Abstract
Patient selection for synthetic lethal–based cancer therapy may be improved by assessment of gene-specific loss of heterozygosity (LOH) and biallelic loss of function (LOF). This report describes SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx), a targeted next-generation sequencing (NGS) panel for detection of LOH and biallelic LOF alterations in 26 target genes focused on DNA damage response pathways, in tumor-only formalin-fixed, paraffin-embedded (FFPE) samples. NGS was performed across all exons of these 26 genes and encompassed a total of 7632 genome-wide single-nucleotide polymorphisms on genomic DNA from 80 FFPE solid tumor samples. The Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing algorithm was optimized to assess tumor purity and copy number based on heterozygous single-nucleotide polymorphisms. SNiPDx demonstrated high sensitivity (95%) and specificity (91%) for LOH detection compared with whole genome sequencing. Positive agreement with local NGS-based testing in the detection of genetic alterations was 95%. SNiPDx detected 93% of biallelic ATM LOF mutations, 100% of ATM single-nucleotide variants and small insertions/deletions, and 100% of all ATM LOH status events identified by orthogonal NGS-based testing. SNiPDx is a novel, clinically feasible test for analysis of allelic status in FFPE tumor samples, which demonstrated high accuracy when compared with other NGS-based approaches in clinical use.
DNA damage response (DDR) and homologous recombination (HR) DNA repair play pivotal roles in the maintenance of genomic integrity and the prevention of the accumulation of genetic alterations.1 The genomic loss of components integral to DDR and HR pathways results in dependence on alternative, error-prone DNA repair mechanisms and thus increased levels of genomic instability.1 The increased dependence of DNA repair–deficient tumors on alternative repair pathways can be targeted through pharmacologic inhibition, resulting in tumor-specific cell death through synthetic lethality (SL).2,3 Poly (ADP-ribose) polymerase inhibitors are canonical agents that were developed and approved based on SL and are indicated for the treatment of patients with HR-deficient cancers, such as BRCA1/2-mutated breast, pancreatic, prostate, and ovarian cancers.2, 3, 4, 5, 6, 7, 8, 9, 10 Small-molecule inhibitors of ataxia-telangiectasia Rad-3-related (ATR) are also in development as SL-based treatments for patients with ataxia-telangiectasia mutated (ATM)–deficient cancers.11, 12, 13, 14
Although alterations affecting one allele (monoallelic) in proto-oncogenes are often sufficient to generate oncogenic phenotypes, tumor suppressor genes, including those that encode DDR- and HR-related proteins, usually require inactivation of both alleles [biallelic loss of function (LOF)].1,15 There are several lines of evidence demonstrating that most genes encoding DDR- and HR-related proteins are not haploinsufficient and require biallelic inactivation for the loss of competent DNA repair.2,16,17 Among biallelic alterations, LOF can often be driven by inactivation of the first allele via germline or somatic mutation events, in combination with inactivating alterations [such as loss of heterozygosity (LOH) through physical deletions or copy number neutral LOH, somatic mutations, rearrangements, or gene promoter methylation] in the second allele.18,19
Published analyses of large cancer cohorts indicate that frequencies of monoallelic and biallelic LOF vary across genes and cancer types (Supplemental Figure S1).16,20 The results shown are based on data generated by The Cancer Genome Atlas (TCGA) Research Network (https://www.cancer.gov/tcga, last accessed June 10, 2020). Across tumor types characterized as part of TCGA, most mutations detected in DDR/HR genes are monoallelic rather than biallelic, with the exception of specific tumor types with high rates of HR deficiency, such as ovarian, breast, pancreatic, and prostate cancer.16,21,22 Most biallelic LOF events are driven by somatic or germline mutations with LOH of the wild-type allele (Supplemental Figure S1B). In addition, biallelic LOF events across various tumor types can result from compound heterozygous mutations (ie, a second inactivating somatic mutation), homozygous deletions (HomDels), and complex rearrangements affecting large regions of the genome.
Evidence is emerging for the clinical significance of biallelic LOF alterations as biomarkers for SL-based pharmaceutical approaches to treatment of patients with cancer.7,16,23,24 Differences in response rates among patients with monoallelic versus biallelic LOF alterations highlight that detection of targeted alterations, without analysis of the wild-type allele, may not be sufficient to accurately predict clinical benefit from SL-based approaches.7 In tumors harboring BRCA1/2 alterations, up to 50%, depending on the cancer type, may be monoallelic (Supplemental Figure S1A). Indeed, it is plausible that the lack of benefit of SL-based agents observed in some studies targeting BRCA1/2 mutant cancers25,26 may be due, in part, to the lack of prospective and accurate assessment of biallelic LOF. Hence, accurate selection of patients who may benefit from SL approaches requires comprehensive detection of LOH to identify tumors harboring biallelic LOF events in target genes.7,24,27
Currently available clinical genomic profiling assays do not systematically and comprehensively report gene-specific LOH and may not be able to classify a substantial subset of alterations as biallelic LOF [according to manufacturers’ technical information (eg, FoundationOne CDx, Foundation Medicine, Inc., Cambridge, MA; and TruSight Oncology 500, Illumina, San Diego, CA)].28,29 Variable purity across clinical samples, coupled with the lack of appropriate markers of allelic imbalance in existing sequencing panels, can lead to indeterminate inference of purity and ploidy, rendering precise LOH assessment challenging. To address this unmet need, SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) was developed and validated as a targeted, next-generation sequencing (NGS)–based multigene panel capable of accurate reporting of monoallelic versus biallelic alterations affecting DDR- and HR-related genes. SNiPDx offers a clinically feasible approach to facilitate the understanding of patient responses to a range of SL treatments.
This article reports on the development of SNiPDx methods and validation of the panel for the detection of monoallelic versus biallelic LOF alterations in formalin-fixed, paraffin-embedded (FFPE) tumor samples, through the comparison of genomic alterations identified with SNiPDx and existing genomic profiling approaches.
Materials and Methods
Allele-Specific LOF Analysis of TCGA Samples
Genomic data for TCGA30 were downloaded from the National Cancer Institute's Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov, last accessed June 10, 2020). Copy number variation estimates used in the analysis were based on the Allele-Specific Copy number Analysis of Tumors (ASCAT) algorithm.31 HomDel was declared when gene-level estimates of total copy number were equal to 0. Somatic single-nucleotide variation and insertion/deletion (InDel) estimates performed by TCGA MC3 Project32 were used to identify LOF mutations in the genes of interest. Germline pathogenic mutations were obtained from the literature.16 Genomic events considered to be biallelic were limited to the following: i) HomDel (defined as ASCAT total copy number estimate of 0); ii) co-occurrence of a mutation and LOH (defined as ASCAT minor copy number estimate of 0); iii) co-occurrence of a somatic and a germline mutation; iv) co-occurrence of two somatic mutations in patients with <1000 somatic mutations; and v) mutation with mutant allele frequency of >90%. All other mutations were considered not biallelic.
SNiPDx Panel Methods
Samples
FFPE samples, including 80 tumor resections and 24 unmatched normal samples from various tissues, were obtained from Avaden Biosciences (Seattle, WA). FFPE tumor biopsies and archival tumors were collected as part of the first-in-human camonsertib Treatment Enabled by SNIPRx (TRESR) study (https://clinicaltrials.gov, identifier NCT04497116, last accessed February 13, 2023). All patients provided consent for tissue acquisition and genomic analysis.
Selection of SNPs for Inclusion in SNiPDx
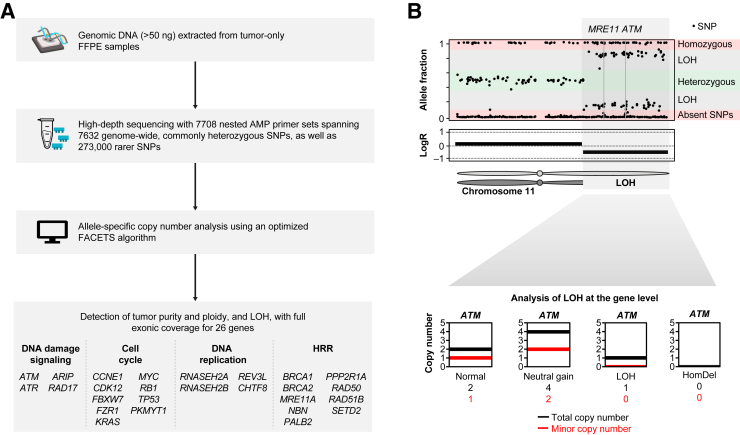
The SNiPDx panel covers full exonic regions of 26 DDR-related genes (Supplemental Table S1). In addition to the 1140 single-nucleotide polymorphisms (SNPs) that overlapped with or were located within 1 megabase (Mb) of the target genes, 6492 SNPs distributed in a genome-wide manner were included to facilitate genome-wide allele-specific copy number analysis (ASCNA). SNPs were selected on the basis of population allele frequencies between 33% and 66% and a low inbreeding coefficient (0 to 0.2), maximizing the number of heterozygous SNPs, as well as having ≥50 bp of unique flanking sequence that contained no other high-frequency SNPs, and GC content 25% to 75%. Furthermore, to ensure adequate numbers of heterozygous SNPs in samples from different genetic ancestries, SNPs at regular intervals from each other were prioritized, reducing the linkage disequilibrium between the proposed SNPs. SNPs in proximity to genes of interest were also added to increase the accuracy of ASCNA. A total of 7708 nested PCR primer pairs were designed to span all selected SNPs on the SNiPDx panel. The total genomic footprint of the SNiPDx panel was 200.6 Kb, with 100.8 Kb covering common SNPs, and a total of 99.8 Kb covering gene coding regions.
