Abstract
Background
Portal vein tumour thrombus (PVTT) is the main pathway of HCC intrahepatic metastasis and is responsible for the poor prognosis of patients with HCC. However, the molecular mechanisms underlying PVTT vascular metastases have not been fully elucidated.
Methods
NDRG1 expression was assessed by immunohistochemistry and immunoblotting in clinical specimens obtained from curative surgery. The functional relevance of NDRG1 was evaluated using sphere formation and animal models of tumorigenicity and metastasis. The relationship between NDRG1 and EpCAM was explored using molecular biological techniques.
Results
NDRG1 protein was upregulated in HCC samples compared to non-tumorous tissues. Furthermore, NDRG1 expression was enhanced in the PVTT samples. Our functional study showed that NDRG1 was required for the self-renewal of tumour-initiating/cancer stem cells (CSCs). In addition, NDRG1 knockdown inhibited the proliferation and migration of PVTT-1 cells in vitro and in vivo. NDRG1 was found to stabilise the functional tumour-initiating cell marker EpCAM through protein–protein interactions and inhibition of EpCAM ubiquitination.
Conclusion
Our findings suggest that NDRG1 enhances CSCs expansion, PVTT formation and growth capability through the regulation of EpCAM stability. NDRG1 may be a promising target for the treatment of patients with HCC and PVTT.
Subject terms: Cancer stem cells, Ubiquitylation
Background
Hepatocellular carcinoma (HCC) is the sixth most prevalent cancer type worldwide, with an incidence of 800,000 new cases per year [1, 2]. Patients are normally diagnosed at a late stage with local and distant metastasis, resulting in a 30% 5-year survival rate [3]. Portal vein tumour thrombus (PVTT), direct portal invasion and pro-coagulant cytokine production by HCC cells, are responsible for the poor prognosis [4, 5]. The average survival time of PVTT patients is 4 months, which is significantly shorter than that of patients without PVTT [4]. Mechanistically, for late-stage HCC development, it was found that the TGF-β–miR-34a-CCL22 axis mediated Treg recruitment by HCC cells, to form an immunosuppressant microenvironment that promotes the colonisation of disseminated HCC cells in the portal vein system [4]. However, the molecular mechanisms underlying PVTT vascular metastasis have not been fully explored.
Tumour-initiating cells (TICs) /cancer stem cells (CSCs) are a subpopulation of biologically distinct cells with an elevated capacity to initiate tumour, resist conventional treatment and metastasis to distant organs [6–8]. Adopting the notion of lineage differentiation from normal cell development, several surface proteins such as CD133, CD44 and EpCAM, have been identified as TICs markers [9–11]. Among these, EpCAM was found to serve as a functional marker that maintains the tumour-initiating cell phenotype. Mechanistically, it has been suggested that the EpCAM intracellular domain (EpICD) is released together with other co-factors, including β-catenin, and transported into the nucleus, which transcriptionally activates key TICs genes, such as c-myc and Sox2 [12, 13]. It is unknown how this complex is modulated in the context of primary HCC. Although the literature reports that EpCAM is highly expressed in HCC [14], it is unclear how EpCAM is regulated during PVTT formation.
N-Myc downstream-regulated 1 (NDRG1) is a stress-responsive protein involved in hormone response, cell growth, migration, lipid metabolism and cancer progression [15–20]. The function of NDRG1 in cancer development is complicated and largely context-dependent. For example, in glioblastoma and colorectal cancer, NDRG1 acts as a major tumour metastasis suppressor while promotes tumour progression in HCC [19–27]. Although several studies have shown that NDRG1 promotes HCC cell proliferation, it remains unclear whether NDRG1 regulates stem-like characteristics and PVTT-related metastasis.
In this study, we screened the protein expression profiles of NDRG1, NDRG2, NDRG3 and NDRG4 in HCC tumour, and found that NDRG1 protein was the only highly expressed in HCC tumour samples. Further analysis revealed significantly enhanced expression in the PVTT samples. Functional studies have shown that NDRG1 is crucial for maintaining HCC TICs characteristics and in vivo tumour initiation and progression. Importantly, we found that one of the key TICs functional markers EpCAM, was stabilised NDRG1 via de-ubiquitination. Taken together, our study provides a fundamental mechanism of NDRG1 in stabilising EpCAM protein and promoting TICs activity in HCC samples.
Methods
Cell culture
Three HCC cell lines (PLC/PRF/5, Huh7 and PVTT-1), murine cell line of hepatoma Hepa1-6 and human embryonic kidney cell HEK-293T were used in this study. PLC/PRF/5, Hepa1-6, and HEK-293T cells were obtained from the American type culture collection (ATCC), whereas Huh7 was acquired from the Japanese Collection of Research Bioresources (JCRB). PVTT-1 was derived from the tissues of patients with PVTT, which has been described previously [28]. All above cells were cultured in DMEM supplemented with 10% foetal bovine serum (FBS) and 1% penicillin–streptomycin at 37 °C in a humidified atmosphere of 5% CO2. Three human cells (PLC/PRF/5, Huh7 and HEK-293T) were recently authenticated using Short Tandem Repeat (STR) profiling. PVTT-1 cell line was verified by western blotting (alpha-fetoprotein and EpCAM for PVTT-1). All cell lines were not contaminated by mycoplasma.
Clinical samples
HCC specimens and corresponding para-tumour tissues were obtained from 48 patients who underwent hepatectomy at the Peking University People’s Hospital. Seven PVTT specimens and the corresponding primary HCC and adjacent non-tumour tissues from seven patients were also obtained from Peking University People’s Hospital. One tissue microarrays were purchased from Shanghai Superbiotek Pharmaceutical Technology Corporation (Cat No.LVC-1201), consisting of 24 cases of PVTT (Supplementary Table S1). The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Research Ethics Committee of the Peking University People’s Hospital. Written informed consent was obtained from all the patients.
Comparison of mRNA levels from public databases
The GEO dataset (GSE14520) was used to evaluate the correlation between NDRG1 family and overall survival (OS) of patients with liver cancer. The cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/) dataset was used to evaluate the mRNA levels of NDRG1 in different TNM stages.
RNA isolation, cDNA synthesis and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from cells or tissues using the TRIzol reagent (Invitrogen, USA). Next, the extracted RNA was reverse-transcribed to single-stranded cDNA using a PrimeScript™ RT-PCR Kit (Takara, Japan). Then, qRT-PCR amplification was performed on the ViiA7 Real-time PCR System (Applied Biosystems, USA) using SYBR Green Real-time PCR Master Mix (Takara, Japan). Relative quantification of mRNA expression was performed using the 2−∆∆CT method, and β-actin was used as an internal control. The gene-specific primers used to detect mRNA amplification are listed in Supplementary Table S2.
Immunohistochemistry for clinical samples
Immunohistochemistry (IHC) was performed on paraffin-embedded slides or tissue microarray (TMA). First, the slides or TMA were de-paraffinized and rehydrated, followed by antigen retrieval using EDTA solution pH 9.0 in a steamer. The endogenous peroxidase activity was blocked by using 3% H2O2. The slides or TMA were incubated with anti-NDRG1 (NBP2-66969, Novus, USA) with 1:600 dilution or anti-EpCAM (66316-1-1 g, Proteintec, China) with 1:200 dilution at 4 °C overnight. The next day, the slides or TMA were washed with PBS and incubated with a secondary antibody for 1 h. The slides or TMA were lightly counterstained with DAB and haematoxylin.
