Abstract
Genomic DNA barcoding has emerged as a sensitive and flexible tool to measure the fates of clonal sub-populations within a heterogeneous cancer cell population. Coupling cellular barcoding with single cell transcriptomics permits the longitudinal analysis of molecular mechanisms with detailed clone-level resolution. Numerous recent studies have employed these tools to track clonal cell states in cancer progression and treatment response. With these new technologies comes the opportunity to examine longstanding questions about the origins and contributions of tumor cell heterogeneity and the roles of selection and phenotypic plasticity in disease progression and treatment.
Keywords: barcoding, tumor heterogeneity, tumor evolution, drug resistance, phenotypic plasticity
Introduction
Heterogeneity is a fundamental property of all cell populations [1] and results from the stochasticity of molecular interactions [2]–[5]. In cancer, as in normal cell development, the inherent phenotypic diversity in a cell population is a substrate for the forces of selection [6]–[8]. Cellular interactions and interactions with the physical tissue environment stabilize particular phenotypes and sculpt the overall population structure [5], [9], [10]. In addition, tumors consist of cells that are genetically heterogeneous, even when derived from a single clone, and genetic instability may contribute to the generation of additional variation with disease progression [11]. A longitudinal understanding of tumor heterogeneity is essential for improving clinical treatments and outcomes [12], [13]. In recent years, cell-based methods have been complemented by high-throughput sequencing and methods for the genomic and transcriptomic analysis of individual single cells. Advances in these areas have also facilitated the development of cellular barcoding methods that permit the tracking of individual cell trajectories with exquisite detail. Barcoding technologies enable the quantification of heterogeneous populations and the evaluation of changes in phenotypic state, in the context of selective pressures arising from the cell population, tissue and environmental interactions, and external perturbations, such as drug treatment.
This review will explore the development and applications of barcoding technology in recent years, including biological questions, techniques, challenges, and outcomes. Specifically, we will delve into the ways that barcoding tools are paving the way for new mechanistic insights into tumor heterogeneity, progression, treatment response and relapse [14].
Overview of Genomic Barcoding Platforms
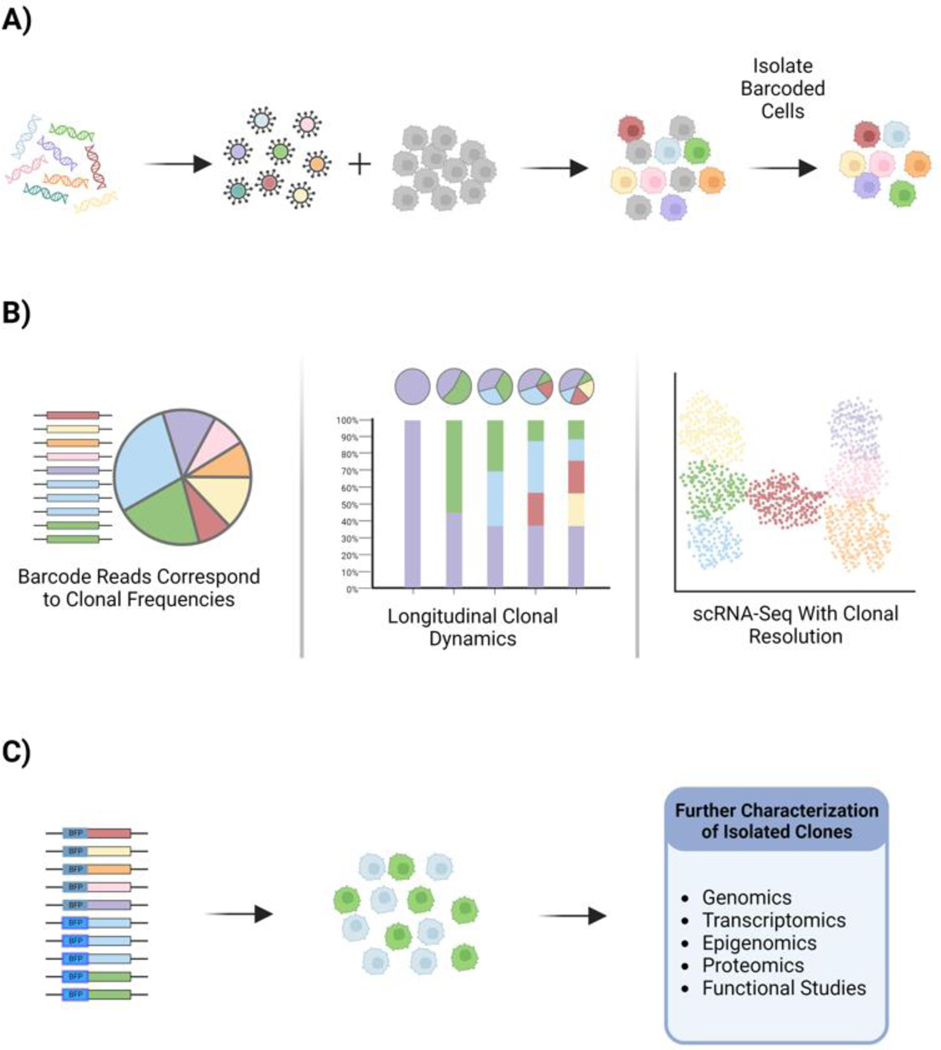
While numerous genomic barcoding technologies are now in use, they share an overall conceptual strategy. A unique nucleic acid sequence is introduced into each founder cell in the population, usually by viral delivery. This label becomes integrated into the genome and is heritable to daughter cells, serving to tag the clone and all of its descendants over many expansions and generations [15], [16] (Figure 1A). Positively-transduced cells are then selected by antibiotic resistance or by FACS using a fluorescent reporter to ensure that the starting cell population consists entirely of barcode-labeled cells (Figure 1A). The number of initial founder cells in this labeled population determines the barcode diversity; a starting population of 1000 cells will include 1000 unique barcode labels. With cell subculturing and expansion, the relative frequency of these 1000 barcodes may change, as some cells proliferate more rapidly than others and some cells are lost from the population. Importantly, it is straightforward to determine the frequency of these 1000 clones in the population by high-throughput sequencing of the ensemble of barcodes. The number of reads of each barcode sequence corresponds to the frequency of that clone in the cell population. In some variations of barcoding technologies, the integrated sequence is also expressed as a transcript that can be detected by standard scRNA-Seq methods allowing detailed transcriptomic analysis of the clonal population [17]–[22] (Figure 1B). Measuring barcode frequencies and barcode-resolved single cell transcriptomes over longitudinal time points enables the study of trajectories of clonal behaviors within an overall heterogeneous tumor cell population. In another extension of the technology, the ClonMapper and CaTCH technologies add additional functionalities to clone tracking studies by enabling the clone-specific expression of a reporter for subpopulation isolation [21], [23], [24] (Figure 1C). In this application, expressed barcodes consist of random sgRNA sequences and a Cas9 transcriptional activator (lacking endonuclease activity) drives the activation of synthetic gene circuits to induce clone-specific fluorescent reporter expression. Transcriptional activation of functional genes has also been demonstrated, enabling clone-specific control of cell fate decisions, such as activation of the apoptosis program by BAX [23].