Anchored Multiplex PCR Library Generation and Sequencing
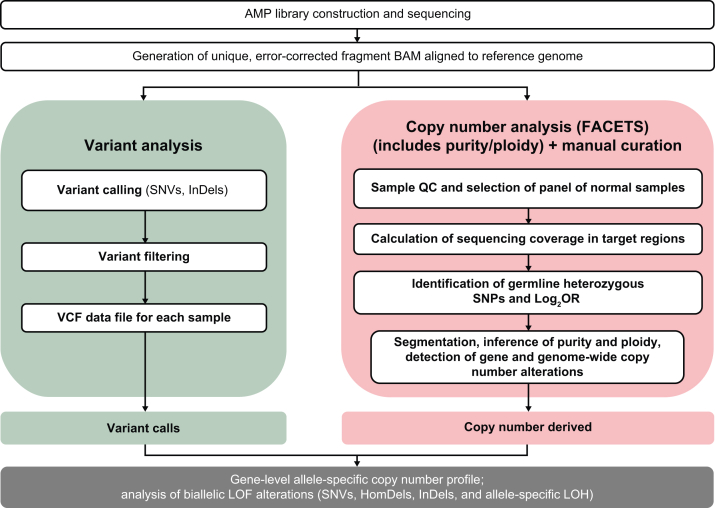
DNA (minimum, 50 ng) was extracted from 10 FFPE slides (5 μm thick) per sample. Anchored multiplex PCR (AMP) was employed for target amplification for NGS, as previously described.33 Genomic DNA underwent end repair (End-Repair Mix, Enzymatics, Beverly, MA), adenylation (Klenow Exo-, Enzymatics; Taq Polymerase, Thermo Fisher Scientific, Waltham, MA), and ligation (T4 DNA Ligase, Enzymatics) with a universal half-functional adapter. Libraries were cleaned using solid-phase reversible immobilization. Target was enriched using two rounds of nested PCR (Platinum Taq Polymerase, Thermo Fisher Scientific). The first PCR (10 cycles) was performed using a primer complementary to the universal adapter and target-specific primers (Operon, Huntsville, AL). After cleanup, the second round of PCR (15 cycles) was executed using a universal adapter primer and a second pool of target-specific primers. These primers were each 5′-tagged sequencing adapters, to generate target amplicons ready for clonal amplification and sequencing (Figure 1). Libraries were quantitated using quantitative PCR (Kapa Biosystems, Woburn, MA) and normalized per manufacturer’s protocol. Amplicons were sequenced on the NovaSeq platform (Illumina, San Diego, CA), according to the manufacturer's standard protocol. Samples with <20 million reads were excluded from analyses.
Figure 1.
SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) workflow. AMP, anchored multiplex PCR; BAM, binary alignment map; FACETS, Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing; HomDel, homozygous deletion; InDel, insertion/deletion; LOF, loss of function; Log2OR, log2 odds ratio; LOH, loss of heterozygosity; QC, quality control; SNP, single-nucleotide polymorphism; SNV, single-nucleotide variant; VCF, variant call format.
Bioinformatics Workflow
Sequence alignment and annotation
AMP libraries were processed using the VariantPlex Pipeline from Archer Analysis Platform version 6.2.8 (ArcherDx, Boulder, CO). Paired-end sequence data were processed using methods developed for AMP to align error-corrected reads (Figure 1).33 Briefly, raw reads were cleaned for quality, trimmed of adapter sequence, and deduplicated to an error-corrected sequence. Reads were mapped to the human GRCh37.p13 (GCA_000001405.1) National Center for Biotechnology Information reference genome using Bowtie 2 version 2.2.9.34 To capture reads that may not be aligned using Bowtie 2 (ie, primarily split reads, to enable structural variant detection), a second round of alignments was performed using Burrows-Wheel Aligner–MEM.35 Binary alignment maps were generated using SAMtools version 1.5 (https://github.com/samtools/samtools). Variants were called using LoFreq version 2.1.1 (http://csb5.github.io/lofreq), Freebayes version 0.9.9 (https://github.com/freebayes/freebayes), and proprietary methods. VariantPlex pipeline parameters were adjusted to accommodate the large footprint of SNiPDx and minimize background noise within variant calls. Variant calls <300 bp from the nearest gene-specific primers within regions of interest in reads with minimum base quality 22, and with minimum allele fraction 0.02, were reported. Genes, transcripts, and consequences of variants were accessed through Alamut Batch (Interactive BioSoftware, Haute-Normandie, France) using database version 1.5-2020.11.25. A variant call format file for each sample was generated using Vcftools version 0.1.11 (https://vcftools.github.io).
Copy Number Analysis from SNiPDx Data
Definition of biallelic versus monoallelic alterations
Alterations in genes were classified as monoallelic versus biallelic based on the presence of single-nucleotide variants (SNVs)/InDels and copy number alterations. Biallelic LOF included the following: i) two somatic pathogenic or likely pathogenic (P/LP) alterations; ii) one germline P/LP alteration and one somatic P/LP alteration; iii) one P/LP alteration and LOH; or iv) complete or partial HomDel of a gene. Variant allele fraction (VAF) estimates for P/LP alterations with LOH were checked for consistency with Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS)–defined tumor purity and allele-specific copy number (asCN) state.36
Selection of SNPs for copy number analysis
All SNPs targeted in the SNiPDx panel were included in the copy number analysis. An additional approximately 273,000 rarer germline SNPs located within 400 bp of SNiPDx panel primers were identified using gnomAD genome version 3.0 and exome version 2.1 (https://gnomad.broadinstitute.org/downloads, last accessed January 21, 2021), and were also included for copy number analysis.
Generation of a baseline data set from normal tissue samples
Aligned sequence data were analyzed from 23 FFPE normal samples, derived from tissue adjacent to tumors that had been resected (colon, n = 10; breast, n = 5; testis, n = 4; ovary, n = 2; and tonsil, n = 2). SNPs with a mean coverage of ≥200× across all samples were selected. To eliminate SNPs coinciding with erroneously mapped reads, only SNPs that were heterozygous (allele fraction 0.4 to 0.6) or homozygous (allele fraction ≤0.1 or ≥0.9) in ≥23 normal samples were included for copy number analysis. Furthermore, only SNPs with average coverage of at least 100× in each normal sample category, and where the SD of coverage in normal samples was <20% of average, were included. To reduce the noise in analyses resulting from loss of sequencing coverage at regions distal from primers (Supplemental Figure S2), data from normal samples were stratified according to DNA fragment size distribution to generate graded panels of normal (PoNs) samples for copy number normalization of tumor sequence data. k-Means clustering (k = 3) was performed to classify data as good (n = 8), fair (n = 12), or poor (n = 3) (Supplemental Figure S3).
Optimization of ASCNA using FACETS and analysis of tumor samples
ASCNA is essential for the accurate detection of loss of the wild-type allele of genes mutated in a given cancer. For the detection of allele-specific gains and amplifications (with corresponding integer copy number), HomDels, LOH caused by heterozygous deletions, and copy number–neutral LOH, an accurate estimation of, and adjustment for, tumor purity and ploidy is fundamental.31,36
FACETS, which was designed for ASCNA of FFPE tumor samples subjected to targeted capture sequencing, was employed.36,37 Curation of asCN calls was performed systematically, with partial automation using standard FACETS parameters for segmentation, inference of sample purity, and ploidy. Samples with <20% tumor purity were excluded from further analysis.
An adaptive PoN selection scheme was added to the standard FACETS workflow to match quality parameters to an FFPE tumor sample analyzed. A PoN with the same insert size distribution class was selected for normalization. Loess regression analysis was used to determine a primer-SNP distance cutoff from the coverage profile of each tumor sample. The distance from primers where the average sequencing depth fell below 500× was used as a cutoff for selection of SNPs to be included in copy number analysis (Supplemental Figure S4). Although all SNPs are useful to derive information on deletion or gain of chromosomal material (as previously demonstrated36), SNPs with VAF 5% to 95% were considered as heterozygous and included in the analysis of allelic imbalances, with results extrapolated to regions without heterozygous SNPs.
Genome-wide major and minor copy numbers were inferred by FACETS.36 Copy number alterations and allelic imbalances in the 26 SNiPDx target genes and other genomic regions were calculated on the basis of the log2R (ie, the log2 ratio of SNP coverage in a tumor sample/coverage in a matched normal sample or PoN) and log2 odds ratio (calculated from the number of reads reporting the alternative allele/number of reads reporting the reference allele), adjusted by tumor purity and ploidy.36 Minor allele (b-allele) fraction for each SNP was defined as the ratio of reads reporting the alternative allele/total number of reads at that position; LOH was determined if the minor allele copy number was 0.
Samples were manually reviewed for technical parameters of sequencing and tumor content, and LOH status per gene was curated for possible mis-segmentations using plots generated from FACETS solutions. Data from each sample were re-baselined, based on a region or chromosome with ploidy 2. FACETS results were then reviewed using the copy number software NexusCopyNumber version 9.0 (BioDiscovery, El Segundo, CA). For each gene, the asCN state was classified according to terms shown in Supplemental Table S2.
Comparison of Genomic Assessment by SNiPDx with Existing Biomarker Profiling Approaches
Comparison of LOH Detection by SNiPDx with a Reference Data Set Derived Using WGS
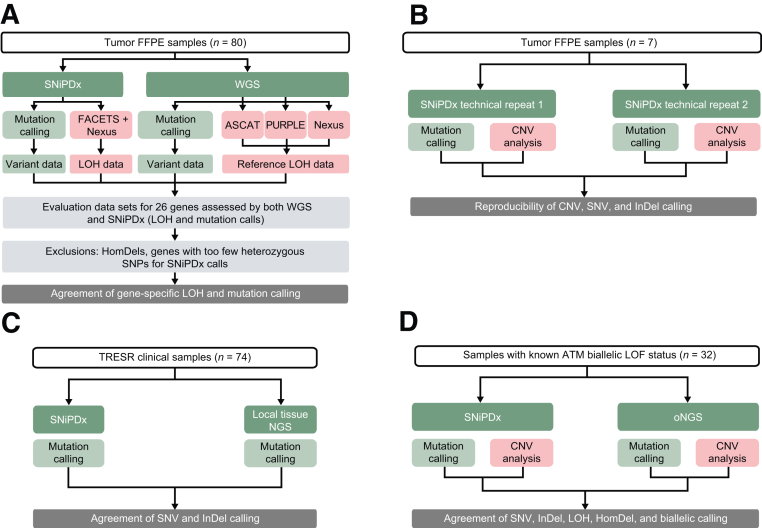
In the absence of a gold standard approach to validate ASCNA from targeted sequencing of FFPE samples, a reference data set of high-confidence LOH events was derived from whole genome sequencing (WGS) in a set of tumor samples. For assay comparisons, SNiPDx and WGS were performed on serial sections. WGS was performed on extracted DNA from 80 tumor samples and 23 unmatched normal FFPE tissue samples (Audubon Bioscience, Houston, TX) (Figure 2A). WGS reads were aligned to the reference genome [University of California, Santa Cruz (UCSC) hg19 genome GCA_000001405.1] across 1.86 million genome-wide SNPs, using the Burrows-Wheel Aligner mapping tool. Duplicate reads were removed.
