Abstract
The development of an ontology facilitates the organization of the variety of concepts used to describe different terms in different resources. The proposed ontology will facilitate the study of cardiothoracic surgical education and data analytics in electronic medical records (EMR) with the standard vocabulary.
Keywords: Ontology, Cardiothoracic Surgery, Common Vocabulary, Education
1. Introduction
The variety of terms typically employed in cardiothoracic surgical education and study can challenge reasoning over different resources due to other representations. We can overcome these challenges with the standard vocabularies that facilitate various concepts' organization as a general idea of something with an ontology [1]. There are several valuable ontologies and terminologies in a medical domain, such as SNOMED CT2 (Systematized Nomenclature of Medicine-Clinical Terms), Unified Medical Language System3 (UMLS), and Cardiovascular Disease Ontology (CVDO)4. But, none of the above cover all terms for clinical study in one place with related codes (e.g., ICD10 and Current Procedural Terminology (CPT) codes are not included in ontologies with all corresponding codes).
2. Methods
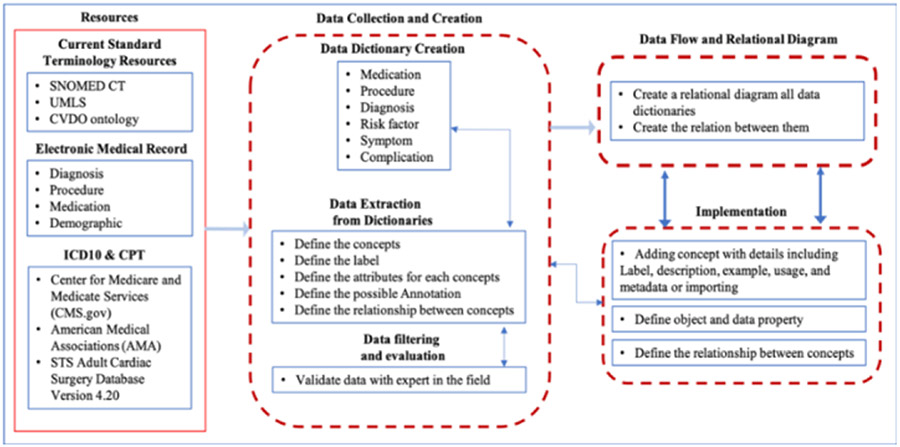
Our systematic approach to the ontology creation is shown in figure 1. In the process of developing the ontology with web protégé [4], we created a new term or imported the term from other resources, we created the definition for each term, and added annotations (e.g., adding comments). Terms are selected from related resources, as shown in figure 1, and with discussion with the experts, we validated the selections.
Figure 1.
shows a systematic approach to the design and development of CTS ontology.
3. Results and Discussion
Our ontology includes descriptions of classes, definitions, and relations. It includes 1041 medication codes in levels of therapeutic, pharmaceutical class, and subclass, ~210 procedure, and ~200 ICD10 codes, 58 risk factors, and the relationships (e.g., “is-a”, “has-a”, “as a result of’). For example, CABG “is-a” surgical procedure “to restore” normal blood flow to an obstructed coronary artery. Statin “is-a” cholesterol-lowering drug “is-taken-by” patients with hyperlipidemia (ICD10 = E78.5) “caused-by” (e.g., obesity).
4. Conclusions
We will deploy our ontology within the BioPortal [2] as a web-services-based portal to enable universal accessibility over the Internet. It allows researchers to access, explore and reuse the resources and products of the related domain [3]. The novelty of this work is filling the gap between different resources with complete ontology for surgical education and data analytics that includes related coeds from EMR.
Footnotes
References
- [1].Gruber TR. A translation approach to portable ontology specifications. Knowledge acquisition. 1993. Jun 1;5(2):199–220. [Google Scholar]
- [2].Noy NF, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic acids research. 2009. Jul 1;37(suppl_2):W170–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Panahiazar M, Sheth AP, Ranabahu A, et al. Advancing data reuse in phyloinformatics using an ontology-driven Semantic Web approach. BMC medical genomics. 2013. Nov;6(3):1–0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Webpage : https://webprotege.stanford.edu, Update. (2022).