Abstract
The fecal microbiome of 55 obese children and adolescents (BMI-SDS 3.2 ± 0.7) and of 25 normal-weight subjects, matched both for age and sex (BMI-SDS − 0.3 ± 1.1) was analysed. Streptococcus, Acidaminococcus, Sutterella, Prevotella, Sutterella wadsworthensis, Streptococcus thermophilus, and Prevotella copri positively correlated with obesity. The inferred pathways strongly associated with obesity concern the biosynthesis pathways of tyrosine, phenylalanine, tryptophan and methionine pathways. Furthermore, polyamine biosynthesis virulence factors and pro-inflammatory lipopolysaccharide biosynthesis pathway showed higher abundances in obese samples, while the butanediol biosynthesis showed low abundance in obese subjects. Different taxa strongly linked with obesity have been related to an increased risk of multiple diseases involving metabolic pathways related to inflammation (polyamine and lipopolysaccharide biosynthesis). Cholesterol, LDL, and CRP positively correlated with specific clusters of microbial in obese patients. The Firmicutes/Bacteroidetes-ratio was lower in obese samples than in controls and differently from the literature we state that this ratio could not be a biomarker for obesity.
Subject terms: Biological techniques, Biomarkers, Diseases, Endocrinology, Medical research
Introduction
During the past decades, the prevalence of childhood obesity has dramatically increased worldwide. In most developed countries (the U.S. at first), being overweight or obese is the most common chronic disease in childhood and adolescence and represents a serious public health problem. It has been reported that obesity affects nearly 107.7 million children and adolescents worldwide1. Several factors have been identified as potential risk factors for pediatric obesity and type 2 diabetes: early nutritional and epigenetic mechanisms, maternal malnutrition and microbiota assessment2.
The adverse consequences of obesity include several conditions, like insulin resistance and type 2 diabetes mellitus3. Both increase the risk for cardiovascular and cerebrovascular morbidity and mortality, which shorten life expectancy.
Different articles describe the microbiome of obese children and adolescents, sometimes with conflictual results. These articles are rather heterogeneous on the procedures for studying the microbiome, ranging from culture technique and PCR targeting only a limited number of taxa to Next-Generation sequencing (NGS)4–7. Moreover, it is known that the microbiome could be affected by different drugs such as Metformin and Liraglutide8,9. Most articles used on NGS are based on the PCR amplification of a single polymorphic region of the 16S-ribosomal subunit gene. Part of the generated conflicting results may also depend on the variegated criteria used to classify the obese population enrolled in the analysis. Indeed, previously published results are based on adult or pediatric populations considering a mix of obese and overweight patients selected based on BMI alone or BMI z-score, but few on BMI-SDS in pediatric cohorts. In the present study, the fecal microbiome was assessed using NGS technology, and the children/adolescent patients were grouped based on BMI-SDS. Since it is known that the analysis of a single or of a couple of 16S-hypervariable regions did not give an exhaustive representation of the microbiome10, we opted for the NGS-sequencing of 7 out of the 9 polymorphic 16S-regions.
Results
Microbiome analysis and the Firmicutes/Bacteroidetes ratio
The 16S-mapped reads obtained from all analyzed microbiome samples were 211,854 ± 56,420 and resulted in the identification of 1797 ± 400 operational taxonomic units (OTUs) in the simple obese patients (OB-G), 1761 ± 509 OTUs in the obese with complications (OBc-G) samples and 1747 ± 602 OTUs in normal weight Healthy Donors (nwHD) used as controls. A visual analysis of the Firmicutes and the Bacteroidetes phyla abundances and their Firmicutes/Bacteroidetes (F/B) ratio indicated that the samples from obese patients had always a lower F/B-ratio value than the group of nwHDs, indicating that F/B ratio is not a biomarker for obesity, differently from what proposed by some authors11,12. These data are confirmed both by the analysis of the consensus of seven 16S-polymorphic regions (F/B-ratio: Obese:1.2 ± 0.8 vs nwHD:1.6 ± 1.8, respectively) and by analyzing the V4-region alone (F/B-ratio: Obese:0.9 ± 0.7 vs nwHD:1.2 ± 1.3, respectively). It is to be stressed that the V4 region is the most studied 16S polymorphic region reported in the literature on metagenomics.
Alpha and beta diversity analysis
Different alpha diversity profiling indices have been used that estimate either the community richness (Chao1-index) or richness and evenness (Shannon or Simpson indexes). It is important to highlight that none of the analyzed alpha diversity indexes reached statistical significance in the comparison between the complete case series of obese patients (55 fullOB) and the 25nwHD. In detail, the trend of Chao1 mean values were slightly higher in pathological samples (143.2 ± 22.3) rather than in nwHDs (141.5 ± 30.1) (Supplementary Fig. 1a). Simpson (Si) and Shannon (Sh) indexes behaved in the opposite way, where both were slightly higher in nwHDs (Si: 0.946 ± 0.014–Sh: 5.057 ± 0.320) rather than fullOB (Si: 0.87 ± 0.10–Sh: 2.83 ± 0.45) (Supplementary Fig. 1a). The Bray–Curtis index used for beta diversity analysis clearly showed statistical significance (PERMANOVA p < 0.02) in the comparison between fullOB and nwHDs, thus indicating a difference in microbiome composition between patients and normal weight controls (Supplementary Fig. 1b).
Comparative analysis: fullOBvsnwHD
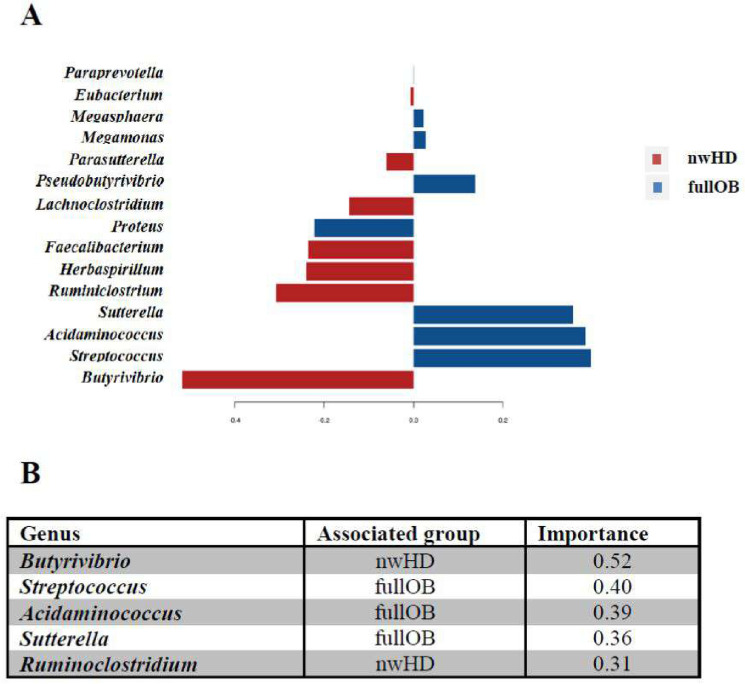
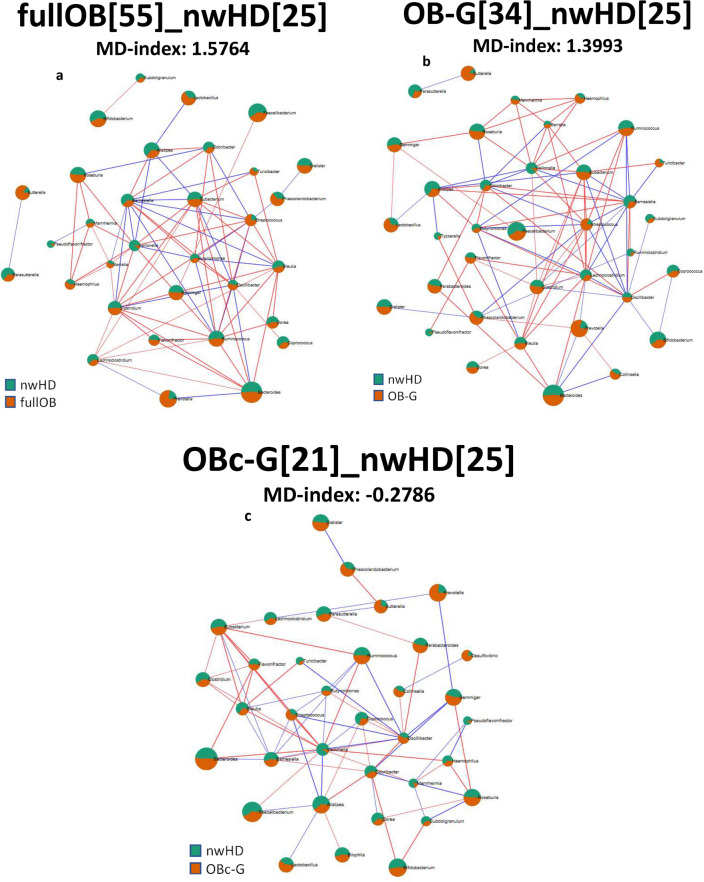
The specific associations between taxa present in fullOB and nwHD were analyzed using the Calypso13,14 package which considers the sparsity (i.e., a dataset with many values equal to zero) and the compositional origin of microbiome data. Specifically, we used the sparse Partial Least Squares-Discriminant Analysis (sPLS-DA)14 that associates the importance of a specific taxon to describe a group of samples (Fig. 1). The results indicated that among the first 5 genera ordered for importance, 3 of them showed a positive correlation with fullOB (Streptococcus, Acidaminococcus and Sutterella with importance scores of 0.40, 0.39, and 0.36, respectively). The relative abundance analysis, using 4 different algorithms within MicrobiomeAnalyst, and the sparse correlation for compositional data (SparCC) analysis confirmed this result (Fig. 2a and Supplementary Table 1). A positive correlation with fullOB was found for Sutterella wadsworthensis and Streptococcus thermophilus, both characterized by 10 times higher abundances in obese patients compared to nwHD (EdgeR log2 fold change values 3.4795 and 3.7707 respectively with p-value < 0.01; see Supplementary Table 1). Prevotella genus and PrevotellA we correlated with full rather than nwHD (Fig. 2a and Supplementary Table 1). The Microbial Dysbiosis index (MD-index) for this comparison is 1.5764 (EdgeR log2 fold change values 3.3504 and 3.3511 respectively with p-value < 0.05; see Fig. 2a) indicating a high imbalance (dysbiosis) in the microbial flora of obese patients compared to controls (nwHD).
