Abstract
Human programmed cell death protein 1 (PD-1) is a checkpoint protein involved in the regulation of immune response. Antibodies are widely used as inhibitors that block the immune checkpoint, preventing strong immune responses. Pembrolizumab is an FDA-approved IgG4 antibody with PD-1 inhibitory ability for the treatment of melanoma. In this study, we investigated the effect of Pembrolizumab on the conformational changes in PD-1 using extensive molecular modeling and simulation approaches. Our study revealed that during the 200 ns simulation, the average values of the solvent accessible surface area, the radius of gyration, and internal hydrogen bonds of PD-1 were 64.46 nm2, 1.38 nm and 78, respectively, while these values of PD-1 in the PD-1/Pembrolizumab complex were 67.29 nm2, 1.39 nm and 76, respectively. The RMSD value of PD-1 gradually increased until 80 ns and maintained its stable conformation at 0.32 nm after 80 ns, while this value of PD-1 in the PD-1/Pembrolizumab complex maintained an increasing trend during 200 ns. The interaction between PD-1 and Pembrolizumab led to a flexible but stable structure of PD-1. PD-1 rotated around the rotation axis of the C’D loop and gradually approached Pembrolizumab. The number of hydrogen bonds involved in the interactions on the C and C’ strands increased from 4 at 100 ns to 7 at 200 ns. The strong affinity of Pembrolizumab for the C’D and FG loops of PD-1 disrupted the interactions between PD-1 and PD-L1. Inhibition of the interaction between PD-1 and PD-L1 increased the T cell activity, and is effective in controlling and curing cancer. Further experimental work can be performed to support this finding.
Keywords: PD-1, pembrolizumab, T cells, cancer, MD simulation
1. Introduction
Cancer treatment impacts people’s lives and health [1]. Conventional approaches to cancer treatment usually involve a combination of surgery and chemotherapy. However, the disadvantage of this approach is that the cancer cannot be completely cured and chemotherapy is often associated with severe side effects and pain [2]. Immunotherapy is a treatment that uses the immune system to fight cancer, not by targeting the tumor itself, but by activating or suppressing the patient’s immune system to eliminate cancer cells [3]. The development of immunotherapy has revolutionized cancer treatment and helped improve patient survival rates [4].
Regulation of T cell and B-cell activity by the activation or inhibition of immune checkpoints is a widely used approach in immunotherapy [5]. Antigen-specific and antigen-nonspecific signals play a critical role in regulating immune cell activity. Antigen-specific signaling refers to the recognition of cancer cells by T cells through the binding of the T cell receptor (TCR), which is expressed on the surface of T cells, to the major histocompatibility complex (MHC) on the surface of cancer cells [6]. In contrast, antigen-nonspecific signaling refers to the synergistic action of the CD28 protein expressed on the surface of T cells with the TCR to regulate T cell activity [7].
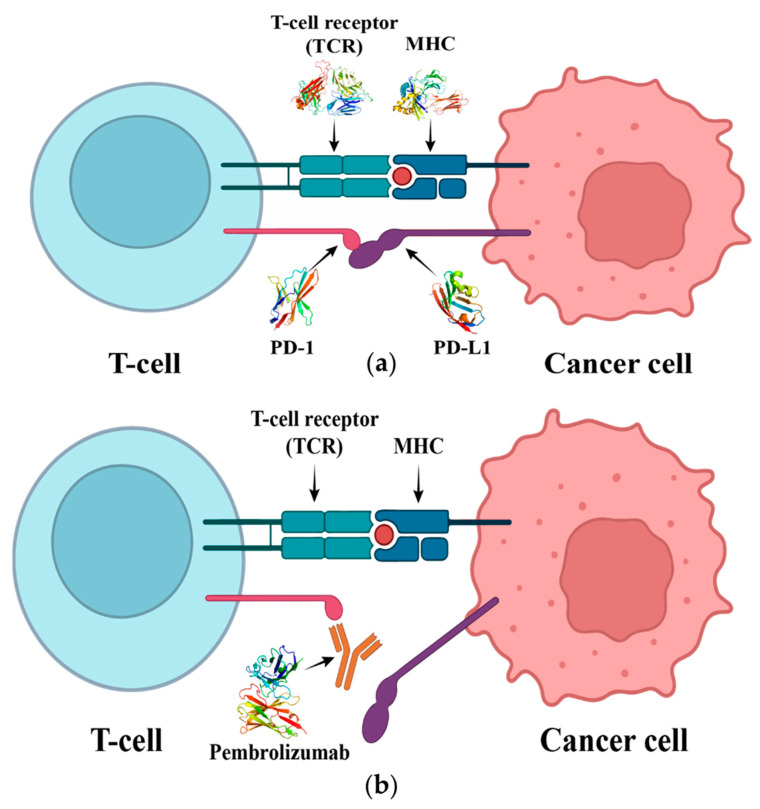
Monoclonal antibody therapies targeting the interaction between Programmed cell death protein 1 (PD-1) and its ligands, Programmed death- ligand 1 (PD-L1) and Programmed death- ligand 2 (PD-L2), have shown remarkable efficacy in treating and curing cancer [8,9]. PD-1 belongs to the CD28 family and is commonly found on the cell membrane surface of activated T cells and B cells [10]. It recognizes endogenous PD-L1 or PD-L2, activating the corresponding signaling pathways to suppress the immune response of T cells [11]. However, PD-L1 and PD-L2 are also commonly present on the surface of some cancer cells, giving them some ability to mount an immune response [12]. PD-1 inhibitors have a high affinity for PD-1 and can compete with endogenous PD-L1 (Figure 1). This leads to an increase in the activity of T cells in destroying cancer cells, which in turn leads to changes in the overall immune system of the patient, possibly causing some autoimmune diseases [13]. Studies of the crystal structure of mouse and human PD-1/PD-L1 have revealed that the PD-1/PD-L1 interaction is based on the CC’ loop and the FG loop of PD-1 [14].
Figure 1.
(a) The interaction of T cell receptor (TCR) with the major histocompatibility complex (MHC). It initiates T cell activation, and the interaction of PD-1 with PD-L1 will lead to the inhibition of T cell. (b) Pembrolizumab blocks the connection between PD-1 and PD-L1, thus preventing T cell inhibition and restoring its activity.
