Abstract
Quantitative optical microscopy—an emerging, transformative approach to single-cell biology—has seen dramatic methodological advancements over the past few years. However, its impact has been hampered by challenges in the areas of data generation, management, and analysis. Here we outline these technical and cultural challenges and provide our perspective on the trajectory of this field, ushering in a new era of quantitative, data-driven microscopy. We also contrast it to the three decades of enormous advances in the field of genomics that have significantly enhanced the reproducibility and wider adoption of a plethora of genomic approaches.
Introduction
As we consider quantitative light microscopy, it is informative to compare it with the enormous success of genomics. The genomic revolution produced a breathtaking transformation in biomedical science, moving from a specialized field focused on single genes to a pervasive framework for understanding the biological and medical world with technologically sophisticated and accessible tools (Topol, 2014). Driving this transformation are the inexpensive next-generation sequencing of genomes, robust platforms for profiling gene expression, the construction of high-quality shared databases and repositories, the development of shared analysis tools, and publications showcasing novel use cases. Major successes include hundreds of genome-wide association studies (Tam et al., 2019; Uffelmann et al., 2021), the realization that most diseases are highly polyallelic (Lappalainen et al., 2019), and the finding that complex and intertwined changes in gene regulatory networks accompany disease, differentiation, and changes in cellular behavior (Chiliński et al., 2021). Innovation in these technologies has also greatly facilitated studying epigenetics (Lake et al., 2018), the 4D regulation of our genome (Zhou et al., 2021), and cell-to-cell heterogeneity via single-cell methodologies (Stuart and Satija, 2019). Three overarching conclusions from all these advances are that (1) there is a synergistic interplay between accessible technology development and scientific progress, (2) understanding and predicting cellular behaviors from genomics alone is challenging and requires augmentation from other sources, and (3) public archives and repositories for storage and dissemination of sequence data have been critical for advancing the field.
Light microscopy provides unique spatial and temporal information that can revolutionize our understanding of the relationships between genotype and pheno-type, but the high-quality production and quantitative analysis of cell and tissue images and the secondary use of this information have lagged behind the genomic sciences, due in part to inadequate funding. The potential of imaging to map, understand, and predict cellular behaviors from cell structure and organization across broad spatial and temporal scales is immense. Since its origins in 19th century pathology, optical microscopy has significantly benefited from breakthrough technologies that recently include high-resolution and high-throughput light microscopy (Bickle, 2010; Mattiazzi Usaj et al., 2016), genome editing for mutations and protein tagging (Wright et al., 2016), new visualization tools for volumetric rendering and interrogation (Lemon and McDole, 2020), and new statistical methods, including artificial intelligence for image analysis and integration (Moen et al., 2019). With genomic technologies that are increasingly capable of resolving or imputing spatial organization (Liu et al., 2020; Takei et al., 2021) and imaging technologies that are increasingly expanding their degree of multiplexing (Lewis et al., 2021), we believe there is an exciting, emerging convergence of approaches that will significantly change the future of quantitative microscopy.
These new imaging technologies promise to transform biomedical research at the cellular level and address fundamental questions from basic to translational science. Can cellular and subcellular organization predict cell behaviors and single-cell gene expression profiles? If not, do they afford more meaningful and useful assessments of cell behaviors when conjoined? Can a richer view of cellular organization lead to better disease diagnoses and prognoses? Over the next decade we see the goal of integrating multiscale image data, both optical and cryoEM (Nogales and Scheres, 2015; Turk and Baumeister, 2020), with other emerging single-cell ‘omics technologies (e.g., single-cell gene expression and proteomics), producing a rich, high replicate, quantitative, integrated atlas for studying and mining cellular phenomena. As these technologies develop and integrate with newer emerging technologies, they will need to be brought to scale, benchmarked, made interoperable, and democratized. Additionally, the large datasets that will be produced will need tools for management and sharing and will require the development of common formats, robust analytical tools, and consensus standards for evaluating analysis quality.
We have begun addressing these challenges through formal and informal conversations across various stakeholders, including relevant funding agencies, NGOs, journals, and research communities. Here, we outline the nature of short- and long-term challenges in quantitative cell imaging for the broad scientific community and provide our perspective on opportunities to enhance the growth of this emerging field in transforming cell biology (Figure 1).
