Summary
The process of metabolic liver zonation is spontaneously established by assigning distributed tasks to hepatocytes along the porto-central blood flow. Hepatocytes fulfil critical metabolic functions, while also maintaining hepatocyte mass by replication when needed. Recent technological advances have enabled us to fine-tune our understanding of hepatocyte identity during homeostasis and regeneration. Subsets of hepatocytes have been identified to be more regenerative and some have even been proposed to function like stem cells, challenging the long-standing view that all hepatocytes are similarly capable of regeneration. The latest data show that hepatocyte renewal during homeostasis and regeneration after liver injury is not limited to rare hepatocytes; however, hepatocytes are not exactly the same. Herein, we review the known differences that give individual hepatocytes distinct identities, recent findings demonstrating how these distinct identities correspond to differences in hepatocyte regenerative capacity, and how the plasticity of hepatocyte identity allows for division of labour among hepatocytes. We further discuss how these distinct hepatocyte identities may play a role during liver disease.
Keywords: Liver regeneration, Hepatocyte identity, Hepatocyte plasticity, Ploidy, Liver stem cells, Metabolic liver zonation
Key points.
-
•
Distinct metabolic hepatocyte identity along the porto-central blood flow is essential for metabolism.
-
•
Hepatocytes can be defined by their location within the lobule, gene expression, cell size and ploidy.
-
•
These factors have been associated with differences in proliferation and contribution to the hepatocyte pool, as well as with assigning metabolic functions.
-
•
During homeostasis, zone 2 hepatocytes rather than a defined subpopulation seem to have increased proliferative potential.
-
•
Hepatocytes across all lobular zones exhibit high plasticity and support liver regeneration on demand.
-
•
An inverse correlation between hepatocyte proliferation and metabolism suggests division of labour between these two essential processes.
-
•
Changes in hepatocyte identity are associated with severe liver disease.
Introduction
As the body’s largest internal organ, the liver is responsible for many vital functions. It metabolises nutrients and xenobiotics and is also an important factory and recycling station.1 The liver controls carbohydrate metabolism, storage (glycogenesis) and release of glucose (glycogenolysis) to maintain homoeostatic glucose levels in the blood. During starvation, the liver can also synthesise glucose from amino acids (AAs), lactate or glycerol (gluconeogenesis).2,3 The liver contributes to lipid metabolism via the production of triglycerides (TGs) and lipoproteins, and via cholesterol synthesis. Bile production and excretion supports fat digestion and vitamin absorption.4 The liver is further responsible for protein synthesis and degradation. Most plasma proteins are produced and recycled in hepatocytes. AAs from recycled proteins are used to produce new plasma proteins or are converted into glucose during gluconeogenesis. In addition, AAs are provided as building blocks for extrahepatic protein synthesis in other tissues, highlighting the importance of the liver in supporting systemic protein homeostasis.5,6 The hepatic recycling station also handles the breakdown of red blood cells and the processing of haemoglobin, including the storage and use of its iron.7 Moreover, the liver breaks down insulin and other hormones, and it performs drug metabolism via cytochrome P450 (CYP) enzymes to modify xenobiotics and enable their excretion via bile or urine. Finally, the liver supports immune defence during sepsis.1,3,4,8 Given these, and many more, vital functions, it is obvious why impaired liver function is associated with so many diseases.
In an industrial factory, assigning the correct task to different workers is critical for success. Likewise, hepatocytes need to be instructed to perform diverse metabolic tasks in the right place and at the right time.9 Given the extended hepatocyte lifespan (200–300 days in rodents10 and 3 years in human11) the liver requires much lower proliferation rates than high-turnover organs that are maintained by constantly cycling tissue stem cell compartments. Yet, lost hepatocytes must be replaced during homeostatic turnover or following injury to maintain a full work force. This requires efficient regenerative mechanisms that enable hepatocytes to proliferate when needed, while maintaining full metabolic capacity of the liver. Therefore, the regenerative response has to be tailored to the site and severity of the injury.12 In recent years, several putative liver stem cells with increased regenerative potential during homeostasis and regeneration have been proposed, but conflicting findings challenge the existence of a bona fide liver stem cell.13 Instead, a modular regenerative response by hepatocytes repairs different types of local or pan-zonal injuries, and the enormous regenerative potential and plasticity of hepatocytes is sufficient to restore liver mass and function. The exception is when hepatocytes are senescent and fail to regenerate, and alternative regenerative mechanisms are then required to refuel the hepatocyte pool.14,15
New hepatocytes must be assigned to the correct metabolic task. Impaired metabolic function is associated with severe hepatic and systemic diseases (reviewed in9). For example, in non-alcoholic steatohepatitis (NASH) impaired lipid metabolism results in TG accumulation and lipotoxicity-induced inflammation and fibrosis. In alcohol-related liver disease, loss of metabolic function leads to toxic ammonia accumulation, causing neurological damage. Another example is small-for-size syndrome (SFSS), a condition where small graft transplantation or large resection leave insufficient amounts of functional hepatocytes to handle the metabolic demand. Many end-stage liver diseases are characterised by insufficient functional hepatocytes or maladaptive repair processes that can have fatal consequences.9
Due to the lack of sufficient liver grafts for transplantation, currently the only cure for end-stage liver disease, regenerative therapies are urgently needed. Current therapeutic concepts focus on promoting hepatocyte proliferation to restore functional liver mass.16 An equally important factor that has received less attention is the spatiotemporal restoration of hepatocyte identity, which is needed to restore liver function.
Here, we summarise the functional, morphological, and anatomical diversity of hepatocytes, and the mechanisms instructing their identity. We highlight the relevance of metabolic zonation and adaptive regenerative principles, as well as the disease-associated consequences of their impairment.
Defining the differences among hepatocytes
The anatomical structure of the liver and its repetitive functional subunits are closely aligned with its physiological tasks. Expression of different metabolic genes in different hepatocytes is tightly coordinated with the liver’s spatiotemporal functional needs. In addition, cell size and ploidy further differentiate hepatocytes from each other.
Lobular liver architecture
The liver is divided into distinct lobes, which each consist of repetitive honeycomb-like substructures termed lobules. Highly oxygenated blood enters the liver via the hepatic artery and the portal vein. The portal vein receives blood from the spleen, pancreas, and gastrointestinal tract, thus making the liver the first-pass organ for most nutrients and xenobiotics. Hepatocytes are the main metabolic workers in the liver, supported by non-parenchymal cells, including phagocytic Kupffer cells, vitamin A-storing and extracellular matrix-depositing hepatic stellate cells, and bile duct-forming biliary epithelial cells (BECs, also termed cholangiocytes). Hepatocytes are lined by fenestrated liver sinusoidal endothelial cells, which allow nutrients, metabolic products, and other molecules to permeate between hepatocytes and the blood. Lymphocytes carry out immune surveillance in the sinusoidal space. The blood leaves the lobules via central veins, which merge into the hepatic veins, and exits the liver to join the inferior vena cava. Canaliculi collect the bile produced by hepatocytes and transport it in the opposite direction to the porto-central blood flow towards the intrahepatic bile ducts, which merge into the extrahepatic bile duct. Meals trigger the release of bile from temporary storage in the gall bladder into the duodenum, to support digestion.1,17 The lobular structure defines the factory’s assembly line, made up of hepatocytes, alongside the porto-central hepatic blood flow.
Functional and anatomical hepatocyte diversity
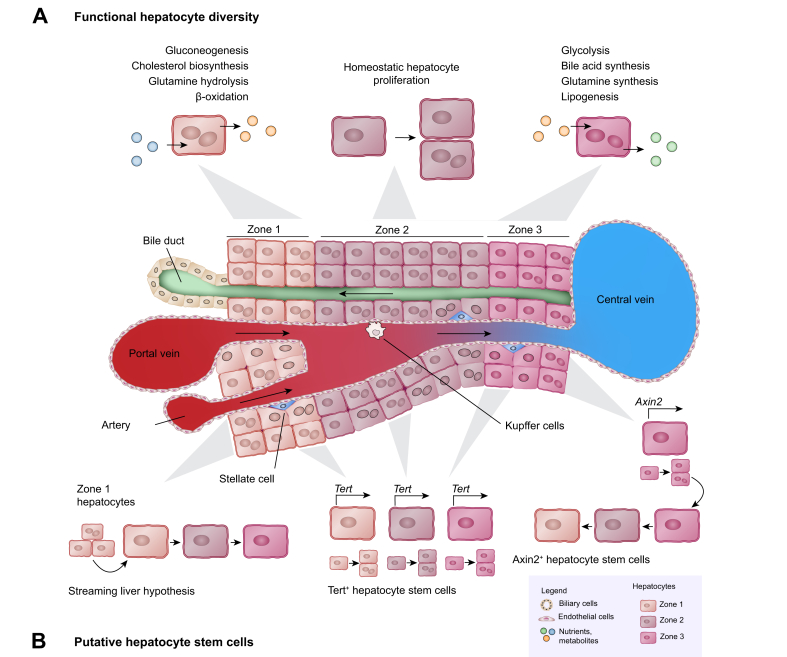
In the classic description of the liver lobule (Fig. 1A), hepatocytes reside in three zones defined according to their location along the porto-central axis, and carry out different metabolic functions.18 Zone 1 (periportal) hepatocytes carry out functions such as oxidative energy metabolism, beta-oxidation, AA catabolism, ureagenesis from AAs, gluconeogenesis, as well as bile acid and bilirubin excretion; while glycolysis, liponeogenesis, ureagenesis from ammonia, and biotransformation are primarily performed by zone 3 (pericentral) hepatocytes. This setup creates a functional “division of labour” so that different hepatocytes specialise in different tasks. Additionally, an even finer understanding of the many differences in gene expression among hepatocytes based on their position in the lobule has recently been revealed through experiments involving spatial sorting of mouse liver cells,19 single-cell RNA sequencing combined with virtual positioning of the captured hepatocytes,20 and spatial transcriptomics.21,22 Human hepatocytes and other liver cell types have also been examined by single-cell RNA sequencing and spatial proteomics, revealing their zonal heterogeneity.23,24 Zonal reprogramming observed by spatial transcriptomics22 and molecular cartography25 combined with perturbation of WNT/β-catenin signalling further indicate that metabolic zonation is dynamic and requires active activation and repression of metabolic gene expression. More work is needed to dissect the interplay of gene regulatory networks driving this complex process.
Fig. 1.
Homeostasis.
(A) Schematic showing a section of the liver lobule from the portal triad to the central vein, with the different cell types as listed in the legend. Hepatocytes residing in the three zones along this axis retain diverse metabolic functions (examples listed). The prevailing view is that hepatocytes proliferate similarly. (B) A recent additional observation was that zone 2 hepatocytes with the least metabolic functions proliferate more during homeostasis. However, zone 1 hepatocytes possibly derived from bile ducts, hepatocytes expressing high levels of telomerase, and pericentral hepatocytes expressing Axin2 have also been reported to drive hepatocyte renewal during homeostasis. Schematic showing the subpopulations of hepatocytes reported to fuel hepatocyte regeneration during homeostasis.
Different metabolic tasks are assigned to hepatocytes based on the spatial needs and the local environment along the hepatic assembly line. In addition, temporal entrainment of metabolic gene expression adds another dimension to the complexity and diversity of hepatocyte identities.26 Spatial zonation principles (reviewed in detail by Ben-Moshe et al.27) include production-line patterns to enable complementary tasks (e.g. neutral bile acid biosynthesis cascade), or spatial segregation of opposing metabolic tasks (e.g. glucose metabolism). In addition, temporal control of metabolic gene expression aligns the availability of metabolic enzymes with their need during diurnal fasting and feeding cycles. Defined spatiotemporal compartmentalisation of metabolic processes, often by strict regulation of key rate-limiting enzymes, enables the liver to assign tasks to the right lobular zone and at the time when they are needed.26 Although hepatocytes may look alike and appear similar, numerous functional and expression profiling studies have revealed their functional and anatomical diversity; this diversity of hepatocytes is a critical prerequisite for maintaining liver function (reviewed in9).
