologically and medically relevant fungus Rhizopus microsporus, the toxin-producing bacterium Mycetohabitans rhizoxinica faces myriad challenges, such as evading the host’s defense mechanisms. However, the bacterial effector(s) that facilitate the remarkable ability of M. rhizoxinica to freely migrate within fungal hyphae have thus far remained unknown. Here, we show that a transcription activator-like (TAL) effector released by endobacteria is an essential symbiosis factor. By combining microfluidics with fluorescence microscopy, we observed enrichment of TAL-deficient M. rhizoxinica in side hyphae. High-resolution live imaging showed the formation of septa at the base of infected hyphae, leading to the entrapment of endobacteria. Using a LIVE/DEAD stain, we demonstrate that the intracellular survival of trapped TAL-deficient bacteria is significantly reduced compared with wild-type M. rhizoxinica, indicative of a protective host response in the absence of TAL proteins. Subversion of host defense in TAL-competent endobacteria represents an unprecedented function of TAL effectors. Our data illustrate an unusual survival strategy of endosymbionts in the host and provide deeper insights into the dynamic interactions between bacteria and eukaryotes.
Keywords: intracellular survival, microfluidics, Mucoromycota, Mycetohabitans, symbiosis, compartmentalization
Highlights
-
•
TAL effectors circumvent host defense in a bacterial-fungal endosymbiosis
-
•
Microfluidics reveal trapping of effector-deficient bacteria in fungal side hyphae
-
•
Unprecedented case of septa formation in Mucoromycota fungi
-
•
Endosymbionts rely on TAL effectors to aid their intracellular lifestyle
Richter et al. show that a transcription activator-like (TAL) effector of bacteria living within a phytopathogenic fungus is an essential symbiosis factor. Microfluidics and live imaging reveal septa formation in fungal side hyphae, which traps TAL effector-deficient bacteria, leading to reduced survival.
Introduction
Endosymbiosis, a symbiotic relationship in which a microbial partner lives within its host, has led to some of the major evolutionary transitions, e.g., the formation of mitochondria and chloroplasts from internalized prokaryotes.1 Endosymbiosis represents the most intimate contact between interacting partners and, in many cases, has reached a point of strict dependency on the symbiotic partner after millions of years of evolution.2 Such obligate relationships are frequently observed between endofungal bacteria and Mucoromycota fungi.3,4
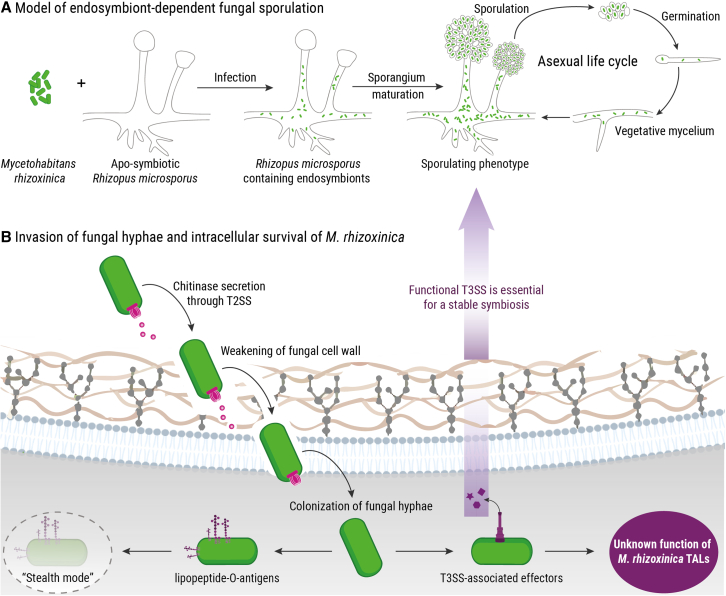
One pertinent example is the intriguing endosymbiosis between the Mucoromycota fungus Rhizopus microsporus and its bacterial symbiont, Mycetohabitans rhizoxinica.5,6,7 R. microsporus is the causative agent of rice seedling blight, a disease that causes severe crop losses in agriculture in Asia.8 The symbiont resides within the fungal hypha, where it produces the highly potent phytotoxin rhizoxin, which is partly tailored by the fungus before being excreted to efficiently stall plant cell division.9 The R. microsporus-M. rhizoxinica symbiosis is unique not only in being a phytotoxin-producing alliance but also because host reproduction through spores is reliant on the presence of endobacteria (Figure 1A). If cured of its endosymbionts by antibiotic treatment, R. microsporus is unable to reproduce vegetatively.10 The dependence of R. microsporus sporulation on M. rhizoxinica ensures that the symbiosis persists over time because the endosymbionts are translocated into the fungal spores during host reproduction.
Figure 1.
Schematic representation of endofungal bacteria and their intracellular lifestyle
(A) The Gram-negative bacterium Mycetohabitans rhizoxinica infects its fungal host Rhizopus microsporus, forming an endosymbiotic relationship. Fungal reproduction through spores is dependent on the presence of endosymbionts. Mature sporangia are absent in apo-symbiotic (endosymbiont-free) fungi.
(B) Entrance of M. rhizoxinica into the fungal hyphae is mediated by the secretion of effectors via a type 2 secretion system (T2SS). Once inside, M. rhizoxinica lives in “stealth mode” with the help of a specialized bacterial lipopeptide O-antigen. Although a functional type 3 secretion system (T3SS) is required for the formation of a stable symbiosis, the molecular function of the T3SS-secreted transcription activator-like (TAL) effectors in M. rhizoxinica is currently unknown.
Multiple investigations have shed light on various factors that are integral to the establishment and maintenance of the R. microsporus-M. rhizoxinica symbiosis. For example, M. rhizoxinica can invade R. microsporus by secreting chitinases via the type II secretion system (T2SS).11 Bacterial invasion is further promoted by a bacterial linear lipopeptide (holrhizin) that reduces surface tension and biofilm formation.12 In order to accommodate the endobacteria, the fungal host responds with upregulation of genes involved in cytoskeletal rearrangements, chitin synthesis, lipid metabolism, and quenching of reactive oxygen species.13,14 Among the factors employed by M. rhizoxinica to persist within the fungal host is a specialized bacterial lipopeptide called O-antigen, which enables the endobacterium to live in “stealth mode.”15 In addition, a bacterial depsipeptide (habitasporin) and a functional type III secretion system (T3SS) are required for the formation of a stable symbiosis (Figure 1B).16,17
Upon initial physical contact with R. microsporus, nearly 60 T3SS-associated effector genes are upregulated in M. rhizoxinica.13 Among them, perhaps the most well known are homologs of transcription activator-like (TAL) effectors from plant-pathogenic Xanthomonas and Ralstonia species.18,19 TAL effectors localize to nuclei of plant hosts, where they bind to specific DNA sequences and act to induce expression of genes that aid bacterial colonization and virulence.20 Although a Mycetohabitans TAL (MTAL) produced by Mycetohabitans sp. B13 is thought to increase the tolerance of R. microsporus to cell membrane stress,21 there is relatively little known regarding the possible functions of MTALs in the interaction between M. rhizoxinica and its fungal host. Here, we show that M. rhizoxinica MTAL1 plays a key role in controlling the intracellular lifestyle of the endobacteria. Furthermore, we present the first real-time view of septa biogenesis in a Mucoromycota fungus, which leads to hyphal trapping of endobacteria incapable of secreting MTAL1.
Results
M. rhizoxinica Δmtal1 does not influence hyphal morphology of R. microsporus
To investigate whether MTAL proteins are important for intracellular survival of Mycetohabitans, we performed a targeted gene deletion using a double-crossover strategy (Figures S1A and S1B).16 We chose to focus on the most abundant mtal gene in Mycetohabitans spp., i.e., mtal1, and deleted this gene in M. rhizoxinica HKI-0454 to generate M. rhizoxinica Δmtal1 (M. rhizoxinica knockout, Figure S1C). M. rhizoxinica Δmtal1 was transformed with pBBR_Ps12_mtal1 (Figure S1D), an expression vector in which mtal1 is under the control of a constitutive promoter, yielding the complemented strain M. rhizoxinica Δmtal1 pBBR-mtal1 (complemented knockout). M. rhizoxinica Δmtal1 was transformed with pBBR_Ps12 to generate M. rhizoxinica Δmtal1 pBBRØ (empty vector control), a control strain containing the relevant empty vector.