Figure 1. Genomic Barcoding.
The general process and outcomes of genomic barcoding are similar among technologies. A) Barcodes are delivered into cells by viral transduction and cells which successfully integrate a barcode are isolated via FACS or antibiotic selection. B) Investigations using barcoded cells measure clonal frequencies and longitudinal clonal dynamics by high-resolution genomic sequencing of the barcode ensemble. Clonally-resolved transcriptome data is measured by scRNA-Seq. C) Some barcoding techniques allow for clone-specific activation of fluorescent reporter genes, allowing for isolation of particular clones and further live cell or molecular characterization.
Lentiviral-based cell barcoding platforms have been used widely in cancer biology and are amenable to in vivo studies in which labeled cells are engrafted. An alternative set of technologies accomplishes clone tracing through genome editing (Table 1) [25], [26]. Here Cas9 target sequences are integrated into the host genome; the entire set of Cas9 targets serves as a barcode label. As Cas9-mediated insertions and deletions accumulate at the targets comprising the barcode, the unique combination of genomic alterations identifies that cell. New edits are added to daughter cells with each round of cell division and subsequent high-resolution sequencing enables reconstruction of the lineage tree. Variations on this method use self-targeting guide RNAs to record information at a particular locus and are able to record a larger number of accumulated edits [27], [28]. These approaches have been used in cultured mammalian cells and in vivo, in zebrafish and mouse models expressing Cas9 to examine developmental lineage relationships.
Table 1.
Overview of cell barcoding approaches.
| Expressed barcodes | Expressed barcode and clone retrieval | Cas9 edited barcodes | Optical barcodes | |
|---|---|---|---|---|
| Library size | + | + | + | - |
| Quantitative | + | + | + | + |
| Multiplexing | + | + | + | + |
| Clonal isolation | - | + | - | + |
| Clone-specific gene expression | - | + | - | - |
| Spatial information | emerging | emerging | - | + |
| In vivo applications | xenograft only | xenograft only | + | + |
Optical barcoding methods employ a different strategy, labeling clones by expressing a unique combination of fluorescent proteins in each. The expression pattern is heritable to all daughter cells within the clonal population and is quantified by fluorescence microscopy or flow cytometry. While early generations of optical barcoding relied on Cre-Lox recombination events to generate diversity of labels (as in the Brainbow/Confetti system), more recent developments have used the lentiviral gene ontology system (LeGO) of modular vectors to label cells with combinations of up to six fluorescent proteins, producing up to 41 detectable hues and combinations [29]–[31]. While the diversity of optical barcodes is much less than that of genetic barcoding, a significant advantage is the ability to acquire spatial information about clonal distributions, as the barcode labels can be visualized by fluorescence microscopy [32], [33].
Biological questions
Broadly speaking, cellular barcoding enables investigations of the clonal relationships among tumor cells and identifies molecular and functional variations among clones that may contribute to disease progression and treatment response. In this section we examine the biological questions that are driving the application of cell barcoding approaches in cancer and the new insights afforded by these methodologies.
Do clones vary in therapeutic resistance and are signatures of resistance detectable in a naïve cell population?
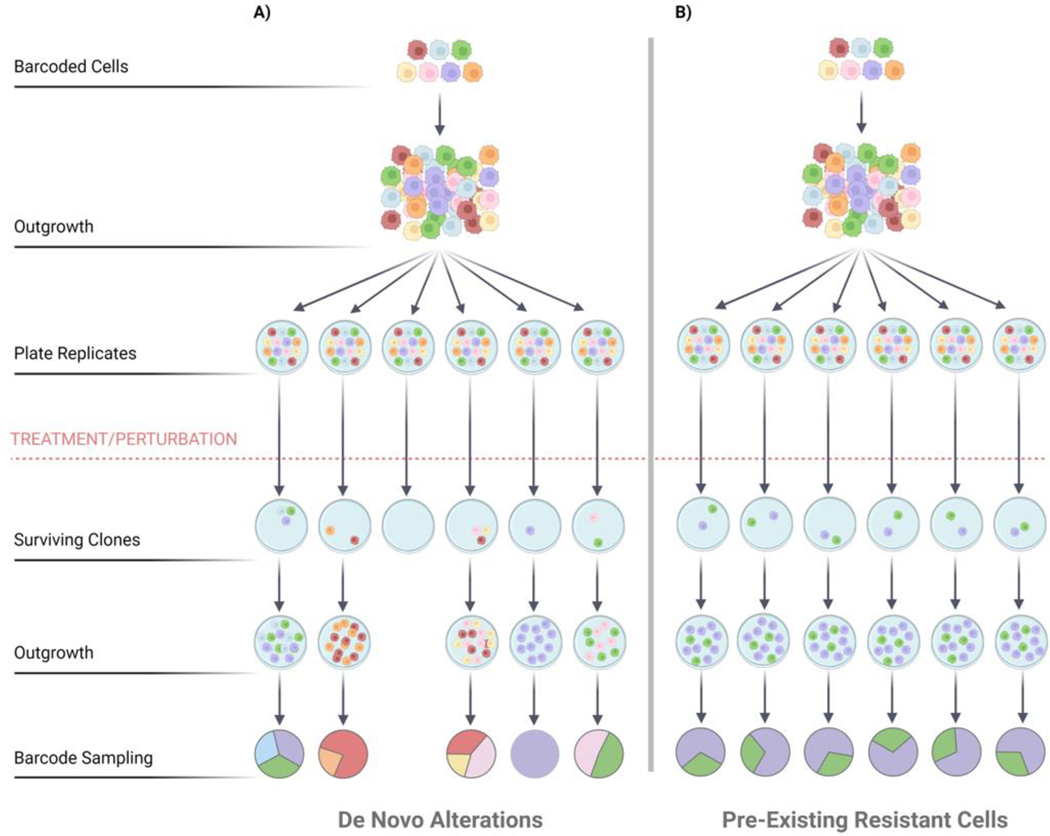
While much work has evaluated the genetic underpinnings of tumor resistance and relapse, in recent years non-genetic mechanisms have also been widely studied. The relative contributions of these processes vary in different tumor settings, drug treatments and drug schedules [34]–[37]. By enabling the quantitation of clonal frequency in parallel with clonal transcriptomic state, barcoded cancer cells shed light on the contributions of selection and non-genetic plasticity to therapeutic response [38], [39]. A unique advantage of working with barcoded cell libraries is ability to compare clonal behaviors across experimental samples (in vitro replicate cultures or in vivo replicate tumors) (Figure 2). The parental barcoded cell population is split into separate replicates, each consisting of a comparable collection of clones. Clones that display similar dynamics across multiple replicates may have a pre-existing genetic mutation that contributes to fitness and serves as a substrate for selection. In contrast, a clone that exhibits differences in abundance (reflecting net survival and proliferation rate) across replicates may have acquired a de novo alteration, which may be genetic or epigenetic [9], [40] (Figure 2). With the advantages of replicate cell libraries and longitudinal quantitative observation, studies of barcoded cells reveal these mechanistic distinctions.