Figure 2.
Overview of the validation of SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) for the detection of genetic alterations. A: Assessment of SNiPDx loss of heterozygosity (LOH) events compared with whole genome sequencing (WGS). B: Assay reproducibility. C: Agreement of single-nucleotide variant (SNV) and insertion/deletion (InDel) calling by SNiPDx and WGS or local next-generation sequencing (NGS)–based profiling. D: Assessment of biallelic loss of function (LOF) calls between SNiPDx panel and orthogonal NGS (oNGS) in clinical samples with known biallelic loss of ataxia-telangiectasia mutated (ATM). ASCAT, Allele-Specific Copy number Analysis of Tumors; CNV, copy number variant; FACETS, Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing; FFPE, formalin fixed, paraffin embedded; HomDel, homozygous deletion; PURPLE, PURity and Ploidy Estimator; SNP, single-nucleotide polymorphism.
SNV and InDel calling
Mutations in SNiPDx genes were identified in a targeted manner from WGS using the MuTect algorithm.38 To ensure confident calling, only coding mutations with six or more supporting reads were included. MuTect and default PoNs reduced the number of recurrent sequencing artifacts and germline mutations identified. Mutations identified by WGS were annotated against canonical transcripts using Ensembl Variant Effect Predictor.39 The mutation set was filtered by consequence and read support to prioritize likely pathogenic mutations in genes also covered by SNiPDx. Specifically, the selected coding variants were supported by at least six reads in samples that had both WGS and SNiPDx data. Using this approach, 36 likely pathogenic mutations were identified from WGS; the possibility that some were germline cannot be excluded.
Allele-specific copy number analysis from WGS
Three existing bioinformatics methods were used for copy number variant analysis of WGS profiles to generate an LOH event reference data set: ASCAT,31 PURity and PLoidy Estimator (PURPLE),40 and NexusCopyNumber. ASCAT and PURPLE were previously optimized for matched-normal, fresh-frozen tissue.31,40 In this study, the algorithms were modified to improve their application to tumor-only DNA from FFPE samples, which is prone to increased DNA fragmentation and base damage,41 and correlates with reduced effective sequencing coverage by nonduplicated reads and increased background noise of coverage profiles. Minor or b-allele fraction (ie, the relative presence of each of the two alternative nucleotides at each SNP profiled) was calculated using a modified form of the ASCAT tool.31 Only reads with a base quality score >27 were included for analysis of WGS profiles by ASCAT. Modifications to the ASCAT algorithms included normalization of the sequencing signal to account for GC content in small and large local windows, plus additional covariates to account for local insert size and local mapping quality in the WGS PoN. The normalization method was built on robust linear regression with normalization, using the glmnet package version 4.1,42 adjusted to correct for local mapping quality and local read length in the panel of normal samples. The elastic network from glmnet (Lasso and Elastic-Net Regularized Generalized Linear Models version 4.1)43 was implemented to deal with extreme noise and correlated covariates. ASCAT was run in tumor-only mode. The panel of 19 unmatched normal samples was used as a comparator in the log2R calculations to normalize coverage signal. Segmentation penalties of P = 50 and P = 100 were used. Copy number profiles were assessed against biological plausibility; solutions without at least two chromosomes with segments of LOH >20 Mb each, or with large HomDels >10 Mb, would be marked as failed quality control. If ASCAT did not find an acceptable solution automatically, plausible purity/ploidy solutions were selected manually, according to the ASCAT sunrise plot with segmentation penalty of P = 100. The algorithm and its modifications were validated through analysis of normal FFPE samples to ascertain whether, as expected, no copy number variants were detected in DNA extracted from normal tissue samples. Samples with no identifiable and plausible solution, or with excessive noise quantified by Nexus (>0.055), were excluded. The PURPLE algorithm was used to perform copy number segmentation, as well as purity and ploidy estimation.40 Samples not meeting quality control criteria from PURPLE were removed from the LOH calling reference data set. The PURPLE algorithm was modified for tumor-only analysis, to enable normalization of the coverage signal (log2R) in a tumor sample against the average coverage in the same regions using a panel of unmatched normal samples. All the other PURPLE parameters were used as standard or default.
Manual curation of gene copy number variation calls was performed using NexusCopyNumber software version 9.0. Each gene of interest was manually inspected to determine the cytogenetic state and event, accounting for events observed on the chromosomal location of the gene of interest. Samples with tumor purity <40% and samples not meeting quality control parameters defined within ASCAT and PURPLE were excluded from the WGS LOH calling reference data set. A higher tumor purity threshold was applied to WGS than to SNiPDx data sets to ensure robust analysis and avoid confounding issues associated with the lower sequencing coverage typical of WGS. LOH predictions were adjusted for tumor purity.
Using a voting system, SNPs that were detected as LOH by at least two of the three bioinformatic approaches were included in the WGS LOH reference data set. Where asCN calls were available and in agreement on gene LOH or heterozygosity from all three WGS analysis approaches, results were also tabulated to generate an even higher confidence reference set of genes. In addition, genes in samples with HomDels were excluded from the reference data set (Figure 2A). Sensitivity and specificity of LOH detection by SNiPDx were assessed using the WGS LOH data set as a reference.
Assay Reproducibility
To investigate the reproducibility of SNiPDx, DNA from seven tumor samples was extracted, sequenced, and analyzed in duplicate. The agreement for the detection of gene-specific LOH status and variants was assessed (Figure 2B).
In Silico Estimation of the Limit of LOH Detection
Simulations of FFPE tumor samples of varying purity were performed on data from four FFPE colorectal cancer samples, which were selected because of their high purity (>45%), making them amenable to experimental in silico dilution. Matched SNiPDx data from normal FFPE tissue from the same patients were also used. FACETS purity estimates from nondiluted tumor samples were assumed to be correct, and used to calculate the dilution proportions. To perform tumor sample dilutions in silico, weighted means of read counts from matched tumor and normal samples were calculated, in proportions that were estimated (from the original tumor purity of the sample) to simulate samples with target purity of 10%, 15%, 20%, 25%, 30%, 35%, and 40%. Data were adjusted for differences in overall sequencing coverage between tumor and normal samples, and noise was added by sampling from normal distribution given the SD of coverage for each SNP in the panel of normal samples (Supplemental Figure S5). Alternative read counts were drawn from the binomial distribution, with the expected allele fraction calculated as a weighted average of allele fraction in the tumor and normal samples. The sampling procedure was repeated 20 times for each dilution of each of the four samples. Estimated tumor purity and the LOH status of each of the SNiPDx genes were reported for 560 FACETS analyses (4 samples × 7 simulated purities × 20 repeats).
Comparison of SNV and InDel Calling in Clinical Tumor Samples
SNiPDx SNV and InDel detection was validated by comparing the accuracy of variant calls from SNiPDx with those in an existing data set, derived using external NGS-based tumor testing as part of screening for patient enrollment from the ongoing phase 1/2a TRESR study (NCT04497116)44 (Figure 2C). Only SNVs or InDels affecting one of the 26 genes included in the SNiPDx assay were considered for comparative analysis. Cases were excluded when FACETS was unable to determine sample purity (often due to low tumor content).
Validation Cohort of Clinical Samples with Known ATM Biallelic LOF
SNiPDx-detected SNVs, InDels, LOH, HomDels, and biallelic calls were assessed in clinical FFPE tumor samples (n = 27) with known ATM biallelic LOF status (Figure 2D). ATM biallelic LOF status was determined previously using an orthogonal, clinical-grade, target capture NGS-based assay with ≥400 key cancer genes including intronic and regulatory regions of selected genes,45 hereon referred to as orthogonal NGS (oNGS). Tumor samples were collected under informed consent as specified in the Internal Review Board–approved protocols. asCN profiles were derived from FACETS analysis, previously optimized for oNGS.36 Matched blood samples were used to identify heterozygous SNPs.
Statistical Analysis
Relationships between tumor purity and ploidy, as determined by different platforms, were evaluated using Pearson correlation coefficients. To compare asCN calls by SNiPDx with existing biomarker profiling approaches, sensitivity and specificity, 95% CIs, and percentage agreement were calculated. Cohen κ scores quantify agreement between binary classifications against reference data, accounting for the probability of agreement by chance, particularly for tasks with class imbalances.46 Cohen κ tests were performed using fsmb version 0.7.3 (https://CRAN.R-project.org/package=fmsb, last accessed October 7, 2022) in R version 4.0.4.
Code Availability
The analysis of SNiPDx data is based on the R package FACETS version 0.6.1 (https://github.com/mskcc/facets, last accessed October 7, 2022). Analysis of WGS data was based on ASCAT and PURPLE algorithms. Version 2.5.2 of ASCAT (https://github.com/VanLoo-lab/ascat, last accessed October 7, 2022) was used. Version 2.51 of PURPLE, relying on AMBER 3.5 and COBALT 1.11 (https://github.com/hartwigmedical/hmftools, last accessed October 7, 2022), was used.
Custom code for selection of SNPs and adaptable selection of PoN for SNiPDx remains proprietary, with a patent application filed.
Results
Design and Optimization of SNiPDx
SNiPDx was developed as a proof-of-concept, NGS-based gene panel for accurate reporting of monoallelic versus biallelic alterations in 26 tumor suppressor DDR genes, including LOH events in the presence of somatic or germline variants, and HomDels (Figure 3). SNiPDx has several key features. First, AMP primer sets (probes) cover all exons of 26 target genes of interest, 7632 genome-wide, commonly heterozygous SNPs, and up to 273,000 rarer SNPs. Second, copy number analysis with SNiPDx enables accurate estimations of sample purity and ploidy, which are essential to inform ASCNA, because of high sequencing coverage (>500×). Third, a custom bioinformatic analysis pipeline generated using the asCN and clonal heterogeneity analysis tool, based on FACETS,36 was further optimized for analysis of FFPE tumor-only tissue samples. Fourth, incorporation of unique molecular identifiers ensures efficient removal of duplicate reads (error correction) for accurate variant detection.
