Abstract
We previously showed the anti-inflammatory effects of kynurenic acid (KYNA) and its brain-penetrable analog N-(2-(dimethylamino)ethyl)-3-(morpholinomethyl)-4-hydroxyquinoline-2-carboxamide (SZR104) both in vivo and in vitro. Here, we identified the cytomorphological effects of KYNA and SZR104 in secondary microglial cultures established from newborn rat forebrains. We quantitatively analyzed selected morphological aspects of microglia in control (unchallenged), lipopolysaccharide (LPS)-treated (challenged), KYNA- or SZR104-treated, and LPS + KYNA or LPS + SZR104-treated cultures. Multicolor immunofluorescence labeling followed by morphometric analysis (area, perimeter, transformation index, lacunarity, density, span ratio, maximum span across the convex hull, hull circularity, hull area, hull perimeter, max/min radii, mean radius, diameter of bounding circle, fractal dimension, roughness, circularity) on binary (digital) silhouettes of the microglia revealed their morphological plasticity under experimental conditions. SZR104 and, to a lesser degree, KYNA inhibited proinflammatory phenotypic changes. For example, SZR104 treatment resulted in hypertrophied microglia characterized by a swollen cell body, enlarged perimeter, increased transformation index/decreased circularity, increased convex hull values (area, perimeter, mean radius, maximum span, diameter of the bounding circle and hull circularity), altered box-counting parameters (such as fractal dimension), and increased roughness/decreased density. Taken together, analysis of cytomorphological features could contribute to the characterization of the anti-inflammatory activity of SZR104 on cultured microglia.
Subject terms: Microglia, Cellular imaging
Introduction
Microglial cells are macrophage-like cells of mesodermal origin that migrate early during the ontogenetic development to the central nervous system (CNS), where they play important physiological and pathophysiological roles1,2. Under physiological conditions in the CNS, most microglia display a ramified morphology3,4. However, inflammatory signals or injury elicit a variety of morphological and functional changes in these cells, reshaping them into an activated, amoeboid form both in vivo5,6 and in vitro7–10. Activated microglia not only exhibits phagocytosis, antigen presentation11,12, and altered gene expression patterns7,8,13 but dynamically remodeled membrane structures such lamellipodia, pseudopodia, filopodia, and podosomes10,14,15.
Kynurenic acid (KYNA), an endogenous antagonist for several neurotransmitter receptors such as ionotropic glutamate receptors16, influences important (patho)physiological processes including neuroinflammation17, neurodegeneration and -protection18–21 as well as some psychiatric diseases22. Pharmacological experiments demonstrated, for example, that KYNA exerts a neuroprotective role in various inflammatory and/or neurodegenerative CNS disorders23. KYNA is not a viable therapeutic drug as it cannot traverse the blood–brain barrier (BBB); however, brain-penetrable analogs could have the potential to ameliorate neurodegenerative or -inflammatory disorders24. One such analog, N-(2-(dimethylamino)ethyl)-3-(morpholinomethyl)-4-hydroxyquinoline-2-carboxamide (SZR104), has been recently shown to cross the BBB25 and successfully applied to pentylenetetrazol-induced epileptiform seizures, significantly decreasing the seizure-evoked field potentials24. Both KYNA and SZR104 exhibit anti-inflammatory properties under in vitro and in vivo conditions as they not only show potent immunosuppressive capacities in an animal model of epilepsy and inhibition of phagocytosis in microglial cultures26 but decreased the levels of the inflammatory marker proteins C–X–C motif chemokine ligand 10 (CXCL10) and C–C motif chemokine receptor 1 (CCR1) after LPS-treatment in microglial cultures27.
This study was, in part, motivated by the apparent similarities in cytomorphological features observed by treatment with SZR104 and with some other well-established, structurally unrelated anti-inflammatory drugs such as rosuvastatin (RST) and aspirin (ASP). Previously, we demonstrated that the proinflammatory LPS and the anti-inflammatory drugs RST and ASP dynamically and conversely altered not only phagocytosis and the expression of some inflammatory genes but the morphology of microglia7,8. Further, we showed the involvement of the actin cytoskeleton in such morphological changes14. Thus, the present study aimed to quantitatively analyze the effects of LPS, KYNA, and SZR104 on selected cytomorphological features (fractal dimension, lacunarity, area, perimeter, transformation index, circularity, hull area, hull perimeter, hull circularity, mean radius, maximum span across the convex hull, diameter of the bounding circle, max/min radii, span ratio, density, roughness) of binary (digital) silhouettes of control (unchallenged) and LPS-challenged microglial cells with or without KYNA or SZR104 treatment. We used these parameters as most have already been successfully used to characterize microglia in vivo28–30. Our study demonstrates that certain morphometric features are characteristic to the anti-inflammatory form, developed as a response to SZR104 treatment, of microglia under in vitro conditions.
Results
Image analysis pipeline
The complete set of CD11b/c-immunopositive cells (a total of 974 microglia) from the control and treated cultures analyzed for this study is shown in Supplementary Fig. S1. The image processing and measurements pipeline is depicted in Supplementary Figs. S2–S4. At the end of the process, CD11b/c-positive microglial images were converted into binary replicas using thresholding procedures implemented by ImageJ and FracLac (Fig. 1). The quantitative cytomorphological parameter values of individual microglial cells from control and experimentally treated cultures are shown in Supplementary Tables S1 and S2.