Figure 2. DNA barcodes enable population replicates.
Quantitation of clonal population structures in replicate studies is useful for the categorization of underlying biological processes that impact changes in population structure. Analysis of barcoded replicates may identify pre-existing variants or de novo changes that arise in many different clones upon treatment or perturbation. A) High resolution sequencing of the barcode ensemble may reveal diverse clonal population structures across replicates, which may result from de novo alterations following drug treatment. B) In contrast, barcode quantitation may find that consistent subsets of clones dominate, suggesting the presence of pre-existing resistant clones in the initial cancer cell population.
Drug resistance is linked to heritable genomic alterations of the cells when a minority of clones in the initial cell population undergo selection and become highly enriched in the population after drug treatment [21], [41]–[45]. One of the first high-complexity cancer cell barcoding systems, the ClonTracer technology, was used to track more than 1 million unique barcode labels in the non–small cell lung cancer cell line HCC827 which harbors an activating epidermal growth factor receptor (EGFR) mutation that confers sensitivity to the EGFR inhibitor erlotinib [41], [42]. Due to the large number of barcodes in this system, the contributions of even very rare clones could be detected. Treatment of replicate barcoded cultures revealed that a small pre-existing subpopulation, representing 0.05% of the initial population, was selected and expanded under erlotinib treatment. Other studies used ClonTracer to pinpoint rare resistant clones in the KCL-22 model of chronic myeloid leukemia that were enriched and swept the population following nilotinib or imatinib treatment [42].
In other studies, clonal selection is not associated with therapeutic resistance, as the barcode diversity is not significantly reduced. Instead, cells from many clones may have the potential to evade the drug through the activation of various non-genetic resistance mechanisms [19], [46]–[48]. For example, an in vivo study of colorectal cancer (CRC) patient-derived cells engrafted barcoded tumor cells into NOD/SCID mice and found no loss of clonal diversity upon chemotherapy treatment. Instead, the barcode composition of individual tumors was unique, and the survivor cells appeared to escape treatment by entering a drug-tolerant persister state that resembles diapause [48].
Which cancer cells contribute to tumor initiation and to seeding of metastatic lesions?
Within a heterogeneous cancer cell population, only a small fraction of cells have the capacity to initiate primary tumor formation and seed secondary tumors sites [12], [46], [49], [50]. Barcoded cell technologies are well-suited to identify and measure these rare initiator cells, as the starting distribution of clones and the population composition at engrafted primary and secondary sites can be quantified. In breast cancer cell lines and patient-derived cell models, barcoding approaches measured frequencies of tumor initiating cells that vary from 1–10%. Likewise, approximately 3% of clones from colorectal cancer cell lines were capable of initiating new tumors in vivo[50], [51]. Tumor initiating frequencies also rely on microenvironmental and immune interactions with specific clones; immunoediting of primary tumor clones has been recently described[52].
