Abstract
Adipogenesis is key to maintaining organism-wide energy balance and healthy metabolic phenotype, making it critical to thoroughly comprehend its molecular regulation in humans. By single-nuclei RNA-sequencing (snRNA-seq) of over 20,000 differentiating white and brown preadipocytes, we constructed a high-resolution temporal transcriptional landscape of human white and brown adipogenesis. White and brown preadipocytes were isolated from a single individual's neck region, thereby eliminating inter-subject variability across two distinct lineages. These preadipocytes were also immortalized to allow for controlled, in vitro differentiation, allowing sampling of distinct cellular states across the spectrum of adipogenic progression. Pseudotemporal cellular ordering revealed the dynamics of ECM remodeling during early adipogenesis, and lipogenic/thermogenic response during late white/brown adipogenesis. Comparison with adipogenic regulation in murine models Identified several novel transcription factors as potential targets for adipogenic/thermogenic drivers in humans. Among these novel candidates, we explored the role of TRPS1 in adipocyte differentiation and showed that its knockdown impairs white adipogenesis in vitro. Key adipogenic and lipogenic markers revealed in our analysis were applied to analyze publicly available scRNA-seq datasets; these confirmed unique cell maturation features in recently discovered murine preadipocytes, and revealed inhibition of adipogenic expansion in humans with obesity. Overall, our study presents a comprehensive molecular description of both white and brown adipogenesis in humans and provides an important resource for future studies of adipose tissue development and function in both health and metabolic disease state.
Keywords: Adipogenesis, White fat, Brown fat, Single-nuclei RNA-seq
Highlights
-
•
Generated comprehensive molecular atlas of human white and brown adipogenesis.
-
•
Mapped modular adipogenic transcriptional dynamics in primary human adipose tissues.
-
•
Provided white- and brown-lineage specific gene signatures for mapping adipogenic progression scores in future single-cell investigations.
-
•
Identified TRPS1 as a novel adipogenic regulator in humans.
1. Introduction
Adipogenesis is a highly orchestrated process in which differentiation of adipose precursor cells (preadipocytes) into mature adipocytes is induced in response to varying metabolic needs such as energy storage during nutrient excess, lipolysis during periods of caloric deficit, or energy expenditure during cold exposure. Thus, adipogenesis is a critical process for maintaining metabolic homeostasis on an organism-wide level, and its dysregulation can contribute to diseases such as obesity, type 2 diabetes, and lipodystrophy. Consequently, there is a need to comprehensively understand the molecular regulation of adipogenic expansion in both health and in the setting of metabolic disease risk.
Adipogenic differentiation is regulated by a network of transcription factors (TFs) and results in the development of distinct types of fat from two distinct kinds of preadipocytes: white adipocytes for energy storage and brown adipocytes for thermogenic energy expenditure. Previous studies of adipogenesis’ regulation have typically employed murine model systems. For example, studies employing the murine 3T3-L1 cell line led to the identification of the core adipogenic transcriptional cascade including factors such as PPARG and C/EBPs [1,2]. Further research focused on identifying core brown versus white fat-selective factors which led to the identification of drivers and effectors of thermogenic phenotypes such as PRDM16, EBFs, and PGC1A and UCP1 [3,4]. More recently, modern transcriptomic investigations have identified multiple auxiliary transcription factors that serve as either positive or negative regulators of adipogenic/thermogenic response in rodents [5]. Overall, studies in rodent models of adipogenesis have offered significant insights into the molecular regulation of adipogenesis. However, their applicability to humans is limited because of the existing differences in the metabolism, physiology, and transcriptomic regulation of adipose tissue between the two species. For example, BAT, which is abundantly and homogeneously present in the interscapular depot in mice, was only found to be present in adult humans over the last decade [6], and its cellular composition is heterogeneous, varying with the sampling depth in a given region [7]. Furthermore, while ADRB3 is an adrenergic receptor believed to mediate murine thermogenesis, there have been conflicting observations of its role in human thermogenesis [8,9]. Additionally, recent studies also highlighted BAT metabolic functions that do not translate from the rodents to the human [10]. Consequently, there has been much interest in understanding the transcriptional control of adipocyte formation in humans [11,12]. A comprehensive understanding of the transcriptional regulation that drives adipogenesis would provide insights into lineage-determining, adipogenic, and thermogenic factors in humans, which may serve as molecular targets for therapeutic stimulation of a healthy metabolic phenotype.
Recently, multiple studies have compared the transcriptomic profiles of human-derived adipose stem cells (ASCs) at multiple stages of adipogenic differentiation using bulk gene expression profiling techniques such as microarray analysis [13,14,15,16,17,18], bulk RNA-seq [19], and RT-qPCR [20]. This has resulted in the identification of novel adipogenic TFs such as KLFs [21], FOXs [22], and GATAs [23]. However, bulk sampling of cells at dense time-intervals during differentiation does not allow detection of heterogeneity caused by asynchronous differentiation and the possibility of multiple lineages existing within the cellular populations of interest. By contrast, single-cell RNA-sequencing (scRNA-seq) overcomes many of these challenges and can provide an unbiased transcriptomic view of complex tissues at an unprecedented resolution [24,25,26]. Within primary adipose tissue, recent investigations utilizing single-cell level measurements have investigated the molecular dynamics of adipocyte development in mice [27,28]. However, single cell studies of primary tissues are not able to thoroughly sample the time course of adipogenesis. In this study, we mapped the transcriptional landscape of human white and brown adipogenesis using a unique, well-controlled, in vitro model system [29,30], which enables isolation of differentiating preadipocytes at multiple well-defined stages of development. In this system, paired white and brown primary preadipocytes were isolated from the neck of a single individual (Methods). This system, therefore, allowed us to measure transcriptional dynamics within and between white and brown lineages, while controlling for inter-individual variation typically associated with transcriptomic profiling of primary human adipose tissue, such as body mass index, genotype, and gender. Preadipocytes from both lineages were isolated and then immortalized to allow for long-term in vitro cell-culture. In vitro differentiated adipocytes were shown to recover gene expression profiles and function of primary human neck BAT and WAT [29,31,30] and the transcriptional profile of the immortalized preadipocytes were highly concordant with primary preadipocytes [32].
In this study, we applied single-nuclei RNA-seq (snRNA-seq) to perform time-resolved transcriptional analysis of differentiating white and brown preadipocytes using this in vitro model system. snRNA-seq was critical to reduce bias in cell recovery because mature adipocytes are typically inefficiently recovered during single cell isolation [11]. We isolated intact nuclei from differentiating white and brown preadipocytes at 5 stages of adipogenesis (Figure 1A). We then defined, lineage-specific adipogenic gene signatures to order single nuclei along a pseudotemporal axis of differentiation to observe the transcriptional dynamics of adipogenesis. Our analyses revealed temporal regulation of distinct gene modules in both white and brown adipogenesis, highlighting the dynamics of biologically relevant functional processes. This pseudotime analysis also allowed us to identify transcription factors (TFs) that are potentially involved in brown and white adipogenesis. Furthermore, we used these lineage-specific gene signatures to explore variation in adipogenesis across distinct metabolic phenotypes and cell-types using publicly accessible genomic datasets.
Figure 1.
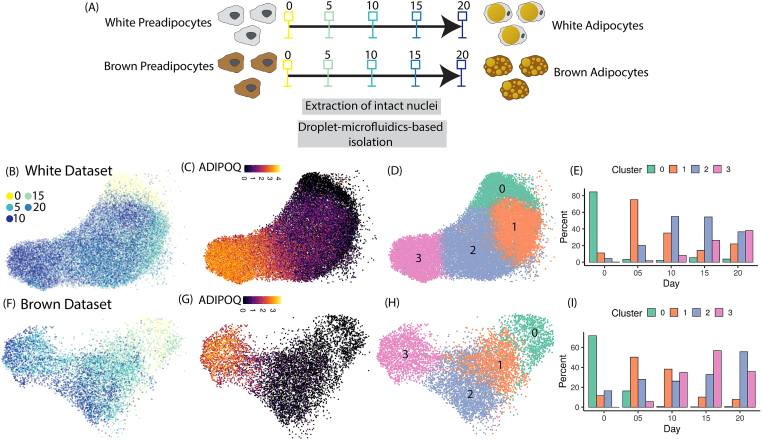
snRNA-seq of differentiating preadipocytes enables high-resolution sampling of adipogenic cellular states (A) Schematic of the experimental design utilized in this study. (B–D) UMAP visualization of white adipogenesis dataset colored by (B) day of collection (C) ADIPOQ gene expression and (D) unsupervised cluster classification. (E) Distribution of nuclei harvested at each experimental time-point with clusters identified in (D). (F–I) Same plots as (B) to (E) but plotted for brown adipogenesis dataset. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Results
2.1. snRNA-seq enables high-resolution sampling of adipogenesis
Cultured white and brown preadipocytes were differentiated into respective mature adipocyte types using an induction cocktail (see Methods). During differentiation, intact nuclei were harvested from white and brown preadipocytes (see Methods) at 5 equally spaced time-points during the 20-day adipogenic induction period (Figure 1A). Isolated nuclei were processed with droplet-based snRNA-seq, for whole-transcriptome analysis (Methods). In total, 25,339 high-quality nuclei were recovered from cells undergoing white adipogenesis and 27,568 high-quality -nuclei were recovered from cells undergoing brown adipogenesis, with ∼2000–6000 genes detected per nucleus (Table 1).
Table 1.
Sequencing metrics for snRNA-seq libraries analyzed in this study.
| Day | White Adipogenesis |
Brown Adipogenesis |
||
|---|---|---|---|---|
| Number of nuclei recovered | Reads per nuclei | Number of nuclei recovered | Reads per nuclei | |
| 00 | 8026 | 174,738 | 6763 | 216,700 |
| 05 | 39,345 | 37,939 | 9107 | 94,812 |
| 10 | 7,229 | 102,155 | 9945 | 83,732 |
| 15 | 4,834 | 174,967 | 3217 | 122,859 |
| 20 | 12,160 | 123,700 | 8085 | 181,758 |
Independent analysis of white preadipocytes at each of the 5 time-points revealed detection of adipocytes as early as day 5 (Fig. S1A), with continuously increasing expression for the mature adipocyte marker gene ADIPOQ and decreasing expression for progenitor marker CD44 (Fig. S1A) over the time course. Integration of these 5 datasets using scVI-tools [33] revealed structuring of nuclei along a continuum of cellular states (Figure 1B), starting from early precursors to mature adipocytes (as marked by ADIPOQ, Figure 1C). Unsupervised clustering revealed four distinct clusters during white adipogenesis (Figure 1D), with the majority of day 0 nuclei grouped in cluster-0 and majority of day 20 nuclei grouped in cluster-3, thereby illustrating the increasing maturity level from cluster-0 to cluster-3. Nuclei harvested at late stages of adipogenesis (day 10 to day 20) were distributed over all clusters (Figure 1E), thereby highlighting the asynchronous behavior of adipogenic differentiation in the in vitro model system.
Single nuclei analysis of brown preadipocytes at day 0 revealed two transcriptionally distinct cell populations, one which was enriched for genes related to a stem cell-like state (Preadipocyte-1) and another which was enriched for genes related to a fibroblast-like state (Preadipocyte-2, [32]. Integrative analysis of all 5 timepoints indicated a differential adipogenic capacity of these two populations (Fig. S1C), with the Preadipocyte-2 cells differentiating into mature adipocytes, and the Preadipcyte-1 population undergoing minimal transcriptional changes over the 20-day time course (Fig. S1C). Pathway analysis of genes that were up regulated in the non-adipogenic population (Preadipocyte-1) showed enrichment of FOXO1 (Fig. S1D), a known repressor of PPARG [34,35], suggesting a possible role of FOXO1 in inhibiting adipogenic response in the Preadipocyte-1 population. We observed similar transcriptional heterogeneity in primary brown adipose tissue, by projecting transcriptional signatures from the Preadipocyte-1 and Preadipocyte-2 populations (Vision [36]; onto a publicly available snRNA-seq database of human brown adipose tissue [37]; Note S2, Fig. S6).
In the brown adipogenic cluster, nuclei harvested across different days were distributed over a continuum of differentiation state (Figure 1F), with a continuously increasing expression of ADIPOQ (Figure 1G). Like white adipogenic response, unsupervised clustering identified 4 clusters during brown adipogenic response (Figure 1H), each with increasing maturity level (Figure 1I). Of note, nuclei harvested on day 20 were primarily grouped in cluster 2 whereas nuclei harvested on day 15 were primarily grouped in cluster 3 (Figure 1I). We attributed this observation to a lower differentiation efficiency on day 20, thereby resulting in reduced number of harvested mature adipocytes (grouped in cluster 3) as compared to day 15. We performed adipogenic signature analysis, in which a score is assigned to each nucleus based on expression of adipogenic genes, that revealed the highest score for day 20 adipocytes in cluster-3 (Fig. S1E). As compared to white adipocytes, differentiated brown adipocytes had up-regulation of the brown-adipocyte-specific marker gene ZIC1 [38] as well as PGC1B, a regulator of hepatic glucose and lipid metabolism [39] and a recognized thermogenic marker [40]; Fig. S1F). Analysis of differential expression followed by transcription factor enrichment analysis between mature white and brown adipocytes (cluster-3 in Figure 1D vs cluster-3 in Figure 1H) identified that the top-ranking transcription factor enriched in brown adipocytes was FOXS1 [41] and FOXC2 [42]. Thus, our experimental strategy allowed us to capture a spectrum of cell-states undergoing differentiation toward white or brown adipocyte lineages.