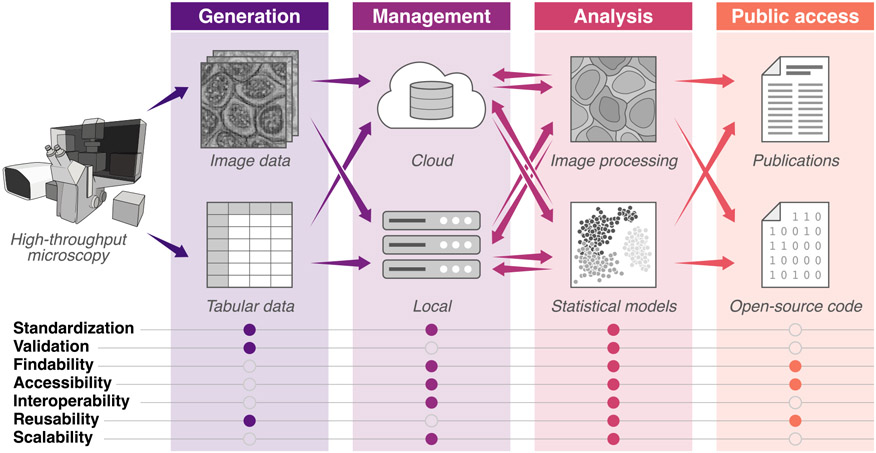
Figure 1. Current image experiment to analysis pipelines have many bottlenecks that hinder making data and tools findable, accessible, interoperable, and reusable (FAIR).
This illustration highlights ways in which the community can build consensus, incentives, and use cases across the imaging pipeline. Information transfer, from data generation through publication, is shown with arrows. Addressing the list of features in filled circles will enable the iterative sharing of data, resources, and tools in robust and easy-to-access repositories, which is critical to the growth of the quantitative cell imaging community and closes the loop between analysis, generation, and discovery.
Quality imaging data
Several recent breathtaking advances in fluorescence microscopy have opened new vistas in cell science. Piercing the diffraction barrier by super-resolution methods has opened the door to new insights into sub-cellular structures (Betzig et al., 2006; Gustafsson, 2000; Hell and Kroug, 1995; Hell and Wichmann, 1994). Light-sheet microscopes have greatly reduced the photodamage intrinsic to light microscopy (Reynaud et al., 2008), allowing vastly longer viewing periods and volumetric imaging. Automation software and robotic platforms bring higher reproducibility and throughput, providing high-content image databases (Bickle, 2010). In addition, a broader spectrum of genetically encoded tags and CRISPR-based gene editing methods for precise endogenous tagging allow multispectral imaging and minimize complications arising from perturbations (Roberts et al., 2017). Deep learning offers the ability to reduce, and even improve, human analysis of images (Moen et al., 2019). An increasing range of commercial, non-profit, grant-supported, and open-source approaches is democratizing access to these state-of-the-art technologies.
While these advances have greatly enhanced the use and potential of light microscopy, several barriers inhibit the large-scale data sharing, reuse, and analysis on the scale of genomic approaches. Sample preparation, user-controlled imaging methods and settings, and instrumentation platforms are far more variable and complex with microscopy than with genomics. Endogenously tagged lines, for example, require quality control to assess structural and functional perturbations as well as off-site genomic alterations. Likewise, cell lines labeled with exogenous fluorescent reporters require similar assessments. Studies using fixed and sectioned tissues, while used for decades, also present reproducibility challenges. A path to address these challenges starts with documented methods and moves toward a goal of community standards that ensure reproducibility both within and across labs. The Human Protein Atlas has developed such guidelines for antibody validation and reproducibility (Uhlen et al., 2016), and the Allen Institute for Cell Science is attempting to realize these goals by using endogenously tagged cell lines with documented quality control (Roberts et al., 2017).
Live cell imaging has the unique ability to explore the temporal domain of cellular processes; however, the challenges of bringing it to scale are significant. In addition to ensuring stable imaging conditions and monitoring phototoxicity during the duration of image capture, the volume of data generated (e.g., terabytes for lattice light sheet) can overwhelm most lab infrastructure for storage.
