Abstract
Background
Eurycomanone (EN) is a diterpenoid compound isolated from the roots of Eurycoma longifolia (E. longifolia). Previous studies have confirmed that E. longifolia can enhance bone regeneration and bone strength. We previously isolated and identified ten quassinoids from E. longifolia, and the result displayed that five aqueous extracts have the effects on promotion of bone formation, among whom EN showed the strongest activity. However, the molecular mechanism of EN on bone formation was unknown, and we further investigated in this study.
Methods
After the verification of purity of extracted EN, following experiments were conducted. Firstly, the pharmacologic action of EN on normal bone mineralization and the therapeutic effect of EN on Dex-induced bone loss using zebrafish larvae. The mineralization area and integral optical density (IOD) were evaluated using alizarin red staining. Then the vital signaling pathways of EN relevant to OP was identified through network pharmacology analysis. Eventually in vitro, the effect of EN on cell viability, osteogenesis activities were investigated in human bone marrow mesenchymal stem cells (hMSCs) and C3H10 cells, and the molecular mechanisms by which applying AKT inhibitor A-443654 in hMSCs.
Results
In zebrafish larvae, the administration in medium of EN (0.2, 1, and 5 μM) dramatically enhanced the skull mineralization area and integral optical density (IOD), and increased mRNA expressions of osteoblast formation genes (ALP, RUNX2a, SP7, OCN). Meanwhile, exposure of EN remarkably alleviated the inhibition of bone formation induced by dexamethasone (Dex), prominently improved the mineralization, up-regulated osteoblast-specific genes and down-regulated osteoclast-related genes (CTSK, RANKL, NFATc1, TRAF6) in Dex-treated bone loss zebrafish larvae. Network pharmacology outcomes showed the MAPK and PI3K-AKT signaling pathways are closely associated with 10 hub genes (especially AKT1), and AKT/GSK-3β/β-catenin was selected as the candidate analysis pathway. In hMSCs and C3H10 cells, results showed that EN at appropriate concentrations of 0.008–5 μM effectively increased the cell proliferation. In addition, EN (0.04, 0.2, and 1 μM) significantly stimulated osteogenic differentiation and mineralization as well as significantly increased the protein phosphorylation of AKT and GSK-3β, and expression of β-catenin, evidencing by the results of ALP and ARS staining, qPCR and western blotting. Whereas opposite results were presented in hMSCs when treated with AKT inhibitor A-443654, which effectively inhibited the pro-osteogenesis effect induced by EN, suggesting EN represent powerful potential in promoting osteogenesis of hMSCs, which may be closely related to the AKT/GSK-3β/β-catenin signaling pathway.
Conclusions
Altogether, our findings indicate that EN possesses remarkable effect on bone formation via activating AKT/GSK-3β/β-catenin signaling pathway in most tested concentrations.
The translational potential of this article
This study demonstrates EN is a new effective monomer in promoting bone formation, which may be a promising anabolic agent for osteoporosis (OP) treatment.
Keywords: Eurycomanone, Bone formation, Zebrafish larvae, Human mesenchymal stem cells, C3H10 cells, AKT/GSK-3β/β-catenin signaling pathway
Graphical abstract
1. Introduction
Bone formation is the process of new bone occurrence and maturation, whereas its abnormal frequently cause serious consequences, and that osteoporosis (OP) and periodontitis are the most obvious [1]. Among them, OP considered as a common chronic bone disease charactered by reducing bone mass and density of bone mineralization, ultimately leading to an increased risk of fracture [2]. With the aging of society, OP has become a widespread global health disease affecting more than 200 million of people [3]. In clinical, anabolic agents are primary drugs applied in promoting bone formation for severe bone loss, and among with whom parathyroid hormone (PTH) analogs (teriparatide and abaloparatide) and sclerostin monoclonal antibody (romosozumab) were approved by US FDA currently [4]. Notably, the above-mentioned anabolic agents can rapidly increase bone mineral density and reduce prevalence of nonvertebral and vertebral fractures, especially suitable for the treatment of women with postmenopausal OP accompanied by high risk of fracture or glucocorticoid induced OP (GIOP) [[5], [6], [7]]. However, there are several concerns that include the risk of developing osteosarcoma (teriparatide) [8], hypercalcemia (abaloparatide) [6], and cardiovascular events (romosozumab) [5]. In order to cope with the above shortcomings, the methods for treatment of OP based on natural products has attracted increasing attention in recent years, by which high throughput screening of natural products is an important mean to obtain active ingredients of anti-osteoporosis drugs.
Eurycoma longifolia (E. longifolia) is a traditional medicinal plant mainly grown in Malaysia, Indonesia and other Southeast Asian countries [9]. Chemical compositions from E. longifolia are varied, and the active components extracted from its leaves, stems and roots are mainly diterpenoids of quassinoid skeleton and canthin-6-one alkaloids, e.g., Eurycomaoside, Eurycolactone, Eurycomalactone, Eurycomanone (EN) and Pasakbumin-B [[10], [11], [12]]. Studies have shown E. longifolia possesses a variety of pharmacologic properties, such as anti-tumor, anti-malaria, improve male sexual dysfunction et al. [13]. A previous study revealed the crude extract of E. longifolia had protective effects on the OP induced by orchidectomy through upregulating osteoprotegerin gene expression [14]. Considering the crude extract of E. longifolia has multitudinous components, it is of great significance to obtain active components of E. longifolia that play a positive role in bone through high throughput screening. Hence in our preliminary study, the isolation and identification of ten quassinoids (EL-1-EL-10) from E. longifolia were conducted. Results shown that most tested concentrations of five water-soluble quassinoids from the roots of E. longifolia (EL-1-EL-5) have the effects on promotion of bone formation, among whom EL-5 (which known as EN) showed the strongest activity [15]. As one of the bioactive components in E. Longifolia, EN play a crucial role in promoting bone formation may due to its functional activity in lipid metabolism and sex hormone metabolism [16,17], while the exact mechanism is worth exploring in depth.
Zebrafish, perceived as bony fish, have been developing rapidly in recent years due to advantages such as small individual size, high reproductive capacity, transparent embryos for easy viewing, high survival rate of larvae, and a short experimental period, rendering them suitable for disease replication and high-throughput drug screening [18]. Notably, zebrafish larvae have been widely applied to bone research due to their highly similar bone structure to that of humans [19]. Like mammals, zebrafish form bone through intramembranous ossification and endochondral ossification. Among them, intramembranous ossification is the dominant form of ossification in zebrafish bones and occurs in the cranial roof, while the chondroosteogenesis occurs on the surface of cartilage and forms in the lower jaw of zebrafish [20]. For zebrafish larvae, the miniralization begins 3–4 dpf (day-post-fertilization), and until day 7, parts of the notochord begin to mineralize and form vertebrae directly [21]. Simultaneously, both osteoblasts and osteoclasts exist in zebrafish larvae [22]. Thus, the zebrafish larvae skull contains sufficient and necessary cells for bone formation and absorption activity, making it easy for researchers to obtain the OP phenotype by using glucocorticoids (GCs) [23]. Above all, with the continuous development and improvement of zebrafish bone research methods, zebrafish larvae are widely used in the study of screening of bone-protecting drugs, especially bone anabolic agents.
Bone marrow mesenchymal stem cells (BMSCs) are multifunctional stem cells, which can be differentiated into various specialized cell types in vitro, such as osteoblasts, chondrocytes and adipose cells [24]. When BMSCs differentiated into osteoblasts, which are responsible for the stimulation of the bone formation process in vivo by producing bone matrix, calcium deposits, and osteoblastic cytokines [25]. Most importantly, decreased quantity and osteogenic differentiation of BMSCs commonly reduced bone formation [26] and increased osteoblast apoptosis, which are closely related to osteoporosis [27]. Besides, C3H10 is a mesenchymal progenitor cells from murine with similar osteogenesis mechanisms action of BMSCs. Recently, BMSCs and C3H10 cells have become common cells for studying bone relevant experiments [28]. It is reported that naringin has a protective effect on Dex-induced bone formation reduction in BMSCs through the PI3K/AKT/mTOR signaling pathway [29], sesamin extracted from Fagara was confirmed to enhanced osteoblastic differentiation of BMSCs via regulating the Wnt/β-catenin pathway [30], and amygdalin derived from bitter apricot kernel promote the differentiation of C3H10 cells to accelerated the fracture healing process by regulating TGF-β/Smad signaling pathway [31]. Taken together, promoting MSCs osteogenesis through natural products may be a promising therapeutic strategy.
Here, we explored the roles of EN on bone formation in zebrafish larvae, and the molecular mechanisms in mesenchymal stem cells. The experimental results revealed that EN represent powerful potential on stimulating bone mineralization and ameliorating the inhibitory effect induced by Dex in zebrafish larvae. Besides, EN possessed active effects on promoting osteogenic differentiation in hMSCs and C3H10 cells, which might be associated with the upregulation of AKT/GSK-3β/β-catenin signaling (see Fig. 1).
Fig.1.
Workflow of experiments of zebrafish larvae, network pharmacology analysis, and validation experiment of mesenchymal stem cells in vitro.
