Abstract
Opsins are universal photosensitive proteins in animals. Vertebrates have a variety of opsin genes for visual and non-visual photoreceptions. Analysis of the gene structures shows that most opsin genes have introns in their coding regions. However, teleosts exceptionally have several intron-less opsin genes that are presumed to have been duplicated by an RNA-based gene duplication mechanism, retroduplication. Among these retrogenes, we focused on the Opn4 (melanopsin) gene responsible for non-image-forming photoreception. Many teleosts have five Opn4 genes including one intron-less gene, which is speculated to have been formed from a parental intron-containing gene in the Actinopterygii. In this study, to reveal the evolutionary history of Opn4 genes, we analyzed them in teleost (zebrafish and medaka) and non-teleost (bichir, sturgeon, and gar) fishes. Our synteny analysis suggests that the intron-less Opn4 gene emerged by retroduplication after the branching of the bichir lineage. In addition, our biochemical and histochemical analyses showed that, in the teleost lineage, the newly acquired intron-less Opn4 gene became abundantly used without substantial changes in the molecular properties of the Opn4 protein. This stepwise evolutionary model of Opn4 genes is quite similar to that of rhodopsin genes in the Actinopterygii. The unique acquisition of rhodopsin and Opn4 retrogenes would have contributed to the diversification of the opsin gene repertoires in the Actinopterygii and the adaptation of teleosts to various aquatic environments.
Keywords: rhodopsin, melanopsin, G protein-coupled receptor (GPCR), retrogene, molecular evolution
Vertebrates utilize light not only for visual functions but also for various non-visual functions. These photoreceptions are generally underlain by photoreceptive proteins, opsins (1, 2, 3). Recent advances in genome analysis have revealed that most vertebrates have multiple opsin genes in their genomes. Opsins share amino acid sequence homology and structural elements including seven transmembrane domains and a chromophore, retinal. Thus, opsin genes are considered to have expanded from a single ancestral gene through gene duplications and mutations, which has led to the diversification of the molecular properties and expression patterns of the proteins that they encode. Opsins can be classified into several groups by phylogenetic relationship analysis. Among mammalian opsins, rhodopsin and Opn4 (melanopsin) are the best-studied opsins responsible for visual and non-visual photoreceptions, respectively. Rhodopsin functions for twilight vision in the rod photoreceptor cells of the retina and belongs to the c-opsin group together with cone visual pigments and Opn3. Rhodopsin binds 11-cis retinal in the dark and activates Gt-type of G protein in a light-dependent manner. By contrast, Opn4 functions for several non-image-forming photoreceptions, such as entrainment of the circadian rhythm and pupillary light reflex, in the intrinsically photosensitive retinal ganglion cells of the retina and belongs to the r-opsin group (4, 5). Opn4 binds 11-cis retinal in the dark and mainly activates Gq-type of G protein in a light-dependent manner.
Analysis of the gene structures of vertebrate opsins has revealed that most vertebrate opsin genes contain introns in their translational regions and several boundary positions of exons and introns are conserved within each opsin group (6). These facts indicate that opsin genes have been diversified mainly by DNA-based gene duplication mechanisms, such as whole-genome duplication, unequal crossing over, or DNA transposons (7, 8). However, it has been reported that a few opsin genes do not contain introns in their coding regions (9, 10, 11). These intron-less (single-exon) genes are supposed to have been acquired by the RNA-based gene duplication mechanism, namely, retroduplication (retrotransposition), in which cDNA was reverse-transcribed from mature mRNA of a parental intron-containing gene and was inserted into another region of the genome (12). In vertebrates, several intron-less opsin genes have been found in fish genomes. In a previous study, we analyzed the evolutionary process of the teleost intron-less rhodopsin gene (13). In general, vertebrates have a single intron-containing rhodopsin gene in their genomes and utilize it in rod cells for visual photoreception. By contrast, teleosts have intron-containing and intron-less rhodopsin genes in their genomes. The intron-less (Rh1) and intron-containing (Exorh) genes are expressed in the rod cells (9) and the pineal gland (14), respectively. Taking account of the phylogenetic relationship of actinopterygian species (ray-finned fishes) (15), we analyzed the rhodopsin genes of non-teleost fishes in the Actinopterygii (Polypteriformes, Acipenseriformes, and Holostei). Our analysis led us to propose an evolutionary scenario in which the intron-less rhodopsin gene emerged from a parental intron-containing one by retroduplication after branching of the Polypteriformes and the intron-containing gene changed its expression pattern to exclusive and abundant distribution in the pineal gland after branching of the Holostei.
Another well-known example of teleost intron-less opsin genes is Opn4. The Opn4 gene was first discovered in the melanophores of Xenopus laevis (16) and was subsequently identified also in the human genome (17). Accumulated information about Opn4 genes has shown that vertebrate Opn4 genes are classified into two lineages, Opn4m (mammalian-like) and Opn4x (Xenopus-like), in the phylogenetic tree and mammals have a single Opn4 gene (Opn4m) due to loss of the Opn4x gene (18). Previous reports about Opn4 in non-mammalian vertebrates showed that Opn4 protein is expressed in several tissues, including the retina, brain, and skin (18), suggesting the possibility that Opn4 protein contributes to various non-visual photoreceptions (4). The genomes of many teleost fishes contain five Opn4 genes, Opn4m1, Opn4m2, Opn4m3, Opn4x1, and Opn4x2, among which Opn4m2 is an intron-less single-exon gene (19). A previous study revealed that zebrafish Opn4 genes show largely non-overlapping expression patterns in the retina and the intron-less Opn4m gene (Opn4m2) is predominantly expressed in the inner nuclear layer (INL) and the photoreceptor cells (19, 20). Thus, it has been supposed that, during the evolutionary process of the Actinopterygii, the Opn4 genes expanded by both retroduplication and teleost-specific whole-genome duplication and functionally differentiated by the diversification of their expression patterns.
In this study, we analyzed the synteny conservation of the Opn4 genes in addition to the rhodopsin genes among actinopterygian species. We also examined the molecular properties of Opn4 protein and the tissue distributions of Opn4 mRNA in actinopterygian species. Our comparison of the diversification processes of intron-containing and intron-less rhodopsin and Opn4 genes suggests a common evolutionary history of these two opsin genes in the Actinopterygii.
Results
Synteny analysis of rhodopsin genes in the Actinopterygii
In our previous study, we obtained the rhodopsin clones from several non-teleost fishes in the Actinopterygii and proposed a stepwise evolutionary model of teleost intron-containing and intron-less rhodopsin genes, that is, the intron-less rhodopsin gene was retroduplicated from an ancestral intron-containing rhodopsin gene after the branching of the Polypteriformes and subsequently the molecular property and tissue distribution of the ancestral intron-containing rhodopsin gene changed to be exclusively utilized for pineal photoreception in teleosts (13). After our analysis, the whole genome assemblies of gray bichir (Polypterus senegalus) and reedfish (Erpetoichthys calabaricus) in the Polypteriformes and sterlet (Acipenser ruthenus) and paddlefish (Polyodon spathula) in the Acipenseriformes were published in the public databases (21, 22). To evaluate our previous model, we examined the genomic locations of these rhodopsin genes and compared the syntenies flanking the rhodopsin genes in the genomes of actinopterygian species based on Actinopterygii evolution (15) (Fig. 1A and Data S1). The genomes of the Acipenseriformes (sterlet and paddlefish), the Holostei (spotted gar (Lepisosteus oculatus)), and the Teleostei (Asian arowana (Scleropages formosus)) contain the intron-less rhodopsin gene in the conserved synteny, whereas the genomes of the Polypteriformes (gray bichir and reedfish) do not contain the rhodopsin gene in the corresponding synteny. These results suggest that an intron-less rhodopsin gene emerged by retroduplication of the parental intron-containing rhodopsin gene and was inserted into the genomic location between adamts9 and magi1 after branching of the Polypteriformes. In addition, the intron-containing rhodopsin gene is found in the conserved synteny of the Asian arowana, spotted gar, gray bichir, and reedfish genomes but is missing in the sterlet and paddlefish genomes. This is consistent with our previous analysis in which we could not obtain the clone of an intron-containing rhodopsin gene in Siberian sturgeon. These findings suggested that the intron-containing gene was lost at the base of the Acipenseriformes. It should be noted that, in the Acipenseriformes, sterlet has one intron-less rhodopsin gene and paddlefish has two intron-less rhodopsin genes, a protein-coding gene and a pseudogene, which emerged by the whole genome duplication. Thus, both species have only one intron-less rhodopsin gene. These synteny analyses supported our evolutionary model of rhodopsin genes in the Actinopterygii (Fig. 1B).
Figure 1.