2.2. Pseudotemporal ordering of differentiating preadipocytes identifies dynamics of extracellular matrix (ECM) remodeling, lipogenesis, and thermogenesis
2.2.1. Lineage-specific gene signatures enable high-resolution ordering of single nuclei
Dense sampling of cellular states with snRNA-seq enabled reconstruction of the adipogenic developmental trajectory by ordering individual nuclei along a pseudo-time axis. To achieve this, we identified a set of genes specific to both white and brown-adipogenesis that defined progression through differentiation and provided a signature score that was used as a proxy for pseudo-time (Note S1). These gene signatures consisted of genes that showed monotonic increase in expression from immature preadipocytes to mature adipocytes (Note S1). Hence, pseudo-temporal scoring based on expression of such genes provided a high dynamic range and resolution of cellular differentiation state assignment.
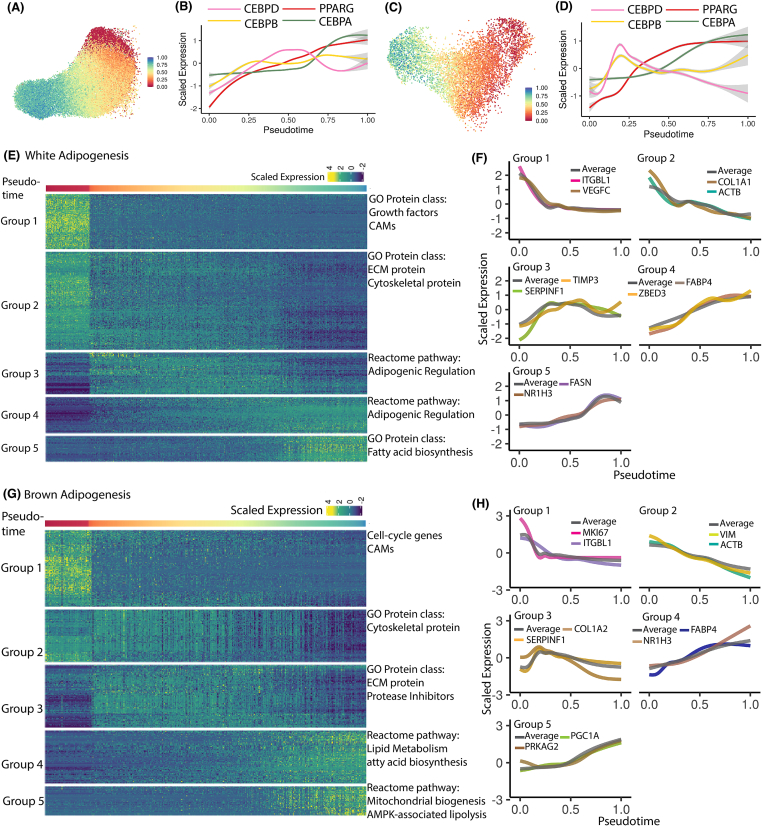
Using the white-/brown-adipogenesis-specific gene signature, cells undergoing adipogenesis were ordered by increasing degree of differentiation (Figure 2A,C). Assessment of expression dynamics in pseudotime of key adipogenic TFs CEBPB, CEBPD, PPARG and CEBPA accurately reflected the core adipogenic signaling cascade (Figure 2B,D), with early induction of CEBPB, followed by sustained expression in response to insulin (see Methods [43], early induction and transient expression of CEBPD, stable increase in expression of PPARG, and late induction of CEBPA, thereby validating our cell-ordering strategy for both white and brown adipocyte development. Next, dynamically regulated genes were identified by binning nuclei in the pseudotemporal space (Fig. S2A and S2B, see Methods, [44]). We reasoned that there would be significant deviation in expression of dynamically regulated genes from either the initial or final stages of adipogenesis, and hence performed differential expression testing for each bin against the first and last pseudotemporal bins (logFC >1 and FDR <0.05) to identify such genes. The bin size in pseudo-temporal space was determined to have a similar number of nuclei in each bin (Fig. S7A), thereby ensuring statistically robust differential gene expression testing. In total, we identified 596 and 454 temporally expressed genes during white and brown adipogenesis respectively. Unsupervised hierarchical gene clustering identified three major expression trends (Figure 2E–H, Tables S1 and S2) which we describe as down-regulated (Module 1 and Module 2), transiently up-regulated (Module 3), and up-regulated (Module 4 and Module 5). Within down-regulated gene modules, Module 1 undergoes immediate down-regulation, whereas Module 2 undergoes a consistent down-regulation (Fig. S2I and S2J). Within up-regulated gene modules, Module 4 has a consistent up-regulation whereas Module 5 has a delayed up-regulation (Fig. S2K and S2L).
Figure 2.
Gene module identification reveals expression dynamics of key biological processes accompanying white and brown adipogenic progression (A) Pseudotemporal ordering of differentiating white preadipocytes. (B) Expression dynamics for core adipogenic transcription factors CEBPs and PPARG during white adipogenesis. (C) and (D) Same plot as (A) and (B) but for brown adipogenesis. (E) Heatmap of expression dynamics for gene modules identified in white adipogenesis. Each column indicates a single nucleus, with columns ordered by increasing pseudotemporal score. Each row depicts a single gene, with rows clustered by unsupervised Louvain clustering. Each gene module is annotated by terms reflecting key biological processes based on Gene Ontology. (F) Smoothed expression dynamics of selected genes from each gene module identified in panel (E) in white adipogenesis dataset. (G) and (H) Same plot as (E) and (F) but for brown adipogenesis dataset. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.2.2. Cell-adhesion disruption is followed by fibrillar to basement-membrane-type ECM remodeling during early adipogenic progression
ECM remodeling during adipogenesis is key to providing the appropriate niche as spindle-shaped preadipocytes transform into spherical, fragile, lipid-laden adipocytes [45,46]. Dysregulation of ECM remodeling is a hallmark of clinical obesity, in which adipose tissue becomes fibrotic because of increased deposition of fibrillar ECM components such as collagen type 1, -3, and -5 [47]. While ECM remodeling is central to healthy adipogenic expansion, the precise dynamics of ECM reorganization and its molecular regulation remains poorly understood. Here, we focused on the differential dynamics across Modules 1, 2, and 3 (Fig. S2I and S2J) to provide novel insights into the temporal regulation of ECM reorganization during human adipogenic progression.
During white adipogenesis, genes undergoing rapid down-regulation upon addition of adipogenic medium (Module 1) primarily included cell adhesion molecules (CAMs) such as ITGB8, ITGA11, and ITGBL1, as well as growth factors such as VEGFA, VEGFC, FGF2, and FGF5 (Figure 2F, Tables S2 and S4). These observations are supported by previous studies which report downregulation of such integrin-associated genes during adipogenesis [48,49,50], and known anti-adipogenic effects of these growth factors [51,52,53]. Notably, CAMs serve as contact points between cells and ECM; thus, down-regulation of CAMs suggests that disruption of cellular-ECM contacts is a very early response to the induction media. Module 2 genes undergoing more gradual down-regulation primarily included ECM structural components (Figure 2F, Tables S2 and S4) such as fibrillar collagen types COL1, COL3, and COL5. This observation is consistent with previous observations of gene- and protein-level ECM remodeling during adipogenesis [45,54], which suggests that degradation of fibrillar ECM components paves the way for basement-membrane-type ECM components such as collagen-4 which are better suited to support spherical adipocytes [55,56]. Indeed, expression of COL4 gradually increased during white adipogenesis in our dataset (Fig. S2C, Table S2). Module 2 also included down-regulated cytoskeletal components such as actin (ACTB), tubulin (TUBB), and vimentin (VIM) (Figure 2F, Tables S2 and S4), in agreement with previous reports [57,58]. Module 3 genes (transient up-regulation) mostly consisted of protease inhibitors such as TIMP3 and SERPINF1 (Figure 2F, Tables S2 and S4). Upregulation of the protease inhibitor TIMP3 was also validated with western-blot analysis (Fig. S9). Protease inhibitors serve as ECM constructive enzymes, antagonizing the ECM degradation activity of metalloproteases such as MMPs, ADAMs, and ADAMTSs [59]. Notably, all such metalloproteases were downregulated during white adipogenesis in our dataset (Fig. S2D). Therefore, our results are consistent with an initial disruption of cell adhesion contacts, followed by metalloprotease activity to promote ECM degradation and a subsequent shift toward ECM regeneration via activity of protease inhibitors. This finding is also consistent with previous studies that have reported functional modulation of adipocyte differentiation via activity of metalloprotease, and its inhibitors [60,61,62,63,59].
Brown adipogenesis recovered similar dynamics of ECM remodeling, with immediate downregulation of integrins and CAMs such as ITGBL1 (Figure 2H), CDH11, CDH13, and CDH2 (Table S1). Similarly, Module 2 included gradually downregulated cytoskeletal components such as ACTB, TUBB, and VIM (Figure 2H, Tables S1 and S5). However, unlike white adipogenesis, fibrillar collagen components such as collagen types −1, −3, and −5 were clustered in Module-3 with initial up-regulation (Figure 2H, Tables S1 and S5), likely providing a fibrillary-type ECM to support early proliferation of brown preadipocytes (Fig. S2E and S2F) until their growth arrest [56]. Finally, several patterns were very similar to those observed during white adipogenesis. Collagen types −1, −3, and −5 are downregulated over time, with a converse increase in expression of basement-membrane-type collagen-4 (Fig. S2G). Likewise, metalloprotease inhibitors were enriched in Module-3 (Figure 2H, Table S5) with consistent downregulation of metalloproteases (Fig. S2H), suggesting a similar shift in enzymatic activity from ECM degeneration to ECM reconstruction.
2.2.3. Differential transcriptional dynamics of adipogenesis, lipogenesis, and thermogenesis
Typically, fat synthesis (or lipogenesis) accompanies later stages of adipogenesis as preadipocytes accumulate lipid droplets characteristic of mature adipocytes. However, previous studies have identified distinct pathways and regulators specific to lipid droplet biogenesis, expansion, and shrinkage during fat cell maturation [64,65], indicating a lipogenesis-specific transcriptional network. Similarly, brown-fat-specific transcriptional networks which regulate thermogenesis have been uncovered [66,4]. Pseudotemporal ordering of single cells provides an opportunity to better understand the relative transcriptional dynamics of these processes during differentiation of white and brown adipose tissue.
In white adipogenesis, Module 4 primarily consisted of genes that regulate lipid mobilization such as PLIN1, FABP4, CD36 (Fig. S8A) and adipogenic transcriptional regulators such as PPARG, MLXIPL, and ZBED3 [67,68] (Fig. S8B), all of which were gradually up-regulated with respect to the pseudotime axis (Figure 2F, Table S2). Module-5, on the other hand, included lipogenic genes such as FASN, ACSL1, GPAM and the lipogenic transcription factor NR1H3 [69]; Figure 2F, Fig. S8C, Table S2). This observation agrees with pathway analysis which revealed enrichment of adipogenic regulation terms in Module-4 (Table S4) and fatty acid biosynthesis terms in Module-5 (Table S4). On average, genes in module 4 were significantly upregulated (logFC > 1) at an earlier pseudotime point than genes in module 5 (Fig. S3A), suggesting a delayed onset of the lipogenic transcriptional response compared to the adipogenic response.
In brown adipogenesis, Module 4 consisted of transcriptional regulators of adipogenic response such as PPARG, FABP4, SREBF1 [70,71] (Figure 2H, Fig. S8D), with pathway analysis identifying enrichment of fatty acid biosynthesis, and lipid metabolism-associated terms (Table S5). Module 5, on the other hand, was enriched for thermogenic terms such as AMPK-associated lipolytic pathways [72,73] and mitochondrial biogenesis pathways (Table S5). Therefore, our results suggest an early onset of adipogenic response during brown fat development, followed by a later thermogenic response.
2.3. High-resolution map of transcription factor dynamics identifies potential regulators of adipogenic and thermogenic response in humans
Differential expression patterns of transcription factor mRNAs can help identify distinct cell-types, -states, or -lineages. Using our snRNA-seq dataset, we specifically characterized and compared the expression dynamics of transcription factors during white and brown fat development.
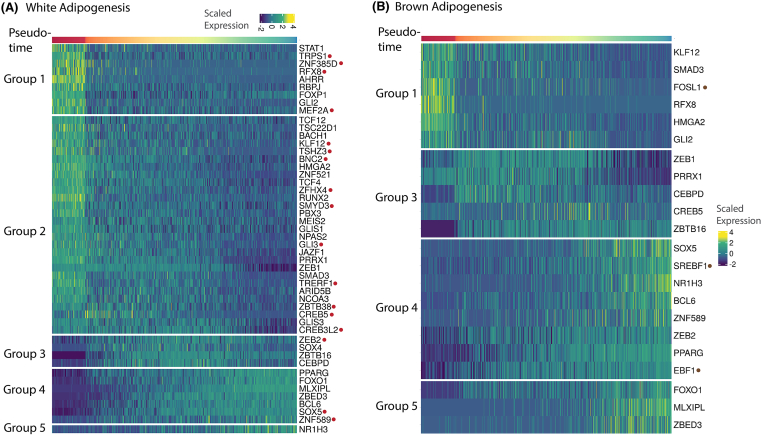
During white adipogenesis, we identified 49 TFs with dynamic gene expression profiles (Figure 3A). As expected, most of these TFs had similar expression dynamics as previously reported in rodents (Figure 3A). This included Module 1 anti-adipogenic TFs such as GLI2 (hedgehog signaling mediator [74,75], RBPJ (Notch signaling mediator [76,77], and AHRR [78], Module 2 anti-adipogenic TFs such as TCF4 & TCF12 (mediator of [79]), and SMAD3 (mediator of TGFB pathway [80]), Module 4 pro-adipogenic TFs such as PPARG, MLXIPL, and ZBED3, and Module 5 pro-lipogenic TF NR1H3. We also identified multiple TFs with dynamic gene expression profiles during white fat development that had not previously been associated with adipogenesis in humans (Figure 3A, highlighted in red), and which may serve as potential adipogenic regulators in humans. This included KLF12, ZEB2, CREB3L2, and MEF2A (Figure 3A); orthologs of these genes KLF8 [81], ZEB1 [82], CREB5 [83], and MEF2D [84] are known regulators of adipogenesis in rodents, thereby suggesting a similar role for these genes in humans. Moreover, transcription factor binding site enrichment analysis demonstrated overrepresentation of RFX8, BNC2, and TSHZ3 binding sites in differentially expressed genes.
