arising from: P. Zhu etal. Nature 10.1038/s41586-021-04062-5 (2021)
In over-wintering annuals of Arabidopsis thaliana grown in temperate regions, prolonged cold exposure in winter, through the physiological process of vernalization, represses the expression of the potent floral repressor FLOWERING LOCUS C (FLC) to enable the transition to flowering in spring1,2. Recently, Zhu et al.3 reported that cold induced nuclear condensation of FRIGIDA (FRI) for FLC repression and that cold-induced antisense RNA COOLAIR promoted FRI condensation during prolonged cold exposure. Here we report that the cold-induced formation of nuclear FRI condensates is independent of COOLAIR.
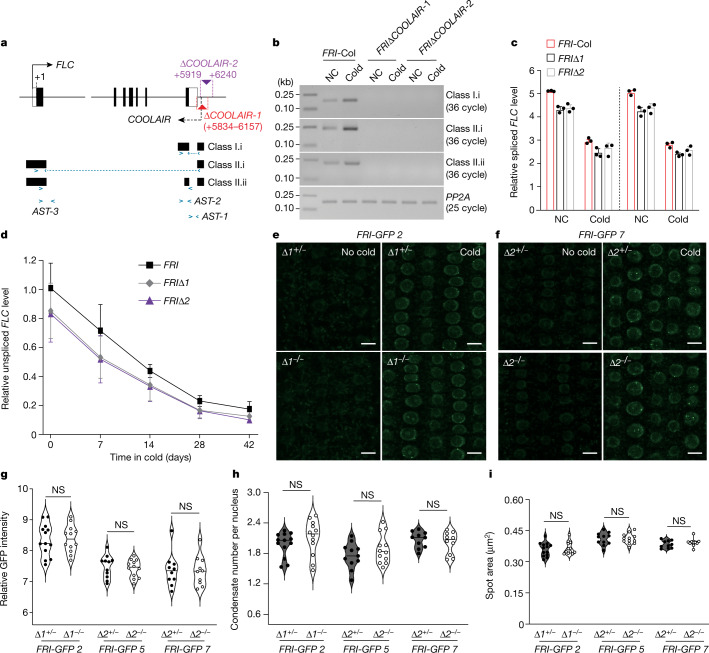
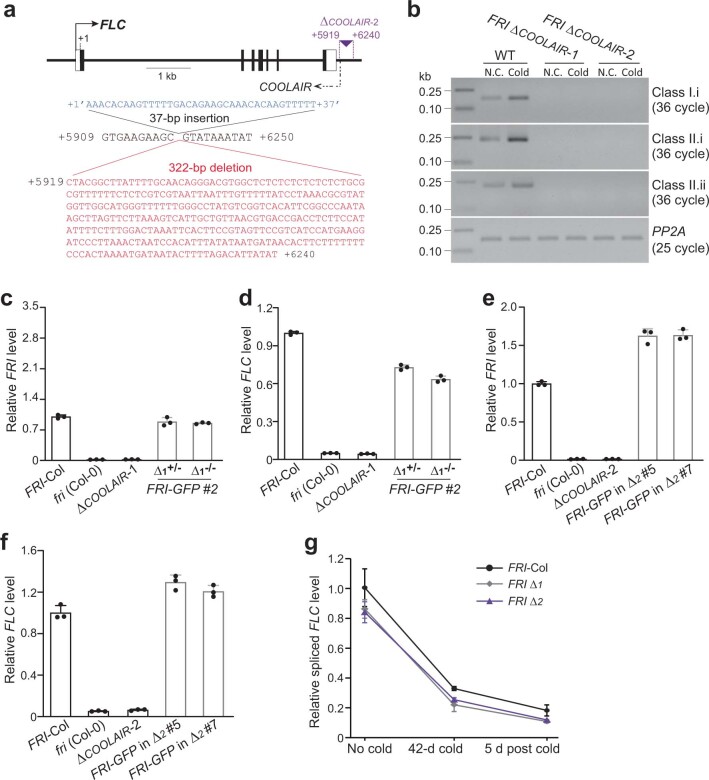
Before exposure to cold, FRI activates FLC expression, and long-term continuous winter cold exposure (typically lasting more than a month) results in FLC repression in FRI-bearing winter annuals grown at high latitudes1,4. Prolonged cold exposure induces the expression of COOLAIR, a group of non-coding antisense RNAs initiating from a region downstream of the 3′ end of FLC that is composed of proximally polyadenylated class I and distally polyadenylated class II transcripts5. COOLAIR expression reaches a high level after around 3 weeks of cold exposure and subsequently declines under constant cold temperature6. Using a CRISPR–Cas9 system, we previously constructed several lines in which a large part of the core COOLAIR promoter region was removed7, resulting in the elimination of both class I and class II COOLAIR transcripts before cold exposure (Fig. 1a,b and Extended Data Fig. 1a,b). Furthermore, consistent with a recent study8, cold induction of COOLAIR expression was eliminated in these core promoter deletion lines (Fig. 1b), partly because the cis-acting cold-responsive elements located in the promoter region have been removed. To examine the role of COOLAIR in FRI condensation, we introduced a functional FRI-GFP into two lines in which the COOLAIR promoter was deleted—ΔCOOLAIR-1 and ΔCOOLAIR-2—in the rapid-cycling accession Col-0 (bearing a loss-of-function fri allele9) by genetic transformation. Subsequently, independent FRI-GFPΔCOOLAIR lines (numbers 2, 5 and 7) were backcrossed to Col-0 and ΔCOOLAIR-1 or ΔCOOLAIR-2, respectively, resulting in F1 progeny of FRI-GFPΔCOOLAIR−/− and FRI-GFPΔCOOLAIR+/−. In these lines, FRI–GFP is fully functional and acts to activate FLC expression before cold exposure (Extended Data Fig. 1c–f).