Figure 1.
Morphological heterogeneity of microglial cells in control and treated cultures. CD11b/c-positive microglial cells from cultures under different treatments were photographed and the digital images (silhouettes) processed for analysis. Scale bar: 100 μm.
Different culturing environments affect microglia morphology
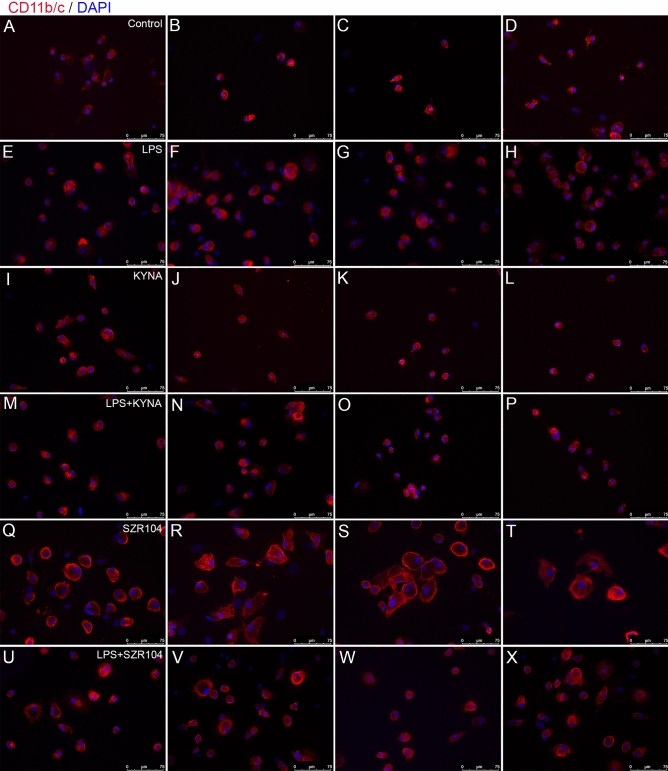
Fluorescent immunohistochemistry analysis showed a mixed population of ameboid and slightly ramified CD11b/c-labeled microglial cells in control (unstimulated) secondary, microglia-enriched cultures (DIV7) (Figs. 1A, 2A–D). Due to the relatively short culturing time, these microglia had only a few short branching processes. As expected, the LPS immunochallenge activated microglia, becoming more reactive and showing a more ameboid body without ramifications (Figs. 1B, 2E–H) compared to control cells. Under the microscope, KYNA-treated cells had relatively small cell bodies, occasionally slightly swollen and barely ramified, sometimes with short filopodia-like processes (Figs. 1C, 2I–L), which did not look significantly different from the controls. Unlike KYNA, its analog SZR104 produced the most profound effects on microglial morphology. SZR104-treated microglia became enlarged and rarely ramified but often displayed filopodia-like processes (Figs. 1E, 2Q–T). Such hypertrophied, swollen cell forms accounted for almost half of the examined microglia in the SZR104-treated group. When SZR104 was added to LPS-challenged cultures (Figs. 1F, 2U–X), the microglial morphology was less hypertrophied and often displayed an intermediate phenotype between the control and LPS-challenged cells, with a less pronounced ameboid shape.
Figure 2.
Immunocytochemical localization of microglial cells in secondary cultures. Secondary, microglia-enriched cultures were prepared and maintained as described in the Materials and Methods. A total of 974 cells were counted under a fluorescence microscope using a 20× or 40× objective. Representative immunocytochemical pictures from unstimulated/control (A–D), LPS- (E–H), KYNA- (I–L), LPS + KYNA- (M–P), SZR104- (Q–T) and LPS + SZR104-treated cultures (U–X). Note the varying levels of morphological heterogeneity of the microglial cells in control (unchallenged) and treated cultures. SZR104-treated cultures included mostly hypertrophied microglia (Q–T). Scale bar: 75 μm.