Figure 1.
The sPLS-DA results between fullOB (blue) vs nwHD (red) associated fecal microbiome. Loading plot from the sPLS-DA applied to the data to discriminate in the microbiome the Obese (fullOB) patient's associated taxa from the ones linked to controls (nwHD). Colors indicate the classes in which the median is maximum for each significant taxa (red) for regular weight Healthy Donors (nwHD) and blue for Obese (fullOB). The negative and positive values indicate positive and negative associations (importance) identified among the statistically significant identified taxa.
Figure 2.
SparCC correlation networks. Taxa are connected by an edge when their correlation meets the p-value (< 0.05) and the correlation thresholds (0.3). The edge size reflects the magnitude of the correlation. These networks show significant positive (red edges) or negative (blue edges) Pearson correlations. The size of the rounded area for each node represents the abundance of that taxon, and the colors show the proportion of the associated group. (a) Green for normal-weight Healthy Donors (25 subjects) and orange for the complete case seriesof obese patients (55 cases). The MD-index was 1.5764, computed at the genus level for comparing the microbiome of obese patients over the normal weight controls. (b) Green for normal-weight Healthy Donors (25 subjects) and orange for the obese patients (34 cases) with no additional complication. The MD-index was 1.3993. (c) Green for normal-weight Healthy Donors (25 subjects) and orange for the obese with complication patients (21 cases). The MD-index was − 0.2786. In the analysis of the microbiome of plain obese patients (OB-G) compared with the microbial flora present in controls, there is a slightly unbalanced overrepresentation of some genera in the Obese samples rather than in control subjects. Whereas obese patients with complications showed an unbalance due to an underrepresentation of some genera confronted with the same population of controls (nwHD). Note that the MD index of a Eubiotic state is equal to 1.
Comparative analyses: OB-G vs nwHD and OBc-G vs nwHD
The analysis of the more relevant genera using supervised Random-Forest indicated that Streptococcus was a genus tightly linked to OB-G, followed by Sutterella, Clostridium, and Lactobacillus (Fig. 2b and Supplementary Table 2). The species that showed a positive association with OB-Group of samples were Sutterella wadsworthensis and Blautiaproducta(OBB error:0.322; Sensitivity: 0.64 and 0.70 of Specificity; see Supplementary Table 2). Both the Sutterella genus and the Sutterella wadsworthensis species have confirmed a stronger association with OB-G rather than OBc-G, considering the number of different methods reaching statistical significance and also the result of the relative abundance analysis (Fig. 2b,c. Supplementary Tables 2 and 3 EdgeR log2 fold change 3.7642 and 4.6938 with p-value < 0.05 and Supplementary Table 3 EdgeR log2 fold change 2.9930 and 4.1985 with p-value < 0.05). The same was true for the Streptococcus genus (EdgeR log2 fold change 2.6028 with p-value < 0.05)and their descendent species (i.e., S. australis, S. salivarius and S. thermophilus. EdgeR log2 fold change 2.4211, 2.1653 and 5.0708 with p-value < 0.05) that were better described in the OB-Gpatients (Supplementary Table 2). Random-forest's supervised analysis indicated that the genera Lactobacillus, Gemminger (OBB error 0.413; Sensitivity: 0.61 and 0.56 of Specificity), and the species Coprococcus comes and Bacteroides massiliensis (OBB error 0.37; Sensitivity: 0.64 and 0.61 of Specificity) showed higher "mean decrease accuracy" and thus a positive association with the OBc-G patients (Supplementary Table 2). More in detail, in relative abundance analysis the Bacteroides massiliensis showed a statistically significant value only in the comparison between OBc-G and nwHD (Supplementary Table 3 EdgeRlog2 fold change 2.9527 with p-value < 0.05), but not in the comparison between OB-G and nwHD (Supplementary Table 2). On the contrary, Coprococcus comes showed a positive association with both OB-G and with OBc-G group of patients (Supplementary Tables 2–3. EdgeRlog2 fold change 2.1999 and LDA: 2.16, both with p-value < 0.05).
It is to be noted that some Bacteroides genera behave in opposite ways, indeed Bacteroides fragilis, Bacteroides plebeius, and Bacteroides thetaiotaomicron (EdgeR log2 fold change 3.7523, zero-inflated Gaussian fit: 0.0189 and EdgerR log2 fold change 2.1903 with p-value < 0.05) showed higher abundances in OB-G patients compared to nwHD. While, on the contrary, Bacteroides faeces and Bacteroides massiliensis (EdgeR log2 fold change 3.1409 and 2.9527 with p-value < 0.05) showed a positive association with OBc-G compared to nwHD (Supplementary Tables 2–3). The MD-index behaves in opposite ways in the OB-G and OBc-G correlation networks with respect to nwHD. Indeed, MD-index computed for OB-G was 1.3993, showing high dysbiosis, while in OBc-G was-0.2786 (Fig. 2b,c, respectively) which still underlinesa dysbiosis but much less evident for OB-G samples.Indeed, in the group of obese patients without any complication (OB-G), there was a slightly unbalanced overrepresentation of some genera concerning the microbiome of control subjects. Whereas in obese patients with complications, some genera were underrepresented concerning controls (nwHD).
Supervised Random Forest analysis and normal weight healthy donors (nwHD)
We applied the Random Forest machine-learning algorithm15 to identify taxa able to discriminate between patients with obesity and normal-weight donors with good classification performances. We already described the correlations between specific taxa and the OB-G or OBc-G. Here we closely analyzed taxa that classify the normal-weight healthy donor group. Alistipes genus (Supplementary Tables 1, 2: zero inflated-Gaussian fit of 0.0027 and 0.0015), different Alistipes species (Supplementary Table 1: A. finegoldii, A. sp. and A. senegalensis Edge R log2 fold change − 1.4927 and − 1.6049 with p-value < 0.05 and zero-inflated Gaussian fit: 0.0376. Supplementary Table 2: A. indistinctus and A. senegalensis. EdgeR log2 fold change − 2.3560 with p-value < 0.05 and 0.0025 of zero-inflated Gaussian fit. Supplementary Table 4: A. onderdonkii. OBB error: 0.262 or 0.319; Sensitivity: 1.00 or 0.65 and 0.72 or 0.77of Specificity), and the Bifidobacterium longum (Supplementary Table 1: EdgeR log2 fold change − 2.6775 with p-value < 0.01. Supplementary Table 2: EdgeR log2 fold change − 2.095 with p-value < 0.05. Supplementary Table 4: OBB error: 0.356 or 0.319; Sensitivity: 0.61 or 0.65 and 0.66 or 0.77 of Specificity) were strongly associated with nwHD. In addition, the genus Akkermansia (Supplementary Table 1: zero-inlated Gaussian fit: 0.0025. Supplementary Table 2: OBB error: 0.386; Sensitivity:0.57 and 0.65 of Specificity) and its descendants Akkermansia muciniphila (Supplementary Table 1a: zero-inlated Gaussian fit: 0.0376. Supplementary Table 2: OBB error: 0.281; Sensitivity: 0.70 and 0.74 of Specificity) were mainly associated with nwHD (Supplementary Tables 1 and 4). Among Blautiaspecies, a different behavior was observed for Blautiawexlerae since it appeared closely linked to nwHD rather than obese samples (Supplementary Table 4: EdgeR log2 fold change − 3.1199 and p-value < 0.01. Supplementary Table 2: OBB error: 0.281; Sensitivity: 0.70 and 0.74 of Specificity) and other Blautiaspecies (B. faecis was linked to nwHD see Supplementary Table 1c, while B. producta was linked to Overall Severe Obese, see Supplementary Table 2) (Supplementary Tables 3 and 4).