Fig. 1. Functional analysis of COOLAIR in cold-mediated nuclear FRI condensation and FLC repression.
a, Schematic of the FLC locus. The A of ATG is indicated (+1), the blue arrows show the primer positions and the dashed lines show the interexon primers. b, COOLAIR expression is eliminated in FRIΔCOOLAIR lines (FRIΔCOOLAIR-1 (FRIΔ1) and FRIΔCOOLAIR-2 (FRIΔ2)). Seedlings were exposed to cold for 14 days or no cold (NC). FRI-Col is a winter-annual reference line. The constitutively expressed PP2A (At1G13320; PROTEIN PHOSPHATASE 2A SUBUNIT A3) was used as an internal control. c,d, Quantification of spliced (c) and unspliced FLC (d) transcripts in cold-treated seedlings. Seedlings were cold-treated for 14 days and two biological replicates were conducted in c. The levels of FLC transcripts were normalized to PP2A. Data are mean ± s.d. of three technical replicates (c) or biological replicates (d). The relative expression to FRI-Col (before cold exposure) is presented in d. e,f, Confocal microscopy images of FRI–GFP in the root tip nuclei of FRI-GFP 2 (e) and FRI-GFP 7 (f) seedlings. Seedlings were treated with cold for 14 days. Scale bars, 10 μm. g, The fluorescence intensity of FRI–GFP in the root tips treated with cold for 14 days. Data points are plotted on bar graphs. h,i, Quantification of FRI–GFP condensates (condensate number per nucleus (h) and spot area (i)) in the nuclei of cold-treated root tips. For g–i, 10–13 seedlings per F1 population were scored and data were analysed using two-tailed t-tests with Welch’s correction; NS, not significant (P > 0.05).
Extended Data Fig. 1. Characterization of COOLAIR promoter deletion lines and functional analysis of FRI-GFP.
a, Genomic sequences around the 322-bp promoter deletion in the ΔCOOLAIR-2 line. Notably, there is a 37-bp insertion at +5919. b, COOLAIR expression is eliminated in the COOLAIR promoter deletion lines. Shown is a biologically-independent repeat experiment of Fig. 1b. c–f, Relative FRI (c,e) and FLC (d,f) expression in the indicated seedlings. Transcript levels of FRI and FLC (spliced) were normalized to PP2A. Relative expression to FRI-Col is presented. Individual data points are shown. Error bars for s.d. of three biological replicates. g, Relative levels of spliced FLC transcripts upon 42-d cold exposure. 5-d post cold, return to warmth for 5 d after 42-d constant cold exposure. Error bars for s.d. of three biological replicates.