Figure 3.
Transcription factor dynamics in white and brown adipogenesis (A) Dynamics of all TFs temporally regulated during white adipogenesis, distributed by their gene module annotation. Highlighted in red are TFs that had not previously been associated with adipogenesis in humans (B) Dynamics of all TFs temporally regulated during brown adipogenesis, distributed by their gene module annotation. Highlighted in brown are TFs that are differentially regulated during brown adipogenesis only. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
While many studies have focused on drivers of adipogenesis, this data provided the opportunity to identify thermogenic TFs based on unique regulation in brown adipogenesis over white adipogenesis, in a model system derived from the same individual. Typically, thermogenic TFs are identified based on differential enrichment of genes in BAT over WAT. While such a strategy is applicable in rodents, in which BAT is predominantly localized in a discrete interscapular depot, it is challenging in humans where BAT is found interspersed within WAT. Moreover, such a strategy provides a tissue-level measurement, with no information on whether identified differences manifest at the early preadipocyte stage or late mature adipocyte stage. Our dataset mitigated this challenge by allowing us to investigate the temporal dynamics of TF expression during fat development across multiple stages of adipogenesis. In total, we identified temporal regulation of 22 TFs, of which 19 TFs were also temporally regulated during white adipogenesis, with similar expression profiles (Figure 3B and Fig. S3B). This included novel TFs RFX8, KLF12, CREB5, ZEB2, SOX5, and ZNF589 identified in the paragraph above (Fig. S3B). Notably, we identified differentially significant regulation of TFs SREBF1, EBF1, and FOSL1 during brown adipogenesis (Figure 3B, Fig. S7), which agrees with their previously demonstrated pro-thermogenic roles [85,4]. Next, we identified TFs with differential activity in brown adipogenesis using binding-site enrichment analysis (Table S3). This included thermogenic regulatory TFs such as SREBF1, KLF15 [86], and TWIST1 [87], along with TFs FOXL1, ZNF117, IRX6, OSR1, and PRRX2, whose involvement in the context of thermogenic response has not been previously characterized. TFs ZNF117 and IRX6 were enriched in up-regulated genes (Module-4 and Module-5, Table S3) along with pro-thermogenic TFs SREBF1 and KLF15, thereby suggesting a potentially similar function for these TFs in human thermogenesis.
2.4. TRPS1 knockdown impairs white adipogenesis
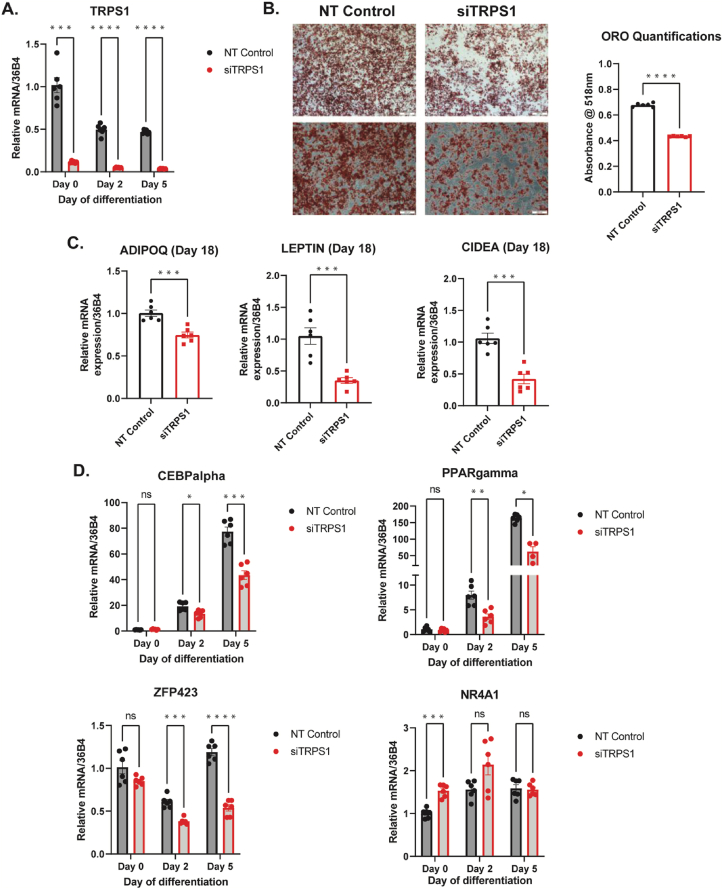
TRPS1 has been shown to be a repressor of GATA-regulated genes, and GATA TFs are known to play important role in fat cell formation [88]. TRPS1 was also identified in our enrichment analysis as one of the TFs expressed early during differentiation (Figure 3A). Hence, we sought to explore the role of TRPS1 on white adipogenesis in vitro, through siRNA-mediated knockdown (KD) of TRPS1 in immortalized white preadipocytes (day −2.5 of differentiation). After siRNA knockdown, we induced differentiation of these preadipocytes (day 0) and evaluated markers of adipogenesis along several time points of adipocyte differentiation (Fig. S10A). First, we confirmed that siRNA against TRPS1 leads to an efficient knockdown (>90% reduction) of TRPS1 expression in adipogenic progenitors on day 0 of differentiation, effect that is maintained on days 2 and 5 after induction of differentiation (Figure 4A). Next, we evaluated whether TRPS1 KD in adipogenic progenitors at the early stages of differentiation (days 0–5) can affect their adipogenic capacity. Oil-Red-O staining demonstrated a significant reduction in the number of mature adipocytes on day 18 of differentiation (Figure 4B), effect which was accompanied by a significant reduction in ADIPOQ, LEP, and CIDEA expression (Figure 4C), suggesting that lack of TRPS1 expression at the early stages of differentiation impairs adipogenesis. To gain insights into the mechanism by which TRPS1 KD impairs adipogenesis, we measured gene expression of established key TFs that regulate early adipocyte differentiation. We observed that TRPS1 KD led to a significant reduction in the expression of the TFs CEBPA, PPARG, and ZFP423 on days 2 and 5 of differentiation (Figure 4D). Notably, expression of NR4A1 was increased on day 0 of differentiation in response to TRPS1 KD (Figure 4D). Previous studies have shown that overexpression of NR4A1 can impair adipogenesis in vitro [89]. Small effects on days 2 and 5 of differentiation were also observed on the expression of CEBPB and CEBPD (Fig. S10B), however expression of GATA2 and GATA3 were not altered in response to TRPS1 KD (Fig. S10C). Overall, these findings suggest that high-resolution measurement of the transcriptional landscape of in vitro WAT and BAT differentiation enables identification of important regulatory drivers of adipogenesis.
Figure 4.
TRPS1 knockdown in human adipogenic progenitors impairs differentiation of mature white adipocytes in vitro. (A) Relative mRNA expression of TRPS1 in white pre-adipocytes during days 0, 2, and 5 of differentiation in response to siRNA-mediated TRPS1 knockdown (KD) or non-targeting control (NT), as quantified by qPCR (B) Oil Red-O-stained images of mature white adipocytes at Day18 of differentiation in response to TRPS1 KD or NT. Scale bar is 200 ums. (C) Relative mRNA expression of Adiponectin (ADIPOQ), Leptin, and CIDEA in mature white adipocytes during Day 18 of differentiation in response to siRNA-mediated TRPS1 knockdown (KD) or non-targeting control (D) Relative mRNA expression of C/EBPα, PPARγ, ZFP423, and NR4A1 in white pre-adipocytes during days 0, 2, and 5 of differentiation in response to siRNA-mediated TRPS1 knockdown (KD) or non-targeting control. Results are expressed as relative mRNA expression (fold-change) normalized to the housekeeping gene (36B4). Gene expression analysis was performed using the ddCT method. Graphs are shown as Mean +/− S.E.M. One-Way ANOVA (4 A, 4D) or unpaired t-test (B, C) were applied. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2.5. Mapping in vitro adipogenesis onto primary human adipose tissues
Utilizing our in-vitro model system, we defined lineage-specific gene signatures that enabled ordering of differentiating white and brown preadipocytes by their degree of differentiation. We also identified gene modules with differing expression dynamics during adipogenesis. To establish the generality of these findings, we used the gene expression data from our in vitro model to investigate transcriptional signatures of adipogenesis in human-derived primary cells and tissues.
We first investigated whether lineage-specific gene signatures defined in our study would recapitulate differences in degree of differentiation between cell-types at the opposite ends of adipogenic spectrum, i.e., preadipocytes and mature adipocytes. We focused on datasets using similar sequencing technique and hence mapped our lineage-specific gene signature scores onto a single-nuclei RNA-seq dataset of primary human WAT and BAT generated in [37]. As characterized in the original study, the snRNA-seq dataset revealed a primary cluster of preadipocytes and adipocytes each in WAT and BAT, as marked by the expression of marker genes DCLK1 and ADIPOQ (Figs. S4A and S4B). Indeed, using lineage-specific gene signatures in WAT and BAT revealed a significantly higher score for mature white and brown adipocytes, as compared to respective preadipocytes (Fig. S4C and S4D). Next, we focused on mapping our lineage-specific gene signature scores onto primary human adipose datasets derived using a slightly different sequencing technique of single-cell RNA-seq. Here, we used the scRNA-seq data of primary human WAT generated by Emont et al., 2022, which we first filtered to minimize sequencing chemistry related, depot-related, and cell-cluster-related biases (Methods). In line with our above observations, human adipocytes had a higher score for white-lineage specific gene signature than precursor cells (Fig. S4E). Overall, our analyses demonstrate the applicability of our lineage-specific gene signatures in recovering large-scale differences in degree of adipogenic differentiation (preadipocytes vs adipocytes) in human-derived in vivo datasets.
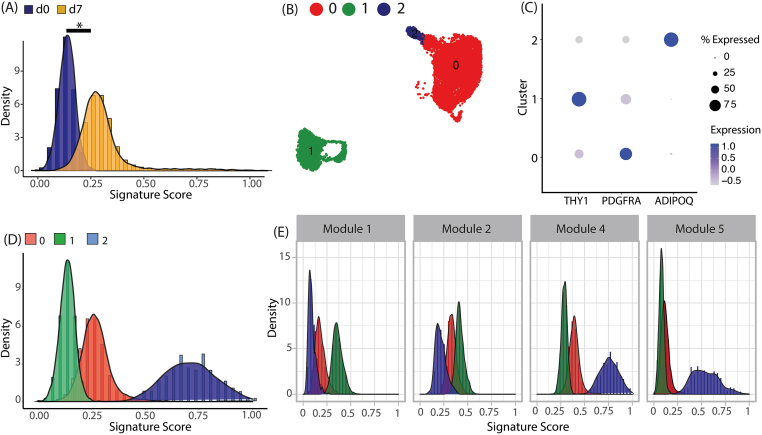
Next, we focused on investigating whether lineage-specific gene signatures defined in our study would recapitulate small-scale differences in degree of adipogenic differentiation. Consequently, we focused on mapping our signature scores onto a white adipogenesis time-course dataset, where scRNA-seq was performed on human-derived mesenchymal stem cells (MSCs) at day 0 and day 7 of adipogenic differentiation (Fig. S4F, Rauch et al., 2019). Indeed, cells isolated on day 7 of differentiation had a significantly higher signature score than cells isolated on day 0 (Figure 5A). Next, unsupervised clustering of the Rauch data identified 3 major clusters, cluster 1 consisting of day 0 MSCs (stem cells marked by THY1), cluster 0 consisting of day 7 differentiating preadipocytes (marked by PDGFRA), and cluster 2 consisting of day 7 mature adipocytes (marked by ADIPOQ, Figure 5B, C). Assessment of cluster-level white adipogenesis signature scores revealed that cluster 2 had the highest signature score, followed by cluster 0, the day 7 preadipocytes and then cluster 1, the day 0 progenitors (Figure 5D). We also defined the list of genes identified in each of the down-regulated (Module 1 and 2) and up-regulated modules (Module 4 and 5) as its own signature, and assigned scores to cells in each cluster based on their combined expression. Scores for downregulated gene modules 1 and 2, which primarily consisted of genes related to cell-adhesion and fibrillar ECM components, were highest in day 0 progenitors (cluster 1), followed by day 7 preadipocytes (cluster 0) and day 7 adipocytes (cluster 2, Figure 5E). For upregulated modules 4 and 5, which consisted of genes related to adipogenic and lipogenic response, day 7 adipocytes (cluster 2) had the highest score (Figure 5E). Additionally, for module 5, which consisted of genes with delayed up-regulation, day 0 progenitors (cluster 1), and day 7 preadipocytes (cluster 1) had similar scores (Figure 5E). This was different from module 4, where day 0 progenitors (cluster 1), and day 7 preadipocytes (cluster 1) had distinct scores, likely because genes in this module were consistently upregulated (Figure 5E). Taken together, these observations indicate that the in vitro derived transcriptional signatures identified in our study correlated well with in vivo differentiation dynamics.