2. Materials and methods
2.1. Solution preparation
EN (chemical structure as shown in Fig. 2a) was provided by the Guangdong Provincial Key Laboratory for Research and Development of Nature Drugs (Zhanjiang, China). The original mass spectra and purity detection of EN are available in supplementary materials. It was solved in dimethyl sulfoxide (DMSO; Solarbio, Beijing, China) to 25 mM and maintained at 4 °C, followed by diluting to the indicated concentrations for experiments using Modified Eagle's Medium alpha (α-MEM; Gibco, Grand Island, NY, USA) or egg water (containing 5.0 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2 and 0.33 mM MgSO4).
Fig. 2.
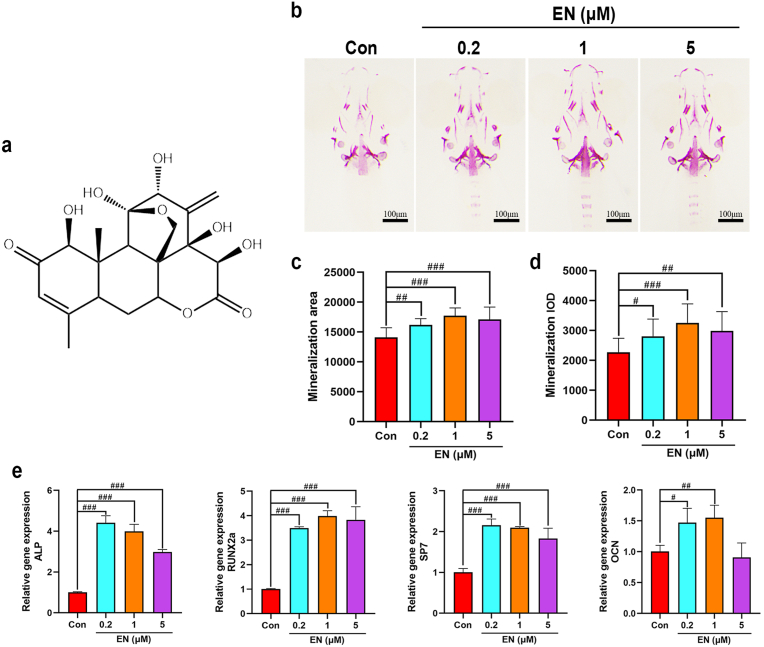
The chemical structure of EN and effects of EN on skull mineralization in zebrafish larvae. (a) Chemical structure of EN. (b) Zebrafish larvae were treated with EN (0.2, 1, and 5 μM), and skull mineralization ability was determined using ARS. (c) Mineralization areaJOTr-D-22-00514R1. (d) Mineralization IOD. (e) The mRNA expression of osteoblast-related genes (ALP, RUNX2a, SP7, and OCN) on 9-dpf in zebrafish larvae. The data are shown as the means ± SD (n = 10 zebrafish larvae/group). #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Con. Scale bar: 100 μm. EN, eurycomanone; ARS, alizarin red S; IOD, integrated optical density; ALP, alkaline phosphatase, RUNX2a, runt-related transcription factor 2a; SP7, Sp7 transcription factor; OCN, osteocalcin; dpf, days after fertilization; Con, Control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. Zebrafish husbandry and experimental designs
Wild-type zebrafish (AB strain) were purchased from Shanghai FishBio Co., Ltd. Company (Shanghai, China), and maintained in a day/night cycle (14 h/10 h, respectively) at 28 °C. Zebrafish larvae were grown in egg water containing methylene blue (0.1 g/100 ml). Zebrafish larvae were randomly divided into 5 groups in 12-well plates at 3 days post fertilization (dpf) for alizarin red S (ARS) staining (n = 12–15 larvae/well/group) and RNA isolation (n = 30 larvae/well/group), and the groups were as follows: (1) Control (Con); (2) EN 0.2 μM; (3) EN 1 μM; (4) EN 5 μM. To investigate the effect of EN in Dex-induced bone loss zebrafish larvae, groups were as follows: (1) Con; (2) Dex10 μM; (3) EN 0.2 μM + Dex 10 μM; (4) EN 1 μM + Dex 10 μM; (5) EN 5 μM + Dex 10 μM. Egg water containing different agents were changed once a day in zebrafish larvae until 9-dpf.
2.3. ARS and quantitative analysis
As a dye that specifically binds calcium salts, ARS is commonly performed to assess bone mineralized matrix deposition, which is a crucial index of bone formation. Zebrafish larvae ARS staining was conducted as previously described [32]. Briefly, zebrafish larvae at 9-dpf were collected, and then fixed in 4% paraformaldehyde solution for 2 h at room temperature. Ethanol (50%) prepared with phosphate-buffered saline (PBS) was added for dehydration 10 min or so. After washing with PBS 3 times, followed by staining with 0.1% (weight/volume) ARS and placed overnight. Next day, a fresh bleach solution (containing 1.5% H2O2 and 1% KOH) was added approximately 30 min in zebrafish larvae, and subsequently soaked in 0.5% KOH and glycerin at ratios of 3:1, 1:1, and 1:3, respectively. Eventually, we photographed zebrafish larvae with a stereo microscope (SOPTOP SZN71, Ningbo, China), and the Image-Pro Plus 6.0 software (IPP; Media Cybernetics, Rockville, MD, USA) was utilized for the quantitative analysis of the mineralization area and integral optical density (IOD).
2.4. RNA isolation and gene expression analysis
Total RNA from each group was extracted in zebrafish larvae using the TRIZOL reagent (Abcam, Austin, TX, USA), whereafter synthesizing the cDNA through the PrimeScript™ RT Master Mix reagent kit (Takara, Dalian, China) on request to manufacturer's instructions. Reverse transcription reaction conditions were as follows: 37 °C for 15 min; 85 °C for 5 s. Subsequently, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was performed using TB Green Premix Ex TaqII (TaKaRa, Dalian, China) to detect the mRNA levels of alkaline phosphatase (ALP), Runt-related transcription factor 2a (RUNX2a), osterix (SP7), osteocalcin (OCN), cathepsin K (CTSK), receptor of nuclear factor-κB (RANKL), nuclear factor of activated T cells 1 (NFATC1), and TNF receptor-associated factor 6 (TRAF6). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was utilized as internal control. Cycling conditions of qRT-PCR were as follows: 95 °C for 5 min for pre-denaturation, followed by 40 cycles at 95 °C for 15 s and 60 °C for 60 s. Relative expression was calculated using the 2−ΔΔCT method. Paired primers of genes were synthesized by Sangon Biotech (Shanghai, China) (Table 1).
Table 1.
Primer sequences of genes used in quantitative real-time-PCR.
| Species | Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|---|
| Zebrafish | ALP | CTGGTGGAAGGTGGACGCATTG | CGTGTCATACTCGCTTGTCAGGAG |
| RUNX2a | AGACTCCGACCTCACGACAACC | GGCAGCACCGAGCACAGAAAG | |
| SP7 | AGGCTTGCTAACACCAACTGGAAG | GGGAAACACTGGAGGTCTGGAAG | |
| OCN | GTCCTGATCTTCTGCTGCCTGATG | GTCACGCTTCACAAACACACCTTC | |
| CTSK | GTGCCTGATGACAGAGTGGGAAAG | AGCCGCATGATCCTTGGTTCTTC | |
| RANKL | ACACTCACGGTCTGAACCTCCTAC | ACACCTCTGCCCTGCTCCATG | |
| NFATC1 | TCTGCCATTGTTGCCGCCATC | TGTCTCGTCTCCACCTGGTTCTAC | |
| TRAF | GGTACAAACTCTGCCTGCGACTG | CCAGCACTGCCAGCCGAATG | |
| GAPDH | CGCTGGTGCTGGTATTGCTCTC | GCCATCAGGTCACATACACGGTTG | |
| Human | COL1 | AAAGATGGACTCAACGGTCTC | CATCGTGAGCCTTCTCTTGAG |
| RUNX2 | AGGCAGTTCCCAAGCATTTCATCC | TGGCAGGTAGGTGTGGTAGTGAG | |
| SP7 | CGGCAAGAGGTTCACTCGTTCG | TGGAGCAGAGCAGGCAGGTG | |
| OCN | AGGGCAGCGAGGTAGTGAAGAG | GCCGATGTGGTCAGCCAACTC | |
| Mouse | COL1 | GACAGGCGAACAAGGTGACAGAG | CAGGAGAACCAGGAGAACCAGGAG |
| RUNX2 | GATGATGACACTGCCACCTCTGAC | TGAGGGATGAAATGCTTGGGAACTG | |
| SP7 | GACTACCCACCCTTCCCTCACTC | TAGACACTAGGCAGGCAGTCAGAC | |
| OCN | CAAGCAGGAGGGCAATAAGGTAGTG | CGGTCTTCAAGCCATACTGGTCTG |
PCR, polymerase chain reaction; ALP, alkaline phosphatase; RUNX2a, runt-related transcription factor 2a; SP7, Sp7 transcription factor; OCN, osteocalcin; CTSK, cathepsin K; RANKL, receptor of nuclear factor-κB; NFATC1, nuclear factor of activated T cells 1; TRAF6, tumor necrosis trapping receptor associated factor 6; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; COL1, collagen type I
2.5. Network pharmacology prediction for EN in osteoporosis
Targets genes and signaling pathways of EN in prevention and treatment of osteoporosis were predicted as previous described [33]. Briefly, target genes related to EN and osteoporosis were retrieved from the following databases: Bioinformatics Analysis Tool for Molecular Mechanism of Traditional Chinese Medicine (BATMAN-TCM; http://bionet.ncpsb.org.cn/batman-tcm/), Online Mendelian Inheritance in Man (OMIM; https://www.omim.org/), DisGeNET (https://www.disgenet.org/home/), Drugbank (https://go.drugbank.com/), GeneCards (https://www.genecards.org/), Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/). Then the original microarray dataset GSE35958 was analyzed with GEO2R to identify differentially expressed genes and generate a volcano plot. After obtaining the Venn diagram of potential osteoporosis of EN, UniProt database (https://www.uniprot.org/) was used to standardize the target genes. Next, the top 10 hub genes were acquired through Search Tool for the Retrieval of Interacting Genes/Proteins database (STRING; https://string-db.org/) and Cytoscape v3.7.2 (https://cytoscape.org/) software (Table 2). Then, the disease and drug component–core targets–signaling pathways/biological process interaction network were constructed and analyzed using Cytoscape v3.7.2 software. Finally, gene ontology (GO) functional annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG; https://www.genome.jp/kegg/) pathway enrichment analysis were conducted by Metascape database and Bioinformatics database (http://www.bioinformatics.com.cn/).
