Abstract
Background
Immune cell activation in early sepsis is beneficial to clear pathogens, but immune cell exhaustion during the inflammatory response induces immunosuppression in sepsis. Here, we studied the relationship between immune cell survival status and the prognosis of sepsis patients.
Methods
Sepsis patients admitted to our hospital with a diagnosis time of less than 24 h were recruited. RNA sequencing technologies were used to study functional alterations in various immune cells in peripheral blood mononuclear cells (PBMCs) from sepsis patients. Flow cytometry and electron microscopy were performed to study cell apoptosis and morphological alterations.
Results
A total of 68 sepsis patients with complete data were enrolled and divided into survival (45 patients) and death (23 patients) groups according to their prognosis. Patients in the death group had significantly increased lactic acid levels compared with those in the survival group, but there was no significant difference in other physiological and coagulation functional indicators between the two groups. Bulk RNA sequencing showed that cell death-related pathways and biomarkers were highly enriched and activated in the PBMCs of the death group than that in the survival group. Signs of mitochondrial damage, autophagosomes, cell surface damage and cell surface pore forming were also more pronounced in PBMCs from the death group under electron microscopy. Further single-cell RNA sequencing revealed that cell death occurred mainly in myeloid cells rather than lymphocytes at the early stage of sepsis; cell death patterns of destructive necrosis and pyroptosis were predominant in neutrophils, and apoptosis, autophagy and ferroptosis with less damage to the surroundings were predominant in monocytes.
Conclusion
Cell death mainly occurs in monocytes and neutrophils in the PBMCs of sepsis at the early stage. The study provides a perspective for the immunotherapy of early sepsis targeting immune cell death.
Keywords: Sepsis, Immune cells, PBMC, Single-cell RNA sequencing, Cell death
1. Introduction
Sepsis is a leading public health concern affecting nearly 20 million people annually worldwide [1]. The definition of sepsis has been updated as life-threatening organ dysfunction caused by a dysregulated host response to infection [2]. A global statistical study showed that the leading causes of sepsis were lower respiratory infections and diarrhoeal diseases [3]. Although the mortality of sepsis has decreased over the years, it remains high due to the diversity of aetiology and susceptible populations, accounting for 1/5 of global deaths in 2017 alone [3]. Meanwhile, sepsis has a poor prognosis, and even if patients survive, they may have severe cognitive impairment and a high risk of recurrent infections [1]. As sepsis is featured by the disequilibrium of hyper-inflammation and immune suppression [4], research on immune dysregulation during sepsis provides more directions or strategies for the prevention and treatment of sepsis.
The immune trajectory during sepsis indicated that the hyper-inflammatory response was the strongest at the early stage of hospitalization [4]. Although many cells and indicators are involved in the regulatory network of sepsis, leukocytes are undoubtedly predominant. Functional and phenotypic alterations of the cells implicated in innate and adaptive immune systems are decisive for the progression of sepsis [5,6]. Using a septic mouse model, studies demonstrated the heterogeneity of neutrophils at the late stage of sepsis, such as in suppression of apoptosis, excessive tissue infiltration and disordered chemotaxis [7]. Decreased expression of human leukocyte antigen-DR (HLA-DR) on circulating monocytes is considered a characteristic event in sepsis [8]. Specific cell surface markers of neutrophils and monocytes are also largely altered in sepsis patients [5]. The changes of T, B and natural killer (NK) lymphocyte numbers in sepsis depend on the origin of the infected bacteria [9]. Immune cells during sepsis have a dynamic change induced by a variety of external and intrinsic factors, which play crucial roles in the occurrence and development of sepsis [10].
The battle between the pathogen and host immune system in sepsis is a race against life. Although numerous immunotherapies targeting host immune response or its related biomarkers for sepsis are under investigation [5,6], there is no specific therapeutic way or medicine for sepsis treatment to date. Sepsis-induced immune cell apoptosis is closely related to immunosuppression, high risks of recurrent infection and poor outcomes in sepsis patients [11,12]. Extensive immune cell death during sepsis further contributes to multiple organ dysfunction syndrome; however, certain cell death types and their related mechanism are far from clear [13]. In this study, using bulk-RNA and single-cell RNA sequencing of peripheral blood mononuclear cells (PBMCs) from sepsis patients, we studied the major cell death types in various immune cells in sepsis at the early stage and provided a perspective for immunotherapy of sepsis focusing on a specific immune cell death pattern.
2. Methods
2.1. Ethics statement
The study was approved by the Ethical Committee of the Xijing Hospital of Fourth Military Medical University, with document number KY20212172 and NCT05229328. All procedures performed relating to human participants complied with the Declaration of Helsinki. Informed consent was signed by all subjects.
2.2. Study population
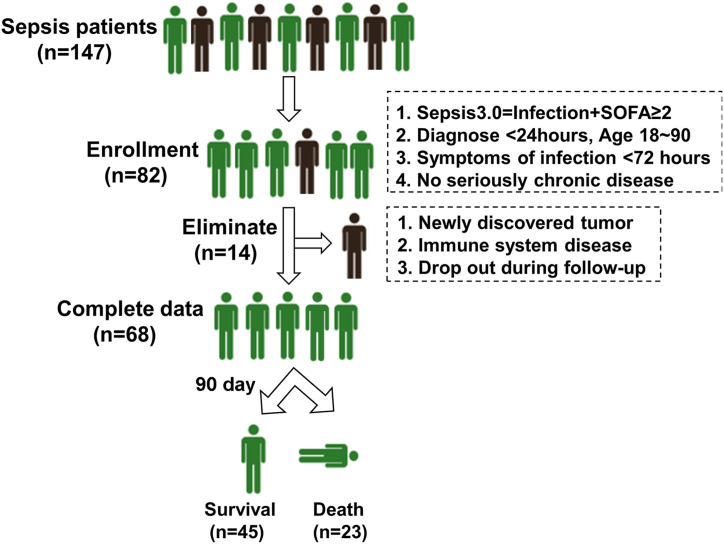
A total of 147 sepsis patients who were admitted to the emergency department and intensive care unit in Xijing Hospital from December 2021 to May 2022 were recruited into this study. The study design is shown in Fig. 1. The inclusion criteria for sepsis patients were Sepsis 3.0 = Infection + SOFA ≥2; symptoms of infection, such as fever <72 h; diagnosis time <24 h (sepsis at the early stage), untreated; age 18–90 with signed informed consent; and no history of seriously chronic diseases. Patients who had newly discovered tumours or immune system diseases or dropped out during follow-up (90 days) were excluded. Sixty-eight patients with complete data were finally enrolled, and their clinical characteristics were collected, including infection sites (respiratory system, digestive system or urogenital system), physiological indexes (sex, age, temperature, heart rate, respiratory rate, systolic blood pressure [SBP] and mean arterial pressure [MAP]), laboratory indicators (white blood cell [WBC], neutrophil, lymphocyte, platelet, prothrombin time [PT], activated partial thromboplastin time [APTT], total bilirubin, alanine aminotransferase [ALT], aspartate aminotransferase [AST], creatinine, cystatin C and lactic acid), and also disease scores (APACHE II, SOFA, qSOFA and NEWS). The details were summarized in Table 1.
Fig. 1.
Study design. Recruitment and elimination of enrolled sepsis patients.
Table 1.
Clinical characteristics of sepsis patients involved (n = 68).