While researchers have been acquiring microscopic images for over a century, quantitative assessment and documentation of image quality and microscope acquisition parameters (e.g., illumination intensity and evenness across the field, magnification, resolution) are rare. Some inexpensive and relatively straight-forward methods to collect, benchmark, and assess image quality are not used widely, and more research is needed. The National Institute of Standards and Technology (NIST), for example, has several efforts for benchmarking optical microscopy methods including fluorescence imaging (https://nist.gov/programs-projects/measurement-assurance-quantitative-cell-imaging-optical-microscopy). Since 2020, the international community of light microscopists and the Quality Assessment and Reproducibility for Instruments & Images in Light Microscopy (QUAREP-LiMi) (Boehm et al., 2021) have been organizing to improve quality assessment and quality control in light microscopy. A key benefit for all stakeholders would be standardized and automated capture of sample and instrument metadata, as well as benchmarked evaluation of microscopy image quality, as DICOM does for medical imaging (https://dicomstandard.org). Ideally, such data would accompany each image file, along with the provenance of image data that specifies any image processing and analysis. Highly qualified data, with good metadata and benchmarks and assurance of data integrity, are necessary standards for sharing of image data to enable analysis with alternative methods and generate new hypotheses.
There are several impediments to standardized image data. The proprietary nature of the software driving the microscopes and accompanying image analysis software produces images in differing formats without details of how images have been processed. In this way, image data—raw and processed—cannot be readily shared, evaluated, and reproduced among labs. One solution is image format conversion software and analysis platforms that can accommodate images from the major manufacturers (e.g., Bio-Formats [Linkert et al., 2010], OMERO [Allan et al., 2012], and ImageIO [https://github.com/AllenCellModeling/aicsimageio]) and the use of microscope-control software (e.g., Micro-Manager [Edelstein et al., 2014]) that interfaces with most microscopes. Another solution is open-source microscope design (Gualda et al., 2013). An additional challenge is that the incorporation of more automation into imaging increases the size and number of image datasets. While most microscope analyses at the cell level tend to rely on a small number (10 s) of captured images, the enormous variance observed within samples dictates going beyond a small, typical image field and argues for larger datasets that sample larger numbers of cells (100–1000 s), both for better science and for statistical analyses. This is already routine in histological studies (Anderson and Badano, 2016; Lin et al., 2021).
The challenge is that the captured data need to be stored, with accompanying metadata, in a manner that can be readily queried and accessed. The increased interest and ability to collect time-lapse imaging of live cells will result in even larger datasets that also need to be transferred, annotated, and stored.
Thus, to produce high quality, shared datasets that can be easily studied and reused, the field will benefit from (1) standardized approaches for image quality and associated metadata, (2) innovative ways of democratizing access to imaging, image analysis platforms, and protocols, and (3) accessible platforms for microscope automation and data management. Our view is that the imaging community will need to support the many researchers actively developing these standards and platforms (http://nature.com/collections/djiciihhjh) and to work with research funders and journals to identify realistic incentives help implement them. The incentives are critical; for example, the OME project has advocated for standard microscopy formats from vendors for many years but is only recently getting traction (Linkert et al., 2010). Cultural change has been slow as well, so we see funder and journal mandates as a more effective mechanism to change the field.
FAIR and compliant imaging data management
Optical microscopy lags behind genomics, and even other biomedical imaging technologies, in the development of general-purpose data repositories. At the core of the challenge is the trade-off between incentives, value, and burden. The case for archiving data for general use is weakened by several factors, including (1) the inherent variety in terms of samples, sample preparation and staining schemes, instrumentation, and data formats, (2) the corresponding lack of sufficiently detailed and standardized metadata schema, (3) the lack of a strong community of data users, and (4) the perceived low value of older or poorly documented datasets.
There are many long-standing repositories and tools, including the Cell Image Library (https://cellimagelibrary.org) and Broad Bioimage Benchmark Collection (https://bbbc.broadinstitute.org) as well as specialized clinical repositories for pathology and ophthalmology and generalpurpose repositories for all types of biomedical imaging, such as The Cancer Imaging Archive (TCIA; https://cancerimagingarchive.net) (Clark et al., 2013). However, the trade-off between value and burden has dramatically changed for microscopy with the emergence of large datasets due to higher-throughput, higher-content, and higher-resolution imaging technologies, analytical methods that require large datasets, and large research programs with the mandate to make their products findable, accessible, interoperable, and reusable (FAIR) (Wilkinson et al., 2016).