Table 2.
Top 10 genes in the protein–protein interaction network ranked by the degree method.
| Rank | UniProt ID | Gene symbol | Protein name | Degree |
|---|---|---|---|---|
| 1 | P12931 | SRC | Proto-oncogene tyrosine-protein kinase Src | 34 |
| 2 | P28482 | MAPK1 | Mitogen-activated protein kinase 1 | 33 |
| 3 | P31749 | AKT1 | RAC-alpha serine/threonine-protein kinase | 28 |
| 4 | P62993 | GRB2 | Growth factor receptor bound protein 2 | 25 |
| 5 | P19793 | RXRA | Retinoid X acid receptor alpha | 24 |
| 5 | Q06124 | PTPN11 | Tyrosine-protein phosphatase non-receptor type 11 | 24 |
| 7 | P05412 | JUN | Transcription factor AP-1 | 23 |
| 8 | Q16539 | MAPK14 | Mitogen-activated protein kinase 14 | 21 |
| 8 | P61586 | RHOA | Transforming protein RhoA | 21 |
| 10 | P45983 | MAPK8 | Mitogen-activated protein kinase 8 | 20 |
ID, identifier
2.6. Cell culture
C3H10 cells were obtained from the key laboratory of research and development of natural drugs (Zhanjiang, China). C3H10 cells were cultured in DMEM medium with 10% fetal bovine serum (Gibco) and 1% penicillin-streptomycin (Solarbio) at 37 °C in an atmosphere with 5% CO2. When cells density reached 90%, they were digested with 0.25% ethylenediaminetetraacetic acid–trypsin (Gibco) and subcultured at a ratio of 1:3. Cells were changed medium fluids every two days. To obtain differentiated cells, C3H10 cells were cultured in osteoblast induction medium (OIM) containing 10 mM β-glycerophosphate (β-GP) and 50 μg/ml ascorbic acid (Vit C).
Primary hMSCs were supplied by the Affiliated Hospital of Guangdong Medical University (Zhan jiang, China). The research was approved by the ethics committee of the hospital. Primary hMSCs were extracted from patients who underwent joint replacement. Briefly, bone marrow samples from clinical surgery were transferred to anti-coagulant tube, and send to the laboratory with low-temperature holding rapidly. Firstly, an equal volume of PBS was added to bone marrow samples, followed by repeated aspiration to fully separate the tissue blocks, and the mixture was slowly added to a cell separation solution (1:1). After centrifugation at 400×g for 20 min in a high-speed refrigerated centrifuge (Thermo Fisher Scientific, Waltham, MA, USA), the liquid was divided into four layers and the interlayer cells were extracted. This was followed by addition of PBS to reach a volume of 10 ml and centrifugation at 250×g for 10 min. Pale red cell precipitates were obtained and cultured in 10-cm culture dishes. Similarly, hMSCs were cultured in α-MEM medium containing 10% Australian fetal bovine serum (Gibco) and 1% penicillin-streptomycin in an atmosphere with 5% CO2 at 37 °C. Avoid moving cells, until 72 h, the medium fluids were first replaced with fresh, and then changed once every 3 days. Primary hMSCs from passages 3–6 was employed in the experiments. In order to detect surface markers, hMSCs were adjusted to 1.0 × 106/ml after trypsin digestion. Murine monoclonal antibodies CD14, CD19, CD44 and CD73 were prepared with PBS solution, and then incubated in cells at room temperature and away from light for 30 min. After centrifugation, cells were washed with PBS for 2–3 times and incubated with FITC-labeled goat anti-rabbit lgG (H + L) secondary antibody under the same conditions. Washing twice in PBS, hMSCs were detection in flow cytometry. Primary hMSCs identification are available in supplementary materials. Besides, to obtain differentiated cells, hMSCs were cultured in OIM containing 10 mM β-GP, 50 μg/ml Vit C, and 10 nM Dex.
2.7. Cell viability analysis
Cell viability was generally determined via the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. To evaluate the effect of EN on the proliferation of C3H10 cells and hMSCs, they were cultured in 96-well plates with the density of 2.5 × 103 and 1.0 × 104 cells/well, respectively. C3H10 cells were treated with 0.008, 0.04, 0.2, 1, 5, and 25 μM EN for 1, 2, 3 days, and hMSCs were exposed to EN at the same concentrations for 1, 3, 7 days. Thereafter, MTT (20 μl; 5 mg/ml) was added at the corresponding time points. 4 h later, adding 150 μl dimethyl sulfoxide (DMSO) to each well after removing the solution, and cells were incubated at room temperature for 30 min. Ultimately, the optical density (OD) was measured with a microplate analyzer (Bio-Rad Laboratories, Hercules, CA, United States) at an absorbance of 490 nm.
2.8. ALP staining and activity assay
ALP is commonly performed to detect the level of early osteoblast differentiation. C3H10 cells and hMSCs were seeded into 24-well plates with the density of 1.0 × 104 and 1.0 × 105 cells/well, respectively. They were treated with 0.04, 0.2, and 1 μM EN for 7 days. For C3H10 cells, we firstly collected the supernatant of each well for the detection of ALP activity with ALP/AKP kit (Nanjing Jiancheng, Nanjing, China) according to the instructions, and OD was detected with a microplate analyzer at an absorbance of 520 nm. ALP activity is calculated as U/L. Besides, BCIP/NBT Kit (Leagene, Beijing, China) was conducted to ALP staining. In accordance with the instructions provided by the manufacturer, cells were washed 2 times with PBS and then fixed in 4% paraformaldehyde for 30 min or so. Washing 2–3 times of PBS again, the prepared ALP solution was added to each well, and followed by incubating under given conditions of normal temperature and away from light for 30 min. Finally, after removing the solution, stereo microscope was performed to capture the images.
While for hMSCs, ALP staining was conducted with BCIP/NBT Kit. Besides, another 24-well plates were lysed overnight with radioimmunoprecipitation assay lysis buffer (RIPA; Beyotime, Shanghai, China) at 4 °C, soon afterwards incubated at 37 °C with the reagents of the ALP/AKP kit about 15 min. After adding coloring solution, the OD value was measured in the same way with a microplate analyzer at 520 nm, followed by the determination of protein concentration with a bicinchoninic acid (BCA; Solarbio) protein quantitation kit (Beyotime). In here, ALP activity is specially counted in U/10 mg protein.
2.9. ARS staining and activity assay
Calcium deposition was assessed by ARS staining and activity assay during osteogenic differentiation. C3H10 cells and hMSCs were inoculated into 12-well plates with the density of 2.0 × 104 and 2.0 × 105 cells/well, respectively. C3H10 cells were treated with EN at different indicated concentrations for 21 days, while EN exposed to hMSCs until 28 days. Being washed 2 times with PBS, cells were subsequently fixed in 70% ethanol for 30 min or so. After rinsing with PBS for 2–3 times, cells were exposed to 0.2% ARS (Sigma, Tokyo, Japan) solution (pH 4.3) and incubated shielding from light at room temperature approximately 30 min. When the mineralization-positive cells were stained red, followed by washing cells twice with PBS again, and photographs were captured under a stereo microscope when the plates were dried. After photographing, 10% cetylpyridine was added to each well, and following by incubating under light protection at room temperature for 30min. Then transferred samples to 96-well plate with 100ul per well, and OD was detected with a microplate analyzer at an absorbance of 562 nm.