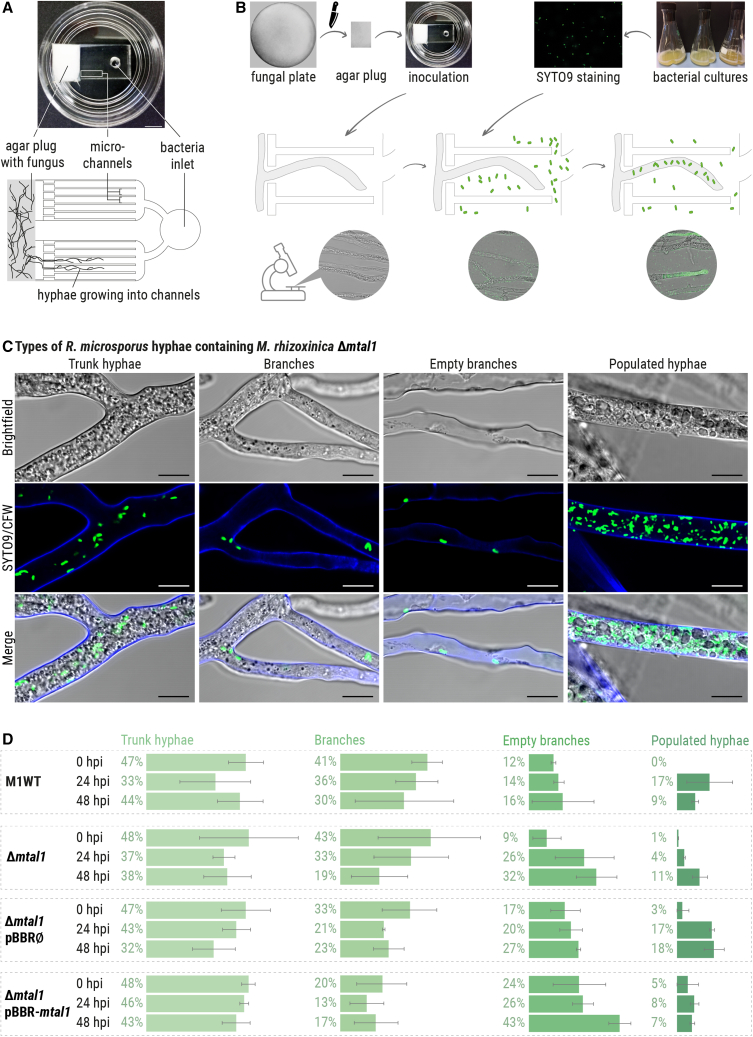
In order to explore specific interactions between intracellular M. rhizoxinica Δmtal1 and fungal hyphae at the cellular level, we utilized a tailor-made microfluidic platform. This bacterial-fungal interaction (BFI) device enables fungal hyphae to be cultured in microchannels, thus allowing imaging of single hyphae over time (Figure 2A).22 First, apo-symbiotic R. microsporus was grown in BFI devices filled with potato dextrose broth (PDB, Figure 2B). After 2 days of incubation, axenic M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, or M. rhizoxinica Δmtal1 pBBR-mtal1 were stained with SYTO9 and then added individually to the bacteria inlet for co-incubation with apo-symbiotic R. microsporus. T3SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT)16 and rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG)9 were also assessed in the assay. Both T3SS-deficient strains were previously reported to have limited ability to reinfect the fungus,16 while deletion of rhiG causes disruption in the biosynthesis of the toxic macrolide rhizoxin, which should not impact the reinfection process.
Figure 2.
Phenotypic observations of R. microsporus co-cultivated with M. rhizoxinica wild type or M. rhizoxinica Δmtal1 in bacterial-fungal interaction (BFI) devices
(A) Photograph and simplified illustration showing the BFI device in a glass Petri dish. The BFI device is made of a patterned poly(dimethylsiloxane) layer bonded to a glass-bottomed dish to form microchannels, which were filled with PDB. Scale bars: 5 mm. Two-dimensional representation showing the narrow entry points into the microchannels that limit the number of hyphae that can enter the device.
(B) Illustration showing the workflow. An agar plug containing apo-symbiotic R. microsporus is placed in direct contact with the microchannels. After 2 days of incubation, hyphae are growing inside the microchannels, and SYTO9-stained bacterial strains are introduced into the microchannels via the “bacteria inlet.” Fungal reinfection is monitored over 48 h, and microscopic images are taken every 24 h.
(C) Microscopic images of R. microsporus (stained with calcofluor white [CFW]) co-cultivated with M. rhizoxinica Δmtal1 (SYTO9-stained) depicting four types of hyphae. Scale bars: 10 μm.
(D) The fungal mycelium area (as a percentage) of each type of hyphae was measured over a 48-h time period of co-incubation in BFI devices. At time point 0, cultures of M. rhizoxinica wild type (M1WT), M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, or M. rhizoxinica Δmtal1 pBBR-mtal1 were stained with SYTO9, individually added to the inlet, and co-incubated with apo-symbiotic R. microsporus. Images were taken at the time of infection (0 h post infection [hpi]), as well as 24 and 48 hpi. n = 3 biological replicates ± SEM (Data S1A–S1D). See also Figures S2, S3A, and S3B.
Using light microscopy, we obtained high-resolution images of individual hyphae growing in the microchannels of the BFI devices. We observed four different types of hyphae, which were broadly classified as follows: (1) trunk hyphae, (2) branches, (3) empty branches, and (4) populated hyphae (Figures 2C, S2, and S3A). Trunk hyphae represent the main vegetative hyphae, which are characterized by their relatively large diameter (approximately 10–15 μm). They appear to be tightly packed with organelles (e.g., nuclei, vacuoles, mitochondria, etc.), which are transported through the hyphae at relatively high speed (Video S1). This hyphal classification has been described for members of the Basidiomycetes and Ascomycetes, but rarely for Mucoromycota.23,24 We also observed smaller side hyphae, which are either filled with cellular components (branches) or are empty (empty branches). Populated hyphae represent another type of side hyphae, which are filled with organelles and are similar in diameter to the trunk hyphae. In addition, they show strong green fluorescence, which is indicative of high bacterial cell numbers (Figure 2C). Comparison of the mycelium area revealed a similar distribution of individual types of hyphae over time and regardless of the infecting M. rhizoxinica strain (Figures 2D and S3B; Data S1).
Endosymbiont-free R. microsporus was co-incubated with M. rhizoxinica Δmtal1 in a microfluidic device. Bacterial cells were stained with SYTO9 and then introduced into the microfluidic device via the bacteria inlet 12 h before image acquisition. Reinfection was monitored over time using fluorescence microscopy. The video was recorded at 1 frame per s (fps). Direction of hyphal growth is from left to right. Scale bars: 10 μm
High bacterial load of M. rhizoxinica Δmtal1 in fungal hyphae
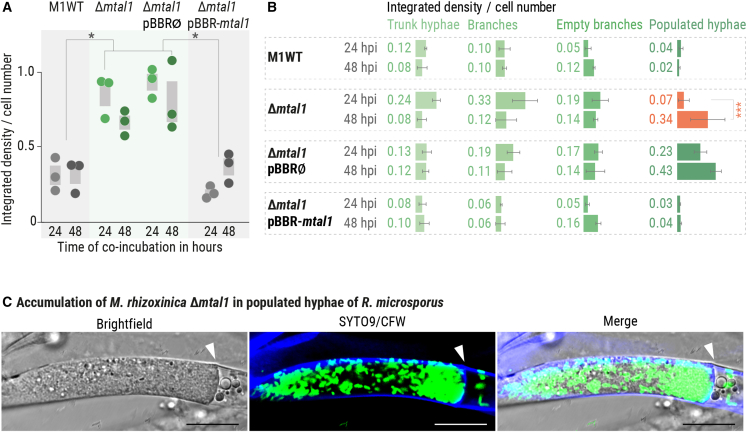
Intrigued by the apparent high cell number of M. rhizoxinica in populated hyphae, we quantified the number of endohyphal bacteria after colonization of R. microsporus by SYTO9-stained M. rhizoxinica in BFI devices. The bacterial load inside the fungal hyphae was measured by calculating the number of fluorescent endohyphal bacteria from the integrated density at 485/498 nm (Figure 3A). In comparison with when apo-symbiotic R. microsporus is colonized by M. rhizoxinica wild type, both M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ show significantly higher cell numbers (p < 0.05) after 1 day (Data S2A–S2D). In contrast, the bacterial load of hyphae colonized by M. rhizoxinica Δmtal1 pBBR-mtal1 is similar to that of M. rhizoxinica wild type (Figure 3A; Data S2A–S2D). As expected, endobacteria were absent from hyphae when the T3SS-deficient M. rhizoxinica strains were co-cultured with the apo-symbiotic fungus (Figures S3C–S3E). In contrast, rhizoxin-deficient bacteria are present in fungal hyphae at a similar level to the wild type (Figures S3C–S3E; Data S2E–S2H).
Figure 3.
Quantification of MTAL1-deficient M. rhizoxinica inside R. microsporus
(A) Apo-symbiotic R. microsporus was individually co-incubated with SYTO9-stained M. rhizoxinica wild type (M1WT), M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, or M. rhizoxinica Δmtal1 pBBR-mtal1 for 48 h. After fluorescence microscopy at 485/498 nm, the integrated density per bacterial cell number was calculated for both time points (24 and 48 hpi). Dots represent three independent replicates (n = 3 biological replicates) ± SEM (gray bars). One-way ANOVA with Tukey’s multiple comparison test (∗p < 0.05, Data S2A–S2D).
(B) The integrated density per cell number was measured for each individual type of hyphae. n = 3 biological replicates ± SEM. One-way ANOVA with Tukey’s multiple comparison test (∗∗∗p < 0.0002, Data S3A–S3D).
(C) Fluorescence microscopy images showing the accumulation of M. rhizoxinica Δmtal1 (green) in populated hyphae of R. microsporus (blue). Septa are indicated by an arrow head. Scale bars: 10 μm. See also Figures S3C–S3E and S4.
The counts of endofungal M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1 at the 24 h time point are not significantly different from those at 48 h (Figure 3A; Data S2A–S2D), suggesting that fungal reinfection takes place within 24 h of their introduction. However, it is important to consider that a potential increase in bacterial cell numbers after 24 h may not have been detected due to dilution of the staining intensity caused by bacterial proliferation and bleaching of SYTO9.25 To minimize SYTO9-bleaching, light exposure was kept to a minimum by using a short exposure time (100 ms). In addition, axenic SYTO9-stained M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1 were shown to remain detectably stained after culturing for 24 and 48 h in refreshed medium (Figure S4).