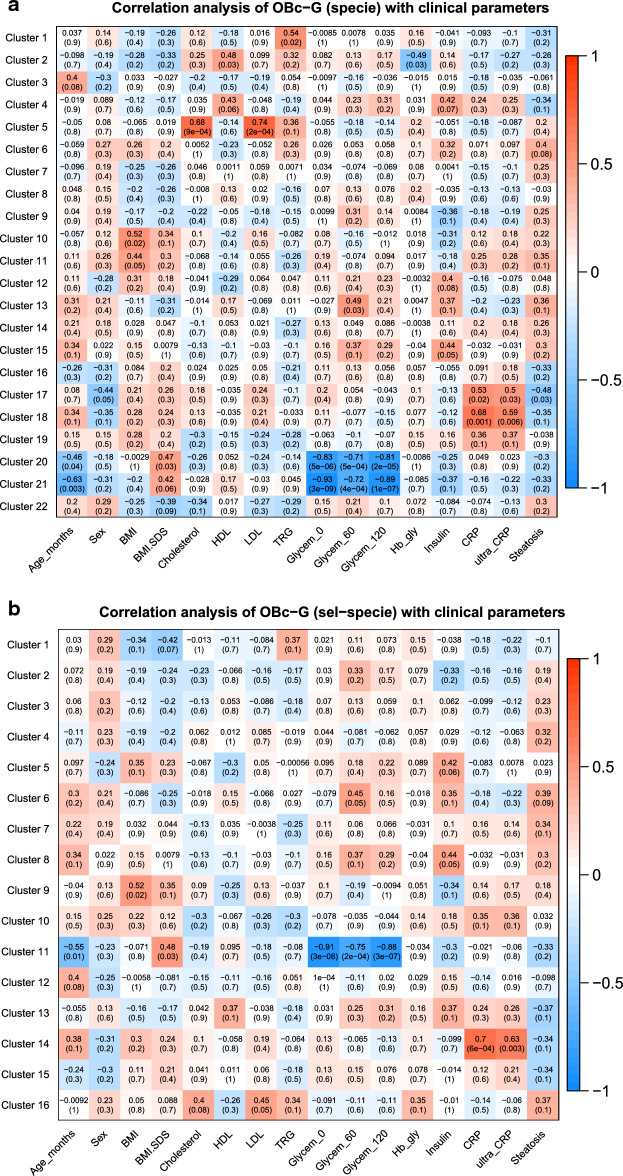
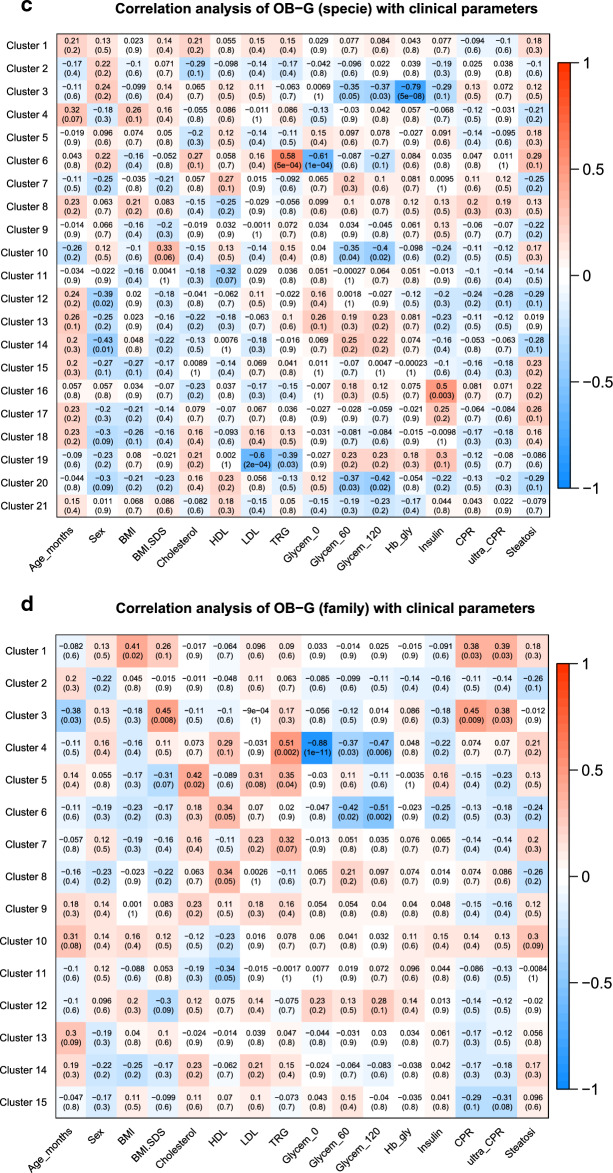
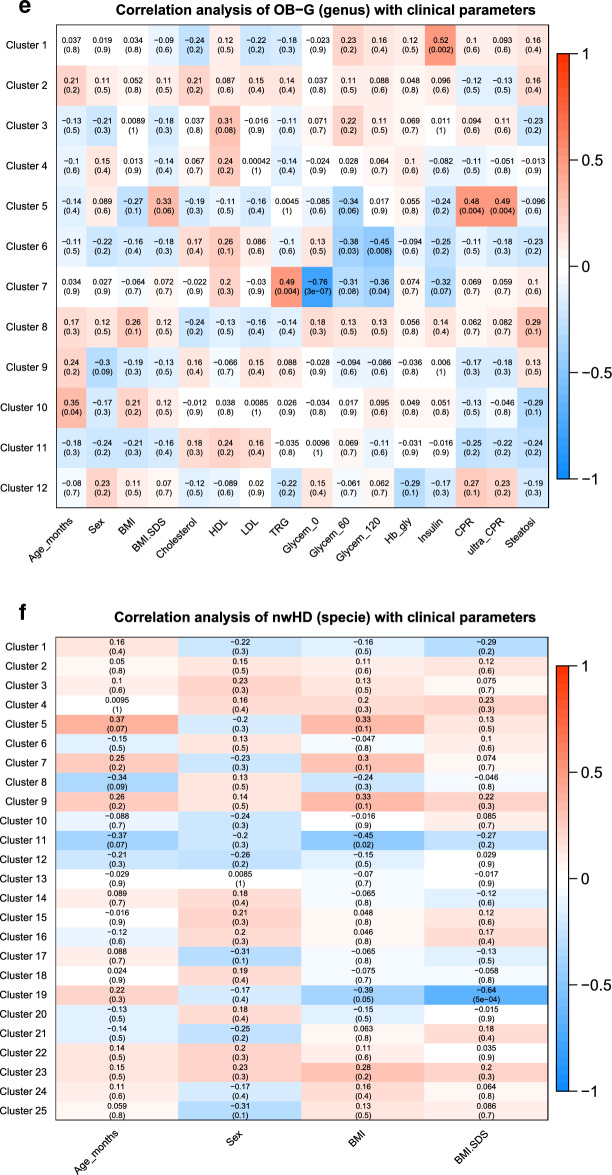
Correlation analysis of taxa clusters with physiological parameters
In the analysis of taxa related to samples of OBc-G, we found interesting positive correlations between some clusters of species and the total cholesterol, LDL levels (Fig. 3a,c and Supplementary Table 5a) and CRP (Fig. 3a,b and Supplementary Table 5a,b). Specifically, the increase of the species enclosed in Cluster 5 is significantly associated with the increase of both total cholesterol and LDL levels (Pearson correlation factor (PC): 0.68 and 0.74 with adj-p-value < 0.05); seemingly the increase of the species enclosed in Clusters 18 (Fig. 3a) and 14 (Fig. 3b), is significantly associated with the increase of CRP, which is a marker of inflammation (PC: 0.59 with adj-p-value 0.04). On the contrary, Glycemia (0′, 60′ and 120′) showed a negative correlation with the species of clusters 20 (PC: − 0.83, − 0.71, − 0.81 with adj-p-values < 0.05), 21 (PC: − 0.93, − 0.72, − 0.89 with adj-p-values < 0.05) (Fig. 3a) and 11 (PC: − 0.91, − 0.75, − 0.88 with adj-p-values < 0.05) (Fig. 3b). In the comparisons of the species analyzed entirely versus the same species analyzed after the feature reduction (Fig. 3a vs. b), we found common species (i) positively associated with CRP and (ii) negatively associated with glycemia (0′, 60′, 120′): specifically, we found (i) Alistipes indistinctus, Clostridium innocuum, Desulfovibrio piger Prevotella ruminicola and Prevotella in common between clusters 18 and 14, while (ii) Acidaminococcus fermentans, Clostridium cocleatum and Clostridium ramosum in common between clusters 20–21 and cluster 11 (see Supplementary Table 5a,b).While the connection of Alistipes indistinctus and Clostridium innocuum with obesity or with clinical parameters related to obesity is not known in the literature, Desulfovibriopiger, Prevotella ruminocula and Prevotella species specie are already known to be associated with inflammation, insulin-resistance, hyperglycemia and type 2-diabetes16. It is important to highlight that Desulfovibrio piger (Clusters 6 for OB-G and Clusters 14, 18 for OBc-G) showed that the increase of the abundance of this species is always associated with the increase in the value of some clinical parameters critical for obesity (such as TRG for OB-G and CRP for OBc-G, respectively).
Figure 3.
Heatmaps of the correlations between taxa clusters and physiological parametersusing WGCNA. The heatmaps show the results of the multivariate clustering analysis considering the physiological parameters and the unadjusted p-values. The red rectangles highlight those correlations that remained statistically significant after the correction for multiple hypotheses with Benjamini–Hochberg. The colored bar aside from the heatmaps shows the color change associated with different Pearson correlation coefficients: the red color indicates positive correlations while the blue color indicates negative correlations. The “sel-specie” refers to the feature reduction step performed before the WGCNA analysis considering the most relevant specie found in our previous analysis (see “Methods” section).
