Abstract
Background and Objective
Midazolam is the preferred clinical probe drug for assessing CYP3A activity. We have previously shown substantial intraindividual variability in midazolam absolute bioavailability and clearance in patients with obesity before and after weight loss induced by gastric bypass or a strict diet. The objective was to describe intraindividual variability in absolute bioavailability and clearance of midazolam in healthy individuals without obesity.
Methods
This study included 33 healthy volunteers [28 ± 8 years, 21% males, body mass index (BMI) 23 ± 2.5 kg/m2] subjected to four pharmacokinetic investigations over a 2-month period (weeks 0, 2, 4, and 8). Semi-simultaneous oral (0 h) and intravenous (2 h later) midazolam dosing was used to assess absolute bioavailability and clearance of midazolam.
Results
At baseline, mean absolute bioavailability and clearance were 46 ± 18% and 31 ± 10 L/h, respectively. The mean coefficient of variation (CV, %) for absolute bioavailability and clearance of midazolam was 26 ± 15% and 20 ± 10%, respectively. Approximately one-third had a CV > 30% for absolute bioavailability, while 13% had a CV > 30% for clearance.
Conclusions
On average, intraindividual variability in absolute bioavailability and clearance of midazolam was low to moderate; however, especially absolute bioavailability showed considerable variability in a relatively large proportion of the individuals.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40262-023-01257-z.
Key Points
| The literature indicates low intraindividual variability in clearance of midazolam, the standard CYP3A probe drug, while absolute bioavailability has not yet been determined. |
| This is the first study to describe intraindividual variability in absolute bioavailability of midazolam in healthy volunteers. |
| On average, there was a low to moderate intraindividual variability in systemic clearance and absolute bioavailability of midazolam in healthy individuals over a 2-month period. However, a relevant number displayed considerable intraindividual variability, which may limit the use of midazolam as a probe drug for CYP3A in certain studies. |
Introduction
To evaluate sources of intra- and interindividual variability in metabolic drug clearance, in vivo probe drug phenotyping is frequently used to assess the activity of a target cytochrome P450 (CYP) enzyme. These phenotyping studies are of high interest both from an industrial perspective, e.g., to quantify drug–drug interactions, and from a clinical perspective, e.g., to quantify the effect of disease states or individualize drug dosing. CYP3A is recognized as the single most important drug-metabolizing subfamily, accounting for around 40% of CYP-mediated drug metabolism, and is frequently investigated in phenotyping studies [1]. CYP3A is abundantly expressed in the liver and the small intestine, accounting for approximately 30% and 80% of the total CYP content in these organs, respectively [2–4]. Accordingly, presystemic metabolism by CYP3A is a crucial factor limiting the oral bioavailability of substrate drugs. Among the four isoenzymes in the CYP3A subfamily, which include CYP3A4, CYP3A5, CYP3A43, and CYP3A7 [5], CYP3A4 is the major contributor to CYP3A-mediated drug metabolism [5, 6]. However, the polymorphic CYP3A5 may contribute significantly in individuals carrying functional CYP3A5 alleles (*1) [7]. There is substantial interindividual variability in CYP3A phenotype, due to environmental factors, drug–drug interactions, and physiological factors, contributing significantly to variability in drug response [8, 9].