Figure 3.
SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) overview and copy number analysis. A: Overview. B: Copy number analysis. Bottom panel: Allele-specific copy number analysis at the locus of one of the 26 genes of interest, the ataxia-telangiectasia mutated (ATM) locus on 11q. AMP, anchored multiplex PCR; FACETS, Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing; FFPE, formalin fixed, paraffin embedded; HomDel, homozygous deletion; HRR, homologous recombination repair; LogR, log2 ratio of SNP coverage in a tumor sample to coverage in a matched normal sample; LOH, loss of heterozygosity; SNP, single-nucleotide polymorphism.
SNiPDx ASCNA
Calculation of Allelic Imbalance in Tumor Samples by SNiPDx
The SNiPDx panel was initially developed using a set of 44 tumor samples, including 9 ovarian cancer, 7 uterine cancer, 6 leiomyosarcoma, 5 lung cancer, 5 breast cancer (3 luminal B and 2 triple negative), 3 bladder cancer, 4 gastric cancer, 4 prostate cancer, and 1 colon cancer. After removal of duplicate reads, median sequencing depth with SNiPDx was 1593× [interquartile range (IQR), 1376× to 1801×] across tumor samples, and 1316× (IQR, 767× to 1964×) across normal samples. On the basis of DNA fragment size distributions, tumor samples were categorized as good (n = 12), fair (n = 22), and poor (n = 10) for normalization to unmatched PoNs.
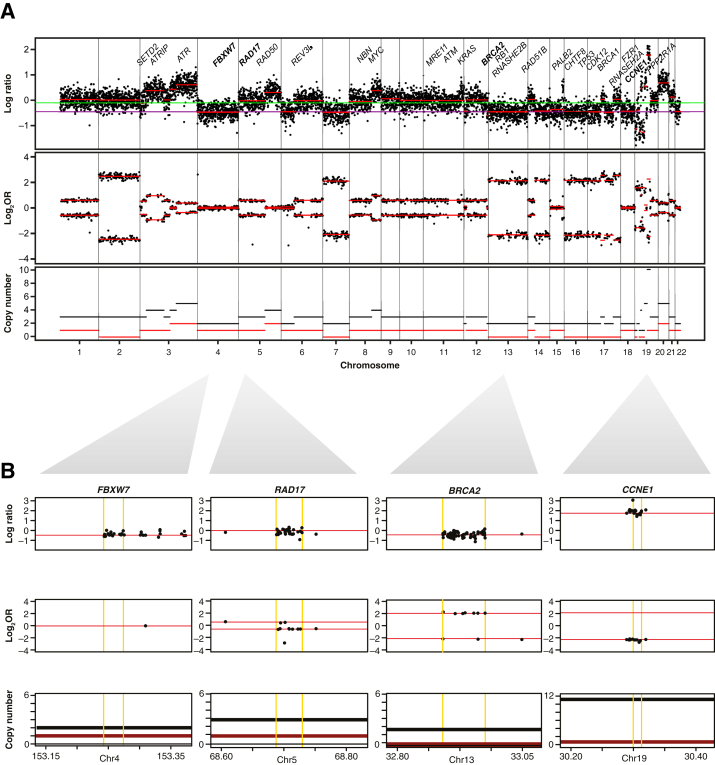
Copy number calculations were performed across all 281,000 SNPs covered by SNiPDx. Heterozygous SNPs inform allelic imbalances, whereas normalized sequencing coverage informs chromosomal gains (in regions of increased local coverage) and losses (otherwise). Homozygous SNPs with VAF of 1, absent SNPs (VAF = 0), and heterozygous SNPs (VAF = 0.5) were predominant in normal tissue samples. SNPs that were deemed as heterozygous in normal samples had a 95% CI within the range of 0.46 to 0.54. Across tumor samples, allelic imbalance was prevalent, with VAFs of heterozygous SNPs ranging between 0.07 and 0.89 (example assay output from tumor sample shown) (Figure 4). A median of 2966 heterozygous SNPs (IQR, 2909 to 3124) per tumor sample were detected by SNiPDx.
Figure 4.
Example of a genome-wide allele-specific copy number analysis (ASCNA) profile from analysis using SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) in a formalin-fixed, paraffin-embedded sample from a 56-year–old woman with ovarian cancer. A: Genome-wide ASCNA profile. B: Example of copy number states identified in FBXW7 (diploid), RAD17 (gain of one copy), BRCA2 [loss of heterozygosity (LOH)], and CCNE1 (amplification). A and B: Within copy number tracks, black lines denote total copy number, and red lines denote minor copy number, with LOH if minor copy number is 0. A: Black dots denote single-nucleotide polymorphisms. Red lines in the log ratio and log2 odds ratio (Log2OR) tracks are a result of segmentation of raw data. In the log ratio track, the purple line denotes log-ratio value inferred at diploid state, and green line denotes median log-ratio level. B: Yellow lines indicate the boundaries of canonical transcripts; their chromosomal coordinates are marked on the horizontal axis. Chr, chromosome.
Copy Number Analysis
FACETS36 allowed for estimation of sample ploidy and purity, which were accounted for in total and minor copy number calculations across all chromosomal locations. Sequencing coverage in the PoNs was used as denominator to calculate the log2R of tumor/normal copy number across all genomic segments (Figure 4). Across all genomic segments analyzed for all tumor samples, the median fraction of the genome with allelic imbalance (log2 odds ratio >0.1) was 39% (IQR, 14% to 58%).
Genome-wide SNP coverage by SNiPDx provided resolution for accurate copy number assessments, including LOH. Of 1092 curated asCN calls (26 genes in 42 samples; two samples analyzed by FACETS were not manually curated and were excluded from comparisons), ASCNA of SNiPDx data (using FACETS and manual curation) detected 533 LOH (allelic imbalance with minor allele = 0) events, including 123 LOH events with copy number loss, 163 copy-neutral LOH events, and 177 LOH events with copy number gain.
Validation of SNiPDx Data in an LOH Reference Data Set, Through Comparison with WGS
Of 79 WGS profiles processed, 34 FFPE tumor biopsy samples passed stringent quality control filters to form a veritable, high-confidence reference data set of LOH/non-LOH events. Reasons for sample attrition during WGS quality control filtering included low tumor fraction (tumor fraction <40%; 33 samples) and high noise score from NexusCopyNumber that captures levels of variation between adjacent coverage probes (additional 13 samples), resulting in inability to infer plausible integer copy number profiles using ASCAT or PURPLE.31,40 Data from 19 unmatched normal samples passed quality control filtering to generate a data set from a PoN for copy number normalization. Median coverage by WGS was 11.9 (IQR, 8.4 to 13.5) across normal tissue samples. Twenty-four WGS samples from the LOH event reference data set were also included in the SNiPDx data set (Supplemental Table S3). The sample data set for comparison of LOH events comprised eight ovarian cancers, four uterine cancers, four breast cancers, two prostate cancers, two leiomyosarcomas, two lung cancers (both adenocarcinoma), one gastric cancer, and one bladder cancer. Median sequencing depth across samples was 1554× (IQR, 1367× to 1807×) by SNiPDx and 18.6× (IQR, 15.7× to 21.6×) by WGS, after removal of duplicate reads.
Comparison of Tumor Purity and Ploidy Assessments in SNiPDx Panel and WGS Data
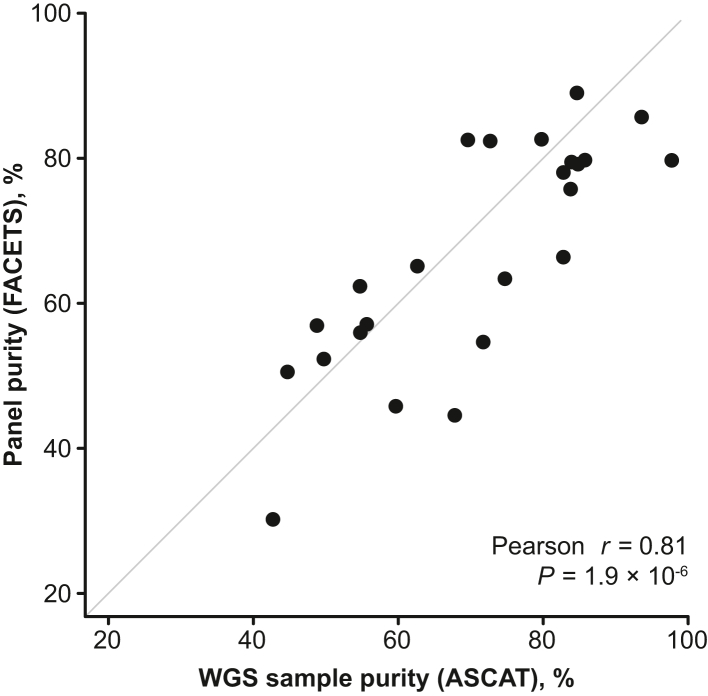
A strong correlation was observed (Pearson r = 0.81, P = 1.9 × 10−6) between tumor purity estimates determined from WGS data (ASCAT) and from SNiPDx panel data (FACETS) (Figure 5). Pearson r for tumor ploidy estimates by WGS and SNiPDx was 0.29 (P = 0.17) (Supplemental Figure S6). Discrepancies between ploidy values may be attributable to more accurate estimates of SNP allele fractions from SNiPDx and differences in assumptions about clonal structure made by different copy number analysis algorithms.
Figure 5.
Comparison of sample purity assessments derived from whole genome sequencing (WGS) data [Allele-Specific Copy number Analysis of Tumors (ASCAT)] and SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) data [Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS)].
