Summary
p97 is a human AAA+ (ATPase associated with diverse cellular activities, also known as valosin-containing protein [VCP]) ATPase, which is involved in diverse cellular processes such as membrane fusion and proteolysis. Lysine-specific methyltransferase of p97 (METTL21D) was identified as a class I methyltransferase that catalyzes the trimethylation of Lys315 of p97, a so-called VCP lysine methyltransferase (VCPKMT). Interestingly, VCPKMT disassembles a single hexamer ring consisting of p97-D1 domain and methylates Lys315 residue. Herein, the structures of S-adenosyl-L-methionine-bound VCPKMT and S-adenosyl-L-homocysteine-bound VCPKMT in complex with p97 N/D1 (N21-Q458) were reported at a resolution of 1.8 Å and 2.8 Å, respectively. The structures revealed the molecular details for the recognition and methylation of monomeric p97 by VCPKMT. Using biochemical analysis, we also investigated whether the methylation of full-length p97 could be sufficiently enhanced through cooperation between VCPKMT and the C terminus of alveolar soft part sarcoma locus (ASPL). Our study provides the groundwork for future structural and mechanistic studies of p97 and inhibitors.
Subject areas: Biochemistry, Molecular biology, Structural biology
Graphical abstract

Highlights
-
•
The crystal structures of VCPKMT and the VCPKMT-p97 N/D1 complex are presented
-
•
The structure explains the recognition and methylation of monomeric p97 by VCPKMT
-
•
A salt bridge between VCPKMT (D44) and p97 (R362) is the primary interaction
-
•
The C terminus of ASPL is sufficient to provide monomeric p97 for VCPKMT
Biochemistry; Molecular biology; Structural biology
Introduction
Methyltransferases (MTases) catalyze the transfer of a methyl group from an S-adenosyl-L-methionine (SAM) to various substrates, such as proteins, DNA, RNA, lipids, and small molecules, which are involved in biosynthesis, cell signaling, protein repair, and gene regulation.1,2,3 Among the five different MTase classes (I–V), the majority of known MTases belong to class I MTases, which are also designated as the SET (Su(var)3–9), enhancer-of-zeste, trithorax) domain proteins.3 Most lysine-specific class I MTases catalyze the transfer to histones, and histone-lysine methyltransferase (DOT1L) has been studied the most.4,5,6,7,8,9,10 A few class I MTases methylate lysine or arginine on non-histones and mainly recognize specific motifs in unstructured polypeptides.3
The p97 adenosine triphosphatase (ATPase) is also known as valosin-containing protein (VCP) in metazoans; and its Cdc48 orthologs in Saccharomyces cerevisiae segregate polyubiquitinated proteins to the proteasomes for degradation.11,12,13 Two ATPase domains, D1 and D2, of p97 form a double hexamer, and the N-terminal domain is in the peripheral region of the first ring consisting of D1 domain.14,15 p97 is engaged in a variety of cellular processes that cooperate with many adaptor proteins through its N-terminal domain.16,17 Lysine-specific methyltransferase of p97 (METTL21D), a lysine-specific MTase (KMT), catalyzes the trimethylation of Lys315 in p97-D1. Therefore, METTL21D is known as VCP lysine methyltransferase (VCPKMT).4,18 In addition, a single hexamer consisting of N and D1 domains was disassembled by VCPKMT.4 In in vitro studies, the movements of VCPKMT-deficient cells, such as proliferation, migration, and invasion, were shown to slow down.4
The alveolar soft part sarcoma locus (ASPL), also known as tether containing a ubiquitin regulatory X (UBX) domain for GLUT4 (TUG), controls the oligomeric status of p97.19 ASPL contains two potential binding sites for p97-N: the suppressor of high-copy PP1 (SHP) box and UBX domain.18,19 Crystallographic studies have revealed that the extended α-helical lariat structure at the N terminus of the UBX domain is critical for hexamer disassembly; thus, the C-terminal domain has been named as the extended UBX domain (eUBX).20 Genetic studies have revealed that the eUBX domain of ASPL can enhance the methylation of full-length p97 by VCPKMT.18 Interestingly, the methylation of disease-related mutant p97 (R155H, R159G, and R191Q) by VCPKMT was not increased by ASPL.18
In this study, we determined the crystal structures of SAM-bound VCPKMT and S-adenosyl-L-homocysteine (SAH)-bound VCPKMT in complex with p97 N/D1 (N21-Q458). Our structures, along with biochemical studies, revealed the binding mode of a class I lysine-specific MTase to the substrate protein and key residues involved in theses interactions.
Results
Overall structure of VCPKMT
Human VCPKMT has been reported as a class I lysine-specific protein MTase, which methylates Lys315 of p97 with high specificity.4,18 To elucidate the mechanistic details, we determined the crystal structures of SAM-bound VCPKMT and SAH-bound VCPKMT in complex with p97 N/D1 (N21-Q458). Detailed statistics related to data collection and refinement are summarized in Table 1. After the model of VCPKMT was refined successfully, omit maps (Fo – Fc) for SAM were obtained (Figure 1A). The asymmetric unit contained one molecule of VCPKMT covering residues 16 to 227, and a SAM molecule, even though full-length VCPKMT was used for crystallographic studies. During this study, the crystal structure of human VCPKMT was deposited in the RCSB Protein Data Bank (RCSB PDB) under the access code of 4LG1 without publication. The overall fold of the structure shown in the topological diagram represents a typical class I MTase fold consisting of a central seven-stranded β sheet (S3 – S9) flanked on both sides by five α helices (H1–H5) and a short helix (H6) (Figures 1A and S1A). The SAM molecule was located in front of the loop between S3 and H2. In the β sheet, S5–S7 and S8 were observed to be parallel to each other, but S9, which is placed between S7 and S8, was antiparallel to the others (Figures 1A and S1A). Interestingly, VCPKMT was noted to have a pair of β strand (S1 and S2) and a helix (H1) at the N terminus as a noble structural feature. Prior to our study, only a few structures of human class I lysine-specific protein MTases had been charaterized.5,21 To gain a deeper understanding of the structural diversity of this enzyme class, we compared the crystal structure of human DOT1L (RCSB PDB code: 1NW3) and human Calmodulin-lysine MTase (RCSB PDB code: 4PWY) to that of VCPKMT (Figure S2). Despite the lack of sequence similarity among the three enzymes, we were able to superimpose their structures based on the typical class I MTase fold which consists of central seven-stranded β sheets. The structure comparison revealed that the N-terminal domain exhibited structural diversity (Figure S2). Furthermore, VCPKMT had the most compact conformation of the N-terminal domain.
Table 1.