Hepatocyte morphology and ploidy
Morphologically, hepatocytes are cuboidal epithelial cells that vary by size and ploidy. In uninjured adult mouse livers, hepatocyte volume ranges from 3,126 μm3 to 10,606 μm3.28 Normal ploidies observed among adult mouse hepatocytes are diploid (2n), tetraploid (4n) and octaploid (8n); and 4n and 8n hepatocytes often have two nuclei (bi-nucleated).28 Higher ploidies (>8n) appear in injured livers, likely due to alterations to cell cycles caused by cellular damage.29 Curiously, at birth, all hepatocytes are diploid, with accumulation of polyploid hepatocytes, mostly 4n and some 8n, occurring during post-natal development (further reviewed in[29], [30], [31]), until polyploid hepatocytes account for ∼90% of hepatocytes in rodent livers and 30% in human livers.[32], [33], [34], [35] In uninjured mouse livers, there is some correlation between hepatocyte size and ploidy and their location within the lobule. 2n mononuclear diploid and 2x2n binuclear tetraploid hepatocytes are more frequent in zone 1 and zone 3, while 2x4n binuclear octaploid hepatocytes are more frequent in zone 2, with 4n mononuclear hepatocytes distributed evenly across the lobule.28 Additionally, when polyploid hepatocytes divide they can give rise to aneuploid hepatocytes, causing genetic variation among hepatocytes in mice as well as humans.32,36 The functional role of ploidy is further reviewed in31 and,37 with some evidence that diploid hepatocytes may be more proliferative, polyploid hepatocytes may be protected against transformation into cancer, while genetic variation due to aneuploidy could allow subsets of hepatocytes to adapt and survive chronic liver injury.11,[37], [38], [39] Recently, a single-nucleus RNA sequencing study found differences between 2n and 4n hepatocytes with regards to their expression of genes associated with metabolism, which shows crosstalk with the hepatocyte’s position within the lobule.40 Thus, although hepatocytes appear similar, their cellular properties and gene expression vary. These differences in gene expression, metabolic function, cell size, and ploidy can be used to specify hepatocyte identity.
Factors that regulate hepatocyte identity during homeostasis
Hepatocyte identity and function are mostly dictated by their position in the liver lobule (Fig. 1A). The porto-central oxygen gradient (pO2: 60–65 mmHg [zone 1]) to 30–35 mmHg [zone 3]), associated hypoxia-inducible factors (HIFs) and interplay with other regulatory mechanisms direct energy-demanding tasks to periportal (high oxygen) hepatocytes and less demanding tasks to pericentral (low oxygen) hepatocytes.41 For example, HIF1α promotes glycolysis and HIF2α suppresses gluconeogenesis in pericentral hepatocytes.41 WNT/β-catenin signalling controlled by the RSPO-LGR4/5-ZNRF3/RNF43 module is a major determinant of hepatocyte identity and metabolic function in the liver.22,[42], [43], [44], [45], [46], [47], [48] WNT2, WNT9B and RSPO3 ligands secreted from central vein endothelial cells and adjacent liver sinusoidal endothelial cells,25,43,45,46,49 as well as distinct expression patterns of regulatory elements of the WNT/β-catenin pathway along the porto-central axis,22,42,44 result in a centro-portal gradient of WNT/β-catenin signalling. Due to high levels of WNT/β-catenin signalling, pericentral hepatocytes express the β-catenin target gene Lgr5, which confers tissue stem cell identity in the gut, but not in the liver.44,50 While activation of WNT/β-catenin signalling reprogrammed periportal into pericentral hepatocytes, pathway blockage induced widespread periportal metabolic enzyme expression.22,42,44,45,51 This indirect regulation of hepatocyte identity is mediated by the competition of HNF4α and β-catenin for binding to TCF. Without nuclear β-catenin, the HNF4α/TCF complex promotes periportal gene expression, whereas nuclear translocation of β-catenin forms a β-catenin/TCF complex that favours pericentral gene expression.52 In addition, periportal Hedgehog signalling contributes to defining hepatocyte identity and metabolic zonation by partially counteracting WNT/β-catenin signalling and regulating glucose homeostasis.53 Higher periportal availability of hormones, such as insulin and glucagon, play important opposing roles in spatial metabolic zonation and interact with the WNT/β-catenin pathway.54 Although a porto-central Yes-associated protein (YAP) expression gradient has been reported,55 the role of YAP/HIPPO signalling in metabolic zonation is unclear. A role for YAP in regulating glutamine synthetase expression56 was questioned by two other studies.57,58 However, YAP promotes SRY-box transcription factor 9 (SOX9) expression in hepatocytes59 and may confer the hybrid hepatocyte-biliary identity of periportal SOX9+ hepatocytes.60 Studying the potential interplay between YAP/HIPPO and WNT/β-catenin signalling may shed further light on the regulation of hepatocyte identity in metabolic liver zonation.
In addition to these signalling pathways, hepatocyte identity is controlled by epigenetic mechanisms. This includes chromatin remodelling, DNA methylation, post-translational modifications of histones, and non-coding RNAs, however, epigenetic regulation of zonal hepatocyte identity and regeneration are not well understood.61 For instance, ARID1A, a component of the SWI/SNF (SWItch/sucrose non-fermentable) chromatin remodelling complex, has been shown to suppress hepatocyte proliferation and regeneration by limiting access of transcription factors to their target genes. These include C/EBPα, HNF4α, and E2F, all of which are known to be involved in regulating zonated gene expression.62 Furthermore, the human liver exhibits differential DNA methylation between the periportal and pericentral zone at binding sites of around 50 transcription factors, which may lead to distinct binding preferences along the centro-portal axis.63 As reviewed elsewhere,64,65 several microRNAs are involved in controlling hepatocyte regenerative identity, for example, by maintaining the hepatocyte quiescent state or promoting cell cycle entry during liver regeneration.
In addition to spatial compartmentalisation, temporal regulation of hepatocyte metabolic gene expression is important to align availability of metabolic enzymes to fasting and feeding cycles. Nutrient availability, as well as interactions between diverse environmental cues and the circadian clock, are the pacemakers responsible for daily fluctuations in virtually all aspects of hepatic function. For example, gluconeogenesis-regulating genes are induced in periportal hepatocytes following prolonged starvation at the end of the resting phase.9,26 Further, the expression of WNT ligands is influenced by diurnal mRNA fluctuations, suggesting that WNT signalling may fluctuate throughout the day.26 Interestingly, core clock genes undergo rhythmic H3 histone acetylation, which can be explained by the rhythmic interaction of p300 with CLOCK.66 Thus, histone acetylation also controls important metabolic functions of the liver in a temporal and spatial manner.
Mechanisms regulating hepatocyte ploidy include insulin signalling through the P13K-AKT pathway, which is influenced by weaning67 and induces formation of binucleate tetraploid hepatocytes during postnatal liver growth.68,69 E2F7 and E2F8 are cell cycle factors expressed at low levels in adult livers but at high levels during the first 7 weeks after birth in mice; they are also responsible for postnatal polyploidisation.38,70 Additionally, miR-122, one of the most abundant microRNAs in the liver, regulates physiological hepatocyte polyploidisation during postnatal growth.71 In injured livers, oxidative stress through induction of ATR/p53/p21 signalling has been reported to induce hepatocyte polyploidy72 (further reviewed in31).
Maintenance of a functional hepatocyte pool during homeostasis
The body maintains a stable liver-to-body weight ratio by tightly controlling mechanisms promoting hepatocyte proliferation and size. This is necessary to adapt the size of the liver and number of hepatocytes to the body`s needs, and the “hepatostat” is set such that the liver regenerates back to 100% of what is required for homeostasis, unlike other solid organs with more limited regenerative potential.73,74 When hepatocytes are lost during homeostatic turnover, they are replaced by proliferation of other hepatocytes. In the absence of injury, hepatocytes in zone 2 have a proliferation rate of 0.042 per week.75 Since the same pathways that induce and maintain metabolic function also control hepatocyte proliferation, restricting unnecessary proliferation while maintaining the metabolic identity of hepatocytes in the different zones is an important balancing act for the liver.22,76 Whether specialised liver stem cells maintain hepatocyte numbers during homeostasis remains controversial.13,77
Controversy around the origin of new hepatocytes during homeostasis
The “streaming liver” model proposed several decades ago hypothesised that hepatocytes are born in the periportal zone possibly from bipotent progenitor cells in the bile ducts, and stream towards the pericentral zone where they die78 (Fig. 1B). This idea followed from observation of “oval cells”,79 putative bipotent progenitor cells thought to originate from bile ducts, that were reported to give rise to new hepatocytes in 2-acetylaminofluorene-damaged rat livers.80,81 While the results of one study using Sox9-CreERT2 lineage tracing in mice supported the streaming liver hypothesis,82 another study using a multicolour fluorescent reporter with Sox9-CreERT2 did not.83 Alternative methods based on tracing of periportal hepatocytes showed that periportal hepatocytes do not expand towards zone 360 under homeostatic conditions, and may reduce over time.84
Recent attention has been devoted to whether subpopulations of hepatocytes are special and function like stem cells. Zone 3 AXIN2+ hepatocytes which experience high WNT signals were labelled by an Axin2-CreERT2 transgene inserted at the endogenous Axin2 locus. These cells were found to self-renew and give rise to progeny hepatocytes that move into zones 2 and 149 (Fig. 1B). However, a subsequent lineage-tracing study using an Axin2-CreERT2 in a BAC transgene did not support this.85 Instead, EdU incorporation suggested that knocking CreERT2 into the endogenous Axin2 locus increases proliferation of hepatocytes due to heterozygous Axin2 deletion.86 Finally, independent validation using the original Axin2-CreERT2 mouse line in a new study did not replicate expansion of the labelled hepatocytes into other zones.87 Alternative zone 3 labelling with other gene promoters also did not show that zone 3 hepatocytes give rise to hepatocytes in other zones during homeostasis.44,50,86
To understand the homeostatic expansion potential of different hepatocytes, a method using AAV8-TBG-Cre activating a multicolour fluorescent reporter was developed to label and trace random samples of hepatocytes throughout the lobule in mice. 3D imaging of labelled cells and their progeny showed that hepatocyte renewal is not driven by zone 3, no hepatocytes formed particularly large clones, and hepatocyte proliferation was distributed throughout the lobule with larger hepatocyte clones more frequently found in zone 2.88 A novel method using a “ProTracer” mouse transgenic line that records cell division demonstrated that there is more proliferation in zone 2 compared to zones 1 and 3 during homeostasis.75 A comprehensive study using several new mouse transgenic lines to label different portions of the lobule also demonstrated that zone 2 hepatocytes expand during homeostasis, with a corresponding contraction of the areas originally occupied by zone 1 or 3 hepatocytes.86
Another putative hepatocyte stem cell population are the high telomerase-expressing hepatocytes. Labelled by Tert-CreERT2, they made up only 2.8% of all hepatocytes, were distributed throughout the lobule, and were observed to give rise to 30% of hepatocytes in mouse livers over 1 year89 (Fig. 1B). However, a random sampling of hepatocytes labelled with AAV8-TBG-Cre did not support homeostatic hepatocyte renewal driven by rare hepatocytes.88 Additionally, sparsely labelled hepatocytes, marked by novel Cre-expressing transgenes from the Hamp2 and Mup3 promoters, also expanded during homeostasis, demonstrating that proliferation is not limited to the high telomerase-expressing population.86
Although there is controversy and unresolved discrepancies between studies (summarised in Table 1), the current consensus is that there is no hepatocyte stem cell uniquely responsible for homeostatic hepatocyte renewal. Nevertheless, hepatocytes do not proliferate equally over time, as results obtained via three independent experimental approaches have led to the same conclusion, namely that zone 2 hepatocytes proliferate more than hepatocytes from the other zones.75,86,88
Table 1.
