Abstract
OBJECTIVE
Advancements in MRI technology have provided improved ways to acquire imaging data and to more seamlessly incorporate MRI into modern pediatric surgical practice. One such situation is image-guided navigation for pediatric neurosurgical procedures, including intracranial catheter placement. Image-guided surgery (IGS) requires acquisition of CT or MR images, but the former carries the risk of ionizing radiation and the latter is associated with long scan times and often requires pediatric patients to be sedated. The objective of this project was to circumvent the use of CT and standard-sequence MRI in ventricular neuronavigation by investigating the use of fast MR sequences on the basis of 3 criteria: scan duration comparable to that of CT acquisition, visualization of ventricular morphology, and image registration with surface renderings comparable to standard of care. The aim of this work was to report image development, implementation, and results of registration accuracy testing in healthy subjects.
METHODS
The authors formulated 11 candidate MR sequences on the basis of the standard IGS protocol, and various scan parameters were modified, such as k-space readout direction, partial k-space acquisition, sparse sampling of k-space (i.e., compressed sensing), in-plane spatial resolution, and slice thickness. To evaluate registration accuracy, the authors calculated target registration error (TRE). A candidate sequence was selected for further evaluation in 10 healthy subjects.
RESULTS
The authors identified a candidate imaging protocol, termed presurgical imaging with compressed sensing for time optimization (PICO). Acquisition of the PICO protocol takes 25 seconds. The authors demonstrated noninferior TRE for PICO (3.00 ± 0.19 mm) in comparison with the default MRI neuronavigation protocol (3.35 ± 0.20 mm, p = 0.20).
CONCLUSIONS
The developed and tested sequence of this work allowed accurate intraoperative image registration and provided sufficient parenchymal contrast for visualization of ventricular anatomy. Further investigations will evaluate use of the PICO protocol as a substitute for CT and conventional MRI protocols in ventricular neuronavigation. https://thejns.org/doi/abs/10.3171/2022.5.JNS22767
Keywords: neuronavigation, ventricular shunt, MRI, CT, image-guided surgery, surgical technique, ultrafast, roentogenic risk, radiation
IMAGE-GUIDED surgery (IGS) allows the operator to match a specific anatomical location on a patient with a corresponding location on medical imaging. IGS is common in neurosurgery because appropriate assessment of position can be vital for successful patient outcomes. Use of IGS in ventricular shunt placement can increase surgical accuracy and reduce the number of cannulation attempts compared with freehand placement,1–5 as well as decrease the shunt failure rate6,7 and improve time until catheter revision.7–9
IGS requires high-resolution CT or MRI. IGS matches locations on the patient to locations on the images by using renderings of the surface of the patient’s scalp. Thus, high-quality image rendering at the skin surface is essential for IGS. However, diagnostic imaging quality of the parenchyma is not as vital for IGS in the setting of shunt placement because imaging is used only to define the catheter entry site and tip destination.
In this work, we developed a fast sequence with noninferior surface registration specifically for ventricular neuronavigation. We tested 13 separate MRI protocols on a single individual, and then we selected 1 protocol (designated presurgical imaging with compressed sensing for time optimization [PICO]) for accuracy and reproducibility testing in 10 participants. We have provided the statistical and methodological framework to assess scan performance, as well as the means to acquire and validate our images (Supplemental Materials), in order to allow other institutions to deploy PICO imaging and validate whether its accuracy and reproducibility are acceptable for use at their site.
Methods
This study was approved by the institutional review board at the University of Southern California/Children’s Hospital Los Angeles (CHLA). All volunteers were personal acquaintances of the investigators, who were recruited and provided consent on the basis of good clinical research practices, including provision of written informed consent before participation in this study.