Comparison of Copy Number Analysis in SNiPDx and WGS Data
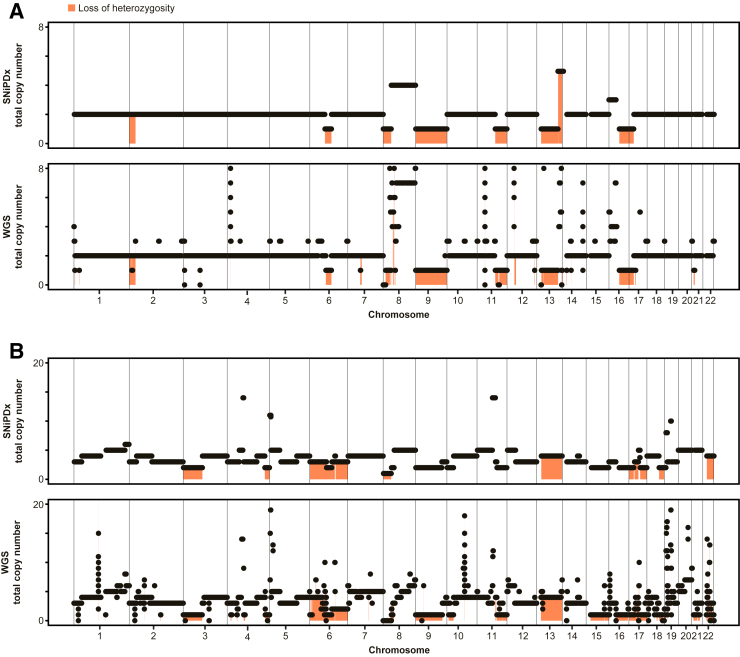
To estimate the accuracy of LOH detection by SNiPDx, a ground truth set of asCN calls from a WGS-derived data set was established. A consensus voting system of LOH detection by ASCAT, PURPLE, and Nexus algorithms was used to determine LOH in the 24 WGS profiles that had matching SNiPDx data. Among the 26 genes per sample (24 samples, 624 asCN calls for comparison in total) (Supplemental Table S4), 225 LOH events (36% of asCN calls) were detected by two or more WGS analysis approaches, of which 96 LOH events were detected by all three algorithms, and 129 were detected by two algorithms. A further 52 asCN calls had weak evidence for LOH (detected by only one algorithm) and were considered non-LOH; these calls were excluded from comparisons where indicated. Among the 288 asCN events that were evaluated by all three methods, the proportion of LOH events was similar: 114 (40%) were deemed LOH. Following exclusion of genes with putative HomDels (n = 11, 2% of calls assessed), and loci where LOH could not be established because of low numbers of heterozygous SNPs covered by SNiPDx (n = 8, 1% of calls assessed), 605 evaluable asCN calls formed the WGS-derived reference data set. The correlation of total gene copy number estimates in 24 validation samples between SNiPDx and WGS was 0.72 (Pearson r, P < 2.2 × 10−16). Two examples comparing ASCNA by SNiPDx and WGS are shown in Figure 6.
Figure 6.
Example comparisons showing copy number analysis by SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) and whole genome sequencing (WGS). A: Luminal type B breast cancer sample with a near-diploid genome (WGS-estimated ploidy of 2.2). B: Ovarian serous carcinoma sample with a whole-genome duplication (WGS-estimated ploidy of 4.2). Black dots and black lines indicate total copy number. Orange shading denotes areas with loss of heterozygosity.
Using standard parameters (without manual curation), SNiPDx demonstrated excellent performance for the detection of LOH events in the ground truth WGS reference data set (200 of 212 LOH events; Cohen κ, 0.85; 95% CI, 0.81–0.89) (Supplemental Table S5). Additional manual curation of SNiPDx data, which was performed with analysts (R.D.D. and J.Z.) blinded to the results of the WGS data set, resulted in SNiPDx detecting 202 of the 212 LOH events detected by WGS (Cohen κ, 0.89; 95% CI, 0.85–0.92) (Table 1). The sensitivity and specificity of LOH detection by SNiPDx were 95% and 91%, respectively. Sensitivity and specificity increased to 99% and 98%, respectively, across 230 asCN calls with LOH agreement by all three WGS algorithms (Cohen κ, 0.97; 95% CI, 0.94–1.00), and to 97% and 99%, respectively, across 235 diploid regions with no sub-clonal alterations (Cohen κ, 0.93; 95% CI, 0.87–0.98). SNiPDx also accurately identified LOH in genes within segments that had small footprints [eg, between 100 kb and 1 Mb (one asCN call) and between 1 Mb and 5 Mb (nine asCN calls)].
Table 1.
Comparison of LOH Events Derived from SNiPDx and WGS Reference Data Set
| Variable | LOH detection by WGS |
|||
|---|---|---|---|---|
| All regions | ||||
| Two or more algorithms (605 LOH events)∗ |
Three-algorithm consensus (230 LOH events)† |
|||
| LOH | Non-LOH | LOH | Non-LOH | |
| LOH events by SNiPDx | ||||
| LOH | 202 | 22 | 93 | 2 |
| Non-LOH | 10 | 371 | 1 | 134 |
| Sensitivity, % | 95 | 99 | ||
| Specificity, % | 91 | 98 | ||
| κ Score (95% CI) | 0.89 (0.85–0.92) | 0.97 (0.94–1.00) | ||
| Variable | Diploid samples with no sub-clonal alterations (by WGS) |
|||
|---|---|---|---|---|
| Two or more algorithms (235 LOH events)∗ |
Three-algorithm consensus (116 LOH events)† |
|||
| LOH | Non-LOH | LOH | Non-LOH | |
| LOH events by SNiPDx | ||||
| LOH | 178 | 1 | 89 | 0 |
| Non-LOH | 5 | 51 | 1 | 26 |
| Sensitivity, % | 97 | 99 | ||
| Specificity, % | 99 | 100 | ||
| κ Score (95% CI) | 0.93 (0.87–0.98) | 0.98 (0.93–1.02) | ||
LOH, loss of heterozygosity; SNiPDx, SyNthetic lethal Interactions for Precision Diagnostics; WGS, whole genome sequencing.
LOH gene calls defined when indicated by at least two WGS algorithms.
LOH calls in the subset of genes and samples where all three WGS algorithms agreed.
SNiPDx Reproducibility
To investigate SNiPDx reproducibility, DNA was extracted twice from seven FFPE tumor blocks and run through separate library preparations and sequencing runs. Across 170 asCN calls, LOH events were determined by SNiPDx in 57, with the remaining 113 asCN calls considered to be non-LOH. LOH detection was 100% reproducible across the seven samples sequenced and analyzed in duplicate (Supplemental Table S6). The pathogenic mutations identified across the technical repeats were found to be identical: 14 of 14 pathogenic mutations were re-called in both replicates, with concordant allele frequency estimates (Pearson r = 0.99).
SNiPDx Limit of Detection
Through in silico dilution experiments where sequencing reads from matched normal FFPE samples were admixed into four high-purity FFPE colorectal cancer samples, it was demonstrated that purity inferences and LOH calls obtained from FACETS were consistent down to a simulated tumor sample purity of 20% (Supplemental Figure S7).
Validation of Mutation Calling by SNiPDx in Clinical Tumor Samples
Agreement of SNV and InDel Detection by SNiPDx and WGS or Local NGS-Based Profiling
Local tumor NGS reports from 74 patients with matched SNiPDx data were included in the analysis. Most frequent NGS tests were FoundationOne/FoundationOne CDx28 (Foundation Medicine; n = 23), OncomineV347 (n = 10), OncoPanel48 (n = 7), and Caris Molecular Intelligence49 (n = 7). The tissue sample tested by SNiPDx included both screening and archival biopsy samples, whereas local NGS testing was always done before screening.
In total, 129 alterations across 11 genes (TP53, n = 48; ATM, n = 21; BRCA1, n = 14; BRCA2, n = 13; KRAS, n = 12; CDK12, n = 6; RB1, n = 5; SETD2, n = 5; NBN, n = 3; PALB2, n = 1; and RAD51B, n = 1) were reported by local tumor NGS profiling of 74 patients. Of 129 alteration events, 122 were detected with SNiPDx (percentage positive agreement with local NGS-based approaches, 95%), including 37 of 39 InDels (percentage positive agreement, 95%) and 85 of 90 SNVs (percentage positive agreement, 94%) (Table 2).
Table 2.
Comparison of InDels and SNVs Derived from SNiPDx and NGS Data from Clinical Samples from the TRESR Trial
| Variable | Local NGS approach |
||
|---|---|---|---|
| Reported | |||
| Total (129 events) | InDels (39 events) | SNVs (90 events) | |
| SNiPDx panel | |||
| Detected | 122 | 37 | 85 |
| Not detected | 7 | 2 | 5 |
| Agreement, % | 95 | 95 | 94 |
InDel, insertion/deletion; NGS, next-generation sequencing; SNiPDx, SyNthetic lethal Interactions for Precision Diagnostics; SNV, single-nucleotide variant.
In all patients in whom an alteration was not detected by SNiPDx (n = 7), the local NGS and SNiPDx analyses were performed on different tumor specimens, some originating from distinct metastatic samples. Five of seven discordant alterations overlapped asCN events (four LOH events and one HomDel), suggesting that the event detected by the local tumor NGS assay was lost in the sample analyzed by SNiPDx. In one case involving a missed TP53 mutation, tumor samples collected for local and SNiPDx analysis were obtained from tumors of distinct histologic types from a patient with biphasic mesothelioma. Finally, SNiPDx analysis did not detect a pathogenic BRCA1 p.E720∗ alteration in a patient with non–small-cell lung cancer with tumor mutational burden of 14 mutations per Mb, likely a tumor of high clonal heterogeneity with many sub-clonal mutations. Consistent with high agreement between local tumor NGS and SNiPDx, 100% agreement was also observed (36 of 36 events) between WGS-identified pathogenic mutations and the same samples run on SNiPDx.
Assessment of Biallelic LOF Calls between SNiPDx Panel and oNGS in Clinical Samples with Known Biallelic Loss of ATM
The ability of SNiPDx to detect ATM biallelic LOF (including mutations, LOH, and HomDels) was assessed by comparison to asCN calls from FFPE samples previously characterized by oNGS. Samples were obtained from patients with esophagogastric (n = 20), prostate (n = 8), breast (n = 3), and pancreatic (n = 1) cancers. Previously characterized ATM alterations included 18 samples with biallelic loss (mutation + LOH, n = 11; and HomDel, n = 7), LOH alone (n = 8), or no ATM alterations (n = 6) (Supplemental Table S7). Five samples, including four with ATM biallelic LOF, failed SNiPDx analysis because of low DNA input material or low purity and were excluded from the comparisons. SNiPDx detected biallelic ATM LOF in 13 of 14 samples with known biallelic LOF (sensitivity, 93%; κ score, 0.93; 95% CI, 0.79–1.07) (Table 3). SNiPDx demonstrated 100% sensitivity for the detection of ATM mutations (κ score, 1) and LOH status (κ score, 0.92; 95% CI, 0.77–1.08). Five of six known ATM HomDels were detected by SNiPDx (sensitivity, 83%; κ score, 0.88; 95% CI, 0.65–1.11). For the one discordant case, ploidy estimates were variable between the two assays (3.96 by SNiPDx and 2.16 by oNGS) (Supplemental Figure S8).