Highlights of recent hepatocyte lineage-tracing results in the mouse liver.
| Tracing method | Hepatocytes labelled at start of tracing | Hepatocytes labelled at end of tracing period in homeostasis or regeneration | Ref. |
|---|---|---|---|
| Sox9-CreERT2 | Zone 1 (SOX9+, next to bile ducts) | Regeneration: Large clones spanning zones 1-3 (12x CCl4) | 60 |
| Mfsd2a-CreERT2 | Zone 1, ∼40% of parenchyma |
Homeostasis: Reduction in by half to ∼20% of lobule (6- to 20-week-old mice, no change 20 to 36 week) Regeneration: Expansion from zones 1 to 2 (PHx and 1x CCl4) and to zone 3 covering nearly all of lobule (10x CCl4) |
84 |
| Gls2-CreERT2 | Zone 1 + ∼1/4 zone 2, ∼60% of lobule |
Homeostasis: Zone 1, ∼40% of lobule (12 months) Regeneration: Zones 1-3, ∼99% of lobule (12x CCl4) |
86 |
| Agr1.2-CreERT2 | Zone 1 + zone 2, 78% of lobule |
Homeostasis: Zone 1 + zone 2 + some of zone 3, 94% of lobule (12 months) Regeneration: Zones 1-3, ∼99% of lobule (12x CCl4) |
86 |
| Agr1.1-CreERT2 | Zones 1-3 except cells next to CV, 94% of lobule | Homeostasis: Zone 1 + zone 2 + zone 3 except cells next to CV, 96% of lobule (12 months) | 86 |
| Cyp1a2-CreERT2 | ∼2/3 of zone 2 + zone 3, ∼60% of lobule |
Homeostasis: Some of zone 1 + zone 2 + zone 3, ∼80% of lobule (12 months) Regeneration: Zones 1-3, ∼67% of lobule (6 weeks DDC) |
86 |
| Oat-CreERT2 | ∼1/4 of zone 2 + zone 3, 46% of lobule | Homeostasis: Some of zone 1 + zone 2 + zone 3; 54% of lobule labelled (6 months) | 86 |
| Axin2-CreERT2 (endogenous locus) | Zone 3 (AXIN2+ subset) | Homeostasis: Large clones spanning zones 3 to 2, sometimes to zone 1 (1 year) | 49 |
| Axin2-CreERT2 (BAC transgenic) | Zone 3 (AXIN2+ subset) |
Homeostasis: Zone 3 only (10 months) Regeneration: Zone 3 + modest expansion to zone 2 (PHx). Upregulation of Axin2 and labelling of regenerating hepatocytes across zones in zonal injury model |
85 |
| Lgr5-CreERT2 (endogenous locus) | Zone 3 (sparse LGR5+ subset) |
Homeostasis: Zone 3 only (18 months) Regeneration: Similar Ki67 positivity as zone 2 hepatocytes (48 h post PHx) |
44 |
| Lgr5-rtTA-IRES-GFP; TetO-Cre | Zone 3 (LGR5+ subset) | Homeostasis: Zone 3 only (18 months) | 50 |
| GS-CreERT2 | Zone 3 (GS+, next to CV), 9.4% of lobule |
Homeostasis: Zone 3 next to CV, 9.9% of lobule (12 months) Regeneration: Zone 3, ∼15% of parenchyma (6 weeks DDC) |
86 |
| Axin2-CreERT2 (endogenous locus) | Zone 3 and neighbouring zone 2, and sparsely in zone 1 (7 days after tamoxifen) | Homeostasis: Similar pattern between 7 days and 365 days tracing | 87 |
| Lgr4-CreERT2 (endogenous locus) | Sparse hepatocytes throughout zones 1-3 | Homeostasis: Throughout lobule with no reported zonal dominance (10 months) | 44 |
| Tert-CreERT2 (knocked into exon 1) | Sparse hepatocytes in zones 1-3 expressing high Tert, 2.8% of parenchyma |
Homeostasis: 30% of parenchyma in 1 year Regeneration: 38% of parenchyma (1-month DDC) |
89 |
| Tert-CreERT2 (knocked into 3’UTR) | Sparse hepatocytes throughout lobule, more in zones 2 and 3 than 1 | Homeostasis: Hepatocytes distributed in all zones, primarily in zone 2 in (7 and 21 days) | 86 |
| AAV8-TBG-Cre | Individual hepatocyte clones (differentiated with multicolour R26-Rainbow reporter), zones 1-3 |
Homeostasis: Broadly distributed hepatocyte division in all zones, more proliferation in zone 2, but no particularly large clones (13 months) Regeneration: Hepatocyte division detected in all uninjured zones (1 to 12x CCl4 and AA) |
88 |
| Hamp2-CreERT2 | Sparse hepatocytes throughout lobule, more in zone 2 than 1 and 3, ∼10% of parenchyma |
Homeostasis: Hepatocytes throughout lobule, more in zone 2 than 1 and 3, ∼25% of parenchyma (12 months) Regeneration: Large hepatocyte clusters primarily in zone 2, ∼17% of parenchyma (6 weeks DDC). Large hepatocyte clusters primarily in zone 2, 13% (12x CCl4). |
86 |
| Mup3-CreERT2 | Very sparse hepatocytes throughout lobule, more in zones 1 and 2 than zone 3, 0.06% of parenchyma | Homeostasis: Very sparse hepatocytes throughout lobule, 0.85% of parenchyma (6 months) Regeneration: Hepatocyte clusters in zones 2 and 3, ∼2.8% of parenchyma (12x CCl4). | 86 |
|
Ki67-Cre-rox-ERT2-rox Activated in hepatocytes though Alb-DreERT2 or AAV8-TBG-Dre |
Proliferating hepatocytes throughout lobule |
Homeostasis: Hepatocytes in zones 1-3, but most frequently in zone 2, less in zone 1 and very little in zone 3 Regeneration: Hepatocytes in zones 1-3, with earlier proliferation zones 1 and 2 (PHx) |
75 |
AAV8-TBG-Cre and AAV8-TBG-Dre are adeno-associated viral vectors expressing Cre or Dre recombinase, respectively. All other lineage-tracing methods employ transgenic mouse lines expressing Cre from the indicated promoters. “∼X%” are numbers estimated from graphs in figures of the respective study when numbers were not provided.
AA, allyl alcohol; CCl4, carbon tetrachloride, DDC, 3,5-diethoxycarbonyl-1,4-dihydrocollidine diet, PHx, 2/3 partial hepatectomy.
Balancing metabolic function and proliferation during homeostasis
To maintain both metabolic function and hepatocyte numbers in the liver, a further division of labour among hepatocytes is required to balance metabolism with proliferation. Spatial transcriptomics profiling identified proliferative hepatocytes mostly in zone 2,22 consistent with the lineage-tracing studies described above.75,86,88 Interestingly, these proliferative hepatocytes showed significantly reduced expression of metabolic genes.22 Given the generally lower expression of metabolic genes in midzonal hepatocytes, along with higher expression of periportal and pericentral metabolic genes in their respective zones, a hypothesis has been proposed that there is a general division of labour between the hepatocytes driving metabolism, and the hepatocytes responsible for maintaining the hepatocyte pool.9 However, hepatocytes in all lobular zones have accessible chromatin in the genes regulating proliferation, as well as periportal and pericentral metabolism.22 This epigenetic configuration may enable hepatocytes to quickly adapt their metabolic identity and proliferative status on demand. For example, hepatocyte metabolic identity can be modified, as illustrated by fate tracing combined with WNT/β-catenin pathway activation. ZNRF3/RNF43 deletion reprogrammed periportal hepatocytes into pericentral hepatocytes. A curious and still unresolved finding was that some hepatocytes with ZNRF3/RNF43 deletion increased metabolic gene expression, whereas others proliferated.22 Collectively, these studies suggest an inverse correlation between metabolic activity and proliferation in hepatocytes.
The RSPO-LGR4/5-ZNRF3/RNF43 module controlling WNT/β-catenin signalling is a growth and size rheostat in the liver. RSPO injections reversibly increased liver size by inducing hepatocyte proliferation, whereas WNT/β-catenin signalling blockade resulted in smaller livers.44,90,91 ZNRF3 and RNF43 cooperate to restrict WNT/β-catenin signalling during homeostasis to prevent uncontrolled proliferation and tumour formation, while enabling metabolic zonation.22,92 While activated YAP signalling reversibly increased liver size,93 its deletion did not affect liver size during homeostasis.57,58,94 Hepatocyte proliferation in zone 2 hepatocytes is mediated by the IGFBP2-mTOR-CCND1 axis.86 Whether mitogenic pathways, such as growth factor and nuclear hormone receptor signalling,74 contribute to zonal hepatocyte identity requires further assessment. Hepatocyte proliferation and the cell cycle are also regulated by DNA methylation.95 For instance, deletion of Dnmt1 in postnatal hepatocytes using Alb-Cre transgenic mice caused global hypomethylation, enhanced the DNA damage response, and induced a senescent state. This was accompanied by liver fibrosis and inflammation, and a reduced hepatocyte proliferative capacity in response to injury. Intriguingly, deletion of Dnmt1 was accompanied by the loss of the perivenous enzyme cytochrome CYP2E1, which is responsible for the bioactivation of carbon tetrachloride (CCl4), thus causing unresponsiveness to this drug.96 Thus, DNA methylation may play a role in regulating hepatocyte identity by constraining hepatocyte proliferation and metabolic gene expression.
Hepatocyte regenerative potential following injury
Liver function depends on the rapid restoration of lost hepatocytes, which requires the regenerative response to be aligned with the site and severity of injury. Diverse local or pan-zonal injuries require different regenerative strategies and spatiotemporal control of repair processes in different zones. In most cases, hepatocytes proliferate to replace lost comrades. Niche signals guide the development of hepatocyte identity to restore metabolic zonation. In addition, hepatocytes and BECs utilise their enormous plasticity to transdifferentiate into one another in special severe injury settings. Directing the right regenerative response to the type of injury is key to minimising the disruption of vital metabolic processes.
Hepatocyte plasticity
One of the ways hepatocytes adapt to the needs of the liver is through plasticity, although not all hepatocytes are the same in this regard. Hepatocytes display remarkable plasticity and under some conditions can transdifferentiate into BECs, epithelial cells of the bile ducts with completely different morphology, gene expression and function59,[97], [98], [99] (Fig. 2A). Curiously, not all hepatocytes transdifferentiate with the same capacity; zone 1 periportal hepatocytes transdifferentiated readily to BECs, but even with activation of NOTCH signalling, which is a strong inducer of biliary fate in hepatocytes, zone 3 hepatocytes did not transdifferentiate.99 While hepatocyte-to-biliary plasticity has been hypothesised to be a transient response (also termed ductular reaction) to injury involving bile ducts,100 a persistent functional biliary tree formed from transdifferentiated hepatocytes in a mouse model developmentally lacking intrahepatic bile ducts. This showed that hepatocyte-to-BEC transdifferentiation can be permanent.98 Which hepatocytes are capable of forming functional bile ducts remains to be further characterised.
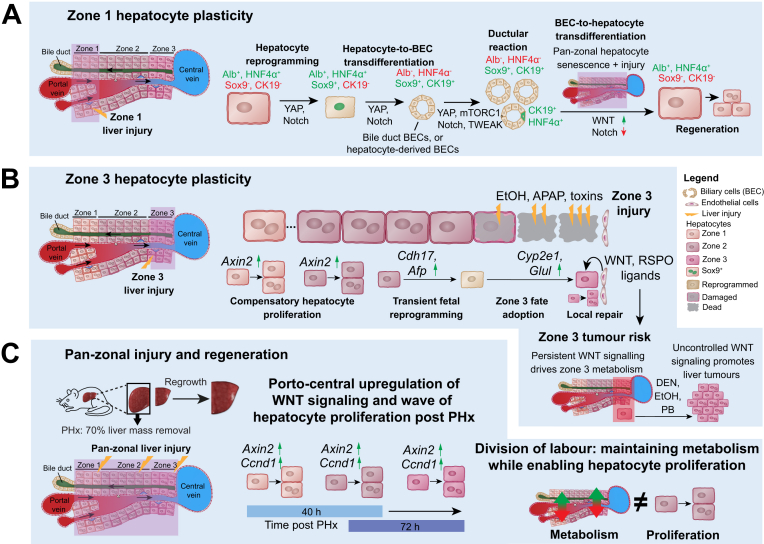
Fig. 2.
Zonal regeneration principles.
(A) Zone 1 hepatocytes show plasticity and can undergo reprogramming to turn on genes normally expressed in BECs, while losing expression of hepatocyte genes. Sometimes hepatocytes can fully transdifferentiate into BECs. BECs are also reported to give rise to hepatocytes when existing hepatocytes in all zones are senescent. (B) Zone 2 and 3 hepatocytes also have plasticity, but different from zone 1. After injury to zone 3, zone 1 and 2 hepatocytes proliferate to repair the damage, and zone 2 hepatocytes are reported to undergo transient fetal reprogramming to adopt a zone 3 fate in response to WNT signalling from zone 3 endothelial cells. Whereas persistent WNT signalling drives zone 3 metabolism, uncontrolled WNT signalling promotes liver tumours, highlighting tumour risk from Zone 3 hepatocytes. (C) However, porto-central upregulation of WNT signalling (indicated by Axin2 upregulation; as well as upregulation of many other factors not shown here74) also drives a wave of hepatocyte proliferation following partial hepatectomy. Additionally, hepatocytes are observed to downregulate metabolism as they proliferate, enabling a division of labour for some hepatocytes to proliferate as others maintain metabolic function. DEN, diethylnitrosamine; BECs, biliary epithelial cells; PHx, partial hepatectomy.