Protocol Development and MRI Acquisition
MRI scans were obtained with a 3-T clinical MR scanner (Philips Achieva) at CHLA. The default MRI protocol used by our institution for preoperative neuronavigation images was designated sequence 1 (S1) and consisted of an approximately 5-minute T1-weighted turbo field echo (TFE) sequence, including TR/TE 7/3.5 msec, flip angle (FA) 8°, matrix 252 × 217, in-plane resolution 1 × 1 mm, and slice thickness 1 mm. A summary of all evaluated sequence parameters is included (Table 1), and the acquisition times for standard diagnostic T1-weighted and T2-weighted scans are supplied in Supplemental Materials.
TABLE 1.
MRI scan sequence parameters
| Sequence | Voxel Dimensions (mm) | Time (mins:sec) | TR (msec) | TE (msec) | FA (°) | Readout | Matrix | CS Factor | Half-Scan | Successful Registration |
|---|---|---|---|---|---|---|---|---|---|---|
| S1 | 1.00 × 1.00 × 1.00 | 5:06 | 7 | 3.5 | 8 | T1-weighted TFE | 252 × 217 | No | No | Yes |
| S2 | 2.98 × 2.98 × 3.00 | 1:42 | 7 | 3.5 | 8 | T1-weighted TFE | 84 × 72 | No | No | Yes |
| S3 | 1.25 × 1.25 × 5.00 | 4:09 | 7 | 3.5 | 8 | T1-weighted TFE | 200 × 177 | No | No | Yes |
| S4 | 1.00 × 1.00 × 1.00 | 2:29 | 7 | 3.5 | 8 | T1-weighted TFE | 252 × 217 | 3 | No | Yes |
| S5* | 2.98 × 2.98 × 3.00 | 0:25 | 7 | 3.5 | 8 | T1-weighted TFE | 84 × 72 | 3 | No | Yes |
| S6 | 2.98 × 2.98 × 3.00 | 1:02 | 7 | 3.5 | 8 | T1-weighted TFE | 84 × 72 | No | 0.6 | Yes |
| S7 | 1.25 × 1.25 × 1.00 | 1:30 | 20 | 6.6 | 15 | T1-weighted TFE | 204 × 180 | No | No | Yes |
| S8 | 1.00 × 1.00 × 5.00 | 1:28 | 20 | 6.6 | 15 | T1-weighted TFE | 200 × 175 | No | No | Yes |
| S9 | 1.24 × 1.25 × 3.00 | 0:46 | 20 | 6.6 | 15 | T1-weighted TFE | 208 × 179 | 3 | No | Yes |
| S10 | 0.99 × 1.00 × 5.00 | 1:01 | 20 | 6.6 | 15 | T1-weighted TFE | 264 × 235 | 3 | No | Yes |
| S11 | 1.25 × 1.25 × 3.00 | 0:22 | 20 | 1.57 | 15 | Spiral | 200 × 200 | No | No | Yes |
| S12 | 1.25 × 1.25 × 3.00 | 1:30 | 20 | 6.6 | 15 | T1-weighted TFE | 208 × 180 | No | No | No |
| S13 | 1.25 × 1.25 × 3.00 | 0:55 | 20 | 6.6 | 15 | T1-weighted TFE | 208 × 180 | No | 0.6 | No |
CS = compressed sense.
This row shows the characteristics of PICO.
The aims of parameter refinement were to reduce the acquisition time and to maintain a high contrast-to-noise ratio (CNR) and signal-to-noise ratio (SNR) on the skin surface. Moderate reductions in image resolution and SNR/CNR in the brain parenchyma itself were considered acceptable trade-offs. A single subject was scanned with all 13 image sequences to evaluate which were suitable for further evaluation in additional volunteers. We acquired baseline (S1) and improved (S5) images from 10 healthy volunteers (age range 25–38 years; 8 males and 2 females), as well as from a pediatric patient (12 years old) to demonstrate proof of concept.
