Abstract
Introduction
Prognostic factors in plasma cell myeloma were proved to be related to signaling pathways and associated transcription factors. RGS1 and mTOR were known to play an important role in the pathogenesis of multiple myeloma. The aim of the study was to evaluate the expression and the prognostic value of RGS1 and mTOR and their relation to clinical as well as other diagnostic criteria in multiple myeloma.
Patients and methods
The present study included 44 denovo Myeloma patients, recruited from the Medical Oncology Department, National Cancer Institute, Cairo University. Detection of RGS1 and mTOR expression was performed using Immunohistochemical staining on bone marrow biopsy sections.
Results
The median age was 51 years with male to female ratio 1.58:1. There was a positive highly statistically significant correlation between RGS1 and mTOR among all studied cases (p value <0.001). Regarding their prognostic value, there was a highly statistically significant association of the expression levels of RGS1 and mTOR with treatment response (p <0.001). Finally, there was a significant influence of RGS1 and mTOR on overall survival probability (p value <0.001 and <0.002 respectively) with better survival for those having low expression.
Conclusion
RGS1 and mTOR were suggested as poor prognostic markers in MM patients, being associated with lower response rate and inferior OS. We recommend considering RGS1 and mTOR as one of the prognostic criteria in different risk stratification and staging classifications. Further trials for RGS1 and mTOR targeting in multiple myeloma are recommended.
Introduction
Plasma cell myeloma (PCM) is ranked the second most common hematological malignancy worldwide, and in South Africa, PCM is more common in African black population than in the white population with a ratio 2:1 [1]. In Egypt, multiple myeloma (MM) accounts for 1.3% of all cancers [2] and the mean age of the disease is 58.5 years [3].
The BM microenvironment (BMM) plays a central role in the pathogenesis of MM [4], where it can provide inflammatory agents as cytokines, growth factors like insulin-like growth factor-1 (IGF-1), and others, which support malignant cell growth, drug resistance and cytotoxicity of healthy cells [5]. Some of the prognostic factors related to the tumor burden are related to the signaling pathways and the associated transcription factors as STAT3 [6].
Regulator of G proteins signaling (RGS) proteins are a member family of intracellular regulatory proteins [7], that are expressed on hematopoietic cells such as T and B lymphocytes [8], natural killer cells [9], and dendritic cells [10]. RGS1 is known to negatively regulate heterotrimeric G protein-coupled receptor (GPCR) signaling pathways, by accelerating intrinsic GTPase activity [11] of G protein α subunits, favoring their inactivation by reassociation with the βγ dimer and thereby inhibiting their downstream activity [12]. On the other hand, an important mechanism of action of RGS1 is the non-GAP signaling, where RGS1 promotes malignancy progression by the competitive action with Gβγ for Gα binding, due to the similarity of the two surfaces of Gα that interact with both Gβγ and RGS proteins. This favors the occurrence of some downstream effects of both Gβγ to Gαs subunits [13].
Generated Gα and Gβγ initiate downstream signaling networks by interacting with several effector molecules as phospholipase C (PLC), phosphoinositide 3- kinase (PI3K), [14], and increasing the activity of mitogen-activated protein kinase (MAPK) signaling pathways through PI3K/Akt and PLCβ. Also, this occurs through interaction with insulin-like growth factor receptor 1 (IGFR1) and c-Src tyrosine kinase [15]. All these factors will promote survival, differentiation, and proliferation of the cells and provide higher resistance to apoptosis [16].
Many studies highlighted the importance of RGS in the autoimmune diseases as multiple sclerosis, Crohn’s disease, ulcerative colitis [17], and undifferentiated spondylo-arthritis, often associated with gastrointestinal lesions [18]. Also, RGS1 gene amplification was detected in various malignancies, including melanoma, non-Hodgkin lymphoma, retinoblastoma, pancreatic cancer, nasopharyngeal carcinoma [19], diffuse large B-cell lymphomas, follicular lymphomas, and multiple myeloma [11]. Roh et al. suggested that RGS1 protein may be a promising prognostic marker for MM risk stratification and a promising target for a new MM therapy as well [11].
The mTOR (mechanistic target of rapamycin) is a serine/threonine protein kinase that is expressed throughout the whole body [20]. mTOR signaling is generally involved in regulating cell survival, growth, metabolism, protein synthesis and autophagy, as well as homeostasis [21]. This signaling pathway is critical in normal hematopoietic cells’ development and function [22], which occurs through regulation of some proteins such as STAT3 [23]. Additionally, mTORC1 (mTOR complex 1) is regulated by several signaling pathways including the PI3K/Akt pathway, the Ras-MAPK pathway, and some other intracellular factors [23,24]. Insulin is the most-characterized growth factor (IGF) that activates mTORCs through the PI3K/Akt/TSC/Rheb pathway [25].
Deregulated mTOR was found to be associated with human growth and metabolic diseases as neuronal degeneration, obesity, and type 2 diabetes [26]. Several studies reported that mTOR is aberrantly overactivated in more than 70% of cancers and mTOR dysfunction contributes to tumorigenesis [23] as in kidney cancer [27], breast cancer, non-small cell lung cancer (NSCLC) [28] and multiple myeloma [29,30].
mTORC is rapamycin sensitive [31]. Accordingly, Rapalogs (rapamycin and its analogs) was found to induce inhibit the tumor progression by inducing tumor cell apoptosis, cell cycle arrest and signal transduction inhibition in many hematological diseases, one of them is multiple myeloma [22,32,33].
The aim of the study was to evaluate the expression of RGS1 and mTOR markers and their relation to the clinical and diagnostic parameters as well as their prognostic value in MM. To the best of our knowledge, this study is one of the few studies that have been done to evaluate the IHC expression of RGS1 and mTOR in BM biopsies of MM patients [11,30,34] and the first study to highlight the role of these markers in Egyptian multiple myeloma patients.
Methods
The present study included 44 de-novo Egyptian Multiple Myeloma cases, above 18 years, attended to the outpatient clinic in the National Cancer Institute (NCI), Cairo University, throughout 18 months. All patients were diagnosed ant treated in the NCI (S1 Appendix and S2 Appendix respectively).
This study was approved by the ethical committee, review board of National Cancer Institute, Cairo University in accordance with Helsinki guidelines for the protection of human subjects. Written informed consent was obtained from all patients (IRB no. IRB00004025).