The immunohistochemical results were evaluated by two independent observers who were blinded to the clinical data. The staining intensity of NDRG1 or EpCAM was scored on a scale of 0 to 3 as follows: 0 (negative, −), 1 (weak, +), 2 (medium, ++), or 3 (strong, +++). The percentage of positive cells was assessed on a scale of 0 (negative), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%). The overall scores were calculated using the formula: IHC score = immunostaining intensity × percentage of positive cells.
Immunofluorescence for clinical samples
PVTT tissue was fixed with paraformaldehyde, embedded in paraffin, and sectioned. The PVTT sections were baked, de-paraffinized and rehydrated. Antigens were repaired using antigen retrieval buffer (pH 9.0) in a microwave oven. The PVTT specimen was incubated with anti-NDRG1 (NBP2-66969, Novus, USA) as well as anti-EpCAM (66316-1-Ig, Proteintec, China) antibodies at 4 °C overnight. After washing, the PVTT slide was incubated with a goat anti-rabbit IgG H&L (Cy3®) (Ab6939, Abcam, USA) and a goat anti-mouse IgG H&L (Alexa Fluor® 488) (Ab150113, Abcam, USA). DAPI was used to counterstain nuclei for 10 min. Fluorescence images were obtained using a confocal laser microscope.
Western blotting
Proteins were extracted from the cells using RIPA lysis buffer supplemented with a protease inhibitor. Proteins were fractionated by SDS-PAGE and transferred to nitrocellulose membranes. These membranes were blocked with 5% non-fat dry milk or BSA and then incubated with specific primary antibodies overnight at 4 °C. The next day, the membranes were washed with TBST five times and incubated with an HRP-conjugated secondary antibody. Protein expression was visualised using a chemiluminescence imaging system. The following primary antibodies were used: NDRG1 (HPA006881, Sigma-Aldrich, USA), EpCAM (2929S, Cell Signalling Technology, USA), γ-tubulin (5886S, Cell Signalling Technology, USA), GAPDH (IMA1052L, Imagentx, China).
Plasmids and lentiviral package
The expression plasmids for NDRG1 overexpression experiments were sub-cloned into a PCDH vector. Human NDRG1 short hairpin RNAs (shRNAs) or mouse NDRG1 shRNAs were sub-cloned into PLKO.1 plasmids, which were purchased from Sigma-Aldrich (USA). Briefly, the lentiviral and packaging plasmids pMD2.G and psPAX2 were transfected into HEK293-T cells. The lentiviral supernatants were collected. NDRG1 overexpression and knockdown cell lines were obtained by antibiotic screening.
Transient transfection
Human NDRG1 siRNA was used for transient transfections. Human NDRG1 siRNA sequences: NDRG1 siRNA1:5’-CGUGAACCCUUGUGCGGAA-3’; NDRG1 siRNA2, 5’-GCAUUAUUGGCAUGGGAAC-3’.
Cell viability and assay
First, the cells were seeded into each well of a 96-well plate and incubated in complete medium. On each day, cell viability assay was performed using CCK-8, according to the manufacturer’s protocol. In total, 10 µl of CCK-8 solution was added to each well containing 100 µL of DMEM. Then, the plates were incubated at 37 °C for 1.5 h, and absorbance was detected at 450 nm.
Colony formation assay
One thousand cells were plated in six-well plates and cultured for 10–14 days. The colonies were fixed with 4% paraformaldehyde and stained with 1% crystal violet. After the cells were washed with PBS three times, the number of colonies was counted.
Sphere formation assay
Cells were plated in ultralow attachment 6-well or 24-well plates in serum-free DMEM/F12 medium, supplemented with 10 ng/ml of epidermal growth factor (EGF, Life Technologies), 5 ng/ml of basic fibroblast growth factor (bFGF, Life Technologies), B27 (Life Technologies), N2 (Life Technologies), 100 U/ml penicillin and 100 ng/ml streptomycin. Fresh sphere medium was added to ultralow attachment plates every 3 days. After the cells were cultured for 1–2 weeks, the primary spheres were formed. The primary spheres were passaged, the secondary spheres were formed after several days of culture. Bright-field images of spheres were captured and the number of spheres was evaluated.
Transwell migrant assay
Transwell inserts were placed in 24-well cell culture plates (Corning). PLC cells transfected with PCDH-NDRG1 or PCDH plasmids were plated in the top chamber, and the lower chamber was filled with 650 µL of complete medium. After 24 h of incubation, the cells that migrated to the underside of the filters were fixed, and stained with 0.1% crystal violet. Data are presented as average cell count in five random fields at 20x magnification. Similarly, the migration properties of PVTT-1 cells transfected with PLKO.1-mock and PLKO.1-shNDRG1 plasmids were also measured.
Protein half-life assay
PLKO.1-shNDRG1 or PLKO.1-mock stably transfected PVTT-1 cells were treated with 50 μg/ml cycloheximide (CHX) for 0, 3, 6 and 9 h separately. The protein was isolated from cells and western blotting was performed to analyse the half-life of the EpCAM protein. The relative levels EpCAM protein at different time points were calculated using ImageJ2x software and γ-tubulin was used as an endogenous control.
Ubiquitination assay
To detect the EpCAM ubiquitination in 293 T cells, cells were transfected with Flag-EpCAM, MYC–Ub, or HA-NDRG1 for 48 h. Next, cells were treated with 10 μM MG132 for 6 h, then the cells were lysed in SDS lysis buffer (0.1% SDS, 150 mM NaCl, 10 mM Tris-HCl (pH = 7.5), 0.5% NP-40) with protease inhibitor mixture, a phosphatase inhibitor, and 10 μM N-ethylmaleimide. The lysates were immunoprecipitated with anti-flag antibody overnight at 4 °C, followed by incubation with Protein A/G agarose beads(MCE) for 2 h at 4 °C. Then the ubiquitination of EpCAM protein was detected by western blotting.
Animal studies
For the xenograft HCC model, the nude mice were randomly divided into three groups: PVTT-1~PLKO.1 (n = 5), PVTT-1 ~ shNDRG1-1 (n = 5), and PVTT-1 ~ shNDRG1-2 (n = 5). It is reported that the incidence of HCC in men is higher than that in women, so male mice were used for the in vivo study. In total, 2.5 × 106 cells were resuspended in 200 μl PBS and injected subcutaneously into the right flank of 6-week-old male Balb/c nude mice. Tumour diameter was measured using an electronic calliper. Tumour volumes were calculated using the formula: V = 1/2 long diameter × (short diameter)2. The tumourigenic abilities of Hepa1-6 cells transfected with PLKO.1 (n = 5), PLKO.1-shNDRG1-1 (n = 5), and PLKO.1-shNDRG1-2 (n = 5) plasmids were also measured.