Figure 5.
Mapping in vitro adipogenesis onto primary huma adipose tissues (A) Distribution of gene signature score for day 0 (d0) and day 7 (d7) cells from Rauch et al. (B) Sub-clustering of Rauch et al. dataset as visualized on UMAP (C) Gene expression for key progenitor and adipocyte marker genes in Rauch et al. sub-clusters (D) Distribution of gene signature score for sub-clusters in Rauch et al. (E) Distribution of module score for sub-clusters in Rauch et al.
2.6. Adipogenic gene signature reveals previously unidentified differences in cell maturation states
Utilization of gene signatures in our dataset enabled ordering of preadipocytes by their degree of differentiation. The applicability of these signatures can also be extended to other primary adipose tissue scRNA-seq/snRNA-seq datasets to investigate differences in degree of adipogenic differentiation that may be driven by factors such as metabolic phenotype, diseased state, BMI etc. This can be achieved by using appropriate signatures (based on the lineage) as input to Vision to assign scores to cells of interest in a given dataset. The assigned score can then be tested for statistical difference, thereby providing comparative, quantitative insights into degree of adipogenic differentiation. Since scores assigned using Vision are specific to a given dataset, it would be inappropriate to compare scores across distinct datasets.
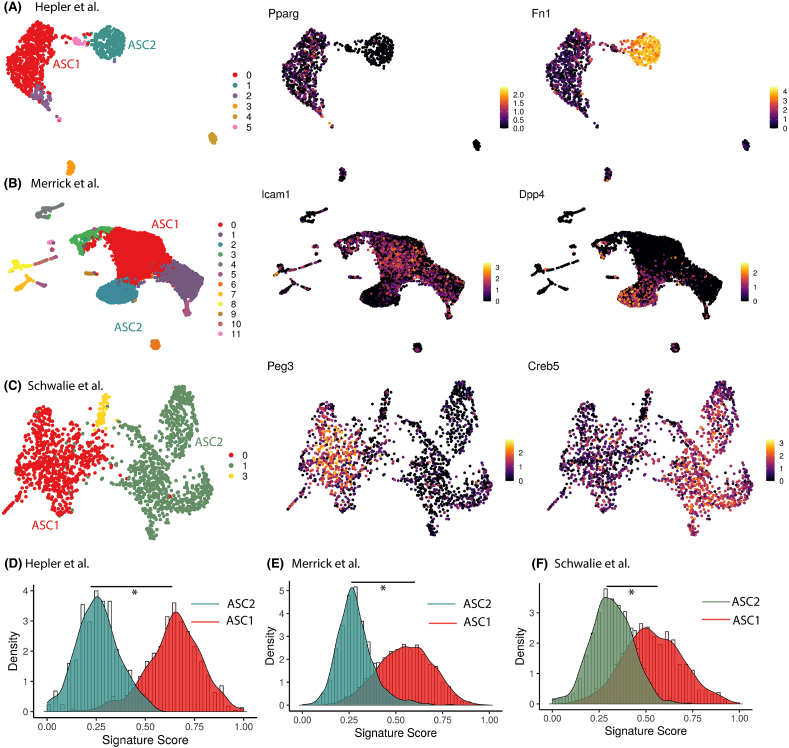
Here, we used these signatures to assess maturation state differences in recently identified murine white adipocyte precursors ASC1 and ASC2 [11,12]. ASC1 and ASC2 were recently identified as two distinct preadipocyte-types in multiple mouse scRNA-seq studies (Figure 6A–C), with in vitro studies reflecting differential
Figure 6.
Cell maturation state assessment for murine white adipocyte precursors ASC1 and ASC2 (A) to (C) UMAP visualization of murine WAT scRNA-seq datasets from different studies colored by cluster (left panel), ASC1 marker expression (middle panel) and ASC2 marker expression (right panel) (D) to (F) Distribution of gene signature score for ASC2 and ASC1 cells from different studies. Score was scaled to vary from 0 to 1.
adipogenic capacity, and hence differential therapeutic capacity, within these cell populations [90,91,92]. However, in vivo studies revealed a transition of ASC2 into ASC1 prior to becoming adipocytes [91], thereby suggesting a less committed progenitor state for ASC2 cells as compared to ASC1. Overall, it remains unclear whether ASC1 and ASC2 cells are distinct cell-types or cells with different maturation states. Hence, we used our gene signatures to determine whether maturation state differed between these cells. We applied our signatures to three distinct scRNA-seq studies that identified ASC1 and ASC2 precursor cells (Figure 6A–C) in murine WAT [90,91,92]. In all three studies, the signature score was significantly higher in ASC1 cells compared to ASC2 (Figure 6D–F), further corroborating the hypothesis that the two precursors are cells at different stages of adipogenic maturation, rather than being two distinct cell-types.
2.7. Differential expression and signature score analyses reveal inhibited adipogenic expansion in obese metabolic phenotype
In healthy adipogenic expansion, new adipocytes are derived from de novo differentiation of adipose progenitors or stem cells (hyperplastic expansion). In obesity, however, dysregulated adipogenesis results in excess fat being stored in existing adipocytes, resulting in their enlarged and inflamed phenotype (hypertrophic expansion; [93]. Here, using our dataset, we investigated which of the genes dynamically regulated during healthy adipogenesis are differentially expressed in lean vs obese individuals.
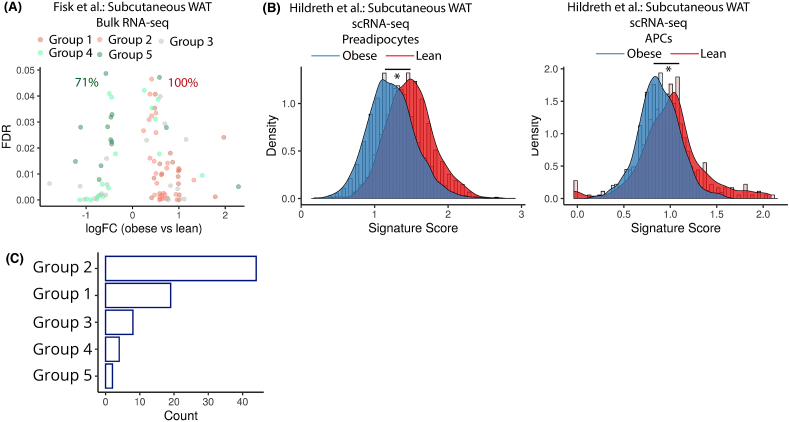
First, we worked with 3 publicly available WAT datasets, 2 of which profiled gene expression using bulk RNA-sequencing [94,95] and the third using microarray [96]. We observed that many dynamically regulated genes in our study were differentially expressed between obese and lean samples (Table S4). Particularly, a majority of Module 1 and Module 2 genes were differentially enriched in obese samples in all 3 studies (Figure 7A and Fig. S5A). This is expected since downregulated genes primarily include CAMs and fibrillar ECM components, which exhibit increased production and accumulation during adipose tissue fibrosis in obesity [47]. Conversely, Module 4 and Module 5 genes, which primarily included adipogenic and lipogenic markers, were preferentially differentially enriched in lean samples in all 3 studies (Figure 7A and Fig. S5A). This observation was consistent with previous reports demonstrating downregulation of adipogenic genes in obesity [97,98], possibly due to hypertrophic adipose tissue expansion, as well as adipocyte dedifferentiation [99].
Figure 7.
Implication of dynamically regulated genes during human adipogenesis in obesity (A) Volcano plot of dynamically regulated genes DE between lean vs obese bulk RNA-seq samples. Each dot is a gene colored by its original gene module annotation. The number in green indicates percent of total DE genes in Module 4 and 5 that are enriched in lean samples. The number in red indicates percent of total DE genes in Module 1 and 2 that are enriched in obese samples. (B) Distribution of adipogenic gene signature scores between lean and obese patient in Preadipocytes (left panel) and Adipocyte precursors (APCs; right panel). (C) Distribution of all dynamically regulated genes during white adipogenesis that are also reported in GWAS datasets according to their annotation using gene modules from the current study. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
At the bulk level, our results suggested there may be adipogenic inhibition phenotypes in samples from individuals with obesity. However, it remains unclear whether similar molecular differences also exist at the preadipocyte level, since cell-type level resolution is lost in bulk measurements. Using a recently published scRNA-seq WAT dataset with comparison of lean vs obese humans [100], we utilized our white lineage gene signature to investigate differences in maturation state at the preadipocyte level. The original study identified two distinct clusters, one of adipocyte precursors (APCs), and another of preadipocytes (Fig. S5B). Preadipocytes are known to be derived from APCs as part of the differentiation process, and this trend was also recovered by our signature analysis (Fig. S5C) Notably, the original study reported a lower fraction of APCs, and a higher fraction of preadipocytes in lean patients, suggesting an accelerated development from APCs to preadipocytes in lean individuals. Our signature analysis revealed a significantly higher maturation score for both preadipocytes and APCs in lean samples (Figure 7B), suggesting existence of adipogenic inhibition amongst the two cell populations associated with obesity.
Besides gene expression profiling, genome-wide association studies have also been critical in linking genetic variants to obesity and metabolic disease risk. Here, we took an integrative approach to further analyze our adipogenic-molecular findings in light of recent GWAS studies and asked whether genes dynamically regulated in human adipogenesis are also linked to metabolic traits associated with increased genetic risk for obesity.
Using publicly available GWAS datasets (see Methods), we identified SNPs that are associated with metabolic traits such as BMI, waist circumference, and hip circumference [101,102]. In total, we analyzed datasets from 19 studies identifying over 1000 SNPs localized within the genic regions of 984 distinct genes. 77 of those 984 genes were temporally regulated during differentiation of white preadipocytes in our dataset. 44 of the 77 genes belonged to Module-2 which was associated with ECM and cytoskeletal remodeling during adipogenesis (Figure 7C). This observation agrees with dysfunctional ECM remodeling being a hallmark of obesity, in which excessive lipid accumulation in adipocytes provokes an excess of deposition of ECM components such as collagens, elastin, and fibronectin in adipose tissue [103,104]. Of the 77 genes, 15 genes were transcription factors that were both temporally regulated during white adipogenesis and associated with metabolic disease risk traits (Fig. S5D). The transcription factor EBF1, which was differentiallyly regulated during brown adipogenesis, was also associated with metabolic disease risk traits. This observation provides further evidence for a potentially thermogenic role of EBF1 [105].
3. Discussion
In this study, we decipher the transcriptional dynamics of human white and brown fat development using an in vitro model system. Utilization of the in vitro model system enables high-resolution sampling of the entirety of adipogenic differentiation spectrum. We ordered differentiating preadipocytes from the white and brown lineages by their progression through differentiation based on lineage-specific adipogenic gene signature scores. After cellular ordering, unsupervised gene clustering revealed five transcriptional modules with distinct expression dynamics that result in the generation of mature white and brown adipocytes. We also validated the applicability of this in vitro model to human-derived primary cells and tissues, by demonstrating the utility of our gene-signatures and -modules to recover differences in degree of adipogenic differentiation on large-scale single cell atlas datasets. Through trajectory comparison, we identified novel TFs potentially involved in regulation of adipogenic or thermogenic transcriptional responses. Of these TFs, we demonstrated the role of a novel transcription factor TRPS1 on adipocyte differentiation and showed that its KD impairs human white adipogenesis in vitro.
Within both white and brown adipogenesis, the early underlying transcriptional program was revealed to be centered around ECM remodeling. Although down-regulation of fibrillar ECM and up-regulation of basement membrane ECM has been demonstrated during 3T3-L1 adipogenesis, the dynamics of ECM reorganization and its regulation during human adipogenesis are not well understood. Our findings provide a high-resolution view into the expression dynamics of specific ECM components during healthy adipose tissue expansion, as well as potential ECM remodeling regulated by an interplay of metalloproteases and its inhibitors. In future investigations, our dataset could be used as a reference for analyses into differential expression dynamics of ECM components or metalloproteases in healthy vs metabolically diseased states.
An important aspect of our model system is the isolation of white and brown preadipocytes from a single individual, and a single anatomical location. Such a paired isolation strategy eliminates the effect of genetic variation, and enables investigation not just within, but also across white and brown lineages. Using our model system, we identified TRPS1, RFX8 and SOX5 as new TFs with potential adipogenic regulatory activity. Moreover, based on differential regulation/enrichment in brown adipogenesis over white adipogenesis, we also identified novel TFs ZNF117 and IRX6 with potential involvement in thermogenic regulation. Further functional investigations focusing on TRPS1 confirmed its role in regulating human white adipogenesis via regulating expression of key adipogenic TFs CEBPA, PPARG, ZFP423, and NR4A1. It remains to be understood whether TRPS1 plays a role in differentially regulating healthy vs diseased adipogenic expansion. Ultimately, identification of novel TFs helps discern specific pharmacological targets for stimulating metabolically healthy, as well as thermogenic fat development.
