Abstract
Simple Summary
A new species of snail-eating snake in the genus Pareas is described, from the Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China, based on one male and three juvenile specimens. The maximum likelihood analyses based on Cytochrome b (Cyt b) and NADH dehydrogenase subunit 4 (ND4) indicated the new taxon is different from its congeners. Morphologically, the new species can be diagnosed from the other species by a combination of seven characters. The recognition of the new species brings the number of described Pareas species to 30.
Abstract
We described a new species of genus Pareas from Baise City, Guangxi Zhuang Autonomous Region, China, based on morphological and molecular evidence. Pareas baiseensis sp. nov. is distinguished from its congeners by the combination of (1) Yellowish-brown body colouration; (2) Frontal subhexagonal to diamond-shaped with its lateral sides converging posteriorly; (3) The anterior pair of chin shields is longer than it is broad; (4) Loreal not in contact with the eye, prefrontal in contact with the eye, two or three suboculars; (5) Rows of 15–15–15 dorsal scales, five rows of mid-dorsal scales keeled at the middle of the body, one vertebral scale row enlarged; (6) 187–191 ventrals, 89–97 subcaudals, all divided, cloacal plate single; (7) Two postocular stripes, the nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. The genetic divergence (uncorrected p-distance) between the new species and other representatives of Pareas ranged from 13.9% to 24.4% for Cytochrome b (Cyt b) and 12.1% to 25.5% for NADH dehydrogenase subunit 4 (ND4). Phylogenetic analyses of mitochondrial DNA gene data recovered the new species from being the sister taxon to (P. boulengeri + P. chinensis) from China.
Keywords: snail-eating snake, Pareas baiseensis sp. nov., taxonomy, phylogeny, morphology
1. Introduction
The family Pareidae Romer, 1956 (Squamata, Serpentes) encompasses four genera [1,2], respectively, namely Aplopeltura Duméril, 1853, Asthenodipsas Peters, 1864, Pareas Wagler, 1830, and Xylophis Beddome, 1878 [3,4]. The Asian snail-eating genus Pareas Wagler, 1830, is widely distributed through the Oriental zoogeographic region and is the most species-diverse genus in the Pareidae. It differs from other Pareidae genera by having 15 rows of dorsal scales at the midbody; all subcaudals divided; preocular and subocular scales present; supralabials usually not in contact with the eye; no anterior single inframaxillary, and three pairs of chin shields [4,5,6]. The morphology of snakes of the genus Pareas is highly conservative, and the morphological differences between the species are subtle and difficult to distinguish. Recent studies have demonstrated that the species diversity of the genus Pareas has been seriously underestimated [4,7,8,9]. Since 2015, based on integrative taxonomic approaches incorporating molecular analyses and morphological comparisons, twelve new species have been described, and seven species have been resurrected [4,6,8,9,10,11,12,13,14,15].
During our field research in the Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China, in November 2022, we collected four specimens of Pareas that differed from all members of the genus. Based on conclusive morphological and molecular evidence, we describe them as a new species.
2. Materials and Methods
2.1. Sampling
The four individuals of snail-eating snakes (one male, three juveniles) were collected from Daleng Township, Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China, in November 2022. The collected specimens were fixed in approximately 95% ethanol and subsequently transferred to 75% ethanol for permanent storage. Liver tissue samples were preserved separately in 95% ethanol. The specimens examined in the present study were preserved and deposited at Anhui Normal University Museum (ANU).
2.2. Morphological Examination
Referring to the following literature, Dowling 1951, Vogel 2015, Wang et al., 2020, and Poyarkov et al., 2022 [4,6,11,16], a total of 21 morphological characters in four specimens of the new species were examined (Table 1). Morphological measurements (all in mm) included: snout-vent length (SVL); tail length (TaL); total length (TL); relative tail length (TaL/TL); head length from snout tip to jaw angles (HL); maximal head width (HW); and eye diameter (ED). Meristic characteristics evaluated were the number of dorsal scale rows counted at one head length behind head (ASR), at mid-body (MSR), namely at SVL/2, and at one head length before vent (PSR); number of enlarged vertebral scale rows (VSE); number of keeled dorsal scale rows at midbody (KMD); number of ventral scales (VEN); number of subcaudal scales (SC); number of cloacal plates (CP); number of supralabials (SL); number of infralabials (IL); number of anterior temporals (At); number of posterior temporals (Pt); number of loreals (LOR); number of preoculars (Preoc); number of suboculars (SoO); and number of postoculars (PoO). We recorded the values for paired head characteristics on both sides of the head (in a left/right order). We measured body and tail lengths with a measuring tape (to the nearest of 1 mm); all other measurements were taken using an electronic slide caliper (to the nearest 0.1 mm).
Table 1.
Samples used for molecular phylogenetic analysis in this study.
| No | Specimen ID | Species | Locality | cyt b | ND4 |
|---|---|---|---|---|---|
| 1 | ANU000220008 = HSR22185 | Pareas baiseensis sp. nov. | Baise, Guangxi, China | OQ054329 | OQ054328 |
| 2 | ANU000220009 = HSR22186 | Pareas baiseensis sp. nov. | Baise, Guangxi, China | OQ054329 | OQ054328 |
| 3 | ANU000220010 = HSR22188 | Pareas baiseensis sp. nov. | Baise, Guangxi, China | OQ054329 | OQ054328 |
| 4 | ANU000220011 = HSR22189 | Pareas baiseensis sp. nov. | Baise, Guangxi, China | OQ054329 | OQ054328 |
| 5 | NMNS 05618 | P. komaii | Taiwan, Taitung, Lijia | KJ642185 | MW287056 |
| 6 | NMNS 05625 | P. komaii | Taiwan, Hualien | MZ712215 | MZ712240 |
| 7 | NMNS 05655 | P. iwasakii | Japan, Okinawa, Ishigaki | KJ642160 | — |
| 8 | NMNS 05654 | P. iwasakii | Japan, Okinawa, Iriomote | MZ712216 | — |
| 9 | NMNS 05594 | P. atayal | Taiwan, Taoyuan, Beiheng | KJ642124 | MW287041 |
