Summary
In glioma modeling, existing organoid protocols lack the ability to replicate glioma cell invasion and interaction with normal brain tissue. Here, we present a protocol for generating in vitro brain disease models using human-induced pluripotent- or embryonic-stem-cell-derived cerebral organoids (COs). We describe steps for forming glioma organoids by co-culturing forebrain organoids with U-87 MG cells. We also detail vibratome sectioning of COs to prevent cell death and enhance contact between U-87 MG cells and cerebral tissues.
Subject areas: Cancer, Neuroscience, Organoids, Tissue Engineering
Graphical abstract

Highlights
-
•
Detailed steps for successful development of forebrain organoid
-
•
Vibratome-sectioned COs enlarge tissue surfaces and accelerate U87 cell invasion
-
•
Organoid-U87 assembloids serve as a drug sensitivity model
-
•
Study communication between glioma cells and COs using organoid-U87 assembloids
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
In glioma modeling, existing organoid protocols lack the ability to replicate glioma cell invasion and interaction with normal brain tissue. Here, we present a protocol for generating in vitro brain disease models using human-induced pluripotent- or embryonic-stem-cell-derived cerebral organoids (COs). We describe steps for forming glioma organoids by co-culturing forebrain organoids with U-87 MG cells. We also detail vibratome sectioning of COs to prevent cell death and enhance contact between U-87 MG cells and cerebral tissues.
Before you begin
Before generating COs, prepare Matrigel-coated well plates to grow hiPSCs or hESCs to an optimal confluency. The culture medium and the steps of generating COs partially refer to Qian et al.,1 and the specific differences will be highlighted in the following sections. The differences here mainly include the process of culturing hiPSCs and hESCs and the medium used to form embryoid bodies. The formula of the medium used after the formation of EBs basically refers to Qian et al.1
Complete mTeSR™ plus medium
Use sterile techniques to prepare complete mTeSR™ Plus medium (Basal Medium + 5X Supplement).
Prepare Matrigel-coated well plates
Timing: 1 h 30 min, to generate 4 coated 6-well plates
-
1.
Thaw Corning® Matrigel® hESC-Qualified Matrix (Cat. 354277) on ice, referring to the recommended aliquot size in the Matrigel® Certificate of Analysis (e.g., 250 μL per tube) to prepare the desired volume of diluted matrix (e.g., 25 mL).
Note: If Corning® Matrigel® hESC-Qualified Matrix (Cat. 354277) is not available, please use CellAdhere™ Laminin-521 for alternatives. After thawing it at 2°C–8°C, dilute CellAdhere™ Laminin-521 in Dulbecco’s phosphate-buffered saline with Ca++ and Mg++ to a final concentration of 5–10 μg/mL.
CRITICAL: Always keep Matrigel on ice and prevent it from gelling.
-
2.
After thawing the aliquot, dilute the Matrigel stock in 100-fold (volume) of cold DMEM/F-12(e.g., 250 μL in 24.75 mL DMEM/F-12 in the 50 mL tube) to prepare the working solution for plate coating.
-
3.
Keep Matrigel aliquots at −20°C for up to 2 months or at −80°C for up to 6 months.
-
4.
To coat 4 coated 6-wells plates, prepare 25 mL of Matrigel working solution by adding 250 μL of Matrigel stock into 24.75 mL of cold DMEM/F-12 in a pre-chilled 50 mL centrifuge tube and mix up gently with pipettes. troubleshooting 1.
-
5.
Add 1 mL Matrigel working solution to each well.
-
6.
Swirl the plate to spread the Matrigel solution to completely cover the surface area.
-
7.
Incubate at 37°C in the settled-down incubator for a minimum of 1 h before using it; cover the plate to prevent water evaporation.
Note: If not used immediately, seal the cultureware and keep at 4°C for up to 1 week.
Grow hiPSCs and hESCs
Timing: 10 min for passaging cells, and 4–5 days for growing to optimal confluency
-
8.
Grow hiPSCs and hESCs in the mTeSR Plus medium on Matrigel-coated 6-well plates and incubate at 37°C with 5% CO2.
-
9.Passaging cells.
-
a.Gently remove the Matrigel working solution in coated 6-well plates. Add 2 mL mTeSR Plus with 10 μΜ Y-27632 to each well and warm the cultureware at 37°C with 5% CO2.
-
b.Remove all medium from the plates and wash cells twice with pre-warmed 1 X DPBS.
-
c.Add 1mL ReLeSR to each well and aspirate it within 1 min, so that cells are exposed to a thin film of liquid, incubate the cells at 37°C for 4–5 min.Note: If ReLeSR is not available, please use Gentle Cell Dissociation Reagent (GCDR) as the alternative. The procedure of using GCDR to passage cells should follow the commercial protocol.
-
d.Add 1 mL of mTeSR Plus with 10 μΜ Y-27632 to each well.
-
e.Detach the colonies by tapping the plates for 30 s.
-
f.Gently pipette cell mixture with P1000 pipette tips 1–2 times when the colonies are at optimal sizes (about 10 μm).
-
g.Plate the cell aggregate mixture on coated-plates at the appropriate density.
-
h.As a recommendation, spilt the mixture at a density of 1:30-1:50, and the specific ratio is determined based on the cell density of the well before subculture. A better reference method is to observe 3–5 aggregates in a field of view under a bright-field microscope with a 4x objective lens, which is the optimal density.
-
i.After 24 h, wash the cells with 1 X DPBS and refresh the medium with mTeSR Plus.
-
a.
-
10.