In order to assign a pseudo-time to differentiating human preadipocytes, we identified gene sets which defined transcriptional signatures specifically associated with white or brown fat development. Scores for these transcriptional signatures could be utilized as a quantitative metric to investigate differences in cellular maturity across varying metabolic conditions, and for different anatomical locations. Such an analysis could help us better understand the differential roles of fat depots towards obesity development and progression. Moreover, our gene signature provides a targeted list of temporally, and biologically relevant genes, which could be specifically profiled using techniques such as spatial transcriptomics, or in situ hybridization/sequencing [106,107], to better understand the spatial organization of adipose tissue development in primary samples. Finally, our gene signature could also be used to understand adipocyte dedifferentiation, and investigate molecular differences between dedifferentiated preadipocytes, and differentiating preadipocytes, at similar stages of maturity.
3.1. Limitations of the study and future directions
Our study design also informs on the future directions for comprehensively investigating molecular regulation of human adipogenesis. In this work, adipogenic transcriptional dynamics were investigated using an immortalized, in vitro system of human white and brown preadipocytes. This dataset will serve as reference to further understand molecular circuitry of fat development in primary adipose tissue samples. Additionally, the currently employed in vitro model system was isolated from a single individual, and from a single anatomical location. And as such, the transcriptional landscape generated here could be made even more comprehensive by isolating similar model systems across multiple individuals and depot locations. In our work, nuclear isolation was performed on days 0, 5, 10, 15, and 20 of differentiation, with sampling of nuclei at intermediate stages due to asynchronous nature of adipogenesis. Further research will likely augment our transcriptional landscape, by honing on specific stages of adipogenesis, for example, very early differentiation time-points, and harvesting nuclei from targeted days of adipogenic induction at higher temporal resolution. During QC of snRNA-seq libraries analyzed in this investigation, we observed varying levels of background mRNA contamination in individual samples. Single-nuclei extraction involves breaking apart the cellular matrix to isolate nuclei, which releases high amounts of debris and cytoplasmic mRNA. During droplet-based single nuclei isolation, this debris gets encapsulated in the droplet along with the nuclei, leading to background mRNA contamination [108,109]. This varying mRNA contamination makes it challenging to identify nuclei that are at similar stages of differentiation but distributed across different days of harvest (different single-nuclei libraries). scRNA-seq dataset integration algorithms do mitigate this challenge partially. However, development of integration strategies taking into account varying background mRNA distribution in snRNA-seq datasets would provide more accurate insights into the gene expression dynamics during differentiation processes.
Overall, our study identifies a new transcription factor TRPS1 for its role in regulating human white adipogenesis, and takes the first steps towards understanding the nature of adipogenic differentiation at a high temporal and cellular resolution in humans. These findings will therefore serve as a resource for multiple efforts into investigating adipose tissue biology in health, as well as disease, ultimately enabling newer therapeutics for improved clinical tackling of a variety of metabolic disorders. Fundamentally, our dataset provides a high-resolution resource for mapping different adipogenic cell-types and states in the human adipose tissue and therefore, serves as a reference for undertakings such as mapping the Human Cell Atlas [110].
4. Methods
4.1. Isolation and immortalization of primary human white and brown fat progenitors
The generation of immortalized human white and brown preadipocytes was described in [29,30] and reproduced here for reference. Briefly, neck fat was collected from a male subject (age 56, BMI 30.8). Isolated SVF cells were subjected to immortalization by overexpression of hTert. Specifically, subcutaneous and subplatysmal neck fat depots were pooled to generate hWAT-SVF, and deep neck fat depots collected from the carotid sheath, longus colli and prevertebral regions were combined for generation of hBAT-SVF. Freshly resected fat depots were collected, minced, and digested using collagenase 1 (2 mg/mL in PBS with the addition of 3.5% BSA; Worthington Biochemical Corporation, Lakewood, NJ), and the SVF was isolated. SVF cells were expanded in culture and split a few times before immortalization. To immortalize, SVF cells were infected with retroviral particles encoding the plasmid pBABE-Hygro-hTERT or pBABE-Neo-hTert (Addgene, Cambridge, MA, USA). Following retrovirus infection, cells were selected with appropriate antibiotics for two weeks. Once drug selection was finished, immortalized cells were allowed to grow in DMEM medium containing 10% FBS. Multiple previous studies have performed transcriptomic and functional analyses that confirm that the cells used in our study are bona fide human brown and white preadipocytes [29,31].
4.2. Preadipocyte culture and adipogenic differentiation
Detailed protocol for maintenance, cryopreservation, and differentiation of white and brown preadipocytes are outlined in a different study [111]. Briefly, for culturing preadipocytes, cells were grown in DMEM medium (Corning, 10-017-CV) supplemented with 10% vol/vol FBS and containing 1% vol/vol Penicillin-Streptomycin (Gibco). Cell culture was maintained at 37 °C in a humidified incubator containing 5% vol/vol CO2. 80% confluent cells were passaged using 0.25% trypsin with 0.1% EDTA (Gibco, 25,200-056) for a 1:3 split in a new 100 mm cell culture dish (Corning).
Prior to adipogenic differentiation, white preadipocytes were allowed to grow up to 100% confluence in a 100 mm cell culture dish (Corning). After 48 h at 100% confluence, growth media was replaced with adipogenic induction media every 48 h for the next 20 days. Induction media was prepared by adding 1 mL FBS, 500 μl Penicillin-Streptomycin, 15 μl human Insulin (0.5 μM, Sigma–Aldrich, I2643-50 MG), 10 μl T3 (2 nM, Sigma–Aldrich,T6397-100 MG), 50 μl Biotin (33 μM, Sigma–Aldrich, B4639-100 MG), 100 μl Pantothenate (17 μM, Sigma–Aldrich, P5155-100G), 1 μl Dexamethasone (0.1 μM, Sigma–Aldrich, D2915-100 MG), 500 μl IBMX (500 μM, Sigma–Aldrich, I7018-100 mg), and 12.5 μl Indomethacin (30 μM, Sigma–Aldrich, I7378-5G) to 48.5 mL DMEM medium and sterile filter.
4.3. Experimental design
Our current study focuses on generating a transcriptional map for the broader course of entire adipogenic differentiation in human WAT and BAT. As a result, snRNA-seq was performed on days 0, 5, 10, 15, and 20 of both white and brown adipogenic differentiation in our study. Although, nuclei are harvested at specific days of differentiation, we can sample nuclei at all stages of adipogenesis due to the asynchronous kinetics of differentiation progression. Additionally, to ensure that the adipocytes are fully differentiated, we opted for a conservative nuclear isolation 20-days post differentiation induction. Indeed, via regulating expression of C/EBPα, PPARγ, ZFP423, and NR4A1 S1E demonstrates that brown adipocytes derived on Day 20 are more mature than brown adipocytes derived on Day 15, thereby enabling sampling of nuclei from fully differentiated adipocytes.
4.4. Nuclei isolation from differentiating preadipocytes and snRNA-seq
Nuclei were isolated from differentiating white and brown preadipocytes using an NP-40 based lysis buffer: To 14.7 mL nuclease-free water (Qiagen), 150 uL of Tris-Hydrochloride (Sigma, T2194), 30 uL of Sodium Chloride (5 M; Sigma, 59222C), 45 uL of Magnesium Chloride (1 M; Sigma, M1028), and 75 uL of NP-40 (Sigma, 74,385) was added. Nuclei harvested on day 0 were isolated from preadipocytes prior to adipogenic differentiation. Two 100 mm dishes were used for nuclei isolation from each preadipocyte type. 500 uL of NP-40 based lysis buffer was added to each 100 mm dish and a cell scraper was employed to release adherent cells from the plates. On day 10, 15, and 20, where cells had visible lipid droplet accumulation, dounce homogenizer was used on scraped out cells to separate the lipids. Cells were then incubated with the lysis buffer for 5 min on ice in a pre-chilled 15 mL falcon tube. Cells were washed with ice-cold PBS supplemented with .2 U/uL RNase Inhibitor (Protector RNase Inhibitor; henceforth called wash buffer) 4 times by centrifuging at 500 rcf for 5 min at 4C. Wash buffer was aspirated after the final round of centrifugation and nuclei were resuspended in the ice-cold wash buffer and filtered using a 40 um cell strainer. Final concentration was adjusted to ∼1000 nuclei/uL using a hemocytometer for downstream sequencing. Nuclei were also stained using 0.08% trypan blue dye to assess nuclear membrane integrity under brightfield imaging. For nuclear isolation on day 10, 15, and 20, the same protocol was implemented as mentioned above with the modification of using 1 mL lysis buffer for each 100 mm dish.
To minimize variation across different days of nuclear isolation during white and brown adipogenesis, preadipocytes derived from the same cryogenic tube were first expanded to culture enough cells for this study. These cells were then seeded at different starting confluency, to allow reaching 100% confluency on staggered days. Upon reaching 100% confluency, cells were maintained for 48 h in growth media, followed by differentiation induction. Post differentiation induction, nuclei were harvested at day 5, 10, 15, and 20 using steps described above. Since differentiation induction was staggered, nuclei collection for each time-point was performed on different days. After preparing nuclei suspension for each day, isolation was performed on the 10× Chromium platform the same day, and libraries prepared as per the manufacturer's protocol using v3 sequencing chemistry. All final libraries were sequenced on the Illumina NovaSeq platform to the following specifications:
4.5. Sequencing data QC and analysis
In total, we had 5 libraries each for the white and brown adipogenesis dataset. For each library, empty droplets were removed using CellBender [109], and doublets were removed using Scrublet [112] or DoubletDetection [113]. Using Seurat, low-quality clusters such as clusters with high MT content, clusters with cellular debris (as marked by the enrichment of translation terms in GO analysis), clusters enriched for empty/doublet barcodes were removed from downstream analysis. Integration of all 5 time points for white and brown dataset was performed using scVI-tools [33]; see Batch Correction and Integration). Post-integration, further QC was performed to remove apoptotic cells. Seurat was used for unsupervised clustering, and differential gene expression analysis. Both intronic and exonic reads were used for all UMI quantification. Transcription factor enrichment analysis was performed using ChEA3. All the analysis scripts used in our study are uploaded here:https://github.com/streetslab/Adipogenesis_time_course
4.6. Batch Correction and Integration
Our experimental design was controlled to limit the variability associated with following parameters:
-
1.
Sequencer Type: All snRNA-seq libraries were sequenced on the Illumina NovaSeq platform.
-
2.
Sequencing Chemistry: All snRNA-seq libraries were prepared using the 10× Genomics v3 chemistry.
-
3.
Cell Culture: Preadipocytes derived from the same cryogenic tube were first expanded to culture enough cells for this study. These cells were then seeded to start cellular culture for harvesting nuclei.
However, due to experimental constraints related to lengthy nuclear isolation protocols, nuclei for only one time point were collected every day. These isolated nuclei were then processed on the 10× Genomics Chromium platform for downstream library preparation the same day, and final libraries were submitted for Illumina Sequencing the next day. Hence, for all 10 time points (5 each for white and brown adipogenesis), nuclear isolation was done on different days, library preparation was done on different days, and Illumina sequencing was done on different days. However, a general consensus in the sequencing community is that batch effects based on flow cells are minimal. Furthermore, technical notes from 10× Genomics demonstrate no batch effects when comparing the same sample run across multiple chips (Tech Note: https://kb.10xgenomics.com/hc/en-us/articles/115003122252-Are-batch-effects-observed-across-Chip-A-). Therefore, in our study, the main technical variable was nuclear isolation of each time point on different days. Hence, for downstream data integration, each time point was defined as a batch.
Integration of all 5 time-points from each lineage in our study was performed using scvi-tools, where each time-point was defined as a batch. In scvi-tools, variable genes from each batch are identified and their intersection is used for integration. Utilization of genes that are commonly variable across batches helps remove batch-specific variation due to batch-specific gene expression. To assess whether integration using scvi-tools worked reasonably well, we referred to the latent space inferred for each white and brown lineage (Figure 1B,F). For white adipogenesis, all five datasets were relatively mixed in latent space, with preadipocytes on one end, and mature adipocytes on another (Figure 1B). For brown adipocytes, scvi-tools was able to correct for nuisance variation due to different days of collection, while maintaining the separation between Preadipocyte-1 and Preadipocyte-2 (Figure 1F and Fig. S1C).
4.7. Pseudo-temporal ordering
4.7.1. Slingshot analysis
For white and brown adipogenesis dataset, the integrated Seurat object was clustered at resolution = 0.4. Slingshot was then used to infer the trajectory using the cluster with the highest contribution from day 0 as the starting cluster. For identifying temporally regulated genes, cells were clustered into 6 equally spaced pseudo-temporal bins. DGE was then performed for each bin against the 1st and last bin, and all genes with logFC $>$ 1 and FDR $<$ 0.05 were considered as temporally regulated. For identifying monotonically increasing genes when cells are ordered using Slingshot, genes were clustered using the ComplexHeatmap package, with k-means clustering algorithm, and the number of clusters set to 5.
4.7.2. Vision analysis
For both white and brown adipogenesis dataset, Vision was used to assign a score to each cell in the integrated Seurat object using the “Hallmark_Adipogenesis” MSigDB signature. This score was used as a proxy for pseudotime. For identifying temporally regulated genes in white adipogenesis dataset, cells were distributed into bins defined using the command cut_points = c (−Inf, seq (0.15, 0.6, 0.15), Inf). For the brown adipogenesis dataset, cells were distributed into bins defined using the command cut_points = c (−Inf, 0.2, 0.3, seq (0.4, 0.6, 0.2), Inf). Temporally regulated genes were identified using the same strategy as defined above. For identifying monotonically increasing genes when cells are ordered using Vision, genes were clustered using the ComplexHeatmap package, with k-means clustering algorithm, and the number of clusters set to 5.