The in vivo limiting dilution assay (LDA) were used to evaluate the relationship between NDRG1 expression and TICs capabilities. Hepa1-6~PLKO.1 and NDRG1-depletion cells (103, 104, 105 and 106) were injected into BALB/c nude mice, respectively. About 25 days later, tumour formation was counted, followed by the calculation of the stem cell frequency [29, 30].
For spontaneous metastasis assays, luciferase-expressing PVTT-1-shNDRG1 and PVTT-1-PLKO.1-mock cells were injected into the spleen of nude mice. Metastatic formation was monitored by the appearance of luciferase activity using the imaging apparatus of IVIS systems.
The animal studies were approved by the Institutional Animal Welfare and Ethics Committee of Peking University People’s Hospital. No statistical method was used to determine the sample size. The researchers recording tumour growth were blinded to the group allocation.
Statistical analysis
Data are shown as mean ± SD from at least three independent experiments. GraphPad Prism5 software was used for statistical analysis. Student’s t test was used to evaluate the statistical significance between the two groups. Statistical significance was set at P < 0.05, **P < 0.01, and ***P < 0.001.
Results
NDRG1 overexpression is prognostic of poor HCC outcome
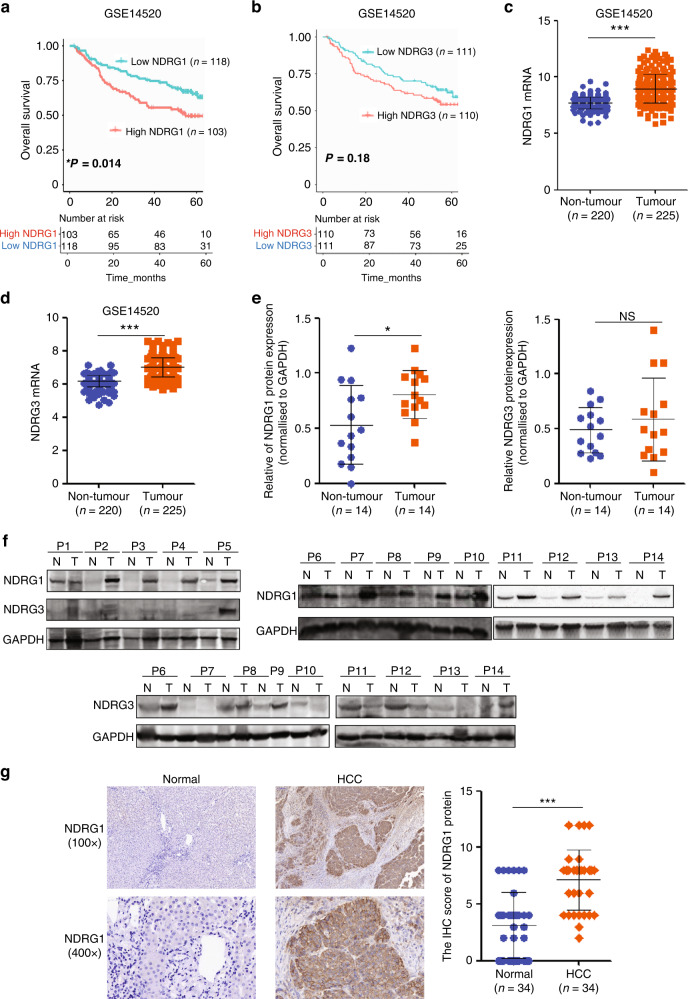
NDRG1 belongs to the NDRG family with three other members sharing a conserved α/β hydrolase-fold motif. Despite the structural similarity among family members, detailed functional overlaps have yet been fully elucidated. To investigate the prognostic significance of NDRG family expression in patients with HCC, data derived from a GEO cohort (GSE14520) was analysed by Kaplan–Meier methods. Patients with high NDRG1 expression were significantly correlated with poor Overall Survival (OS) (*P = 0.014) (Fig. 1a), whereas high NDRG2 expression indicated good survival in HCC (*P = 0.033) (Supplementary Fig. 1a). NDRG3 and NDRG4 mRNA levels were not significantly correlated with OS (Fig. 1b and Supplementary Fig. 1b). Data from TCGA also showed high NDRG1 expression indicated bad survival in HCC (Supplementary Fig. 2a).
Fig. 1. NDRG1 expression is upregulated in HCC and indicates the unfavourable prognosis of patients with HCC.
a Kaplan–Meier analysis showing the overall survival of 221 patients with HCC with high (red; n = 103) or low NDRG1 expression (blue; n = 118) from the GEO dataset (GSE14520) (*P = 0.014). b The overall survival rates of 221 patients with HCC were compared between the NDRG3 high (n = 110) and NDRG3low (n = 111) groups (P = 0.18). c, d The mRNA levels of NDRG1 and NDRG3 in HCC tissues (n = 220) compared with non-tumorous liver tissues (n = 225) from the dataset GSE14520. e, f Western blot analysis for NDRG1 or NDRG3 expression in HCC tissues (T; n = 14) compared to non-tumorous tissues (N; n = 14). g Representative pictures and quantitative evaluation of NDRG1 staining intensity in primary HCC (n = 34) and matched non-tumorous liver tissues (n = 34).
The mRNA levels of NDRG1, NDRG3, and NDRG4 were higher in HCC malignant liver samples than in non-tumorous samples (Fig. 1c, d and Supplementary Figs. 1d and 2b), whereas NDRG2 mRNA was decreased in cancerous tissues (Supplementary Fig. 1c). Proteins are the executors of cell function; therefore, NDRG family protein levels were detected by western blotting. Intriguingly, only NDRG1 was strongly upregulated in HCC samples (Fig. 1e, f). Compared to non-tumorous tissues, the protein profiles of NDRG2, NDRG3, and NDRG4 did not change significantly (Fig. 1e, f and Supplementary Fig. 1e, f).
Among them, NDRG1 is interesting because of its complicated and highly context-dependent functions in cancer development, including its implications for stem cell maintenance [15, 19–27]. We then confirmed the induction of NDRG1 in HCC IHC samples compared with that in their adjacent tissues (Fig. 1g). Taken together, these data strongly support that NDRG1 expression is specifically elevated in HCC.
NDRG1 expression is high in PVTT compared to primary HCC and normal liver tissues
During PVTT progression, HCC tumour cells colonised within the portal veins to initial tumour thrombosis. Based on this finding, we further explored the expression of NDRG1 in IHC samples from patients with PVTT. NDRG1 protein levels were found to be induced in primary HCC samples and dramatically elevated in paired PVTT samples (Fig. 2a). Western blot data confirmed the serial expression enhancement of NDRG1 in paired normal adjacent tissues, HCC primary tumour and PVTT tissues (Fig. 2b). Interestingly, we found that NDRG1 was not only highly expressed in PVTT samples but also significantly enhanced in primary tumours with thrombosis when compared to the tumours without vascular invasion (Fig. 2c). NDRG1 expression was positively correlated with advances in TNM staging (Fig. 2d, e). In summary, the correlation between NDRG1 expression and PVTT indicates that NDRG1 might affect the late-stage HCC tumour progression.