Change medium daily. Monitor the confluency of cells daily to prevent non-specific differentiation and potential contamination. troubleshooting 2.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse Monoclonal Anti-SOX2 (1:100) | Santa Cruz | Cat#sc-365823; RRID: AB_2064130 |
| Goat Monoclonal Anti-SOX2 (1:500) | R&D | Cat#AF2018; RRID: AB_355110 |
| Rabbit Polyclonal Anti-TBR2 (1:500) | Abcam | Cat# ab23345, RRID: AB_778267 |
| Rat Monoclonal Anti-CTIP2 (1:100) | Abcam | Cat# ab18465, RRID: AB_2064130 |
| Rabbit Polyclonal Tubulin β 3 (TUJ1) (1:500) | Abcam | Cat#ab18207, RRID: AB_444319 |
| Mouse Monoclonal BRN2 (1:300) | Millipore | Cat#MABD51, RRID: AB_11212125 |
| Rabbit Polyclonal SATB2 (1:100) | Abcam | Cat#ab34735, RRID: AB_2301417 |
| Chicken Polyclonal Glial Fibrillary Acidic Protein (GFAP) (1:2000) | Abcam | Cat#ab4674, RRID: AB_304558 |
| Rabbit polyclonal to Ki67 | Abcam | Cat# 15580 |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor® 568) | Abcam | Cat# ab175472 |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150077 |
| Goat Anti-Rat IgG H&L (Alexa Fluor® 647) | Abcam | Cat# ab150159 |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 488) | Abcam | Cat# ab150073 |
| Goat Anti-Chicken IgY H&L (Alexa Fluor® 647) | Abcam | Cat# ab150171 |
| Chemical peptides and recombinant proteins | ||
| Matrigel Basement Membrane Matrix, LDEV-free | Corning | Cat# 354277 Cat# 354230 |
| CellAdhere™ Laminin-521 | Stem Cell | Cat# 200-0117 |
| Dorsomorphin | Stem Cell Technologies | Cat# 72102 |
| A83-01 | Stem Cell Technologies | Cat# 2022 |
| SB-431542 | Stem Cell Technologies | Cat# 72232 |
| CHIR-99021 | Stem Cell Technologies | Cat# 72052 |
| Wnt-3a | MCE | Cat# HY-P70453 |
| Y-27632 dihydrochloride | MCE | Cat# HY-10583 |
| BDNF | Stem Cell Technologies | Cat# 78005 |
| GDNF | Stem Cell Technologies | Cat# 78058 |
| Ascorbic acid | MCE | Cat# HY-B0166 |
| cAMP | Sigma-Aldrich | Cat# A9501 |
| Penicillin–streptomycin | Gibco | Cat# 15140122 |
| 2-Mercaptoethanol | Gibco | Cat# 21985023 |
| N-2 supplement | Gibco | Cat# 17502048 |
| B-27 supplement | Gibco | Cat# 17504044 |
| Insulin | MCE | Cat# HY-P0035 |
| DMEM | Corning | Cat# 10-013-CV |
| DMEM/F-12 | Gibco | Cat# 11330032 |
| D-PBS | Gibco | Cat# 14190144 |
| Neurobasal medium | Gibco | Cat# 21103049 |
| KnockOut Serum Replacement | Gibco | Cat# 10828028 |
| GlutaMAX supplement | Gibco | Cat# 35050061 |
| MEM non-essential amino acids | Gibco | Cat# 11140050 |
| Mtesr Plus | Stem Cell Technologies | Cat# 100-0276 |
| Relesr | Stem Cell Technologies | Cat# 05872 |
| Accutase | Sigma-Aldrich | Cat# A6964 |
| Gentle Cell Dissociation Reagent | Stem Cell Technologies | Cat# 07174 |
| FBS | Hyclone | Cat# SH30084.03 |
| Normal Donkey Serum | Jackson | Cat# 017-000-121 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| Penicillin/streptomycin/amphotericin B, sterile solution | Leagene | Cat# CA0077 |
| Sucrose | Sigma-Aldrich | Cat# S7903 |
| Bacterial and virus strains | ||
| VSVG-Lentai-hEF1a-PuroR-CMV-mRuby3 | Hanbio | / |
| SFFV-luciferasr-IRES2-EGFP | Hanbio | / |
| Experimental models: Cell lines | ||
| DXR0109B | ATCC | Cat# ACS-1023 |
| H9 | Wicell | Cat# Wa09 |
| U-87 MG | Mingzhoubio | Cat# MZ-2455 |
| Others | ||
| CO2 incubators | Esco | Cat# CCL-170T-8 |
| Biological safety cabinet | Esco | Cat# 2010655 |
| Six-well tissue culture dishes | Biofil | Cat# TCP011006 |
| Sterile filter pipette tips | Kirgen | Cat# KG1313; KG1212; KG1111 |
| Sterile microcentrifuge tubes | Axgen | Cat# MCT-150-C-S |
| U-bottom ultralow attachment plates, 96 well | Corning | Cat# CLS7007 |
| Conical tubes, 15 mL | NEST | Cat# 601002 |
| Ultralow attachment plates, 6 well | Corning | Cat# CLS73471 |
| Tissue culture dish, 60 mm | Biofil | Cat# TCD010060 |
| Stericup 0.2-μm filter unit | Biofil | Cat# FPV103150, FPV103250, FPV103500 |
| Orbital shaker | NEST | Cat# 105008 |
| Serological pipettes, 10 mL | NEST | Cat# 327001 |
| 1 channel Mechanical Pipette | Eppendorf | Cat# 3120000020, 3120000011, 3120000062 3120000046 |
| 8-channel Mechanical Pipette | Eppendorf | Cat# 3122000051 |
| Electronic Pipette Controller | Eppendorf | Cat# 4430000018 |
| Software | ||
| GraphPad Prism | https://www.graphpad.com/features | |
| Adobe Illustrator | https://www.adobe.com/products/illustrator.html | |
| Fiji | https://fiji.sc/ | |
| Adobe Premiere Pro | https://www.adobe.com/cn/products/premiere.html | |
Materials and equipment
EB Formation Medium
| Reagent | Final concentration | Amount | Storage conditions |
|---|---|---|---|
| mTeSR Plus | N/A | 50 mL | 4°C |
| Y-27632 Rock Inhibitor [10 mM] | 50 μM | 500 μL | −20°C |
| Total | N/A | 50 mL | 4°C |
Mix all these reagents on the day of use. If the hiPSCs or hESCs are cultured in other culture systems, such as E8, we recommend using the cell culture medium that cultured the cells with the addition of 50 μM rock inhibitor as the medium for EB formation.
Storage condition and storage time: The prepared culture medium should be stored at 4°C and used up within two weeks.
U-87 MG cells culture medium
| Reagent | Final concentration | Amount | Storage conditions |
|---|---|---|---|
| DMEM | N/A | 45 mL | 4°C |
| FBS | 10% | 5 mL | −20°C |
| Total | N/A | 50 mL | 4°C |
Storage condition and storage time: The prepared culture medium should be stored at 4°C and used up within two weeks.
30% sucrose solution
| Reagent | Final concentration | Amount | Storage conditions |
|---|---|---|---|
| Sucrose | 30% | 15 g | RT |
| D-PBS | N/A | 50 mL | RT |
| Total | N/A | 50 mL | 4°C |
RT: Room temperature.
Storage condition and storage time: The prepared sucrose solution should be stored at 4°C and used up within one month.
Permeabilization Buffer
| Reagent | Final concentration | Amount | Storage conditions |
|---|---|---|---|
| Triton X-100 | 0.1% | 500 μL | RT |
| D-PBS | N/A | 500 mL | RT |
| Total | N/A | 500 mL | 4°C |
Storage condition and storage time: The prepared permeabilization buffer should be stored at 4°C and used up within one month.