Pembrolizumab, an IgG4 antibody with PD-1 inhibitory activity, was approved by the FDA in 2014 for the treatment of melanoma [15]. The study of the crystal structure of the PD-1/Pembrolizumab complex provided a basis for the exploration of protein–protein interaction (PPI) [16]. However, the knowledge gained from this analysis is limited. The use of molecular dynamics (MD) simulations provides a unique perspective that can be used to further refine our investigations. Conventional crystallization methods provide only a static snapshot of the PPI, whereas molecular dynamics simulations represent a dynamic process that can reflect possible conformational changes in the protein [17,18]. In addition, free energy analyzes based on molecular dynamics simulations can help identify key residues involved in PPI [19]. These data are important for the development of small molecules or peptide inhibitors targeting PD-1. In this article, we present a comprehensive analysis of the interaction mechanism between PD-1 and Pembrolizumab. To this end, molecular dynamics simulations of the PD-1/Pembrolizumab complex were performed using the GROMACS program for a duration of 200 ns. A number of computational methods were used to determine the plausible binding mechanism between PD-1 and Pembrolizumab. Our goal is to contribute to future research on PD-1 and the interaction methods of the antibody Pembrolizumab.
2. Results
2.1. Structure Analysis of PD-1 Complex
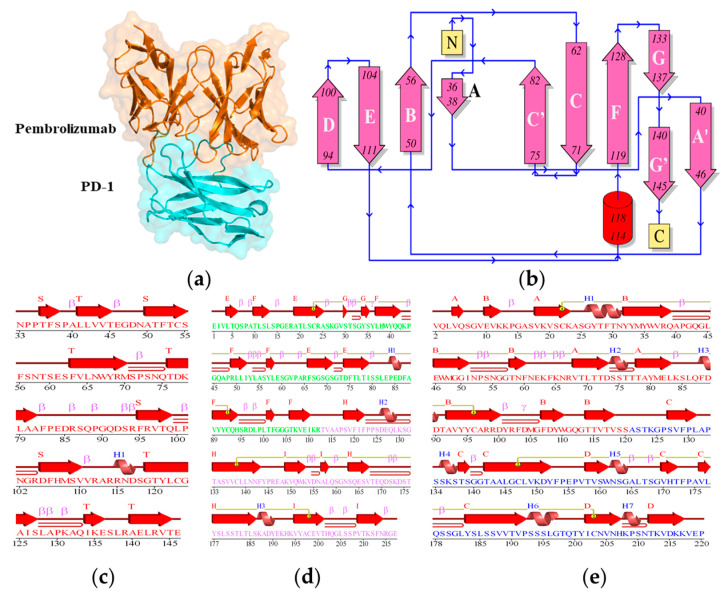
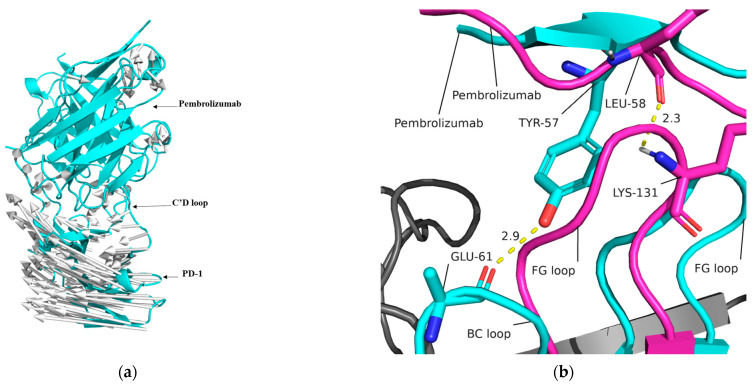
The complex structure of PD-1 and Pembrolizumab was obtained from the Protein Data Bank (PDB: 5B8C) and missing atoms were added to the structure. Pembrolizumab consists of light and heavy chains. PD-1 is a β-sandwich structure with nine loops, of which the FG loop and the C’D loop play the major part in the interaction of PD-1 with Pembrolizumab. The PD-1 has 2 β-sheets, 3 β-hairpins, 2 β-bulges, 10 strands, 1 α-helix, 14 β-turns, 1 γ-turn, and 1 disulphide bond. The light chain of Pembrolizumab has 3 β-sheets, 5 β-hairpins, 3 β-bulges, 12 strands, 1 α-helix, 15 β-turns, 1 γ-turn, and 1 disulphide bond, whereas the heavy chain of Pembrolizumab has 2 β-sheets, 4 β-hairpins, 4 β-bulges, 11 strands, 3 α-helices, 10 β-turns, 1 γ-turn, and 1 disulphide bond. The total secondary structural elements present in the light and heavy chains of Pembrolizumab and PD-1 are shown in Figure 2. It was found that the PD-1 forms hydrogen bonds such as Ser60-Tyr34, Glu61-Tyr57 and Leu128-Tyr53, and van der Waals interactions such as Glu61-Gly33 and Glu61-Tyr34 with the light chain of Pembrolizumab. A salt-bridge Arg86-Arg96 was present between PD-1 and light chain of Pembrolizumab. Several strong hydrogen bonds, van der Waals interactions, and a salt bridge existed between PD-1 and heavy chain of Pembrolizumab (Table 1).
Figure 2.
(a) The crystal structure, and (b) the topology of the PD-1/Pembrolizumab complex. The secondary structure elements of (c) the PD-1, (d) the light-chain of Pembrolizumab and (e) the heavy-chain of Pembrolizumab.
Table 1.
