Abstract
Simple Summary
Next-generation sequencing (NGS) is a cutting-edge technology that enables rapid, high-throughput sequencing of DNA and RNA. Researchers and clinicians can identify genetic mutations, gene fusions, and other alterations that may drive cancer growth. This is particularly important in precision oncology as it is applied in the context of Molecular Tumor Boards (MTBs). The latter are multidisciplinary teams of experts who use NGS and bioinformatics tools to analyze patients’ genetic profiles and develop personalized treatment recommendations for cancer patients. Thus, a crucial process for MTB decision-making is the analysis, compilation, and presentation of high-dimensional sequencing data, which are used for both preparation of and case presentation to all stakeholders. MIRACUM-Pipe precisely addresses these requirements and offers an easy-to-use, one-prompt standardized solution to analyze NGS data, including quality control, variant calling, copy number estimation, annotation, visualization, and report generation.
Abstract
(1) Background: Next-generation sequencing (NGS) of patients with advanced tumors is becoming an established method in Molecular Tumor Boards. However, somatic variant detection, interpretation, and report generation, require in-depth knowledge of both bioinformatics and oncology. (2) Methods: MIRACUM-Pipe combines many individual tools into a seamless workflow for comprehensive analyses and annotation of NGS data including quality control, alignment, variant calling, copy number variation estimation, evaluation of complex biomarkers, and RNA fusion detection. (3) Results: MIRACUM-Pipe offers an easy-to-use, one-prompt standardized solution to analyze NGS data, including quality control, variant calling, copy number estimation, annotation, visualization, and report generation. (4) Conclusions: MIRACUM-Pipe, a versatile pipeline for NGS, can be customized according to bioinformatics and clinical needs and to support clinical decision-making with visual processing and interactive reporting.
Keywords: molecular tumor board, next-generation sequencing, pipeline, precision oncology, bioinformatics, computational biology, software, workflow, somatic variant calling
1. Introduction
Molecular precision oncology aims to manipulate or influence a defined, direct, or indirect tumor-specific molecular target. The principle and clinical efficacy of highly effective “molecular targeting” has meanwhile been demonstrated for numerous compounds in large phase III trials [1,2,3,4]. However, the use of approved targeted agents currently remains almost exclusively within histologically defined entities and is often dependent on the evidence of a predictive biomarker. In the concept of molecular oncology, a tumor is not defined exclusively by its histological features but primarily by its molecular (genetic) tumor profile. According to molecular oncology, this profile and its associated biomarkers can be similar across entities and require the same molecular treatment. For example, many targeted compounds with promising results are currently in biomarker-stratified, tumor-agnostic phase I/II trials, providing a glimpse of the future molecular approval landscape. Unfortunately, only a few molecularly driven phase I/II trials are currently available (especially in Europe), so molecular drugs outside of regulatory approval are frequently used “off-label” in individual therapeutic trials. These molecularly driven “off-label” therapy trials are within the scope of Molecular Tumor Boards (MTBs). The main aim of MTBs is to provide therapeutic or diagnostic indications, usually based on genomic analysis, for cancer patients. To this end, data on specific and recurrent molecular mechanisms are collected from many individual patient cases to provide scientific and clinical evidence for the efficacy of therapeutic approaches targeting these mechanisms [5,6,7,8,9]. Eventually, this will bring highly evident molecular therapies into routine clinical care. MTBs consist of a multidisciplinary team that combines medical and scientific expertise with translational oncology, molecular biology, and bioinformatics [10,11]. There are several approaches to expanded molecular genetic analysis, each identifying a different spectrum of genetic alterations. In the context of MTBs, either targeted combinations of genes are studied using targeted NGS (tNGS) approaches to identify specific targets for which there are approved drugs or extended genetic diagnostics are performed using whole exome sequencing (WES), which covers all protein-coding genes, accounting for approximately 1% of the total genome. With the increasing complexity of genetic data available for individual patients, molecular alterations need to be interpreted according to predefined standards and ultimately reviewed in a multidisciplinary MTB. The therapeutic interpretation of molecular data as well as the assessment of pathogenic mutations require not only a high level of scientific interdisciplinarity but also a continuous literature search and poses a great challenge to the MTB team. Due to the complexity of the results, the need for software or supporting tools to interpret and present the results is tremendously high [12,13,14].
To support transparent data integration and decision-making across the MTBs we have developed MIRACUM-Pipe, an automated analysis workflow for NGS, that produces reliable and reproducible results across different facilities.
2. Materials and Methods
MIRACUM-Pipe is a workflow that combines many individual tools to create a seamless sequence for comprehensive analysis, annotation, and reporting of NGS data. The workflow mainly consists of several parts: (i) quality control, (ii) alignment, (iii) variant calling, (iv) copy number variation estimation, (v) RNA fusion detection, (vi) annotation, and (vii) reporting and visualization. MIRACUM-Pipe currently supports three different run-modes tailored to the following:
WES analysis, i.e., a patient-matched tumor–normal pair of sequencing samples (DNA only),
tNGS analysis, i.e., tumor-only sequencing on a hybrid capture-based gene panel (DNA and optional RNA),
Tumor-only analysis, i.e., a single tumor sample from WES (DNA only).
Depending on the selected mode different tools, workflows, and parameter settings are used, and the differences are highlighted in the following.
2.1. Quality Control
Preprocessing and quality control are performed with FastQC [https://www.bioinformatics.babraham.ac.uk/projects/fastqc; accession: 26 May 2023], Trimmomatic [15], SAMtools [16,17,18], and bedtools [19] to obtain various quality metrics:
mean base quality,
mean coverage over (exonic) target region,
library size,
insert size length.
2.2. Alignment
The preprocessed sequencing samples are aligned using the Burrows–Wheeler aligner (BWA-MEM) [20] to the reference genome hg19 from UCSC. To obtain high confidence alignments we applied the Genome Analysis Toolkit (GATK) [21] indel realignment, and conducted base quality score recalibration, and duplication removal following the GATK Best Practice recommendations [22,23].
2.3. Variant Calling
Somatic variants for WES tumor–normal pairs are identified and filtered for false positives with VarScan2 to obtain high-confidence variant calls. VarScan2 separates the identified variants into somatic, germline, and loss of heterozygosity (LOH). The latter means that a variant is heterozygous in the germline but evolved to a homozygous variant in the tumor.