Interestingly enough, in the clusters of taxa found negatively correlated with glycemia (Fig. 3d,e, and Supplementary Table 5d,e) we found that the family Oxalobacteraceae and two genera descending from it, namely Herbaspirillum and Oxalobacter, were already known to be associated with a decreased of insulin-resistance and glycemia17.
Finally, in the analysis performed on the nwHD group, BMI-SDS showed a negative association with five species, enclosed in Cluster 19 (PC: − 0.64 with adj-p-value = 0.05) (Fig. 3f and Supplementary Table 5f). This data strongly suggests that BMI-SDS could represent the most sensible clinical parameter to correctly classify children and adolescents as normal weight, overweight, and obese subjects and that the species belonging to this cluster could be considered protective against obesity.
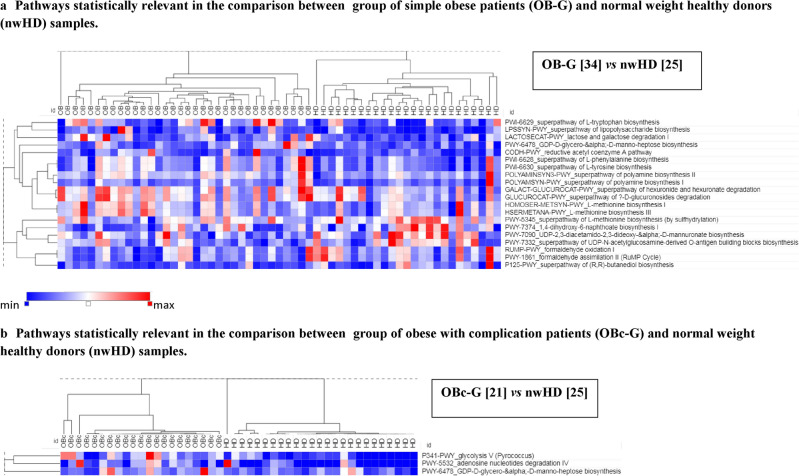
Inferring functional (metabolic) pathways characterizing OB-G and OBc-G
At least 20 different metabolic pathways, inferred with PICRUSt2 and present in the MetaCycdatabase18, showed statistical significance in the comparison between OB-G vs nwHD (Fig. 4a). Different pathways involved in amino acid biosynthesis showed a positive correlation with obesity in pediatric patients. Thus, pathways involving the phenylalanine (PWY-6628) and the tyrosine (PWY-6630) aromatic amino acids were among the entries with the highest statistical importance. In addition, tryptophan (PWY-6629) and methionine biosynthesis (HSERMETANA-PWY and HOMOSER-METSYN-PWY) showed higher abundances in OB-G, while an additional pathway (PWY-5345) for methionine biosynthesis showed an opposite behavior. More, different pathways involved in polyamine biosynthesis known to play a role in bacterial pathogenicity and biofilm formation showed higher abundances in the microbiome of OB-G. Among them, the POLYAMINSYN3-PWY is the one showing the higher importance of all pathways analyzed by Random-Forest analysis (mean decrease accuracy 0.006). Others like POLYAMSYN-PWY and the pro-inflammatory lipopolysaccharide19 pathway (LPSSYN-PWY) showed higher abundances in OB-G compared to lean subjects. More, the second in order of importance in the Random-Forest analysis (mean decrease accuracy 0.004) was the super-pathway of (R,R)-butanediol biosynthesis (P125-PWY) which was overabundant only in nwHDs.
Figure 4.
Heatmaps of the significant inferred pathways identified with PICRUSt2. (a) Heat-map of the comparisons group of Obese with no complication (OB-G) vs controls (nwHD) and (b) group of Obese with complication (OBc-G) vs controls (nwHD).
Obesity in adolescents is often associated with clinical complications such as insulin resistance, hyperglycemia, dyslipidemia, and hypertension, which together are termed "metabolic syndrome". Regarding this issue, we found 3 pathways more abundant in OBc-G with respect to nwHD: PWY-341 (glycolysis V), PWY-5532 (adenosine nucleotides degradation), and PWY-6478 (GDP-d-glycero-alpha-d-manno-heptose biosynthesis) (Fig. 4b). The heptose-sugars are components of bacterial cell surface common in the pro-inflammatory lipopolysaccharide (LPS)20.
Discussion
Microbiome studies might be hampered by different technical problems related to the methods used for their analysis21 not evident to the average readers. The 16S gene is structured in nine variable regions useful to define microbial taxonomy22–24. Primer pairs design is to hybridize in the conserved sequence regions, so it is clear that the choice of their sequence directly influences the taxa composition of the microbiome under analysis. Less important but still relevant in the microbiome definition might be the use of different 16S-ribosomal sequence reference databases. More, additional parameters show only marginal effects in the resulting microbiome composition. Indeed methods of clustering the 16S sequences, bioinformatic pipeline, and the parameters used in the analysis might slightly modify the microbiome composition21. All these issues might be potential biases to complicate the comparison of microbiome biomarker (taxa or Firmicutes/Bacteroidetes-ratio) composition in different publications, even for the same pathology. Due to the not ideal primer pairs design that usually targets a single 16S variable region, it is likely to have bacterial taxa in the analyzed microbiome might be under-represented. Thus, to overcome these problems, a possibility is to increase the number of the variable regions to be studied and to use different 16S-ribosomal sequences reference databases, bioinformatic pipelines, and parameters, thus improving the definition of biomarkers associated with the pathological microbiome. Our aim was not only an academical re-analysis of taxa associated with obesity but with the introduction of the analysis of multiple 16S-variable regions in the microbiome analysis together with the use of rarely used bioinformatic pipelines and parameters try to define which biomarkers were still associated with the obese patients. We believe that comparing the biomarkers defined using different primer pairs, chemistry, and methods with the ones already shown in published reports indicates the taxa that are more strictly associated with the pathology.
The relative abundance analysis using the complete cohort of fecal samples of obese patients (fullOB) compared to nwHD confirmed that Acidaminococcus, Sutterella, Streptococcus, Prevotella, Lactobacillus, and some Bacteroides species correlated with obesity, as shown by different published articles (see a meta-analysis)25. Indeed, the Acidaminococcus genus was reported to be significantly associated with obesity in adult Hispanic subjects26 and with pro-inflammatory diets27. In our data, Acidaminococcus was better associated with the group of patients with Severe Obesity (OB-SO-G) (Supplementary Table 4). Therefore, it was not astonishing that others have found such an increase in patients with type 2 diabetes (T2D)28. Sutterella genus was already described to correlate positively with obesity in obese Chinese children29, but others found the opposite in adults30. In line with this data, Sutterella wadsworthenis was reported to be positively associated with insulin resistance31. Our data showed taxa strongly associated with the microbiome of Obese patients (fullOB, OB-G, and OBc-G) in the comparisons with the fecal flora of normal weight control subjects (nwHD) (Supplementary Tables 1–4).
Streptococcus genus was already shown to be correlated in adult cases with BMI32. In this study, Streptococcus descendant like S.thermophilus was found to be associated with obesity also in our data. These also indicated a positive Prevotellaassociation between and its descendants in obesity. Indeed, the role of the Prevotell and Prevotella copri in obesity is still debated since beneficial and detrimental roles in health have been described for both taxa33,34. Prevotellaceae and the genus Prevotella have been associated with inflammation and insulin resistance25,35. It is also of note that Prevotellacoprishowed to be associated with an altered glucose metabolism leading to glucose intolerance and reduced insulin sensitivity due to the presence of the LeuBgene31. It is also known that Bacteroides and Prevotella genera have a negative correlation with serum leptin levels and a positive correlation with GHrelin serum levels36. Since leptin is known to inhibit hunger while GHrelin increases the drive to eat, the effect of these taxa on obesity is consistent36. Indeed, the leptin sensitizer butendiol (produced in P125-PWY pathways) was found to have a lower abundance in our simple-obese patients than in controls. Thus, the mechanism inducing obesity relative to the P125-PWY might induce a less efficient regulation of appetite by leptin37.
Obesity has been related to an increased risk of multiple diseases involving oxidative stress and inflammation38, and Prevotella species have been already described as being more abundant in obese patients with an inflammatory condition26. Along this line, in our obese patients with complications, at least ten different species were found to correlate positively with standard CRP levels, a marker of chronic inflammation. Among them, we found Prevotella ruminicola, Prevotella sp., the genus Mitsuokella and Desulfovibriopiger. Indeed, Desulfovibrio descendant species are known to produce endotoxins and favor alteration of gut permeability leading to the induction of pro-inflammatory responses39–41. Thisalso applies to the genus Collinsella, which we have shown with greater abundance in severely obese patients39. Furthermore, in our data, Bacteroides correlated with obesity, as described by others who have found a higher abundance in obesity and a positive correlation with BMI42. Some authors have found an inverse correlation between obesity and Bacteroides thetaiotamicron43 compared to us. This discrepancy is intriguing since this specie is known to produce high amounts of small-chain fatty acids (acetate and propionate), and the overproduction of acetate is known to induce hepatic de novo lipogenesis and increase adiposity44.
Our data showed that descendant species belonging to the Gordonibacter genus have a positive correlation with plasma levels of total and LDL cholesterol. Along this line, species belonging to this genus were already known to have a positive association with total cholesterol plasma level45.