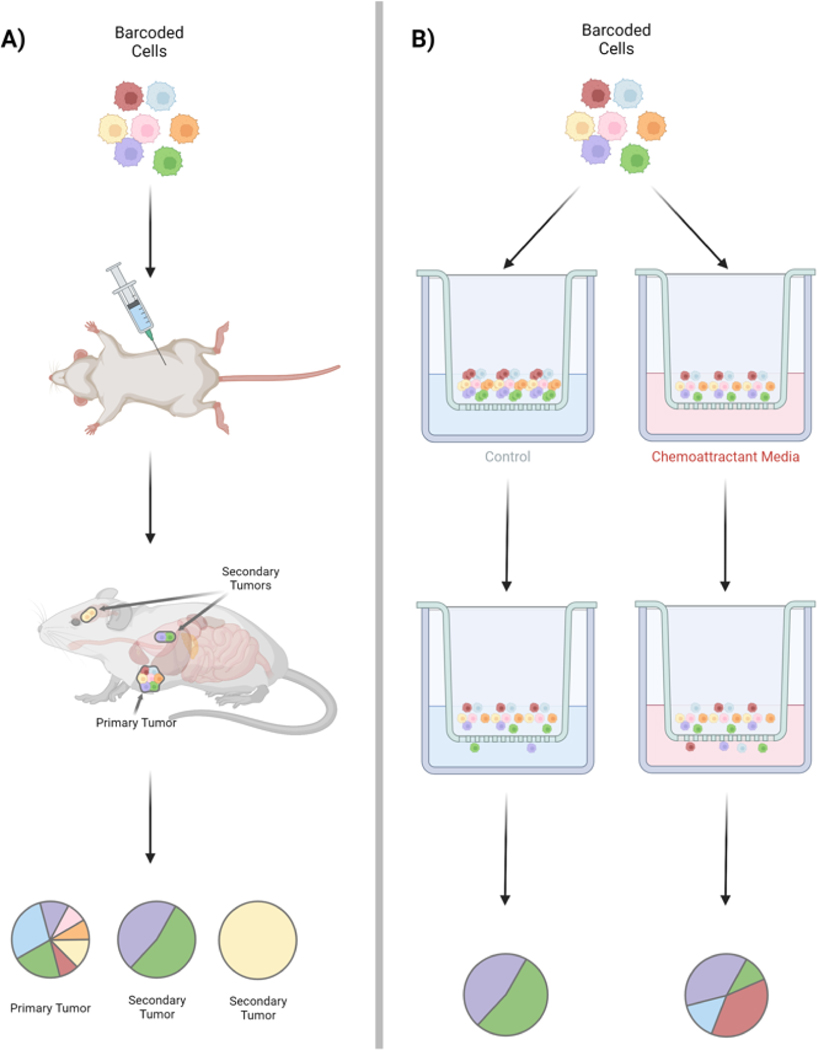
Barcoding methods provide an opportunity to quantify heterogeneity of metastatic potential through direct quantitative comparison of the clonal composition at the primary site with secondary lesions [50], [53]–[55] (Figure 3). Previous approaches have relied on genomic sequencing of the primary and metastatic tumors to compare mutational profiles; targeting sequencing and comparison of the barcode distributions is considerably less costly and less laborious. One recent study investigated metastasis using the human KRAS-mutant lung adenocarcinoma cell line (A549) in an orthotopic xenograft setting [56]. Here a Cas9-based barcoding method was utilized in which a target site was cleaved by Cas9 and an inheritable allele inserted which could then be tracked through future generations. This allowed for single-cell sequencing of individual cells and their progeny, in addition to rates of metastasis for tumor populations, the heritability of these metastatic phenotypes, and tissue routes of metastasis [56]. Metastatic seeding in this model was primarily by pre-existing, heritable subpopulations with distinct clonal transcriptomic signatures of invasion that were detectable in vitro and amplified further in vivo. These studies also revealed a new role for KRT17 as a suppressor of metastatic populations. Thus, tissue interactions and the interplay of clonal populations with the microenvironment appears critical.
Figure 3. Uncovering the dynamics of metastasis with cell barcoding.
Barcoding techniques provide a platform for investigation of tumor phenotypes such as metastasis in vivo. A) Barcoded cells may be implanted into mice to form primary tumors and secondary tumors can be isolated and assessed for clonal frequency and cellular characteristics. B) Complementary in vitro studies may also be performed, such as transwell migration of barcoded cells with or without chemoattractant or ECM changes. Cells which migrate through the transwell can be identified by their barcode and the relationship to clonal identity and gene expression state can assessed.
The longitudinal dynamics of metastatic seeding have also been examined through barcoded cell models [57], [58] and interestingly these differ depending on the primary tumor context of the overall tumor population. When primary tumors were resected in barcoded mouse models, the clonal diversity of metastatic lesions was reduced, indicating that many disseminated cells are incapable of seeding in this context [47]. This highlights a role for clonal interactions, an aspect of tumor biology that has historically been understudied and can now be approached through barcoding technologies. In particular, optical barcoding has illuminated spatial heterogeneities in clonal growth and metastasis dynamics, identifying differences in clonal behaviors at the edge and tumor core [59].
How do cellular barcodes provide detailed quantitative information about tumor clonal dynamics in vivo?
To measure the frequency of clones within a tumor population, barcoded cells are subjected to deep sequencing. This approach only requires sequencing a short region of the genome (i.e., the barcodes), which allows for high-depth coverage of the barcode and enables high-resolution quantitation of individual lineages. These data, combined theoretical population genetics, can serve to measure the distribution of fitness effects and identify clones that are expanding faster than expected by neutral stochastic drift. For example, the role of 11 tumor suppressor pathways was assessed in barcoded cells by using CRISPR/Cas9 genome editing to introduce mutations and then measuring clonal expansion in mouse models of lung cancer[60]. Mutations in SETD2 and LKB1 proved to have the largest fitness effect and resulted in the largest tumors[61]. In murine glioblastoma models, quantitation of barcoded cell dynamics revealed that intratumor heterogeneity resulted primarily from the stochastic fates of cells in a stem cell hierarchy [51]. Within these populations, rare treatment-resistant clones could be identified that displayed deviations from these dynamics [51].
Cell fates and clonal dynamics of acute myeloid leukemia have been elucidated with the single-cell profiling and lineage tracing technology (SPLINTR), which utilizes expressed barcodes [19]. While malignant clonal dominance is a clone-intrinsic property of leukemia cells, increased transcriptional heterogeneity was also a consistent feature of clonal fitness in these cells [19].
To examine clonal patterns in breast cancer recurrence, a conditional mouse model of mammary tumorigenesis MMTV-rtTA;TetO-neu was labeled by lentiviral-mediated cell barcoding [62]. Approximately half of the recurrent tumors in this study were marked by clonal dominance, with just one or two clones sweeping the population. Transcriptomic analysis revealed that these dominant clones demonstrated de novo acquisition of Met amplification. The other half of the recurrent tumors were polyclonal, with no obvious reduction in clonal diversity compared to the primary tumors and characterized by alterations in the IL-6 -Jak/Stat3 pathway [62]. In this model then, recurrence can proceed through multiple distinct routes. Future work may investigate how tumor-intrinsic factors and microenvironmental conditions contribute to the particular clonal dynamics displayed by any individual tumor.
Can clonally-resolved screening assays improve therapy development and evaluation?