VarScan2 is similarly used for tumor-only WES samples. The only difference is that no somatic, germline, nor LOH can be distinguished due to the lack of an appropriate control sample. Therefore, the mpileup routines from VarScan2 are used followed by the false positive filter algorithm.
In the case of tNGS, Mutect2 from GATK is used to identify variants followed by the implemented filtering routine to filter and remove false positives. Mutect2 offers the possibility to supply known platform specific sequencing artifacts and a known germline resource to further limit false positive variant calls and help identify potential germline variants. This is particularly useful when no control sample has been analyzed to identify variants that are unique to the tumor.
2.4. Copy Number Variation Calling
Copy number variations (CNVs) are identified with Control-FREEC [24,25]. A matched control sample together with GC-content is used for normalization. If no control sample is available, only GC-content is used. Additionally, the tool Sequenza [26] is used to bioinformatically infer tumor purity and ploidy, and with the help of the scarHRD R package [27,28] the HRD score of the sample is inferred. Tumor purity and ploidy results are further used as input parameters for Control-FREEC.
2.5. RNA Fusion Calling
RNA fusions are called with the tool FusionCatcher [29] in case RNA sequencing samples are available in tNGS mode.
2.6. Annotation
Identified variants are annotated with the tools ANNOVAR [30,31] and SnpEff [32], covering basic gene annotations from either RefSeq or Ensembl, curated databases like gnomAD [33], ClinVar [34], or InterVar [35], and functional annotation sources contained in dbNSFP [36,37] including 38 prediction scores, e.g., REVEL [38] and 8 conservation scores. SnpEff is used to infer the canonical transcript of the alteration.
In addition, cancer genes from OncoKB [39] as well as known hotspot variants [40,41] are highlighted.
Furthermore, the following sample-specific complex biomarkers are calculated.
tumor mutational burden (TMB),
homologous recombination deficiency (HRD) score,
microsatellite instability (MSI),
bioinformatic tumor purity, and
ploidy.
For TMB calculation, all somatic and protein-coding variants are used. HRD score, consisting of the sum of large-scale transitions (LST) [42], number of telomeric allelic imbalances (TAI) [43], and loss of heterozygosity (HRD-LOH) [44], is inferred, as described above with the R package scarHRD [27] and the Sequenza [26] output as well as ploidy and tumor purity values are also inferred, as described above. Microsatellite status is identified with MSIsensor-pro [45] for tumor–normal pairs and MSIsensor2 [46] for tumor-only cases.
2.7. Functional Enrichment Analysis
Functional enrichment analysis with Fisher’s exact test is performed based on either all identified variants, or all genes affected by CNVs to obtain insights into altered pathways. As a signaling pathway source, the Molecular Signatures Database (MSigDB) [47] with the Hallmark gene sets [48] is used.
2.8. Mutational Signature Analysis
A mutational signature analysis is conducted with the R package YAPSA [49] based on the COSMIC signature V2 [50,51,52].
2.9. Reporting and Visualization
MIRACUM-Pipe reports all protein-coding and protein-altering variants fulfilling all quality metrics, the variant allele frequency (VAF) and population frequency cutoffs. As default cutoffs, a VAF above 5% and a population frequency below 0.1% are set. However, not only simple variants, such as SNVs and InDels, are reported but also all findings regarding to quality metrics, complex biomarkers, CNVs, and RNA fusions, if applicable. This information is presented in the form of an interactive PDF report that includes hyperlinks to curated online data sources, such as Genome Nexus [53], MetaKB [54] and VarSome [55], which provide background information on variants. Alternatively, the results are written in a data format that can be directly imported into cBioPortal [56,57].
3. Results
The results, as well as the appropriate selection and presentation of the MIRACUM-Pipe tools, were carefully chosen and adapted through close collaboration with clinicians and members of the MTBs, based on a comprehensive stakeholder analysis [13].
3.1. MIRACUM-Pipe
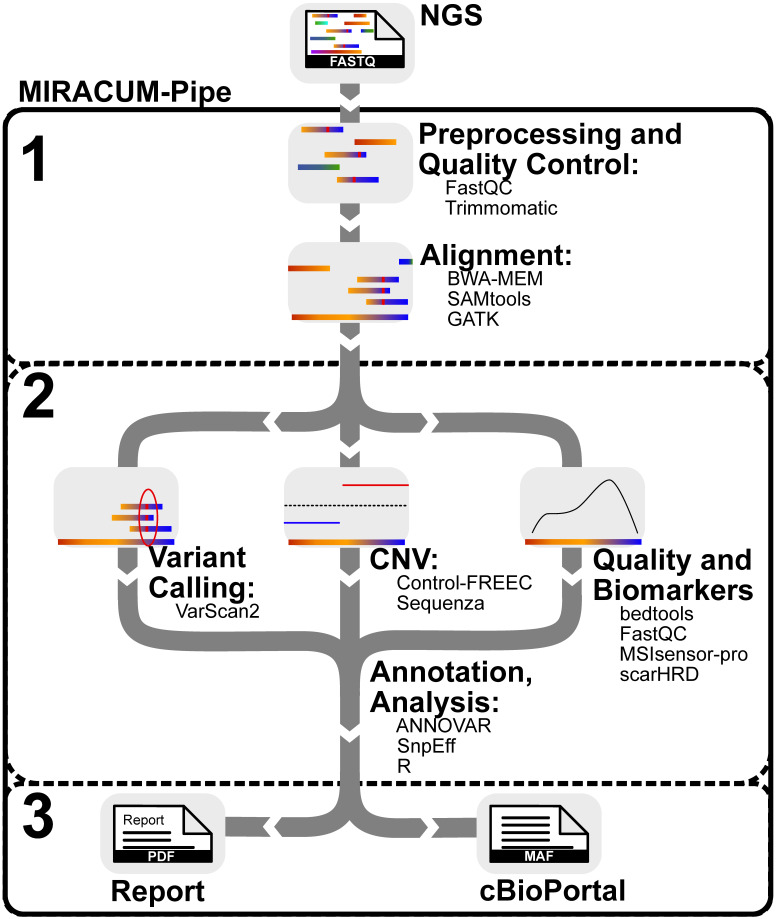
MIRACUM-Pipe incorporates tools for detecting SNVs, InDels, LOH variants, CNVs, and RNA fusions as well as for determining quality and statistics. Various functional prediction and annotation databases are integrated to annotate the identified variants automatically. The workflow is designed as a fully automated one-prompt solution from the raw sequencing files to the interactive PDF report containing quality assessments, the identified and annotated genetic variations as well as a gene set enrichment analysis of the SNVs and CNVs, respectively. MIRACUM-Pipe consists of bash and R scripts to perform NGS data processing, basic annotation, complex functional annotations, and downstream analysis of the results. The pipeline is divided into three main parts, as shown in Figure 1, namely (1) preprocessing, quality control, and alignment; (2) analysis, annotation, and interpretation, including variant calling and copy number calling; and (3) assembly of results into a PDF report and an input format for visualization in cBioPortal. In addition to identifying genetic alterations, another focus is annotating identified variants and other results from various available database sources to facilitate their interpretation.