Crystallographic statistics for data collection and refinement
| SeMet VCPKMT (peak) | VCPKMT (+SAM) | VCPKMT in complex with VCP ND1 (+SAH) | |
|---|---|---|---|
| PDB ID | 8HL6 | 8HL7 | |
| Data collection | |||
| Wavelength (Å) | 0.9791 | 1.0000 | 1.0000 |
| Space group | P3221 | P3121 | P43212 |
| Total reflections | 412,136 | 613,125 | 336,520 |
| Unique reflections | 19,355 | 23,428 | 26,103 |
| Cell dimensions | |||
| a, b, c (Å) | 98.8, 98.8, 96.2 | 58.1, 58.1, 129.3 | 60.7, 60.7, 473.9 |
| (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 28.56–2.80 (2.95–2.80) | 64.66–1.80 (1.90–1.80) | 30.00–2.80 (2.95–2.80) |
| Rpima | 0.036 (0.255) | 0.023 (0.683) | 0.058 (0.310) |
| CC1/2 | 0.998 (0.924) | 0.999 (0.405) | 0.974 (0.890) |
| I/σ I | 18.4 (4.4) | 17.2 (0.9) | 8.8 (2.3) |
| Completeness (%) | 99.8 (99.9) | 96.2 (81.5) | 99.8 (100.0) |
| Multiplicity | 20.1 (20.7) | 9.4 (4.6) | 12.7 (10.7) |
| MR-SAD phasing | Figure of merit 0.29 |
||
| Refinement | |||
| PDB ID | |||
| Resolution (Å) | 32.75–1.80 (1.88–1.80) | 29.57–2.80 (2.91–2.80) | |
| No. reflections | 23,334 (2,394) | 23,283 (2,297) | |
| Rwork/Rfreeb | 0.193/0.226 (0.335/0.366) | 0.218/0.261 (0.319/0.418) | |
| No. atoms | |||
| Protein | 1,638 | 4,600 | |
| Ligand/ion | 27 | 70 | |
| Water | 98 | 10 | |
| B-factors | |||
| Protein | 49.1 | 66.26 | |
| Ligand | 37.0 | 61.60 | |
| Water | 52.8 | 45.55 | |
| R.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.009 | |
| Bond angles (°) | 0.923 | 1.34 | |
| Ramachandran plot (%) | |||
| Favored | 96.19 | 92.35 | |
| Allowed | 3.81 | 7.65 | |
| Outliers | 0 | 0 | |
Values in parentheses refer to the highest resolution shell.
Rpim = Σhkl|[1/(N-1)]1/2 Σi|Ii(hkl)- |/Σ hkl Σ iIi(hkl).
Rwork = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where Rfree was calculated for a randomly chosen 5% of reflections that were not used for structure refinement, and Rwork was calculated for the remaining reflections.
Figure 1.
Overview of the Valosin-Containing Protein Lysine Methyltransferase (VCPKMT)
(A) Front view of VCPKMT. The stick model represents the S-adenosyl-L-methionine (SAM) molecule, overlaid with polder Fo – Fc difference maps contoured at 3.0 times the RMSD (green).
(B) Zoomed view of the VCPKMT active site and SAM molecule, highlighting key interacting residues shown as stick model representations. Water molecules are depicted as red balls, and hydrogen bonds are represented by black dotted lines. The stick model representation of the SAM molecule is consistent with that shown in Figure 1A.
Next, we analyzed the binding pocket for SAM in our structure. The narrow channel formed in front of the methyl group of SAM is a plausible substrate-binding site for a targeted lysine (Figure S1B). The entrance of the SAM-binding pocket is wide at the adenine group of SAM, but it becomes narrower toward the center of the pocket. The inside of the SAM-binding pocket is a conserved acidic pocket formed by the GxGxG motif in motif I, whereas the other side of the pocket is more hydrophobic for binding the adenine group and the methionine moieties (Figures S1B and S3). The detailed interactions are shown in Figure 1B. Sequence analysis indicated that VCPKMT contains conserved sequence motifs denoted as motif I (residues 71–79, located between S3 and H2), motif post I (residues 92–96, located in S4), and motif II (residues 143–147, located between S6 and H4) (Figure S3). Motif I containing Gly75 along with post I containing Asp96 forms the back side of the pocket, and motif II containing Tyr147 and Tyr148 forms the left side and front of the pocket. Interestingly, two tryptophan residues, Trp43 and Trp126, contributed to the hydrophobic environment of SAM (Figure 1B). Specifically, Trp43, Try147, and Tyr148 were observed to generate a hydrophobic environment for the methyl moiety of SAM (Figures 1B and S1B). The indole ring of Trp126 forms a π–π stacking interaction with the adenosyl moiety of SAM, and the hydroxyl group of Ser151 forms a hydrogen bonding with the N6 atom of SAM (Figure 1B). Furthermore, the amide of Trp126 forms a hydrogen bond with the N1 atom of SAM (Figure 1B). Interestingly, despite not being part of the motifs, Y148 forms a π–interaction with the adenosyl moiety of SAM. These additional interactions between Y148 and SAM are likely to contribute to the stability of the protein-SAM complex as well as the specificity of SAM toward its target protein, along with other key residues mentioned above.
Overall structure of p97 N-D1 in complex with VCPKMT
To gain insights into the structural details of the VCPKMT-p97 complex, the crystal structure of the SAH-bound VCPKMT in complex with p97 N/D1 (N21-Q458) was determined at a resolution of 2.8 Å. First, we focused on the reconstitution and purification of the VCPKMT-p97 complex. We used a pull-down method to ensure that VCPKMT with a C-terminal His-tag could capture the non-tagged p97 during immobilized metal affinity chromatography. Although we screened three fragments of p97 for crystallographic studies (D1, N/D1, or N/D1/D2), the N-D1 fragment (residues N21–Q458) in complex with VCPKMT could produce crystals. The structure of the complex was solved by molecular replacement using wild-type p97 N/D1 (N21-Q458) as a phasing model. However, the N and D1 domains had to be used as separated models, indicating that the arrangement between the N and D1 domains differs from that of the wild type. The asymmetric unit contained a copy of N-D1 and a copy of VCPKMT. The structure was successfully refined, converging to Rfree and Rwork of 0.260 and 0.214, respectively (Table 1). The final models of the VCPKMT contained 207 residues from Phe16 to Lys222, and the following two loops were observed to be disordered: one was located between H1 and S3 (residues 60–64) and the other was located between S5 and S6 (residues 131–133). The linker of p97 N/D1 (residues 190–199) and C terminus of the D1 domain (residues 433–444) was observed to be completely disordered (Figures 2A and 2B). During iterative structure refinement, we found that the electron density maps of Lys315 represented trimethylation (Figures 2C and S4). The ε-ammonium group of Lys315 was observed to have a spherical shape in the polder maps (Fo – Fc), even though the electron density maps of SAH were not clear because of resolution limit (Figure 2C). The degree of methylation on Lys315 was not clear at this resolution; however it is known that VCPKMT catalyzes preferencially the trimethylation of p97 both in vitro and in vivo.4,18 The successful refinement of the trimethylated lysine was achieved through iterative refinement processes, which revealed the presence of positive difference electron density maps (Fo – Fc) around the trimethylated lysine (Figure 2C). The shape of the 2Fo – Fc difference electron density maps was a distinguished lysine residue from the other lysine residues (Figure S4). The substrate loop (residues 305–323) between S15 and H9 in the D1 domain was found to be surrounded by the three loops of VCPKMT. The first loop is between S2 and H1, the second is motif II between S6 and H4, and the third is between S7 and H5 (Figures 2A and 2B). The interface area of the complex between VCPKMT and p97 was 1158.7 Å2. The details of these interactions are discussed in subsequent sections.