Preclinical drug evaluation has often measured the sensitivity of cell lines and primary human cells, without considering the heterogeneity of cellular responses. While advances in organoid culture and biomimetic models now incorporate key features of the tissue environment to increase cell diversity, traditional cell culture models may still harbor heterogeneous subpopulations. Designing in vitro studies to capture this subpopulation information may provide new insights. A large-scale drug screening effort of 578 human cancer cell lines was performed using the PRISM platform of nucleic acid barcoding with pooled screening, to assess the potential for drug-repurposing of non-oncological agents. [63]. Researchers screened 4,518 drugs from the Drug Repurposing Hub in a 2-stage dosing protocol. Six non-oncological treatments demonstrated cytotoxic effects on cancer cells; however, the genomic characteristics of the cell lines suggests these cytotoxic effects are likely variable. In some cases, CRISPR/Cas9 loss-of-function and gain-of-function screening shed additional light on molecular pathways of some drug targets and with further contributions this type of dataset may become a valuable tool in preclinical testing and therapeutic development.
Other studies have employed barcoded cells to explore the effects of treatment schedules on the evolution of drug resistance in heterogeneous cancer cell populations [64]. An extensive characterization of the spectrum of cell responses in the triple negative breast cancer MDA-MB-231 cell line compared 696 treatment conditions, all administering concurrent or sequential crizotinib and navitoclax with different intervals in exposure window and recovery time. Quantitation of barcode dynamics and gene expression states with expressed barcodes revealed that navitoclax selects for pre-existing resistant clones and that the two drugs in combination at low doses resulted in a similar distribution of survivor barcodes to a single high dose of crizotinib. Importantly, the response of subpopulations depends on the history of prior drug exposure, a phenomenon which has been highlighted in numerous studies and can now be directly measured using barcode dynamics [47], [48]. Future studies in the field will likely focus on the molecular mechanisms by which clonal subpopulations interact and may mutually impact therapeutic response and treatment resistance.
Technical considerations
Despite the rapid growth of new platforms for genomic barcoding, there remain some technological considerations and limitations. Understanding these aspects in the experimental design phase will be important for expanding the tools to explore new models and questions.
The size of the barcode library is a key parameter in these studies; in genomic barcoding, the theoretical diversity of the library is extraordinarily high (for a random nucleic acid barcode of 20 nucleotides, the number of possible unique sequences exceeds one trillion). What then are the practical considerations that constrain the initial labeling of a cell population? First the cell population must be sufficiently large to capture the underlying biological variation; a low number of cells in the starting population may represent only a snapshot of the potential cell diversity. Second, the size of the founder cell population must be scaled to the size of the barcode library. If there are more cells than unique sequences in the barcode library, multiple cells will be labeled by the same barcode. Finally, there must be careful attention to the manner in which the cells and the barcodes are combined in the initial labeling event. For many of the studies described here, lentiviral transduction has been the means for barcode integration. This should be carried out at a low multiplicity of infection (MOI) to ensure that most cells receive one and only one barcode by Poisson statistics. For many protocols an MOI of 0.2 is recommended and some methods recommend 0.1 or less. The rationale here is that a low MOI results in fewer doubly-barcoded cells, as well as more cells receiving no barcode—however the unlabeled cells may be easily eliminated by antibiotic selection or FACS for a reporter of transduction. Thus, the starting population will consist entirely of barcoded cells, with few doubly integrated cells.
A further consideration is that clones are identified by relatively short nucleic acid sequences and mutations to the barcode sequence may be introduced in the sequencing step or earlier during PCR amplification of the barcode region. This presents a challenge in distinguishing whether two highly similar barcodes in a dataset represent distinct clones or are derived from the same clonal population before and after a mutation event. (This is sometimes called “barcode collision”.) One solution is to perform deep sequencing of the initial barcode-labeled cell population to generate a “whitelist” of verified barcode labels, although for very high diversity libraries it may not be possible to completely exclude the possibility of rare clones with highly similar sequence.
Integrating transcribed barcode labels with the 10x Genomics Chromium platform for single-cell transcriptomic measurements introduces new technical considerations, as only partial barcode data may be captured in an individual cell. In addition, single-cell experimental design needs to consider the starting diversity of the barcoded population and the expected frequency of clones in the dataset, in order to obtain sufficient individual cell measurements of each.
The genotypic and phenotypic heterogeneity among barcoded cells reflects the population status at the instantiation of labeling. A cell population that is genetically homogenous at the time of barcode introduction will include many clones with similar genotype, although they are tagged with different barcodes. In a population with high genetic heterogeneity, it is possible that low frequency genotypes may be lost. All protocols for barcode delivery and integration introduce the potential for clonal selection, as some clones may be less amenable or less robust to viral delivery or transfection. One practical biological challenge is to monitor the potential loss of barcode diversity with long-term passaging of cell lines in vitro and to understand how various culture conditions impact this process. Similarly, in xenograft studies it is necessary to measure the variation among clones in rates of engraftment, immune escape, and proliferation rates in vivo. An absence of concordance among clonal-specific behaviors in vitro and in vivo may identify new considerations for improving the physiological relevance of cell culture models.
Some experimental settings are not amenable to labeling by exogenous barcode sequences and a retrospective method of identifying clonal relationships is preferred (examples may include archival clinical samples). mtDNA lineage tracking has emerged as an attractive alternative to tracking somatic mutation frequencies via nuclear genomic sequencing [65]–[67]. The mitochondrial genome undergoes significant mutagenesis, at a rate one to two orders of magnitude greater than the mutation rate observed in nuclear genomes [68]. In addition, mtDNA is devoid of histones and nucleosomes and thus accessible to standard chromatin accessibility protocols. Due to the small size of the mtDNA genome, deep mitochondrial sequencing can be performed at low cost and even low frequency somatic mutations can be quantified.
Concluding Remarks
Understanding the behavior of different clones in disease progression and in response to environmental perturbations will be an essential step in integrating cellular heterogeneity into clinical approaches. Pinpointing aggressive clones that are linked to recurrence and resistance is necessary to improve monitoring and treatment. The growing adoption of barcoding methodologies presents unique opportunities for the cancer biology field. Once a barcoded cell population has been generated for a tumor cell line or primary cell population, it becomes possible to compare the behavior of specific clones across different experimental settings, in different culture conditions or microenvironments, and under different therapeutic pressures (Outstanding Questions). Barcoded cell models may therefore be a key to the facilitation of sharing and synergy across laboratories, as the barcodes serve as a stable index for identifying a particular clone over time. We envision the generation of databases that compile clonal cell behaviors and subpopulation behaviors within each cancer cell model. These resources could serve as tools to integrate multidimensional characterization of individual clones, encompassing transcriptomic and genomic information about specific clonal subpopulations, and also drug sensitivity, migration rate, invasion rate, measurements of physical parameters and many more properties that can now be evaluated at clonal resolution. The tremendous potential for incorporating cellular barcoding into existing experimental workflows is therefore catalyzing a shift towards a deeper understanding of heterogeneous cancer cells.