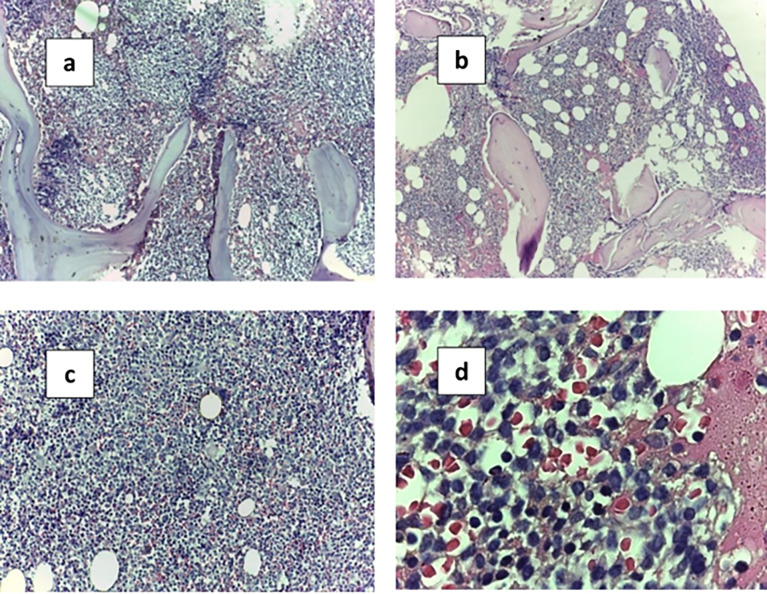
The BM trephine biopsy was performed at diagnosis by using trephine biopsy needles from the posterior superior iliac crest. Biopsy cores were prepared and stained for microscopic examination (Fig 1).
Fig 1. BMB H&E sections with plasma cells infiltration.
A. Para trabecular and patchy infiltration by plasma cells with power magnification (10x). B. Interstitial infiltration by plasma cells with power magnification (10x). C. Interstitial infiltration by plasma cells with power magnification (40x). D. Plasma cells infiltration power magnification (100X Oil emersion lens).
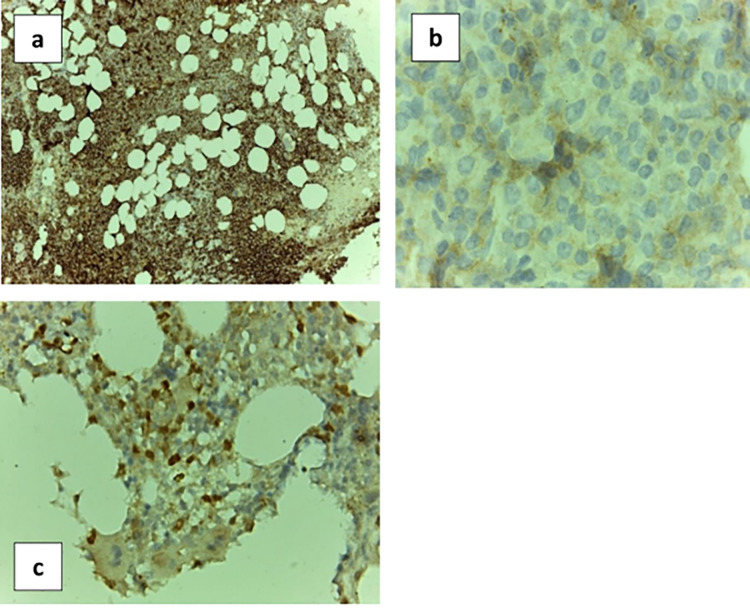
Immunohistochemical (IHC) staining was done by monoclonal Mouse IgG Anti-Human CD138 Clone (MI15 Ready-to-Use (Dako Omnis) Code GA642), monoclonal rabbit IgG Anti-Human mTOR (7C10, #2983, from Cell Signaling Technology, Beverly, MA) (1:250), and polyclonal rabbit IgG Anti-Human RGS1 protein (NBP1-68645, Novus Biologicals, Littleton, Colorado, USA) (1:250) (Fig 2).
Fig 2. BMB IHC sections with plasma cells infiltration.
A. Positive immunohistochemical staining CD138 with power magnification (10x). B. Positive immunohistochemical staining mTOR (100x). C. Positive immunohistochemical staining RGS1 (40x).
Interpretation of the immunohistochemical stain analysis. Analysis of whole tissue section slides under an Olympus light microscope was performed and histopathological features were evaluated. Plasma cell ratio in bone marrow was confirmed by correlation with the PC counts in Giemsa-stained bone marrow aspirations and CD138 IHC to confirm the presence and actual percentage of plasma cells.
Immunostaining for mTOR was cytoplasmic. IHC was scored with regards to both the extent and intensity of staining. The intensity was graded from 0 to 2 (‘0’ for absent staining, ‘1’ for dim expression, and ‘2’ for moderate to strong expression). The percentage of tumor with positive staining was graded from 1 to 3 (“1” represents <10% tumor positivity, “2” represents 10–50% tumor positivity, and “3” for > 50% positivity). For comparison with clinical variables, the expression level of the biomarker was semi-quantitated by multiplying the scores of the staining intensity and the percentage of positive tumor [30]. Then we classified the patients into two groups according to the best scoring level by using Receiver operating characteristics (ROC) curve analysis. Correlations of mTOR to clinical variables and other investigations were done.
Regarding RGS1, three major RGS1 IHC expression patterns were observed: negative, predominantly cytoplasmic and both cytoplasmic and nuclear. Subsequently, the relative percentage of simultaneous cytoplasmic and nuclear RGS1-positive cells were counted and analyzed relative to the overall number of tumor cells [11]. The optimal cut-off value of RGS1 IHC was evaluated using a ROC curve analysis and correlations were done with different investigational and clinical variables.
Data was analyzed using IBM SPSS statistics (V. 26.0, IBM Corp., USA, 2019) (S3 Appendix).
Results
The demographic, clinical and laboratory data of the studied groups are represented in Table 1. The studied cases were 27/44 males (61.4%) and 17/44 females (38.6%) with male to female ratio 1.58:1. Their age ranged from 25 to 82 years with a mean value 52.23 ± 11.06 and a median value 51. The cut off levels of defining end organ damage were set according to the updated IMWG criteria 2016 [35].