2.10. Real-time quantitative PCR (qRT-PCR)
In the light of the instructions of manufacturer, total cell RNA was extracted with TRIZOL reagent, and cDNA was synthesized using the PrimeScript™ RT Master Mix reagent kit. Next, TB Green Premix EX Taq II was put to use for real-time detection. Gene expression levels of collagen type I (COL1), RUNX2, SP7, OCN in hMSCs and C3H10 cells were normalized to those of the housekeeping gene GAPDH. Primer sequences used in real-time PCR are shown in Table 1. Relative expression levels of mRNA was calculated with the 2−ΔΔCT method.
2.11. Western blotting
Western blotting analysis of C3H10 cells and hMSCs were conducted after 10 days and 14 days, respectively. Briefly, cells were extracted with a mixture lysis solution composed of RIPA plus phenylmethylsulfonylfluoride (PMSF; Beyotime) at the ratio of 100:1, and we measured the protein concentration with BCA assays according to the instructions. After separation of samples (10–30 μg protein) through 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (activated with methanol 10 min in advance), followed by the incubation with 5% fat-free milk for 2–3 h. Above membranes were cut according to the size of protein molecule we needed, and put into the corresponding primary antibody solution overnight at 4 °C. Antibodies were used against: COL1 (1:1000), RUNX2 (1:1000), SP7 (1:1000) and OCN (1:500) (all from Abcam); protein kinase B (AKT; 1:1000) and phosphorylated-AKT (p-AKT; 1:1000) (all from Signalway Antibody, College Park, MD, USA); GSK-3β (1:500; BD Biosciences, Beijing, China); phospho-GSK-3β (Ser9) (1:500) and β-catenin (1:500) (all from Cell Signaling Technology, Danvers, Massachusetts, USA); and GAPDH (1:2000; Abway, Shanghai, China). Subsequently, aforementioned membranes were further incubated with secondary antibodies of goat anti-rabbit/mouse IgG H&L (horseradish peroxidase) (1:2000; ORIGENE, Beijing, China) in circumstances of room temperature for 2 h. An ultra-hypersensitive electrochemiluminescence kit (Beyotime) was performed to visualize the signals, and imaging results of membrane proteins were quantified via the ImageJ software (National Institutes of Health, Bethesda, MD, USA).
2.12. Statistical analysis
Data from each group were expressed as mean ± standard deviation. One-way analysis of variance (ANOVA) was performed to analyze the differences between groups. When the variance between groups was equal, the least significant difference post hoc test (P > 0.05); otherwise, Dunnett's post hoc test will be used. P-values <0.05 was considered statistically significant. SPSS 26.0 software (IBM Corporation, Armonk, NY, USA) was used for statistical analysis.
3. Results
3.1. EN enhanced bone formation in zebrafish larvae
Given that mineralization is considered as a vital index in the process of bone formation, ARS staining in here was performed to investigate the effect of EN (0.2, 1 and 5 μM) in zebrafish larvae. Our results showed that, in comparison to the control group, both mineralization area and IOD of zebrafish larvae treated with diverse concentrations of EN were observably increased, and 1 μM EN shows the peak induction (Fig. 2b–d). Besides, treatment with EN, zebrafish larvae showed a markedly increased in osteogenic genes (ALP, RUNX2, SP7, and OCN), which as shown in Fig. 2e.
3.2. EN alleviated the inhibition in dex-induced zebrafish larvae bone loss
To further examine the effect of EN on secondary bone loss, zebrafish larvae were co-treated with EN (0.2, 1, and 5 μM) and Dex (10 μM) until 9-dpf. Results showed that 10 μM Dex caused a significant reduction in the skull mineralization compared with control. Besides, Dex obviously promoted the mRNA expression levels of osteoblasts-related genes of ALP,RUNX2a, SP7 (except for OCN) and osteoclasts-specified genes of CTSK, RANKL, NFATC1, TRAF6. Conversely, EN has direct protective effects against bone loss, which significantly improved the mineralization and IOD. Intriguingly, EN prominently up-regulated the levels of osteogenesis genes, but markedly reduced those of osteoclast-related genes to different degrees (Fig. 3). Above results indicated that EN reverses inhibition induced by Dex in zebrafish larvae bone loss.
Fig. 3.
Effects of EN on Dex-induced zebrafish larvae bone loss. (a) Zebrafish larvae were treated with EN (0.2, 1, and 5 μM) combined with 10 μM Dex, and skull mineralization ability was determined using ARS. (b) Mineralization area. (c) Mineralization IOD. (d) The mRNA expression of osteoblast-related genes (ALP, RUNX2a, SP7, and OCN) on 9-dpf in Dex-induced zebrafish larvae. (e) The mRNA expression of osteoclast-related genes (CTSK, RANKL, NFATC1, and TRAF6) on 9-dpf in Dex-induced zebrafish larvae. The data are shown as the means ± SD (n = 13 zebrafish larvae/group). ###P < 0.001 vs. Con; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. Dex. Scale bar: 100 μm. EN, eurycomanone; Dex, dexamethasone; ARS, alizarin red S; IOD, integrated optical density; ALP, alkaline phosphatase, RUNX2a, runt-related transcription factor 2a; SP7, Sp7 transcription factor; OCN, osteocalcin; CTSK, cathepsin K; RANKL, receptor of nuclear factor-κB; NFATC1, nuclear factor of activated T cells 1; TRAF, TNF receptor-associated factor 6; dpf, days after fertilization; Con, control. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.3. Network pharmacological analysis for EN in osteoporosis
As presented in Fig. 4a–d, 387 target genes related to EN and 4009 target genes related to osteoporosis were identified respectively, among which 158 target genes of EN in osteoporosis were obtained. Deleting 30 free targets, 128 target genes were identified. As showed in Fig. 4e, there are 396 connections among 128 targets in the protein–protein interaction network. Further analysis of the target genes determined the top 10 hub genes as follows: SRC proto-oncogene, non-receptor tyrosine kinase (SRC), mitogen-activated protein kinase 1 (MAPK1), AKT serine/threonine kinase 1 (AKT1), growth factor receptor bound protein 2 (GRB2), retinoid X receptor alpha (RXRA), protein tyrosine phosphatase non-receptor type 11 (PTPN11), Jun proto-oncogene, AP-1 transcription factor subunit (JUN), MAPK14, and MAPK8 (Table 2 and Fig. 4f). Based on the above findings, we further performed a disease and drug component–core targets–signaling pathways/biological process to visualize the relationship between the target genes of EN in osteoporosis (Fig. 4g). In Fig. 4h–k, GO functional annotation and KEGG pathway enrichment analysis related to target genes revealed that most of the target genes were related to biological process including inflammatory response, oxidative stress and the MAPK cascade, and signaling pathways such as the MAPK signaling pathway (has04010), phosphatidylinositol-3-kinase (PI3K)-AKT signaling pathway (hsa04151), T helper 17 cell differentiation (hsa04659), osteoclast differentiation (hsa04380), interleukin 17 (IL17) signaling pathway (hsa04657), forkhead box O (FOXO) signaling pathway (hsa04068). Ultimately, MAPK signaling pathway and PI3K-AKT signaling pathway were chose for verification in hMSCs.
Fig. 4.
Network pharmacological analysis for EN related to osteoporosis. Venn diagram of (a) EN targets and (b) OP-related genes. (c) Volcano plot of DEGs obtained from GSE35958. (d) Venn diagram of target genes of EN in OP. (e) PPI network of EN targets in OP. (f) Venn diagram of hub genes of EN in OP. (g) Target–signaling pathway/biological process interaction network of EN in OP. Note: left single node, EN; right single node, OP; middle nodes in grid arrangement, overlapping EN-OP targets; upward side nodes in circular arrangement, biological process; downward side nodes in circular arrangement, signaling pathways. (h) Biological process (BP). (i) Molecular function (MF). (j) Cellular component (CC). (k) KEGG pathway. DEGs, differentially expressed genes; EN, eurycomanone; OP, osteoporosis; PPI, protein–protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
3.4. EN promoted cells proliferation in vitro
To probe into the function of EN on proliferation, C3H10 cells and hMSCs were exposed to EN (0.008–25 μM) for 3 days and 7 days respectively, followed by adding MTT at the corresponding time points. For C3H10 cells, 25 μM EN significantly inhibited cell proliferation on days 1, but there was no significant change in other concentrations. On days 2, EN at concentrations of 0.008–5 μM promoted cell viability, but only at 0.04 μM and 5 μM showed statistically significant. On days 3, EN at concentrations of 0.04–5 μM markedly promoted cell growth, but the optimal EN were 0.04 ∼ 1 μM (Fig. 5a).
Fig. 5.