4.7.3. Identifying lineage-specific gene signatures
Once monotonically increasing genes were identified using both Vision and Slingshot, the intersection of the two was taken to define lineage-specific gene signatures. These signatures were used as input to Vision to assign a score to differentiating white and brown preadipocytes, and used as a proxy for pseudotime.
4.7.4. Gene module clustering
For identifying temporally regulated genes in white dataset, when cells are ordered using lineage-specific gene signatures, cells were distributed into bins defined using the command cut_points = c (−Inf, 0.1, seq (0.3, 0.8, 0.1), Inf). DGE was then performed for each bin against the 1st and last bin, and all genes with logFC $>$ 1 and FDR $<$ 0.05 were considered as temporally regulated. Clustering for genes was performed using Seurat with resolution set to 0.5. For the brown adipogenesis dataset, the same steps were used with cut_points defined using the command cut_points = c (−Inf, 0.15, 0.225, seq (0.3, 0.7, 0.2), Inf).
4.8. TRPS1 knockdown analysis during white adipogenesis
4.8.1. siRNA-mediated knockdown in vitro in human adipocytes
White preadipocytes were transiently transfected with 10 nM of siRNAs targeting human TRPS1 or a non-targeting control (NT Control) (Dharmacon) using the transfection reagent Dharmafect in OptiMEM. In detail, 100,000 cells were counted and seeded in wells with transfection mix (Dharmafect + siRNA) in growth medium. Cells were incubated with transfection mix for 48–60 h prior to induction of differentiation.
4.8.2. RNA extraction – reverse transcription
RNA was extracted using the Trizol/Chloroform extraction method. In brief, cells were lysed in Trizol and the aqueous phase was collected after chloroform addition and centrifugation. RNA was pelleted using isopropanol and was subsequently treated using DNase I (NEB) for removal of genomic DNA. After DNase treatment, RNA was pelleted using the sodium acetate – ethanol method. Subsequently, RNA was reverse-transcribed to cDNA using the Applied Biosystems cDNA kit.
4.8.3. qPCR gene expression analysis
cDNA was diluted at a concentration of 5ng/uL and qPCR was performed in a QuantStudio 6 (Applied Biosystems) using SYBR green. Validated primers for 36B4, TRPS1, ADIPOQ, LEPTIN, CIDEA, C/EBPA, C/EBPB, C/EBPD, GATA2, GATA3, ZNF423, and NR4A1 were used. Gene expression analysis was performed using the ddCT method. Results are expressed as relative mRNA expression (fold-change) normalized to the housekeeping gene (36B4).
4.9. Western-blot analysis for TIMP3 during white adipogenesis
White preadipocytes undergoing differentiation were lysed at different time points using RIPA Buffer (Boston BioProducts Inc., Ashland, MA) containing cOmplete protease inhibitor cocktail (Sigma–Aldrich, Dallas, TX). Lysates diluted in Laemmli Buffer (Bio-Rad, Hercules, CA) containing BoltTM Sample Reducing Agent (Thermo Fisher Scientific, Waltham, MA) and boiled at 95 °C for 10 min. Proteins were separated using a 4–20% TGX Stain Free Gel (Bio-Rad, Hercules, CA). Gels were imaged using the ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA) before transferring using Power Blotter System (Thermo Fisher Scientific, Waltham, MA). Membranes were blocked for 1 h using Western Blocking Reagent (Roche, Basel, Switzerland) and probed overnight at 4 °C with anti-TIMP3 (1:2000, ab276134; Abcam, Cambridge, MA) primary antibodies and 30 min at room temperature with HRP-coupled secondary antibodies (1:20,000, 7074; Cell Signaling Technologies, Beverly, MA). Proteins were detected using SuperSignal West Femto ECL substrate (Thermo Fisher Scientific, Waltham, MA).
4.10. GWAS analysis
The GWAS dataset was downloaded from the GWAS catalog (gwas_catalog_v1.0-associations_e100_r2021-04-20. tsv) and subset to metabolic traits defined in Locke et al. [101] and Shungin et al. [102]. The catalog was further subset to SNPs that were mapped to a single gene.
4.11. Bulk RNA-seq and microarray analysis
RNA-seq datasets were downloaded from the GEO Accession Viewer using Accession# GSE25401 and GSE162653. Microarray data was downloaded from the journal's website (oby22950-sup-0007-TableS1.xlsx). Differential expression analysis for RNA-seq datasets was performed using the DESeq package in R.
4.12. Adipogenic signature scoring analysis in publicly available scRNA-seq/snRNA-seq datasets
Vision was used to assign lineage-specific adipogenic signature scores to cells in publicly available scRNA-seq datasets to characterize maturation states. Datasets were downloaded from accession numbers mentioned in the original manuscript.
QC was performed on Rauch et al. data to filter low-quality clusters and non-adipogenic clusters prior to signature analysis (Fig. S4F and G). We filtered cluster 3 which was derived from the day-0 cells but was composed of <1% of the total cells. Furthermore, it did not contain any differentially expressed genes compared to cluster 1 which was primarily composed of day-0 cells as well, indicating that cluster 3 might be an artifact of cellular debris.
Additionally, we identified a second cluster of cells (cluster 4, Fig. S4G) with enrichment for metallothioneins. Metallothioneins have previously not been reported in the context of adipogenic differentiation, but rather are associated with osteogenesis in mesenchymal stem cells (MSCs, Rauch et al. also used bone-marrow derived MSCs, [114,115]. Additionally, Rauch et al. reported that MSCs are more similar to osteoblasts than adipocytes in their undifferentiated state (Rauch et al., 2019). Existing literature also suggests a role of metallothioneins in regulating the metal ion pool necessary for bone growth [116]. Taken together, our findings suggested an osteoblast-like state for MSCs in this cluster, and hence this cluster was removed prior to downstream adipogenic gene signature analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35GM124916. This publication was also supported by grant numbers 2019-002454 and 2022-246198 from the Chan Zuckerberg Initiative DAF, an advised fund of Silicon Valley Community Foundation and grant numbers R01DK102898 and K01DK125608 from the National Institutes of Health. A.S. is a Chan-Zuckerberg Biohub Investigator and a Pew Scholar in the Biomedical Sciences, supported by the Pew Charitable Trusts. A.G. is supported by the University of California at Berkeley Lloyd Fellowship in Bioengineering. We would also like to thank Norma Neff and the CZ Biohub Sequencing Team for lending support with sequencing snRNA-seq libraries.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101746.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The human adipogenesis data (raw and normalized counts, integrated embedding, cell type annotations, and pseudotemporal ordering) is publicly available and can be downloaded via cellxgene: https://cellxgene.cziscience.com/collections/b1912131-de29-4f13-b528-164b45d69cfd. Raw sequence data from this study has been submitted to the NCBI database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/dbgap) under accession number phs002461. v2. p1. The scripts to reproduce the work presented in this study can be found here: https://github.com/streetslab/Adipogenesis_time_course.
References
- 1.Ma Xiuquan, Lee Paul, Chisholm Donald J., James David E. Control of adipocyte differentiation in different fat depots; implications for pathophysiology or therapy. Front Endocrinol. 2015;6:1. doi: 10.3389/fendo.2015.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosen Evan D., Spiegelman Bruce M. Review what we talk about when we talk about fat. Cell. 2014;156(1–2):20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harms Matthew, Seale Patrick. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19(10):1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 4.Shapira Suzanne N., Seale Patrick. Transcriptional control of Brown and beige fat development and function. Obesity. 2019;27(1):13–21. doi: 10.1002/oby.22334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mota de Sá Paula, Allison J. Richard, Hang Hardy, Stephens Jacqueline M. Transcriptional regulation of adipogenesis. Compr Physiol. 2017;7(2):635–674. doi: 10.1002/cphy.c160022. [DOI] [PubMed] [Google Scholar]
- 6.Betz Matthias J., Enerbäck Sven. Human Brown adipose tissue: what we have learned so far. Diabetes. 2015;64(7):2352–2360. doi: 10.2337/db15-0146. [DOI] [PubMed] [Google Scholar]
- 7.Cypess Aaron M., White Andrew P., Vernochet Cecile, Schulz Tim J., Xue Ruidan, Sass Christina A., et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck Brown fat. Nat Med. 2013;19(5):635–639. doi: 10.1038/nm.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blondin Denis P., Nielsen Soren, Kuipers Eline N., Severinsen Mai C., Jensen Verena H., Miard Stéphanie, et al. Human Brown adipocyte thermogenesis is driven by β2-AR stimulation. Cell Metabol. 2020;32(2) doi: 10.1016/j.cmet.2020.07.005. 287–300.e7. [DOI] [PubMed] [Google Scholar]
- 9.Cero Cheryl, Lea Hannah J., Zhu Kenneth Y., Shamsi Farnaz, Tseng Yu-Hua, Aaron M., et al. β3-Adrenergic receptors regulate human Brown/beige adipocyte lipolysis and thermogenesis. JCI Insight. 2021;6(11) doi: 10.1172/jci.insight.139160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Xuejiao, Cervantes Christopher, Liu Feng. Common and distinct regulation of human and mouse Brown and beige adipose tissues: a promising therapeutic target for obesity. Protein Cell. 2017;8(6):446–454. doi: 10.1007/s13238-017-0378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deutsch Alana, Feng Daorong, Pessin Jeffrey E., Shinoda Kosaku. The impact of single-cell genomics on adipose tissue research. Int J Mol Sci. MDPI AG. 2020 doi: 10.3390/ijms21134773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rondini Elizabeth A., Granneman James G. Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochem J. 2020;477(3):583–600. doi: 10.1042/BCJ20190467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caserta F., Tchkonia T., Civelek V.N., Prentki M., Brown N.F., McGarry J.D., et al. Fat depot origin affects fatty acid handling in cultured rat and human preadipocytes. Am J Physiol Endocrinol Metab. 2001;280(2):E238–E247. doi: 10.1152/ajpendo.2001.280.2.E238. [DOI] [PubMed] [Google Scholar]
- 14.Ross Sarah E., Erickson Robin L., Gerin Isabelle, DeRose Paul M., Bajnok Laszlo, Longo Kenneth A., et al. Microarray analyses during adipogenesis: understanding the effects of wnt signaling on adipogenesis and the roles of liver X receptor α in adipocyte metabolism. Mol Cell Biol. 2002;22(16):5989–5999. doi: 10.1128/MCB.22.16.5989-5999.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satish Latha, Michael Krill-Burger J., Gallo Phillip H., Etages Shelley Des, Liu Fang, Philips Brian J., et al. Expression analysis of human adipose-derived stem cells during in vitro differentiation to an adipocyte lineage. BMC Med Genom. 2015;8(1):1–12. doi: 10.1186/s12920-015-0119-8. 2015 8:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tchkonia Tamara, Giorgadze Nino, Pirtskhalava Tamar, Thomou Thomas, DePonte Matthew, Koo Ada, et al. Fat depot-specific characteristics are retained in strains derived from single human preadipocytes. Diabetes. 2006;55(9):2571–2578. doi: 10.2337/db06-0540. [DOI] [PubMed] [Google Scholar]
- 17.Tchkonia Tamara, Tchoukalova Yourka D., Giorgadze Nino, Pirtskhalava Tamar, Karagiannides Iordanes, Armour Forse R., et al. Abundance of two human preadipocyte subtypes with distinct capacities for replication, adipogenesis, and apoptosis varies among fat depots. Am J Physiol Endocrinol Metab. 2005;288(1):E267–E277. doi: 10.1152/ajpendo.00265.2004. [DOI] [PubMed] [Google Scholar]
- 18.Urs Sumithra, Smith Colton, Campbell Brett, Saxton Arnold M., Taylor James, Zhang Bing, et al. Gene expression profiling in human preadipocytes and adipocytes by microarray analysis. J Nutr. 2004;134(4):762–770. doi: 10.1093/jn/134.4.762. [DOI] [PubMed] [Google Scholar]
- 19.Ehrlund Anna, Mejhert Niklas, Björk Christel, Andersson Robin, Kulyté Agné, Åström Gaby, et al. Transcriptional dynamics during human adipogenesis and its link to adipose morphology and distribution. Diabetes. 2017;66(1):218–230. doi: 10.2337/db16-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambele, Anyasi Melvin, Dessels Carla, Durandt Chrisna, Pepper Michael Sean. Genome-wide analysis of gene expression during adipogenesis in human adipose-derived stromal cells reveals novel patterns of gene expression during adipocyte differentiation. Stem Cell Res. 2016;16(3):725–734. doi: 10.1016/j.scr.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Wu Zeni, Wang Suqing. Role of kruppel-like transcription factors in adipogenesis. Dev Biol. 2013;373(2):235–243. doi: 10.1016/j.ydbio.2012.10.031. [DOI] [PubMed] [Google Scholar]