Antibody Diluent Solution
| Reagent | Final concentration / Dilution | Amount |
|---|---|---|
| Mouse Monoclonal Anti-SOX2 | 1:100 | N/A |
| Goat Monoclonal Anti-SOX2 | 1:500 | N/A |
| Rabbit Polyclonal Anti-TBR2 | 1:500 | N/A |
| Rat Monoclonal Anti-CTIP2 | 1:100 | N/A |
| Rabbit Polyclonal Tubulin β 3 (TUJ1) | 1:500 | N/A |
| Mouse Monoclonal BRN2 | 1:300 | N/A |
| Rabbit Polyclonal SATB2 | 1:100 | N/A |
| Chicken Polyclonal Glial Fibrillary Acidic Protein (GFAP) | 1:1000 | N/A |
| Rabbit polyclonal to Ki67 | 1:100 | N/A |
| Donkey Anti-Mouse IgG H&L (Alexa Fluor® 568) | 1:500 | N/A |
| Goat Anti-Rabbit IgG H&L (Alexa Fluor® 488) | 1:500 | N/A |
| Goat Anti-Rat IgG H&L (Alexa Fluor® 647) | 1:500 | N/A |
| Donkey Anti-Rabbit IgG H&L (Alexa Fluor® 488) | 1:500 | N/A |
| Goat Anti-Chicken IgY H&L (Alexa Fluor® 647) | 1:500 | N/A |
Storage condition and storage time: Antibody solution should be stored at 4°C and used up within one month.
Step-by-step method details
Generating embryoid bodies (EBs)
Timing: 30 min, to generate a 96-well plate of EBs
This procedure demonstrates the method for generating embryoid bodies from hiPSCs and hESCs.
-
1.Grow cells at 70%–80% confluency.
-
a.The healthy condition of cells is crucial for generating organoids, in short, cells confluency should reach 70%–80% and appear as flat and smooth colonies (Figure 1A).
-
a.
-
2.
Detach colonies into single cell by Accutase and incubate cells at 37°C with 5% CO2 for 4–5 min.
-
3.
Add 1 mL EB formation medium, that is, mTeSR Plus with 50 μΜ Y-27632, into each well to stop the dissociation.
Note: mTeSR Plus used as the EB formation medium is because hiPSCs and hESs are maintained in this medium for a long time. If cells are cultured in other medium, such as Essential 8 medium, it’s better to use the medium that culturing cells to form EBs.
-
4.
Collect cells in the 15 mL centrifuge tube.
-
5.
Centrifuge at 300 × g for 5 min, remove the supernatant and keep the cell pellet.
-
6.Resuspend the cells in 1 mL EB formation medium and calculate the live cell density.
-
a.Resuspend the respective number of cells in the appropriate volume of EB formation medium.
-
b.The generation of single EB requires 1500 cells/well.
-
c.The total volume of each well is 150 μL.
-
d.For a 96-well plate, resuspend cells in 15 mL EB formation medium at the total number of 150,000.
-
a.
-
7.
Transfer the cell suspension to multichannel reagent reservoirs, then aliquot 150 μL into each well of an ultra-low cell attachment U bottom 96-well plate by using the multichannel pipette.
Optional: Centrifuge the plate at 480 × g for 5 min at room temperature. EBs can form naturally in the wells, this step can help EBs gather faster without any impact on EBs formation.
-
8.
Incubate EBs at 37°C with 5% CO2 for 24h and avoid disturbance.
-
9.
At Day 1 (24 h after EBs generation), observed the successfully formed EBs under a bright field microscope. EB presents a spherical shape and smooth peripheral. Some dead cell debris that appeared around EBs will not affect the formation of organoids. troubleshooting 3.
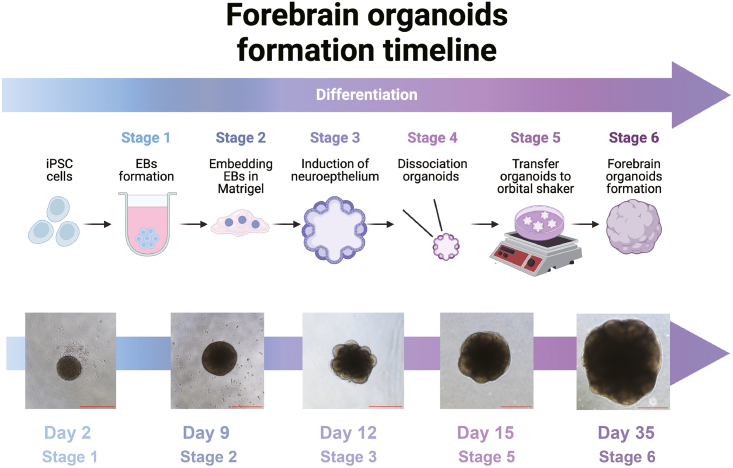
Figure 1.
EBs embedded in Matrigel to form neuroepithelium, related to step 28
(A) Figure A shows a smooth and round hiPSCs colony.
(B) At Day 8, EBs appear spherical with a transparent edge and smooth surface.
(C) Figures show the procedure of making ‘cookies’ when embedding EBs in Matrigel. (Scale bar: 500 μm).
Induction of forebrain organoids
Timing: 6 days
This procedure is to generate organoid of forebrain regions.
-
10.
On Day1, remove half volume of EB formation medium and replace with Medium 1, that is, the medium used at the First stage (Table 1).
-
11.
Half-half refresh the medium daily from Day 2 to Day 5.
-
12.
On Day 5 and Day 6, refresh half volume of the medium with the Medium 2 (Table 1).
Table 1.