Outstanding Questions.
The implementation of personalized medicine in cancer has been limited, as tumor heterogeneity and real-time monitoring introduce significant challenges. How can cellular barcoding data provide insight regarding the potential trajectories of subpopulation responses that may aid in clinical decision making?
Preclinical drug development and screening relies on in vitro measurements in cancer cell models; inherent heterogeneity within these cell populations may mask specific subpopulation responses to the therapeutic agent. Can barcoded cancer cells be a tool for the discovery and testing of new therapies with subpopulation-specific activity?
Can we envision the establishment of databases and repositories that compile clonal subpopulation behaviors within and across cancer models, to promote sharing and synergy across laboratories?
Highlights.
Barcoding technologies enable the high-resolution quantification of heterogeneous cancer cell populations, in the context of selective pressures arising from the cell population, microenvironmental interactions and external perturbations, such as drug treatment.
By enabling the quantitation of clonal frequency in parallel with clonal transcriptomic state, barcoded cancer cells shed light on the contributions of selection and non-genetic plasticity to resistance mechanisms.
There is a notable interest in characterization of the qualities and clonal dynamics of cells comprising metastatic lesions. Cellular barcoding can be used to study these characteristics of metastasis and compare heterogeneous cancer cells both in vivo and in vitro.
Acknowledgments
The authors thank Sui Huang, Arja Kaipainen and Aziz Al’Khafaji for ongoing discussions and are grateful for funding support through the NIH (1R01CA226258, 1U01CA25354 and 1R01CA255536 to AB) and DOD CDMRP (W81XWH-18-1-0420). KH is supported by an NSF Graduate Research Fellowship.
Footnotes
The authors report no financial conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- [1].Elsasser WM, “Outline of a theory of cellular heterogeneity.,” Proc Natl Acad Sci U S A, vol. 81, no. 16, pp. 5126–5129, Aug. 1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].“The Origin of Phenotypic Heterogeneity in a Clonal Cell Population In Vitro,” PLOS ONE, vol. 2, no. 4, p. e394, Apr. 2007, doi: 10.1371/journal.pone.0000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].“Role of network-mediated stochasticity in mammalian drug resistance | Nature Communications.” https://www.nature.com/articles/s41467-019-10330-w (accessed Jan. 23, 2023). [DOI] [PMC free article] [PubMed]
- [4].Kim E, Kim J-Y, Smith MA, Haura EB, and Anderson ARA, “Cell signaling heterogeneity is modulated by both cell-intrinsic and -extrinsic mechanisms: An integrated approach to understanding targeted therapy,” PLOS Biology, vol. 16, no. 3, p. e2002930, Mar. 2018, doi: 10.1371/journal.pbio.2002930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Brock A, Chang H, and Huang S, “Non-genetic heterogeneity--a mutation-independent driving force for the somatic evolution of tumours,” Nat Rev Genet, vol. 10, no. 5, pp. 336–342, May 2009, doi: 10.1038/nrg2556. [DOI] [PubMed] [Google Scholar]
- [6].Neildez-Nguyen TMA et al. , “Epigenetic gene expression noise and phenotypic diversification of clonal cell populations,” Differentiation, vol. 76, no. 1, pp. 33–40, Jan. 2008, doi: 10.1111/j.1432-0436.2007.00219.x. [DOI] [PubMed] [Google Scholar]
- [7].Sonnenschein C and Soto AM, “Over a century of cancer research: Inconvenient truths and promising leads,” PLoS Biol, vol. 18, no. 4, p. e3000670, Apr. 2020, doi: 10.1371/journal.pbio.3000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kulkarni P et al. , “Addressing the genetic/nongenetic duality in cancer with systems biology,” Trends Cancer, pp. S2405-8033(22)00265–5, Jan. 2023, doi: 10.1016/j.trecan.2022.12.004. [DOI] [PubMed]
- [9].Greaves M and Maley CC, “Clonal evolution in cancer,” Nature, vol. 481, no. 7381, Art. no. 7381, Jan. 2012, doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McGranahan N and Swanton C, “Biological and Therapeutic Impact of Intratumor Heterogeneity in Cancer Evolution,” Cancer Cell, vol. 27, no. 1, pp. 15–26, Jan. 2015, doi: 10.1016/j.ccell.2014.12.001. [DOI] [PubMed] [Google Scholar]
- [11].Nowell PC, “The clonal evolution of tumor cell populations,” Science, vol. 194, no. 4260, pp. 23–28, Oct. 1976, doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- [12].Marusyk A and Polyak K, “Tumor heterogeneity: causes and consequences,” Biochim Biophys Acta, vol. 1805, no. 1, p. 105, Jan. 2010, doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zeng AGX et al. , “A cellular hierarchy framework for understanding heterogeneity and predicting drug response in acute myeloid leukemia,” Nat Med, vol. 28, no. 6, pp. 1212–1223, Jun. 2022, doi: 10.1038/s41591-022-01819-x. [DOI] [PubMed] [Google Scholar]
- [14].Serrano A, Berthelet J, Naik SH, and Merino D, “Mastering the use of cellular barcoding to explore cancer heterogeneity,” Nat Rev Cancer, vol. 22, no. 11, Art. no. 11, Nov. 2022, doi: 10.1038/s41568-022-00500-2. [DOI] [PubMed] [Google Scholar]
- [15].Navin N et al. , “Inferring tumor progression from genomic heterogeneity,” Genome Res, vol. 20, no. 1, pp. 68–80, Jan. 2010, doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang Y et al. , “Clonal evolution in breast cancer revealed by single nucleus genome sequencing,” Nature, vol. 512, no. 7513, Art. no. 7513, Aug. 2014, doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Biddy BA et al. , “Single-cell mapping of lineage and identity in direct reprogramming,” Nature, vol. 564, no. 7735, pp. 219–224, Dec. 2018, doi: 10.1038/s41586-018-0744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Weinreb C, Rodriguez-Fraticelli A, Camargo F, and Klein AM, “Lineage tracing on transcriptional landscapes links state to fate during differentiation,” Science, vol. 367, no. 6479, p. eaaw3381, Feb. 2020, doi: 10.1126/science.aaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Fennell KA et al. , “Non-genetic determinants of malignant clonal fitness at single-cell resolution,” Nature, vol. 601, no. 7891, Art. no. 7891, Jan. 2022, doi: 10.1038/s41586-021-04206-7. [DOI] [PubMed] [Google Scholar]
