Summary
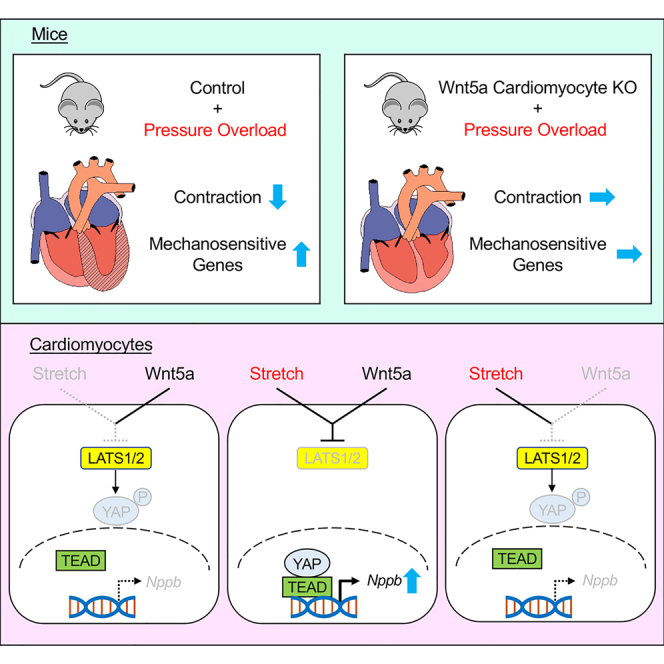
Non-canonical Wnt signaling activated by Wnt5a/Wnt11 is required for the second heart field development in mice. However, the pathophysiological role of non-canonical Wnt signaling in the adult heart has not been fully elucidated. Here we show that cardiomyocyte-specific Wnt5a knockout mice exhibit improved systolic function and reduced expression of mechanosensitive genes including Nppb when subjected to pressure overload. In cultured cardiomyocytes, Wnt5a knockdown reduced Nppb upregulation induced by cyclic cell stretch. Upstream analysis revealed that TEAD1, a transcription factor that acts with Hippo pathway co-activator YAP, was downregulated both in vitro and in vivo by Wnt5a knockdown/knockout. YAP nuclear translocation was induced by cell stretch and attenuated by Wnt5a knockdown. Wnt5a knockdown-induced Nppb downregulation during cell stretch was rescued by Hippo inhibition, and the rescue effect was canceled by knockdown of YAP. These results collectively suggest that Wnt5a-YAP signaling axis mediates mechanotransduction in cardiomyocytes and contributes to heart failure progression.
Subject areas: Cardiovascular medicine, Molecular physiology
Graphical abstract
Highlights
-
•
Wnt5a deletion in cardiomyocytes attenuates TAC-induced cardiac dysfunction
-
•
Myocyte Wnt5a is required for TAC-induced upregulation of mechanosensitive genes
-
•
Wnt5a is required for stretch-induced YAP nuclear translocation in cardiomyocytes
-
•
Wnt5a-YAP signaling axis mediates mechanotransduction in cardiac myocytes
Cardiovascular medicine; Molecular physiology
Introduction
Wnts constitute a large family of cysteine-rich secreted proteins that elicit evolutionarily conserved intracellular signaling and control diverse processes during development.1 Wnt signaling also plays critical roles in various physiological and pathological processes in adult organisms including stem cell self-renewal/differentiation, degenerative diseases, and carcinogenesis.2 The initially identified intracellular signaling pathway activated by Wnts was a β-catenin-dependent or canonical pathway. In canonical Wnt pathway, Wnt stimulation induces cytosolic β-catenin stabilization and nuclear translocation, where it binds to T cell factor/Lymphoid enhancer factor (Tcf/Lef) and activates Tcf/Lef-dependent transcription.2,3 Subsequently β-catenin-independent or non-canonical Wnt pathway was also identified. There are at least two non-canonical Wnt pathways called planar cell polarity (PCP) pathway and Ca2+ pathway, the former is mediated by small G proteins such as RhoA and Rac and regulates cell polarity and migration whereas the latter is mediated by intracellular Ca2+ mobilization and stimulates cell migration and inhibits canonical Wnt pathway.3,4,5 Wnt5a and Wnt11 are two major Wnt ligands that preferentially activate non-canonial Wnt signaling, although the selectivity of canonical versus non-canonical Wnt signaling appears to be determined by distinct set of receptors rather than the Wnt ligands themselves.3,6
Previous studies have shown that canonical Wnt signaling is required for heart development and involved in cardiac hypertrophy and remodeling in the adult.7,8 It was also reported that non-canonical Wnt signaling is required for normal heart development: inhibition of Wnt11 in frogs disrupts heart formation,9 and combined deletion of Wnt5a/Wnt11 genes in mice results in loss of second heart field progenitors and single-chambered heart.10 However, the pathophysiological role of non-canonical Wnt signaling in the adult heart remains elusive.
Here, we show that cardiomyocyte-specific Wnt5a knockout mice exhibit improved systolic function and reduced expression of mechanosensitive genes when subjected to pressure overload. We also show that Wnt5a mediates mechanotransduction in cardiac myocytes through YAP activation. These results suggest that Wnt5a-YAP signaling axis mediates mechanotransduction in cardiac myocytes and contributes to the transition to heart failure.
