Abstract
Species differences in the host factor ANP32A/B result in the restriction of avian influenza virus polymerase (vPol) in mammalian cells. Efficient replication of avian influenza viruses in mammalian cells often requires adaptive mutations, such as PB2-E627K, to enable the virus to use mammalian ANP32A/B. However, the molecular basis for the productive replication of avian influenza viruses without prior adaptation in mammals remains poorly understood. We show that avian influenza virus NS2 protein help to overcome mammalian ANP32A/B-mediated restriction to avian vPol activity by promoting avian vRNP assembly and enhancing mammalian ANP32A/B-vRNP interactions. A conserved SUMO-interacting motif (SIM) in NS2 is required for its avian polymerase-enhancing properties. We also demonstrate that disrupting SIM integrity in NS2 impairs avian influenza virus replication and pathogenicity in mammalian hosts, but not in avian hosts. Our results identify NS2 as a cofactor in the adaptation process of avian influenza virus to mammals.
NS2 acts as a cofactor in avian influenza virus adaptation to mammalian hosts.
INTRODUCTION
Host restriction limits cross-species transmission of avian influenza virus (AIV) from migratory aquatic birds to mammals (1). However, some highly pathogenic AIVs (e.g., H5N1 and H7N9) have overcome this host barrier and are able to infect humans, resulting in a serious disease with a high mortality rate and posing a huge threat to human welfare (2, 3). In addition, it is not uncommon for a human host to be infected with multiple different subtypes of AIVs, e.g., H9N2, H6N1, H7N7, H10N8, and H5N6, concurrently (4–8). The AIV must overcome multiple host range barriers to effectively infect and spread among mammals. A major species barrier is the restriction of avian virus polymerase (vPol) in mammalian cells (9), which is the driving force for the emergence of adaptive mutations (10). Two most well-known mammalian adaptation mutations, E627K and D701N in PB2, increase the replication, pathogenicity, and transmittance of AIVs in mammals (11–16). Recent studies have identified that species-specific differences in proteins of the acidic nuclear phosphoprotein 32 (ANP32) family determine the restriction of avian vPol in mammalian cells (17).
The host factor ANP32A/B regulates the synthesis of vRNA from the complementary RNA (cRNA) of AIV (18). Compared with human ANP32A/B (huANP32A/B), chicken ANP32A (chANP32A) has a unique insertion of 33 amino acids that enables it to efficiently support the vPol activity of both avian- and human-adapted influenza viruses. However, huANP32A/B only supports the vPol activity of human-adapted influenza virus and cannot support that of the AIV due to the absence of this insertion (17). In addition, further studies have revealed that the avian hosts carry up to three chANP32A isoforms. These three isoforms differ only in the composition of the 33 amino acid inserts but support the activity of avian-signature vPol to differing extents (19, 20). A hydrophobic SUMO-interacting motif (SIM)–like sequence in the 33–amino acid insertion of chANP32A is required to selectively support avian vPol activity (21), suggesting that SUMO-dependent function is important for the function of avian vPol. In addition, results from our laboratory and others have demonstrated that huANP32A and huANP32B equally support the polymerase activity of human-adapted IAV and that loss of huANP32A and huANP32B simultaneously results in failure of IAV replication (22, 23). The chicken ANP32B (chANP32B) has natural mutations at positions 129 and 130, and it has lost its ability to support the activity of influenza A vPol (22, 24). Swine ANP32A (swANP32A) is able to support avian vPol activity to a greater extent than other mammalian ANP32A/B, as a result of a recently found unique 106V/156S signature, which explains why pigs can act as intermediate hosts for the cross-species transmission of AIV (25, 26). In addition, human ANP32 proteins, including ANP32A, ANP32B, and ANP32E, have recently been identified as cofactors of influenza B vPol (27).
Mammalian ANP32A/B proteins, with the exception of swANP32A, provide poor support for avian vPol activity, so to achieve high levels of replication in mammalian cells, AIVs require mutations to adapt to mammalian ANP32A/B (17, 22, 25, 26). However, some human isolates of AIVs H9N2, H5N1, and H7N9 were able to establish productive infections in humans without the emergence of previously identified adaptive mutations (28–30). Avian isolates of multiple subtypes of AIV are even able to establish productive infections in mammalian hosts and are even pathogenic for mice without prior adaptation (10, 31–33). These studies suggest that the polymerases of certain subtypes of AIVs can use mammalian ANP32A/B to some extent for productive replication in the process of cross-species transmission into mammals. This contradicts the conclusions drawn from cell-based vPol reconstitution assays that mammalian ANP32A/B poorly support avian vPol activity (17, 22, 25, 26). It has been demonstrated that during transcription and replication, the regulatory events that occur in the IAV replication process are quite different from those that occur in cell-based vPol reconstitution assays (34). With the exceptions of the trimeric polymerase complex and nucleoprotein (NP), the contributions of other IAV proteins produced during viral replication to avian vPol activity are largely unknown.
In this study, we show that the IAV NS2 promotes avian-signature vPol activity supported by mammalian ANP32A/B, but not chANP32A. NS2 has a relatively limited effect on the activity of mammalian-signature vPol supported by both mammalian ANP32A/B and chANP32A. NS2 proteins derived from IAVs isolated from a range of hosts are conserved in this avian-signature polymerase-enhancing function. In addition, we demonstrate that NS2 exerts this function by facilitating the assembly of avian viral ribonucleoprotein (vRNP) and enhancing the interaction of mammalian ANP32A/B with avian vRNP in mammalian cells. NS2 harbors a highly conserved SIM, which is required for its avian-signature polymerase-enhancing properties. Disruption of the integrity of this SIM in NS2 has deleterious effects on the replication and pathogenicity of AIVs in mammalian hosts, but not in avian hosts. Together, our data suggest a model in which the adaptation of avian-signature vPol to mammalian ANP32A/B relies on the help of the SIM supplied by IAV-encoded NS2 protein, providing previously unknown insights into the adaptation of AIV to mammals.
RESULTS
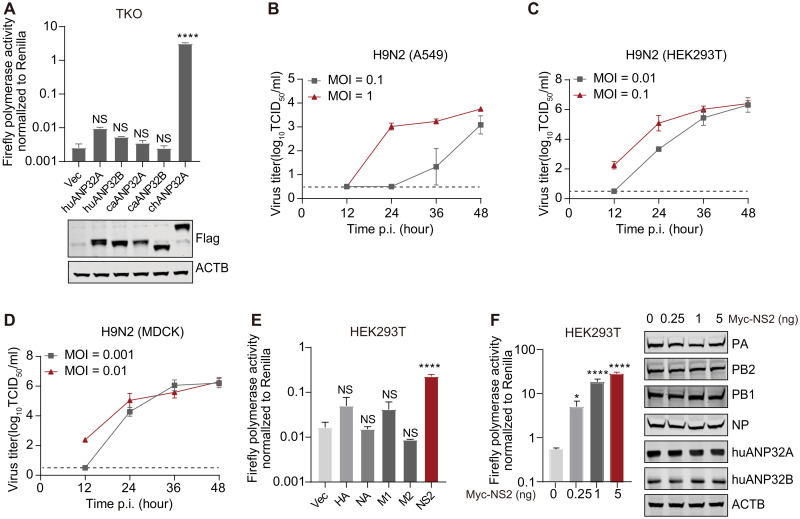
NS2 regulates the activity of the avian H9N2 virus vPol
Host factor ANP32 proteins determine the function of IAVs polymerase, and mammalian cells are nonpermissive for AIVs’ vPol because of the restriction from their ANP32A/B (17, 19–26, 35). However, many H9N2, H5N1, or H7N9 AIVs can establish productive infection in humans without prior adaptation (28–30, 33). To investigate the mechanism by which the avian vPol can function efficiently in mammal cells, we used huANP32A/B/E triple knockout (TKO) human embryonic kidney (HEK) 293T cells (27) to establish an ANP32-dependent mini replicon vPol reporter system. We found that huANP32A/B and canine ANP32A/B (caANP32A/B) poorly supported the vPol activity of the AIVs A/chicken/Zhejiang/B2013/2012(H9N2) (Fig. 1A), which was consistent with previous results (17, 22, 25, 26). However, when the replication ability of H9N2 in mammalian cells was evaluated, the results suggested that the H9N2 virus could establish productive replication in mammalian cell lines, including A549 cells, HEK293T cells, and Madin-Darby canine kidney (MDCK) cells (Fig. 1, B to D). These data indicate that huANP32A/B or caANP32A/B support the vPol activity of the avian H9N2 virus to a degree sufficient to support productive replication under infection conditions, which is inconsistent with the results of vPol reconstitution assays in TKO cells (Fig. 1A).
Fig. 1. NS2 enhances H9N2 vPol activity in human cells.
(A) vPol reconstitution assay in TKO cells to compare the effect of each ANP32-Flag construct on avian H9N2 vPol activity. The accompanying WBs show expression of Flag-tagged ANP32 constructs. Vec, empty vector control; hu, human; ca, canine; ch, chicken. (B) A549 cells, (C) HEK293T cells, or (D) MDCK cells were infected with H9N2 virus, and the supernatant was collected to detect the H9N2 growth kinetics using TCID50 assays. Data are shown as means ± SD (n = 3) and are representative of three independent experiments. MOI, multiplicity of infection. (E) vPol reconstitution assay to compare the effect of different influenza viral proteins on H9N2 vPol activity in HEK293T cells. (F) vPol reconstitution assay in HEK293T cells to compare the effect of increasing doses of H9N2-NS2 on H9N2 vPol activity. WB analysis shown right (RNP proteins of H9N2 and endogenous huANP32A/B). ACTB indicates β-Actin. In (A), (E), and (F), data shown are firefly activity normalized to Renilla, plotted as means ± SD (n = 3), and are one representative of three independent experiments. P values were determined using one-way analysis of variance (ANOVA) followed by a Dunnett’s multiple comparisons test. NS, not significant; *P < 0.05 and ****P < 0.0001.
As the vPol reconstitution assay involves only the RNP(s) required for viral transcription and replication, we hypothesized that besides these RNP(s), other viral proteins produced under infection conditions could regulate the supporting function of mammalian ANP32A/B toward avian vPol activity and could account for the efficient replication of AIV in mammalian cells. To investigate this hypothesis, a vPol reconstitution assay was performed in HEK293T cells to assess whether other viral proteins produced during infection process, including HA, NA, M1, M2, and NS2, have an impact on the vPol activity of the H9N2 virus. As shown in Fig. 1E, H9N2-NS2 significantly enhanced the vPol activity of avian H9N2 virus in HEK293T cells. In addition, we found that this enhancing effect of NS2 toward the vPol activity of the avian H9N2 virus was dose dependent (Fig. 1F). Together, these data identified NS2 as a positive regulator of the vPol activity of the avian H9N2 virus in human cells.
