Abstract
Background
B-raf inhibitors (BRAFi) are effective for BRAF-mutated papillary (PTC) and anaplastic (ATC) thyroid carcinomas, although acquired resistance impairs tumour cells’ sensitivity and/or limits drug efficacy. Targeting metabolic vulnerabilities is emerging as powerful approach in cancer.
Methods
In silico analyses identified metabolic gene signatures and Hif-1α as glycolysis regulator in PTC. BRAF-mutated PTC, ATC and control thyroid cell lines were exposed to HIF1A siRNAs or chemical/drug treatments (CoCl2, EGF, HGF, BRAFi, MEKi and diclofenac). Genes/proteins expression, glucose uptake, lactate quantification and viability assays were used to investigate the metabolic vulnerability of BRAF-mutated cells.
Results
A specific metabolic gene signature was identified as a hallmark of BRAF-mutated tumours, which display a glycolytic phenotype, characterised by enhanced glucose uptake, lactate efflux and increased expression of Hif-1α-modulated glycolytic genes. Indeed, Hif-1α stabilisation counteracts the inhibitory effects of BRAFi on these genes and on cell viability. Interestingly, targeting metabolic routes with BRAFi and diclofenac combination we could restrain the glycolytic phenotype and synergistically reduce tumour cells’ viability.
Conclusion
The identification of a metabolic vulnerability of BRAF-mutated carcinomas and the capacity BRAFi and diclofenac combination to target metabolism open new therapeutic perspectives in maximising drug efficacy and reducing the onset of secondary resistance and drug-related toxicity.
Subject terms: Cancer metabolism, Targeted therapies
Background
Thyroid carcinomas, and especially the most frequent form (i.e., papillary, PTC), represent prototypic examples of how distinct oncogenic lesions drive the activation of different signalling pathways. Indeed, tumours carrying somatic mutations in BRAF and RET genes (constituting the BRAF-like subtype) exhibit a marked ERK-induced transcriptional signature which allows them to escape from the canonical ERK inhibitory feedback [1, 2]. On the other hand, tumours carrying either mutations in RAS genes or PPARG/PAX8 fusions (i.e., the RAS-like subtype) display a more pronounced induction of the PI3K/AKT signalling [1, 2]. The differential activation of signalling pathways is reflected in the lower sensitivity of BRAF-like tumours to the ERK-mediated inhibitory feedback loop and, from the clinical point of view, in higher tumour aggressiveness [1]. In the last decade, the alteration of distinct oncogenes has been associated with the different capacity of tumour cells to undergo metabolic reprogramming, which has emerged as a driving tumorigenesis force having a strong impact on drug responsiveness and tumour aggressiveness [3–6]. Indeed, the substantially higher glucose demand (even in normoxia) [7] is associated with increased metastatic capacity, reduced drug sensitivity [8–10] and poor prognosis [11]. Hence, identifying mutation-specific metabolic patterns is the first step to design new effective treatments to restrain tumour cells’ growth and aggressiveness.
Among the others, BRAFV600E mutation in thyroid, melanoma and colorectal carcinomas has been associated with increased expression of key glycolysis-related genes [9, 12–14]. Accordingly, B-raf inhibition by vemurafenib (VMR) in melanoma cell lines, xenograft models and primary melanomas potently suppresses glucose uptake [15–19]. Few recent studies have used The Cancer Genome Atlas (TCGA) data [1], from the thyroid carcinoma (THCA) cohort, to detect gene signatures of glucose transporters (i.e., Gluts), glycolysis and lipid metabolism genes associated with multiple clinicopathological features of PTCs [20–27]. The overexpression of specific Gluts was associated with tumour dedifferentiation [22] and increased mortality in PTC patients [20], although Gluts and glycolysis signatures were oppositely correlated to tumour differentiation [22]. Moreover, Ma et al. defined a risk score for dedifferentiation based on the expression levels of metabolic genes, which is higher in patients carrying BRAFV600E mutation [21]. Higher expression of lactate dehydrogenase A (LDHA) in PTCs in patients carrying BRAFV600E mutation has finally been related to aggressive clinicopathological features [24, 25].
Here we report—for the first time—a comprehensive investigation of all metabolic genes in TCGA-THCA cohort [1], focusing on a comparative analysis between BRAF- and RAS-like tumours. This analysis revealed subtype-specific signatures of metabolic genes in PTC, leading to the identification of Hif-1α as one the main transcriptional contributors to the metabolic rewiring of BRAF-driven tumours. However, among thyroid cancers, the high metastatic potential and the poorer prognosis of the anaplastic form (ATC)—of which 20–25% of cases carry BRAFV600E mutation—indicate the clinical need to design more effective therapeutic regimens to treat this aggressive tumour type. Thus, here we further report the capacity of B-raf inhibitors to restrain the glycolytic phenotype of both BRAF-mutated PTCs and ATCs. However, the effectiveness of B-raf inhibitors is known to decrease within 6–8 months [28–30] from starting therapy due to acquired resistance. Hence, we sought to exploit the glycolytic dependency of BRAF-driven tumours as a metabolic vulnerability and to adopt new therapeutic approaches to maximise drug efficacy (even at low doses) and avoid—or delay—the onset of secondary resistance and drug-related toxicity. In this regard, in line with recent reports on BRAF-mutated melanoma [10, 31], we found that diclofenac, a non-steroidal anti-inflammatory drug (NSAID), is able to impair the glycolytic flux of tumour cells and—synergistically with B-raf inhibitors (i.e., either vemurafenib or dabrafenib)—to reduce the viability of papillary and anaplastic thyroid cancer cells.
Materials and methods
TCGA data selection, processing and analysis
Whole-exome and RNA-Seq data were downloaded in March 2019 from the TCGA portal and analysed as previously described [32]. Briefly, according to the presence of specific somatic mutations, THCA patients were stratified using Exome-Seq data files (.maf format). All files retrieved from each consortium were merged. Afterwards, using a customised computational pipeline in R language, BRAF- and RAS-mutant samples were identified, and the entire cohort was subdivided into BRAF- and RAS-like tumours. Then, RNA-Seq data were used to identify patients having RET/PTC and PAX8/PPARG rearrangements to be further included in the BRAF-like and RAS-like subgroup, respectively. A total of 448 samples were analysed in this work: 327 BRAF-like PTCs 71 marked as RAS-like tumours and 50 healthy samples. The analysis of gene expression—and the identification of differentially expressed genes (DEGs)— was carried out by comparing transcriptome data (normalised values) of BRAF- vs RAS-like PTCs and of tumour vs normal samples (paired analysis). Genes differentially expressed both between the two tumour subtypes and vs healthy counterparts are indicated as BRAFup, BRAFdown, RASup or RASdown. Differential expression analysis was carried out using generalised linear models (gLM) implemented in the EdgeR (Bioconductor package, version 3.17.10; RRID:SCR_006442; [33]). Multiple corrections were performed applying the false discovery rate (FDR) method. The FDR threshold for differentially expressed genes was set to ≤0.05. A comprehensive list of human metabolic or metabolism-related genes was retrieved in PhosphoSitePlus database http://www.phosphosite.org/psrSearchAction.do) by selecting “containing metabolism” in the section named “Protein type”, as described in ref. [34]. Moreover, the correlation analysis on gene expression data (TCGA) was performed using linear models implemented in R language (lmp function). Correlations between the selected genes were reported in the graphs as Pearson’s coefficient (r) and P values were computed by one-way ANOVA in R. DNA Methylation data (THCA) of BRAF- and RAS-mutant samples were retrieved from the TCGA repository using the TCGABiolinks (RRID:SCR_017683) Bioconductor package in March 2019 [35]. Collected data were produced by Illumina Methylation 450k microarray platform. To perform a reliable differential expression analysis, a visual inspection of probe intensity distributions was carried out, and only probes mapping in CpG islands, shores and shelves were selected for further analyses. Beta values were transformed in M values by applying the formula M = log2(Beta/(1-Beta)). Differential DNA methylation analysis between BRAF- and RAS-mutant samples was performed using the limma package in R language [36]. Nominal P values were adjusted by applying the Benjamini-Hochberg method [37]. Probeset IDs were annotated on the base of the IlluminaHumanMethylation450kanno.ilmn12.hg19 annotation. Methylation data—related to probe intensities on the whole gene body—were downloaded in September 2021 from cBioportal (https://www.cbioportal.org/) for THCA cohort and were used to generate plots. Over-represented Transcription Factor Binding Site Motifs were searched using PScan online resources [38], using Jaspar 2018 matrix. Motifs’ search was carried out in a 1000-bp window upstream the transcriptional starting site of selected genes, as previously described [39]. All the heatmaps, scatter, violin and box plots were generated using Ggplot2-based scripts in R language and custom scripts, available upon request.
Gene set enrichment analysis and LINCS L1000 data manipulation
Gene Set Enrichment Analysis (GSEA; RRID:SCR_003199) was performed by clusterProfiler R package (RRID:SCR_016884; 35). The GSEA was carried out by using the list of differentially expressed genes ranked by logFC and the KEGG pathways as gene sets. The transcriptome-wide effect of treatment with vemurafenib on cancer cell lines was evaluated by retrieving gene expression data from the L1000 Connectivity Map perturbation profiles of the Broad Institute LINCS (GEO ID: GSE70138; [36, 37]). Differential expression analysis on L1000 data for vemurafenib treatment on tumour cell lines was performed by non-parametric Wilcoxon test, comparing gene expression profiles of treated and plate-specific untreated samples as a control. All the computations carried out on the LINCS L1000 data were performed by compiling tailored R scripts. Ggplot2-based scripts in R language were used to generate the related images.
Cell cultures
Human papillary, anaplastic thyroid carcinoma and normal thyroid follicular epithelial cell lines (i.e., BCPAP, 8505c and Nthy-ori 3-1, respectively)—kindly provided by Profs. Alfredo Fusco and Massimo Santoro (Medicina Molecolare e Biotecnologie Mediche, Università degli Studi di Napoli Federico II, Napoli, Italy)—were available in the laboratory from a previous study [40]. All cell lines—routinely analysed for Mycoplasma contamination—were “myco-free”. BCPAP were cultured in DMEM, whereas 8505c and Nthy-ori 3-1 in RPMI. Culture media were supplemented with South American foetal bovine serum (FBS, 10%), glutamine and antibiotics, and cell cultures were maintained in a humidified atmosphere of 5% CO2 at 37 °C. Media, sera and antibiotics were purchased from Thermo Fisher Scientific (Waltham, Massachusetts, USA). Cell lines were used for following assays between the 4th and the 16th cell passage.
Cell treatments and multidrug combination analysis
Before treatments, all cell lines were starved for 16–18 h. For evaluating the effects of MAPK activation on metabolic genes, Nthy-ori 3-1 were treated with human recombinant Epidermal Growth Factor (EGF; 100 ng/mL; Thermo Fisher Scientific, Waltham, Massachusetts, USA) or Hepatocyte Growth Factor (HGF; 100 ng/mL; Thermo Fisher Scientific, Waltham, Massachusetts, USA) proteins for 3 or and 6 h, as previously described [40].
BCPAP were treated with PLX4032 5 µM (Selleckchem, USA, i.e., a vemurafenib analogue) at different time points: 3, 6 and 24 h for the analysis of metabolic genes and Erk phosphorylation levels, 24 h for assessing the expression levels of Hif-1α and metabolic genes—in presence or absence of Cobalt(II) chloride hexahydrate 125 µM (CoCl2·6H2O; Sigma Aldrich, St Louis, MO, USA)—and 72 h for all viability assays. Moreover, BCPAP cells were exposed for 72 h to increasing PLX4032 concentrations (from 0.5 to 25 µM) for determining LC50, glucose uptake and lactate secretion (0.5, 1, 5 µM). To assess Hif-1α stabilisation and the effects on metabolic genes, BCPAP were also treated with CoCl2 250 µM for 24 h. Furthermore, BCPAP cells were treated with diclofenac sodium salt (Sigma Aldrich, St Louis, MO, USA; 50 µM and 100 µM) and mRNA levels of metabolic genes (3 and 6 h) and Hif-1α protein levels (24 h) have been analysed. The treatment with increasing doses of diclofenac (5, 25, 50 µM) for 72 h was performed for assessing cell viability—in presence/absence of PLX4032 (0.5, 1, 5 µM) or dabrafenib (Selleckchem, USA, 0.05, 0.1, 0.5 µM)—glucose uptake and lactate secretion.
Similarly, for viability assays 8505c cells were exposed for 72 h at PLX4032 (0.5, 1, 5, 10, 25 µM), diclofenac (5, 50, 100 µM), dabrafenib (0.05, 0.25, 0.5 µM) and trametinib (1, 5, 50 nM), alone or in combination. Particularly, for determining LC50 of the drugs, cells were exposed to increasing concentrations of PLX4032 (from 0.5 to 25 µM) or trametinib (MedChemExpress, USA; from 0.1 to 2000 nM). The levels of Erk phosphorylation were analysed after treatment with PLX4032 (5 and 10 µM for increasing time points from 30 min to 24 h), trametinib (50 nM for 30 min) or their combination. The expression of metabolic genes was evaluated at 3, 6 and 24 h after treatment with PLX4032 10 µM or trametinib 50 nM, whereas glucose uptake and lactate secretion were assessed 72 h after treatment with increasing doses of PLX4032 (1, 5 and 10 µM) or trametinib (1, 50 and 50 nM). The treatment of 8505c with diclofenac was also performed (100 µM for 3 and 6 h) for analysing the effects on metabolic genes, and at increasing concentrations (5, 50, 100 µM for 72 h) for evaluating glucose uptake and lactate secretion.