| Survival (n = 45) | Death (n = 23) | χ2/t/Z | p-value | |
|---|---|---|---|---|
| Infection site | ||||
| Respiratory system n (%) | 15 (33.3) | 8 (34.8) | 0.505 | 0.888 |
| Digestive system n (%) | 21 (46.7) | 12 (52.2) | ||
| Urogenital system n (%) | 9 (20) | 3 (13) | ||
| Physiological index | ||||
| Female n (%) | 20 (44.4) | 8 (34.8) | 0.587 | 0.603 |
| Age year | 64.5 ± 15.3 | 66.5 ± 12.8 | 0.542 | 0.590 |
| Temperature °C | 36.70 (36.5–37.9) | 36.50 (36.4–36.9) | 1.345 | 0.179 |
| HR beats per minute | 101.3 ± 24.4 | 106.8 ± 20.9 | 0.919 | 0.362 |
| RR breaths per minute | 24 (21.5–26.5) | 23 (22–27) | 0.052 | 0.958 |
| SBP mmhg | 105 (89.5–125) | 98 (92–126) | 0.266 | 0.790 |
| MAP mmhg | 80.3 ± 15.9 | 80.9 ± 13.4 | 0.140 | 0.889 |
| Laboratory results | ||||
| WBC × 109/L | 9.8 (7.2–16.1) | 12.5 (6∼23.4) | 0.369 | 0.712 |
| Neutrophil % | 85.9 (80.3–90) | 89.2 (82.2–93.5) | 1.783 | 0.075 |
| Lymphocyte % | 8 (4.5–13.2) | 5.4 (3.3–12.8) | 1.239 | 0.215 |
| Platelet × 109/L | 131 (70.5–186) | 72 (57–181) | 1.582 | 0.114 |
| PT s | 14 (12.9–15.8) | 13.9 (12.5–16.1) | 0.272 | 0.785 |
| APTT s | 35.8 (31.2–39.7) | 40.9 (31.4–44.8) | 1.439 | 0.150 |
| Total bilirubin umol/L | 26 (17.7–42.3) | 33.2 (24.4–79.2) | 1.290 | 0.197 |
| ALT U/L | 30 (19–57) | 34 (19–84) | 0.318 | 0.750 |
| AST U/L | 42 (27∼65.5) | 35 (27–68) | 0.078 | 0.938 |
| Creatinine umol/L | 78 (62–153) | 134 (76–255) | 1.491 | 0.136 |
| Cystatin C mg/L | 1.4 (1.3–2.1) | 1.8 (1.4–3) | 1.789 | 0.074 |
| Lactic acid mmol/L | 1.5 (1∼2.3) | 2.7 (1.3–6.3) | 2.530 | 0.011* |
| Disease score | ||||
| APACHE II | 13 (10–16) | 15 (11–17) | 0.833 | 0.405 |
| SOFA | 6 (5–8) | 9 (6–11) | 2.167 | 0.030* |
| quick SOFA | 2 (2–3) | 2 (2–3) | 0.815 | 0.415 |
| NEWS | 5.9 ± 3 | 7.6 ± 4 | 1.966 | 0.054 |
Abbreviations: HR, heart rate; RR, respiratory rate; SBP, systolic blood pressure; MAP, mean arterial pressure; WBC, white blood cell; PT, prothrombin time; APTT, activated partial prothrombin time; ALT, alanine aminotransferase; AST, aspartate aminotransferase; APACHE II, acute physiology and chronic health evaluation II; SOFA, sequential organ failure assessment; NEWS, national early warning score.
2.3. Blood sample collection
Blood samples were taken to obtain plasma and PBMCs once the subjects agreed to be enrolled. A total of 8 ml venous blood samples were drawn from the cubital vein of patients and healthy volunteers into EDTA vacutainer tubes and processed within 1 h after collection. After transferring the anticoagulant blood samples into Lymphocyte Separation Tube for Human Peripheral Blood (DAKEWE BIOTECH, Shenzhen, China), density gradient sedimentation (400 g, 15 min, 20 °C) was applied to separate blood samples into layers. Following centrifugation, the top layer was plasma, PBMCs were below plasma and the separation liquid was at the bottom. Plasma and PBMC samples were then collected and stored at −80 °C or liquid nitrogen for further use.
2.4. Bulk RNA sequencing
PBMC samples from the first 14 sepsis patients (10 survivals and 4 death) were collected and sent to Gene Denovo Biotechnology Co. (Guangzhou, China) to conduct bulk RNA (both mRNA and microRNA) sequencing. Total RNAs were extracted through TRIzol reagent kit (Invitrogen, Carlsbad, CA, USA). For transcriptome sequencing, Oligo (dT) beads were used to enrich eukaryotic mRNAs that were then broken into fragments and reverse transcribed into cDNAs. After purification, end repairing and poly (A) tail adding, cDNA fragments were ligated to Illumina sequencing adapters and sequenced through Illumina Novaseq 6000. For microRNA sequencing, the RNAs at 18–30 nt were enriched by polyacrylamide gel electrophoresis, followed by addition of 3′ adapters and enrichment again the RNA molecules at 36–44 nt. After ligating 5′ adapters to the RNAs, amplification was conducted and the PCR products at 140–160 bp were selected to construct a cDNA library, which was then sequenced using the Illumina Novaseq 6000 platform.
2.5. Single-cell RNA sequencing
We also randomly collected PBMC samples of three sepsis patients at the early stage and three healthy volunteers to perform single-cell RNA sequencing by Gene Denovo Biotechnology Co. Trypan blue was used to detect sample quantity and cell viability. Chromium (10 × Genomics) instrument was used for single-cell RNA sequencing. Gene libraries were sequenced using the Illumina Novaseq 6000. Moreover, raw gene expression matrices generated using CellRanger (version 5.0.0) in each sample were combined in R (version 3.5.3) mapping to GRCh38 and converted to a Seurat object using the Seurat R package (version 3.0.1). Exclusive criteria for the cells were: >17,000 UMIs, >3700 or <400 expressed genes or >20% UMIs from mitochondrial genome. TSNE with resolution set to 0.5 was applied to study the deviations among all detected cells. Here, we mainly studied the changes in the functional signals of early cell death (Necrosis, Necroptosis, Pyroptosis, Apoptosis, Autophagy and Ferroptosis) in different types of immune cells in sepsis.
2.6. Functional analysis of differentially expressed genes (DEGs)
Differential expression analysis for mRNAs and microRNAs was conducted using DESeq2 software and edgeR software, respectively, between the two groups. The transcripts or microRNAs with the parameters of fold change ≥2 and P value less than 0.05 were regarded as significant DEGs or DE microRNAs (differentially expressed microRNAs). Screened DEGs were mapped to Gene Ontology (GO) terms in the GO database that mainly includes three ontologies: molecular function, cellular component and biological process, to evaluate significantly enriched GO terms of DEGs. Pathway enrichment analysis was achieved by Kyoto Encyclopedia of Genes and Genomes (KEGG) database to identify significantly enriched KEGG annotation of DEGs.
2.7. Luminex assay
Luminex detection was performed to assess the concentration of cytokines and chemokines in plasma using Human XL Cytokine Luminex Performance Panel Premixed Kit (#FCSTM18–30, R&D Systems, Minneapolis, MN, USA). The factors detected were TNF-α, IL-6, IL-1β, CXCL1, CCL2, CCL4, CXCL10, GM-CSF, 1L-1 ra, IL-8, IL-10 and IL-15. Quality control: the standard curve was fitted to a 5-parameter logistic model; coefficient of variance ≤20%; and recovery ≥80%.
2.8. ELISA
ELISA kits for the content detection of plasma apoptosis and pyroptosis-related factors (CASP 1, CASP 8, GSDMD, NFKBIA, BID, BAX, BCL2L1 and BCL2) were purchased from Shanghai JiangLai Biotechnology Co., Ltd., China. All detection was followed the manufacturer instructions.
2.9. Flow cytometry
The cell apoptotic state of PMBC samples was detected using the FITC Annexin V Apoptosis Detection Kit I (#556547, BD Pharmingen, San Diego, CA) according to the manufacturer's instructions. Briefly, cells (1 × 106 cells per group) were resuspended with 1 × binding buffer (100 μl), followed by addition of 5 μl Annexin V-FITC and 10 μl PI. After incubation for 30 min at 4 °C in the dark, the fluorescence in the samples was detected with a COULTER EPICS XL flow cytometer.
2.10. Electron microscopy
For electron microscopy, the PMBC samples were fixed in electron microscopy fixatives (Servicebio, Wuhan, China). Specimens were attached to metallic stubs, sputter-coated with gold for 30 s, and then observed and photographed under the scanning electron microscope (HITACHI SU8100, Tokyo, Japan). For transmission electron microscopy, the cells were pre-embedded in agarose. After dehydration, resin penetration and embedding, the specimens were ultrathin sectioned into 60–80 nm slices with a Leica EM UC7 Ultramicrotome (Wetzlar, Germany) and placed onto 150-mesh cuprum grids with formvar film, followed by uranium acetate and lead citrate staining. The samples were observed under the Transmission Electron Microscope (HITACHI HT7800).
2.11. Western blotting
Protein extraction was performed in RIPA lysis buffer (GenStar, Beijing, China). The protein concentration was quantified with the BCA Protein Assay Kit (Zhonghuihecai Biopharmaceutical Technology, Shaanxi, China). After SDS-PAGE and transferring onto PVDF membranes, the membranes were incubated with diluted primary antibodies and the corresponding secondary ones. Protein bands were visualized using the ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA). ImageJ software was applied for densitometry analyses. Each experiment was repeated at least three times. The primary antibodies used were anti-CASP 3 (#9662 S, Cell Signaling Technology, CST, Shanghai, China). anti-BAX (#2772 S, CST), anti-CASP 5 (#46680 S, CST), anti-cleaved GSDMD (#36425 S, CST), anti-BCL2 (#3498 S, CST) and anti-Beta actin (#66009-I-IG, Proteintech, Wuhan, China).