Several probe drugs have been suggested as CYP3A metrics, such as erythromycin, alfentanil, alprazolam, and triazolam [10]. Still, midazolam is recognized as the gold standard method for assessing CYP3A phenotype in vivo [11–13]. In addition to being a selective CYP3A substrate in vivo [14, 15], midazolam has a high sensitivity to CYP3A inhibitors and inducers [16–18], making it a preferred CYP3A probe drug in clinical trials and drug research. Previous studies have also indicated a low intraindividual (within-subject) variability in midazolam clearance following intravenous administration (coefficient of variation (CV) < 20%) in healthy volunteers [19, 20], which is a prerequisite for phenotyping metrics used to study changes in enzyme activity over time. However, in a recent study performed by our research group, midazolam was used as a probe drug to investigate changes in CYP3A activity in patients with severe obesity before and after weight loss induced by gastric bypass or a strict diet [21]. In this study, more than 60% of the patients had an intraindividual CV > 30% for midazolam absolute bioavailability during a 9-week period with three study investigations, and almost half of the patients also displayed a CV > 30% for clearance [21]. In view of this finding, we wanted to investigate if the large intraindividual variability was attributed to an actual change in CYP3A activity and not variability in midazolam pharmacokinetics in this specific patient population. To which extent midazolam actually reflects CYP3A activity in all patient populations is a controversial subject, given its highly variable extraction ratio [22–24]. Accordingly, changes in midazolam clearance depend not only on changes in CYP3A activity (intrinsic metabolic capacity) but also for example on alterations in hepatic blood flow and protein binding. High intraindividual variability in midazolam clearance challenges the use of midazolam to study changes in CYP3A phenotype over time. Also, to the best of our knowledge, within-subject variability in midazolam absolute bioavailability has not yet been investigated in healthy volunteers. Therefore, the objective of this study was to describe intraindividual variability in absolute bioavailability and clearance of midazolam in healthy individuals during a 2-month period.
Methods
Study Participants and Study Design
This study was an open, prospective, single-armed, and single-center study performed at Oslo University Hospital, Rikshospitalet, Norway. The participants were recruited from the Greater Oslo Region, and healthy individuals with no underlying disease, aged 18 years or above, and with body mass index (BMI) < 30 kg/m2 were eligible for inclusion in the study. Pregnant or nursing mothers were not eligible to participate in the study. Exclusion criteria also included conditions anticipated to interfere with gastrointestinal or hepatic drug disposition, and treatment with substances that may influence midazolam pharmacokinetics. Individuals carrying functional CYP3A5*1 alleles were excluded after the baseline investigation (week 0). The study was approved by the Regional Committee for Medical and Health Research Ethics (2021/255289/REK) and performed in accordance with the Declaration of Helsinki and Good Clinical Practice. Written informed consent was collected before inclusion.
Study Visits and Procedures
All individuals were subjected to four 6-h pharmacokinetic investigations: weeks 0, 2, 4, and 8 (± 7 days). On the study days, the participants fasted at least 2 h prior to the pharmacokinetic investigations except for water. Following baseline blood sampling, the participants first received 1.5 mg oral midazolam syrup (Midazolam Ratiopharm), followed by 1.0 mg intravenous midazolam (Midazolam B. Braun) 2 h later. The intravenous midazolam was administered via a separate peripheral venous catheter and flushed with 10 mL saline. A standardized meal was served within 30 min after the intravenous midazolam administration for all participants. Blood samples were collected from a peripheral venous catheter before (0 h) and 0.25, 0.5, 1, 1.5, 2, 2.25, 2.5, 3, 3.5, 4, 5, and 6 h after oral midazolam administration. The blood samples were drawn in 4 mL K2 EDTA vacutainers and centrifuged for 10 min at 4 °C (1800g). Plasma was separated into Cryovials and frozen at − 80 °C until analysis.
Clinical Chemistry Analyses
Fasting blood sample assessments of alanine aminotransferase (ALT), aspartate aminotransferase (AST), gamma-glutamyl transferase (GGT), total cholesterol, C-reactive protein (CRP), albumin, hemoglobin, and hemoglobin A1c (HbA1c) were performed at baseline as standard safety analyses. An analysis of CYP3A variant alleles was also performed in association with the first study investigation, and the following variant alleles were assessed: CYP3A5, the null allele *3 (rs776746), and CYP3A4, the reduced function allele *22 (rs35599367). Clinical chemistry analyses and genotyping were performed on fresh blood samples according to the hospital’s routine procedure.