The effects obtained by drug combinations were assessed by analyzing viability data with SynergyFinder (https://www.synergyfinder.org/; RRID:SCR_019318) [41]—based on HSA reference—and by comparing the observed and expected drug combination responses (i.e., positive values denote synergy). Moreover, cNMF algorithm [42] implemented in SynergyFinder was used for the estimation of outlier measurements.
Cell transfection
BCPAP cells were transfected with two different HIF1A-targeting or scrambled siRNAs (20 nM) designed from IDT (Coralville, Iowa, USA) by Lipofectamine 3000 (Life Technologies, Carlsbad, CA, USA), in culture medium without antibiotics and serum, according to manufacturer’s instructions. After 24 h, transfected cells were used for further analysis. The efficiency of Hif-1α was estimated by assessing its mRNA (by qPCR) and protein expression levels (by western blot).
RNA extraction, RT-PCR and qPCR
Total RNA—isolated using TRIzol Reagent (Life Technologies, Carlsbad, CA, USA) as described in ref. [38]—was used for cDNA synthesis by High-Capacity cDNA Reverse Transcription kit (Invitrogen, Carlsbad, CA, USA), according to manufacturer’s instructions. Expression analysis was performed by relative qPCR assays. Gene-specific primers were designed using the Oligo 4.0 program (listed in Supplementary Table S1). The iTaq Universal Sybr Green Supermix (Bio-Rad, Hercules, CA, USA) was used for qPCR on CFX Connect Detection System (Bio-Rad, Hercules, CA, USA), according to manufacturer instructions, as previously described [39]. Relative mRNA expression was measured by the 2-ΔΔCt method. The PPIA gene was selected as house-keeping gene and cells treated with the vehicles were used as reference samples. All reactions were performed in duplicates in at least three independent experiments. Student’s t test (one sample or two samples test; two-tailed) was used for assessing statistical significance of differences for normally distributed data. For each assay, SEM and statistical significance are specifically reported in figures and legends.
Western blot
Cell lysates were extracted by RIPA buffer and blotted (30–50 mg protein/sample) with anti-Hif-1α (1:4000; Proteintech, Manchester, UK; RRID:AB_10732601), anti-pErk (1:1000; Cell Systems, Kirkland, WA, USA; RRID:AB_331646), anti-Hsp90 (1:5000; Origene, Rockville, Maryland, USA; RRID: AB_11141759), anti-Pfkfb3 (1:1000; ABclonal, Woburn, MA, USA; RRID: AB_2863158), anti-Ldha (1:1000; ABclonal, Woburn, MA, USA; RRID:AB_2758572), anti-Glut1 (1:1000; CUSABIO TECHNOLOGY LLC, Houston, USA; CSB-PA021546ESR2HU), anti-Mct4 (1:1000; CUSABIO TECHNOLOGY LLC, Houston, USA; CSB-PA021410LA01HU) antibodies and secondary anti-IgG (mouse and rabbit) antibodies (1:5000; Bio-Rad, Hercules, CA, USA; RRID: AB_11125547 and RRID: AB_2617112). The autoradiographs included in the figures are representative of at least three independent experiments. Densitometric data analysis was performed by calculating pixel density with GelQuant.NET software (http://biochemlabsolutions.com/) and displayed as mean values ± SEM of different experiments. Intensity values were normalised using the expression signals of Hsp90 and compared to signals detected in reference samples, and Student’s t test (two samples test; two-tailed) was used for assessing the statistical significance of differences.
Cell viability analysis and growth curves
Viability assays were performed on BCPAP (RRID:CVCL_0153) and 8505c (RRID:CVCL_1054) cells by CellTiter-Glo Kit (Promega, Madison, Wisconsin, USA), according to the manufacturer instructions. Cells were plated and treated in 96-well white opaque plates. Luminescence was measured on VICTOR Multilabel Plate Reader (Perkin Elmer, USA). The effect of each treatment on cell viability was estimated after 72 h as the percentage of luminescence (means ± SEM of four independent experiments) compared to control cells, as specified in figure legends. The luminescence of control cells was set to 100% and Student’s t test (one sample or two samples test; two-tailed) was used for assessing statistical significance of differences. Moreover, for each cell line and treatment, cell growth curves were obtained using a standard curve of cell number and plotting viability values at different time points (i.e., 0 h, 12 h, 24 h, 48 h, 72 h). Then, the area under the curves (AUC) has been calculated as described in ref. [10].
Glucose uptake and lactate secretion assays
Glucose uptake analysis was performed on Nthy-ori 3-1, BCPAP and 8505c cells using the Glucose Uptake Colorimetric Assay Kit (Sigma Aldrich, St Louis, MO, USA), according to manufacturer instructions. Cells were plated in 96-well plates, and the effect of each treatment/co-treatment was estimated after 72 h. Cells were serum-starved for 14 h and then glucose-starved for 40 min by KRPH buffer 2% BSA. Insulin (1 mM) stimulation was performed for 40 min and a negative control was prepared by incubating a parallel sample without insulin and 2-DG. Colorimetric detection of 2-DG6P was performed on VICTOR Multilabel Plate Reader (Perkin Elmer, USA). OD values were corrected for dilution factor and by subtracting background (blank sample) and negative control values. Standard curves were plotted using 2-DG6P standards. For the analysis of the total glucose uptake, each value (pmol) was normalised for the related AUC and plotted. The insulin-induced glucose uptake was calculated for each sample as the difference between insulin-stimulated and insulin-free cells. Student’s t test (one sample or two samples test; two-tailed) was used for assessing the statistical significance of differences.
L-lactic acid content in cell culture supernatant was measured by L-lactic acid/lactate (LA) Colorimetric Assay Kit (Elabscience, USA), according to manufacturer instructions. The supernatant samples were collected from Nthy-ori 3-1, BCPAP and 8505c plated in 96-well plates, treated or not for 72 h and centrifuged (at 10,000×g for 5 min, 4 °C). Colorimetric detection of secreted lactate was performed on VICTOR Multilabel Plate Reader (Perkin Elmer, USA). OD values were corrected considering background (blank sample), the dilution factor and standard concentration (mmol/L). Lactate concentration (mmol/L) was calculated by using standard curves, the obtained values were normalised for the related AUC and plotted. The effect of each treatment was estimated as the percentage of lactate secreted by control cells (set to 100%) and Student’s t test (one sample or two samples test; two-tailed) was used for assessing statistical significance of differences.
Results
Identification of subtype-specific metabolic gene signatures in BRAF-like PTCs
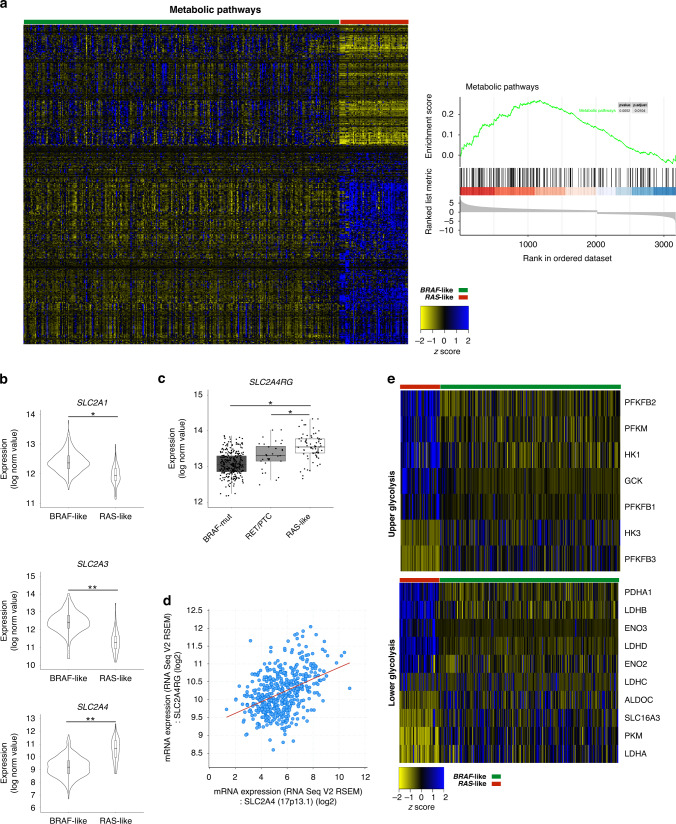
To investigate whether BRAF-like papillary thyroid carcinomas display specific expression signatures of metabolic genes, we took advantage of TCGA whole-exome and transcriptome data to stratify tumours in the BRAF- and RAS-like subtypes and to identify differentially expressed genes (DEGs). The overall comparison of expression data from the two distinct PTC subtypes explored by GSEA indicated “Metabolic pathways” as the only enriched process (Fig. 1a). Accordingly, metabolic and metabolism-related genes rank among the top DEGs (~31%, i.e., 459/1462, gene IDs retrieved in March 2019 from PhosphoSitePlus database) and their clustering is evident between the two PTC subtypes (Fig. 1a and Supplementary File S1). Using the KEGG database, we focused on four main processes, i.e., carbohydrate, lipid, amino acid and nucleotide metabolism, which include 246 DEGs (Supplementary Fig. S1). To identify those specifically altered in distinct PTC subtypes, we selected genes differentially expressed between BRAF- and RAS-like PTCs and deregulated even vs the related healthy samples (i.e., 133 genes) and we labelled them BRAFup and BRAFdown or RASup and RASdown genes (Supplementary Fig. S1A and Supplementary File S1). In addition, taking advantage of RNA-Seq data from our previously published [2] independent cohort of PTC patients (n = 22), we validated ~55% of them (i.e., 73/133 genes) as differentially expressed between tumour subgroups (Supplementary File S1). Notably, BRAF-like tumours displayed major expression changes in several genes accounting for energy metabolism (i.e., 116 genes), whereas only 20 were identified as altered in RAS-like PTCs. Particularly, focusing on the expression of membrane glucose transporters, we observed that, despite SLC2A1 gene (alias Glut1) is reported as the most abundant channel expressed in primary thyroid tumours and cell lines [43, 44], SLC2A3 gene (alias Glut3) display comparable expression levels and both genes show higher expression in BRAF- vs RAS-like tumours (Supplementary Fig. S1B), as confirmed by RNA-Seq data of healthy thyroids on GTEx database (Supplementary Fig. S1C). However, both SLC2A1 (FDR < 0.01) and SLC2A3 (FDR < 0.001) genes—associated with higher glucose uptake and oncogenic growth [12, 45, 46]—are markedly overexpressed in BRAF- (vs RAS)-like tumours (Fig. 1b). Differently from BRAF-like PTCs, RAS-like tumours show higher levels of the inducible transporter SLC2A4 (alias Glut4; Fig. 1b) and of its main regulator, the transcription factor SLC2A4RG (Fig. 1c), whose expression levels are positively correlated across tumour samples (Fig. 1d). Then, considering the processes following glucose internalisation and phosphorylation we focused on pentose phosphate pathway (PPP) and glycolysis. Except for TKT gene, all PPP-related genes are not differentially expressed, whereas the expression of genes encoding enzymes of upper and lower glycolysis is largely affected between tumour subtypes, also compared to the healthy counterparts (Fig. 1e). In particular, we identified a specific signature of glycolysis-related genes in BRAF-like tumours, which display the overexpression of SLC2A1, SLC2A3, HK3, PFKFB3, PKM, LDHA and SLC16A3 (alias Mct4, the membrane channel regulating lactate efflux). Notably, also LDHA/LDHB expression ratio—measured within each tumour sample—shows a marked increase only in BRAF-like tumours, whereas LDHB is modestly upregulated in the RAS-like subtype (Supplementary Fig. S2A, S2B). Thus, in contrast with the glycolytic phenotype of BRAF-like PTCs, RAS-like tumours display expression signatures compatible with oxidative metabolism [47], characterised by the usage of extracellular lactate to produce ATP (via OXPHOS). Conversely, the expression of SLC16A1 and SLC16A7 genes (Supplementary Fig. S2C, D)—encoding MCT1 and MCT2 lactate transporters, respectively—as well as of GJA1 (data not shown) encoding connexin 43 and responsible of lactate diffusion [48], does not differ between the two tumour subtypes. Thus, according to the flux control coefficients reported by Tanner and colleagues [49], transcriptome data indicate that BRAF-like tumours have a marked increase of the entire glycolytic flux—from glucose uptake to lactic fermentation and excretion—compared to RAS-like sample (Supplementary Fig. S3). Moreover, we also found that TCA cycle, FAO and OXPHOS genes display a subtype-specific expression pattern (Supplementary Figs. S1A and S4). The overexpression in BRAF-like tumours of specific genes encoding enzymes responsible for anaplerotic TCA cycle reactions (Supplementary Fig. S4A, B and Supplementary File S1) possibly contributes to replenishing the oxaloacetate pool from pyruvate and aspartate. Glutamine influx is another anaplerotic process enhanced in BRAF-like PTCs, which may contribute to replenishing TCA intermediates. Indeed, glutamine transporter (SLC16A5) and enzymes converting glutamine to glutamate (GLS and GLS2) are specifically overexpressed in BRAF-like PTCs (Supplementary Fig. S4A, B). Moreover, GOT1—regulating one of the main cataplerotic reactions—is downregulated only in this subtype (Supplementary Fig. S4A, B). In line with the high glycolytic flux and low amount of intermediates feeding the TCA cycle, BRAF-like tumours display a mild downregulation of OXPHOS genes compared both to RAS-like and healthy samples (Supplementary Fig. S4C, D and Supplementary File S1). Taken together, transcriptome data indicate that BRAF-like PTCs undergo extensive metabolic rewiring with subtype-specific expression patterns indicative of enhanced glucose uptake and glycolytic flux and reduced ATP production via OXPHOS. Thus, the subtype-specific expression patterns of metabolic genes strongly suggest a differential metabolic rewiring between subtypes in line with the lower thyroid differentiation score (TDS [1]) of BRAF-like tumours (TDS markers are reported in Supplementary Fig. S4E). Finally, confirming the subtype-specific reprogramming of metabolic genes we further disclosed that BRAF-like PTCs display specific patterns of expression also for genes involved in amino acid metabolism (Supplementary Fig. S5A, B), sterol and lipid biosynthesis (Supplementary Fig. S5C, D), purine and pyrimidine biosynthesis (Supplementary Fig. S6A, B).