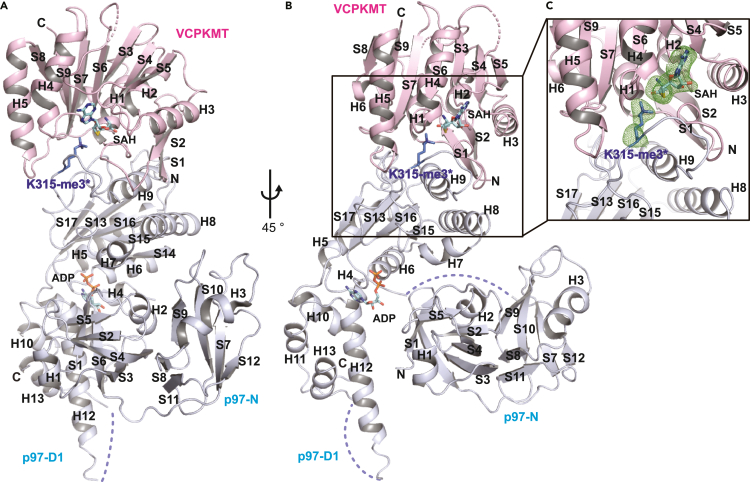
Figure 2.
Overview of the VCPKMT in Complex with p97-N/D1
(A) Front view of the complex. The color codes are as follows: VCPKMT (pink) and p97 (blue). The stick model representations of S-adenosyl-L-homocysteine (SAH) molecule in VCPKMT and adenine dinucleotide phosphate (ADP) molecule in p97 are shown with the same color code as that of SAM molecule in Figure 1A. The side chain of the trimethylated lysine is depicted as a stick model representation. The dotted lines represent the disordered loop in p97.
(B) Side view of the complex, with a 45-degree counterclockwise rotation from Figure 2A.
(C) Zoomed view of the VCPKMT active site. The stick model representations of s-adenosyl-l-homocysteine (SAH) molecule and the side chain of trimethylated lysine are superimposed with polder Fo – Fc difference maps contoured at 3.0 times the RMSD (green).
Biochemical analysis of binding interface between p97 and VCPKMT
VCPKMT has been characterized as a lysine-specific MTase that methylates Lys315 in p97-D1. To determine the kinetic parameters of VCPKMT, we used two enzymes, namely SAH nucleosidase and adenine deaminase.22 As these two enzymes are 100-fold more active than general MTase (kcat≒1 min−1), the kinetics of coupled enzyme reactions can be used to determine the kinetic parameters of MTase (kcat = 4.12 ± 0.10 s−1 for SAH nucleosidase and kcat = 35.2 ± 0.92 s−1 for adenine deaminase).22 The following kinetic parameters were determined for the trimethylation of the p97-N/D1 (N21-Q458) as a substrate: Vmax = 29.412 nmol min−1 mg−1, Km = 3.697 μM, and kcat = 0.759 min−1 (Figure 3B). Protein-interfaces-surfaces-and-sssemblies (PISA) analysis revealed that VCPKMT bound tightly to p97-D1, which was also confirmed by the Michaelis constant.23 The tight binding affinity was mainly attributed to three salt bridges (Figure 3A). First, the carboxyl group of Aps44 in VCPKMT is in proximity of the guanidinium group of Arg362 in p97, and the distances are 2.7 Å and 3.4 Å, respectively. Next, the guanidinium group of Arg18 in VCPKMT also forms hydrogen bonds with Thr330 and Asp333 in p97, and the distances of those bonds are 2.6 Å and 2.3 Å, respectively. The third salt bridge includes the corresponding hydrogen bonds between Glu174/Asp214 in VCPKMT and Arg359 in p97, and the distances of those bonds are 2.4 Å and 2.6 Å, respectively. In addition to the three salt bridges, the interaction between VCPKMT and p97 is facilitated by a perpendicular pi-pi interaction between Y210 of VCPKMT and F360 of p97. Y210 is a highly conserved residue in VCPKMT, with the exception of Acyrthosiphon pisum, which has phenylalanine on that position (Figure S3). Furthermore, R359 and F360 in p97 are part of the second region of homology (SRH), located within the nucleotide-binding site of the adjacent subunit in the p97 hexamer (Figure S7).24,25,26,27 Next, we investigated the contribution of each salt bridge in the methylation of p97-N/D1 (N21-Q458) using an in vitro assay (Figure 3C). Interestingly, most of the methylation activity was abolished by a single mutation in VCPKMT, Asp44. A total of 90% of the activity was lost upon the mutation of Asp44, but approximately 80% of the activity was retained in the case of other mutations, including R18A, D214A, and E174A. The results obtained from the in vitro assay correlated with those from the nickel-nitrilotriacetic acid (Ni-NTA) pull-down assay, which utilized a C-terminal His-tagged VCPKMT (Figure 3D). The binding affinity of VCPKMT for p97-N/D1 (N21-Q458) was abolished by a single mutation in its sequence, specifically D44A. The presence of GST-tagged p97 was not detected by Coomassie blue staining. The other VCPKMT mutations, namely R18A, D214A, and E174A, did not affect the band intensity in the SDS-PAGE analysis. The mutants underwent additional further analysis to assess their abilities to disassemble the p97-N/D1 (N21-Q458) hexamer. This analysis involved size-exclusion chromatography combined with multi-angle light scattering (SEC-MALS) and native PAGE (Figure S5 and S6). The results indicated that D44A mutant of VCPKMT has experienced a significant loss in its ability to disassemble the p97-N/D1 (N21-Q458) hexamer, while its MTase activity, which remains to be fully understood, is still retained. Thus, our results have clearly indicated that the binding affinity between the VCPKMT and p97 mainly originates from a salt bridge between Asp44 of VCPKMT and Arg362 of p97. Furthermore, we identified an acidic residue, D144, that stabilizes the positive-charged ε-ammonium group of Lys315 in the substrate loop of p97. Previous studies have demonstrated that the activity of VCPKMT is abolished by the mutation of D144V.4
Figure 3.
Structural Details and Mutational Binding Analysis of the Interface
(A) The key interaction residues of the interface are depicted as stick model representations, with hydrogen bonds represented by black dotted lines. The color codes used are consistent with those in Figure 2.
(B) The initial reaction rate was plotted using eight different concentrations of the substrate p97-N/D1 to analyze the Michaelis-Menten kinetics. The data are presented as mean values with the standard errors obtained from three independent measurements.
