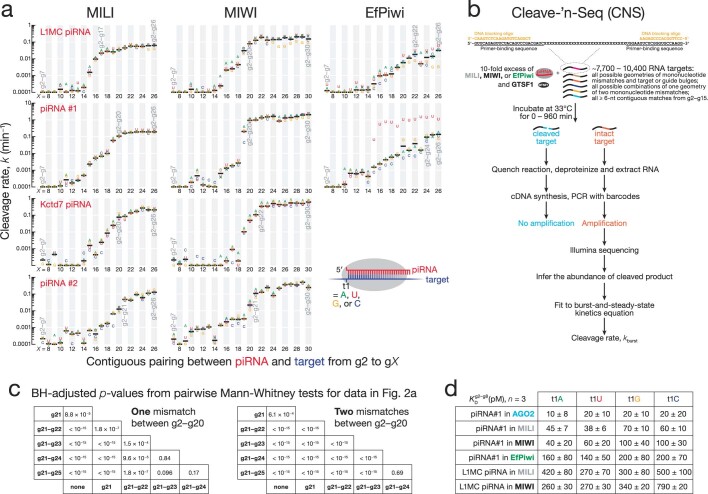
Extended Data Fig. 3. Pairing to piRNA 3′ end is dispensable for PIWI slicing.
a, MILI, MIWI, and EfPiwi pre-steady-state cleavage rates for targets of piRNAs contiguously paired from nucleotide g2. Data are for targets with all possible identities of nucleotide t1.b, Overview of Cleave-’n-Seq. c, Benjamini-Hochberg corrected p-values for post hoc pairwise, two-tailed Mann-Whitney tests for difference in pre-steady-state cleavage rate of targets in Fig. 2a. Kruskal-Wallis test (one-way ANOVA on ranks) p-values are < 10−15 for data with one and two mismatches. d, MILI, MIWI, EfPiwi, and mouse AGO2, binding affinities (KD) for a g2–g8 match with different t1 nucleotide identities. Mean and standard deviation from three independent trials are shown.