| 10 | CAS 235254 | P. victorianus | Myanmar, Chin, Nat Ma Taung N.P. | MW438300 | MW438302 |
| 11 | KIZ 014167 | P. monticola | China, Tibet (Xizang), Motuo | MK135109 | MK805374 |
| 12 | ZMMU R-16631 | P. monticola | Myanmar, Sagaing, Ban Mauk | MW438296 | MW438301 |
| 13 | CAS235359 | P. andersonii | Myanmar, Chin, Nat Ma Taung N.P. | MT968772 | MW287040 |
| 14 | ZMMU R-16628 | P. macularius | Laos, Xaisomboun, Long Tien | MT968770 | MZ712241 |
| 15 | ZMMU R-16629 | P. macularius | Myanmar, Sagaing, Ban Mauk | MT968771 | MW287057 |
| 16 | MZMU1293 | P. modestus | India, Mizoram, Aizawl, Tanhril | MT968773 | — |
| 17 | ZMMU R-13451 | P. margaritophorus | Vietnam, Binh Puoc, Bu Gia Map N.P. | KJ642195 | MW287058 |
| 18 | ZMMU NAP-09759 | P. margaritophorus | Thailand, Ratchaburi, Suan Phueng | MZ712217 | MZ712243 |
| 19 | KIZ 09966 | P. boulengeri | China, Hubei, Jiannan | JF827678 | JF827656 |
| 20 | GP 2923 | P. boulengeri | China, Guizhou, Jiangkou | MK135090 | MK805355 |
| 21 | CIB 010140 | P. chinensis | China, Sichuan, Tianquan | JF827691 | JF827669 |
| 22 | HM 2007-S001 | P. stanleyi | China, Guangxi, Guilin | JN230704 | JN230705 |
| 23 | CAS 248147 | P. vindumi | Myanmar, Kachin, Lukpwi | MW287080 | MW287059 |
| 24 | CHS 656 | P. nigriceps | China, Yunnan, Gaoligongshan N.R. | MK201455 | — |
| 25 | BNHS 3575 | P. kaduri | India, Arunachal Pradesh, Lohit, Kamlang W.S. | MT188734 | — |
| 26 | BNHS 3574 | P. kaduri | India, Arunachal Pradesh, Lohit, Kamlang W.S. | MW026190 | — |
| 27 | ZMMU NAP-09088 | P. hamptoni | Vietnam, Lao Cai, Bat Xat | MW287079 | MW287053 |
| 28 | YPX 18219 | P. hamptoni | Myanmar, Kachin | MK135077 | MK805342 |
| 29 | ZMMU R-16478 | P. geminatus | Thailand, Chiang Mai | MW287074 | MW287050 |
| 30 | KIZ-XL1 | P. xuelinensis | China, Yunnan, Lancang, Xuelin | MW436709 | — |
| 31 | NMNS 05637 | P. formosensis | Taiwan, Nantou | MW287060 | MW287042 |
| 32 | ZMMU R-16333 | P. formosensis | Vietnam, Gia Lai, Kon Chu Rang N.R. | MW287066 | MW287048 |
| 33 | GP 4122 | P. niger | China, Yunnan, Kunming | MK135084 | MK805349 |
| 34 | AUP 01573 | P. berdmorei berdmorei | Thailand, Chiang Mai | MZ712218 | MZ712244 |
| 35 | ZMMU R-13753-2 | P. berdmorei unicolor | Vietnam, Dong Nai, Ma Da N.R. | MZ712224 | MZ712250 |
| 36 | ZMMU R-16802 | P. kuznetsovorum | Vietnam, Phu Yen, Song Hinh | MZ712232 | MZ712258 |
| 37 | CAS 247982 | P. carinatus tenasserimicus | Myanmar, Tanintharyi, Yaephyu | MZ712233 | MZ712259 |
| 38 | KIZ 011972 | P. carinatus | Malaysia (peninsular) | MK135111 | MK805376 |
| 39 | ZMMU R-16393 | P. abros | Vietnam, Quang Nam, Song Thanh N.P. | MZ712235 | MZ712262 |
| 40 | ZMMU R-14788 | P. abros | Vietnam, Thua Thien-Hue, A Roang | MZ712237 | MZ712264 |
| 41 | ZMMU R-13656 | P. temporalis | Vietnam, Lam Dong, Cat Loc | MZ712238 | MZ712265 |
| 42 | FK 2626 | P. nuchalis | Brunei, Brunei Darussalam, Belait | MZ603794 | U49311 |
| 43 | LSUHC 7248 | Aplopeltura boa | Malaysia, Sabah, Sepilok | KC916746 | U49312 |
| 44 | FMNH 241296 | Asthenodipsas laevis | Malaysia, Penang, Pulau Pinang | KX660468 | KX660596 |
| 45 | LSUHC 9098 | Asthenodipsas lasgalenensis | Malaysia, Pahang, Fraser’s Hill | KC916755 | MZ712267 |
| 46 | FMNH 273617 | Asthenodipsas borneensis | Malaysia, Sarawak, Kelabit Highlands, Bario | KX660469 | KX660597 |
| 47 | BNHS 3376 | Xylophis captaini | India | MK340914 | MK340912 |
| 48 | BNHS 3582 | Xylophis perroteti | India | MN970042 | MN970046 |
2.3. Molecular Phylogeny
Total genomic DNA was extracted from an ethanol-preserved liver or muscle tissue using Tissue DNA Kits (Takara Biotechnology (Dalian) Co., Ltd., Dalian, China). We amplified the fragments of cytochrome b (cyt b; primer L14910, H16064; Queiroz et al., 2002) and NADH dehydrogenase subunit 4 (ND4; primer ND4F, ND4LEUR; Salvi et al. 2013) mtDNA genes, using the Polymerase Chain Reaction (PCR) [17,18]. The PCR products were sequenced at Shanghai Map Biotech Co., Ltd. The raw sequences were stitched using SeqMan in the DNAstar software package [19]. In addition to sequences of 26 species of the genus Pareas, six outgroup taxa [3,4,11,19,20,21,22,23,24] were downloaded from GenBank (Details on taxonomy, localities, GenBank accession numbers in Table 1) and aligned with the newly generated sequences using the software MEGA X [25].
A maximum likelihood (ML) tree was reconstructed using RaxML v7.2.6 using the GTRGAMMA model with 1000 ultrafast bootstrap (BS) replicates [26,27]. We also calculated the pairwise distances (p-distances) among ingroup taxa using the neighbour-joining method in MEGA X [25,28].
The electronic version of this article in Portable Document Format (PDF) will represent a published work according to the International Commission on Zoological Nomenclature (ICZN), and hence the new names contained in the electronic version are effectively published under that Code from the electronic edition alone. This published work and the nomenclatural acts it contains have been registered in ZooBank, the online registration system for the ICZN. The ZooBank LSIDs (Life Science Identifiers) can be resolved, and the associated information can be viewed through any standard web browser by appending the LSID to the prefix http://zoobank.org/ (accessed on 8 June 2023). The LSID for this publication is urn:lsid:zoobank.org:pub:73634CF1-1ACA-4C07-B8A2-3929E2306558.
3. Results
3.1. Phylogenetic Relationship
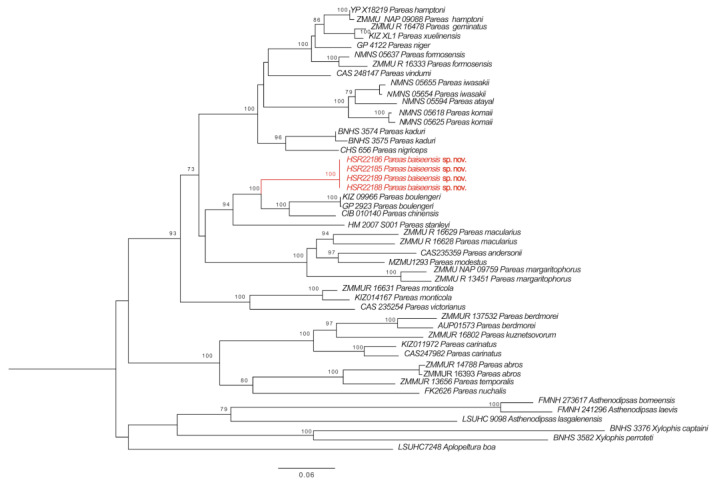
The newly generated sequences of Cyt b and ND4 genes of four specimens shared one haplotype for each gene. The sequences were submitted to GenBank (accession numbers, OQ054328 for Cyt b, OQ054329 for ND4). The p-distances based on fragments of Cyt b between the new species and other species of the genus Pareas varied from 13.9% (Pareas boulengeri) to 24.4% (Pareas carinatus) (Table 2), and those of ND4 varied from 12.1% (Pareas boulengeri) to 25.5% (Pareas berdmorei) (Table 3). Phylogenetic analyses of mitochondrial DNA data recovered the new species to be the sister taxon to (P. boulengeri + P. chinensis) from China (Figure 1). Combined with morphological data, the specimens from Baise, Guangxi, China, are considered to be a new species.
Table 2.