When the bacterial load was quantified for each individual type of hyphae, we noticed that the cell number of M. rhizoxinica Δmtal1 in populated hyphae is significantly higher (p < 0.0002) after 2 days of co-incubation than after 1 day (Figure 3B; Data S3A–S3D). Populated hyphae also contain a higher number of M. rhizoxinica Δmtal1 pBBR∅ after 48 h, reaching a similar cell number as M. rhizoxinica Δmtal1. Genetic complementation restores wild-type behavior, with M. rhizoxinica Δmtal1 pBBR-mtal1 distribution across the different types of hyphae being similar to that of M. rhizoxinica wild type (Figure 3B; Data S3A–S3D). Accumulation of M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ in populated hyphae is clearly visible in microscopic images (Figures 3C and S2B). In contrast, accumulation of M. rhizoxinica wild type, M. rhizoxinica Δmtal1 pBBR-mtal1, and M. rhizoxinica ΔrhiG in populated hyphae was only occasionally observed. As expected, populated hyphae are absent in R. microsporus co-cultivated with T3SS-deficient M. rhizoxinica because these strains are unable to effectively colonize fungal hyphae (Figure S3D; Data S3E–S3H). Taken together, these results reveal that MTAL1-deficient M. rhizoxinica reaches higher cell densities than M. rhizoxinica wild type in R. microsporus-populated hyphae.
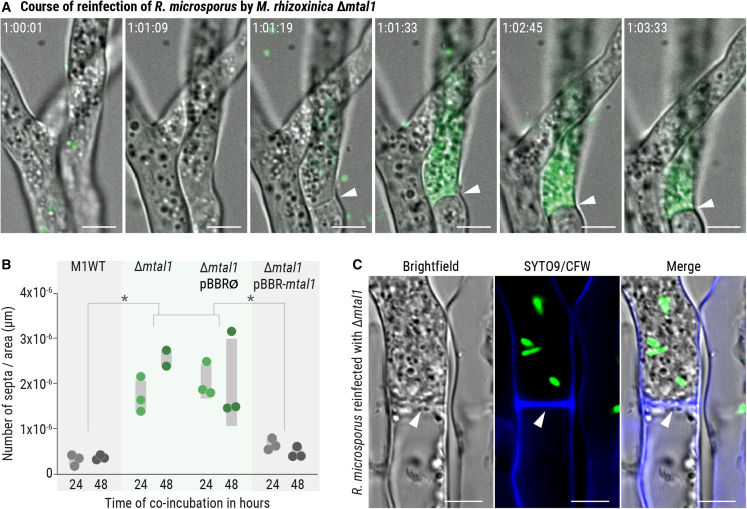
M. rhizoxinica Δmtal1 colonization of R. microsporus causes hyphal septation
To visualize the process by which M. rhizoxinica Δmtal1 reaches high cell densities in fungal hyphae, the colonization of R. microsporus by M. rhizoxinica Δmtal1 was monitored in real time using a BFI device (Figure 4A; Video S1). First, M. rhizoxinica Δmtal1 penetrates and enters selected hyphae of R. microsporus. Following colonization, the bacterial cells accumulate in high numbers (strong green fluorescent signal), and a septum is formed at the base of these hyphae in a relatively short amount of time (approximately 3 min). As a result, the cytoplasmic flow between colonized hyphae and the remaining fungal mycelium is abolished, leading to the physical containment of bacteria (Video S1).
Figure 4.
Formation of septa after colonization of R. microsporus by MTAL1-deficient M. rhizoxinica
(A) Course of colonization of R. microsporus by M. rhizoxinica Δmtal1. Colonization was monitored over time using fluorescence microscopy (Video S1). Arrowheads indicate the formation of septa. Scale bars: 5 μm.
(B) The number of septa per hyphal area (in μm) was calculated for M. rhizoxinica wild type (M1WT), M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1. Dots represent three independent replicates (n = 3 biological replicates) ± SEM (gray bars). One-way ANOVA with Tukey’s multiple comparison test (∗p < 0.05, Data S4A–S4D).
(C) Fluorescence microscopy images showing the formation of septa (indicated by an arrow head) in R. microsporus (blue stain) colonized by M. rhizoxinica Δmtal1 (green). Scale bars: 5 μm. See also Figures S5A and S5B.
Intrigued by our observation of septa formation in R. microsporus, we analyzed the number of septa formed after fungal reinfection by M. rhizoxinica strains in a BFI device. The data were normalized by dividing the septa number by the mycelium area (in μm) that was analyzed (Figure 4B). Although septa formation occurs occasionally in hyphae containing M. rhizoxinica wild type, we observed a significant increase in septa formation in hyphae containing M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ after 24 h of co-incubation (p < 0.05, Figure 4B; Data S4A–S4D). This increase in septa formation is maintained at the 48 h time point. In contrast, the number of septa in hyphae colonized by M. rhizoxinica Δmtal1 pBBR-mtal1, T3SS-deficient M. rhizoxinica strains (M. rhizoxinica ΔsctC and M. rhizoxinica ΔsctT), and M. rhizoxinica ΔrhiG is comparable to the wild type (Figures 4B, S5A, and S5B; Data S4E–S4H).
To confirm that the observed septa are true septa, fungal mycelium containing SYTO9-stained M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, or M. rhizoxinica Δmtal1 pBBR-mtal1 was counter-stained with calcofluor white, a fungal cell-wall-specific dye. Fluorescence microscopy images reveal that septa are indeed composed of fungal cell wall material (Figures 4C and S5B). This formation of septa may represent a mechanism to limit hyphal damage caused by MTAL1-deficient M. rhizoxinica, considering that R. microsporus is known to produce septa to wall off old or injured hyphae.23,26
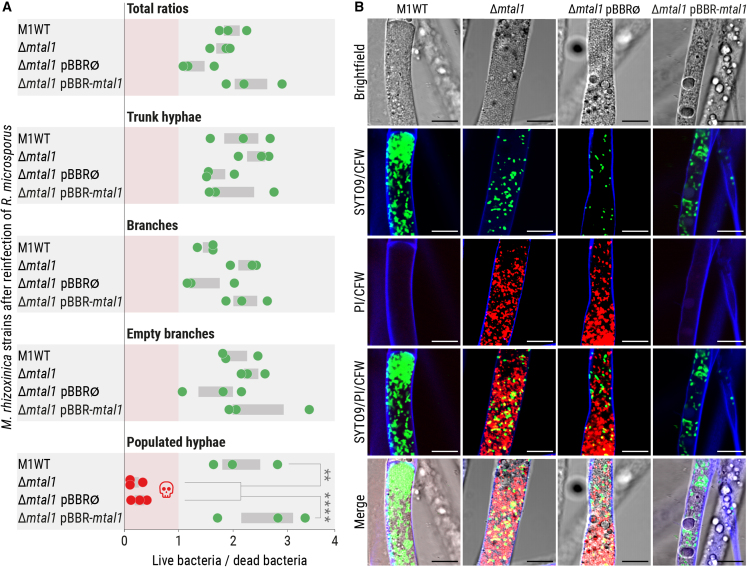
Survival of trapped M. rhizoxinica Δmtal1 is reduced in R. microsporus-populated hyphae
Based on the observation that septa formation stops the cytoplasmic flow between populated hyphae and the remaining fungal mycelium, we investigated whether trapped bacteria are alive in these hyphae. Using a bacterial LIVE/DEAD stain (SYTO9/propidium iodide [PI]) we observed that the majority of bacteria in trunk hyphae, branches, and empty branches are alive independent of MTAL1 production because M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ have ratios of live vs. dead that are similar to those of M. rhizoxinica wild type and M. rhizoxinica Δmtal1 pBBR-mtal1 (Figure 5A; Data S5). In contrast, the survival of M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ in populated hyphae is significantly reduced compared with M. rhizoxinica wild type and M. rhizoxinica Δmtal1 pBBR-mtal1 (p < 0.002, Figure 5A; Data S5). These results were confirmed by microscopic images showing mainly dead (red stained) M. rhizoxinica Δmtal1 and M. rhizoxinica Δmtal1 pBBR∅ in populated hyphae, while the majority of M. rhizoxinica wild type and M. rhizoxinica Δmtal1 pBBR-mtal1 are alive (green stained) in these hyphae (Figure 5B). These results suggest decreased M. rhizoxinica survival in the absence of MTAL1 (Figure 6).
Figure 5.
Viability test of M. rhizoxinica following colonization of R. microsporus
(A) Viability of M. rhizoxinica wild type (M1WT), M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1 inside R. microsporus after 72 h of co-cultivation in a bacterial-fungal interaction (BFI) device. Co-cultures were stained with LIVE/DEAD BacLight fluorescent dyes inside the BFI device. Following fluorescence microscopy, the integrated density was calculated for both live (SYTO9-stained) and dead bacteria (propidium iodide-stained [PI]) using Fiji. The ratio (live/dead) was plotted for the total number of bacteria and for each individual type of hyphae. Dots represent three independent replicates (n = 3 biological replicates) ± SEM (gray bars). One-way ANOVA with Tukey’s multiple comparison test (∗∗p < 0.002, ∗∗∗∗p < 0.0001, Data S5).
(B) Microscopic images of R. microsporus colonized by M. rhizoxinica wild type (M1WT), M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, or M. rhizoxinica Δmtal1 pBBR-mtal1 stained with LIVE/DEAD BacLight fluorescent dyes. Scale bars: 10 μm. See also Figure S5C.
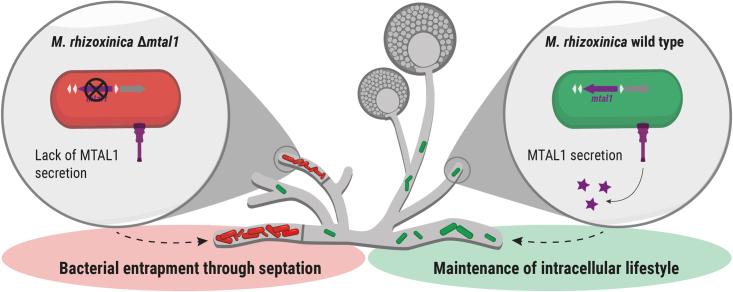
Figure 6.