Fig. 1. BRAF-like PTC subtype displays a specific signature of metabolic genes.
a Left panel. Heatmap showing normalised expression data (RNA-Seq from THCA; n = 398) of genes flagged as “metabolism” or “metabolism-related” (PhosphoSitePlus database) in BRAF- (green) vs RAS-like (red) PTCs. Right panel. The enrichment score curve (in green) of genes belonging to the “Metabolic pathway” obtained as output of the Gene Set Enrichment Analysis (GSEA) on the genes differentially expressed (P value=0.0052; P-adjusted=0.0104) between BRAF- and RAS-like tumours (THCA cohort, TCGA). The vertical black lines indicate gene position in the ranked list, whereas horizontal bars in graded colour (upregulated in red; downregulated in blue) indicate the rank-ordered, non-redundant list of genes. b Violin plots illustrating normalised expression values (log2 scale) of SLC2A1, SLC2A3 and SLC2A4 – encoding glucose transporters —in BRAF- (n = 327) and RAS-like (n = 71) PTCs (THCA cohort). *FDR ≤ 0.05, **FDR ≤ 0.01, ***FDR ≤ 0.001. c Box plots displaying normalised expression values (log2 scale) of SLC2A4RG—encoding the main transcription regulator of Glut4—in BRAF-like (BRAFV600E and RET/PTC) and RAS-like tumours. *FDR ≤ 0.05. d Scatter plot of normalised expression data (log2 scale) showing the correlation trend between SLC2A4 and SLC2A4RG overall THCA cohort (generated in cBioPortal); Spearman’s correlation: 0.39 (P = 3.98e-19), Pearson’s correlation: 0.41 (P = 3.01e-21). e Heatmaps showing normalised expression data of genes involved in upper and lower glycolysis in the BRAF- (green) vs RAS-like (red) PTCs.
Analysis of DNA methylation and transcription factors as contributors to the metabolic reprogramming of BRAF-like PTCs
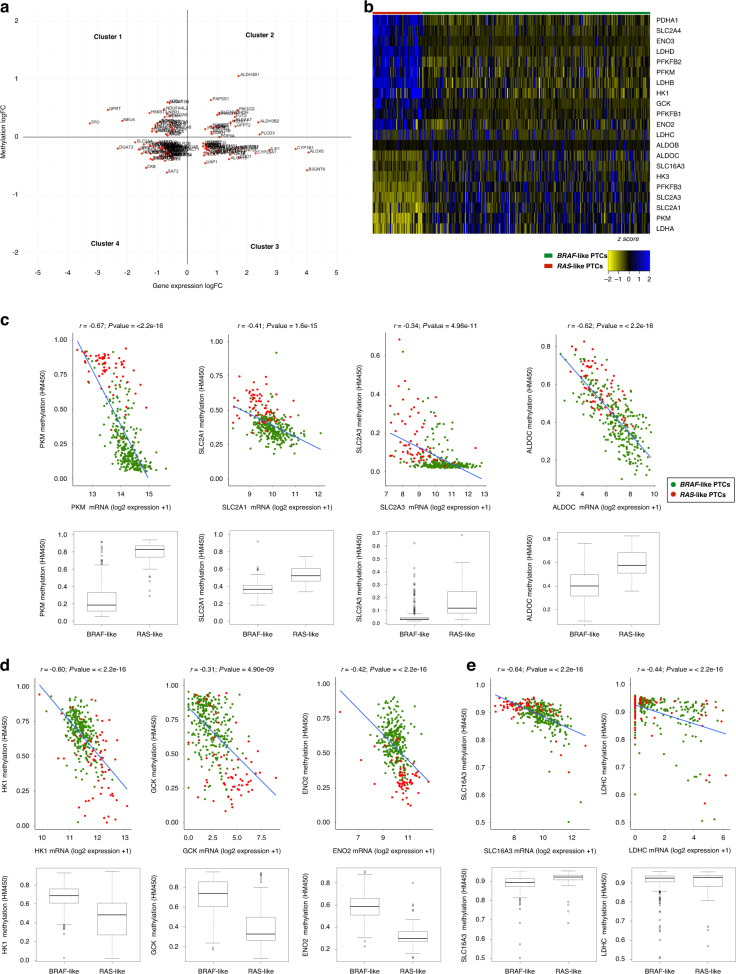
To identify the mechanisms underlying the specific metabolic signatures of BRAF-like PTCs, we first analysed DNA methylation, the main epigenetic alteration participating in the transcriptional regulation of metabolic genes in cancer [50–52]. Taking advantage of TCGA methylation data available for PTC (THCA dataset), we identified differentially methylated probes (DMPs) in CpG islands. Focusing on CpG islands of all metabolic genes differentially expressed between BRAF- and RAS-like tumours (i.e., 459 genes) we found that about 30% of them display significant methylation changes (i.e., 144 genes, of which 48 hyper- and 96 hypomethylated; Supplementary Files S1 and S2). Thus, considering the mean methylation values and expression levels of each independent gene, we classified these metabolic genes into four clusters (Fig. 2a), focusing on those belonging to cluster 1 (i.e., 28 genes characterised by CpG islands’ hypomethylation paralleled by overexpression in BRAF-like tumours) and cluster 3 (i.e., 37 genes hypermethylated and downregulated in BRAF-like tumours; Supplementary File S2).
Fig. 2. PTC subtypes do not display marked differences in DNA methylation of glucose transporters and glycolytic genes.
a Scatter plot of expression and methylation data (logFC; THCA datasets) of genes differentially expressed between BRAF- and RAS-like PTCs. Four distinct clusters of genes have been identified (clockwise from upper left) as altered in BRAF-like tumours: hypermethylated and downregulated (cluster 1); hypermethylated and overexpressed (cluster 2); hypomethylated and overexpressed (cluster 3); hypomethylated and downregulated (cluster 4). b Heatmap showing normalised expression data (RNA-Seq datasets from THCA cohort; n = 398) of genes encoding glucose transporters and glycolysis-related proteins (selected according to KEGG database) in BRAF- (green) vs RAS-like (red) PTCs. c–e Scatter plots showing the correlation—by linear regression analysis—between normalised expression (log2 scale) and methylation data (logFC; THCA cohort) of 9 selected genes differentially expressed between PTC subtypes. These genes were selected as showing a good (0.30 > r < 0.49) or strong (0.5 > r < 0.7) correlation (Pearson’s coefficient) and distinguished in significantly hypomethylated and overexpressed (c), hypermethylated and downregulated in BRAF-like subtype (d) and highly correlated, but not differentially methylated, between tumour subtypes (e).
Interestingly, using KEGG pathway classification, we observed that lipid and nucleic acid metabolism are enriched for differentially methylated genes (Supplementary Fig. S7A and Supplementary File S1). However, CpG islands of genes encoding glucose transporters and glycolysis-related enzymes do not display—except PFKFB3 (Supplementary Fig. S7A)— significant methylation changes between tumour subtypes. Hence, for these genes, we extended the analysis of methylation from promoters to gene bodies. Moreover, we used gene expression data of each independent tumour sample to measure the correlation index for all these genes. Interestingly, for 9 out of 21 genes differentially expressed between PTC subtypes (Fig. 2b) we measured good (0.30 > r < 0.49) or strong (0.5 > r < 0.7) correlation (Pearson’s coefficient) between global gene methylation and expression. Using this approach, we could define three different classes of genes: (1) significantly hypomethylated—and overexpressed—in BRAF-like tumours (Fig. 2c) and, conversely, (2) hypermethylated—and downregulated—in the same tumour subtype (Fig. 2d), and finally (3) highly correlated but not differentially methylated among tumour subtypes (Fig. 2e). The finding that altered expression of genes responsible for glucose uptake and glycolysis in BRAF-like tumours is not supported, at least convincingly, by their differential methylation, makes these tumours possibly not amenable for epigenetic drugs.
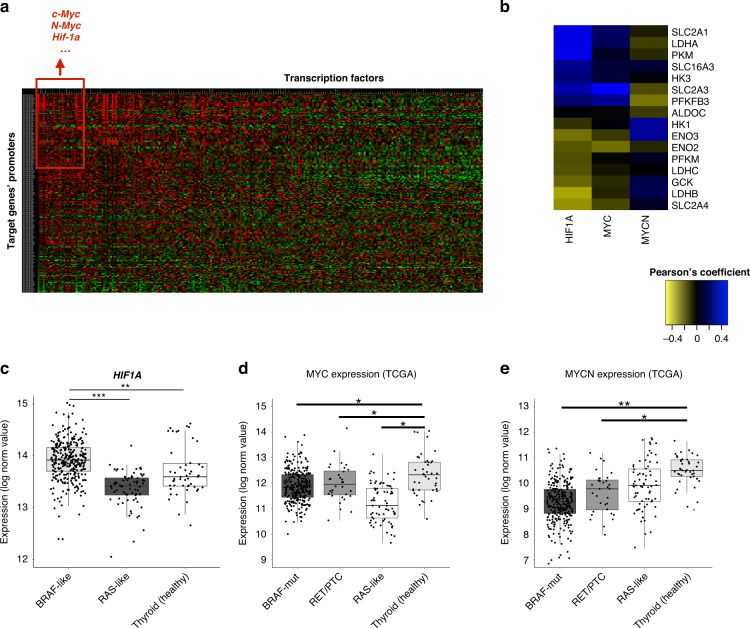
Thus, we investigated transcription factors (TFs) as the most likely candidates by screening the promoters of metabolism-related DEGs and searching for TFs motifs’ enrichment. Interestingly, c-Myc, n-Myc and Hif-1α were the three top-ranked TFs with the highest number of predicted binding sites (Fig. 3a), in line with the notion that c-Myc overexpression in various cancers [53–55] positively modulates the expression of genes involved in glucose metabolism [56–59]. In addition, Hif-1α and c-Myc crosstalk has been well documented [55, 56], and the glycolytic phenotype of BRAFV600E-mutated melanoma is known to be directly linked to the increase of Hif-1α and c-Myc TFs, which regulate the expression of metabolic genes [9, 60]. Combining TCGA RNA-Seq data and co-expression network data implemented in cBioPortal (https://www.cbioportal.org/), we used a guilt-by-association approach to determine which among top-ranked TFs is most likely to regulate the expression of the glucose transporters and other glycolysis-related genes, which we identified as specifically perturbed in BRAF-like tumours. Using this approach, we found that HIF1A gene has the highest correlation coefficients (Fig. 3b)—especially with genes reported as glycolysis drivers [35]—and is significantly overexpressed in BRAF- (vs RAS-like) PTCs (Fig. 3c). Conversely, MYC and MYCN genes display very low correlation (Fig. 3b) and show only a very modest induction in the BRAF- (vs RAS-like) tumour subgroup and are not differentially expressed in tumours vs healthy samples’ comparison (Fig. 3d, e). Overall, these data suggest Hif-1α as a potential regulator of energy metabolism genes specifically altered in BRAF-like PTCs.