SZR104 elicits cytomorphological changes associated with the anti-inflammatory phenotype
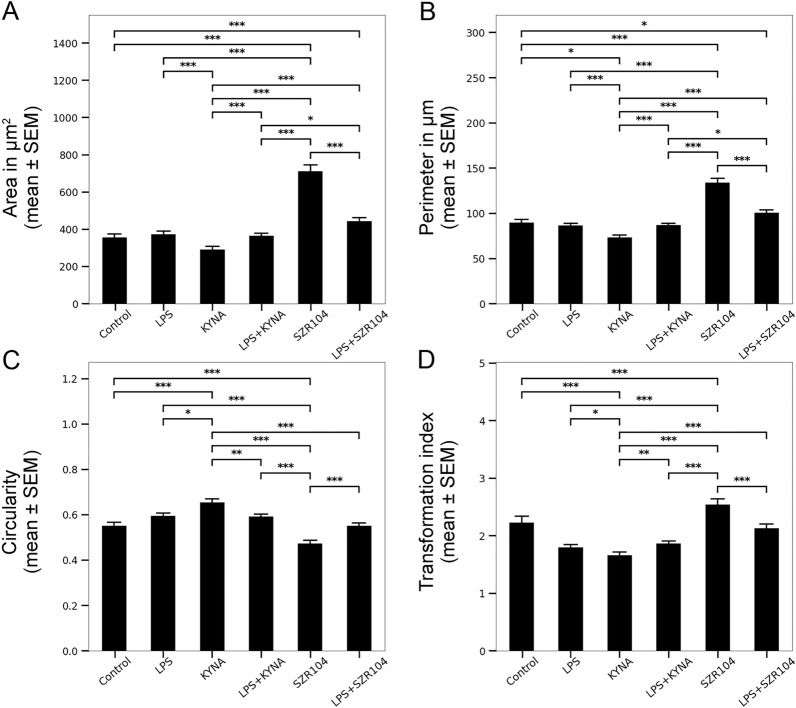
Quantitative analysis on the binary silhouettes of the microglia revealed that several parameters changed significantly with the different treatments (Supplementary Tables S1, S2). According to the cell area and perimeter values, as well as their derived parameters, the circularity and transformation index, microglia treated with SZR104 showed the most obvious changes (Fig. 3). For example, while control microglia had area values of 356.08 ± 18.10 μm2, perimeter values of 89.72 ± 3.43 μm, circularity values of 0.55 ± 0.02, and transformation index values of 2.23 ± 0.11, SZR104-treated cells had area values of 710.79 ± 34.92 μm2, perimeter values of 134.19 ± 4.73 μm, circularity values of 0.47 ± 0.01 and transformation index values of 2.54 ± 0.10, respectively (Fig. 3A–D, Supplementary Tables S1, S2). The significantly increased area, perimeter, and transformation index values, together with the significantly decreased circularity value, strongly indicate SZR1040’s inhibitory effects on the development of a proinflammatory morphology in microglia.
Figure 3.
Morphological heterogeneity of microglial cells under various conditions as evidenced by the area, perimeter, circularity, and transformation index values. CD11b/c-positive microglial cells from the control and differently treated cultures were photographed, digitized, and quantitatively analyzed according to their morphological characteristics. Area (A; μm2), perimeter (B; μm), circularity (C) and transformation index (D) values.
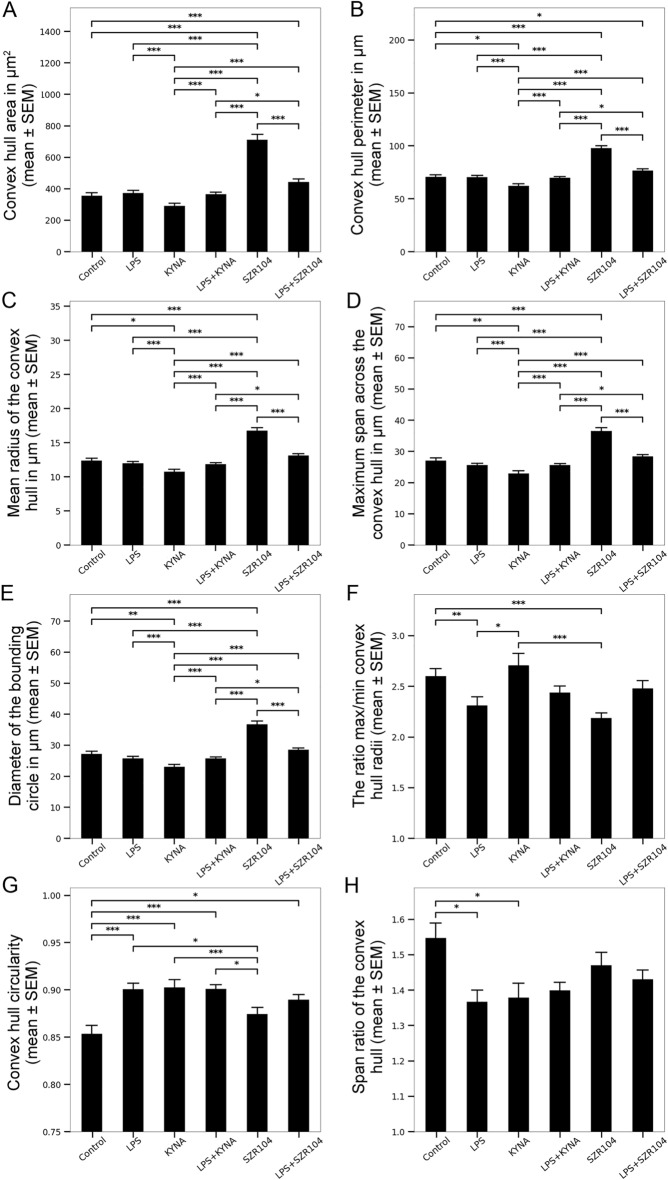
When convex hull parameters were examined using FracLac, a more differentiated pictures emerged. The area of the convex hull (Fig. 4A), the perimeter (Fig. 4B), the mean radius (Fig. 4C), the maximum span across it (Fig. 4D), and the diameter of the bounding circle (Fig. 4E) consistently showed the largest significant differences between controls and SZR104-treated cells. While the maximum/minimum ratio of the convex hull, also showed significantly different values between control (2.60 ± 0.08) and SZR104-treated cells (2.19 ± 0.05; Fig. 4F), other dimensionless hull values, such as the convex hull circularity and the span ratio of the convex hull, showed no significant differences between these two groups (Fig. 4G, H). Most convex hull parameter values for SZR104-treated microglia indicated an increased cell size, a measure often indicative of hypertrophied and swollen cell bodies.