Schematic model of the intracellular survival of endofungal bacteria
The endosymbiotic bacterium Mycetohabitans rhizoxinica secretes transcription activator-like effector 1 (MTAL1) via the type 3 secretion system, which is essential for its survival within R. microsporus (green). Absence of MTAL1 leads to entrapment of M. rhizoxinica through fungal septation and subsequent bacterial cell death (red).
It should be considered that LIVE/DEAD staining using SYTO9 and PI may underestimate bacterial viability. SYTO9 (live stain) can stain live cells with different efficiencies,25 while PI (dead stain) has a strong binding affinity to extracellular nucleic acids, a major component of bacterial biofilms.27 Because M. rhizoxinica is a known biofilm producer,28 we performed LIVE/DEAD stains of axenic cultures before co-incubation in BFI devices. The majority of the cells stained green with negligible red staining (Figure S5C), indicating that underestimation of bacterial viability due to PI binding to extracellular nucleic acids is unlikely.
Discussion
Both pathogenic and symbiotic bacteria can invade and control their eukaryotic hosts by employing effector molecules.29,30 One prominent example is the TAL effectors, a family of highly homologous proteins secreted by the T3SS that have been implicated in the virulence of numerous plant pathogens.31,32,33 In recent years, TALs have become popular as biotechnological tools due to their ability to manipulate DNA in a site-directed manner.34 Consequently, TAL homologs have been identified in a wide range of bacterial species, including M. rhizoxinica (MTAL1, MTAL2, and MTAL3).35 In this study, we combined genetic and phenotypic studies to functionally characterize MTAL1 of the fungal endosymbiont M. rhizoxinica. Targeted gene knockouts resulted in MTAL1-deficient bacteria that became trapped in side hyphae through the formation of septa, signifying that MTAL1 is an important symbiosis factor for the Rhizopus-Mycetohabitans interaction.
By means of a tailor-made microfluidic device,22 we show that the R. microsporus mycelium is composed of four phenotypically distinct types of hyphae (trunk hyphae, branches, empty branches, and populated hyphae). These fungal hyphae types have been rarely observed among the Mucoromycota,23 and this is the first time that different cell types have been described for R. microsporus. The vegetative main hyphae, or so-called trunk hyphae,36,37 are important for transporting intracellular material across the mycelium at relatively high speeds. This allows R. microsporus and fungi in general to cope well with heterogeneous distributions of soil nutrients in their natural habitat.24 In addition, cytoplasmic flow enables the free migration of M. rhizoxinica throughout the host mycelium.38 Hyphae serving as dispersal vectors for motile bacteria are an important feature of BFI ecology39 and are thought to promote colonization of new ecological niches.40
Fluorescence microscopy combined with microfluidics was used to gain a deeper understanding of the interaction between MTAL1-deficient M. rhizoxinica and R. microsporus. We have demonstrated (1) efficient fungal reinfection by MTAL1 knockout strains within 1 day and (2) accumulation and trapping of these strains in side hyphae after 2 days. These infected side hyphae (populated hyphae) are then walled off from the remaining mycelium via the formation of septa, which abrogates cytosolic exchange. In this way, M. rhizoxinica, which normally moves freely within the mycelium,10 becomes physically restricted (Figure 6). The observed increase in bacterial density may be caused by the ongoing cell division of MTAL1-deficient M. rhizoxinica while trapped. Alternatively, it is plausible that MTAL1-deficient bacteria are redistributed inside the fungal mycelium after reinfection and subsequently accumulate in side hyphae, as total bacterial numbers reach a maximum after 1 day and a significant increase within populated hyphae is observed after 2 days. We propose that the spatial separation of MTAL1-deficient symbionts in walled-off hyphae may allow the fungal host to impede interactions with non-beneficial symbionts.41 Although such compartmentalization in fungi is only described for arbuscular mycorrhizal fungi, its benefits to the host are well documented in a wide range of host-microbe mutualisms (e.g., insects and bacteria, legumes and rhizobia, and squid and Vibrio).41 For example, mycorrhiza form partial compartments in the host plant’s root tissue (arbuscules). The host uses these compartments to monitor the quality of the symbiont and subsequently regulate symbiont success.42 A similar regulatory mechanism could be used by R. microsporus to protect itself against colonization by MTAL1-deficient bacteria. Because the majority of trapped MTAL1-deficient bacteria are dead, it could be that bacterial cell death results from a host-produced toxic factor. Alternatively, entrapment may result in an insufficiency of the host-derived metabolites and lipids on which M. rhizoxinica depends.14,43
Our visualization of septa formation by R. microsporus is one of the most surprising results because vegetative hyphae of Mucoromycota generally lack regular septa.23 Interestingly, R. microsporus has been observed to produce septa as separators between mycelia and spore-forming structures.23,26 Thus, given that R. microsporus grown submerged in liquid medium struggles to produce mature sporangia, it could be that populated hyphae represent abortive sporangia due to the set-up of the microfluidics devices. However, considering that the persistence of the symbiosis is dependent on spores containing healthy endobacteria,10 we would expect the majority of the trapped MTAL1-deficient endobacteria to be alive (as seen in the rare instances of trapped wild-type bacteria). Since the majority of MTAL1-deficient bacteria are dead, we reason that it is unlikely that populated hyphae represent abortive sporangia. Instead, it is possible that septa are formed as a response to limit hyphal damage caused by MTAL1-deficient bacteria because Mucoromycota fungi can form septa to wall off old or injured hyphae.23,26 In addition, induced host suicide by bacteria is a mechanism that is well described in plant-pathogen interactions. For example, TALs from X. campestris induce a suicide gene in resistant plants, causing programmed cell death in the host and thereby preventing the pathogen from spreading.44 Another mechanism to restrict the horizontal propagation of harmful cytoplasmic components in filamentous fungi is vegetative (heterokaryon) incompatibility.45 In Neurospora crassa, hyphal fusion of two heterokaryon-incompatible individuals leads to septa formation and subsequent death of the resulting compartments.46 This behavior appears strikingly similar to the formation of septa by R. microsporus after colonization by MTAL1-deficient M. rhizoxinica. Although it remains to be investigated whether populated hyphae are dead and/or damaged, it is nonetheless an interesting proposition that MTAL1-deficient endobacteria may trigger a response similar to nonself-recognition in Mucormycota fungi.
Notably, the MTAL1 proteins described in this study are the first TAL-related proteins whose absence causes an increase in host colonization. In contrast, TAL proteins of plant pathogens induce susceptibility genes, which leads to a dramatic increase in bacterial cell numbers in the host.47 Thus, MTAL1 might represent DNA-binding proteins with a function different from TALs34 because multiple unrelated DNA-binding proteins are known to aid host colonization by various pathogens and symbionts.29 It is conceivable that R. microsporus perceives M. rhizoxinica lacking MTAL1 not as an endosymbiont but as a pathogen. Indeed, M. rhizoxinica likely was initially a parasite before becoming a mutualist,14,48,49 with R. microsporus having acquired resistance against the potent phytotoxin (rhizoxin) produced by M. rhizoxinica.49 Thus, MTAL1 proteins might represent an important factor for the maintenance of the mutualistic relationship in this BFI,21 and their absence may cause a reversion from a mutualistic to a parasitic lifestyle. Our data designating MTAL1 as a symbiosis factor in the R. microsporus-M. rhizoxinica relationship illuminate the “sliding scale” nature of BFIs, wherein symbionts can act as mutualists or pathogens depending on the genetic context.50
In summary, we show that MTAL1-deficient endosymbionts successfully recolonize the host but become trapped in side hyphae due to the formation of septa. These observations represent an unprecedented case in which bacterial colonization induces biogenesis of septa in a Mucoromycota fungus, a likely defense strategy of the host. Based on microscopic investigations, it appears that MTAL1-deficient mutants are perceived as pathogens in the fungal cytosol, in accordance with a plausible antagonism-mutualism shift in the evolution of this microbial symbiosis. Our results not only offer insight into the dynamic interactions between bacteria and fungi but also open new avenues of research into endobacterial control of fungal host physiology.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Mycetohabitans rhizoxinica | This study | HKI-454 |
| Electrocompetent Escherichia coli TOP10 | Invitrogen | CAT#C404052 |
| Chemicals, peptides, and recombinant proteins | ||
| Potato Dextrose Agar | Becton, Dickinson & Co. | CAT#213400 |