Fig. 3. Identification of transcription factors potentially contributing to the metabolic rewiring in BRAF-like PTCs.
a Heatmap showing the enrichment of predicted transcription factors’ binding sites in the putative promoters (window of 1 kb up- and downstream the transcription start site, TSS) of metabolic genes differentially expressed between BRAF- and RAS-like PTCs. Red squares indicate higher enrichment scores for TFs’ predicted binding. b Heatmap showing Pearson’s coefficients between the expression values of top-ranked TFs—having the highest number of predicted binding sites in the promoter of metabolic genes—and the glucose transporters and other glycolytic genes differentially expressed between BRAF- and RAS-like PTCs (THCA cohort). c Box plot comparing normalised HIF1A expression values (log2 scale) in BRAF-, RAS-like PTCs and in healthy thyroid samples. ***FDR ≤ 0.001. d, e Box plots comparing normalised MYC (D) and MYCN (E) expression values (log2 scale) in BRAF-mutated, RET/PTC, RAS-like PTCs and in healthy thyroid samples (THCA cohort). *FDR ≤ 0.05, **FDR ≤ 0.01.
Hif-1α modulates the transcription of genes responsible of the glycolytic phenotype in BRAF-mutated PTC cells
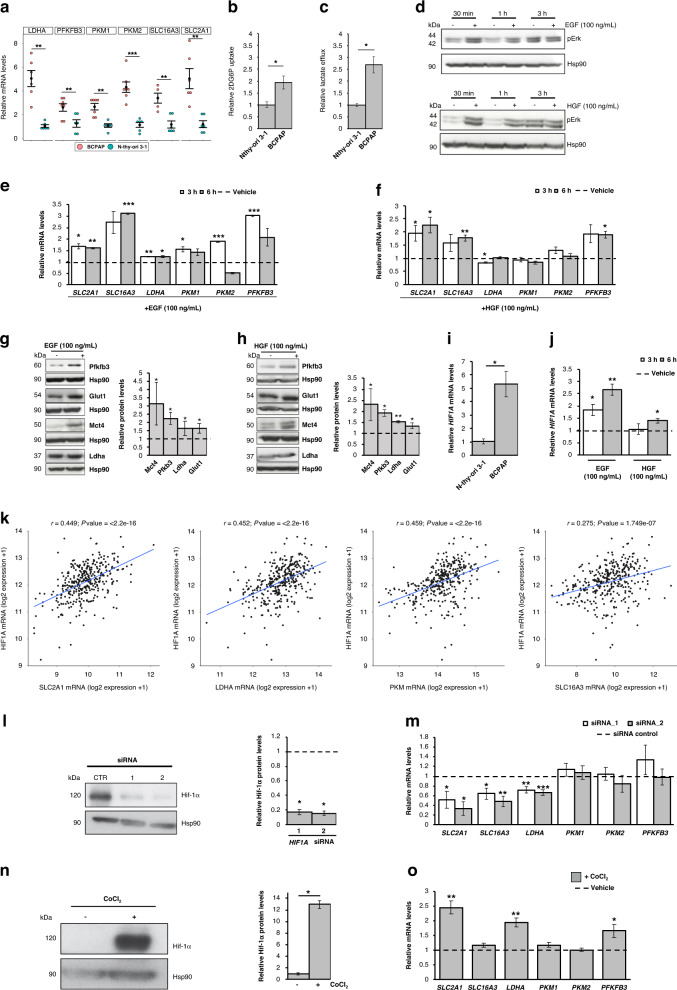
The analysis of transcriptome data from TCGA overall indicates a glycolytic phenotype in BRAF-like PTCs. Thus, to experimentally validate this metabolic gene signature, we compared a BRAFV600E-mutated papillary (BCPAP) tumour cell line and the BRAF wild-type normaloid thyroid follicular epithelial cell line (Nthy-ori 3-1). For the expression analysis, we selected glycolysis-related genes (i.e., SLC2A1, SLC16A3, PFKFB3, PKM1, PKM2 and LDHA) (i) being the most overexpressed in BRAF- vs RAS-like tumours, (ii) displaying the highest glycolytic flux’s coefficients according to Tanner and colleagues [49] and (iii) being also unfavourable prognostic markers across multiple tumour types according to The Human Protein Atlas database (https://www.proteinatlas.org/). Notably, BCPAP cells display the overexpression of selected genes (Fig. 4a) and increased glucose uptake (Fig. 4b) and lactate efflux (Fig. 4c) compared to Nthy-ori 3-1 cells, confirming the high glycolytic phenotype of BRAF-mutated tumours. Afterwards, we evaluated if MAPK activation alone—in non-tumoral thyroid cells—is sufficient to transcriptionally induce the glycolytic genes. Interestingly, most of them are overexpressed upon short-term treatment with two distinct MAPK activators, ie., epidermal and hepatocyte growth factors (EGF and HGF, respectively, Fig. 4d–h). Moreover, in line with the finding that HIF1A is overexpressed in BRAF-like tumour samples, BRAF-mutated cells display increased levels of HIF1A compared to Nthy-ori 3-1 cells (Fig. 4i). Moreover, this TF is also induced by EGF and HGF in normaloid cells (Fig. 4j). Interestingly, we found that all (except PFKFB3) selected metabolic genes—upregulated in BCPAP cells and induced by MAPK stimulation (Fig. 4a, e–h)—display robust co-expression with HIF1A in the THCA cohort (Fig. 4k). Thus, to understand if Hif-1α regulates the expression of these metabolic genes, we used two distinct siRNAs targeting HIF1A mRNA to knock it down in BRAF-mutated tumour cells (Fig. 4l and Supplementary Fig. S7B). Interestingly, as reported in Fig. 4m, tumour cells depleted for Hif-1α display a significantly reduced expression of SLC2A1, LDHA and SLC16A3. Moreover, tumour cells cultured in the presence of CoCl2—a known hypoxia-mimicking chemical inducing Hif-1α protein stabilisation (Fig. 4n and Supplementary Fig. S7C)—display a marked induction of SLC2A1, LDHA and PFKFB3 (Fig. 4o), further indicating that this transcription factor contributes to positively modulate glycolysis-related genes altered in BRAF-like PTCs.
Fig. 4. BRAF-mutated PTC cells display a glycolytic phenotype mediated by Hif-1α.
a Relative mRNA quantification (qPCR) of selected metabolic genes in BCPAP compared to normaloid thyroid cell line (Nthy-ori 3-1). Data are reported as mean ± SEM vs Nthy-ori 3-1 cells of at least six independent experiments. PPIA was used as reference gene. **P value ≤0.01 and ***P value ≤0.001. b Relative colorimetric detection of 2-DG6P uptake in BCPAP compared to Nthy-ori 3-1 cells. Corrected values (pmol) were normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. *P value ≤ 0.05. c Relative colorimetric detection of l-lactic acid content in the cell culture supernatant of BCPAP compared to Nthy-ori 3-1 cells. Lactate concentration (mmol/L) was normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. *P value ≤0.05. d Representative autoradiographs of the western blot analysis for pErk in Nthy-ori 3-1 treated or not with Egf (upper panel) or Hgf (lower panel) 100 ng/mL at multiple time points. Hsp90 was used as a loading control. e, f Relative mRNA quantification (qPCR) of selected metabolic genes in Nthy-ori-3-1 cells upon EGF (100 ng/mL; e) and HGF (100 ng/mL; f) treatment (3, 6 h). Data are reported as mean ± SEM vs control cells (vehicle-treated; dotted line) of three independent experiments. PPIA was used as reference gene. *P value ≤0.05, **P value ≤0.01 and ***P value ≤0.001. g, h Representative autoradiographs of the western blot analysis for selected metabolic enzymes and transporters (i.e., Pfkfb3, Glut1, Mct4 and Ldha) in Nthy-ori 3-1 treated or not with EGF (g) or HGF (h) 100 ng/mL at multiple time points. Hsp90 was used as a loading control. Bar graphs (right part of each panel) report relative protein levels normalised on Hsp90 expression (pixel density analysis of western blots). Data are reported as mean ± SEM vs control cells (i.e., treated with the vehicle) of three independent experiments. *P value ≤0.05, **P value ≤0.01. i, j Relative mRNA quantification (qPCR) of HIF1A gene in BCPAP vs Nthy-ori 3-1 (i) and in BCPAP treated with EGF and HGF (100 ng/Ml, j) for 3 and 6 h vs control cells (i.e., treated with the vehicles; dotted line). Data are reported as mean ± SEM of three independent experiments. PPIA was used as a reference. *P value ≤0.05 and **P value ≤0.01. k Scatter plot of normalised expression data (log2 scale) showing the correlation—by linear regression analysis—of HIF1A and selected glycolysis-related genes in THCA cohort (expression data downloaded from cBioPortal). Pearson correlation coefficient (r) and P value (P) are shown. l Representative autoradiographs of western blot analysis (left panel) of Hif-1α protein levels in BCPAP transfected with two different HIF1A siRNAs. Hsp90 was used as a loading control. Bar graphs (right panel) report relative Hif-1α levels normalised on Hsp90 expression (pixel density analysis of western blots). Data are reported as mean ± SEM vs control cells (i.e., BCPAP transfected with scrambled siRNAs; dotted line) of three independent experiments. *P value ≤0.05. m Relative mRNA quantification (qPCR) of selected metabolic genes upon HIF1A silencing in BCPAP. Data are reported as mean ± SEM vs control cells (scrambled siRNAs; dotted line) of at least three independent experiments. PPIA was used as reference. *P value ≤0.05, **P value ≤0.01 and ***P value ≤0.001. n Representative autoradiographs of western blot analysis (left panel) of Hif-1α protein levels in BCPAP treated with CoCl2 (250 µM, 24 h). Hsp90 was used as a loading control. Bar graphs (right panel) report relative Hif-1α levels normalised on Hsp90 expression (pixel density analysis of western blots). Data are reported as mean ± SEM vs control cells (i.e., treated with the vehicle) of three independent experiments. *P value ≤0.05. o Relative mRNA quantification (qPCR) of selected metabolic genes in BCPAP treated with CoCl2 (250 µM, 24 h). Data are reported as mean ± SEM vs control cells (BCPAP treated with the vehicle; dotted line) of at least four independent experiments. PPIA was used as a reference. *P value ≤0.05 and **P value ≤0.01.
Hif-1α-mediated glycolytic phenotype of BRAF-mutated PTC cells is attenuated by BRAFi
The inhibition of oncogenic B-raf by VMR has been shown to impair glucose uptake and glycolysis in BRAF-mutated melanoma [9]. In the same tumour type, BRAFV600E-mediated regulation of glycolysis has been demonstrated to occur at the transcriptional level [9]. Hence, to address if the VMR-mediated transcriptional effect on glycolysis-related genes is common to other BRAF-mutated tumours, we analysed public RNA-Seq datasets from LINCS L1000 Project. Interestingly, we disclosed that VMR treatment has a marked transcriptional effect—for several metabolic genes—that is shared across multiple BRAF-mutated tumour cell lines. Indeed, as reported in Fig. 5a, VMR reverts the expression of a large fraction of genes involved in glucose uptake, glycolysis, TCA cycle and OXPHOS, identified as hallmarks of BRAF-like tumours (Supplementary File S1). This analysis, other than corroborating the anti-glycolytic effect of VMR also in other BRAF-mutated tumour types, uncovered also a previously unrecognised repressive effect on genes belonging to lipid (Supplementary Fig. S7D), nucleotide (Supplementary Fig. S7E, F) and amino acid metabolism pathways (Supplementary Fig. S7G).