The main residues participated in the hydrogen bonds, Van Der Waals and salt bridges in the crystal structure of PD-1/Pembrolizumab complex.
| Complex | Hydrogen Bonds | Van Der Waals | Salt Bridges |
|---|---|---|---|
| PD-1—Light chain of Pembrolizumab | Ser60-Tyr34, Glu61-Tyr57 and Leu128-Tyr53 | Glu61-Gly33 and Glu61-Tyr34 | Arg86-Arg96 |
| PD-1—Heavy chain of Pembrolizumab | Asn66-Arg102, Gln75-Thr30, Thr76-Tyr101, Asp77-Ser54, Lys78-Tyr101, Lys78-Thr33, Ser87-Arg99, Ser87-Asn59, Ser87-Thr35 and Gly90-Thr58 |
Gln75-Thr28, Lys78-Arg99, Lys78-Arg102, Arg86-Tyr33, Asp85-Tyr101, Ser87-Ser95, Ser87-Leu98, Ser87-Leu100, Gly90-Gly57 and Gly90-Asn59 | Asp85-Arg99 |
2.2. Structure Conformation of PD-1/Pembrolizumab Complex
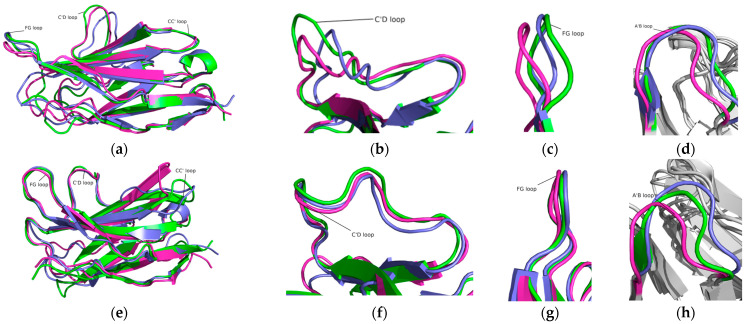
To investigate the structural conformations, the simulated structures of the PD-1 and PD-1/Pembrolizumab complexes were analyzed at 100 ns, 150 ns, and 200 ns (Figure 3). It can be seen that the BC loop, the C’D loop, and the FG loop of PD-1 in the PD-1/Pembrolizumab complex have almost no conformational differences at the three time scales, while some other loops have large conformational differences. All loops in PD-1 show strong fluctuations and divergences, while these loops in the PD-1/Pembrolizumab complex are stable and show the least divergence. The C’D loop of PD-1 in the PD-1/Pembrolizumab complex was found to be relatively distant from the β-sheet and in contact with the Pembrolizumab heavy chain. The FG loop of PD-1 in the PD-1/Pembrolizumab complex shows almost no changes, whereas some deviations in the FG loop have been reported in the case of PD-1. The AB loop in PD-1 and the PD-1/Pembrolizumab complex are not stable. The results suggest that the binding of PD-1 with the antibody Pembrolizumab leads to stabilization of the C’D and FG loops. This strong affinity of Pembrolizumab for the C’D and FG loops of PD-1 leads to the disruption of the PD-1 and PD-L1 interactions. In addition, the FG and A’B loops experience a shift of ~6.5 Å and ~15.5 Å, respectively, during the 200 ns MD simulations.
Figure 3.
Structural dynamics of the PD-1 and PD-1/Pembrolizumab complex at 100 ns (green), 150 ns (slate), and 200 ns (magenta). (a) PD-1, (b) C’D loop in PD-1, (c) FG loop in PD-1, and (d) A’B loop in PD-1. (e) PD-1 in the PD-1/Pembrolizumab complex, (f) C’D loop in the PD-1/Pembrolizumab complex, (g) FG loop in the PD-1/Pembrolizumab complex, and (h) A’B loop in the PD-1/Pembrolizumab complex.
2.3. Interaction of Pembrolizumab and PD-1
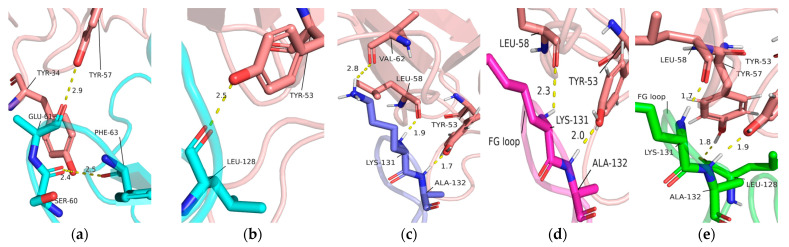
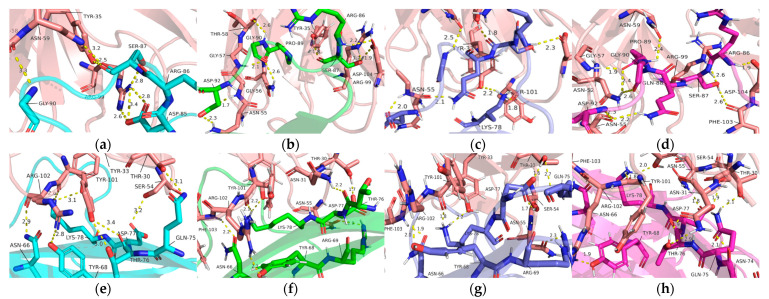
To investigate the interaction mechanism of PD-1 with Pembrolizumab, the simulated structures of the PD-1/Pembrolizumab complex were analyzed at 100 ns, 150 ns, and 200 ns. Interactions occur mainly at three different sites on PD-1, namely in the FG loop, in the C’D loop and near the CC’ loop. Residues Ser60, Glu61 and Phe63 in the BC loop are only involved in the interaction between PD-1 and the light chain of Pembrolizumab with Tyr34 and Tyr57 in the initial state (Figure 4a). Residues Leu128, Lys131, and Ala132 of the FG loop in PD-1 form hydrogen bonds with residues Tyr53, Tyr57, and Leu58, Val62 of the Pembrolizumab light chain during different time scales (Figure 4b–e).
Figure 4.
(a) Interaction details of light chain of Pembrolizumab (tints) with BC loop at initial state. Interaction details of light chain of Pembrolizumab (tints) with FG loop of PD-1 at (b) initial state (cyan), (c) 100 ns (green), (d) 150 ns (slate), and (e) 200 ns (magenta) during MD simulations.
The C’D loop contributes to a large number of hydrogen bonds in the interaction of PD-1 with Pembrolizumab. The C’D loop residues such as Arg86, Ser87, Gln88, Pro89, Gly90 and Asp92 form hydrogen bonds with residues Thr35, Asn52, Asn55, Gly57, Thr58, Asn59, Arg99, Phe103, and Asp104 at 200 ns of Pembrolizumab (Figure 5a–d). CC’ loop regions are also involved in the interaction with the heavy chain of Pembrolizumab. The C strand and C’ strand regions allow more interactions with PD-1 Asn66, Tyr68, Arg69, Asn74, Gln75, Thr76, Asp77, and Lys78 due to the presence of side chain Thr30, Asn31, Tyr33, Ser54, Asn55, Tyr101, Arg102, Phe103, which may contribute to the stability of PD-1 (Figure 5e–h).