The establishment of the EMBL-EBI Bio Image Archive (https://www.ebi.ac.uk/bioimage-archive/) and the associated Image Data Resource (https://idr.openmicroscopy.org) reflect the European Union’s wider investment in life science data management as part of the ERIC and Elixir programs. The Global BioImaging network (https://globalbioimaging.org), initiated in 2015, is bringing the international microscopy communities together to support capacity-building and to harmonize infrastructure, while targeted programs such as the Allen Institute for Cell Science (https://alleninstitute.org/what-we-do/cell-science), the Human Protein Atlas (https://proteinatlas.org), the Chan-Zuckerberg Biohub (Cho et al., 2021) (https://czbiohub.org), the HHMI-Janelia Farm’s Advanced Imaging Center (https://janelia.org/open-science/advanced-imaging-center-aic), and several NIH Programs (e.g., Human Biomolecular Atlas Program [Snyder et al., 2019], Human Tumor Atlas Network [Rozenblatt-Rosen et al., 2020], 4DNucleome [Dekker et al., 2017], and Kidney Precision Medicine Program [Ferkowicz et al., 2021]) are building, demonstrating, and disseminating full workflows for large-scale microscopy.
Still, many challenges remain that lead to trade-offs. Resources come at a cost and drive the economics of what to store, where to store it, and how to make it accessible. At one extreme, there is a desire to share all data. However, technologies like lightsheet microscopy can produce terabytes of data daily, creating a bottleneck for its upload, storage, transfer, and analysis. While tools such as BioFormats (https://openmicroscopy.org/bio-formats) are a boon for the interoperability of microscopy data formats, the lack of standards around how to record annotations, metadata associated with instruments, experimental conditions, and data pre-processing and how to report data quality hamper the sharing of imaging datasets and the accompanying raw data. Traditional data repositories are centralized on premises, though the rapid emergence of affordable cloud solutions make the potential for distributed repositories and data workbenches more practical. It is likely several economic models will emerge for supporting cloud repositories, potentially including fees for accessing and analyzing data, credits provided by funders, cloud provider-paid (for data deemed likely to generate use of their compute service; e.g., AWS OpenData), and tiered storage and access. If data users need to payfordata, it will potentially push data portals to provide interfaces that allow smarter identification of subsets of data based on metadata, curation, or modeling.
The research community, in partnership with funders and journals, could play an important role over the next five years to shape the trajectory of microscopy data management. We believe there needs to be a good balance of incentives and mandates for (1) data generators to prepare and upload interoperable datasets, (2) data managers to make data and tools FAIR, and (3) repositories to avoid siloing on data types or specific use cases. Beyond ensuring resources are FAIR, access to data repositories needs to be equitable, with sufficient annotation and documentation to assess quality and reproducibility. Support for algorithm sharing and testing needs to be bolstered, and a common understanding of what should be stored and for how long needs to emerge. The stakeholders, by working together, could address these challenges in a stepwise fashion, enabling insights awaiting discovery from broadly accessible secondary analyses of microscopy data.
Robust and interoperable tools for image analysis
The dramatic expansion of cell imaging from its initial focus on the careful production of small numbers of images probing a specific cell biological question to the production of high-volume datasets for mining and analysis is introducing many new opportunities and challenges. Recently, artificial intelligence and machine learning approaches have taken center stage in this effort because they enable processing, de-noising, compression, classification, visualization, integration, and comparison of large datasets. The key opportunity is the development of accessible, interoperable tools to analyze and interpret complex image data to extract new biological insights.
Many image analysis tools are developed in-house with the initial purpose of addressing lab and context-specific questions. Thus, each analysis pipeline tends to be unique. This decentralized analysis landscape—one in which each PI generates, manages, and analyzes data—does not scale and enable a community to easily share and modify tools. It disincentivizes the development and dissemination of efficient and modular code, the benchmarking of tools, and trust in data quality.