Not all hepatocytes that acquire a biliary gene signature undergo full transdifferentiation into BECs. Following diverse injuries, often involving cholestasis, YAP signalling also drives hepatocyte reprogramming via SOX9 induction. These reprogrammed zone 1 hepatocytes exhibited reduced metabolic function, upregulated tissue stem cell markers and increased proliferative capacity, contributing to regeneration.59,60,100,101 Arid1a supports hepatocyte reprogramming by facilitating binding of YAP to genes associated with a liver progenitor cell state.102 However, the upstream regulatory signals promoting YAP signalling in zone 1 remain elusive. A recent study showed that Kupffer cell-derived IL6 induced SOX9 in hepatocytes via STAT3 signalling,103 indicating that several pathways collectively induce reprogramming via SOX9 induction.
Hepatocyte reprogramming has also been observed after zone 3 injury (Fig. 2B). Following ablation of pericentral hepatocytes, neighbouring zone 2 hepatocytes upregulated WNT/β-catenin signalling and acquired zone 3 identity to restore metabolic zonation.85,104,105 This fate adoption is likely induced by locally confined secretion of WNT and RSPO ligands from the central vein endothelium and neighbouring sinusoidal endothelial cells.25,43 Transient fetal reprogramming of peri-injury hepatocytes, which subsequently adopted a zone 3 fate and contributed to local repair, suggests that hepatocytes in this zone have high plasticity104 (Fig. 2B). During pan-zonal regeneration induced by partial hepatectomy, hepatocytes across the liver underwent transient fetal reprogramming to acquire proliferative potential.106,107 Hepatocytes also underwent an EMT (epithelial-mesenchymal transition)-like response to enable them to grow in a TGFβ-enriched microenvironment.108 Injury-dependent upregulation of AXIN2 in hepatocytes across the lobule suggests that hepatocytes can acquire a regenerative signature on demand.85 In response to cholestatic injury, BECs upregulate WNT ligands.57,94,109,110 To which extent this mediates biliary repair or induction of BEC markers in hepatocytes,110,111 hepatocyte-mediated regeneration57 or even BEC-to-hepatocyte transdifferentiation,112 requires further investigation.
When hepatocyte-mediated regeneration is impaired, following chronic injury and induction of hepatocyte senescence, BECs can transdifferentiate into zone 1 hepatocytes which subsequentially proliferate and repopulate other lobular zones[112], [113], [114], [115], [116], [117], [118] (Fig. 2A), and the new hepatocytes are reported to persist long-term and through chronic injury.113,119 Several markers have been proposed to mark BEC subsets with liver progenitor cell (LPC) potential, including CD24/CD133,119 FOXL1[120], [121], [122] and TWEAK/FN14.123 It was further suggested that BEC-to-hepatocyte transdifferentiation occurs through a CK19+/HNF4α+ bi-phenotypic state113 and that Notch and WNT/β-catenin signalling,124,125 as well as TET1/YAP signalling126 are involved in LPC fate decisions. However, definitive dual-recombinase lineage-tracing studies labelling BEC subsets with LPC potential have been missing. Only recently, such studies identified bipotential CK19+/HNF4α+ transient LPCs (TLPCs) that emerge from BECs following severe liver injury and transdifferentiate into hepatocytes or differentiate back into BECs. Mechanistically, NOTCH and WNT/β-catenin signalling orchestrate the stepwise BEC-TLPC-hepatocyte transdifferentiation process. The appearance of CK19+/HNF4α+ BECs in most human liver disease indications correlates with the degree of hepatocyte senescence.112 Whether CK19+/HNF4α+ BECs in patients contribute to liver regeneration, as well as the mechanisms linking hepatocyte senescence to the induction of TLPCs, remain to be studied. It is also possible that the HNF4α- LPCs described above additionally contribute to the formation of new hepatocytes during liver regeneration.
Inverse correlation between homeostatic hepatocyte proliferation and metabolic function
Following liver injury, hepatocytes re-enter the cell cycle to restore the hepatocyte pool and liver mass. The signalling mechanisms regulating hepatocyte proliferation after liver injury are well reviewed elsewhere (73,74 as examples). In brief, redundancies between multiple signalling pathways ensure that hepatocytes proliferate to restore liver mass. For example, HGF (ligand of the MET receptor) and ligands of EGFR are direct mitogens on hepatocytes. While MET or EGFR signalling alone can compensate for one another, blocking both signalling pathways results in deficient liver regeneration, with respect to original liver mass, and animal death after partial hepatectomy.[127], [128], [129] Another example of redundancies that secure liver regrowth following partial hepatectomy is insulin-mTORC1 activation, which can compensate for loss of WNT/β-catenin signalling.130
In addition to restoring liver mass, hepatic metabolism and hepatocyte identity need to be adapted to the changing energy demands of regenerating hepatocytes. At the same time, vital liver functions must be maintained. Several studies suggest that hepatocyte proliferation leads to metabolic remodelling and that hepatocytes across the liver can adapt dynamically to changes during regeneration106,107,[131], [132], [133], [134], [135] (Fig. 2C). Following partial hepatectomy, a subset of hepatocytes transiently acquired an early-postnatal-like gene expression programme to proliferate, while other hepatocytes became metabolically hyperactive to compensate for any temporary deficits in liver function.106 Interestingly, some hepatocytes retained chromatin landscapes and transcriptomics of metabolically active uninjured hepatocytes, while others showed changing chromatin and expression signatures suggesting transient fetal reprogramming.107 Moreover, other liver injury models suggest division of labour by confining different hepatocyte identities across the lobule. During two distinct phases of regeneration, macrophage-derived WNT ligands were required for functional compensation and induction of metabolic gene expression, whereas endothelium-derived WNT ligands were more important for inducing hepatocyte proliferation.134 It is conceivable that the porto-central wave of hepatocyte proliferation following partial hepatectomy, induced by a preceding wave of WNT/β-catenin signalling induction, also contributes to maintaining liver function by distributing proliferation events cross the lobule over time.
Proliferation of specialised hepatocytes during liver regeneration
In addition to plasticity and division of labour to meet the needs of a regenerating liver, the intrinsic regenerative potential of hepatocytes is another important factor. A prevailing theory over the last several decades is that all hepatocytes have similar regenerative potential (Fig. 3A). A recent update to this theory is that hepatocytes transiently undergo reprogramming (EMT, or fetal gene expression) as they proliferate.104,108 Challenging the prevailing theory, several recent studies demonstrated that hepatocyte regenerative potential is heterogenous, with some hepatocytes shown to be more regenerative than others. However, a complicating factor in these experiments is that the results may depend on the type of injury. Zone 1 SOX9+ hepatocytes next to the bile ducts expanded throughout the lobule, giving rise to large clones when CCl4 was applied repeatedly to damage zone 3 hepatocytes60 (Fig. 3B). However, proliferation during regeneration in the CCl4 model was not limited to SOX9+ hepatocytes. Zone 2 hepatocytes labelled by AAV8-TBG-Cre or CreERT2 expressed from the Hamp2 or Mup3 promoters also proliferated and clonally expanded.86,88 Additionally, injury to zone 1 by allyl alcohol injection or 3,5-diethoxycarbonyl-1,4-dihydrocollidine (DDC) diet was compensated by proliferation of hepatocytes in zones 2 and 3.85,86,88 A recent study shows that zone 2 hepatocytes traced by Igfbp2-CreER proliferate more during regeneration after partial hepatectomy and during pregnancy. Additionally, they can replace zone 3 and 1 hepatocytes after CCl4- or DDC-induced injury, and their proliferation is nutritionally regulated136 (Fig. 3C).
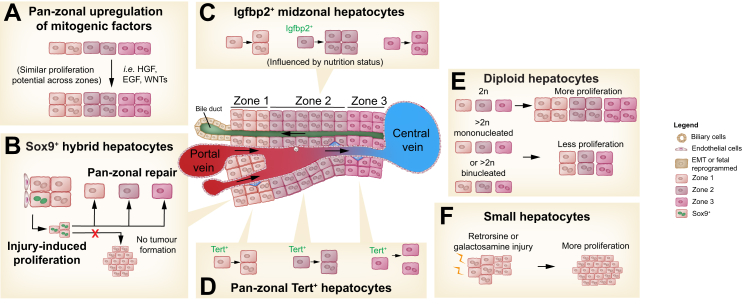
Fig. 3.
Models of hepatocyte regenerative potential.
(A) Schematic illustrating the long-prevailing hypothesis that all hepatocytes are regenerative and have similar regenerative potential, regardless of location, cell ploidy or other characteristics. (B) Alternatively, Sox9+ hepatocytes in zone 1 next to the portal vein or bile ducts are reported to be especially regenerative hepatocytes and can replenish all zones with hepatocytes without giving rise to tumours. (C) Very recent data showing Lgfbp2+ zone 2 hepatocytes fuel hepatocyte regeneration during partial hepatectomy and liver growth in pregnancy. Their proliferation is affected by nutrition status. (D) Hepatocytes expressing high levels of telomerase (Tert+) found in all zones are reported to be more regenerative, functioning like stem cells after injury. (E) Diploid hepatocytes are reported to be more proliferative than polyploid hepatocytes. (F) Small hepatocytes found in retrorsine- or galactosamine-injured rat livers have been reported to fuel regeneration. EMT, epithelial-mesenchymal transition.
Rare distributed high telomerase-expressing hepatocytes were observed to be highly regenerative, giving rise to 38% of the parenchyma after 1 month of DDC-induced injury89 (Fig. 3D). However, ∼80% of hepatocytes randomly labelled by AAV8-TBG-Cre gave rise to expanding hepatocyte clones in mouse livers subjected to repeated CCl4 injury, indicating that proliferative hepatocytes are relatively abundant.88 Additionally, hepatocytes traced from the Hamp2 and Mup3 promoters readily gave rise to hepatocyte clusters after CCl4 and DDC injury.86
Whether hepatocyte regenerative potential is linked to ploidy is controversial. Livers enriched for diploid hepatocytes showed a regeneration advantage over normal adult mouse livers with ∼90% polyploid hepatocytes38 (Fig. 3E). However, polyploid hepatocytes marked by a multicolour reporter system appeared to be as proliferative as diploid hepatocytes in mouse models of hepatocyte transplantation and liver regeneration.137
An interesting but less well understood population of hepatocytes are the small hepatocytes found in retrorsine-treated rat livers[138], [139], [140] and the small Thy1+ hepatocytes observed in rat livers following galactosamine-induced injury141 (Fig. 3F). These small hepatocytes were reported to have robust regenerative properties. While it was speculated that they arise from putative progenitor cells in the bile ducts,142 a number of different approaches demonstrated that hepatocytes are the source of small hepatocytes.[143], [144], [145], [146] These small hepatocytes could be further characterised by lineage-tracing experiments.
As summarised in Table 1, several studies have recently demonstrated a unique set of proliferative hepatocytes during liver regeneration. It remains to be resolved whether these populations are each uniquely regenerative, or whether there is overlap between them. Additionally, investigating whether increased proliferative potential is due to the hepatocytes’ unique identity, or due to the influences of their surrounding environment, which is affected by the type of injury, is important to understanding the extent to which different hepatocyte identities translate into regenerative potential.
Heterogeneity among hepatocytes during liver disease
Injury-related differences in hepatocyte identity are also relevant during liver disease. Many of the CYP450 enzymes, which are responsible for metabolising drugs and xenobiotics, are expressed in pericentral hepatocytes. Associated toxic metabolites therefore predominantly damage hepatocytes in this zone.147 Examples include alcohol148 and paracetamol,149 which damage pericentral hepatocytes and are responsible for the majority of fatal acute liver injury cases. Cholangiopathies, including primary biliary cirrhosis or primary sclerosing cholangitis, often promote damage in periportal hepatocytes due to accumulation of cytotoxic bile in this area.150,151 Such cholestatic injury also activates YAP signalling and thereby reprogrammes periportal hepatocytes, reducing the amount of metabolically active workers in this zone.57,94,102 Mutations in the periportal hepatocyte-enriched genes HSD17B13152 and PNPLA3153 are associated with NASH, although the role of the spatially confined proteins in the pathophysiology of NASH remains to be fully elucidated. Steatosis in adults with non-alcoholic fatty liver disease (NAFLD) usually begins in pericentral hepatocytes and then extends into other lobular zones during disease progression.154 Lipotoxicity-induced hepatocyte injury further leads to a loss of hepatocyte identity and reprogramming, causing loss of metabolic function and the induction of profibrotic and proinflammatory factors.155 The exact spatiotemporal mechanisms responsible for TG accumulation in patients with NAFLD remain to be further elucidated. The consequences associated with loss of hepatocyte identity in zone 1 or 2 were illustrated by the severe metabolic defects in patients with monogenic liver disease (reviewed in9). For example, mutations in a single enzyme (AGXT), predominantly expressed in periportal hepatocytes, results in a life-threatening condition, primary hyperoxaluria type 1.26,156 How loss of zonal hepatocyte identity and function contribute to the many detrimental symptoms of chronic liver disease (e.g. visceral bleeding, hepatic encephalopathy, cachexia and ascites) requires further assessment.