Bioanalytical Assay
Midazolam plasma concentrations were determined by a validated ultrahigh-performance liquid chromatography tandem mass spectrometry (UHPLC–MS/MS) method at the Department of Pharmacy, University of Oslo, Norway as previously reported [25]. In brief, 100 μL of plasma was added to 200 μL of 95% acetonitrile and 5% methanol and deuterated internal standard to 5 ng/mL (midazolam-d6). The samples were vortex-mixed briefly, followed by 1 h of storage at − 20 °C. Samples were centrifuged for 10 min at 2272g at 4 °C, and the supernatant (50 μL) was added to 50 μL mobile phase A before injection (5 μL) into the UPLC–MS/MS system [Vanquish UPLC coupled to an Altis triple quadrupole mass spectrometer (Thermo-Fisher, Waltham, MA)]. The analytical column was an Aquity HSS T3 C18, 2.1 × 50 mm column (Waters, Milford, MA), with an Aquity HSS T3 VanGuard 2.1 mm × 5 mm Pre-column (Waters, Milford, MA). Mobile phase A consisting of 5% acetonitrile and 10 mM ammonium formate and mobile phase B consisting of 90% acetonitrile and 10% methanol were delivered in a gradient flow rate of 0.4 mL/min. Calibrators and quality control (QC) samples were prepared in blank plasma and analyzed in each analytical run. Ten calibrators in the range 0.25–100 ng/mL were applied. Lower limit of quantification (LLOQ) was 0.25 ng/mL, and upper limit of quantification (ULOQ) was 100 ng/mL. All study samples from each individual were analyzed in the same analytical run to minimize the bioanalytical variation. Within-series and between-series performance were assessed with resulting CV of < 4.3% and < 10.4%, and the mean accuracy ranged from 93 to 101% and 93 to 102 %, respectively.
Population Pharmacokinetic Modeling
Given the semi-simultaneous dosing of midazolam, a previously developed and validated population pharmacokinetic model of midazolam was used as Bayesian prior for predictions of absolute bioavailability and area under the curve (AUC) was used for determination of clearance in the present study [21]. The population pharmacokinetic modeling was performed using the nonparametric adaptive grid approach implemented in Pmetrics (version 1.5.2) for R (version 4.1.1) [26, 27]. This pharmacokinetic model is a catenary three-compartment model with absorption lag-time and first-order elimination from the central compartment and parameterized to determine individual absolute bioavailability. The model was developed using data with semi-simultaneous oral and intravenous dosing design with rich pharmacokinetic profiles (18 samples over 24 h) [21]. The predictive performance for determination of absolute bioavailability and AUC when applying the dosing design in the present study (2 h between oral and intravenous midazolam dosing and 6-h sampling) was investigated in the validation dataset of the previously developed model before the study start by comparing the results from the full 24-h data (reference) with the results obtained with the 6-h truncated data. The results are presented in Supplementary Table S1.
A total of 1737 midazolam concentrations corresponding to 134 unique pharmacokinetic profiles were available from a total of 38 individuals in the present study. The diagnostic plots of observed versus individual predicted concentration plots and residual plots were considered, with no major bias (Supplementary Fig. S1). Detailed results from the adapted population pharmacokinetic modeling, as well as a visual predictive check and an observed versus population predicted concentration plot, are presented in Supplementary Figs. S2 and S3 and Supplementary Tables S2 and S3.
Statistical Analysis and Calculations
No strict sample size calculation has been performed due to the descriptive nature of this study. However, on the basis of the literature on midazolam absolute bioavailability, we planned to include 30 individuals to ensure a good descriptive power of the intraindividual variability in midazolam pharmacokinetics. Intraindividual variability was determined by calculating CV% of absolute bioavailability and clearance of midazolam. Posterior individual parameter values obtained from the population pharmacokinetic model were used to describe midazolam pharmacokinetics. Absolute bioavailability, maximum plasma concentrations (Cmax) after oral midazolam administration, and time to reach Cmax (Tmax) were obtained directly from the model predictions, while clearance was calculated using dose and model derived AUC from zero to infinity. The function makeAUC() in the Pmetrics package for R was used to calculate the model-derived area under the concentration versus time curve from zero to infinity (AUC0-inf). Clearance was calculated according to Eq. (1).
| 1 |
Visual inspection of plots and the Shapiro–Wilk test were used to assess the normality of the data. A linear mixed-effects model was used to estimate changes over time and potential diurnal variation. The pharmacokinetic parameter was treated as dependent variable, and visit (weeks 0, 2, 4, and 8), time of day (morning or afternoon), and their interaction were treated as fixed effects. The unique patient identifier was used as a random effect (individual intercepts), to account for individual variability. Contrast analyses were performed for parameters of interest. The confidence intervals were adjusted using the Tukey method. Student’s t-test was used to compare means between two groups. Data from the first pharmacokinetic investigation (week 0) in individuals carrying functional CYP3A5*1 alleles were included in a separate cross-sectional analysis investigating the impact of genotype on absolute bioavailability and clearance of midazolam. All statistical analyses were performed using R, and a P value < 0.05 was considered statistically significant [26].