IAV NS2 selectively promotes avian-signature vPol activity when supported by huANP32A/B, but not by chANP32A
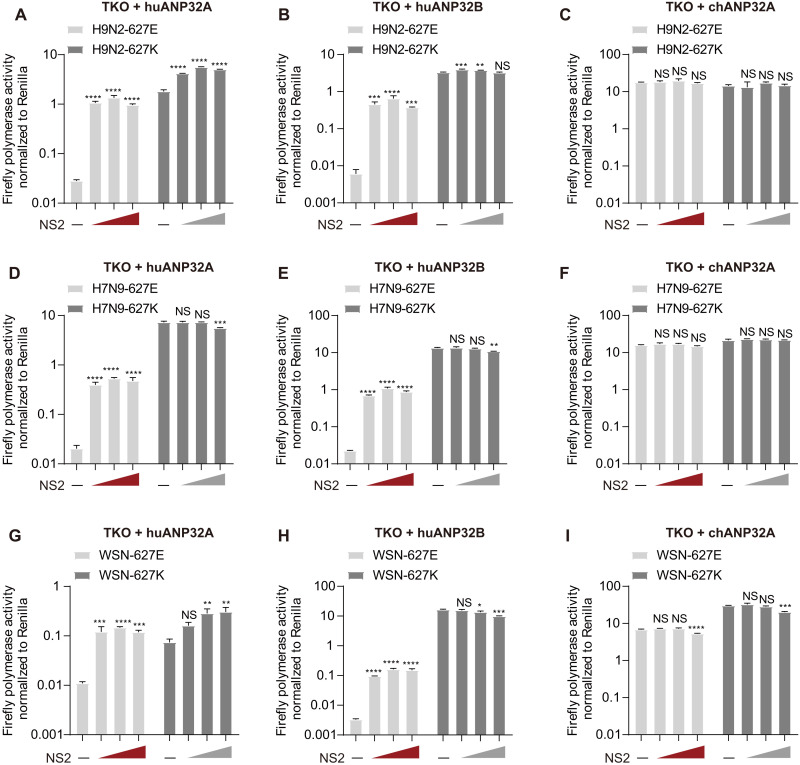
We and other laboratories independently found that both huAP32A and huANP32B play important roles in supporting the vPol activity and both exhibited a limited supporting function toward avian vPol activity (22, 23, 25, 26). We next evaluated the effect of H9N2-NS2 on the avian and mammalian-signature (PB2-627K) vPol activity of the H9N2 virus supported by ANP32 proteins. To do this, we established an ANP32-dependent vPol reconstitution assay in TKO cells. We found that NS2 markedly enhanced the avian H9N2 vPol activity when supported by huANP32A (~47.01-fold increase) or huANP32B (~105.99-fold increase), but not by chANP32A. However, NS2 had limited effects on the activity of the mammalian-signature H9N2 (PB2-627K) vPol when supported by huANP32A, huANP32B, or chANP32A (Fig. 2, A to C). We obtained consistent results using the NS2 and vPol from the genetic background of the avian influenza H7N9AH13 virus (Fig. 2, D to F).
Fig. 2. NS2 selectively enhances avian-signature vPol activity when supported by huANP32/B, but not by chANP32A.
(A to C) vPol reconstitution assay to compare the effect of increasing doses of H9N2-NS2 (0 to 5 ng) on the activities of H9N2 and H9N2 (PB2-627K) vPols in TKO cells reconstituted with huANP32A (A), huANP32B (B), or chANP32A (C). TKO cells were transfected with H9N2-PB2, H9N2-PB1, H9N2-PA, H9N2-NP, different ANP32s as indicated, a mini-genome reporter, and a Renilla expression control together with or without NS2. Luciferase activity was measured 24 hours later. (D to F) Similar to (A) to (C) but using NS2 and RNP proteins from avian influenza H7N9AH13. (G to I) Similar to (A) to (C) but using NS2 and RNP proteins from the A/WSN/33 virus. In (A) to (I), data shown are firefly activity normalized to Renilla plotted as means ± SD (n = 3) and are one representative of three independent experiments. P values were determined using one-way ANOVA followed by a Dunnett’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To further investigate the general role of NS2 in enhancing huANP32A/B-mediated avian vPol activity, we used a human influenza H1N1WSN polymerase to investigate the effect of H1N1WSN-NS2 on vPol activity of avian signature (PB2-627E and WSN-627E) when supported by different ANP32 proteins. We found that H1N1WSN-NS2 also selectively enhanced the WSN-627E vPol activity when supported by huANP32A (~12.43-fold increase) or huANP32B (~48.10-fold increase), but not by chANP32A. H1N1WSN-NS2 also had a limited effect on the mammalian-signature (PB2-627K and WSN-627 K) vPol activity when supported by huANP32A, huANP32B, or chANP32A (Fig. 2, G to I). These data suggest that the role of NS2 in selectively promoting the avian-signature vPol activity when supported by huANP32A/B may not be virus strain specific. To obtain additional evidence for this, we performed vPol reconstitution assays in TKO cells to test whether the NS2 proteins from IAVs isolated from different host species have a similar role in promoting avian- signature vPol activity when supported by huANP32A/B. As shown in fig. S1 (A to E), all NS2s of IAVs isolated from different taxa (including human, bird, pig, dog, and equine) have the ability to promote avian H9N2 and H7N9 (PB2-627E) vPol activity when supported by huANP32A/B. However, the NS2 of influenza B virus (IBV) did not have this function.
The main known function of NS2 is to mediate the nuclear export of vRNP, which requires the nuclear export signal (NES) of NS2 and its binding to M1 (36–38). To test whether the avian-signature polymerase-enhancing properties of NS2 depend on its role in vRNP nuclear export, we generated a panel of NS2 mutants (fig. S2A), including those lacking NES or harboring a W78S point mutation that impairs the NS2-M1 interaction. The performance of these NS2 mutants was then evaluated in a vPol reconstitution assay in TKO cells. These NS2 mutants had the same positive effect on avian H9N2 vPol activity when supported by huANP32A/B as did wild-type (WT) NS2 (fig. S2B), suggesting that the avian-signature polymerase-enhancing properties of NS2 is independent of its role in the mediation of vRNP nuclear export. Together, these results suggest that IAV NS2 can overcome the limitations of avian-signature vPol in human cells due to species-specific differences in the ANP32 protein.
NS2 promotes avian-signature vPol activity when supported by ANP32A/B from different mammalian species
Previous studies have confirmed that ANP32A/B is a critical factor for the function of influenza A vPols from different species (17, 22–24). Mammalian ANP32A/B proteins, including caANP32A/B, equine ANP32A/B (eqANP32A/B), mouse ANP32B (muANP32B), and swANP32B, lack a 33-residue insertion present in chANP32A, meaning that avian-signature vPol activity is restricted in most mammalian cells (17, 22, 25, 26). However, swANP32A contains a unique sequence feature that enables it to support avian vPol activity to a higher degree than other mammalian ANP32A/B (25, 26). In addition, muANP32A is inactive in supporting IAV polymerase activity, and only muANP32B supports IAV replication in mice (22, 23).
To examine the effect of NS2 on the activity of avian vPol when supported by caANP32A/B, eqANP32A/B, muANP32B, or swANP32A/B, we performed a vPol reconstitution assay in TKO cells reconstituted with those ANP32A and ANP32B proteins. As shown in Fig. 3 (A to D), H9N2-NS2 markedly enhanced the activity of the H9N2 virus vPol when supported by the above-mentioned mammalian ANP32A/B but had a relatively limited effect on the activity of the mammalian-signature (PB2-627K) vPol. We obtained consistent results using NS2 and vPol from the avian H7N9AH13 virus (Fig. 3, E to H). These data suggest that NS2 can compensate for the defects of avian-signature vPol in mammalian cells, making it an indispensable cofactor in the adaptation of avian-signature polymerase to mammalian hosts.
Fig. 3. NS2 enhances avian-signature vPol activity when supported by ANP32A/B from different mammalian species.
(A to D) vPol reconstitution assay comparing the effect of H9N2-NS2 on H9N2 and H9N2 (PB2-627K) vPols activities in TKO cells reconstituted with ANP32A/B from dog (A), equine (B), pig (C), and mouse (D). TKO cells were transfected with H9N2-PB2, H9N2-PB1, H9N2-PA, H9N2-NP, different ANP32s as indicated, a mini-genome reporter, and a Renilla expression control together with or without NS2. Luciferase activity was measured 24 hours later. (E to H) Similar to (A) to (D) but using NS2 and RNP proteins from avian influenza H7N9AH13. In (A) to (H), data shown are firefly activity normalized to Renilla plotted as means ± SD (n = 3) and are one representative of three independent experiments. P values were determined using one-way ANOVA followed by an unpaired Student’s t test. ***P < 0.001 and ****P < 0.0001.
NS2 enhances avian vRNP assembly and the interaction between avian vRNP and huANP32A/B to selectively promote avian vPol activity in human cells
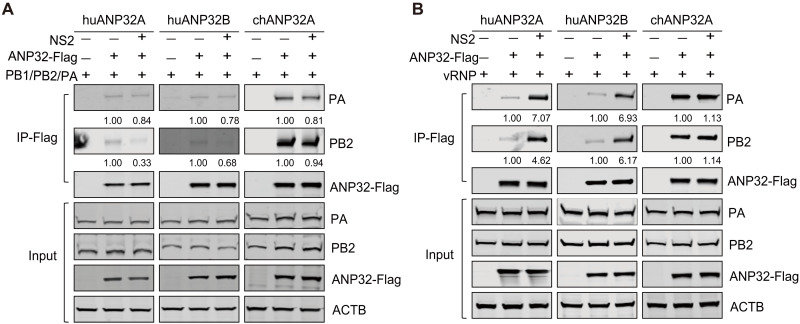
Upon nuclear import, viral polymerase subunits (PA/PB1/PB2) assemble into the trimeric polymerase complex, which, together with genomic RNA and NP, further assemble into vRNPs. Previous reports have shown that assembly of the trimeric viral polymerase is unaffected by the identity of amino acid 627 in the PB2 [K627 versus E627 (9)]. In contrast, vRNP assembly defects are associated with the restriction of avian-signature vPol activity in mammalian cells (9). To gain insight into the mechanism by which NS2 enhances avian-signature vPol activity in mammalian cells, we first confirmed these observations by monitoring vRNP assembly in TKO cells reconstituted with huANP32A/B or chANP32A. In the presence of huANP32A/B, the amount of H9N2 polymerase (PB2-627E and PA) that coprecipitated with NP was less than that of mammalian-signature polymerase (PB2-627K and PA). However, in the presence of chANP32A, the coprecipitation of H9N2 (PB2-627E and PA) was increased (Fig. 4A). This species-specific effect of ANP32 protein on avian-signature vRNP assembly may determine the restriction of avian-signature vPol activity in mammalian cells.