Effects of EN on cell proliferation and osteogenic differentiation in C3H10 cells. (a) Viability of C3H10 cells treated with EN at different concentrations (0.008–25 μM). (b–c) ALP staining and activity assays were measured after treating C3H10 cells with EN (0.04, 0.2, and 1 μM) for 7 days. (d–e) ARS staining and activity assays were performed after 21 days. (f) The mRNA expression levels of osteoblast-related genes (COL1, RUNX2, SP7, and OCN) in C3H10 cells were detected on day 10. (g–h) The protein expression levels of COL1, RUNX2, SP7, and OCN were detected by western blotting on day 10. #P < 0.05, ##P < 0.01, ###P < 0.001 vs. Con; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM. The data are shown as the means ± SD (n = 3). Scale bar: 100 μm. EN, eurycomanone; C3H10 cells, mouse mesenchymal stem cell line; ALP, alkaline phosphatase; ARS, alizarin red S; COL1, collagen type I; RUNX2, runt-related transcription factor 2; SP7, Sp7 transcription factor; OCN, osteocalcin; Con, Control; OIM, osteogenic induction medium. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
For hMSCs, results showed that EN in the concentration of 25 μM inhibited the cell growth in the same way with C3H10 cells on days 1, whereas other concentrations showed a slight proliferative effect. Since days 3, EN at concentrations between 0.04 and 1 μM prominently promoted proliferation, which increased over time. In addition, EN at concentrations of 0.008 μM and 5 μM also showed a stimulative effect on days 7, but to a lesser extent than 0.04 to 1 uM. In summary, EN at concentrations from 0.04 to 1 μM were best selected for subsequent experiments (Fig. 6a).
Fig. 6.
Effects of EN on cell proliferation and osteogenic differentiation in hMSCs. (a) Viability of hMSCs treated with EN at different concentrations (0.008–25 μM). (b–c) ALP staining and activity assays were measured after treating hMSCs with EN (0.04, 0.2, and 1 μM) for 7 days. (d–e) ARS staining and activity assays were performed after 28 days. (f) The mRNA expression levels of osteoblast-related genes (COL1, RUNX2, SP7, and OCN) in hMSCs were detected on day 14. (g–h) The protein expression levels of COL1, RUNX2, SP7, and OCN were detected by western blotting on day 14. #P < 0.05, ###P < 0.001 vs. Con; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM. The data are shown as the means ± SD (n = 3). Scale bar: 100 μm. EN, eurycomanone; hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; ARS, alizarin red S; COL1, collagen type I; RUNX2, runt-related transcription factor 2; SP7, Sp7 transcription factor; OCN, osteocalcin; Con, Control; OIM, osteogenic induction medium. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.5. EN increased the osteogenic differentiation in C3H10 and hMSCs
To determine whether EN (0.04, 0.2, and 1 μM) plays a part in the process of osteogenic differentiation, cells were consistently cultured in OIM for 7 days. Subsequently, ALP staining and activity assays were carried out. For C3H10 cells, results showed that OIM group led to a significant secretion of ALP compared with control. Additionally, EN further induced osteogenic differentiation, with higher ALP activity than OIM (Fig. 5b and c). For hMSCs, results were similarly to C3H10 cells. OIM stimulated the production of ALP, While EN extremely stimulated the expression of ALP, and 0.2 μM had the best promoting effect (Fig. 6b and c).
Given the stimulative effect of EN on osteogenic differentiation in C3H10 cells and hMSCs, we further evaluated the direct influence of EN on mineralization. The results were similar to those described above. Treatment with OIM effectively promoted the formation of mineralized nodules, whereas cells treated with EN (0.04, 0.2, and 1 μM) showed more mineralized nodules than those observed after culture in OIM (Fig. 5d-e 6 d-e).
3.6. EN increased the expression of osteoblast differentiation-related genes in C3H10 and hMSCs
To better explore the role of EN in osteoblast differentiation, we furthermore investigated the expression of osteoblast-related genes (COL1, RUNX2, SP7, and OCN) in C3H10 cells cultured in OIM for 10 days by qPCR and western blotting (WB), and hMSCs for 14 days. Results indicated that the mRNA and proteins levels from osteogenic genes were dramatically increased at the presence of EN (0.04, 0.2, and 1 μM) compared with those recorded in cells treated only with OIM (Fig. 5f-h,6f-h).
3.7. EN activated the AKT/GSK-3β/β-catenin signaling pathway in C3H10 and hMSCs
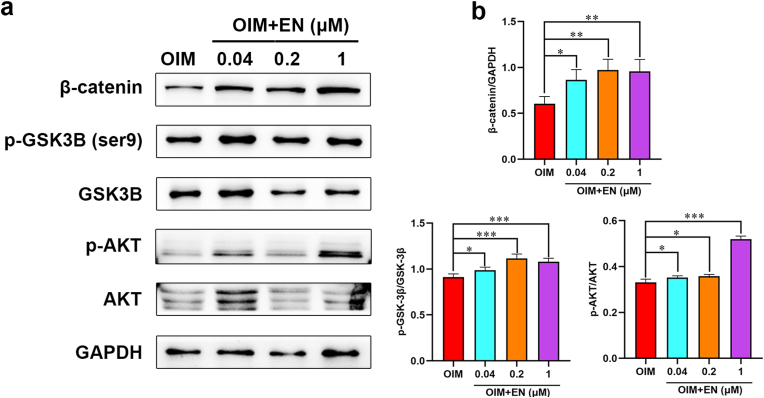
To explore the underlying mechanism of osteogenic differentiation based on network pharmacology, protein expression levels related to AKT/GSK-3β/β-catenin signaling pathway were detected by WB after C3H10 cells treated with EN (0.04, 0.2, and 1 μM) for 10 days, and hMSCs for 14days. As shown in Fig. 7, Fig. 8, the ratio of p-AKT/AKT, GSK-3β (Ser9)/GSK-3β and the expression of β-catenin were significantly promoted. These results suggested that EN may play an osteogenic role through the upregulation of AKT/GSK-3β/β-catenin signaling pathway.
Fig. 7.
Effect of EN on AKT/GSK-3β/β-catenin pathway in C3H10 cells. (a) Protein levels of AKT/GSK-3β/β-catenin pathway in C3H10 cells were detected by western blotting on day 10: (b) β-catenin/GAPDH, p-GSK-3β (ser9)/GSK-3β, p-AKT/AKT. The data are shown as the means ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM. EN, eurycomanone (p-) AKT (phosphorylated-) protein kinase B (p-) GSK-3β (ser9) (phosphorylated-) glycogen synthase kinase 3β at ser9; hMSCs, human mesenchymal stem cells; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OIM, osteogenic induction medium.
Fig. 8.
Effect of EN on AKT/GSK-3β/β-catenin pathway in hMSCs. (a) Protein levels of AKT/GSK-3β/β-catenin pathway in hMSCs were detected by western blotting on day 14: (b) β-catenin/GAPDH, p-GSK-3β (ser9)/GSK-3β, p-AKT/AKT. The data are shown as the means ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM. EN, eurycomanone (p-) AKT (phosphorylated-) protein kinase B (p-) GSK-3β (ser9) (phosphorylated-) glycogen synthase kinase 3β at ser9; hMSCs, human mesenchymal stem cells; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OIM, osteogenic induction medium.
3.8. EN improved the intervention of AKT inhibitor A-443654 in hMSCs
To further validate the action of EN (0.2 μM) on AKT/GSK-3β/β-catenin signaling pathway, AKT inhibitor A-443654 was exposed to hMSCs. As shown in Fig. 9, Fig. 10, EN significantly promoted the differentiation, mineralization and the expression of osteoblasts-related genes (COL1, RUNX2, SP7 and OCN), even the levels of COL1, RUNX2, p-AKT/AKT, p-GSK-3β (ser9)/GSK-3β and β-catenin. Nevertheless, A-443654 significantly reverse the above phenomena, markedly inhibit the osteogenesis of EN in hMSCs, which indicate that EN may play an osteogenic role through the activation of AKT/GSK-3β/β-catenin signaling pathway.
Fig. 9.
Effects of EN on the intervention with AKT inhibitor A-443654 in hMSCs. (a–b) ALP staining and activity assays were measured after treating hMSCs with EN (0.2 μM) and A-443654 (41 nM) for 7 days. (c–d) ARS was performed on day 28. (e) After exposed to EN and A-443654 for 14 days, the mRNA expression levels of osteoblast-related genes (COL1, RUNX2, SP7, and OCN) in hMSCs were detected. (f-g) The protein expression levels of COL1 and RUNX2 were detected by western blotting on day 14. ##P < 0.01, ###P < 0.001 vs. Con; ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM; ◇P < 0.05, ◇◇P < 0.01, ◇◇◇P < 0.001 vs. A-443654; ◆P < 0.05, ◆◆P < 0.01, ◆◆◆P < 0.001 vs. EN. The data are shown as the means ± SD (n = 3). EN, eurycomanone (p-) AKT (phosphorylated-) protein kinase B; hMSCs, human mesenchymal stem cells; ALP, alkaline phosphatase; ARS, alizarin red S; COL1, collagen type I; RUNX2, runt-related transcription factor 2 (p-) GSK-3β (ser9) (phosphorylated-) glycogen synthase kinase 3β at ser9; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; Con, Control; OIM, osteogenic induction medium. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Fig. 10.