Results
Pressure overload-induced cardiac dysfunction is attenuated in cardiomyocyte-specific Wnt5a knockout mice
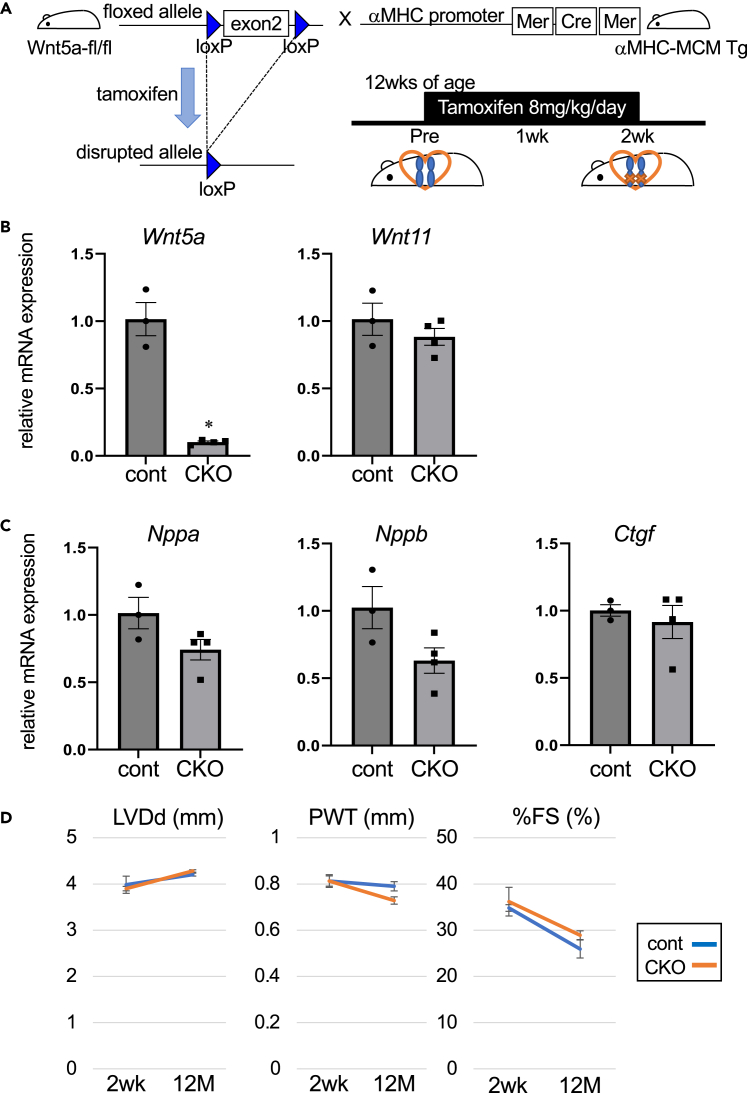
To explore the pathophysiological role of Wnt5a in the adult heart, we generated tamoxifen-inducible Wnt5a cardiomyocyte-specific knockout (CKO) mice by crossing Wnt5a floxed mice11 with Myh6-Cre/Esr1 transgenic mice expressing tamoxifen-inducible Cre under the control of α-myosin heavy chain promoter12 (Figure 1A). Intraperitoneal tamoxifen administration (8 mg/kg/day) was started at 12 weeks of age, which resulted in 90% reduction in Wnt5a expression level in the heart of CKO mice compared to control mice after 2 weeks of tamoxifen injection (Figure 1B). The expression levels of Wnt11 in the hearts of CKO mice were comparable to those in control mice (Figure 1B). The expression levels of Nppa (encoding atrial natriuretic peptide), Nppb (encoding brain natriuretic peptide), and Ctgf (encoding connective tissue growth factor) genes that are known to be upregulated in stressed hearts were not significantly different between control and CKO mice (Figure 1C). Echocardiographic parameters both at 2 weeks and 12 months after initial tamoxifen injection were also comparable between control and CKO mice (Figure 1D).
Figure 1.
Generation of cardiomyocyte-specific Wnt5a knockout mice
(A) Schematic illustration depicting generation of cardiac-specific Wnt5a knockout mice and the timing of tamoxifen treatment.
(B and C) Relative mRNA levels of Wnt5a, Wnt11, Nppa, Nppb, and Ctgf at day 14 after initial tamoxifen injection. ∗sp < 0.05 versus cont, n = 3 for cont and n = 4 for CKO.
(D)Echcardiographic evaluation of left ventricular dimension and contraction at 14 days and 12 months after initial tamoxifen injection. LVDd, left ventricular dimension at diastole; PWT, posterior wall thickness; %FS, percentile fractional shortening. n = 4 for cont and n = 3 for CKO.
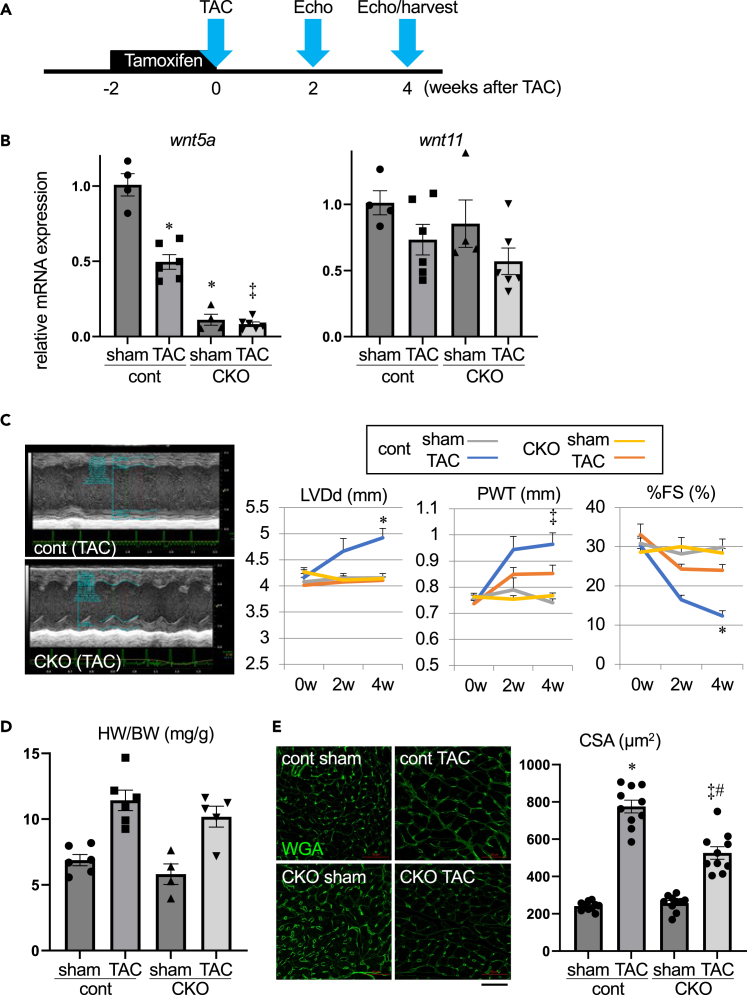
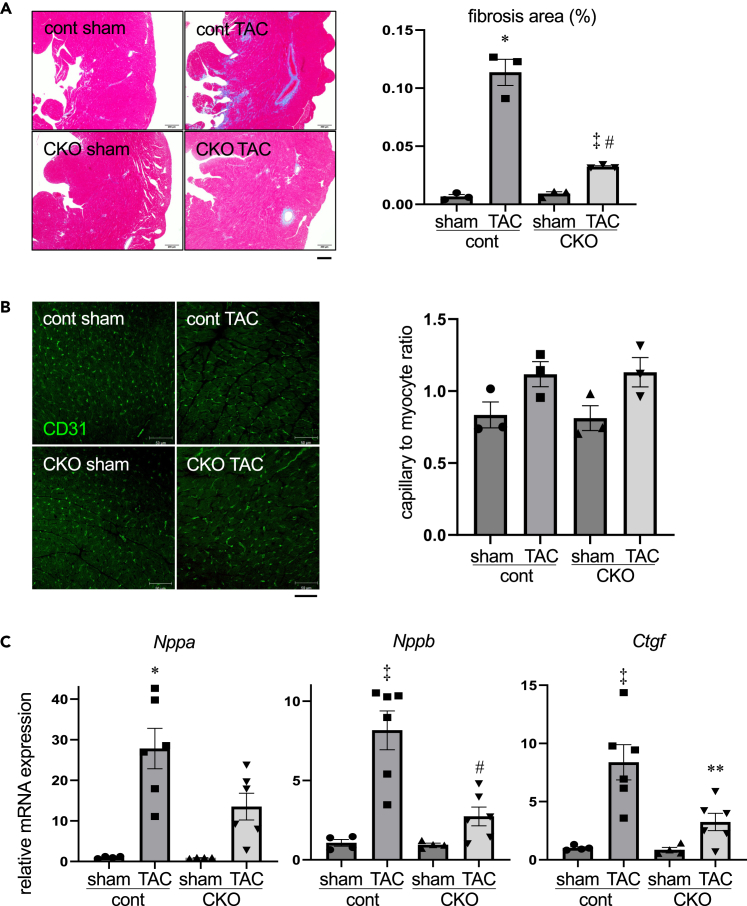
We next performed transverse aortic constriction (TAC) at 2 weeks after initial tamoxifen injection (Figure 2A). At 4 weeks after TAC, Wnt5a expression was significantly downregulated in control hearts, and the low levels of Wnt5a in CKO hearts remained unchanged (Figure 2B). The expression levels of Wnt11 were not altered by TAC both in control and CKO hearts (Figure 2B). Echocardiography revealed that chamber dilatation, increased wall thickness, and contractile dysfunction of left ventricle induced by TAC were attenuated in CKO hearts compared to control hearts (Figure 2C). Although the heart weight/body weight ratio was not significantly different between control and CKO hearts 4 weeks after TAC (Figure 2D), histological analysis revealed that TAC-induced cardiomyocyte hypertrophy was significantly reduced in CKO hearts as evidenced by decreased myocyte cross sectional area (CSA) (Figure 2E). Histological analysis also revealed that TAC-induced interstitial fibrosis was attenuated in CKO hearts (Figure 3A), and myocardial capillary density did not differ between control and CKO hearts (Figure 3B). TAC significantly increased the expression levels of Nppa, Nppb, and Ctgf genes, and Wnt5a deletion resulted in the reduction of Nppb expression levels in response to TAC (Figure 3C). These observations indicate that pressure overload-induced cardiac dysfunction is attenuated in the heart of Wnt5a CKO mice.