A recent report showed that different taxa belonging to the Phylum of the Firmicutes were found in the microbiome of adult obese or overweight patients30. Also, our data showed that Firmicutes descendants Acidaminococcus spp., Lachnospiraceae, Mitsuokella, Ruminococcus spp., Streptococcaceae, and Streptococcus are among the taxa that shared higher abundance values in obese patients. One exception is represented by the Odoribacter genus, although belonging to Firmicutes, which showed a higher relative abundance in normal-weight healthy (nwHD) donors. Indeed, the fact that this genus associates better with nwHDs is not surprising due to the anti-inflammatory potential of this microorganism46. Although the Firmicutes/Bacteroidetes-ratio was believed to be a biomarker for obesity11,12, its role in this condition was found to be contradictory47 since some studies reported a positive correlation between F/B-ratio and the BMI values48, others, like our work, found no correlation or showed an opposite trend48–51 and essentially a dominance of the Bacteroides genus in obesity52. About the inferred metabolic pathways associated with obesity, it is interesting to point out that the abundance of aromatic amino acids (tyrosine, phenylalanine, and tryptophan) has already been reported to be associated with obesity and insulin resistance48. In particular, tyrosine was shown to be the more prevalent amino acid associated with insulin resistance in obese children53. Tryptophan has also been implicated in the pathogenesis of metabolic disorders such as obesity54. Indeed increased levels of tryptophan have been linked to over-nutrition and might be responsible for obesity-related inflammation pathways55. Pro-inflammatory conditions are even supported by the pathways LPSSYN-PWY lipopolysaccharide and PWY-6478, both involved in LPS synthesis and assembly19. These pathways, along with PWY-341 (glycolysis V), are known to play a central role in promoting a pro-inflammatory environment that supports the production of inflammatory mediators by macrophages, thus contributing to insulin resistance. Furthermore, pyruvate, the final product of glycolysis, is metabolized into acetyl-CoA, which is essential for cholesterol and lipid synthesis56. All these conditions are linked to obesity and its complications. In addition, it was shown that the transfer of stools from lean donor recipients into metabolic syndrome patients increased insulin sensitivity of the latter and the abundance of 16 different taxa, including Oxalobacter formigenes. This is in line with the negative correlation found between fasting plasma glucose and Taxa Cluster 1, 2, and 3 shown in Supplementary Table 3a17.
The main limitation of our study is not many pediatric patients enrolled compared to the number of obese subjects in our region.The major strengths include: (i) the mono-centric recruitment, with strict patients selection; (ii) the use of BMI-SDS to define pediatric obesity (in detail, all overweight patients have been excluded); (iii) similar protocols of selection have been used for classifying regular weight healthy donors; (iv) the analysis of multiple 16S-polymorphic regions to define taxonomy and also (v) the use of different algorithms to analyze the microbiome composition among different groups. Furthermore,it is important to highlight thatour results have not been influenced by the use of drugs known to interfere with microbiota composition, such as metformin and liraglutide8,9.
Methods
Patients
Patients were stratified into different groups based on the BMI-SDS values obtained comparing WHO growing curves corrected for sex and month age following the references shown https://www.who.int/tools/growth-reference-data-for-5to19-years/indicators. https://www.who.int/tools/growth-reference-data-for-5to19-years/indicators. The patients were defined as overweight if the BMI-SDS values had a standard deviation (SD): > + 1 SD < + 2; obese when > + 2 SD < + 3, and severely obese SD: > + 3. Monogenic or syndromic obesity were ruled out in all patients. Thus, we evaluated the fecal-microbiome of a total of 55 obese children and adolescents (fullOB) recruited at Giannina Gaslini Institute in Genoa, Italy, between February 2016, and October 2021 (mean age 13.1 ± 2.9, median 13.0, 36% female; BMI-SDS 3.2 ± 0.7). Inclusion criteria were: Caucasian subjects living in Northern Italy, personal history negative for acute or chronic gastrointestinal diseases, and/or antibiotics or probiotics administration in the previous month. All patients were negative for autoimmune disease screening (i.e celiac and thyroid diseases). Among these fullOB samples, 32 patients were grouped based on BMI-SDS as severely obese (mean age 13.5 ± 3.5, median 13.6, 34% female; BMI-SDS 3.6 ± 0.5), while 22 of them were grouped as obese (mean age 13.2 ± 2.4, median 12.9, 36% female; BMI-SDS 2.6 ± 0.2) (see Tables 1, 2).
Table 1.
Simple obese (OB-G) patients.
| Code | Age | Sex | BMI at onset | BMI-SDS | Obeisty subgroup | Total cholesterol | HDL | LDL | TRG | Glycemia 0' | Glycemia 60' | Glycemia 120 | Glicated Hb | Total insulin | CRP | Ultrasensitive CRP | HepaticSteatosis | Metformin therapy | Other clinical infos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5D | 12.8 | F | 37.8 | 3.39 | OB-SO-G | 184 | 40 | ND | ND | 87 | 199 | 174 | 5.6 | 1364 | 0.23 | 2.22 | No | No | Systolic hypertension |
| 6D | 8.8 | F | 31.6 | 3.62 | OB-SO-G | 180 | 35 | 120 | ND | 99 | 142 | 136 | 4.5 | 331 | 0.61 | 4.93 | Yes | No | |
| 7D | 15.8 | M | 36.3 | 3.16 | OB-SO-G | ND | ND | ND | ND | 96 | 117 | 118 | ND | 727 | 0.23 | ND | ND | No | |
| 24D | 15.3 | F | 34.1 | 2.79 | OB-O-G | 177 | 43 | 110 | 93 | 88 | 92 | 89 | 4.87 | 627 | 0.23 | 1.36 | Yes | No | Sodium butyrrate |
| 27D | 9.8 | F | 25.2 | 2.54 | OB-O-G | 160 | 70 | 86 | 55 | 95 | 193 | 132 | 5.28 | 606 | 0.65 | 6.25 | ND | No | |
| 39D | 16.3 | F | 33.4 | 2.65 | OB-O-G | 151 | 40 | 95 | 88 | 89 | 89 | 109 | 5.03 | 316 | 0.23 | 0.79 | Yes | No | |
| 51D | 10.2 | M | 24.6 | 2.64 | OB-O-G | 186 | 66 | 104 | 58 | 90 | 136 | 119 | 5.36 | 558 | 0.23 | 2.83 | No | No | |
| 67D | 11.1 | M | 34.3 | 3.56 | OB-SO-G | 163 | 68 | 97 | 69 | 83 | 104 | 104 | 5.04 | 665 | 0.67 | 6.07 | No | No | Hepatomegaly |
| 79D | 13.8 | F | 39.8 | 3.29 | OB-SO-G | 147 | 47 | 92 | 71 | 104 | 183 | 148 | 5.32 | 1166 | 0.97 | 9.73 | No | Yes | |
| 83D | 12.3 | M | 34 | 3.29 | OB-SO-G | 128 | 47 | 74 | 65 | 98 | 152 | 130 | 5.17 | 244 | 0.54 | 5.11 | ND | No | Obesity and asma |
| 85D | 8.8 | M | 34.6 | 4.4 | OB-SO-G | 130 | 58 | 64 | 114 | 95 | ND | ND | 4.56 | ND | 0.23 | 1.3 | Yes | No | |
| 108D | 10.9 | M | 29 | 3.12 | OB-SO-G | 117 | 44 | 71 | 39 | 85 | 113 | 118 | 5.35 | 302 | 0.23 | 1.95 | No | No | Severe Obesity, Asma and OSAS |