Effect of EN on AKT/GSK-3β/β-catenin pathway at the present of AKT inhibitor A-443654 in hMSCs. (a) Protein levels of AKT/GSK-3β/β-catenin pathway in hMSCs were detected by western blotting on day 14: (b) β-catenin/GAPDH, p-GSK-3β (ser9)/GSK-3β, p-AKT/AKT. The data are shown as the means ± SD (n = 3). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001 vs. OIM; ◇◇◇P < 0.001 vs. A-443654; ◆◆P < 0.01, ◆◆◆P < 0.001 vs. EN. EN, eurycomanone (p-) AKT (phosphorylated-) protein kinase B (p-) GSK-3β (ser9) (phosphorylated-) glycogen synthase kinase 3β at ser9; hMSCs, human mesenchymal stem cells; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; OIM, osteogenic induction medium.
4. Discussion
Osteoporosis is a chronic disease that endangers bone health [34]. The development of anti-osteoporosis drugs, especially anabolic agents, has become a hot topic at home and abroad. Commonly used drugs in clinic for promoting bone formation have different side effects and simultaneously bring economic burden to patients [35]. Natural products from plants are active ingredients with a variety of pharmacological effects, which are performed to prevent and treat various diseases [36]. At present, various studies have proved that plant drugs have active anti-osteoporosis effects, such as Cistanche deserticola (total glycosides and polysaccharides) [37], Astragalus [38], Hibicus syriacus L. [39], Piper betleleaves [40] and Fructus Ligustri Lucidi [41]. Hence, seeking novel bone formation drugs through natural products represents a promising treatment strategy of OP in clinical, since whom are multi-component, more safety and possess extensive targets. In this study, EN is a diterpenoid compound isolated from the roots of E. longifolia. E. longifolia have proven to be a medical plant employed in the prevention and treatment of androgen deficiency osteoporosis [14], but it's not clear which monomer is responsible for the beneficial effects. According to this, we extracted ten quassinoids from E. longifolia, and we initially found that five aqueous had a positive effect on bone formation in previous studies, while the exact mechanism is unknown. We choose EN for further experiments on account of EN showed the strongest activity among them. Herein, we explored the effects of EN on bone mineralization in zebrafish larvae and osteogenic differentiation potential in hMSCs and C3H10 cells, and further explored its mechanism of action with the interference of AKT inhibitor A-443654.
In this study, our preliminary results showed that the existing of EN contributed to the increase of mineralization and IOD of skull in zebrafish larvae by ARS staining, suggesting that EN has a strong osteogenic effect on zebrafish larvae skull. Besides, further PCR study found that EN dramatically improve the expression levels of osteogenic genes (COL 1、RUNX2、SP7 and OCN) in zebrafish larvae. COL1 is a crucial protein of bone formation in early stage [42]. RUNX2 is responsible for proliferation and differentiation of early osteoblasts, which involved in the expression of SP7 [43] and directly regulates OCN [44]. SP7 is an essential transcription factor in osteogenic differentiation [45]. OCN, a marker of late osteogenic differentiation, promotes the formation of mineralized nodules, ultimately completing the process of osteogenic differentiation [46]. Thus, the above genes play significant contributory roles in the process of osteogenesis, and our results suggested that EN promote bone formation may be via activating the transcription of osteoblast-related genes.
Dex, a kind of glucocorticoids (GCs), as strong anti-inflammatory agent and immunosuppressant, which administrated with long-term or high dose results in secondary osteoporosis. Research has shown that the GCs-induced zebrafish bone loss was rapidly applied to the researches of osteoporosis in 2006 [18], whose main nosogenesis is bone formation disorder [23]. In this study, Dex in the concentration of 10 μM showing a strong inhibition on mineralization area and IOD in zebrafish. Our previous research found that treated with Dex (5 μM–20 μM) for 6 days, the mineralization area and IOD decreased in a concentration dependent manner, while among them, 10 μM Dex is the optimal experimental concentration to induce the establishment of bone formation inhibition of zebrafish larvea, because it can not only significantly inhibit the skull formation of zebrafish larvae, but has no observably effect on its activity [32] It's astoundingly that Dex observably increased the mRNA expression of osteoclast-specific genes (CTSK, RANKL, NFATC1, and TRAF6), but also surprisingly increased the levels of osteogenic genes except OCN. Whereas EN markedly altered the changes of the phenomenon. We further found that Dex-induced bone loss zebrafish larvae that had been treated with EN exhibited an elevation in expressions of osteogenic genes (COL 1, RUNX2, SP7, and OCN), which also showed the reduction levels of osteoclast-related genes (CTSK, RANKL, NFATC1, and TRAF6). It's noteworthy that, EN exhibited more potent effects than Dex in osteogenic gene expression and not surprisingly down-regulated osteoclast genes. Nevertheless, the most puzzling issue is the elevated expression of osteogenic genes caused by Dex, which is not in line with our expectations. Hence, it is necessary to discuss the effects of GCs on osteoblasts in zebrafish larvae. Previous research has reported that stimulation by GCs up-regulated the expression of osteogenic genes in the early stage of prednisone-induced osteoporosis (consistent with our results), while an opposite trend was observed in later stages [47]. In other words, the increased expression of osteogenic genes observed in the early stage may be a compensatory effect following exposure to Dex. This suggests that treatment with Dex may induce significantly different gene expression levels in various developmental stages of zebrafish larvae. We therefore speculated that, Dex simultaneously promoted both osteogenic and osteoclast genes expression in the early osteogenic differentiation process of zebrafish larvae. But after osteoclast activation, the enhancement of bone resorption activity is far greater than the rate of bone formation, resulting in a greatly increased bone conversion rate, followed by the inhibition of bone formation and aggravated bone loss. Due to this results, we draw a conclusion that EN has efficacious effect on inhibition of bone loss, which may be a candidate drug for OP.
Network pharmacology is a subject with systems biology [48], which has become an effective adjunct tool in recent years, assisting researchers to understand the mechanisms of drugs through systematic pharmacological analysis [49]. This new approach is important in drug discovery, and particularly suitable for research on multi-component herbal medicine [50]. Therefore, researchers can analyze the relationship between drugs and diseases through network pharmacology, so as to predict the mechanism of action of drugs, and providing a new approach to exploring drug targets. Undoubtedly, we further performed a network pharmacologic analysis to explore the mechanism of EN promoting bone formation. Identification and validation of target genes of EN in OP showed that EN is close related to SRC, MAPK1, and AKT1, which are the top three genes in the protein–protein interaction network. Additionally, GO function annotation and KEGG pathway enrichment analysis showed that the MAPK and PI3K-AKT signaling pathways are closely associated with the 10 hub genes (especially AKT1), indicating that they may be key signaling of EN in promoting osteogenic differentiation. Considering the high frequency of AKT1 and the closely relationship between AKT1 and bone related signaling pathways, we ultimately targeted AKT to choose the AKT/GSK-3β/β-catenin signaling pathway and verify in C3H10 cells and hMSCs. AKT (a downstream target of PI3K), namely protein Kinase B, is a protein serine/threonine kinase, including three differentially encoded subtypes: AKT1, AKT2, and AKT3, whose activity is regulated by phosphorylation and interacting proteins [51]. While the phosphorylation of AKT (p-AKT) affects a serious of activities in cells, especially cell growth, proliferation, differentiation, and motility [52]. Nowadays, increasing evidence has confirmed that AKT is an indispensable target for bone formation, and AKT1 is the major AKT isoform in bone [52]. Previous studies have proven that mice with AKT1 knockout exhibited of bone formation disorders, and AKT1 deficiency similarly inhibited osteoblast activities [53]. Surprisingly, natural product of Astragaloside IV (AS-IV) contributes to the coupling of osteogenesis and angiogenesis in BMSCs through AKT/GSK-3β/β-catenin pathway [54]. So, we speculate that AKT/GSK-3β/β-catenin pathway may be an important target pathway for natural products to promoting bone formation.
As we expected, the effects of EN for osteogenesis were evident both in C3H10 cells and hMSCs. Our results demonstrated that EN significantly promoted the proliferation, osteogenic differentiation and mineralization, effectively increased the levels of mRNA and protein of osteoblast-related key markers (COL1, RUNX2, SP7, and OCN) in C3H10 cells and hMSCs. However, it seems that EN has a stronger osteogenic ability to hMSCs than C3H10 cells, possibly due to the stronger differentiation ability of primary hMSCs from 3 to 6 generations compared with C3H10 cells. Further western blotting analysis showed that EN remarkably promoted the ratio of p-AKT/AKT, p-GSK-3β (ser9)/GSK-3β and upregulated the protein expression of β-catenin. From this we speculated that EN may be play a critical role in osteogenesis via AKT/GSK-3β/β-catenin pathway. In support of this view, the same osteogenic experiments were carried out at the present of AKT inhibitor A-443654, and we found that A-443654 effectively reduced the osteogenic promotion of EN in hMSCs. As a cell-signaling growth factor, AKT plays a critical role in stimulating central nodes downstream of cytokines and other cells [55]. The activation of p-AKT further phosphorylates GSK-3β at ser9 (GSK-3β inactivation), thereby the prevention of the phosphorylation and degradation of β-catenin and facilitating entry into the nucleus [56]. Taken together, our above findings suggested that EN expedited osteogenic differentiation by activating the AKT/GSK-3β/β-catenin signaling pathway (Fig. 11).