Uncorrected p-distance based on a fragment of Cyt b among the genus Pareas.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. P. baiseensis sp. nov. | — | ||||||||||||||
| 2. P. komaii | 0.194–0.200 | 0.017 | |||||||||||||
| 3. P. iwasakii | 0.192–0.194 | 0.078–0.083 | 0.010 | ||||||||||||
| 4. P. atayal | 0.205 | 0.085–0.093 | 0.069–0.072 | — | |||||||||||
| 5. P. victorianus | 0.189 | 0.187–0.192 | 0.194–0.198 | 0.194 | — | ||||||||||
| 6. P. monticola | 0.203–0.206 | 0.180–0.187 | 0.174–0.180 | 0.174–0.178 | 0.144–0.152 | 0.040 | |||||||||
| 7. P. andersonii | 0.207 | 0.194–0.195 | 0.199–0.205 | 0.201 | 0.209 | 0.191–0.192 | — | ||||||||
| 8. P. macularius | 0.187–0.194 | 0.181–0.194 | 0.191–0.197 | 0.189–0.193 | 0.192–0.193 | 0.173–0.176 | 0.135–0.145 | 0.114 | |||||||
| 9. P. modestus | 0.194 | 0.176–0.180 | 0.191–0.194 | 0.180 | 0.192 | 0.182–0.188 | 0.117 | 0.102–0.118 | — | ||||||
| 10. P. margaritophorus | 0.200–0.205 | 0.186–0.193 | 0.185–0.195 | 0.188–0.192 | 0.208–0.212 | 0.193–0.197 | 0.151–0.159 | 0.135–0.146 | 0.139–0.141 | 0.047 | |||||
| 11. P. boulengeri | 0.139–0.141 | 0.178–0.181 | 0.173 | 0.181 | 0.193 | 0.185–0.188 | 0.198 | 0.183–0.186 | 0.191–0.193 | 0.190–0.192 | 0.002 | ||||
| 12. P. chinensis | 0.146 | 0.181–0.185 | 0.177–0.181 | 0.187 | 0.176 | 0.182–0.186 | 0.192 | 0.176–0.179 | 0.188 | 0.187–0.192 | 0.089–0.090 | — | |||
| 13. P. stanleyi | 0.160 | 0.172–0.177 | 0.182–0.187 | 0.191 | 0.190 | 0.189–0.194 | 0.206 | 0.186–0.194 | 0.194 | 0.188–0.194 | 0.155 | 0.153 | — | ||
| 14. P. vindumi | 0.192 | 0.150 | 0.145–0.148 | 0.149 | 0.176 | 0.178–0.182 | 0.207 | 0.189–0.191 | 0.194 | 0.195–0.198 | 0.182 | 0.175 | 0.192 | — | |
| 15. P. nigriceps | 0.169 | 0.162 | 0.159–0.161 | 0.157 | 0.191 | 0.188–0.191 | 0.188 | 0.183–0.193 | 0.164 | 0.178–0.19 | 0.169 | 0.162 | 0.190 | 0.123 | — |
| 16. P. kaduri | 0.196–0.207 | 0.155–0.164 | 0.149–0.156 | 0.147–0.159 | 0.186–0.193 | 0.189–0.194 | 0.203–0.212 | 0.200–0.210 | 0.192–0.195 | 0.199–0.215 | 0.192–0.202 | 0.188–0.199 | 0.202–0.208 | 0.125–0.135 | 0.097–0.104 |
| 17. P. hamptoni | 0.184–0.187 | 0.141–0.147 | 0.134–0.139 | 0.139 | 0.182–0.183 | 0.186–0.189 | 0.214 | 0.188–0.194 | 0.194 | 0.192–0.200 | 0.166–0.169 | 0.181 | 0.183–0.184 | 0.115–0.116 | 0.125–0.126 |
| 18. P. geminatus | 0.200 | 0.152–0.153 | 0.135–0.139 | 0.142 | 0.192 | 0.194–0.198 | 0.219 | 0.192–0.199 | 0.206 | 0.199–0.202 | 0.169–0.170 | 0.187 | 0.193 | 0.122 | 0.132 |
| 19. P. xuelinensis | 0.201 | 0.147 | 0.133–0.138 | 0.137 | 0.186 | 0.194–0.197 | 0.213 | 0.195–0.198 | 0.202 | 0.196–0.200 | 0.165–0.166 | 0.185 | 0.192 | 0.124 | 0.125 |
| 20. P. formosensis | 0.189–0.195 | 0.147–0.154 | 0.133–0.148 | 0.145–0.149 | 0.175–0.184 | 0.188–0.200 | 0.212–0.214 | 0.193–0.201 | 0.198–0.203 | 0.192–0.196 | 0.165–0.173 | 0.171–0.181 | 0.193–0.195 | 0.115–0.120 | 0.125–0.133 |
| 21. P. niger | 0.187 | 0.146–0.149 | 0.133–0.138 | 0.142 | 0.176 | 0.187–0.190 | 0.207 | 0.189–0.191 | 0.188 | 0.196–0.198 | 0.174–0.175 | 0.176 | 0.194 | 0.110 | 0.126 |
| 22. P. berdmorei | 0.227–0.230 | 0.235–0.240 | 0.230–0.245 | 0.230–0.236 | 0.220–0.224 | 0.212–0.223 | 0.231–0.39 | 0.216–0.227 | 0.233–0.237 | 0.228–0.241 | 0.231–0.232 | 0.233–0.248 | 0.249–0.250 | 0.238–0.242 | 0.224–0.229 |
| 23. P. kuznetsovorum | 0.235 | 0.239–0.241 | 0.236–0.241 | 0.229 | 0.226 | 0.221–0.225 | 0.233 | 0.216–0.222 | 0.238 | 0.227–0.232 | 0.220–0.222 | 0.230 | 0.249 | 0.231 | 0.239 |
| 24. P. carinatus | 0.229–0.243 | 0.236–0.242 | 0.231–0.245 | 0.229–0.236 | 0.226–0.228 | 0.223–0.227 | 0.225–0.237 | 0.216–0.228 | 0.237–0.245 | 0.226–0.242 | 0.216–0.221 | 0.227–0.231 | 0.243–0.244 | 0.238–0.243 | 0.229–0.231 |
| 25. P. abros | 0.243–0.244 | 0.228–0.232 | 0.225–0.233 | 0.226–0.227 | 0.242–0.244 | 0.227–0.229 | 0.234–0.235 | 0.246–0.242 | 0.231–0.232 | 0.241–0.250 | 0.233–0.234 | 0.238 | 0.255–0.257 | 0.244–0.245 | 0.232–0.236 |
| 26. P. temporalis | 0.239 | 0.240 | 0.227–0.234 | 0.234 | 0.246 | 0.218–0.231 | 0.233 | 0.231–0.233 | 0.231 | 0.242–0.248 | 0.224–0.225 | 0.219 | 0.242 | 0.251 | 0.241 |
| 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | 26 | 27 | ||||
| 1. P. baiseensis sp. nov. | |||||||||||||||
| 2. P. komaii | |||||||||||||||
| 3. P. iwasakii | |||||||||||||||
| 4. P. atayal | |||||||||||||||
| 5. P. victorianus | |||||||||||||||
| 6. P. monticola | |||||||||||||||
| 7. P. andersonii | |||||||||||||||
| 8. P. macularius | |||||||||||||||
| 9. P. modestus | |||||||||||||||
| 10. P. margaritophorus | |||||||||||||||
| 11. P. boulengeri | |||||||||||||||
| 12. P. chinensis | |||||||||||||||
| 13. P. stanleyi | |||||||||||||||
| 14. P. vindumi | |||||||||||||||
| 15. P. nigriceps | |||||||||||||||
| 16. P. kaduri | 0.015 | ||||||||||||||
| 17. P. hamptoni | 0.125–0.136 | 0.005 | |||||||||||||
| 18. P. geminatus | 0.139–0.151 | 0.083–0.084 | — | ||||||||||||
| 19. P. xuelinensis | 0.133–0.141 | 0.078–0.082 | 0.027 | — | |||||||||||
| 20. P. formosensis | 0.128–0.141 | 0.072–0.074 | 0.087–0.096 | 0.080–0.088 | 0.041 | ||||||||||
| 21. P. niger | 0.122–0.132 | 0.056–0.060 | 0.076 | 0.073 | 0.071–0.079 | — | |||||||||
| 22. P. berdmorei | 0.242–0.251 | 0.227–0.246 | 0.245–0.255 | 0.244–0.251 | 0.241–0.248 | 0.230–0.236 | 0.071 | ||||||||
| 23. P. kuznetsovorum | 0.228–0.233 | 0.230–0.231 | 0.247 | 0.243 | 0.230–0.232 | 0.223 | 0.121–0.125 | — | |||||||
| 24. P. carinatus | 0.221–0.244 | 0.231–0.232 | 0.237–0.242 | 0.237–0.241 | 0.233–0.240 | 0.229–0.233 | 0.130–0.149 | 0.128–0.133 | 0.080 | ||||||
| 25. P. abros | 0.248–0.257 | 0.231–0.236 | 0.231–0.232 | 0.230–0.231 | 0.231–0.236 | 0.226–0.227 | 0.208–0.218 | 0.206–0.209 | 0.216–0.230 | 0.015 | |||||
| 26. P. temporalis | 0.249–0.252 | 0.237–0.238 | 0.248 | 0.246 | 0.234–0.240 | 0.236 | 0.203–0.209 | 0.196 | 0.194–0.197 | 0.127–0.132 | — | ||||
| 27. P. nuchalis | 0.253–0.26 | 0.250 | 0.264 | 0.260 | 0.240–0.248 | 0.254 | 0.210 | 0.203 | 0.206–0.2 | 0.212–0.21 | 0.201 | — |
Table 3.