(C) The data presented in the figure represent the mean relative methyltransferase activity of both the wild-type and mutant VCPKMT, along with the standard errors obtained from three independent measurements. The specific activity of the wild-type VCPKMT was set as the reference value at 100%.
(D) An in vitro His-tag pull-down assay was performed using VCPKMT-His and GST-p97.
Structural changes in p97 and VCPKMT upon forming heterodimeric complex
One of the notable features of VCPKMT is its ability to disrupt the p97 single hexamer composed of the D1 domain. However, it was found that the double hexamer of p97-D1/D2 presented challenges in terms of disassembly and methylation by VCPKMT.4,18 In order to comprehend how VCPKMT disrupts the hexamer of p97-D1, we superimposed the structure of the heterodimer complex formed by VCPKMT and p97-N/D1 onto the hexamer of the p97-N/D1 (Figure 4). Given the interest in the conformation state of the N-terminal domain within the heterodimer, we compared the structure of the heterodimer with two conformations of p97-N/D1. These conformations correspond to an N-terminal domain in a down conformation (PDB entry: 4KOD) and an up conformation (PDB entry: 4KO8).24 The movement of p97-N involves a 111.1° rotation from the down conformation and 168.0° rotation from the up conformation, occurring around a hinge located at residues from Glu187 to Val201 in the N-D1 linker region (Figures 4C and 4D).28 N-D1 linker of p97 is found to be disordered in the structure of the heterodimer, indicating significant conformational changes in the N-terminal domain of p97. Furthermore, it is noteworthy that the key residues responsible for the formation of the p97 hexamer are situated at the interface of the heterodimer, implying that the binding of VCPKMT leads to the disruption of the p97 hexamer (Figures 4A and 4B). The substrate loop (residues 305–323) between S15 and H9 in p97-D1 is also stretched out to the binding pocket of VCPKMT (Figures 4A and 4B). To address the conformational changes in VCPKMT upon binding to p97, the apo structure was compared to that of the complex (Figure 5). The most striking conformational changes were observed in the two loops that capture the substrate loop containing Lys315 of p97. The first loop between S2 and H1, which is the right side of the substrate loop, is moved to the substrate loop of p97 by a distance of 9.0 Å, and the second loop between S7 and H5, which is the left side of the substrate loop, also approaches the substrate loop of the p97 at a distance of 2.6 Å. The side chains of the two tyrosine residues of the VCPKMT, Tyr147 and Tyr148, swing out to form a hydrogen bond between Try147 of VCPKMT and His317 of p97. Tyr417 of VCPKMT is a highly conserved residue in motif II of the class I MTase. The trimethylated lysine is located within a hydrophobic environment composed of V38, W43, Y147, and Y148, following a conformational change of VCPKMT.
Figure 4.
Comparison of the Heterodimer (Comprising VCPKMT and p97-N/D1) with Two Conformations of p97-N/D1 Hexamer (PDB Entry: 4KOD and 4KO8)
(A) Superposition of the p97-D1 domains from the p97-N/D1 hexamer (PDB entry: 4KOD) and the heterodimer comprising VCPKMT and p97-ND1. The position of the p97-N domain is the same as its position in the individual structures. The color codes are as following: p97-N domain (green) and D1 domain (light orange) from the hexamer, p97-N domain (marine), D1 domain (light blue), and VCPKMT (light pink) from the heterodimer. The cartoon representations of the N-D1 linker are shown in red. The key residues involved in the formation of the p97 hexamer are highlighted in bold, with the D1 domain in the hexamer shown in orange and the D1 domain in the heterodimer shown in slate color.
(B) Superposition of the p97-D1 domains from the p97-N/D1 hexamer (PDB entry: 4KO8) and the heterodimer comprising VCPKMT and p97-ND1. The position of the p97-N domain is the same as its position in the individual structures. The color codes are as following: p97-N domain (dark gray) and D1 domain (light green) from the hexamer, p97-N domain (marine), D1 domain (light blue), and VCPKMT (light pink) from the heterodimer. The cartoon representations of the N-D1 linker are shown in red. The key residues involved in the formation of the p97 hexamer are highlighted in bold, with the D1 domain in the hexamer shown in deep teal and the D1 domain in the heterodimer shown in slate color.
(C) The p97-N domain is rotated upon VCPKMT binding to the p97-D1 domain (PDB entry: 4KOD) along the axis indicated by a black arrow which lies on the N-D1 linker. Domain movements of p97-N domain were analyzed by using the Dyndom server.29 The color codes used are consistent with those in Figure 4A.
(D) The p97-N domain is rotated upon VCPKMT binding to the p97-D1 domain (PDB entry: 4KO8) along the axis indicated by a black arrow which lies on the N-D1 linker. Domain movements of p97-N domain were analyzed by using the Dyndom server.29 The color codes used are consistent with those in Figure 4B.
Figure 5.
Superposition of the unbound VCPKMT and the p97-bound VCPKMT from the heterodimer comprising VCPKMT and p97-N/D1
The color codes used for the heterodimer are consistent with those in Figure 2. The unbound VCPKMT is represented as a gray cartoon. The conformational changes of VCPKMT upon binding to p97 are indicated by black dotted arrows. Hydrogen bonds are represented by black dotted lines.
Enhancement in in vitro methylation for full-length p97 upon assistance by ASPL
ASPL, which contains a tandem ubiquitin-like (UBL) domains, an 8-residue SHP box, and an eUBX domain, was reported to disassemble a p97 hexamer through a direct interaction19 (Figure 6A). In vitro studies have revealed that the methylation of full-length p97 by VCPKMT was enhanced upon the addition of ASPL, which directly interacts with the MTase.18 Interestingly, ASPL did not show a stimulatory effect when disease-related mutants of p97 (R155H, R159H, and R191Q) were used.18 Structural studies of the p97:ASPL complex demonstrated that the α-helical lariat preceding the UBX domain directly disrupted D1:D1 protomer interactions, resulting in the dissociation of the p97 hexamer.20 We screened six fragments of ASPL to identify the minimal domain that enhances the methylation of full-length p97 by VCPKMT. First, we confirmed using a Ni-NTA pull-down assay that VCPKMT did not bind to ASPL (Figure S8). Next, using in vitro enzyme and Ni-NTA pull-down assays, we found that the minimal domains of ASPL enhanced the methylation (Figure 6B). Although the SHP box has been recognized as a binding motif to the N-terminal domain of p97, we were unable to determine the impact of the SHP box in ASPL.30 This might be attributed to the proteolysis of the relevant fragment (ASPL-C1). Previous studies have indicated that the N-terminal domains (residues 1–290) and disordered C-terminal amino acids (residues 499–553) do not exhibit binding to full-length p97.19,20 Expanding on these findings, our study demonstrated that the C-terminal fragment, consisting of the eUBX domain and an additional N-terminal 27 residues (residues 291–498), significantly enhanced the methylation of the full-length p97, akin to the behavior observed with full-length ASPL. We also successfully demonstrated the formation of a ternary complex involving p97, the ASPL-C3 fragment, and VCPKMT, as confirmed by pull-down assays using a C-terminal His-tagged full-length p97, non-tagged VCPKMT, and non-tagged ASPL (Figure 6C). Furthermore, we have demonstrated that ASPL can enhance the methylation of full-length p97 by assisting in the disassembly of the full-length p97 hexamer. However, this enhancement was not observed when the full-length p97 was replaced with the p97-N/D1 hexamer (N21-Q458) (Figure 6B). Therefore, while ASPL is not an essential component for the methylation of p97-N/D1 hexamer (N21-Q458), it appears to be a prerequisite for the methylation of full-length p97 as it provides monomeric p97 to VCPKMT.