Fig. 4. NS2 promotes the avian vRNP assembly in TKO cells reconstituted with huANP32A/B, but not with chANP32A.
(A) Comparison of avian (H9N2-PB2-627E) and mammalian-signature (H9N2-PB2-627K) vRNP assembly in TKO cells reconstituted with chANP32A or huANP32A/B. TKO cells were transfected with the indicated plasmids for 24 hours; cell lysates were immunoprecipitated with anti-Flag M2 Magnetic Beads and then analyzed by Western blotting. (B) Comparison of the effect of H9N2-NS2 on the H9N2 vRNP assembly in TKO cells reconstituted with either huANP32A/B or chANP32A. TKO cells were transfected with the indicated plasmids. After anti-Flag precipitation at 24 hours after transfection, the indicated proteins were analyzed by Western blotting. (C) Comparison of the effect of H9N2 NS2 on the H9N2 vRNP assembly in TKO cells without reconstitution of ANP32 proteins. Experiments were performed as in (B), except that the ANP32 constructs were not transfected. Following anti-Flag precipitation at 24 hours after transfection, the indicated proteins were analyzed by Western blotting. (D to G) GST affinity isolation analysis of the complex formation of NS2 with PA (D), PB1 (E), PB2 (F), or NP (G). In (A) to (G), three independent experiments were performed, with consistent results in each experiment.
Next, to confirm whether the enhanced avian-signature vPol activity conferred by NS2 results from enhanced binding of the viral polymerase to the NP in mammalian cells, we assessed the effect of NS2 on the H9N2 vRNP assembly in TKO cells reconstituted with either huANP32A/B or chANP32A. Immunoprecipitation assays showed that when TKO cells were reconstituted with huANP32A/B, the amounts of the H9N2 polymerase subunits polymerase acidic protein (PA) and PB2 coprecipitated by NP increased in the presence of NS2, whereas when chANP2A was present, coprecipitation of PB2 and PA by NP was not affected by NS2 (Fig. 4B). These results confirm that the increased avian-signature vPol activity conferred by NS2 correlates with increased vRNP assembly in mammalian cells.
To further investigate the role of NS2 in facilitating vRNA assembly, the assembly process of H9N2 vRNP was monitored in TKO cells without reconstitution of any ANP32 proteins. In this case, vPol functions poorly during replication and transcription, resulting in a stable level of vRNA. However, to our surprise, NS2 was still able to promote the assembly of avian-signature vRNP (Fig. 4C). In addition, we used glutathione S-transferase (GST) pull-down analysis to demonstrate that NS2 associates with H9N2 RNP(s), including PB1, PB2, and NP, but not PA (Fig. 4, D to G). These data strongly suggest that, such as chANP32A, NS2 can compensate for the defective assembly of avian vRNP in mammalian cells and rescue avian-signature vPol activity in these cells through its association with RNP(s).
Although there are many unknowns regarding the specific molecular mechanism by which the ANP32 protein supports the vPol activity of the IAV, current evidence suggests that the interaction of ANP32 with the trimeric polymerase complex is critical for its function in supporting polymerase activity (18, 19, 21–24, 39, 40). In addition, recent studies have shown that a unique feature (106V/156S) of swANP32A allows the protein to bind avian-signature trimeric polymerase complexes more strongly than other mammalian ANP32 proteins and enabling it to support avian-signature vPol activity to a greater extent (25, 26). Therefore, we propose that NS2 promotes the avian-signature vPol activity in mammalian cells by enhancing the interaction between mammalian ANP32 proteins and the trimeric polymerase complex. To our surprise, H9N2-NS2 did not affect the interaction between huANP32A/B and the trimeric H9N2 vPol complex, and neither was the interaction between chANP32A and the trimeric polymerase complex affected by NS2 (Fig. 5A). However, when we explored whether NS2 affected the interaction between huANP32A/B and the H9N2 vRNP, we found that the amount of PA and PB2 coprecipitated by huANP32A/B was increased in the presence of NS2. NS2 did not affect the interaction between chANP32A and the vRNP (Fig. 5B).
Fig. 5. NS2 promotes the interaction of H9N2 vRNP with huANP32A/B, but not with chANP32A.
(A) Effect of H9N2-NS2 on the binding of ANP32 proteins to H9N2 polymerase trimeric complexes. TKO cells were transfected with the indicated plasmids for 24 hours; cell lysates were immunoprecipitated with anti-Flag M2 Magnetic Beads and then analyzed by Western blotting. (B) Effect of H9N2-NS2 on the interaction between ANP32 proteins and H9N2 vRNP. Experiments were performed as in (A), except that the NP and vRNA luciferase reporter were included in the transfection. In (A) and (B), three independent experiments were performed, with consistent results in each experiment.
To further confirm that the enhanced avian-signature vPol activity conferred by NS2 results from increased vRNP formation and vRNP-ANP32 interactions, we next performed similar experiments using muANP32B. As shown in fig. S3A, H9N2-NS2 enhanced the assembly of H9N2 vRNP in TKO cells reconstituted with muANP32B. In addition, H9N2-NS2 also enhanced the interaction between muANP32B and the H9N2 vRNP (fig. S3B). Together, these data suggest that the rescue of avian-signature vPol activity by NS2 in mammalian cells correlates with its ability to promote the assembly process of avian vRNP and mammalian ANP32A/B-vRNP interactions.
The SIM of NS2 determines its avian-signature polymerase-enhancing properties
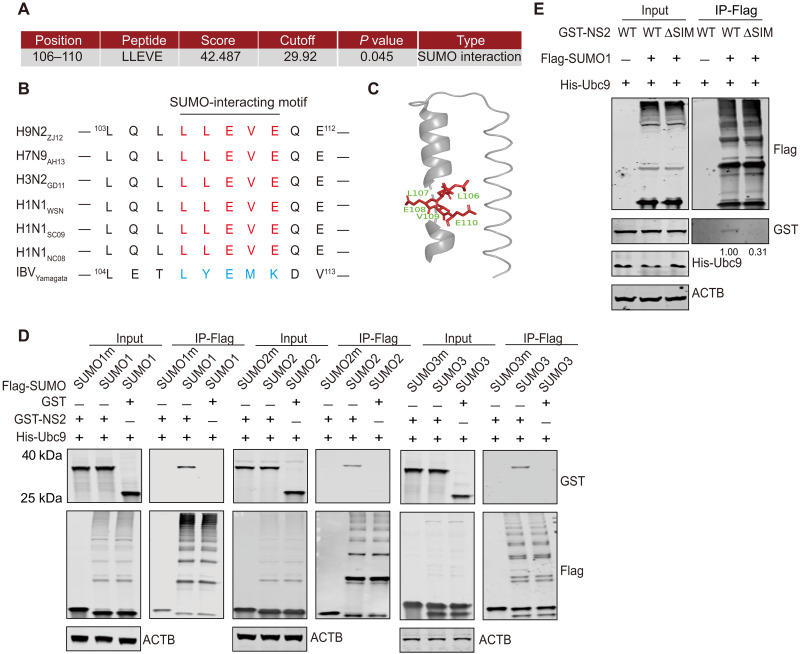
Unlike mammalian ANP32A/B, chANP32A contains a 33-residue insertion, which allows it to effectively support the avian vPol activity (17). Further evidence suggests that the presence of a SIM-like sequence in this 33-residue insertion is critical for this supporting function (21), suggesting that a SUMO-dependent function plays an important role in chANP32A-mediated avian vPol function. Therefore, we hypothesized that the SUMO-dependent function of NS2 might be related to its ability to promote avian-signature vPol activity in mammalian cells. To our surprise, when we used the in silico prediction program GPS(Group-based Prediction System)-SUMO (41), we found that H9N2-NS2 contains an SIM (106LLEVE110) (Fig. 6A). A comparison of the amino acid sequence of this region (amino acids 106 to 110) with those from NS2 proteins from other viruses revealed that this SIM was highly conserved among IAVs isolated from different host species, but not IBV (Fig. 6B). In addition, the crystal structure of the NS2 C-terminal domain (37) showed that the SIM was located on the C2 helix (Fig. 6C). The presence of a SIM in NS2 suggested that this protein can interact noncovalently with SUMO1/2/3 or SUMO1/2/3-conjugated proteins. To gain more evidence for this, we performed SUMO binding assays in HEK293T cells by cotransfecting GST-tagged NS2 with His-Ubc9 and Flag-SUMO1/2/3. As shown in Fig. 6D, GST-NS2, but not GST, could be coprecipitated by Flag-tagged SUMO1/2/3. However, the mutants defective in SUMO conjugation (SUMOm) failed to precipitate NS2, suggesting that NS2 specifically interacts noncovalently with SUMO1/2/3-conjugated proteins. Furthermore, the deletion of this SIM resulted in a failure of NS2 binding to Flag-SUMO1–conjugated proteins, further confirming that LLEVE is a bona fide SIM (Fig. 6E).
Fig. 6. Identification of a conserved SIM in NS2.
(A) Candidate SIM in H9N2-NS2 was predicted using the GPS-SUMO software. (B) Sequence alignment of NS2-SIMs of IAVs isolated from different host species and IBV. (C) Position of SIM in the structure of the NS2 C-terminal domain. (D) In vivo SUMO binding assays. HEK293T cells were transfected with plasmids expressing GST-tagged NS2 from H9N2 (1 μg) together with 1 μg of Flag-tagged SUMO1/SUMO1m (left), SUMO2/SUMO2m (middle), and SUMO3/SUMO3m (right) and His-tagged Ubc9 (1 μg). At 24 hours after transfection, cell lysates were prepared for anti-Flag precipitation and the indicated proteins were analyzed by Western blotting. (E) In vivo SUMO binding assays. HEK293T cells were transfected with plasmids expressing GST-tagged NS2 or GST-tagged NS2 lacking SIM (ΔSIM) from H9N2 (1 μg) together with 1 μg of Flag-tagged SUMO1 and His-tagged Ubc9 (1 μg). At 24 hours after transfection, cell lysates were prepared for anti-Flag precipitation and the indicated proteins were analyzed by WB. In (D) and (E), three independent experiments were performed, with comparable results in each experiment.