Figure 1.
Schematic workflow of MIRACUM-Pipe. MIRACUM-Pipe is divided into three main parts: (1) preprocessing, quality control, and alignment, (2) analysis, annotation, and interpretation, subdivided into variant calling (VC) copy number variations (CNV) calling, and quality assessment and biomarker calculation, and (3) assembly of results.
Performance, Usability, and Configuration
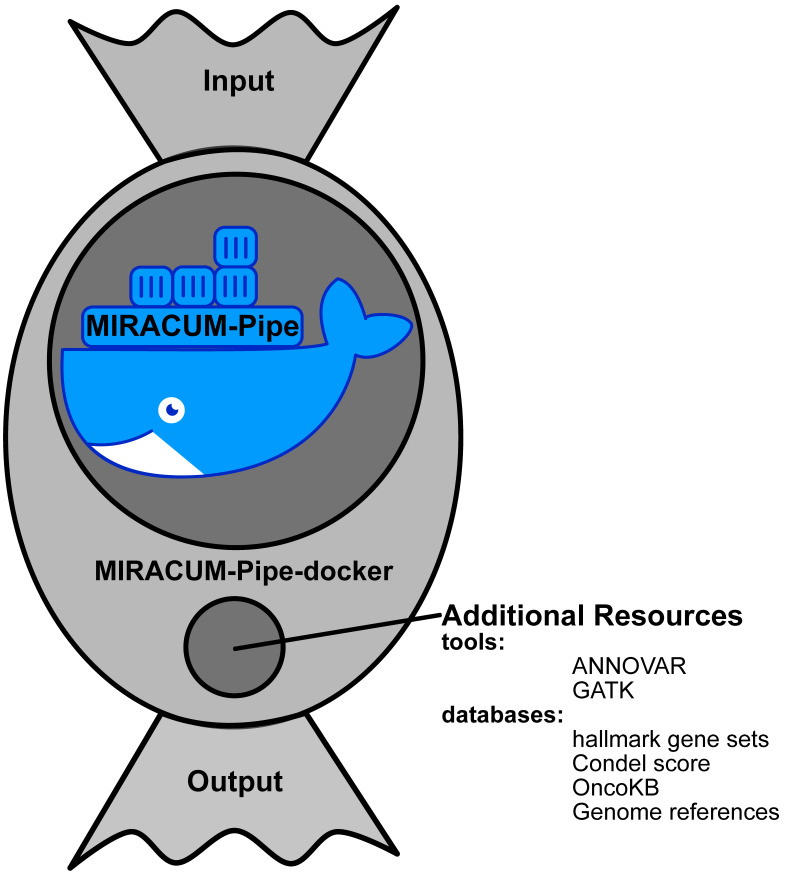
The pipeline is intended to operate on a high-performance computing (HPC) cluster with a minimum number of eight cores, 150 GB of RAM, and 500 GB of hard drive space. After alignment, variant calling and annotation and copy number analysis is run in parallel by default (see Figure 1), saving processing time and distributing resources evenly. As a benchmark example, a WES dataset, consisting of a tumor and a matched germline sample with 100 million paired-end reads each, was analyzed within 12 h on a computer cluster with two 18-Core Intel Xeon E5-2697v4 processors (2.3–3.6 GHz) having 1 TB of RAM. We provide a Docker container to make the installation flawless and error-free, while allowing MIRACUM-Pipe to be distributed to other sites. This container includes a shell script that implements the pipeline processes, certain tools, R libraries, and several databases (Supplementary Material Table S1). Since MIRACUM-Pipe requires additional user-specific databases and files (Supplementary Material Table S2), we designed an environment wrapped around the Docker container (MIRACUM-Pipe-docker). The environment projects the structure inside the Docker container onto the host system, providing an easy solution to add additional databases and tools. It also serves as an interface for data input and output. In addition, the wrapper takes care of the correct Docker syntax when starting the pipeline, and the user can run it as a simple command line tool. This simplifies the application and setup of MIRACUM-Pipe. Due to existing license restrictions, some tools cannot be delivered within the Docker container. To address these issues, our software is split into two GitHub repositories; one for the pipeline itself, which is intended to be used as a Docker container: MIRACUM-Pipe, and another one for its application and setup: MIRACUM-Pipe-docker. The implementation scheme is shown in Figure 2.
Figure 2.
MIRACUM-Pipe implementation scheme with the two available repositories, namely MIRACUM-Pipe and MIRACUM-Pipe-docker. The MIRACUM-Pipe repository builds the Docker container with all necessary source code and most tools, while MIRACUM-Pipe-docker serves as a wrapper to provide additional tools and databases.
3.2. MIRACUM-Pipe Results
Results are provided both as an interactive PDF report and as a machine-readable csv-based file that can be seamlessly imported into cBioPortal.