Another major challenge is the quality of in-house code, which is usually not readily FAIR. In contrast, professional code is well documented, maintained, hardened, updated (as software languages evolve), and packaged to be easily used by others. It also runs more efficiently, reducing computational expense. The requirements and demands for professional code development make it difficult to produce in most labs, which lack both incentives and appropriate personnel (e.g., software engineers and web designers) to produce it. Thus, there is a need to develop appropriate standards and create incentives or other mechanisms to develop high-quality shared software. At present, some large, openly accessible platforms are taking on that challenge. A collaboration between NIST and NCATS produced a web-based platform for data and software sharing where image data can be organized and operated on with plug-in analytical tools and where image processing and analysis provenance can be saved, facilitating data sharing (Bajcsy et al., 2018) (https://isg.nist.gov/deepzoomweb/home). CellProfiler (https://cellprofiler.org) and ImageJ/Fiji (https://imagej.github.io/software/fiji) are examples of long-standing image analysis platforms that are openly accessible and accommodate community tools. The napari platform is a community-built, open-source platform designed for large datasets (https://napari.org), and the Allen Cell Segmenter is an example of a high-quality 3D image analysis platform, with tutorials, that can stand alone or be plugged into larger platforms (http://www.allencell.org/segmenter). In contrast, pipeline languages and cloud containers have already moved genomic analysis to be more FAIR, a vision outlined for imaging a decade ago (Swedlow, 2012).
Once developed, analysis tools require large datasets for assessing accuracy and generalizability. This is particularly important for tools using artificial intelligence, including deep neural nets, where there is a need for ground truth training sets and benchmarks for assessing accuracy and biological validation. In the end, the value of the analysis depends on the quality and quantity of available data. In this light, there is a major need for well-validated, annotated datasets. While small collections (Ljosa et al., 2012) and individual large datasets exist, the deep learning revolution demands major efforts here to produce a large collection of large, annotated datasets. Perhaps datasets meeting standards could be given a special designation or funding incentives given for groups to generate or synthesize these datasets.
Finally, there are several kinds of data produced routinely—datasets developed for reuse or mining, and smaller one-off datasets used for validation or further analysis—each requiring some unique solutions. Establishing common standards for metadata and quality assessments will fortify trust and catalyze data reuse both within and across the scientific community (Hammer et al., 2021). Finally, the use of these datasets might be enhanced through competitions, which would drive development and bring in new investigators; they could also be used for cross comparison of tools (e.g., segmentation or classification). The Human Protein Atlas has shown how useful this kind of effort can be (Ouyang et al., 2019).
Cultural barriers
Over the past 20 years, genomics transformed from a molecule-based field of study to one highly dependent on computational approaches. Microscopy is just beginning this transformation, evolving from its qualitative, single-molecule origins into a highly quantitative, computational discipline producing and analyzing large, complex datasets with sophisticated statistical methods. In this light, we believe the future of imaging will be greatly accelerated by research organizations embracing and adapting to this new computational landscape (Way et al., 2021). This will require quantitatively focused and collaboratively oriented scientists to catalyze it. The transformation will also need to be supplemented with training in computational and statistical methods at the undergraduate, graduate, and professional levels. Exposing researchers to image analysis early on can help spur a cultural change toward quantitation of images, as well as broader computational fluency, bringing a greater diversity and integration of expertise from across the sciences. Bioimage analysis is a fantastic way to introduce students to computation because the results of algorithms can be seen concretely using public datasets. Furthermore, researchers entering the field can take advantage of the explosion of excellent online training materials for various bioimaging software packages (https://bioimagingna.org; https://youtube.com/c/neubias).
Outlook
Quantitative microscopy is likely to change significantly in the next decade and move beyond a method for studying spatial organization of biological systems toward a quantitative approach for tracking tens to hundreds of specific biomolecules, subcellular structures, and cells in space and time. However, as covered here, there are substantial barriers to progress throughout the workflow, particularly in data generation, management, and analysis. In addressing these challenges, there are lessons to be learned from genomics and medical imaging that can be tailored as solutions to accelerate wider adoption.