| 120D | 16.7 | F | 33 | 2.58 | OB-O-G | 112 | 38 | 71 | 80 | 115 | 191 | 178 | 5.4 | ND | 0.23 | 1.34 | No | No | Pre-diabetes with insulin resistance. OSAS—hypercapnia |
| 125D | 17.8 | F | 35.5 | 2.9 | OB-O-G | 130 | 40 | 72 | 50 | 83 | 92 | 107 | 4.96 | 488 | 0.23 | 0.93 | No | Yes | Asma. dicoflor therapy. polycystic ovary syndrome (PCOS) |
| 135D | 10.6 | M | 47 | 4.33 | OB-SO-G | 127 | 47 | 68 | 80 | 84 | ND | 101 | 4.95 | ND | 1.42 | 14.45 | No | No | Niaprazine, aripiprazole and depakin therapy |
| 137D | 10.4 | M | 23 | 2.26 | OB-O-G | 165 | 56 | 104 | 55 | 115 | 169 | 189 | 5.75 | 601 | 0.23 | 2.07 | No | No | |
| 138D | 15.9 | M | 43.7 | 3.76 | OB-SO-G | 182 | 54 | 109 | 99 | 94 | 155 | 135 | 5.21 | 804 | 0.23 | 1.39 | Yes | No | Gallbladder stones |
| 140D | 10.6 | M | 36.2 | 3.8 | OB-SO-G | 157 | 69 | 79 | 157 | ND | ND | ND | 5.43 | ND | 0.68 | 6.48 | Yes | No | |
| 146D | 12.5 | M | 29.2 | 2.7 | OB-O-G | 140 | 52 | 79 | 82 | 84 | 135 | 115 | 5.4 | 1128 | 0.48 | 4.63 | ND | No | Essential hypertension under therapy |
| 148D | 17.5 | F | 44.7 | 3.83 | OB-SO-G | 157 | 53 | 94 | 74 | 86 | 118 | 127 | 5.10 | 552 | 0.23 | 3.28 | ND | No | Severe obesity |
| 149D | 12.8 | M | 31.4 | 2.98 | OB-O-G | 178 | 64 | 108 | 43 | 93 | ND | ND | ND | ND | 0.23 | 0.71 | Yes | No | |
| 151D | 14 | M | 42.2 | 3.66 | OB-SO-G | 109 | 58 | 53 | 42 | 93 | 131 | 137 | 5.19 | 289 | 0.47 | 4.24 | Yes | No | Severe obesity and hepatic steatosis |
| 154D | 11.4 | M | 27.2 | 2.74 | OB-O-G | 177 | 42 | 122 | 133 | 88 | 140 | 122 | 5.4 | 748 | 0.23 | 1.5 | Yes | No | Celiac. obesity and steatosis |
| 156D | 13.2 | M | 38 | 3.46 | OB-SO-G | 113 | 53 | 56 | 46 | 87 | 183 | 139 | 5.34 | 2000 | 0.83 | 7.88 | Yes | No | insulin resistance (2000). Mental retardation, q21.1 duplication |
| 160D | 9.8 | F | 24.9 | 2.49 | OB-O-G | 176 | 65 | 97 | 64 | 89 | 187 | 114 | 5.02 | 787 | 0.23 | 2.07 | ND | No | |
| 161D | 10.6 | F | 26 | 2.48 | OB-O-G | 172 | 68 | 96 | 50 | 104 | ND | ND | 4.74 | ND | 0.23 | 0.25 | ND | No | Hypertension |
| 162D | 12.5 | M | 39.8 | 3.65 | OB-SO-G | 157 | 38 | 111 | 87 | 85 | 119 | 91 | 5.17 | 765 | 0.23 | 3.33 | No | No | Obesity + mild OSAS |
| 168D | 11.2 | M | 50.4 | 4.24 | OB-SO-G | 149 | 46 | 101 | 89 | 94 | 116 | 118 | 4.9 | 340 | 1.22 | 12.3 | Yes | No | Severe obesity and hepatic steatosis |
| 170D | 12.1 | M | 37.5 | 3.57 | OB-SO-G | 119 | 61 | 59 | 54 | 102 | 156 | 133 | 4.81 | 858 | 0.23 | 2.8 | Yes | No | Severe obesity and RPM. Glucose intollerance. Known chromosome deletion Sigmacillin i.m.therapy |
| 179D | 16.2 | F | 37.9 | 3.18 | OB-SO-G | 148 | 43 | 104 | 75 | 94 | 119 | 108 | 4.26 | 326 | 0.23 | 1.03 | ND | No | Depression |
| 180D | 5.8 | M | 26.6 | 4.94 | OB-SO-G | 135 | 41 | 90 | 89 | 104 | 105 | 138 | 5.35 | 215 | 1.37 | 12.38 | No | No | Severe obesity and moderate OSAS |
| 184D | 16.5 | M | 43 | 3.75 | OB-SO-G | 133 | 34 | 88 | 75 | 99 | 134 | 119 | 5.5 | 929 | 0.9 | 8.4 | Yes | No | Severe obesity and OSAS |
| 186D | 9.8 | M | 29.1 | 3.45 | OB-SO-G | 130 | 33 | 85 | 74 | 87 | 117 | 122 | 5.38 | 690 | 0.23 | 0.75 | No | No | Hypertension |
| 195D | 14.5 | M | 27.7 | 2.25 | OB-SO-G | 210 | 39 | 131 | 136 | 97 | 202 | 127 | 4.97 | 1259 | 0.23 | 0.36 | Yes | No |
Table 2.
Obese with complication (OBc-G) patients.
| Code | Age (years) | Sex | BMI at onset | BMI-SDS | Obeisty subgroup | Total cholesterol | HDL | LDL | TRG | Glycemia 0' | Glycemia 60' | Glycemia 120 | Glicated Hb | Total insulin | CRP | Ultrasensitive CRP | Hepatic steatosis | Metformin therapy | Other clinical infos |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4V | 14.7 | F | ND | ND | ND | 194 | 34 | ND | ND | 82 | 194 | 184 | 5.25 | 2797 | 0.61 | 6 | Si | No | |
| 59D | 11.3 | M | 28 | 2.89 | OBc-O-G | 188 | 31 | 115 | 219 | 108 | 105 | 129 | 5.52 | 221 | 0.23 | ND | No | No | Language and learning difficulties |
| 80D | 11.5 | F | 27.3 | 2.45 | OBc-O-G | 176 | 56 | 105 | 108 | 101 | 148 | 138 | ND | 1404 | 0.23 | 0.14 | No | No | |
| 84D | 14.1 | M | 30 | 2.59 | OBc-O-G | 134 | 38 | 85 | 103 | 88 | 204 | 148 | 5.02 | 338 | 0.23 | 1.42 | Yes | Yes | Macrogol therapy |
| 86D | 6.1 | F | 28 | 4.28 | OBc-SO-G | 161 | 44 | 103 | 162 | ND | ND | ND | 4.64 | ND | 0.23 | 0.7 | ND | No | |
| 106D | 15.2 | M | 28.6 | 2.23 | OBc-O-G | 109 | 39 | 68 | 59 | 92 | 165 | 133 | 5.4 | 576 | 0.23 | 1.42 | Yes | No | Hypertension |
| 109D | 13.8 | M | 28.4 | 2.42 | OBc-O-G | 159 | 41 | 105 | 106 | 91 | 161 | 133 | 6.12 | 643 | 0.23 | 1.39 | No | No | |
| 112D | 13.7 | M | 32.8 | 2.98 | OBc-O-G | 277 | 34 | 210 | 262 | 97 | 121 | 124 | 5.88 | 842 | 0.23 | 2.91 | Yes | No | |
| 117D | 16.4 | F | 42.8 | 3.63 | OBc-SO-G | 161 | 47 | 106 | 115 | 89 | 172 | 152 | 5.36 | 861 | 0.76 | 6.64 | No | No | |
| 118D | 13.9 | M | 29 | 2.49 | OBc-O-G | 160 | 50 | 91 | 93 | 88 | 192 | 172 | 5.79 | 1869 | 0.82 | 7.35 | ND | No | Hypertension |
| 119D | 16.9 | M | 30 | 2.23 | OBc-O-G | 170 | 45 | 99 | 171 | 82 | 232 | 150 | 4.83 | 1746 | 0.23 | 0.64 | Yes | No | |
| 126D | 19.0 | F | 36.6 | 3.05 | OBc-SO-G | 135 | 34 | 83 | 163 | 89 | 113 | 124 | 4.96 | 865 | 0.23 | 3.56 | No | No | Estro-progestinic therapy |
| 141D | 13.0 | M | 26.7 | 2.3 | OBc-O-G | 170 | 38 | 112 | 154 | 94 | 133 | 121 | 5.48 | 876 | 0.23 | 2.3 | Yes | No | |
| 153D | 14.3 | M | 42.6 | 3.68 | OBc-SO-G | 111 | 36 | 66 | 56 | 78 | 112 | 113 | 5.62 | 1117 | 0.93 | 8.72 | No | Glucophage | Hypertension therapy with amlodipin, GHD |
| 157D | 12.8 | M | 25.2 | 2.16 | OBc-O-G | 197 | 36 | 116 | 338 | 83 | 154 | 133 | 5.82 | 892 | 0.23 | 0.98 | ND | No | |
| 166D | 14.8 | F | 41.2 | 3.51 | OBc-SO-G | 158 | 31 | 115 | 167 | 96 | 172 | 154 | 5.11 | 1892 | 0.23 | 2.57 | No | No | Hypertension |
| 169D | 10.8 | F | 37.4 | 3.65 | OBc-SO-G | 170 | 40 | 114 | 81 | 100 | 175 | 141 | 5.38 | 849 | 0.6 | 6.51 | ND | No | |
| 174D | 17 | M | 38.3 | 3.34 | OBc-SO-G | 140 | 39 | 101 | 65 | 108 | 158 | 151 | 4.96 | 1085 | 0.85 | 7.39 | Yes | No | Hypertension |
| 176D | 18.0 | F | 41.4 | 3.56 | OBc-SO-G | 192 | 39 | 139 | 151 | 99 | 136 | 110 | 5.44 | 625 | 1.38 | 11.49 | ND | No | |
| 182D | 12.8 | M | 50.6 | 4.04 | OBc-SO-G | 175 | 34 | 122 | 120 | 93 | 112 | 127 | 5.21 | 168 | 0.62 | 6.24 | Yes | No | Moderate OSAS |
| 189D | 13.0 | M | 45.3 | 3.86 | OBc-SO-G | 148 | 33 | 79 | 229 | 90 | 151 | 140 | 5.25 | 1600 | 0.7 | 6.3 | Yes | No | Mild OSAS |
Patients and relevant clinical features: Sex (M:male, F:female), BMI and BMI-SDS at onset, TRG: triglycerids, Obesity group (OB-O-G: Obese in Simple obesity, OB-SO-G: Severe Obese in Simple obesity, OBc-O*G: Obese in obesity with complication, OBc-SO-G: Severe Obese in obesity with complication. CRP (C-reactive protein): mg/dL (< = 0.23:negative value).