Table 1. Demographic, clinical and laboratory data of the studied groups.
| Number = 44 (%) | |
|---|---|
| Age | |
| median(range) | 51 (25–82) |
| Gender | |
| Female | 27 (61.4%) |
| Male | 17 (38.6%) |
|
CRAB criteria
| |
| Hemoglobin (n = 44) | |
| < 10 g/dl | 35 (79.5) |
| ≥ 10 g/dl | 9 (20.5) |
| median(range) | 8.65 (5.3–16) |
| Serum Ca (n = 44) | |
| ≤ 11 mg/dl | 33(75.0) |
| > 11 mg/dl | 11(25.0) |
| median(range) | 9.4 (3.9–14.3) |
| Serum Creatinine (n = 44) | |
| ≤ 2 mg/dl | 27 (61.4) |
| > 2 mg/dl | 17 (38.6) |
| median(range) | 1.2 (0.4–7.8) |
| Osteolytic bony lesions (n = 44) | |
| Negative | 6 (13.6) |
| Positive | 38 (86.3) |
| Other Biochemical laboratory profile | |
| β2 microglobulin | |
| < 3.5 mg/l | 1 (2.3) |
| ≥ 3.5 mg/l | 97.7) |
| Albumin (gm/dl) | |
| median(range) | 3 (2–9) |
| LDH (IU/L) | |
| median(range) | 310 (114–1045) |
| Bone marrow examination | |
| BMA plasma cells (%) | |
| median(range) | 30 (15.0–84.0) |
| BMB CD 138 (%) | |
| median(range) | 80 (20–100) |
| BMB pattern of distribution | |
| Diffuse | 28 (63.6) |
| Interstitial | 16 (36.4 |
| Serum protein electrophoresis and immunofixation | |
| IgA Kappa | 5 (11.4) |
| IgA lambda | 5 (11.4) |
| IgG kappa | 22 (50.0) |
| IgG lambda | 12 (27.3) |
| Free light chain (FLC) | |
| K-FLC | 3 (6.8) |
| L-FLC | 5 (11.4) |
| No FLC | 36 (81.8) |
| Response to treatment (n = 44) | |
| Responders | 16 (36.4) |
| Non-responders | 28 (63.6) |
Plasma cells were examined from BMA smears and the percentage ranged from 15% to 84%. Immunohistochemistry CD138 showed increased levels in all cases, correlating with the PC counts. The % of CD138 positivity ranged from 20% to 100% infiltration with a diffuse or interstitial pattern of infiltration. M band was detected in SPE in all patients. Immunofixation revealed either IgA or IgG bands. Free light chain was present in 8/44 patients (18.2%).
Immunohistochemical staining of RGS1 and mTOR in MM patients
A. RGS1 and mTOR expression distribution in MM patients (n = 44)
RGS1 expression was estimated in all the patients and the % of expression ranged from 0.0% to 100% with a mean value 46.57 ± 23.88 and a median value 50%. As regards the mTOR expression, every patient had a different score level according to intensity and % of expression. Accordingly, the patient fell into 1 of 7 categories from score 0 to 6 as shown in Table 2.
Table 2. RGS1 and mTOR expression distribution in the studied patients.
| Marker expression | Number = 44 (%) |
|---|---|
| RGS1 | |
| >35% | 28 (63.6) |
| ≤35% | 16 (36.3) |
| Median (range) | 50 (0.0–100.0) |
| mTOR score level | |
| 0 | 7 (15.9) |
| 1 | 10 (22.7) |
| 2 | 5 (11.3) |
| 3 | 11(25.0) |
| 4 | 3 (6.8) |
| 5 | 0 (0.0) |
| 6 | 8 (18.2) |
B. Receiver operating characteristics (ROC) curve for determination of the best cut-off values for RGS1 and mTOR
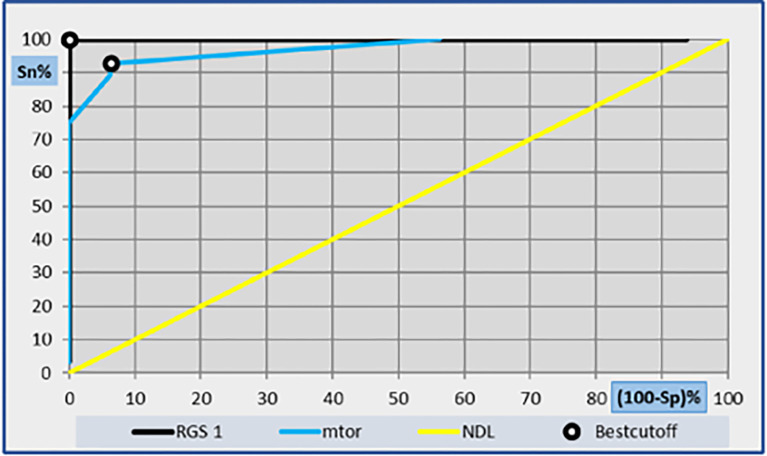
ROC curve was done for calculation of RGS1 and mTOR cut offs as regards sensitivity and specificity with 95% confidence interval. As regards RGS1, the area under the ROC curve (AUC) was 0.938 with the best cut-off value 35% with 100% specificity and 100% sensitivity. As regards mTOR, the AUC was 0.533 and the best cut-off score value was 1 with 93.8 specificity and 92.9 sensitivity as shown in Fig 3.
Fig 3. ROC curve analysis showing the diagnostic performance of RGS1 and mTOR for discriminating of the best cutoff value.
C. Comparative analysis of RGS1 and mTOR with the studied patients
Association between RGS1 expression at cutoff value 35% and MM patients as regarding demographic & laboratory data
There was a highly statistically significant association between RGS1 expression and LDH levels. Where, 13/16 (81.3%) of those having low RGS1 were having low LDH levels as well. While 26/28 (92.9%) of those having high RGS1 expression were having high LDH levels as well (P <0.001). OR = 56.3 (95CI: 8.35–380.1); Significant positive risk (Table 3).