3.2.1. Interactive PDF Report
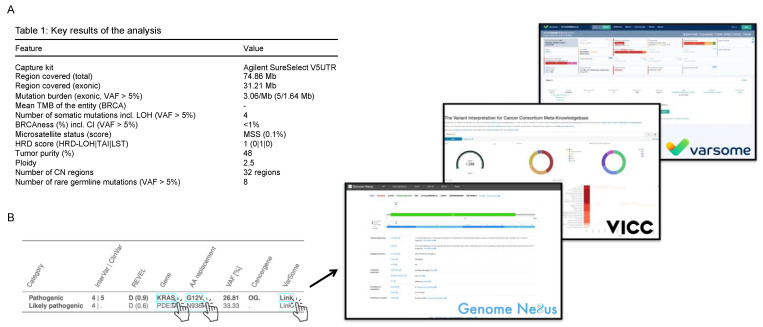
All identified and annotated variants and copy number variations are automatically compiled into a PDF report. All key results are presented on the first page of the report. These include the TMB, HRD score, and BRCAness (Figure 3A). In addition, the mutations, ranked according to the ACMG classification (InterVar/ClinVar [34,35]), are presented in a tabular form. Further information about a gene can be obtained from hyperlinks to the Genome Nexus database [53], and information about an amino acid exchange can be obtained from the Variant Interpretation for Cancer Consortium Meta-Knowledgebase (VICC) [54], or from the VarSome [55] database (Figure 3B). Quality anomalies are also mentioned on the first page of the report to better assess the results. A detailed presentation of the assessed quality metrics is shown in the report in tabular form with corresponding reference ranges (Figure 4A).
Figure 3.
Representations from the first page of the interactive PDF report. (A) The most important analysis results are concisely presented. (B). Listing of variants and their corresponding link to the database of Genome Nexus [53] (https://www.genomenexus.org/variant/chr12:g.25398284C%3EA; accession: 26 May 2023), the Variant Interpretation for Cancer Consortium Meta-Knowledgebase (VICC) [54] (https://search.cancervariants.org/#KRAS%20G12V; accession: 26 May 2023) and VarSome [55] (https://varsome.com/variant/hg19/chr12%3A25398284%3AC%3AA?; accession: 26 May 2023). The variants are also assigned to the categories of pathogenicity. Further parameters are the REVEL Score [38], the information on the variant allele frequency (VAF), and the assignment to cancer genes (tumor suppressor genes (TSGs) or oncogenes (OGs)).
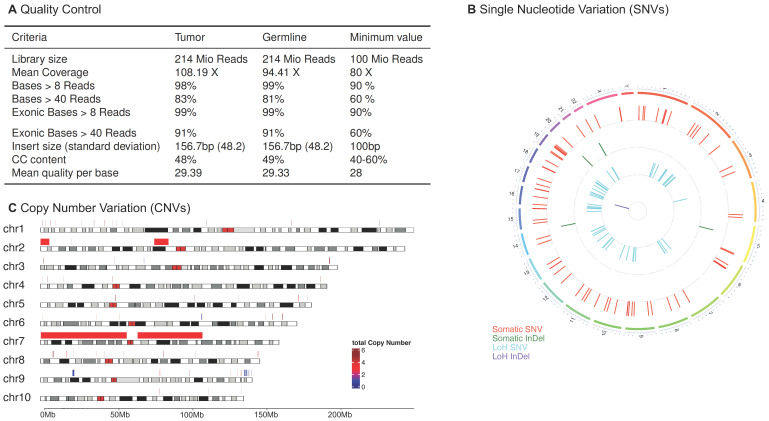
Figure 4.
Visual representations of the three key analysis results of MIRACUM-Pipe: (A) Quality control (QC) parameters: minimum reference values are provided to assess the sufficiency of the NGS results for the other analyses. The table has been modified to be in English for illustrative purposes, whereas the output language of MIRACUM-Pipe is German, as the report is addressed to clinical experts in Germany. (B) Circos plot [58] visualizing the different types of identified variants. Somatic single nucleotide variants (SNVs) are shown in red, somatic insertions and deletions (InDels) in green, loss of heterozygosity (LOH) SNVs in cyan, and (LOH) InDels in purple. Variants are aligned to their actual position on the chromosomes. (C) Selection of an ideogram [59] showing copy number variations (CNVs) in the first ten chromosomes of a patient’s tumor genome. The figure briefly assesses which regions are likely to be amplified (red-shaded regions) or lost (blue-shaded regions), indicating increased or decreased activity of genes within that region. A complete table of genes with CNVs is provided in the full report.
In addition, the report includes the total number of SNVs and InDels, which are further categorized as homozygous or heterozygous variants, and LOH. Mutations are labeled as tumor suppressor genes (TSGs), oncogenes (OGs), or cancer hotspots. They are only considered if they occur in a protein-coding sequence with a population frequency (MAF) below 0.1% and a sample-specific VAF above 5%. However, these thresholds can be adjusted within the pipeline. The identified variants are displayed in a Circos plot (Figure 4B). To better understand the biological processes affected by the variants, a functional enrichment is calculated on hallmark gene sets [48], where terms with the highest significance are reported. In addition to the classical gene set enrichment, the variants are checked against five cancer-associated signaling pathways, namely PI3K-AKT-mTOR, RAF-MEK-ERK, DNA Damage Response, Cell Cycle and Tyrosine Kinases, that play a role in known cancer processes. The lists of genes involved in these pathways were obtained from Qiagen (https://geneglobe.qiagen.com/us/knowledge/pathways; accession: 26 May 2023). The COSMIC mutational signatures, e.g., the BRCAness signature (AC3, DNA damage), which according to Alexandrov et al. [51] provide insight into the selection of therapeutic options, such as PARP inhibitors, calculated using the R package YAPSA [49]. Copy number variations are visualized as an ideogram highlighting the altered regions above the chromosomes (Figure 4C) and explicitly reported for TSGs and Ogs. For a better insight into the processes altered by the chromosomal instabilities, a functional enrichment based on the hallmark gene sets is performed. Furthermore, the quality criteria and coverage are reported. The reports’ appendix lists all detectable somatic variants, LOH, and germline variants that meet all the criteria. Finally, all tools and databases used are listed, including version information.
All provided information related to complex biomarkers, genetic alterations, as well as quality abnormalities, provide clinicians with all the facts necessary to make a comprehensive assessment of the results and prepare a treatment recommendation for discussion within the MTB.