Results
Participant characteristics
Between 1 June 2021 and 5 October 2021, 45 healthy volunteers were screened for eligibility. Seven individuals were found to be ineligible due to obesity (n = 1), tachycardia (n = 1), systolic murmur (n = 1), Crohn’s disease (n = 1), and vasovagal syncope (n = 3), leaving 38 individuals that were included in the study. Five individuals were heterozygote carriers of the CYP3A5*1 allele and were excluded after the investigation at week 0. One individual withdrew after the study visit at week 2, and another did not show for the week 2 investigation. Accordingly, 31 individuals completed all four study visits. Baseline characteristics are presented in Table 1. The individuals (97% Caucasians) had a mean age of 28 ± 8 years and a mean BMI of 23 ± 2.5 kg/m2, and 79% were women.
Table 1.
Patient characteristics at baseline
| Healthy volunteers (n = 33) | |
|---|---|
| Sex (female/male) | 26/7 |
| Age (years) | 28 ± 8 |
| Ethnicity (Caucasian/other) | 32/1 |
| Body weight (kg) | 69 ± 9 |
| BMI (kg/m2) | 23 ± 2.5 |
| Hemoglobin (g/dL) | 13 ± 1.1 |
| AST (U/L)a | 22 ± 11 |
| ALT (U/L) | 21 ± 9.1 |
| Gamma-glutamyl transferase (U/L) | 17 ± 7.3 |
| HbA1c (mmol/mol) | 31 ± 2.2 |
| Total cholesterol (mmol/L) | 4.2 ± 0.81 |
| C-reactive protein (mg/L) | 1.5 ± 2.5 |
| Albumin (g/L) | 45 ± 3.0 |
| Genotype | |
| CYP3A4*1/*1 | 31 (94%) |
| CYP3A4*1/*22 | 2 (6%) |
Data are presented as mean ± SD or number (%)
ALT alanine aminotransferase, AST aspartate aminotransferase, BMI body mass index, HbA1c hemoglobin A1c
aAST is missing for two individuals at baseline due to hemolysis. C-reactive protein and albumin are missing for two individuals due to technical errors
Intraindividual variability
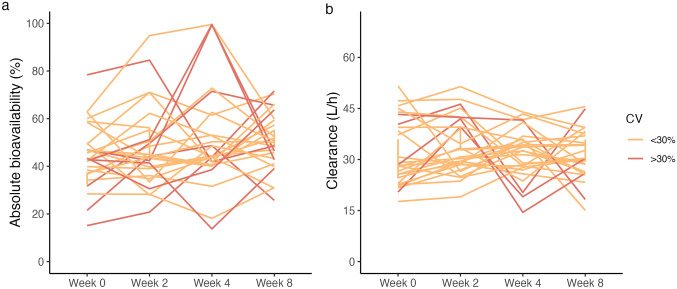
Mean CV% for midazolam absolute bioavailability and clearance was 26 ± 15% and 20 ± 10%, respectively. Approximately one-third (n = 10) had a CV > 30% for absolute bioavailability, ranging from 6 to 69% (Supplementary Table S4). The intraindividual variability was lower for clearance (Supplementary Table S5), with CV values ranging from 7 to 41%. Individual plots for the midazolam absolute bioavailability and clearance are shown in Fig. 1. The mixed model analysis showed no statistically significant differences in absolute bioavailability or clearance of midazolam between the four study investigations and no diurnal variation (all P > 0.05). Further, no correlation between absolute bioavailability and clearance was observed (R = 0.10, P = 0.26) (Supplementary Fig. S4). Pharmacokinetic data from the four study visits are presented in Table 2.