Fig. 11.
Diagram of hypothesized molecular effects of EN contributing to bone.
Above all, we have preliminarily demonstrated the effect of EN that stimulates bone mineralization of zebrafish larvae and osteogenic differentiation of mesenchymal stem cells. Nonetheless, there are some limitations of this study. On the one hand, there is no experiment in cellular study by using Dex or Dex + EN, as well as no data to show the effect of EN on osteoclastogenesis in vitro though we have result in zebrafish. On the other hand, contradictory effect of Dex in osteogenic markers and mineralization. In this zebrafish larvae study, we think that Dex inhibited mineralization, but promote osteogenic differentiation. This contradictory effect could be one of the limitations regarding Dex-induced bone loss, which will be addressed thoroughly in mice in our further study.
5. Conclusion and future perspectives
Collectively, our study demonstrated that EN promoted bone mineralization and ameliorated the inhibitory effect induced by Dex in zebrafish larvae, which may be attributed to the up-regulation of AKT/GSK-3β/β-catenin pathway in stem cells during osteogenic differentiation. Our study highlights EN as a potential anabolic agent in bone formation, which sheds light on a new drug for preventing OP in clinical.
Credit author statement
YTZ and HBL conceived and designed the experiments. ZQY performed the integrated network pharmacology analysis. LC and XW guided the experimental design and contributed to the data analysis. LC provide research funding. YTZ, HBL, LMK, JXL, and JYH conducted the experiments. YTZ and ZQY wrote the manuscript. LC reviewed the manuscript. XW provided the eurycomanone. HSJ and BW provided the hMSCs. YTZ, HBL, and ZQY contributed equally to this work and share first authorship.
Declaration of competing interest
All authors disclosed no relevant relationship, and no potential conflict of interest was reported by the authors.
Acknowledgements
This work was funded by the Science and Technology Plan Project of Guangdong Province, China (grant no. 2020B151520052), and Discipline Construction Project of Guangdong Medical University, China (grant no. 4SG23002G).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jot.2023.05.006.
Contributor Information
Xin Wu, Email: woodbirdlhb@163.com.
Liao Cui, Email: cuiliao@163.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Esfahanian V., Shamami M.S., Shamami M.S. Relationship between osteoporosis and periodontal disease: review of the literature. J Dent. 2012;9(4):256–264. [PMC free article] [PubMed] [Google Scholar]
- 2.Anastasilakis A.D., Polyzos S.A., Makras P. Therapy of endocrine disease: denosumab vs bisphosphonates for the treatment of postmenopausal osteoporosis. Eur J Endocrinol. 2018;179(1):R31–R45. doi: 10.1530/EJE-18-0056. [DOI] [PubMed] [Google Scholar]
- 3.Lewiecki E.M. Safety of long-term bisphosphonate therapy for the management of osteoporosis. Drugs. 2011;71(6):791–814. doi: 10.2165/11585470-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 4.Haas A.V., LeBoff M.S. Osteoanabolic agents for osteoporosis. J Endocr Soc. 2018;2(8):922–932. doi: 10.1210/js.2018-00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalyan S. Romosozumab treatment in postmenopausal osteoporosis. N Engl J Med. 2017;376(4):395–396. doi: 10.1056/NEJMc1615367. [DOI] [PubMed] [Google Scholar]
- 6.Martin T.J., Seeman E. Abaloparatide is an anabolic, but does it spare resorption? J Bone Miner Res. 2017;32(1):11–16. doi: 10.1002/jbmr.3042. [DOI] [PubMed] [Google Scholar]
- 7.Tanaka T., Takao-Kawabata R., Takakura A., Shimazu Y., Nakatsugawa M., Ito A., et al. Teriparatide relieves ovariectomy-induced hyperalgesia in rats, uggesting the involvement of functional regulation in primary sensory by PTH-mediated signaling. Sci Rep. 2020;10(1):5346. doi: 10.1038/s41598-020-62045-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Subbiah V., Madsen V.S., Raymond A.K., Benjamin R.S., Ludwig J.A. Of mice and men: divergent risks of teriparatide-induced osteosarcoma. Osteoporos Int. 2010;21(6):1041–1045. doi: 10.1007/s00198-009-1004-0. [DOI] [PubMed] [Google Scholar]
- 9.Cs M., George A., Ashok G., Choudhary Y.K. Effect of herbal extract Eurycoma longifolia (Physta((R))) on female reproductive hormones and bone biochemical markers: an ovariectomised rat model study. BMC Complement Med Ther. 2020;20(1):31. doi: 10.1186/s12906-020-2814-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohd E.N., Mohamed N., Muhammad N., Naina M.I., Shuid A.N. Eurycoma longifolia: medicinal plant in the prevention and treatment of male osteoporosis due to androgen deficiency. Evid Based Complement Alternat Med. 2012;2012:125761. doi: 10.1155/2012/125761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teh C.H., Murugaiyah V., Chan K.L. Developing a validated liquid chromatography-mass spectrometric method for the simultaneous analysis of five bioactive quassinoid markers for the standardization of manufactured batches of Eurycoma longifolia Jack extract as antimalarial medicaments. J Chromatogr A. 2011;1218(14):1861–1877. doi: 10.1016/j.chroma.2011.02.014. [DOI] [PubMed] [Google Scholar]
- 12.Li Z., Ruan J.Y., Sun F., Yan J.J., Wang J.L., Zhang Z.X., et al. Relationship between structural characteristics and plant sources along with pharmacology research of quassinoids. Chem Pharm Bull (Tokyo) 2019;67(7):654–665. doi: 10.1248/cpb.c18-00958. [DOI] [PubMed] [Google Scholar]
- 13.Thu H.E., Mohamed I.N., Hussain Z., Jayusman P.A., Shuid A.N. Eurycoma Longifolia as a potential adoptogen of male sexual health: a systematic review on clinical studies. Chin J Nat Med. 2017;15(1):71–80. doi: 10.1016/S1875-5364(17)30010-9. [DOI] [PubMed] [Google Scholar]
- 14.Shuid A.N., El-arabi E., Effendy N.M., Razak H.S., Muhammad N., Mohamed N., et al. Eurycoma longifolia upregulates osteoprotegerin gene expression in androgen- deficient osteoporosis rat model. BMC Compl Alternative Med. 2012;12:152. doi: 10.1186/1472-6882-12-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao H.B., Zhong Y.T., Zhou D.H., Xie Q.J., Zhang Z.P., Wu Y.M., et al. Quassinoids from Eurycoma longifolia and their bone formation evaluation inzebrafish, C3H10 cells and silico. Chem Biol Interact. 2022:367. doi: 10.1016/j.cbi.2022.110140. [DOI] [PubMed] [Google Scholar]
- 16.Low B.S., Choi S.B., Abdul W.H., Das P.K., Chan K.L. Eurycomanone, the major quassinoid in Eurycoma longifolia root extractincreases spermatogenesis by inhibiting the activity of phosphodiesterase and aromatase in steroidogenesis. J Ethnopharmacol. 2013;149(1):201–207. doi: 10.1016/j.jep.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 17.Lahrita L., Hirosawa R., Kato E., Kawabata J. Isolation and lipolytic activity of eurycomanone and its epoxy derivative from Eurycoma longifolia. Bioorg Med Chem. 2017;25(17):4829–4834. doi: 10.1016/j.bmc.2017.07.032. [DOI] [PubMed] [Google Scholar]
- 18.Barrett R., Chappell C., Quick M., Fleming A. A rapid, high content, in vivo model of glucocorticoid-induced osteoporosis. Biotechnol J. 2006;1(6):651–655. doi: 10.1002/biot.200600043. [DOI] [PubMed] [Google Scholar]
- 19.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496(7446):498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tonelli F., Bek J.W., Besio R., De Clercq A., Leoni L., Salmon P., et al. Zebrafish: a resourceful vertebrate model to investigate skeletal disorders. Front Endocrinol. 2020;11:489. doi: 10.3389/fendo.2020.00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dietrich K., Fiedler I.A., Kurzyukova A., López-Delgado A.C., McGowan L.M., Geurtzen K., et al. Skeletal biology and disease modeling in zebrafish. J Bone Miner Res. 2021;36(3) doi: 10.1002/jbmr.4256. [DOI] [PubMed] [Google Scholar]
- 22.Kwon R.Y., Watson C.J., Karasik D. Using zebrafish to study skeletal genomics. Bone. 2019;126:37–50. doi: 10.1016/j.bone.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luderman L.N., Unlu G., Knapik E.W. Zebrafish developmental models of skeletal diseases. Curr Top Dev Biol. 2017;124:81–124. doi: 10.1016/bs.ctdb.2016.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Veldhuis-Vlug A.G., Rosen C.J. Clinical implications of bone marrow adiposity. J Intern Med. 2018;283(2):121–139. doi: 10.1111/joim.12718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karsenty G. Transcriptional control of skeletogenesis. Annu Rev Genom Hum Genet. 2008;9:183–196. doi: 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- 26.Lian F., Zhao C., Qu J., Lian y, Cui Y., Shan L., et al. Icariin attenuates titanium particle-induced inhibition of osteogenic differentiation and matrix mineralization via miR-21-5p. Cell Biol Int. 2018;42(8):931–939. doi: 10.1002/cbin.10957. [DOI] [PubMed] [Google Scholar]