Uncorrected p-distance based on a fragment of ND4 among the genus Pareas.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. P. baseensis sp. nov | — | |||||||||||||||||||||
| 2. P. komaii | 0.208 | — | ||||||||||||||||||||
| 3. P. ata yal | 0.202 | 0.076 | — | |||||||||||||||||||
| 4. P. victorianus | 0.184 | 0.189 | 0.183 | — | ||||||||||||||||||
| 5. P. monticola | 0.186–0.193 | 0.205–0.211 | 0.189–0.198 | 0.127–0.139 | 0.056 | |||||||||||||||||
| 6. P. andersonii | 0.180 | 0.195 | 0.189 | 0.183 | 0.186–0.194 | — | ||||||||||||||||
| 7. P. macularius | 0.186–0.189 | 0.201–0.207 | 0.195–0.196 | 0.196–0.204 | 0.195–0.215 | 0.120 | 0.103 | |||||||||||||||
| 8. P. margaritophorus | 0.189 | 0.201–0.210 | 0.196–0.201 | 0.189–0.201 | 0.193–0.213 | 0.139–0.147 | 0.147–0.153 | 0.071 | ||||||||||||||
| 9. P. boulengeri | 0.121 | 0.190 | 0.184 | 0.180 | 0.178–0.181 | 0.179 | 0.181–0.196 | 0.177–0.187 | — | |||||||||||||
| 10. P. chinensis | 0.122 | 0.192 | 0.021 | 0.184 | 0.174–0.184 | 0.182 | 0.193–0.201 | 0.190–0.208 | 0.101 | — | ||||||||||||
| 11. P. stanleyi | 0.162 | 0.186 | 0.195 | 0.194 | 0.192–0.200 | 0.179 | 0.179–0.183 | 0.195–0.204 | 0.148 | 0.166 | — | |||||||||||
| 12. P. vindumi | 0.148 | 0.142 | 0.139 | 0.154 | 0.174–0.175 | 0.161 | 0.178 | 0.172–0.184 | 0.156 | 0.157 | 0.165 | — | ||||||||||
| 13. P. hamptoi | 0.174–0.177 | 0.172–0.175 | 0.163 | 0.169–0.175 | 0.177–0.186 | 0.189 | 0.196–0.204 | 0.189–0.201 | 0.178–0.181 | 0.165 | 0.204 | 0.128–0.131 | 0.006 | |||||||||
| 14. P. geminatus | 0.178 | 0.172 | 0.160 | 0.172 | 0.183–0.192 | 0.179 | 0.186–0.207 | 0.199–0.208 | 0.181 | 0.181 | 0.197 | 0.134 | 0.053–0.056 | — | ||||||||
| 15. P. formosensis | 0.171 | 0.157–0.174 | 0.156–0.171 | 0.174–0.183 | 0.190–0.205 | 0.188–0.198 | 0.199–0.211 | 0.195–0.201 | 0.189–0.199 | 0.174–0.183 | 0.189–0.194 | 0.123–0.139 | 0.076–0.100 | 0.082–0.107 | 0.051 | |||||||
| 16. P. niger | 0.180 | 0.162 | 0.160 | 0.184 | 0.189–0.198 | 0.188 | 0.195–0.204 | 0.184–0186 | 0.177 | 0.171 | 0.191 | 0.136 | 0.089–0.091 | 0.101 | 0.082–0.101 | — | ||||||
| 17. P. berdmorei | 0.242–0.255 | 0.224 | 0.211–0.221 | 0.211–0.230 | 0.204–0.216 | 0.203–0.212 | 0.225–0.234 | 0.213–0.227 | 0.215–0.227 | 0.224–0.228 | 0.236–0.239 | 0.201–0.213 | 0.205–0.215 | 0.207–0.218 | 0.208–0.222 | 0.222–0.230 | 0.066 | |||||
| 18. P. kuznetsovorum | 0.228 | 0.213 | 0.211 | 0.215 | 0.195–0.202 | 0.214 | 0.216–0.237 | 0.202–0.219 | 0.199 | 0.218 | 0.224 | 0.193 | 0.189 | 0.198 | 0.187–0.219 | 0.213 | 0.140–0.142 | — | ||||
| 19. P. carinatus | 0.224–0.231 | 0.211–0.216 | 0.210–0.211 | 0.205–0.207 | 0.204–0.218 | 0.229–0.230 | 0.230–0.242 | 0.211–0.218 | 0.210–0.221 | 0.227–0.228 | 0.224–0.225 | 0.198–0.204 | 0.196–0.210 | 02101–0.211 | 0.198–0.227 | 0.199–0.213 | 0.137–0.150 | 0.144–0.151 | 0.053 | |||
| 20. P. abros | 0.218–0.221 | 0.219–0.222 | 0.224 | 0.202 | 0.205–0.219 | 0.202–0.203 | 0.221–0.224 | 0.205–0.216 | 0.211–0.215 | 0.211–0.215 | 0.216–0.218 | 0.193 | 0.193–0.196 | 0.184 | 0.186–0.207 | 0.199–0.202 | 0.177–0.199 | 0.201 | 0.189–0.201 | 0.003 | ||
| 21. P. temporalis | 0.208 | 0.196 | 0.199 | 0.181 | 0.186–0.196 | 0.198 | 0.218 | 0.195–0.208 | 0.202 | 0.187 | 0.201 | 0.187 | 0.195 | 0.187 | 0.193–0.201 | 0.181 | 0.186–0.195 | 0.190 | 0.189–0.192 | 0.095–0.098 | — | |
| 22. P. nuchalis | 0.228 | 0.236 | 0.233 | 0.234 | 0.208–0.210 | 0.197 | 0.221–0.233 | 0.211–0.234 | 0.222 | 0.224 | 0.228 | 0.199 | 0.207–0.210 | 0.202 | 0.198–0.215 | 0.211 | 0.178–0.181 | 0.195 | 0.187–0.189 | 0.172–0.175 | 0.172 | — |
Figure 1.