2.12. Quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
Total RNA was isolated from PBMCs using the TRIzol Reagent kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. RNA was quantified at 260 nm using a spectrophotometer, and only samples with an OD 260/280 ratio between 1.8 and 2.0 were used. First-strand cDNA was synthesized in a volume of 20 μL using 1 μg total RNA and PrimeScript® RT reagent (Takara, Shiga, Japan). MiRNA qRT-PCR Stater Kit (#C107R-1, RiboBio, Guangzhou, China) was used for the RT and qPCR for miRNA. The primer pairs of miRNA were synthesized by RiboBio. Further quantitative PCR reaction was performed using cDNA templates, primers, PCR Mix (#11141ES60, Yeasen), and SYBR Green Mix (#11201ES08, Yeasen). Specific primer pairs were designed by Tsingke Biotechnology (Beijing, China) and are listed in Supplementary Table 1.
2.13. Statistical analysis
The clinical data were collected and recorded by trained staff. IBM SPSS Statistics 28.0 was used for statistical analysis, and GraphPad Prism 9 was used for mapping. The data were evaluated by the Shapiro-Wilk test to determine the normal distribution and were expressed as mean ± standard deviation (SD) or median with quartile. A Student's t-test or Mann–Whitney test was used to compare differences between groups. Qualitative data were represented by the constituent ratio. Fisher's exact probability method was used to compare rates between groups. The threshold for statistical significance was set at p < 0.05.
3. Results
3.1. Clinical characteristics of sepsis patients
A total of 147 sepsis patients were admitted to our hospital, and 82 patients who met the inclusion criteria were enrolled. After follow-up, 14 patients were eliminated and 68 patients with complete data were chosen, including 45 who had survived and 23 who were dead (Fig. 1). To explore the relationship between prognosis and clinical parameters in patients with sepsis, we tested for differences in infection sites, physiological indexes, laboratory indicators, and disease scores between the two groups (Table 1). The results exhibited no significant difference in the infection sites, sex, age, heart rate, respiratory rate, SBP, MAP, immune cells, PT, APTT, total bilirubin, ALT, AST, creatinine and cystatin C of sepsis patients between the survival and death group. However, the lactic acid level in the death group was significantly increased compared with that of survival patients, suggesting more severe cytotoxicity and bodily injury. Meanwhile, the SOFA scores of septic patients who died were significantly higher than those of patients who survived, but the other scoring system showed no significant difference between them.
3.2. Cell death-related pathways are highly activated in the death group of sepsis patients
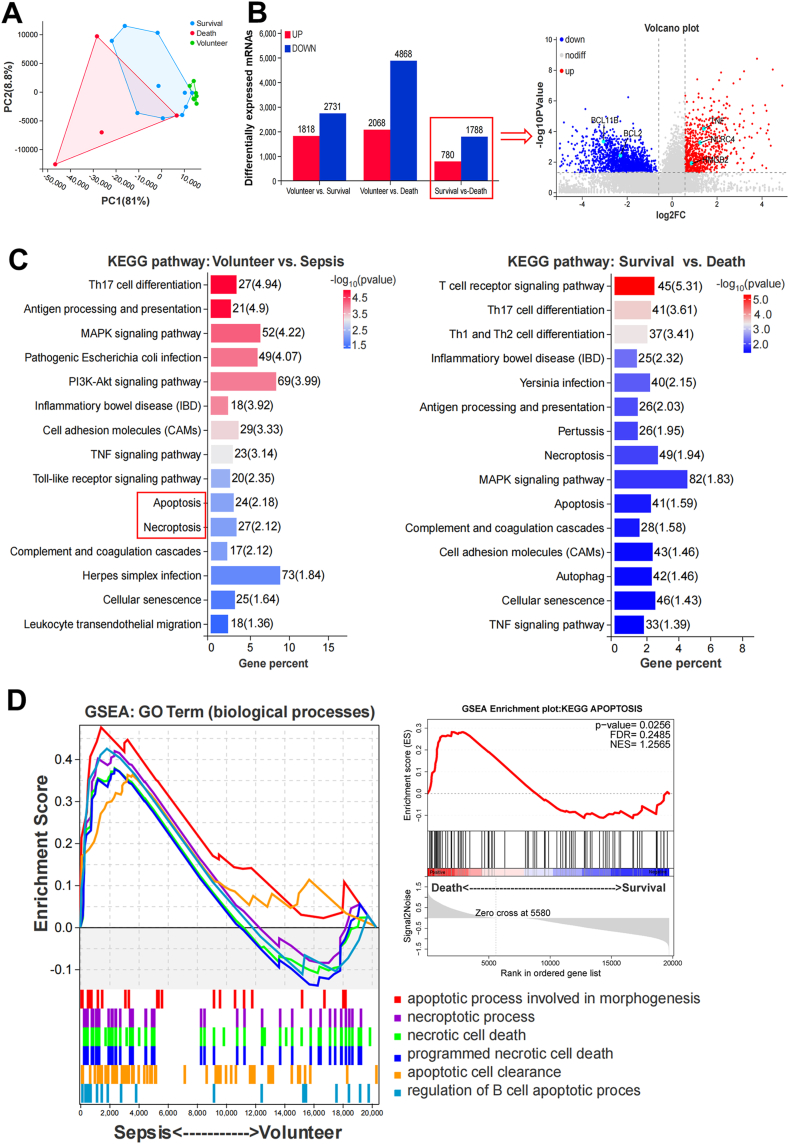
To further study the functional cellular differences in sepsis patients between the survival and death groups, we collected PBMC samples from the first 14 sepsis patients (Table S2) and performed transcriptome sequencing. The PCA plot showed two sample clusters in the survival and death groups (Fig. 2A). There were 2568 DEGs in the death group compared to the survival group, including 780 upregulated and 1788 downregulated genes (Fig. 2B). Significant DEGs between the two groups are shown in a volcano plot. Further functional enrichment analysis of the DEGs between the volunteer and sepsis groups as well as the survival and death groups showed that pathways related to the immune or infection response, apoptotic processes, and cell death were significantly enriched in both KEGG database and GO-biological processes ontology (Fig. 2C, Figure S1). In addition, we conducted KEGG functional enrichment analysis on the upregulated (780) and downregulated (1788) DEGs in deaths compared to those in survivors, and found that upregulated genes were mainly enriched in the pathways of cell cycle, cellular senescence, and cell death-related signaling cascades; moreover, downregulated genes were mainly enriched in the pathways of hematopoietic cell lineage, Th17 cell differentiation, Th1 and Th2 cell differentiation, autoimmune thyroid disease, rheumatoid arthritis, primary immunodeficiency, and systemic lupus erythematosus (Figure S2). We then performed GSEA analysis of several cell death-related pathways from GO-biological processes and found that genesets of cell apoptosis, necrosis and necroptosis were largely enriched in the sepsis group compared to the volunteer group; meanwhile, the geneset of the KEGG apoptosis pathway was also highly enriched in the death group versus the survival group (Fig. 2D).
Fig. 2.
mRNA sequencing of PBMCs from sepsis patients at the early stage (Volunteer n = 7, Survival n = 10, Death n = 4). A Sample correlation via PCA plot. B Bar chart and volcano plot showed a total of 2568 significant DEGs between survival and death groups. C KEGG pathway function enrichment of the screened DEGs (p < 0.05). D GESA indicated that genesets of cell necrosis, necroptosis, and apoptosis were significantly enhanced in the sepsis group (p < 0.05).