Fig. 2. NDRG1 expression is upregulated in PVTT samples compared to primary cancer and para-cancerous tissues.
a Representative IHC images and quantitative evaluation showing different NDRG1 expression level in non-tumorous liver tissue (N), primary tumour nodules (T), and PVTT (n = 31 per group). b Images and quantification of western blotting for NDRG1 in specimens from 4 HCC patients with PVTT. γ-tubulin was shown as a loading control. c The mRNA expression of NDRG1 in HCC group with vascular invasion (n = 109) or without vascular invasion (n = 206) from the TCGA dataset. d, e The NDRG1 level in different TNM stages from TCGA and GSE14520 datasets.
NDRG1 promotes HCC cell proliferation
Several studies have reported that NDRG1 promotes tumour proliferation [20, 25, 26]. To verify this notion in our system, we used the medium NDRG1 expressing primary HCC cell line PLC/PRF/5 as the research model. Upon ectopic expression, the proliferation rate of PLC-NDRG1 OE cells was significantly enhanced in vitro (Fig. 3a–c). Consistently, when NDRG1 was knocked down in the PVTT patient-derived cell line, the in vitro proliferation rate of the cells was impaired (Fig. 3d–f). To further explore the impact of NDRG1 on HCC tumour in vivo proliferation, we used the high NDRG1 expression cell lines PVTT-1 and Hepa1-6 as research models. As expected, we observed a consistent decrease tumour volume in both tumour types when NDRG1 was knocked down. (Fig. 3g and Supplementary Fig. 3). Thus, we showed that NDRG1 is able to promote HCC tumour cell proliferation in vitro and carcinogenesis in vivo.
Fig. 3. NDRG1 promotes tumour proliferation and carcinogenesis in HCC.
a Expression plasmids for NDRG1 gene were made in a PCDH vector. Data from western blotting showed the successful overexpression of NDRG1 (NDRG1 OE) in PLC/PRF5 cells. b Growth curves of PLC/PRF/5 cells stably expressing PCDH-NDRG1 or PCDH vector. c The clony formation assay were performed to determine the proliferation of PLC-NDRG1 OE and PLC-PCDH cells. d Expression plasmids for NDRG1 shRNAs were made in a PLKO.1 lentiviral vector. Successful knockdown of NDRG1 (shNDRG1#1, shNDRG1#2) in PVTT-1 cells was also confirmed. e Growth curves of PVTT-1 cells stably expressing PLKO.1-shNDRG1 or PLKO.1-mock. f The colony formation of PVTT-1 cells stably expressing PLKO.1-shNDRG1 or PLKO.1-mock. g Suppression of NDRG1 protein reduced tumour initiation induced by PVTT-1 cells in nude mice. 2.5 million PLKO.1-mock or PLKO.1-NDRG1 shRNA stably transfected PVTT-1 cells were injected into the dorsal flank of 6-week-old nude mice subcutaneously and tumour incidence was checked 5 days after injection. Black arrows showed the tumour masses.
NDRG1 enhances tumour-initiating capacity and metastasis in HCC tumours
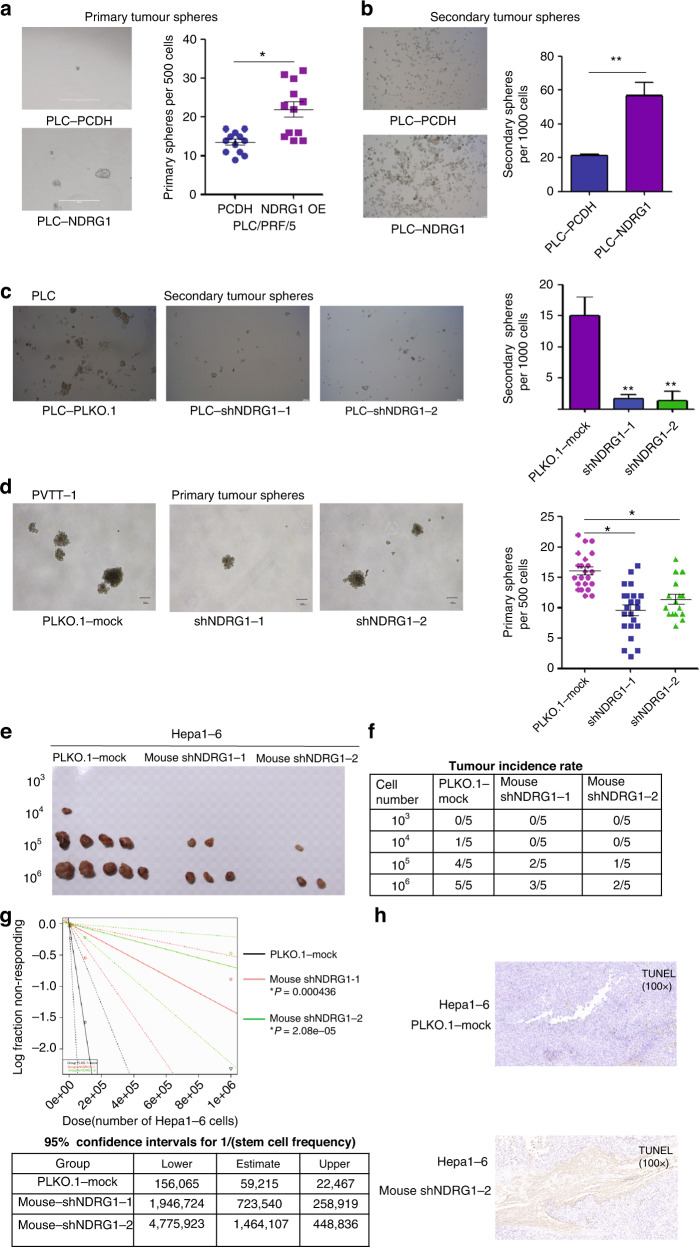
As mentioned above, NDRG1 expression is tightly correlated with late tumour progression and low differentiation signature, and we hypothesised that NDRG1 might affect TICs maintenance in HCC tumours. Conventionally, TICs are defined by their ability to form spheres under serum-free and non-adherent culture conditions. We performed the sphere formation assay by manipulating NDRG1 expression. C-myc, Sox2 and Nanog have been identified as master transcription factors that maintain TIC fate. Thus, we also tested the expression of these major transcription factors in PVTT-1 and PLC cell lines using NDRG1 manipulation.
In a sphere formation assay performed with PLC/PRF/5 cells, NDRG1 overexpression significantly increased the number and size of primary spheres and their secondary passages, whereas NDRG1 knockdown significantly attenuated the capacity of tumour sphere formation (Fig. 4a–c). In addition, fewer primary spheres were observed in the PVTT-1 cells with NDRG1 knockdown (Fig. 4d). The in vivo limiting dilution assay (LDA) in mice was performed to evaluate the relationship between NDRG1 depletion and TICs abilities. The CSCs frequencies of Hepa1-6-shNDRG1 cells were significantly lower than Hepa1-6-PLKO.1-mock cells, suggesting NDRG1 knockdown inhibits the in vivo tumour-initiating potential of HCC cells (Fig. 4e–g). Compared to control group, the apoptosis ratio was significantly increased in grafts from Hepa1-6-shNDRG1-2 cells (Fig. 4h).