Comparison of rhodopsin gene repertoires in the Actinopterygii. A, synteny of orthologous genes flanking the intron-containing rhodopsin gene (blue triangle with vertical lines) and the intron-less rhodopsin gene (pink triangle) in actinopterygian species and Xenopus tropicalis. One of the two intron-less rhodopsin genes in paddlefish is a pseudogene (pink triangle with horizontal stripes) which contains two stop codons in the coding region. The genes flanking the rhodopsin loci are shown by white triangles. Gene names are indicated above the X. tropicalis genes, intraflagellar transport 122 homolog (ift122) (gene number 1 shown by white triangle), H1.8 linker histone (h1-8) (gene number 2), plexin D1 (plxnd1) (gene number 3), ADAM metallopeptidase with thrombospondin type 1 motif 9 (adamts9) (gene number 4) and membrane-associated guanylate kinase, WW and PDZ domain containing 1 (magi1) (gene number 5). The animals of the NCBI assembly accession number: X. tropicalis, GCF_000004195.4; gray bichir (P. senegalus), GCF_016835505.1; reedfish (E. calabaricus), GCF_900747795.1; sterlet (A. ruthenus), GCF_010645085.1; paddlefish (P. spathul), GCF_017654505.1; spotted gar (L. oculatus), GCF_000242695.1; Asian arowana (S. formosus), GCF_900964775.1. Detailed gene information is shown in Data S1. B, evolutionary model of teleost intron-containing and intron-less rhodopsin genes in the Actinopterygii. Fishes in the Polypteriformes have only one rhodopsin gene (Rh1), which contains introns. After branching of the Polypteriformes, the intron-less rhodopsin gene (Rh1-2) emerged by retroduplication. The intron-containing rhodopsin gene (Rh1-1) was lost in the Acipenseriformes. The ancestor of the Teleostei duplicated the intron-less rhodopsin gene (Rh1-2, Rh1) by a whole genome duplication and changed the expression pattern of the intron-containing rhodopsin gene (Exorh (Rh1-1)) for pineal photoreception (13). The nomenclature of rhodopsin genes we use refers to that in the previous studies (36, 49, 50, 51).
Synteny analysis of Opn4 genes in the Actinopterygii
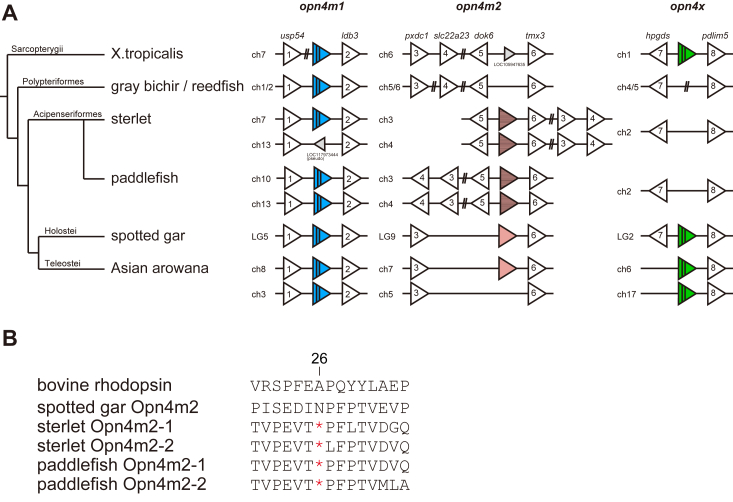
Next, we compared the repertoires of Opn4 genes among actinopterygian species. The Xenopus tropicalis genome contains two intron-containing Opn4 genes (Opn4m and Opn4x) and the Asian arowana genome contains four intro-containing Opn4 genes (Opn4m1, Opn4m3, Opn4x1, and Opn4x2) and one intron-less Opn4 gene (Opn4m2). Thus, we searched for intron-containing and intron-less Opn4 genes in the genomes of non-teleost fishes in the Actinopterygii (Fig. 2A and Data S1). The spotted gar genome contains an Opn4m2 protein-coding gene and the sterlet and paddlefish genomes contain two Opn4m2 pseudogenes (Fig. 2B). These intron-less Opn4m genes are located upstream of tmx3 in the genomes. By contrast, Opn4m2 gene is missing in the corresponding synteny of the gray bichir and reedfish genomes. Thus, we speculate that the intron-less Opn4m gene emerged by retroduplication of the parental intron-containing Opn4m gene (Opn4m) after the branching of the Polypteriformes. The parental intron-containing Opn4m gene is located in the conserved synteny, between usp54 and ldb3, of the Asian arowana, spotted gar, sterlet, paddlefish, gray bichir, and reedfish genomes, among which the Asian arowana and paddlefish genomes have two intron-containing Opn4m genes because of the whole genome duplication. The other intron-containing Opn4 gene, Opn4x, is identified in the spotted gar genome. The location of Opn4x gene in the genome is conserved in upstream of pdlim5 among Asian arowana, spotted gar, and X. tropicalis. By contrast, Opn4x gene is missing in the corresponding synteny of the sterlet, paddlefish, gray bichir, and reedfish genomes. This suggests that Opn4x gene was independently lost in the Polypteriformes and Acipenseriformes lineages. Therefore, these synteny analyses showed that the repertoires of Opn4 genes in non-teleost fishes have been diversified by retroduplication, whole genome duplication, and gene loss including pseudogenization.
Figure 2.
Comparison of Opn4 gene repertoires in the Actinopterygii. A, synteny of orthologous genes flanking the Opn4m1 gene (blue triangle with vertical lines), Opn4m2 gene (pink triangle), and Opn4x gene (green triangle with vertical lines) in actinopterygian species and X. tropicalis. The intron-less Opn4m2 genes in the Acipenseriformes are pseudogenes (pink triangle with horizontal stripes) which contain a stop codon at the same position in their coding regions (B). The genes flanking the Opn4 loci are shown by white triangles. Gene names are indicated above the X. tropicalis genes, ubiquitin specific peptidase 54 (usp54) (gene number 1 shown by white triangle), LIM domain binding 3 (ldb3) (gene number 2), PX domain containing 1 (pxdc1) (gene number 3), solute carrier family 22 member 23 (slc22a23) (gene number 4), docking protein 6 (dok6) (gene number 5), thioredoxin related transmembrane protein 3 (tmx3) (gene number 6), hematopoietic prostaglandin D synthase (hpgds) (gene number 7) and PDZ, and LIM domain 5 (pdlim5) (gene number 8). The animals of the NCBI assembly accession number: X. tropicalis, GCF_000004195.4; gray bichir (P. senegalus), GCF_016835505.1; reedfish (E. calabaricus), GCF_900747795.1; sterlet (A. ruthenus), GCF_010645085.1; paddlefish (P. spathul), GCF_017654505.1; spotted gar (L. oculatus), GCF_000242695.1; Asian arowana (S. formosus), GCF_900964775.1. Detailed gene information is shown in Data S1. B, comparison of deduced amino acid residues between those encoded by the intron-less Opn4m2 gene of spotted gar and the intron-less Opn4m2 pseudogenes of sterlet and paddlefish. The sequences of Opn4m2 are aligned with the sequence of bovine rhodopsin (BAB83621.1). Both sterlet and paddlefish have two intron-less Opn4m2 pseudogenes (Opn4m2-1 and Opn4m2-2), all of which share a nonsense mutation site at position 26 (based on the bovine rhodopsin numbering system). Gene IDs of these sequences are as follows: spotted gar Opn4m2, 102695598; sterlet Opn4m2-1, 117400101; sterlet Opn4m2-2, 117394593; paddlefish Opn4m2-1, 121307772. We found the sequence of paddlefish Opn4m2-2 using blast search in the whole-genome shotgun sequence, JADDYA010000170.1 (127199–127727).
Analysis of the molecular properties of Opn4 proteins
Next, we analyzed the molecular properties of Opn4 proteins among actinopterygian species. We obtained full-length Opn4 cDNAs from gray bichir in the Polypteriformes (Opn4m), Siberian sturgeon (Acipenser baerii) in the Acipenseriformes (Opn4m1), spotted gar in the Holostei (Opn4m1, Opn4m2, and Opn4x), and medaka in the Teleostei (Opn4m1, Opn4m2, Opn4m3, Opn4x1, and Opn4x2). To improve the expression level of Opn4 proteins, we truncated the C-terminus according to our previous study on mouse Opn4 (23) and added the Rho1D4 epitope sequence as a purification tag. It has been reported that the truncation of the C-terminus does not affect the photoreaction of cephalopod rhodopsins, which are closely related to vertebrate Opn4 in the phylogenetic tree of opsins (24, 25). In addition, it should be noted that the photochemical properties of the C-terminal truncated protein of mouse Opn4 (23) are consistent with the spectral sensitivities of Opn4-expressing ganglion cells that were measured by the electrophysiological recording in the mouse retina (26).