3.2.2. Visualization in cBioPortal
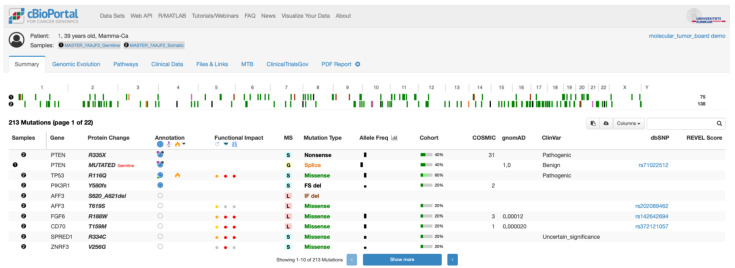
MIRACUM-Pipe reports all findings not only as an interactive PDF report but also in a format for seamless import into cBioPortal. cBioPortal is an open platform for exploring, visualizing, and analyzing multidimensional cancer genomics data and thus supports the translation of large data sets into biological insights and clinical applications [56,57]. For this reason, we have extended cBioPortal according to the needs of the MTB stakeholders [13] within the context of the MIRACUM consortium [60,61,62]. The upload is performed as follows: variants are exported in the mutation annotation format, the copy number variations as discrete copy number values and segmented data, and RNA fusions as structural variants. Additionally, all complex biomarkers and the results of the mutational signature analysis are stored as patient-specific clinical attributes. With all this information at hand, clinicians make extensive use of the features provided by cBioPortal to visualize the mutational landscape (Figure 5) as well as specific variants, including related information on therapeutic options from, e.g., OncoKB [39] (Figure S1). The latter was extended after information on the approval status of a given drug based on the European Medicines Agency (EMA). The extended cBioPortal even offers the possibility to search directly for clinical trials involving the patient’s genetic alterations through an extension of ClincalTrials.gov (Figure S2) [60]. Furthermore, the clinician can use cBioPortal’s powerful virtual trial options to identify similar patients who have already been discussed with the option of documenting therapy recommendations based on the genetic findings generated by MIRACUM-Pipe, thus creating evidence for further similar individual therapeutic trials.
Figure 5.
Visualized summary of the mutational landscape in cBioPortal, hosted for the MTB at the Comprehensive Cancer Center Freiburg (CCCF) depicting all SNVs with appropriate structural and additional clinically relevant information. Added features like REVEL score, ClincalTrials.gov, MTB documentation or PDF report can be seen in additional columns and in the tab bar.
4. Discussion
WES analysis is a high-throughput technology that allows the simultaneous sequencing of thousands of genes in a single experiment. In the context of an MTB, WES analysis can provide clinicians with a wealth of information about a patient’s cancer, including potential driver mutations, actionable targets, and drug resistance mechanisms. Therefore, standardized analysis pipelines and visualization strategies are needed to assist all stakeholders involved in an MTB in handling the complex data that will ultimately lead to therapy recommendations for personalized oncology. In this study, we present the design and use of MIRACUM-Pipe for the analysis of NGS and tNGS data and the associated visualization capabilities of these data.
4.1. Use and Insights of MIRACUM-Pipe in the Context of an MTB
In general, WES provides a comprehensive tumor genome analysis, identifying known and novel mutations in cancer cells. This information can help clinicians understand the molecular drivers of the tumor and select appropriate treatment options. A WES analysis generates a large amount of data and interpreting the results can be challenging. Clinicians may need specialized training to interpret the data and use it effectively to guide treatment decisions. MIRACUM-Pipe has been developed to support this in a mostly automated way. It combines a thorough selection of useful tools, some with appropriate adaptations, and a dedicated annotation process with various databases. However, the greatest advantage of MIRACUM-Pipe is that all codes and analysis processes are presented transparently and can be adapted flexibly if required. Another advantage of MIRACUM-Pipe is the compilation of the results into a PDF report and an input format for the visualization in cBioPortal. The PDF report can document the analysis and be added to the corresponding medical records. Visualization tools, such as cBioPortal, can significantly impact the MTB by providing a clear and concise representation of complex genomic data. cBioPortal can help to identify patterns and relationships within the data that may not be apparent in a text-based format. Visualization tools help the MTB to communicate findings more effectively to other team members and the patients. The interactive PDF report and cBioPortal facilitate discussion and decision-making processes by presenting data in a clear and concise manner. Other existing pipelines, e.g., [63,64,65,66], do not yet provide a comparatively in-depth and clearly structured presentation of genetic variants, quality metrics, complex biomarkers, CNVs, and RNA fusions in single outputs.
4.2. Limitations and Future Directions
The overall flexibility and scalability of the current implementation as a single Docker container is not ideal. Not all features of a current HPC cluster can be used, such as automated queuing and resource distribution. Therefore, we will implement the pipeline in Nextflow’s workflow language [67] in the future. Using a Nextflow workflow offers the possibility of greater flexibility, scalability, and reproducibility, and with Nextflow’s software containers, it becomes easier to exchange tools and incorporate new tools. This modular approach also facilitates the implementation of multiple tools for the same task, such as streamlining the integration of new methodologies. Another technical challenge in using WES data is the quality of the sequencing data. WES analysis requires high-quality DNA samples, and the quality of sequencing data can be affected by several technical factors, such as sequencing depth and coverage. This has to be taken into account in the analysis and has to be manually adjusted in MIRACUM-Pipe accordingly. Similarly, the cost of WES and its analysis must be considered and may currently be a barrier to its widespread use in clinical practice. However, even these technological challenges will dissolve or adapt, so the use of analysis pipelines and visualization tools, such as MIRACUM-Pipe and cBioPortal, will continue to be beneficial.
Although some of the tools used in MIRACUM-Pipe have been developed using machine learning and artificial intelligence (AI) methods, MTB therapy recommendation currently relies on expert knowledge due to the lack of large enough and well-stratified patient cohorts. One approach to compensate for this lack is few-shot learning, which combines individual patient information with additional data from in vitro screening [68]. This situation will change in the future. Current initiatives, such as PM4Onco (Personalized Medicine for Oncology, Medical Informatics Initiative: https://www.medizininformatik-initiative.de/en/node/801, accession: 26 May 2023) aim to harmonize MTB reporting across hospitals to generate a multi-center cohort of genomic and clinical information of MTB cases on which AI methods can be applied for therapy recommendations and response or resistance. Standardized reporting tools, such as MIRACUM-Pipe, will be instrumental in this as they can provide standardized, machine-readable output reports.
5. Conclusions
Next-generation sequencing, particularly WES, is increasingly being used to identify new therapeutic options. However, no common standards for analysis strategies, depth, or medical implementation have been established so far. To overcome these problems and to enable easy-to-use analysis, data interoperability and reuse, we have developed MIRACUM-Pipe. This pipeline can be easily adapted to integrate or merge future databases, analysis tools, and workflows. Furthermore, its visualization capabilities as an interactive PDF report and its integration with cBioPortal facilitate the understanding of the complex data for clinicians and all stakeholders in the MTB.