Figure 4.
Morphological heterogeneity of microglial cells under various conditions as evidenced by convex hull parameters (different aspects of the convex polygons, with all interior angles < 180°, containing the whole cell shape). Various convex hull measurements (A–H) were performed in each group in terms of area (μm2) and distance (μm). Convex hull area (A), convex hull perimeter (B), mean radius of the convex hull (C), maximum span across the convex hull (D), diameter of the bounding circle (E), the ration max/min convex hull radii (F), convex hull circularity (G) and span ratio of the convex hull (H) were quantitatively analyzed.
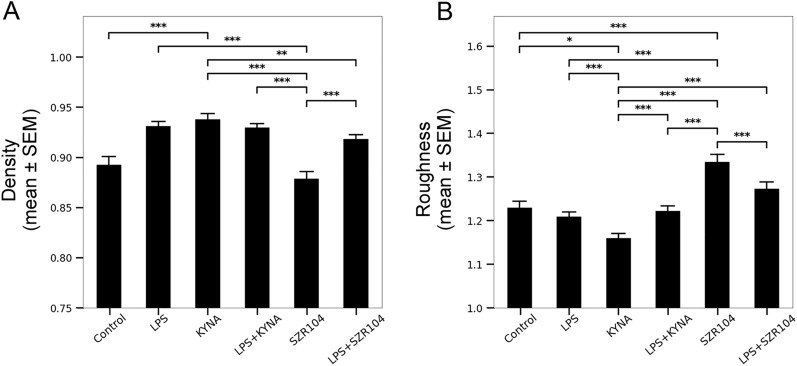
Density and roughness, dimensionless parameters defined as the ratios of cell areas or perimeters to convex hull areas or perimeters, showed the largest differences between control and SZR104-treated microglia (Fig. 5A, B). A decreased density is indicative of a less compact cell, an obvious characteristic of SZR104-treated microglia (Fig. 5A). In these cells, the density was the smallest (0.878695 ± 0.007411) while the roughness was the largest (1.334733 ± 0.017267), indicating that this analog affected cell morphology the most. Interestingly, the roughness value was significantly higher in microglia treated with SZR104 than in any other group (Fig. 5B). Taken together, both density and roughness values indicated that SZR104 inhibited the proinflammatory morphology of microglia.
Figure 5.
Morphological heterogeneity of microglial cells under various conditions as evidenced by density (A) and roughness (B) values. Density and roughness are two additional, dimensionless parameters that can be derived from the area, the perimeter, and the convex hull measurements.
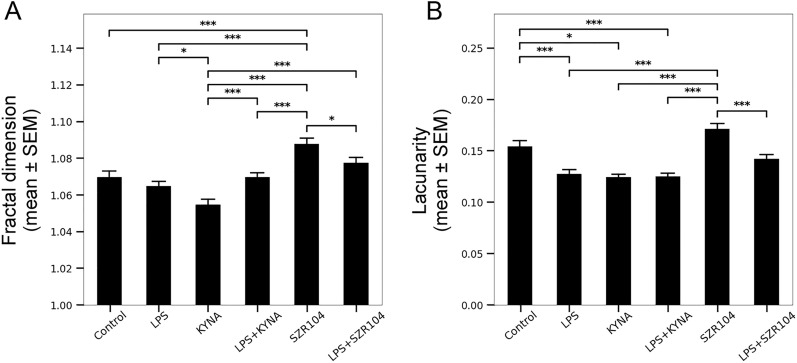
Finally, fractal dimension and lacunarity, determined by the box counting method, were analyzed to determine the heterogeneity and complexity of the cell morphology (Fig. 6). Activation of microglia by LPS and KYNA was evidenced by lower fractal dimension (Fig. 6A) and lacunarity (Fig. 6B), a measure of translational and rotational invariance and increased heterogeneity, respectively. The fractal dimension showed a significant difference (p < 0.001) between control (1.069746 ± 0.003361) and SZR104-treated cells (1.087849 ± 0.003153; Fig. 6A), indicating an increase in pattern complexity. Interestingly, this parameter differed the most between cells treated either with KYNA or its analog SZR104. Whereas KYNA treatment tended to lead to a simpler geometry, SZR104 treatment resulted in more complex cell forms (Figs. 1, 6A). Increased lacunarity was observed in SZR104-treated microglia (Fig. 6B); in fact, SZR104 treatment resulted in the largest lacunarity values among experimental groups (0.171496 ± 0.005049), even if not significantly different from control microglia (0.154234 ± 0.005571). Taken together, fractal dimension and lacunarity parameters indicated a strong anti-inflammatory effect of SZR104 on the microglial cells. All cytomorphological parameters that could collectively point to the anti-inflammatory activity of SZR104 are summarized in Table 1.
Figure 6.
Morphological heterogeneity of microglial cells under various conditions as evidenced by the fractal dimension (A) and lacunarity (B) parameters. Morphological complexity was analyzed using fractal analysis to obtain fractal dimension and lacunarity (translational and rotational invariance) values. Fractal dimension and lacunarity were measured by the box counting method.
Table 1.