Figure 6.
In vitro Methylation of Full-length p97 by VCPKMT with The Assistance of ASPL
(A) Schematic diagrams presenting the domains of ASPL.
(B) The data represent the mean relative methyltransferase activity of wild-type VCPKMT, along with the standard error, obtained from three independent measurements. The components of each reaction are summarized in the table below. The specific activity using p97-N/D1 (N21-Q458) as a substrate was designed as the reference value and set at 100%.
(C) An in vitro His-tag pull-down assay was performed using full-length p97-His, non-tagged VCPKMT, and non-tagged ASPL.
Discussion
In this study, our structures revealed how VCPKMT can disrupt a single hexamer of p97 N/D1 (N21-Q458) and methylate a Lys315 of p97, as supported by biochemical experiments. Most lysine-specific MTases catalyze the addition of a methyl group on histones; however, a few lysine-specific MTases target lysine or arginine in the substrate protein. In humans, the histone-specific DOT1L is the most studied MTase. DOT1L transfers a methyl group from cofactor SAM to H3 substrate.5 Similarly, VCPKMT catalyzes the transfer of a methyl group from cofactor SAM to Lys315 of p97. The binding mode of MTase to the substrate protein is quite different; however auxiliary modification for both MTases is required prior to methylation. While DOT1L has the ability to bind to ubiquitinated histone H3,6,7,8,9,10 the full-length p97 requires monomerization by ASPL prior to its binding with VCPKMT. Surprisingly, a salt bridge between Asp44 of VCPKMT and Arg362 of p97 mainly contributes to the interaction between the MTase and p97. In addition, the interaction between the heterodimer composed of VCPKMT and p97-N/D1 involves the contribution of R359 and F360 in p97, which are located within the nucleotide-binding site of the neighboring subunit in the p97 hexamer and are recognized as the SRH. Furthermore, The binding of VCPKMT induces a significant conformational change in p97-N/D1 (N21-Q458). The conformation of p97-N in the heterodimer is similar to the up conformation, even though the p97-D1 domain contains adenosine-5′-diphosphate (ADP). During the review process of our manuscript, a publication reported the structure of the ternary complex involving p97-N/D1 (M1-G481), VCPKMT, and ASPL (V313-S500).31 Intriguingly, in that structure, the conformation of p97-N resembles a down conformation bound to ASPL, while p97-D1 contains adenosine-5′-triphosphate (ATP) (Figure S9). However, the mechanism by which the p97-N/D1 linker in our structure displays high flexibility, resulting in the repositioning and conformational changes of p97-N, remains unclear. We also investigated how full-length p97 could be methylated by VCPKMT with the assistance of ASPL. Recent structural studies have shown that ASPL disrupts the D1:D1 protomer interactions to disassemble the double hexamer p97.20 Our biochemical studies revealed that the C terminus (P291-A498), including the eUBX domain with an N-terminal coiled-coil domain of ASPL, is a minimal region that forms a ternary complex and enhances the methylation of full-length p97 by VCPKMT. The previous in vitro assays demonstrated that the addition of ASPL does not enhance the methylation of p97 mutants (R155H, R159G, and R191Q) by VCPKMT.18 These p97 mutants (R155H, R159G, and R191Q) exhibit lower ADP binding affinity at the D1 domain compared with the wile type p97, as they disrupt the intersubunit communication within the p97 hexamer, resulting in elevated ATPase activities in the D1 and D2 domains.24 This effect is attributed to the fixed side chain conformations of R359, F360, and R362, particularly F360, which are part of the SRH motifs and remain unchanged regardless of the nucleotide type (ATP or ADP) in p97 mutants.24 The conformational changes of the SRH motifs induced by the mutation on p97 may hinder the binding of VCPKMT to p97, while ASPL facilitates the interaction by providing monomeric forms of the p97 mutants. The hypothesized function of downregulating ATPase activity in the D1 domain, as confirmed by previous studies, may be a potential role in regulating p97 against disease-related mutations.18 Further research is needed to identify additional protein factors that regulate the activity of the D2 domain and the overall chaperone activities of p97. Additionally, conducting further structural studies on methylated p97 subsequent to the dissociation of ASPL and VCPKMT is crucial in order to obtain a comprehensive understanding of the functionalities associated with methylated p97.
Limitations of the study
Structural and biochemical analyses have elucidated the molecular mechanism underlying the recognition and disassembly of the p97-N/D1 hexamer. However, further investigation is needed to understand the physiological function of methylation on Lys315 in p97. Additionally, the role of ASPL in p97 methylation remains elusive.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Escherichia coli: DH10B | Invitrogen | Cat# EC0113 |
| Escherichia coli: Rosetta 2 | Novagen | Cat# 71402 |
| Critical commercial assays | ||
| SaltRx 1 | Hampton Research | Cat# HR2-107 |
| SaltRx 2 | Hampton Research | Cat# HR2-109 |
| Index | Hampton Research | Cat# HR2-144 |
| PEG/Ion | Hampton Research | Cat# HR2-126 |
| PEG/Ion 2 | Hampton Research | Cat# HR2-098 |
| The Protein Complex Suite | Qiagen | Cat# 130715 |
| The Classic Suite | Qiagen | Cat# 130701 |
| The Classic II Suite | Qiagen | Cat# 130723 |
| Deposited data | ||
| hVCPKMT | This work | PDB:8HL6 |
| hVCPKMT-p97-N/D1 (N21-Q458) complex | This work | PDB:8HL7 |
| Oligonucleotides | ||
| 5′-agatatacatatggcttctggagccgat | This work | FLp97_F |
| 5′-cgcggatccgccatacaggtcatcatc | This work | FLp97_R |
| 5′-agatatacatatgaaccgtcccaatcggtta | This work | p97ND1_F |
| 5′-cgcggatccctggctcaaggcccaccg | This work | p97ND1_His_R |
| 5′-cgcggatccttactggctcaaggcccaccg | This work | p97ND1_non-R |
| 5′-cgcggatccaaccgtcccaatcggtta | This work | p97ND1_GST_F |
| 5′-cgcgaattcctggctcaaggcccaccg | This work | p97ND1_GST_R |
| 5′-agatatacatatggccgacacgctggaatcg | This work | FLKMT_F |
| 5′-ctgttctcgaggctaggaaatttagactt | This work | FLKMT_R |
| 5′-agatatacatatgtttgtgcgcgtgcttgag | This work | 16KMT_F |
| 5′-cgcggatccgccgacacgctggaatcg | This work | FLKMT_BH_F |
| 5′-cgcgaattcttagctaggaaatttagactt | This work | FLKMT_ER_R |
| 5′-cgcgaattcatggcggccccggca | This work | FLASPL_F |
| 5′-cgcaagcttttacctcttgctggccgg | This work | FLASPL_R |
| 5′-cgcaagcttttaggcagctgcgggtgc | This work | ASPL_240_R |
| 5′-cgcgaattcccctgctgcgagcac | This work | 223_ASPL_F |
| 5′-cgcgaattcgggggccctcctgggccc | This work | 253_ASPL_F |