Medium for different stages of COs
| Stage | Medium | Container used | Reference |
|---|---|---|---|
| EB formation (Day 0 - Day 1) | mTeSR Plus with 50 μΜ Y-27632 | Ultra-low attachment 96-well plate | / |
| First (Day 2 – Day 4) | Medium 1: DMEM/F12(1:1)basic(1X), KOSR(Knockout Serum Replacement) (20%), GlutaMAX(1X), MEM NEAA(MEM Non-Essential Amino Acids Solution)(1X), Penicillin-Streptomycin (10,0 U/mL), 2-Mercaptoethanol (0.1 mM), Dorsomorphin (2μm), A-83 (2μm), |
Qian et al.1 | |
| Second (Day 5 – Day 7) | Medium 2: DMEM/F12(1:1)basic (1X), GlutaMAX (1X), MEM NEAA(MEM Non-Essential Amino Acids Solution) (1X), Penicillin-Streptomycin (100 U/mL), N-2 Supplement (1X), CHIR-99021 (1μm), SB-431542 (1μm), Wnt3a Protein (2μm), |
Ultra-low attachment 96-well plate | |
| Third (Day 8– Day 14) | Medium 2: DMEM/F12(1:1)basic (1X), GlutaMAX (1X), MEM NEAA(MEM Non-Essential Amino Acids Solution) (1X), Penicillin-Streptomycin (100 U/mL), N-2 Supplement (1X), CHIR-99021 (1μm), SB-431542 (1μm), Wnt3a Protein (2μm), |
Ultra-low attachment 6-well plate ,Embedding Matrigel(354230) (Mix 50% Medium 2) | |
| Forth (Day 15 – Day 34) | Medium 3:DMEM/F12(1:1)basic (50%), Neurobasal Medium (50%), GlutaMAX (1X), MEM NEAA(MEM Non-Essential Amino Acids Solution) (1X), 2-Mercaptoethanol (0.1 mM), Penicillin-Streptomycin (80 U/mL), Penicillin / Streptomycin / Amphotericin B, sterile solution (0.2X) N-2 Supplement (1X), B-27 Supplement (1X), Ascorbic acid (0.2 mM), cAMP (0.5 mM), Insulin(2.5 μg/mL), BDNF (20 ng/mL), GDNF (20 ng/mL), |
60 mm TC-treated Cell Culture Dish, Orbital Shaker 100rpm | |
| Forth + Matrigel (Day 35 – Day 69) | Medium 3 + Matrigel: DMEM/F12(1:1)basic (50%), Neurobasal Medium (50%), GlutaMAX (1X), MEM NEAA(MEM Non-Essential Amino Acids Solution) (1X), 2-Mercaptoethanol (0.1 mM), Penicillin-Streptomycin (80 U/mL), Penicillin / Streptomycin / Amphotericin B, sterile solution (0.2X) N-2 Supplement (1X), B-27 Supplement (1X), Ascorbic acid (0.2 mM), cAMP (0.5 mM), Insulin(2.5 μg/mL), BDNF (20 ng/mL), GDNF (20 ng/mL), Matrigel(354230) (1%) |
60 mm TC-treated Cell Culture Dish, Orbital Shaker 100rpm | |
| Fifth (Day 70 – ∞) | Medium 4: Neurobasal Medium (1X), GlutaMAX (1X), MEM NEAA(MEM Non-Essential Amino Acids Solution) (1X), 2-Mercaptoethanol (0.1 mM), Penicillin-Streptomycin (80 U/mL), Penicillin / Streptomycin / Amphotericin B, sterile solution (0.2X), B-27 Supplement (1X), Ascorbic acid (0.2 mM), cAMP (0.5 mM), BDNF (20 ng/mL), GDNF (20 ng/mL), |
60 mm TC-treated Cell Culture Dish, Orbital Shaker 100rpm |
Storage condition and storage time: The prepared culture medium should be stored at 4°C and used up within two weeks. We recommend mixing small molecule compounds at the day before use to achieve their maximum effect.
Notice: The formula of Medium 2 refers to Qian et al.,1 the only difference is that Medium 2 contains WNT3A. It was mentioned in Qian et al. 20162 that WNT3A and CHIR99021 activate the same downstream signaling pathway. They considered that WNT3A had little effect in it, so they deleted it in their article at 2018. But we found that the success rate of forebrain region induction is higher with the addition of WNT3A, which may be caused by the insufficient concentration of CHIR99021.
-
13.
On Day 7, EB appears spherical with a transparent and smooth surface. The sphere edge is brighter than the interior with a clear interface in brightness between the two regions (Figure 1Β).
Note: The quality of healthy EBs is crucial for successful outcome.
Generation of neuroepithelium of organoids
Timing: 1 h for embedding EBs in Matrigel, 5 days for generation of neuroepithelium of organoids, and 1 h for mechanically dissociating organoids from Matrigel
This procedure is to generate neuroepithelium buds of organoids.
-
14.
On Day 7, thaw an aliquot of Matrigel (Cat. 354230) on ice.
-
15.
Wet 600 μL centrifuge tubes with the Medium 2 for later use.
-
16.
Cut the P200 pipette tips to widen the openings to 1.5–2 mm in diameter, and transfer EBs into pre-wet tubes.
-
17.
Let the EBs settle down and remove all the supernatant.
-
18.
Add 67 μL Medium 2 to each tube.
-
19.
Place the tubes on ice.
Note: Each tube containing 20–30 EBs.
-
20.
Add 100 μL Matrigel and mix with medium and EBs on ice by using a cut tip.
-
21.
Pipette the suspension up and down.
-
22.
Using the cut tip, transfer the EBs-Matrigel mixture to the ultra-low-attachment 6-well plate to make a ‘cookie’ (Figure 1C).
-
23.
Use P10 pippette tips to distribute EBs in the cookie, and place the cookie in the middle of each well.
CRITICAL: The whole process should be performed on ice.
-
24.
Incubate the EBs-Matrigel cookies at 37°C with 5% CO2 for 30 min.
-
25.
Gently add 4 mL pre-warmed Medium 2 into each well, incubate EBs at 37°C with 5% CO2.
-
26.
Completely refresh the Medium 2 each day from Day 8 to Day 14. To avoid disrupting EB-Matrigel complex, add pre-warmed medium gently. troubleshooting 4.
-
27.
On Day 14, pipette the cookies by using 15 mL serological pipettes at a volume rate of 2 mL per s. More details can be referred to Qian et al.1
-
28.
Observe whether the organoids are completely detached from the Matrigel under a bright field microscope; if not, repeat step 27 for 1–2 times.
Growing organoids of forebrain region
Timing: long term culture, approximately 45 days
This procedure describes the method for long-term cultivation of organoids.
-
29.
Transfer the detached organoids in 60 mm cell culture dishes for long term culture.
-
30.
Each 60 mm cell culture dish contains 10–15 organoids.
-
31.
Completely replace Medium 2 with 5 mL Medium 3 each dish (Table 1).
-
32.
Place an orbital shaker in the incubator.
-
33.Place dishes on the orbital shaker at the speed of 85–95 rpm.
-
a.Place organoids on the orbital shaker for long-term culture.
-
b.Change Medium completely every two days.
-
a.
-
34.
On Day 35, refresh the medium with Medium 3 containing supplementary Matrigel (Cat. 354230) at the dilution ratio 1: 100 in order to create an extracellular matrix coating on the surface of organoids.
Note: COs can be maintained for at least 6 months. The scheme of organoid pattern is shown in Figure 2.
Figure 2.
Scheme of organoid pattern with bright field microscope images, related to step 34
Figure 1 shows the formation time line of forebrain organoids. The bright field microscope images of different stages organoids are shown respectively. (Scale bar: 500 μm).