- [20].Oren Y et al. , “Cycling cancer persister cells arise from lineages with distinct programs,” Nature, vol. 596, no. 7873, pp. 576–582, Aug. 2021, doi: 10.1038/s41586-021-03796-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gutierrez C et al. , “Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment,” Nat Cancer, vol. 2, no. 7, pp. 758–772, Jul. 2021, doi: 10.1038/s43018-021-00222-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Johnson KE et al. , “Integrating transcriptomics and bulk time course data into a mathematical framework to describe and predict therapeutic resistance in cancer,” Phys Biol, vol. 18, no. 1, p. 016001, Nov. 2020, doi: 10.1088/1478-3975/abb09c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Al’Khafaji AM, Deatherage D, and Brock A, “Control of Lineage-Specific Gene Expression by Functionalized gRNA Barcodes,” ACS Synth Biol, vol. 7, no. 10, pp. 2468–2474, Oct. 2018, doi: 10.1021/acssynbio.8b00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Umkehrer C et al. , “Isolating live cell clones from barcoded populations using CRISPRa-inducible reporters,” Nat Biotechnol, vol. 39, no. 2, pp. 174–178, Feb. 2021, doi: 10.1038/s41587-020-0614-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, and Shendure J, “Whole organism lineage tracing by combinatorial and cumulative genome editing,” Science, vol. 353, no. 6298, p. aaf7907, Jul. 2016, doi: 10.1126/science.aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Spanjaard B et al. , “Simultaneous lineage tracing and cell-type identification using CRISPR/Cas9-induced genetic scars,” Nat Biotechnol, vol. 36, no. 5, pp. 469–473, Jun. 2018, doi: 10.1038/nbt.4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kalhor R, Mali P, and Church GM, “Rapidly evolving homing CRISPR barcodes,” Nat Methods, vol. 14, no. 2, Art. no. 2, Feb. 2017, doi: 10.1038/nmeth.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Perli SD, Cui CH, and Lu TK, “Continuous genetic recording with self-targeting CRISPR-Cas in human cells,” Science, vol. 353, no. 6304, p. aag0511, Sep. 2016, doi: 10.1126/science.aag0511. [DOI] [PubMed] [Google Scholar]
- [29].Livet J et al. , “Transgenic strategies for combinatorial expression of fluorescent proteins in the nervous system,” Nature, vol. 450, no. 7166, pp. 56–62, Nov. 2007, doi: 10.1038/nature06293. [DOI] [PubMed] [Google Scholar]
- [30].Schepers AG et al. , “Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas,” Science, vol. 337, no. 6095, pp. 730–735, Aug. 2012, doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- [31].Mohme M et al. , “Optical Barcoding for Single-Clone Tracking to Study Tumor Heterogeneity,” Mol Ther, vol. 25, no. 3, pp. 621–633, Mar. 2017, doi: 10.1016/j.ymthe.2016.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tiede S et al. , “Multi-color clonal tracking reveals intra-stage proliferative heterogeneity during mammary tumor progression,” Oncogene, vol. 40, no. 1, pp. 12–27, Jan. 2021, doi: 10.1038/s41388-020-01508-4. [DOI] [PubMed] [Google Scholar]
- [33].Berthelet J et al. , “The site of breast cancer metastases dictates their clonal composition and reversible transcriptomic profile,” Sci Adv, vol. 7, no. 28, p. eabf4408, Jul. 2021, doi: 10.1126/sciadv.abf4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Neftel C et al. , “An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma,” Cell, vol. 178, no. 4, pp. 835–849.e21, Aug. 2019, doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yuan S, Norgard RJ, and Stanger BZ, “Cellular Plasticity in Cancer,” Cancer Discov, vol. 9, no. 7, pp. 837–851, Jul. 2019, doi: 10.1158/2159-8290.CD-19-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Marjanovic ND et al. , “Emergence of a High-Plasticity Cell State during Lung Cancer Evolution,” Cancer Cell, vol. 38, no. 2, pp. 229–246.e13, Aug. 2020, doi: 10.1016/j.ccell.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Foo J et al. , “Roadmap on plasticity and epigenetics in cancer,” Phys Biol, vol. 19, no. 3, Apr. 2022, doi: 10.1088/1478-3975/ac4ee2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Huang S, “Reconciling Non-Genetic Plasticity with Somatic Evolution in Cancer,” Trends Cancer, vol. 7, no. 4, pp. 309–322, Apr. 2021, doi: 10.1016/j.trecan.2020.12.007. [DOI] [PubMed] [Google Scholar]
- [39].Pillai M and Jolly MK, “Systems-level network modeling deciphers the master regulators of phenotypic plasticity and heterogeneity in melanoma,” iScience, vol. 24, no. 10, p. 103111, Sep. 2021, doi: 10.1016/j.isci.2021.103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jones PA and Gonzalgo ML, “Altered DNA methylation and genome instability: A new pathway to cancer?,” Proc Natl Acad Sci U S A, vol. 94, no. 6, pp. 2103–2105, Mar. 1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Hata AN et al. , “Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition,” Nat. Med, vol. 22, no. 3, pp. 262–269, Mar. 2016, doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bhang HC et al. , “Studying clonal dynamics in response to cancer therapy using high-complexity barcoding,” Nat Med, vol. 21, no. 5, pp. 440–448, May 2015, doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- [43].Hinohara K et al. , “KDM5 histone demethylase activity links cellular transcriptomic heterogeneity to therapeutic resistance,” Cancer Cell, vol. 34, no. 6, pp. 939–953.e9, Dec. 2018, doi: 10.1016/j.ccell.2018.