The field can move forward effectively by working in partnership with funders and journals to define better baseline standards and benchmarks to ensure rigor, reproducibility, and reuse(Figure 2).Toachievethese goals, there needs to be a balance of incentives and mandates in several areas, including comprehensively annotating and sharing data in standardized ways and professionalizing the development of code that supports lasting FAIR practices. The dissemination and reuse of imaging data will be greatly enhanced by a more standardized approach to the development and distribution of well-documented and hardened tools, workflows from published papers, tutorials, and hands-on work-shops. The development and sharing of high-quality benchmarked datasets, along with the democratization of in silico training datasets, will also empower individual researchers, entire labs, and new trainees to reuse and improve upon one another’s methods. Facilitating the transfer of innovative tools to new users will enable iterative improvements that generate robust analyses and future models that will carry the entire field forward.
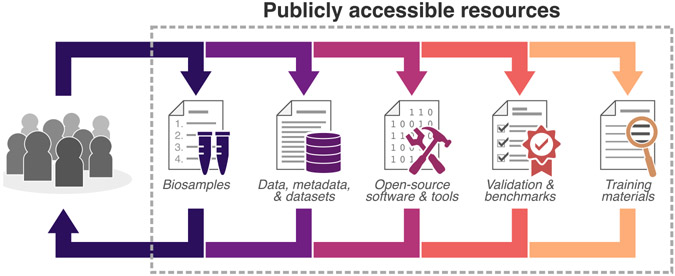
Figure 2. Community-established standards enable findable, accessible, interoperable, and reusable (FAIR) resources.
The consistent use of dynamic standards, benchmarks, and annotations for data generation and the sharing of these resources in repositories is critical to developing a quantitative cell imaging community. Supporting publicly available resources positions the community to build upon shared expertise and develop resources that will empower and engage a new wave of investigators and lower the barrier for productive collaborations.
There is already significant movement in this direction. The Network of European Bioimage Analysts (NEUBIAS; https://eubias.org/neubias), QUAREP (https://quarep.org), and BioImaging North America (BINA; https://bioimagingna.org) are examples of efforts that aim to build stronger communities of users and promote shared resources in image processing and analysis.
If the hurdles we describe can be overcome, the result will be a dramatic change in the way that images are used (and reused) in biology. As imaging becomes routinely higher-throughput and higher-resolution, automated feature extraction from images, particularly via deep learning, offers the potential to use images as raw material for discovery and insight. One may even envision autonomous (smart or learning) microscopes that merge with edge computing to allow on-camera data processing and smart data acquisition for truly data-driven quantitative imaging. From identifying how genes functionally impact each other in pathways to predicting the response of cells to previously untested compounds, and from modeling the localization of proteins and cellular machinery in silico to identifying causal relationships among them, there is much to be gained by treating microscopy as a data science. We think investments in this field will make linking cell morphology to pheno-types as computable as genomes and transcriptomes.
There is much to share and learn in this rapidly evolving field, and we hope the desire of stakeholders to work together will catalyze change during this transformative time.
ACKNOWLEDGMENTS
Illustration credit: Jessica S. Yu
N.B. and R.H. thank the Allen Institute for Cell Science founder, Paul G. Allen, for his vision, encouragement, and support. A.E.C. is supported by the National Institute of General Medical Sciences of the National Institutes of Health (R35 GM122547). E.L. is supported by the Knut and Alice Wallenberg Foundation, Sweden (KAW 2021.0346).
Any mention of commercial products by or within NIST web pages is for information only; it does not imply recommendation or endorsement by NIST.