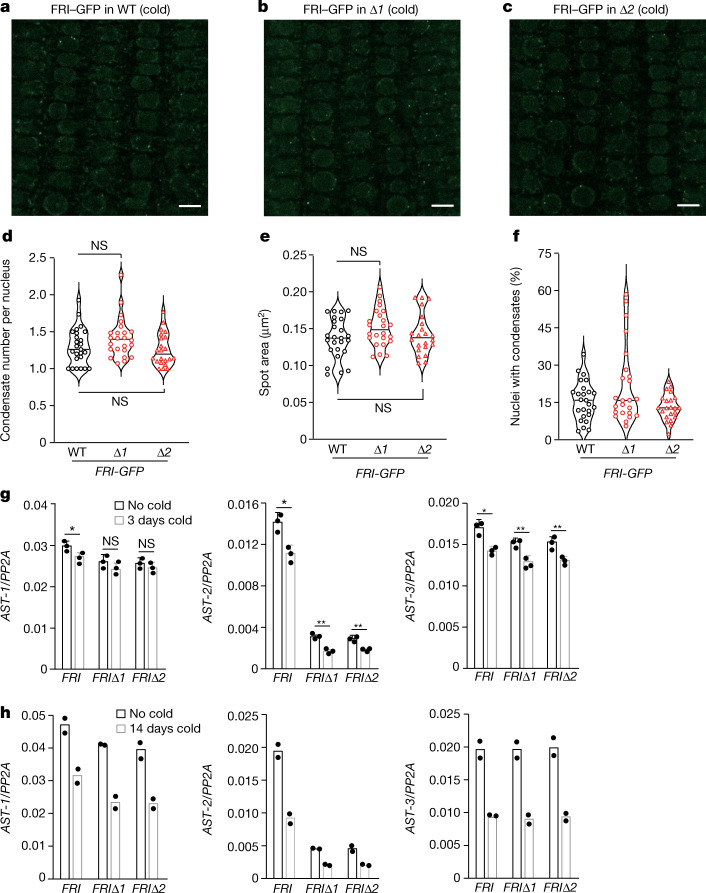
We next determined whether the loss of COOLAIR expression might reduce nuclear FRI condensation. We measured the fluorescence intensity of FRI–GFP in the root tips and the size and number of FRI–GFP condensates in root tip nuclei of cold-treated FRI-GFPΔCOOLAIR+/− and FRI-GFPΔCOOLAIR−/− seedlings and found that there was no difference between these two genotypes (Fig. 1e–i). Furthermore, we crossed a FRI-GFP line3 to both ΔCOOLAIR-1 and ΔCOOLAIR-2 and obtained homozygous FRI-GFPΔCOOLAIR lines. Subsequently, we measured the size and number of FRI–GFP condensates in root tip nuclei in these lines after cold exposure and found that there was no statistically significant difference (Fig. 2a–f). Together, these results show that the cold-induced formation of nuclear FRI condensates is independent of COOLAIR expression, given that, before and during cold exposure, COOLAIR expression (including class I and class II transcripts) was eliminated in both the ΔCOOLAIR-1 and ΔCOOLAIR-2 lines.
Fig. 2. Characterization of nuclear FRI–GFP condensation and antisense transcription at FLC in the absence of COOLAIR expression.
a–c, Confocal microscopy images of FRI–GFP in root-tip nuclei from the indicated seedlings (WT (a), ΔCOOLAIR-1 (b) and ΔCOOLAIR-2 (c)) exposed to cold for 14 days. Scale bars, 10 μm. d–f, Quantification of FRI–GFP condensates (number (d), spot area (e) and percentage of nuclei with condensates (f)) in the nuclei of cold-treated root tips. 22–25 seedlings per line were scored. For d and e, statistical analysis was performed using two-tailed t-tests with Welch’s correction. g,h, Quantification of ASTs at FLC in the indicated seedlings exposed to cold for 3 days (g) and 14 days (h). The examined regions with ASTs are indicated in Fig. 1a. AST-3 is known as CAS (convergent antisense transcript). For g, data are mean ± s.d. of three biological replicates. For g, statistical analysis was performed using two-tailed t-tests; *P < 0.05, **P < 0.01. Data points in h denote two biological replicates.