Figure 5.
Interaction details of heavy chain of Pembrolizumab (tints) with C’D loop at (a) initial state (cyan), (b) 100 ns (green), (c) 150 ns (slate), and (d) 200 ns (magenta). The C strand and C’ strand region of PD-1 at (e) initial state (cyan), (f) 100 ns (green), (g) 150 ns (slate), and (h) 200 ns (magenta) during MD simulations.
2.4. Structure Deviations and Compactness
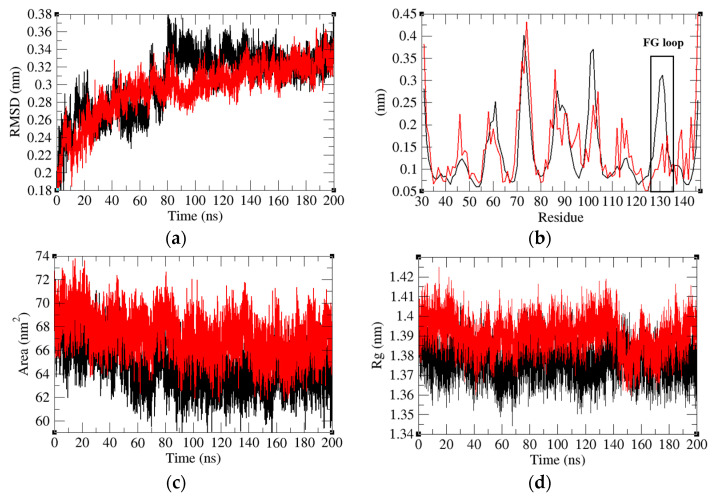
The root mean square deviation (RMSD) is one of the most important properties to determine whether a protein is stable and close to its experimental structure. To better determine the interaction mechanism of PD-1 with Pembrolizumab, we first determined the RMSD values of PD-1 and PD-1 in the PD-1/Pembrolizumab complex. The RMSD value of PD-1 fluctuates upward during the first 80 ns and maintains a stable average value. The average RMSD values during the stable region (150–200 ns) of PD-1 and PD-1 in the PD-1/Pembrolizumab complex were 0.32 nm and 0.33 nm, respectively (Figure 6b). It was found that PD-1 has rather random fluctuations and the binding of Pembrolizumab leads to the minimization of the average fluctuations. The root mean square fluctuation (RMSF) is also commonly used to assess flexibility. The results show that there is a peak at Lys131 of PD-1, the part of the FG loop, while PD-1 in the PD-1/Pembrolizumab complex remains in a lower steady state. This suggests that binding to Pembrolizumab makes the FG loop more stable.
Figure 6.
(a) Root mean square deviations, (b) root mean square fluctuations, (c) solvent accessible surface area, and (d) radius of Gyration of PD-1 (black) and PD-1 in the PD-1/Pembrolizumab complex (red).
A protein’s solvent accessible surface area (SASA) refers to the area of its surface that interacts with its solvent molecules. The average SASA of PD-1 and PD-1 in the PD-1/Pembrolizumab complex was 64.46 nm2 and 67.29 nm2, respectively (Figure 6c). Binding of Pembrolizumab to the loop of PD-1 leads to an increase in the SASA of PD-1. An increase in SASA may lead to the suppression of the effect of PD-1. In addition, the mean radius of gyration (Rg) values of PD-1 and PD-1 in the PD-1/Pembrolizumab complex were found to be 1.38 nm and 1.39 nm, respectively (Figure 6d). The Rg is a parameter linked to the tertiary structural volume of a protein and has been applied to obtain insight into the stability of the protein in a biological system. The Rg values suggest that the binding of Pembrolizumab could slightly relax the structure of PD-1.
2.5. Analysis of Intermolecular and Intramolecular Hydrogen Bonds
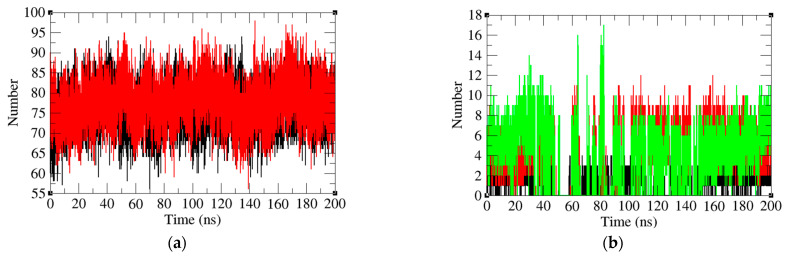
Hydrogen bond analysis is a widely used method to study protein–protein interactions. The average number of hydrogen bonds in PD-1 and PD-1 in the PD-1/Pembrolizumab complex was reported to be 78 and 76, respectively. The Rg results indicate that the PD-1 has a less compact structure (Figure 7a). This might be due to the fact that PD-1 has more loops that can move freely. The average number of hydrogen bonds between Pembrolizumab and three specific regions of the PD-1 protein, namely, the FG loop, the CD loop, and the C and C’ strands were found to be 1, 4, and 4, respectively. (Figure 7b). The number of hydrogen bonds on FG loop and C’D loop remained stable after 80 ns time scale. In contrast, the number of hydrogen bonds on the C strand and C’ strand starts to increase at 150 ns, and eventually increases from 4 at 100 ns to 7 at 200 ns. This may indicate that the number of amino acids involved in the C strand and C’ strand to bind to Pembrolizumab increases.
Figure 7.
(a) Intramolecular hydrogen bond number of PD-1 (black) and PD-1 in PD-1/Pembrolizumab complex (red). (b) Hydrogen bond number between Pembrolizumab and the FG loop (black), the C strand and the C’ strand (green) and C’D loop (red).