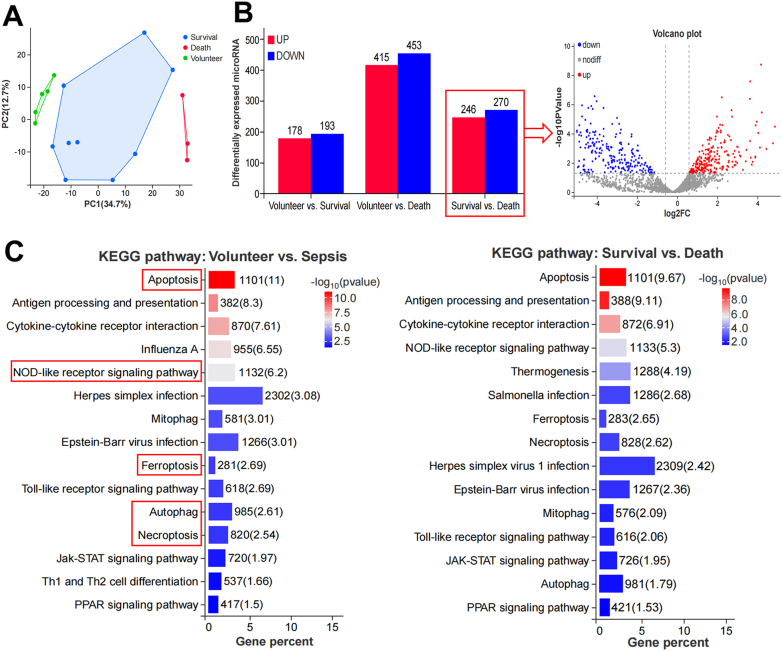
Moreover, we conducted microRNA sequencing of PBMC samples from sepsis patients (survival and death, Table S2) and healthy volunteers. The distinct distribution of the involved samples was shown in the PCA plot (Fig. 3A). Compared to the volunteers, 178 DE microRNAs were upregulated and 193 were downregulated in the survival group and 415 were upregulated and 453 were downregulated in the death group. Compared to the survival group, there were 246 microRNAs upregulated and 270 downregulated in the death group (Fig. 3B). Further KEGG and GO functional analysis of the significant DE microRNAs showed that pathways of cell death, programmed cell death and immune responses were markedly enriched between volunteer vs. sepsis and survival vs. death (Fig. 3C, Figure S3), which agreed with the results of mRNA sequencing. We also withdrew some key regulatory genes and their targeted microRNAs of various cell death pathways from the sequencing results to undergo heatmap expression analysis (Figure S4). Unsurprisingly, pyroptosis-related CASP 4 [14], autophagy-related SAT1 [15] and ferroptosis-related ATG7 [16] were significantly upregulated in the death group, while apoptosis-related BCL2 [17,18] was remarkably downregulated compared to other groups (Figure S4A). Their corresponding microRNAs were oppositely expressed (Figure S4B), all of which were validated by qRT-PCR (Figure S4C). The results here indicate that cell death-related pathways are highly activated in the septic death group compared to the septic survival and healthy volunteers.
Fig. 3.
MicroRNA sequencing of PBMCs from sepsis patients at the early stage (Volunteer n = 5, Survival n = 9, Death n = 3). A Sample correlation between groups. B Bar chart and volcano plot showed 516 significant DE microRNAs between survival and death groups. C KEGG pathway functional enrichment analysis of the screened DE microRNAs (p < 0.05).
3.3. Morphological alteration of PBMCs from sepsis patients in the survival and death groups
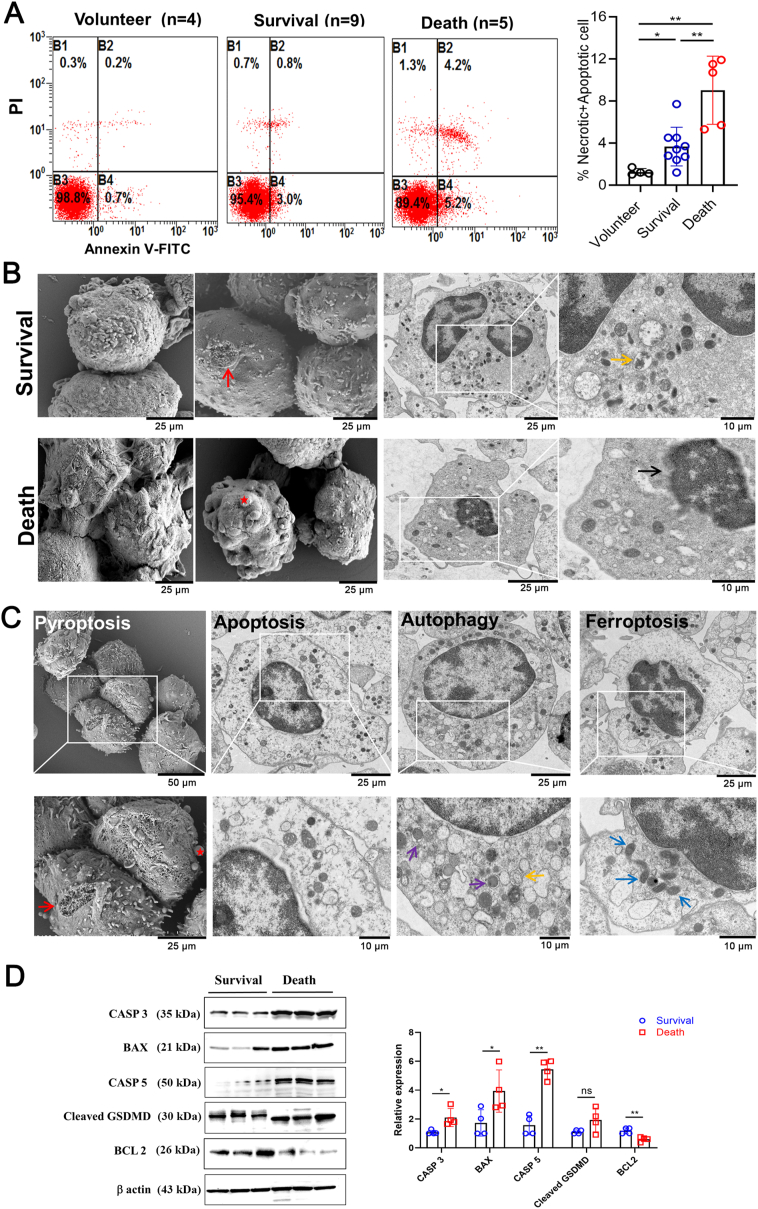
In order to validate the changes in the activation of cell death-related pathways among groups, we then carried out flow cytometry of PBMCs to detect cell necrotic and apoptotic death. The results showed that the percentage of apoptotic and necrotic cells were significantly increased in the death group compared to both the survival group and volunteers (Fig. 4A). Morphological changes in PBMCs of septic survival and death groups were then observed through TEM and SEM. Generally, in the survival group, most of the cell membranes were intact with occasional damage or pore formation. Meanwhile, there was local oedema in the cytoplasm, accompanied by mitochondrial pyknosis and a small amount of autolysosomes. In the death group, the cells were irregularly formed, with bulges and obvious pore-forming. Cell damage was evident, shown by condensed chromatin and blurred nuclear membrane. A small number of mitochondria were pyknotic and scattered in the cytoplasm (Fig. 4B). Further analysis of the cells in death group revealed some typical morphological changes in various cell death patterns (Fig. 4C). SEM showed obvious bulges and pore-forming on the cell membranes (Pyroptosis). From TEM, the cell nucleus was irregularly shaped with a blurred nuclear membrane and widened nuclear gap (Apoptosis); more autophagosomes and autolysosomes appeared (Autophagy); and some mitochondria were pyknotic with high electron density and cristae dilatation (Ferroptosis). We also detected several biomarkers of cell apoptosis and pyroptosis in PBMCs from the survival and death groups through western blotting. The results showed that the relative expression levels of BAX, CASP 3, and CASP 5 were significantly increased and BCL2 was decreased in the death group compared to the survival group. Cleaved-GSDMD had a nonsignificant increasing tendency (Fig. 4D, Figure S5). The experimental findings were in line with the above sequencing results.
Fig. 4.