Zhu et al.3 reported that, in the cold, the FRI protein enriched class II.ii COOLAIR transcripts and that the FRI–class II.ii interaction was closely connected with cold-induced FRI condensation. Using a transgenic FLC terminator exchange (TEX) line in which the COOLAIR promoter was replaced with an RBCS3B (encoding Rubisco small subunit 3B) terminator, Zhu et al.3 found that the size and number per nucleus of FRI–GFP condensates were reduced in the TEX line compared with those in the non-transgenic background (Col-0). In the ΔCOOLAIR lines that we used in this study, the class II transcripts are eliminated before cold exposure and in the cold. Thus, we conclude that the FRI-class II.ii interaction and COOLAIR expression are not involved in cold-induced FRI condensation. The cause for the discrepancy between the two studies is unclear.
In addition to the COOLAIR transcripts initiated downstream of the 3′ end of FLC, there are other antisense transcripts (ASTs) initiated within the FLC locus (see, for example, ref. 10). We examined these ASTs in FRI-Col (a reference winter-annual line11) and FRIΔCOOLAIR seedlings, and found that they were at low levels before cold exposure (Fig. 2g). After cold exposure for 3 days, the expression of ASTs in all three examined regions declined in FRI-Col, and was apparently reduced in two examined regions in the FRIΔCOOLAIR seedlings (Fig. 2g); cold exposure for 14 days strongly suppressed the expression of ASTs in both the FRI-Col and FRIΔCOOLAIR lines (Fig. 2h). Thus, in contrast to COOLAIR, the expression of ASTs is repressed along the early phase of long-term cold exposure or vernalization. The function of ASTs in the vernalization-mediated FLC repression remains to be seen.
Cold induction of COOLAIR expression in the early phase of vernalization was reported to mediate FLC repression5,6. We measured the levels of FLC transcripts (both spliced and unspliced) in the cold-treated FRIΔCOOLAIR seedlings, and found that loss of COOLAIR expression in either FRIΔCOOLAIR-1 or FRIΔCOOLAIR-2 had no effect on the progression of transcriptional shutdown of FLC during cold exposure or on post-cold stable silencing of FLC (Fig. 1c,d and Extended Data Fig. 1g), consistent with observations in a recent study8. Notably, in our vernalization study, like several other studies reporting a role of COOLAIR for FLC repression5,6, seedlings were exposed to a constant cold temperature, the mechanisms uncovered through which may not fully represent FLC regulation by winter cold in the fields with fluctuating cold temperatures.
In summary, our study shows that the cold-induced formation of nuclear FRI condensates is independent of COOLAIR. Moreover, our vernalization study with constant cold temperature shows that COOLAIR is not involved in FLC repression by prolonged cold exposure. Thus, more in-depth experiments would be required to resolve the role of COOLAIR in vernalization.
Methods
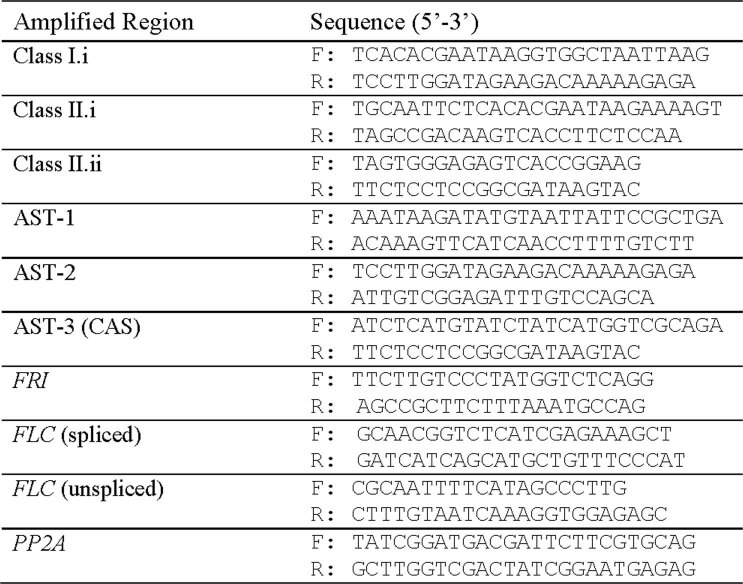
Arabidopsis thaliana FRI-Col, Col-0, ΔCOOLAIR-1 and FRIΔCOOLAIR-1 were described previously7. Treatment of the seedlings with constant cold and quantification of the expression of genes of interest using quantitative PCR were performed as previously described12. COOLAIR expression was examined by semiquantitative PCR, after reverse transcription using transcript-specific primers (5′-TGGTTGTTATTTGGTGGTGTGAA-3′ for class I; and 5′-GCCCGACGAAGAAAAAGTAG-3′ for class II10). A list of the PCR primers is provided in Extended Data Table 1.