In this way, MIRACUM-Pipe can support physicians in making personalized therapy recommendations in oncology and unify the standardization of personalized oncology efforts in the German healthcare system.
Acknowledgments
The authors are grateful to the many members of the MIRACUM team and the Molecular Tumor Board Freiburg team who have actively participated in the development of MIRACUM-Pipe. The authors further express their gratitude to Silke Kowar for her technical collaboration and to Geritt Batt for his support with the computing infrastructure management, and to the high throughput sequencing unit team of the DKFZ Genomics and Proteomics Core Facility, Heidelberg, for providing the sequencing services.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15133456/s1, Figure S1: Illustration of cBioPortal and OncoKB. Figure S2: Integration of ClinicalTrials.gov to simplify the search study search. Supplementary Table S1: Tools, R libraries, and databases included in the MIRACUM-Pipe Docker container. Supplementary Table S2: Additional Resources.
Author Contributions
Conceptualization, P.M., H.B., G.A. and M.B.; methodology P.M., M.E.H., A.B., T.P., V.S., R.M., J.C., P.U., N.R. and R.S.; software, P.M., M.E.H., A.B., T.P., V.S., J.C., P.U., R.S., N.R., H.B. and G.A.; validation, P.M., H.B., A.L.I. and M.B.; resources, M.B.; data curation, P.M., M.E.H., T.P., R.M. and M.B.; writing—original draft preparation, P.M. and M.B.; writing—review and editing, P.M., M.E.H., A.B., T.P., V.S., R.M., J.C., P.U., N.R., R.S., A.L.I., H.B., G.A. and M.B.; supervision, M.B. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Please add “The study was conducted in accordance with the Declaration of Helsinki, and approved by the local institutional review board of the Medical Center—University of Freiburg (protocol code 369/19). All patients gave written informed consent.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The two repositories are available on GitHub and can be downloaded from https://github.com/AG-Boerries/MIRACUM-Pipe (accession: 26 May 2023) and https://github.com/AG-Boerries/MIRACUM-Pipe-docker (accession: 26 May 2023).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was funded by the German Federal Ministry of Education and Research (BMBF) by MIRACUM within the Medical Informatics Funding Scheme (01ZZ1801B to P.M. and M.B., 01ZZ1801A to J.C. and P.U.). EkoEstMed–FKZ 01ZZ2015 to G.A. A.L.I. is supported by a Mildred-Scheel-Professorship Grant by the German Cancer Aid (#70114112). H.B. acknowledges funding by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy—EXC 22167-390884018.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Maemondo M., Inoue A., Kobayashi K., Sugawara S., Oizumi S., Isobe H., Gemma A., Harada M., Yoshizawa H., Kinoshita I., et al. Gefitinib or Chemotherapy for Non–Small-Cell Lung Cancer with Mutated EGFR. N. Engl. J. Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 2.Robert C., Karaszewska B., Schachter J., Rutkowski P., Mackiewicz A., Stroiakovski D., Lichinitser M., Dummer R., Grange F., Mortier L., et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N. Engl. J. Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 3.Solomon B.J., Mok T., Kim D.-W., Wu Y.-L., Nakagawa K., Mekhail T., Felip E., Cappuzzo F., Paolini J., Usari T., et al. First-Line Crizotinib versus Chemotherapy in ALK-Positive Lung Cancer. N. Engl. J. Med. 2014;371:2167–2177. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 4.Abou-Alfa G.K., Sahai V., Hollebecque A., Vaccaro G., Melisi D., Al-Rajabi R., Paulson A.S., Borad M.J., Gallinson D., Murphy A.G., et al. Pemigatinib for previously treated, locally advanced or metastatic cholangiocarcinoma: A multicentre, open-label, phase 2 study. Lancet Oncol. 2020;21:671–684. doi: 10.1016/S1470-2045(20)30109-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsimberidou A.M., Fountzilas E., Nikanjam M., Kurzrock R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat. Rev. 2020;86:102019. doi: 10.1016/j.ctrv.2020.102019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tamborero D., Dienstmann R., Rachid M.H., Boekel J., Baird R., Braña I., De Petris L., Yachnin J., Massard C., Opdam F.L., et al. Support systems to guide clinical decision-making in precision oncology: The Cancer Core Europe Molecular Tumor Board Portal. Nat. Med. 2020;26:992–994. doi: 10.1038/s41591-020-0969-2. [DOI] [PubMed] [Google Scholar]
- 7.Horak P., Heining C., Kreutzfeldt S., Hutter B., Mock A., Hüllein J., Fröhlich M., Uhrig S., Jahn A., Rump A., et al. Comprehensive Genomic and Transcriptomic Analysis for Guiding Therapeutic Decisions in Patients with Rare Cancers. Cancer Discov. 2021;11:2780–2795. doi: 10.1158/2159-8290.CD-21-0126. [DOI] [PubMed] [Google Scholar]
- 8.Stenzinger A., Edsjö A., Ploeger C., Friedman M., Fröhling S., Wirta V., Seufferlein T., Botling J., Duyster J., Akhras M., et al. Trailblazing precision medicine in Europe: A joint view by Genomic Medicine Sweden and the Centers for Personalized Medicine, ZPM, in Germany. Semin. Cancer Biol. 2022;84:242–254. doi: 10.1016/j.semcancer.2021.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Westphalen C.B., Bokemeyer C., Büttner R., Fröhling S., Gaidzik V.I., Glimm H., Hacker U.T., Heinemann V., Illert A.L., Keilholz U., et al. Conceptual framework for precision cancer medicine in Germany: Consensus statement of the Deutsche Krebshilfe working group ‘Molecular Diagnostics and Therapy’. Eur. J. Cancer. 2020;135:1–7. doi: 10.1016/j.ejca.2020.04.019. [DOI] [PubMed] [Google Scholar]
- 10.Hoefflin R., Geißler A.-L., Fritsch R., Claus R., Wehrle J., Metzger P., Reiser M., Mehmed L., Fauth L., Heiland D.H., et al. Personalized Clinical Decision Making Through Implementation of a Molecular Tumor Board: A German Single-Center Experience. JCO Precis. Oncol. 2018;2:1–16. doi: 10.1200/PO.18.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoefflin R., Lazarou A., Hess M.E., Reiser M., Wehrle J., Metzger P., Frey A.V., Becker H., Aumann K., Berner K., et al. Transitioning the Molecular Tumor Board from Proof of Concept to Clinical Routine: A German Single-Center Analysis. Cancers. 2021;13:1151. doi: 10.3390/cancers13051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hinderer M., Boerries M., Haller F., Wagner S., Sollfrank S., Acker T., Prokosch H.-U., Christoph J. Supporting molecular tumor boards in molecular-guided decision-making—The current status of five German university hospitals. Stud. Health Technol. Inform. 2017;236:48–54. doi: 10.3233/978-1-61499-759-7-48. [DOI] [PubMed] [Google Scholar]