| Glycerol | Roth | CAT#6967.1 |
| Yeast Extract | Becton, Dickinson & Co. | CAT#288620 |
| K2HPO4 | Roth | CAT#P749.2 |
| KH2PO4 | Roth | CAT#3904.1 |
| C6H7NaO | Roth | CAT#HN13.3 |
| (NH4)2SO4 | Roth | CAT#9212.1 |
| Mg2SO4 | Roth | CAT#P027.1 |
| Standard Nutrient Agar I | Merck | CAT#1.03864.0500 |
| Phusion® High-Fidelity PCR Master Mx | New England Biolabs | CAT#M0530L |
| Apramycin | Sigma | CAT#A2024 |
| SpeI | New England Biolabs | CAT#R3133L |
| KpnI | New England Biolabs | CAT#R3142L |
| OneTAQ® Quick-Load® 2x Master Mix | New England Biolabs | CAT#M0486L |
| Ethidium bromide | New England Biolabs | CAT#E1510 |
| NdeI | New England Biolabs | CAT#R0111L |
| AflII | New England Biolabs | CAT#R0520L |
| BstBI | New England Biolabs | CAT#R0519L |
| Chloramphenicol | Roth | CAT#3886.1 |
| Potato Dextrose Broth | Becton, Dickinson & Co. | CAT#254920 |
| NaCl | Roth | CAT#3957.1 |
| SYTO™ 9 | Invitrogen | CAT#S34854 |
| LIVE/DEAD™ BacLight™ Bacterial Viability Kit | Invitrogen | CAT#L7012 |
| Propidium iodide | Invitrogen | CAT#P21493 |
| Calcofluor white | Sigma | CAT#18909 |
| GeneRuler™ DNA Ladder Mix | Thermo Scientific | CAT#SM0333 |
| T4 DNA Ligase | New England Biolabs | CAT#M0202L |
| Ciprofloxacin | Sigma | CAT#17850 |
| Critical commercial assays | ||
| Monarch® DNA Gel Extraction Kit | New England Biolabs | CAT#T1020L |
| NEBuilder® HiFi DNA Assembly Cloning Kit | New England Biolabs | CAT#E2621L |
| MasterPure™ DNA Purification Kit | Biozym | CAT#160502 |
| Monarch® Plasmid Miniprep Kit | New England Biolabs | CAT#T1010L |
| Experimental models: Organisms/strains | ||
| Rhizopus microsporus | Jena Microbial Resource Collection | ATCC 62417 |
| Oligonucleotides | ||
| See Data S6 for a list of oligonucleotides | N/A | |
| Recombinant DNA | ||
| Plasmid pZU17 | This paper | N/A |
| Plasmid pBBR-mtal1 | This paper | N/A |
| Plasmid pBBR∅ | This paper | N/A |
| Plasmid pGL42 | Lackner et al.16,43 | N/A |
| Software and algorithms | ||
| Zen Blue | Zeiss | N/A |
| Zen Black | Zeiss | N/A |
| Fiji | Schindelin et al.51 | N/A |
| GraphPad Prism 9.0 | GraphPad Software | N/A |
| Other | ||
| M. rhizoxinica genome | Lackner et al.16,43 | Genbank Accession #GCA_000198775.1 |
| NanoDrop™ One | Thermo Scientific | N/A |
| Eporator Electroporator | Eppendorf | N/A |
| Quantum ST5 Xpress Gel Imager | Vilber Lourmat | N/A |
| Bacterial-Fungal-Interaction devicesDevices | This paper | N/A |
| CASY Cell Counter | OMNI Life Science | N/A |
| Fluorescence Spinning Disc Microscope | Zeiss | Axio Observer platform with Cell Observer SD |
| Confocal Laser Scanning Microscope | Zeiss | LSM 710 |
| Fluorescence Widefield Microscope | Zeiss | Elyra7 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Christian Hertweck (Christian.Hertweck@leibniz-hki.de).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
The fungal strain Rhizopus microsporus ATCC62417 harboring the endobacteria Mycetohabitans rhizoxinica was used in this study.48 Endobacteria from R. microsporus ATCC62417 were eliminated by continuous antibiotic treatment10 and the apo-symbiotic fungal strain was named ATCC62417/S. Absence of endobacteria was confirmed by fluorescence microscopy and an absence of rhizoxin extracts of the fungal mycelium.52 Both R. microsporus strains (ATCC62417 and ATCC62417/S) were cultivated on Potato Dextrose Agar (PDA; Becton, Dickinson & Company, Sparks, MD, USA) at 30 °C. Bacterial endosymbionts were isolated from R. microsporus mycelium as previously reported.10 Axenic cultures of M. rhizoxinica were grown at 30 °C in MGY M9 medium (10 g/L glycerol, 1.25 g/L yeast extract, M9 salts: 40 mM K2HPO4, 14 mM KH2PO4, 2.2 mM C6H7NaO7, 7.5 mM (NH4)2SO4, and 0.8 mM Mg2SO4) or Standard I Nutrient Agar (Merck, Darmstadt, Germany) supplemented with 1% glycerol.
Method details
Generation of M. rhizoxinica Δmtal1 strains
To investigate the role of MTAL1 in the symbiosis, the mtal1 gene (RBRH_01844) was deleted using a double crossover strategy as previously described.16
Using a proofreading polymerase, the upstream and downstream regions of the gene of interest were amplified. Primers were designed to contain 20 bp overlap with the gene of interest as well as a 20 bp overlap with an antibiotic resistance cassette (apramycin). The apramycin cassette was amplified from pJI773 using primers carrying the same 20 bp.
The knockout vector pGL42a was used to generate a M. rhizoxinica Δmtal1::ApraR deletion mutant. pGL42a was double-digested with the restriction enzymes SpeI and KpnI (New England Biolabs, Ipswich, MA, USA). The linear vector was gel-purified (Monarch® DNA Gel Extraction Kit, New England Biolabs) and quantified using a NanoDrop™ (Thermo Fisher Scientific).
Equimolar amounts of three PCR products and linear pGL42a were mixed with NEBuilder® 2X Master Mix (NEBuilder® HiFi DNA Assembly Cloning Kit, New England Biolabs) and incubated at 60 °C for 1 hr following the manufacturer’s recommendations. The new plasmid pZU17 (Δmtal1) was introduced into E. coli by chemical transformation. Transformants were selected on Standard I Nutrient Agar (Merck) supplemented with 50 μg/mL apramycin and 1% glycerol (pZU17).
Competent M. rhizoxinica HKI-454 (M1WT) cells were transformed with pZU17 via electroporation. Transformants were grown on standard nutrient agar containing 50 μg/mL of apramycin. Colonies were subsequently passaged onto agar plates containing double selection medium16 until the correct knockout constructs were observed using colony PCR. Colony PCRs were carried out in 12.0 μL final volumes containing: 5 μL of high-fidelity OneTaq® Quick-Load® 2X Master Mix (New England Biolabs), appropriate forward and reverse control primers (both 0.4 μM, Data S6A), and 5 μL colony suspension. The following thermocycling conditions were used for amplification: 96 °C / 3 min, 1 cycle; 96 °C / 10 s, 58 °C / 15 s, 68 °C / 1 min, 30 cycles; 68 °C / 5 min, 1 cycle; 16 °C / hold. The resulting PCR products were visualized on an agarose gel (Figure S1E). Primers were designed to span the two recombination sites, yielding amplicons A and B in M. rhizoxinica Δmtal1 strains and amplicons C and D in M. rhizoxinica wild type strains (Figures S1A and S1B).
Generation of genetically complemented M. rhizoxinica Δmtal1 strains
In order to genetically complement the knockout strain M. rhizoxinica Δmtal1, genomic DNA from M. rhizoxinica (M1WT) was isolated using the MasterPure™ DNA Purification Kit (Biozym Scientific, Hessisch Oldendorf, Germany) following the manufacturer's recommendations. The mtal1 gene was amplified by PCR with the primer pairs listed in Data S6B using Phusion® High-Fidelity PCR Master Mix with HF Buffer (New England Biolabs). The PCR product was gel-purified with the Monarch DNA Gel Extraction Kit (New England Biolabs). The purified amplicon was cloned into the NdeI/AflII restricted pBBR_Ps12_gfp downstream of the promoter Ps12 with the NEBuilder® 2X Master Mix (NEBuilder® HiFi DNA Assembly Cloning Kit, New England Biolabs), yielding pBBR-mtal1 (Figure S1D). The reaction mixture was subsequently used to transform chemically competent E. coli TOP10 cells (Invitrogen™, One Shot™, Carlsbad, CA, USA).
To generate an empty vector control, pBBR_Ps12_gfp was digested with the restriction enzyme BstBI. The resulting linear vector lacking gfp was self-circularized using T4 DNA Ligase (New England Biolabs) to yield pBBR∅ and used to transform chemically competent E. coli TOP10 cells (Invitrogen™, One Shot™). All plasmids were purified from E. coli TOP10 overnight cultures using the Monarch Plasmid Miniprep Kit (New England Biolabs) and verified by restriction digest and Sanger sequencing using the primers cmr_seq_fw and BBR_seq_rv (Data S6B).
The new plasmids (pBBR-mtal1 or pBBR∅) were introduced into competent M. rhizoxinica Δmtal1 cells via electroporation. Transformants were grown on standard nutrient agar containing 50 μg/mL chloramphenicol and 50 μg/mL apramycin. Colonies containing the respective plasmids were observed using colony PCR and control primers (Figure S1E; Data S6B).
Inoculation of bacterial-fungal interaction devices with fungus
Bacterial-fungal interaction (BFI) devices were prepared as previously described.22 Before the BFI devices were inoculated, apo-symbiotic (endosymbiont-free) R. microsporus ATCC62417/S was sub-cultured on PDA plates for two days at 30 °C. A piece of young, growing mycelium (app. 1 cm3) was cut from the agar plate and placed upside down in front of the microchannels of a BFI device filled with Potato Dextrose Broth (PDB; Becton, Dickinson & Company). The agar plug was positioned with the growth direction of the fungus facing the microchannels. We paid special attention to the size of the agar plug to minimize variation between experiments. After two days of incubation at 30 °C, the hyphae had grown far enough into the microchannels to perform subsequent reinfection experiments.
Bacterial inoculation into bacterial-fungal interaction devices
Bacterial over-night cultures (500 μL) were harvested in an Eppendorf tube and resuspended in 0.5 mL NaCl (0.85%) containing SYTO9 (5 nM final concentration, Invitrogen). Following incubation in the dark for 5 min, stained cells were washed and resuspended in 0.5 mL PDB medium. Cells were counted using a CASY Cell Counter (OMNI Life Science, Bremen, Germany) and then added to the bacteria inlet of the BFI device (100 μL), which is connected with the microchannels containing the cured fungus (Figures 2A and 2B).