Table 3. The association of RGS1expression and other parameters.
| Clinicopathological features | RGS1 | Total No. (%) |
P value |
||
|---|---|---|---|---|---|
| ≤ 35 (N = 16) | >35 (N = 28) | ||||
| No. (%) | No. (%) | ||||
| LDH (IU/L) | ≤ 250 | 13 (81.3) | 2 (7.1) | 15 (34.1) | <0.001 * |
| >250 | 3 (18.8) | 26 (92.9) | 29 (65.9) | ||
| Clinicopathological features | mTOR score level |
Total
No. (%) |
P
value |
||
| > 1 (n = 27) | ≤ 1 (n = 17) | ||||
| No. (%) | No. (%) | ||||
| LDH (IU/L) | ≤ 250 | 2 (7.4) | 13 (76.5) | 15 (34.1) | <0.001 ** |
| >250 | 25 (92.6) | 4 (23.5) | 29 (65.9) | ||
| FLC | Kappa-LC | 2 (5.4) | 1 (14.3) | 3 (6.8) | 0.074 ** |
| Lambda-LC | 4 (10.8) | 1 (14.3) | 5 (11.4) | ||
| No FLC | 31 (83.8) | 5 (71.4) | 36 (81.8) | ||
** P-value < 0.05: Significant; P-value < 0.01: Highly significant (Chi-square test).
* OR = 56.3 (95CI: 8.35–380.1); Sig. Pos. Risk.
On the contrary, there was no statistically significant association between RGS1 expression and demographic and other chemical laboratory data (S4 Table (1) in S4 Appendix).
Association of mTOR expression at cutoff score value 1 and MM patients regarding demographic & laboratory data
There was a highly statistically significant association between mTOR expression and LDH levels. Where, 25/27 (92.6%) patients of those having mTOR expression level >1 were having high LDH levels as well. While 13/17 (76.5%) patients of those having expression level of mTOR ≤1 were having low LDH levels as well (P <0.001). On the contrary, there was no statistically significant association between mTOR expression and other demographic and laboratory data) (S4 Table (2) in S4 Appendix).
Moreover, there was a borderline statistically significant association between mTOR expression and FLC (P value = 0.074). While there was no statistically significant association between mTOR expression and SPE (P value = 0.597).
Association of RGS1 and mTOR expression and response to treatment
There was a highly statistically significant association between the expression of RGS1 as well as mTOR expression and the response to treatment among our patients (Table 4).
Table 4. Association of RGS1 expression and mTOR score level and response to treatment.
| Response | RGS1 | P value | |
|---|---|---|---|
| ≤ 35 no = 16 (%) |
>35 no = 28 (%) |
||
| Responders | 16 (100) | 0 (0.0) | <0.001 * |
| Non responders | 0 (0.0) | 28 (100) | |
| mTOR score level | |||
|
>1
no = 27 (%) |
≤1
no = 17 (%) |
||
| Responders | 1 (3.7) | 15 (88.2) | <0.001 * |
| Non responders | 26 (96.3) | 2 (11.8) | |
*p-value < 0.01: Highly significant (Chi-square test).
The relation between mTOR and RGS1 expression in MM patients
There was a positive highly statistically significant correlation between mTOR expression and RGS1 expression among all studied cases (P-value <0.001) (Table 5).
Table 5. The relation between RGS1 and mTOR expression in MM patients.
| RGS.1 | Total | P value | ||||
|---|---|---|---|---|---|---|
| ≤35 | >35 | |||||
| mTOR score level | >1 | Count (%) | 1 (3.7) | 26 (96.3) | 27 (61.3) | <0.001 * |
| ≤1 | Count (%) | 15 (88.2) | 2 (11.8) | 17 (38.6) | ||
| Total | Count (%) | 16 (100) | 28 (100) | 44 (100) | ||
*P-value < 0.01: Highly significant (Chi-square test).
D. Survival analysis; overall survival (OS)
Median follow up time of the studied group was 20.5 months ranging from 6 month to 44 months. Overall survival probability was estimated for the whole group; and the survival estimate at 1 year was 38.6% and at 2 years was 38.6% with a median estimate of 20.5 months.
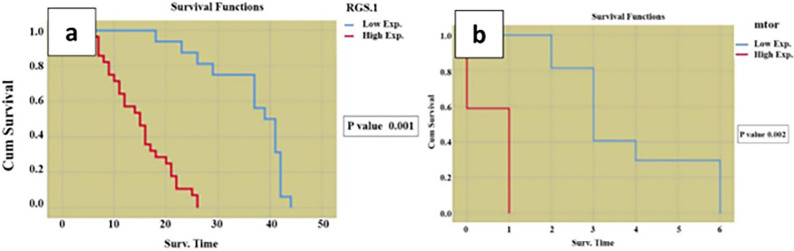
Relationship between OS and RGS1 as well as mTOR expression (Fig 4)
Fig 4.
Kaplan- Meier analysis for the relation between (a) RGS1 and (b) mTOR results and overall survival among the studied.
Results revealed that there was a significant influence of RGS1 expression and mTOR expression on overall survival probability (p value 0.001 and 0.002 respectively) with better survival for those having low expression.
Relationship between OS and different studied parameters
P value of the relationship between demographic, clinical and laboratory data with the OS (S4 Table (3) in S4 Appendix). Results showed that there was a statistically significant difference in the OS among MM patients in our study and LDH with a better overall survival for those having LDH level < 250 IU/L (p = 0.003). Multivariate analysis was done for the statistically significant variables in the univariate level as shown in S4 Table (4) in S4 Appendix, it revealed that RGS1 and mTOR remained an independent prognostic factors of the OS in MM patients with p value = 0.05 and p value = 0.01 respectively. However, LDH lost its prognostic significance in the OS.
Discussion
MM is characterized by a wide heterogeneity in outcome ranging from few months to more than 10 years [36]. For this reason, various prognostic factors and staging systems have been developed trying to predict the outcome of the disease [37]. The present study included 44 de-novo Multiple Myeloma cases with male to female ratio 1.58:1. Their age ranged from 25 years till 82 years with a median age 51 years. This prevalence is nearly like previous studies done in Egyptian multiple myeloma patients, where the mean age was 58.5 with a male to female ratio 1.5:1 and like some previous studies as well [3,38–40]. Most of our patients usually visit the clinics in advanced stage. So, our results reflect a more advanced stage according to IMWG, ISS and mSMART staging systems [41,42].
Several recent reviews have described the multiple roles of GPCR signaling in cancer [43]. RGS1 was shown to have a role in the proliferation, invasion, and metastasis of malignancies [44]. Previous studies have reported the upregulation of RGS1 expression in MM [11,45]. Also, mTOR shares some signaling pathways with RGS1 as the PI3K/Akt cell cycling pathway, which contributes to the initiation and maintenance of cancer as in Non-Hodgkin lymphomas [46,47]. Review of literature reveals a few studies on MM cell lines to study the dysregulation of m-TOR pathway [22,48], and nowadays, mTOR signaling has become a target for treatment to increase overall survival [29].