SZR104 promotes microglia polarization toward anti‐inflammatory (M2) phenotype.
| Cytomorphological features analyzed in automated image analysis workflow | Change after SZR104 as compared to control | p | Change after SZR104 as compared to LPS challenge | p | Change toward anti-inflammatory phenotype |
|---|---|---|---|---|---|
| Area | Up | *** | Up | *** | Yes |
| Perimeter | Up | *** | Up | *** | Yes |
| Circularity | Down | *** | Down | *** | Yes |
| Transformation index | Up | *** | Up | *** | Yes |
| Convex hull area | Up | *** | Up | *** | Yes |
| Convex hull perimeter | Up | *** | Up | *** | Yes |
| Mean radius of the convex hull | Up | *** | Up | *** | Yes |
| Maximum span across the convex hull | Up | *** | Up | *** | Yes |
| Diameter of the bounding circle | Up | *** | Up | *** | Yes |
| The ratio max/min convex hull radii | Down | *** | Down | N.S. | Likely |
| Convex hull circularity | Up | N.S. | Up | ** | Yes |
| Span ratio of the convex hull | Down | N.S. | Down | N.S. | Likely |
| Density | Down | N.S. | Down | *** | Yes |
| Roughness | Up | *** | Up | *** | Yes |
| Fractal dimension | Up | *** | Up | *** | Yes |
| Lacunarity | Up | N.S. | Up | *** | Yes |
The anti-inflammatory potential (M2 polarization) of SZR104 was classified according to the changes it caused on selected cytomorphological features of cultured microglial cells [see refs.7,8,28,29,57,60]. Significant changes (p) are indicated as: *** (p < 0.005), ** (p < 0.05). N.S.: non-significant.
Discussion
Microglial morphology under physiological and pathophysiological states31,32 have been extensively analyzed both in vivo28 and in vitro7,8,10. Under in vivo conditions, microglial morphology is greatly influenced by the age of the host33 and the surrounding tissue cytoarchitecture34 resulting in a characteristic spatial arrangement of their branches that are sometimes difficult to precisely reconstruct. While microglial cells also show a wide range of morphological differentiation in vivo28,29, their ramified processes are flattened to a monolayer during culturing and thus analyzing their digital silhouettes easily10,35.
The morphological changes in microglia occurring in response to various physiological and pathological conditions are well documented. Upon activation, sedentary, ramified microglia became reactive, often displaying ameboid shape and a unique set of immunoreactive markers1,12. Microglial hypertrophy has been studied extensively in vivo in several mammalian species including mice36, rats37, and humans38. When activated by physiological stimuli such as salt stress37, diabetic induction39, high fat diet36, neuronal trauma40, infection41 or neurodegenerative stimuli38, microglia became hypertrophied. Interestingly, both ameboid and ramified microglia can become hypertrophic42. Although hypertrophic microglia are sometimes indicative of neuronal tissue injury36,43, this microglial phenotype is dominant during physiological activation as well37. Moreover, a recent study observed dystrophic but not hypertrophied microglia in Alzheimer’s disease38.
The quantitative morphology parameters for SZR104 consistently showed an anti-inflammatory drug in action. While the ramified, surveillant phenotype of the microglia with a relatively small cell body and long primary and thin secondary branches display a high fractal dimension as well as high lacunarity and perimeter values together with low density and typically larger hull density values, the ameboid, activated, phagocytosis-capable microglia is characterized by small, roundish cell bodies without branches that would be associated to low fractal dimension, lacunarity and perimeter values as well as typically lower hull density values. This latter form corresponds to the activated, proinflammatory phenotype, capable of avid phagocytosis and proinflammatory cytokine production. Conversely, microglia treated with SZR104 show an increased area, perimeter, and transformation index values, as well as a decreased circularity value and high complex hull, density, and roughness values, all indicative of hypertrophied and swollen cell bodies. These findings further support that SZR104 inhibited the proinflammatory morphology in microglia induced by LPS. Previous in vivo and in vitro studies suggested that SZR104 is an anti-inflammatory agent17,26,27,44. In an animal model of epilepsy, for example, we showed that SZR104 potently inhibited both the synthesis of the proinflammatory IL-6 in the hippocampus during pilocarpine-induced seizures and the proinflammatory morphological transformation of microglia following status epilepticus26. Moreover, we also showed that SZR104 strongly inhibited microglial phagocytosis26 and the synthesis of the inflammatory marker proteins CXCL10 and CCR1 after LPS-treatment in microglial cultures27. Furthermore, our university-wide KYNA research project (GINOP 2.3.2-15-2016-00034) demonstrated that SZR104 not only inhibited tumor necrosis factor-α (TNF-α) synthesis, a molecule known for its proinflammatory function, and increased the expression of the anti-inflammatory gene TNF-Stimulated Gene-6 in U-937 cells after immunochallenge17, but also protected against sepsis-associated neutrophil activation and brain mitochondrial dysfunction in rats44. Thus, our previous in vivo and in vitro results and the present in vitro data seem to support the anti-inflammatory effects of SZR104.