IGS Registration
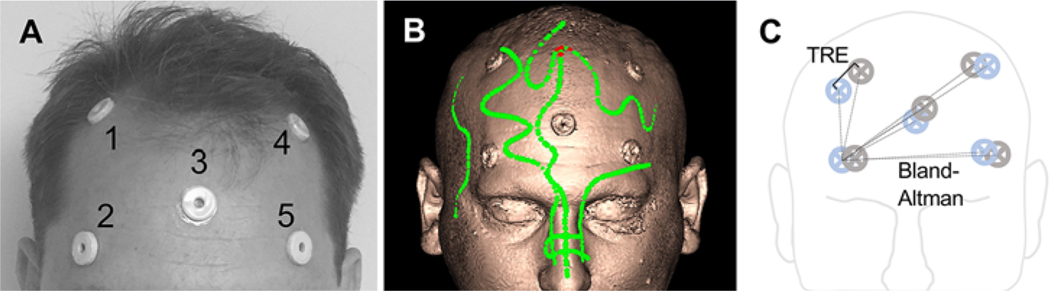
We placed 5 self-adhesive fiducial markers on each participant before MRI (Fig. 1A). During IGS registration (StealthStation S7 surgical navigation system, Medtronic), we affixed a noninvasive electromagnetic reference tracker (AxiEM, Medtronic) to the forehead at a location above the left eyebrow at the midpupillary line. The participant was supine, with their head positioned neutral on a donut headrest. The surgeon performing registration was blinded to the scan sequence and performed standard surface matching by using points from roughly similar locations on all scans. The surgeon avoided the fiducial markers during surface mapping because the markers would later be used to evaluate registration (Fig. 1C). The surgeon repeated the registration process up to 3 times to evaluate the reliability of registration. The sequence was deemed unreliable if a sequence failed to register during at least 2 of these attempts.
FIG. 1. Quantification of registration accuracy.
A: The locations of the 5 target markers applied to the head of a human subject. B: Representative surface rendering from the baseline S1 sequence obtained with neuronavigation software. Green points represent data from surface-mapping registration. The fiducial markers are seen on the image. C: A schematic of the target markers in image space (blue) and real space (gray). TRE (solid line) is shown as the 3D distance between image−real space fiducial center pairs (performed for all marker pairs). Bland-Altman analysis was used to compare the differences in distance from a single image space marker to each adjacent marker pair (dashed lines). Bland-Altman analysis was performed over all image space points of origin. Figure is available in color online only.
Digitizing the Real Space and Image Space Locations of the Fiducial Markers
There were 2 sets of locations, one in imaging space and the other in real space. To assess the real space location of each fiducial marker, we placed the handheld tracer probe on the center of each fiducial marker on the patient and collected the corresponding digitized coordinates of the probe location from the StealthStation system. To assess the image space location of each fiducial marker, we placed the digital cursor on the center of each fiducial marker on the images and collected the corresponding coordinates of the cursor location from the imaging system.
Statistical Analysis of Fiducial Marker Locations
We used target registration error (TRE), root mean square error (RMSE), and Bland-Altman analysis to assess the accuracy and precision of imaging sequence registration. TRE is the 3D Euclidian distance between a target marker in real space and its equivalent marker in imaging space (Fig. 1C), where the ideal value would be 0. Mean TRE and standard error of the mean were calculated using all fiducial reference points. To identify protocols with suitable accuracy compared with the control (S1) protocol, we compared the data from S1 and all other scans by using 1-way analysis of variance with Dunnett post hoc analysis. We used the unpaired 2-tailed t-test (p < 0.05) to compare the mean TRE values of the control (S1) and PICO (S5) scans. We also calculated the RMSE for each participant as a descriptive statistic of image accuracy.
We assessed differences in the long distances across the scalp with Bland-Altman analysis. To perform Bland-Altman analysis, the location was obtained from a single fiducial in real space, and the differences in distances to neighboring fiducials were calculated in real and image space. Supplemental Materials include details on the TRE, RMSE, and Bland-Altman analyses, as well as a spreadsheet to enable others to perform these calculations and assess scan performance. Table 1 contains the defined parameters for PICO imaging. These parameters are not proprietary and can be deployed on most modern 3-T MR scanners.