Slice organoids to prevent cell death and sustain health
Timing: 1 h for slicing a dish of organoids
This procedure is set for preventing hypoxia and cell death of organoid, by exposing organoid sections into culture medium.
-
35.Preparation before vibrating sectioning.
-
a.Place the vibratome (Leica VT 1200S) into the hood.
-
b.Expose the vibratome to UV sterilization for at least 1 h to prevent contamination.
-
c.Collect forebrain organoids at Day 45 from the orbital shaker and immerse organoids in the 3% (w/v) ultra-low-melting-point agarose dissolved in DMEM/F-12.
-
d.Keep the agarose mixture at 45°C to maintain a liquid phase to avoid solidifying.
-
e.Embed up to 15 organoids in agarose within a disposable plastic base mold (1.5 cm wide, square).
-
f.Move the agarose blocks on ice for 5–10 min to solidify.
-
a.
Note: When embedding organoids, first lay a thin layer of agarose solution in the block. Then quickly use a Pasteur pipette to place the organoids into the block, and finally drop agarose solution to make all organoids sink to the bottom of the block, to keep the organoids in the same plane within the block. troubleshooting 5.
-
36.
Perform vibrating sectioning by Leica VT 1200S in ice-cold DMRM/F-12 containing 1 X Penicillin-Streptomycin-Amphotericin B.
-
37.
Set vibratome as 0.12 mm/s in speed and 1 mm in amplitude.
Note: For users who have no access to this vibratome, we recommend using Leica VT1000S to perform vibratome with same parameters. Or use spring scissors to cut COs, refer to Momoko et al..
-
38.
Collect and separate 500 μm-thick sections from agarose by gentle pipetting.
Note: Only the middle sections are collected. Each organoid can be considered as an irregular sphere with a diameter of 1.2–1.5 mm, so each organoid can collect 1–2 middle sections. The first and last sections of each organoid cut with a vibratome should be discarded, as it cannot be guaranteed that they are 500 μm-thick.
-
39.
Transfer organoid sections to the ultra-low-attachment 6-well plate with the Medium 3 supplemented with 1% Matrigel(354230).
-
40.
Maintain organoid sections in a static state at 37°C with 5% CO2 for 1 h. troubleshooting 6.
-
41.
After 1 h, transfer organoid sections to 60 mm cell culture dishes on the orbital shaker at 37°C with 5% CO2 for long term culture.
Note: COs should be sliced once a month from Day 45 to avoid hypoxia following the same procedure as described in steps 35–41.
Validation of forebrain organoids
Timing: 1 week
The purpose of this step is to confirm the success of the forebrain pattern and the formation of the ventricular zone (VZ) and cortical plate (CP).
-
42.
On Day 1, collect Day 60 COs in a 35 mm dish without medium and fixed in 4% paraformaldehyde (PFA) pH7.4 at temperature for 20 min.
-
43.
Rinse COs in phosphate buffered saline (PBS) 3 times for 10 min each at room temperature.
-
44.
Gently transfer COs in 1.5 mL microcentrifuge tubes with 30% sucrose solution.
-
45.
Incubate COs in 30% sucrose solution at 4°C overnight (8–12 h).
-
46.
On Day 2, transfer each CO to a 7∗7∗5 mm disposable plastic base mold and embed in optimal cutting temperature compound (OCT) for supporting tissue integrity during cryosectioning.
Note: COs embedded in OCT should stay at 4°C overnight (8–12 hrs) before being transferred to −80°C or −20°C, to completely infiltrate the tissue.
-
47.
On Day 3, before cryosectioning, thaw the mold with the frozen COs for 30 min in a cryostat chamber set to −20°C.
-
48.
Remove COs from mold and set on the specimen stage with OCT.
-
49.
Collect 10 μm sections and place two sections each glass slide.
-
50.
Allow slides to dry at temperature for at least 1 h.
-
51.
Rinse slides with PBS 3 times for 10 min to remove OCT.
-
52.
Use a pap-pen or liquid blocker pen to draw a border around the sections on the slide and allow to dry.
-
53.
Block and permeabilize the tissues by adding blocking solution (PBS supplemented with 5% normal donkey serum and 0.1% Triton X-100).
-
54.
Block for 1 h at room temperature or 4°C overnight (8–12 h) in a humidified chamber.
-
55.
Stain with an appropriate primary antibody in blocking solution overnight (8–12 h) in a humidified chamber at 4°C.
-
56.
Rinse slides with PBST (PBS supplemented with 0.1% Triton X-100) twice and PBS twice for 10 min each time.
-
57.
Stain with the secondary antibody in blocking solution for 2 h in humidified chamber at room temperature.
-
58.
Rinse slides with PBST (PBS supplemented with 0.1% Triton X-100) twice and PBS twice for 10 min each time.
-
59.
Stain with DAPI (1ug/mL in PBS) for 5–10 min at room temperature.
-
60.
Remove the DAPI solution and add glycerol to samples on the slides.
-
61.
Mount the slides with cover glass and seal them using clear nail polish.
-
62.
Store the slides in store at −80°C or imaging immediately using confocal microscopy. Forebrain organoids successfully develop VZ, CP and astrocytes at Day 60 (Figure 3).
Figure 3.
VZ, CP, neuron and glia formation of day 60 organoids, related to step Multiple antibodies were used to verified the formation of forebrain organoids, related to step 62
(A) including TUJ1, a marker for neurons of early stage; GFAP, a marker for astrocytes.
(B) SOX2, a marker for neurons in ventricular zone; TBR2, a marker for neurons in subventricular zone; CTIP2, a marker for neurons cortical plate.
(C) BRN2, a marker for neurons in layers 2/3 of CP; SATB2, a marker for neurons from layers 2–4 of CP. Forebrain organoids at Day 60 show appropriate glia formations and polarized rosettes. (Scale bar: 300 μm).
Transfection of fluorescent protein in U-87 MG cell line
Timing: 1.5 week
This procedure is to produce U-87 MG cell line with a fluorescent protein which can be visualized its activity under the fluorescence microscope.
-
63.
Plate U-87 MG into 24-well plate at a density of 1 × 105 cells/mL one day in advance using Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). 50%–70% confluence should be reached on the following day.
-
64.
Before infection, calculate the volume of virus based on MOI and resuspend the melted virus in the 250ul cell culture medium.
Example: If the virus titer is 1 × 108 IU/mL and you want to deliver 1 × 105 cells, you need 10 μL volume of virus to achieve an MOI of 10.
-
65.