RGS1 expression was estimated in all the patients and the % of expression ranged from 0.0% to 100% with a median value 50%. The cutoff value was chosen by using ROC curve. High RGS1 expression (>35%) was detected in 28/44 patients (63.6%), while 16/44 patients (36.3%) showed low expression (≤35%). In a study done by Roh et al., to study the RGS1 expression in multiple myeloma BMB sections, they classified their patients into high versus low expression with a cutoff value 7 which is equivalent to 60%-70% expression, they found that this is the significant cutoff value in their population [11]. This may be related to the larger number of patients in their study and ethnic variation between the Asian and Egyptian populations. Also, 59.5% of their studied group was classified as a standard risk category according to IMWG risk stratification, while our study included all stages of MM.
RGS1 expression showed a highly statistically significant association with LDH levels. Also, Roh et al., found a borderline correlation between RGS1 expression and LDH levels (p value = 0.060) [11]. This may be related to the advanced stage of the disease in our studied population. On the contrary, there is no statistically significant association between RGS1 expression and other demographic and laboratory data. Moreover, there was no statistically significant association between RGS1 expression and neither BMA plasma cells percentage, nor BMB CD 138 nor BMB infiltration pattern. Similar results were found in Roh et al. [11].
As regards mTOR expression, in accordance with Stockwin et al., patients were categorized into 7 scores according to the mTOR expression as regards the intensity of the marker and the percentage of expression. Patients with score level 6 in our study were 18.7% compared to the 25.8% in Stockwin et al., however, Stockwin et al. performed IHC staining with mTOR and phospho-mTOR (p-mTOR) on multiple myeloma BMB sections, but in our study, we studied only the mTOR [30]. So, this may reflect a relative higher expression of the marker in their study. Accordingly, in our study, a ROC curve was done and score level 1 was chosen. According to the scoring system, 17/44 (38.6%) patients were having a score level ≤1 and 27/44 (61.4%) were having a score level >1.
Near to our results, Sebestyen et al. verified that mTOR activity was presented among DLBCL patients in 62% of the studied population, by positive immunostaining for p-S6 the most sensitive marker of mTOR activity [49]. Also, Schedel et al., examined the expression of mTOR and its phosphorylated (active) counterpart found [50]. The results suggested that mTOR is highly active in different cancer types, as various permanent cell lines of bladder cancer and head & neck squamous cell carcinoma.
Like RGS1, mTOR showed a highly statistically significant association with LDH levels. This may be related to the advanced stage of the disease. Also, there was a borderline statistically significant association with FLC assay. On the contrary, there is no statistically significant association with other parameters.
Like our results, Stockwin et al. found no significant association with the age of their patients [30]. However, they found a significant correlation between mTOR expression and male gender in the MM patients (p = 0.04) and Vajpayee et al. found a significant correlation between mTOR expression and male gender in diffuse large B-cell lymphoma (DLBDL) patients (85% vs. 46%, p < 0.01) [46]. However, the former study showed a male to female ratio 1:0.72 and the latter a male to female ratio 1.89:1 and this is different from our studied population ratio which is 1.58:1.
Regarding the response to treatment, there was a highly statistically significant association between the expression of both RGS1 and mTOR with the response to treatment among our patients. This can be explained by the poor prognostic association between RGS1 and different cancer diseases, supporting the proliferation and invasion of cancer cells (11,46,53). Vajpayee et al. studied the mTOR marker among the diffuse large B cell lymphoma patients and revealed that the high mTOR expression showed a trend toward advanced clinical stage than others (78% vs. 54%, p = 0.08) [46].
Also, there was highly statistically significant positive association between RGS1 and mTOR expression in MM patients, and both markers showed inferior prognostic significance in MM. Where, both RGS1 and mTOR showed an inferior overall survival influence in MM, with better survival for those having low marker expression levels. Both RGS1 and mTOR remained an independent prognostic factor of the OS in MM patients after multivariate analysis.
Like our results, RGS1 protein expression was significantly associated with unfavorable prognosis of the patients in DLBCL [51] and in melanoma [44]. Also, similar mTOR results were found in previous studies among DLBCL patients [46,49].
This association and prognostic significance can be explained by sharing some receptors and common signaling pathways by both RGS1 and mTOR, that are involved in the pathogenesis of MM as well, like IGFR1, PIK3/AKT, MAPK [52–54]. So, higher expressions may be associated with hyperactivity in the signaling pathways and myeloma progression as well.
Gβγ, which is activated by RGS1, has been shown to increase the activity of MAPK signaling pathways through PI3K/Akt and PLCβ, as well as through IGFR1 [53]. Similarly, the mTORC is regulated by the IGF, PI3K/Akt/mTOR pathway, the Ras-MAPK pathway, and some other intracellular factors [25,55–57].
Akt, which is commonly found to be hyperactive in cancers, is an important substrate of mTORC [58]. The cytokines secreted in BMM which are contributed to the pathogenesis of MM as IL6, VEGF and IGF activate their respective receptors expressed on MM cells through an important pathway, PI3K/AKT/mTOR, and promote tumor growth as well as resistance development to existing therapies [59,60].
Since RGS1 & mTOR were shown to play a significant role in disease pathogenesis, they may be a new target for immunotherapy in many diseases. RGS1 was shown to be a novel target for inflammatory diseases as Rheumatoid arthritis [61], melanoma [62] and cervical cancer [63].
Regarding mTOR, Rapamycin was the first mTOR inhibitor, and was initially used as an immunosuppressive drug in the field of solid organ transplantation [33]. Moreover, there was a strong rationale for mTOR inhibitors (mTORi) effects in many hematological diseases due to the associated hyperactivation of mTOR, as in acute leukemia, lymphoma, multiple myeloma, Waldenström macroglobulinemia, and GVHD [22,32]. However, preclinical trials are needed to confirm these findings.
Conclusion
RGS1 and mTOR were found to be poor prognostic markers in MM patients, being associated with lower response rate and inferior OS. Further studies should be done on larger population to assess the association with other prognostic markers. From these findings, we could recommend considering RGS1 and mTOR as one of the prognostic criteria in the different risk stratification and staging classifications. Further trials for RGS1 and mTOR targeting in multiple myeloma are recommended.
Limitations
The study enrolled a relatively small number of cases which reduced the capability to identify certain associations.