Live-cell imaging of bacterial-fungal interactions
A fluorescence spinning disc microscope (Axio Observer microscope-platform equipped with Cell Observer SD, Zeiss, Oberkochen, Germany) was used to visualize BFIs at 0 h, 24 h, and 48 h after bacterial inoculation. For each time point, 16 images were taken at random positions of the microchannels, including both ends of the BFI device (N = 16 technical replicates). Brightfield images were captured using a laser intensity of 7.1 V and an exposure time of 100 ms. Images of bacterial cells (M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, M. rhizoxinica Δmtal1 pBBR-mtal1, M. rhizoxinica ΔsctT, M. rhizoxinica ΔsctC, and M. rhizoxinica ΔrhiG), stained with SYTO9, were captured at 485/498 nm with an exposure time of 100 ms. As a reference, wild-type R. microsporus ATCC62417, naturally containing endosymbionts, was also analyzed, without additional bacterial cultures being added through the inlet. Each reinfection experiment was performed three times independently (N = 3 biological replicates).
Imaging of bacterial survival inside of bacterial-fungal interaction devices
To test the bacterial survival rate in BFI devices, cells were stained with LIVE/DEAD BacLight fluorescent dyes (Invitrogen) after 72 h of bacterial-fungal co-incubation. The fungal cell wall was counter-stained with calcofluor white (Sigma-Aldrich, St. Louis, MI, USA). To stain fungal and bacterial cells inside the BFI device, the PDB medium was removed through the bacteria inlet and replaced with 100 μL staining solution (10 nM SYTO9, 60 nM propidium iodide (Invitrogen), 1 μM calcofluor white). Fluid replacement was repeated three times, before the devices were incubated in the dark for 15 min. Following staining, fluorescent dyes were replaced with PDB medium by flushing the devices four times. Fluorescence microscopy was carried out using a fluorescence spinning disc microscope (Axio Observer microscope-platform equipped with Cell Observer SD, Zeiss). Images of living bacterial cells were captured at 485/498 nm, dead bacterial cells were captured at 493/636 nm, and the fungal cell wall was captured at 380/475 nm using the Zeiss-Zen software.
Quantification of bacterial cells inside of fungal hyphae
Fluorescent images, containing spatial calibration meta data, were imported to Fiji51 and the same spatial scale was applied to all images using the global scale function. Prior to image analyses, all images were converted to 16-bit gray-scale, which was propagated to all channels (brightfield, 485/498 nm, 493/636 nm). The hyphal area and integrated density (ID; product of area and mean grey value) of all channels were measured using the freehand tool and measuring tool implemented in Fiji. According to the brightfield images, four types of hyphae were defined (Figure 2C) and the number of septa observed was counted. Subsequent calculations and data visualization were carried out using MS Excel and GraphPad Prism 9.0. To correct for background autofluorescence, raw ID values at 0 hours post infection (hpi) were subtracted from raw ID values at 24 hpi and 48 hpi. Corrected ID values were divided by the number of bacterial cells (cells/mL) inoculated in the bacteria inlet and then plotted. The same approach was applied to correct for the number of septa observed. The corrected number of septa was divided by the mycelium area analyzed (in μm).
Imaging of axenic bacterial cultures
To test whether stained bacteria remain detectably stained, axenic M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅ or M. rhizoxinica Δmtal1 pBBR-mtal1 were grown at 30 °C in 2 mL MGY M9 medium. When the exponential phase was reached, bacterial cells (1 mL) were stained with 5 nM SYTO9 for 5 min. Fluorescence microscopy was carried out using a fluorescence wide-field microscope (ELYRA 7, Zeiss). Images of bacterial cells were captured at 485/498 nm using the Zeiss-Zen software. Stained cells were centrifuged, the cell pellets were resuspended in 2 mL fresh MGY M9 medium, and the bacterial cultures were incubated at 30 °C. After 24 h of incubation, 20 μL of cells were used for fluorescence imaging at 485/498 nm. Sub-cultures (1 mL) of the 24-hour-old cultures were centrifuged, the cell pellets were resuspended in 2 mL fresh MGY M9 medium, and the bacterial cultures were incubated at 30 °C. After 24 h of incubation, 20 μL of the 48-hour-old cultures were used for fluorescence imaging at 485/498 nm.
Imaging of fungal septa
Fungal hyphae were harvested from one-week old bacterial-fungal co-cultures, stained with 5 nM SYTO9 and 1 μM calcofluor white, and fixed on glass slides. Fluorescent microscopy was performed using a confocal laser scanning microscope (LSM 710 equipped with a Cell Observer SD, Zeiss). Images of bacterial cells were captured at 485/498 nm and the fungal cell wall was captured at 380/475 nm using the Zeiss-Zen software.
Quantification and statistical analysis
Raw data from reinfection experiments and bacterial survival experiments were processed with MS Excel. GraphPad Prism 9.0 was used for statistical analysis and graphing. One-way analysis of variance (ANOVA) was used to study the relationship between different M. rhizoxinica strains (M. rhizoxinica wild type, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 control, or M. rhizoxinica Δmtal1 pBBR-mtal1) and fungal physiology (e.g. mycelium area, reinfection efficiency, septation rate, bacterial survival) following fungal reinfection using the Tukey HSD test function. P-values with p<0.05 were considered statistically significant. The Brown-Forsythe test was used to test for equal variance and a p value with <0.05 was considered significant.
Acknowledgments
We thank S. Lindner for assistance with image analyses. I.R. is grateful for financial support from the European Union’s Horizon 2020 Research and Innovation Program under the Marie Skłodowska-Curie grant agreement no. 794343. Financial support by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 2051 (Cluster of Excellence “Balance of the Microverse”) Project-ID 390713860 and the SFB 1127 ChemBioSys, Project-ID 239748522, and the Leibniz Award (to C.H.); the JSMC to Z.U.; and the Swiss National Science Foundation in the form of an Ambizione Career Grant (PZ00P2_168005) to C.E.S. is gratefully acknowledged. We thank the Microverse Imaging Center and Aurélie Jost for providing microscope facility support for data acquisition. The ELYRA 7 (used for producing images of axenic bacterial cultures) was funded by the Free State of Thuringia with grant number 2019 FGI 0003. The Microverse Imaging Center is funded by the DFG under Germany’s Excellence Strategy—EXC 2051, Project-ID 390713860.
Author contributions
I.R. conceived the idea, developed the study design, interpreted the data, and wrote the manuscript. P.W. designed plasmids and generated complemented strains. Z.U. conceived the idea and generated the mtal1 knockout strain. C.E.S. designed and manufactured microfluidic devices and revised the manuscript. J.K. assisted with plasmid design. E.M.M. assisted in data interpretation and manuscript revision. N.M. conceived the idea. I.F. assisted with method development of image analysis. F.H. assisted in data interpretation and manuscript revision. C.H. conceived the idea and drafted and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cub.2023.05.028.
Supplemental information
(A and B) (A) Approximate probabilities (p) of Browns-Forsythe test and (B) one-way analysis of variance (ANOVA) for the mycelium area of each type of hyphae following co-cultivation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of individual types of hyphae at specific time points (0, 24, or 48 h post infection [hpi]) across all four M. rhizoxinica strains using the Tukey HSD post hoc test.
(D) Comparison of types of hyphae from individual M. rhizoxinica strains over time (0 vs. 24 hpi and 0 vs. 48 hpi) using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the mycelium area of each type of hyphae following co-cultivation of apo-symbiotic R. microsporus with M. rhizoxinica wild type (M1WT), type 3 secretion system knockout strains (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT) or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of individual types of hyphae at specific time points (0, 24, or 48 h post infection [hpi]) across all four M. rhizoxinica strains using the Tukey HSD post hoc test.
(H) Comparison of types of hyphae from individual M. rhizoxinica strains over time (0 vs. 24 hpi and 0 vs. 48 hpi) using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the bacterial load of Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1) after colonization of apo-symbiotic Rhizopus microsporus. Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) The number of endohyphal bacteria in each M. rhizoxinica strain (indicated by green fluorescence) 24 h post infection (hpi) was compared with the number of endohyphal bacteria 48 h after co-cultivation (24 vs. 48 hpi).
(D) Comparison of endohyphal bacteria at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the bacterial load of M. rhizoxinica wild type (M1WT), T3SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG) following colonization of apo-symbiotic R. microsporus. Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) The number of endohyphal bacteria in each M. rhizoxinica strain (indicated by green fluorescence) 24 h post infection (hpi) was compared with the number of endohyphal bacteria 48 hours after co-cultivation (24 vs. 48 hpi).
(H) Comparison of endohyphal bacteria at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the bacterial load in individual types of hyphae of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of types of hyphae from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(D) Comparison of individual types of hyphae at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the bacterial load in individual types of hyphae of apo-symbiotic R. microsporus co-incubated with M. rhizoxinica wild type (M1WT), T2SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of types of hyphae from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(H) Comparison of individual types of hyphae at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the number of septa formed per mycelium area following co-incubation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of septa from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(D) Comparison of septa at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the number of septa formed per mycelium area following co-incubation of apo-symbiotic R. microsporus with M. rhizoxinica wild type (M1WT), T2SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of septa from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(H) Comparison of septa at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the ratio of live vs. dead endohyphal bacteria following co-incubation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of overall ratios of live/dead bacteria inside the fungal hyphae between M1WT, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1. 72 h post infection.
(D) The ratios of live/dead bacteria inside the fungal hyphae was compared between the different types of hyphae of individual strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A) Primers used for generation of M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1).
(B) Primers used for generation of genetically complemented M. rhizoxinica Δmtal1 (Δmtal1 pBBR-mtal1) or control M. rhizoxinica Δmtal1 (Δmtal1 pBBR∅).