- 22.Gerin Isabelle, Bommer Guido T., Lidell Martin E., Cederberg Anna, Enerback Sven, McDougald Ormond A. On the role of FOX transcription factors in adipocyte differentiation and insulin-stimulated glucose uptake. J Biol Chem. 2009;284(16):10755–10763. doi: 10.1074/jbc.M809115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong Q., Dalgin G., Xu H., Ting C.N., Leiden J.M., Hotamisligil G.S. Function of GATA transcription factors in preadipocyte-adipocyte transition. Science. 2000;290(5489):134–138. doi: 10.1126/science.290.5489.134. [DOI] [PubMed] [Google Scholar]
- 24.Birnbaum Kenneth D. Power in numbers: single-cell RNA-seq strategies to dissect complex tissues. Annu Rev Genet. 2018 doi: 10.1146/annurev-genet-120417-031247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han Xiaoping, Wang Renying, Zhou Yincong, Fei Lijiang, Sun Huiyu, Lai Shujing, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172(5):1091–1107.e17. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Trapnell Cole. Defining cell types and states with single-cell genomics. Genome Res. 2015 doi: 10.1101/gr.190595.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burl Rayanne B., Ramseyer Vanesa D., Rondini Elizabeth A., Pique-Regi Roger, Lee Yun Hee, Granneman James G. Deconstructing adipogenesis induced by β3-adrenergic receptor activation with single-cell expression profiling. Cell Metabol. 2018;28(2) doi: 10.1016/j.cmet.2018.05.025. 300–309.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sárvári Anitta Kinga, Laila Van Hauwaert Elvira, Kruse Markussen Lasse, Gammelmark Ellen, Marcher Ann Britt, Ebbesen Morten Frendø, et al. Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metabol. 2021;33(2) doi: 10.1016/j.cmet.2020.12.004. 43–453.e5. [DOI] [PubMed] [Google Scholar]
- 29.Kriszt Rókus, Arai Satoshi, Itoh Hideki, Lee Michelle H., Goralczyk Anna G., Min Ang Xiu, et al. Optical visualisation of thermogenesis in stimulated single-cell Brown adipocytes. Sci Rep. 2017;7(1) doi: 10.1038/s41598-017-00291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xue Ruidan, Lynes Matthew D., Dreyfuss Jonathan M., Shamsi Farnaz, Schulz Tim J., Zhang Hongbin, et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human Brown and white preadipocytes. Nat Med. 2015;21(7):760–768. doi: 10.1038/nm.3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Chih-Hao, Lundh Morten, Fu Accalia, Kriszt Rókus, Huang Tian Lian, Lynes Matthew D., et al. CRISPR-engineered human Brown-like adipocytes prevent diet-induced obesity and ameliorate metabolic syndrome in mice. Sci Transl Med. 2020;12(558) doi: 10.1126/scitranslmed.aaz8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta Anushka, Shamsi Farnaz, Altemose Nicolas, Dorlhiac Gabriel F., Cypess Aaron M., White Andrew P., et al. Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res. 2022 doi: 10.1101/gr.275509.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gayoso Adam, Lopez Romain, Xing Galen, Boyeau Pierre, Amiri Valeh Valiollah Pour, Hong Justin, et al. A Python library for probabilistic analysis of single-cell omics data. Nat Biotechnol. 2022;40(2):163–166. doi: 10.1038/s41587-021-01206-w. [DOI] [PubMed] [Google Scholar]
- 34.Chen Junye, Lu Yi, Tian Mengyuan, Huang Qiren. Molecular mechanisms of foxo1 in adipocyte differentiation. J Mol Endocrinol. 2019 doi: 10.1530/JME-18-0178. [DOI] [PubMed] [Google Scholar]
- 35.Fan Wu Qiang, Imamura Takeshi, Sonoda Noriyuki, Sears Dorothy D., Patsouris David, Kim Jane J., et al. FOXO1 transrepresses peroxisome proliferator-activated receptor γ transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J Biol Chem. 2009;284(18):12188–12197. doi: 10.1074/jbc.M808915200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeTomaso David, Jones Matthew G., Subramaniam Meena, Ashuach Tal, Ye Chun J., Nir Yosef. Functional interpretation of single cell similarity maps. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-12235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Wenfei, Dong Hua, Balaz Miroslav, Slyper Michal, Drokhlyansky Eugene, Colleluori Georgia, et al. 2020. Single-nucleus RNA-seq reveals a new type of Brown adipocyte regulating thermogenesis. [DOI] [Google Scholar]
- 38.Sharp Louis Z., Shinoda Kosaku, Ohno Haruya, Scheel David W., Tomoda Emi, Ruiz Lauren, et al. “Human BAT possesses molecular signatures that resemble beige/brite cells.” edited by hironori waki. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0049452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medina-Gomez Gema, Gray Sarah, Vidal-Puig Antonio. Adipogenesis and lipotoxicity: role of peroxisome proliferator-activated receptor γ (PPARγ) and PPARγcoactivator-1 (PGC1) Publ Health Nutr. 2007;10(10A):1132–1137. doi: 10.1017/S1368980007000614. [DOI] [PubMed] [Google Scholar]
- 40.Hussain, Faiz Mohammed, Roesler Anna, Lawrence Kazak. Regulation of adipocyte thermogenesis: mechanisms controlling obesity. FEBS J. 2020;287(16):3370–3385. doi: 10.1111/febs.15331. [DOI] [PubMed] [Google Scholar]
- 41.Heglind Mikael, Anna Cederberg, Aquino Jorge, Lucas Guilherme, Ernfors Patrik, Enerbäck Sven. Lack of the central nervous system- and neural crest-expressed forkhead gene Foxs1 affects motor function and body weight. Mol Cell Biol. 2005;25(13):5616–5625. doi: 10.1128/MCB.25.13.5616-5625.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cederberg Anna, Gronning Line M., Ahrén Bo, Taskén Kjetil, Carlsson Peter, Enerbäck Sven. FOXC2 is a winged helix gene that counteracts obesity, hypertriglyceridemia, and diet-induced insulin resistance. Cell. 2001;106(5):563–573. doi: 10.1016/s0092-8674(01)00474-3. [DOI] [PubMed] [Google Scholar]
- 43.MacDougald O.A., Cornelius P., Liu R., Lane M.D. Insulin regulates transcription of the CCAAT/enhancer binding protein (C/EBP) α, β, and δ genes in fully-differentiated 3T3-L1 adipocytes. J Biol Chem. 1995;270(2):647–654. doi: 10.1074/jbc.270.2.647. [DOI] [PubMed] [Google Scholar]
- 44.Steier Z., McIntyre L.L., Lutes L.K., Huang T.S., Robey E.A., Yosef N., et al. Single-cell multi-omic analysis of thymocyte development reveals NFAT as a driver of CD4/CD8 lineage commitment. bioRxiv. 2021 doi: 10.1101/2021.07.12.452119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mariman E.C.M., Wang Ping. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell Mol Life Sci. 2010 doi: 10.1007/s00018-010-0263-4. Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nakajima Ikuyo, Muroya Susumu, Tanabe Ryo Ichi, Chikuni Koichi. Extracellular matrix development during differentiation into adipocytes with a unique increase in type V and VI collagen. Biol Cell Auspices Eur Cell Biol Org. 2002;94(3):197–203. doi: 10.1016/s0248-4900(02)01189-9. [DOI] [PubMed] [Google Scholar]
- 47.DeBari Megan K., Abbott Rosalyn D. Adipose tissue fibrosis: mechanisms, models, and importance. Int J Mol Sci. 2020;21(17) doi: 10.3390/ijms21176030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morandi E.M., Verstappen R., Zwierzina M.E., Geley S., Pierer G., Ploner C. ITGAV and ITGA5 diversely regulate proliferation and adipogenic differentiation of human adipose derived stem cells. Sci Rep. 2016;6(1):1–14. doi: 10.1038/srep28889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spurgin Stephen B., Vishvanath Lavanya, Macpherson Karen A., Hepler Chelsea, Shao Mengle, Gupta Rana K. Identification of Itgbl1: a novel regulator of adipogenesis. 2016. https://utswmed-ir.tdl.org/handle/2152.5/3261
- 50.Ullah Mujib, Michael Sittinger, Ringe Jochen. Extracellular matrix of adipogenically differentiated mesenchymal stem cells reveals a network of collagen filaments, mostly interwoven by hexagonal structural units. Matrix Biol: J Int Soc Matrix Biol. 2013;32(7–8):452–465. doi: 10.1016/j.matbio.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Côté Julie Anne, Lessard Julie, Pelletier Mélissa, Marceau Simon, Lescelleur Odette, Fradette Julie, et al. Role of the TGF-β pathway in dedifferentiation of human mature adipocytes. FEBS Open Bio. 2017;7(8):1092–1101. doi: 10.1002/2211-5463.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Karaman Sinem, Hollmén Maija, Yoon Sun Young, Alkan H. Furkan, Alitalo Kari, Wolfrum Christian, et al. Transgenic overexpression of VEGF-C induces weight gain and insulin resistance in mice. Sci Rep. 2016;6 doi: 10.1038/srep31566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Sooho, Ahn Chihoon, Bong Naeun, Choe Senyon, Lee Dong Kun. “Biphasic effects of FGF2 on adipogenesis.” edited by marià alemany. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0120073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ojima Koichi, Oe Mika, Nakajima Ikuyo, Muroya Susumu, Nishimura Takanori. Dynamics of protein secretion during adipocyte differentiation. FEBS Open Bio. 2016;6(8):816–826. doi: 10.1002/2211-5463.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Al Hasan Mohammad, Martin Patricia E., Shu Xinhua, Patterson Steven, Bartholomew Chris. Type III collagen is required for adipogenesis and actin stress fibre formation in 3T3-L1 preadipocytes. Biomolecules. 2021;11(2):1–17. doi: 10.3390/biom11020156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mor-Yossef Moldovan Lisa, Lustig Maayan, Naftaly Alex, Mardamshina Mariya, Geiger Tamar, Amit Gefen, et al. Cell shape alteration during adipogenesis is associated with coordinated matrix cues. J Cell Physiol. 2019;234(4):3850–3863. doi: 10.1002/jcp.27157. [DOI] [PubMed] [Google Scholar]
- 57.Spiegelman Bruce M., Farmer Stephen R. Decreases in tubulin and actin gene expression prior to morphological differentiation of 3T3 adipocytes. Cell. 1982;29(1):53–60. doi: 10.1016/0092-8674(82)90089-7. [DOI] [PubMed] [Google Scholar]
- 58.Yang Wulin, Guo Xiangxiang, Thein Shermaine, Xu Feng, Sugii Shigeki, Baas Peter W., et al. Regulation of adipogenesis by cytoskeleton remodelling is facilitated by acetyltransferase MEC-17-dependent acetylation of α-tubulin. Biochem J. 2013;449(3):606–612. doi: 10.1042/BJ20121121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lilla Jennifer, Stickens Dominique, Werb Zena. Metalloproteases and adipogenesis: a weighty subject. Am J Pathol. 2002 doi: 10.1016/S0002-9440(10)61100-5. American Society for Investigative Pathology Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernot Denis, Barruet Emilie, Poggi Marjorie, Bonardo Bernadette, Alessi Marie-Christine, Peiretti Franck. Down-regulation of tissue inhibitor of metalloproteinase-3 (TIMP-3) expression is necessary for adipocyte differentiation. J Biol Chem. 2010;285(9):6508–6514. doi: 10.1074/jbc.M109.078444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bouloumié A., Sengenès C., Portolan G., Galitzky J., Lafontan M. Adipocyte produces matrix metalloproteinases 2 and 9: involvement in adipose differentiation. Diabetes. 2001;50(9):2080–2086. doi: 10.2337/diabetes.50.9.2080. [DOI] [PubMed] [Google Scholar]
- 62.Chavey Carine, Mari Bernard, Monthouel Marie-Noëlle, Bonnafous Stéphanie, Anglard Patrick, Van Obberghen Emmanuel, et al. Matrix metalloproteinases are differentially expressed in adipose tissue during obesity and modulate adipocyte differentiation. J Biol Chem. 2003;278(14):11888–11896. doi: 10.1074/jbc.M209196200. [DOI] [PubMed] [Google Scholar]
- 63.Fenech Matthew, Gavrilovic Jelena, Turner Jeremy. Effect of tissue inhibitor of metalloproteinases 3 on DLK1 shedding in cultured human pre-adipocytes and implications for adipose tissue remodelling. Lancet. 2015;385(Suppl 1):S35. doi: 10.1016/S0140-6736(15)60350-6. [DOI] [PubMed] [Google Scholar]
- 64.Meex Ruth C.R., Schrauwen Patrick, Hesselink Matthijs K.C. Modulation of myocellular fat stores: lipid droplet dynamics in health and disease. Am J Physiol Regul Integr Comp Physiol. 2009;297(4):R913–R924. doi: 10.1152/ajpregu.91053.2008. [DOI] [PubMed] [Google Scholar]
- 65.Yu Jinhai, Li Peng. “The size matters: regulation of lipid storage by lipid droplet dynamics.” science China. Life Sci. 2017;60(1):46–56. doi: 10.1007/s11427-016-0322-x. [DOI] [PubMed] [Google Scholar]
- 66.Seale Patrick, Bryan Bjork, Yang Wenli, Kajimura Shingo, Chin Sherry, Kuang Shihuan, et al. PRDM16 controls a Brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hurtado Del Pozo Carmen, Vesperinas-García Gregorio, Rubio Miguel Ángel, Corripio-Sánchez Ramón, Torres-García Antonio J., Jesus Obregon Maria, et al. ChREBP expression in the liver, adipose tissue and differentiated preadipocytes in human obesity. Biochim Biophys Acta, Mol Cell Biol Lipids. 2011;1811(12):1194–1200. doi: 10.1016/j.bbalip.2011.07.016. [DOI] [PubMed] [Google Scholar]