Results
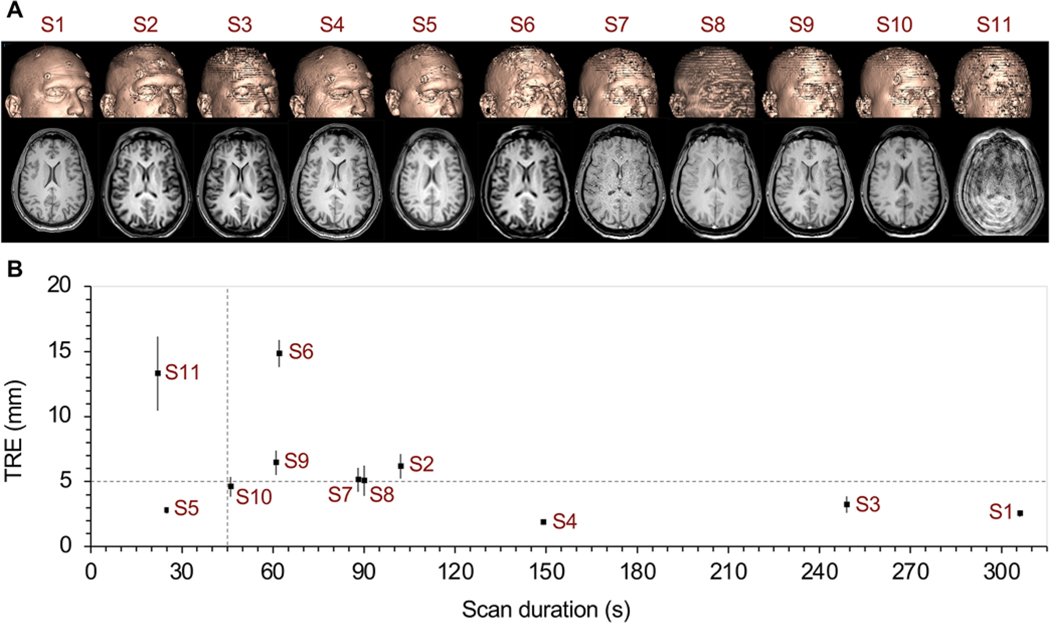
Figure 2A shows 3D surface renderings and corresponding axial cross-sectional images of the experimental sequences obtained for parameter optimization (S1–S11). All scans obtained during the parameter refinement phase (Fig. 2) were tested on the same individual with identical fiducial positions. Scan sequences S12 and S13 were not included due to their inability to yield successful registration on the neuronavigation platform.
FIG. 2. Initial protocol screen.
A: 3D surface rendering and axial images at the level of the lateral ventricles, displayed for all successfully registered sequences (S1–S11) in a single individual. B: TRE plotted as a function of scan duration. Dashed lines are included as references. The horizontal line at TRE of 5 mm represents twice the TRE value of the baseline S1 scan (2.5 mm). The vertical line at 45 seconds was the goal for scan duration. Figure is available in color online only.
Qualitative visual inspection of the reconstructed skin surfaces (Fig. 2A) revealed that sequences S1, S2, S4, and S5 demonstrated reasonable surface smoothness; sequences S3, S6, S7, and S11 showed relatively poor surface SNR; and sequences S8, S9, and S10 had sufficient surface SNR but an uneven surface appearance due to thicker slices. All scans except one retained sufficient SNR and CNR in the parenchyma for discrimination of ventricular boundaries. The exception was S11, where spiral acquisition introduced an artifact and degraded the scan quality.