We expressed the recombinant Opn4 proteins in cultured cells and reconstituted the corresponding photo-pigments (Fig. 3, A–D). To measure the absorption maximum (λmax) of these Opn4 proteins, we added 11-cis retinal to the Opn4 protein-containing cell membranes and solubilized them with 1% dodecyl maltoside (DDM). We irradiated the extracts with yellow light (>500 nm) in the presence of 30 mM hydroxylamine and calculated their difference spectra by subtracting the spectrum after light irradiation from that before irradiation. Based on these spectra, we estimated λmax of gray bichir Opn4m (473 nm) and Siberian sturgeon Opn4m1 (479 nm) proteins (Fig. 3, A and B). In addition, the estimated absorption maximum (λmax) of spotted gar Opn4m1 (482 nm) and Opn4m2 (477 nm) proteins were red-shifted compared to that of spotted gar Opn4x protein (461 nm) (Fig. 3C). Likewise, λmax of medaka Opn4m1 (487 nm), Opn4m2 (482 nm), and Opn4m3 (480 nm) proteins were red-shifted compared to those of medaka Opn4x1 (463 nm) and Opn4x2 (474 nm) proteins (Fig. 3D). We also conducted affinity column chromatography to remove other extract components from DDM-solubilized samples of spotted gar Opn4 and medaka Opn4 proteins and obtained their absorption spectra (Fig. S1, A and B), whose spectral peaks were comparable to the values estimated from Figure 3, C and D. However, we could not obtain the absorption spectrum of gray bichir Opn4m or Siberian sturgeon Opn4m1 protein after the purification procedure, probably because of their low expression levels in mammalian cultured cells. Taken together, our analysis showed that λmax values of Opn4 proteins from actinopterygian species have a common feature, that is, λmax of Opn4m protein (470–480 nm) is red-shifted compared to that of Opn4x protein (450–470 nm). This is consistent with λmax of Opn4 proteins from other vertebrate species previously reported (19, 23, 27, 28).
Figure 3.
Molecular characteristics of Opn4 proteins. A and B, difference absorption spectra of gray bichir Opn4m protein (A) and Siberian sturgeon Opn4m1 protein (B). The Opn4-expressing cell membranes were mixed with 11-cis retinal and solubilized with 1% DDM. The absorption spectra of the extracts were measured before and after yellow light (>500 nm) irradiation in the presence of 30 mM hydroxylamine. Difference spectra were calculated by subtracting the spectrum after light irradiation from that before irradiation. λmax of gray bichir Opn4m protein and Siberian sturgeon Opn4m1 protein were estimated to be at 473 nm and 479 nm, respectively. C, difference absorption spectra of spotted gar Opn4m1 protein (blue), Opn4m2 protein (red) and Opn4x protein (green). λmax of Opn4m1, Opn4m2 and Opn4x protein were estimated to be 482 nm, 477 nm, and 461 nm, respectively. D, difference absorption spectra of medaka Opn4m1 protein (blue), Opn4m2 protein (red), Opn4m3 protein (light blue), Opn4x1 protein (green) and Opn4x2 protein (light green). λmax of Opn4m1, Opn4m2, Opn4m3, Opn4x1 and Opn4x2 protein were estimated to be 487 nm, 482 nm, 480 nm, 463 nm, and 474 nm, respectively. E–H, light-induced changes of intracellular Ca2+ level by Opn4 protein. The Ca2+ level in the HEK293S cells expressing gray bichir Opn4m protein (E), Siberian sturgeon Opn4m1 protein (F), and spotted gar Opn4 protein (Opn4m1, Opn4m2 or Opn4x) (G) or medaka Opn4 protein (Opn4m1, Opn4m2, Opn4m3, Opn4x1 or Opn4x2) (H) was measured using aequorin-based luminescent assay. Ca2+-dependent luminescence change of aequorin was triggered by light irradiation from blue (450 nm) LED for 5 s. Data are presented as the means ± deviations of two independent experiments compared with those of mock-transfected cells.
We also analyzed the ability of Opn4 proteins from actinopterygian species to activate G protein. Opn4 protein generally activates Gq-type of G protein to increase the intracellular Ca2+ concentration in a light-dependent manner. Thus, we measured the change in the intracellular Ca2+ level based on aequorin bioluminescence. We expressed the C-terminal truncated gray bichir, Siberian sturgeon, spotted gar, or medaka Opn4 protein with the Rho1D4 epitope sequence in cultured cells and observed an increase in the luminescence from aequorin induced by blue light irradiation (Fig. 3, E–H), indicating that blue light irradiation can induce the activation of Gq-type of G protein by these Opn4 proteins. We also prepared two constructs, one of which has the full-length sequence with the Rho1D4 epitope sequence and the other of which has the truncated C-terminus without the Rho1D4 epitope sequence, of spotted gar Opn4m1, Opn4m2, and Opn4x. Our analysis confirmed that these two constructs showed a light-dependent elevation in the Ca2+ level comparably to the construct which has the truncated C-terminus with the Rho1D4 epitope sequence (Fig. S1, C–E). Our data showed that these Opn4 proteins can trigger the Ca2+ signaling pathway in the cells, although it needs to be confirmed experimentally by using intact cells. Therefore, we concluded that Opn4 proteins encoded by both intron-containing and intron-less Opn4 genes from actinopterygian species share common spectral and biochemical properties.
Analysis of the expression patterns of Opn4 mRNA in the retina
To elucidate whether Opn4 genes changed their mRNA distribution patterns during the evolutionary process in the Actinopterygii, we conducted in situ hybridization analysis in the retinas of non-teleost fishes: gray bichir, Siberian sturgeon, and spotted gar. Our analysis showed that the gray bichir intron-containing Opn4 gene (Opn4m) was expressed abundantly in the cells (presumed horizontal cells) abutting the outer plexiform layer (OPL) (Fig. 4, A and F). We also detected gray bichir Opn4m mRNA in a small number of the presumed amacrine cells and bipolar cells or Müller cells on the INL and retinal ganglion cells. Likewise, the Siberian sturgeon intron-containing Opn4 gene (Opn4m1) showed high expression in the cells abutting the OPL and a small number of the cells on the INL and ganglion cell layer (GCL) (Fig. 4, B and G). Additionally, Siberian sturgeon Opn4m1 mRNA was detected in the retinal pigment epithelium (RPE) cells. Next, we analyzed the expression patterns of the spotted gar Opn4 genes. The transcript of the spotted gar intron-containing Opn4 gene (Opn4m1) was distributed abundantly in the presumed horizontal cells and in a subset of cells in the INL and GCL, which covered the distribution areas of gray bichir Opn4m and Siberian sturgeon Opn4m1 mRNAs (Fig. 4, C and H). In addition, we could detect spotted gar Opn4m1 mRNA signals in the RPE cells in the fundus of the retina (Fig. S2). On the other hand, the hybridization signals of mRNA of the spotted gar intron-less Opn4m gene (Opn4m2) were restricted to a quite small number of the cells abutting the OPL (Fig. 4, D and I). The spotted gar Opn4x gene (Opn4x) was expressed weakly in the OPL and in a sparse population of the cells in the ONL and GCL, which is similar to the expression patterns of spotted gar Opn4m gene (Opn4m1) (Fig. 4, E and J).
Figure 4.
Distribution of Opn4 mRNA in the retina of non-teleost fishes in the Actinopterygii. Distribution of the transcripts of gray bichir Opn4m (A and F), Siberian sturgeon Opn4m1 (B and G) and spotted gar Opn4m1 (C and H), Opn4m2 (D and I) and Opn4x (E and J) in the retina. These sections were hybridized with antisense probes (A–E) or corresponding sense probes (F–J). Scale bar: 20 μm.