The maximum likelihood (ML) phylogenetic relationship trees based on concatenated Cyt b and ND4 fragments. Numbers near each node indicate the bootstrap support (The branch where the P. baiseensis sp. nov. is located is marked in red).
3.2. Taxonomic Account
Pareas baiseensis sp. nov. WU, GONG, HUANG, and XU
http://zoobank.org/984D3880-9849-46DB-A488-7D6542478AB7 (accessed on 8 June 2023)
Figure 2, Figure 3 and Figure 4
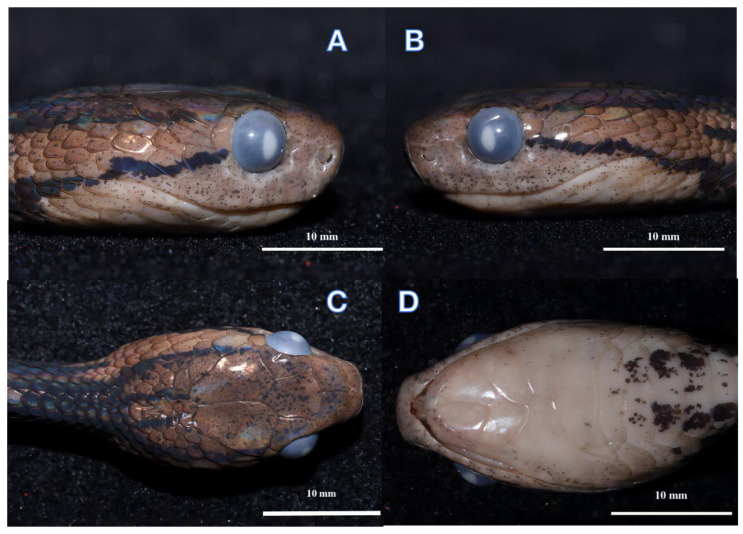
Figure 2.
Pareas baiseensis sp. nov. in life. (A) Holotype male (ANU20220011); (B) Paratype juvenile (ANU20220014).
Figure 3.
Holotype male (ANU20220011) of Pareas baiseensis sp. nov. in preservative. Right (A), Left (B), Ventral (C), Dorsal (D) views of the head.
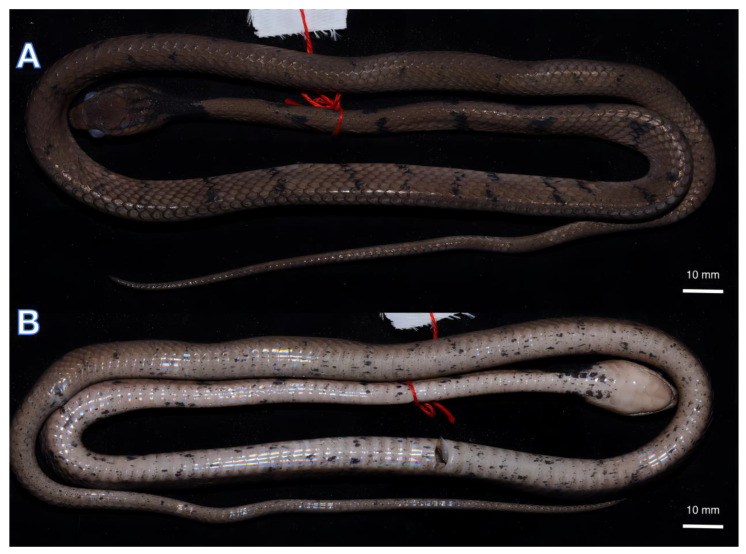
Figure 4.
Dorsal (A) Vntral (B) views of the preserved holotype of Pareas baiseensis sp. nov. (ANU 20220011).
Holotype. ANU20220011 (collection number HSR22185), an adult male (Figure 2A), was found in the Daleng Township, Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China (23.73521N, 106.39544E (DD); ca 781 m a.s.l.). The specimen was collected by Jiaxiang Wu and Yongjin Liu on 25 November 2022 and deposited at Anhui Normal University Museum.
Paratypes. ANU20220012 (collection number HSR22186), juvenile; ANU20220013 (collection number HSR22188, Figure 2B), juvenile; ANU20220014 (collection number HSR22189), juvenile, all with the same collecting information as the holotype.
3.3. Diagnosis
Pareas baiseensis sp. nov. is distinguished from all other Pareas by a combination of the following characteristics: (1) Yellow-brown body colouration; (2) Frontal subhexagonal to diamond-shaped with its lateral sides converging posteriorly; (3) The anterior pair of chin shields is longer than it is broad; (4) The loreal is not in contact with the eye, prefrontal in contact with the eye, two or three suboculars; (5) Rows of 15–15–15 dorsal scales, five rows of mid-dorsal scales keeled at the middle of the body, one vertebral scale row enlarged; (6) 187–191 ventrals, 89–97 subcaudals, all divided, cloacal plates single; (7) Two postocular stripes, the nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines.
3.4. Comparisons
Pareas baiseensis sp. nov. differs from P. margaritophorus, P. macularius, P. modestus and P. andersonii by having a light brown dorsum with irregular dark bands (vs. uniform grey to black to dark colouration, and with bicolored spots in P. margaritophorus, P. macularius and P. andersonii); 9/9 infralabials (vs. 7–8 infralabials); one vertebral scale row enlarged (vs. not enlarged); a higher number of ventrals (187–191 vs. 133–173); and a higher number of subcaudals (89–97 vs. 35–54). [4,6,7,8,28,29].
Pareas baiseensis sp. nov. differs from P. nigriceps, P. niger, and P. stanleyi by two or three suboculars (vs. one or suboculars fused with postoculars); the dorsal surface of the head is light brown with dark brown spots (vs a large black area on the back of the head); nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. (vs. nuchal area no collar); 9/9 infralabials (vs. 7 or 8 infralabials); a higher number of ventrals (187–191 vs. 151–184); and a higher number of subcaudals (89–97 vs. 48–77) [4,6,7,9,29,30,31].
Pareas baiseensis sp. nov. differs from P. abros, P. kuznetsovorum, P. temporalis, P. berdmorei, P. nuchalis and P. carinatus by frontal subhexagonal with lateral sides converging posteriorly (vs. frontal hexagonal with lateral sides parallel to body axis); and the anterior pair of chin shields is longer than it is broad (vs. anterior pair of chin shields broader than long or slightly longer) [4,6,7,20,29,30,31,32].
Pareas baiseensis sp. nov. differs from P. vindumi, P. victorianus, and P. monticola by the loreal not contacting the eye (vs. the loreal contacting the eye); two or three suboculars (vs. one or suboculars fused with postoculars); five slightly keeled dorsal scale rows at midbody (vs. smooth or 7–11 keeled dorsal scale rows at midbody); and the nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. (vs. nuchal area no collar) [6,7,10,29].