Figure 2.
Pressure overload-induced cardiac dysfunction is attenuated in cardiomyocyte-specific Wnt5a knockout mice
(A) Timeline depicting tamoxifen treatment, transverse aortic constriction (TAC), echocardiography, and tissue harvest.
(B) Relative mRNA levels of Wnt5a and Wnt11 at 4 weeks after TAC. ∗p < 0.001 versus sham-control, ‡p < 0.001 versus TAC-control, n = 4, 6, 4, and 6 for each group.
(C) Echcardiographic evaluation of left ventricular dimension and contraction at 2 weeks and 4 weeks after TAC. ∗p < 0.01 versus other groups, ‡p < 0.01 versus sham-cont and sham-CKO, n = 4, 5, 4, and 5 for each group.
(D) Heart weight (HW) (mg)/body weight (BW) (g) ratio at 4 weeks after TAC. n = 6, 6, 4, and 5 for each group.
(E) Cross sectional area (CSA) of myocytes at 4 weeks after TAC. Left panel shows the wheat germ agglutinin (WGA) staining of sarcolemma. Scale bar, 50μm. ∗p < 0.001 versus sham-cont, ‡p < 0.001 versus sham-CKO, #p < 0.001 versus TAC-cont. n = 10 for each group.
Figure 3.
Pressure overload-induced cardiac fibrosis and stress-responsive gene expression are attenuated in cardiomyocyte-specific Wnt5a knockout mice
(A) Interstitial fibrosis at 4 weeks after TAC. Left panel shows the Masson’s trichrome staining of connective tissues. Right panel shows the relative fibrosis area. Scale bar, 200μm. ∗p < 0.001 versus sham-cont, ‡p < 0.05 versus sham-CKO, #p < 0.001 versus TAC-cont. n = 3 for each group.
(B) Capillaries in the myocardium 4 weeks after TAC. Left panel shows the CD31 staining of capillaries. Right panel shows capillary to myocyte ratio. Scale bar, 50μm. n = 3 for each group.
(C) Relative mRNA levels of Nppa, Nppb, and Ctgf at 4 weeks after TAC. ∗p < 0.05 versus sham-cont, ‡p < 0.001 versus sham-cont, #p < 0.001 versus TAC-cont, ∗∗p < 0.005 versus TAC-cont. n = 4, 6, 4, and 6 for each group.
Cardiomyocyte-specific Wnt5a deletion diminishes the pressure overload-induced upregulation of genes related to “cellular response to mechanical stimuli”
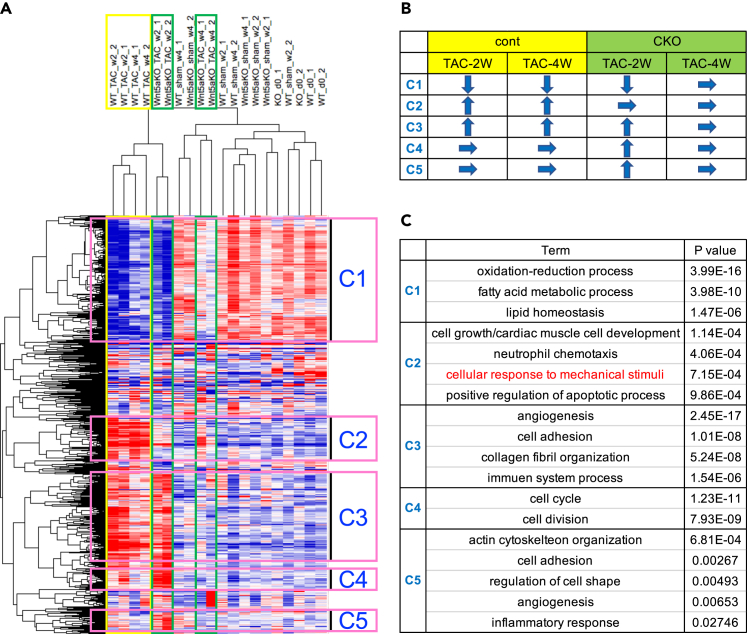
To gain mechanistic insights into how cardiomyocyte-specific Wnt5a deletion blunts the response to pressure overload, we carried out RNA-seq analysis for mRNA samples obtained from control and Wnt5a CKO hearts at 3 different time points (at the end of tamoxifen treatment, 2 weeks after sham/TAC operation, and 4 weeks after sham/TAC operation). Heatmap of the differentially expressed genes identified 5 clusters of genes whose expression patterns in response to pressure overload were altered by Wnt5a deletion in myocytes (Figure 4A and Table S1). The differential expression patterns of genes in each cluster are shown in Figure 4B. For instance, expressions of genes in cluster 1 are downregulated both in 2 weeks and 4 weeks after TAC in control hearts but they are downregulated only at 2 weeks after TAC in CKO hearts. We then focused on genes in “cellular response to mechanical stimuli” category in cluster 2 (Figure 4C). Genes in this cluster are upregulated in control hearts but not in Wnt5a CKO hearts both at 2 weeks and 4 weeks after TAC, and these differential expression patterns of genes are consistent with our observation that cardiomyocyte-specific Wnt5a deletion blunts the response to pressure overload. Based on the results of RNA-seq analysis, we hypothesized that Wnt5a is involved in mechanosensing in cardiac myocytes.