Data and code availability
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.
References
- 1.Wernegreen J.J. Endosymbiosis. Curr. Biol. 2012;22:R555–R561. doi: 10.1016/j.cub.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 2.Sachs J.L., Skophammer R.G., Regus J.U. Evolutionary transitions in bacterial symbiosis. Proc. Natl. Acad. Sci. USA. 2011;108:10800–10807. doi: 10.1073/pnas.1100304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pawlowska T.E., Gaspar M.L., Lastovetsky O.A., Mondo S.J., Real-Ramirez I., Shakya E., Bonfante P. Biology of fungi and their bacterial endosymbionts. Annu. Rev. Phytopathol. 2018;56:289–309. doi: 10.1146/annurev-phyto-080417-045914. [DOI] [PubMed] [Google Scholar]
- 4.Bonfante P., Desirò A. Who lives in a fungus? The diversity, origins and functions of fungal endobacteria living in Mucoromycota. ISME J. 2017;11:1727–1735. doi: 10.1038/ismej.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Partida-Martinez L.P., Hertweck C. Pathogenic fungus harbours endosymbiotic bacteria for toxin production. Nature. 2005;437:884–888. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 6.Partida-Martinez L.P., Hertweck C. A gene cluster encoding rhizoxin biosynthesis in Burkholderia rhizoxina, the bacterial endosymbiont of the fungus Rhizopus microsporus. ChemBioChem. 2007;8:41–45. doi: 10.1002/cbic.200600393. [DOI] [PubMed] [Google Scholar]
- 7.Estrada-de Los Santos P., Palmer M., Chávez-Ramírez B., Beukes C., Steenkamp E.T., Briscoe L., Khan N., Maluk M., Lafos M., Humm E., et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes. 2018;9 doi: 10.3390/genes9080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lackner G., Hertweck C. Impact of endofungal bacteria on infection biology, food safety, and drug development. PLoS Pathog. 2011;7:e1002096. doi: 10.1371/journal.ppat.1002096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scherlach K., Busch B., Lackner G., Paszkowski U., Hertweck C. Symbiotic cooperation in the biosynthesis of a phytotoxin. Angew. Chem. Int. Ed. Engl. 2012;51:9615–9618. doi: 10.1002/anie.201204540. [DOI] [PubMed] [Google Scholar]
- 10.Partida-Martinez L.P., Monajembashi S., Greulich K.O., Hertweck C. Endosymbiont-dependent host reproduction maintains bacterial-fungal mutualism. Curr. Biol. 2007;17:773–777. doi: 10.1016/j.cub.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 11.Moebius N., Üzüm Z., Dijksterhuis J., Lackner G., Hertweck C. Active invasion of bacteria into living fungal cells. eLife. 2014;3:e03007. doi: 10.7554/eLife.03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niehs S.P., Scherlach K., Hertweck C. Genomics-driven discovery of a linear lipopeptide promoting host colonization by endofungal bacteria. Org. Biomol. Chem. 2018;16:8345–8352. doi: 10.1039/c8ob01515e. [DOI] [PubMed] [Google Scholar]
- 13.Lastovetsky O.A., Krasnovsky L.D., Qin X., Gaspar M.L., Gryganskyi A.P., Huntemann M., Clum A., Pillay M., Palaniappan K., Varghese N., et al. Molecular dialogues between early divergent fungi and bacteria in an antagonism versus a mutualism. mBio. 2020;11:e02088-20. doi: 10.1128/mBio.02088-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lastovetsky O.A., Gaspar M.L., Mondo S.J., LaButti K.M., Sandor L., Grigoriev I.V., Henry S.A., Pawlowska T.E. Lipid metabolic changes in an early divergent fungus govern the establishment of a mutualistic symbiosis with endobacteria. Proc. Natl. Acad. Sci. USA. 2016;113:15102–15107. doi: 10.1073/pnas.1615148113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leone M.R., Lackner G., Silipo A., Lanzetta R., Molinaro A., Hertweck C. An unusual galactofuranose lipopolysaccharide that ensures the intracellular survival of toxin-producing bacteria in their fungal host. Angew. Chem. Int. Ed. Engl. 2010;49:7476–7480. doi: 10.1002/anie.201003301. [DOI] [PubMed] [Google Scholar]
- 16.Lackner G., Moebius N., Hertweck C. Endofungal bacterium controls its host by an hrp type III secretion system. ISME J. 2011;5:252–261. doi: 10.1038/ismej.2010.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niehs S.P., Scherlach K., Dose B., Uzum Z., Stinear T.P., Pidot S.J., Hertweck C. A highly conserved gene locus in endofungal bacteria codes for the biosynthesis of symbiosis-specific cyclopeptides. Proc. Natl. Acad. Sci. USA. 2022;1:pgac152. doi: 10.1093/pnasnexus/pgac152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Juillerat A., Bertonati C., Dubois G., Guyot V., Thomas S., Valton J., Beurdeley M., Silva G.H., Daboussi F., Duchateau P. BurrH: a new modular DNA binding protein for genome engineering. Sci. Rep. 2014;4:3831. doi: 10.1038/srep03831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lange O., Wolf C., Dietze J., Elsaesser J., Morbitzer R., Lahaye T. Programmable DNA-binding proteins from Burkholderia provide a fresh perspective on the TALE-like repeat domain. Nucleic Acids Res. 2014;42:7436–7449. doi: 10.1093/nar/gku329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholze H., Boch J. TAL effectors are remote controls for gene activation. Curr. Opin. Microbiol. 2011;14:47–53. doi: 10.1016/j.mib.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Carter M.E., Carpenter S.C.D., Dubrow Z.E., Sabol M.R., Rinaldi F.C., Lastovetsky O.A., Mondo S.J., Pawlowska T.E., Bogdanove A.J. A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host. Proc. Natl. Acad. Sci. USA. 2020;117:17122–17129. doi: 10.1073/pnas.2003857117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stanley C.E., Stöckli M., van Swaay D., Sabotič J., Kallio P.T., Künzler M., deMello A.J., Aebi M. Probing bacterial-fungal interactions at the single cell level. Integr. Biol. (Camb) 2014;6:935–945. doi: 10.1039/c4ib00154k. [DOI] [PubMed] [Google Scholar]
- 23.Dijksterhuis J., Samson R.A. In: Food Spoilage Microorganisms. Blackburn C.d.W., editor. Woodhead Publishing; 2006. Zygomycetes; pp. 415–436. [DOI] [Google Scholar]
- 24.Fricker M.D., Tlalka M., Bebber D., Takagi S., Watkinson S.C., Darrah P.R. Fourier-based spatial mapping of oscillatory phenomena in fungi. Fungal Genet. Biol. 2007;44:1077–1084. doi: 10.1016/j.fgb.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 25.Stiefel P., Schmidt-Emrich S., Maniura-Weber K., Ren Q. Critical aspects of using bacterial cell viability assays with the fluorophores SYTO9 and propidium iodide. BMC Microbiol. 2015;15:36. doi: 10.1186/s12866-015-0376-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore-Landecker E. Encyclopedia of Life Sciences (ELS) John Wiley & Sons, Ltd.; 2008. Zygomycota and glomeromycota. [DOI] [Google Scholar]
- 27.Rosenberg M., Azevedo N.F., Ivask A. Propidium iodide staining underestimates viability of adherent bacterial cells. Sci. Rep. 2019;9:6483. doi: 10.1038/s41598-019-42906-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uzum Z., Silipo A., Lackner G., De Felice A., Molinaro A., Hertweck C. Structure, genetics and function of an exopolysaccharide produced by a bacterium living within fungal hyphae. Chembiochem. 2015;16:387–392. doi: 10.1002/cbic.201402488. [DOI] [PubMed] [Google Scholar]
- 29.Bierne H., Cossart P. When bacteria target the nucleus: the emerging family of nucleomodulins. Cell. Microbiol. 2012;14:622–633. doi: 10.1111/j.1462-5822.2012.01758.x. [DOI] [PubMed] [Google Scholar]
- 30.Vlisidou I., Hapeshi A., Healey J.R.J., Smart K., Yang G., Waterfield N.R. The Photorhabdus asymbiotica virulence cassettes deliver protein effectors directly into target eukaryotic cells. eLife. 2019;8:e46259. doi: 10.7554/eLife.46259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boch J., Scholze H., Schornack S., Landgraf A., Hahn S., Kay S., Lahaye T., Nickstadt A., Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 32.Kay S., Bonas U. How Xanthomonas type III effectors manipulate the host plant. Curr. Opin. Microbiol. 2009;12:37–43. doi: 10.1016/j.mib.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 33.de Lange O., Schreiber T., Schandry N., Radeck J., Braun K.H., Koszinowski J., Heuer H., Strauß A., Lahaye T. Breaking the DNA-binding code of Ralstonia solanacearum TAL effectors provides new possibilities to generate plant resistance genes against bacterial wilt disease. New Phytol. 2013;199:773–786. doi: 10.1111/nph.12324. [DOI] [PubMed] [Google Scholar]
- 34.Perez-Quintero A.L., Szurek B. A decade decoded: spies and hackers in the history of TAL effectors research. Annu. Rev. Phytopathol. 2019;57:459–481. doi: 10.1146/annurev-phyto-082718-100026. [DOI] [PubMed] [Google Scholar]
- 35.Stella S., Molina R., Bertonatti C., Juillerrat A., Montoya G. Expression, purification, crystallization and preliminary X-ray diffraction analysis of the novel modular DNA-binding protein BurrH in its apo form and in complex with its target DNA. Acta Crystallogr. F Struct. Biol. Commun. 2014;70:87–91. doi: 10.1107/S2053230X13033037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmieder S.S., Stanley C.E., Rzepiela A., van Swaay D., Sabotič J., Nørrelykke S.F., deMello A.J., Aebi M., Künzler M. Bidirectional propagation of signals and nutrients in fungal networks via specialized hyphae. Curr. Biol. 2019;29:217–228.e4. doi: 10.1016/j.cub.2018.11.058. [DOI] [PubMed] [Google Scholar]
- 37.Hickey P.C., Jacobson D., Read N.D., Glass N.L. Live-cell imaging of vegetative hyphal fusion in Neurospora crassa. Fungal Genet. Biol. 2002;37:109–119. doi: 10.1016/s1087-1845(02)00035-x. [DOI] [PubMed] [Google Scholar]