Fig. 1.
Intraindividual variability in midazolam pharmacokinetics. Individual plots for a absolute bioavailability and b clearance of midazolam
Table 2.
Midazolam pharmacokinetic variables and parameters at the four study visits
| Week 0 (n = 33) | Week 2 (n = 32) | Week 4 (n = 32) | Week 8 (n = 32) | |
|---|---|---|---|---|
| Absolute bioavailability (%) | 46 ± 18 | 48 ± 19 | 52 ± 21 | 51 ± 14 |
| Clearance (L/h) | 31 ± 9.9 | 33 ± 8.9 | 32 ± 7.9 | 31 ± 7.7 |
| Cmax, oral (µg/L) | 6.3 ± 2.4 | 6.5 ± 2.2 | 6.9 ± 2.3 | 7.2 ± 2.9 |
| Tmax, oral (h) | 0.97 ± 0.47 | 0.90 ± 0.32 | 1.1 ± 0.46 | 0.93 ± 0.32 |
| AUC (µg h/L) | 62 ± 23 | 57 ± 20 | 58 ± 15 | 61 ± 23 |
Data are presented as mean ± SD
AUC area under the curve, Cmax maximum plasma concentrations, Tmax time to reach Cmax
Interindividual variability
There was a seven- and four-fold difference in absolute bioavailability and clearance of midazolam between individuals, respectively. Females had lower absolute bioavailability compared with males: − 21% (95% CI − 33 to − 9), while there was no difference in clearance [0.02 L/h (− 4.3 to 4.3)]. Individuals carrying one functional CYP3A5*1 allele had similar absolute bioavailability [13% (95% CI − 8 to 31)] and clearance [− 0.64 L/h (95% CI − 6 to 4)] as individuals homozygous for the nonfunctional CYP3A5*3 allele (week 0).
Discussion
This is the first study to describe intraindividual variability in absolute bioavailability of midazolam in healthy volunteers. The main findings were that intraindividual variability in absolute bioavailability was moderate, and as expected, the variability was more extensive than for clearance. Assuming that these parameters actually reflect intestinal and hepatic CYP3A activity, the findings suggest that CYP3A activity is relatively stable over time in most healthy individuals. However, a considerable intraindividual variability was observed in a relevant proportion of the healthy individuals. Given that a low intraindividual variability is a prerequisite for phenotyping metrics used to study changes in the activity of a target enzyme, the findings in the present study should be taken into consideration in phenotyping studies using midazolam to study changes in CYP3A activity over time.
The wider range in intraindividual variability for absolute bioavailability may be attributed to numerous factors influencing the absorption and presystemic metabolism of midazolam, such as gastric and intestinal transit time, gastrointestinal motility, and splanchnic blood flow [28, 29]. Some individuals displayed a large intraindividual variability, and a CV above 40% was observed in three study participants, and one individual had a CV of 69%. We could not identify any subject-dependent factors (such as gender) or other covariates that could explain the high variability in these individuals. In addition, we observed no correlation between absolute bioavailability and clearance of midazolam, i.e., low clearance values did not necessarily correspond to high absolute bioavailability values and vice versa. Thus, it appears that intestinal extraction is of greater importance for the observed variability. Similar to our findings, a recent study reported significant intraindividual variability in midazolam exposure in several of the study participants and low intraindividual variability in others (mean CV was 11%) [30]. Furthermore, previous studies have reported a low mean intraindividual variability in midazolam clearance [19, 20, 31], which is in agreement with the results in this study. Kharasch et al. reported a similar mean CV of midazolam clearance (19 ± 12%) over 21 days as in the present study [20]. Kashuba et al. also investigated the CV of midazolam clearance six times over a 3-month period. Median CV in the 20 individuals (10 males) was 10%, ranging from 5 to 22% [19].