Functional SIMs are characterized by a hydrophobic core preceded or followed by negatively charged residues, and replacement of these hydrophobic residues with glycine impairs SIM-mediated function, as evidenced in previous reports (21). The same approach was used here to generate a set of NS2 mutants to explore the contribution of the SIM in NS2 toward its avian polymerase-enhancing function. The NS2 protein expression level was not markedly affected by these amino acid changes in NS2-SIM (Fig. 7A). As expected, substitution of these hydrophobic residues in this motif for glycines (SIMLLGG, SIMVEGG, and SIMGGGG) severely impairs the interaction between Flag-SUMO1–conjugated proteins and NS2, while substitution at the nonhydrophobic position E108 (SIMEA) in this SIM did not (Fig. 7B). Next, the effects of these NS2 mutants on avian vRNP assembly and avian vRNP-ANP32A interactions were assessed in TKO cells reconstituted with either huANP32A or chANP2A. Unexpectedly, in TKO cells reconstituted with huANP32A, NS2 mutants in which hydrophobic residues in the SIM were replaced with glycines lost the ability to promote avian vRNP assembly, while SIMEA retained this ability (fig. S4A). In contrast, WT and mutant NS2 did not affect avian vRNP assembly in the presence of chANP32A (fig. S4A). Avian vRNP-huANP32A interactions were enhanced in the presence of WT NS2 and SIMEA, but not in NS2 mutants with glycine substitutions in the SIM (fig. S4B). None of the NS2 constructs affected the avian vRNP-chANP32A interaction (fig. S4B).
Fig. 7. The SIM in NS2 determines its avian-signature polymerase-enhancing function.
(A) Panel of generated NS2 mutants. The accompanying WBs show the effect of NS2 SIM changes on protein expression levels in these mutants. HEK293T cells were transfected with equal amount of indicted GST-tagged H9N2-NS2 constructs. At 24 hours after transfection, cell lysates were prepared with radioimmunoprecipitation assay lysis buffer and subjected to SDS–polyacrylamide gel electrophoresis, followed by analysis with Western blotting. (B) Effects of H9N2-NS2 SIM mutations on H9N2-NS2 binding to Flag-tagged SUMO1. Experiments were performed as in Fig. 6E. Three independent experiments were performed, with comparable results in each experiment. (C to E) vPol reconstitution assay to compare the effect of H9N2-NS2 SIM mutations on H9N2 vPol activity in TKO cells reconstituted with huANP32A (C), huANP32B (D), and chANP32A (E). TKO cells were transfected with H9N2-PB2, H9N2-PB1, H9N2-PA, H9N2-NP, different ANP32s as indicated, a mini-genome reporter, and a Renilla expression control together with increasing dose of different H9N2-NS2. Luciferase activity was measured 24 hours later. For all assays, data were firefly activity–normalized to Renilla and plotted as fold change to empty vector (0 ng of NS2 constructs). Data are presented as the means ± SD (n = 3) and are one representative of three independent experiments. Significance was determined by two-way ANOVA. ****P < 0.0001.
Subsequent vPol reconstitution assays were performed in TKO cells to assess the effect of NS2 mutants on avian-signature vPol activity in TKO cells reconstituted with huANP32A, huANP32B or chANP32A. As shown in Fig. 7 (C and D), the NS2-SIMEA mutant was even more potent than WT-NS2 in promoting avian H9N2 vPol activity supported by huANP32A/B, whereas the NS2-SIMLLGG, NS2-SIMVEGG, and NS2-SIMGGGG mutants almost completely lost this ability. Notably, both WT NS2 and these mutants had only a limited effect on the avian H9N2 vPol activity supported by chANP32A (Fig. 7E). We confirmed these observations using NS2 and RNP proteins from an H7N9 AIV genetic background. Similarly, H7N9-NS2 mutants in which the hydrophobic residues in the SIM were substituted with glycines lost the avian-signature polymerase-enhancing properties, while NS2-SIMEA did not (fig. S5, A to C). None of the H7N9-NS2 mutants had more than a limited effect on H7N9 (PB2-627E) vPol activity when supported by chANP32A (fig. S5D).
The above data indicate that the SIM in NS2 is required for its avian-signature polymerase-enhancing function. Therefore, we next assessed the prevalence of this motif among IAVs. The SIM (106LLEVE110) was highly conserved in avian isolates of H1-H16 influenza viruses (table S1). In addition, we focused on several subtypes of AIVs capable of cross-species transmission, including H5N1, H7N9, H5N6, H5N8, and H9N2. The analysis showed that their NS2 SIMs were similarly well conserved (table S1).
Previous studies have shown that NS2 can be SUMOylated (42). To assess the contribution of the SUMOylation of NS2 to its avian-signature polymerase-enhancing properties, we generated an NS2 mutant with lysine-to-arginine loss-of-function substitutions at all six lysine residues of the protein (termed NS2-K0) (fig. S6A). Unexpectedly, NS2-K0 still retained the ability to promote the avian-signature vPol activity when supported by huANP32A/B (fig. S6, B and C). However, similar to WT NS2, NS2-K0 had only a very limited effect on chANP32A-mediated avian-signature vPol function (fig. S6D). Meanwhile, the NS2 mutant lacking SIM (NS2-ΔSIM) lost its avian-signature polymerase-enhancing properties (fig. S6, B to D). These data suggest that NS2 selectively relies on its SIM to exert its avian-signature polymerase-enhancing function.
SIM of NS2 is crucial for efficient replication of AIV in mammalian cells
Our vPol reconstitution assays identified NS2 as a cofactor necessary for the adaptation of avian-signature polymerase to mammalian hosts. To determine whether the species-specific effect on the avian-signature vPol activity conferred by NS2 manifested as species-specific changes in the replication of AIV in mammalian cells, we first intended to rescue H9N2 AIVs carrying different mutant SIMs on NS2, which cause defects in promoting avian-signature polymerase activity in mammalian cells. Unfortunately, we were unable to rescue the mutant avian influenza H9N2 viruses bearing NS2 SIMLLGG, SIMVEGG, or SIMGGGG mutations, which have almost completely lost their avian-signature polymerase-enhancing properties. However, the mutant avian influenza H9N2 virus with NS2 bearing the SIME110G mutation was successfully rescued. We first demonstrated that the E110G mutation in NS2 impaired its ability to bind SUMO1-conjugated proteins (fig. S7A). In line with this, the vPol reconstitution assays in TKO cells further demonstrated that the ability of NS2-E110G to promote H9N2 vPol activity supported by huANP32A/B was impaired (fig. S7, B and C). Mechanistically, we found that E110G mutation in NS2 impaired its ability to promote avian vRNP assembly and avian vRNP/huANP32A interaction in huANP32A reconstituted TKO cells (fig. S7, D and E). Furthermore, E110G mutations did not affect avian vRNP assembly and avian vRNP-chANP32A interaction in chANP32A reconstituted TKO cells (fig. S7, D and E). Together, these experiments demonstrated that the E110G mutation, similar to the NS2 SIMLLGG, SIMVEGG or SIMGGGGG mutations, affects this newly identified function of NS2 in a species-specific manner.
We next performed multicycle replication assays to compare the replication ability of WT (SIMWT) and mutant (SIME110G) AIV H9N2 in different cell lines (Fig. 8, A to D). The replication kinetics and titer of the H9N2-SIME110G virus in primary chicken embryo fibroblasts (CEFs) were comparable to those of the H9N2-SIMWT virus (Fig. 8A), which is consistent with the fact that chANP32A-mediated avian-signature vPol function was not greatly affected by the NS2. However, replication of the H9N2-SIME110G virus was significantly impaired in A549 and MDCK cells (Fig. 8, B and C). Notably, the replication defect of H9N2-SIME110G virus in MDCK cells was completely rescued upon overexpression of chANP32A in these cells (Fig. 8D). These data suggest that the species-specific effect on the avian-signature vPol activity conferred by NS2 also affects the replication of the AIV in a species-specific fashion. Consistent results were obtained when multicycle replication assays were performed using the two highly pathogenic AIVs H7N9HN17 (Fig. 8, E to H) and H5N6GD16 (Fig. 8, I to L). In addition, to obtain more evidence to support the general role of NS2 in promoting the function of AIV polymerase in mammalian cells, we used a recombinant A/WSN/33 virus encoding PB2-627E (WSN-627E), which shows substantial replication defects in mammalian cells due to host restrictions as previously described (9), as an avian-like virus in the following experiment. As we expected, the WSN-627E-SIME110G virus showed replication defects in A549 and MDCK cells (Fig. 8, N and O), but no change in viral titers was detected from primary CEFs (Fig. 8M) and MDCK-chANP32A cells (Fig. 8P).
Fig. 8. SIM of NS2 promotes AIV replication in mammalian cells but not in avian cells.
(A to D) Comparison of the replication abilities of H9N2-SIMWT and H9N2-SIME110G virus in indicated cell lines. Primary CEFs (A), A549 cells (B), MDCK cells (C), or MDCK-chANP32A cells (MDCK cells stably expressing chANP32A) (D) were infected with the H9N2-SIMWT or the H9N2-SIME110G virus. Cell supernatants were collected at the described time points post-infection (p.i.), and virus titers were determined using 50% tissue culture infective dose (TCID50) assays. (E to H) Comparison of the replication abilities of H7N9-SIMWT and H7N9-SIME110G virus in indicated cell lines. Experiments were performed as in (A) to (D). (I to L) Comparison of the replication abilities of H5N6-SIMWT and H5N6-SIME110G virus in the indicated cell lines. Experiments were performed as in (A) to (D). (M to R) Comparison of the replication abilities of the avian-signature WSN-PB2-627E virus (WSN-627E, SIMWT) and the avian-signature WSN-PB2-627E virus with NS2 SIM harboring an E110G mutation (WSN-627E, SIME110G) in the indicated cell lines. Experiments were performed as in (A) to (D). In (A) to (R), dotted line denotes the limit of detection based on the dilution factor in our assays. Graphs shown are of one representative (means ± SD, n = 3). Asterisks indicate significant differences between SIMWT and SIME110G, and statistical significance was calculated per time point by unpaired Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
To further support that the AIV polymerase is dependent on the NS2-SIM for effective function in different mammalian cells, we performed viral replication experiments with an avian virus (WSN-627E) in two different mouse cell lines, MLE-12 (mouse lung epithelium-12) and NIH3T3 (mouse embryonic fibroblasts), and the results showed that the WSN-627E-SIME110G virus had a replication defect in both MLE-12 and NIH3T3 cells (Fig. 8, Q and R). Together, these results confirm the essential role of the SIM of NS2 in promoting the adaptation of AIV to mammalian cells.