Supporting information
(DOCX)
(DOCX)
(DOCX)
Tables: • S4 Table (1): The association of RGS1expression and other parameters
• S4 Table (2): The association of mTOR and other parameters
• S4 Table (3): Overall Survival (OS) of the studied patients and its relation to other prognostic factors
• S4 Table (4): Multivariate analysis for OS (cox regression model)
(DOCX)
Acknowledgments
We thank all our colleagues in the National Cancer Institute, Cairo University.
Abbreviations
- ABMT
autologous bone marrow transplant
- AC
adenylyl cyclase
- AUC
area under the curve
- BMM
Bone marrow microenvironment
- CR
complete response
- GPCR
G protein-coupled receptor
- IGF
insulin growth factor
- IGF-1
insulin-like growth factor-1
- IHC
Immunohistochemistry
- LDH
lactate dehydrogenase
- MAPK
mitogen-activated protein kinase
- MM
multiple myeloma
- mTOR
mechanistic target of rapamycin
- NSCLC
non-small cell lung cancer
- PCM
Plasma cell myeloma
- PI3K
phosphoinositide 3- kinase
- PLC
phospholipase C
- Rapalogs
rapamycin and its analogs
- RGS
Regulator of G proteins signaling
- ROC
receiver operating characteristics
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The author(s) received no specific funding for this work.
References
- 1.Zamagni E, Tacchetti P and Cavo M (2018) Imaging in multiple myeloma: How? When?. Blood.; 133:7. [DOI] [PubMed] [Google Scholar]
- 2.Ibrahim A, Khaled H and Mikhail N (2014) Cancer Incidence in Egypt: Results of the National Population-Based Cancer Registry Program. Journal of Cancer Epidemiology. Hindawi Publishing Corporation; p 1–18. doi: 10.1155/2014/437971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Husseiny NM, Kasem N, El Azeeim HA, Mattar MW (2014) Multiple myeloma: a descriptive study of 217 Egyptian patients. Ann Hematol.; doi: 10.1007/s00277-013-1849-3 [DOI] [PubMed] [Google Scholar]
- 4.Smedt ED, Lui H, Maes K, Veirman KD, Menu E, Vanderkerken K, et al. (2018) The Epigenome in Multiple Myeloma: Impact on Tumor Cell Plasticity and Drug Response Oncol. doi: 10.3389/fonc.2018.00566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gilbert LC, Chen H, Lu X, Nanes MS (2013) Chronic low dose tumor necrosis factor-α (TNF) suppresses early bone accrual in young mice by inhibiting osteoblasts without affecting osteoclasts. Bone.; doi: 10.1016/j.bone.2013.06.002 [DOI] [PubMed] [Google Scholar]
- 6.Chong PS, Chng WJ, and de Mel S (2019) STAT3: A Promising Therapeutic Target in Multiple Myeloma. Cancers journal.; doi: 10.3390/cancers11050731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Senese NB, Kandasamy R, Kochan KE, Traynor JR (2020) Regulator of G-Protein Signaling (RGS) Protein Modulation of Opioid Receptor Signaling as a Potential Target for Pain Management. Front. Mol. Neurosci.; doi: 10.3389/fnmol.2020.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Agenès F, Bosco N, Mascarell L, Fritah S, Ceredig R (2005) Differential expression of regulator of G-protein signalling transcripts and in vivo migration of CD4+ naïve and regulatory T cells. Immunology.; doi: 10.1111/j.1365-2567.2005.02146.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kveberg L, Ryan JC, Rolstad B, Inngjerdingen M (2005) Expression of regulator of G protein signaling proteins in natural killer cells, and their modulation by Ly49A and Ly49D. Immunology.; doi: 10.1111/j.1365-2567.2005.02174.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y, Yan H, Frost P, Gera J, Lichtenstein A (2005) Mammalian target of rapamycin inhibitors activate the AKT kinase in multiple myeloma cells by up-regulating the insulin-like growth factor receptor/insulin receptor substrate-1/phosphatidylinositol 3-kinase cascade. Mol Cancer Ther.; doi: 10.1158/1535-7163.MCT-05-0068 [DOI] [PubMed] [Google Scholar]
- 11.Roh J, Shin SJ, Lee AN, Yoon DH, Suh C, Park CJ, et al. (2017) RGS1 expression is associated with poor prognosis in multiple myeloma. Journal of Clinical Pathology.; doi: 10.1136/jclinpath-2016-203713 [DOI] [PubMed] [Google Scholar]
- 12.Hurst JH and Hooks SB (2009) Regulator of G-protein signaling (RGS) proteins in cancer biology. Biochem Pharmacol.; doi: 10.1016/j.bcp.2009.06.028 [DOI] [PubMed] [Google Scholar]
- 13.Tang W, Tu Y, Nayak SK, Woodson J, Jehl M, Ross EM (2006) Gbetagamma inhibits Galpha GTPase-activating proteins by inhibition of Galpha-GTP binding during stimulation by receptor. J Biol Chem.; doi: 10.1074/jbc.M510573200 [DOI] [PubMed] [Google Scholar]
- 14.Senarath K, Payton JL, Kankanamge D, Siripurapu P, Tennakoon M, Karunarathne A (2018) Gγ identity dictates efficacy of Gβγ signaling and macrophage migration. J Biol Chem.; doi: 10.1074/jbc.RA117.000872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gulati S, Jin H, Masuho I, Orban T, Cai Y, Pardon E, et al. (2018) Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat Commun.; doi: 10.1038/s41467-018-04432-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Urnukhsaikhan E, Cho H, Mishig-Ochir T, Seo YK, Park JK (2016) Pulsed electromagnetic fields promote survival and neuronal differentiation of human BM-MSCs. Life Sci.; doi: 10.1016/j.lfs.2016.02.066 [DOI] [PubMed] [Google Scholar]
- 17.Gourraud PA (2011) International Multiple Sclerosis Genetics Consortium (IMSGC). When is the absence of evidence, evidence of absence? Use of equivalence-based analyses in genetic epidemiology and a conclusion for the KIF1B rs10492972*C allelic association in multiple sclerosis. Genet Epidemiol.; doi: 10.1002/gepi.20592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orlando A, Renna S, Perricone G, Cottone M (2009) Gastrointestinal lesions associated with spondyloarthropathies. World J Gastroenterol.; doi: 10.3748/wjg.15.2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtin JA, Fridlyand J, Kageshita T, Patel HN, Busam KJ, Kutzner H, et al. (2005) Distinct sets of genetic alterations in melanoma. N Engl J Med.; doi: 10.1056/NEJMoa050092 [DOI] [PubMed] [Google Scholar]