The findings in the present study are in contrast to the large intraindividual variability that we observed in patients with obesity before and after weight loss induced by gastric bypass or a strict diet [21]. This discrepancy may be explained by major physiological changes occurring in the patients with obesity following the intervention. In view of the results in the present study, we hypothesize that the large intraindividual variability observed in the patients with obesity previously was attributed to within-subject variability in midazolam absolute bioavailability and clearance rather than actual intraindividual variability in CYP3A activity. Thus, it seems that other processes such as alterations in hepatic blood flow or protein binding may influence midazolam pharmacokinetics more than previously assumed, particularly in specific patient populations. Given that our study population was a homogeneous group of young Caucasians, the intraindividual variability may be different in other more heterogeneous populations.
The major strengths of this study include that we could determine both absolute bioavailability and clearance of midazolam in more than 30 individuals, and that over 95% of the participants completed all four study visits. Also, the bioanalytical within-series CV was below 5%, suggesting that the bioanalytical variability should not have contributed significantly to the observed intraindividual variability in this study. Limitations of the present study include the semi-simultaneous administration of midazolam, with a short time period between oral and intravenous dosing. However, we used a previously validated population pharmacokinetic model [21] that was developed on a large dataset in which there were 4 h between the oral and intravenous dose and blood samples for determination of midazolam plasma concentrations collected over 24 h. Using this dataset as a Bayesian prior, the absolute bioavailability and AUC of midazolam were accurately predicted. Food intake has been reported to increase clearance and reduce AUC of intravenously administered drugs with a moderate to high hepatic extraction ratio due to an increase in hepatic blood flow [32]. As such, we cannot rule out that the standardized meal that was served in close approximation to the intravenous midazolam dose to some degree has influenced the results. Also, external factors, such as food intake prior to the 2-h fasting and physical activity, were not controlled. To minimize any potential influence by circadian variations, we aimed for the individuals to have their study visit at the same time of the day each time (~ 2:00 p.m.), but 15 individuals had one or two study investigations that started in the morning (~ 9:00 a.m.). While the diurnal variation in clearance appears to be small [33, 34], oral bioavailability of midazolam has been shown to display 24-h variation, with a relative difference of ~ 30% between peak and trough levels [33]. However, time of day had no impact on absolute bioavailability and clearance of midazolam in the mixed effects model analysis. Thus, we do not believe that diurnal variation has influenced the results significantly. Finally, we did not measure albumin, ALT, and CRP over time. However, as the participants were healthy individuals, we do not suspect this influenced the results.
In conclusion, intraindividual variability in midazolam absolute bioavailability and clearance was low to moderate on average; however, a relevant proportion of the individuals displayed high intraindividual variability. Altogether, the results indicate minor variability in CYP3A activity over time in healthy individuals. The considerable intraindividual variability observed in some of the individuals limits the use of midazolam as a sole metric to study changes in CYP3A activity over time.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgments
The authors would like to thank the participants in the study. We also thank Kristin Sandnes, Cristell Melby Magnussen, May Ellen Lauritsen, Merethe Larsen Bjerke, and Edmar Agustin at the Clinical Research Unit, Department of Pharmacology, Oslo University Hospital.
Funding
Open access funding provided by University of Oslo (incl Oslo University Hospital).
Declarations
Funding
Internal funds at the University of Oslo and Oslo University Hospital.
Conflict of Interest
The authors have no conflict of interest to declare.
Availability of Data, Code, and Material
Data sharing requires Ethical Committee approval according to Norwegian regulations. Contact the corresponding author with requests.
Ethics Approval
The study was approved by the Regional Committee for Medical and Health Research Ethics (255289) and complied with the Declaration of Helsinki.
Consent to Participate
Informed consent was obtained from all individual participants included in the study.
Consent for Publication
Not applicable.
Author Contributions
KEK, IR, and AÅ designed the study. KEK, OMD, NH, ES, HKZ, and IR performed the research. KEK, OMD, and IR analyzed the data. KEK wrote the manuscript. All authors contributed to critically reviewing the manuscript and gave their final approval for submission.