Our previous in vitro studies also showed that hypertrophic phenotypes can also be of anti-inflammatory nature, as observed in RST- and ASP-activated hypertrophic microglia7,8. We found that these two widely used pleiotropic drugs displaying anti-inflammatory properties, affected microglia very similarly after LPS immunochallenge in culture7,8. When secondary microglia were cultured for a week and treated with SZR104, RST, or ASP cells displayed a distinct morphological characteristic with transformation index values significantly higher than those in the control (unchallenged) or LPS-treated microglia cultures. This similarity was observed irrespectively to the purity of the microglial cultures used as microglia-enriched secondary cultures for KYNA and SZR104 had, on average, 78% of purity, while pure microglial cultures for RST and ASP had > 99% purity7,8. When treated with SZR104, RST or ASP, microglial cells acted very similarly and became enlarged, often with two-fold increase in their area, and developed short filopodia and pseudopodia, effectively increasing their perimeter. The combined treatment of LPS with either SZR104, RST7 or ASP8 resulted in a similarly elevated area, perimeter, and transformation index values of the cultured microglia.
Phenotypic plasticity of microglia is a feature of the normal structural remodeling that accompanies microglial activation37, where hypertrophy of cytoplasm is a characteristic feature of the activated microglia12,36. Based on our present quantitative measurements, we assume that the hypertrophic enlargement of the cell somata and filopodia formation exemplify anti-inflammatory processes in microglia. The cytomorphological changes elicited by SZR104 were like those of other anti-inflammatory drugs such as RST and ASP and point out to the involvement of actin cytoskeleton14.
The synthetic SZR104 is a structural analog of the endogenous KYNA. It must be noted that despite their chemical similarity, they are not necessarily functional analogs in every aspect; indeed, they have different physiological, biochemical, pharmacological, or cytomorphological properties (for example, while KYNA is an endogenous compound, SZR104 is a synthetic molecule; KYNA cannot pass the blood–brain barrier, SZR104 can; both have anti-inflammatory properties, etc.). Studies on the relationship between molecular and biological similarity showed that cell morphology changes and chemical structure differences contain valuable information for the prediction of biological activity45–47. While the extensive molecular structure modifications of SZR104 certainly attributed to its improved anti-inflammatory characteristics as compared to that of KYNA, the mode of action is still undetermined.
Collectively, our quantitative morphology data strongly indicate that SZR104 is a more potent anti-inflammatory activator of microglia than the physiologic KYNA. As it can also cross the BBB, SZR104 could be a potential target molecule to explore the endogenous KYNA pathway when applied in vivo. In conclusion, our cytomorphology-based analysis can provide useful information for predicting a measure of biological activity, such as the anti-inflammatory nature, of a KYNA analog.
Materials and methods
Animals
All animal experiments were carried out in strict compliance with the European Council Directive (86/609/EEC) and EC regulations (O.J. of EC No. L 358/1, 18/12/1986) regarding the care and use of laboratory animals for experimental procedures and followed relevant Hungarian and local legislation requirements. Our experimental protocols were approved by the Institutional Animal Welfare Committee of the University of Szeged (II./1131/2018). Pregnant Sprague–Dawley rats (190–210 g) were maintained under standard housing conditions and fed ad libitum.
Reagents
KYNA was purchased from Sigma-Aldrich (Steinheim, Germany). The KYNA analog SZR104 was synthesized in house at the Institute of Pharmaceutical Chemistry, University of Szeged, Hungary. For this study, we further optimized the previously published synthesis of the amide and aminoalkylated derivatives48,49. The direct amidation was carried out in ethanol at reflux, using 1,2 equivalent of amine, 6 h reaction time (yield: 78%). The subsequent aminoalkylation was also optimized as the 3 equivalent aqueous formaldehyde previously described was replaced by a 1,2 equivalent of paraformaldehyde resulting in a yield of 89%50. KYNA and SZR104 were dissolved in phosphate buffered saline (PBS) and added to the cell cultures at a final concentration of 1 μM. Unless stated otherwise, all reagents were purchased from Sigma (St. Louis, MO, USA).
Preparation of high-purity microglial cultures
Mixed primary neuron/glia cultures were established from newborn rats of both sexes. The animals were decapitated in accordance with ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines/experimental-procedures) and AVMA Guidelines for the Euthanasia of Animals (https://www.avma.org/sites/default/files/2020-02/Guidelines-on-Euthanasia-2020.pdf; see Section M3.7 for explanation). The brains were quickly removed and cleared from the meninges. The forebrains were minced with scissors and incubated in 9 mL of Dulbecco’s Modified Eagle’s Medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 1 g/L of d-glucose, 110 mg/L of Na-pyruvate, 4 mM L-glutamine, 3.7 g/L of NaHCO3, 10,000 U/mL of penicillin G, 10 mg/mL of streptomycin sulfate, and 25 μg/mL of amphotericin B, supplemented with 1 mL 2.5% trypsin (Invitrogen) for 10 min at 37 °C, followed by centrifugation at 1000 × g for 10 min at room temperature (RT)10,51,52. The pellet was resuspended in 10 mL DMEM containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen) and filtered using a sterile filter (100 µm pore size; Greiner Bio-One Hungary Kft., Mosonmagyaróvár, Hungary), to eliminate the tissue fragments that resisted dissociation. The filtered cell suspension was centrifuged for 10 min at 1000 × g at RT and the pellet resuspended in 5 mL DMEM/10% FBS; then, the primary mixed cells were seeded in the same medium on poly-L-lysine-coated culture flasks (75 cm2; 107 cells/flask). The number of collected cells was determined in a Bürker chamber after trypan blue staining. The cultures were maintained at 37 °C in a humidified air atmosphere supplemented with 5% CO2. The medium was changed the following day and on the fourth day.