Fig. 4. NDRG1 enhances tumour-initiating capacity in HCC tumours.
a, b NDRG1 overexpression significantly increased the number and size of primary spheres and their secondary passages. c NDRG1 knockdown significantly attenuated the capacity of tumour sphere formation. d NDRG1 knockdown significantly attenuates sphere-forming property in sphere cells derived from PVTT-1 cells. e, f Hepa1-6-shNDRG1 and Hepa1-6-PLKO.1-mock cells (103, 104, 105 and 106) were subcutaneously injected into BALB/c nude mice for observation of tumour growth. Tumour sizes and tumour incidence rate were demonstrated. g Liver CSCs frequency was analysed by extreme limiting dilution analysis (ELDA) software. A log-fraction plot of the limiting dilution model. The slope of the line was the log-active cell fraction and the dotted lines showing the 95% confidence interval (top). The estimated frequency and its 95% confidence intervals displayed in table (bottom). h Representative images showing IHC staining of TUNEL on tumours derived from Hepa1-6-shNDRG1-2 and control cells.
Intriguingly, NDRG1 ectopic expression enhanced the expression of CSCs genes, including c-myc, Sox2 and Nanog (Supplementary Fig. 4a). NDRG1 knockdown diminished the induction of transcription factors in PVTT-1 cells (Supplementary Fig. 4d). Protein levels of c-myc and Sox2 were inhibited in PLC-shNDRG1 cells compared to control group; however, Nanog protein was not affected by NDRG1 manipulations (Supplementary Fig. 4b, c).
TICs are characterised by enhanced cellular motility and distal metastasis. Thus, we explored HCC cell migration in different NDRG1 manipulations. NDRG1 knockdown eliminated the migration ability for more than 100-folds in PVTT cells, while ectopic expression promoted cell motility (Supplementary Fig. 5a, b). Further analysis in orthotropic HCC model confirmed that NDRG1 knockdown hampered primary tumour metastasis (Supplementary Fig. 5c, d). Taken together, we found NDRG1 was potent in mediating HCC cell migration and tumour metastasis.
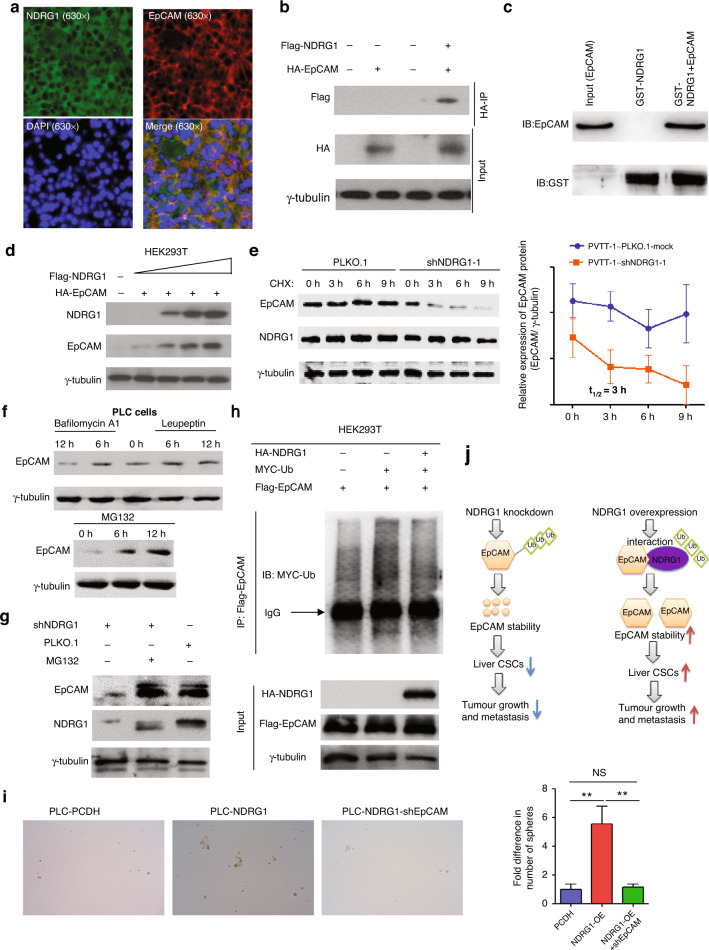
NDRG1 directly maintains EpCAM expression by promoting the protein stability
EpCAM is a major functional marker of TICs that potently activates TICs transcriptional factors, such as Sox2 and c-myc. As we found that NDRG1 could modulate Sox2 and c-Myc expression in HCC cells, we hypothesised that NDRG1 might modulate TICs transcription factors and the phenotype by directly modulating EpCAM. Upon shRNA-mediated NDRG1 knockdown, we observed a decrease in the surface membrane bond EpCAM without a noticeable change in the expression of other TICs markers, such as CD133 and CD44 (Fig. 5a). We further validated the overall EpCAM expression by western blotting. We found that EpCAM protein levels increased with NDRG1 ectopic expression and decresed with shRNA- or siRNA-mediated NDRG1 knockdown (Fig. 5b–d). Moreover, to test if EpCAM expression is aligned with NDRG1 levels, we sorted EpCAM-positive and -negative HCC cells and tested NDRG1 expression in each population. Encouragingly, in Huh7 cell lines, NDRG1 expression was found to be significantly lower in EpCAM-negative cells (Fig. 5e). To probe the detailed mechanism by which NDRG1 controls EpCAM expression, we first tested whether this regulation occurs at the transcriptional level. Unexpectedly, there was no major difference in RNA when NDRG1 was knocked down using siRNA or shRNA (Fig. 5f, g). In the same experimental batch, EpCAM declined at the protein level, along with NDRG1 knockdown (Fig. 5b, c). Hence, we assumed that NDRG1 mediates EpCAM expression post-translation. To validate our findings in specimens from patients with HCC, both NDRG1 and EpCAM protein levels were evaluated in 34 tumour tissues. EpCAM levels were elevated in tumour tissues with high expression levels of NDRG1 (Fig. 5h). EpCAM protein expression is highly correlated with NDRG1 expression in HCC samples.
Fig. 5. EpCAM is a functional target of NDRG1 in HCC.
a PLKO.1-mock or PLKO.1-NDRG1 shRNA stably transfected PVTT-1 cells were subjected to flow cytometry analysis to quantify EpCAM, CD44 or CD133 positive cells. b–d EpCAM protein were increased in PVTT-1 cells transfected with NDRG siRNAs or shRNAs, while opposite effects were observed in NDRG1 overexpressed cells. e NDRG1 protein level was determined by western blot in EpCAM-positive and negative cells isolated from Huh7. γ-tubulin was used as a loading control. f, g The mRNA and protein levels of NDRG1 in PVTT-1 cells transfected with NDRG1 siRNAs. h Correlation graphs between NDRG1 and EpCAM protein in 34 patients with HCC. Representative IHC images of low or high NDRG1/EpCAM expression in HCC tissues (left). Pearson correlation analysis of NDRG1 and EpCAM protein expressions in 34 HCC samples (r = 0.501, **P = 0.0025, n = 34) (right).