Figure 4.
Cardiomyocyte-specific Wnt5a deletion diminishes the pressure overload-induced upregulation of genes related to “cellular response to mechanical stimuli”
(A) Heatmap of RNA-seq transcriptome analysis showing differentially expressed genes between TAC-cont and TAC-CKO at 2 and 4 weeks after the operation by hierarchical clustering analysis. Yellow rectangles show TAC-cont, green rectangles show TAC-CKO, and gene clusters are shown by pink rectangles.
(B) Schematic illustration of the expression patterns of genes in 5 clusters.
(C) Gene ontology terms of differentially expressed genes in 5 clusters. Gene names in each gene ontology term are shown in Table S1. Gene names in each GO term are shown in Table S1.
Wnt5a is required for the responses to mechanical stimuli in cardiac myocytes
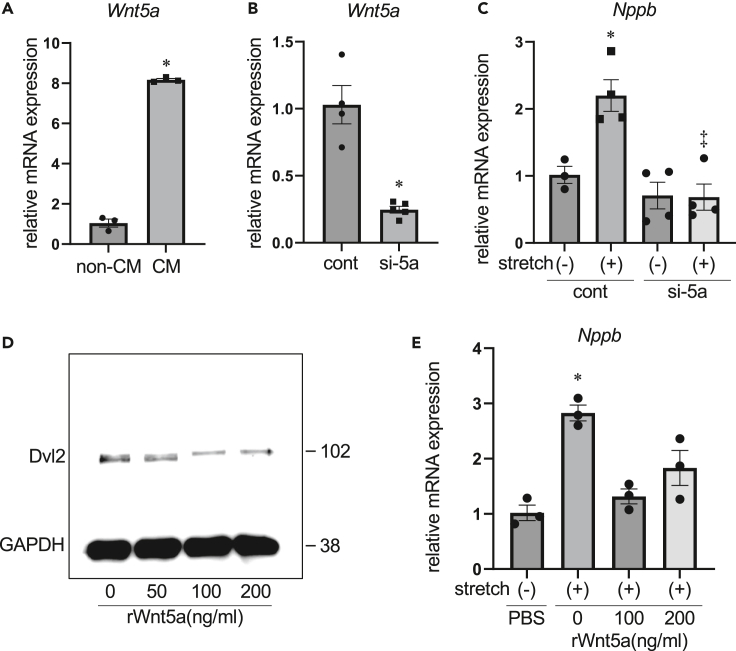
To test the above-mentioned hypothesis, we performed experiments in neonatal rat ventricular cardiomyocytes (NRVCs) subjected to cyclic stretch, where cultured cardiac myocytes seeded on deformable silicon dishes were uniaxially stretched for 8 h (20% in length, 30 cycles/minute). Wnt5a was predominantly expressed in myocyte fraction compared to non-myocyte fraction (Figure 5A), and siRNA-mediated knockdown of Wnt5a gene reduced the expression of endogenous Wnt5a approximately by 75% (Figure 5B). Cyclic stretch for 8 h significantly upregulated Nppb expression levels in NRVCs, and this stretch-induced Nppb upregulation was diminished by knockdown of Wnt5a (Figure 5C). Treatment of cultured NRVCs with recombinant Wnt5a protein13 induced Disheveled2 (Dvl2) phosphorylation at the concentration of 100 and 200 ng/mL as indicated by reduced electrophoretic mobility (Figure 5D). However, supplementation of Wnt5a protein (100 and 200 ng/mL) did not increase but rather decreased Nppb expression following cyclic stretch (Figure 5E). These observations suggest that Wnt5a is required for but does not enhance mechanotransduction-like responses in cardiac myocytes.
Figure 5.
Wnt5a is required for the responses to mechanical stimuli in cardiac myocytes
(A) Relative mRNA levels of Wnt5a in cardiomyocyte (CM) fraction and non-CM fraction. ∗p < 0.01 versus non-CM. n = 3 for each group.
(B) Relative mRNA levels of Wnt5a in cardiac myocytes following siRNA-mediated Wnt5a knockdown. ∗p < 0.05 versus cont. n = 4 for cont and n = 5 for si-Wnt5a (si-5a).
(C) Relative mRNA levels of Nppb in cardiac myocytes after 8 h of cyclic stretch. ∗p < 0.005 versus stretch (−)-cont, ‡p < 0.001 versus stretch (+)-cont. n = 3, 4, 4, and 4 for each group.
(D) Western blot analysis of Disheveled2 (Dvl2) in cardiac myocytes treated with recombinant Wnt5a protein (50, 100, and 200 ng/mL) for 3 h.
(E) Relative mRNA levels of Nppb in cardiac myocytes treated with recombinant Wnt5a protein (100 and 200 ng/mL) during 8 h of stretch. ∗p < 0.005 versus stretch (−)-cont. n = 3 for each group.
Wnt5a mediates mechanotransduction in cardiac myocytes through YAP activation
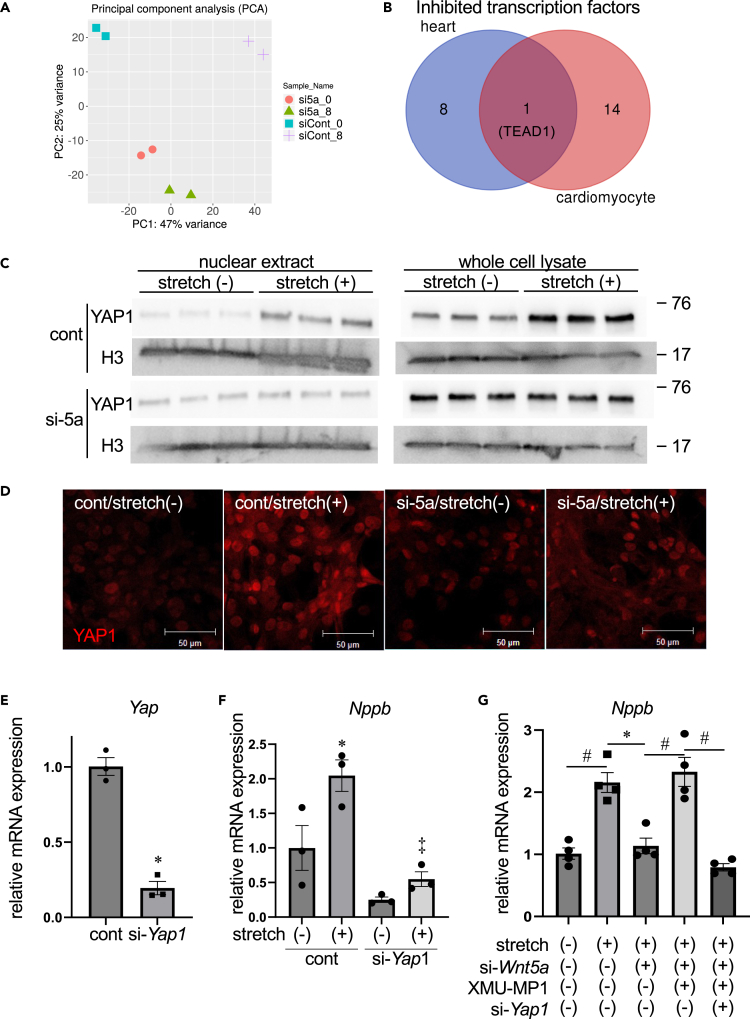
To examine the mechanism by which Wnt5a mediates mechanotransduction in cardiomyocytes, we carried out RNA-seq analysis in NRVCs. Principal component analysis demonstrated that siRNA-mediated knockdown of Wnt5a affected a large scale of transcriptional regulation in NRVCs at baseline before cyclic stretch (Figure 6A, blue squares versus orange circles), and the contribution of PC1 to changes in gene expression levels because of cyclic stretch was relatively small in the Wnt5a knockdown group (Figure 6A, compare the changes from blue squares to purple crosses with those from orange circles to green triangles). Moreover, pathway analysis suggested that Wnt5a knockdown significantly downregulated transcription factor binding in NRVCs compared to control cells after 8 h of cell stretch (Table S2). We therefore carried out upstream analysis of transcription factors and found that Wnt5a knockdown inhibited 15 transcription factors in NRCMs during 8 h of cyclic stretch (Table S3). We also performed upstream analysis of transcription factors in the hearts and found that 9 transcription factors were inhibited in Wnt5a CKO hearts compared to control hearts (Table S4). When we compared the results of upstream analysis of transcription factors between in vivo and in vitro experiments, TEA domain transcription factors 1 (TEAD1) was the only transcription factor inhibited in both settings (Tables S3, S4, and Figure 6B). TEAD regulates gene expression through interaction with yes-associated protein (YAP) and its paralog, transcriptional coactivator with a PDZ-binding motif (TAZ).14 YAP/TAZ is a transcriptional co-activator in the Hippo pathway, and Mst1/2-Lats1/2 kinase cascade phosphorylates YAP/TAZ to induce its cytoplasmic retention and inhibition of YAP/TAZ activity.14 Consistently, YAP1 activity was shown to be downregulated by Wnt5a deletion in NRVCs (Table S3). Moreover, Wnt5a was previously shown to activate YAP/TAZ.15,16,17 Based on these data and previous reports, we hypothesized that Wnt5a mediates mechanotransduction in cardiac myocytes through YAP activation.
Figure 6.
Wnt5a mediates mechanotransduction in cardiac myocytes through YAP activation
(A) Principal component analysis of control and Wnt5a-knocked down cardiac myocytes at baseline and 8 h after stretch.