The preparation of secondary, microglia-enriched cultures from mixed primary forebrain cultures followed a slightly modified version of our previous protocol51. Briefly, after seven days of culture, the microglial cells in the primary cultures were shaken off using a platform shaker (120 rpm for 20 min) at 37 °C, collected from the supernatant by centrifugation (3000 × g for 8 min at RT), resuspended in 4 mL of DMEM/10% FBS and plated in the same medium on poly-l-lysine-coated coverslips (15 × 15 mm; 2 × 105 cells/coverslip) for immunocytochemistry51. The number of collected cells was determined in a Bürker chamber after trypan blue staining. After allowing the cells to adhere to the poly-l-lysine-coated surface for 30 min, the supernatant which may contain floating cells was carefully removed and cell culture medium (DMEM/10% FBS) was added to the cells. On the sixth day of subcloning, the culture medium was replaced, and the microglia were treated for 24 h with either LPS (20 ng/mL), KYNA (1 μM), or SZR104 (1 μM) alone, or with a combination of LPS + KYNA or LPS + SZR104. LPS treatment served as immunochallenge. Microglial cells seeded on coverslips were fixed in 0.05 M PBS (pH 7.4 at RT) containing 4% formaldehyde for 10 min at RT and stored at – 20 °C until use.
Fluorescent immunocytochemistry
For the identification of microglial cells, we used the anti-CD11b/c antibody (clone OX42) as it recognizes a common epitope shared between CD11b and CD11c (integrin αM and αX chains) and reacts with all monocytes and macrophages53,54, identifying resident microglia in the CNS10. Light microscopic immunofluorescence labeling was performed as previously described51. Fixed secondary cultures on coverslips were washed three times for 5 min each in 0.05 M PBS. After permeabilization and blocking of nonspecific sites for 30 min at 37 °C in 0.05 M PBS containing 5% normal goat serum, 1% heat-inactivated FBS albumin, and 0.05% Triton X-100, the cells on the coverslips were incubated overnight at 4 °C with the microglia-specific mouse anti-CD11b/c monoclonal primary antibody (1:100 final dilution; Abcam, Cambridge, England). The cultured cells were then washed four times for 10 min each at RT in 0.05 M PBS and incubated with the goat anti-mouse Alexa Fluor 568 fluorochrome-conjugated secondary antibody (final dilution: 1/1000; Invitrogen, Carlsbad, CA, USA, without Triton X-100) in the dark for 3 h at RT. The cells on the coverslip were washed four times for 10 min each in 0.05 M PBS at RT, rinsed in distilled water, and mounted on microscope slides covered with Prolong Diamond Antifade with DAPI (ThermoFisher). To confirm the specificity of the secondary antibody, omission control experiments (i.e., staining without the primary antibody) were performed. In these experiments, no immunocytochemical signals were observed.
Image analysis
Digital images were captured by a Leica DMLB epifluorescence microscope using a Leica DFC7000 T CCD camera (Leica Microsystems CMS GmbH, Wetzlar, Germany) and the LAS X Application Suite X (Leica). For each culture, 15–20 randomly selected microscope fields per coverslip from ≥ 8 coverslips were counted and analyzed using the computer programs ImageJ (version 1.47; developed at the U.S. National Institutes of Health by W. Rasband, available at https://imagej.net/Downloads;55) and the plugin FracLac for ImageJ (https://imagej.nih.gov/ij/plugins/fraclac/fraclac.html;56,57). The complete set of CD11b/c-immunopositive cells analyzed for this study from the control and treated cultures is shown in Supplementary Fig. S1. For the measurements, CD11b/c-positive microglial images were converted into binary replicas using thresholding procedures implemented in ImageJ and FracLac. The image processing and measurements pipeline is depicted in Supplementary Figs. S2, S3. Digital images in tagged image file formats (.tif) were opened in ImageJ (Supplementary Fig. S2A). The measurements were performed on segmented (binary) and processed images of selected individual cells (400 × 400 pixel cropped images) identified by both the DAPI-labeled cell nuclei (Supplementary Fig. 2SB) and the CD11b/c-positive cytoplasm (Supplementary Fig. S2C). To this end, each fluorescence image obtained was processed as follows: the labeled cytoplasmic and nuclear compartments of individual microglial cells were identified on the two-channel image (Supplementary Fig. S2A), but the blue (Supplementary Fig. S2B) and the red (Supplementary Fig. S2C) color channel images were converted to 8-bit grayscale images and threshold separately selecting the following sequence of menus: Image → Color → Split channels/Merge channels/Channels Tool…, Image → Type → 8-bit (Supplementary Figs. S2D, E, F, S3G). Since the Alexa Fluor 568 fluorochrome-conjugated secondary antibody-labeled cytoplasm showed a weaker staining in the location of the cell nuclei, merging their binary silhouettes (cleared foreground pixels of threshold color channels) with the binary silhouettes of the corresponding cell nuclei proved beneficial in many cases, resulting in complementary silhouettes (Supplementary Fig. S3H, I, J). For this reason, two color channels (red and blue) were used to extract the binary silhouettes. Thresholding was performed separately on the images of the two-channels (after channel splitting and converting to 8-bit grayscale images; Supplementary Fig. S2D, E), since transforming them together into 8-bit grayscale images would lead to them canceling out each other's intensity values; further, after binarization, the cells could become connected to neighboring ones due to the parallel use of two-channels.