2.6. Secondary Structural Analysis
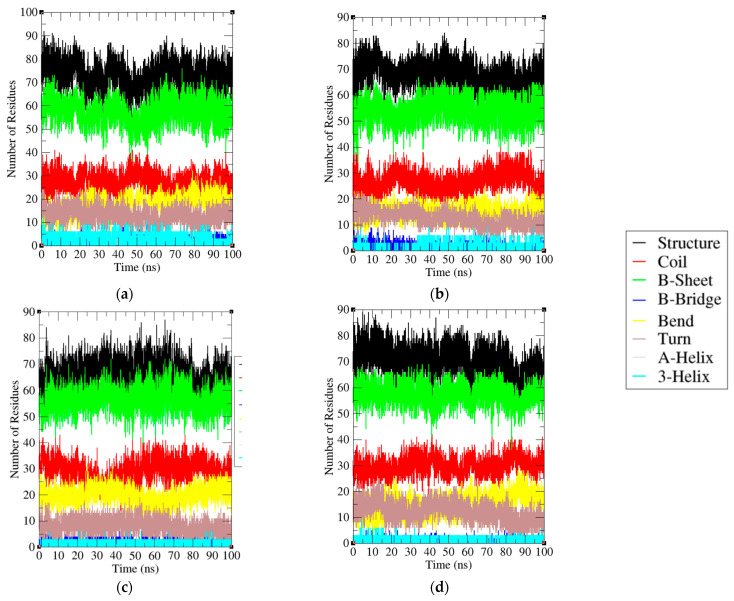
It was found that the average residues involved in the PD-1 total structure, coil, β-sheet, β-bridge, bend, and turn are 57%, 25%, 49%, 1%, 17%, and 7%, respectively. On other hand, the average residues involved in the PD-1/Pembrolizumab total structure, coil, β-sheet, β-bridge, bend, and turn are 61%, 25%, 50%, 0%, 13%, and 11%, respectively (Table 2). A secondary structure analysis of PD-1 and PD-1/Pembrolizumab complex showed that the interaction with Pembrolizumab resulted in a decrease in bends and β-bridges and an increase in the total amount of secondary structure, β-sheet and turn. The increased percentage of amino acids involved in secondary structure formation enhances the stability of the protein structure, leading to inhibition of the interaction between PD-1 and PD-L1. The secondary structure of the light and heavy chains of Pembrolizumab is also shown (Figure 8a–d).
Table 2.
Percentage of average secondary structure elements present in PD-1, and PD-1 in PD-1/Pembrolizumab complex during MD simulations.
| Protein | Structure | Coil | β-Sheet | β-Bridge | Bend | Turn | α-Helix | 3-Helix |
|---|---|---|---|---|---|---|---|---|
| PD-1 | 57% | 25% | 49% | 1% | 17% | 7% | 0% | 1% |
| PD-1/Pembrolizumab complex | 61% | 25% | 50% | 0% | 13% | 11% | 0% | 1% |
Figure 8.
Secondary structure of (a) Light chain of Pembrolizumab, (b) Heavy chain of Pembrolizumab, (c) PD-1, and (d) PD-1 in PD-1/Pembrolizumab complex during 200 ns MD simulation. Total structure, coil, β-Sheet, β-Bridge, Bend, Turn, α-Helix and 3-Helix are represented by black, red, green, blue, yellow, brown, silver and cyan, respectively.
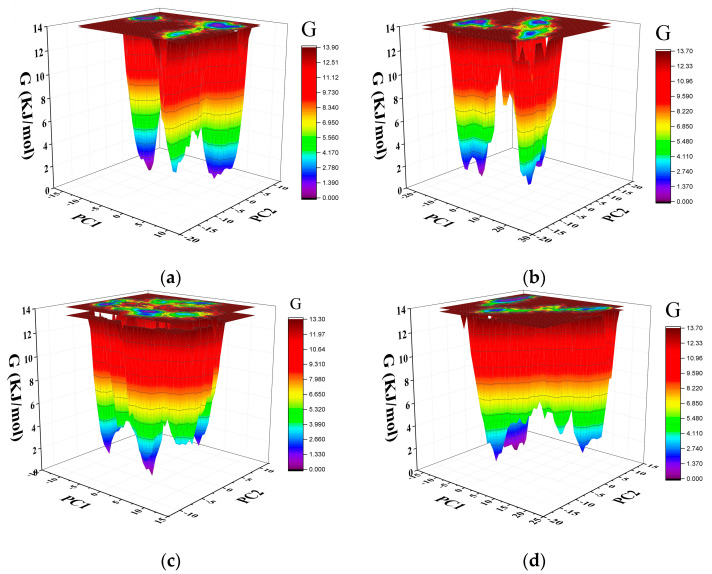
2.7. Gibbs Free Energy Landscape
The Gibbs free energy (GFE) landscape provides information about protein folding and states. The Gibbs free energy landscape was computed using gmx covar, gmx anaeig, and gmx sham using the projections of their own first (PC1) and second (PC2) eigenvectors, respectively. The closer the color is to blue, the lower the free energy. Multiple peaks of low free energy indicate that the protein can switch between different conformations. The GFE landscape of Pembrolizumab light chain, Pembrolizumab heavy chain, PD-1 in the PD-1/Pembrolizumab complex and PD-1 were plotted separately (Figure 9a–d). PD-1 has a larger area of lower free energy regions, and PD-1/Pembrolizumab complex have multiple peaks. This suggests that PD-1 may undergo structural changes more readily.
Figure 9.
The GFE landscape of (a) Pembrolizumab light chain, (b) Pembrolizumab heavy chain, (c) PD-1, and (d) PD-1 in PD-1/Pembrolizumab complex.