| 5′-cgcgaattcccccagcaggagcaggag | This work | 291_ASPL_F |
| 5′-cgcaagcttttaggcggccctggacat | This work | ASPL_498_R |
| 5′-cgcaagcttttacacctttgggtagcg | This work | ASPL_391_R |
| 5′-gctgctggtaccatgaataaagaagcgcta | This work | AD_F |
| 5′-gctgcttctagattattgcagtgatatgtg | This work | AD_R |
| 5′-gctgctggtaccatgaaaatcggcatcatt | This work | SN_F |
| 5′-gctgcttctagattagccatgtgcaagttt | This work | SN_R |
| Recombinant DNA | ||
| Plasmid: modified pET26b-NdeI-p97(1-806)-BamHI with C-terminal 6xHis-tag | This work | Genbank ID: NM_007126 |
| Plasmid: modified pET26b-NdeI-p97(21-458)-BamHI (non-tag) | This work | Genbank ID: NM_007126 |
| Plasmid: modified pET26b-NdeI-p97(21-458)-BamHI with C-terminal 6xHis-tag | This work | Genbank ID: NM_007126 |
| Plasmid: pGEX-6P-1-GST-BamHI-p97(21-458)-EcoRI | This work | Genbank ID: NM_007126 |
| Plasmid: pET26b-NdeI-VCPKMT(1-229)-XhoI with C-terminal 6xHis-tag | This work | Synthesized gene |
| Plasmid: pET26b-NdeI-VCPKMT(16-229)-XhoI with C-terminal 6xHis-tag | This work | Synthesized gene |
| Plasmid: modified pET26b -6xHis-tag-TEV protease recognition site, ENLYFQG-BamHII-VCPKMT(1-229)-EcoRI | This work | Synthesized gene |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli-TEV protease recognition site, ENLYFQG -EcoRI-ASPL(1-553)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli-TEV protease recognition site, ENLYFQG -EcoRI-ASPL(1-240)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli -TEV protease recognition site, ENLYFQG -EcoRI-ASPL(223-553)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli -TEV protease recognition site, ENLYFQG -EcoRI-ASPL(253-553)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli -TEV protease recognition site, ENLYFQG -EcoRI-ASPL(291-498)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: modified pET26b -6XHis-tag-Thiredoxin1 from E. coli -TEV protease recognition site, ENLYFQG -EcoRI-ASPL(253-391)-HindIII | This work | Genbank ID: NM_024083 |
| Plasmid: pCOLD1-6xHis-tag-KpnI-Adenine deaminase from Bacillus subtilis-XbaI | This work | Genbank ID: BAA07052 |
| Plasmid: pCOLD1-6xHis-tag-KpnI-MTA/SAH nucleosidase from E. coli-XbaI | This work | Genbank ID: AAC38291 |
| Software and algorithms | ||
| HKL2000 suite | Otwinowski and Minor,32 | https://hkl-xray.com |
| CCP4 Program Suite | https://www.ccp4.ac.uk | |
| PHENIX Suite | https://phenix-online.org/ | |
| Coot | Emsley et al.33 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PyMOL | PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC | https://pymol.org/2/ |
| Origin | Origin Lab, USA | https://www.originlab.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fullfilled by the lead contact, Prof. Jin Kuk Yang (jinkukyang@ssu.ac.kr).
Materials availability
Plasmids used in this study are available from the lead contact.
Method details
Cloning
The details pertaining to the plasmids used in this study are summarized in key resources table. Briefly, the gene was amplified using PCR and cloned into the corresponding vector. The nucleotide sequences of the inserts were confirmed by sequencing (Bionics, Korea).
Protein expression and purification
All proteins were expressed in Escherichia coli Rosetta2 (DE3) strain (Novagen, USA) and purified using immobilized metal affinity chromatography. Proteins were further purified by size-exclusion chromatography (Superdex-200, Cytiva, USA) pre-equilibrated with a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM sodium chloride, 0.1 mM Tris(2-carboxyethyl)phosphine hydrochloride, and 5 % (v/v) glycerol.
Crystallization and structure determination
Initial crystallization screening was performed using the sitting drop vapor diffusion method at 295 K using crystallization reagent kits supplied by Hampton Research (USA) and Qiagen (Germany). The best crystal of hVCPKMT in complex with p97 N/D1 was grown at 295 K using the hanging drop vapor diffusion method by mixing 1.0 μL of protein solution with an equal amount of reservoir solution containing 20 % (w/v) polyethylene glycol (PEG) 3350, 0.1M Na-HEPES (pH 7.5), 2 % (v/v) Tacsimate (pH 7.0), and 3 % (w/v) 6-aminohexanoic acid along with the micro seed crystals after a few days later. The best crystal of the hVCPKMT was grown at 295 K with the same method, but the composition of the reservoir solution was 25 % (w/v) PEG 3350, 0.1 M Na-HEPES (pH 7.5), 0.2 M sodium chloride, and 3 % (w/v) sucrose. Selenomethionine (SeMet) hVCPKMT (full-length) was crystallized by applying a microseeding technique using native VCPKMT crystals. The crystal of hVCPKMT in the presence of SAM was grown at 295 K using the same method, but the composition of the reservoir solution was 20 % PEG 1000, 0.1 M Na/K phosphate (pH 6.2), and 0.2M sodium chloride. The concentrations of PEG and glycerol were gradually increased by adding cryo-solution before the crystal was directly flash-cooled in liquid nitrogen for data collection. The SAD data for SeMet VCPKMT were collected at the K-edge of Se on an ADSC Quantum Q315R CCD detector at the beam line 5C of the Pohang Accelerator Laboratory (Republic of Korea), and the native data were collected at beam line 5C of the Pohang Accelerator Laboratory (Republic of Korea). The X-ray diffraction data were indexed, integrated, and scaled using MOSFLM and SCALA in the CCP4 program suite. The Phaser EP in the CCP4 program suite was used to locate anomalous scatters, for phase calculation, and for density modification. The Phenix suite was used for the initial model building. SAD phasing produced high-quality density maps covering the most of the VCPKMT (F16 – F227), except for 15 residues at the N-terminus and the two residues at the C-terminus. The crystal structure of SAM-bound VCPKMT was determined by the molecular replacement, using the apo-VCPKMT model obtained in this study as a search model. The structure including SAM was further refined to 0.193 of Rwork and 0.226 of Rfree at a resolution of 1.8 Å. The initial dataset from a crystal consisting of hVCPKMT in complex with p97 N/D1 showed low completeness because of the blind region caused by the long crystallographic axis (473.9 Å). Therefore, the mounting loop was bent perpendicular to the rotation axis, and the completeness and resolution of the dataset were then improved.34 The initial model of hVCPKMT in complex with p97 N-D1 was determined by molecular replacement with Phaser in the CCP4 program suite using the hVCPKMT model (this study) and p97 N-D1 model (PDB: 1S3S). Subsequent manual rebuilding and refinement were performed iteratively using Coot and phenix.refine in Phenix suite. Figures were prepared using PyMOL (PyMOL Molecular Graphics System, Version 2.3.2 Schrödinger, LLC).