Next, we analyzed the distribution patterns of Opn4 mRNAs in the eyes of medaka and zebrafish in the Teleostei. Among the distribution patterns of medaka Opn4m mRNAs in the retina (Fig. 5), the transcript of the intron-containing Opn4m gene (Opn4m1) was abundant in the cells (presumed horizontal cells) abutting OPL and a small number of the cells on the inner side of the INL (Fig. 5, A and F). The transcript of the other intron-containing Opn4m gene (Opn4m3) was distributed in the INL and GCL (Fig. 5, C and H). On the other hand, the hybridization signals of mRNA of the intron-less Opn4m gene (Opn4m2) were observed only in a smaller number of the cells (presumed horizontal cells) abutting OPL (Fig. 5, B and G). These results showed that the distribution patterns of the medaka Opn4m mRNAs in the retina were similar to those of the spotted gar Opn4m mRNAs. Our double fluorescence in situ hybridization analysis confirmed that the expression signals of the horizontal cell marker gene (gja10b encoding connexin 52.6) are merged with several Opn4m-positive cells abutting the OPL in the medaka retina (Fig. 6, A–I) (29). In addition, to compare the distribution patterns of Opn4m mRNAs between medaka and zebrafish in teleosts, we conducted in situ hybridization analysis in the zebrafish retina. Zebrafish Opn4m2 mRNA showed abundant distribution in the presumed horizontal cells and the photoreceptor cells (Fig. 5, L and Q), whereas Opn4m1 mRNA was expressed in a subset of the cells in the INL (Fig. 5, K and P) and Opn4m3 mRNA was expressed in the OPL and a small number of cells on the inner side of the INL (Fig. 5, M and R). These results for zebrafish Opn4m genes are consistent with a previous report (19, 20, 30). Thus, the expression patterns of Opn4m mRNAs are diversified among teleost species. In addition, the comparison of the localization of Opn4m2 mRNA in the retina among spotted gar, medaka, and zebrafish showed that the expression patterns of the intron-less Opn4m gene changed to be more abundant in zebrafish. We also analyzed the distribution patterns of Opn4x mRNAs in the retinas of medaka (Fig. 5, D, E, I, and J) and zebrafish (Fig. 5, N, O, S, and T). In both teleosts, Opn4x1 and Opn4x2 mRNAs were detected in the INL, and the signals of Opn4x2 mRNA were more abundant than those of Opn4x1 mRNA. In addition, we confirmed that the horizontal cell marker gene (gja10b) is expressed in several Opn4x2-positive cells abutting the OPL in the medaka retina (Fig. 6, J–L). The comparison of the localization of Opn4x mRNAs in the retina among spotted gar, medaka, and zebrafish showed that the exclusive expression of the Opn4x genes in the INL was conserved among the actinopterygian species.
Figure 5.
Distribution of Opn4 mRNA in the retina of medaka and zebrafish.A–J, distribution of the transcripts of medaka Opn4m1 (A and F), Opn4m2 (B and G), Opn4m3 (C and H), Opn4x1 (D and I) and Opn4x2 (E and J) in the retina. K–T, distribution of the transcripts of zebrafish Opn4m1 (K and P), Opn4m2 (L and Q), Opn4m3 (M and R), Opn4x1 (N and S) and Opn4x2 (O and T) in the retina. These sections were hybridized with antisense probes (A–E, K–L) or corresponding sense probes (F–J, P–T). Scale bar: 20 μm.
Figure 6.
Distribution of Opn4 mRNA in the horizontal cells of medaka.A–L, distribution of the transcripts of medaka Opn4m1 (A–C), Opn4m2 (D–F), Opn4m3 (G–I), and Opn4x2 (J–L) and horizontal cell marker (gja10b) in the retina. Nuclei were stained with Hoechst (blue). White arrowheads indicate the colocalization of Opn4 genes and gja10b in some cells in the OPL. Scale bar: 20 μm.
Analysis of the expression patterns of Opn4 mRNA in the brain
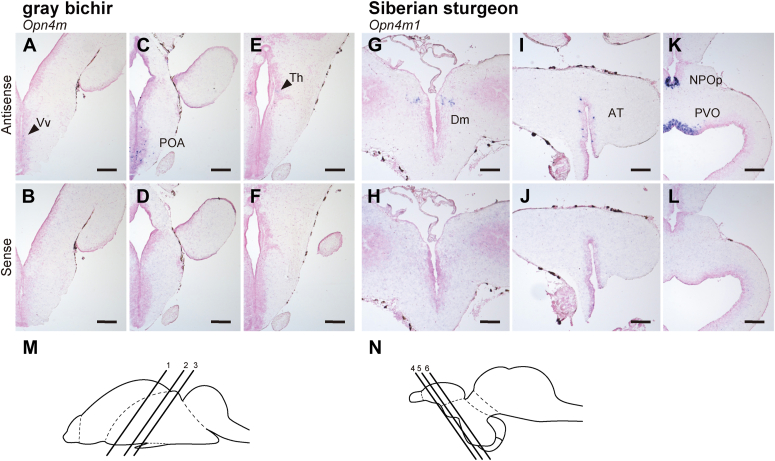
Finally, we analyzed the mRNA distribution of Opn4 genes within the brains of non-teleost fishes (Fig. 7). We observed the expression signals of these Opn4 genes in several areas of the telencephalon, diencephalon, and mesencephalon. In the gray bichir brain, Opn4m mRNA was expressed in the ventral part of the ventral telencephalic area (Vv) (Fig. 7, A and B), in the preoptic area (POA), and in a small number of neurons in the thalamus (Th) of the diencephalon (Fig. 7, C–F). In the Siberian sturgeon brain, Opn4m1 mRNA was expressed in some parts of the medial part of the dorsal telencephalic area (Dm) and anterior tuberal region (AT) (Fig. 7, G–J). Moreover, we detected strong hybridization signals of Siberian sturgeon Opn4m1 mRNA in cerebrospinal fluid-contacting neurons in the parvocellular preoptic nucleus (NPOp) and paraventricular organ (PVO) of the hypothalamus (Fig. 7, K and L). In the spotted gar telencephalon, diencephalon, and mesencephalon, three Opn4 genes showed their characteristic expression patterns (Figs. 8 and S3). Opn4m1 mRNA was distributed in the inner cell layer of the olfactory bulb (OB), POA, the optic tectum (OT), and the periventricular nucleus of the hypothalamus (PNH) (Fig. 8, A–D). The hybridization signal of spotted gar Opn4x mRNA was detected in the dorsal habenula (dHb) (Fig. 8, I–L). The transcripts of the intron-containing Opn4 genes, Opn4m1 and Opn4x, were also expressed in the ventral part of the ventral telencephalic area (Vv) in the telencephalon (Fig. 8, B and J). By contrast, we could not detect the hybridization signals of the transcript of the intron-less Opn4 gene (Opn4m2) in the anterior region of the spotted gar brain (Fig. 8, E–H).
Figure 7.
Distribution of Opn4 mRNA in the brain of gray bichir and Siberian sturgeon. (A–L) Distribution of the transcripts of gray bichir Opn4m (A–F) and Siberian sturgeon Opn4m1 (G–L) in the brain. These sections were hybridized with antisense probes (A, C, E, G, I, and K) or corresponding sense probes (B, D, F, H, J, and L). Scale bar: 200 μm. M, N, Schematic drawings of the gray bichir brain (M) and Siberian sturgeon brain (N) in lateral view. Numbered lines indicate the positions of cross sections shown in A and B (1), C and D (2), E and F (3), G and H (4), I and J (5) or K and L (6).
Figure 8.
Distribution of Opn4 mRNA in the brain of spotted gar. (A–L) Distribution of the transcripts of spotted gar Opn4m1 (A–D), Opn4m2 (E–H), and Opn4x (I–L) in the brain. These coronal sections were hybridized with antisense probes. The sections which were hybridized with the corresponding sense probes are shown in Fig. S3. Scale bar: 200 μm. (M) Schematic drawing of the spotted gar brain in dorsal view. Numbered lines indicate the positions of cross sections shown in (A, E, and I) (1), (B, F, and J) (2), (C, G, and K) (3) or (D, H, and L) (4).
To compare the expression patterns of Opn4 mRNAs between teleost and non-teleost species, we also analyzed the distribution of Opn4 mRNAs in the brains of medaka and zebrafish (Figs. 9 and S4). We observed the distribution of Opn4m mRNAs in a large area within the medaka brain (Fig. 9, A–C). Notably, the transcript of the medaka intron-less Opn4m gene (Opn4m2) was expressed in OB, Dm, dorsal part of the ventral telencephalic area (Vd), supracommisural part of the ventral telencephalic area (Vs), Vv, POA, Th, OT, ventral tuberal region (VT), anterior tuberal region (AT), posterior tuberal region (PT), nucleus recessus posterioris (NRP), inferior lobe (IL), and cerebellar corpus (CCe) (Fig. 9B). Medaka Opn4m1 mRNA was detected in Vd, Vs, Vv, POA, VT, AT, and PT. The expression signals of the other intron-containing Opn4m gene (Opn4m3) in medaka were observed in Vv, POA, dHb, Th, OT, VT, AT, PT, and IL. In the zebrafish brain, Opn4m2 mRNA was detected only in the pineal gland (Pin) (Fig. 9G), whereas the intron-containing Opn4 genes (Opn4m1 and Opn4m3) were expressed in several parts of the brain including Dm, POA, the ventral habenula (vHb), Th, AT and the ventral zone of the periventricular hypothalamus (Hv) (Fig. 9, F and H). A previous study reported the significantly low expression level of the Opn4 gene in the zebrafish brain (6), which is consistent with our results. The comparison of the localization of Opn4m2 mRNA in the brain among spotted gar, medaka, and zebrafish showed that the expression patterns of the intron-less Opn4m gene changed to be more abundant in some teleost lineages. Moreover, the Opn4x genes showed different expression patterns in the brain between medaka and zebrafish. In the rostral surface of Dm, the expression signals of medaka Opn4x2 mRNA, but not Opn4x1, were observed (Fig. 9, D and I). By contrast, in the corresponding region of the zebrafish brain, the hybridization signals of Opn4x1 mRNA, but not Opn4x2, were detected (Fig. 9, E and J). Medaka Opn4x1 and Opn4x2 mRNAs were also detected in POA, AT, and PT, although zebrafish Opn4x1 and Opn4x2 mRNAs were not observed in regions other than Dm. Collectively, these results indicated that the expression patterns of Opn4 genes in the brain were diversified among the actinopterygian species.