Cell apoptosis and morphological changes of cell death pattern in PBMCs from sepsis patients and healthy volunteers. A Flow cytometry of PBMCs showing cell apoptosis and necrosis rate. B Electron microscopy images of PBMCs in sepsis patients from survival and death groups (SEM, scale bar = 25 μm; TEM, scale bar = 25 and 10 μm). C Representative electron microscope images of PBMCs showing typical cell death morphology of pyroptosis, apoptosis, autophagy, and ferroptosis in death group. (SEM, scale bar = 50 and 25 μm; TEM, scale bar = 25 and 10 μm). Red arrow: pore formation; Red star: bulges; Yellow arrow: autolysosomes; Black arrow: blurred nuclear membrane; Purple arrow: autophagosomes; Blue arrow: mitochondrial pyknosis. D Western blotting of several key molecules related to cell apoptosis and pyroptosis in PBMCs of sepsis patients. *p < 0.05, **p < 0.01, ns = no significance. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
3.4. Plasma levels of inflammatory cytokines and cell death indicators are elevated in septic death group
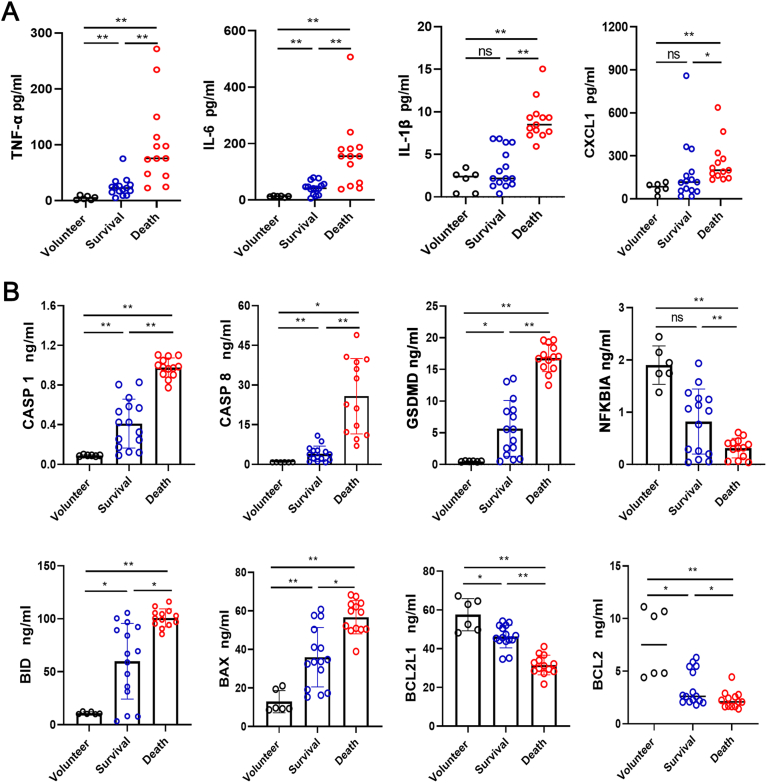
Plasma levels of inflammatory cytokines and chemokines in sepsis patients (survival and death) and healthy volunteers were evaluated through the Luminex assay. The results demonstrated that the concentrations of the inflammatory cytokines TNF-α, IL-6, IL-1β, and the chemokine CXCL1 were significantly increased in the death group compared to the survival and volunteer groups (Fig. 5A). Other cytokines like IL-1 ra, IL-8, and IL-15, and chemokines like CCL2, CCL4, and CXCL10 showed similar results in the plasma (Figure S6). Moreover, ELISA was performed to detect some cell death markers in the plasma samples between the groups. Compared to the survival, plasma levels of CASP 1, CASP 8, GSDMD, BID and BAX were significantly elevated in the death group, combined with significantly decreased levels of NFKBIA, BCL2L1 and BCL2 (Fig. 5B).
Fig. 5.
Plasma levels of inflammatory factors and key molecules involved in cell death pathways in sepsis patients (Volunteer n = 6, Survival n = 15, Death n = 13). A Luminex assay showed the levels of inflammatory factors TNF-α, IL-6, IL-1β, and chemokine CXCL1 between groups. B ELISA results showed the plasma levels of CASP 1, CASP 8, GSDMD, NFKBIA, BID, BAX, BCL2L1, and BCL2 between groups. *p < 0.05, **p < 0.01, ns = no significance.
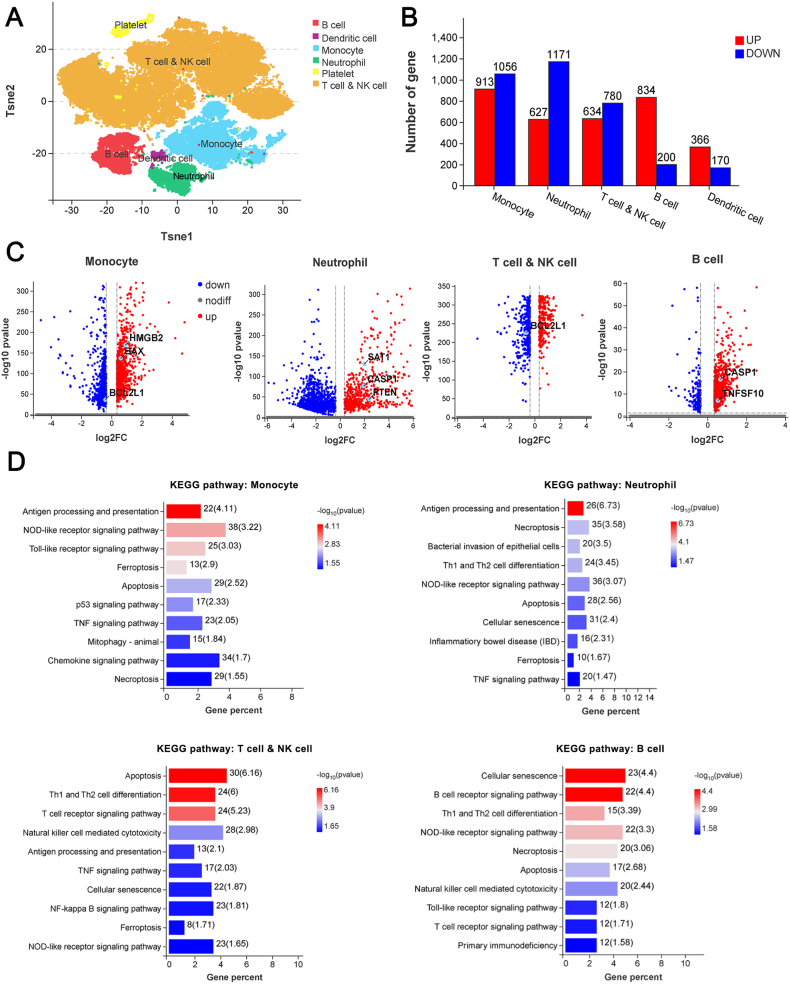
3.5. Cell death occurs mainly in myeloid cells rather than lymphocytes in patients with sepsis at the early stage
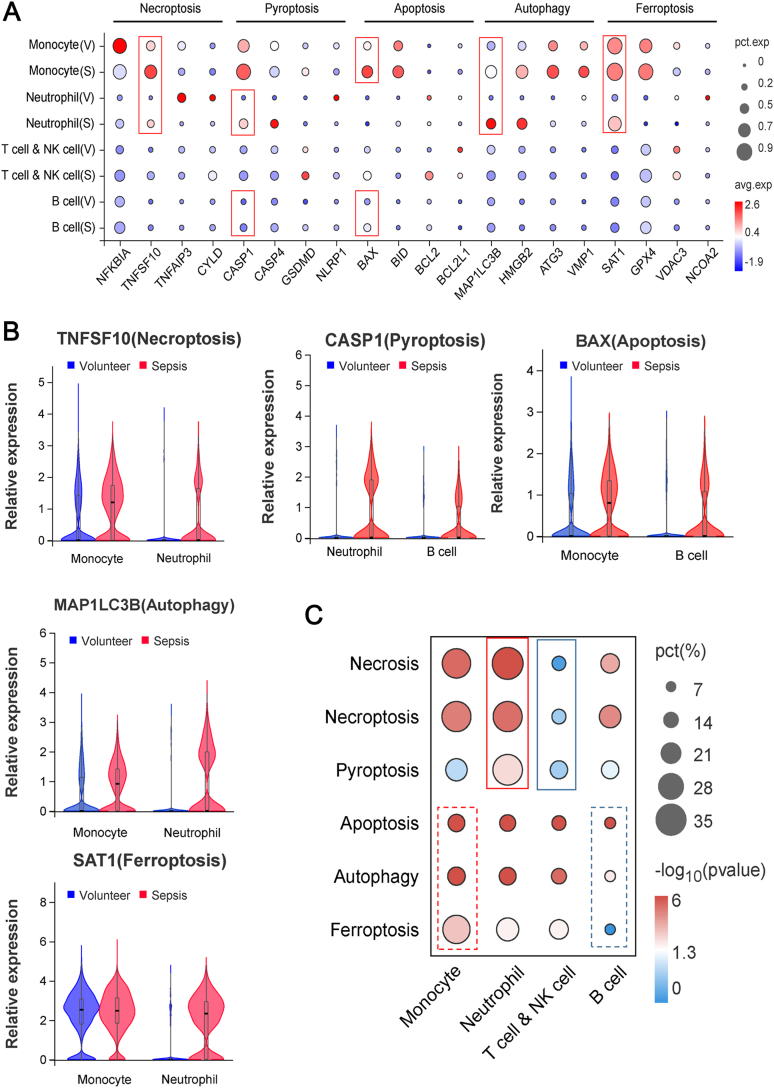
To further clarify the cell types and death patterns that predominate in the early development of sepsis, we collected PBMCs from three healthy volunteers and sepsis patients (Table S3) to conduct single-cell RNA sequencing. There were five main annotated immune cell groups (monocytes, neutrophils, T cells and NK cells, B cells, and dendritic cells), and the significant DEGs were mostly concentrated in the first four cell groups (Fig. 6A and B). Volcano plots showed an overview of the significant DEGs in monocytes, neutrophils, T and NK cells and B cells (Fig. 6C). KEGG functional enrichment analysis indicated that in the peripheral immune cells of patients with sepsis at the early stage, cellular signaling related to apoptosis, necroptosis, ferroptosis and also pyroptosis was significantly enriched (Fig. 6D, Table S4). Similar results were also observed by GO term annotation (Figure S7). We further studied the interaction between immune cells and the cell death-related pathways in sepsis. At the early stage of sepsis, key molecules of necroptosis (TNFSF10), pyroptosis (CASP 1), apoptosis (BAX), autophagy (MAP1LC3B) and ferroptosis (SAT1) were all significantly increased compared with those in volunteers (Fig. 7A). Among them, TNFSF10, MAP1LC3B and SAT1 were mainly significantly increased in monocytes and neutrophils of sepsis patients, CASP 1 was mainly increased in neutrophils and B cells and BAX was increased in monocytes and B cells (Fig. 7B). Moreover, we studied the cell types with the most significantly-altered death pattern based on the quantity of DEGs in GO terms related to the above five death patterns in various immune cells. The results showed that the necrosis, necroptosis, and pyroptosis pathways were significantly enriched in neutrophils but less altered in T and NK cells; while apoptosis, autophagy and ferroptosis patterns showed the most obviously increase in monocytes and less changes in B cells (Fig. 7C). Therefore, the above findings suggest that peripheral immune cell death occurs mainly in myeloid cells, not lymphocytes, in sepsis patients at the early stage. Meanwhile, neutrophil deaths are mainly destructive necrosis and pyroptosis, while apoptosis and autophagy which have little effects on the surrounding tissues are the main patterns for monocyte deaths.