REFERENCES
- Allan C, Burel J-M, Moore J, Blackburn C, Linkert M, Loynton S, Macdonald D, Moore WJ, Neves C, Patterson A, et al. (2012). OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9, 245–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson N, and Badano A (2016). Technical Performance Assessment of Digital Pathology Whole Slide Imaging Devices - Guidance for Industry and Food and Drug Administration Staff (U.S. Department of Health and Human Services, Food and Drug Administration; ). [Google Scholar]
- Bajcsy P, Chalfoun J, and Simon M (2018). Web Microanalysis of Big Image Data (Springer; ). [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, and Hess HF (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Bickle M. (2010). The beautiful cell: high-content screening in drug discovery. Anal. Bioanal. Chem 393, 219–226. [DOI] [PubMed] [Google Scholar]
- Boehm U, Nelson G, Brown CM, Bagley S, Bajcsy P, Bischof J, Dauphin A, Dobbie IM, Eriksson JE, Faklaris O, et al. (2021). QUAREP-LiMi: a community endeavor to advance quality assessment and reproducibility in light microscopy. Nat. Methods 18, 1423–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiliński M, Sengupta K, and Plewczynski D (2021). From DNA human sequence to the chromatin higher order organisation and its biological meaning: Using biomolecular interaction networks to understand the influence of structural variation on spatial genome organisation and its functional effect. Semin. Cell Dev. Biol S1084-9521(21)00211-1. 10.1016/j.semcdb.2021.08.007. [DOI] [PubMed] [Google Scholar]
- Cho NH, Cheveralls KC, Brunner A-D, Kim K, Michaelis AC, Raghavan P, Kobayashi H, Savy L, Li JY, Canaj H, et al. (2021). OpenCell: proteome-scale endogenous tagging enables the cartography of human cellular organization. bioRxiv. 10.1101/2021.03.29.437450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M, et al. (2013). The Cancer Imaging Archive (TCIA): maintaining and operating a public information repository. J. Digit. Imaging 26, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B, et al. ; 4D Nucleome Network (2017). The 4D nucleome project. Nature 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, and Stuurman N (2014). Advanced methods of microscope control using μManager software. J. Biol. Methods 1, e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferkowicz MJ, Winfree S, Sabo AR, Kamocka MM, Khochare S, Barwinska D, Eadon MT, Cheng Y-H, Phillips CL, Sutton TA, et al. ; Kidney Precision Medicine Project (2021). Large-scale, three-dimensional tissue cytometry of the human kidney: a complete and accessible pipeline. Lab. Invest 101, 661–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualda EJ, Vale T, Almada P, Feijó JA, Martins GG, and Moreno N (2013). OpenSpinMicroscopy: an open-source integrated microscopy platform. Nat. Methods 10, 599–600. [DOI] [PubMed] [Google Scholar]
- Gustafsson MGL (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc 198, 82–87. [DOI] [PubMed] [Google Scholar]
- Hammer M, Huisman M, Rigano A, Boehm U, Chambers JJ, Gaudreault N, North AJ, Pimentel JA, Sudar D, Bajcsy P, et al. (2021). Towards community-driven metadata standards for light microscopy: tiered specifications extending the OME model. Nat. Methods 18, 1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell SW, and Kroug M (1995). Ground-state-depletion fluorscence microscopy: A concept for breaking the diffraction resolution limit. Appl. Phys. B 60, 495–497. [Google Scholar]
- Hell SW, and Wichmann J (1994). Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett 19, 780–782. [DOI] [PubMed] [Google Scholar]
- Lake BB, Chen S, Sos BC, Fan J, Kaeser GE, Yung YC, Duong TE, Gao D, Chun J, Kharchenko PV, and Zhang K (2018). Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat. Biotechnol 36, 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Scott AJ, Brandt M, and Hall IM (2019). Genomic Analysis in the Age of Human Genome Sequencing. Cell 177, 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon WC, and McDole K (2020). Live-cell imaging in the era of too many microscopes. Curr. Opin. Cell Biol 66, 34–42. [DOI] [PubMed] [Google Scholar]
- Lewis SM, Asselin-Labat M-L, Nguyen Q, Berthelet J, Tan X, Wimmer VC, Merino D, Rogers KL, and Naik SH (2021). Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 18, 997–1012. [DOI] [PubMed] [Google Scholar]