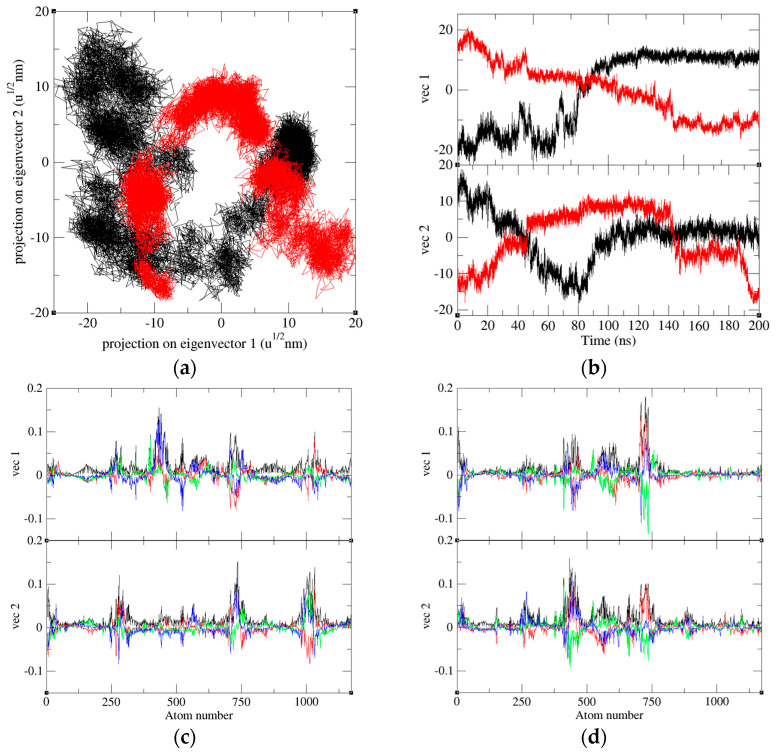
2.8. Principal Component Analysis
PCA is a powerful statistical analysis technique that can show trends, jumps, clusters and outliers by reducing the dimensionality of a data set with a high number of dimensions. The 2D projections of trajectories onto eigenvectors at a different time scale are reported. Interaction of PD-1 with Pembrolizumab results in significant changes in the collective motion of PD-1. During the 100–200 ns MD simulations, the eigenvalues of PD-1 and PD-1 in the PD-1/Pembrolizumab complex were found to be 535.66 nm2 and 408.96 nm2, respectively (Figure 10). The eigenvector trajectory of PD-1 was consistently higher than that of PD-1 in PD-1/Pembrolizumab. The eigenvector 1 and eigenvector 2 of PD-1 and PD-1 in PD-1/Pembrolizumab complex were analyzed and recorded to understand atomic fluctuations and intramolecular collective motions, respectively. The results show that the atomic fluctuations in PD-1 occur in a wider range. In contrast, most atoms of PD-1 in the PD-1/Pembrolizumab complex are relatively stable, with only a few specific loops undergoing large deformations. Most of them are involved in the interaction of PD-1 with Pembrolizumab. This means that the presence of Pembrolizumab makes the structure of PD-1 more stable.
Figure 10.
The plot of eigenvectors with eigen components. (a) The 2D projections of trajectories on eigenvectors of PD-1 (black) and PD-1 in the PD-1/Pembrolizumab complex (red). (b) The projections of trajectories on eigenvectors of PD-1 (black) and PD-1 in complex (red) at different time scale. Eigenvector components were further calculated for (c) PD-1 and (d) PD-1 in the PD-1/Pembrolizumab complex. The eigenvector components pertaining to the total, x-axis, y-axis, and z-axis were represented by the colors black, red, green, and blue, respectively.
2.9. Mechanism of PD-1 and Pembrolizumab Interaction
The 200 ns simulation revealed a displacement of PD-1 and Pembrolizumab. By comparing the structure at 200 ns with the initial state, it was observed that PD-1 rotated as a whole around the axis of the C’D loop in relation to Pembrolizumab (as shown in Figure 11a). Additionally, it was observed that after 200 ns of simulation, the FG loop of PD-1 occupied a binding site on Pembrolizumab, which is the binding site of the BC loop of PD-1 at the starting state (Figure 11b).
Figure 11.
(a) The rotation of PD-1 during MD simulation. The arrows (silver) describe the rotation direction. (b) The FG loop at 200 ns (magenta) occupy the binding site of BC loop at initial state (cyan) during the rotation.
3. Discussion
The immune checkpoint inhibitor agents could reactivate cytotoxic T cells to work against cancer cells. The FDA approval of PD-1 inhibitors (Pembrolizumab) have diversified the clinical activity toward a wide variety of solid tumors including lung cancer, renal cell cancer, and ovarian cancer [20]. A comprehensive analysis of the interactions of the immune checkpoint blockers can provide a better understanding of their therapeutic mechanisms of action [21]. We investigated the effect of Pembrolizumab on the conformational changes in PD-1 using extensive molecular modeling and simulation approaches [22]. During the 200 ns simulation, the relative position and binding sites of PD-1 changed significantly after Pembrolizumab binding. The structural conformation of PD-1 and PD-1 in the PD-1/Pembrolizumab complex shows the interaction with Pembrolizumab makes the loops of the PD-1 binding domain more stable. By comparing the average value of intramolecular hydrogen bonds, the radius of gyration and SASA of PD-1 and PD-1 in the PD-1/Pembrolizumab complex, it was found that the binding of Pembrolizumab makes PD-1 more flexible. By comparing the structures of PD-1 and PD-1/Pembrolizumab complex, we propose that the alteration in the binding site is a consequence of the rotation of PD-1 along the C’D loop axis. CC’ loop region is an essential binding region of PD-1 for Pembrolizumab. The number of hydrogen bonds between the CC’ loop region of PD-1 and Pembrolizumab gradually increased during the simulation. This might be due to the rotation of PD-1. At the same time, the rotation of PD-1 causes the FG loop to occupy the binding site belonging to the BC loop. This also allows the FG loop to bind more stably to Pembrolizumab.
4. Materials and Methods
4.1. Molecular Modeling
The crystal structure of the PD-1/Pembrolizumab complex (PDB: 5B8C) was obtained from the Protein Data Bank [23]. The missing atoms in the structure of PD-1 were modeled using Modeller [24]. The complete protocols are available in previous publications [25,26,27,28,29]. Structural analysis was performed using PDBsum [30] and numerous modules of MD simulations. Discovery Studio [31] and Pymol [32] were used to visualize and draw the structure.
4.2. Molecular Dynamics Simulations
Molecular dynamics simulations were performed on PD-1 and PD-1/Pembrolizumab complex using GROMACS 2018.2 using a standard protocol [33,34,35,36]. Atomic positions were first constrained using a GROMOS96 43a1 force field file. Energy minimization was achieved using the steepest descent algorithm. Two levels of equilibration were performed: NVT (constant number of particles, volume and temperature) and NPT (constant number of particles, pressure and temperature). The details of the methodology of MD are explained in previous publications [37,38].
4.2.1. Root Mean Square Deviation
Root mean square deviation (RMSD) is a fundamental property extensively employed in the analysis of protein structures. It serves as a valuable metric to quantify the differences between the experimental and simulated protein structures obtained from molecular dynamics simulations. The RMSD value enables the assessment of the accuracy and reliability of the MD simulation results, and thereby aids in the interpretation of the structural dynamics and conformational changes that occur during the simulation.
| (1) |
where M denotes the mass of protein and denotes the atomic mass of i. denotes the location of atom i at moment t. denotes the location of atom i in reference structure.