Table 3.
Comparison of Alteration Events in Samples with Biallelic Loss of ATM, Derived from SNiPDx and oNGS
| Variable | LOH detection by oNGS |
|||||||
|---|---|---|---|---|---|---|---|---|
| Biallelic LOF |
Mutation status (SNVs + InDels) |
LOH status |
HomDel status |
|||||
| Biallelic | Not biallelic | Mutant | WT | LOH | Non-LOH | HomDel | Not HomDel | |
| Calls by SNiPDx | ||||||||
| Detected | 13 | 0 | 8 | 0 | 16 | 1 | 5 | 0 |
| Not detected | 1 | 13 | 0 | 19 | 0 | 10 | 1 | 17 |
| Sensitivity, % | 93 | 100 | 100 | 83 | ||||
| κ Score (95% CI) | 0.93 (0.79–1.07) | 1 | 0.92 (0.77–1.08) | 0.88 (0.65–1.11) | ||||
ATM, ataxia-telangiectasia mutated; HomDel, homozygous deletion; InDel, insertion/deletion; LOF, loss of function; LOH, loss of heterozygosity; oNGS, orthogonal next-generation sequencing; SNiPDx, SyNthetic lethal Interactions for Precision Diagnostics; SNV, single-nucleotide variant; WT, wild type.
Discussion
The field of precision oncology is evolving from its original focus on developing targeted therapeutics for oncogenes to target a broader profile, including LOF mutations in tumor suppressor genes, which account for a large proportion of all cancer mutations. SL is a validated approach to target tumors with LOF mutations in DDR/HR-deficient genes, but it requires knowledge of the allelic status of target genes (namely, biallelic LOF). NGS-based tumor profiling is a promising diagnostic tool to aid the development of SL-based therapies that target the DDR and to identify patients with cancer who are most likely to respond to SL-targeted therapies.24 This study reports on the development and testing of SNiPDx, a target gene panel assay that accurately detects biallelic inactivation of 26 SL-therapy target genes and therefore may facilitate the identification of patients who might benefit from these innovative therapeutic approaches. Although this proof-of-concept panel incorporates analysis across a limited number of genes, AMP technology allows for the addition of more genes for novel SL approaches in future iterations of SNiPDx.
In this study, SNiPDx exhibited reproducibility and accuracy in the detection of monoallelic and biallelic genomic alterations, demonstrating its potential for use in a real-world clinical diagnostic setting. Types of biallelic inactivation detected by SNiPDx included those occurring through HomDels, and mutations in conjunction with LOH of the wild-type allele or compound heterozygotes. In lieu of a validated gold standard approach, a WGS-derived reference data set was generated for the assessment of biallelic LOF, and the performance of SNiPDx for the analysis of allelic status in FFPE tumor samples was compared with this as well as with a variety of other NGS-based approaches.
LOH detection by SNiPDx was reproducible in tumor samples sequenced and analyzed in duplicate, and LOH events detected by SNiPDx displayed excellent agreement with the WGS-derived reference data set, despite differences in the ploidy inferences observed. Discrepancies in ploidy estimates may be explained by differences in sequencing coverage between the two assay methods, and it could be posited that SNiPDx may be better powered than WGS to discern even modest allele imbalance in tumors, and by differences in the algorithm assumptions employed to ascertain purity and ploidy. Consistent with the performance of SNiPDx against a WGS reference data set, SNiPDx also accurately detected the major mechanism of biallelic LOF events (deleterious mutations coupled with inactivation of the wild-type allele via LOH), as detected by alternative clinically validated methods. Variant detection by SNiPDx also showed high percentage agreement (95%) with clinical NGS approaches from Clinical Laboratory Improvement Amendments–certified or College of American Pathologists–accredited laboratories. In all cases of disagreement, the most parsimonious explanations for the discrepancies were related to sample collection chronology (ie, distinct samples from different time points in the patients' journeys) and/or heterogeneity between samples analyzed by SNiPDx and other NGS methods for each patient.
In-depth analysis of LOF alterations affecting ATM, as one example of an SL target, revealed that SNiPDx compared favorably with oNGS. The single instance of discordance stemmed from different ploidy levels being inferred by SNiPDx and oNGS, highlighting the challenges associated with current approaches to ASCNA. Hence, it was anticipated that even with the enhanced SNP coverage by SNiPDx compared with other methods, which allows for a higher resolution and more accurate ploidy estimation than most targeted sequencing assays, a small minority of samples may yield uncertain results.
Assessment by SNiPDx relies on a publicly available bioinformatics pipeline. The methods described here for the detection and validation of LOH events could be adapted for use in other clinical assays. Although expert manual curation and interpretation of SNiPDx data are required for detection of genomic alterations with the highest accuracy, automated analysis with default parameters still resulted in accurate mutation and LOH detection. An optimized SNiPDx workflow provides a new strategy to overcome challenges associated with obtaining accurate asCN calls from tumor biopsies. Although existing comprehensive genomic profiling approaches [eg, OncoPanel (Dana-Faber Institute, Boston, MA)48 and Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT; Memorial Sloan Kettering Cancer Center, New York, NY)45] are capable of providing the information to define biallelic inactivation of specific genes, including LOH assessment, this information is not systematically included in reports; hence, SNiPDx may help address this unmet need.
Limitations
This study has several limitations. Comparisons with existing NGS-based methods were performed on a small number of archival samples. Further validation studies with larger sample cohorts are warranted. For the assessment of the performance of SNiPDx against a US Food and Drug Administration–authorized tumor-normal multigene sequencing assay, work focused on the detection of mutation, HomDel, LOH, and allelic status of ATM; hence, further analyses will be required to define the performance for other genes included in the SNiPDx panel. SNiPDx cannot detect all mechanisms of biallelic loss, including structural rearrangements and epigenetic silencing via promoter methylation. Further work is required to establish the minimum tumor purity for delivery of accurate ASCNA results by SNiPDx.
Conclusion
SNiPDx constitutes a clinically feasible platform for the assessment of biallelic alterations affecting key DDR/HR-related genes, and prospective testing to identify patients who may be eligible for SL therapies targeting these genes will assess the utility of SNiPDx as a companion diagnostic. Further validation of SNiPDx in the context of prospective clinical trials of SL therapies is therefore warranted.
Acknowledgments
We thank the patients, their families, and all investigators involved in the TRESR study; Theresa Zhang, Parham Nejad, and Amanda Rode for invaluable contributions; and Annie Macpherson, Ph.D., for medical writing support, and Richard McDonald, B.Sc., for editorial support (both of Onyx, London, UK), supported by Repare Therapeutics, according to Good Publication Practice guidelines.50
Footnotes
Development of the SNiPDx assay was supported by Repare Therapeutics; this study was supported in part by the Breast Cancer Research Foundation (J.S.R.-F.), by Susan G. Komen through a Komen Scholar Leadership grant (J.S.R.-F.), by an NIH/National Cancer Institute (NCI) P50 CA247749 01 grant (J.S.R.-F.), and by the NCI Cancer Center support grant P30 CA08748 (J.S.R.-F.).
Disclosures: I.M.S., M.Z., A.V., M.K., and V.R. are employees of Repare Therapeutics. D.G. was an employee of, and stockholder in, Repare Therapeutics at the time of the study. I.M.S., M.Z., M.K., D.G., V.R., and J.S.R.-F. declare stock in Repare Therapeutics. R.A.R., J.Z., N.M., R.D.D., and V.J. are employees of Invitae. S.H. is an employee of Tempus. J.S.R.-F. also declares personal/consultancy fees from Goldman Sachs, Repare Therapeutics, Paige.AI and Eli Lilly; is a member of the scientific advisory boards for Repare Therapeutics, VolitionRx, Paige.AI, and Personalis; is a member of the Board of Directors of Grupo Oncoclínicas; and is an ad hoc member of the scientific advisory boards of AstraZeneca, MSD, and Daiichi Sankyo, outside the scope of this study. N.R. declares research funding from Repare Therapeutics, Invitae, and Pfizer. P.S. and D.M. have nothing to disclose. D.G., I.M.S., M.Z., M.K., R.D.D., V.R., and J.S.R.-F. have contributed to the patent application PCT/CA2022/050655, pertaining to the detection of loss of heterozygosity and biallelic loss of function alterations in tumor-only formalin-fixed, paraffin-embedded samples, pending to Repare Therapeutics Inc.
Current address of S.H., Tempus, Raleigh, NC.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2023.02.004.
Author Contributions
D.G., I.M.S., A.V., V.J., R.A.R., and R.D.D. designed the study, developed SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx) assay, and acquired and analyzed data; P.S. developed SNiPDx assay and analyzed data; J.Z., N.M., S.H., and D.M. acquired and analyzed data; M.K. conceptualized and designed the study and interpreted data; M.Z. conceptualized and designed the study; N.R. reviewed and interpreted data; V.R. designed the study and developed SNiPDx assay; J.S.R.-F. provided samples, designed, coordinated, and supervised the study, and interpreted data; and all authors contributed to the development of the manuscript and reviewed the final version. J.S.R.-F. is the guarantor of this work and, as such, had full access to all of the data utilized in the study and takes responsibility for the integrity of the data, as well as the accuracy of the data analysis and interpretation of the findings.
Supplemental Data
Biallelic loss of function (LOF) genomic alterations affecting genes of interest in The Cancer Genome Atlas data set. A: Frequency of monoallelic versus biallelic LOF for alterations. B: Mechanisms of biallelic LOF. LOH, loss of heterozygosity.
Sequencing coverage profiles in 23 normal samples profiled using SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx), with respect to distance to nearest primer, dependent on insert size distribution for a given sample. A: Unnormalized coverage values. B: Coverage values normalized by median coverage for each sample.
k-Means (k = 3) analysis of insert size distributions across tumor and normal formalin-fixed, paraffin-embedded samples. A: Clustering of insert size distributions. B: Average densities of insert sizes according to sample category.