Figure 9.
Distribution of Opn4 mRNA in the brain of medaka and zebrafish.A–E, distribution of the transcripts of medaka Opn4m1 (A), Opn4m2 (B), Opn4m3 (C), Opn4x1 (D), and Opn4x2 (E) in the brain. F–J, distribution of the transcripts of zebrafish Opn4m1 (F), Opn4m2 (G), Opn4m3 (H), Opn4x1 (I), and Opn4x2 (J) in the brain. These sagittal sections were hybridized with antisense probes. The sections which were hybridized with the corresponding sense probes are shown in Fig. S4. Scale bar: 200 μm.
Discussion
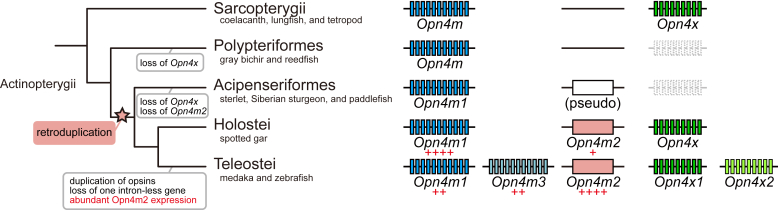
In this study, to assess the diversification process of the intron-containing and intron-less Opn4 genes in the Actinopterygii, we analyzed the synteny of the Opn4 genes in both teleost and non-teleost fishes. Based on our analysis, we propose an evolutionary model of Opn4 genes in the Actinopterygii (Fig. 10). The ancestor of the Actinopterygii had two Opn4 genes, Opn4m and Opn4x, both of which contained introns. After branching of the Polypteriformes, the intron-less Opn4m gene (Opn4m2) emerged by retroduplication of the parental intron-containing Opn4m gene (Opn4m). The common ancestor of the Holostei and the Teleostei had three Opn4 genes (Opn4m1, Opn4m2, and Opn4x), and subsequently, in the Teleostei lineage, the intron-containing Opn4 genes (Opn4m1 and Opn4x) duplicated by whole genome duplication. In addition, our analysis of the molecular properties of Opn4 proteins showed that Opn4 proteins maintained the sensitivity to blue light and the coupling to Gq-type G protein during the diversification process in the Actinopterygii. Also, the comparison of the distribution patterns of Opn4 mRNA showed that the newly acquired intron-less Opn4m gene (Opn4m2) was expressed in a quite-restricted area of the retina in spotted gar, whereas the orthologous gene was expressed in a large area of the anterior region of the brain in medaka and expressed abundantly in the horizontal cells and the photoreceptor cells of the retina in zebrafish. This showed that the intron-less Opn4m gene expanded its expression areas in the retina or brain in the Teleostei lineage. As mentioned in Figure 1, it can be speculated that the intron-less rhodopsin gene (Rh1-2) emerged after the branching of the Polypteriformes and became predominantly used in the retina without substantial changes of the molecular property of the rhodopsin protein in the Teleostei lineage. Therefore, our analysis of the rhodopsin and Opn4 genes in the Actinopterygii suggested that the teleost intron-less rhodopsin and Opn4 genes coincidently experienced similar evolutionary events after their emergence by retroduplication.
Figure 10.
Evolutionary model of teleost intron-containing and intron-less Opn4 genes in the Actinopterygii. It can be speculated that the ancestor of the Actinopterygii had two Opn4 genes, Opn4m and Opn4x, both of which contained introns. After branching of the Polypteriformes, the intron-less Opn4m gene (Opn4m2) emerged by retroduplication of the parental intron-containing Opn4m gene (Opn4m). However, this newly acquired intron-less Opn4m gene was pseudogenized in the Acipenserformes. In addition, the Opn4x gene was lost independently in the Polypteriformes and the Acipenserformes. The common ancestor of the Holostei and the Teleostei had three Opn4 genes (Opn4m1, Opn4m2 and Opn4x) and subsequently, in the Teleostei lineage, the intron-containing Opn4 genes (Opn4m1 and Opn4x) duplicated by a whole genome duplication. In addition, an increase of the cells which express the intron-less Opn4m gene (Opn4m2) in the retina or brain occurred in several teleost species.
The comparison of the synteny of the rhodopsin and Opn4 genes also unveiled a loss of the opsin genes in several lineages. In the Acipenseriformes, paddlefish has one intron-less rhodopsin protein-coding gene together with one intron-less rhodopsin pseudogene which was speculated to have been pseudogenized after a whole genome duplication in this lineage. In addition, sterlet and paddlefish have one or two intron-containing Opn4 protein-coding gene(s) together with two intron-less Opn4 pseudogenes. These pseudogenes share the same synteny and nonsense mutation position (Fig. 2B). This suggests that the intron-less Opn4 genes of sterlet and paddlefish were pseudogenized in their common ancestor before the divergence of the Acipenseridae and Polyodontidae in the Acipenseriformes (153 Mya) (21). These gene losses are thought to be driven by the functional redundancy of opsins. The intron-containing rhodopsin gene (Rh1) in gray bichir and the intron-less rhodopsin gene (Rh1-2) in Siberian sturgeon exhibit similar expression patterns, exclusive and abundant expression in the photoreceptor cells of the retina, and encode proteins that have similar spectral and biochemical properties (13). The intron-containing Opn4m gene also has generally conserved expression patterns in gray bichir and Siberian sturgeon, although the Siberian sturgeon Opn4m gene (Opn4m1) shows several specific expressions such as in the RPE cells of the retina and the cerebrospinal fluid-contacting neurons of the brain. Also, Opn4 proteins encoded by the intron-containing Opn4m genes in gray bichir and Siberian sturgeon share spectral and biochemical properties. These functional redundancies of opsins would have led to a random loss of the intron-containing or the intron-less opsin gene in the Acipenseriformes.
In our results of in situ hybridization analysis, we observed that teleost Opn4 genes show more widespread expression patterns in the brain than non-teleost Opn4 genes. However, we found that Opn4 genes in the Actinopterygii show several common distribution patterns in the retina and the brain, such as expression in the telencephalon and diencephalon. For example, all actinopterygian species we analyzed speculated to express the transcripts of the Opn4 genes in the horizontal cells of the retina. These results suggest that the Opn4-mediated photoreception in the horizontal cells is well conserved among the Actinopterygii (30). In addition, in all actinopterygian species analyzed in this study, we detected the expression signals of the Opn4 genes in the POA, where several other opsin genes such as the Opn3/tmt-opsins and Opn5 genes in the Actinopterygii have been reported to be expressed (31, 32). Also, several studies showed that Opn4 proteins in the zebrafish brain are involved in the regulation of locomotor activity in the larva (33, 34). Further comparative investigations will reveal the functional similarities and differences of the Opn4 genes among actinopterygian species and lead to the understanding of the physiological relevance of the presence of multiple Opn4 genes in the Actinopterygii.
In addition to the rhodopsin and Opn4 genes in the Actinopterygii, it has been reported that Cyprinodontiformes fishes have a retrogene (S180r) of long wavelength sensitive (LWS) cone visual pigments (11, 35). For example, guppy (Poecilia reticulate) has four LWS opsin genes, among which one is a retrogene (two exons) duplicated from a parental LWS opsin gene (six exons). The sequence comparison of the LWS opsins encoded by the retrogenes in the Cyprinodontiformes shows that guppy LWS-4 protein in the Poeciliidae has a histidine residue at position 181 (based on the bovine rhodopsin numbering system), whereas sheepshead minnow (Cyprinodon variegatus) LWS-4 protein in the Cyprinodontidae has a tyrosine residue at this position (Fig. S5) (36). Most LWS opsins in vertebrates have His181, which participates in the binding of Cl- to induce the red shift of λmax (37). By contrast, the opsins in the Glires, such as mouse (Mus musculus) in the Rodentia and rabbit (Oryctolagus cuniculus) in the Lagomorpha, have Tyr181, which is responsible for their blue-shifted λmax (38, 39, 40). This H181Y mutation has rarely been found in the opsins of species other than the Glires. Thus, it can be speculated that this unique mutation in the retrogene results in a blue shift of λmax of the LWS-4 protein and contributes to the expansion of the spectral sensitivities covered by the LWS proteins in the Cyprinodontidae.