(B) Venn diagram showing the number of shared and exclusive transcription factors downregulated by Wnt5a knockout in the heart (blue) and Wnt5a knockdown in cultured cardiac myocytes (orange). See also Tables S3 and S4.
(C) Western blot analysis of YAP in nuclear extract (left) and whole cell lysate (right) in cultured cardiac myocytes.
(D) Immunohistochemistry of YAP1 in cultured cardiac myocytes. Scale bar, 50μm.
(E) Relative mRNA levels of Yap1 in cardiac myocytes following siRNA-mediated Yap1 knockdown. ∗p < 0.01 versus cont. n = 6 for each group.
(F) Relative mRNA levels of Nppb in cardiac myocytes. ∗p < 0.05 versus stretch (−)-cont, ‡p < 0.005 versus stretch (+)-cont. n = 3 for each group.
(G) Relative mRNA levels of Nppb in cardiac myocytes. #p < 0.01, ∗p < 0.05. n = 4 for each group.
To test this hypothesis, nuclear localization of YAP after cyclic stretch was examined in NRVCs. Western blot analysis of NRVC nuclear extracts revealed that cyclic stretch induced nuclear translocation of YAP in control cells but not in Wnt5a knockdown cells (Figure 6C, left panel). In western blot analysis of NRVC whole cell lysate, total protein amount of YAP was increased by cyclic stretch in control cells but not in Wnt5a knockdown cells (Figure 6C, right panel). YAP immunostaining also revealed stretch-induced nuclear retention of YAP in control cells but not in Wnt5a knockdown cells (Figure 6D). These results indicate that cyclic stretch in NRVCs induces nuclear retention of YAP in a Wnt5a-dependent manner.
To test whether YAP is involved in mechanotransduction in cardiac myocytes, the effect of Yap1 knockdown in NRVCs was examined. siRNA-mediated knockdown of Yap1 gene reduced the expression of endogenous Yap1 approximately by 80% (Figure 6E). Upregulation of Nppb gene in NRVCs induced by cyclic stretch was attenuated by Yap1 knockdown (Figure 6F), indicating that stretch-induced Nppb upregulation is dependent on YAP. To further confirm that Wnt5a-YAP signaling axis mediates mechanotransduction in cardiac myocytes, we attempted to activate YAP independently of Wnt5a in NRVCs. For this purpose, we used XMU-MP-1, which pharmacologically inhibits Mst1/2 kinases and thereby promotes YAP nuclear translocation.18 Wnt5a knockdown attenuated cyclic stretch-induced Nppb upregulation, and XMU-MP1 treatment reversed this inhibitory effect of Wnt5a knockdown, suggesting that Wnt5a signaling affects Nppb expression upstream or independently of Mst1/2. Of note, this upregulation of Nppb expression induced by XMU-MP-1 in Wnt5a knockdown cells was canceled by additional knockdown of Yap1 (Figure 6G). Collectively, these results suggest that Wnt5a mediates mechanotransduction in cardiomyocytes through YAP activation.
Discussion
In the present study we explored the pathophysiological role of cardiomyocyte-derived Wnt5a in the adult heart and found that (i) Wnt5a CKO mice exhibit improved systolic function in response to pressure overload and (ii) Wnt5a mediates cell stretch-induced upregulation of Nppb in cultured cardiac myocytes through activation of transcriptional co-factor YAP. These findings suggest that non-canonical Wnt5a-YAP signaling axis mediates mechanotransduction in cardiac myocytes and contributes to the transition to heart failure in response to pressure overload.
Wnt5a-YAP signaling axis mediates mechanotransduction in cardiac myocytes
The ability of cells to respond to the alterations in external physical stimuli is crucial for the maintenance of tissues such as muscle and bone that are exposed to varying degrees of mechanical stress.19 YAP was originally identified as a protein that associates with Src family of tyrosine kinase Yes. Subsequently YAP was shown to act as a co-activator of TEAD family of transcription factors downstream of Hippo signaling pathway, where upstream kinase MST1/2 phosphorylates and activates LATS1/2, which in turn phosphorylates and inactivates YAP by inducing cytoplasmic retention and proteasome-dependent degradation.14 It was also shown that YAP is an effector of cellular mechanosensing pathway and translocates to the nucleus in response to various mechanical stimuli.20 Furthermore, it was also shown that Wnt5a activates YAP in a β-catenin-independent but LATS1/2-dependent fashion,16 collectively suggesting the possibility that Wnt5a-YAP signaling axis is involved in cellular responses to mechanical stimuli. However, to the best of our knowledge, this is the first report demonstrating that endogenous Wnt5a mediates mechanotransduction through YAP activation. Wnt5a was necessary for the activation of YAP and the upregulation of Nppb gene expression in response to cell stretch in cardiac myocytes. However, treatment with Wnt5a protein did not increase but rather decreased Nppb expression in the presence of cyclic stretch. Although the precise reasons for such apparent discrepancies are not clear at present, we speculate that appropriate levels of Wnt5a signaling may be required for the integrity of mechanosensing apparatus, and that both reduced and enhanced Wnt5a signaling may result in the impairment of mechanotransduction and YAP activation. The exact mechanistic basis for Wnt5a-mediated mechanotransduction remains to be elucidated.