Remove the preceding medium and wash once with the DPBS. Then U-87 MG are inoculated with VSVG-Lentai-hEF1a-PuroR-CMV-mRuby3 lentivirus at multiplicities of infection (MOI) of 0, 10, 20, 40, 80 with or without the 5ug/mL polybrene.
-
66.
Refresh the culture medium after 12–16h incubation in 37°C.
-
67.
Check the fluorescence via microscopy after 72h to choose one appropriate MOI for subsequent experiments, which ensures the high lentiviral transfection efficiency with minimum damage.
-
68.
After choosing an appropriate MOI for infection, prepare uninfected U-87 MG cells in 6-well plate 24h in advance.
-
69.
Repeat the procedure 61–63 at an MOI of 10.
Note: In fact, both MOI of 10 and 20 are relatively appropriate. Viruses within this range of MOI will cause cells to express strong red fluorescent protein without affecting cell viability.
-
70.
After 24h, change the medium completely for U-87 MG and incubate under the same conditions for 48h.
-
71.
Next, select stably transfected U-87 MG cells by changing medium supplemented with puromycin (5ug/mL) until the non-transfected cells die off. This procedure lasts for 1 week.
Organoids slices co-culture with U-87 MG cells
Timing: 1 day
This step is to generate glioma organoids with high reproducibility.
-
72.
Prepare the co-culture medium: mix the differential organoid medium with U-87 MG culture medium at the volume ratio of 1: 1.
Note: Co-culture medium can prevent cell death from both organoids and U-87 MG cells.
-
73.
On day 0, transfer each vibratome-sectioned Day 60 forebrain organoids in a well of ultra-low-attachment 24-well plate. troubleshooting 7.
-
74.
Add 1 mL co-culture medium to each well.
-
75.
Rinse the U-87 MG cells twice with PBS (1x) to remove FBS in the dishes.
-
76.
Add 3 mL of 0.25% Trypsin-EDTA solution to each 10 cm dish.
-
77.
Incubate U-87 MG cells at 37°C with 5% CO2 for 5 min.
-
78.
Add 3 mL of U-87 MG culture medium to stop the digestion, and pipette the suspension to completely detach the cells from the culture dish.
-
79.
Transfer the cell suspension to a 15mL centrifuge tube.
-
80.
Centrifuge Cells for 5 min at 300 × g.
-
81.
Remove the supernatant. Resuspend the cells with 1 mL co-culture medium and calculate the live cell density.
-
82.Resuspend the respective number of cells in the appropriate volume of co-culture medium.
-
a.The generation of a single organoid slide in co-culture with U-87 MG cells require seeding of 10ˆ5 U-87 cells in each well. Load each well with one organoid slide (approximately 3–4 mm in diameter).
-
b.Load each well with 1 mL co-culture medium and kept consistent across wells.
-
c.For a 24-well plate, resuspend cells in total 24 mL co-culture medium at the total number of 2.4∗106 cells per well.
-
a.
-
83.
Add 1 mL cell suspension into each well.
-
84.
Incubate organoids co-cultured with U-87 MG cells at 37°C with 5% CO2 for 24 h under static culture.
-
85.
On Day 1, U-87 MG cells successfully invade into forebrain organoids, which can be observed under the fluorescence microscope (Figure 4). troubleshooting 8 and 9.
Figure 4.
Timeline of forebrain organoids co-cultured with U-87 MG, related to step 85
U87 cells carried mRuby fluorescent proteins invaded in the organoids after 24 h and formed the special structures in organoids. On the first day of co-culture, U87 cells are observed to diffuse and begin to aggregate in the organoid under a fluorescent microscope (see zoom-in image of Day 1); as the days increased, U87 cells show a stronger preference for clustering together in the organoids. (Scale bars: Expect for the last panel, scale bar represents 500 μm. Scale bar of the last panel images represent 300 μm).
Validation of organoid-U87 assembloids
Timing: 1 week
This procedure is to verify that U87 cells have successfully invaded the organoids and that the cell-cell interactions between U87 cells and organoids can be observed under confocal microscopy.
-
86.
On the third day of co-culture, fix the assembloids in 4% PFA at room temperature for 20 min.
-
87.
Rinse the assembloids for three time with PBS (1x), each time lasts for 10 min.
-
88.
Transfer the assembloids to a 1.5 mL centrifuge tube for subsequent immunofluorescence staining.
-
89.
Block and permeabilize the tissues by adding blocking solution (PBS supplemented with 5% normal donkey serum and 0.1% Triton X-100). Block for 1 h at room temperature or 4°C overnight (8–12 h) in the tubes.
-
90.
Stain with an appropriate primary antibody in blocking solution for 1–2 day at 4°C.
-
91.
Rinse the assembloids with PBST (PBS supplemented with 0.1% Triton X-100) twice and PBS twice for 10 min each time.
-
92.
Stain with the secondary antibody in blocking solution for 1 day at 4°C.
-
93.
Rinse assembloids with PBST (PBS supplemented with 0.1% Triton X-100 ) twice and PBS twice for 10 min each time.
-
94.
Stain with DAPI (1ug/mL in PBS) for at least 3 h at room temperature.
-
95.
Using FOCM (ultrafast optical clearing method) to perform tissue clearing on assembloids. Ultrafast optical clearing method refers to Zhu et al.3
-
96.FOCM reagent preparing:
-
a.Dissolve urea and D-mannitol in DMSO.
-
b.Stir overnight (8–12 h) at room temperature (25°C) until completely dissolved.
-
c.Add glycerol and stir further.
-
d.Store the reagent at room temperature for several months and gently shake before use.
-
e.Transfer the assembloids to the glass bottom cell culture dishes.
-
a.
-
97.
Remove all the liquid and add 100 μL FOCM reagent on the dish.
-
98.
Culture organoids in FOCM reagent for at least 15 min until sent for confocal scanning (Figure 5 & Methods video S1). troubleshooting 10.
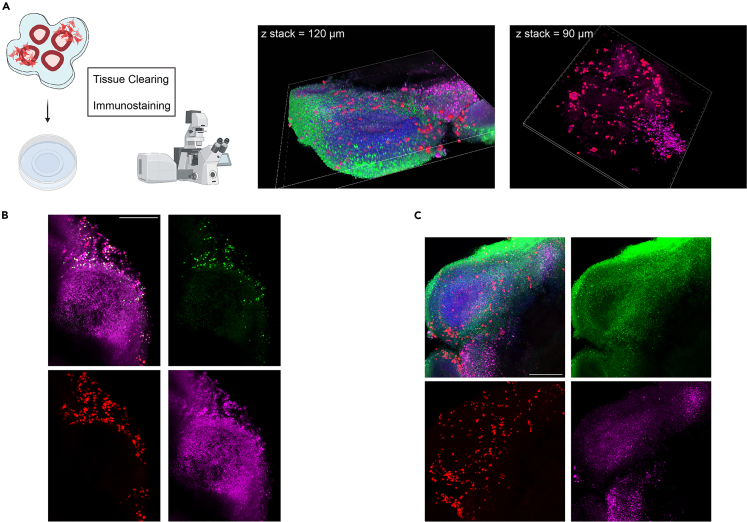
Figure 5.