Fig. 6.
Single cell sequencing of PBMCs from healthy volunteers and sepsis (n = 3). A Sample cell annotation showed mainly five types of immune cells. B The number of significant DEGs of the 5 immune cells between volunteer and sepsis. C Volcanic maps of differential gene distribution between volunteer and sepsis in monocyte, neutrophil, T cell and NK cell, and B cell. D KEGG enrichment pathway with significant differences in the four immune cells in disease state (p < 0.05).
Fig. 7.
Single-cell RNA sequencing revealed the changes of cell death patterns in various immune cells in sepsis. A Expression difference of the key molecules of five cell death signaling (necroptosis, pyroptosis, apoptosis, autophagy, and ferroptosis) in the four immune cells in sepsis at the early stage. B Expression changes of the biomarkers related to the five cell death signaling in immune cells (p < 0.05). C Comparison of the significant degree of functional changes in different cell death pathways of the four immune cells in sepsis at the early stage. Bubble plots were drawn according to the percentage of significant DEGs in the related GO term.
4. Discussion
Although the morbidity and mortality of sepsis is decreasing as the development of medical techniques and growing awareness, there is still no effective specific treatment for sepsis. Sepsis has a poor prognosis. Multiple organ dysfunction syndrome caused by sepsis easily leads to human death. Severe cognitive impairment and a high risk of recurrent infections are also the main threats to sepsis patients who survive [1]. The questions are: Are there any physiological or biological differences between sepsis patients who survive and those who do not? What can those differences bring to the therapeutic insight into sepsis? Is there any possibility of preventing sepsis from occurring? Hence, we enrolled sepsis patients admitted to our hospital and carried out a 90-day follow-up study of them to record their survival state. The clinical characteristics of the sepsis patients between the survival and death group informed that the lactic acid levels were significantly higher in the death group, combined with an elevated SOFA score, relative to the survival group. An increased lactic acid level via anaerobic glycolysis is an indicator for cytotoxicity and tissue damage [19,20]. Nowadays, serum lactate has been incorporated into the latest definition of septic shock [21]. Elevated blood lactate levels are closely associated with high mortality and severity of sepsis [22,23]. A retrospective observational study also demonstrated that blood lactate level was strongly correlated to SOFA scores at the early stage of intensive care unit stay [24], which is consistent with the current study.
Next, bulk RNA sequencing revealed more enrichment of the pathways related to cell death of PBMCs in the death group of sepsis at both the mRNA and microRNA levels. Morphological observation and molecular detection of PBMCs also confirmed the activation of diverse cell death patterns in the death group. Except for PBMCs, plasma levels of proinflammatory factors and chemokines in the death group were significantly higher than those in the survival group. Similarly, cell death-related molecules in the plasma were also significantly increased in the death group compared to the survival and healthy volunteers. All these results indicated that larger and more severe cell death occurred in sepsis patients who died than in those who survived, which is, of course, logical and realistic. Research has shown that programmed cell death is the body's resistance to infection, including apoptosis, necroptosis, pyroptosis and NETosis. These cell death pathways are activated independently and also engage in crosstalk with each other, to disrupt intercellular niches and coordinate appropriate innate immune responses [25]. Excessive defence can cause irreversible damage and even death. As programmed cell death is crucial for the development and homeostasis of eukaryotic organisms, its dysregulation leads to the occurrence of a wide range of diseases, including infection and sepsis, making the cell death-related regulators as potential therapeutic targets for them [26,27].
Back to the current work, cell death signaling in sepsis patients was studied in relation to their prognosis, which provided a better understanding of septic pathological development and cellular fate at the early stage. The number of differentially expressed mRNA and miRNAs was quantitatively larger in PBMCs from the deaths versus those from the survivors, both compared to those in healthy volunteers, suggesting a more violate immune responses in the death group than the survival one. In addition, we found that upregulated genes in deaths compared to those in survivors were mainly enriched in the pathways of cell cycle, cellular senescence, and cell death-related signaling cascades, suggesting that cell death-related molecules were highly expressed in the immune cells of sepsis patients with poor prognosis.
Several cytokines have been studied as biomarkers in sepsis, including IL-6, IL-1, IL-10 and IL-8, to regulate the body's immune responses [28]. Concurrence of initial immune activation and chronic immunosuppression during sepsis can lead to immune cell death [29]. Inversely, uncontrolled immune cell death is also considered as a main incentive for marked immunosuppression [11]. Above results revealed the occurrence of diverse cell death patterns in the PMBCs from septic death group. Due to the existence of a wide range of immune cells in PBMCs, increasing researches use single-cell RNA sequencing technology to analyze the global landscape of immune cell alterations [[30], [31], [32]]. Miguel Reyes et al. identified a unique CD14+ monocyte state and its surface markers using single-cell RNA sequencing [10]. Linfeng Tao et el. Found new co-diagnostic genes for sepsis and metabolic syndrome through single-cell data analysis and machine learning algorithms [33]. Recently, Wenfeng Xie also found SPI1-mediated autophagy of peripheral blood monocytes during sepsis based on single-cell RNA sequencing [34].
Nowadays, increasing studies began to use bulk RNA sequencing combined with single-cell RNA sequencing technology to screen core molecules and important signaling pathways involved in the pathological changes of diseases. Bulk RNA sequencing is used with large sample sizes to discover biological phenomena, and then single-cell RNA sequencing technique is adopted to dig deeper into the relevant core molecules and signaling pathways [35,36]. Here, we further conduced single-cell RNA sequencing and found that most DEGs were in monocytes, neutrophils, T and NK cells and B cells. Pathway enrichment analysis suggested that the changes of cell death type in immune cells were not exactly the same. Monocytes and neutrophils at the first line of defence showed the most severe cell death in sepsis at the early stage. Among them, necrosis, necroptosis and pyroptosis were significantly enriched in neutrophils, while apoptosis, autophagy and ferroptosis were in monocytes. Neutrophils receive signals from antigen presenting cells activated by infection through their own functional receptors, and exert functions to eradicate pathogen by phagocytosis and NET formation [37]. Monocytes/macrophages recognize and clear pathogens to respond to signals from infected cells [38]. Recently, researchers found that monocyte distribution width is an effective indicator for the diagnosis of early sepsis [39,40]. Since macrophages have high plasticity, different macrophage death mechanisms have been studied in response to sepsis, like pyroptosis and necroptosis [41]. Inhibition of macrophage pyroptosis by gasdermin D inhibitor prolonged the survival of a sepsis mouse model [42]. Meanwhile, neutrophil pyroptosis also plays a crucial role in sepsis [43], suggesting a potential therapeutic targeting pyroptotic death in sepsis. Besides, studies revealed that excessive cell death drives sepsis, which mainly consists of T and B cell apoptosis in the substantial immune tissues [13]. However, cell death of T and B cells in the adaptive immune system showed less changes in PBMCs in the current study. It seemed that peripheral immune cell death occurs mainly in myeloid cells not lymphocytes in sepsis patients at the early stage. Therefore, further studies on the changes of the survival status of different immune cell type in sepsis are needed to provide a scientific basis for the development of precise intervention for sepsis.