- 38.Desirò A., Salvioli A., Ngonkeu E.L., Mondo S.J., Epis S., Faccio A., Kaech A., Pawlowska T.E., Bonfante P. Detection of a novel intracellular microbiome hosted in arbuscular mycorrhizal fungi. ISME J. 2014;8:257–270. doi: 10.1038/ismej.2013.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kohlmeier S., Smits T.H., Ford R.M., Keel C., Harms H., Wick L.Y. Taking the fungal highway: mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 2005;39:4640–4646. doi: 10.1021/es047979z. [DOI] [PubMed] [Google Scholar]
- 40.Simon A., Hervé V., Al-Dourobi A., Verrecchia E., Junier P. An in situ inventory of fungi and their associated migrating bacteria in forest soils using fungal highway columns. FEMS Microbiol. Ecol. 2017;93 doi: 10.1093/femsec/fiw217. [DOI] [PubMed] [Google Scholar]
- 41.Chomicki G., Werner G.D.A., West S.A., Kiers E.T. Compartmentalization drives the evolution of symbiotic cooperation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190602. doi: 10.1098/rstb.2019.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Limpens E., Geurts R. Plant-driven genome selection of arbuscular mycorrhizal fungi. Mol. Plant Pathol. 2014;15:531–534. doi: 10.1111/mpp.12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lackner G., Moebius N., Partida-Martinez L., Hertweck C. Complete genome sequence of Burkholderia rhizoxinica, an endosymbiont of Rhizopus microsporus. J. Bacteriol. 2011;193:783–784. doi: 10.1128/JB.01318-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Römer P., Hahn S., Jordan T., Strauss T., Bonas U., Lahaye T. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- 45.Paoletti M. Vegetative incompatibility in fungi: from recognition to cell death, whatever does the trick. Fungal Biol. Rev. 2016;30:152–162. doi: 10.1016/j.fbr.2016.08.002. [DOI] [Google Scholar]
- 46.Jacobson D.J., Beurkens K., Klomparens K.L. Microscopic and ultrastructural examination of vegetative Incompatibility in partial diploids heterozygous at het loci in Neurospora crassa. Fungal Genet. Biol. 1998;23:45–56. doi: 10.1006/fgbi.1997.1020. [DOI] [PubMed] [Google Scholar]
- 47.Verdier V., Triplett L.R., Hummel A.W., Corral R., Cernadas R.A., Schmidt C.L., Bogdanove A.J., Leach J.E. Transcription activator-like (TAL) effectors targeting OsSWEET genes enhance virulence on diverse rice (Oryza sativa) varieties when expressed individually in a TAL effector-deficient strain of Xanthomonas oryzae. New Phytol. 2012;196:1197–1207. doi: 10.1111/j.1469-8137.2012.04367.x. [DOI] [PubMed] [Google Scholar]
- 48.Lackner G., Möbius N., Scherlach K., Partida-Martinez L.P., Winkler R., Schmitt I., Hertweck C. Global distribution and evolution of a toxinogenic Burkholderia-Rhizopus symbiosis. Appl. Environ. Microbiol. 2009;75:2982–2986. doi: 10.1128/AEM.01765-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmitt I., Partida-Martinez L.P., Winkler R., Voigt K., Einax E., Dölz F., Telle S., Wöstemeyer J., Hertweck C. Evolution of host resistance in a toxin-producing bacterial-fungal alliance. ISME J. 2008;2:632–641. doi: 10.1038/ismej.2008.19. [DOI] [PubMed] [Google Scholar]
- 50.Pérez-Brocal V., Latorre A., Moya A. In: Between Pathogenicity and Commensalism. Dobrindt U., Hacker J.H., Svanborg C., editors. Springer; 2013. Symbionts and pathogens: what is the difference? pp. 215–243. [DOI] [PubMed] [Google Scholar]
- 51.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scherlach K., Partida-Martinez L.P., Dahse H.M., Hertweck C. Antimitotic rhizoxin derivatives from a cultured bacterial endosymbiont of the rice pathogenic fungus Rhizopus microsporus. J. Am. Chem. Soc. 2006;128:11529–11536. doi: 10.1021/ja062953o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Endosymbiont-free R. microsporus was co-incubated with M. rhizoxinica Δmtal1 in a microfluidic device. Bacterial cells were stained with SYTO9 and then introduced into the microfluidic device via the bacteria inlet 12 h before image acquisition. Reinfection was monitored over time using fluorescence microscopy. The video was recorded at 1 frame per s (fps). Direction of hyphal growth is from left to right. Scale bars: 10 μm
(A and B) (A) Approximate probabilities (p) of Browns-Forsythe test and (B) one-way analysis of variance (ANOVA) for the mycelium area of each type of hyphae following co-cultivation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of individual types of hyphae at specific time points (0, 24, or 48 h post infection [hpi]) across all four M. rhizoxinica strains using the Tukey HSD post hoc test.
(D) Comparison of types of hyphae from individual M. rhizoxinica strains over time (0 vs. 24 hpi and 0 vs. 48 hpi) using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the mycelium area of each type of hyphae following co-cultivation of apo-symbiotic R. microsporus with M. rhizoxinica wild type (M1WT), type 3 secretion system knockout strains (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT) or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of individual types of hyphae at specific time points (0, 24, or 48 h post infection [hpi]) across all four M. rhizoxinica strains using the Tukey HSD post hoc test.
(H) Comparison of types of hyphae from individual M. rhizoxinica strains over time (0 vs. 24 hpi and 0 vs. 48 hpi) using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the bacterial load of Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1) after colonization of apo-symbiotic Rhizopus microsporus. Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) The number of endohyphal bacteria in each M. rhizoxinica strain (indicated by green fluorescence) 24 h post infection (hpi) was compared with the number of endohyphal bacteria 48 h after co-cultivation (24 vs. 48 hpi).
(D) Comparison of endohyphal bacteria at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the bacterial load of M. rhizoxinica wild type (M1WT), T3SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG) following colonization of apo-symbiotic R. microsporus. Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) The number of endohyphal bacteria in each M. rhizoxinica strain (indicated by green fluorescence) 24 h post infection (hpi) was compared with the number of endohyphal bacteria 48 hours after co-cultivation (24 vs. 48 hpi).
(H) Comparison of endohyphal bacteria at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the bacterial load in individual types of hyphae of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of types of hyphae from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(D) Comparison of individual types of hyphae at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the bacterial load in individual types of hyphae of apo-symbiotic R. microsporus co-incubated with M. rhizoxinica wild type (M1WT), T2SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of types of hyphae from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(H) Comparison of individual types of hyphae at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the number of septa formed per mycelium area following co-incubation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of septa from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(D) Comparison of septa at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(E and F) (E) Approximate probabilities (p) of Brown-Forsythe test and (F) one-way analysis of variance (ANOVA) for the number of septa formed per mycelium area following co-incubation of apo-symbiotic R. microsporus with M. rhizoxinica wild type (M1WT), T2SS-deficient M. rhizoxinica (M. rhizoxinica ΔsctC or M. rhizoxinica ΔsctT), or rhizoxin-deficient M. rhizoxinica (M. rhizoxinica ΔrhiG). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(G) Comparison of septa from individual M. rhizoxinica strains over time (24 vs. 48 h post infection [hpi]) using the Tukey HSD post hoc test.
(H) Comparison of septa at specific time points (24 or 48 hpi) across all four M. rhizoxinica strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A and B) (A) Approximate probabilities (p) of Brown-Forsythe test and (B) one-way analysis of variance (ANOVA) for the ratio of live vs. dead endohyphal bacteria following co-incubation of apo-symbiotic Rhizopus microsporus with Mycetohabitans rhizoxinica wild type (M1WT), M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1), M. rhizoxinica Δmtal1 control (M. rhizoxinica Δmtal1 pBBR∅), or complemented M. rhizoxinica Δmtal1 (M. rhizoxinica Δmtal1 pBBR-mtal1). Homogeneous data (non-significant Brown-Forsythe) are shown in black numbers and non-homogeneous data (significant Brown-Forsythe) are highlighted in red numbers.
(C) Comparison of overall ratios of live/dead bacteria inside the fungal hyphae between M1WT, M. rhizoxinica Δmtal1, M. rhizoxinica Δmtal1 pBBR∅, and M. rhizoxinica Δmtal1 pBBR-mtal1. 72 h post infection.
(D) The ratios of live/dead bacteria inside the fungal hyphae was compared between the different types of hyphae of individual strains using the Tukey HSD post hoc test. p values with p < 0.05 were considered statistically significant (highlighted in gray).
(A) Primers used for generation of M. rhizoxinica transcription activator-like effector (MTAL) knockout strain (M. rhizoxinica Δmtal1).
(B) Primers used for generation of genetically complemented M. rhizoxinica Δmtal1 (Δmtal1 pBBR-mtal1) or control M. rhizoxinica Δmtal1 (Δmtal1 pBBR∅).
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. The accession numbers for the datasets are listed in the key resources table.
-
•
Microscopy data reported in this paper will be shared by the lead contact upon request.