In conclusion, we propose an evolutionary model of the teleost intron-less Opn4 gene, that is, the intron-less Opn4 gene emerged after branching of the Polypteriformes and became abundantly used in the Teleostei lineage. This stepwise evolutionary model is common with that of the teleost intron-less rhodopsin gene, which would have contributed to the different repertoires of the opsin genes and the diversity of the photoreceptive functions in the Actinopterygii. It should be noted that the intron-containing and intron-less genes of rhodopsin and Opn4 show several similar expression patterns in the retinas of actinopterygian species, such as rhodopsin genes in the rod cells and Opn4m genes in the cells abutting the OPL in the spotted gar retina. This suggests the possibility that these retrogenes were formed by the reverse transcription of mRNAs including the upstream regions of their parental genes (12). In fact, a previous study reported that the upstream sequences of intron-containing and intron-less rhodopsin genes of zebrafish share several common cis-elements responsible for the transcriptional regulation of the retinal genes (41). Further comparative analysis of the cis-regulation mechanisms between the intron-containing and intron-less opsin genes based on the accumulated genomic information will reveal the detailed evolutionary process of rhodopsin and Opn4 genes in the Actinopterygii.
Experimental procedures
Animals and ethics statement
Gray bichir (P. senegalus: ∼15 cm), Siberian sturgeon (A. baerii: ∼20 cm), and spotted gar (L. oculatus: ∼7 cm) were purchased from a local pet shop. Zebrafish (Danio rerio) was purchased from Kamihata Fish Industries Ltd. Medaka (Oryzias latipes) was maintained and bred at Kyoto University. The use of animals in these experiments was in accordance with guidelines established by the Ministry of Education, Culture, Sports, Science and Technology of Japan. The protocols in this paper were approved by the Animal Care and Use Committee of Kyoto University (permit number: H2919, H3015, 201913, 202012 and 202109).
Isolation of cDNA encoding opsin
To isolate the clones of gray bichir, spotted gar, zebrafish, and medaka Opn4, we searched them in the genomic databases and identified their full-length ORF sequences. The clones of gray bichir Opn4m (accession no. XM_039768409.1), spotted gar Opn4m1 (XM_006630896.2), Opn4m2 (LC743797) and Opn4x (XM_015340274.1), zebrafish Opn4m1 (GQ925715.1), Opn4m2 (GQ925716.1), Opn4m3 (GQ925717.1), Opn4x1 (GQ925718.1) and Opn4x2 (GQ925719.1) and medaka Opn4m1 (XM_023963602.1), Opn4m2 (XM_011486336.3), Opn4m3 (XM_020712260.2), Opn4x1 (XM_011481794.2), and Opn4x2 (XM_023958407.1) were isolated by PCR from the first strand cDNA of the eyes or brain as shown in our previous studies (13, 42). To isolate the clone of Siberian sturgeon Opn4m1, we first searched it in the transcriptome data deposited at the NCBI Sequence Read Archive (BioProject accession ID: SRX2426785). We successfully identified sequence reads including the start codon and stop codon of Opn4m1 ORF. Based on these sequence reads, we performed RT-PCR on eye RNA and isolated the full-length ORF sequence of Siberian sturgeon Opn4m1 (LC743796).
Preparation of Opn4 recombinant proteins
To improve the expression level of Opn4 recombinant proteins, we conducted C-terminus truncation according to our previous study on mouse Opn4 (23). We truncated 199 amino acid residues from the C-terminus of gray bichir Opn4m, 186 residues from Siberian sturgeon Opn4m1, 192 residues from spotted gar Opn4m1, 110 residues from spotted gar Opn4m2, 189 residues from spotted gar Opn4x, 205 residues from zebrafish Opn4m1, 125 residues from zebrafish Opn4m2, 184 residues from zebrafish Opn4m3, 198 residues from zebrafish Opn4x1, 132 residues from zebrafish Opn4x2, 167 residues from medaka Opn4m1, 96 residues from medaka Opn4m2, 157 residues from medaka Opn4m3, 79 residues from medaka Opn4x1, and 216 residues from medaka Opn4x2, which correspond to the deletion region of mouse Opn4 shown in our previous paper (23). The C-terminal truncated Opn4 cDNAs were tagged with the epitope sequence of the anti-bovine rhodopsin monoclonal antibody Rho1D4 (ETSQVAPA) at the C-terminus and were introduced into the mammalian expression vector pCAGGS (43). We also prepared the full-length cDNAs with the Rho1D4 epitope sequence and the C-terminal truncated cDNAs without the Rho1D4 epitope sequence for spotted gar Opn4m1, Opn4m2, and Opn4x. The plasmid DNA was transfected into HEK293S cells, which were maintained at 37 °C with 5% CO2, using the calcium phosphate method as previously described (23). 11-cis retinal was added to the medium (final concentration: 5 μM) 24 h after transfection. The cells were kept in the dark before they were collected at 48 h after transfection. The reconstituted pigments were extracted from cell membranes with 1% DDM in Buffer A (50 mM HEPES, 140 mM NaCl, pH 6.5). To remove other extract components from the DDM-solubilized sample, the elution was incubated with Rho1D4-conjugated agarose overnight and washed with 0.02% DDM and 20% glycerol (v/v) in Buffer A. The Opn4 pigments were eluted with 0.02% DDM and 20% glycerol in Buffer A containing the synthetic peptide that corresponds to the C-terminus of bovine rhodopsin. We carried out all procedures on ice under dim red light.
Spectroscopic measurements
UV/Vis absorption spectra were recorded using a spectrophotometer (UV2400, UV2450 or UV2600, Shimadzu) and an optical cell (width, 2 mm; light path, 1 cm). The sample temperature was maintained using a temperature controller (RTE-210, NESLAB) at 0 ± 0.1 °C. The sample was irradiated with light which was generated by a 1-kW tungsten halogen lamp (Master HILUX-HR, Rikagaku Seiki) and passed through an optical filter (Y-52, AGC Techno Glass).
Ca2+ level measurement in cultured cells
Ca2+ levels in cultured cells were measured using an aequorin-based bioluminescence assay (44, 45). To identify the effect of the C-terminus modifications on G protein signaling, we additionally prepared full-length cDNAs with 1D4 epitope (FL-1D4) and C-terminus truncated cDNAs without 1D4 epitope (CT-no1D4) encoding spotted gar Opn4m1, Opn4m2, and Opn4x proteins and were introduced into the mammalian expression vector pCAGGS. HEK293S cells were seeded in 96-well plates at a density of 60,000 cells/well in medium (D-MEM/F12 containing 10% FBS). After incubation for 24 h, the plasmid DNA was transfected into the cells (100 ng/well) using the polyethyleneimine transfection method. After incubation for 6 h, 11-cis retinal was added to the medium (final concentration: 5 μM). After overnight incubation, the medium was replaced with the equilibration medium (CO2-independent medium containing coelenterazine h and 10% FBS). Following equilibration for 2 h at room temperature, luminescence from the cells was measured using a microplate reader (SpectraMax L, Molecular Devices). The cells were irradiated for 5 s with blue LED light (450 nm) to trigger the change of the luminescence.
In situ hybridization
Tissue sample preparation and in situ hybridization were performed according to a previous paper (32) with slight modifications. The fish were euthanized by immersion in 0.04% MS-222 and immediate decapitation. After the eyes and brains were dissected from those fish, they were fixed overnight in PBS containing 4% PFA at 4 °C. The tissues were then cryoprotected by soaking overnight in PBS containing 20% sucrose, embedded in OCT compound, and frozen at −80 °C. Frozen tissues were sliced into 16-μm sections and affixed to glass slides. They were stored at −20 °C until use.
Digoxigenin-labeled riboprobes of Opn4 genes were synthesized from full-length cDNAs flanked by T7 and T3 promoter sequences which were inserted into pBluescript II KS (+) or pTA2 (TOYOBO). All of the following procedures were performed at room temperature (RT) unless otherwise noted. Tissue sections were sequentially immersed in 4% PFA in PBS for 15 min, 100% methanol for 30 min, PBS for 5 min, Proteinase K (0.5 μg/ml)/Tris-EDTA buffer (50 mM Tris-HCl, 5 mM EDTA, pH 7.6) for 15 min, 4% PFA in PBS for 15 min, dimethyl dicarbonate-treated water for 30 s, acetylation buffer (0.27% (v/v) acetic anhydride, 100 mM triethanolamine, pH 8.0) for over 10 min, PBS for 5 min, and hybridization buffer (750 mM NaCl, 75 mM sodium citrate, 0.4 mg/ml yeast RNA, 0.1 mg/ml heparin sodium, 1 × Denhardt's solution, 0.1% (v/v) Tween, 0.1% (w/v) CHAPS, 5 mM EDTA, 70% (v/v) formamide) at 65 °C for 3 h. Digoxigenin-labeled riboprobes (final concentration: 0.17 μg/ml) diluted in hybridization buffer were then applied to tissue sections and incubated at 65 °C for approximately 40 h.