Quantitative Comparison
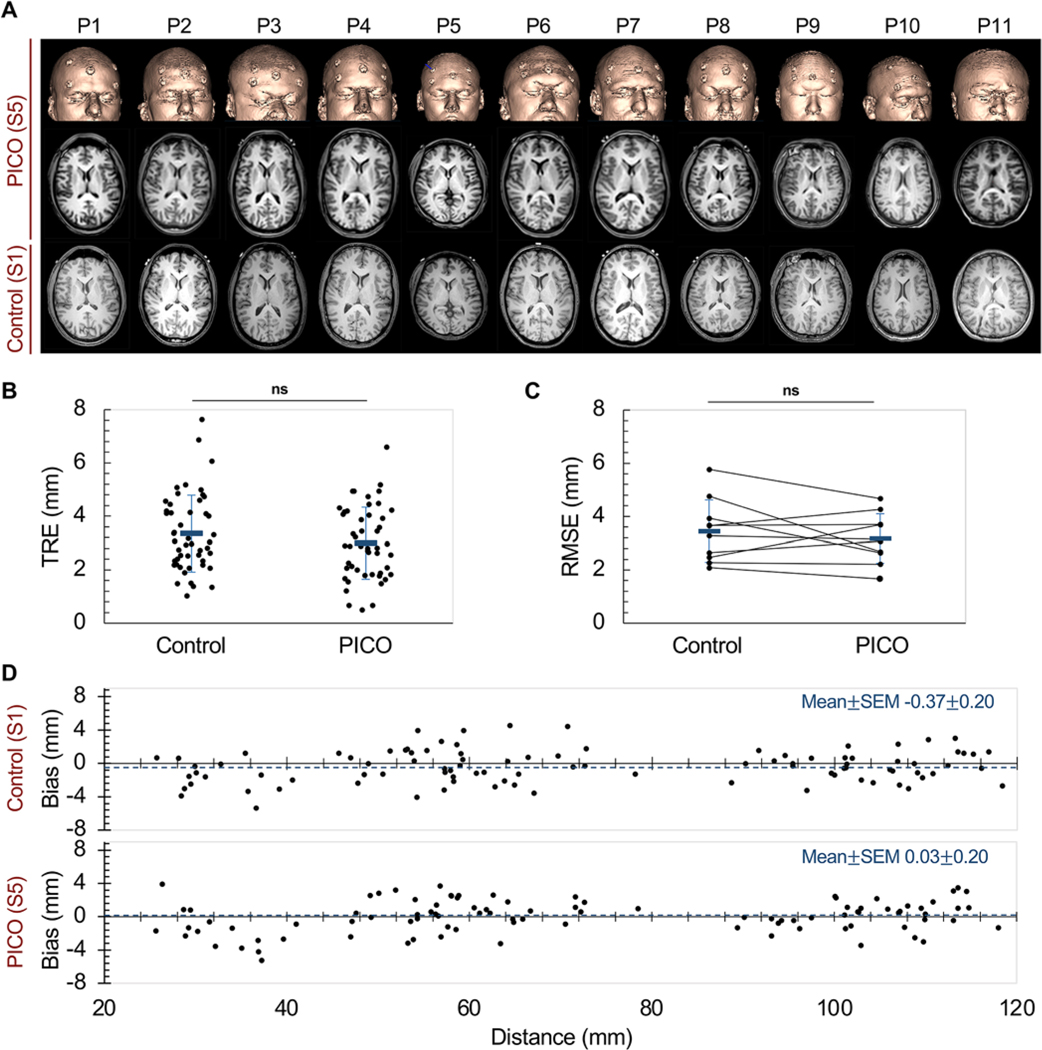
We imaged our 11 participants with the baseline neuronavigation scan (S1) and PICO (S5). Figure 3A shows StealthStation-generated PICO surface renderings, as well as corresponding axial PICO cross-sectional images at the level of the foramen of Monro and baseline neuronavigation sequence (S1) cross-sectional images at an equivalent plane.
FIG. 3. Implementation of PICO in multiple participants.
A: 3D surface rendering and axial images over the lateral ventricles obtained with PICO (S5) for each of the 10 participants (P1–P10) and 1 pediatric patient (P11), displayed alongside the corresponding S1 axial images. B: TRE was independently calculated for each fiducial for participants 1–10 (black dots). Random jitter was used in the x-dimension to separate values for visual identification. Mean ± SD TRE is shown in blue. C: RMSE values, averaged over all fiducials for each participant, are shown. Black lines connect data from individual participants. Mean ± SD is RMSE shown in blue. D: Bland-Altman analysis comparing bias between S1 and S5. Bias (i.e., differences in the distances between pairs consisting of an image space fiducial and a neighboring real space−image space fiducial) was plotted against the corresponding image–image space distance. The horizontal line (blue) indicates mean bias. Figure is available in color online only.