Immunofluorescence validation of the sectioned assembloids at Day 3, related to step 98
(A) First, the organoid-U87 cell assembloids on Day 3 are fixed and immune-fluorescence stained. Prior to performing confocal microscopy, tissue clearing is required for assembloids to facilitate laser penetration through the tissue. The 3D graph in the middle column shows a 120 μm thick structure of assembloid, with green representing TUJ1, red representing U87 cells, and magenta representing GFAP. The 3D graph in the left column shows the invasion of U87 cells in a 90-micron thick section of the organoid-U87 assembloid, with red representing U87 cells and magenta representing GFAP. These results indicate that U87 cells can infiltrate to at least 120 μm thick and distribute throughout the organoid slices after co-culturing for three days.
(B) Two-dimensional immunofluorescence images of organoid-U87 assembloids: Green represents Ki67, a marker for cell proliferation, red represents U87, and magenta represents MAP2
(C) Green represents TUJ1, red represents U87, and magenta represents GFAP. The scale bars for both B and C are 300 μm.
The drug sensitivity (e.g., TMZ) of U-87 MG glioma organoid models
Timing: 2 days
This procedure is to test the temozolomide (TMZ) sensitivity, a widely used clinical drug for glioma cancer, of U-87 MG glioma organoid models (Figure 6B).
-
99.Generate U-87 MG cells stably expressing luciferase by lentiviral infection of the SFFV-luciferasr-IRES2-EGFP construct.
-
a.U-87 MG/Luciferase cells, which have been transfected with SFFV-luciferasr-IRES2-EGFP, are transfected with VSVG-Lentai-hEF1a-PuroR-CMV-mRuby3 lentivirus, refer to step 63–71.
-
b.Since that, U-87 MG/Luciferase/mRuby3 cells are generated.
-
a.
-
100.
Transfer Organoids co-cultured with U-87 MG/Luciferase/mRuby3 cells of Day 2 to an ultra-low-attachment 24-well cell culture plate.
-
101.Place organoids in individual wells in a co-culture medium containing TMZ or vehicle control.
-
a.Respective concentration of TMZ: 200 μM, 500 μM, and 1000 μM is diluted in co-culture medium.
-
a.
-
102.
Incubate Organoids at 37°C with 5% CO2 for 48 h.
Note: Do not refresh the medium within 48 h.
-
103.
Take images of each organoid on Day 0 and Day 2 (Figure 6A).
-
104.On Day 2, prepare D-Luciferin Potassium Salt Bioluminescent Substrate solution.
-
a.Prepare 200X Luciferin stock solution (30 mg/mL) in sterile water. Mix the solution by pipetting repeatedly until the solute is completely dissolved.
-
b.Aliquot the luciferin stock solution and store it in −80°C.
-
c.Prepare Luciferin working solution (150 mg/mL final concentration) in co-culture medium. 1 mL for each well.
-
a.
-
105.
Remove all the medium in wells. Add 1 mL luciferin working solution to each well.
-
106.
Incubate organoids at 37°C with 5% CO2 for 10–20 min before imaging.
Note: Incubation time will influence the signal.
-
107.
Check the in vitro bioluminescence with Tecan Spark Cyto. Analyze the signal among each group (Figure 6C). troubleshooting 11.
Note: To obtain multiple luciferase readings from the same drug-treated organoids at different time points, after each measurement of luciferase activity, completely remove the luciferin working solution and wash the organoid-U87 assembloids twice with the co-culture medium. Then, add back the medium at step 105 into the corresponding wells. Repeat steps 105–107 when performing the next measurement.
-
108.
Further application of U-87 MG glioma organoids: Although we have only demonstrated one of the applications of U-87 MG glioma organoids, which is to test the drug sensitivity of U87 cells fused with organoid tissues, other types of cell activity in the cerebral organoids, such as neurons and astrocytes, can also be monitored during drug treatment, to predict potential side effects or adverse reactions of new drugs in clinical use. In addition to drug-related applications, this model can serve as a mechanism research model for glioma cells invasion and interaction with other types of cells in the brain.
Figure 6.
TMZ drug sensitivity of U-87 MG glioma organoids, related to step 99–107
(A) Figures show the changes of the density and the fluorescent intensity of U-87 MG glioma organoids under TMZ treatment after 48 h.
(B) To more accurately measure drug sensitivity, U87 cells were transfected by SFFV-luciferasr-IRES2-EGFP and VSVG-Lentai-hEF1a-PuroR-CMV-mRuby3. The U87 cells used for testing drug sensitivity will stably express the luciferase protein. After co-culturing with organoid slices and drug treatment for 48 h, luciferin substrate was added and allowed to react with the expressed luciferase from U87 cells for 10–20 min. Luciferase activity was measured via microplate reader to quantitate the activity of U87 cells.
(C) And the histogram shows the drug sensitivity of U-87 MG glioma organoids exposed to different concentration of TMZ, measured by the fold change of luciferase activity. The histogram was shown as the mean ± SEM (n = 4). Statistical significance (∗∗∗P < 0.001, ∗∗P < 0.01, ∗P < 0.05) was assessed by one-way ANOVA followed by Bonferroni’s multiple comparison tests. (Scale bar: 500 μm).
Expected outcomes
This protocol describes the generation of forebrain organoids and the method of preventing hypoxia by vibratome section. The vibratome sectioned forebrain organoids are easily invaded by U-87 MG cells, which helps the formation of glioma organoids. Our glioma organoids can be formed within 24 h, and a large number of glioma organoids can be reproduced by vibratome section method. A large number of U-87 MG invaded into the brain organoids could be observed under the fluorescence microscope on the second day after co-culture. On the third day of co-culture, using tissue clearing and immunofluorescence staining, U87 cells can be observed to form structures in the organoids and interact with other types of cells through confocal scanning of sectioned organoid-U87 assembloids. Following our protocol, the formation of forebrain organoids can be confirmed by bright field microscopy and immunofluorescence staining in the early stage, and the invasion of U-87 MG can be observed in real time under a fluorescence microscope when co-culturing with forebrain organoids. The organoid-U87 assembloids can be used to study the communication mechanisms between glioblastoma cells and cells from brain organoids. In addition, we found that on the fifth day of U87 cell invasion into the organoids, they tended to cluster rather than diffusing uniformly throughout the forebrain organoids. This phenomenon is consistent with the behavior of real glioblastomas, where cancer cells prefer to cluster and form solid tumors inside the patient’s brain.