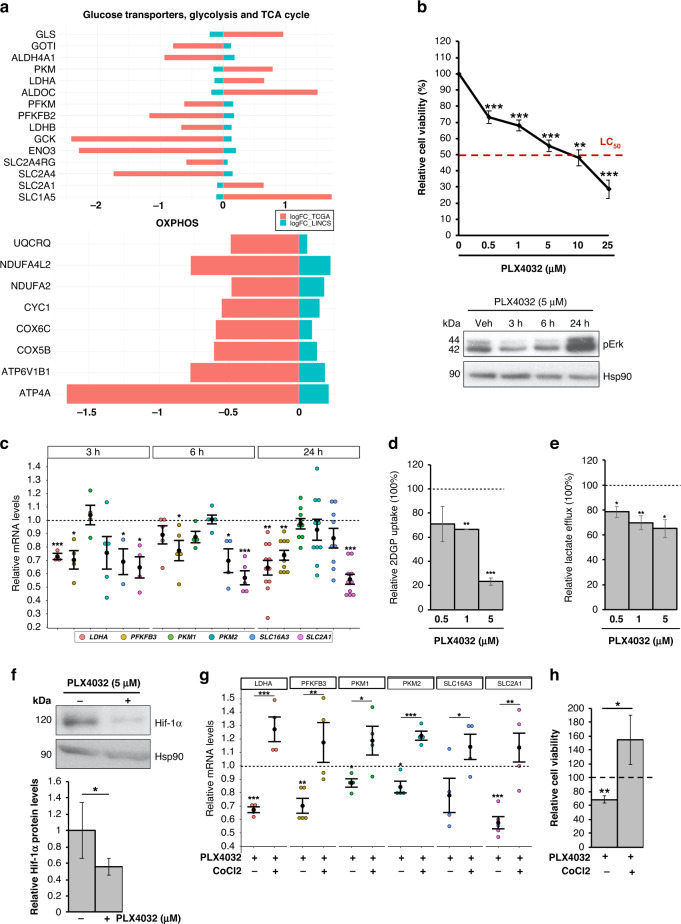
Fig. 5. BRAFi counteract the glycolytic phenotype and Hif-1α-modulated transcription signature of BRAF-mutant PTC cells.
a Bar graphs indicate the expression levels (logFC) of genes involved in energy metabolism—i.e., glucose transport, glycolysis, TCA cycle (upper panel) and OXPHOS (lower panel)—and differentially expressed between BRAF- and RAS-like tumours, whose expression is reverted upon treatment with vemurafenib (blue bars) in multiple BRAF-mutated tumour cell lines from the LINCS 1000 Project. TCGA data (THCA cohort) for the same genes are indicated as red bars (logFC). b Relative cell viability (percentage; upper panel) in BCPAP cells upon treatment with PLX4032 treatment (0.5, 1, 5, 10 and 25 µM)—a vemurafenib analogue—for 72 h. Red dotted line indicates the median lethal concentration (lethal concentration 50%, LC50). Data are reported as mean ± SEM vs control cells (i.e., BCPAP treated with Veh; set to 100% of viability) of at least four independent experiments. **P value ≤0.01 and ***P value ≤0.001. Representative autoradiographs (lower panel) of western blot analysis of Erk phosphorylation (i.e., pErk) levels in BCPAP treated with PLX4032 (5 µM; 3, 6 and 24 h). Hsp90 was used as a loading control. c Relative mRNA quantification (qPCR) of selected metabolic genes in BCPAP treated with PLX4032 (5 µM; 3, 6 and 24 h). Data are reported as mean ± SEM vs control cells (i.e., treated with the vehicle; dotted line) of at least four independent experiments. *P value ≤0.05, **P value ≤0.01 and ***P value ≤0.001. d Relative colorimetric detection of total 2-DG6P uptake in BCPAP treated with different concentrations of PLX4032 for 72 h. Corrected values (pmol) were normalised for the related AUC and data are reported as mean ± SEM of at least three independent experiments. The effect of each treatment was estimated as the percentage of glucose uptake of control cells (i.e., BCPAP treated with the vehicle, set to 100%; dotted line). **P value ≤0.01 and ***P value ≤0.01. e Relative colorimetric detection of l-lactic acid content in cell culture supernatant of BCPAP treated with different concentrations of PLX4032 for 72 h. Lactate concentration (mmol/L) was normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. The effect of each treatment was estimated as the percentage of lactate secreted by control cells (i.e., BCPAP treated with the vehicle, set to 100%; dotted line). *P value ≤0.05 and **P value ≤0.01. f Representative autoradiographs of western blot analysis (upper panel) of Hif-1α protein levels in BCPAP treated with PLX4032 (5 µM; 24 h). Hsp90 was used as a loading control. Bar graphs (lower panel) report relative Hif-1α levels normalised on Hsp90 expression (pixel density analysis of western blots). Data are reported as mean ± SEM vs control cells (i.e., BCPAP treated with the vehicle) of three independent experiments. *P value ≤ 0.05. g Relative mRNA quantification (qPCR) of selected metabolic genes in BCPAP upon PLX4032 treatment (5 µM) alone—vs cells treated with the vehicle—or in combination with CoCl2 (125 µM)—vs cells treated with CoCl2 alone—for 24 h. Data are reported as mean ± SEM vs control cells (dotted line) of at least four independent experiments. PPIA was used as reference. *P value ≤0.05, **P value ≤0.01 and ***P value ≤0.001. h Relative cell viability in BCPAP upon PLX4032 treatment (5 µM) alone—vs cells treated with the vehicle—or in combination with CoCl2 (125 µM)—vs cells treated with CoCl2 alone—for 72 h. Data are reported as mean ± SEM vs control cells (dotted line, set to 100% of viability) of four independent experiments. *P value ≤0.05 and **P value ≤0.01.
To experimentally verify the transcriptional effect of VMR on metabolic genes in the context of thyroid carcinomas, we evaluated if PLX4032—a VMR analogue—could repress the expression of glycolysis-related genes in a BRAF-mutated PTC cell line. We first evaluated BCPAP cells’ viability at increasing drug doses (Fig. 5b, upper panel). Notably, we assessed that PLX4032 concentrations lower than LC50 are sufficient to inhibit ERK phosphorylation even at early time points (3, 6 h; Fig. 5b, lower panel). However, in line with the report from Montero-Conde et al. [61], MAPK pathway inhibition by PLX4032 induces a rapid rebound of pErk (24 h; Fig. 5b, lower panel). As we measured a marked reduction of tumour cells viability 72 h upon PLX4032 treatment we explored the possibility that this cytotoxic effect may be due to metabolic restraining. Interestingly, a transcriptional repression of the selected metabolic genes (Fig. 5c) and a reduction of glucose uptake (Fig. 5d and Supplementary Fig. S8A) and lactate efflux (Fig. 5e) was observed in BRAF-mutated PTC cells. Notably, in line with our finding that Hif-1α contributes to the glycolytic phenotype of BRAF-mutated cells, PLX4032 treatment also impaired Hif-1α levels (Fig. 5f). Accordingly, Hif-1α stabilisation is able to rescue both the expression of glycolysis-related genes (Fig. 5g) and tumour cells’ viability (Fig. 5h) induced by B-raf inhibition. These findings indicate that the attenuation of the glycolytic phenotype induced by B-raf inhibition is mediated—at least in part—by the transcription factor Hif-1α, which controls the expression of a network of genes involved in energy metabolism and specifically altered in BRAF-like tumour samples.
The antitumoral effect of BRAFi on BRAF-mutated PTC and ATC cells is potentiated by the combination with diclofenac
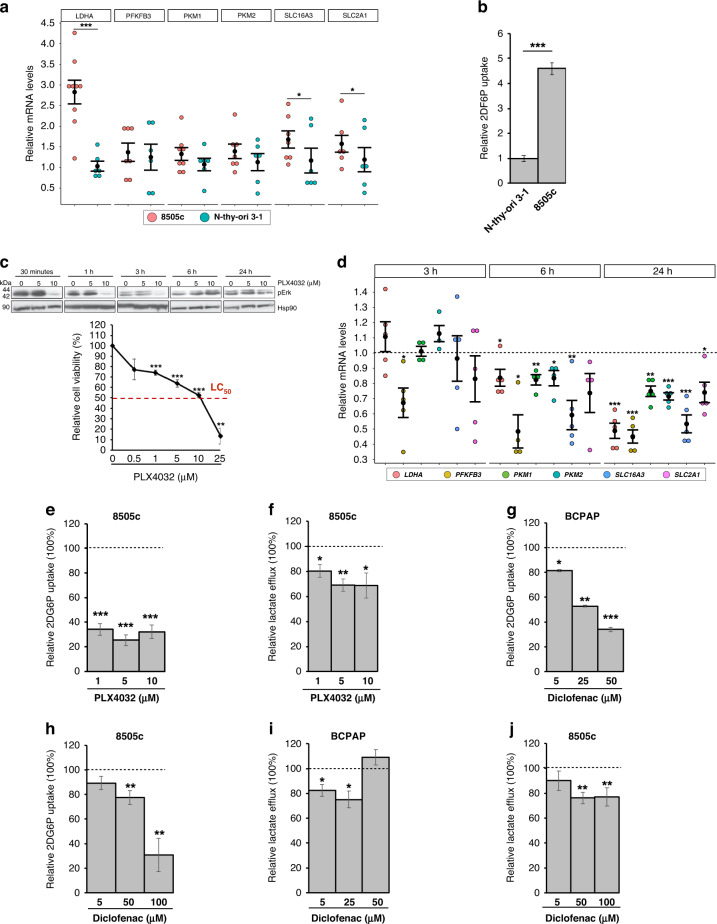
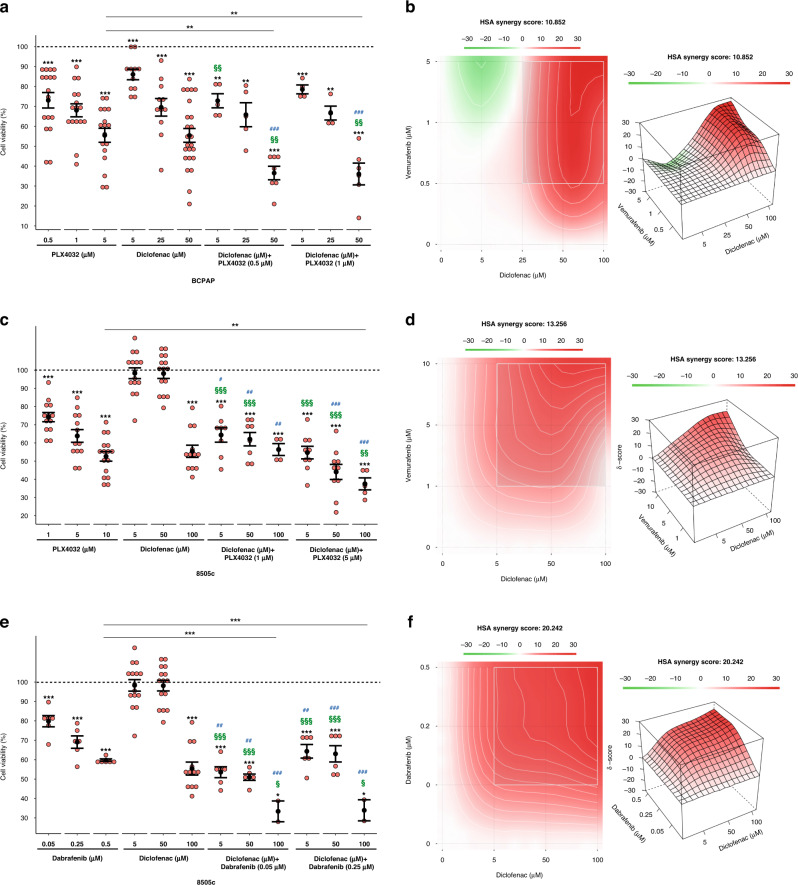
As 20–25% of the aggressive ATC cases display BRAFV600E mutation, we sought to verify, likewise reported in BRAF-driven PTCs, the presence of a BRAF-mediated glycolytic phenotype in this tumor and if B-raf inhibitors are able to restrain it. Notably, the highly aggressive anaplastic BRAF-mutated tumour cells (i.e., 8505c)—compared to normal thyroid cells—display higher levels of multiple metabolic genes (Fig. 6a) and increased glucose uptake (Fig. 6b). Moreover, in line with the notion that B-raf inhibition by vemurafenib is effective on BRAFV600E-mutated ATCs [62–65], PLX4032 markedly reduces pErk and cell viability of ATC cells (Fig. 6c), likewise the treatment with trametinib, an FDA-approved MEK inhibitor (MEKi) for ATC (Supplementary Fig. S8B). Noteworthy, we found that it impairs the glycolytic phenotype also of these aggressive tumour cells, by transcriptionally repressing glycolysis-related genes (Fig. 6d) and reducing glucose uptake (Fig. 6e and Supplementary Fig. S8A) and lactate efflux (Fig. 6f). Further validating the MAPK-dependent modulation of the metabolic phenotype and of glycolytic genes, similar results were obtained using trametinib (Supplementary Fig. S8C, D). Considering that the combination of BRAFi and MEKi is currently used in the therapy of BRAF-mutated ATC, 8505c cells were treated with PLX4032 and trametinib combination. Interestingly, despite the marked suppression of pErk (Supplementary Fig. S8E), tumour cell viability was not significantly impaired by BRAFi and MEKi combination compared to the single drug treatments (Supplementary Fig. S8F). Accordingly, no synergistic effect has been disclosed for PLX4032 and trametinib combination (Supplementary Fig. S8G), further indicating the need to identify other drugs able to maximise the BRAFi effects. Thus, we sought to exploit metabolic pathways as possible targets for new promising anti-cancer strategies. Indeed, several trials based on the simultaneous targeting of multiple pathways are currently evaluating the effects of combined therapies in BRAF-mutated tumours, including in PTC and especially ATC. In this context, non-steroidal anti-inflammatory drugs (NSAIDs)—commonly used as COX inhibitors—have been proposed as VMR-sensitisers in BRAF-mutated melanoma [10]. Hence, we exploited the glycolytic dependency of BRAF-driven thyroid carcinomas using combinations of NSAIDs and BRAFi. To this aim, we selected diclofenac - reported to restrain glycolytic flux in melanoma [10, 31, 66]—as a candidate drug. Notably, diclofenac has been recently reported as a possible inhibitor of Glut1, Mct4 and Ldha [67]. Hence, we treated BRAF-mutated PTC and ATC cells with different doses of diclofenac. We found that such a treatment has no effect on MAPK signalling (Supplementary Fig. S9A), as well as on the expression of both HIF1A and glycolysis-related genes at multiple time points in both tumour cell lines (Supplementary Fig. S9B–F). However, more interestingly, diclofenac is able to impair glucose uptake (Fig. 6g, h, in PTC and ATC, respectively, and Supplementary Fig. S9G) and lactate excretion (Fig. 6i, j, in PTC and ATC, respectively) in both the BRAF-mutated tumour cell lines. Thus, the effect of diclofenac on the energy metabolism is not due to transcriptional repression—as it occurs for B-raf inhibition by PLX4032—but rather it may depend on the inhibition of key glycolytic enzymes. In line with the ability of these drugs to target distinct pathways/processes, we assessed that diclofenac acts synergistically with PLX4032 in reducing cell viability in both tumour types (Fig. 7a–d). We further confirmed the synergistic effect of NSAID and BRAFi combining diclofenac and dabrafenib (DBR), a B-raf inhibitor currently approved by FDA in combination with trametinib for BRAF-mutated ATCs (Fig. 7e, f for ATC and Supplementary Fig. S9H for PTC), further confirming the synergistic effect of NSAID and BRAFi. Taken together, our results suggest that diclofenac potentiates the cytotoxic effect of BRAFi, possibly by inhibiting tumour glycolytic phenotype. Notably, the combination of diclofenac with low doses of BRAFi allows obtaining the same (or more pronounced) effect on cell viability reached by high doses of PLX4032. For instance, in BCPAP cells the co-treatment with diclofenac 50 µM causes a stronger reduction of cell viability even with five to tenfold lower doses of BRAFi (Fig. 7a). Likewise, in 8505c cells the combination of diclofenac 100 µM induces a more marked impairment of cell viability even with two to tenfold lower doses of PLX4032 and DBR (Fig. 7c, e). Thus, the finding that BRAFi/diclofenac combinations exert a potent anti-tumour activity even at very low BRAFi doses in both PTC and ATC BRAF-mutated cell lines (Fig. 7a–f) strongly suggests the possibility to delay—or reduce—the onset of acquired resistance, limiting the risk of unwanted effects.