- 20.Maiese K (2014) Cutting through the complexities of mTOR for the treatment of stroke. Curr Neurovasc Res.; doi: 10.2174/1567202611666140408104831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe R, Wei L, Huang J (2011) mTOR signaling, function, novel inhibitors, and therapeutic targets. J Nucl Med.; doi: 10.2967/jnumed.111.089623 [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Chen X, Cassady K, Zou Z, Yang S, Wang Z, et al. (2021) The Role of mTOR Inhibitors in Hematologic Disease: From Bench to Bedside. Front Oncol.; doi: 10.3389/fonc.2020.611690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tian T, Li X, Zhang J (2019) mTOR Signaling in Cancer and mTOR Inhibitors in Solid Tumor Targeting Therapy. Int J Mol Sci.; doi: 10.3390/ijms20030755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D (2012) PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Curr Oncol Rep.; doi: 10.1007/s11912-012-0227-y [DOI] [PubMed] [Google Scholar]
- 25.Avruch J, Hara K, Lin Y, Liu M, Long X, Ortiz-Vega S, et al. (2006) Insulin and amino-acid regulation of mTOR signaling and kinase activity through the Rheb GTPase. Oncogene.; doi: 10.1038/sj.onc.1209882 [DOI] [PubMed] [Google Scholar]
- 26.Mossmann D, Park S, Hall MN (2018) mTOR signalling and cellular metabolism are mutual determinants in cancer. Nat Rev Cancer.; doi: 10.1038/s41568-018-0074-8 [DOI] [PubMed] [Google Scholar]
- 27.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. (2014) A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov.; doi: 10.1158/2159-8290.CD-13-0929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Balko JM, Giltnane JM, Wang K, Schwarz LJ, Young CD, Cook RS, et al. (2014) Molecular profiling of the residual disease of triple-negative breast cancers after neoadjuvant chemotherapy identifies actionable therapeutic targets. Cancer Discov.; doi: 10.1158/2159-8290.CD-13-0286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koldehoff M, Beelen DW, Elmaagacli AH (2014) Inhibition of mTOR with everolimus and silencing by vascular endothelial cell growth factor-specific siRNA induces synergistic antitumor activity in multiple myeloma cells. Cancer Gene Ther.; doi: 10.1038/cgt.2014.27 [DOI] [PubMed] [Google Scholar]
- 30.Stockwin W, Johnson P, Vajpayee N (2016) Immunohistochemical Expression of mTOR in Multiple Myeloma: Retrospective Analysis of 31 Cases, a Clinicopathological Study. Ann Clin Lab Sci.; 46:2. [PubMed] [Google Scholar]
- 31.Laplante M, Sabatini DM (2012) mTOR signaling in growth control and disease. Cell. 13;149(2):274–93. doi: 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin HG, Wu GZ, Wu GH, Bao YG (2018) Combining the mammalian target of rapamycin inhibitor, rapamycin, with resveratrol has a synergistic effect in multiple myeloma. Oncol Lett.; doi: 10.3892/ol.2018.8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelchtermans J, Chang J, Glaberson W, DeFreitas M, Alba-Sandoval M, Chandar J (2020) A Pediatric Case of Sirolimus-Associated Pneumonitis After Kidney Transplantation. J Pediatr Pharmacol Ther.; doi: 10.5863/1551-6776-25.5.459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, et al. (2016) Sestrin2 is a leucine sensor for the mTORC1 pathway. Science.; doi: 10.1126/science.aab2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bergstrom DJ, Kotb R, Louzada ML, Sutherland HJ, Tavoularis S, Venner CP (2020) Myeloma Canada Research Network Consensus Guideline Consortium. Consensus Guidelines on the Diagnosis of Multiple Myeloma and Related Disorders: Recommendations of the Myeloma Canada Research Network Consensus Guideline Consortium. Clin Lymphoma Myeloma Leuk.; doi: 10.1016/j.clml.2020.01.017 [DOI] [PubMed] [Google Scholar]
- 36.Perrot A, Lauwers-Cances V, Tournay E, Hulin C, Chretien ML, Royer B, et al. (2019) Development and Validation of a Cytogenetic Prognostic Index Predicting Survival in Multiple Myeloma. J Clin Oncol.; doi: 10.1200/JCO.18.00776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Kwok-Shing Ng P, Kucherlapati M, Chen F, Liu Y, Tsang YH, et al. (2017) A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell.; doi: 10.1016/j.ccell.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar SK, Rajkumar SV, Dispenzieri A, Lacy MQ, Hayman SR, Buadi FK, et al. (2008) Improved survival in multiple myeloma and the impact of novel therapies. Blood.; doi: 10.1182/blood-2007-10-116129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerecke C, Fuhrmann S, Strifler S, Schmidt-Hieber M, Einsele H, Knop S. The Diagnosis and Treatment of Multiple Myeloma. Dtsch Arztebl Int. 2016; doi: 10.3238/arztebl.2016.0470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Naymagon L and Abdul-Hay M (2016) Novel agents in the treatment of multiple myeloma: a review about the future. J Hematol Oncol.; doi: 10.1186/s13045-016-0282-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moreau P, San Miguel J, Sonneveld P, Mateos MV, Zamagni E, Avet-Loiseau H, et al. (2017) ESMO Guidelines Committee. Multiple myeloma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol.; doi: 10.1093/annonc/mdx096 [DOI] [PubMed] [Google Scholar]
- 42.Rajkumar SV (2016) Updated Diagnostic Criteria and Staging System for Multiple Myeloma. Am Soc Clin Oncol Educ Book.; doi: 10.1200/EDBK_159009 [DOI] [PubMed] [Google Scholar]
- 43.Spiegelberg BD, Hamm HE (2007) Roles of G-protein-coupled receptor signaling in cancer biology and gene transcription. Curr Opin Genet Dev.; doi: 10.1016/j.gde.2006.12.002 [DOI] [PubMed] [Google Scholar]