- 27.Liu H., Yi X., Tu S.T., Cheng C., Luo J. Kaempferol promotes BMSC osteogenic differentiation and improves osteoporosis by downregulating miR-10a-3p and upregulating CXCL12. Mol Cell Endocrinol. 2021;520 doi: 10.1016/j.mce.2020.111074. [DOI] [PubMed] [Google Scholar]
- 28.Zhao F., Ma X.L., Qiu W.X., Wang P., Zhang R., Chen Z.H., et al. Mesenchymal MACF1 facilitates SMAD7 nuclear translocation to drive. Bone Formation. Cells. 2020;9(3) doi: 10.3390/cells9030616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X., Zhou G. Protective effects of naringin on glucocorticoid-induced osteoporosis through regulating the PI3K/Akt/mTOR signaling pathway. Am J Transl Res. 2021;13(6):6330–6341. [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Z.P., Zhang Z.F., Yang Y.F., Yang Y. Sesamin promotes osteoblastic differentiation and protects rats from osteoporosis. Med Sci Mon Int Med J Exp Clin Res. 2019;25:5312–5320. doi: 10.12659/MSM.915529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying J., Ge Q., Hu S., Luo C., Lu F., Yu Y., et al. Amygdalin promotes fracture healing through TGF-beta/smad signaling in mesenchymal stem cells. Stem Cell Int. 2020;2020 doi: 10.1155/2020/8811963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo S., Yang Y., Chen J., Zhong Z., Huang H., Zhang J., et al. Tanshinol stimulates bone formation and attenuates dexamethasone-induced inhibition of osteogenesis in larval zebrafish. J Orthop Translat. 2016;4:35–45. doi: 10.1016/j.jot.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Chen Z., Liao H., Zhong Y., Hua J., Su M., et al. Anti-osteogenic effect of danshensu in ankylosing spondylitis: an in vitro study based on integrated network pharmacology. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.772190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang X.X., Chen W.K., Gu C., Liu H., Hou M.Z., Qin W.J., et al. Melatonin suppresses bone marrow adiposity in ovariectomized rats by rescuing the imbalance between osteogenesis and adipogenesis through SIRT1. Journal of orthopaedic translation. 2023:38. doi: 10.1016/j.jot.2022.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skjødt M.K., Frost M., Abrahamsen B. Side effects of drugs for osteoporosis and metastatic bone disease. British journal of clinical pharmacology. 2019;85(6) doi: 10.1111/bcp.13759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leung P.C., Siu W.S. Herbal treatment for osteoporosis: a current review. Journal of Traditional and Complementary Medicine. 2013;3(2) doi: 10.4103/2225-4110.110407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang F., Tu P., Zeng K., Jiang Y. Total glycosides and polysaccharides of Cistanche deserticola prevent osteoporosis by activating Wnt/β-catenin signaling pathway in SAMP6 mice. J Ethnopharmacol. 2021;271 doi: 10.1016/j.jep.2021.113899. [DOI] [PubMed] [Google Scholar]
- 38.Sun N.Y., Liu X.L., Gao J., Wu X.H., Dou B. AstragalosideIV modulates NGF-induced osteoblast differentiation via the GSK-3β/β-catenin signalling pathway. Mol Med Rep. 2021;23(1) doi: 10.3892/mmr.2020.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karunarathne W., Molagoda I., Lee K.T., Choi Y.H., Jin C.Y., Kim G.Y. Anthocyanin-enriched polyphenols from Hibiscus syriacus L. (Malvaceae) exert anti-osteoporosis effects by inhibiting GSK-3β and subsequently activating β-catenin. Phytomedicine. 2021;91 doi: 10.1016/j.phymed.2021.153721. [DOI] [PubMed] [Google Scholar]
- 40.Mishra R., Das N., Varshney R., Juneja K., Sircar D., Roy P. Food Funct; 2021. Betel leaf extract and its major component hydroxychavicol promote osteogenesis and alleviate glucocorticoid-induced osteoporosis in rats. [DOI] [PubMed] [Google Scholar]
- 41.Liu H., Guo Y., Zhu R., Wang L., Chen B., Tian Y., et al. Fructus Ligustri Lucidi preserves bone quality through induction of canonical Wnt/β- catenin signaling pathway in ovariectomized rats. Phytother Res. 2021;35(1):424–441. doi: 10.1002/ptr.6817. [DOI] [PubMed] [Google Scholar]
- 42.Lepe-Balsalobre E., Santotoribio J.D., Nunez-Vazquez R., Garcia-Morillo S., Jimenez-Arriscado P., Hernandez-Arevalo P., et al. Genotype/phenotype relationship in Gaucher disease patients. Novel mutation in glucocerebrosidase gene. Clin Chem Lab Med. 2020;58(12):2017–2024. doi: 10.1515/cclm-2020-0306. [DOI] [PubMed] [Google Scholar]
- 43.Molagoda I., Karunarathne W., Choi Y.H., Park E.K., Jeon Y.J., Lee B.J., et al. Fermented oyster extract promotes osteoblast differentiation by activating the wnt/beta-catenin signaling pathway, leading to bone formation. Biomolecules. 2019;9(11) doi: 10.3390/biom9110711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsubara T., Kida K., Yamaguchi A., Hata K., Ichida F., Meguro H., et al. BMP2 regulates Osterix through Msx2 and Runx2 during osteoblast differentiation. J Biol Chem. 2008;283(43):29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carnovali M., Ottria R., Pasqualetti S., Banfi G., Ciuffreda P., Mariotti M. Effects of bioactive fatty acid amide derivatives in zebrafish scale model of bone metabolism and disease] Pharmacol Res. 2016;104:1–8. doi: 10.1016/j.phrs.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 46.Komori T. Functions of osteocalcin in bone, pancreas, testis, and muscle. Int J Mol Sci. 2020;21(20) doi: 10.3390/ijms21207513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He H., Wang C., Tang Q., Yang F., Xu Y. Possible mechanisms of prednisolone-induced osteoporosis in zebrafish larva. Biomed Pharmacother. 2018;101:981–987. doi: 10.1016/j.biopha.2018.02.082. [DOI] [PubMed] [Google Scholar]
- 48.Yang L., Li H., Yang M., Zhang W., Li M., Xu Y., et al. Exploration in the mechanism of kaempferol for the treatment of gastric cancer based on network pharmacology. BioMed Res Int. 2020;2020 doi: 10.1155/2020/5891016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang W., Bai Y., Wang Y., Xiao W. Polypharmacology in drug discovery: a review from systems pharmacology perspective. Curr Pharmaceut Des. 2016;22(21):3171–3181. doi: 10.2174/1381612822666160224142812. [DOI] [PubMed] [Google Scholar]
- 50.Zhai L., Ning Z.W., Huang T., Wen B., Liao C.H., Lin C.Y., et al. Cyclocarya paliurus leaves tea improves dyslipidemia in diabetic mice: a lipidomics-based network pharmacology study] Front Pharmacol. 2018;9:973. doi: 10.3389/fphar.2018.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gu Z., Wang L., Yao X., Long Q., Lee K., Li J., et al. ClC-3/SGK1 regulatory axis enhances the olaparib-induced antitumor effect in human stomach adenocarcinoma. Cell Death Dis. 2020;11(10):898. doi: 10.1038/s41419-020-03107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Q.W., Cheng C., Hackett C., Feldman M., Houseman B.T., Nicolaids T., et al. Akt and autophagy cooperate to promote survival of drug-resistant glioma. Sci Signal. 2010;3(147) doi: 10.1126/scisignal.2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kawamura N., Kugimiya F., Oshima Y., Ohba S., Lkeda T., Saito T., et al. Akt1 in osteoblasts and osteoclasts controls bone remodeling. PLoS One. 2007;2(10) doi: 10.1371/journal.pone.0001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang F., Qian H.J., Kong L.C., Wang W.B., Wang X.Y., Xu Z., al Wet. Accelerated bone regeneration by Astragaloside IV through stimulating the coupling of osteogenesis and angiogenesis. Int J Biol Sci. 2021;17(7):1821–1836. doi: 10.7150/ijbs.57681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manning B.D., Cantley L.C. AKT/PKB signaling: navigating downstream. Cell. 2007;129(7):1261–1274. doi: 10.1016/j.cell.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng W., Yao M., Wu M., Yang J., Yao D., Wang L. Secretory clusterin promotes hepatocellular carcinoma progression by facilitating cancer stem cell properties via AKT/GSK-3beta/beta-catenin axis. J Transl Med. 2020;18(1):81. doi: 10.1186/s12967-020-02262-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.