After hybridization, the tissue sections were washed in 1 × SSC buffer (150 mM NaCl, 15 mM sodium citrate, pH 7.0) containing 50% formamide at 65 °C for 15 min and 1 h, followed by washing in 0.2 × SSC buffer at 65 °C for 1 h and MABT (100 mM maleate, 150 mM NaCl, 0.1% Tween 20, pH 7.5) three times for 30 min each. After washing, tissue sections were soaked in blocking buffer (1% (w/v) bovine serum albumin, 10% (v/v) sheep normal serum, 0.08% (v/v) Triton-X100 in PBS) for 30 min and then incubated with alkaline phosphatase-conjugated anti-digoxigenin Fab fragment (1:2000 dilution; Roche) overnight at 4 °C.
Tissue sections were then rinsed three times for 30 min each with MABT and twice for 5 min each with AP reaction buffer (100 mM Tris-HCl, 50 mM MgCl2, 100 mM NaCl, and 0.1% Tween 20, pH 9.5) for 5 min each. Color development was performed with 50 μg/ml nitro blue tetrazolium, 175 μg/ml 5-bromo-4-chloro-3-indolyl phosphate, 1 mM levamisole hydrochloride, and 5% (w/v) polyvinyl alcohol in AP buffer at 28 °C for 2 days. Tissue sections were counterstained with Nuclear Fast Red, dehydrated with 70 and 100% Tissue Dehydration Solution (FUJIFILM Wako Pure Chemical), and cleared by immersion in G-NOX (Genostaff). After clearing, the tissue sections were coverslipped with PARAmount mounting medium (FALMA).
Fluorescence in situ hybridization
The mRNA of medaka opn4 and gja10b were detected using signal amplification by exchanging reaction (SABER) with single-strand DNA probes and branches extended by primer exchanging reaction according to the original paper (46) with slight modifications. Probe sequences were designed using OligoMiner software (47). The synthesized single strand DNA probes and branches were confirmed to be 400 to 700 nucleotides in length and purified using DNA Clean & Concentrator-25 (Zymo Research).
The tissue sections were immersed in PBST (PBS containing 0.1% Tween 20), treated with 0.05 mg/ml pepsin in 0.01 M HCl at 37 °C for 20 min, and washed with PBST three times for 5 min each. The following hybridization and washing steps were repeated once for probes and twice for branch DNAs: The tissue sections were immersed in Whyb (0.3 M NaCl, 30 mM sodium citrate, 40% deionized formamide, pH 7.0) buffer for 10 min. Probes or branches diluted with Hyb1 (Whyb buffer containing 10% dextran sulfate) buffer were applied onto the sections at 1 μg/ml and incubated for 16 h. After hybridization, the tissue sections underwent a series of washes with Whyb for 30 min twice, 2 × SSC buffer containing 0.1% Tween 20 for 5 min twice. These steps were performed at 43 °C for probes and 37 °C for branches. After these iterations, the tissue sections were rinsed twice with PBST at RT, incubated with oligo DNAs labeled by fluorophore diluted in PBST at 0.1 μM at 37 °C for 1 h, rinsed twice with PBST again, and counterstained with 1 μg/ml Hoechst 33342 in PBST. Finally, coverslips were mounted on glass slides with homemade PVA/glycerol mounting medium.
Confocal fluorescence images were collected with a laser scanning confocal microscope system, Zeiss LSM 780 (Carl Zeiss Microscopy GmbH, Oberkochen, Germany) with 405, 488, and 561 nm laser lines at the Central Research Laboratory, Okayama University Medical School. Confocal z-stack images were acquired at 0.555 μm intervals. Confocal fluorescence images were manipulated by ZEN 2012 SP1 black edition (64 bit, version 8.1) and ImageJ (48). DNA sequences of probes, branches, hairpins, and oligos labeled by fluorophore, and combinations of applied probes and branches in the above procedure were summarized in Table S1.
Data availability
All data are available in the main text or the supplementary materials. The sequences reported in this paper have been deposited in the GenBank database (accession numbers LC743796 and LC743797).
Supporting information
This article contains supporting information (37, 38, 39, 40).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Prof. R. S. Molday for the generous gift of a Rho1D4-producing hybridoma and Prof. J. Nathans for providing the HEK293S cell line. We also thank Dr E. Nakajima for a critical reading of the manuscript.
Author contributions
C. F., K. S., H. O., T. Y. Conceptualization; C. F., K. S., T. Y. Investigation; C. F., K. S., T. Y. Writing - Original Draft; C. F., K. S., H. O., T. Y. Writing - Review & Editing; C. F., K. S., H. O., T. Y. Visualization; C. F., K. S., T. Y. Funding acquisition; T. Y. Supervision; T. Y. Project administration.
Funding and additional information
This work was supported in part by Japan Society for the Promotion of Science (JSPS) Grants-in-Aid for Scientific Research to C. F. (22J20019) and K. S. (20K08885), a grant from the Okayama Medical Foundation to K. S., grants from the Takeda Science Foundation to K. S. and T. Y., a grant from Fujiwara Natural History Foundation to C. F., Japan Science and Technology Agency (JST) CREST to T. Y. (JPMJCR1753) and Japan Agency for Medical Research and Development (AMED) CREST to T. Y. (22gm1510007). C. F. is supported by a JSPS Research Fellowship for Young Scientists.
Reviewed by members of the JBC Editorial Board. Edited by Kirill Martemyanov
Supporting information
References
- 1.Terakita A. The opsins. Genome Biol. 2005;6:213. doi: 10.1186/gb-2005-6-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shichida Y., Matsuyama T. Evolution of opsins and phototransduction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2009;364:2881–2895. doi: 10.1098/rstb.2009.0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porter M.L., Blasic J.R., Bok M.J., Cameron E.G., Pringle T., Cronin T.W., et al. Shedding new light on opsin evolution. Proc. Biol. Sci. 2012;279:3–14. doi: 10.1098/rspb.2011.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayak S.K., Jegla T., Panda S. Role of a novel photopigment, melanopsin, in behavioral adaptation to light. Cell Mol. Life Sci. 2007;64:144–154. doi: 10.1007/s00018-006-5581-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Do M.T., Yau K.W. Intrinsically photosensitive retinal ganglion cells. Physiol. Rev. 2010;90:1547–1581. doi: 10.1152/physrev.00013.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bellingham J., Foster R.G. Opsins and mammalian photoentrainment. Cell Tissue Res. 2002;309:57–71. doi: 10.1007/s00441-002-0573-4. [DOI] [PubMed] [Google Scholar]
- 7.Lamb T.D. Evolution of phototransduction, vertebrate photoreceptors and retina. Prog. Retin. Eye Res. 2013;36:52–119. doi: 10.1016/j.preteyeres.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 8.Pantzartzi C.N., Pergner J., Kozmik Z. The role of transposable elements in functional evolution of amphioxus genome: the case of opsin gene family. Sci. Rep. 2018;8:2506. doi: 10.1038/s41598-018-20683-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fitzgibbon J., Hope A., Slobodyanyuk S.J., Bellingham J., Bowmaker J.K., Hunt D.M. The rhodopsin-encoding gene of bony fish lacks introns. Gene. 1995;164:273–277. doi: 10.1016/0378-1119(95)00458-i. [DOI] [PubMed] [Google Scholar]
- 10.Bellingham J., Whitmore D., Philp A.R., Wells D.J., Foster R.G. Zebrafish melanopsin: isolation, tissue localisation and phylogenetic position. Brain Res. Mol. Brain Res. 2002;107:128–136. doi: 10.1016/s0169-328x(02)00454-0. [DOI] [PubMed] [Google Scholar]
- 11.Ward M.N., Churcher A.M., Dick K.J., Laver C.R., Owens G.L., Polack M.D., et al. The molecular basis of color vision in colorful fish: four long wave-sensitive (LWS) opsins in guppies (Poecilia reticulata) are defined by amino acid substitutions at key functional sites. BMC Evol. Biol. 2008;8:210. doi: 10.1186/1471-2148-8-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaessmann H., Vinckenbosch N., Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat. Rev. Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiyabu C., Sato K., Utari N.M.L., Ohuchi H., Shichida Y., Yamashita T. Evolutionary history of teleost intron-containing and intron-less rhodopsin genes. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mano H., Kojima D., Fukada Y. Exo-rhodopsin: a novel rhodopsin expressed in the zebrafish pineal gland. Brain Res. Mol. Brain Res. 1999;73:110–118. doi: 10.1016/s0169-328x(99)00242-9. [DOI] [PubMed] [Google Scholar]