- 44.Rangel J, Nosrati M, Leong SP, Haqq C, Miller JR 3rd, Sagebiel RW, et al. (2008) Novel role for RGS1 in melanoma progression. Am J Surg Pathol.; doi: 10.1097/PAS.0b013e31816fd53c [DOI] [PubMed] [Google Scholar]
- 45.Croonquist PA, Linden MA, Zhao F, Van Ness BG (2003) Gene profiling of a myeloma cell line reveals similarities and unique signatures among IL-6 response, N-ras-activating mutations, and coculture with bone marrow stromal cells. Blood.; doi: 10.1182/blood-2003-04-1227 [DOI] [PubMed] [Google Scholar]
- 46.Vajpayee N, Thakral C, Gopaluni S, Newman N, Gajra A (2012) Activation of mammalian target of rapamycin in diffuse large B-cell lymphoma: a clinicopathological study. Leuk Res.; doi: 10.1016/j.leukres.2012.07.016 [DOI] [PubMed] [Google Scholar]
- 47.Gulati S, Jin H, Masuho I, Orban T, Cai Y, Pardon E, et al. (2018) Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat Commun.; doi: 10.1038/s41467-018-04432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng H, Zou Y, Ross JS, Wang K, Liu X, Halmos B, et al. (2015) RICTOR Amplification Defines a Novel Subset of Patients with Lung Cancer Who May Benefit from Treatment with mTORC1/2 Inhibitors. Cancer Discov.; doi: 10.1158/2159-8290.CD-14-0971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sebestyén A, Sticz TB, Márk A, Hajdu M, Timár B, Nemes K, et al. (2012) Activity and complexes of mTOR in diffuse large B-cell lymphomas—a tissue microarray study. Mod Pathol.; doi: 10.1038/modpathol.2012.141 [DOI] [PubMed] [Google Scholar]
- 50.Schedel F, Pries R, Thode B, Wollmann B, Wulff S, Jocham D, et al. (2011) mTOR inhibitors show promising in vitro activity in bladder cancer and head and neck squamous cell carcinoma. Oncol Rep.; doi: 10.3892/or.2011.1146 [DOI] [PubMed] [Google Scholar]
- 51.Carreras J, Kikuti YY, Beà S, Miyaoka M, Hiraiwa S, Ikoma H, et al. (2017) Clinicopathological characteristics and genomic profile of primary sinonasal tract diffuse large B cell lymphoma (DLBCL) reveals gain at 1q31 and RGS1 encoding protein; high RGS1 immunohistochemical expression associates with poor overall survival in DLBCL not otherwise specified (NOS). Histopathology.; doi: 10.1111/his.13106 [DOI] [PubMed] [Google Scholar]
- 52.Bieghs L, Johnsen HE, Maes K, Menu E, Van Valckenborgh E, Overgaard MT, et al. (2016) The insulin-like growth factor system in multiple myeloma: diagnostic and therapeutic potential. Oncotarget.; doi: 10.18632/oncotarget.8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gulati S, Jin H, Masuho I, Orban T, Cai Y, Pardon E, et al. (2018) Targeting G protein-coupled receptor signaling at the G protein level with a selective nanobody inhibitor. Nat Commun.; doi: 10.1038/s41467-018-04432-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Urnukhsaikhan E, Cho H, Mishig-Ochir T, Seo YK, Park JK (2016) Pulsed electromagnetic fields promote survival and neuronal differentiation of human BM-MSCs. Life Sci.; doi: 10.1016/j.lfs.2016.02.066 [DOI] [PubMed] [Google Scholar]
- 55.Willems L, Tamburini J, Chapuis N, Lacombe C, Mayeux P, Bouscary D (2012) PI3K and mTOR signaling pathways in cancer: new data on targeted therapies. Curr Oncol Rep.; doi: 10.1007/s11912-012-0227-y [DOI] [PubMed] [Google Scholar]
- 56.Rad E, Murray JT, Tee AR (2018) Oncogenic Signaling through Mechanistic Target of Rapamycin (mTOR): A Driver of Metabolic Transformation and Cancer Progression. Cancers (Basel).; doi: 10.3390/cancers10010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu ZZ, Xia ZG, Wang AH, Wang WF, Liu ZY, Chen LY, et al. (2013) Activation of the PI3K/AKT/mTOR pathway in diffuse large B cell lymphoma: clinical significance and inhibitory effect of rituximab. Ann Hematol.; doi: 10.1007/s00277-013-1770-9 [DOI] [PubMed] [Google Scholar]
- 58.Inoki K, Li Y, Zhu T, Wu J, Guan KL (2002) TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol.; doi: 10.1038/ncb839 [DOI] [PubMed] [Google Scholar]
- 59.Ramakrishnan V, Kumar S (2018) PI3K/AKT/mTOR pathway in multiple myeloma: from basic biology to clinical promise. Leuk Lymphoma.; doi: 10.1080/10428194.2017.1421760 [DOI] [PubMed] [Google Scholar]
- 60.Di Marzo L, Desantis V, Solimando AG, Ruggieri S, Annese T, Nico B, et al. (2016) Microenvironment drug resistance in multiple myeloma: emerging new players. Oncotarget.; doi: 10.18632/oncotarget.10849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu X, Tang J, Zeng G, Hu X, Bao P, Wu J, et al. (2019) RGS1 silencing inhibits the inflammatory response and angiogenesis in rheumatoid arthritis rats through the inactivation of Toll-like receptor signaling pathway. J Cell Physiol. Nov;234(11):20432–20442. doi: 10.1002/jcp.28645 Epub 2019 Apr 22. . [DOI] [PubMed] [Google Scholar]
- 62.Kashani-Sabet M, Nosrati M, Miller JR 3rd, Sagebiel RW, Leong SPL, Lesniak A, et al. (2017) Prospective validation of molecular prognostic markers in cutaneous melanoma: a correlative analysis of E1690. Clin Cancer Res.;23:6888–92. doi: 10.1158/1078-0432.CCR-17-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang S, Wang H, Liu J, Tao T, Zeng Z, Wang M (2022) RGS1 and related genes as potential targets for immunotherapy in cervical cancer: computational biology and experimental validation. J Transl Med. Jul 25;20(1):334. doi: 10.1186/s12967-022-03526-0 ; PMCID: PMC9310486. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
(DOCX)
Tables: • S4 Table (1): The association of RGS1expression and other parameters
• S4 Table (2): The association of mTOR and other parameters
• S4 Table (3): Overall Survival (OS) of the studied patients and its relation to other prognostic factors
• S4 Table (4): Multivariate analysis for OS (cox regression model)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.