References
- 1.Saravanakumar A, et al. Physicochemical properties, biotransformation, and transport pathways of established and newly approved medications: a systematic review of the top 200 most prescribed drugs vs. the FDA-approved drugs between 2005 and 2016. Clin Pharmacokinet. 2019;58(10):1281–1294. doi: 10.1007/s40262-019-00750-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paine MF, et al. The human intestinal cytochrome P450 "pie". Drug Metab Dispos. 2006;34(5):880–886. doi: 10.1124/dmd.105.008672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Achour B, Barber J, Rostami-Hodjegan A. Expression of hepatic drug-metabolizing cytochrome p450 enzymes and their intercorrelations: a meta-analysis. Drug Metab Dispos. 2014;42(8):1349–1356. doi: 10.1124/dmd.114.058834. [DOI] [PubMed] [Google Scholar]
- 4.Grangeon A, et al. Determination of CYP450 expression levels in the human small intestine by mass spectrometry-based targeted proteomics. Int J Mol Sci. 2021;22(23):12791. doi: 10.3390/ijms222312791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daly AK. Significance of the minor cytochrome P450 3A isoforms. Clin Pharmacokinet. 2006;45(1):13–31. doi: 10.2165/00003088-200645010-00002. [DOI] [PubMed] [Google Scholar]
- 6.Rendic S, Guengerich FP. Survey of human oxidoreductases and cytochrome p450 enzymes involved in the metabolism of xenobiotic and natural chemicals. Chem Res Toxicol. 2015;28(1):38–42. doi: 10.1021/tx500444e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuehl P, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27(4):383–391. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 8.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Lenoir C, et al. Influence of inflammation on cytochromes p450 activity in adults: a systematic review of the literature. Front Pharmacol. 2021;12:733935. doi: 10.3389/fphar.2021.733935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann N, Haefeli WE, Mikus G. CYP3A activity: towards dose adaptation to the individual. Expert Opin Drug Metab Toxicol. 2016;12(5):479–497. doi: 10.1517/17425255.2016.1163337. [DOI] [PubMed] [Google Scholar]
- 11.Fuhr U, Jetter A, Kirchheiner J. Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the "cocktail" approach. Clin Pharmacol Ther. 2007;81(2):270–283. doi: 10.1038/sj.clpt.6100050. [DOI] [PubMed] [Google Scholar]
- 12.Chung E, et al. Comparison of midazolam and simvastatin as cytochrome P450 3A probes. Clin Pharmacol Ther. 2006;79(4):350–361. doi: 10.1016/j.clpt.2005.11.016. [DOI] [PubMed] [Google Scholar]
- 13.Keller GA, et al. In vivo phenotyping methods: cytochrome p450 probes with emphasis on the cocktail approach. Curr Pharm Des. 2017;23(14):2035–2049. doi: 10.2174/1381612823666170207100724. [DOI] [PubMed] [Google Scholar]
- 14.de Jonge H, et al. Impact of CYP3A5 genotype on tacrolimus versus midazolam clearance in renal transplant recipients: new insights in CYP3A5-mediated drug metabolism. Pharmacogenomics. 2013;14(12):1467–1480. doi: 10.2217/pgs.13.133. [DOI] [PubMed] [Google Scholar]
- 15.Yu KS, et al. Effect of the CYP3A5 genotype on the pharmacokinetics of intravenous midazolam during inhibited and induced metabolic states. Clin Pharmacol Ther. 2004;76(2):104–112. doi: 10.1016/j.clpt.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Backman JT, Olkkola KT, Neuvonen PJ. Rifampin drastically reduces plasma concentrations and effects of oral midazolam. Clin Pharmacol Ther. 1996;59(1):7–13. doi: 10.1016/S0009-9236(96)90018-1. [DOI] [PubMed] [Google Scholar]
- 17.Krishna G, et al. Effects of oral posaconazole on the pharmacokinetic properties of oral and intravenous midazolam: a phase I, randomized, open-label, crossover study in healthy volunteers. Clin Ther. 2009;31(2):286–298. doi: 10.1016/j.clinthera.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 18.Olkkola KT, Backman JT, Neuvonen PJ. Midazolam should be avoided in patients receiving the systemic antimycotics ketoconazole or itraconazole. Clin Pharmacol Ther. 1994;55(5):481–485. doi: 10.1038/clpt.1994.60. [DOI] [PubMed] [Google Scholar]