Next, we investigated whether protein degradation was directly mediated through protein–protein interactions. Using immunofluorescence assay, the localisation of NDRG1 and EpCAM was found to be tightly overlapped in PVTT-1 and PLC/PRF/5 cells (Fig. 6a and Supplementary Fig. 6). In addition, the direct protein interaction between EpCAM and NDRG1 was confirmed through immunoprecipitation and GST-pull down experiments (Fig. 6b, c). Next, we examined whether NDRG1 affected EpCAM protein stability. EpCAM degradation was specifically tested by inhibiting protein synthesis using cycloheximide (CHX). We found that NDRG1 expression is required for long-term EpCAM stabilisation, and loss of NDRG1 expression resulted in significant EpCAM degradation (Fig. 6d, e).
Fig. 6. NDRG1 interacts with and increases EpCAM protein stability.
a Subcellular localisation of NDRG1 and EpCAM in PVTT tissues. b NDRG1 interacted with EpCAM in HEK293 cells. 6ug Flag-NDRG1 and (or) 6ug HA-EpCAM plasmids were transfected into HEK293 cells in 10-cm cell culture dishes. 48 h after transfection. Cell lysates were immunoprecipitated with anti-HA beads and probed with anti-Flag antibody. Input cell lysates were utilised as controls. c GST-pull down was performed to detect whether NDRG1 interacted with EpCAM. d NDRG1 increased EpCAM protein stability in vitro. In total, 0.5 µg HA-EpCAM and increasing amounts (0, 0.5, 1.0, 1.5 µg) of NDRG1 constructs were transfected into HEK293T cells. EpCAM protein level was determined by western blot 48 h after transfection. e NDRG1 knockdown promoted degradation of EpCAM protein in PVTT-1 cells. PVTT-1 cells transfected with shNDRG1 or shCtrl were treated with 100 μg/ml cycloheximide (CHX) for the indicated times, harvested and cell lysates were immunoblotted with the indicated antibodies. f EpCAM ubiquitination was inhibited by MG132 treatment in PLC cells; compared with the control group, EpCAM protein was not changed significantly in PLC cells treated with lysosomal inhibitor including leupeptin and bafilomycin A1. g The inhibition of NDRG1 knockdown on EpCAM could be rescued by MG132, a proteasome inhibitor. h NDRG1 inhibited ubiquitination of EpCAM. 293 T cells were transfected with the indicated plasmids, the cell lysates were incubated with Flag antibody and the ubiquitylation level of Flag-EpCAM was detected by western blotting with an anti-MYC antibody. i Knockdown of EpCAM could diminish the improved self-renewal capability of PLC cells after NDRG1 overexpression. j A proposed model how NDRG1 regulates EpCAM in HCC. Left panel: Knockdown of NDRG1 promotes the ubiquitination and degradation of EpCAM, therefore leads to the self-renewal of liver CSCs, ultimately resulting in tumour suppression. Right panel: Overexpression of NDRG1 promotes EpCAM stability, therefore leading to the self-renewal of liver CSCs, ultimately resulting in tumour progression.
Proteasome–ubiquitination and lysosome–proteolysis are two major signalling pathways involved in cellular protein degradation. To narrow down the possible EpCAM degradation pathway, we treated cells with the proteasome inhibitor MG132 and the lysosome inhibitor bafilomycin A1 or leupeptin. Interestingly, EpCAM degradation is inhibited only when cells were treated with a proteasome inhibitor (Fig. 6f, g). Furthermore, we observed a reduction of EpCAM ubiquitination following NDRG1 overexpression (Fig. 6h). Knockdown of EpCAM could suppress the improved self-renewal capability of PLC/PRF/5 cells after NDRG1 overexpression (Fig. 6i). These results demonstrate that NDRG1 maintains EpCAM protein expression by inhibiting ubiquitination and degradation (Fig. 6j). Collectively, we found that NDRG1 can directly stabilise EpCAM through direct protein–protein interactions.
EpCAM expression is high in PVTT compared to primary HCC and normal liver tissues
Considering the high levels of NDRG1 in PVTT tissues, we speculated that the expression of EpCAM in PVTT differed from that in the corresponding primary tumours. Hence, we examined EpCAM protein levels in PVTT, primary HCC, and normal liver tissues using IHC. Our data showed that EpCAM protein was higher in PVTT tissues than in primary HCC tissues (Supplementary Fig. 7a, b).
Discussion
HCC is a major cause of cancer-related death in developing countries. Frequent intrahepatic and extrametastases of HCC are responsible for unsatisfactory clinical prognosis of HCC patients. In 30% of patients with HCC metastasis, malignant cells migrate and colonise the portal vein and form PVTT, leading to intractable ascites, and oesophageal variceal bleeding which results in organ failure [4, 31]. Thus, symptoms are a major obstacle to clinical treatment. Although numerous factors are found to contribute to HCC initiation, development and metastasis, the detailed mechanism of PVTT formation has not been widely studied.
In this study, we aimed to identify genes involved in PVTT formation. Using clinical samples, we identified one promising candidate NDRG1, which was enhanced in PVTT samples compared with primary HCC samples. NDRG1 has been reported paradoxically to mediate malignancy development in different cancer types. Our functional showed that, NDRG1 was able to stabilise the functional TICs marker EpCAM through direct protein–protein interaction and inhibition of EpCAM ubiquitination.
In this study, we screened the spectrum of NDRG family proteins in HCC samples. NDRG1 protein was elevated in HCC tumours compared to non-tumour tissues, whereas NDRG2, NDRG3, and NDRG4 expression did not change significantly in HCC tumours compared to non-tumour tissues. Furthermore, we analysed the relationship between NDRG1 expression and PVTT formation. Data from TCGA database analysis showed that NDRG1 mRNA was significantly enhanced in primary HCC with PVTT compared to tumours without PVTT. Our findings were consistent with those of previous studies, which showed that NDRG1 high levels in primary HCC [32–34]. We not only detected the expression of NDRG1 in primary HCC but also in metastatic HCC. We showed that NDRG1 protein were higher in PVTT tissues than in primary HCC and normal liver tissues. Taken together, aberrant activation of NDRG1 appears to be frequent in patients with HCC and PVTT and is an attractive candidate target for its treatment. Since many patients with HCC and PVTT have lost the opportunity for surgery, we could not analyse the expression of NDRG1 in a large number of PVTT specimens.
NDRG1 acts as a suppressor of tumour metastasis and growth in glioblastoma, colorectal, cancer and breast cancer [21–24]. Paradoxically, NDRG1 stimulates oncogenesis in cancers of the oesophageal squamous cell, liver, and stomach [25, 26, 35]. Although it has been reported that NDRG1 promote the proliferation of HCC cell lines derived from primary HCC tissues, the correlation between NDRG1 and PVTT-1 cells derived from metastatic HCC specimens has not been studied. Using loss-of-function experiments, we found that NDRG1 inhibition suppressed the growth of PVTT-1 cells both in vitro and in vivo. Using gain-function experiments, we found that the overexpression of NDRG1 enhanced the growth of PLC cells in vitro and in vivo.