Cardiomyocyte-derived Wnt5a contributes to the transition to heart failure in response to pressure overload
In this study we show that Wnt5a CKO mice exhibit improved systolic function in response to pressure overload, suggesting that cardiomyocyte-derived Wnt5a contributes to heart failure progression. Although the precise mechanism for Wnt5a-mediated cardiac dysfunction is not clear at present, we speculate that activation of mechanosensitive genes contributes to the progression of heart failure following pressure overload. Responses of the heart to increased load is generally recognized as adaptive processes. However, sustained mechanical stress results in maladaptive responses of the heart, leading to adverse remodeling and contractile dysfunction of the heart.21 Thus, it is possible that appropriate levels of reduction in mechanosensation and mechanoresponse inhibits or delays the transition from adaptive to maladaptive cardia hypertrophy following pressure overload. Of note, it was previously reported that neutrophil-specific Wnt5a deletion also protects the heart from pressure overload-induced dysfunction,22 suggesting that Wnt5a derived from both myocytes and non-myocytes plays a significant role in heart failure progression.
YAP is a downstream effector of Hippo pathway, and several studies have investigated the role of Hippo signaling in pressure overload-induced heart failure. Cardiac-specific homozygous deletion of YAP results in dilated cardiomyopathy-like phenotype at baseline and heterozygous cardiac-specific YAP knockout mice exhibit exacerbated contractile dysfunction after TAC, suggesting that YAP is cardioprotective.23 However, YAP activation by cardiac-specific deletion of WW45 (also called Salvador), a scaffold protein required for LATS1/2 activation by MST1/2, results in enhanced contractile dysfunction after TAC, suggesting that YAP activation in this case is deleterious for the heart.24 Moreover, although both inactivation (heterozygous deletion of YAP) and activation (homozygous deletion of WW45) of YAP promotes heart failure in response to pressure overload, combined deletion of YAP (heterozygous) and WW45 (homozygous) in cardiac myocytes is protective against pressure overload-induced cardiac dysfunction.24 These results collectively suggest that YAP must be maintained at appropriate levels for the heart to maintain normal function. It is therefore possible that pressure overload-induced YAP activation is decreased by Wnt5a deletion to “appropriate levels for the heart”, leading to maintained contractile function in response to pressure overload. Future studies will elucidate how mechanotransduction mediated by Wnt5a-YAP signaling axis contributes to the regulation of cardiac function.
There are some discrepancies between our present study and several previous literature. Although we found that Wnt5a gene expression is downregulated in response to pressure overload (Figure 1B), it was previously reported that Wnt5a is upregulated in the myocardium of patients with heart failure and in the heart of mice following TAC.25,26 The precise reasons for such discrepancies are not clear at present. On the other hand, consistent with our present study, it was previously reported that simultaneous in vivo knockdown of Wnt5a and Wnt11 using shRNA-Wnt5a/Wnt11-AAV9 vector exhibits cardioprotective effects (e.g., improved contraction and reduced fibrosis) after TAC.26 However, it should be noted that in vivo knockdown by shRNA vectors affect all cell types in the heart whereas Wnt5a gene was knocked out in a cardiomyocyte-specific manner in the present study.
Limitations of the study
Several limitations exist in the present study. First, although we were able to show that Wnt5a is necessary to activate YAP and induce Nppb gene expression in response to cell stretch, the detailed mechanistic basis for Wnt5a-mediated mechanotransduction remains unresolved. Second, the observation that Wnt5a CKO mice exhibit improved systolic function and reduced expression of mechanosensitive genes in response to pressure overload does not directly demonstrate that reduced mechanotransduction signaling induced by Wnt5a knockout plays a causal role in functional improvement. It is also possible that other Wnt5a-mediated signaling plays a role in pressure overload-induced contractile dysfunction.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD31 | BioLegend | Cat# 102414; RRID: AB_493408 |
| Dvl2 | CST | Cat# 3216; RRID: AB_2093338 |
| GAPDH | CST | Cat# 2118; RRID: AB_561053 |
| Histone H3 | CST | Cat# 4499; RRID: AB_10544537 |
| YAP | CST | Cat# 14074; RRID: AB_2650491 |
| Chemicals | ||
| tamoxifen | SIGMA | Cat# T5648 |
| wheat germ agglutinin | Invitrogen | W11261 |
| XMU-MP-1 hydrochloride | SIGMA | SML2233 |
| Deposited Data | ||
| Raw and processed RNA-seq data | This paper | GEO: GSE200667 |
| Experimental models: organisms/strains | ||
| Mouse: Wnt5aflox | (Sato et al.11) | N/A |
| Mouse: Myh6-Cre/Esr1Tg | JACKSON | #005657 |
| Oligonucleotides and siRNA | ||
| Primers and siRNA sequences | This paper | N/A |
| are listed in Table S5 | ||
| Software and algorithms | ||
| cellSens standard 1.11 | OLYMPUS | N/A |
| Image J bundled with Java 1.8.0 | NIH | N/A |
| ZEN | ZEISS | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Ichiro Shiojima (shiojima@hirakata.kmu.ac.jp).
Materials availability
This study did not generate new unique reagents.
Experimetal model and study participant details
Animals
Wnt5a floxed mice (Wnt5afl/fl)11 and Myh6-Cre/Esr1 transgenic mice12 were housed and genotyped with primers listed in Table S5. To generate cardiomyocyte-specific Wnt5a knockout mice, Wnt5a floxed mice were crossed with Myh6-Cre/Esr1 transgenic mice, and Wnt5afl/fl; Myh6-Cre/Esr1Tg (Wnt5a CKO) mice and Wnt5afl/fl (control) mice were treated with intraperitoneal injection of tamoxifen (8 mg/kg/day) for 14 days beginning at 12 weeks of age. Transverse aortic constriction (TAC) was performed in male mice at 14 weeks of age. All experimental procedures involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) at Kansai Medical University (21–108).