Example of sample-specific copy number normalization calculations in a leiomyosarcoma sample from an 81-year–old man. A: Using a cutoff of 500× sequencing coverage, single-nucleotide polymorphisms within 168 bp from the primer were selected for inclusion. Gray line denotes sequencing coverage cutoff of 500×. B: Sample-specific insert size distribution. Vertical red lines correspond to quantiles, and blue line corresponds to mean. C: Matching of the sample-specific insert size distribution (from B) to typical insert size distributions derived from the panel of normal samples (PoNs). In this example, the fair PoN profile most resembled the tumor sample and was hence selected for normalization. FFPE, formalin fixed, paraffin embedded.
In silico limit of detection experiments. A: List of tumor-normal sample pairs and their characteristics. B: The algorithm used for sampling of read counts to simulate tumor samples with target purity values. FACETS, Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing; ID, identifier.
Ploidy estimates derived from whole genome sequencing [WGS; Allele-Specific Copy number Analysis of Tumors (ASCAT)] and SyNthetic lethal Interactions for Precision Diagnostics [SNiPDx; Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS)].
In silico experiments simulating low-purity tumor samples by diluting pure tumor samples with read counts from matched normal samples. A: Inference of tumor purity in diluted samples by Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS). Red lines correspond to median of inferred purity; blue lines delineate upper and lower quartiles. B: Accuracy of loss of heterozygosity detection in in silico diluted samples inferred by FACETS. For each simulated purity value, the number in parentheses represents the number of times (of a total of 20) FACETS identified a solution.
A and B: Discordant ataxia-telangiectasia mutated (ATM) biallelic loss of function called by SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx; A) and orthogonal next-generation sequencing (B). Within copy number tracks, black lines denote total copy number, and red lines denote minor copy number, with loss of heterozygosity if minor copy number is 0. A:Black dots denote single-nucleotide polymorphisms. Red lines in the log ratio and log2 odds ratio tracks are a result of segmentation of raw data. In the log ratio track, the purple line denotes log-ratio value inferred at diploid state, and green line denotes median log-ratio level.
References
- 1.Brown J.S., O'Carrigan B., Jackson S.P., Yap T.A. Targeting DNA repair in cancer: beyond PARP inhibitors. Cancer Discov. 2017;7:20–37. doi: 10.1158/2159-8290.CD-16-0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Farmer H., McCabe N., Lord C.J., Tutt A.N., Johnson D.A., Richardson T.B., Santarosa M., Dillon K.J., Hickson I., Knights C., Martin N.M., Jackson S.P., Smith G.C., Ashworth A. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 3.Bryant H.E., Schultz N., Thomas H.D., Parker K.M., Flower D., Lopez E., Kyle S., Meuth M., Curtin N.J., Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 4.Tutt A.N.J., Garber J.E., Kaufman B., Viale G., Fumagalli D., Rastogi P., et al. Adjuvant olaparib for patients with BRCA1- or BRCA2-mutated breast cancer. N Engl J Med. 2021;384:2394–2405. doi: 10.1056/NEJMoa2105215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swisher E.M., Lin K.K., Oza A.M., Scott C.L., Giordano H., Sun J., Konecny G.E., Coleman R.L., Tinker A.V., O'Malley D.M., Kristeleit R.S., Ma L., Bell-McGuinn K.M., Brenton J.D., Cragun J.M., Oaknin A., Ray-Coquard I., Harrell M.I., Mann E., Kaufmann S.H., Floquet A., Leary A., Harding T.C., Goble S., Maloney L., Isaacson J., Allen A.R., Rolfe L., Yelensky R., Raponi M., McNeish I.A. Rucaparib in relapsed, platinum-sensitive high-grade ovarian carcinoma (ARIEL2 part 1): an international, multicentre, open-label, phase 2 trial. Lancet Oncol. 2017;18:75–87. doi: 10.1016/S1470-2045(16)30559-9. [DOI] [PubMed] [Google Scholar]
- 6.Mateo J., Carreira S., Sandhu S., Miranda S., Mossop H., Perez-Lopez R., et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015;373:1697–1708. doi: 10.1056/NEJMoa1506859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abida W., Patnaik A., Campbell D., Shapiro J., Bryce A.H., McDermott R., Sautois B., Vogelzang N.J., Bambury R.M., Voog E., Zhang J., Piulats J.M., Ryan C.J., Merseburger A.S., Daugaard G., Heidenreich A., Fizazi K., Higano C.S., Krieger L.E., Sternberg C.N., Watkins S.P., Despain D., Simmons A.D., Loehr A., Dowson M., Golsorkhi T., Chowdhury S., Triton Investigators Rucaparib in men with metastatic castration-resistant prostate cancer harboring a BRCA1 or BRCA2 gene alteration. J Clin Oncol. 2020;38:3763–3772. doi: 10.1200/JCO.20.01035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poveda A., Floquet A., Ledermann J.A., Asher R., Penson R.T., Oza A.M., Korach J., Huzarski T., Pignata S., Friedlander M., Baldoni A., Park-Simon T.-W., Sonke G.S., Lisyanskaya A.S., Kim J.-H., Filho E.A., Vergote I., Rowe P., Pujade-Lauraine E. Final overall survival (OS) results from SOLO2/ENGOT-ov21: a phase III trial assessing maintenance olaparib in patients (pts) with platinum-sensitive, relapsed ovarian cancer and a BRCA mutation. J Clin Oncol. 2020;38:6002. [Google Scholar]
- 9.Moore K., Colombo N., Scambia G., Kim B.G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., Gourley C., Banerjee S., Oza A., González-Martín A., Aghajanian C., Bradley W., Mathews C., Liu J., Lowe E.S., Bloomfield R., DiSilvestro P. Maintenance olaparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2018;379:2495–2505. doi: 10.1056/NEJMoa1810858. [DOI] [PubMed] [Google Scholar]
- 10.Chi J., Chung S.Y., Parakrama R., Fayyaz F., Jose J., Saif M.W. The role of PARP inhibitors in BRCA mutated pancreatic cancer. Therap Adv Gastroenterol. 2021;14 doi: 10.1177/17562848211014818. 17562848211014818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmitt A., Knittel G., Welcker D., Yang T.P., George J., Nowak M., Leeser U., Büttner R., Perner S., Peifer M., Reinhardt H.C. ATM deficiency is associated with sensitivity to PARP1- and ATR inhibitors in lung adenocarcinoma. Cancer Res. 2017;77:3040–3056. doi: 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 12.Yap T.A., O'Carrigan B., Penney M.S., Lim J.S., Brown J.S., de Miguel Luken M.J., Tunariu N., Perez-Lopez R., Rodrigues D.N., Riisnaes R., Figueiredo I., Carreira S., Hare B., McDermott K., Khalique S., Williamson C.T., Natrajan R., Pettitt S.J., Lord C.J., Banerji U., Pollard J., Lopez J., de Bono J.S. Phase I trial of first-in-class ATR inhibitor M6620 (VX-970) as monotherapy or in combination with carboplatin in patients with advanced solid tumors. J Clin Oncol. 2020;38:3195–3204. doi: 10.1200/JCO.19.02404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yap T.A., Tan D.S.P., Terbuch A., Caldwell R., Guo C., Goh B.C., Heong V., Haris N.R.M., Bashir S., Drew Y., Hong D.S., Meric-Bernstam F., Wilkinson G., Hreiki J., Wengner A.M., Bladt F., Schlicker A., Ludwig M., Zhou Y., Liu L., Bordia S., Plummer R., Lagkadinou E., de Bono J.S. First-in-human trial of the oral ataxia telangiectasia and RAD3-related (ATR) inhibitor BAY 1895344 in patients with advanced solid tumors. Cancer Discov. 2021;11:80–91. doi: 10.1158/2159-8290.CD-20-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roulston A., Zimmermann M., Papp R., Skeldon A., Pellerin C., Dumas-Bérube É., Dumais V., Dorich S., Fader L.D., Fournier S., Li L., Leclaire M.-E., Yin S.Y., Chefson A., Alam H., Yang W., Fugère-Desjardins C., Vignini-Hammond S., Skorey K., Mulani A., Rimkunas V., Veloso A., Hamel M., Stocco R., Mamane Y., Li Z., Young J.T.F., Zinda M., Black W.C. RP-3500: a novel, potent, and selective ATR inhibitor that is effective in preclinical models as a monotherapy and in combination with PARP inhibitors. Mol Cancer Ther. 2022;21:245–256. doi: 10.1158/1535-7163.MCT-21-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lodish H., Berk A., Matsudaira P., Kaiser C.A., Krieger M., Scott M.P., Zipursky L., Darnell J. Section 23.2: the genetic basis of cancer. In Molecular Cell Biology, ed 5. New York, NY: W.H. Freeman. 2003:943–951. [Google Scholar]
- 16.Riaz N., Blecua P., Lim R.S., Shen R., Higginson D.S., Weinhold N., Norton L., Weigelt B., Powell S.N., Reis-Filho J.S. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8:857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mutter R.W., Riaz N., Ng C.K., Delsite R., Piscuoglio S., Edelweiss M., Martelotto L.G., Sakr R.A., King T.A., Giri D.D., Drobnjak M., Brogi E., Bindra R., Bernheim G., Lim R.S., Blecua P., Desrichard A., Higginson D., Towers R., Jiang R., Lee W., Weigelt B., Reis-Filho J.S., Powell S.N. Bi-allelic alterations in DNA repair genes underpin homologous recombination DNA repair defects in breast cancer. J Pathol. 2017;242:165–177. doi: 10.1002/path.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao A., Guo M. Epigenetic based synthetic lethal strategies in human cancers. Biomark Res. 2020;8:44. doi: 10.1186/s40364-020-00224-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jia P., Zhao Z. Characterization of tumor-suppressor gene inactivation events in 33 cancer types. Cell Rep. 2019;26:496–506.e3. doi: 10.1016/j.celrep.2018.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demeulemeester J., Dentro S.C., Gerstung M., Van Loo P. Biallelic mutations in cancer genomes reveal local mutational determinants. Nat Genet. 2022;54:128–133. doi: 10.1038/s41588-021-01005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nguyen L., W M Martens J., Van Hoeck A., Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11:5584. doi: 10.1038/s41467-020-19406-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Srinivasan P., Bandlamudi C., Jonsson P., Kemel Y., Chavan S.S., Richards A.L., et al. The context-specific role of germline pathogenicity in tumorigenesis. Nat Genet. 2021;53:1577–1585. doi: 10.1038/s41588-021-00949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swisher E.M., Kristeleit R.S., Oza A.M., Tinker A.V., Ray-Coquard I., Oaknin A., Coleman R.L., Burris H.A., Aghajanian C., O'Malley D.M., Leary A., Welch S., Provencher D., Shapiro G.I., Chen L.M., Shapira-Frommer R., Kaufmann S.H., Goble S., Maloney L., Kwan T., Lin K.K., McNeish I.A. Characterization of patients with long-term responses to rucaparib treatment in recurrent ovarian cancer. Gynecol Oncol. 2021;163:490–497. doi: 10.1016/j.ygyno.2021.08.030. [DOI] [PubMed] [Google Scholar]