The replication and virulence of the AIVs H5N6 with NS2-SIME110G mutations are attenuated in mice, but not in chickens
To determine the importance of avian-signature polymerase-enhancing properties of NS2 on the replication capacity and virulence of AIV in mice, the H5N6GD16 AIV was selected for the following in vivo experiments. H5N6GD16 was selected because it has been widely detected in wild birds and poultry, and because, in recent years, it has also been reported to infect humans, posing a huge threat and causing high mortality in human populations (43). To compare the replication ability of the WT H5N6GD16 (H5N6-SIMWT) virus and the mutant H5N6GD16 (H5N6-SIME110G) virus in mice, two groups of five 6-week-old female BALB/c mice were inoculated with these viruses at an infectious dose of either 102 50% egg infectious dose (EID50) or 104 EID50, respectively. The nasal turbinates and lungs of the mice were collected on days 3, 5, and 7, and the viral titer was determined in MDCK-chANP32A cells using median tissue culture infectious dose (TCID50) assays. The replication ability of the H5N6-SIME110G virus in the lungs of mice was significantly reduced at an infectious doses of both 102 EID50 (Fig. 9A) and 104 EID50 (Fig. 9B) on day 5 post-infection (p.i.) and day 7 p.i. At the low infectious dose, the replication ability of H5N6GD16 in nasal turbinate was relatively low, and under these conditions, the viral titers in the nasal turbinates of mice infected with the H5N6-SIME110G virus were similar to those of mice infected with the H5N6-SIMWT virus (Fig. 9C). However, when mice were inoculated with 104 EID50 of the H5N6 virus, the viral titers in the nasal turbinates of those infected with the H5N6-SIME110G virus were lower (P = 0.0513 by t test) than of those infected with the H5N6-SIMWT virus on day 5 p.i. (Fig. 9D). Groups of 6-week-old female BALB/c mice were then inoculated with serial dilutions of the H5N6-SIMWT virus or the H5N6-SIME110G virus, and the 50% mouse lethal-dose (MLD50) values of these viruses were determined and the subjects were monitored for mortality for 2 weeks. As shown in Fig. 9 (E and F), the MLD50 of the H5N6-SIMWT virus was 3.22 log10EID50, whereas the MLD50 of the H5N6-SIME110G virus was reduced approximately 19.05-fold (MLD50 = 4.50 log10EID50). These results suggest that the replication and virulence of the H5N6 avian virus in mice also depend on the avian-signature polymerase-enhancing properties of the NS2 SIM.
Fig. 9. SIM of NS2 affects the replication and virulence of the AIV H5N6 in mice but not in chickens.
(A to D) Replication of the H5N6-SIMWT or H5N6-SIME110G virus in mice. Five 6-week-old BALB/c mice were inoculated intranasally with the H5N6-SIMWT virus or the H5N6-SIME110G mutant virus at an infectious dose of either 102 EID50 (A and C) or 104 EID50 (B and D), respectively. Lungs (A and B) and nasal turbinates (C and D) of mice were collected on day 3 p.i., day 5 p.i., and day 7 p.i. for virus titration using TCID50 assays. Experiments were repeated twice with consistent results. Bars represent mean values of the replicates within one representative experiment (n = 5, ±SD). Dotted line denotes the limit of detection based on the dilution factor in our assays. (E and F) MLD50 for mice infected with the H5N6-SIMWT virus (E) or the H5N6-SIME110G virus (F). (G) Mortality of chickens inoculated with the H5N6-SIMWT virus or the H5N6-SIME110G virus. (H) Viral titers in the organs of chickens inoculated with the H5N6-SIMWT virus or the H5N6-SIME110G virus. Three chickens were inoculated intranasally with 106 EID50 of the H5N6-SIMWT virus or the H5N6-SIME110G mutant virus in a 0.1-ml volume. The organs—including the brains, lungs, kidneys, spleens, pancreas, duodenums, liver, tracheae, and ceca—were collected on day 3 p.i. for viral titration in MDCK-chANP32A cells using TCID50 assays. Bars represent mean values of the replicates within one representative experiment (n = 3, ± SD). In (A) to (D) and (H), significance was determined using an unpaired Student’s t test. *P < 0.05 and **P < 0.01.
Next, to determine whether the replication and pathogenicity of H5N6 AIV in chickens were affected by the NS2-SIME110G mutation, an intravenous pathogenicity index (IVPI) test was performed using the H5N6-SIMWT and H5N6-SIME110G viruses according to the recommendations of the Office International Des Epizooties (44). Groups of 10 6-week-old SPF chickens were inoculated intravenously with 0.1 ml of a 1:10 dilution of bacterium-free allantoic fluid containing either H5N6-SIMWT or H5N6-SIME110G virus and were observed for signs of disease or death for 10 days. All chickens inoculated with either the WT or the mutant virus died within 48 hours after inoculation, yielding an IVPI value of 3 (Fig. 9G). To compare the abilities of the H5N6-SIMWT and H5N6-SIME110G viruses to replicate in chickens, two groups of three 6-week-old SPF chickens were inoculated intranasally with 106 EID50 of either H5N6-SIMWT or H5N6-SIME110G virus in a 0.1-ml volume. The chickens were killed on day 3 p.i., and their organs—including the brains, lungs, kidneys, spleens, pancreas, duodenums, liver, tracheae, and ceca—were collected for viral titration using TCID50 assays in MDCK-chANP32A cells. Both WT and H5N6-SIME110G virus replicated systemically with comparable titers in chickens (Fig. 9H). These results indicated that NS2 SIM function is not required for replication of avian IAV in chickens. Together, these results suggest that the replication and virulence of the AIV in mice, but not in chickens, are also dependent on the help of the NS2 proteins with a functional SIM.
DISCUSSION
Mammalian ANP32A/B efficiently support the activity of mammalian-signature polymerase but do not support avian vPol due to the species-specific differences in ANP32 proteins (17, 19–26, 35). Therefore, AIV requires adaptive mutations, such as PB2-E627K, to adapt to mammalian ANP32 proteins and establish productive replication when jumping from birds to mammals (17, 22, 24). However, in some cases, certain isolates of AIV, such as H7N9 and H5N1, infect humans to establish productive infections and can be fatal without acquiring previously identified adaptive mutations (45, 46). The molecular mechanisms by which AIVs adapt to mammals without prior adaptation are currently unknown. Furthermore, the molecular mechanisms underlying the early-stage replication process of nonadapted AIVs before the emergence of adaptive mutations also remain to be elucidated. In this study, we show that NS2 selectively enhances the activity of the avian-signature vPol when supported by mammalian ANP32A/B but not chANP32A, thereby eliminating the restriction of avian-signature polymerase in mammalian cells. NS2 not only can promote H9N2 vRNP formation but also can enhance the interaction between H9N2 vRNP and huANP32A/B (but not the H9N2 vRNP-chANP32A interaction). The presence of a SIM within the C-terminal tail of NS2 is required to confer its avian-signature polymerase-enhancing ability. NS2 has a relatively limited effect on the activity of mammalian-signature (PB2-627K) vPol supported by mammalian ANP32A/B or chANP32A. Thus, we have identified NS2 as a potential alternative strategy for the AIV to overcome the restriction of polymerase and thus better adapt when it crosses the species barrier from birds to mammals (Fig. 10).
Fig. 10. The working mechanism for NS2 in promoting AIV polymerase in human cells.
Avian polymerase functions efficiently in avian cells due to high levels of vRNP assembly and strong chANP32A-vRNP interaction. However, in human cells, avian polymerase functions poorly due to limited vRNP assembly and weak huANP32A/B-vRNP interactions. Under infection conditions, the viral protein NS2 relies on its SIM to compensate for the defects in vRNP assembly and huANP32A/B-vRNP interactions, thereby greatly enhancing avian vPol activity and the replication capacity of AIVs. Impairment of SIM in NS2 caused it to lose its ability to promote vRNP assembly and huANP32A/B-vRNP interactions, thereby losing its avian polymerase-enhancing properties. Overall, we have identified a previously unknown mechanism by which AIVs use the SIM in NS2 to help their polymerase adapt to mammalian ANP32A/B.
The main known function of NS2 is to mediate the nuclear export of vRNPs, a process that requires the combined participation of its NES and M1 (36, 37). In addition, NS2 regulates the transcription and replication process of the influenza A/WSN/33 virus (34) and is also required to for the production of small viral RNAs (47). A previous study found that adaptative mutations in NS2 promoted the adaptation of the AIV H5N1 to human cells (48). Here, we discovered a novel function of NS2 in species-specific support of AIV replication in mammals in vitro and in vivo. Our results indicate that NS2 from avian and mammalian IAVs can selectively enhance avian-signature vPol activity when supported by mammalian ANP32A/B, but not by chANP32A. However, the mammalian-signature vPol activity supported by mammalian ANP32A/B was limited affected by the NS2. Furthermore, we demonstrate that the avian-signature polymerase-enhancing function of NS2 is independent of its role in the vRNP nuclear export by generating a panel of NS2 mutants with defects in mediating vRNP nuclear export.