We differentiated patients with simple obesity (OB-G), regardless of grade, from patients who had complicated obesity (OBc-G), regardless of grade, but who had at least three out of five of these characteristics HDL < 5th centile, triglyceride values above the 95th centile, blood pressure systolic and/or diastolic above 90° centile, impaired fasting glycemia, impaired glucose tolerance after 2 h from the meal, hepatic Steatosis (see Table 1). It is also noted that patients in the OBc-G have on average a higher ultrasensitive CRP than patients with uncomplicated obesity. More in detail, as reported in Table 1, 34 patients had a diagnosis with simple obesity (OB-Group, OB-G) (mean age 12.6 ± 2.9, median 12.4, 35% female, BMI-SDS: 3.3 ± 0.7), while 21 classify as Obese with complications (OBc-G) (mean age 14.0 ± 2.8, median 13.9, 38% of them of the female gender, BMI-SDS: 3.1 ± 0.7). It is also to be stressed that among the 34 simple-obese (OB-G) patients, 22 were classifiable as severely obese (OB-SO-G) (BMI-SDS: 3.6 ± 0.5), while 12 were obese (OB-O-G) (BMI-SDS: 2.6 ± 0.20). While among the 21 obese with complication (OBc-G) patients, 10 of them were severely obese with complications (OBc-SO-G) (BMI-SDS: 3.7 ± 0.3), and the rest were obese with complications (OBc-O-G) (BMI-SDS: 2.5 ± 0.28) (see Table 1). As a control, we have analyzed the metagenome of fecal samples from 25 normal-weight subjects (nwHD) matched both for age and sex (mean age 12.1 ± 3.0, median 12.7, 40% female, BMI-SDS: − 0.3 ± 1.1).Written informed consent was obtained by patients and caregivers.
Research was performed in accordance with the Declaration of Helsinki.
The study was approved by the local Ethical Committee of Liguria Region (approval letter, enclosed) and by Giannina Gaslini Institute (authorization letter enclosed).
Fecal microbiota analysis
DNA extraction from fecal samples was performed as reported57 and it was used for the 16S amplification reaction performed with Ion 16S™ Metagenomics Kit (Thermo-Fisher Scientific). This method allows the PCR-amplification of 7 out of 9 informative 16S polymorphic regions58. Then up to 16 differently bar-coded libraries were automatically loaded onto an Ion-520-chip by the Ion-Chef and sequenced by the GeneStudio-S5-system (Thermo-Fisher Scientific). Data analysis was performed with the Ion-Reporter™ suite (v 5.18.0.2) using the curated-Greengenes (v13.5) and the MicroSEQ ID 16S-rRNA reference library (v2013.1) databases using standard parameters.
Data analysis
Compositional/functional profiling and comparative-analysis of microbiome data were performed with Microbiome Analyst and Calypso web-tools13,14,59,60. All p-values have been adjusted to correct for multiple hypotheses, using Benjamini and Hochberg false discovery rate (FDR < 0.05), unless differently specified. Sparse Correlations for Compositional data (SparCC)61 was applied after data-filtering to remove low-quality or uninformative features to study the network of correlation among taxa from the microbiome of obese patients and nwHD controls. In addition, we computed the Microbial Dysbiosis index (MD-index) as the logarithm of the sum of all taxa that increase their abundance over the sum of all taxa that decrease it57. WGCNA identified groups of taxa, or modules that were present across a set of clinical conditions, computing a similarity measure, such as Pearson's correlation coefficient, to calculate the relationship between pairs of taxa. These relationships are then used to construct a weighted network of taxa clustered to identify highly interconnected microorganisms, which are assumed to have a similar biological function or to be commonly regulated. For all the analysis Pearson's correlation coefficients were computed and its associated p-values were corrected for multiple comparisons using False Discovery Rate (FDR).The Multivariate clustering methodology based on the weighted correlation network analysis (WGCNA) was used to verify correlations of taxa clusters with the clinical parameters characterizing OB-G,OBc-G and nwHD62. In particular, we considered the following list of clinical parameters: sex, age (in months), BMI, BMI-SDS, serum levels of total HDL, LDL and cholesterol, triglycerides (TRG), glycemia-0', glycemia-60', glycemia-120' during oral glucose tolerance test, glycated-HbA1c, total-insulin, ultrasensitive CRP (C-reactive protein), liver steatosis by ultrasound. For the normal weight healthy control group, namely nwHD we considered a subselection of the above-mentioned clinical parameters, which are sex, age, BMI and BMI-SDS. The red and blu colors in the WGCNA heatmaps indicate respectively the identified positive or negative Pearson correlations: a positive correlation means that the abundance increase of that specific cluster(s) of taxa is associated with the increase of a specific clinical or metabolic parameter while a negative correlation is associated with a decrease of that specific clinical or metabolic parameter. Furthermore, for the species of OB-G and OBc-G samples groups we proceed with the analysis of the complete data set, but also with a manual selection of the species (indicated by the label “sel-specie”), namely a feature reduction, considering only those species that we found relevant in both supervised random forest and relative abundance analyses. The metagenome functional content was predicted using PICRUSt263 from the biom file, to get the KEGG Orthology (KO) terms table and the inferred MetaCycpathways18. These data were analyzed with the Shotgun-data-profiling module of Microbiome Analyst to identify a list of the most significant pathways able to discriminate cases (OB-G or OBc-G) from controls (nwHD). The abundance of the pathways between the groups was also analyzed with the Wilcoxon test and the statistically significant pathways were clustered, considering the Pearson correlation measureand plotted using Morpheus tool (Morpheus, https://software.broadinstitute.org/morpheus).
Informed consent
Informed consent was obtained from all individual participants or their families included in the study.
Supplementary Information
Acknowledgements
We would like to thank Umberto Buelli and Gianni Berretta, SIA—Istituto Giannina Gaslini for helping with the computer network support.
Author contributions
M.S.: wrote the manuscript and researched data, C.B.: selected patients, produced data, researched data and wrote and edited the manuscript, A.L.V.: selected patients, collected and organizeclinical information, researched data, reviewed the manuscript, E.D.M.: produced data, researched data and edited the manuscript, Gi.Pi.: collected clinical information, N.M.: reviewed the manuscript, Gi.Pa.: collected clinical data, F.N.: collected samples, M.B.: collected samples, M.M.: revised manuscript, G.d'A.: collected clinical and metabolic data, wrote the manuscript, reviewed the manuscript, R.B.: planned the research and received funds, wrote the manuscript and researched data.
Funding
Funding support was provided by Ricerca Corrente grant from the Ministry of Health (RB) and from Compagnia di San Paolo—2019.0880—ID ROL 32612 (RB).
Data availability
Raw 16S rRNA gene reads were deposited at the short read archive (SRA_BioProject ID PRJNA794317).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Margherita Squillario, Carola Bonaretti, Alberto La Valle and Roberto Biassoni.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-36533-2.