In addition to loss of zonal hepatocyte identity and hepatocyte cell death associated with acute liver failure or chronic liver disease, insufficient functional hepatic mass either following hepatic transplantation or after resection, termed small-for-size syndrome (SFSS), can be fatal. SFSS is associated with a liver remnant or graft to body weight ratio <0.8% or a remnant to total liver volume of less than 25%–30%, where the liver is too small to handle post-operative metabolic demands.157 Accelerating hepatocyte proliferation and restoring liver function are therefore considered therapeutic options to promote liver regeneration and prevent complications associated with SFSS.16 Treatment with RSPO ligands44,158 or tetravalent FZD-LRP antibodies25 enhanced WNT/β-catenin signalling, accelerated liver regeneration and increased metabolic function in diverse injury models. While this may offer therapeutic potential for patients with acute or chronic liver injury, balancing WNT/β-catenin pathway activation will likely be important to prevent metabolic reprograming of periportal hepatocytes into pericentral hepatocytes and the associated loss of periportal metabolic gene expression.22
Maladaptive repair in the liver is another complication associated with loss of hepatocyte identity. Persistent injury and induction of a pro-proliferative programme in hepatocytes not only reduces the availability of metabolically active hepatocytes, but also increases the risk of tumour formation. Many patients with chronic liver disease are at an increased risk of developing hepatocellular carcinoma (HCC) or cholangiocellular carcinoma.159,160 While HCC is more frequent in patients with zone 3 injury, such as NASH or alcohol-related liver disease,160 cholangiocellular carcinoma risk is dramatically increased in patients with persistent periportal injury, e.g. in those with primary sclerosing cholangitis.159 Likewise, the fibrotic ‘wound healing’ response is tightly linked to zonal injury and changes in hepatocyte identity via reprogramming.155,159,160 Activating mutations in the WNT/β-catenin pathway are frequently found in liver tumours and hepatocytes with high WNT/β-catenin activity are highly susceptible to malignant transformation in experimental tumour models.50,90,91,85 Constant pericentral WNT/β-catenin signalling, which is required to ensure metabolic zonation and function, depends on tight control mechanisms to prevent unwanted proliferation. Redundant regulatory mechanisms, such as the two ubiquitin ligases ZNRF3 and RNF43,22,92 therefore balance WNT/β-catenin activity and prevent tumour formation. Frequent mutations in HCC in downstream pathway nodes, such as AXIN1-inactivating or β-catenin-stabilising mutations, highlight the risk associated with uncontrolled WNT/β-catenin signalling in hepatocytes91 (Fig. 2C).
In addition, chronically damaged livers can induce significant changes to their hepatocyte composition. Whole exome and ultra-deep sequencing from the non-dysplastic tissue in livers of patients with cirrhosis showed recurrent mutations in several genes that promote clonal expansion.161 During regeneration, the polyploid hepatocyte population has been shown to shift from predominantly cellular polyploid to nuclear polyploid,31 possibly via pathological processes. For example, highly polyploid mononucleated hepatocytes observed in mouse models of NAFLD arise via activation of the ATR-p53-p21 pathway from oxidative stress, delaying the S to G2 transition and leading to endoreplication.72 Telomere deprotection162 and infection by HBV or HCV163,164 were also reported to promote polyploidisation via unknown mechanisms. The consequences of these changes are not fully understood and may involve altered metabolism or a pre-disposition to cancer.31,161
Conclusions and future directions
Despite differences in gene expression, hepatocytes appear to be the same cell type and can adopt gene expression characteristics of each zone depending on their surrounding environment. As described in previous sections, changes, for example in porto-central WNT/β-catenin activity or oxygen gradient, can significantly affect hepatocyte identity.9,41,91 Additionally, lineage-tracing data have shown that hepatocytes in the different zones can adopt the gene expression programme of the new zone they reside in.49,60,84,86 Transient changes in hepatocyte identity are required when hepatocytes re-enter the cell cycle in response to injury, while others increase metabolic function to maintain hepatic metabolism.25,104,106,107,134 Unfortunately, persistent changes in hepatocyte identity are associated with most liver diseases and are often accompanied by severe hepatic and systemic consequences.9 More research is required to dissect how spatiotemporal hepatocyte identity is controlled during liver homeostasis, regeneration, and disease. This may enable the design of novel therapies that restore locally confined liver function to help patients with various acute or chronic liver diseases. Likewise, understanding how persisting changes in hepatocyte identity trigger inflammation, fibrosis and cancer could help prevent the vicious cycle associated with maladaptive liver repair.
One important area for future research is the development of biomarkers to monitor pericentral metabolic function. The LiMAX exhalation test can measure CYP1A2 function as a predictor of post-operative liver function.165 However, the majority of routine liver function tests only measure the function of periportal hepatocytes (e.g. serum albumin detection, prothrombin time or lactate dehydrogenase measurements) or assess hepatocyte damage (e.g. measurement of transaminases).166 Expanding the possibilities to non-invasively measure the function of hepatocytes in different lobular zones could not only help to better classify disease stages (e.g. in NASH and alcohol-related liver disease changes in hepatocyte identity extend from zone 3 into zone 1 with disease progression), but also to target therapies to the right patient population. Ultimately, they could also serve as an early predictor for severe systemic complications like hepatic encephalopathy.
Another area where spatial differences in hepatocyte identity are still mostly ignored is in vitro assay. Mechanistic studies and pharmacokinetic assessments in primary hepatocytes or hepatocyte-derived cell lines are key components of most research projects and drug discovery campaigns.167 Although partially zonated metabolism has been established in vitro,[168], [169], [170] complex in vitro systems reflecting the full repertoire of differential hepatocyte identities in the liver do not exist. While primary hepatocytes retain some metabolic function in short-term experiments, prolonged culture or use of hepatocyte-like cell systems dramatically changes their metabolic identity. Moreover, 3D culture systems and organoids are not yet capable of reconstructing the complex diversity of hepatocyte identity and metabolic zonation found in a liver.171 Improving ex vivo systems by modulating hepatocyte identity to the zone that is studied would significantly advance the possibilities to study metabolism outside of the liver.
Financial support
This work was supported by a Novartis Postdoctoral Fellowship Grant.
Conflict of interest
The authors declare no conflicts of interest that pertain to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgement
The authors thank Alan Abrams for generating graphics artwork used for generating the Figures.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2023.100779.
Contributor Information
Feng Chen, Email: feng-2.chen@novartis.com.
Jan S. Tchorz, Email: jan.tchorz@novartis.com.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Trefts E., Gannon M., Wasserman D.H. The liver. Curr Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han H.S., Kang G., Kim J.S., Choi B.H., Koo S.H. Regulation of glucose metabolism from a liver-centric perspective. Exp Mol Med. 2016;48:e218. doi: 10.1038/emm.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petersen M.C., Vatner D.F., Shulman G.I. Regulation of hepatic glucose metabolism in health and disease. Nat Rev Endocrinol. 2017;13:572–587. doi: 10.1038/nrendo.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen P., Leray V., Diez M., Serisier S., Le Bloc'h J., Siliart B., et al. Liver lipid metabolism. J Anim Physiol Anim Nutr (Berl) 2008;92:272–283. doi: 10.1111/j.1439-0396.2007.00752.x. [DOI] [PubMed] [Google Scholar]
- 5.Häussinger D., Lamers W.H., Moorman A.F. Hepatocyte heterogeneity in the metabolism of amino acids and ammonia. Enzyme. 1992;46:72–93. doi: 10.1159/000468779. [DOI] [PubMed] [Google Scholar]
- 6.Hou Y., Hu S., Li X., He W., Wu G. Amino acid metabolism in the liver: nutritional and physiological significance. Adv Exp Med Biol. 2020;1265:21–37. doi: 10.1007/978-3-030-45328-2_2. [DOI] [PubMed] [Google Scholar]
- 7.Meynard D., Babitt J.L., Lin H.Y. The liver: conductor of systemic iron balance. Blood. 2014;123:168–176. doi: 10.1182/blood-2013-06-427757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strnad P., Tacke F., Koch A., Trautwein C. Liver - guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 9.Martini T., Naef F., Tchorz J.S. Spatiotemporal metabolic liver zonation and consequences on pathophysiology. Annu Rev Pathol. 2023;18:439–466. doi: 10.1146/annurev-pathmechdis-031521-024831. [DOI] [PubMed] [Google Scholar]
- 10.Bucher N.L., Malt R.A. Little, Brown; 1971. Regeneration of liver and kidney. [Google Scholar]
- 11.Heinke P., Rost F., Rode J., Trus P., Simonova I., Lazar E., et al. Diploid hepatocytes drive physiological liver renewal in adult humans. Cell Syst. 2022;13:499–507 e412. doi: 10.1016/j.cels.2022.05.001. [DOI] [PubMed] [Google Scholar]
- 12.Stanger B.Z. Cellular homeostasis and repair in the mammalian liver. Annu Rev Physiol. 2015;77:179–200. doi: 10.1146/annurev-physiol-021113-170255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun T., Annunziato S., Tchorz J.S. Prometheus revisited: liver homeostasis and repair. Aging (Albany NY) 2020;12:4685–4687. doi: 10.18632/aging.102957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gadd V.L., Aleksieva N., Forbes S.J. Epithelial plasticity during liver injury and regeneration. Cell Stem Cell. 2020;27:557–573. doi: 10.1016/j.stem.2020.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Li W., Li L., Hui L. Cell plasticity in liver regeneration. Trends Cell Biol. 2020;30:329–338. doi: 10.1016/j.tcb.2020.01.007. [DOI] [PubMed] [Google Scholar]
- 16.Greenbaum L.E., Ukomadu C., Tchorz J.S. Clinical translation of liver regeneration therapies: a conceptual road map. Biochem Pharmacol. 2020;175 doi: 10.1016/j.bcp.2020.113847. [DOI] [PubMed] [Google Scholar]
- 17.Saxena R., Theise N.D., Crawford J.M. Microanatomy of the human liver-exploring the hidden interfaces. Hepatology. 1999;30:1339–1346. doi: 10.1002/hep.510300607. [DOI] [PubMed] [Google Scholar]
- 18.Jungermann K., Katz N. Functional hepatocellular heterogeneity. Hepatology. 1982;2:385–395. doi: 10.1002/hep.1840020316. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Moshe S., Shapira Y., Moor A.E., Manco R., Veg T., Bahar Halpern K., et al. Spatial sorting enables comprehensive characterization of liver zonation. Nat Metab. 2019;1:899–911. doi: 10.1038/s42255-019-0109-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halpern K.B., Shenhav R., Matcovitch-Natan O., Toth B., Lemze D., Golan M., et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature. 2017;542:352–356. doi: 10.1038/nature21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrandt F., Andersson A., Saarenpaa S., Larsson L., Van Hul N., Kanatani S., et al. Spatial Transcriptomics to define transcriptional patterns of zonation and structural components in the mouse liver. Nat Commun. 2021;12:7046. doi: 10.1038/s41467-021-27354-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun T., Annunziato S., Bergling S., Sheng C., Orsini V., Forcella P., et al. ZNRF3 and RNF43 cooperate to safeguard metabolic liver zonation and hepatocyte proliferation. Cell Stem Cell. 2021;28:1822–1837 e1810. doi: 10.1016/j.stem.2021.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Guilliams M., Bonnardel J., Haest B., Vanderborght B., Wagner C., Remmerie A., et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell. 2022;185:379–396 e338. doi: 10.1016/j.cell.2021.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu S., Liu S., Bian Y., Poddar M., Singh S., Cao C., et al. Single-cell spatial transcriptomics reveals a dynamic control of metabolic zonation and liver regeneration by endothelial cell Wnt2 and Wnt9b. Cell Rep Med. 2022;3 doi: 10.1016/j.xcrm.2022.100754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Droin C., Kholtei J.E., Bahar Halpern K., Hurni C., Rozenberg M., Muvkadi S., et al. Space-time logic of liver gene expression at sub-lobular scale. Nat Metab. 2021;3:43–58. doi: 10.1038/s42255-020-00323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ben-Moshe S., Itzkovitz S. Spatial heterogeneity in the mammalian liver. Nat Rev Gastroenterol Hepatol. 2019;16:395–410. doi: 10.1038/s41575-019-0134-x. [DOI] [PubMed] [Google Scholar]