Previous studies have shown that the restriction of avian-signature vPol activity is associated with impaired vRNP assembly in mammalian cells (9, 19, 49). Recent evidence suggests that the species-specific differences in ANP33A determine the restriction of avian-signature vPol activity in mammalian cells (17). The restriction of avian-signature vPol activity in human cells could be compensated for by expression of chANP32A via unknown mechanisms (17, 19), and it is unclear whether chANP32A functions to support avian-signature vPol activity by promoting vRNP assembly in mammalian cells. Here, we provide evidence that the defects of avian-signature vRNP assembly in human cells could be compensated for by expression of chANP32A or NS2. However, when TKO cells are reconstituted with chANP32A, the avian vRNP assembly process is unaffected by NS2 expression. These data suggest that NS2 promotes avian vRNP assembly in a species-specific manner. On the basis of our findings, we propose that NS2 mediates the enhancement of avian vPol activity through a mechanism common to chANP32A. To exclude the possibility that the failure of avian vRNP assembly in TKO cells reconstituted with huANP32A/B was due to a limiting amount of vRNA due to low avian vPol activity when supported by huANP32A/B in the absence of NS2, the avian vRNP assembly process was assessed in TKO cells without reconstitution of any ANP32 proteins. In this case, the avian polymerase was not functional and vRNA levels were stable. However, we still found that NS2 promoted the assembly of avian vRNP. These results suggest that the defects in avian vRNP formation do not arise directly from intrinsic differences in vPol activity. Furthermore, in addition to polymerase directing genome replication and transcription in the form of vRNP, genomic segments are also packaged into virions in the form of vRNP. Therefore, we hypothesize that NS2 likely promotes AIV replication in mammalian cells not only by enhancing avian polymerase activity but also through other mechanisms remaining to be determined.
Previous studies have shown that the polymerase-supporting function of ANP32 is associated with its association with vPol (18, 19, 21–24, 39, 40). However, avian-signature vPol activity is poorly supported by the mammalian ANP32A, a fact which cannot be explained by differences in the interaction of ANP32A with vPol, because the binding of ANP32A to vPol is independent of PB2-627 identity (21). Here, we found that NS2 promoted the avian-signature vPol activity supported by mammalian ANP32A/B without affecting the interaction between mammalian ANP32A/B and vPol. In contrast, we found that NS2 enhanced the interaction between mammalian ANP32 protein and avian-signature vRNP. The avian-signature vPol activity supported by chANP32A was affected by NS2 only to a limited extent, which corresponds to the fact that NS2 protein had only a limited effect on the chANP32A-vRNP interaction.
SUMOylation is an important posttranslational modification in which SUMO molecules can be covalently attached to the lysine of the target protein (50). In mammals, only three SUMO isoforms, SUMO1, SUMO2, and SUMO3, are associated with SUMOylation. SUMOylation typically targets lysines located within the consensus motif (ҨKxD/E, where Ҩ represents hydrophobic amino acid and x represents any amino acid) for covalent modification. In addition, SUMO and target proteins can interact noncovalently through SIMs (51, 52). Current studies have shown that multiple proteins of the IAV, including NS1, M1, NP, PB1, and PB2, can be modified by SUMOylation (53–57). In this study, we found that NS2 contains a previously identified conserved SIM sequence in its C-terminal tail. NS2 can noncovalently bind to SUMO1/2/3-conjugated proteins through this SIM. Disruption of the integrity of this SIM by replacing these hydrophobic residues in this motif for glycines (SIMLLGG, SIMVEGG, and SIMGGGG) resulted in the loss of binding ability of the NS2 protein to SUMO1-conjugated proteins. Moreover, NS2 mutants with impaired SIM integrity failed to enhance avian vRNP assembly and the binding of avian vRNP to mammalian ANP32A/B, thereby almost completely losing their avian-signature polymerase-enhancing properties. We were unable to rescue recombinant AIVs H9N2 with NS2 bearing SIMLLGG, SIMVEGG, or SIMGGGG mutations, which have almost completely lost their avian-signature polymerase-enhancing properties. Single SIM-E110G mutation is already sufficient to affect the replication of AIV in a species-specific manner.
Previous studies have shown that the presence of SIM typically enhances the SUMOylation of its own proteins (58). Here, we provided evidence that the SUMOylation of NS2 is not required for its avian-signature polymerase-enhancing ability by generating a lysine-free mutant of NS2, suggesting that the SIM-mediated function of NS2 is independent of its SUMOylation. Furthermore, the failure of SUMO conjugation–deficient mutants to bind to NS2 suggests that NS2 may interact with specific SUMO-conjugated viral proteins or other host factors through its SIM to exert its polymerase-enhancing function, which remains to be elucidated in future studies.
The mechanism underlying the early stages of AIV replication in mammalian cells, before the emergence of adaptative mutations, has long remained unknown. In most cases, the emergence of adaptative mutations was still required for better adaptation. Here, we provide evidence that nonadapted avian vPol can use mammalian ANP32A/B with the help of NS2 to replicate productively in the early stages before the emergence of adaptive mutations. Notably, SIM (106LLEVE110) is highly conserved in avian isolates of H1-H16 IAVs. On the basis of our findings, we hypothesize that the replication advantage conferred by the avian-signature polymerase-enhancing properties of NS2 may facilitate the adaptation process by accelerating the acquirement of adaptive mutations. Adaptative mutations in NS2 have been reported previously. The adaptive mutation M16I in H5N1 NS2 was shown to compensate for the replication defect of H5N1 in human cells (48). Considering that the NS2s from IAVs isolated from different host species are conserved in this avian-signature polymerase-enhancing function, we reason that the M16I mutation only positively regulates this function. Because we demonstrate that the SIM in NS2 determines this function, it is possible that M16I might enhance the binding of NS2 to SUMO-conjugated proteins, an area that requires further investigation.
To conclude, our study identified NS2 as a cofactor in the adaptation of the AIV to mammalian hosts. We uncovered a previously unknown mechanism by which AIVs use the SIM in NS2 to assist in crossing the species barrier from birds to mammals.
MATERIALS AND METHODS
Ethics statements
This study was conducted in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People’s Republic of China. The protocol for in vivo studies was approved by the Committee on the Ethics of Animal Experiments of the Harbin Veterinary Research Institute (HVRI) of the Chinese Academy of Agricultural Sciences (CAAS). Nine-day-old embryonated chicken eggs and chicken blood were obtained from the National Poultry Laboratory Animal Resource Center.
Biosafety statement and facility
All experiments using live H5N6 and H7N9 viruses were conducted within the enhanced animal biosafety level 3 (ABSL3+) facility at the HVRI of CAAS, which is approved for such use by the Ministry of Agriculture and Rural Affairs of China and the China National Accreditation Service for Conformity Assessment. Details of the facility and the biosafety and biosecurity measures used have been previously reported (59).
Cells lines and constructs
HEK293T cells, TKO cells (ANP32A, ANP32B, and ANP32E TKO HEK293T cell lines), MDCK cells, MDCK-chANP32A cells (MDCK cells stably expressing chANP32A), MLE-12, NIH3T3 (mouse embryonic fibroblast cells), and primary CEFs were cultured in Dulbecco’s modified Eagle’s medium(DMEM; Gibco) containing 10% fetal bovine serum and 1% penicillin-streptomycin (Gibco). Preparation of TKO cells has been previously described (27). All cells were maintained at 37°C and 5% CO2.
Preparation of the pCAGGS expression plasmids encoding the RNP(s) of H7N9 (A/Anhui/01/2013;H7N9AH13), H9N2 AIV (A/chicken/Zhejiang/B2013/2012) and H1N1 human IAV strain A/WSN/1933 (WSN) has been described in previous studies (22, 25). pCAGGS expression plasmids encoding ANP32A and ANP32B from different species were kept in our laboratory, and preparation was described in previous studies (22, 25, 27). The open reading frames (ORFs) of NS2 form IAVs isolated from different host species were cloned into the pCAGGS vector with a GST tag or Myc tag at the N terminus. Site-directed mutants and deletion mutants of NS2 were generated with polymerase chain reaction (PCR) and identified with DNA sequencing. The full-length ORFs of human SUMO1, SUMO2, and SUMO3 were cloned into the primary CEF vector with a Flag tag at the N terminus. To create the pCAGGS expression plasmids encoding for mutant SUMO1, SUMO2, and SUMO3, the required mutations were introduced with PCR using primers containing GG-to-AA mutations at the C terminus of SUMO1/2/3. The ORFs of human Ubc9 were cloned into the pCAGGS vectors with an N-terminal His6 tag. Development of the mini-genome reporters of IAVs and IBVs and Renilla luciferase expression plasmids (pRL-TK) have been described previously (22, 25, 27).
vPol reconstitution assays
Cells plated in 24-well plates were cotransfected with pPolI-luc (40 ng), Renilla luciferase expression plasmid (5 ng), and four RNP expression plasmids such as pCAGGS-PB1 (20 ng), pCAGGS-PB2 (20 ng), pCAGGS-PA (10 ng), and pCAGGS-NP (40 ng) together with the indicated amount of the plasmids of interest. After 24 hours of transfection, cells were lysed and luciferase activity was measured using the Dual-Glo Luciferase Assay system (Promega) with a Berthold Centro LB 960 Microplate Luminometer.
Immunoprecipitations and GST pull-down
After transfection with the indicated plasmids for 24 hours, transfected cells in a 25-cm2 flask were lysed with 1 ml of lysis buffer [50 mM Hepes-NaOH (pH 7.9), 100 mM NaCl, 50 mM KCl, 0.25% NP-40, and 1 mM dithiothreitol]. Cell lysate (100 μl) were saved as an input control, and the rest of the cell lysate was immunoprecipitated with anti-Flag M2 Magnetic Beads (Sigma-Aldrich, M8823) at 4°C overnight. The precipitated immune complexes were washed three times with lysis buffer and then eluted with 3× Flag peptides. Immunoprecipitated proteins were then analyzed by Western blotting.
For the GST pull-down assay, transfected cells in a 25-cm2 flask were lysed with 1 ml of lysis buffer as above. Cell lysate (100 μl) was saved as an input control, and the remaining cell lysate was incubated with Glutathione MagBeads (GenScript, L00327) overnight at 4°C. The beads were then washed three times with 1× phosphate-buffered saline (PBS) and boiled with sample loading buffer prior Western blot (WB) analysis.
Western blots
WB analysis was performed using the standard described methods (22). The following antibodies were used: rabbit anti-GST (1:2000 for WB; GenScript, A00097), rabbit anti-Flag (1:2000 for WB; Sigma-Aldrich, F7425), rabbit anti-V5 (1:2000 for WB; Proteintech, 14440-1-AP), mouse anti-His (1:5000 for WB; Proteintech, 66005-1-Ig), rabbit anti-ACTB (β-Actin) (1:10,000 for WB; Abclonal, AC026), mouse anti-ACTB (1:2000 for WB; Abclonal, AC004), rabbit anti-Flag (1:5000 for WB; Sigma-Aldrich, F7425), rabbit anti-IAV PB2 (1:5000 for WB; GeneTex, GTX125926), mouse anti-human ANP32A(1:5000 for WB; Proteintech, 67687-1-Ig), rabbit anti-human ANP32B (1:5000 for WB; Proteintech, 10843-1-AP), mouse anti-IAV PA (1:5000 for WB; prepared in our laboratory), mouse anti-IAV PB2 (1:1000 for WB; prepared in our laboratory), and mouse anti-IAV NP (1:1000 for WB; prepared in our laboratory).