Fig. 6. BRAF-mutated PTC and ATC cells display a similar glycolytic phenotype modulated by BRAFi and diclofenac.
a Relative mRNA quantification (qPCR) of selected metabolic genes in 8505c compared to Nthy-ori 3-1. Data are reported as mean ± SEM vs Nthy-ori 3-1 cells of at least six independent experiments. PPIA was used as reference. *P value ≤0.05 and ***P value ≤0.001. b Relative colorimetric detection of total 2-DG6P uptake in 8505c compared to Nthy-ori 3-1 cells. Corrected values (pmol) were normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. ***P value ≤0.01. c Representative autoradiographs of western blot analysis (upper panel) of Erk phosphorylation (i.e., pErk) levels in 8505c cells treated with PLX4032 (5 µM and 10 µM; 30 min, 1, 3, 6 and 24 h). Hsp90 was used as a loading control. Relative cell viability (percentage; lower panel) in 8505c cells upon treatment with PLX4032 treatment (0.5, 1, 5, 10 and 25 µM)—a vemurafenib analogue—for 72 h. Red dotted line indicates the median lethal concentration (lethal concentration 50%, LC50). Data are reported as mean ± SEM vs control cells (i.e., BCPAP treated with the vehicle; set to 100% of viability) of at least four independent experiments. **P value ≤0.01, ***P value ≤0.001. d Relative mRNA quantification (qPCR) of selected metabolic genes in 8505c treated with PLX4032 (10 µM; 3, 6 and 24 h). Data are reported as mean ± SEM vs control cells (i.e., 8505c treated with the vehicle; dotted line) of at least four independent experiments. *P value ≤0.05, **P value ≤0.01 and ***P value ≤0.001. e Relative colorimetric detection of total 2-DG6P uptake in 8505c treated with different concentrations of PLX4032 for 72 h. Corrected values (pmol) were normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. The effect of each treatment was estimated as the percentage of glucose uptake of control cells (i.e., 8505c treated with the vehicle, set to 100%; dotted line). **P value ≤0.01 and ***P value ≤0.01. f Relative colorimetric detection of l-lactic acid content in cell culture supernatant of 8505c treated with different concentrations of PLX4032 for 72 h. Lactate concentration (mmol/L) was normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. The effect of each treatment was estimated as the percentage of lactate secreted by control cells (i.e., 8505c treated with the vehicle, set to 100%; dotted line). *P value ≤0.05 and **P value ≤0.01. g, h Relative colorimetric detection of total 2-DG6P uptake in BCPAP (g) and 8505c (h) treated with different concentrations of diclofenac for 72 h. Corrected values (pmol) were normalised for the related AUC and data are reported as mean ± SEM of at least three independent experiments. The effect of each treatment was estimated as the percentage of glucose uptake of control cells (i.e., cells treated with the vehicle, set to 100%; dotted line). *P value ≤0.05, **P value ≤0.01 and ***P value ≤ 0.01. i, j Relative colorimetric detection of l-lactic acid content in cell culture supernatant of BCPAP (i) and 8505c (j) treated with different concentrations of diclofenac for 72 h. Lactate concentration (mmol/L) was normalised for the related AUC and data are reported as mean ± SEM of at least four independent experiments. The effect of each treatment was estimated as the percentage of lactate secreted by control cells (i.e., cells treated with the vehicle, set to 100%; dotted line). *P value ≤0.05 and **P value ≤0.01.
Fig. 7. The combination of BRAFi and diclofenac synergistically reduces the cell viability of BRAF-mutated PTC and ATC cells.
a Relative cell viability in BCPAP cells upon single (PLX4032 and diclofenac) and combined treatment with the two drugs at different doses for 72 h. Data are reported as mean ± SEM vs control cells (i.e., BCPAP treated with the vehicles—indicated as dotted line—are set to 100% of viability) of at least four independent experiments. **P value ≤0.01 and ***P value ≤0.001 vs control cells or BCPAP treated with the maximum dose of PLX4032 (5 µM). ###P value ≤0.001 vs BCPAP treated with the same doses of PLX4032 alone. §§P value ≤ 0.01 vs BCPAP treated with the same doses of diclofenac alone. b Heatmap showing—in 2D (left panel) and 3D (right panel)—the HSA synergy scores (i.e., positive values in red denote synergy), analysed by SynergyFinder tool, for the combination of PLX4032 and diclofenac in BCPAP. c Relative cell viability in 8505c cells upon single (PLX4032 and diclofenac) and combined treatment with the two drugs at different doses for 72 h. Data are reported as mean ± SEM vs control cells (i.e., 8505c treated with the vehicles—indicated as dotted line—are set to 100% of viability) of at least four independent experiments. ***P value ≤0.001 vs control cells or 8505c treated with the maximum dose of PLX4032 (10 µM). #P value ≤0.05, ##P value ≤0.01 and ###P value ≤0.001 vs 8505c treated with the same doses of PLX4032 alone. §§P value ≤0.01 and §§§P value ≤0.01 vs 8505c treated with the same doses of diclofenac alone. d Heatmap showing—in 2D (left panel) and 3D (right panel)—the HSA synergy scores (i.e., positive values in red denote synergy), analysed by SynergyFinder tool, for the combination of PLX4032 and diclofenac in 8505c. e Relative cell viability in 8505c cells upon single (dabrafenib or diclofenac) and combined treatment with the two drugs at different doses for 72 h. Data are reported as mean ± SEM vs control cells (i.e., 8505c treated with the vehicles—indicated as dotted line—are set to 100% of viability) of at least three independent experiments. *P value ≤0.05 and ***P value ≤0.001 vs control cells or 8505c treated with the maximum dose of DBR (0.5 µM). ##P value ≤0.01 and ###P value ≤0.001 vs 8505c treated with the same doses of dabrafenib alone. §P value ≤0.05 and §§§P value ≤0.01 vs 8505c treated with the same doses of diclofenac alone. f Heatmap showing—in 2D (left panel) and 3D (right panel)—the HSA synergy scores (i.e., positive values in red denote synergy), analysed by SynergyFinder tool, for the combination of dabrafenib and diclofenac in 8505c.
Discussion
Metabolic reprogramming of tumour cells is among the ten hallmarks of cancer [68] and has been proposed as one of the driver mechanisms able to increase the metastatic potential and reduce drug sensitivity, leading to poor prognosis in multiple cancer types [3–11, 43, 69, 70]. Hence, targeting metabolism is emerging as a promising strategy to improve current therapies and overcome drug resistance. In line with independent studies reporting the association of metabolic gene signatures and clinicopathological features in papillary thyroid carcinomas [20–27], our work provides evidence of a strong perturbation of metabolic genes between the two distinct tumour subtypes (i.e., BRAF- and RAS-like PTCs). Our results indicate that these tumour subtypes largely differ for the expression of key genes encoding glucose, lactate and glutamine transporters, glycolytic enzymes as well as genes involved in the metabolism of lipids, amino acids and nucleic acids. Particularly, the finding that the altered expression of energy metabolism genes does not associate with a differential methylation pattern makes this metabolic route not amenable for epigenetic drug treatment, at least in the context of PTCs. However, the identification of Hif-1α—overexpressed in BRAF-like tumours—as one of the orchestrators of metabolic processes in these tumours, likewise in melanoma [9], opens new intriguing therapeutic perspectives. Indeed, together with c-Myc, Hif-1α is known to simultaneously induce glycolytic genes and inhibitors of mitochondrial metabolism [60, 71–73], to reduce mitochondrial biogenesis [74, 75] and to play a key role in metabolic reprogramming in many tumour types [69, 72, 74–76]. Interestingly, the modulation of Hif-1α expression can affect the sensitivity to radiotherapy in thyroid neoplasia and to chemotherapy in multiple tumours [77], making it a promising therapeutic target for many cancer types, including thyroid carcinomas [78].