- 28.Morales-Navarrete H., Segovia-Miranda F., Klukowski P., Meyer K., Nonaka H., Marsico G., et al. A versatile pipeline for the multi-scale digital reconstruction and quantitative analysis of 3D tissue architecture. Elife. 2015;4 doi: 10.7554/eLife.11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gentric G., Desdouets C. Polyploidization in liver tissue. Am J Pathol. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- 30.Duncan A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Donne R., Saroul-Ainama M., Cordier P., Celton-Morizur S., Desdouets C. Polyploidy in liver development, homeostasis and disease. Nat Rev Gastroenterol Hepatol. 2020;17:391–405. doi: 10.1038/s41575-020-0284-x. [DOI] [PubMed] [Google Scholar]
- 32.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gandillet A., Alexandre E., Holl V., Royer C., Bischoff P., Cinqualbre J., et al. Hepatocyte ploidy in normal young rat. Comp Biochem Physiol A Mol Integr Physiol. 2003;134:665–673. doi: 10.1016/s1095-6433(02)00374-4. [DOI] [PubMed] [Google Scholar]
- 34.Guidotti J.E., Bregerie O., Robert A., Debey P., Brechot C., Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278:19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- 35.Kudryavtsev B.N., Kudryavtseva M.V., Sakuta G.A., Stein G.I. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:387–393. doi: 10.1007/BF02915139. [DOI] [PubMed] [Google Scholar]
- 36.Duncan A.W., Hanlon Newell A.E., Smith L., Wilson E.M., Olson S.B., Thayer M.J., et al. Frequent aneuploidy among normal human hepatocytes. Gastroenterology. 2012;142:25–28. doi: 10.1053/j.gastro.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilkinson P.D., Duncan A.W. Differential roles for diploid and polyploid hepatocytes in acute and chronic liver injury. Semin Liver Dis. 2021;41:42–49. doi: 10.1055/s-0040-1719175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson P.D., Delgado E.R., Alencastro F., Leek M.P., Roy N., Weirich M.P., et al. The polyploid state restricts hepatocyte proliferation and liver regeneration in mice. Hepatology. 2019;69:1242–1258. doi: 10.1002/hep.30286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S., Zhou K., Luo X., Li L., Tu H.C., Sehgal A., et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018;47:390. doi: 10.1016/j.devcel.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richter M.L., Deligiannis I.K., Yin K., Danese A., Lleshi E., Coupland P., et al. Single-nucleus RNA-seq2 reveals functional crosstalk between liver zonation and ploidy. Nat Commun. 2021;12:4264. doi: 10.1038/s41467-021-24543-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kietzmann T. Metabolic zonation of the liver: the oxygen gradient revisited. Redox Biol. 2017;11:622–630. doi: 10.1016/j.redox.2017.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benhamouche S., Decaens T., Godard C., Chambrey R., Rickman D.S., Moinard C., et al. Apc tumor suppressor gene is the "zonation-keeper" of mouse liver. Dev Cell. 2006;10:759–770. doi: 10.1016/j.devcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 43.Ma R., Martinez-Ramirez A.S., Borders T.L., Gao F., Sosa-Pineda B. Metabolic and non-metabolic liver zonation is established non-synchronously and requires sinusoidal Wnts. Elife. 2020;9 doi: 10.7554/eLife.46206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planas-Paz L., Orsini V., Boulter L., Calabrese D., Pikiolek M., Nigsch F., et al. The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size. Nat Cell Biol. 2016;18:467–479. doi: 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- 45.Preziosi M., Okabe H., Poddar M., Singh S., Monga S.P. Endothelial Wnts regulate beta-catenin signaling in murine liver zonation and regeneration: a sequel to the Wnt-Wnt situation. Hepatol Commun. 2018;2:845–860. doi: 10.1002/hep4.1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rocha A.S., Vidal V., Mertz M., Kendall T.J., Charlet A., Okamoto H., et al. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 2015;13:1757–1764. doi: 10.1016/j.celrep.2015.10.049. [DOI] [PubMed] [Google Scholar]
- 47.Sekine S., Lan B.Y., Bedolli M., Feng S., Hebrok M. Liver-specific loss of beta-catenin blocks glutamine synthesis pathway activity and cytochrome p450 expression in mice. Hepatology. 2006;43:817–825. doi: 10.1002/hep.21131. [DOI] [PubMed] [Google Scholar]
- 48.Tan X., Behari J., Cieply B., Michalopoulos G.K., Monga S.P. Conditional deletion of beta-catenin reveals its role in liver growth and regeneration. Gastroenterology. 2006;131:1561–1572. doi: 10.1053/j.gastro.2006.08.042. [DOI] [PubMed] [Google Scholar]
- 49.Wang B., Zhao L., Fish M., Logan C.Y., Nusse R. Self-renewing diploid Axin2(+) cells fuel homeostatic renewal of the liver. Nature. 2015;524:180–185. doi: 10.1038/nature14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ang C.H., Hsu S.H., Guo F., Tan C.T., Yu V.C., Visvader J.E., et al. Lgr5(+) pericentral hepatocytes are self-maintained in normal liver regeneration and susceptible to hepatocarcinogenesis. Proc Natl Acad Sci U S A. 2019;116:19530–19540. doi: 10.1073/pnas.1908099116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., Mowry L.E., Nejak-Bowen K.N., Okabe H., Diegel C.R., Lang R.A., et al. beta-catenin signaling in murine liver zonation and regeneration: a Wnt-Wnt situation. Hepatology. 2014;60:964–976. doi: 10.1002/hep.27082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gougelet A., Torre C., Veber P., Sartor C., Bachelot L., Denechaud P.D., et al. T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology. 2014;59:2344–2357. doi: 10.1002/hep.26924. [DOI] [PubMed] [Google Scholar]
- 53.Matz-Soja M., Aleithe S., Marbach E., Bottger J., Arnold K., Schmidt-Heck W., et al. Hepatic Hedgehog signaling contributes to the regulation of IGF1 and IGFBP1 serum levels. Cell Commun Signal. 2014;12:11. doi: 10.1186/1478-811X-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng X., Kim S.Y., Okamoto H., Xin Y., Yancopoulos G.D., Murphy A.J., et al. Glucagon contributes to liver zonation. Proc Natl Acad Sci U S A. 2018;115:E4111–E4119. doi: 10.1073/pnas.1721403115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fitamant J., Kottakis F., Benhamouche S., Tian H.S., Chuvin N., Parachoniak C.A., et al. YAP inhibition restores hepatocyte differentiation in advanced HCC, leading to tumor regression. Cell Rep. 2015;10:1692–1707. doi: 10.1016/j.celrep.2015.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cox A.G., Hwang K.L., Brown K.K., Evason K., Beltz S., Tsomides A., et al. Yap reprograms glutamine metabolism to increase nucleotide biosynthesis and enable liver growth. Nat Cell Biol. 2016;18:886–896. doi: 10.1038/ncb3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Planas-Paz L., Sun T., Pikiolek M., Cochran N.R., Bergling S., Orsini V., et al. YAP, but not RSPO-LGR4/5, signaling in biliary epithelial cells promotes a ductular reaction in response to liver injury. Cell Stem Cell. 2019;25:39–53 e10. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 58.Verboven E., Moya I.M., Sansores-Garcia L., Xie J., Hillen H., Kowalczyk W., et al. Regeneration defects in yap and taz mutant mouse livers are caused by bile duct disruption and cholestasis. Gastroenterology. 2021;160:847–862. doi: 10.1053/j.gastro.2020.10.035. [DOI] [PubMed] [Google Scholar]
- 59.Yimlamai D., Christodoulou C., Galli G.G., Yanger K., Pepe-Mooney B., Gurung B., et al. Hippo pathway activity influences liver cell fate. Cell. 2014;157:1324–1338. doi: 10.1016/j.cell.2014.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Font-Burgada J., Shalapour S., Ramaswamy S., Hsueh B., Rossell D., Umemura A., et al. Hybrid periportal hepatocytes regenerate the injured liver without giving rise to cancer. Cell. 2015;162:766–779. doi: 10.1016/j.cell.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monga S.P., Sadler K.C. An epigenetic perspective on liver regeneration. Epigenomics. 2020;12:381–384. doi: 10.2217/epi-2020-0010. [DOI] [PubMed] [Google Scholar]
- 62.Sun X., Chuang J.C., Kanchwala M., Wu L., Celen C., Li L., et al. Suppression of the SWI/SNF component Arid1a promotes mammalian regeneration. Cell Stem Cell. 2016;18:456–466. doi: 10.1016/j.stem.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brosch M., Kattler K., Herrmann A., von Schonfels W., Nordstrom K., Seehofer D., et al. Epigenomic map of human liver reveals principles of zonated morphogenic and metabolic control. Nat Commun. 2018;9:4150. doi: 10.1038/s41467-018-06611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lauschke V.M., Mkrtchian S., Ingelman-Sundberg M. The role of microRNAs in liver injury at the crossroad between hepatic cell death and regeneration. Biochem Biophys Res Commun. 2017;482:399–407. doi: 10.1016/j.bbrc.2016.10.084. [DOI] [PubMed] [Google Scholar]
- 65.Yi P.S., Zhang M., Xu M.Q. Role of microRNA in liver regeneration. Hepatobiliary Pancreat Dis Int. 2016;15:141–146. doi: 10.1016/s1499-3872(15)60036-4. [DOI] [PubMed] [Google Scholar]
- 66.Etchegaray J.P., Lee C., Wade P.A., Reppert S.M. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 67.Margall-Ducos G., Celton-Morizur S., Couton D., Bregerie O., Desdouets C. Liver tetraploidization is controlled by a new process of incomplete cytokinesis. J Cell Sci. 2007;120:3633–3639. doi: 10.1242/jcs.016907. [DOI] [PubMed] [Google Scholar]
- 68.Celton-Morizur S., Merlen G., Couton D., Desdouets C. Polyploidy and liver proliferation: central role of insulin signaling. Cell Cycle. 2010;9:460–466. doi: 10.4161/cc.9.3.10542. [DOI] [PubMed] [Google Scholar]
- 69.Celton-Morizur S., Merlen G., Couton D., Margall-Ducos G., Desdouets C. The insulin/Akt pathway controls a specific cell division program that leads to generation of binucleated tetraploid liver cells in rodents. J Clin Invest. 2009;119:1880–1887. doi: 10.1172/JCI38677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandit S.K., Westendorp B., Nantasanti S., van Liere E., Tooten P.C., Cornelissen P.W., et al. E2F8 is essential for polyploidization in mammalian cells. Nat Cell Biol. 2012;14:1181–1191. doi: 10.1038/ncb2585. [DOI] [PubMed] [Google Scholar]
- 71.Hsu S.H., Delgado E.R., Otero P.A., Teng K.Y., Kutay H., Meehan K.M., et al. MicroRNA-122 regulates polyploidization in the murine liver. Hepatology. 2016;64:599–615. doi: 10.1002/hep.28573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gentric G., Maillet V., Paradis V., Couton D., L'Hermitte A., Panasyuk G., et al. Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest. 2015;125:981–992. doi: 10.1172/JCI73957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Michalopoulos G.K. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 74.Michalopoulos G.K., Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 75.He L., Pu W., Liu X., Zhang Z., Han M., Li Y., et al. Proliferation tracing reveals regional hepatocyte generation in liver homeostasis and repair. Science. 2021:371. doi: 10.1126/science.abc4346. [DOI] [PubMed] [Google Scholar]