Methods details
Transverse aortic constriction (TAC) and echocardiography
TAC was performed essentially as described27 except that a 27 gauge needle was used in this study. For echocardiography we used the Vevo 2100 High Resolution Imaging system (VisualSonics, Toronto, Canada) with a 15–50 MHz transducer mounted on an integrated rail system. Mice were anesthetized using 2–3% isoflurane and moved to a biofeedback warming station that measured heart rate. Standard imaging planes, M-mode, and functional calculations were obtained according to American Society of Echocardiography guidelines. The LV parasternal short-axis view was used to derive fractional shortening, ejection fraction, and ventricular dimensions.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using RNeasy Mini-kit (QIAGEN), and concentration and quality of extracted RNA was determined by Spectrophotometer ND-1000 (Nano Drop). cDNA was prepared using SuperScript III reverse transcriptase (Invitrogen). qRT-PCR was performed using the SYBR green PCR Kit (QIAGEN) and the Rotor Gene Q machine. β-actin gene was used as an internal control. Primers for qRT-PCR are listed in Table S5.
Histological analysis
Hearts were fixed in 4% paraformaldehyde, embedded either in paraffin or OCT compound. Sections were stained with Masson’s trichrome for detection of fibrosis, Alexa Fluor 488-conjugated wheat germ agglutinin to evaluate cross sectional area of myocytes, and Alexa Fluor 488-conjugated CD31 to measure capillary density.
RNA-seq analysis
Total RNA was extracted using miRNeasy Mini Kit (QIAGEN), and library was prepared using a TruSeq Stranded mRNA sample preparation kit (Illumina, San Diego, CA). For heart tissues, sequencing was performed on HiSeq 2500 platform in a 75-base single-end mode. Generated reads were mapped to the mouse (mm10) reference genome using TopHat v2.1.1 in combination with Bowtie2 ver. 2.2.8 and SAMtools ver. 0.1.18. For neonatal rat ventricular cardiomyocytes, sequencing was performed in a 101-base single-end mode and generated reads were mapped to the rat reference genome sequences (rn6). Fragments per kilobase of exon per million mapped fragments (FPKMs) were calculated using Cuffdiff 2.2.1. Raw data concerning this study was submitted under Gene Expression Omnibus (GEO) accession number GSE200667.
Hierarchical clustering was performed using Cluster 3.028 and JAVA Treeview.29 Gene ontology analysis was performed using DAVID.30 Pathway analysis was performed with iDEP (http://ge-lab.org/idep/). For upstream analysis of transcription factors, QIAGEN’s Ingenuity Pathway Analysis was applied. Statistical thresholds were determined through the calculation of the activation z score of gene sets composed of randomly chosen perturbed genes with random sign of fold change that do not lead to significant results on average. (Ingenuity Downstream Effects Analysis, whitepaper). Adjusted p values (adj. Pval) and Activated z-scores were used to identify significant pathways and upstream regulators. The adj. P val indicates significance, while z-scores were used to define activation or inhibition. The strongest predicted activation corresponds to z scores ≥2, and the strongest predicted inhibition corresponds to z scores ≤ −2.
Preparation of neonatal rat cardiac myocytes
Neonatal rat cardiac myocytes and non-myocytes were prepared from 1-day-old Wister rats. In brief, ventricles were incubated in 0.25% trypsin-EDTA solution (Gibco) for overnight at 4°C and then digested with collagenase type II (Worthington). The dispersed cells were then incubated in 10-cm culture dishes for 70 minutes to remove non-myocytes. The unattached viable cells were rich in cardiomyocytes (90–95%), and attached cells were used as non-cardiac myocytes fraction. All experimental procedures involving neonatal rats were approved by IACUC at Kansai Medical University (21–108).
Cyclic stretch of cultured cardiac myocytes
Cardiac myocytes were seeded on stretch chambers coated with laminin (Invitrogen) and cultured for 3 days. Cells were then subjected to uniaxial cyclic stretch (20% in length, 30 cycles/minute) for 8 hours with cell stretch system (STB-1400-10; STREX, Japan).
siRNA-mediated knockdown and Wnt5a stimulation
For Wnt5a and/or Yap1 knockdown in cultured cardiac myocytes, siRNA was transfected using Lipofectamine RNAiMAX reagent (Invitrogen 13778150) approximately 60 hours before starting cyclic stretch. For Wnt5a stimulation, cells were treated with recombinant Wnt5a protein (100 ng/ml)13 during 8 hours of stretch.
Western blot analysis
Total protein was extracted and protein concentrations were measured using a BCA assay kit (Takara). Nuclear extract was prepared using Cell fractionation kit (CST #9038). The proteins were separated by SDS-PAGE and transferred to PVDF membranes. The membranes were blocked in 5% nonfat milk in TBST and incubated with diluted primary antibodies (1:1000) overnight at 4°C. After washing with TBST, the membranes were incubated with Horseradish peroxidase anti-rabbit (CST) secondary antibody for 2 hours at room temperature. Signals were detected with FUSION SOLOS (VILBER).
Immunocytochemistry
Cells were fixed for 10 min at room temperature in PBS containing 4% (w/v) paraformaldehyde and permeabilized in PBS containing 0.1% (w/v) Triton X-100 and 3% (w/v) BSA for 10 min. Cells were incubated with primary antibodies overnight at 4°C. In case of unconjugated primary antibodies, cells were incubated with Alexa Fluor secondary antibodies (Invitrogen, 1:200) for 1 hour at room temperature. Images were obtained using a confocal microscope (LSM700, Carl-Zeiss, Jena, Germany).
Quantification and statistical analysis
Data are shown as mean ± SE. Statistical analysis were made by the non-parametric Mann-Whitney U test for 2 groups comparison or the ANOVA for more than three groups comparison. When appropriate, 2-way ANOVA followed by post-hoc tests were used for the comparison of four groups with two different interventions. Statistical significance was assumed if p < 0.05. All statistical analyses were performed with SPSS (Version 19.0, SPSS Inc).
Acknowledgments
We thank Yoshiko Karatsu for technical assistance and Tomoki Kitawaki for advices in statistical analyses. This work was supported by Grant-in-Aid for Scientific Research (18H02814) from the Japan Society for the Promotion of Science.
Author contributions
Conceptualization, M.I. and I.S.; Investigation, H.K, M.I., K.W., and K.H.; Resources, O.T, K.K., S.M., T.H., S.T., and A.K.; Formal Analysis, S.N, D.M., D.O., and I.K.; Writing – Original Draft, H.K, M.I., and I.S.; Writing – Review and Editing, M.I., S.N., O.D., A.K., and I.S.; Visualization, H.K, M.I., K.H., S.N., and I.S.; Supervision, M.I. and I.S.; Funding Acquisition, I.S.
Declaration of interests
The authors declare no competing interests.
Published: June 15, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107146.
Contributor Information
Masayoshi Iwasaki, Email: iwasakim@hirakata.kmu.ac.jp.
Ichiro Shiojima, Email: shiojima@hirakata.kmu.ac.jp.
Supplemental information
Data and code availability
-
•
Raw and processed RNAseq data have been deposited at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). Accession number is listed in the key resources table.