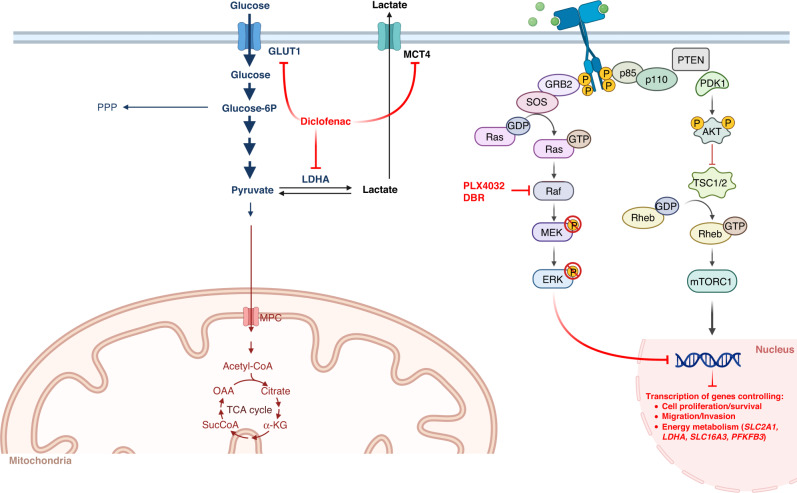
Overall, our findings indicate the possibility to interfere with energy metabolism and—in the spirit of precision oncology—to specifically target tumour cells. Noteworthy, the identification of subtype-specific signatures of metabolic genes reinforces the need to stratify thyroid carcinoma patients based on their background of somatic mutations. This approach is particularly relevant as BRAF-mutated PTCs and ATCs can be selectively targeted by BRAFi—i.e., vemurafenib and dabrafenib (combined with trametinib in ATC)—whose efficacy has been proven both in BRAFV600E-positive metastatic melanoma [79] and in non-melanoma cancers [65, 80]. However, despite sharing the same—and most frequent—BRAF mutation (i.e., BRAFV600E), different tumour types display a heterogeneous responsiveness to BRAFi-based therapies. An interesting hypothesis to explain such heterogeneity is that drug effectiveness may depend on a different metabolic vulnerability, reported as the Achilles’ heel of BRAF-driven melanoma [8–10]. In line with this, both in PTC and ATC, (i) the overexpression of key glucose transporters and glycolytic enzymes (identified herein as hallmark of papillary and anaplastic BRAF-mutated tumours), (ii) their transcriptional repression and (iii) the impairment of glucose uptake and glycolytic flux upon inhibition of MAPK pathway (by targeting B-raf and/or MEK1/2 proteins), indicate that thyroid carcinomas behave similarly to melanoma cells. Although we did not evaluate if impairing the glycolytic flux induces a switch toward OXPHOS in BRAF-mutated tumour cells, our results encouraged us to exploit energy metabolism as an additional process to target together with the MAPK pathway. Drug combinations to simultaneously target multiple pathways/processes are under active evaluation in several BRAF-mutated tumours, and especially in the most aggressive forms, such as anaplastic tumours. In particular, NSAIDs have been recently proposed as BRAFi-sensitisers in melanoma [10, 31]. Our work indicates that papillary and anaplastic thyroid carcinomas can be included in the growing list of tumours amenable to treatment with combinatorial approaches targeting both canonical pathways (MAPK) and new metabolic ones. Indeed, we report that diclofenac—a well-studied and clinically safe and approved drug—restrains the glycolytic phenotype of BRAF-mutated papillary and anaplastic thyroid carcinoma cells, opening new relevant clinical perspectives for these tumours. Moreover, the finding that diclofenac treatment has no effect on the transcriptional modulation of either HIF1A or glycolytic genes suggests that its activity on glucose uptake and glycolytic flux is independent from MAPK pathway (Fig. 8). The synergistic effect of diclofenac and PLX4032 on tumour cells viability can be explained by the simultaneous repression of the metabolism-shaping TF Hif-1α (and its metabolic network) and by the inhibition of Ldha enzyme, Glut1 and Mct4 transporters (schematized in Fig. 8). Noteworthy, the repressive effect of diclofenac on tumour metabolism is evident even at low concentrations (25 μM and 50 μM for PTC and ATC cells, respectively), which are below the physiologically relevant plasmatic levels (0.15–105 mg/l, corresponding to ∼50–350 μM) [10, 81, 82] achieved at the recommended doses to treat inflammatory states [83]. Moreover, several independent reports about intravenous administration of diclofenac, showing twofold higher plasma concentrations (vs oral administration) [81, 84–86] and 3-times higher steady-state levels in synovial fluid, suggest that similar drug concentrations may be eventually reached in tumours [10]. More interestingly, diclofenac treatment becomes synergic with PLX4032 and DBR even at very low doses of BRAFi (i.e., 0.5 μM for PTC and 1 μM for ATC), which show only a modest impact on tumour cells viability (20–25% decrease). Noteworthy, our results clearly show that the combination with diclofenac outperforms even the first-line treatment approved for BRAF-mutated ATC, i.e., BRAFi plus MEKi (trametinib) combination. This finding acquires higher relevance in light of the urgent need to optimise therapeutic regimens for improving drug sensitivity and overcoming resistance onset and tumour relapse. In this regard, the identification of new pharmacological targets, as well as different drug combinations, appears promising to evaluate new therapeutic strategies. For instance, Ldha inhibition has been recently proposed as an attractive strategy for novel approaches in oncology, especially for targeting tumours highly addicted to aerobic glycolysis (reviewed in [87, 88]). However, because of tumour heterogeneity, Ldha inhibition may be more effective—in some cancers—if combined with other drugs targeting also OXPHOS-addicted cells [88]. Therefore, it would be relevant to test new drug combinations (BRAFi and LDHAi) in BRAF-mutated tumours, including—but not limiting to—thyroid carcinomas. Moreover, considering our results and the potential of NSAIDs to target multiple glycolysis-related proteins, it would be interesting to test these drugs in tumours where Ldha inhibition has proven successful in preclinical studies, such as pancreatic and lung cancers. Until now, few studies in glioma [89] and melanoma [10, 31] have evaluated the metabolic effect of the combinatorial use of diclofenac in cancer therapies, whereas most of the studies have focused on its anti-angiogenic, immunomodulatory and pro-apoptotic properties (reviewed in ref. [90]). In conclusion, our results suggest that the combinatorial use of BRAFi and diclofenac is likely to represent a new possible therapeutic approach to treat BRAF-mutated papillary and aggressive anaplastic thyroid carcinomas. We envision that diclofenac, having an established role in oncological practice for the treatment of cancer-related pain, may eventually be considered as an active component of new therapies targeting tumour metabolism in BRAF-mutated cancers.
Fig. 8. Schematic model of BRAFi and diclofenac effects on metabolic processes.
Schematic representation of the possible molecular mechanism underlying the synergism between diclofenac and vemurafenib/dabrafenib (PLX4032 and DBR) in BRAF-mutated papillary and anaplastic thyroid carcinomas. Created with Biorender.com.
Supplementary information
Supplementary Figures, legends and Table
Acknowledgements
We thank all members of our team for their critical input and suggestions.
Author contributions
MA: conceptualisation, formal analysis, investigation, methodology, validation, visualisation, writing—original draft, writing—review and editing. SC: conceptualisation, formal analysis, investigation, methodology, validation, visualisation, writing—review and editing. CP: formal analysis, investigation, validation, visualisation, writing—review and editing. AF: formal analysis, data curation and visualisation. AC: resources, supervision, writing—review and editing. VC: conceptualisation, data curation, funding acquisition, investigation, methodology, project administration, resources, supervision, visualisation, writing— original draft, writing—review and editing.
Funding
This work was supported by My First AIRC Grant (MFAG) Programme (grant n. 23453) and SATIN-POR CAMPANIA FESR 2014/2020 to VC.
Data availability
All data generated or analysed during this study are included either in this article or in additional files, and the links to public datasets are reported in the manuscript.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
This manuscript has not been previously published and is not under consideration for publication elsewhere.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Marianna Aprile, Simona Cataldi.
Contributor Information
Marianna Aprile, Email: marianna.aprile@igb.cnr.it.
Valerio Costa, Email: valerio.costa@igb.cnr.it.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02282-2.
References
- 1.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–90. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costa V, Esposito R, Ziviello C, Sepe R, Bim LV, Cacciola NA, et al. New somatic mutations and WNK1-B4GALNT3 gene fusion in papillary thyroid carcinoma. Oncotarget. 2015;6:11242–51. doi: 10.18632/oncotarget.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer’s Achilles’ heel. Cancer Cell. 2008;13:472–82. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fendt SM, Frezza C, Erez A. Targeting metabolic plasticity and flexibility dynamics for cancer therapy. Cancer Discov. 2020;10:1797–807. doi: 10.1158/2159-8290.CD-20-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xia L, Oyang L, Lin J, Tan S, Han Y, Wu N, et al. The cancer metabolic reprogramming and immune response. Mol Cancer. 2021;20:28. doi: 10.1186/s12943-021-01316-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warburg O, Wind F, Negelein E. The metabolism of tumors in the body. J Gen Physiol. 1927;8:519–30. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haq R, Shoag J, Andreu-Perez P, Yokoyama S, Edelman H, Rowe GC, et al. Oncogenic BRAF regulates oxidative metabolism via PGC1α and MITF. Cancer Cell. 2013;23:302–15. doi: 10.1016/j.ccr.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parmenter TJ, Kleinschmidt M, Kinross KM, Bond ST, Li J, Kaadige MR, et al. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4:423–33. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummer C, Faerber S, Bruss C, Blank C, Lacroix R, Haferkamp S, et al. Metabolic targeting synergizes with MAPK inhibition and delays drug resistance in melanoma. Cancer Lett. 2019;442:453–63. doi: 10.1016/j.canlet.2018.11.018. [DOI] [PubMed] [Google Scholar]
- 11.Som P, Atkins HL, Bandoypadhyay D, Fowler JS, MacGregor RR, Matsui K, et al. A fluorinated glucose analog, 2-fluoro-2-deoxy-D-glucose (F-18): nontoxic tracer for rapid tumor detection. J Nucl Med. 1980;21:670–5. [PubMed] [Google Scholar]
- 12.Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science. 2009;325:1555–9. doi: 10.1126/science.1174229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee MH, Lee SE, Kim DW, Ryu MJ, Kim SJ, Kim SJ, et al. Mitochondrial localization and regulation of BRAFV600E in thyroid cancer: a clinically used RAF inhibitor is unable to block the mitochondrial activities of BRAFV600E. J Clin Endocrinol Metab. 2011;96:E19–E30. doi: 10.1210/jc.2010-1071. [DOI] [PubMed] [Google Scholar]
- 14.Cesi G, Walbrecq G, Zimmer A, Kreis S, Haan C. ROS production induced by BRAF inhibitor treatment rewires metabolic processes affecting cell growth of melanoma cells. Mol Cancer. 2017;16:102. doi: 10.1186/s12943-017-0667-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bollag G, Hirth P, Tsai J, Zhang J, Ibrahim PN, Cho H, et al. Clinical efficacy of a RAF inhibitor needs broad target blockade in BRAF-mutant melanoma. Nature. 2010;467:596–9. doi: 10.1038/nature09454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Søndergaard JN, Nazarian R, Wang Q, Guo D, Hsueh T, Mok S, et al. A. Differential sensitivity of melanoma cell lines with BRAFV600E mutation to the specific Raf inhibitor PLX4032. J Transl Med. 2010;8:39. doi: 10.1186/1479-5876-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baudy AR, Dogan T, Flores-Mercado JE, Hoeflich KP, Su F, van Bruggen N, et al. FDG-PET is a good biomarker of both early response and acquired resistance in BRAFV600 mutant melanomas treated with vemurafenib and the MEK inhibitor GDC-0973. EJNMMI Res. 2012;2:22. doi: 10.1186/2191-219X-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McArthur GA, Puzanov I, Amaravadi R, Ribas A, Chapman P, Kim KB, et al. Marked, homogeneous, and early [18F]fluorodeoxyglucose-positron emission tomography responses to vemurafenib in BRAF-mutant advanced melanoma. J Clin Oncol. 2012;30:1628–34. doi: 10.1200/JCO.2011.39.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carlino MS, Saunders CA, Haydu LE, Menzies AM, Martin Curtis C, Jr, Lebowitz PF, et al. [18]F-labelled fluorodeoxyglucose-positron emission tomography (FDG-PET) heterogeneity of response is prognostic in dabrafenib treated BRAF mutant metastatic melanoma. Eur J Cancer. 2013;49:395–402. doi: 10.1016/j.ejca.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 20.Chai YJ, Yi JW, Oh SW, Kim YA, Yi KH, Kim JH, et al. Upregulation of SLC2 (GLUT) family genes is related to poor survival outcomes in papillary thyroid carcinoma: analysis of data from The Cancer Genome Atlas. Surgery. 2017;161:188–94. doi: 10.1016/j.surg.2016.04.050. [DOI] [PubMed] [Google Scholar]
- 21.Ma B, Jiang H, Wen D, Hu J, Han L, Liu W, et al. Transcriptome analyses identify a metabolic gene signature indicative of dedifferentiation of papillary thyroid cancer. J Clin Endocrinol Metab. 2019;104:3713–25. doi: 10.1210/jc.2018-02686. [DOI] [PubMed] [Google Scholar]
- 22.Suh HY, Choi H, Paeng JC, Cheon GJ, Chung JK, Kang KW. Comprehensive gene expression analysis for exploring the association between glucose metabolism and differentiation of thyroid cancer. BMC Cancer. 2019;19:1260. doi: 10.1186/s12885-019-6482-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M, Sun T, Wen S, Zhang T, Wang X, Cao Y, et al. Characteristics of lipid metabolism-related gene expression-based molecular subtype in papillary thyroid cancer. Acta Biochim Biophys Sin. 2020;52:1166–70. doi: 10.1093/abbs/gmaa092. [DOI] [PubMed] [Google Scholar]