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Caiado F et al. , “Lineage tracing of acute myeloid leukemia reveals the impact of hypomethylating agents on chemoresistance selection,” Nat Commun, vol. 10, p. 4986, Nov. 2019, doi: 10.1038/s41467-019-12983-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Yu C et al. , “High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines,” Nat Biotechnol, vol. 34, no. 4, pp. 419–423, Apr. 2016, doi: 10.1038/nbt.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Echeverria GV et al. , “Resistance to neoadjuvant chemotherapy in triple negative breast cancer mediated by a reversible drug-tolerant state,” Sci Transl Med, vol. 11, no. 488, p. eaav0936, Apr. 2019, doi: 10.1126/scitranslmed.aav0936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Merino D et al. , “Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer,” Nat Commun, vol. 10, no. 1, Art. no. 1, Feb. 2019, doi: 10.1038/s41467-019-08595-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Rehman SK et al. , “Colorectal Cancer Cells Enter a Diapause-like DTP State to Survive Chemotherapy,” Cell, vol. 184, no. 1, pp. 226–242.e21, Jan. 2021, doi: 10.1016/j.cell.2020.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Klein CA, “Parallel progression of primary tumours and metastases,” Nat Rev Cancer, vol. 9, no. 4, Art. no. 4, Apr. 2009, doi: 10.1038/nrc2627. [DOI] [PubMed] [Google Scholar]
- [50].Nguyen LV et al. , “DNA barcoding reveals diverse growth kinetics of human breast tumour subclones in serially passaged xenografts,” Nat Commun, vol. 5, p. 5871, Dec. 2014, doi: 10.1038/ncomms6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lan X et al. , “Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy,” Nature, vol. 549, no. 7671, pp. 227–232, Sep. 2017, doi: 10.1038/nature23666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Baldwin LA et al. , “DNA barcoding reveals ongoing immunoediting of clonal cancer populations during metastatic progression and immunotherapy response,” Nat Commun, vol. 13, no. 1, Art. no. 1, Nov. 2022, doi: 10.1038/s41467-022-34041-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Seth S et al. , “Pre-existing Functional Heterogeneity of Tumorigenic Compartment as the Origin of Chemoresistance in Pancreatic Tumors,” Cell Rep, vol. 26, no. 6, pp. 1518–1532.e9, Feb. 2019, doi: 10.1016/j.celrep.2019.01.048. [DOI] [PubMed] [Google Scholar]
- [54].Howard GR, Jost TA, Yankeelov TE, and Brock A, “Quantification of long-term doxorubicin response dynamics in breast cancer cell lines to direct treatment schedules,” PLoS Comput Biol, vol. 18, no. 3, p. e1009104, Mar. 2022, doi: 10.1371/journal.pcbi.1009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].West JB, Dinh MN, Brown JS, Zhang J, Anderson AR, and Gatenby RA, “Multidrug Cancer Therapy in Metastatic Castrate-Resistant Prostate Cancer: An Evolution-Based Strategy,” Clin Cancer Res, vol. 25, no. 14, pp. 4413–4421, Jul. 2019, doi: 10.1158/1078-0432.CCR-19-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Quinn JJ et al. , “Single-cell lineages reveal the rates, routes, and drivers of metastasis in cancer xenografts,” Science, vol. 371, no. 6532, p. eabc1944, Feb. 2021, doi: 10.1126/science.abc1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Singh SK et al. , “Identification of human brain tumour initiating cells,” Nature, vol. 432, no. 7015, pp. 396–401, Nov. 2004, doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- [58].Zhang M et al. , “Identification of Tumor-initiating Cells in a p53 Null Mouse Model of Breast Cancer,” Cancer Res, vol. 68, no. 12, pp. 4674–4682, Jun. 2008, doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].van der Heijden M et al. , “Spatiotemporal regulation of clonogenicity in colorectal cancer xenografts,” Proc Natl Acad Sci U S A, vol. 116, no. 13, pp. 6140–6145, Mar. 2019, doi: 10.1073/pnas.1813417116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rogers ZN et al. , “A quantitative and multiplexed approach to uncover the fitness landscape of tumor suppression in vivo,” Nat Methods, vol. 14, no. 7, Art. no. 7, Jul. 2017, doi: 10.1038/nmeth.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Rogers ZN et al. , “Mapping the in vivo fitness landscape of lung adenocarcinoma tumor suppression in mice,” Nat Genet, vol. 50, no. 4, Art. no. 4, Apr. 2018, doi: 10.1038/s41588-018-0083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Walens A et al. , “Adaptation and selection shape clonal evolution of tumors during residual disease and recurrence,” Nature Communications, vol. 11, 2020, doi: 10.1038/s41467-020-18730-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Corsello SM et al. , “Discovering the anti-cancer potential of non-oncology drugs by systematic viability profiling,” Nat Cancer, vol. 1, no. 2, pp. 235–248, Feb. 2020, doi: 10.1038/s43018-019-0018-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Patwardhan GA, Marczyk M, Wali VB, Stern DF, Pusztai L, and Hatzis C, “Treatment scheduling effects on the evolution of drug resistance in heterogeneous cancer cell populations,” NPJ Breast Cancer, vol. 7, p. 60, May 2021, doi: 10.1038/s41523-021-00270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Xu J et al. , “Single-cell lineage tracing by endogenous mutations enriched in transposase accessible mitochondrial DNA,” Elife, vol. 8, p. e45105, Apr. 2019, doi: 10.7554/eLife.45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Velten L et al. , “Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics,” Nat Commun, vol. 12, no. 1, Art. no. 1, Mar. 2021, doi: 10.1038/s41467-021-21650-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Penter L, Gohil SH, and Wu CJ, “Natural Barcodes for Longitudinal Single Cell Tracking of Leukemic and Immune Cell Dynamics,” Front Immunol, vol. 12, p. 788891, Jan. 2022, doi: 10.3389/fimmu.2021.788891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Ludwig LS et al. , “Lineage Tracing in Humans Enabled by Mitochondrial Mutations and Single-Cell Genomics,” Cell, vol. 176, no. 6, pp. 1325–1339.e22, Mar. 2019, doi: 10.1016/j.cell.2019.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]