Moreover, since severe COVID-19 patients showed similar clinic symptoms of sepsis, researchers suggested treating severe COVID-19 disease as a sepsis syndrome [44]. Studies found that SARS-coronavirus infection stimulated pyroptotic death of human THP-1 macrophages as well as PBMCs from COVID-19 patients [45,46]. Cell apoptosis of human airway epithelial cells was also found to be elevated induced by SARS-coronavirus infection [47], but the necroptosis of immune cells after this infection was rare or had less evidence [48]. The effects of pathogen infection–induced cell death on the host are two-sided. Excessive cell death can not only limit the infection but also trigger more severe inflammatory and immune responses [48]. Therefore, precisely controlling immune cell death during infection and sepsis remains challenging, but it has great significance for future treatment and improvement of the prognosis of sepsis.
In the current study, we focused on the relationship between the changes of immune cell functions and sepsis prognosis. We enrolled the patients with strict inclusion criteria. Bulk RNA and single-cell RNA sequencing were conducted to analyze the DEGs and their enriched signaling pathways in various immune cells during sepsis. The sequencing results were further validated by a variety of techniques, including flow cytometry, scanning and transmission electron microscopy, WB assay, luminex assay, and qRT-PCR. However, disadvantages still existed: the study was a single-center study; the sample size of sequencing data was small; and the included population was heterogeneous. Hence, further studies with larger population and in-depth clinical verification are still needed.
5. Conclusions
PBMCs in the septic death group incurred more severe cell damage and diverse patterns of cell death than those in the survival group. Elevated plasma levels of inflammatory cytokines and cell death-related indicators were detected in the dead sepsis patients compared to those who survived. Moreover, through single-cell RNA sequencing of PBMCs, we found that cell death patterns occurred mainly in myeloid cells (neutrophils and monocytes) rather than lymphocytes (T and B cells) in septic patients at the early stage. Neutrophil deaths are mainly destructive necrosis and pyroptosis, and monocyte deaths are mainly apoptosis and autophagy that had little effect on the surroundings. The study shed light on the various mechanisms of immune cell death in early sepsis, which may provide novel insight for the development of sepsis-related therapeutics.
Ethics statement
The study was approved by the Ethical Committee of the Xijing Hospital of Fourth Military Medical University, with document number KY20212172 and NCT05229328. All procedures performed relating to human participants complied with the Declaration of Helsinki. Informed consent was signed by all subjects.
Author contribution statement
Shanshou Liu: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Chujun Duan: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Jiangang Xie, Jinxin Zhang, Xu Luo, Qianmei Wang and Xiaojun Zhao: Contributed reagents, materials, analysis tools or data; Performed the experiments.
Xiaoli Liang and Ran Zhuang: Contributed reagents, materials, analysis tools or data.
Wei Zhao: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Wen Yin: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This study was supported by the National Natural Science Foundation of China (No. 81871587), Basic research program of Natural Science in Shaanxi Province (No. 2020JQ-466), Basic research project of the Logistics Support Department of the Chinese Military Commission (No. BWS21J002), Medical Science and Technique Foundation for Fostering Young Scientist (No. 18QNP026), and Funds provided by Fourth Military Medical University (No. 2020rcfczr).
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the subjects in our clinical cohorts for participating and donating blood for our study. We are also grateful to the clinical research coordinators and fellows who conducted patient enrollment and performed blood collections. In addition, the authors thank medical statistical expert Yuhai Zhang from the Department of Statistics, the Fourth Military Medical University, for assisting us to carefully review the statistical analysis of the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17764.
Contributor Information
Wei Zhao, Email: zhaowei0624@fmmu.edu.cn.
Wen Yin, Email: xjyyyw@126.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Prescott H.C., Angus D.C. Enhancing recovery from sepsis: a review. JAMA. 2018;319:62–75. doi: 10.1001/jama.2017.17687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singer M., Deutschman C.S., Seymour C.W., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., van der Poll T., Vincent J.L., Angus D.C. The third international consensus definitions for sepsis and septic shock (Sepsis-3) JAMA. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudd K.E., Johnson S.C., Agesa K.M., Shackelford K.A., Tsoi D., Kievlan D.R., Colombara D.V., Ikuta K.S., Kissoon N., Finfer S., Fleischmann-Struzek C., Machado F.R., Reinhart K.K., Rowan K., Seymour C.W., Watson R.S., West T.E., Marinho F., Hay S.I., Lozano R., Lopez A.D., Angus D.C., Murray C.J.L., Naghavi M. Global, regional, and national sepsis incidence and mortality, 1990-2017: analysis for the Global Burden of Disease Study. Lancet. 2020;395:200–211. doi: 10.1016/S0140-6736(19)32989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Poll T., Shankar-Hari M., Wiersinga W.J. The immunology of sepsis. Immunity. 2021;54:2450–2464. doi: 10.1016/j.immuni.2021.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Rimmele T., Payen D., Cantaluppi V., Marshall J., Gomez H., Gomez A., Murray P., Kellum J.A., Workgroup A.X. Immune cell phenotype and function in sepsis. Shock. 2016;45:282–291. doi: 10.1097/SHK.0000000000000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hotchkiss R.S., Monneret G., Payen D. Sepsis-induced immunosuppression: from cellular dysfunctions to immunotherapy. Nat. Rev. Immunol. 2013;13:862–874. doi: 10.1038/nri3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qi X., Yu Y., Sun R., Huang J., Liu L., Yang Y., Rui T., Sun B. Identification and characterization of neutrophil heterogeneity in sepsis. Crit. Care. 2021;25:50. doi: 10.1186/s13054-021-03481-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukaszewicz A.C., Grienay M., Resche-Rigon M., Pirracchio R., Faivre V., Boval B., Payen D. Monocytic HLA-DR expression in intensive care patients: interest for prognosis and secondary infection prediction. Crit. Care Med. 2009;37:2746–2752. doi: 10.1097/CCM.0b013e3181ab858a. [DOI] [PubMed] [Google Scholar]
- 9.Holub M., Kluckova Z., Helcl M., Prihodov J., Rokyta R., Beran O. Lymphocyte subset numbers depend on the bacterial origin of sepsis. Clin. Microbiol. Infect. 2003;9:202–211. doi: 10.1046/j.1469-0691.2003.00518.x. [DOI] [PubMed] [Google Scholar]
- 10.Reyes M., Filbin M.R., Bhattacharyya R.P., Billman K., Eisenhaure T., Hung D.T., Levy B.D., Baron R.M., Blainey P.C., Goldberg M.B., Hacohen N. An immune-cell signature of bacterial sepsis. Nat. Med. 2020;26:333–340. doi: 10.1038/s41591-020-0752-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao C., Yu M., Chai Y. Pathological alteration and therapeutic implications of sepsis-induced immune cell apoptosis. Cell Death Dis. 2019;10:782. doi: 10.1038/s41419-019-2015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Tulzo Y., Pangault C., Gacouin A., Guilloux V., Tribut O., Amiot L., Tattevin P., Thomas R., Fauchet R., Drenou B. Early circulating lymphocyte apoptosis in human septic shock is associated with poor outcome. Shock. 2002;18:487–494. doi: 10.1097/00024382-200212000-00001. [DOI] [PubMed] [Google Scholar]