Extended Data Table 1.
Primers used in the study
FRIpro:FRI-GFP was constructed by cloning a 4.8 kb genomic FRI fragment (2.5 kb promoter plus the 2.3 kb entire coding region without the stop codon) upstream of the GFP-coding region in the binary vector pMDC11013. Microscopy analysis and image quantification of nuclear FRI–GFP condensates were performed as follows. Root tips of the seedling samples were imaged using the Zeiss LSM900 confocal microscope with a ×40/1 NA water objective and an Airyscan detector of GaAsP-PMT. GFP fluorescence was excited at a wavelength of 488 nm (argon ion laser and laser power 8.0%), and detected at 490–620 nm in lambda mode. All of the images were obtained with a pixel size of 0.119 μm, and exported using the ZEN3.1 software (Zeiss) for quantitative analysis. The number of spots (with an area of larger than 0.1 μm2) per nucleus and the spot area in the F1 seedlings were obtained by outlining the spots using Graphics from ZEN3.1. Similarly, the fluorescent spots with an area of larger than 0.05 μm2 were scored in the root tips of the seedlings bearing the homozygous FRI-GFP3.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this Article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41586-023-06189-z.
Supplementary information
Source data
Acknowledgements
We thank P. Zhu and C. Dean for providing FRI-GFP seeds. We acknowledge support from the National Natural Science Foundation of China (grant nos 31830049 and 31721001) and the Peking-Tsinghua Center for Life Sciences.
Extended data figures and tables
Author contributions
Y.H. conceived the study. Z.Z., X.L. and Y.Y. conducted the experiments. Y.H. wrote the paper with help from Z.Z. and X.L.
Data availability
Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhicheng Zhang, Xiao Luo
Extended data
is available for this paper at 10.1038/s41586-023-06189-z.
Supplementary information
The online version contains supplementary material available at 10.1038/s41586-023-06189-z.
References
- 1.Bouche F, Woods DP, Amasino RM. Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol. 2017;173:27–35. doi: 10.1104/pp.16.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat. Rev. Genet. 2012;13:627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 3.Zhu P, Lister C, Dean C. Cold-induced Arabidopsis FRIGIDA nuclear condensates for FLC repression. Nature. 2021;599:657–661. doi: 10.1038/s41586-021-04062-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antoniou-Kourounioti RL, et al. Temperature sensing is distributed throughout the regulatory network that controls FLC epigenetic silencing in vernalization. Cell Syst. 2018;7:643–655. doi: 10.1016/j.cels.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swiezewski S, Liu F, Magusin A, Dean C. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature. 2009;462:799–802. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 6.Csorba T, Questa JI, Sun Q, Dean C. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc. Natl Acad. Sci. USA. 2014;111:16160–16165. doi: 10.1073/pnas.1419030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo X, Chen T, Zeng XL, He DW, He YH. Feedback regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5′ FLC promoter region in Arabidopsis. Mol. Plant. 2019;12:285–288. doi: 10.1016/j.molp.2019.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Jeon M, et al. Vernalization-triggered expression of the antisense transcript COOLAIR is mediated by CBF genes. eLife. 2023;12:e84594. doi: 10.7554/eLife.84594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johanson U, et al. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, et al. Natural temperature fluctuations promote COOLAIR regulation of FLC. Genes Dev. 2021;35:888–898. doi: 10.1101/gad.348362.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee I, Michaels SD, Masshardt AS, Amasino RM. The late-flowering phenotype of FRIGIDA and luminidependens is suppressed in the Landsberg erecta strain of Arabidopsis. Plant J. 1994;6:903–909. doi: 10.1046/j.1365-313X.1994.6060903.x. [DOI] [Google Scholar]
- 12.Xu G, Tao Z, He Y. Embryonic reactivation of FLOWERING LOCUS C by ABSCISIC ACID-INSENSITIVE 3 establishes the vernalization requirement in each Arabidopsis generation. Plant Cell. 2022;34:2205–2221. doi: 10.1093/plcell/koac077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper.