Graphical abstract
Schematic diagram displays the potential mechanism of MAT2A-induced ferroptosis in osteosarcoma.
Keywords: Osteosarcoma, Ferroptosis, MAT2A, miR-26b-5p, STAT3/SLC7A11
Highlights
-
•
MAT2A depletion dramatically inhibited tumor progression of osteosarcoma in vivo.
-
•
miR-26b-5p/MAT2A regulates tumor malignant progression by controlling p-STAT3 and SLC7A11 expressions.
-
•
miR-26b-5p/MAT2A triggers ferroptosis in osteosarcoma cells by increasing intracellular ferrous iron levels and inhibiting the STAT3/SLC7A11 axis.
Abstract
Osteosarcoma (OS) is the most frequent primary malignant bone tumor. Ferroptosis, a form of regulated cell death, is a key tumor suppression mechanism. Although methionine adenosyltransferase II alpha (MAT2A) has been reported to inhibit several tumor cells, it is unclear whether inhibition of MAT2A in OS cells can reduce ferroptosis. CCK-8, flow cytometry, and Transwell assays were performed to evaluate cell viability, cell apoptosis/cycle, and cell migration, respectively. The levels of ferrous iron and glutathione (GSH) levels in cells were measured to evaluate the degree of cell ferroptosis. Western blot analysis was performed to detect protein levels of MAT2A, p-STAT3 (Ser727)/STAT3, and solute carrier family 7 member 11 (SLC7A11) in OS cells. MAT2A was significantly upregulated in OS specimens and high MAT2A expression was associated with a poorer prognosis in OS patients. shRNA targeting MAT2A significantly increased OS cell apoptosis, triggered cell cycle arrest in the G2 phase, and attenuated migration ability in vitro. MAT2A depletion dramatically inhibited tumor progression of OS in vivo. Overexpression of MAT2A rescued the tumor inhibition caused by miR-26b-5p. MAT2A knockdown promoted OS cell ferroptosis. miR-26b-5p/MAT2A regulates tumor malignant progression and OS cell ferroptosis by controlling p-STAT3 and SLC7A11 expressions. Taken together, our study displayed that miR-26b-5p/MAT2A triggers ferroptosis in OS cells by increasing intracellular ferrous iron levels and inhibiting the STAT3/SLC7A11 axis. Our results reveal a MAT2A-mediated ferroptosis defense mechanism used by OS cells and propose a potential ferroptosis-inducing strategy for the treatment of OS patients.
1. Introduction
Osteosarcoma (OS) is one of the most frequent primary malignant bone tumors for children and adolescents, which is characterized by a high propensity for local invasion and metastasis, as well as recurrence after therapy [1]. Although combining surgery with chemotherapy has immensely improved the outcomes of OS patients, the prognosis of OS with metastatic or recurrent is still unsatisfactory [2]. Targeted therapy has proven to be a promising therapeutic strategy against human malignancies that are often used in combination with chemotherapy, and biomarker development has increased the number of patients who benefit from targeted therapies in recent years [3]. However, OS is a very heterogeneous tumor, both at the intra- and inter-tumor level, with no identified driver mutation. Consequently, efforts to improve treatments using targeted therapies have faced this lack of specific OS targets [4].
Cells undergo ferroptosis as part of their apoptosis, invasion, and metastasis processes. Divalent iron or ester oxygenase is present during ferroptosis, which catalyzes the reduction of lipid-derived unsaturated fatty acids from cell membranes, causing lipid peroxidation and induces cell death [5], [6]. Ferroptosis terminates in mitochondrial dysfunction and toxic lipid peroxidation [6]. In addition to breast cancer, and colorectal cancer [7], [8], ferroptosis also exerts anticancer effects on OS [9], [10]. Such as, KDM4A regulates SLC7A11 transcription and OS cell ferroptosis by controlling H3K9me3 demethylation in the promoter region of SLC7A11, suggesting that KDM4A activity may be a potential therapeutic target for OS treatment [11]. STAT3 inhibitor reactivated ferroptosis in the OS cells and consequently increased sensitivity to cisplatin [12]. The in-depth study of ferroptosis is likely to reveal other molecular targets that could be used to treat patients with OS.
Cell metabolism provides energy and material basis for cellular signaling transduction [13]. Methionine adenosyltransferase (MAT) catalyzes the synthesis of S-adenosyl methionine (SAM) from methionine and ATP, and SAM is an essential amino acid in animals that plays a crucial role in human cancer owing to its activities in epigenetic regulation, especially DNA methylation [14]. The metabolic enzyme methionine adenosyltransferase 2α (MAT2A) was identified as a synthetic lethal target in MTAP-deleted cancers, which accounts for 15% of all cancer types [15]. MAT2A inhibitors substantially reduce SAM and demonstrate antiproliferative activity in MTAP-deleted cancer cells and tumors[16], such as osteosarcomas. Anti-cancer therapies are less effective when MAT2A is expressed in cancer cells [17]. Activation of MAT2A-related pathways was proved to be carcinogenic progress to promote proliferation and metastasis in various cancers [18], [19]. An increasing number of studies have implicated the vital role of metabolic pathways in the regulation of ferroptosis [20]. However, whether MAT2A regulates ferroptosis in osteosarcoma cells or not, and its underlying mechanism remains largely elusive.
Emerging evidence suggested that MAT2A was verified as a target gene of several miRNAs, which formed miRNAs/MAT2A axis were involved in cancer progression [21], [22], [23]. miRNAs exhibit functions by binding to the 3′-untranslated regions of target mRNAs and suppressing their expression[24]. Some studies have revealed a relationship between miRNAs and ferroptosis. In radioresistant cells, miR-7-5p inhibited ferroptosis via downregulating mitoferrin and thus reducing iron levels. Furthermore, miR-9 and miR-137 enhanced ferroptosis via reduction of intracellular GSH levels. Moreover, miR-6852 inhibited growth of lung cancer cells via promoting ferroptosis[24].The overexpression or inhibition of certain miRNAs plays a significant role in limiting the invasion and migration of OS cells, miRNA mimics and antagonists may help improve the efficacy of conventional chemotherapy agents in the treatment of metastatic OS [25]. Earlier in this study, a series of online databases and differential analysis, miR-26b-5p was found be abnormally expressed in OS. miR-26b-5p is known to be involved in the progression of multiple cancers, for example, breast cancer[26], stomach cancer[27], esophageal cancer[28], liver cancer[29] and osteosarcoma [30], [31]. miR-26b-5p inhibits or promotes cancer progression. miR-26b-5p/CCNB1 axis promotes the tumorigenesis and metastasis of osteosarcoma[30], and accelerates chemo-resistance of osteosarcoma[31]. Current studies have shown that ferroptosis is closely related to OS and could reduce chemotherapy resistance to a certain extent, which has great therapeutic potential[32]. Based on the above evidences, we hypothesized that miR-26b-5p might be involved in the progression of osteosarcoma by regulating ferroptosis, but the detailed mechanism between mir and ferroptosis is still unclear.
Studies have shown that ferroptosis-related gene signature associates with immunity and predicts prognosis accurately in patients with osteosarcoma[33]. In order to further elucidate the relationship between MAT2A and the occurrence, development and prognosis of osteosarcoma, and whether knocking out MAT2A could be a drug target for the treatment of osteosarcoma in the future, and explore novel therapies for OS, we conducted this study. The present study aims to investigate the role and underlying mechanisms of MAT2A in regulating ferroptosis and osteosarcoma progression.
2. Materials and methods
2.1. Patient samples
The study was approved by the Ethics Committee of Zhongshan Hospital, Fudan University (Y2014-185) according to the Declaration of Helsinki. Written informed consent was obtained from all participants or their guardians (for children younger than 18 years). Two cohorts of patients involving 76 OS tissues and 21 paired normal tissues were obtained from Zhongshan Hospital, Fudan University. The clinicopathological features of 76 OS patients are listed in Table 1. These data do not contain identity-related information. None of these patients had received preoperative chemotherapy or radiation therapy. The cancer and para-cancerous specimens were divided into two parts: one part was frozen immediately in a −80 °C refrigerator and the other was fixed in 4% paraformaldehyde that was subsequently embedded in paraffin for immunohistochemistry experiments.
Table 1.
Relationship between MAT2A expression and tumor characteristics in patients with osteosarcoma.
| Features | No. of patients | MAT2A expression |
p value | |
|---|---|---|---|---|
| low | high | |||
| All patients | 76 | 33 | 43 | |
| Age (years) | 0.037* | |||
| <26 | 38 | 21 | 17 | |
| ≥26 | 38 | 12 | 26 | |
| Gender | 0.36 | |||
| Male | 51 | 24 | 27 | |
| Female | 25 | 9 | 16 | |
| Grade malignancy | 0.0008*** | |||
| G2 | 41 | 25 | 16 | |
| G3 | 35 | 8 | 27 | |
| T Infiltrate | 0.827 | |||
| T1 | 13 | 6 | 7 | |
| T2 | 63 | 27 | 36 | |
| lymphatic metastasis (N) | 0.849 | |||
| N0 | 74 | 32 | 42 | |
| N1 | 2 | 1 | 1 | |
| Stage | 0.0008*** | |||
| 1 | 41 | 25 | 16 | |
| 2/4 | 35 | 8 | 27 | |
2.2. Cell culture and treatments
Normal osteoblasts (hFOB 1.19) and OS cell lines (MNNG/HOS and U2OS) were purchased from American Type Culture Collection (ATCC) (Manassas, VA, USA). Culture medium was Roswell Park Memorial Institute 1640 (RPMI-1640) (Corning, USA)) with 10% fetal bovine serum (FBS) (Gibco), and 1% penicillin/streptomycin (Invitrogen). All cells were maintained in a humidified environment at 5% CO2 and 37 ◦C.
Until 70–80% of confluence, cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Briefly, overexpressing (OE) plasmid, a negative control (NC) vector, and short hairpin RNA (shRNAs) targeting MAT2A and miR-26b-5p mimics or NC were purchased from Guangdong RiboBio LTD.(China), Briefly, cells were transfected with the plasmids using Lipofectamine 2000 (Invitrogen, China). Transfection solution was stood for 20 min and applied in each well. Transfected cells were harvested at 48 h for the following experiments. Expression levels of target genes were determined by immunoblotting with corresponding antibodies.
In some cases, OS cells (1.0×106/per well) were plated into a 6-well plated and cultured in complete media in the presence or absence of MAT2A inhibitor (FIDAS-5) (10 μM) (Merck Millipore, USA) and/or the iron chelator deferoxamine (DFO, 100 μM) for 24 h.
2.3. Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA (Invitrogen) was extracted using TRIzol reagent (Invitrogen, USA) and their relative complementary deoxyribose nucleic acid (cDNA) was synthesized by Primescript RT Reagent (TaKaRa, Japan). QRT-PCR was performed by using StepOne Plus Real-Time PCR system (Applied Biosystems, USA) with SYBR® Premix Ex Taq™ Reagent (TaKaRa). QRT-PCR was performed at 95 °C for 5 min, 95 °C for 15 s, 58 °C for 30 s, and 74 °C for 30 s, for a total of 40 cycles. The following primers were used for qRT-PCR: MAT2A, F: 5ʹ-ATGAACGGACAGCTCAACGG-3′, R: 5ʹ-CCAGCAAGAAGGATCATTCCAG-3′; miRNA-26b-5p, F: 5ʹ-CCTGTGGAGATTGATGGGGT-3′, R: 5ʹ-TCTCTGGGCCTCTGACATTC-3′; U6, F: 5ʹ-CTCGCTTCGGCAGCACA-3′, R: 5ʹ-AACGCTTCACGAATTTGCGT-3′; GAPDH, F: 5ʹ-GGGAGCCAAAAGGGTCAT-3ʹ, R: 5ʹ-GAGTCCTTCCACGATACCAA-3′.
2.4. Western blotting analysis
The cells were lysed and homogenized using RIPA Lysis Buffer (Beyotime, China) with a protease inhibitor. The protein concentration of the samples was measured using the BCA Protein Assay Kit (Beyotime, China). Proteins were denatured in boiling water for five minutes after adding them to loading buffer (Beyotime, China). Each protein sample (40 μg) was loaded onto an SDS-PAGE 10% gel and transferred to the PVDF membranes (Millipore, USA). The membranes were blocked with 5% skim milk for 2 h, and incubated with the primary antibodies, including anti-MAT2A antibody (1:2500, Abcam, ab154343), anti-SLC7A11 antibody (1:5000; Cell Signaling Technology, 12691), and anti-STAT3 antibody (1: 1000; Affinity Bioscience, AF6294,), anti-p-STAT3 (7 0 5) antibody (1: 1000; Affinity Bioscience, AF3293), at 4 °C overnight. The membranes were subsequently incubated with the secondary antibody for 2 h at room temperature. Finally, the bands were exposed using the electrochemiluminescence (ECL) and analyzed by Image Software (NIH, USA).
2.5. Cell counting kit-8 assay
To measure cell viability, 1×104 cells per well were seeded in a 96-well plate 1 day before treatment. After indicated treatments, each well was replaced with a fresh medium containing Cell Counting Kit-8 (CCK8) reagent (Sigma). Cell viability was then analyzed every 24 hr. The absorbance values of the samples were read at 450 nm using a fluorescence microplate reader (Thermo Fisher, China) according to the protocol. Cell viability (%) = (OD value of tested cells/OD value of control cells) × 100 %.
2.6. Transwell migration assay
Cells were suspended in a serum-free medium with 1.0×105/ml. Transwell chamber without Matrigel pre-coating was inserted in the 24-well plates, containing 200 μL of suspension in the apical chamber and 500 μL of medium with 10% FBS in the basolateral chamber. At 48 h, chambers were taken out, and penetrating cells were fixed with 5% paraformaldehyde for 20 min and stained with 1% crystal violet at room temperature for 10 min. Penetrating cells in five randomly selected fields of each sample were captured for counting using a microscope (Nikon Corporation) under ×200 magnification.
2.7. Cell apoptosis analysis
After indicated treatments, cells were digested and collected by centrifugation at 1000 r/min for 5 min. Cells were resuspended and washed briefly in 1 mL of PBS twice, and then resuspended in 500 μL of 1X binding buffer containing 5 μL of propidium iodide (Beyotime). After incubated for 5 min in the dark at room temperature, cell apoptosis of each group was immediately detected under the FACSCanto II flow cytometer (BD Biosciences, USA) following the manufacturer’s instructions.
2.8. Cell cycle analysis
Cells were seeded in 10 cm dishes (1.5×106 cells/dish) for 24 h of adherence and were synchronized by 10 µM Nocodazole for 8 h. After indicated treatments, the cells were washed with PBS and fixed with 70% ice-cold ethanol overnight at −20 °C. Fixed cells were washed in PBS and stained with Propidium iodide/RNase Staining Buffer for 15 min at room temperature, followed by analysis using a flow cytometer (BD Biosciences).
2.9. Dual-luciferase reporter gene assay
Until 50–70% of confluence, cells were co-transfected with 2 μg pMiR-report vector-MAT2A 3′UTR and miRNA-26b-5p mimics using Lipofectamine 2000. Transfected cells were lysed at 48 h for luciferase activity determination using the Dual-luciferase Reporter Assay System. The luciferase reporter vectors were provided by GenePharma (Shanghai, China). All experiments were repeated three times.
2.10. RNA sequencing
After indicated treatments, total RNA of each group was extracted by TRIzol (Invitrogen) reagent according to the manufacturer’s instructions. RNA quality was assessed using Fragment Analyzer System (Agilent) and RNA Kit (Agilent). RNA samples were polyA-enriched, and libraries were prepared using the Illumina TruSeq_Stranded RNA kit. HiSeq 4000 single-end (150 bp) RNA sequencing runs (3 lanes) with a depth of approximately 20–30 million reads per sample were performed on an Illumina’s Hi-Seq 2500 device at the Genomic Technologies Facility of the University of Lausanne.
The differentially expressed genes between OS patients and normal samples, upon knock downing MAT2A in MNNG/HOS cells, were defined by the cutoff of FDR <0.05 with at least a 1.5-fold change. The expression data of MAT2A, MAT1A, and GNMT were obtained from RNA sequencing data in OS tissues.
2.11. In vivo experiments
Nude mice (4–5 weeks) were purchased from the Shanghai Experimental Animal Center of the Chinese Academy of Sciences, Shanghai, and housed in a pathogen-free facility in the Experimental Animal Centre of Zhongshan Hospital, Fudan University. All animal experiments were in accordance with the Institutional Animal Care and Use Committee Guide (IACUC) of Zhongshan Hospital, Fudan University (2018–014). 1×106 MNNG/HOS cells in the presence of shMAT2A or shCtrl in 0.1 mL PBS were injected subcutaneously into unilateral flank areas of nude mice. Tumor growth was determined for 47 days using calipers fitted with a vernier scale. Then, tumor-bearing mice were sacrificed. The tumors formed were surgically removed and weighed. The length (L) and width (W) were measured with calipers, and tumor volumes were calculated using the following formula: (L*W2)/2. Tumor tissues were fixed with histological tissue fixative (Sigma), embedded in paraffin, and stained with hematoxylin and eosin and KI67 detection.
2.12. Histology and immunohistochemistry.
OS tissues and xenograft tissue samples were collected and immediately fixed in 10% neutral-buffered formalin (ThermoFisher Scientific) overnight. After being washed once with PBS, samples were transferred into 70% ethanol and submitted to the Research Histology Core Laboratory at MD Anderson Cancer Center for embedding and hematoxylin and eosin staining. The antibody used for IHC staining was anti-MAT2A (1:2500, Abcam, ab154343). Images were obtained at 400× magnification using a microscope (Olympus, BX43).
2.13. Ferrous iron assay
Cellular ferrous iron levels were detected using the Cell Ferrous colorimetric Kit (E-BC-K881-M; Elabscience, China). According to the protocol, the cells were incubated with FerroOrange for 10 min at 37 °C. Fluorescence intensity (Ex: 590 nm, Em: 600 nm) was assessed using a fluorescence microplate reader (Thermo Fisher, China). The ferrous iron (Fe2+) levels were finally expressed as a ratio to the fluorescence intensity value of the control.
2.14. GSH assay
GSH levels were assayed using the GSH colorimetric Kit (E-BC-K030-M; Elabscience, China). The culture medium from the wells was carefully removed, and 100 μL of prepared 1× GSH Reagent was added, followed by incubation at room temperature for 30 min. Next, 100 μL of reconstituted Luciferin Detection Reagent was added to each well and mixed briefly on a plate shaker. The harvested cells were crushed by sonication to obtain the supernatant used for measuring the GSH levels in a fluorescence microplate reader (Thermo Fisher, China). A standard curve for GSH concentration was generated and the levels of GSH were expressed as a ratio to the absorbance value of the control cells assessed at 405 nm.
2.15. Statistical analysis
To determine whether significant genes were enriched in specific functions, we performed an overrepresentation analysis of Gene Ontology biological process gene sets using the enricher function of the clusterProfiler package v.3.10.1. Data are presented as means ± SD (n = 3). One-way ANOVA and Tukey's multiple comparisons test were used to analyze the data. All statistical analyses were performed using GraphPad Prism 8. Statistical significance was set at p < 0.05.
3. Results
3.1. Differential gene analysis and identification of MAT2A in OS
To uncover the dependent genes regulating OS progression, we performed differential gene expression analysis by subjecting eight OS tissue samples to bulk RNA-sequencing (RNA-seq). In the principal component analysis (PCA), OS tumor tissues clustered separately from their normal counterparts, suggesting reshaping of the OS transcriptome (Fig. 1A). We identified 2897 differentially expressed genes (DEGs); of these, 1757 were upregulated and 1140 were downregulated in OS tissues when compared with the para-tumor normal tissues (absolute fold change >1; adjusted P < 0.02) (Fig. 1B). The analysis of DEGs revealed that MAT2A was significantly upregulated in OS (∣logFC∣= 0.925843; P < 0.01), but MAT1A and GNMT (encoding glycine N-methyltransferase) were downregulated (Fig. 1C). This implies a strong link exists between MAT2A and OS progression. Based on this initial result, we next focused on the function of MAT2A in OS. We further assessed MAT2A expression in OS cells and found a higher level of MAT2A in two OS cell lines (MNNG/HOS and U2OS) when compared with the control cell line hFOB1.19 (Fig. 1D). Three shRNAs (shMAT2A-1, shMAT2A-2, and shMAT2A-3) were selected to knockdown MAT2A expression in MNNG/HOS cells. We found that the shMAT2A-1-expressing vector was the best at reducing MAT2A expression and had the most significant effect on cell viability (Fig. 1E and F). Therefore, we chose shMAT2A-1 for subsequent experiments. These results showed that MAT2A was a key gene in osteosarcoma.
Fig. 1.
Identification of key gene MAT2A in osteosarcoma (OS) samples. (A) Principal component analysis (PCA) of 8 normal samples (red circles) and 8 tumor samples (green circles), performed using all genes detected by RNA sequencing. (B) Volcano plot showing upregulated genes more expressed in OS samples (red points) and down-regulated genes (blue points) when compared with normal samples, and genes not significantly differentially expressed (gray). MAT2A, MAT1A, and GNMT were tagged in the heat map. P-value < 0.01, while the vertical dashed lines depict an absolute log2(fold change) threshold = 1. (C) The heatmap shows the expression profile of three genes (MAT2A, MAT1A, and GNMT) between 8 pairs of OS tumor samples and para-tumor normal samples after RNA sequencing. (D) The expression of MAT2A in hFOB1.19, MNNG/HOS, and U2OS cells were assessed by qRT-PCR. (E) The mRNA levels of MAT2A in MNNG/HOS cells were verified by qRT-PCR after transfections with three shRNAs targeting MAT2A (shMAT2A-1, shMAT2A-2, and shMAT2A-3) and shControl (shCtrl) plasmids. (F) Cell viability of MNNG/HOS cells was detected by CCK-8 assay after transfections with three shRNAs targeting MAT2A (shMAT2A-1, shMAT2A-2, and shMAT2A-3) and shControl (shCtrl) plasmids. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.2. Evaluating the effect of MAT2A expression in clinically diverse patients with OS
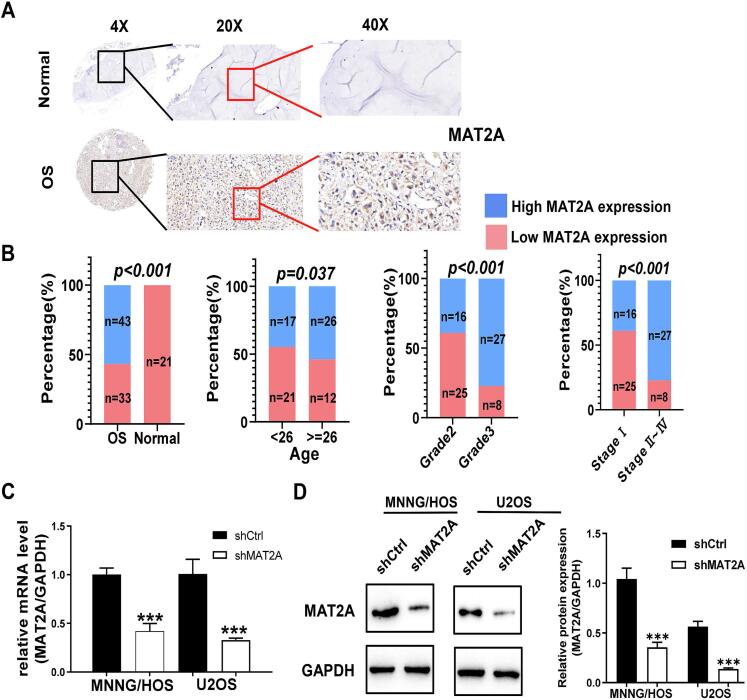
As shown in Fig. 2A, IHC detected a higher expression of MAT2A in OS tissues. We found that MAT2A expression in 76 tumor tissue samples from patients with OS was significantly higher than that in 21 normal tissue samples (P < 0.001; Fig. 2B). Furthermore, MAT2A expression was significantly different in patients with OS who differed in age, grade of malignancy, and pathological stage (Table 1 and Fig. 2B). Importantly, we observed that MAT2A expression in patients with OS was positively correlated with the degree of their tumor malignancy. Thus, MAT2A is related to the occurrence, development, and prognosis of OS, and could potentially be used as a drug target for the treatment of OS. Last, we showed that shMAT2A transfection could reduce the mRNA and protein levels of MAT2A in MNNG/HOS and U2OS cells (Fig. 2C and D).
Fig. 2.
High expression of MAT2A in osteosarcoma (OS) tissues may predict a higher degree of OS malignancy. (A) MAT2A was significantly highly expressed in OS tissues using IHC staining. Scale bar, 50 µm. (B) The high expression of MATA2A was closely related to the higher age, grade malignancy, and pathological stage. (C-D) MAT2A was significantly inhibited by shRNA in MNNG/HOS and U2OS and cells using qRT-PCR (C) and western blot (D) assays. The histogram on the right in represents the results of the statistical analysis. Data represent the mean ± S.D. from three independent experiments. **, p < 0.01; ***, p < 0.001.
3.3. Knockdown of MAT2A inhibited the malignant progression of OS cells in vitro
Moreover, the knockdown of MAT2A significantly reduced cell viability, increased apoptosis, and induced cell cycle arrest in the G2 phase (Fig. 3A – C). Additionally, MAT2A silencing decreased OS cell migration when compared with the cells transfected with shCtrl vectors (Fig. 3D). These findings strongly indicate that MAT2A knockdown inhibited the malignant progression of OS cells in vitro, showing that it was a definite oncogenic factor in OS.
Fig. 3.
MAT2A shRNA transfection decreases MAT2A expression, suppresses cell growth and migration, and induces cell apoptosis, and cell cycle arrest. (A) CCK-8 assays showed that the knockdown of MAT2A remarkably suppressed the viability of MNNG/HOS and U2OS cells. (B) The apoptosis rate of MNNG/HOS and U2OS cell lines was significantly increased after MAT2A shRNA transfection. (C) Knockdown of MAT2A arrested cell cycle at the G2/M period. (D) The migration abilities of MNNG/HOS and U2OS cell lines was significantly inhibited by knockdown of MAT2A. Scale bar, 50 µm. Data represent the mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
3.4. Evaluating the effect of MAT2A expression in a mouse OS xenograft model
We next injected nude mice subcutaneously with MNNG/HOS cells to establish xenograft tumors. Consistent with our in vitro observations, the tumor volume and weight were markedly decreased in the shMAT2A-transfected MNNG/HOS xenograft mouse model, compared to untransfected controls (Fig. 4A – C). In accordance, MAT2A expression was downregulated in xenograft tumors formed using shMAT2A-MNNG/HOS cells (Fig. 4D). Knockdown of MAT2A in tumor xenografts inhibited tumor growth in vivo as determined by H&E (Fig. 4E) and IHC staining for Ki67 expression (Fig. 4F). These in vivo experiments provide solid evidence for the carcinogenic effects of MAT2A in OS.
Fig. 4.
MAT2A silences its anticancer effect in vivo. 2 × 106 MKNNG/HOS cells infected with the lentivirus vector of shMAT2A or shCtrl in 100 μL PBS, then subcutaneously injected in the rear flank of nude mice (n = 3). Representative images of tumors formed. Scale bar, 10 mm. (A-C) The tumor size (mm3) and the weight on day 47 were measured. (D) The protein expressions of MAT2A in 3 cases of tumor tissues were determined by WB. The histogram on the lower represents the results of the statistical analysis. (E, F) Hematoxylin and eosin and immunohistochemical staining of KI67 in tumor xenografts. Scale bar, 50 μm. The experiment was repeated thrice independently with similar results. Data represent the mean ± S.D. from three independent experiments. *, p < 0.05; **, p < 0.01;
3.5. Identification of upstream regulatory factors of MAT2A and exploration of the mechanisms that regulate OS progression
To better understand how MAT2A regulates OS carcinogenesis, we used four databases (TarBase, TargetScan, miRDB, and StarBase) to predict the sequences of upstream MAT2A-regulating miRNAs. We found three overlapping miRNAs: miR-26a-5p, miR-26b-5p, and miR-340-5p (Fig. 5A); and their levels were upregulated in MNNG/HOS cells after shMAT2A transfection (Fig. 5B). Because miR-26b-5p was the most highly upregulated, it was chosen for use in subsequent studies to explore how MAT2A is regulated in OS. The interaction between miR-26b-5p and MAT2A was validated using a dual-luciferase reporter assay (Fig. 5C). We found that miR-26b-5p was upregulated but MAT2A was downregulated in eight tumor samples from patients with OS, compared with their para-tumor normal tissues (Fig. 5D). Interestingly, when miR-26b-5p was overexpressed, MAT2A expression was significantly downregulated in OS cells (Fig. 5E). Moreover, the overexpression of miR-26b-5p inhibited the proliferation and migration of MNNG/HOS and U2OS cells. The introduction of MAT2A into these cells not only caused an obvious increase in their viability and migration capacity but also restored OS tumor growth (Fig. 5F and G). Together, these findings indicate that miR-26b-5p confers an anticancer effect by acting as an upstream regulatory factor of MAT2A and downregulating its expression in OS tumor cells.
Fig. 5.
MiR-26b-5p reversed the regulatory effects of MAT2A on cell viability and migratory potentials of osteosarcoma (OS). (A) Venn diagram showing the overlapping miRNAs targeting MAT2A among four online predicted databases (Tarbase, TargetScan, miRDB, and Starbase). (B) qRT-PCR indicated that the mRNA levels of miR-26a-5p, miR-26b-5p, and miR-340-5p were significantly upregulated in MNNG/HOS cells after shMAT2A transfection. (C) Potential binding sites of MAT2A to miR-26b-5p (upper). Dual-luciferase reporter gene assay showed that MAT2A could bind to miR-26b-5p (bottom). (D) The mRNA levels of miR-26b-5p and MAT2A in 8 pairs of OS samples and para-tumor normal samples were determined using qRT-PCR. (E) Transfection of miR-26b-5p mimics in MNNG/HOS and U2OS cells downregulated MAT2A expression. The histogram on the right represents the results of the statistical analysis from WB. (F) Overexpression of MAT2A facilitates cell viability, but miR-26b-5p overexpression inhibited cell viability in MNNG/HOS and U2OS cells while was further upregulated by co-overexpression of MAT2A and miR-26b-5p. (G) The inhibited migration of miR-26b-5p on MNNG/HOS and U2OS cells was partially reversed by MAT2A overexpression. Scale bar, 50 μm. Data represent the mean ± S.D. from three independent experiments from three mice. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
3.6. The miR-26b-5p/MAT2A axis mediated ferroptosis through the STAT3/SLC7A11 pathway in OS cells
Over-representation analyses were performed against the Gene Ontology (GO) gene sets from the TARGET-OS database to investigate the global effects of MAT2A on gene expression programs. There was a clearly distinguishable difference between OS patients with low MAT2A expression and those with high MAT2A expression in their upregulated mRNA metabolism genes (Fig. 6A). RNA-seq analysis of OS samples and the GO metabolic analysis of the genes showed a striking enrichment in metabolism-associated biological processes, especially regulation of DNA metabolism (Fig. 6B). We further performed RNA-seq transcriptome and gene enrichment analysis and found that the signaling pathways involved Foxo signaling pathway, Glutathone metabolism, p53 signaling pathway, MAPK signaling pathway and ferroptosis pathway (Fig. 6C). Further correlation analysis showed that there was indeed a significant positive correlation between MAT2A and ferroptosis (Fig. 6D). These data suggest that the ferroptosis plays a role in OS.
Fig. 6.
Identification of ferroptosis pathway in osteosarcoma (OS). (A) GO metabolic analysis for the differently expressed genes in OS patients with high or low MAT2A expression from the TARGET dataset. (B) GO metabolic analysis of the genes from the RNA-seq data between OS and normal samples. Top annotation clusters are shown according to their enrichment scores, “regulation of DNA metabolic process” was the most significantly enriched. (C) Lollipop chart displaying the KEGG enrichment analyses, based on the RNA-seq data between normal and OS group. (D) Correlation analysis of MAT2A and ferroptosis pathway based our RNA-seq data.
In addition, cell metabolism is believed to play an important role in ferroptosis regulation [34], [35]. Therefore, we treated OS cells with a MAT2A inhibitor FIDAS-5 (10 μM in DMSO) to further explore MAT2A's involvement in ferroptosis sensitivity. To determine the potential binding mode of FIDAS-5 with MAT2A, we performed dock studies using the MAT2A crystal structure. The results showed a binding model of FIDAS-5 with MAT2A (Estimated Free Energy of Binding = −6.18 kcal/mol) (Fig. 7A and B). The suppression of MAT2A protein levels in OS cells by FIDAS-5 was first verified by western blotting (Fig. 7C). And, the level of Fe2+ was markedly increased in OS cells treated with FIDAS-5 (Fig. 7D), compared with OS cells treated with DMSO alone, which was accompanied by a dramatic drop in the abundance of glutathione (GSH) (Fig. 7E). Transient exposure to FIDAS-5 (24 h) significantly reduced the viability of OS cells. Importantly, the ferroptosis inhibitor ferrostatin or the iron chelator deferoxamine (DFO) could reverse FIDAS-5-induced cell growth inhibition (Fig. 7F). Together, our findings prove that miR-26b-5p may promote the ferroptosis of OS by inhibiting MAT2A.
Fig. 7.
The miR-26b-5p/MAT2A axis mediated ferroptosis through STAT3/SLC7A11 pathway in osteosarcoma (OS). (A) Chemical structures of FIDAS-5 and protein structure of MAT2A. (B) Computational molecular docking analysis to investigate the interaction of FIDAS-5 binding to MAT2A. The green molecule represents FIDAS-5. The Estimated Free Energy of Binding of FIDAS-5 with MAT2A protein is −6.18 kcal/mol. (C) Western blot analysis of MAT2A expression in MNNG/HOS and U2OS cells treated with FIDAS-5 (10 μM) dissolved in DMSO for 24 h. The histogram on the lower represents the results of the statistical analysis. (D) FIDAS-5 treatment (24 h) induced ferrous iron (Fe2+) accumulation in MNNG/HOS and U2OS cells. (E) Relative abundance of GSH was measured by GSH colorimetric Kit in MNNG/HOS and U2OS cells treated with FIDAS-5 (10 μM) or DMSO for 24 h. (F) MNNG/HOS and U2OS cells were treated with FIDAS-5 (10 μM) for 24 h and then were treated with DFO (100 μM), and then cell viability was determined. Deferoxamine (DFO) rescues cells from FIDAS-5-induced cell inactivation. (G) Venn diagram showing 85 key overlapping genes, including STAT3 and SLC7A11, which are contained in both 4625 DEGs and 259 ferroptosis-related genes, simultaneously. (H) Western blotting analysis shows the expression of p-STAT3, STAT3, and SLC7A11 in MNNG/HOS and U2OS cells treated with FIDAS-5 (10 μM) or DMSO for 24 h. The histogram on the lower represents the results of the statistical analysis. Data represent the mean ± S.D. from three independent experiments. **, p < 0.01; ***, p < 0.001. (I) Genome browser display of STAT3-binding events on promoters of SLC7A11 gene and MAT2A gene, data from previous reported ChIP-seq data (GSE117164). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We next studied the signaling pathway that implicates MAT2A in ferroptosis regulation. On MAT2A knockdown, we found 4625 DEGs in OS cells. A comparison of these DEGs with 259 ferroptosis-related genes (from previously published articles) revealed 85 overlapping genes (Fig. 7G). The top two DEGs, STAT3 and SLC7A11, were selected from the overlapping gene list. Notably, exposure to FIDAS-5 markedly reduced the protein levels of phosphorylated (p)-STAT3 and SLC7A11 in OS cells (Fig. 7H). Based on the ChIP-seq data from the GSE117164 cohort, we found the potential links between STAT3 and SLC7A11, and MAT2A genes, suggesting STAT3 might mediate ferroptosis through transcriptional regulation of SLC7A11 and MAT2A in OS. Furthermore, we selected three OS-related GEO datasets (GSE36001, GSE65071 and GSE65071) to analyze the expression of miR-26b-5p/MAT2A/STAT3/SLC7A11 pathway and clinical prognostic features (Supplementary Fig. S1). These results demonstrate that MAT2A depletion suppressed OS tumor development through ferroptosis activation via the STAT3/SLC7A11 signaling pathway.
4. Discussion
Through the STAT3/SLC7A11 pathway, we demonstrated a key role for MAT2A in regulating OS cell ferroptosis. There is an association between high levels of MAT2A expression and a poorer prognosis in patients with OS. Depleting MAT2A significantly increased tumor cell apoptosis and induced tumor cell cycle arrest in the G2 phase, so OS cells became less viable and migratory in vitro. MAT2A targeting significantly inhibited tumor growth in vivo, as we found, suggesting that MAT2A activity may be a potential therapeutic target for OS treatment.
Metastasis was the key issue affecting the prognosis of patients with OS. Our results highlighted that MAT2A plays an oncogenic role in the progression of OS, highly expressed miR-26b-5p restrained MAT2A expression, thereby inhibiting cell growth and migration in OS cells, and MAT2A depletion significantly promotes ferroptosis in vivo.
A growing volume of evidence has demonstrated that a diverse number of metabolic pathways ultimately converge in ferroptosis [20]. Ferroptosis is a recently identified metabolic stress-induced non-apoptotic form of regulated cell death that is caused by cystine depletion and overproduction of lipid-based reactive oxygen species (ROS), particularly lipid hydroperoxide, in an iron-dependent manner [36]. Cellular iron, lipid, and glutaminolysis metabolism have been deemed the major metabolic processes impacting cell vulnerability to ferroptosis [20]. Our data suggested that the MAT2A-mediated anticancer effect is associated with cell metabolism involved in ferroptosis, and linked MAT2A function to ferroptosis in OS. However, the underlying mechanisms by which MAT2A-mediated ferroptosis remain poorly understood. Glutathione peroxidase 4 (GPX4) utilizes reduced glutathione (GSH) to reduce lipid hydroperoxides to lipid alcos, to protect cells against membrane lipid peroxidation and inhibit ferroptosis [37]. At that time, researchers found that cystine deprivation causes cell death with a unique microscopic morphology and that restoration of GSH promotes cell growth [38]. Insufficient cystine supply induces metabolic stress and provokes ferroptosis cell death in tumor cells [36]. Here, MAT2A deletion by FIADS-5 treatment in OS cells results in increased Fe2+ content and decreased GSH synthesis and cell viability. Furthermore, desferrioxamine (DFO) can reduce the content of free iron directly. Importantly, FIADS-5-induced cell death could be largely suppressed by the ferroptosis inhibitor ferrostatin (DFO), suggesting activation of ferroptosis in tumor cells is detrimental to cell survival.
Solute carrier family 7 member 11 (SLC7A11), one catalytic subunit of the cystine/glutamate antiporter system Xc−, is the major transporter of extracellular cysteine [7], [39]. Correspondingly, cystine depletion block SLC7A11-mediated cystine uptake to induce ferroptosis [36]. SLC7A11, which transports cystine into the cells, enhances GSH synthesis, further promoting the inhibition of ferroptosis by GPX4. Signal transducer and activator of transcription 3 (STAT3) belongs to the STAT family and is a vital transcription factor involved in inflammation and tumor progression [40]. Classical STAT3 activation involves the phosphorylation of STAT3 at Tyr705 (p-STAT3 (7 0 5)), which interacts with and inhibits P53 [41]. As described above, it is critical to inactivate STAT3 to halt tumor progression. More importantly, STAT3 overexpression, and SLC7A11 overexpression, were shown to rescue OS cell ferroptosis [42]. Fortunately, the ferroptosis-related genes STAT3 and SLC7A11 were significantly altered by MAT2A in the present study. Our study displayed that FIADS-5 treatment reduced p-STAT3 and SLC7A11 expressions in OS cells, suggesting MAT2A depletion suppressed tumorigenesis by facilitating ferroptosis at least partly through STAT3/SLC7A11 signaling pathway.
In cell line studies, ferroptosis can be specifically induced by treatment with MAT2A inhibitor FIDAS-5. Currently, our ability to study the clinical relevance of ferroptosis in cancer biology is hindered by a lack of established assays for characterizing ferroptosis in tissue and tumor samples. To our knowledge ferroptosis has not previously been identified in tumor samples in vivo. In our study, we conducted cell ferrous colorimetry analysis on tumor cells, the gold standard assay for characterizing cell ferroptosis, and provided definitive evidence of ferroptosis in tumors with MAT2A depletion.
This study extends the results of previous studies and has many innovations. Similar to previous study, which showed that activation of MAT2A-ACSL3 pathway protects cells from ferroptosis in gastric cancer [43], our study also explores the effects of MAT2A associated pathways and ferroptosis on cancer. The biggest highlight and difference of this study is that we selected targeted MAT2A based on RNA sequencing data of collected clinical samples in our hospital. However, previous study achieved the goal through in vitro and in vivo experiments and relevant molecular analyses. The previous study linked MAT2A with the downstream ACSL3 signaling pathway, which differ from miR-26b-5p/MAT2A/STAT3 signaling axis in our study. This finding may reveal several novel targets for inhibiting osteosarcoma progression via ferroptosis pathway.
Previous findings have also suggested that miR-26b-5p is involved in the progression of osteosarcoma. For example, CircPVT1 promotes the tumorigenesis and metastasis of osteosarcoma via mediation of miR-26b-5p/CCNB1 axis[30]. Circular RNA_ANKIB1 accelerates chemo-resistance of osteosarcoma via binding microRNA-26b-5p and modulating enhancer of zeste homolog 2[31]. In the present study, the role and mechanism of MAT2A in regulating the progression of ferroptosis and osteosarcoma were discussed, and the epigenetic mechanism mediated by miR-26b-5p/MAT2A was identified, which further enhanced the study of miR-26b-5p on the progression of OS tumors and provided new ideas for the treatment of OS.Although inhibition of MAT2A and FIDAS-5 specifically induces ferroptosis in cell lines, our capacity to research the clinical relevance of ferroptosis in cancer biology limits ferroptosis in tissue and tumor samples. To our knowledge, ferroptosis has never been detected in live tumor specimens. In our study, we performed a colorimetric iron test on tumor cells, which is the gold standard test for describing cellular ferroptosis and provided clear evidence of tumor ferroptosis following MAT2A deficiency.
Our data cast light on the potential role of the miR-26b-5p/MAT2A-STAT3/SLC7A11 signaling axis in the efficacy of ferroptosis-mediated cancer treatment (Graphical Abstract). Specific MAT2A inhibitors could induce ferroptosis, which suggests a novel cancer therapy strategy for OS. We would like to explore the synergistical anti-tumor effect of the combination of Specific MAT2A inhibitors and ferroptosis inducers in the future. These advances may pave the way for novel treatments requisite for patients with OS in need of new therapies.
CRediT authorship contribution statement
Shuchi Xia: Investigation, Formal analysis, Methodology, Conceptualization. Yun Liang: Investigation, Formal analysis, Methodology, Conceptualization. Yuqing Shen: Validation. Wuxue Zhong: Validation. Yiqun Ma: Validation, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
Funding: This work was supported by the National Natural Science Foundation of China (Grant No.82002841).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbo.2023.100490.
Contributor Information
Wuxue Zhong, Email: zhongwuxue@163.com.
Yiqun Ma, Email: mayiqun090@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplementary Fig. 1.
References
- 1.Chen C., Xie L., Ren T., Huang Y., Xu J., Guo W. Immunotherapy for osteosarcoma: Fundamental mechanism, rationale, and recent breakthroughs. Cancer Lett. 2021;500:1–10. doi: 10.1016/j.canlet.2020.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Simpson E., Brown H.L. Understanding osteosarcomas. Understanding osteosarcomas. Jaapa. 2018;31(8):15–19. doi: 10.1097/01.JAA.0000541477.24116.8d. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Liu R., Wang W., Wang C., Zhang N., Shao X., He Q., Ying M. Advances in targeted therapy for osteosarcoma based on molecular classification. Pharmacol Res. 2021;169:105684. doi: 10.1016/j.phrs.2021.105684. [DOI] [PubMed] [Google Scholar]
- 4.Corre I, Verrecchia F, Crenn V, Redini F and Trichet V. The Osteosarcoma Microenvironment: A Complex But Targetable Ecosystem. Cells 2020; 9. [DOI] [PMC free article] [PubMed]
- 5.Jiang M., Wang Z., He X., Hu Y., Xie M., Jike Y., Bo Z., Qin W., Wang F.u. A Risk-Scoring Model Based on Evaluation of Ferroptosis-Related Genes in Osteosarcoma. J Oncol. 2022;2022:1–17. doi: 10.1155/2022/4221756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y., Zhao B., Zhou L., Zhang Z., Shen Y., Lv H., AlQudsy L.H.H., Shang P. Ferroptosis, a novel pharmacological mechanism of anti-cancer drugs. Cancer Lett. 2020;483:127–136. doi: 10.1016/j.canlet.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Li Z, Chen L, Chen C, Zhou Y, Hu D, Yang J, Chen Y, Zhuo W, Mao M, Zhang X, Xu L, Wang L and Zhou J. Targeting ferroptosis in breast cancer. Biomarker research 2020; 8: 58-58. [DOI] [PMC free article] [PubMed]
- 8.Shen L.D., Qi W.H., Bai J.J., Zuo C.Y., Bai D.L., Gao W.D., Zong X.L., Hao T.T., Ma Y., Cao G.C. Resibufogenin inhibited colorectal cancer cell growth and tumorigenesis through triggering ferroptosis and ROS production mediated by GPX4 inactivation. Anat Rec (Hoboken) 2021;304:313–322. doi: 10.1002/ar.24378. [DOI] [PubMed] [Google Scholar]
- 9.Lv H.-H., Zhen C.-X., Liu J.-y., Shang P. PEITC triggers multiple forms of cell death by GSH-iron-ROS regulation in K7M2 murine osteosarcoma cells. Acta Pharmacol Sin. 2020;41(8):1119–1132. doi: 10.1038/s41401-020-0376-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin H., Chen X., Zhang C., Yang T., Deng Z., Song Y., Huang L., Li F., Li Q., Lin S., Jin D. EF24 induces ferroptosis in osteosarcoma cells through HMOX1. Biomed Pharmacother. 2021;136:111202. doi: 10.1016/j.biopha.2020.111202. [DOI] [PubMed] [Google Scholar]
- 11.Chen M., Jiang Y., Sun Y. KDM4A-mediated histone demethylation of SLC7A11 inhibits cell ferroptosis in osteosarcoma. Biochem Biophys Res Commun. 2021;550:77–83. doi: 10.1016/j.bbrc.2021.02.137. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q., Wang K. The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin. Cell Biol Int. 2019;43(11):1245–1256. doi: 10.1002/cbin.11121. [DOI] [PubMed] [Google Scholar]
- 13.Chen D.-P., Ning W.-R., Jiang Z.-Z., Peng Z.-P., Zhu L.-Y., Zhuang S.-M., Kuang D.-M., Zheng L., Wu Y. Glycolytic activation of peritumoral monocytes fosters immune privilege via the PFKFB3-PD-L1 axis in human hepatocellular carcinoma. J Hepatol. 2019;71(2):333–343. doi: 10.1016/j.jhep.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Kargbo R.B. Methionine Adenosyltransferase Inhibitors for the Treatment of Cancer. ACS Med Chem Lett. 2021;12(2):180–181. doi: 10.1021/acsmedchemlett.1c00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo J., Yang Y., Buettner R., Rosen S.T. Targeting the methionine-methionine adenosyl transferase 2A- S -adenosyl methionine axis for cancer therapy. Curr Opin Oncol. 2022;34:546–551. doi: 10.1097/CCO.0000000000000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kalev P., Hyer M.L., Gross S., Konteatis Z., Chen C.-C., Fletcher M., Lein M., Aguado-Fraile E., Frank V., Barnett A., Mandley E., Goldford J., Chen Y., Sellers K., Hayes S., Lizotte K., Quang P., Tuncay Y., Clasquin M., Peters R., Weier J., Simone E., Murtie J., Liu W., Nagaraja R., Dang L., Sui Z., Biller S.A., Travins J., Marks K.M., Marjon K. MAT2A Inhibition Blocks the Growth of MTAP-Deleted Cancer Cells by Reducing PRMT5-Dependent mRNA Splicing and Inducing DNA Damage. Cancer Cell. 2021;39(2):209–224.e11. doi: 10.1016/j.ccell.2020.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Wang Z., Yip L.Y., Lee J.H.J., Wu Z., Chew H.Y., Chong P.K.W., Teo C.C., Ang H.-K., Peh K.L.E., Yuan J.u., Ma S., Choo L.S.K., Basri N., Jiang X., Yu Q., Hillmer A.M., Lim W.T., Lim T.K.H., Takano A., Tan E.H., Tan D.S.W., Ho Y.S., Lim B., Tam W.L. Methionine is a metabolic dependency of tumor-initiating cells. Nat Med. 2019;25(5):825–837. doi: 10.1038/s41591-019-0423-5. [DOI] [PubMed] [Google Scholar]
- 18.Chu P.-Y., Wu H.-J., Wang S.-M., Chen P.-M., Tang F.-Y., Chiang E.-P. MAT2A Localization and Its Independently Prognostic Relevance in Breast Cancer Patients. Int J Mol Sci. 2021;22(10):5382. doi: 10.3390/ijms22105382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y., Yang H., Zhao J., Wan P., Hu Y.e., Lv K., Hu YiRen, Yang X.i., Ma M. Yang X and Ma M. Activation of MAT2A-RIP1 signaling axis reprograms monocytes in gastric cancer. J Immunother Cancer. 2021;9(2):e001364. doi: 10.1136/jitc-2020-001364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng J., Conrad M. The Metabolic Underpinnings of Ferroptosis. Cell Metabolism. 2020;32(6):920–937. doi: 10.1016/j.cmet.2020.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Chen H., Gu B., Zhao X., Zhao Y., Huo S., Liu X., Lu H. Circular RNA hsa_circ_0007364 increases cervical cancer progression through activating methionine adenosyltransferase II alpha (MAT2A) expression by restraining microRNA-101-5p. Bioengineered. 2020;11(1):1269–1279. doi: 10.1080/21655979.2020.1832343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lo T.-F., Tsai W.-C., Chen S.-T., Avila M.A. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One. 2013;8(9):e75628. doi: 10.1371/journal.pone.0075628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Y.-W., Du Q.-R., He Y.-J., Chen W.-S., Jiang W.-Y., Gui Q.i., Xu C.-C., Wang W., Cheng H.-Y., Lu X.-J. Circ_0044516 Regulates miR-136/MAT2A Pathway to Facilitate Lung Cancer Development. J Immunol Res. 2021;2021:1–12. doi: 10.1155/2021/5510869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., Wang L., Li H., Zhang L., Zheng X., Cheng W. Crosstalk between noncoding RNAs and ferroptosis: new dawn for overcoming cancer progression. Cell Death Dis. 2020;11:580. doi: 10.1038/s41419-020-02772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soghli N., Ferns G.A., Sadeghsoltani F., Qujeq D., Yousefi T., Vaghari-Tabari M. MicroRNAs and osteosarcoma: Potential targets for inhibiting metastasis and increasing chemosensitivity. Biochem Pharmacol. 2022;201:115094. doi: 10.1016/j.bcp.2022.115094. [DOI] [PubMed] [Google Scholar]
- 26.Du Q., Yuan Z., Huang X., Huang Y., Zhang J., Li R. miR-26b-5p suppresses chemoresistance in breast cancer by targeting serglycin. Anticancer Drugs. 2022;33:308–319. doi: 10.1097/CAD.0000000000001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu T., Xie M., Jing X., Jiang H., Wu X., Wang X., Shu Y. Loss of miR-26b-5p promotes gastric cancer progression via miR-26b-5p-PDE4B/CDK8-STAT3 feedback loop. J Transl Med. 2023;21:77. doi: 10.1186/s12967-023-03933-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yin X., Tian M., Zhang J., Tang W., Feng L., Li Z., Zheng C., Liu C., Yan L., Yu X., Li B. MiR-26b-5p in small extracellular vesicles derived from dying tumor cells after irradiation enhances the metastasis promoting microenvironment in esophageal squamous cell carcinoma. Cancer Lett. 2022;541:215746. doi: 10.1016/j.canlet.2022.215746. [DOI] [PubMed] [Google Scholar]
- 29.Khosla R., Hemati H., Rastogi A., Ramakrishna G., Sarin S.K., Trehanpati N. miR-26b-5p helps in EpCAM+cancer stem cells maintenance via HSC71/HSPA8 and augments malignant features in HCC. Liver Int. 2019;39(9):1692–1703. doi: 10.1111/liv.14188. [DOI] [PubMed] [Google Scholar]
- 30.Huang S.-X., Mei H.-B., Liu K., Tang J., Wu J.-Y., Zhu G.-H., Ye W.-H. CircPVT1 promotes the tumorigenesis and metastasis of osteosarcoma via mediation of miR-26b-5p/CCNB1 axis. J Bone Miner Metab. 2022;40(4):581–593. doi: 10.1007/s00774-022-01326-6. [DOI] [PubMed] [Google Scholar]
- 31.Tang JinShan, Duan G., Wang YunQing, Wang B., Li WenBo, Zhu ZiQiang. Circular RNA_ANKIB1 accelerates chemo-resistance of osteosarcoma via binding microRNA-26b-5p and modulating enhancer of zeste homolog 2. Bioengineered. 2022;13(3):7351–7366. doi: 10.1080/21655979.2022.2037869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Du S., Wang S., Ye K. Ferroptosis in osteosarcoma: A promising future. Front Oncol. 2022;12:1031779. doi: 10.3389/fonc.2022.1031779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lei T., Qian H.u., Lei P., Hu Y. Ferroptosis-related gene signature associates with immunity and predicts prognosis accurately in patients with osteosarcoma. Cancer Sci. 2021;112(11):4785–4798. doi: 10.1111/cas.15131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J.-Y., Nam M., Son H.Y., Hyun K., Jang S.Y., Kim J.W., Kim M.W., Jung Y., Jang E., Yoon S.-J., Kim J., Kim J., Seo J., Min J.-K., Oh K.-J., Han B.-S., Kim W.K., Bae K.-H., Song J., Kim J., Huh Y.-M., Hwang G.-S., Lee E.-W., Lee S.C. Polyunsaturated fatty acid biosynthesis pathway determines ferroptosis sensitivity in gastric cancer. Proceedings of the National Academy of Sciences. 2020;117(51):32433–32442. doi: 10.1073/pnas.2006828117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mao C., Liu X., Zhang Y., Lei G., Yan Y., Lee H., Koppula P., Wu S., Zhuang L.i., Fang B., Poyurovsky M.V., Olszewski K., Gan B. DHODH-mediated ferroptosis defence is a targetable vulnerability in cancer. Nature. 2021;593(7860):586–590. doi: 10.1038/s41586-021-03539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dixon S., Lemberg K., Lamprecht M., Skouta R., Zaitsev E., Gleason C., Patel D., Bauer A., Cantley A., Yang W., Morrison B., Stockwell B. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang W., SriRamaratnam R., Welsch M., Shimada K., Skouta R., Viswanathan V., Cheah J., Clemons P., Shamji A., Clish C., Brown L., Girotti A., Cornish V., Schreiber S., Stockwell B. Regulation of ferroptotic cancer cell death by GPX4. Cell. 2014;156(1-2):317–331. doi: 10.1016/j.cell.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eagle H. Amino Acid Metabolism in Mammalian Cell Cultures. Science. 1959;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- 39.Wang X., Xu S., Zhang L., Cheng X., Yu H., Bao J., Lu R. Vitamin C induces ferroptosis in anaplastic thyroid cancer cells by ferritinophagy activation. Biochem Biophys Res Commun. 2021;551:46–53. doi: 10.1016/j.bbrc.2021.02.126. [DOI] [PubMed] [Google Scholar]
- 40.Yu H., Pardoll D., Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9(11):798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pham T.-H., Park H.-M., Kim J., Hong J.T., Yoon D.-Y. STAT3 and p53: Dual Target for Cancer Therapy. Biomedicines. 2020;8(12):637. doi: 10.3390/biomedicines8120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo Y.i., Gao X.u., Zou L., Lei M., Feng J., Hu Z., Gil G. Bavachin Induces Ferroptosis through the STAT3/P53/SLC7A11 Axis in Osteosarcoma Cells. Oxid Med Cell Longev. 2021;2021:1–14. doi: 10.1155/2021/1783485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma M., Kong P., Huang Y., Wang J., Liu X., Hu Y., Chen X., Du C., Yang H. Activation of MAT2A-ACSL3 pathway protects cells from ferroptosis in gastric cancer. Free Radic Biol Med. 2022;181:288–299. doi: 10.1016/j.freeradbiomed.2022.02.015. [DOI] [PubMed] [Google Scholar]