- 24.Ban EJ, Kim D, Kim JK, Kang SW, Lee J, Jeong JJ, et al. Lactate dehydrogenase a as a potential new biomarker for thyroid cancer. Endocrinol Metab. 2021;36:96–105. doi: 10.3803/EnM.2020.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hou X, Shi X, Zhang W, Li D, Hu L, Yang J, et al. LDHA induces EMT gene transcription and regulates autophagy to promote the metastasis and tumorigenesis of papillary thyroid carcinoma. Cell Death Dis. 2021;12:347. doi: 10.1038/s41419-021-03641-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu F, Xu H, Li Z, Huang Y, Huang X, Li Y, et al. Glycolysis-based genes are potential biomarkers in thyroid cancer. Front Oncol. 2021;11:534838. doi: 10.3389/fonc.2021.534838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen S, Luo Y, Wu W, Zhang T, Yang Y, Ji Q, et al. Identification of lipid metabolism-related genes as prognostic indicators in papillary thyroid cancer. Acta Biochim Biophys Sin. 2021;53:1579–89. doi: 10.1093/abbs/gmab145. [DOI] [PubMed] [Google Scholar]
- 28.Fedorenko IV, Paraiso KH, Smalley KS. Acquired and intrinsic BRAF inhibitor resistance in BRAF V600E mutant melanoma. Biochem Pharm. 2011;82:201–9. doi: 10.1016/j.bcp.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez JN, Wang T, Cohen MS. BRAF and MEK inhibitors: use and resistance in BRAF-mutated cancers. Drugs. 2018;78:549–66. doi: 10.1007/s40265-018-0884-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi H, Moriceau G, Kong X, Lee MK, Lee H, Koya RC, et al. Melanoma whole-exome sequencing identifies (V600E)B-RAF amplification-mediated acquired B-RAF inhibitor resistance. Nat Commun. 2012;3:724. doi: 10.1038/ncomms1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottfried E, Lang SA, Renner K, Bosserhoff A, Gronwald W, Rehli M, et al. New aspects of an old drug-diclofenac targets MYC and glucose metabolism in tumor cells. PLoS ONE. 2013;8:e66987. doi: 10.1371/journal.pone.0066987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Federico A, Rienzo M, Abbondanza C, Costa V, Ciccodicola A, Casamassimi A. Pan-cancer mutational and transcriptional analysis of the integrator complex. Int J Mol Sci. 2017;18:936. doi: 10.3390/ijms18050936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang HB, Fan J, Lin R, Elf S, Ji Q, Zhao L, et al. Metabolic rewiring by oncogenic BRAF V600E links ketogenesis pathway to BRAF-MEK1 signaling. Mol Cell. 2015;59:345–58. doi: 10.1016/j.molcel.2015.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–7. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stathias V, Turner J, Koleti A, Vidovic D, Cooper D, Fazel-Najafabadi M, et al. LINCS Data Portal 2.0: next generation access point for perturbation-response signatures. Nucleic Acids Res. 2020;48:D431–D439. doi: 10.1093/nar/gkz1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niepel M, Hafner M, Mills CE, Subramanian K, Williams EH, Chung M, et al. A multi-center study on the reproducibility of drug-response assays in mammalian cell lines. Cell Syst. 2019;9:35–48.e5. doi: 10.1016/j.cels.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costa V, Conte I, Ziviello C, Casamassimi A, Alfano G, Banfi S, et al. Identification and expression analysis of novel Jakmip1 transcripts. Gene. 2007;402:1–8. doi: 10.1016/j.gene.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Aprile M, Cataldi S, Ambrosio MR, D’Esposito V, Lim K, Dietrich A, et al. PPARγΔ5, a naturally occurring dominant-negative splice isoform, impairs PPARγ function and adipocyte differentiation. Cell Rep. 2018;25:1577–1592.e6. doi: 10.1016/j.celrep.2018.10.035. [DOI] [PubMed] [Google Scholar]
- 40.Esposito R, Esposito D, Pallante P, Fusco A, Ciccodicola A, Costa V. Oncogenic properties of the antisense lncRNA COMET in BRAF- and RET-driven papillary thyroid carcinomas. Cancer Res. 2019;79:2124–35. doi: 10.1158/0008-5472.CAN-18-2520. [DOI] [PubMed] [Google Scholar]
- 41.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 3.0: an interactive analysis and consensus interpretation of multi-drug synergies across multiple samples. Nucleic Acids Res. 2022;50:W739–43. doi: 10.1093/nar/gkac382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ianevski A, Giri AK, Gautam P, Kononov A, Potdar S, Saarela J, et al. Prediction of drug combination effects with a minimal set of experiments. Nat Mach Intell. 2019;1:568–77. doi: 10.1038/s42256-019-0122-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Coelho RG, Fortunato RS, Carvalho DP. Metabolic reprogramming in thyroid carcinoma. Front Oncol. 2018;8:82. doi: 10.3389/fonc.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nahm JH, Kim HM, Koo JS. Glycolysis-related protein expression in thyroid cancer. Tumour Biol. 2017;39:1010428317695922. doi: 10.1177/1010428317695922. [DOI] [PubMed] [Google Scholar]
- 45.Birsoy K, Possemato R, Lorbeer FK, Bayraktar EC, Thiru P, Yucel B, et al. Metabolic determinants of cancer cell sensitivity to glucose limitation and biguanides. Nature. 2014;508:108–12. doi: 10.1038/nature13110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onodera Y, Nam JM, Bissell MJ. Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J Clin Invest. 2014;124:367–84. doi: 10.1172/JCI63146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol. 2019;9:1143. doi: 10.3389/fonc.2019.01143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Payen VL, Mina E, Van Hée VF, Porporato PE, Sonveaux P. Monocarboxylate transporters in cancer. Mol Metab. 2020;33:48–66. doi: 10.1016/j.molmet.2019.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanner LB, Goglia AG, Wei MH, Sehgal T, Parsons LR, Park JO, et al. Four key steps control glycolytic flux in mammalian cells. Cell Syst. 2018;7:49–62.e8. doi: 10.1016/j.cels.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaelin WG, Jr, McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miranda-Gonçalves V, Lameirinhas A, Henrique R, Jerónimo C. Metabolism and epigenetic interplay in cancer: regulation and putative therapeutic targets. Front Genet. 2018;9:427. [DOI] [PMC free article] [PubMed]
- 52.Mehta A, Patel BM. Therapeutic opportunities in colon cancer: focus on phosphodiesterase inhibitors. Life Sci. 2019;230:150–61. doi: 10.1016/j.lfs.2019.05.043. [DOI] [PubMed] [Google Scholar]
- 53.Shim H, Dolde C, Lewis BC, Wu CS, Dang G, Jungmann RA, et al. c-Myc transactivation of LDH-A: implications for tumor metabolism and growth. Proc Natl Acad Sci USA. 1997;94:6658–63. doi: 10.1073/pnas.94.13.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, et al. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J Biol Chem. 2000;275:21797–800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 55.Gordan JD, Thompson CB, Simon MC. HIF and c-Myc: sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell. 2007;12:108–13. doi: 10.1016/j.ccr.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol. 2007;27:7381–93. doi: 10.1128/MCB.00440-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–64. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 58.Dang CV, Le A, Gao P. MYC-induced cancer cell energy metabolism and therapeutic opportunities. Clin Cancer Res. 2009;15:6479–83. doi: 10.1158/1078-0432.CCR-09-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Qu Y, Yang Q, Liu J, Shi B, Ji M, Li G, et al. c-Myc is required for BRAFV600E-induced epigenetic silencing by H3K27me3 in tumorigenesis. Theranostics. 2017;7:2092–107. doi: 10.7150/thno.19884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao D, Hou M, Guan YS, Jiang M, Yang Y, Gou HF. Expression of HIF-1alpha and VEGF in colorectal cancer: association with clinical outcomes and prognostic implications. BMC Cancer. 2009;9:432. doi: 10.1186/1471-2407-9-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, et al. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3:520–33. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rosove MH, Peddi PF, Glaspy JA. BRAF V600E inhibition in anaplastic thyroid cancer. N Engl J Med. 2013;368:684–5. doi: 10.1056/NEJMc1215697. [DOI] [PubMed] [Google Scholar]
- 63.Marten KA, Gudena VK. Use of vemurafenib in anaplastic thyroid carcinoma: a case report. Cancer Biol Ther. 2015;16:1430–3. doi: 10.1080/15384047.2015.1071734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prager GW, Koperek O, Mayerhoefer ME, Muellauer L, Wrba F, Niederle B, et al. Sustained response to vemurafenib in a BRAFV600E-mutated anaplastic thyroid carcinoma patient. Thyroid. 2016;26:1515–6. doi: 10.1089/thy.2015.0575. [DOI] [PubMed] [Google Scholar]
- 65.Subbiah V, Puzanov I, Blay JY, Chau I, Lockhart AC, Raje NS, et al. Pan-cancer efficacy of vemurafenib in BRAFV600-mutant non-melanoma cancers. Cancer Discov. 2020;10:657–63. doi: 10.1158/2159-8290.CD-19-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Albano F, Arcucci A, Granato G, Romano S, Montagnani S, De Vendittis E, et al. Markers of mitochondrial dysfunction during the diclofenac-induced apoptosis in melanoma cell lines. Biochimie. 2013;95:934–45. doi: 10.1016/j.biochi.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 67.Pandey SK, Yadav S, Goel Y, Temre MK, Singh VK, Singh SM. Molecular docking of anti-inflammatory drug diclofenac with metabolic targets: Potential applications in cancer therapeutics. J Theor Biol. 2019;21:117–25. doi: 10.1016/j.jtbi.2019.01.020. [DOI] [PubMed] [Google Scholar]
- 68.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Koukourakis MI, Giatromanolaki A, Sivridis E, Bougioukas G, Didilis V, Gatter KC, et al. Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br J Cancer. 2003;89:877–85. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang X, Zhao H, Li Y, Xia D, Yang L, Mam Y, et al. The role of YAP/TAZ activity in cancer metabolic reprogramming. Mol Cancer. 2018;17:134. [DOI] [PMC free article] [PubMed]
- 71.Klaus A, Fathi O, Tatjana TW, Bruno N, Oskar K. Expression of hypoxia-associated protein HIF-1α in follicular thyroid cancer is associated with distant metastasis. Pathol Oncol Res. 2018;24:289–96. doi: 10.1007/s12253-017-0232-4. [DOI] [PubMed] [Google Scholar]
- 72.Marín-Hernández A, Gallardo-Pérez JC, Ralph SJ, Rodríguez-Enríquez S, Moreno-Sánchez R. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem. 2009;9:1084–101. doi: 10.2174/138955709788922610. [DOI] [PubMed] [Google Scholar]
- 73.Semenza GL. Hypoxia-inducible factors: mediators of cancer progression and targets for cancer therapy. Trends Pharmacol Sci. 2012;33:207–14. doi: 10.1016/j.tips.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gottlieb E, Tomlinson IPM. Mitochondrial tumour suppressors: a genetic and biochemical update. Nat Rev Cancer. 2005;5:857–66. doi: 10.1038/nrc1737. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller K, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–20. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 76.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–85. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 77.Kumar SM, Yu H, Edwards R, Chen L, Kazianis S, Brafford P, et al. Mutant V600E BRAF increases hypoxia inducible factor-1alpha expression in melanoma. Cancer Res. 2007;67:3177–84. doi: 10.1158/0008-5472.CAN-06-3312. [DOI] [PubMed] [Google Scholar]
- 78.Wang JZ, Zhu W, Han J, Yang X, Zhou R, Lu HC, et al. The role of the HIF-1α/ALYREF/PKM2 axis in glycolysis and tumorigenesis of bladder cancer. Cancer Commun. 2021;41:560–75. doi: 10.1002/cac2.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–16. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hyman DM, Puzanov I, Subbiah V, Faris JE, Chau I, Blay JY, et al. Vemurafenib in multiple nonmelanoma cancers with BRAF V600 mutations. N Engl J Med. 2015;373:726–36. doi: 10.1056/NEJMoa1502309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rordorf CM, Choi L, Marshall P, Mangold JB. Clinical pharmacology of lumiracoxib: a selective cyclo-oxygenase-2 inhibitor. Clin Pharmacokinet. 2005;44:1247–66. doi: 10.2165/00003088-200544120-00004. [DOI] [PubMed] [Google Scholar]
- 82.Davies NM, Anderson KE. Clinical pharmacokinetics of diclofenac: therapeutic insights and pitfalls. Clin Pharmacokinet. 1997;33:184–213. doi: 10.2165/00003088-199733030-00003. [DOI] [PubMed] [Google Scholar]
- 83.FDA, Prescribing Information “diclofenac Sodium Enteric-coated Tablets. 2018; https://www.accessdata.fda.gov/drugsatfda_docs/label/2009/019201s038lbl.pdf.
- 84.Willis JV, Kendall MJ, Flinn RM, Thornhill DP, Welling PG. The pharmacokinetics of diclofenac sodium following intravenous and oral administration. Eur J Clin Pharm. 1979;16:405–10. doi: 10.1007/BF00568201. [DOI] [PubMed] [Google Scholar]
- 85.Fowler PD, Shadforth MF, Crook PR, John VA. Plasma and synovial fluid concentrations of diclofenac sodium and its major hydroxylated metabolites during long-term treatment of rheumatoid arthritis. Eur J Clin Pharm. 1983;25:389–94. doi: 10.1007/BF01037953. [DOI] [PubMed] [Google Scholar]
- 86.Scott G, Rordorf C, Reynolds C, Kalbag J, Looby M, Milosavljev S, et al. Pharmacokinetics of lumiracoxib in plasma and synovial fluid. Clin Pharmacokinet. 2004;43:467–78. doi: 10.2165/00003088-200443070-00003. [DOI] [PubMed] [Google Scholar]
- 87.Claps G, Faouzi S, Quidville V, Chehade F, Shen S, Vagner S, et al. The multiple roles of LDH in cancer. Nat Rev Clin Oncol. 2022;19:749–62. doi: 10.1038/s41571-022-00686-2. [DOI] [PubMed] [Google Scholar]
- 88.Comandatore A, Franczak M, Smolenski RT, Morelli L, Peters GJ, Giovannetti E. Lactate Dehydrogenase and its clinical significance in pancreatic and thoracic cancers. Semin Cancer Biol. 2022;86:93–100. doi: 10.1016/j.semcancer.2022.09.001. [DOI] [PubMed] [Google Scholar]
- 89.Chirasani SR, Leukel P, Gottfried E, Hochrein J, Stadler K, Neumann B, et al. Diclofenac inhibits lactate formation and efficiently counteracts local immune suppression in a murine glioma model. Int J Cancer. 2013;132:843–53. doi: 10.1002/ijc.27712. [DOI] [PubMed] [Google Scholar]
- 90.Pantziarka P, Sukhatme V, Bouche G, Meheus L, Sukhatme VP. Repurposing drugs in oncology (ReDO)-diclofenac as an anti-cancer agent. Ecancermedicalscience. 2016;10:610. doi: 10.3332/ecancer.2016.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures, legends and Table
Data Availability Statement
All data generated or analysed during this study are included either in this article or in additional files, and the links to public datasets are reported in the manuscript.