Influenza virus infection
IAVs containing NS2-SIMWT or NS-SIME110G were made with the genome from H1N1 human IAV(WSN; provided by Y. Kawaoka), H9N2 avian IAV (A/chicken/Zhejiang/B2013/2012; provided by Z. Li), H7N9 avian IAV (A/Chicken/Hunan/S1220/2017; kept in our laboratory), and H5N6 avian IAV (A/duck/Guangdong/S1330/2016; kept in our laboratory). Virus rescue was performed by transfecting TKO cells with a 12-plasmid rescue system. Briefly, TKO cells were cotransfected with eight pPol1 plasmids encoding all eight segments and expression plasmids encoding RNP(s) and chANP32A. Twenty-four hours after transfection, supernatants were harvested and injected into 9-day-old specific pathogen–free embryonated eggs for virus propagation. Viral stocks were harvested after 48 hours of incubation at 35°C for and titrated in MDCK-chANP32A cell lines (MDCK cells stably expressing chANP32A) using standard methods. Viral genotypes were confirmed by DNA sequencing.
To determine virus growth curves, the indicated cells were infected with each virus diluted in Opti-MEM and replaced 1 to 2 hours later with Opti-MEM supplemented with tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (0 to 2 μg/ml). Cell supernatants were harvested at the indicated time points after infection, and the viral titers were determined in MDCK-chANP32A cells using TCID50 assay.
Mouse study
To determine the viral replication in mice, the indicated dose of H5N6-SIMWT or H5N6-SIME110G was diluted in PBS and used for intranasal inoculation of five 6-week-old female BALB/c mice. The mice were euthanized for the collection of nasal turbinate and lungs on day 3 p.i., day 5 p.i., and day 7 p.i., respectively. Viral titers were then determined in MDCK-chANP32A cells using TCID50 assays.
To determine the MLD50 value, groups of five BALB/c mice that were age-matched were intranasally inoculated with 101 to 106 EID50 of virus in a volume of 50 μl. Then, the inoculated mice were monitored for mortality for 2 weeks and the MLD50 values were calculated by the Reed and Muench methods (60).
Chicken study
To determine the virulence of WT H5N6 and their mutants in chickens, an IVPI was performed according to the method described in the “Diagnostic Manual for Avian Influenza”. Ten 6-week-old chickens were inoculated intravenously with the virus, after which the mortality was monitored for 10 days. To determine the ability of H5N6 virus to replicate in chickens, three chickens were inoculated intranasally with 106 EID50 of tested viruses in a 0.1-ml volume. The chickens were euthanized on day 3 p.i.., and their organs—including the brains, lungs, kidneys, spleens, pancreas, duodenums, liver, tracheae, and ceca—were collected for viral titration in MDCK-chANP32A cells using TCID50 assays.
Statistical analysis
Quantitative data are presented as means ± SD as indicated in the figure legends. Statistical differences were analyzed with an unpaired Student’s t test, two-way analysis of variance (ANOVA) or one-way ANOVA followed by a Dunnett’s multiple comparisons test or a Tukey’s multiple comparisons test using GraphPad Prism 7.0 software. Statistical parameters are reported in the figures and figure legends (NS, not significant; P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001).
Acknowledgments
We thank Y. Kawaoka and Z. Li for providing plasmids.
Funding: This work was supported by the National Key R&D Program of China (2021YFD1800200 to H.C.), the National Natural Science Foundation of China (31521005 to H.C.), the National Natural Science Foundation of China (32002275 to H.Z.), the Natural Science Foundation of Heilongjiang Province of China (YQ2020C021 to H.Z.), and the Natural Science Foundation of Heilongjiang Province of China (TD2022C006 to X.W.).
Author contributions: X.W. conceived and supervised the study and revised the manuscript. L.S., H.C., and X.W. analyzed the data and designed the study. L.S. wrote the original draft. L.S., H.K., M.Y.., L.N., Y.Q., and Y.Z. performed the experiments. Z.Z. and H.Z. provided reagents and resources.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
REFERENCES AND NOTES
- 1.Long J. S., Mistry B., Haslam S. M., Barclay W. S., Host and viral determinants of influenza A virus species specificity. Nat. Rev. Microbiol. 17, 67–81 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Su S., Gu M., Liu D., Cui J., Gao G. F., Zhou J., Liu X., Epidemiology, evolution, and pathogenesis of H7N9 influenza viruses in five epidemic waves since 2013 in China. Trends Microbiol. 25, 713–728 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Neumann G., Chen H., Gao G. F., Shu Y., Kawaoka Y., H5N1 influenza viruses: Outbreaks and biological properties. Cell Res. 20, 51–61 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen H., Yuan H., Gao R., Zhang J., Wang D., Xiong Y., Fan G., Yang F., Li X., Zhou J., Zou S., Yang L., Chen T., Dong L., Bo H., Zhao X., Zhang Y., Lan Y., Bai T., Dong J., Li Q., Wang S., Zhang Y., Li H., Gong T., Shi Y., Ni X., Li J., Zhou J., Fan J., Wu J., Zhou X., Hu M., Wan J., Yang W., Li D., Wu G., Feng Z., Gao G. F., Wang Y., Jin Q., Liu M., Shu Y., Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: A descriptive study. Lancet 383, 714–721 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Pan Y., Cui S., Sun Y., Zhang X., Ma C., Shi W., Peng X., Lu G., Zhang D., Liu Y., Wu S., Yang P., Wang Q., Human infection with H9N2 avian influenza in northern China. Clin. Microbiol. Infect. 24, 321–323 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Wei S. H., Yang J. R., Wu H. S., Chang M. C., Lin J. S., Lin C. Y., Liu Y. L., Lo Y. C., Yang C. H., Chuang J. H., Lin M. C., Chung W. C., Liao C. H., Lee M. S., Huang W. T., Chen P. J., Liu M. T., Chang F. Y., Human infection with avian influenza A H6N1 virus: An epidemiological analysis. Lancet Respir. Med. 1, 771–778 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jonges M., Welkers M. R., Jeeninga R. E., Meijer A., Schneeberger P., Fouchier R. A., de Jong M. D., Koopmans M., Emergence of the virulence-associated PB2 E627K substitution in a fatal human case of highly pathogenic avian influenza virus A(H7N7) infection as determined by Illumina ultra-deep sequencing. J. Virol. 88, 1694–1702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu W., Shi J., Cui P., Yan C., Zhang Y., Wang C., Zhang Y., Xing X., Zeng X., Liu L., Tian G., Suzuki Y., Li C., Deng G., Chen H., Novel H5N6 reassortants bearing the clade 2.3.4.4b HA gene of H5N8 virus have been detected in poultry and caused multiple human infections in China. Emerg. Microbes Infect. 11, 1174–1185 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehle A., Doudna J. A., An inhibitory activity in human cells restricts the function of an avian-like influenza virus polymerase. Cell Host Microbe 4, 111–122 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang L., Jiang L., Li J., Zhao Q., Wang J., He X., Huang S., Wang Q., Zhao Y., Wang G., Sun N., Deng G., Shi J., Tian G., Zeng X., Jiang Y., Liu L., Liu J., Chen P., Bu Z., Kawaoka Y., Chen H., Li C., Low polymerase activity attributed to PA drives the acquisition of the PB2 E627K mutation of H7N9 avian influenza virus in mammals. MBio 10, 10.1128/mBio.01162-19, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Subbarao E. K., London W., Murphy B. R., A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J. Virol. 67, 1761–1764 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Almond J. W., A single gene determines the host range of influenza virus. Nature 270, 617–618 (1977). [DOI] [PubMed] [Google Scholar]
- 13.Van Hoeven N., Pappas C., Belser J. A., Maines T. R., Zeng H., Garcia-Sastre A., Sasisekharan R., Katz J. M., Tumpey T. M., Human HA and polymerase subunit PB2 proteins confer transmission of an avian influenza virus through the air. Proc. Natl. Acad. Sci. U.S.A. 106, 3366–3371 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steel J., Lowen A. C., Mubareka S., Palese P., Transmission of influenza virus in a mammalian host is increased by PB2 amino acids 627K or 627E/701N. PLOS Pathog. 5, e1000252 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mok C. K., Lee H. H., Lestra M., Nicholls J. M., Chan M. C., Sia S. F., Zhu H., Poon L. L., Guan Y., Peiris J. S., Amino acid substitutions in polymerase basic protein 2 gene contribute to the pathogenicity of the novel A/H7N9 influenza virus in mammalian hosts. J. Virol. 88, 3568–3576 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao Y., Zhang Y., Shinya K., Deng G., Jiang Y., Li Z., Guan Y., Tian G., Li Y., Shi J., Liu L., Zeng X., Bu Z., Xia X., Kawaoka Y., Chen H., Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLOS Pathog. 5, e1000709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long J. S., Giotis E. S., Moncorge O., Frise R., Mistry B., James J., Morisson M., Iqbal M., Vignal A., Skinner M. A., Barclay W. S., Species difference in ANP32A underlies influenza A virus polymerase host restriction. Nature 529, 101–104 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugiyama K., Kawaguchi A., Okuwaki M., Nagata K., pp32 and APRIL are host cell-derived regulators of influenza virus RNA synthesis from cRNA. eLife 4, e08939 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker S. F., Ledwith M. P., Mehle A., Differential splicing of ANP32A in birds alters its ability to stimulate RNA synthesis by restricted influenza polymerase. Cell Rep. 24, 2581–2588.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domingues P., Eletto D., Magnus C., Turkington H. L., Schmutz S., Zagordi O., Lenk M., Beer M., Stertz S., Hale B. G., Profiling host ANP32A splicing landscapes to predict influenza A virus polymerase adaptation. Nat. Commun. 10, 3396 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingues P., Hale B. G., Functional insights into ANP32A-dependent influenza a virus polymerase host restriction. Cell Rep. 20, 2538–2546 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang H., Zhang Z., Wang Y., Wang M., Wang X., Zhang X., Ji S., Du C., Chen H., Wang X., Fundamental contribution and host range determination of ANP32A and ANP32B in influenza a virus polymerase activity. J. Virol. 