- 19.Kashuba AD, et al. Quantification of 3-month intraindividual variability and the influence of sex and menstrual cycle phase on CYP3A activity as measured by phenotyping with intravenous midazolam. Clin Pharmacol Ther. 1998;64(3):269–277. doi: 10.1016/S0009-9236(98)90175-8. [DOI] [PubMed] [Google Scholar]
- 20.Kharasch ED, et al. Intraindividual variability in male hepatic CYP3A4 activity assessed by alfentanil and midazolam clearance. J Clin Pharmacol. 1999;39(7):664–669. doi: 10.1177/00912709922008290. [DOI] [PubMed] [Google Scholar]
- 21.Kvitne KE, et al. Short- and long-term effects of body weight loss following calorie restriction and gastric bypass on CYP3A-activity—a non-randomized three-armed controlled trial. Clin Transl Sci. 2022;15(1):221–233. doi: 10.1111/cts.13142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers JF, et al. An evaluation of the suitability of intravenous midazolam as an in vivo marker for hepatic cytochrome P4503A activity. Clin Pharmacol Ther. 2003;73(3):153–158. doi: 10.1067/mcp.2003.23. [DOI] [PubMed] [Google Scholar]
- 23.Kirwan C, MacPhee I, Philips B. Using drug probes to monitor hepatic drug metabolism in critically ill patients: midazolam, a flawed but useful tool for clinical investigation of CYP3A activity? Expert Opin Drug Metab Toxicol. 2010;6(6):761–771. doi: 10.1517/17425255.2010.482929. [DOI] [PubMed] [Google Scholar]
- 24.Klotz U, Ziegler G. Physiologic and temporal variation in hepatic elimination of midazolam. Clin Pharmacol Ther. 1982;32(1):107–112. doi: 10.1038/clpt.1982.133. [DOI] [PubMed] [Google Scholar]
- 25.Egeland EJ, et al. Chronic inhibition of CYP3A is temporarily reduced by each hemodialysis session in patients with end-stage renal disease. Clin Pharmacol Ther. 2020;108(4):866–873. doi: 10.1002/cpt.1875. [DOI] [PubMed] [Google Scholar]
- 26.R Foundation for Statistical Computing. R: a language and environment for statistical computing 2018, Vienna, Austria.
- 27.Neely M, et al. Accurate Detection of outliers and subpopulations with Pmetrics a nonparametric and parametric pharmacometric modeling and simulation packagefor R. Thera Drug Monit. 2012;34:467–476. doi: 10.1097/FTD.0b013e31825c4ba6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abuhelwa AY, et al. Food, gastrointestinal pH, and models of oral drug absorption. Eur J Pharm Biopharm. 2017;112:234–248. doi: 10.1016/j.ejpb.2016.11.034. [DOI] [PubMed] [Google Scholar]
- 29.Jamei M, et al. Population-based mechanistic prediction of oral drug absorption. AAPS J. 2009;11(2):225–237. doi: 10.1208/s12248-009-9099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthaei J, et al. Inherited and acquired determinants of hepatic CYP3A activity in humans. Front Genet. 2020;11:944. doi: 10.3389/fgene.2020.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kharasch ED, et al. Menstrual cycle variability in midazolam pharmacokinetics. J Clin Pharmacol. 1999;39(3):275–280. doi: 10.1177/009127009903900311. [DOI] [PubMed] [Google Scholar]
- 32.Zou P. Does food affect the pharmacokinetics of non-orally delivered drugs? A review of currently available evidence. AAPS J. 2022;24(3):59. doi: 10.1208/s12248-022-00714-0. [DOI] [PubMed] [Google Scholar]
- 33.van Rongen A, et al. Population pharmacokinetic model characterizing 24-hour variation in the pharmacokinetics of oral and intravenous midazolam in healthy volunteers. CPT Pharmacomet Syst Pharmacol. 2015;4(8):454–464. doi: 10.1002/psp4.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomalik-Scharte D, et al. Population pharmacokinetic analysis of circadian rhythms in hepatic CYP3A activity using midazolam. J Clin Pharmacol. 2014;54(10):1162–1169. doi: 10.1002/jcph.318. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.