We calculated and compared TRE values for S1 and PICO images from participants 1 to 10 (Fig. 3B). The mean TRE values (Fig. 3B) were 3.35 ± 0.20 mm and 3.00 ± 0.19 mm for S1 and PICO, respectively (p = 0.20). The mean RMSE values were 3.45 ± 0.37 mm and 3.17 ± 0.29 mm for S1 and PICO, respectively (p = 0.56) (Fig. 3C). The Bland-Altman analysis of S1 and PICO demonstrated comparable biases of −0.37 ± 0.21 mm and 0.03 ± 0.20 mm, respectively (p = 0.80). Thus, PICO was noninferior to S1 in terms of all 3 metrics.
Qualitative Comparison
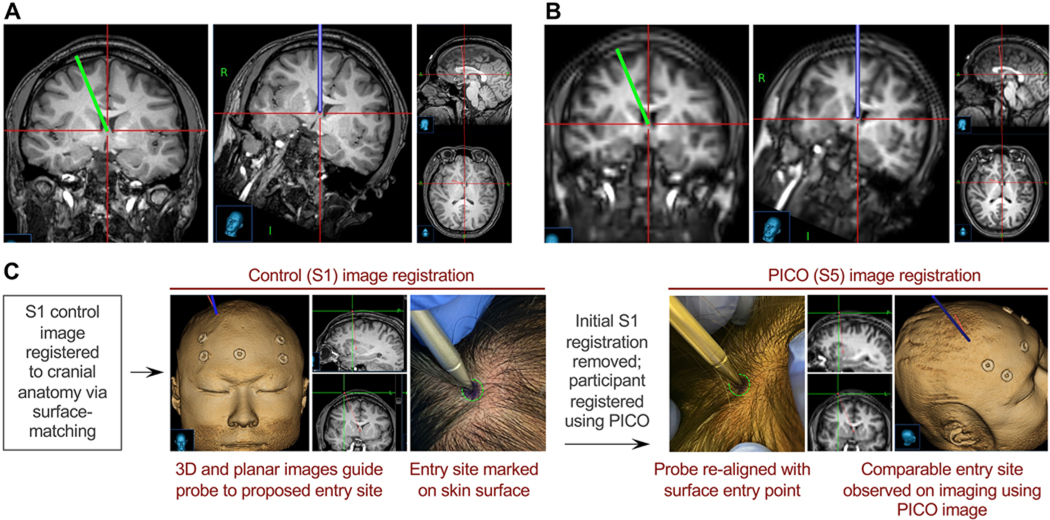
We reviewed all S1 and PICO images for image warping or distortion that may lead to malpositioning of a ventricular catheter. The coregistered images revealed equivalent trajectories in all cases, with representative images shown in Fig. 4. A standard frontal-approach trajectory from Kocher’s point was planned with baseline S1 imaging and then transferred to the coregistered PICO sequence without any changes to the trajectory position or angle. Inspection revealed the same clinically appropriate catheter plan.
FIG. 4. Qualitative assessment of catheter trajectory using PICO.
A: A standard frontal-approach ventricular catheter trajectory was planned with baseline S1 neuronavigation imaging and displayed via coronal and trajectory-approach views. Sagittal and axial images of the target site at the foramen of Monro are shown. B: The PICO image coregistered to S1 was used to display the trajectory planned with S1. Coronal and trajectory-approach views, as well as sagittal and axial images of the target, can be used to visually assess the suitability of the planned trajectory. C: Schematic of a qualitative experiment used to demonstrate correspondence between the appropriate scalp entry points, as planned with surface-registered S1 or PICO. Figure is available in color online only.