Site directed mutagenesis
hVCPKMT mutants were generated using the QuikChange Site-Directed Mutagenesis Kit (Stratagem, USA). The expression and purification of the mutants were performed following the same protocol used for the wile-type strain.
Preparation of the enzyme for methyltransferase assay
The genes encoding SAH nucleosidase from Escherichia coli strain B (ATCC 11303, USA) and adenine deaminase from Bacillus subtilis strain 168 (ATCC 23857D-5, USA) were amplified from the genomic DNA using PCR. The amplified gene was cloned into the pCOLD1 vector (Takara, Japan) using the restriction enzymes, Kpn I and Xba I, both of which were expressed in the Rosetta2 (DE3) strain (Novagen, USA), and purified using a HisTrap column (Cytiva, USA). The eluted protein was further dialyzed into the storage buffer containing 50 mM Tris-HCl (pH 7.5), 5 mM magnesium chloride, 50 mM potassium chloride, and 20 % (v/v) glycerol using a desalting column (Cytiva, USA).
Michaelis-Menten kinetics
The in vitro methyltransferase activity of VCPKMT was assessed by measuring the decrease in absorbance at 265 nm in a reaction coupled with SAH nucleosidase (EC 3.2.2.9) and adenine deaminase (EC 3.5.4.2), as previously described in the literature.22 In this assay, the SAH, which is produced by VCPKMT, is enzymatically hydrolyzed by SAH nucleosidase, resulting in the formation of S-ribosylhomocysteine and adenine. Furthermore, recombinant adenine deaminase converts the adenine generated from SAH to hypoxanthine and ammonia through a deamination reaction. This deamination process leads to a measurable decrease in absorbance at 265 nm, which can be continuously monitored during the assay. The reaction mixture contained 60 μM s-adenosyl-L-methionine (SAM), 2.76 μM VCPKMT, 208 μM SAH nucleosidase, 0.1 μM adenine deaminase, and 1050 μM manganese sulfate, along with eight different concentrations of p97 N/D1 (N21 - Q458) in 50 mM Tris-HCl (pH 7.5), 5 mM magnesium chloride, 50 mM potassium chloride, and 10 % (v/v) glycerol. The reaction was initiated by adding p97 to the mixture and was monitored at 265 nm and 310 K using a UV-3101PC UV-Visible spectrophotometer (Shimadzu, Japan) in thermostatted 1-cm quartz cuvette for 90 seconds. Three independent measurements were carried out.
In vitro relative methyltransferase assay
The overall experimental procedure was the same as that used for assessing the Michaelis-Menten kinetics, with the addition of 2.4 μM p97 at the start of the reaction. When ASPL was included in the assay, 7.2 μM of ASPL was added into the mixture along with p97. The reaction was monitored at 265 nm and 310 K using a UV-3101PC UV-Visible spectrophotometer (Shimadzu, Japan). Three independent measurements were conducted.
In vitro His-tag pull-down assay
The Ni-NTA resin (Cytiva, USA) was mixed with purified proteins, and incubated overnight at 277 K. Afterward, the resin was washed six times with the binding buffer (20 mM Tris-HCl (pH 8.0), 0.1 mM TCEP, 100 mM sodium chloride, 5 % (v/v) glycerol, and 20 mM imidazole). the elutes were assessed using SDS-PAGE with the glycine buffer system.
Size-exclusion chromatography combined with multi-angle light scattering (SEC-MALS)
Individual purified VCPKMT and p97-N/D1 (N21-Q458) were mixed at 2:1 ratio, ensuring an excess amount of VCPKMT was presented. The mixture was then incubated at 277 K for two hours before being subjecting to size-exclusion chromatography column. A total of 100 μl of protein solution was loaded onto a Superdex 200 Increase 10/300 GL column (Cytiva, USA), which has been pre-equilibrated with a buffer containing 20mM Tris-HCl, 0.1mM TCEP, and 200mM NaCl. The elution of the protein was carried out at a flow rate of 0.5 ml/min and was subsequently applied to inline DAWN Heleos II MALS and Optilab T-Rex differential refractive index detector (Wyatt Technology, USA). The resulting data were analyzed using the ASTRA 6 software package (Wyatt Technology, USA) and the final results were visualized using Origin software (OriginLab, USA).
Native PAGE
A total of 50 μl of the same protein mixture, consisting of VCPKMT and p97-N/D1 (N21-Q458), intended for SEC-MALS analysis, was mixed with the sampling buffer. The sampling buffer consisted of 625 mM Tris-HCl (pH 8), 20% v/v glycerol, and 0.02% (w/v) bromophenol blue. Subsequently, 10 μl of resulting mixture was loaded onto Mini-PROTEAN TGX Precast Gel (Bio-rad, USA). The gel was then subjected to electrophoresis at 70 V for 2 hours in ice-cold running buffer composed of 192 mM glycine and 25 mM Tris. Finally, the gel was analyzed by staining with Coomassie Bleu R-250.
Quantification and statistical analysis
The kinetic assays in the manuscript were conducted with three replicates, and data are presented as the mean with the standard error. Statistical details of experiments, where applicable, can be found in figure legends.
Acknowledgments
This research was supported by the Basic Science Research Program of the National Research Foundation of Korea (NRF) (2021R1A6A1A10044154 and 2022R1C1C1004221 to W.K.; 2019R1F1A1063268 and 2017R1D1A1B03035446 to J.K.Y.). We thank the staff at Beamline 5C of the Pohang Accelerator Laboratory in the Republic of Korea for their assistance in X-ray data collection. Additionally, we would also like to express our gratitude to the staff at the Korea Basic Science Institute for their contributions in conducting SEC-MALS analyses.
Author contributions
Conceptualization, W.K. and J.K.Y.; Investigation, N.T.Q., S.K, J.K., S.J., W.K., and J.K.Y.; Writing - Original Draft, W.K. and J.K.Y.; Writing - Review & Editing, W.K. and J.K.Y.; Supervision, W.K. and J.K.Y.; Funding Acquisition, W.K. and J.K.Y.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 26, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107222.
Contributor Information
Wonchull Kang, Email: wonchullkang@ssu.ac.kr.