- Lin J-R, Wang S, Coy S, Tyler M, Yapp C, Chen Y-A, Heiser CN, Lau KS, Santagata S, and Sorger PK (2021). Multiplexed 3D atlas of state transitions and immune interactions in colorectal cancer. bioRxiv. 10.1101/2021.03.31.437984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linkert M, Rueden CT, Allan C, Burel J-M, Moore W, Patterson A, Loranger B, Moore J, Neves C, Macdonald D, et al. (2010). Metadata matters: access to image data in the real world. J. Cell Biol 189, 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Yang M, Deng Y, Su G, Enninful A, Guo CC, Tebaldi T, Zhang D, Kim D, Bai Z, et al. (2020). High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665–1681.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljosa V, Sokolnicki KL, and Carpenter AE (2012). Annotated high-throughput microscopy image sets for validation. Nat. Methods 9, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiazzi Usaj M, Styles EB, Verster AJ, Friesen H, Boone C, and Andrews BJ (2016). High-Content Screening for Quantitative Cell Biology. Trends Cell Biol. 26, 598–611. [DOI] [PubMed] [Google Scholar]
- Moen E, Bannon D, Kudo T, Graf W, Covert M, and Van Valen D (2019). Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E, and Scheres SHW (2015). Cryo-EM: A Unique Tool for the Visualization of Macromolecular Complexity. Mol. Cell 58, 677–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang W, Winsnes CF, Hjelmare M, Cesnik AJ, Åkesson L, Xu H, Sullivan DP, Dai S, Lan J, Jinmo P, et al. (2019). Analysis of the Human Protein Atlas Image Classification competition. Nat. Methods 16, 1254–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Kržič U, Greger K, and Stelzer EHK (2008). Light sheet-based fluorescence microscopy: more dimensions, more photons, and less photodamage. HFSP J. 2, 266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B, Haupt A, Tucker A, Grancharova T, Arakaki J, Fuqua MA, Nelson A, Hookway C, Ludmann SA, Mueller IA, et al. (2017). Systematic gene tagging using CRISPR/Cas9 in human stem cells to illuminate cell organization. Mol. Biol. Cell 28, 2854–2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, Ashenberg O, Cerami E, Coffey RJ, Demir E, et al. ; Human Tumor Atlas Network (2020). The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 181,236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder MP, Lin S, Posgai A, Atkinson M, Regev A, Rood J, Rozenblatt-Rosen O, Gaffney L, Hupalowska A, Satija R, et al. ; HuBMAP Consortium (2019). The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, and Satija R (2019). Integrative single-cell analysis. Nat. Rev. Genet 20, 257–272. [DOI] [PubMed] [Google Scholar]
- Swedlow JR (2012). Innovation in biological microscopy: current status and future directions. BioEssays 34, 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takei Y, Yun J, Zheng S, Ollikainen N, Pierson N, White J, Shah S, Thomassie J, Suo S, Eng CL, et al. (2021). Integrated spatial genomics reveals global architecture of single nuclei. Nature 590, 344–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam V, Patel N, Turcotte M, Bossé Y, Paré G, and Meyre D (2019). Benefits and limitations of genome-wide association studies. Nat. Rev. Genet 20, 467–484. [DOI] [PubMed] [Google Scholar]
- Topol EJ (2014). Individualized medicine from prewomb to tomb. Cell 157, 241–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk M, and Baumeister W (2020). The promise and the challenges of cryo-electron tomography. FEBS Lett. 594, 3243–3261. [DOI] [PubMed] [Google Scholar]
- Uffelmann E, Huang QQ, Munung NS, de Vries J, Okada Y, Martin AR, Martin HC, Lappalainen T, and Posthuma D (2021). Genome-wide association studies. Nat. Rev. Methods Primer 1, 1–21. [Google Scholar]
- Uhlen M, Bandrowski A, Carr S, Edwards A, Ellenberg J, Lundberg E, Rimm DL, Rodriguez H, Hiltke T, Snyder M, and Yamamoto T (2016). A proposal for validation of antibodies. Nat. Methods 13, 823–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Way GP, Greene CS, Carninci P, Carvalho BS, de Hoon M, Finley SD, Gosline SJC, Lê Cao KA, Lee JSH, Marchionni L, et al. (2021). A field guide to cultivating computational biology. PLoS Biol. 19, e3001419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten JW, da Silva Santos LB, Bourne PE, et al. (2016). The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AV, Nuñez JK, and Doudna JA (2016). Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 164, 29–44. [DOI] [PubMed] [Google Scholar]
- Zhou T, Zhang R, and Ma J (2021). The 3D Genome Structure of Single Cells. Annu Rev Biomed Data Sci 4, 21–41. [DOI] [PubMed] [Google Scholar]