- 13.Buechner P., Hinderer M., Unberath P., Metzger P., Boeker M., Acker T., Haller F., Mack E., Nowak D., Paret C., et al. Requirements Analysis and Specification for a Molecular Tumor Board Platform Based on cBioPortal. Diagnostics. 2020;10:93. doi: 10.3390/diagnostics10020093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinderer M., Boerries M., Boeker M., Neumaier M., Loubal F.-P., Acker T., Brunner M., Prokosch H.-U., Christoph J. Implementing Pharmacogenomic Clinical Decision Support into German Hospitals. Stud. Health Technol. Inform. 2018;247:870–874. doi: 10.3233/978-1-61499-852-5-870. [DOI] [PubMed] [Google Scholar]
- 15.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danecek P., Bonfield J.K., Liddle J., Marshall J., Ohan V., Pollard M.O., Whitwham A., Keane T., McCarthy S.A., Davies R.M., et al. Twelve years of SAMtools and BCFtools. GigaScience. 2021;10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonfield J.K., Marshall J., Danecek P., Li H., Ohan V., Whitwham A., Keane T., Davies R.M. HTSlib: C library for reading/writing high-throughput sequencing data. Gigascience. 2021;10:giab007. doi: 10.1093/gigascience/giab007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014;47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 20131303.3997 [Google Scholar]
- 21.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., Del Angel G., Rivas M.A., Hanna M., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boeva V., Popova T., Bleakley K., Chiche P., Cappo J., Schleiermacher G., Janoueix-Lerosey I., Delattre O., Barillot E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeva V., Zinovyev A., Bleakley K., Vert J.-P., Janoueix-Lerosey I., Delattre O., Barillot E. Control-free calling of copy number alterations in deep-sequencing data using GC-content normalization. Bioinformatics. 2010;27:268–269. doi: 10.1093/bioinformatics/btq635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Favero F., Joshi T., Marquard A.M., Birkbak N.J., Krzystanek M., Li Q., Szallasi Z., Eklund A.C. Sequenza: Allele-specific copy number and mutation profiles from tumor sequencing data. Ann. Oncol. 2014;26:64–70. doi: 10.1093/annonc/mdu479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sztupinszki Z., Diossy M., Krzystanek M., Reiniger L., Csabai I., Favero F., Birkbak N.J., Eklund A.C., Syed A., Szallasi Z. Migrating the SNP array-based homologous recombination deficiency measures to next generation sequencing data of breast cancer. NPJ Breast Cancer. 2018;4:16. doi: 10.1038/s41523-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team . R: A Language and Environment for Statistical Computing. R Core Team; Vienna, Austria: 2022. [(accessed on 26 May 2023)]. Available online: https://www.R-project.org/ [Google Scholar]
- 29.Nicorici D., Şatalan M., Edgren H., Kangaspeska S., Murumägi A., Kallioniemi O., Virtanen S., Kilkku O. FusionCatcher—A tool for finding somatic fusion genes in paired-end RNA-sequencing data. bioRxiv. 2014:011650. doi: 10.1101/011650. [DOI] [Google Scholar]
- 30.Wang K., Li M., Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. 2015;10:1556–1566. doi: 10.1038/nprot.2015.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cingolani P., Platts A., Wang L.L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M., Maglott D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42:980–985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q., Wang K. InterVar: Clinical Interpretation of Genetic Variants by the 2015 ACMG-AMP Guidelines. Am. J. Hum. Genet. 2017;100:267–280. doi: 10.1016/j.ajhg.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu X., Jian X., Boerwinkle E. dbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X., Li C., Mou C., Dong Y., Tu Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020;12:103. doi: 10.1186/s13073-020-00803-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ioannidis N.M., Rothstein J.H., Pejaver V., Middha S., McDonnell S.K., Baheti S., Musolf A., Li Q., Holzinger E., Karyadi D., et al. REVEL: An Ensemble Method for Predicting the Pathogenicity of Rare Missense Variants. Am. J. Hum. Genet. 2016;99:877–885. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakravarty D., Gao J., Phillips S., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H., et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017;1:1–16. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang M.T., Bhattarai T.S., Schram A.M., Bielski C.M., Donoghue M.T., Jonsson P., Chakravarty D., Phillips S., Kandoth C., Penson A., et al. Accelerating Discovery of Functional Mutant Alleles in Cancer. Cancer Discov. 2018;8:174–183. doi: 10.1158/2159-8290.CD-17-0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang M.T., Asthana S., Gao S.P., Lee B.H., Chapman J.S., Kandoth C., Gao J., Socci N.D., Solit D.B., Olshen A.B., et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat. Biotechnol. 2015;34:155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popova T., Manié E., Rieunier G., Caux-Moncoutier V., Tirapo C., Dubois T., Delattre O., Sigal-Zafrani B., Bollet M., Longy M., et al. Ploidy and Large-Scale Genomic Instability Consistently Identify Basal-like Breast Carcinomas with BRCA1/2 Inactivation. Cancer Res. 2012;72:5454–5462. doi: 10.1158/0008-5472.CAN-12-1470. [DOI] [PubMed] [Google Scholar]