Pareas baiseensis sp. nov. differs from P. hamptoni, P. kaduri, P. geminatus, P. xuelinensis, P. komaii, P. atayal, P. iwasakii, and P. formosensis by nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. (vs. nuchal area no collar); five slightly keeled dorsal scale rows at midbody (vs. smooth or 5–13 keeled dorsal scale rows at midbody); and two or three suboculars (vs. one or suboculars fused with postoculars) [6,7,9,10,11,12,29,30,31,32,33,34].
Pareas baiseensis sp. nov. differs from Pareas dulongjiangensis by the nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines (vs. two brownish-black longitudinal stripes running on each side of the neck leaving a pale central portion); the loreal not contacting the eye (vs. loreal contacting the eye); absence of preoculars (vs. preoculars being present); two or three suboculars (vs. suboculars fused with postoculars); a higher number of ventrals (187–191 vs. 182); and a higher number of subcaudals (89–97 vs. 76) [13].
Pareas baiseensis sp. nov. differs from Pareas tigerinus by the dorsal surface of the head, which is light brown with dark brown spots (vs. dorsal surface of head solid black or reddish-brown); two anterior temporals (vs. one anterior temporal); two or three suboculars (vs. suboculars fused with postoculars); a higher number of ventrals (187–191 vs. 160–171); and a higher number of subcaudals (89–97 vs. 62–64) [14].
Pareas baiseensis sp. nov. differs from Pareas yunnanensis by the dorsal surface of the head, which is light brown with dark brown spots (vs. dorsal surface of head is black); sides of the head with two lateral postorbital stripes (vs. no or one or two indistinct large black spots on each side of the head, no stripe on each side of the head); a higher number of infralabials (9 vs. 6–8); five slightly keeled dorsal scale rows at midbody (vs. 5–7 rows of middorsal scales keeled on the middle part of the body); a higher number of ventrals (187–191 vs. 169–175); and a higher number of subcaudals (89–97 vs. 59–65) [13].
Pareas baiseensis sp. nov. differs from P. chinensis by five slightly keeled dorsal scale rows at midbody (vs. smooth or seven keeled dorsal scale rows at midbody); one vertebral scale row enlarged (vs. three vertebral scale rows enlarged); two or three suboculars (vs. one subocular); nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. (vs. nuchal area no collar); a higher number of ventrals (187–191 vs. 169–180); and a higher number of subcaudals (89–97 vs. 69–76) [4,6,7,9,29,35].
Pareas baiseensis sp. nov. differs from P. boulengeri by the loreal not contacting the eye (vs. the loreal contacting the eye); one vertebral scale row enlarged (vs. not enlarged); five slightly keeled dorsal scale rows at midbody (vs. smooth); two or three suboculars (vs. suboculars fused with postoculars); nuchal area forming a dark black four-pointed fork collar with the middle tines shorter than the outside tines. (vs. nuchal area no collar); a higher number of ventrals (187–191 vs. 164–187); and a higher number of subcaudals (89–97 vs. 63–78) [4,6,9,29,36].
3.5. Description of Holotype
An adult male, SVL 428 mm, TaL 151 mm, TL 579 mm, TaL/TL ratio 0.26; body slender, compressed; head elongate, clearly distinct from neck; snout round in dorsal view; eye slightly enlarged, pupil vertical and slightly elliptical; rostral slightly visible in dorsal view; frontal subhexagonal to diamond-shaped with its lateral sides converging posteriorly; nasal scale single; two prefrontals large, in contact with the eye; single loreal not in contact with the eye; temporals 2 + 3/3 + 3; 1/1 supraocular; 1/1 preocular; 4/4 suboculars; 1/1 postoculars; 8/8 supralabial scales; 9/9 infralabials; 191 (+1 preventral) ventrals; 15–15–15 dorsal scale rows, five rows of mid-dorsal scales keeled at the middle of the body; 97 subcaudals; cloacal plate single (Table 4).
Table 4.
Measurements (mm) and pholidosis for the holotype and paratypes of Pareas baiseensis sp. nov.; for abbreviations, see the Section 2.
| Voucher Number | ANU2022008 (Holotype) |
ANU2022009 (Paratype) |
ANU2022010 (Paratype) |
ANU2022011 (Paratype) |
|---|---|---|---|---|
| collection number |
HSR22185 | HSR22186 | HSR22188 | HSR22189 |
| SEX | male | juvenile | juvenile | juvenile |
| SVL | 428 | 174 | 171 | 185 |
| TaL | 151 | 56 | 51 | 67 |
| TL | 579 | 230 | 222 | 252 |
| TaL/TL | 0.26 | 0.24 | 0.23 | 0.27 |
| HL | 19.6 | 11.1 | 10.9 | 12.3 |
| HW | 10.6 | 6.2 | 6.0 | 6.5 |
| ED | 4.1 | 2.6 | 2.6 | 2.5 |
| ASR | 15 | 15 | 15 | 15 |
| MSR | 15 | 15 | 15 | 15 |
| PSR | 15 | 15 | 15 | 15 |
| VSE | 1 | 1 | 1 | 1 |
| KMD | 5 | 5 | 5 | 5 |
| VEN | 191 | 190 | 187 | 189 |
| SC | 97 | 91 | 89 | 96 |
| CP | entire | entire | entire | entire |
| SL | 8/8 | 8/8 | 8/8 | 8/8 |
| IL | 9/9 | 9/9 | 9/9 | 9/9 |
| At | 2/3 | 2/2 | 2/2 | 2/2 |
| Pt | 3/3 | 3/3 | 3/4 | 3/2 |
| LOR | 1/1 | 1/1 | 1/1 | 1/1 |
| Preoc | 2/2 | 2/2 | 2/2 | 2/2 |
| SoO | 3/3 | 2/2 | 2/2 | 2/2 |
| PoO | 1/1 | 1/1 | 1/1 | 1/1 |
Colouration: In life, the dorsal surface of the head is light brown with dark brown spots. The dorsum is brown with dark-brown speckling, and there are 34 irregular black cross-bands on the lateral sides of the body from neck to vent. The ventral is creamish–yellow with a few small black spots, the background colour gradually darkens to the rear, and the subcaudal scales are light brown. The sides of the head have two lateral postorbital stripes: the upper stripe extends from the temporal area backward extension to the dorsal scales of the neck, where it joins a large black collar around the nape, forming a dark black Ψ-shaped chevron pattern overall; the lower stripe extends backwards past the 9th supralabial, and at the throat contacting the four-pointed fork collar with the middle tines shorter than the outside tines. There are two black lines on the back of the parietal that extend back to the neck, with two lateral postorbital stripes; together, they form a dark black four-pointed fork collar.
In the preserved state, the colouration still resembles the specimen in life, but the dorsum colour fades to yellowish-brown (Figure 3 and Figure 4).
3.6. Variation
Measurements and scalation features of the type series (n = 4) are presented in Table 4. There is a certain variation observed in the number of ventrals, subcaudals, and temporals: ventrals (187–190, n = 4); subcaudals (89–96, n = 4); temporals (2 + 2, 2 + 3, 2 + 4, n = 4). Numerous irregular black cross-bands on the lateral sides of the body from neck to vent (32–40 bands, n = 4). The coloration features among the members of the type series were very similar.
3.7. Distribution
This species is currently only known from onelocality, Daleng Township, Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China. We found the snakes between 10:00 pm to 1:00 am after light rain in November 2022. The habitat environment was a well-preserved subtropical evergreen broad-leaved forest at elevations of 750–790 m.