- 76.Tchorz J.S. The conundrum of the pericentral hepatic niche: WNT/-Catenin signaling, metabolic zonation, and many open questions. Gene Expr. 2020;20:119–124. doi: 10.3727/105221620X16007982788168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monga S.P. No zones left behind: democratic hepatocytes contribute to liver homeostasis and repair. Cell Stem Cell. 2020;26:2–3. doi: 10.1016/j.stem.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 78.Zajicek G., Oren R., Weinreb M., Jr. The streaming liver. Liver. 1985;5:293–300. doi: 10.1111/j.1600-0676.1985.tb00252.x. [DOI] [PubMed] [Google Scholar]
- 79.Farber E. Similarities in the sequence of early histological changes induced in the liver of the rat by ethionine, 2-acetylamino-fluorene, and 3'-methyl-4-dimethylaminoazobenzene. Cancer Res. 1956;16:142–148. [PubMed] [Google Scholar]
- 80.Evarts R.P., Nagy P., Nakatsukasa H., Marsden E., Thorgeirsson S.S. In vivo differentiation of rat liver oval cells into hepatocytes. Cancer Res. 1989;49:1541–1547. [PubMed] [Google Scholar]
- 81.Evarts R.P., Nagy P., Marsden E., Thorgeirsson S.S. A precursor-product relationship exists between oval cells and hepatocytes in rat liver. Carcinogenesis. 1987;8:1737–1740. doi: 10.1093/carcin/8.11.1737. [DOI] [PubMed] [Google Scholar]
- 82.Furuyama K., Kawaguchi Y., Akiyama H., Horiguchi M., Kodama S., Kuhara T., et al. Continuous cell supply from a Sox9-expressing progenitor zone in adult liver, exocrine pancreas and intestine. Nat Genet. 2011;43:34–41. doi: 10.1038/ng.722. [DOI] [PubMed] [Google Scholar]
- 83.Tarlow B.D., Finegold M.J., Grompe M. Clonal tracing of Sox9+ liver progenitors in mouse oval cell injury. Hepatology. 2014;60:278–289. doi: 10.1002/hep.27084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pu W., Zhang H., Huang X., Tian X., He L., Wang Y., et al. Mfsd2a+ hepatocytes repopulate the liver during injury and regeneration. Nat Commun. 2016;7 doi: 10.1038/ncomms13369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun T., Pikiolek M., Orsini V., Bergling S., Holwerda S., Morelli L., et al. AXIN2(+) pericentral hepatocytes have limited contributions to liver homeostasis and regeneration. Cell Stem Cell. 2020;26:97–107.e106. doi: 10.1016/j.stem.2019.10.011. [DOI] [PubMed] [Google Scholar]
- 86.Wei Y., Wang Y.G., Jia Y., Li L., Yoon J., Zhang S., et al. Liver homeostasis is maintained by midlobular zone 2 hepatocytes. Science. 2021:371. doi: 10.1126/science.abb1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.May S., Muller M., Livingstone C.R., Skalka G.L., Walsh P.J., Nixon C., et al. Absent expansion of AXIN2+ hepatocytes and altered physiology in Axin2CreERT2 mice challenges the role of pericentral hepatocytes in homeostatic liver regeneration. J Hepatol. 2023;78:1028–1036. doi: 10.1016/j.jhep.2023.01.009. [DOI] [PubMed] [Google Scholar]
- 88.Chen F., Jimenez R.J., Sharma K., Luu H.Y., Hsu B.Y., Ravindranathan A., et al. Broad distribution of hepatocyte proliferation in liver homeostasis and regeneration. Cell Stem Cell. 2020;26:27–33 e24. doi: 10.1016/j.stem.2019.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin S., Nascimento E.M., Gajera C.R., Chen L., Neuhofer P., Garbuzov A., et al. Distributed hepatocytes expressing telomerase repopulate the liver in homeostasis and injury. Nature. 2018;556:244–248. doi: 10.1038/s41586-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Annunziato S., Sun T., Tchorz J.S. The RSPO-LGR4/5-ZNRF3/RNF43 module in liver homeostasis, regeneration, and disease. Hepatology. 2022;76:888–899. doi: 10.1002/hep.32328. [DOI] [PubMed] [Google Scholar]
- 91.Russell J.O., Monga S.P. Wnt/beta-Catenin signaling in liver development, homeostasis, and pathobiology. Annu Rev Pathol. 2018;13:351–378. doi: 10.1146/annurev-pathol-020117-044010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Belenguer G., Mastrogiovanni G., Pacini C., Hall Z., Dowbaj A.M., Arnes-Benito R., et al. RNF43/ZNRF3 loss predisposes to hepatocellular-carcinoma by impairing liver regeneration and altering the liver lipid metabolic ground-state. Nat Commun. 2022;13:334. doi: 10.1038/s41467-021-27923-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Camargo F.D., Gokhale S., Johnnidis J.B., Fu D., Bell G.W., Jaenisch R., et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 94.Pepe-Mooney B.J., Dill M.T., Alemany A., Ordovas-Montanes J., Matsushita Y., Rao A., et al. Single-cell analysis of the liver epithelium reveals dynamic heterogeneity and an essential role for YAP in homeostasis and regeneration. Cell Stem Cell. 2019;25:23–38 e28. doi: 10.1016/j.stem.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Arechederra M., Berasain C., Avila M.A., Fernandez-Barrena M.G. Chromatin dynamics during liver regeneration. Semin Cell Dev Biol. 2020;97:38–46. doi: 10.1016/j.semcdb.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 96.Kaji K., Factor V.M., Andersen J.B., Durkin M.E., Tomokuni A., JU Marquardt, et al. DNMT1 is a required genomic regulator for murine liver histogenesis and regeneration. Hepatology. 2016;64:582–598. doi: 10.1002/hep.28563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Michalopoulos G.K., Barua L., Bowen W.C. Transdifferentiation of rat hepatocytes into biliary cells after bile duct ligation and toxic biliary injury. Hepatology. 2005;41:535–544. doi: 10.1002/hep.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Schaub J.R., Huppert K.A., Kurial S.N.T., Hsu B.Y., Cast A.E., Donnelly B., et al. De novo formation of the biliary system by TGFbeta-mediated hepatocyte transdifferentiation. Nature. 2018;557:247–251. doi: 10.1038/s41586-018-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yanger K., Zong Y., Maggs L.R., Shapira S.N., Maddipati R., Aiello N.M., et al. Robust cellular reprogramming occurs spontaneously during liver regeneration. Genes Dev. 2013;27:719–724. doi: 10.1101/gad.207803.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tarlow B.D., Pelz C., Naugler W.E., Wakefield L., Wilson E.M., Finegold M.J., et al. Bipotential adult liver progenitors are derived from chronically injured mature hepatocytes. Cell Stem Cell. 2014;15:605–618. doi: 10.1016/j.stem.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tanimizu N., Nishikawa Y., Ichinohe N., Akiyama H., Mitaka T. Sry HMG box protein 9-positive (Sox9+) epithelial cell adhesion molecule-negative (EpCAM-) biphenotypic cells derived from hepatocytes are involved in mouse liver regeneration. J Biol Chem. 2014;289:7589–7598. doi: 10.1074/jbc.M113.517243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li W., Yang L., He Q., Hu C., Zhu L., Ma X., et al. A homeostatic arid1a-dependent permissive chromatin state licenses hepatocyte responsiveness to liver-injury-associated YAP signaling. Cell Stem Cell. 2019;25:54–68.e55. doi: 10.1016/j.stem.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 103.Li L., Cui L., Lin P., Liu Z., Bao S., Ma X., et al. Kupffer-cell-derived IL-6 is repurposed for hepatocyte dedifferentiation via activating progenitor genes from injury-specific enhancers. Cell Stem Cell. 2023;30:283–299.e289. doi: 10.1016/j.stem.2023.01.009. [DOI] [PubMed] [Google Scholar]
- 104.Ben-Moshe S., Veg T., Manco R., Dan S., Papinutti D., Lifshitz A., et al. The spatiotemporal program of zonal liver regeneration following acute injury. Cell Stem Cell. 2022;29:973–989 e910. doi: 10.1016/j.stem.2022.04.008. [DOI] [PubMed] [Google Scholar]
- 105.Zhao L., Jin Y., Donahue K., Tsui M., Fish M., Logan C.Y., et al. Tissue repair in the mouse liver following acute carbon tetrachloride depends on injury-induced wnt/beta-catenin signaling. Hepatology. 2019;69:2623–2635. doi: 10.1002/hep.30563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chembazhi U.V., Bangru S., Hernaez M., Kalsotra A. Cellular plasticity balances the metabolic and proliferation dynamics of a regenerating liver. Genome Res. 2021;31:576–591. doi: 10.1101/gr.267013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen T., Oh S., Gregory S., Shen X., Diehl A.M. Single-cell omics analysis reveals functional diversification of hepatocytes during liver regeneration. JCI Insight. 2020;5 doi: 10.1172/jci.insight.141024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Oh S.H., Swiderska-Syn M., Jewell M.L., Premont R.T., Diehl A.M. Liver regeneration requires Yap1-TGFbeta-dependent epithelial-mesenchymal transition in hepatocytes. J Hepatol. 2018;69:359–367. doi: 10.1016/j.jhep.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Itoh T., Kamiya Y., Okabe M., Tanaka M., Miyajima A. Inducible expression of Wnt genes during adult hepatic stem/progenitor cell response. FEBS Lett. 2009;583:777–781. doi: 10.1016/j.febslet.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 110.Okabe H., Yang J., Sylakowski K., Yovchev M., Miyagawa Y., Nagarajan S., et al. Wnt signaling regulates hepatobiliary repair following cholestatic liver injury in mice. Hepatology. 2016;64:1652–1666. doi: 10.1002/hep.28774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Thompson M.D., Awuah P., Singh S., Monga S.P. Disparate cellular basis of improved liver repair in beta-catenin-overexpressing mice after long-term exposure to 3,5-diethoxycarbonyl-1,4-dihydrocollidine. Am J Pathol. 2010;177:1812–1822. doi: 10.2353/ajpath.2010.100173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pu W., Zhu H., Zhang M., Pikiolek M., Ercan C., Li J., et al. Bipotent transitional liver progenitor cells contribute to liver regeneration. Nat Genet. 2023 doi: 10.1038/s41588-023-01335-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Deng X., Zhang X., Li W., Feng R.X., Li L., Yi G.R., et al. Chronic liver injury induces conversion of biliary epithelial cells into hepatocytes. Cell Stem Cell. 2018;23:114–122.e113. doi: 10.1016/j.stem.2018.05.022. [DOI] [PubMed] [Google Scholar]
- 114.Espanol-Suner R., Carpentier R., Van Hul N., Legry V., Achouri Y., Cordi S., et al. Liver progenitor cells yield functional hepatocytes in response to chronic liver injury in mice. Gastroenterology. 2012;143:1564–1575.e1567. doi: 10.1053/j.gastro.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 115.Manco R., Clerbaux L.A., Verhulst S., Bou Nader M., Sempoux C., Ambroise J., et al. Reactive cholangiocytes differentiate into proliferative hepatocytes with efficient DNA repair in mice with chronic liver injury. J Hepatol. 2019;70:1180–1191. doi: 10.1016/j.jhep.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 116.Raven A., Lu W.Y., Man T.Y., Ferreira-Gonzalez S., O'Duibhir E., Dwyer B.J., et al. Cholangiocytes act as facultative liver stem cells during impaired hepatocyte regeneration. Nature. 2017;547:350–354. doi: 10.1038/nature23015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rodrigo-Torres D., Affo S., Coll M., Morales-Ibanez O., Millan C., Blaya D., et al. The biliary epithelium gives rise to liver progenitor cells. Hepatology. 2014;60:1367–1377. doi: 10.1002/hep.27078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Russell J.O., Lu W.Y., Okabe H., Abrams M., Oertel M., Poddar M., et al. Hepatocyte-Specific beta-catenin deletion during severe liver injury provokes cholangiocytes to differentiate into hepatocytes. Hepatology. 2019;69:742–759. doi: 10.1002/hep.30270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lu W.Y., Bird T.G., Boulter L., Tsuchiya A., Cole A.M., Hay T., et al. Hepatic progenitor cells of biliary origin with liver repopulation capacity. Nat Cell Biol. 2015;17:971–983. doi: 10.1038/ncb3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sackett S.D., Li Z., Hurtt R., Gao Y., Wells R.G., Brondell K., et al. Foxl1 is a marker of bipotential hepatic progenitor cells in mice. Hepatology. 2009;49:920–929. doi: 10.1002/hep.22705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shin S., Upadhyay N., Greenbaum L.E., Kaestner K.H. Ablation of Foxl1-Cre-labeled hepatic progenitor cells and their descendants impairs recovery of mice from liver injury. Gastroenterology. 2015;148:192–202 e193. doi: 10.1053/j.gastro.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shin S., Walton G., Aoki R., Brondell K., Schug J., Fox A., et al. Foxl1-Cre-marked adult hepatic progenitors have clonogenic and bilineage differentiation potential. Genes Dev. 2011;25:1185–1192. doi: 10.1101/gad.2027811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jakubowski A., Ambrose C., Parr M., Lincecum J.M., Wang M.Z., Zheng T.S., et al. TWEAK induces liver progenitor cell proliferation. J Clin Invest. 2005;115:2330–2340. doi: 10.1172/JCI23486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Boulter L., Govaere O., Bird T.G., Radulescu S., Ramachandran P., Pellicoro A., et al. Macrophage-derived Wnt opposes Notch signaling to specify hepatic progenitor cell fate in chronic liver disease. Nat Med. 2012;18:572–579. doi: 10.1038/nm.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Minnis-Lyons S.E., Ferreira-Gonzalez S., Aleksieva N., Man T.Y., Gadd V.L., Williams M.J., et al. Notch-IGF1 signaling during liver regeneration drives biliary epithelial cell expansion and inhibits hepatocyte differentiation. Sci Signal. 2021:14. doi: 10.1126/scisignal.aay9185. [DOI] [PubMed] [Google Scholar]