- 24.Setton J., Zinda M., Riaz N., Durocher D., Zimmermann M., Koehler M., Reis-Filho J.S., Powell S.N. Synthetic lethality in cancer therapeutics: the next generation. Cancer Discov. 2021;11:1626–1635. doi: 10.1158/2159-8290.CD-20-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robson M., Im S.A., Senkus E., Xu B., Domchek S.M., Masuda N., Delaloge S., Li W., Tung N., Armstrong A., Wu W., Goessl C., Runswick S., Conte P. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 26.Golan T., Hammel P., Reni M., Van Cutsem E., Macarulla T., Hall M.J., Park J.O., Hochhauser D., Arnold D., Oh D.Y., Reinacher-Schick A., Tortora G., Algül H., O'Reilly E.M., McGuinness D., Cui K.Y., Schlienger K., Locker G.Y., Kindler H.L. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med. 2019;381:317–327. doi: 10.1056/NEJMoa1903387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carreira S., Porta N., Arce-Gallego S., Seed G., Llop-Guevara A., Bianchini D., Rescigno P., Paschalis A., Bertan C., Baker C., Goodall J., Miranda S., Riisnaes R., Figueiredo I., Ferreira A., Pereira R., Crespo M., Gurel B., Nava Rodrigues D., Pettitt S.J., Yuan W., Serra V., Rekowski J., Lord C.J., Hall E., Mateo J., de Bono J.S. Biomarkers associating with PARP inhibitor benefit in prostate cancer in the TOPARP-B Trial. Cancer Discov. 2021;11:2812–2827. doi: 10.1158/2159-8290.CD-21-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frampton G.M., Fichtenholtz A., Otto G.A., Wang K., Downing S.R., He J., et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31:1023–1031. doi: 10.1038/nbt.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagahashi M., Shimada Y., Ichikawa H., Kameyama H., Takabe K., Okuda S., Wakai T. Next generation sequencing-based gene panel tests for the management of solid tumors. Cancer Sci. 2019;110:6–15. doi: 10.1111/cas.13837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weinstein J.N., Collisson E.A., Mills G.B., Shaw K.R., Ozenberger B.A., Ellrott K., Shmulevich I., Sander C., Stuart J.M. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Loo P., Nordgard S.H., Lingjærde O.C., Russnes H.G., Rye I.H., Sun W., Weigman V.J., Marynen P., Zetterberg A., Naume B., Perou C.M., Børresen-Dale A.L., Kristensen V.N. Allele-specific copy number analysis of tumors. Proc Natl Acad Sci U S A. 2010;107:16910–16915. doi: 10.1073/pnas.1009843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ellrott K., Bailey M.H., Saksena G., Covington K.R., Kandoth C., Stewart C., Hess J., Ma S., Chiotti K.E., McLellan M., Sofia H.J., Hutter C., Getz G., Wheeler D., Ding L. Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Syst. 2018;6:271–281.e7. doi: 10.1016/j.cels.2018.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J., Pao W., Engelman J.A., Iafrate A.J., Le L.P. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 34.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. [Preprint] [DOI] [Google Scholar]
- 36.Shen R., Seshan V.E. FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res. 2016;44:e131. doi: 10.1093/nar/gkw520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mandelker D., Zhang L., Kemel Y., Stadler Z.K., Joseph V., Zehir A., et al. Mutation detection in patients with advanced cancer by universal sequencing of cancer-related genes in tumor and normal DNA vs guideline-based germline testing. JAMA. 2017;318:825–835. doi: 10.1001/jama.2017.11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cameron D.L., Baber J., Shale C., Papenfuss A.T., Valle-Inclan J.E., Besselink N., Cuppen E., Priestley P. GRIDSS, PURPLE, LINX: unscrambling the tumor genome via integrated analysis of structural variation and copy number. bioRxiv. 2019 doi: 10.1101/781013. [Preprint] [DOI] [Google Scholar]
- 41.Srinivasan M., Sedmak D., Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 43.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization paths for Cox's proportional hazards model via coordinate descent. J Stat Softw. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap T.A., Silverman I.M., Fontana E., Lee E., Spigel D., Højgaard M., Lheureux S., Mettu N., Carneiro B.A., Carter L., Plummer R., Schonhoft J.D., Ulanet D., Nejad P., Manley P., Reis-Filho J.S., Xu Y., Rimkunas V., Koehler M., Rosen E. Genomic and pathologic determinants of response to RP-3500, an ataxia telangiectasia and Rad3-related inhibitor (ATRi), in patients (pts) with DNA damage repair (DDR) loss-of-function (LOF) mutant tumors in the Phase 1/2 TRESR trial. Cancer Res. 2022;82 Abstract CT030. [Google Scholar]
- 45.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., Brannon A.R., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McHugh M.L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 47.Luthra R., Patel K.P., Routbort M.J., Broaddus R.R., Yau J., Simien C., Chen W., Hatfield D.Z., Medeiros L.J., Singh R.R. A targeted high-throughput next-generation sequencing panel for clinical screening of mutations, gene amplifications, and fusions in solid tumors. J Mol Diagn. 2017;19:255–264. doi: 10.1016/j.jmoldx.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 48.Garcia E.P., Minkovsky A., Jia Y., Ducar M.D., Shivdasani P., Gong X., Ligon A.H., Sholl L.M., Kuo F.C., MacConaill L.E., Lindeman N.I., Dong F. Validation of OncoPanel: a targeted next-generation sequencing assay for the detection of somatic variants in cancer. Arch Pathol Lab Med. 2017;141:751–758. doi: 10.5858/arpa.2016-0527-OA. [DOI] [PubMed] [Google Scholar]
- 49.Herzog T.J., Spetzler D., Xiao N., Burnett K., Maney T., Voss A., Reddy S., Burger R., Krivak T., Powell M., Friedlander M., McGuire W. Impact of molecular profiling on overall survival of patients with advanced ovarian cancer. Oncotarget. 2016;7:19840–19849. doi: 10.18632/oncotarget.7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeTora L.M., Toroser D., Sykes A., Vanderlinden C., Plunkett F.J., Lane T., Hanekamp E., Dormer L., DiBiasi F., Bridges D., Baltzer L., Citrome L. Good Publication Practice (GPP) guidelines for company-sponsored biomedical research: 2022 update. Ann Intern Med. 2022;175:1298–1304. doi: 10.7326/M22-1460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Biallelic loss of function (LOF) genomic alterations affecting genes of interest in The Cancer Genome Atlas data set. A: Frequency of monoallelic versus biallelic LOF for alterations. B: Mechanisms of biallelic LOF. LOH, loss of heterozygosity.
Sequencing coverage profiles in 23 normal samples profiled using SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx), with respect to distance to nearest primer, dependent on insert size distribution for a given sample. A: Unnormalized coverage values. B: Coverage values normalized by median coverage for each sample.
k-Means (k = 3) analysis of insert size distributions across tumor and normal formalin-fixed, paraffin-embedded samples. A: Clustering of insert size distributions. B: Average densities of insert sizes according to sample category.
Example of sample-specific copy number normalization calculations in a leiomyosarcoma sample from an 81-year–old man. A: Using a cutoff of 500× sequencing coverage, single-nucleotide polymorphisms within 168 bp from the primer were selected for inclusion. Gray line denotes sequencing coverage cutoff of 500×. B: Sample-specific insert size distribution. Vertical red lines correspond to quantiles, and blue line corresponds to mean. C: Matching of the sample-specific insert size distribution (from B) to typical insert size distributions derived from the panel of normal samples (PoNs). In this example, the fair PoN profile most resembled the tumor sample and was hence selected for normalization. FFPE, formalin fixed, paraffin embedded.
In silico limit of detection experiments. A: List of tumor-normal sample pairs and their characteristics. B: The algorithm used for sampling of read counts to simulate tumor samples with target purity values. FACETS, Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing; ID, identifier.
Ploidy estimates derived from whole genome sequencing [WGS; Allele-Specific Copy number Analysis of Tumors (ASCAT)] and SyNthetic lethal Interactions for Precision Diagnostics [SNiPDx; Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS)].
In silico experiments simulating low-purity tumor samples by diluting pure tumor samples with read counts from matched normal samples. A: Inference of tumor purity in diluted samples by Fraction and Allele-Specific Copy Number Estimates from Tumor Sequencing (FACETS). Red lines correspond to median of inferred purity; blue lines delineate upper and lower quartiles. B: Accuracy of loss of heterozygosity detection in in silico diluted samples inferred by FACETS. For each simulated purity value, the number in parentheses represents the number of times (of a total of 20) FACETS identified a solution.
A and B: Discordant ataxia-telangiectasia mutated (ATM) biallelic loss of function called by SyNthetic lethal Interactions for Precision Diagnostics (SNiPDx; A) and orthogonal next-generation sequencing (B). Within copy number tracks, black lines denote total copy number, and red lines denote minor copy number, with loss of heterozygosity if minor copy number is 0. A:Black dots denote single-nucleotide polymorphisms. Red lines in the log ratio and log2 odds ratio tracks are a result of segmentation of raw data. In the log ratio track, the purple line denotes log-ratio value inferred at diploid state, and green line denotes median log-ratio level.