4. Discussion
The phylogenetic results of Poyarkov et al. (2022) support the genus Pareas sensu lato being divided into two subgenera (Pareas sensu stricto and Eberhardtia) and six species groups; the subgenus Pareas sensu stricto includes two species groups (P. carinatus and P. nuchalis groups), the subgenus Eberhardtia includes four species groups (P. chinensis, P. hamptoni, P. monticola and P. margaritophorus groups). The members of the subgenus Eberhardtia differ from the members of the subgenus Pareas by the following combination of morphological characters: frontal subhexagonal to diamond-shaped with its lateral sides converging posteriorly; the anterior pair of chin shields is longer than it is broad; a single thin elongated subocular; and the ultrastructure of dorsal scales not ravine-like, having pore and arc structures, with arcs connecting to each other forming characteristic lines [4,8,37,38,39]. Our phylogenetic results support Pareas baiseensis sp. nov. belongs to the P. chinensis species groups in the subgenus Eberhardtia, but Pareas baiseensis sp. nov. have two or three suboculars, which is inconsistent with the diagnostic characteristic of subgenus Eberhardtia. Therefore, we propose deleting the diagnostic characteristic of a single thin elongated subocular.
The discovery of Pareas baiseensis sp. nov. increases the number of species of the P.chinensis species group to four species (Pareas baiseensis sp. nov., P.boulengeri, P. stanleyi, P.chinensis) in China. Among them, P.boulengeri is the most widely distributed, which is distributed in Guizhou, Sichuan, Yunnan, Chongqing, Henan, Hubei, Hunan, Guangxi, Guangdong, Jiangsu, Zhejiang, Anhui, Jiangxi, Fujian, Shaanxi, and Gansu. P. stanleyi is distributed in Fujian, Zhejiang, Jiangxi, Guizhou, Sichuan, Hunan and Guangxi; The distribution of P.chinensis in China is limited to the western and southern marginal mountains of the Sichuan Basin [6,7,14,15]; Pareas baiseensis sp. nov. is currently known only from the locality investigated, but Baise City is close to the borders of Yunnan and Vietnam, and this species may also occur in these adjacent areas.
The description of Pareas baiseensis sp. nov. from southern China brings the total number of recognized Pareas species to 30, of which 24 occur in China [2,4,14,15]. The genus Pareas has an ancient origin and poor migration ability, and the morphological difference between different species are subtle [9,21,40,41,42]; a large range of intensive sampling is helpful in discovering cryptic species, especially for some widely distributed types with unclear internal relationships, so sampling should be increased.
5. Conclusions
A new species of Pareas, Pareas baiseensis sp. nov., is described based on four specimens collected from the Youjiang District, Baise City, Guangxi Zhuang Autonomous Region, China, since Baise City is close to the borders of Yunnan and Vietnam and this species may also occur in these adjacent areas. However, their discovery is largely accidental, which makes it difficult for us to make accurate judgments on the distribution and population status of this new species. Further investigations will be necessary to assess the distribution and population status of this species.
Acknowledgments
We are grateful to Ruyi Huang, Jing Yu, Yi Zhang, Zhangbo Cui and Xinge Wang for their help in the study. We are also grateful for the anonymous reviewers’ useful and constructive comments on our manuscript.
Author Contributions
Conceptualization, Y.G., J.W., P.L. and Y.X.; methodology, all authors; data curation, Y.G., D.Y., Y.L. and J.W.; writing—original draft preparation, Y.G. and S.H.; writing—review and editing, Y.G., P.L. and S.H.; visualization, P.L., Y.L., S.L. and D.Y.; supervision, S.H. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study received ethical review and approval from the Animal Ethics Committees at Anhui Normal University (project number AHNU-ET2021025). All sampling and procedures involving live snakes were performed in accordance with the Wild Animals Protection Law of the People’s Republic of China and approved by the Animal Ethics Committees at Anhui Normal University (project number AHNU-ET2021025).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the Postdoctoral Research Program of the Department of Human Resources and Social Security of Anhui Province (2020B422), Doctoral Research Starting Foundation of Anhui Normal University (752017), National Natural Science Foundation of China (31471968, 32001222).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Deepak V., Ruane S., Gower D.J. A new subfamily of fossorial colubroid snakes from the Western Ghats of peninsular India. J. Nat. Hist. 2018;52:2919–2934. doi: 10.1080/00222933.2018.1557756. [DOI] [Google Scholar]
- 2.Uetz P., editor. The Reptile Database. [(accessed on 10 June 2023)]. Available online: http://www.reptile-database.org. [Google Scholar]
- 3.Narayanan S., Mohapatra P.P., Balan A., Das S., Gower D.J. A new species of Xylophis beddome, 1878 (Serpentes: Pareidae) from the southern western Ghats of India. Vertebr. Zool. 2021;71:219–230. doi: 10.3897/vz.71.e63986. [DOI] [Google Scholar]
- 4.Poyarkov N.A., Nguyen T.V., Pawangkhanant P., Yushchenko P.V., Brakels P., Nguyen L.H., Nguyen H.N., Suwannapoom C., Orlov N., Vogel G. An integrative taxonomic revision of slug-eating snakes (Squamata: Pareidae: Pareineae) reveals unprecedented diversity in Indochina. PeerJ. 2022;10:e12713. doi: 10.7717/peerj.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grossmann W., Tillack F. On the taxonomic status of Asthenodipsas tropidonotus (Van Lidth de Jeude, 1923) and Pareas vertebralis (Boulenger, 1900) (Serpentes: Colubridae: Pareatinae) Russ. J. Herpetol. 2003;10:175–190. doi: 10.30906/1026-2296-2003-10-3-175-190. [DOI] [Google Scholar]
- 6.Wang P., Che J., Liu Q., Li K., Jin J.Q., Jiang K., Shi L., Guo P. A revised taxonomy of Asia snail-eating snakes Pareas (Squamata, Pareidae): Evidence from morphological comparison and molecular phylogeny. ZooKeys. 2020;939:45–64. doi: 10.3897/zookeys.939.49309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L., Chen Z., Suwannapoom C., Nguyen T.V., Poyarkov N.A., Vogel G. A new species of the Pareas hamptoni complex (Squamata Serpentes: Pareidae) from the Golden Triangle. Taprobanica. 2020;9:174–193. doi: 10.47605/tapro.v9i2.230. [DOI] [Google Scholar]
- 8.Vogel G., Nguyen T.V., Lalremsanga H.T., Biakzuala L., Hrima V., Poyarkov N.A. Taxonomic reassessment of the Pareas margaritophorus-macularius species complex (Squamata, Pareidae) Vertebr. Zool. 2020;70:547–569. doi: 10.26049/VZ70-4-2020-02. [DOI] [Google Scholar]
- 9.Wu Y.H., Hou S.B., Yuan Z.Y., Jiang K., Huang R.Y., Wang K., Liu Q., Yu Z.B., Zhao H.P., Zhang B.L., et al. DNA barcoding of Chinese snakes reveals hidden diversity and conservation needs. Mol. Ecol. Resour. 2023;23:1124–1141. doi: 10.1111/1755-0998.13784. [DOI] [PubMed] [Google Scholar]