We also qualitatively assessed catheter planning, wherein we registered the S1 images, planned a catheter trajectory, and marked the skin of the participant. The participant was reregistered by using the PICO sequence, the probe was held to the ink mark, and the trajectory was planned. The 2 sets of tip coordinates were comparable (difference < 0.5 mm) (Fig. 4C).
Discussion
Advancements in MRI hardware have improved the contrast, signal resolution, and speed of MR acquisition. We have presented a new imaging sequence that can be obtained in the same amount of time as CT and provides accuracy equivalent to gold-standard MRI. This work focused on MR sequences that were modified for ventricular catheter neuronavigation, with optimization of existing technology for a clinical need and a rigorous quantitative testing. This framework can be reused as future imaging technologies become available.
The product of this optimization is the PICO images, which retain the qualities needed by neurosurgeons: accurate location information and adequate image quality to delineate ventricular morphology and to guide catheter placement to a target within these fluid spaces. The potential benefits to be gained from widespread implementation of accelerated imaging for neuronavigation include reduced delivery of unnecessary radiation, improved healthcare efficiency in the scanning environment, and avoidance of unnecessary procedural sedation. Instead of working to improve CT and minimize radiation to as low as reasonably achievable (i.e., ALARA), we focused on accelerating the acquisition of MRI without degrading image quality in the setting of IGS for shunt placement. Although our primary objective was improved pediatric imaging, approximately 25% of adults have claustrophobia associated with MRI.10 The faster imaging protocol that we have proposed also limits the time adult patients spend while uncomfortable in the scanner. Thus, both children and adults may benefit from our approach.
The measured TRE values for S1 and PICO (3.35 ± 0.20 and 3.00 ± 0.19 mm, respectively) are not statistically different from each other. Moreover, our TRE values are equivalent to the accepted values for registration accuracy reported in the literature (0.7–6 mm,11 5.35 ± 1.64 mm,12 and 1.9–4.6 mm), depending on the combination of registration methods (e.g., fiducial, surface, implanted screws) and imaging platforms (MRI, CT).
The present study included only a demonstration of frontal catheter trajectories, with participants registered in a supine position. Follow-up work should assess the ability of PICO to provide accurate placement of both frontal and posterior catheter trajectories. Due to the use of modified parameters, we do not anticipate PICO to be particularly more motion sensitive than the baseline S1 approach. We acquired the PICO scan without motion artifact in a single pediatric patient, and although further validation of registration accuracy is ongoing in this population, this patient serves as a successful proof of concept. In practice, patients are generally capable of holding still for brief periods, and thus rapid imaging modalities such as PICO can be acquired during brief periods of compliance.
This is the first implementation and multisubject assessment of a scan protocol adjusted for specific variables of interest for ventricular neuronavigation. The PICO protocol may be refined over time, as this demonstration may prompt the development of complementary or superior protocols. This work and discussion highlight the impact of surgical application–specific protocol/sequence design with available modern MRI technology.
Conclusions
We have presented PICO, the result of refinement of standard T1-weighted MRI parameters, with moderately reduced in-plane resolution and compressed sensing for k-space acquisition. The result was fast 25-second image acquisition compatible with standard neuronavigation platforms, with confirmed noninferior registration accuracy in 10 separate subjects and moderate reduction in parenchymal detail that is not expected to have a significant clinical impact (parenchymal detail was superior to CT). This protocol refinement can be adopted/implemented at most modern institutions with a 3-T clinical scanner.
Supplementary Material
Acknowledgments
We thank the Norris Comprehensive Cancer Center in Los Angeles, CHLA, Concern Foundation for Cancer Research, and Tri Delta. This research was conducted through the University of Southern California/CHLA Summer Oncology Research Fellowship Program and was supported in part by a National Cancer Institute R25 grant (no. CA225513), the Norris Comprehensive Cancer Center in Los Angeles, CHLA, Concern Foundation for Cancer Research, and Tri Delta. Additional funding support was provided by CHLA Core Pilot Program (PID 000013525).