- 15.Betancur R.R., Wiley E.O., Arratia G., Acero A., Bailly N., Miya M., et al. Phylogenetic classification of bony fishes. BMC Evol. Biol. 2017;17:162. doi: 10.1186/s12862-017-0958-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Provencio I., Jiang G., De Grip W.J., Hayes W.P., Rollag M.D. Melanopsin: an opsin in melanophores, brain, and eye. Proc. Natl. Acad. Sci. U. S. A. 1998;95:340–345. doi: 10.1073/pnas.95.1.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provencio I., Rodriguez I.R., Jiang G., Hayes W.P., Moreira E.F., Rollag M.D. A novel human opsin in the inner retina. J. Neurosci. 2000;20:600–605. doi: 10.1523/JNEUROSCI.20-02-00600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bellingham J., Chaurasia S.S., Melyan Z., Liu C., Cameron M.A., Tarttelin E.E., et al. Evolution of melanopsin photoreceptors: discovery and characterization of a new melanopsin in nonmammalian vertebrates. PLoS Biol. 2006;4:e254. doi: 10.1371/journal.pbio.0040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davies W.I., Zheng L., Hughes S., Tamai T.K., Turton M., Halford S., et al. Functional diversity of melanopsins and their global expression in the teleost retina. Cell Mol. Life Sci. 2011;68:4115–4132. doi: 10.1007/s00018-011-0785-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matos-Cruz V., Blasic J., Nickle B., Robinson P.R., Hattar S., Halpern M.E. Unexpected diversity and photoperiod dependence of the zebrafish melanopsin system. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du K., Stock M., Kneitz S., Klopp C., Woltering J.M., Adolfi M.C., et al. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 2020;4:841–852. doi: 10.1038/s41559-020-1166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bi X., Wang K., Yang L., Pan H., Jiang H., Wei Q., et al. Tracing the genetic footprints of vertebrate landing in non-teleost ray-finned fishes. Cell. 2021;184:1377–1391.e1314. doi: 10.1016/j.cell.2021.01.046. [DOI] [PubMed] [Google Scholar]
- 23.Matsuyama T., Yamashita T., Imamoto Y., Shichida Y. Photochemical properties of mammalian melanopsin. Biochemistry. 2012;51:5454–5462. doi: 10.1021/bi3004999. [DOI] [PubMed] [Google Scholar]
- 24.Naito T., Nashima-Hayama K., Ohtsu K., Kito Y. Photoreactions of cephalopod rhodopsin. Vis. Res. 1981;21:935–941. doi: 10.1016/0042-6989(81)90195-4. [DOI] [PubMed] [Google Scholar]
- 25.Ashida A., Matsumoto K., Ebrey T.G., Tsuda M. A purified agonist-activated G-protein coupled receptor: truncated octopus Acid Metarhodopsin. Zoolog. Sci. 2004;21:245–250. doi: 10.2108/zsj.21.245. [DOI] [PubMed] [Google Scholar]
- 26.Emanuel A.J., Do M.T. Melanopsin tristability for sustained and broadband phototransduction. Neuron. 2015;85:1043–1055. doi: 10.1016/j.neuron.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun L., Kawano-Yamashita E., Nagata T., Tsukamoto H., Furutani Y., Koyanagi M., et al. Distribution of mammalian-like melanopsin in cyclostome retinas exhibiting a different extent of visual functions. PLoS One. 2014;9 doi: 10.1371/journal.pone.0108209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukamoto H., Kubo Y., Farrens D.L., Koyanagi M., Terakita A., Furutani Y. Retinal attachment instability is diversified among mammalian melanopsins. J. Biol. Chem. 2015;290:27176–27187. doi: 10.1074/jbc.M115.666305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klaassen L.J., de Graaff W., van Asselt J.B., Klooster J., Kamermans M. Specific connectivity between photoreceptors and horizontal cells in the zebrafish retina. J. Neurophysiol. 2016;116:2799–2814. doi: 10.1152/jn.00449.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng N., Tsunenari T., Yau K.W. Intrinsic light response of retinal horizontal cells of teleosts. Nature. 2009;460:899–903. doi: 10.1038/nature08175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato K., Yamashita T., Haruki Y., Ohuchi H., Kinoshita M., Shichida Y. Two UV-sensitive photoreceptor proteins, Opn5m and Opn5m2 in ray-finned fish with distinct molecular properties and broad distribution in the retina and brain. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato K., Nwe K.N., Ohuchi H. The Opsin 3/Teleost multiple tissue opsin system: mRNA localization in the retina and brain of medaka (Oryzias latipes) J. Comp. Neurol. 2021;529:2484–2516. doi: 10.1002/cne.25106. [DOI] [PubMed] [Google Scholar]
- 33.Fernandes A.M., Fero K., Arrenberg A.B., Bergeron S.A., Driever W., Burgess H.A. Deep brain photoreceptors control light-seeking behavior in zebrafish larvae. Curr. Biol. 2012;22:2042–2047. doi: 10.1016/j.cub.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dekens M.P.S., Fontinha B.M., Gallach M., Pflugler S., Tessmar-Raible K. Melanopsin elevates locomotor activity during the wake state of the diurnal zebrafish. EMBO Rep. 2022;23 doi: 10.15252/embr.202051528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang C.H. Correlated expression of the opsin retrogene LWS-R and its host gene in two poeciliid fishes. Zool Stud. 2022;61:e16. doi: 10.6620/ZS.2022.61-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin J.J., Wang F.Y., Li W.H., Wang T.Y. The rises and falls of opsin genes in 59 ray-finned fish genomes and their implications for environmental adaptation. Sci. Rep. 2017;7 doi: 10.1038/s41598-017-15868-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z., Asenjo A.B., Oprian D.D. Identification of the Cl(-)-binding site in the human red and green color vision pigments. Biochemistry. 1993;32:2125–2130. doi: 10.1021/bi00060a001. [DOI] [PubMed] [Google Scholar]
- 38.Sun H., Macke J.P., Nathans J. Mechanisms of spectral tuning in the mouse green cone pigment. Proc. Natl. Acad. Sci. U. S. A. 1997;94:8860–8865. doi: 10.1073/pnas.94.16.8860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Davies W.I., Wilkie S.E., Cowing J.A., Hankins M.W., Hunt D.M. Anion sensitivity and spectral tuning of middle- and long-wavelength-sensitive (MWS/LWS) visual pigments. Cell Mol. Life Sci. 2012;69:2455–2464. doi: 10.1007/s00018-012-0934-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamashita T., Nakamura S., Tsutsui K., Morizumi T., Shichida Y. Chloride-dependent spectral tuning mechanism of L-group cone visual pigments. Biochemistry. 2013;52:1192–1197. doi: 10.1021/bi3016058. [DOI] [PubMed] [Google Scholar]
- 41.Asaoka Y., Mano H., Kojima D., Fukada Y. Pineal expression-promoting element (PIPE), a cis-acting element, directs pineal-specific gene expression in zebrafish. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15456–15461. doi: 10.1073/pnas.232444199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato K., Yamashita T., Kojima K., Sakai K., Matsutani Y., Yanagawa M., et al. Pinopsin evolved as the ancestral dim-light visual opsin in vertebrates. Commun. Biol. 2018;1:156. doi: 10.1038/s42003-018-0164-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niwa H., Yamamura K., Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 44.Bailes H.J., Lucas R.J. Human melanopsin forms a pigment maximally sensitive to blue light (lambdamax approximately 479 nm) supporting activation of G(q/11) and G(i/o) signalling cascades. Proc. Biol. Sci. 2013;280 doi: 10.1098/rspb.2012.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakai K., Ikeuchi H., Fujiyabu C., Imamoto Y., Yamashita T. Convergent evolutionary counterion displacement of bilaterian opsins in ciliary cells. Cell Mol. Life Sci. 2022;79:493. doi: 10.1007/s00018-022-04525-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kishi J.Y., Lapan S.W., Beliveau B.J., West E.R., Zhu A., Sasaki H.M., et al. SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat. Met. 2019;16:533–544. doi: 10.1038/s41592-019-0404-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beliveau B.J., Kishi J.Y., Nir G., Sasaki H.M., Saka S.K., Nguyen S.C., et al. OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E2183–E2192. doi: 10.1073/pnas.1714530115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Met. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morrow J.M., Lazic S., Chang B.S. A novel rhodopsin-like gene expressed in zebrafish retina. Vis. Neurosci. 2011;28:325–335. doi: 10.1017/S0952523811000010. [DOI] [PubMed] [Google Scholar]
- 50.Sukeena J.M., Galicia C.A., Wilson J.D., McGinn T., Boughman J.W., Robison B.D., et al. Characterization and Evolution of the Spotted Gar Retina. J. Exp. Zool B Mol. Dev. Evol. 2016;326:403–421. doi: 10.1002/jez.b.22710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beaudry F.E.G., Iwanicki T.W., Mariluz B.R.Z., Darnet S., Brinkmann H., Schneider P., et al. The non-visual opsins: eighteen in the ancestor of vertebrates, astonishing increase in ray-finned fish, and loss in amniotes. J. Exp. Zool B Mol. Dev. Evol. 2017;328:685–696. doi: 10.1002/jez.b.22773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. The sequences reported in this paper have been deposited in the GenBank database (accession numbers LC743796 and LC743797).