Limitations
This protocol established a reproducible method for generating glioma organoids by co-culturing U-87 MG and COs. However, using a glioma stem cell line instead of GSCs derived from patients may result in different performance of glioma stem cells invading brain organoids from different patients. In addition, glioma organoids generated by this method lack microglia which plays an important role in glioma in patients. Although our protocol can maintain the survival of U87 cells in the COs for at least 9 days, the brain organoids will eventually be invaded and collapsed by U87 cells after 9 days, rendering the collapsed brain organoids unsuitable as a scaffold for U87 cells survival. Therefore, our model has limitations, and using smaller numbers of U87 cells to invade the organoids may prolong the survival of the model.
Troubleshooting
Problem 1
Matrigel doesn’t dissolve properly or there are visible clumps (related to before you begin: step 4).
Potential solution
Matrigel has probably become gelling. It’s better to thaw the Matrigel on ice overnight (8–12 h) rather than at 2°C–8°C. Try to thaw another aliquot to coat the plate.
Problem 2
There is differentiation and contamination when culturing hiPSCs and hESs (related to before you begin: step 10).
Potential solution
If the proportion of differentiated cells in the well is less than 10%, mark the position with a marker pen, and scrape off the cells at the marked position with a P1000 pipette tip. During the process of culturing cells, regular detection of mycoplasma is required to ensure the healthy growth of cells. If there is bacterial, fungal, mycoplasma contamination, it is necessary to check the cell room environment, medium and consumables.
Problem 3
EBs do not form after 24 h (related to step 9).
Potential solution
There are many reasons why EBs cannot be formed at this step, the first possible reason is the poor quality of hiPSCs. When using hiPSCs to generate EBs, pay attention to the fact that hiPSCs cannot be over-fused at this step, which will have a negative impact on the generation of EBs. A second possible explanation is that the EBs were not well settled down during the 24 h of formation. A potential solution to guarantee the successful generation of EBs is to ensure the health of the hiPSCs before seeding the EBs, which includes verifying that colonies are flat and smooth and not fused with others.
Problem 4
Organoids embedded in Matrigel do not form neuroepithelium structure (related to step 26).
Potential solution
Organoids embedded in Matrigel do not form neuroepithelium buds (Figure 1) or extend cells in Matrigel showing the failure differentiation of COs. The possible reason is the poor quality of EBs before Day 7. Healthy EBs at Day 7 should perform smooth and round morphology, with transparent edge.
Problem 5
Organoids cannot lie in the same plane or sink at the bottom of the block when they are embedded in the agarose (related to step 35).
Potential solution
The operation time is too long, resulting in partial solidification of the agarose. The best solution is to take out the agarose solution from the water bath after all the preparations are ready, and then complete the embedding process within 30 s.
Problem 6
Organoids slices cannot be easily separated from the agarose (related to step 40).
Potential solution
During the process of vibratome sectioning, organoid slices will automatically detach from the agarose. If the organoid slices do not detach from the agarose, try gently suctioning them out using a Pasteur pipette.
Problem 7
Why is it necessary to use vibratome-sliced COs for invasive experiments (related to step 73)?
Potential solution
Vibratome-sectioned forebrain organoids with a flat surface are more likely to fuse with U-87 MG, compared to unsliced organoids. Using ultra-low-attachment 24-well plate prevents U-87 MG cells from adherence.
Problem 8
The cells did not successfully invade the organoids, or only a small number of cells invaded (related to step 85).
Potential solution
It is possible to extend the time for COs fusion with cells under static culture or increase the number of cells, but this is not recommended as an excessive amount of U87 cells can lead to broken-apart organoids. Instead, we recommend using older COs, specifically those that are at least day 60 or older, as they are more easily invaded by U87 cells.
Problem 9
Organoids start to break apart after invasion in a few days (related to step 85).
Potential solution
The reason for organoids breaking apart is that there are too many cells invading the organoids. If the organoid slices are smaller (less than 3 mm in diameter), try using less cells to perform invasion.
Problem 10
Why did the sectioned organoids not become transparent after being treated with FOCM reagent for 15 min (related to step 98)?
Potential solution
The transparency is related to the amount of time the organoids stay in the FOCM reagent and the length of time they are fixed in 4% PFA. The longer the fixed time, the more likely they are to become transparent. It is recommended to try extending the fixed time to around 1 h, but be careful, as a fixed time that is too long can affect the efficiency of antibody binding.
Problem 11
How to control the consistency of organoid models when they are used for drug screening? If different focus plane affects the drug sensitivity analysis (related to step 107)?
Potential solution
When using brain organoids as a screening model for glioblastoma, it is necessary to use hiPSCs or hESCs from the same batch and induce the organelles at the same time. However, even with this, there can be individual differences between organelles from the same batch. When selecting organelles to establish the glioblastoma organelle model, we choose organoid slices of similar diameter (between 3–4mm in diameter) and with larger rosette structures. The size of the rosette reflects to some extent the maturity of organoids development.
Although organoids are randomly distributed in the medium, the U-87 MG/Luciferase/mRuby3 cells will react with the substrate luciferin throughout the system regardless of their location. By measuring the bioluminescence intensity of the entire well, the activity of cells carrying luciferase can be quantified.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Shaohua Ma, ma.shaohua@sz.tsinghua.edu.cn.
Materials availability
This study did not generate any unique reagents.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (grant number: 61971255 and 82111530212) and the Natural Science Foundation of Guangdong Province (grant number: 2021B1515020092), Shenzhen Bay Laboratory Fund (grant number: SZBL2020090501014), and Shenzhen Science and Technology Innovation Commission (grant number: WDZC20200821141349001, KCXFZ20200201101050887, RCYX20200714114736146, KCXFZ20201221173207022).
Author contributions
S.M. conceived and supervised the work. Y.F. wrote the manuscript and generated the figures. Y.F., H.Z., and J.T. performed the experiments. Y.F., H.Z., J.T., and S.M. edited the final manuscript. X.L. and R.T. revised and advised on this work.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xpro.2023.102346.
Data and code availability
This study did not generate any datasets or codes.
References
- 1.Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., Ming G.L. Generation of human brain region–specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu X., Huang L., Zheng Y., Song Y., Xu Q., Wang J., Si K., Duan S., Gong W. Ultrafast optical clearing method for three-dimensional imaging with cellular resolution. Proc. Natl. Acad. Sci. USA. 2019;116:11480–11489. doi: 10.1073/pnas.1819583116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any datasets or codes.