ABBREVIATIONS
- CHLA
Children’s Hospital Los Angeles
- CNR
contrast-to-noise ratio
- FA
flip angle
- IGS
image-guided surgery
- PICO
presurgical imaging with compressed sensing for time optimization
- RMSE
root mean square error
- SNR
signal-to-noise ratio
- TFE
turbo field echo
- TRE
target registration error
Footnotes
Disclosures
The authors report no conflict of interest concerning the materials or methods used in this study or the findings specified in this paper.
Supplemental Information
Online-Only Content
Supplemental material is available with the online version of the article.
Supplemental Materials. https://thejns.org/doi/suppl/10.3171/2022.5.JNS22767.
References
- 1.Hermann EJ, Capelle HH, Tschan CA, Krauss JK. Electromagnetic-guided neuronavigation for safe placement of intraventricular catheters in pediatric neurosurgery. J Neurosurg Pediatr. 2012; 10(4): 327–333. [DOI] [PubMed] [Google Scholar]
- 2.Levitt MR, O’Neill BR, Ishak GE, et al. Image-guided cerebrospinal fluid shunting in children: catheter accuracy and shunt survival. J Neurosurg Pediatr. 2012; 10(2): 112–117. [DOI] [PubMed] [Google Scholar]
- 3.Lind CRP, Tsai AMC, Lind CJ, Law AJJ. Ventricular catheter placement accuracy in non-stereotactic shunt surgery for hydrocephalus. J Clin Neurosci. 2009; 16(7): 918–920. [DOI] [PubMed] [Google Scholar]
- 4.Toma AK, Camp S, Watkins LD, Grieve J, Kitchen ND. External ventricular drain insertion accuracy: is there a need for change in practice? Neurosurgery. 2009; 65(6): 1197–1201. [DOI] [PubMed] [Google Scholar]
- 5.Wilson TJ, Stetler WR Jr, Al-Holou WN, Sullivan SE. Comparison of the accuracy of ventricular catheter placement using freehand placement, ultrasonic guidance, and stereotactic neuronavigation. J Neurosurg. 2013; 119(1): 66–70. [DOI] [PubMed] [Google Scholar]
- 6.Dickerman RD, McConathy WJ, Morgan J, et al. Failure rate of frontal versus parietal approaches for proximal catheter placement in ventriculoperitoneal shunts: revisited. J Clin Neurosci. 2005; 12(7): 781–783. [DOI] [PubMed] [Google Scholar]
- 7.Khan NR, DeCuypere M, Vaughn BN, Klimo P. Image guidance for ventricular shunt surgery: an analysis of ventricular size and proximal revision rates. Neurosurgery. 2019; 84(3): 624–635. [DOI] [PubMed] [Google Scholar]
- 8.Janson CG, Romanova LG, Rudser KD, Haines SJ. Improvement in clinical outcomes following optimal targeting of brain ventricular catheters with intraoperative imaging. J Neurosurg. 2014; 120(3): 684–696. [DOI] [PubMed] [Google Scholar]
- 9.Coulson NK, Chiarelli PA, Su DK, et al. Ultrasound stylet for non-image-guided ventricular catheterization. J Neurosurg Pediatr. 2015; 16(4): 393–401. [DOI] [PubMed] [Google Scholar]
- 10.Koh SAS, Lee W, Rahmat R, Salkade PR, Li H. Interethnic variation in the prevalence of claustrophobia during MRI at Singapore General Hospital: does a wider bore MR scanner help? Proc Singap Healthc. 2017; 26(4): 241–245. [Google Scholar]
- 11.Stieglitz LH, Fichtner J, Andres R, et al. The silent loss of neuronavigation accuracy: a systematic retrospective analysis of factors influencing the mismatch of frameless stereotactic systems in cranial neurosurgery. Neurosurgery. 2013; 72(5): 796–807. [DOI] [PubMed] [Google Scholar]
- 12.Mongen MA, Willems PWA. Current accuracy of surface matching compared to adhesive markers in patient-to-image registration. Acta Neurochir (Wien). 2019; 161(5): 865–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.