- 43.Birkbak N.J., Wang Z.C., Kim J.-Y., Eklund A.C., Li Q., Tian R., Bowman-Colin C., Li Y., Greene-Colozzi A., Iglehart J.D., et al. Telomeric Allelic Imbalance Indicates Defective DNA Repair and Sensitivity to DNA-Damaging Agents. Cancer Discov. 2012;2:366–375. doi: 10.1158/2159-8290.CD-11-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abkevich V., Timms K.M., Hennessy B.T., Potter J., Carey M.S., Meyer L.A., Smith-McCune K., Broaddus R., Lu K.H., Chen J., et al. Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br. J. Cancer. 2012;107:1776–1782. doi: 10.1038/bjc.2012.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jia P., Yang X., Guo L., Liu B., Lin J., Liang H., Sun J., Zhang C., Ye K. MSIsensor-pro: Fast, Accurate, and Matched-normal-sample-free Detection of Microsatellite Instability. Genom. Proteom. Bioinform. 2020;18:65–71. doi: 10.1016/j.gpb.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niu B., Ye K., Zhang Q., Lu C., Xie M., McLellan M.D., Wendl M.C., Ding L. MSIsensor: Microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics. 2013;30:1015. doi: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hübschmann D., Jopp-Saile L., Andresen C., Krämer S., Gu Z., Heilig C.E., Kreutzfeldt S., Teleanu V., Fröhling S., Eils R., et al. Analysis of mutational signatures with yet another package for signature analysis. Genes Chromosom. Cancer. 2020;60:314–331. doi: 10.1002/gcc.22918. [DOI] [PubMed] [Google Scholar]
- 50.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Campbell P.J., Stratton M.R. Deciphering Signatures of Mutational Processes Operative in Human Cancer. Cell Rep. 2013;3:246–259. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alexandrov L.B., Nik-Zainal S., Wedge D.C., Aparicio S.A.J.R., Behjati S., Biankin A.V., Bignell G.R., Bolli N., Borg A., Børresen-Dale A.-L., et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nik-Zainal S., Davies H., Staaf J., Ramakrishna M., Glodzik D., Zou X., Martincorena I., Alexandrov L.B., Martin S., Wedge D.C., et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Bruijn I., Li X., Sumer S.O., Gross B., Sheridan R., Ochoa A., Wilson M., Wang A., Zhang H., Lisman A., et al. Genome Nexus: A Comprehensive Resource for the Annotation and Interpretation of Genomic Variants in Cancer. JCO Clin. Cancer Inform. 2022;6:e2100144. doi: 10.1200/CCI.21.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wagner A.H., Variant Interpretation for Cancer Consortium. Walsh B., Mayfield G., Tamborero D., Sonkin D., Krysiak K., Deu-Pons J., Duren R.P., Gao J., et al. A harmonized meta-knowledgebase of clinical interpretations of somatic genomic variants in cancer. Nat. Genet. 2020;52:448–457. doi: 10.1038/s41588-020-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Aguilera M.A., Meyer R., Massouras A. VarSome: The human genomic variant search engine. Bioinformatics. 2019;35:1978–1980. doi: 10.1093/bioinformatics/bty897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B.E., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gu Z., Eils R., Schlesner M. gtrellis: An R/Bioconductor package for making genome-level Trellis graphics. BMC Bioinform. 2016;17:169. doi: 10.1186/s12859-016-1051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Unberath P., Mahlmeister L., Reimer N., Busch H., Boerries M., Christoph J. Searching of Clinical Trials Made Easier in cBioPortal Using Patients’ Genetic and Clinical Profiles. Appl. Clin. Inform. 2022;13:363–369. doi: 10.1055/s-0042-1743560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reimer N., Unberath P., Busch H., Börries M., Metzger P., Ustjanzew A., Renner C., Prokosch H.-U., Christoph J. Studies in Health Technology and Informatics. IOS Press; Amsterdam, The Netherlands: 2021. Challenges and Experiences Extending the cBioPortal for Cancer Genomics to a Molecular Tumor Board Platform; pp. 139–143. [DOI] [PubMed] [Google Scholar]
- 62.Ustjanzew A., Desuki A., Ritzel C., Dolezilek A.C., Wagner D.-C., Christoph J., Unberath P., Kindler T., Faber J., Marini F., et al. cbpManager: A web application to streamline the integration of clinical and genomic data in cBioPortal to support the Molecular Tumor Board. BMC Med. Inform. Decis. Mak. 2021;21:358. doi: 10.1186/s12911-021-01719-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Garcia M., Juhos S., Larsson M., Olason P.I., Martin M., Eisfeldt J., DiLorenzo S., Sandgren J., De Ståhl T.D., Ewels P., et al. Sarek: A portable workflow for whole-genome sequencing analysis of germline and somatic variants [version 2; peer review: 2 approved] F1000Research. 2020;9:63. doi: 10.12688/f1000research.16665.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marriott H., Kabiljo R., Al Khleifat A., Dobson R.J., Al-Chalabi A., Iacoangeli A. DNAscan2: A versatile, scalable, and user-friendly analysis pipeline for human next-generation sequencing data. Bioinformatics. 2023;39:btad152. doi: 10.1093/bioinformatics/btad152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sithara A.A., Maripuri D.P., Moorthy K., Ganesh S.S.A., Philip P., Banerjee S., Sudhakar M., Raman K. iCOMIC: A graphical interface-driven bioinformatics pipeline for analyzing cancer omics data. NAR Genom. Bioinform. 2022;4:lqac053. doi: 10.1093/NARGAB/LQAC053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmed Z., Renart E.G., Mishra D., Zeeshan S. JWES: A new pipeline for whole genome/exome sequence data processing, management, and gene-variant discovery, annotation, prediction, and genotyping. FEBS Open Bio. 2021;11:2441–2452. doi: 10.1002/2211-5463.13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Di Tommaso P., Chatzou M., Floden E.W., Barja P.P., Palumbo E., Notredame C. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 2017;35:316–319. doi: 10.1038/nbt.3820. [DOI] [PubMed] [Google Scholar]
- 68.Ma J., Fong S.H., Luo Y., Bakkenist C.J., Shen J.P., Mourragui S., Wessels L.F.A., Hafner M., Sharan R., Peng J., et al. Few-shot learning creates predictive models of drug response that translate from high-throughput screens to individual patients. Nat. Cancer. 2021;2:233–244. doi: 10.1038/s43018-020-00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The two repositories are available on GitHub and can be downloaded from https://github.com/AG-Boerries/MIRACUM-Pipe (accession: 26 May 2023) and https://github.com/AG-Boerries/MIRACUM-Pipe-docker (accession: 26 May 2023).