NDRG1 has been reported to mediate the self-renewal of CSCs, depending on the cancer type. NDRG1 maintains CSCs stemness in lung cancer, as well as in osteosarcoma, whereas NDRG1 acts as a suppressor of stem-like characteristics in colon cancer [36, 37]. We hypothesised that NDRG1 functions in a specific manner. Using a sphere formation assay, we found that NDRG1 knockdown inhibited the stem-like properties of HCC cells, whereas NDRG1 overexpression promoted the stem-like features of HCC cells. TICs are characterised by enhanced cellular motility and distal metastasis. Thus, we explored HCC cell migration in different NDRG1 manipulations. Our data showed that NDRG1 was potent in mediating HCC cell migration and metastasis. Taken together, these results suggest that NDRG1 is an oncogene in HCC.
To explore the mechanism by which NDRG1 affects CSCs and PVTT development, we analysed well-established surface markers such as EpCAM, CD133 and CD44 in PVTT-1 cells with NDRG1 knockdown. Our findings showed that knockdown of NDRG1 could inhibit the ratio of EpCAM protein, but had no significant effect on the rate of CD133 and CD44 protein expression. Our western data showed similar results. In addition, NDRG1 protein levels were much higher in EpCAM+ HCC cells than in EpCAM- HCC cells. Although a series of molecules, such as c-Myc and CD44, regulated by NDRG1 have been reported in different tumour types [36, 38], we first demonstrated that NDRG1 could trigger the activation of the stemness-related protein EpCAM.
As a major form of post-translational modifications, ubiquitin plays an important role in the development and progression of HCC. However, ubiquitination of the EpCAM protein in many tumours has not been evaluated. We first showed that NDRG1 inhibition suppressed EpCAM protein levels by driving its ubiquitin–proteasome destruction. An obvious issue that remains to be clarified the mechanism by which NDRG1 affects EpCAM ubiquitination. Immunofluorescence data showed that NDRG1 located with EpCAM in the cell membrane of the PVTT specimen. Co-IP and GST-pull down data showed that NDRG1 interacted with EpCAM protein. Collectively, NDRG1 promoted EpCAM protein stability by interacting with EpCAM protein. A limitation of this study is that we could not identify a direct or indirect connection between NDRG1 and EpCAM ubiquitination.
Significantly, we not only found that NDRG1 protein was highly expressed in PVTT tissues but also that the expression of EpCAM in PVTT tissues was higher than that in primary HCC and normal tissues. Thus, the NDRG1-EpCAM axis may be a potential therapeutic target for treating patients with HCC and PVTT.
TICs are a small subpopulation of cancer cells capable of initiating tumours in nude mice, with a limited number of cells. In addition to the conventional serial dilution tumour in vivo formation, TICs has also been characterised with forming spheres in non-adherent culture conditions. Multiple reports have indicated that EpCAM modulates TICs activity by directly activating the master TICs transcription factors Sox2 and c-Myc. In our study, we proved that as a main stabiliser for EpCAM, NDRG1 is sufficient and required for maintaining TICs characteristics, including limited cell tumour initiation, sphere formation, and distal metastasis. In conclusion, our study provides a detailed molecular insight into NDRG1 modulating EpCAM, which drives TICs maintenance, cancer progression and PVTT-associated metastasis in HCC tumours.
Conclusions
In conclusion, our present results demonstrate that NDRG1 protein is increased in PVTT tissues and is associated with metastatic potential of HCC. NDRG1 facilitates CSCs properties of HCC cells by promoting EpCAM protein stability. The NDRG1-EpCAM axis may be a promising candidate for treating patients with HCC and PVTT.
Supplementary information
Acknowledgements
We would like to thank the Prof. Hongyang Wang (Shanghai Eastern Hepatobiliary Surgery Hospital) for providing technical support.
Author contributions
QC and JZ designed the work and provided material support. QC and SN acquired the main data. LZ performed several molecular biology experiments. CZ and SJ performed several cell experiments. YH provided bioinformatics analysis. QC, LZ and SN drafted the manuscript. JZ revised the manuscript.
Funding
This work was supported by National Natural Science Foundation of China (81502581, 82172693, 81872508) and the China Postdoctoral Science Foundation (2022M711911).
Data availability
All results presented in the article are available upon request from the corresponding author.
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The experiments involved in clinical specimens were performed in accordance with the Declaration of Helsinki and were approved by the Research Ethics Committee of Peking University People’s Hospital. Informed consent was obtained from all participants. In addition, the animal studies were approved by the Institutional Animal Welfare and Ethics Committee of Peking University People’s Hospital.
Consent for publication
Not applicable.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Qian Cheng, Shanglei Ning.
Contributor Information
Qian Cheng, Email: chengqian@bjmu.edu.cn.
Jiye Zhu, Email: gandanwk@vip.sina.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s41416-023-02278-y.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Loh JJ, Li TW, Zhou L, Wong TL, Liu X, Ma VW, et al. FSTL1 secreted by activated fibroblasts promotes hepatocellular carcinoma metastasis and stemness. Cancer Res. 2021;81:5692–705. doi: 10.1158/0008-5472.CAN-20-4226. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA A Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Yang P, Li QJ, Feng Y, Zhang Y, Markowitz GJ, Ning S, et al. TGF-β-miR-34a-CCL22 signaling-induced treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell. 2012;22:291–303. doi: 10.1016/j.ccr.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan X, Li Y, Yi X, Chen G, Jin S, et al. Epigenome-wide DNA methylation profiling of portal vein tumor thrombosis (PVTT) tissues in hepatocellular carcinoma patients. Neoplasia. 2020;22:630–43. doi: 10.1016/j.neo.2020.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng H, Pomyen Y, Hernandez MO, Li C, Livak F, Tang W, et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–40. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–8. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling S, Shan Q, Zhan Q, Ye Q, Liu P, Xu S, et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut. 2020;69:1322–34. doi: 10.1136/gutjnl-2019-319616. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–24. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang KH, Ma S, Lee TK, Chan YP, Kwan PS, Tong CM, et al. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology. 2012;55:807–20. doi: 10.1002/hep.24739. [DOI] [PubMed] [Google Scholar]