- 10.Liu S., Rao D.Q. A new species of the genus Pareas (Squamata, Pareidae) from Yunnan, China. ZooKeys. 2021;1011:121–138. doi: 10.3897/zookeys.1011.59029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogel G. New montane species of the genus Pareas Wagler, 1830 (Squamata: Pareatidae) from Northern Myanmar. Taprobanica. 2015;7:1–7. doi: 10.47605/tapro.v7i1.148. [DOI] [Google Scholar]
- 12.You C.W., Poyarkov N.A., Lin S.M. Diversity of the snail-eating snakes Pareas (Serpentes, Pareatidae) from Taiwan. Zool. Scr. 2015;44:349–361. doi: 10.1111/zsc.12111. [DOI] [Google Scholar]
- 13.Bhosale H., Phansalkar P., Sawant M., Gowande G., Patel H., Mirza Z.A. A new species of snail-eating snakes of the genus Pareas Wagler, 1830 (Reptilia: Serpentes) from eastern Himalayas, India. Eur. J. Taxon. 2020;729:54–73. doi: 10.5852/ejt.2020.729.1191. [DOI] [Google Scholar]
- 14.Liu S., Yang M.J., Rao J.Q., Guo Y.H., Rao D.Q. A New Species of Pareas Wagler, 1830 (Squamata, Pareidae) from Northwestern Yunnan, China. Taxonomy. 2023;3:169–182. doi: 10.3390/taxonomy3020013. [DOI] [Google Scholar]
- 15.Liu S., Zhang D.R., Poyarkov N.A., Hou M., Wu L., Rao D.Q., Nguyen T.V., Vogel G. Resurrection of Pareas yunnanensis (Vogt, 1922) with description of a new species of Pareas from Yunnan Province, China (Squamata, Pareidae) Eur. J. Taxon. 2023;860:1–26. doi: 10.5852/ejt.2023.860.2045. [DOI] [Google Scholar]
- 16.Dowling H.G. A proposed standard system of counting ventrals in snakes. Br. J. Herpetol. 1951;1:97–99. [Google Scholar]
- 17.Queiroz A.D., Lawson R., Lemos-Espinal J.A. Phylogenetic relationships of North American garter snakes (Thamnophis) based on four mitochondrial genes: How much DNA is enough? Mol. Phylogenetics Evol. 2002;22:315–329. doi: 10.1006/mpev.2001.1074. [DOI] [PubMed] [Google Scholar]
- 18.Salvi D., Harris D.J., Kaliontzopoulou A., Carretero M.A., Pinho C. Persistence across Pleistocene ice ages in Mediterranean and extra-Mediterranean refugia: Phylogeographic insights from the common wall lizard. BMC Evol. Biol. 2013;13:147. doi: 10.1186/1471-2148-13-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burland T.G. DNASTAR’s Lasergene sequence analysis software. Methods Mol. Biol. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 20.Kraus F., Brown W.M. Phylogenetic relationships of colubroid snakes based on mitochondrial DNA sequences. Zool. J. Linn. Soc. 1998;122:455–487. doi: 10.1111/j.1096-3642.1998.tb02159.x. [DOI] [Google Scholar]
- 21.Guo Y.H., Wu Y.K., He S.P., Shi H.T., Zhao E.M. Systematics and molecular phylogenetics of Asian snail-eating snakes (Pareatidae) Zootaxa. 2011;3001:57–64. doi: 10.11646/zootaxa.3001.1.4. [DOI] [Google Scholar]
- 22.Loredo A.I., Wood P.L., Quah E.S.H., Anuar S., Greer L.F., Ahmad N., Grismer L.L. Cryptic speciation within Asthenodipsas vertebralis (Boulenger, 1900) (Squamata: Pareatidae), the description of a new species from Peninsular Malaysia, and the resurrection of A. tropidonotus (Lidth de Jue, 1923) from Sumatra: An integrative taxonomic analysis. Zootaxa. 2013;3664:505–524. doi: 10.11646/zootaxa.3664.4.5. [DOI] [PubMed] [Google Scholar]
- 23.Figueroa A., McKelvy A.D., Grismer L.L., Bell C.D., Lailvaux S.P. A species-level phylogeny of extant snakes with description of a new colubrid subfamily and genus. PLoS ONE. 2016;11:e0161070. doi: 10.1371/journal.pone.0161070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deepak V., Narayanan S., Das S., Rajkumar K.P., Easa P.S., Sreejith K.A., Gower D.J. Description of a new species of Xylophis Beddome, 1878 (Serpentes: Pareidae: Xylophiinae) from the Western Ghats, India. Zootaxa. 2020;4755:231–250. doi: 10.11646/zootaxa.4755.2.2. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 27.Minh B.Q., Nguyen M.A.T., von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K., Nei M., Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Suntrarachun S., Chanhome L., Hauser S., Sumontha M., Kanya K. Molecular phylogenetic support to the resurrection of Pareas macularius from the synonymy of Pareas margaritophorus (Squamata: Pareidae) Trop. Nat. Hist. 2020;20:182–190. [Google Scholar]
- 30.Vogel G., Nguyen T.V., Zaw T., Poyarkov N.A. A new species of the Pareas monticola complex (Squamata, Serpentes, Pareidae) from Chin Mountains with additions to the Pareas fauna of Myanmar. J. Nat. Hist. 2021;54:2577–2612. doi: 10.1080/00222933.2020.1856953. [DOI] [Google Scholar]
- 31.Boulenger G.A. Proceedings of the Zoological Society of London. Vol. 69. Blackwell Publishing Ltd.; Oxford, UK: 1900. Descriptions of new reptiles and batrachians from Borneo; pp. 182–187. [Google Scholar]
- 32.Guo K.J., Deng X.J. A new species of Pareas (Serpentes: Colubridae: Pareatinae) from the Gaoligong Mountains, southwestern China. Zootaxa. 2008;1:53–60. doi: 10.11646/zootaxa.2008.1.5. [DOI] [Google Scholar]
- 33.Stuebing R.B., Inger R.F., Lardner B. A Field Guide to the Snakes of Borneo. Natural History Publication; Kota Kinabalu, Malaysia: 2014. 310p [Google Scholar]
- 34.Maki M. Monograph of the Snakes of Japan. Dai-ichi Shobo; Tokyo, Japan: 1931. 240p [Google Scholar]
- 35.Maki M. A new subspecies, Amblycephalus formosensis iwasakii, belonging to Amblycephalidae from Ishigaki-jima. Trans. Nat. Hist. Soc. 1937;27:217–218. [Google Scholar]
- 36.Barbour T. Some Chinese vertebrates: Amphibia and reptilia. Mem. Mus. Comp. Zoölogy. 1912;40:125–136. [Google Scholar]
- 37.Angel M.F. Liste de reptiles récémment déterminés et entrés dans les collections et description d’une nouvelle espèce du genre Amblycephalus. Bull. Du Muséum Natl. D’histoire Nat. 1920;2:112–114. [Google Scholar]
- 38.Smith M.A. The Fauna of British India Ceylon and Burma, Including the Whole of the Indo-Chinese Sub-Region. Reptilia and Amphibia. Vol. III. Serpentes. Vol. III. Taylor and Francis; London, UK: 1943. [Google Scholar]
- 39.Taylor E.H. The serpents of Thailand and adjacent waters. Univ. Kans. Sci. Bull. 1965;45:609–1096. [Google Scholar]
- 40.Li J.N., Liang D., Wang Y.Y., Guo P., Huang S., Zhang P. A large-scale systematic framework of Chinese snakes based on a unified multilocus marker system. Mol. Phylogenetics Evol. 2020;148:106807. doi: 10.1016/j.ympev.2020.106807. [DOI] [PubMed] [Google Scholar]
- 41.Pyron R.A., Burbrink F.T., Wiens J.J. A phylogeny and revised classification of Squamata, including 4161 species of lizards and snakes. BMC Evol. Biol. 2013;13:93. doi: 10.1186/1471-2148-13-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaher H., Murphy R.W., Arredondo J.C., Graboski R., Grazziotin F.G. Correction: Large-scale molecular phylogeny, morphology, divergence-time estimation, and the fossil record of advanced caenophidian snakes (squamata: Serpentes) PLoS ONE. 2019;14:e0217959. doi: 10.1371/journal.pone.0217959. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.