4.2.2. Root Mean Square Fluctuation
The assessment of the fluctuations of atoms in a protein is crucial in the analysis of MD simulations. The root mean square fluctuation (RMSF) is a widely employed metric for quantifying the displacement of a specific atom or group of atoms in the protein structure, relative to a reference structure, based on MD simulation results.
| (2) |
where T denotes the simulation duration. denotes the location of atom i at moment j. denotes the location of atom i in reference structure.
4.2.3. Radius of Gyration
The atomic distribution of the target protein can be assessed using the radius of gyration (Rg), which quantifies the distance between each atom in the protein and its center of mass during rotation. In addition, Rg is frequently employed as an indicator of the compactness of a protein.
| (3) |
where denotes the distance between atom i and the center of mass of the target protein, while represents its mass.
4.3. Principal Component Analysis
Principal component analysis (PCA) is a technique for reducing the dimensionality of large data sets, thereby increasing interpretability while minimizing information loss. The diagonalized covariance matrix is the most appropriate decomposition method.
| (4) |
C represents covariance matrix, N represents the total number of Cα atoms. ri represents the Cartesian coordinate of ith Cα atom. <ri> represents the average Cartesian coordinate in all time steps.
4.4. Gibbs Free Energy Landscape
The GFE landscape shows the conformational changes of the PD-1 and PD-1/Pembrolizumab complex.
| (5) |
Principal component 1 (PC1) represents the linear combination of the variables that extract the maximum variance. Principal component 2 (PC2) represents the vertical linear combination with PC1. kB represents the Boltzmann constant; T represents temperature.
5. Conclusions
This research has contributed to a comprehensive understanding of the dynamic binding mechanism between PD-1 and Pembrolizumab. In this study, 200 ns molecular dynamics simulations were carried out to evaluate the dynamic binding between PD-1 and Pembrolizumab. The results showed that PD-1 became more stable and flexible after interaction with Pembrolizumab. In addition, the number of residues involved in the PD-1/Pembrolizumab interaction on the C and C’ strands in PD-1 gradually increased with the simulation time. This resulted in a change in the conformation of the protein complex and the relative interaction of PD-1 and Pembrolizumab. In addition, the number of intermolecular hydrogen bonds between PD-1 and Pembrolizumab increased, resulting in a more stable interaction between PD-1 and Pembrolizumab.
Acknowledgments
We would like to acknowledge the computational facility provided at Xi’an Jiaotong-Liverpool University, China.
Author Contributions
Conceptualization, F.I.K. and S.W.; methodology, F.I.K. and S.W.; software, F.I.K. and S.W.; validation, F.I.K. and S.W.; formal analysis, F.I.K. and S.W.; investigation, F.I.K. and S.W.; resources, F.I.K. and S.W.; data curation, F.I.K. and S.W.; writing—original draft preparation, S.W.; writing—review and editing, F.I.K.; visualization, F.I.K.; supervision, F.I.K.; project administration, F.I.K.; funding acquisition, F.I.K. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the XJTLU Research Development Fund (RDF-22-02-090).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Feain I.J., Court L., Palta J.R., Beddar S., Keall P. Innovations in radiotherapy technology. Clin. Oncol. R Coll. Radiol. 2017;29:120–128. doi: 10.1016/j.clon.2016.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Waldmann T.A. Immunotherapy: Past, present and future. Nat. Med. 2003;9:269–277. doi: 10.1038/nm0303-269. [DOI] [PubMed] [Google Scholar]
- 4.Couzin-Frankel J. Cancer Immunotherapy. American Association for the Advancement of Science; Washington, DC, USA: 2013. [Google Scholar]
- 5.Wu M., Huang Q., Xie Y., Wu X., Ma H., Zhang Y., Xia Y. Improvement of the anticancer efficacy of PD-1/PD-L1 blockade via combination therapy and PD-L1 regulation. J. Hematol. Oncol. 2022;15:24. doi: 10.1186/s13045-022-01242-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sewell A.K. Why must T cells be cross-reactive? Nat. Rev. Immunol. 2012;12:669–677. doi: 10.1038/nri3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fife B.T., Pauken K.E. The role of the PD-1 pathway in autoimmunity and peripheral tolerance. Ann. N. Y. Acad. Sci. 2011;1217:45–59. doi: 10.1111/j.1749-6632.2010.05919.x. [DOI] [PubMed] [Google Scholar]
- 8.Robert C., Ribas A., Wolchok J.D., Hodi F.S., Hamid O., Kefford R., Weber J.S., Joshua A.M., Hwu W.-J., Gangadhar T.C. Anti-programmed-death-receptor-1 treatment with pembrolizumab in ipilimumab-refractory advanced melanoma: A randomised dose-comparison cohort of a phase 1 trial. Lancet. 2014;384:1109–1117. doi: 10.1016/S0140-6736(14)60958-2. [DOI] [PubMed] [Google Scholar]
- 9.Yi M., Zheng X., Niu M., Zhu S., Ge H., Wu K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer. 2022;21:28. doi: 10.1186/s12943-021-01489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gong J., Chehrazi-Raffle A., Reddi S., Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: A comprehensive review of registration trials and future considerations. J. Immunother. Cancer. 2018;6:8. doi: 10.1186/s40425-018-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patsoukis N., Wang Q., Strauss L., Boussiotis V.A. Revisiting the PD-1 pathway. Sci. Adv. 2020;6:eabd2712. doi: 10.1126/sciadv.abd2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han Y., Liu D., Li L. PD-1/PD-L1 pathway: Current researches in cancer. Am. J. Cancer Res. 2020;10:727. [PMC free article] [PubMed] [Google Scholar]