Jin Kuk Yang, Email: jinkukyang@ssu.ac.kr.
Supplemental information
Data and code availability
-
•
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 8HL6 (VCPKMT), and 8HL7 (VCPKMT-p97N/D1 complex) and are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Feoli A., Viviano M., Cipriano A., Milite C., Castellano S., Sbardella G. Lysine methyltransferase inhibitors: where we are now. RSC Chem. Biol. 2022;3:359–406. doi: 10.1039/d1cb00196e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelraheem E., Thair B., Varela R.F., Jockmann E., Popadic D., Hailes H.C., Ward J.M., Iribarren A.M., Lewkowicz E.S., Andexer J.N., et al. Methyltransferases: Functions and Applications. Chembiochem. 2022 doi: 10.1002/cbic.202200212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubert H.L., Blumenthal R.M., Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Biochem. Sci. 2003;28:329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kernstock S., Davydova E., Jakobsson M., Moen A., Pettersen S., Mælandsmo G.M., Egge-Jacobsen W., Falnes P.Ø. Lysine methylation of VCP by a member of a novel human protein methyltransferase family. Nat. Commun. 2012;3:1038. doi: 10.1038/ncomms2041. [DOI] [PubMed] [Google Scholar]
- 5.Min J., Feng Q., Li Z., Zhang Y., Xu R.M. Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell. 2003;112:711–723. doi: 10.1016/s0092-8674(03)00114-4. [DOI] [PubMed] [Google Scholar]
- 6.Jang S., Kang C., Yang H.S., Jung T., Hebert H., Chung K.Y., Kim S.J., Hohng S., Song J.J. Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase. Genes Dev. 2019;33:620–625. doi: 10.1101/gad.323790.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anderson C.J., Baird M.R., Hsu A., Barbour E.H., Koyama Y., Borgnia M.J., McGinty R.K. Structural Basis for Recognition of Ubiquitylated Nucleosome by Dot1L Methyltransferase. Cell Rep. 2019;26:1681–1690.e5. doi: 10.1016/j.celrep.2019.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yao T., Jing W., Hu Z., Tan M., Cao M., Wang Q., Li Y., Yuan G., Lei M., Huang J. Structural basis of the crosstalk between histone H2B monoubiquitination and H3 lysine 79 methylation on nucleosome. Cell Res. 2019;29:330–333. doi: 10.1038/s41422-019-0146-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Worden E.J., Hoffmann N.A., Hicks C.W., Wolberger C. Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L. Cell. 2019;176:1490–1501.e12. doi: 10.1016/j.cell.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valencia-Sánchez M.I., De Ioannes P., Wang M., Vasilyev N., Chen R., Nudler E., Armache J.P., Armache K.J. Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination. Mol. Cell. 2019;74:1010–1019.e6. doi: 10.1016/j.molcel.2019.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia D., Tang W.K., Ye Y. Structure and function of the AAA+ ATPase p97/Cdc48p. Gene. 2016;583:64–77. doi: 10.1016/j.gene.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer H., Weihl C.C. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. J. Cell Sci. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erzberger J.P., Berger J.M. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 14.DeLaBarre B., Brunger A.T. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat. Struct. Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee S., Bartesaghi A., Merk A., Rao P., Bulfer S.L., Yan Y., Green N., Mroczkowski B., Neitz R.J., Wipf P., et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeung H.O., Kloppsteck P., Niwa H., Isaacson R.L., Matthews S., Zhang X., Freemont P.S. Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 2008;36:62–67. doi: 10.1042/BST0360062. [DOI] [PubMed] [Google Scholar]
- 17.Buchberger A., Schindelin H., Hänzelmann P. Control of p97 function by cofactor binding. FEBS Lett. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- 18.Cloutier P., Lavallée-Adam M., Faubert D., Blanchette M., Coulombe B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orme C.M., Bogan J.S. The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment. J. Biol. Chem. 2012;287:6679–6692. doi: 10.1074/jbc.M111.284232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arumughan A., Roske Y., Barth C., Forero L.L., Bravo-Rodriguez K., Redel A., Kostova S., McShane E., Opitz R., Faelber K., et al. Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers. Nat. Commun. 2016;7 doi: 10.1038/ncomms13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnani R., Dirk L.M.A., Trievel R.C., Houtz R.L. Calmodulin methyltransferase is an evolutionarily conserved enzyme that trimethylates Lys-115 in calmodulin. Nat. Commun. 2010;1:43. doi: 10.1038/ncomms1044. [DOI] [PubMed] [Google Scholar]
- 22.Dorgan K.M., Wooderchak W.L., Wynn D.P., Karschner E.L., Alfaro J.F., Cui Y., Zhou Z.S., Hevel J.M. An enzyme-coupled continuous spectrophotometric assay for S-adenosylmethionine-dependent methyltransferases. Anal. Biochem. 2006;350:249–255. doi: 10.1016/j.ab.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 24.Tang W.K., Xia D. Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. J. Biol. Chem. 2013;288:36624–36635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karata K., Inagawa T., Wilkinson A.J., Tatsuta T., Ogura T. Dissecting the role of a conserved motif (the second region of homology) in the AAA family of ATPases. Site-directed mutagenesis of the ATP-dependent protease FtsH. J. Biol. Chem. 1999;274:26225–26232. doi: 10.1074/jbc.274.37.26225. [DOI] [PubMed] [Google Scholar]
- 26.Davies J.M., Brunger A.T., Weis W.I. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 27.Zhang X., Shaw A., Bates P.A., Newman R.H., Gowen B., Orlova E., Gorman M.A., Kondo H., Dokurno P., Lally J., et al. Structure of the AAA ATPase p97. Mol. Cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 28.Qi G., Lee R., Hayward S. A comprehensive and non-redundant database of protein domain movements. Bioinformatics. 2005;21:2832–2838. doi: 10.1093/bioinformatics/bti420. [DOI] [PubMed] [Google Scholar]
- 29.Veevers R., Hayward S. Methodological improvements for the analysis of domain movements in large biomolecular complexes. Biophys. Physicobiol. 2019;16:328–336. doi: 10.2142/biophysico.16.0_328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le L.T.M., Kang W., Kim J.Y., Le O.T.T., Lee S.Y., Yang J.K. Structural Details of Ufd1 Binding to p97 and Their Functional Implications in ER-Associated Degradation. PLoS One. 2016;11 doi: 10.1371/journal.pone.0163394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrović S., Roske Y., Rami B., Phan M.H.Q., Panáková D., Heinemann U. Structural remodeling of AAA+ ATPase p97 by adaptor protein ASPL facilitates posttranslational methylation by METTL21D. Proc. Natl. Acad. Sci. USA. 2023;120 doi: 10.1073/pnas.2208941120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dauter Z. Data-collection strategies. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1703–1717. doi: 10.1107/s0907444999008367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 8HL6 (VCPKMT), and 8HL7 (VCPKMT-p97N/D1 complex) and are publicly available as of the date of publication.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.