- 126.Aloia L., McKie M.A., Vernaz G., Cordero-Espinoza L., Aleksieva N., van den Ameele J., et al. Epigenetic remodelling licences adult cholangiocytes for organoid formation and liver regeneration. Nat Cell Biol. 2019;21:1321–1333. doi: 10.1038/s41556-019-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tsagianni A., Mars W.M., Bhushan B., Bowen W.C., Orr A., Stoops J., et al. Combined systemic disruption of MET and epidermal growth factor receptor signaling causes liver failure in normal mice. Am J Pathol. 2018;188:2223–2235. doi: 10.1016/j.ajpath.2018.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lopez-Luque J., Caballero-Diaz D., Martinez-Palacian A., Roncero C., Moreno-Caceres J., Garcia-Bravo M., et al. Dissecting the role of epidermal growth factor receptor catalytic activity during liver regeneration and hepatocarcinogenesis. Hepatology. 2016;63:604–619. doi: 10.1002/hep.28134. [DOI] [PubMed] [Google Scholar]
- 129.Paranjpe S., Bowen W.C., Mars W.M., Orr A., Haynes M.M., DeFrances M.C., et al. Combined systemic elimination of MET and epidermal growth factor receptor signaling completely abolishes liver regeneration and leads to liver decompensation. Hepatology. 2016;64:1711–1724. doi: 10.1002/hep.28721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu S., Cao C., Poddar M., Delgado E., Singh S., Singh-Varma A., et al. Hepatocyte beta-catenin loss is compensated by Insulin-mTORC1 activation to promote liver regeneration. Hepatology. 2022 doi: 10.1002/hep.32680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bangru S., Arif W., Seimetz J., Bhate A., Chen J., Rashan E.H., et al. Alternative splicing rewires Hippo signaling pathway in hepatocytes to promote liver regeneration. Nat Struct Mol Biol. 2018;25:928–939. doi: 10.1038/s41594-018-0129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Caldez M.J., Van Hul N., Koh H.W.L., Teo X.Q., Fan J.J., Tan P.Y., et al. Metabolic remodeling during liver regeneration. Dev Cell. 2018;47:425–438 e425. doi: 10.1016/j.devcel.2018.09.020. [DOI] [PubMed] [Google Scholar]
- 133.Huang J., Rudnick D.A. Elucidating the metabolic regulation of liver regeneration. Am J Pathol. 2014;184:309–321. doi: 10.1016/j.ajpath.2013.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Walesky C.M., Kolb K.E., Winston C.L., Henderson J., Kruft B., Fleming I., et al. Functional compensation precedes recovery of tissue mass following acute liver injury. Nat Commun. 2020;11:5785. doi: 10.1038/s41467-020-19558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang A.W., Wangensteen K.J., Wang Y.J., Zahm A.M., Moss N.G., Erez N., et al. TRAP-seq identifies cystine/glutamate antiporter as a driver of recovery from liver injury. J Clin Invest. 2018;128:2297–2309. doi: 10.1172/JCI95120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lin Y.-H., Wei Y., Wang Y., Pagani C.A., Li L., Zhu M., et al. IGFBP2 expressing midlobular hepatocytes preferentially contribute to liver homeostasis and regeneration. bioRxiv. 2022 doi: 10.1016/j.stem.2023.04.007. 2022.2010.2012.511675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Matsumoto T., Wakefield L., Tarlow B.D., Grompe M. In Vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell. 2020;26:34–47 e33. doi: 10.1016/j.stem.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gordon G.J., Coleman W.B., Hixson D.C., Grisham J.W. Liver regeneration in rats with retrorsine-induced hepatocellular injury proceeds through a novel cellular response. Am J Pathol. 2000;156:607–619. doi: 10.1016/S0002-9440(10)64765-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Best D.H., Coleman W.B. Liver regeneration by small hepatocyte-like progenitor cells after necrotic injury by carbon tetrachloride in retrorsine-exposed rats. Exp Mol Pathol. 2010;89:92–98. doi: 10.1016/j.yexmp.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 140.Katayama S., Tateno C., Asahara T., Yoshizato K. Size-dependent in vivo growth potential of adult rat hepatocytes. Am J Pathol. 2001;158:97–105. doi: 10.1016/S0002-9440(10)63948-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kon J., Ichinohe N., Ooe H., Chen Q., Sasaki K., Mitaka T. Thy1-positive cells have bipotential ability to differentiate into hepatocytes and biliary epithelial cells in galactosamine-induced rat liver regeneration. Am J Pathol. 2009;175:2362–2371. doi: 10.2353/ajpath.2009.080338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen Y.H., Chang M.H., Chien C.S., Wu S.H., Yu C.H., Chen H.L. Contribution of mature hepatocytes to small hepatocyte-like progenitor cells in retrorsine-exposed rats with chimeric livers. Hepatology. 2013;57:1215–1224. doi: 10.1002/hep.26104. [DOI] [PubMed] [Google Scholar]
- 143.Best D.H., Coleman W.B. Treatment with 2-AAF blocks the small hepatocyte-like progenitor cell response in retrorsine-exposed rats. J Hepatol. 2007;46:1055–1063. doi: 10.1016/j.jhep.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Best D.H., Coleman W.B. Bile duct destruction by 4,4'-diaminodiphenylmethane does not block the small hepatocyte-like progenitor cell response in retrorsine-exposed rats. Hepatology. 2007;46:1611–1619. doi: 10.1002/hep.21876. [DOI] [PubMed] [Google Scholar]
- 145.Avril A., Pichard V., Bralet M.P., Ferry N. Mature hepatocytes are the source of small hepatocyte-like progenitor cells in the retrorsine model of liver injury. J Hepatol. 2004;41:737–743. doi: 10.1016/j.jhep.2004.07.029. [DOI] [PubMed] [Google Scholar]
- 146.Pichard V., Aubert D., Ferry N. Direct in vivo cell lineage analysis in the retrorsine and 2AAF models of liver injury after genetic labeling in adult and newborn rats. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 148.Massey V.L., Arteel G.E. Acute alcohol-induced liver injury. Front Physiol. 2012;3:193. doi: 10.3389/fphys.2012.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yoon E., Babar A., Choudhary M., Kutner M., Pyrsopoulos N. Acetaminophen-induced hepatotoxicity: a comprehensive update. J Clin Transl Hepatol. 2016;4:131–142. doi: 10.14218/JCTH.2015.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Karlsen T.H., Folseraas T., Thorburn D., Vesterhus M. Primary sclerosing cholangitis - a comprehensive review. J Hepatol. 2017;67:1298–1323. doi: 10.1016/j.jhep.2017.07.022. [DOI] [PubMed] [Google Scholar]
- 151.Trauner M., Meier P.J., Boyer J.L. Molecular pathogenesis of cholestasis. N Engl J Med. 1998;339:1217–1227. doi: 10.1056/NEJM199810223391707. [DOI] [PubMed] [Google Scholar]
- 152.Abul-Husn N.S., Cheng X., Li A.H., Xin Y., Schurmann C., Stevis P., et al. A protein-truncating HSD17B13 variant and protection from chronic liver disease. N Engl J Med. 2018;378:1096–1106. doi: 10.1056/NEJMoa1712191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Speliotes E.K., Butler J.L., Palmer C.D., Voight B.F., Consortium G., Consortium M.I., et al. PNPLA3 variants specifically confer increased risk for histologic nonalcoholic fatty liver disease but not metabolic disease. Hepatology. 2010;52:904–912. doi: 10.1002/hep.23768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brunt E.M. Pathology of nonalcoholic fatty liver disease. Nat Rev Gastroenterol Hepatol. 2010;7:195–203. doi: 10.1038/nrgastro.2010.21. [DOI] [PubMed] [Google Scholar]
- 155.Loft A., Alfaro A.J., Schmidt S.F., Pedersen F.B., Terkelsen M.K., Puglia M., et al. Liver-fibrosis-activated transcriptional networks govern hepatocyte reprogramming and intra-hepatic communication. Cell Metab. 2021;33:1685–1700.e1689. doi: 10.1016/j.cmet.2021.06.005. [DOI] [PubMed] [Google Scholar]
- 156.Wang X., Danese D., Brown T., Baldwin J., Sajeev G., Cook E.E., et al. Primary hyperoxaluria type 1 disease manifestations and healthcare utilization: a multi-country, online, chart review study. Front Med. 2021;8 doi: 10.3389/fmed.2021.703305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Tucker O.N., Heaton N. The 'small for size' liver syndrome. Curr Opin Crit Care. 2005;11:150–155. doi: 10.1097/01.ccx.0000157080.11117.45. [DOI] [PubMed] [Google Scholar]
- 158.Zhang Z., Broderick C., Nishimoto M., Yamaguchi T., Lee S.J., Zhang H., et al. Tissue-targeted R-spondin mimetics for liver regeneration. Sci Rep. 2020;10 doi: 10.1038/s41598-020-70912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Deeb M., Karlsen T.H., Hirschfield G.M. The 6 C's of primary sclerosing cholangitis. J Hepatol. 2020;73:1255–1256. doi: 10.1016/j.jhep.2020.06.033. [DOI] [PubMed] [Google Scholar]
- 160.Zhu C., Tabas I., Schwabe R.F., Pajvani U.B. Maladaptive regeneration - the reawakening of developmental pathways in NASH and fibrosis. Nat Rev Gastroenterol Hepatol. 2021;18:131–142. doi: 10.1038/s41575-020-00365-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Zhu M., Lu T., Jia Y., Luo X., Gopal P., Li L., et al. Somatic mutations increase hepatic clonal fitness and regeneration in chronic liver disease. Cell. 2019;177:608–621.e612. doi: 10.1016/j.cell.2019.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lazzerini Denchi E., Celli G., de Lange T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes Dev. 2006;20:2648–2653. doi: 10.1101/gad.1453606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Toyoda H., Bregerie O., Vallet A., Nalpas B., Pivert G., Brechot C., et al. Changes to hepatocyte ploidy and binuclearity profiles during human chronic viral hepatitis. Gut. 2005;54:297–302. doi: 10.1136/gut.2004.043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ahodantin J., Bou-Nader M., Cordier C., Megret J., Soussan P., Desdouets C., et al. Hepatitis B virus X protein promotes DNA damage propagation through disruption of liver polyploidization and enhances hepatocellular carcinoma initiation. Oncogene. 2019;38:2645–2657. doi: 10.1038/s41388-018-0607-3. [DOI] [PubMed] [Google Scholar]
- 165.Stockmann M., Lock J.F., Malinowski M., Niehues S.M., Seehofer D., Neuhaus P. The LiMAx test: a new liver function test for predicting postoperative outcome in liver surgery. HPB (Oxford) 2010;12:139–146. doi: 10.1111/j.1477-2574.2009.00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Lala V., Zubair M., Minter D.A. StatPearls. Treasure Island (FL); 2022. Liver function tests. [PubMed] [Google Scholar]
- 167.Jin M., Yi X., Liao W., Chen Q., Yang W., Li Y., et al. Advancements in stem cell-derived hepatocyte-like cell models for hepatotoxicity testing. Stem Cell Res Ther. 2021;12:84. doi: 10.1186/s13287-021-02152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sato A., Kadokura K., Uchida H., Tsukada K. An in vitro hepatic zonation model with a continuous oxygen gradient in a microdevice. Biochem Biophys Res Commun. 2014;453:767–771. doi: 10.1016/j.bbrc.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 169.Kang Y.B.A., Eo J., Mert S., Yarmush M.L., Usta O.B. Metabolic patterning on a chip: towards in vitro liver zonation of primary rat and human hepatocytes. Sci Rep. 2018;8:8951. doi: 10.1038/s41598-018-27179-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tonon F., Giobbe G.G., Zambon A., Luni C., Gagliano O., Floreani A., et al. In vitro metabolic zonation through oxygen gradient on a chip. Sci Rep. 2019;9 doi: 10.1038/s41598-019-49412-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kaluthantrige Don F., Huch M. Organoids, where we stand and where we go. Trends Mol Med. 2021;27:416–418. doi: 10.1016/j.molmed.2021.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.