- 13.Francisco L.M., Sage P.T., Sharpe A.H. The PD-1 pathway in tolerance and autoimmunity. Immunol. Rev. 2010;236:219–242. doi: 10.1111/j.1600-065X.2010.00923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi D., Zhou S., Liu X., Zhao C., Liu H., Yao X. Understanding the structural and energetic basis of PD-1 and monoclonal antibodies bound to PD-L1: A molecular modeling perspective. Biochim. Biophys. Acta BBA-Gen. Subj. 2018;1862:576–588. doi: 10.1016/j.bbagen.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Sun X., Yan X., Zhuo W., Gu J., Zuo K., Liu W., Liang L., Gan Y., He G., Wan H. PD-L1 nanobody competitively inhibits the formation of the PD-1/PD-L1 complex: Comparative molecular dynamics simulations. Int. J. Mol. Sci. 2018;19:1984. doi: 10.3390/ijms19071984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horita S., Nomura Y., Sato Y., Shimamura T., Iwata S., Nomura N. High-resolution crystal structure of the therapeutic antibody pembrolizumab bound to the human PD-1. Sci. Rep. 2016;6:35297. doi: 10.1038/srep35297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollingsworth S.A., Dror R.O. Molecular dynamics simulation for all. Neuron. 2018;99:1129–1143. doi: 10.1016/j.neuron.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klepeis J.L., Lindorff-Larsen K., Dror R.O., Shaw D.E. Long-timescale molecular dynamics simulations of protein structure and function. Curr. Opin. Struct. Biol. 2009;19:120–127. doi: 10.1016/j.sbi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Karplus M., McCammon J.A. Molecular dynamics simulations of biomolecules. Nat. Struct. Biol. 2002;9:646–652. doi: 10.1038/nsb0902-646. [DOI] [PubMed] [Google Scholar]
- 20.Alsaab H.O., Sau S., Alzhrani R., Tatiparti K., Bhise K., Kashaw S.K., Iyer A.K. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: Mechanism, combinations, and clinical outcome. Front. Pharmacol. 2017;8:561. doi: 10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee H.T., Lee S.H., Heo Y.S. Molecular interactions of antibody drugs targeting PD-1, PD-L1, and CTLA-4 in immuno-oncology. Molecules. 2019;24:1190. doi: 10.3390/molecules24061190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baig M.H., Ahmad K., Roy S., Ashraf J.M., Adil M., Siddiqui M.H., Khan S., Kamal M.A., Provazník I., Choi I. Computer aided drug design: Success and limitations. Curr. Pharm. Des. 2016;22:572–581. doi: 10.2174/1381612822666151125000550. [DOI] [PubMed] [Google Scholar]
- 23.Bernstein F.C., Koetzle T.F., Williams G.J., Meyer E.F.J., Brice M.D., Rodgers J.R., Kennard O., Shimanouchi T., Tasumi M. The protein data bank: A computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/S0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 24.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2016;54:5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan F.I., Nizami B., Anwer R., Gu K.R., Bisetty K., Hassan M.I., Wei D.Q. Structure prediction and functional analyses of a thermostable lipase obtained from Shewanella putrefaciens. J. Biomol. Struct. Dyn. 2017;35:2123–2135. doi: 10.1080/07391102.2016.1206837. [DOI] [PubMed] [Google Scholar]
- 26.Khan F.I., Wei D.Q., Gu K.R., Hassan M.I., Tabrez S. Current updates on computer aided protein modeling and designing. Int. J. Biol. Macromol. 2016;85:48–62. doi: 10.1016/j.ijbiomac.2015.12.072. [DOI] [PubMed] [Google Scholar]
- 27.Khan F.I., Govender A., Permaul K., Singh S., Bisetty K. Thermostable chitinase II from thermomyces lanuginosus SSBP: Cloning, structure prediction and molecular dynamics simulations. J. Theor. Biol. 2015;374:107–114. doi: 10.1016/j.jtbi.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 28.Khan F.I., Song H., Hassan F., Tian J., Tang L., Lai D., Juan F. Impact of amino acid substitutions on the behavior of a photoactivatable near infrared fluorescent protein PAiRFP1. Spectrochim. Acta. Part A Mol. Biomol. Spectrosc. 2021;253:119572. doi: 10.1016/j.saa.2021.119572. [DOI] [PubMed] [Google Scholar]
- 29.Khan F.I., Lan D., Durrani R., Huan W., Zhao Z., Wang Y. The lid domain in lipases: Structural and functional determinant of enzymatic properties. Front. Bioeng. Biotechnol. 2017;5:16. doi: 10.3389/fbioe.2017.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laskowski R.A., Jabłońska J., Pravda L., Vařeková R.S., Thornton J.M. PDBsum: Structural summaries of PDB entries. Protein Sci. A Publ. Protein Soc. 2018;27:129–134. doi: 10.1002/pro.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biovia D.S. Discovery Studio Modeling Environment. 2017. [(accessed on 6 June 2023)]. Release. Available online: https://www.bibsonomy.org/bibtex/23b2245c892c43eb0c25b9e7096a7737a/salotz.
- 32.DeLano W.L. The PyMOL Molecular Graphics System. 2002. [(accessed on 6 June 2023)]. Available online: http://www.pymol.org.
- 33.Khan F.I., Hassan F., Anwer R., Juan F., Lai D. Comparative analysis of bacteriophytochrome Agp2 and its engineered photoactivatable NIR fluorescent proteins PAiRFP1 and PAiRFP2. Biomolecules. 2020;10:1286. doi: 10.3390/biom10091286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan F.I., Gupta P., Roy S., Azum N., Alamry K.A., Asiri A.M., Lai D., Hassan M.I. Mechanistic insights into the urea-induced denaturation of human sphingosine kinase 1. Int. J. Biol. Macromol. 2020;161:1496–1505. doi: 10.1016/j.ijbiomac.2020.07.280. [DOI] [PubMed] [Google Scholar]
- 35.Khan F.I., Hassan F., Ali H., Lai D. Mechanism of pH-induced conformational changes in MurE ligase obtained from salmonella enterica serovar typhi. J. Biomol. Struct. Dyn. 2021;39:1898–1905. doi: 10.1080/07391102.2020.1739560. [DOI] [PubMed] [Google Scholar]
- 36.Hess B., Kutzner C., Van Der Spoel D., Lindahl E. GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 37.Khan F.I., Kang T., Ali H., Lai D. Remdesivir strongly binds to RNA-dependent RNA polymerase, membrane protein, and main protease of SARS-CoV-2: Indication from molecular modeling and simulations. Front. Pharmacol. 2021;12:710778. doi: 10.3389/fphar.2021.710778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan F.I., Ali S., Chen W., Anjum F., Shafie A., Hassan M., Lai D. High-resolution MD simulation studies to get mechanistic insights into the urea-induced denaturation of human sphingosine kinase 1. Curr. Top. Med. Chem. 2021;21:2839–2850. doi: 10.2174/1568026621666211105095731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.