- 68.Xu Haoying, Yang Yanlei, Fan Linyuan, Deng Luchan, Fan Junfen, Li Di, et al. Lnc13728 facilitates human mesenchymal stem cell adipogenic differentiation via positive regulation of ZBED3 and downregulation of the WNT/β-Catenin pathway. Stem Cell Res Ther. 2021;12(1):176. doi: 10.1186/s13287-021-02250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schultz Joshua R., Tu Hua, Luk Alvin, Repa Joyce J., Medina Julio C., Li Leping, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14(22):2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shao Wei, Espenshade Peter J. Expanding roles for SREBP in metabolism. Cell Metab. 2012 doi: 10.1016/j.cmet.2012.09.002. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Talebi Ali, Jonas Dehairs, Rambow Florian, Rogiers Aljosja, Nittner David, Derua Rita, et al. Sustained SREBP-1-dependent lipogenesis as a key mediator of resistance to BRAF-targeted therapy. Nat Commun. 2018;9(1):1–11. doi: 10.1038/s41467-018-04664-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmadian Maryam, Abbott Marcia J., Tang Tianyi, Carolyn S., Hudak S., Kim Yangha, et al. Desnutrin/ATGL is regulated by AMPK and is required for a Brown adipose phenotype. Cell Metabol. 2011;13(6):739–748. doi: 10.1016/j.cmet.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Herzig Sébastien, Shaw Reuben J. Nature reviews molecular cell biology. Nature Publishing Group; 2018. AMPK: guardian of metabolism and mitochondrial homeostasis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fontaine Coralie, Cousin Wendy, Plaisant Magali, Dani Christian, Pascal Peraldi. Hedgehog signaling alters adipocyte maturation of human mesenchymal stem cells. Stem Cell. 2008;26(4):1037–1046. doi: 10.1634/stemcells.2007-0974. [DOI] [PubMed] [Google Scholar]
- 75.Shi Yu, Long Fanxin. Hedgehog signaling via Gli2 prevents obesity induced by high-fat diet in adult mice. Elife. 2017;6 doi: 10.7554/eLife.31649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bi Pengpeng, Shan Tizhong, Liu Weiyi, Yue Feng, Yang Xin, Liang Xin Rong, et al. Inhibition of notch signaling promotes browning of white adipose tissue and ameliorates obesity. Nat Med. 2014;20(8):911–918. doi: 10.1038/nm.3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shan Tizhong, Liu Jiaqi, Wu Weiche, Xu Ziye, Wang Yizhen. Roles of notch signaling in adipocyte progenitor cells and mature adipocytes. J Cell Physiol. 2017 doi: 10.1002/jcp.25697. Wiley-Liss Inc. [DOI] [PubMed] [Google Scholar]
- 78.Ishihara Yasuhiro, Tsuji Mayumi, Vogel Christoph F.A. Suppressive effects of aryl-hydrocarbon receptor repressor on adipocyte differentiation in 3T3-L1 cells. Arch Biochem Biophys. 2018;642:75–80. doi: 10.1016/j.abb.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hrckulak Dusan, Janeckova Lucie, Lanikova Lucie, Kriz Vitezslav, Horazna Monika, Babosova Olga, et al. Wnt effector TCF4 is dispensable for wnt signaling in human cancer cells. Genes. 2018;9(9) doi: 10.3390/genes9090439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choy Lisa, Skillington Jeremy, Derynck Rik. Roles of autocrine TGF-β receptor and smad signaling in adipocyte differentiation. J Cell Biol. 2000;149(3):667–681. doi: 10.1083/jcb.149.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee Haemi, Kim Hyo Jung, Lee Yoo Jeong, Lee Min-Young, Choi Hyeonjin, Lee Hyemin, et al. Krüppel-like factor KLF8 plays a critical role in adipocyte differentiation. PLoS One. 2012 doi: 10.1371/journal.pone.0052474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gubelmann Carine, Schwalie Petra C., Raghav Sunil K., Röder Eva, Delessa Tenagne, Kiehlmann Elke, et al. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. Elife. 2014;3:1–30. doi: 10.7554/eLife.03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reusch J.E., Colton L.A., Klemm D.J. CREB activation induces adipogenesis in 3T3-L1 cells. Mol Cell Biol. 2000;20(3):1008–1020. doi: 10.1128/mcb.20.3.1008-1020.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Meihang, Liu Zhenjiang, Zhang Zhenzhen, Liu Guannv, Sun Shiduo, Sun Chao. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol Chem. 2015;396(3):235–244. doi: 10.1515/hsz-2014-0241. [DOI] [PubMed] [Google Scholar]
- 85.Sanchez-Gurmaches Joan, Tang Yuefeng, Jespersen Naja Zenius, Wallace Martina, Martinez Calejman Camila, Gujja Sharvari, et al. Brown fat AKT2 is a cold-induced kinase that stimulates ChREBP-mediated de novo lipogenesis to optimize fuel storage and thermogenesis. Cell Metabol. 2018;27(1):195–209.e6. doi: 10.1016/j.cmet.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yamamoto Ken-Ichi, Sakaguchi Masakiyo, Medina Reinhold J., Niida Aya, Sakaguchi Yoshihiko, Miyazaki Masahiro, et al. Transcriptional regulation of a Brown adipocyte-specific gene, UCP1, by KLF11 and KLF15. Biochem Biophys Res Commun. 2010;400(1):175–180. doi: 10.1016/j.bbrc.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 87.Seale P. Transcriptional control of Brown adipocyte development and thermogenesis. Int J Obes. 2010;34(Suppl 1):S17–S22. doi: 10.1038/ijo.2010.178. [DOI] [PubMed] [Google Scholar]
- 88.Tong Qiang, Tsai Judy, Gökhan S., Hotamisligil GATA transcription factors and fat cell formation. Drug News Perspect. 2003;16(9):585–588. doi: 10.1358/dnp.2003.16.9.829340. [DOI] [PubMed] [Google Scholar]
- 89.Zhang Yang, Federation Alexander J., Kim Soomin, O'Keefe John P., Lun Mingyue, Xiang Dongxi, et al. Targeting nuclear receptor NR4A1–dependent adipocyte progenitor quiescence promotes metabolic adaptation to obesity. J Clin Investig. 2018;128(11):4898–4911. doi: 10.1172/JCI98353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hepler Chelsea, Shan Bo, Zhang Qianbin, Henry Gervaise H., Shao Mengle, Vishvanath Lavanya, et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. Elife. 2018;7 doi: 10.7554/eLife.39636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Merrick David, Alexander Sakers, Irgebay Zhazira, Okada Chihiro, Calvert Catherine, Morley Michael P., et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science. 2019;364(6438) doi: 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schwalie Petra C., Dong Hua, Zachara Magda, Russeil Julie, Daniel Alpern, Akchiche Nassila, et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature. 2018;559(7712):103–108. doi: 10.1038/s41586-018-0226-8. [DOI] [PubMed] [Google Scholar]
- 93.Jo Junghyo, Gavrilova Oksana, Pack Stephanie, Jou William, Mullen Shawn, Sumner Anne E., et al. “Hypertrophy and/or hyperplasia: dynamics of adipose tissue growth.” edited by Jason A. Papin. PLoS Comput Biol. 2009;5(3) doi: 10.1371/journal.pcbi.1000324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fisk Helena L., Childs Caroline E., Miles, Robert Ayres Elizabeth A., Noakes Paul S., Paras-Chavez Carolina, Kuda Ondrej, et al. Dysregulation of endocannabinoid concentrations in human subcutaneous adipose tissue in obesity and modulation by omega-3 polyunsaturated fatty acids. Clin Sci. 2021;135(1):185–200. doi: 10.1042/CS20201060. [DOI] [PubMed] [Google Scholar]
- 95.Zhou Qiuzhong, Fu Zhenzhen, Gong Yingyun, Seshachalam Veerabrahma Pratap, Jia Li, Ma Yizhe, et al. Metabolic health status contributes to transcriptome alternation in human visceral adipose tissue during obesity. Obesity. 2020;28(11):2153–2162. doi: 10.1002/oby.22950. [DOI] [PubMed] [Google Scholar]
- 96.Arner Erik, Mejhert Niklas, Kulyté Agné, Balwierz Piotr J., Pachkov Mikhail, Cormont Mireille, et al. Adipose tissue microRNAs as regulators of CCL2 production in human obesity. Diabetes. 2012;61(8):1986–1993. doi: 10.2337/db11-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Dubois Severine G., Heilbronn Leonie K., Smith Steven R., Albu Jeanine B., Kelley David E., Ravussin Eric, et al. Decreased expression of adipogenic genes in obese subjects with type 2 diabetes. Obesity. 2006;14(9):1543–1552. doi: 10.1038/oby.2006.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nadler S.T., Stoehr J.P., Schueler K.L., Tanimoto G., Yandell B.S., Attie A.D. The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc Natl Acad Sci U S A. 2000;97(21):11371–11376. doi: 10.1073/pnas.97.21.11371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Song Tongxing, Kuang Shihuan. Adipocyte dedifferentiation in health and diseases. Clin Sci. 2019;133(20):2107–2119. doi: 10.1042/CS20190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hildreth Andrew D., Ma Feiyang, Wong Yung Yu, Sun Ryan, Pellegrini Matteo, Timothy E., et al. Single-cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat Immunol. 2021;22(5):639–653. doi: 10.1038/s41590-021-00922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Locke Adam E., Kahali Bratati, Berndt Sonja I., Justice Anne E., Pers Tune H., Day Felix R., et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518(7538):197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shungin Dmitry, Winkler Thomas W., Croteau-Chonka Damien C., Ferreira Teresa, Locke Adam E., Mägi Reedik, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518(7538):187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lin De, Chun Tae-Hwa, Kang Li. Adipose extracellular matrix remodelling in obesity and insulin resistance. Biochem Pharmacol. 2016;119:8–16. doi: 10.1016/j.bcp.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ruiz-Ojeda, Javier Francisco, Méndez-Gutiérrez Andrea, Aguilera Concepción María, Plaza-Díaz Julio. Extracellular matrix remodeling of adipose tissue in obesity and metabolic diseases. Int J Mol Sci. 2019;20(19) doi: 10.3390/ijms20194888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Angueira Anthony R., Shapira Suzanne N., Ishibashi Jeff, Sampat Samay, Sostre-Colón Jaimarie, Emmett Matthew J., et al. Early B cell factor activity controls developmental and adaptive thermogenic gene programming in adipocytes. Cell Rep. 2020;30(9) doi: 10.1016/j.celrep.2020.02.023. 2869–2878.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Asp Michaela, Joseph Bergenstråhle, Lundeberg Joakim. Spatially resolved transcriptomes-next generation tools for tissue exploration. Bioessays: News Rev Mol Cell Dev Biol. 2020;42(10) doi: 10.1002/bies.201900221. [DOI] [PubMed] [Google Scholar]
- 107.Burgess Darren J. Spatial transcriptomics coming of age. Nat Rev Genet. 2019;20(6) doi: 10.1038/s41576-019-0129-z. 317–317. [DOI] [PubMed] [Google Scholar]
- 108.Alvarez Marcus, Rahmani Elior, Jew Brandon, Garske Kristina M., Miao Zong, Benhammou Jihane N., et al. Enhancing droplet-based single-nucleus RNA-seq resolution using the semi-supervised machine learning classifier DIEM. Sci Rep. 2020;10(1):1–16. doi: 10.1038/s41598-020-67513-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Fleming Stephen J., Marioni John C., Babadi Mehrtash. CellBender remove-background: a deep generative model for unsupervised removal of background noise from scRNA-seq datasets. bioRxiv. 2019 [Google Scholar]
- 110.Regev Aviv, Teichmann Sarah A., Lander Eric S., Amit Ido, Benoist Christophe, Birney Ewan, et al. The human cell atlas. Elife. 2017;6 doi: 10.7554/eLife.27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shamsi Farnaz, Tseng Yu Hua. Protocols for generation of immortalized human Brown and white preadipocyte cell lines. Methods Mol Biol. 2017;1566:77–85. doi: 10.1007/978-1-4939-6820-6_8. Humana Press Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wolock Samuel L., Lopez Romain, Klein Allon M. Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Systems. 2019;8(4) doi: 10.1016/j.cels.2018.11.005. 281–291.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gayoso Adam, Shor Jonathan, Carr Ambrose J., Sharma Roshan, Dana Pe’er. 2019. JonathanShor/DoubletDetection: HOTFIX: correct Setup.py installation. [DOI] [Google Scholar]
- 114.Li Santie, Kim Myeong-Ji, Lee Sung-Ho, Jin Litai, Cong Weitao, Jeong Hye-Gwang, et al. Metallothionein 3 promotes osteoblast differentiation in C2C12 cells via reduction of oxidative stress. Int J Mol Sci. 2021;22(9) doi: 10.3390/ijms22094312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Liu An-Ling, Zhang Zhong-Ming, Zhu Bi-Feng, Liao Zhao-Hui, Zhu Liu. Metallothionein protects bone marrow stromal cells against hydrogen peroxide-induced inhibition of osteoblastic differentiation. Cell Biol Int. 2013;28(12):905–911. doi: 10.1016/j.cellbi.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 116.De-Ugarte Laura, Balcells Susana, Nogues Xavier, Grinberg Daniel, Diez-Perez Adolfo, Garcia-Giralt Natalia. Pro-osteoporotic miR-320a impairs osteoblast function and induces oxidative stress. PLoS One. 2018;13(11) doi: 10.1371/journal.pone.0208131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The human adipogenesis data (raw and normalized counts, integrated embedding, cell type annotations, and pseudotemporal ordering) is publicly available and can be downloaded via cellxgene: https://cellxgene.cziscience.com/collections/b1912131-de29-4f13-b528-164b45d69cfd. Raw sequence data from this study has been submitted to the NCBI database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/dbgap) under accession number phs002461. v2. p1. The scripts to reproduce the work presented in this study can be found here: https://github.com/streetslab/Adipogenesis_time_course.