- 13.Cheng Z., Abrams S.T., Toh J., Wang S.S., Wang Z., Yu Q., Yu W., Toh C.H., Wang G. The critical roles and mechanisms of immune cell death in sepsis. Front. Immunol. 2020;11:1918. doi: 10.3389/fimmu.2020.01918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng J., Song X., Yan G., Wang H., Li H., Lou D. Dendrobine suppresses endoplasmic reticulum stress-induced apoptosis through upregulating microRNA miR-381-3p to decrease caspase-4. Bioengineered. 2021;12:4452–4463. doi: 10.1080/21655979.2021.1956672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao Y., Li C., Wang H., Liu Y. LINC00265 targets miR-382-5p to regulate SAT1, VAV3 and angiogenesis in osteosarcoma. Aging (Albany NY) 2020;12:20212–20225. doi: 10.18632/aging.103762. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Liu K., Hong D., Zhang F., Li X., He M., Han X., Zhang G., Xu G., Stonehouse N.J., Jiang Z., An W., Guo L. MicroRNA-106a inhibits autophagy process and antimicrobial responses by targeting ULK1, ATG7, and ATG16L1 during mycobacterial infection. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.610021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H., Dong B., Zhang N., Liu S., Zhao H. microRNA-182 negatively influences the neuroprotective effect of apelin against neuronal injury in epilepsy. Neuropsychiatric Dis. Treat. 2020;16:327–338. doi: 10.2147/NDT.S238826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye S., Xiong F., He X., Yuan Y., Li D., Ye D., Shi L., Lin Z., Zhao M., Feng S., Zhou B., Weng H., Hong L., Ye H., Gao S. DNA hypermethylation-induced miR-182 silence targets BCL2 and HOXA9 to facilitate the self-renewal of leukemia stem cell, accelerate acute myeloid leukemia progression, and determine the sensitivity of BCL2 inhibitor venetoclax. Theranostics. 2023;13:77–94. doi: 10.7150/thno.77404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kling D.E., Cavicchio A.J., Sollinger C.A., Madoff L.C., Schnitzer J.J., Kinane T.B. Lactic acid is a potential virulence factor for group B Streptococcus. Microb. Pathog. 2009;46:43–52. doi: 10.1016/j.micpath.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 20.Bakker J., Postelnicu R., Mukherjee V. Lactate: where are we now? Crit. Care Clin. 2020;36:115–124. doi: 10.1016/j.ccc.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 21.Gauer R., Forbes D., Boyer N. Sepsis: diagnosis and management. Am. Fam. Physician. 2020;101:409–418. [PubMed] [Google Scholar]
- 22.Weinberger J., Klompas M., Rhee C. What is the utility of measuring lactate levels in patients with sepsis and septic shock? Semin. Respir. Crit. Care Med. 2021;42:650–661. doi: 10.1055/s-0041-1733915. [DOI] [PubMed] [Google Scholar]
- 23.Kushimoto S., Akaishi S., Sato T., Nomura R., Fujita M., Kudo D., Kawazoe Y., Yoshida Y., Miyagawa N. Lactate, a useful marker for disease mortality and severity but an unreliable marker of tissue hypoxia/hypoperfusion in critically ill patients. Acute Med. Surg. 2016;3:293–297. doi: 10.1002/ams2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen T.C., van Bommel J., Woodward R., Mulder P.G., Bakker J. Association between blood lactate levels, Sequential Organ Failure Assessment subscores, and 28-day mortality during early and late intensive care unit stay: a retrospective observational study. Crit. Care Med. 2009;37:2369–2374. doi: 10.1097/CCM.0b013e3181a0f919. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen I., Rayamajhi M., Miao E.A. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 2017;17:151–164. doi: 10.1038/nri.2016.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kist M., Vucic D. Cell death pathways: intricate connections and disease implications. EMBO J. 2021;40 doi: 10.15252/embj.2020106700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gibellini L., Moro L. Programmed cell death in health and disease. Cells. 2021;10 doi: 10.3390/cells10071765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kany S., Vollrath J.T., Relja B. Cytokines in inflammatory disease. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20236008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nedeva C. Inflammation and cell death of the innate and adaptive immune system during sepsis. Biomolecules. 2021;11 doi: 10.3390/biom11071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang T., Zhang X., Liu Z., Yao T., Zheng D., Gan J., Yu S., Li L., Chen P., Sun J. Single-cell RNA sequencing reveals the sustained immune cell dysfunction in the pathogenesis of sepsis secondary to bacterial pneumonia. Genomics. 2021;113:1219–1233. doi: 10.1016/j.ygeno.2021.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C., Li Y., Li S., Chen M., Hu Y. Proteomics combined with RNA sequencing to screen biomarkers of sepsis. Infect. Drug Resist. 2022;15:5575–5587. doi: 10.2147/IDR.S380137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y., Wang C., Lu Q., Zhang C., Hu L., Ling J., Chen M., Hu Y. Screening of potential immune-related genes expressed during sepsis using gene sequencing technology. Sci. Rep. 2023;13:4258. doi: 10.1038/s41598-022-23062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao L., Zhu Y., Liu J. Identification of new co-diagnostic genes for sepsis and metabolic syndrome using single-cell data analysis and machine learning algorithms. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1129476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie W., Zou S., Dong C., Yang C. SPI1-mediated autophagy of peripheral blood monocyte cells as a mechanism for sepsis based on single-cell RNA sequencing. Int. Immunopharm. 2023;117 doi: 10.1016/j.intimp.2023.109909. [DOI] [PubMed] [Google Scholar]
- 35.Li X., Liao Z., Deng Z., Chen N., Zhao L. Combining bulk and single-cell RNA-sequencing data to reveal gene expression pattern of chondrocytes in the osteoarthritic knee. Bioengineered. 2021;12:997–1007. doi: 10.1080/21655979.2021.1903207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun D., Guan X., Moran A.E., Wu L.Y., Qian D.Z., Schedin P., Dai M.S., Danilov A.V., Alumkal J.J., Adey A.C., Spellman P.T., Xia Z. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2022;40:527–538. doi: 10.1038/s41587-021-01091-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 38.Narasimhan P.B., Marcovecchio P., Hamers A.A.J., Hedrick C.C. Nonclassical monocytes in health and disease. Annu. Rev. Immunol. 2019;37:439–456. doi: 10.1146/annurev-immunol-042617-053119. [DOI] [PubMed] [Google Scholar]
- 39.Agnello L., Bivona G., Vidali M., Scazzone C., Giglio R.V., Iacolino G., Iacona A., Mancuso S., Ciaccio A.M., Lo Sasso B., Ciaccio M. Monocyte distribution width (MDW) as a screening tool for sepsis in the Emergency Department. Clin. Chem. Lab. Med. 2020;58:1951–1957. doi: 10.1515/cclm-2020-0417. [DOI] [PubMed] [Google Scholar]
- 40.Hausfater P., Robert Boter N., Morales Indiano C., Cancella de Abreu M., Marin A.M., Pernet J., Quesada D., Castro I., Careaga D., Arock M., Tejidor L., Velly L. Monocyte distribution width (MDW) performance as an early sepsis indicator in the emergency department: comparison with CRP and procalcitonin in a multicenter international European prospective study. Crit. Care. 2021;25:227. doi: 10.1186/s13054-021-03622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robinson N., Ganesan R., Hegedus C., Kovacs K., Kufer T.A., Virag L. Programmed necrotic cell death of macrophages: focus on pyroptosis, necroptosis, and parthanatos. Redox Biol. 2019;26 doi: 10.1016/j.redox.2019.101239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rathkey J.K., Zhao J., Liu Z., Chen Y., Yang J., Kondolf H.C., Benson B.L., Chirieleison S.M., Huang A.Y., Dubyak G.R., Xiao T.S., Li X., Abbott D.W. Chemical disruption of the pyroptotic pore-forming protein gasdermin D inhibits inflammatory cell death and sepsis. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aat2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L., Sun B. Neutrophil pyroptosis: new perspectives on sepsis. Cell. Mol. Life Sci. 2019;76:2031–2042. doi: 10.1007/s00018-019-03060-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kocak Tufan Z., Kayaaslan B., Mer M. COVID-19 and sepsis. Turk. J. Med. Sci. 2021;51:3301–3311. doi: 10.3906/sag-2108-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi C.S., Nabar N.R., Huang N.N., Kehrl J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Dis. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodrigues T.S., de Sa K.S.G., Ishimoto A.Y., Becerra A., Oliveira S., Almeida L., Goncalves A.V., Perucello D.B., Andrade W.A., Castro R., Veras F.P., Toller-Kawahisa J.E., Nascimento D.C., de Lima M.H.F., Silva C.M.S., Caetite D.B., Martins R.B., Castro I.A., Pontelli M.C., de Barros F.C., do Amaral N.B., Giannini M.C., Bonjorno L.P., Lopes M.I.F., Santana R.C., Vilar F.C., Auxiliadora-Martins M., Luppino-Assad R., de Almeida S.C.L., de Oliveira F.R., Batah S.S., Siyuan L., Benatti M.N., Cunha T.M., Alves-Filho J.C., Cunha F.Q., Cunha L.D., Frantz F.G., Kohlsdorf T., Fabro A.T., Arruda E., de Oliveira R.D.R., Louzada-Junior P., Zamboni D.S. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218 doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu N., Wang W., Liu Z., Liang C., Wang W., Ye F., Huang B., Zhao L., Wang H., Zhou W., Deng Y., Mao L., Su C., Qiang G., Jiang T., Zhao J., Wu G., Song J., Tan W. Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells. Nat. Commun. 2020;11:3910. doi: 10.1038/s41467-020-17796-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee S., Channappanavar R., Kanneganti T.D. Coronaviruses: innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41:1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.