93, e00174-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Staller E., Sheppard C. M., Neasham P. J., Mistry B., Peacock T. P., Goldhill D. H., Long J. S., Barclay W. S., ANP32 proteins are essential for influenza virus replication in human cells. J. Virol. 93, e00217-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long J. S., Idoko-Akoh A., Mistry B., Goldhill D., Staller E., Schreyer J., Ross C., Goodbourn S., Shelton H., Skinner M. A., Sang H., McGrew M. J., Barclay W., Species specific differences in use of ANP32 proteins by influenza A virus. eLife 8, e45066 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Li H., Wang W., Wang Y., Han G. Z., Chen H., Wang X., A unique feature of swine ANP32A provides susceptibility to avian influenza virus infection in pigs. PLOS Pathog. 16, e1008330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peacock T. P., Swann O. C., Salvesen H. A., Staller E., Leung P. B., Goldhill D. H., Zhou H., Lillico S. G., Whitelaw C. B. A., Long J. S., Barclay W. S., Swine ANP32A supports avian influenza virus polymerase. J. Virol. 94, e00312-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Z., Zhang H., Xu L., Guo X., Wang W., Ji Y., Lin C., Wang Y., Wang X., Selective usage of ANP32 proteins by influenza B virus polymerase: Implications in determination of host range. PLOS Pathog. 16, e1008989 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Q. M., Ito M., Muramoto Y., Hoang P. V., Vuong C. D., Sakai-Tagawa Y., Kiso M., Ozawa M., Takano R., Kawaoka Y., Pathogenicity of highly pathogenic avian H5N1 influenza A viruses isolated from humans between 2003 and 2008 in Northern Vietnam. J. Gen. Virol. 91, 2485–2490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manz B., de Graaf M., Mogling R., Richard M., Bestebroer T. M., Rimmelzwaan G. F., Fouchier R. A. M., Multiple natural substitutions in avian influenza a virus PB2 facilitate efficient replication in human cells. J. Virol. 90, 5928–5938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song W., Qin K., Human-infecting influenza A (H9N2) virus: A forgotten potential pandemic strain? Zoonoses Public Health 67, 203–212 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Yin X., Deng G., Zeng X., Cui P., Hou Y., Liu Y., Fang J., Pan S., Wang D., Chen X., Zhang Y., Wang X., Tian G., Li Y., Chen Y., Liu L., Suzuki Y., Guan Y., Li C., Shi J., Chen H., Genetic and biological properties of H7N9 avian influenza viruses detected after application of the H7N9 poultry vaccine in China. PLOS Pathog. 17, e1009561 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui P., Zeng X., Li X., Li Y., Shi J., Zhao C., Qu Z., Wang Y., Guo J., Gu W., Ma Q., Zhang Y., Lin W., Li M., Tian J., Wang D., Xing X., Liu Y., Pan S., Zhang Y., Bao H., Liu L., Tian G., Li C., Deng G., Chen H., Genetic and biological characteristics of the globally circulating H5N8 avian influenza viruses and the protective efficacy offered by the poultry vaccine currently used in China. Sci. China Life Sci. 65, 795–808 (2022). [DOI] [PubMed] [Google Scholar]
- 33.Guo J., Wang Y., Zhao C., Gao X., Zhang Y., Li J., Wang M., Zhang H., Liu W., Wang C., Xia Y., Xu L., He G., Shen J., Sun X., Wang W., Han X., Zhang X., Hou Z., Jin X., Peng N., Li Y., Deng G., Cui P., Zhang Q., Li X., Chen H., Molecular characterization, receptor binding property, and replication in chickens and mice of H9N2 avian influenza viruses isolated from chickens, peafowls, and wild birds in eastern China. Emerg. Microbes Infect. 10, 2098–2112 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robb N. C., Smith M., Vreede F. T., Fodor E., NS2/NEP protein regulates transcription and replication of the influenza virus RNA genome. J. Gen. Virol. 90, 1398–1407 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Bi Z., Ye H., Wang X., Fang A., Yu T., Yan L., Zhou J., Insights into species-specific regulation of ANP32A on the mammalian-restricted influenza virus polymerase activity. Emerg. Microbes Infect. 8, 1465–1478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O’Neill R. E., Talon J., Palese P., The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 17, 288–296 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akarsu H., Burmeister W. P., Petosa C., Petit I., Muller C. W., Ruigrok R. W., Baudin F., Crystal structure of the M1 protein-binding domain of the influenza A virus nuclear export protein (NEP/NS2). EMBO J. 22, 4646–4655 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paterson D., Fodor E., Emerging roles for the influenza A virus nuclear export protein (NEP). PLOS Pathog. 8, e1003019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang F., Sheppard C. M., Mistry B., Staller E., Barclay W. S., Grimes J. M., Fodor E., Fan H., The C-terminal LCAR of host ANP32 proteins interacts with the influenza A virus nucleoprotein to promote the replication of the viral RNA genome. Nucleic Acids Res. 50, 5713–5725 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrique L., Fan H., Walker A. P., Keown J. R., Sharps J., Staller E., Barclay W. S., Fodor E., Grimes J. M., Host ANP32A mediates the assembly of the influenza virus replicase. Nature 587, 638–643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Q., Xie Y., Zheng Y., Jiang S., Liu W., Mu W., Liu Z., Zhao Y., Xue Y., Ren J., GPS-SUMO: A tool for the prediction of sumoylation sites and SUMO-interaction motifs. Nucleic Acids Res. 42, W325–W330 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pal S., Santos A., Rosas J. M., Ortiz-Guzman J., Rosas-Acosta G., Influenza A virus interacts extensively with the cellular SUMOylation system during infection. Virus Res. 158, 12–27 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Bi Y., Chen Q., Wang Q., Chen J., Jin T., Wong G., Quan C., Liu J., Wu J., Yin R., Zhao L., Li M., Ding Z., Zou R., Xu W., Li H., Wang H., Tian K., Fu G., Huang Y., Shestopalov A., Li S., Xu B., Yu H., Luo T., Lu L., Xu X., Luo Y., Liu Y., Shi W., Liu D., Gao G. F., Genesis, evolution and prevalence of H5N6 avian influenza viruses in China. Cell Host Microbe 20, 810–821 (2016). [DOI] [PubMed] [Google Scholar]
- 44.Manual of Diagnostic Tests and Vaccines for Terrestrial Animal (Office International des Epizooties, Paris, 2011).
- 45.Zhao D., Liang L., Li Y., Jiang Y., Liu L., Chen H., Phylogenetic and pathogenic analyses of avian influenza A H5N1 viruses isolated from poultry in Vietnam. PLOS ONE 7, e50959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi J., Deng G., Ma S., Zeng X., Yin X., Li M., Zhang B., Cui P., Chen Y., Yang H., Wan X., Liu L., Chen P., Jiang Y., Guan Y., Liu J., Gu W., Han S., Song Y., Liang L., Qu Z., Hou Y., Wang X., Bao H., Tian G., Li Y., Jiang L., Li C., Chen H., Rapid evolution of H7N9 highly pathogenic viruses that emerged in China in 2017. Cell Host Microbe 24, 558–568.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez J. T., Varble A., Sachidanandam R., Zlatev I., Manoharan M., Garcia-Sastre A., tenOever B. R., Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc. Natl. Acad. Sci. U.S.A. 107, 11525–11530 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Manz B., Brunotte L., Reuther P., Schwemmle M., Adaptive mutations in NEP compensate for defective H5N1 RNA replication in cultured human cells. Nat. Commun. 3, 802 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Labadie K., Dos Santos Afonso E., Rameix-Welti M. A., van der Werf S., Naffakh N., Host-range determinants on the PB2 protein of influenza A viruses control the interaction between the viral polymerase and nucleoprotein in human cells. Virology 362, 271–282 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Geiss-Friedlander R., Melchior F., Concepts in sumoylation: A decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Song J., Durrin L. K., Wilkinson T. A., Krontiris T. G., Chen Y., Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U.S.A. 101, 14373–14378 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hecker C. M., Rabiller M., Haglund K., Bayer P., Dikic I., Specification of SUMO1- and SUMO2-interacting motifs. J. Biol. Chem. 281, 16117–16127 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Xu K., Klenk C., Liu B., Keiner B., Cheng J., Zheng B. J., Li L., Han Q., Wang C., Li T., Chen Z., Shu Y., Liu J., Klenk H. D., Sun B., Modification of nonstructural protein 1 of influenza A virus by SUMO1. J. Virol. 85, 1086–1098 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo J., Chen J., Li Y., Li Y., Deng G., Shi J., Liu L., Chen H., Li X., SUMOylation of matrix protein M1 and filamentous morphology collectively contribute to the replication and virulence of highly pathogenic H5N1 avian influenza viruses in mammals. J. Virol. 96, e0163021 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han Q., Chang C., Li L., Klenk C., Cheng J., Chen Y., Xia N., Shu Y., Chen Z., Gabriel G., Sun B., Xu K., Sumoylation of influenza A virus nucleoprotein is essential for intracellular trafficking and virus growth. J. Virol. 88, 9379–9390 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li J., Liang L., Jiang L., Wang Q., Wen X., Zhao Y., Cui P., Zhang Y., Wang G., Li Q., Deng G., Shi J., Tian G., Zeng X., Jiang Y., Liu L., Chen H., Li C., Viral RNA-binding ability conferred by SUMOylation at PB1 K612 of influenza A virus is essential for viral pathogenesis and transmission. PLOS Pathog. 17, e1009336 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang G., Zhao Y., Zhou Y., Jiang L., Liang L., Kong F., Yan Y., Wang X., Wang Y., Wen X., Zeng X., Tian G., Deng G., Shi J., Liu L., Chen H., Li C., PIAS1-mediated SUMOylation of influenza A virus PB2 restricts viral replication and virulence. PLOS Pathog. 18, e1010446 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M., Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y., Zhang Q., Kong H., Jiang Y., Gao Y., Deng G., Shi J., Tian G., Liu L., Liu J., Guan Y., Bu Z., Chen H., H5N1 hybrid viruses bearing 2009/H1N1 virus genes transmit in guinea pigs by respiratory droplet. Science 340, 1459–1463 (2013). [DOI] [PubMed] [Google Scholar]
- 60.Reed L. J., A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27, 493–497 (1938). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1