References
- 1.Farpour-Lambert NJ, et al. Childhood obesity is a chronic disease demanding specific health care—A position statement from the Childhood Obesity Task Force (COTF) of the European Association for the Study of Obesity (EASO) Obes. Facts. 2015;8:342–349. doi: 10.1159/000441483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soderborg TK, Borengasser SJ, Barbour LA, Friedman JE. Microbial transmission from mothers with obesity or diabetes to infants: An innovative opportunity to interrupt a vicious cycle. Diabetologia. 2016;59:895–906. doi: 10.1007/s00125-016-3880-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lascar N, et al. Type 2 diabetes in adolescents and young adults. Lancet Diabetes Endocrinol. 2018;6:69–80. doi: 10.1016/S2213-8587(17)30186-9. [DOI] [PubMed] [Google Scholar]
- 4.Gao X, et al. A study of the correlation between obesity and intestinal flora in school-age children. Sci. Rep. 2018;8:14511. doi: 10.1038/s41598-018-32730-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu HJ, et al. Obesity alters the microbial community profile in Korean adolescents. PLoS ONE. 2015;10:e0134333. doi: 10.1371/journal.pone.0134333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollister EB, Foster BA, Dahdouli M, Ramirez J, Lai Z. Characterization of the stool microbiome in hispanic preschool children by weight status and time. Child Obes. 2018;14:122–130. doi: 10.1089/chi.2017.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.López-Contreras BE, et al. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatr. Obes. 2018;13:381–388. doi: 10.1111/ijpo.12262. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Q, Hu N. Effects of metformin on the gut microbiota in obesity and Type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 2020;13:5003–5014. doi: 10.2147/DMSO.S286430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreira GV, et al. Liraglutide modulates gut microbiota and reduces NAFLD in obese mice. J. Nutr. Biochem. 2018;62:143–154. doi: 10.1016/j.jnutbio.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Fuks G, et al. Combining 16S rRNA gene variable regions enables high-resolution microbial community profiling. Microbiome. 2018;6:17. doi: 10.1186/s40168-017-0396-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 12.Verdam FJ, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 2013;21:E607–E615. doi: 10.1002/oby.20466. [DOI] [PubMed] [Google Scholar]
- 13.Zakrzewski M, Proietti C, Ellis JJ, et al. Calypso: A user-friendly web-server for mining and visualizing microbiome-environment interactions. Bioinformatics. 2017;33:782–783. doi: 10.1093/bioinformatics/btw725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.LêCao KA, et al. MixMC: A multivariate statistical framework to gain insight into microbial communities. PLoS ONE. 2016;11:e0160169. doi: 10.1371/journal.pone.0160169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breiman L. Random forests. Mach. Learn. 2001;45:5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 16.Doumatey AP, et al. Gut microbiome profiles are associated with type 2 diabetes in urban Africans. Front. Cell Infect. Microbiol. 2020;10:63. doi: 10.3389/fcimb.2020.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vrieze A, et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology. 2012;143:913–967. doi: 10.1053/j.gastro.2012.06.031. [DOI] [PubMed] [Google Scholar]
- 18.Caspi R, et al. The MetaCyc Database of metabolic pathways and enzymes. Nucleic Acids Res. 2018;46:D633–D639. doi: 10.1093/nar/gkx935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiu SF, et al. Helicobacter pylori GmhB enzyme involved in ADP-heptose biosynthesis pathway is essential for lipopolysaccharide biosynthesis and bacterial virulence. Virulence. 2021;12:1610–1628. doi: 10.1080/21505594.2021.1938449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holst O, Zähringer U, Brade H, Zamojski A. Structural analysis of the heptose/hexose region of the lipopolysaccharide from Escherichia coli K-12 strain W3100. Carbohydr. Res. 1991;215:323–335. doi: 10.1016/0008-6215(91)84031-9. [DOI] [PubMed] [Google Scholar]
- 21.Abellan-Schneyder I, Matchado MS, Reitmeier S, Sommer A, Sewald Z, Baumbach J, List M, Neuhaus K. Primer, pipelines, parameters: Issues in 16S rRNA gene sequencing. mSphere. 2021;6:e01202–e1220. doi: 10.1128/mSphere.01202-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woese CR, Kandler O, Wheelis ML. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. U. S. A. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J. Clin. Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boudewijns M, Bakkers JM, Sturm PDJ, Melchers WJG. 16S rRNA gene sequencing and the routine clinical microbiology laboratory: A perfect marriage? J. Clin. Microbiol. 2006;44:3469–3470. doi: 10.1128/JCM.01017-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinart M, et al. gut microbiome composition in obese and non-obese persons: A systematic review and meta-analysis. Nutrients. 2021;14:12. doi: 10.3390/nu14010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan RC, et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019;20:219. doi: 10.1186/s13059-019-1831-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng J, et al. Dietary inflammatory potential in relation to the gut microbiome: Results from a cross-sectional study. Br. J. Nutr. 2020;124:931–942. doi: 10.1017/S0007114520001853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agarwal DM, et al. Disruptions in oral and nasal microbiota in biomass and tobacco smoke associated chronic obstructive pulmonary disease. Arch. Microbiol. 2021;203:2087–2099. doi: 10.1007/s00203-020-02155-9. [DOI] [PubMed] [Google Scholar]
- 29.Chen X, et al. Characterization of the gut microbiota in Chinese children with overweight and obesity using 16S rRNA genesequencing. PeerJ. 2021;9:e11439. doi: 10.7717/peerj.11439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palmas V, et al. Gut microbiota markers associated with obesity and overweight in Italian adults. Sci. Rep. 2021;11:5532. doi: 10.1038/s41598-021-84928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 32.Naderpoor N, et al. Faecal microbiota are related to insulin sensitivity and secretion in overweight or obese adults. J. Clin. Med. 2019;8:452. doi: 10.3390/jcm8040452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cani PD. Human gut microbiome: Hopes, threats and promises. Gut. 2018;67:1716–1725. doi: 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ley RE. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol. 2016;13:69–70. doi: 10.1038/nrgastro.2016.4. [DOI] [PubMed] [Google Scholar]
- 35.Aranaz P, et al. A predictive regression model of the obesity-related inflammatory status based on gut microbiota composition. Int. J. Obes. (Lond.) 2021;45:2261–2268. doi: 10.1038/s41366-021-00904-4. [DOI] [PubMed] [Google Scholar]
- 36.Queipo-Ortuño MI, et al. Gut microbiota composition in male rat models under different nutritional status and physical activity and its association with serum leptin and ghrelin levels. PLoS ONE. 2013;8:e65465. doi: 10.1371/journal.pone.0065465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isoda M, et al. Leptin sensitizing effect of 1,3-butanediol and its potential mechanism. Sci. Rep. 2021;11:17691. doi: 10.1038/s41598-021-96460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fernández-Navarro T. Different intestinal microbial profile in over-weight and obese subjects consuming a diet with low content of fiber and antioxidants. Nutrients. 2017;9:551. doi: 10.3390/nu9060551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, et al. Glucose but not fructose alters the intestinal paracellular permeability in association with gut inflammation and dysbiosis in mice. Front. Immunol. 2021;12:742584. doi: 10.3389/fimmu.2021.742584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beerens H, Romond C. Sulfate-reducing anaerobic bacteria in human feces. Am. J. Clin. Nutr. 1977;30:1770–1776. doi: 10.1093/ajcn/30.11.1770. [DOI] [PubMed] [Google Scholar]
- 41.Weglarz L, et al. Desulfovibriodesulfuricans lipopolysaccharides induce endothelial cell IL-6 and IL-8 secretion and E-selectin and VCAM-1 expression. Cell. Mol. Biol. Lett. 2003;8:991–1003. [PubMed] [Google Scholar]
- 42.Patil DP, et al. Molecular analysis of gut microbiota in obesity among Indian individuals. J. Biosci. 2012;37:647–657. doi: 10.1007/s12038-012-9244-0. [DOI] [PubMed] [Google Scholar]
- 43.Liu R, et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 2017;23:859–868. doi: 10.1038/nm.4358. [DOI] [PubMed] [Google Scholar]
- 44.Samuel BS, Gordon JI. A humanized gnotobiotic mouse model of host-archaeal-bacterial mutualism. Proc. Natl. Acad. Sci. USA. 2006;103:10011–10016. doi: 10.1073/pnas.0602187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kang K, et al. Distinctive gut microbial dysbiosis between chronic alcoholic fatty liver disease and metabolic-associated fatty liver disease in mice. Exp. Ther. Med. 2021;21:418. doi: 10.3892/etm.2021.9862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliphant K, Allen-Vercoe E. Macronutrient metabolism by the human gut microbiome: Major fermentation by-productsand their impact on host health. Microbiome. 2019;7:91. doi: 10.1186/s40168-019-0704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters WA, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magne F, et al. The firmicutes/bacteroidetes ratio: A relevant marker of gut dysbiosis in obese patients? Nutrients. 2020;12:1474. doi: 10.3390/nu12051474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Duncan SH, Lobley GE, Holtrop G, Ince J, Johnstone AM, Louis P, Flint HJ. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 2008;32:1720–1724. doi: 10.1038/ijo.2008.155. [DOI] [PubMed] [Google Scholar]
- 50.Schwiertz A, Taras D, Schäfer K, Beijer S, Bos NA, Donus C, Hardt PD. Microbiota and SCFA in lean and overweight healthy subjects. Obesity. 2010;18:190–195. doi: 10.1038/oby.2009.167. [DOI] [PubMed] [Google Scholar]
- 51.Tims S, Derom C, Jonkers DM, Vlietinck R, Saris WH, Kleerebezem M, de Vos WM, Zoetendal EG. Microbiota conservation and BMI signatures in adult monozygotic twins. ISME J. 2013;7:707–717. doi: 10.1038/ismej.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo K, et al. Plasma metabolomics and lipidomics. Differentiate obese individuals by peripheral neuropathy status. J. Clin. Endocrinol. Metab. 2022;107:1091–1109. doi: 10.1210/clinem/dgab844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Raybould HE, Zumpano DL. Microbial metabolites and the vagal afferent pathway in the control of food intake. Physiol. Behav. 2021;240:113555. doi: 10.1016/j.physbeh.2021.113555. [DOI] [PubMed] [Google Scholar]
- 54.Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut. 2021;70:1174–1182. doi: 10.1136/gutjnl-2020-323071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lischka J, et al. Tryptophan metabolism is associated with BMI and adipose tissue mass and linked to metabolic disease in pediatric obesity. Nutrients. 2022;14:286. doi: 10.3390/nu14020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Niekerk G, Engelbrecht AM. Inflammation-induced metabolic derangements or adaptation: An immunometabolic perspective. Cytokine Growth Factor Rev. 2018;43:47–53. doi: 10.1016/j.cytogfr.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 57.Biassoni R, et al. Gut Microbiota in T1DM-onset pediatric patients: Machine-learning algorithms to classify microorganisms as disease linked. J. Clin. Endocrinol. Metab. 2020;105:dgaa407. doi: 10.1210/clinem/dgaa407. [DOI] [PubMed] [Google Scholar]
- 58.Wang S, Sun B, Tu J, Lu Z. Improving the microbial community reconstruction at the genus level by multiple 16S rRNA regions. J. Theor. Biol. 2016;398:1–8. doi: 10.1016/j.jtbi.2016.03.016. [DOI] [PubMed] [Google Scholar]
- 59.Dhariwal A, et al. MicrobiomeAnalyst—A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017;45:W180–188. doi: 10.1093/nar/gkx295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chong J, Liu P, Zhou G, Xia J. Using microbiome analyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020;15:799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 61.Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012;8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Douglas GM, et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38:685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw 16S rRNA gene reads were deposited at the short read archive (SRA_BioProject ID PRJNA794317).