-
•
This manuscript does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 2.Nusse R., Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–999. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 3.Kikuchi A., Yamamoto H., Sato A. Selective activation mechanisms of Wnt signaling pathways. Trends Cell Biol. 2009;19:119–129. doi: 10.1016/j.tcb.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Kikuchi A., Yamamoto H., Sato A., Matsumoto S. Wnt5a: its signalling, functions and implication in diseases. Acta Physiol. 2012;204:17–33. doi: 10.1111/j.1748-1716.2011.02294.x. [DOI] [PubMed] [Google Scholar]
- 5.Nishita M., Enomoto M., Yamagata K., Minami Y. Cell/tissue-tropic functions of Wnt5a signaling in normal and cancer cells. Trends Cell Biol. 2010;20:346–354. doi: 10.1016/j.tcb.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 6.van Amerongen R., Mikels A., Nusse R. Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 2008;1:re9. doi: 10.1126/scisignal.135re9. [DOI] [PubMed] [Google Scholar]
- 7.Gessert S., Kühl M. The multiple phases and faces of wnt signaling during cardiac differentiation and development. Circ. Res. 2010;107:186–199. doi: 10.1161/CIRCRESAHA.110.221531. [DOI] [PubMed] [Google Scholar]
- 8.Bergmann M.W. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ. Res. 2010;107:1198–1208. doi: 10.1161/CIRCRESAHA.110.223768. [DOI] [PubMed] [Google Scholar]
- 9.Pandur P., Läsche M., Eisenberg L.M., Kühl M. Wnt-11 activation of a non-canonical Wnt signalling pathway is required for cardiogenesis. Nature. 2002;418:636–641. doi: 10.1038/nature00921. [DOI] [PubMed] [Google Scholar]
- 10.Cohen E.D., Miller M.F., Wang Z., Moon R.T., Morrisey E.E. Wnt5a and Wnt11 are essential for second heart field progenitor development. Development. 2012;139:1931–1940. doi: 10.1242/dev.069377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato A., Kayama H., Shojima K., Matsumoto S., Koyama H., Minami Y., Nojima S., Morii E., Honda H., Takeda K., Kikuchi A. The Wnt5a-Ror2 axis promotes the signaling circuit between interleukin-12 and interferon-gamma in colitis. Sci. Rep. 2015;5 doi: 10.1038/srep10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sohal D.S., Nghiem M., Crackower M.A., Witt S.A., Kimball T.R., Tymitz K.M., Penninger J.M., Molkentin J.D. Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen-inducible Cre protein. Circ. Res. 2001;89:20–25. doi: 10.1161/hh1301.092687. [DOI] [PubMed] [Google Scholar]
- 13.Kurayoshi M., Yamamoto H., Izumi S., Kikuchi A. Post-translational palmitoylation and glycosylation of Wnt-5a are necessary for its signalling. Biochem. J. 2007;402:515–523. doi: 10.1042/BJ20061476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng Z., Moroishi T., Guan K.L. Mechanisms of Hippo pathway regulation. Genes Dev. 2016;30:1–17. doi: 10.1101/gad.274027.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y., Liang Y., Zhu X., Wang M., Gui Y., Lu Q., Gu M., Xue X., Sun X., He W., et al. The signaling protein Wnt5a promotes TGFbeta1-mediated macrophage polarization and kidney fibrosis by inducing the transcriptional regulators Yap/Taz. J. Biol. Chem. 2018;293:19290–19302. doi: 10.1074/jbc.RA118.005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park H.W., Kim Y.C., Yu B., Moroishi T., Mo J.S., Plouffe S.W., Meng Z., Lin K.C., Yu F.X., Alexander C.M., et al. Alternative Wnt Signaling Activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo C., Balsa E., Perry E.A., Liang J., Tavares C.D., Vazquez F., Widlund H.R., Puigserver P. H3K27me3-mediated PGC1alpha gene silencing promotes melanoma invasion through WNT5A and YAP. J. Clin. Invest. 2020;130:853–862. doi: 10.1172/JCI130038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan F., He Z., Kong L.L., Chen Q., Yuan Q., Zhang S., Ye J., Liu H., Sun X., Geng J., et al. Pharmacological targeting of kinases MST1 and MST2 augments tissue repair and regeneration. Sci. Transl. Med. 2016;8:352ra108. doi: 10.1126/scitranslmed.aaf2304. [DOI] [PubMed] [Google Scholar]
- 19.Jaalouk D.E., Lammerding J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 2009;10:63–73. doi: 10.1038/nrm2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panciera T., Azzolin L., Cordenonsi M., Piccolo S. Mechanobiology of YAP and TAZ in physiology and disease. Nat. Rev. Mol. Cell Biol. 2017;18:758–770. doi: 10.1038/nrm.2017.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Opie L.H., Commerford P.J., Gersh B.J., Pfeffer M.A. Controversies in ventricular remodelling. Lancet. 2006;367:356–367. doi: 10.1016/s0140-6736(06)68074-4. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y., Sano S., Oshima K., Sano M., Watanabe Y., Katanasaka Y., Yura Y., Jung C., Anzai A., Swirski F.K., et al. Wnt5a-Mediated Neutrophil Recruitment Has an Obligatory Role in Pressure Overload-Induced Cardiac Dysfunction. Circulation. 2019;140:487–499. doi: 10.1161/CIRCULATIONAHA.118.038820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byun J., Del Re D.P., Zhai P., Ikeda S., Shirakabe A., Mizushima W., Miyamoto S., Brown J.H., Sadoshima J. Yes-associated protein (YAP) mediates adaptive cardiac hypertrophy in response to pressure overload. J. Biol. Chem. 2019;294:3603–3617. doi: 10.1074/jbc.RA118.006123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeda S., Mizushima W., Sciarretta S., Abdellatif M., Zhai P., Mukai R., Fefelova N., Oka S.I., Nakamura M., Del Re D.P., et al. Hippo Deficiency Leads to Cardiac Dysfunction Accompanied by Cardiomyocyte Dedifferentiation During Pressure Overload. Circ. Res. 2019;124:292–305. doi: 10.1161/CIRCRESAHA.118.314048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abraityte A., Vinge L.E., Askevold E.T., Lekva T., Michelsen A.E., Ranheim T., Alfsnes K., Fiane A., Aakhus S., Lunde I.G., et al. Wnt5a is elevated in heart failure and affects cardiac fibroblast function. J. Mol. Med. 2017;95:767–777. doi: 10.1007/s00109-017-1529-1. [DOI] [PubMed] [Google Scholar]
- 26.Zou Y., Pan L., Shen Y., Wang X., Huang C., Wang H., Jin X., Yin C., Wang Y., Jia J., et al. Cardiac Wnt5a and Wnt11 promote fibrosis by the crosstalk of FZD5 and EGFR signaling under pressure overload. Cell Death Dis. 2021;12:877. doi: 10.1038/s41419-021-04152-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.deAlmeida A.C., van Oort R.J., Wehrens X.H. Transverse aortic constriction in mice. J. Vis. Exp. 2010 doi: 10.3791/1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Hoon M.J.L., Imoto S., Nolan J., Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 29.Saldanha A.J. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Raw and processed RNAseq data have been deposited at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). Accession number is listed in the key resources table.
-
•
This manuscript does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.