- 11.Asai R, Tsuchiya H, Amisaki M, Makimoto K, Takenaga A, Sakabe T, et al. CD44 standard isoform is involved in maintenance of cancer stem cells of a hepatocellular carcinoma cell line. Cancer Med. 2019;8:773–82. doi: 10.1002/cam4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani SK, Zhang H, Diab A, Pascuzzi PE, Lefrançois L, Fares N, et al. EpCAM‑regulated intramembrane proteolysis induces a cancer stem cell‑like gene signature in hepatitis B virus‑infected hepatocytes. J Hepatol. 2016;65:888–98. doi: 10.1016/j.jhep.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Münz M, Kieu C, Mack B, Schmitt B, Zeidler R, Gires O. The carcinoma-associated antigen EpCAM upregulates c-myc and induces cell proliferation. Oncogene. 2004;23:5748–58. doi: 10.1038/sj.onc.1207610. [DOI] [PubMed] [Google Scholar]
- 14.Nio K, Yamashita T, Okada H, Kondo M, Hayashi T, Hara Y, et al. Defeating EpCAM(+) liver cancer stem cells by targeting chromatin remodeling enzyme CHD4 in human hepatocellular carcinoma. J Hepatol. 2015;63:1164–72. doi: 10.1016/j.jhep.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Bae DH, Jansson PJ, Huang ML, Kovacevic Z, Kalinowski D, Lee CS, et al. The role of NDRG1 in the pathology and potential treatment of human cancers. J Clin Pathol. 2013;66:911–7. doi: 10.1136/jclinpath-2013-201692. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Tchou-Wong KM, Costa M. Egr-1 mediates hypoxia-inducible transcription of the NDRG1 gene through an overlapping Egr-1/Sp1 binding site in the promoter. Cancer Res. 2007;67:9125–33. doi: 10.1158/0008-5472.CAN-07-1525. [DOI] [PubMed] [Google Scholar]
- 17.Lane DJ, Saletta F, Suryo Rahmanto Y, Kovacevic Z, Richardson DR. N-myc downstream regulated 1 (NDRG1) is regulated by eukaryotic initiation factor 3a (eIF3a) during cellular stress caused by iron depletion. PLoS ONE. 2013;8:e57273. doi: 10.1371/journal.pone.0057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merlot AM, Porter GM, Sahni S, Lim EG, Peres P, Richardson DR. The metastasis suppressor, NDRG1, differentially modulates the endoplasmic reticulum stress response. Biochim Biophys Acta Mol Basis Dis. 2019;1865:2094–110. doi: 10.1016/j.bbadis.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 19.Sevinsky CJ, Khan F, Kokabee L, Darehshouri A, Maddipati KR, Conklin DS. NDRG1 regulates neutral lipid metabolism in breast cancer cells. Breast Cancer Res. 2018;20:55. doi: 10.1186/s13058-018-0980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Villodre ES, Hu X, Eckhardt BL, Larson R, Huo L, Yoon EC, et al. NDRG1 in aggressive breast cancer progression and brain metastasis. J Natl Cancer Inst. 2022;114:579–91. doi: 10.1093/jnci/djab222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ito H, Watari K, Shibata T, Miyamoto T, Murakami Y, Nakahara Y, et al. Bidirectional regulation between NDRG1 and GSK3b controls tumor growth and is targeted by differentiation inducing factor-1 in glioblastoma. Cancer Res. 2020;80:234–48. doi: 10.1158/0008-5472.CAN-19-0438. [DOI] [PubMed] [Google Scholar]
- 22.Mi L, Zhu F, Yang X, Lu J, Zheng Y, Zhao Q, et al. The metastatic suppressor NDRG1 inhibits EMT, migration and invasion through interaction and promotion of caveolin-1 ubiquitylation in human colorectal cancer cells. Oncogene. 2017;36:4323–35. doi: 10.1038/onc.2017.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama Y, Ono M, Kawahara A, Yokoyama T, Basaki Y, Kage M, et al. Tumor growth suppression in pancreatic cancer by a putative metastasis suppressor gene Cap43/NDRG1/Drg-1 through modulation of angiogenesis. Cancer Res. 2006;66:6233–42. doi: 10.1158/0008-5472.CAN-06-0183. [DOI] [PubMed] [Google Scholar]
- 24.Bandyopadhyay S, Pai SK, Hirota S, Hosobe S, Takano Y, Saito K, et al. Role of the putative tumor metastasis suppressor gene Drg-1 in breast cancer progression. Oncogene. 2004;23:5675–81. doi: 10.1038/sj.onc.1207734. [DOI] [PubMed] [Google Scholar]
- 25.Ai R, Sun Y, Guo Z, Wei W, Zhou L, Liu F, et al. NDRG1 overexpression promotes the progression of esophageal squamous cell carcinoma through modulating Wnt signaling pathway. Cancer Biol Ther. 2016;17:943–54. doi: 10.1080/15384047.2016.1210734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu WJ, Chua MS, So SK. Suppressing N-Myc downstream regulated gene 1 reactivates senescence signaling and inhibits tumor growth in hepatocellular carcinoma. Carcinogenesis. 2014;35:915–22. doi: 10.1093/carcin/bgt401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berghoff AS, Liao Y, Karreman MA, Ilhan-Mutlu A, Gunkel K, Sprick MR, et al. Identification and characterization of cancer cells that initiate metastases to the brain and other organs. Mol Cancer Res. 2021;19:688–701. doi: 10.1158/1541-7786.MCR-20-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang T, Hu HS, Feng YX, Shi J, Li N, Guo WX, et al. Characterisation of a novel cell line (CSQT-2) with high metastatic activity derived from portal vein tumour thrombus of hepatocellular carcinoma. Br J Cancer. 2010;102:1618–26. doi: 10.1038/sj.bjc.6605689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang Y, Liu S, Li N, Guo W, Shi J, Yu H, et al. 14-3-3ζ promotes hepatocellular carcinoma venous metastasis by modulating hypoxia-inducible factor-1α. Oncotarget. 2016;7:15854–67. doi: 10.18632/oncotarget.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2016;347:70–8. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Zhu P, Luo J, Wang J, Liu Z, Wu W, et al. LncRNA HAND2-AS1 promotes liver cancer stem cell self-renewal via BMP signaling. EMBO J. 2019;38:e101110. doi: 10.15252/embj.2018101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R, et al. NDRG1 as a biomarker for metastasis, recurrence and of poor prognosis in hepatocellular carcinoma. Cancer Lett. 2011;310:35–45. doi: 10.1016/j.canlet.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 33.Dang H, Chen L, Tang P, Cai X, Zhang W, Zhang R, et al. LINC01419 promotes cell proliferation and metastasis in hepatocellular carcinoma by enhancing NDRG1 promoter activity. Cell Oncol. 2020;43:931–47. doi: 10.1007/s13402-020-00540-6. [DOI] [PubMed] [Google Scholar]
- 34.Akiba J, Ogasawara S, Kawahara A, Nishida N, Sanada S, Moriya F, et al. N-myc downstream regulated gene 1 (NDRG1)/Cap43 enhances portal vein invasion and intrahepatic metastasis in human hepatocellular carcinoma. Oncol Rep. 2008;20:1329–35. [PubMed] [Google Scholar]
- 35.Chang X, Xu X, Xue X, Ma J, Li Z, Deng P, et al. N-myc downstream-regulated gene 1 promotes tumor inflammatory angiogenesis through JNK activation and autocrine loop of interleukin-1α by human gastric cancer cells. J Biol Chem. 2013;288:25025–37. doi: 10.1074/jbc.M113.472068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Zhou Y, Tao F, Chai S, Xu X, Yang Y, et al. N-myc downstream regulated gene 1(NDRG1) promotes the stem-like properties of lung cancer cells through stabilized c-Myc. Cancer Lett. 2017;401:53–62. doi: 10.1016/j.canlet.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 37.Zhao T, Meng Y, Wang Y, Wang W. NDRG1 regulates osteosarcoma cells via mediating the mitochondrial function and CSCs differentiation. J Orthop Surg Res. 2021;16:364. doi: 10.1186/s13018-021-02503-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wangpu X, Yang X, Zhao J, Lu J, Guan S, Lu J, et al. The metastasis suppressor, NDRG1, inhibits “stemness” of colorectal cancer via down-regulation of nuclear beta-catenin and CD44. Oncotarget. 2015;6:33893–911. doi: 10.18632/oncotarget.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All results presented in the article are available upon request from the corresponding author.