In most cases, a few simple ImageJ commands were sufficient to clean the binary silhouettes (Analyze→ Analyze Particles: Size: depending on the circumstances, Show: Masks, Exclude on edges; Process→ Binary: Fill holes, Watershed); not all commands were necessarily applied in each case (see58 for details, and the documentation web pages for ImageJ at http://rsb.info.nih.gov/ij/docs/index.html); however, in some cases manual editing was required to separate the microglial cells in the binarized images (Freehand selections, Edit → Cut, Wand tool, Edit→ Selection→ Make Inverse). The ImageJ Macro Recorder was used to generate macro codes to apply commands and automate image processing in some cases. For instance, Plugins→ Macros → Record… →’used commands’ → Create→ Run provided a cell profile formed by a continuous, filled set of pixels (Supplementary Fig. S3H, I, J). These final, clean, and merged binary silhouettes (Supplementary Fig. S3K) were used to measure most analyzed parameters (such as area, perimeter, lacunarity and convex hull measurements), but before measuring fractal dimension, we applied the Process→ Binary → Outline commands on the images (Supplementary Fig. S3K; see also the black contour lines enclosing the dark gray areas in Supplementary Fig. S3L). Using the FracLac plugin, Box Counting (BC) measurements were performed without selecting “Legacy model”. The process involved the following commands: “Use binary”, “Lock white as background”. In “Grid design” section, the Scaling Method of “Power series” with base 2 and exponent 2 was selected, and “Db” (to measure the fractal dimension by box counting) was chosen in the corresponding dialog box. Convex hull measurements were performed with the following settings: in “Grid design”, the number of grid orientations was set to 0, and in Graphics options, “Hull and circle” and “metrics” as well as “bounding circle” and “convex hull” were selected (black edges of convex polygons enclosing the dark gray areas and white concavities and black bounding circles enclosing the light gray parts as well) (Supplementary Figs. 3L, S4). The full set of binary silhouettes used in this study is shown in Fig. 1.
The quantitative parameters used to analyze microglial morphology (described in detail in28,29,58–60) were grouped into four classes: (1) fractal dimension and lacunarity (computed using box counting software); (2) area (number of pixels in the filled shape of the binary image, μm2), perimeter (μm), transformation index (determined according to Fujita et al.61 using the following formula: [perimeter of the cell (μm)]2/4π [cell area (μm2)]), and circularity (calculated as [4π × cell area (μm2)]/cell perimeter (μm)2]; note that transformation index = 1/circularity where the circularity value of a circle = 1); (3) convex hull area (smallest convex polygon containing the whole cell shape, μm2), convex hull perimeter (single outline of the convex hull, μm), convex hull circularity ([4π × convex hull area]/[convex hull perimeter]2; this value is 1 for a circle), mean radius of the convex hull (mean length, μm) from the center of mass of the convex hull to an exterior point, maximum span across the convex hull (maximum distance between two points across the convex hull, μm), diameter of the bounding circle (the smallest circle that encloses the convex hull, μm), max/min radii (ratio of the largest to the smallest radius from the center of mass of the convex hull to an exterior point), and span ratio of the convex hull (ratio of the major to the minor axes of the convex hull, as a measure of cell shape/elongation); and 4) density (ratio of the cell area to its convex hull area), and roughness (ratio of cell perimeter to convex hull perimeter) were analyzed using ImageJ and FracLac28,29,55–57,59,60,62,63.
Statistical analysis
A total of 974 microglia, identified by the presence of the microglia-specific marker CD11b/c, were quantitatively analyzed: 148 controls, 131 LPS-treated, 107 KYNA-treated, 215 LPS + KYNA-treated, 164 SZR104-treated, and 209 LPS + SZR104-treated microglia. The statistical analysis on the measured cytomorphological parameters of the cells was performed in a Colab notebook of the Google Colaboratory (https://colab.research.google.com) running Python version 3.7.1364,65. The non-parametric Kruskal–Wallis H-test was performed using the function (scipy.stats.kruskal) of the SciPy scientific library (https://scipy.org)66 to determine whether there was a difference between the six groups with regard to the given parameters (Supplementary Table S1, S2). Mann–Whitney U post-hoc testing was performed using normal approximation, continuous distributions, and continuity correction since n1 > 20 and n2 > 20. The Bonferroni correction method was also applied for multiple comparisons (for controlling Type I error; https://scikit-image.org; see ref.67 for details) to indicate which groups are different. Python software libraries Pandas (ver. 1.4.4, https://pandas.pydata.org) and NumPy (ver. 1.23.0, https://numpy.org) were also used for basic data manipulation, formatting, and processing68,69. Values are indicated as the mean ± SEM. p < 0.05 was considered significant; *, **, and *** denote p < 0.05, p < 0.01, and p < 0.005, respectively.
Ethics declarations
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. Experimental procedures were carried out in strict compliance with the European Communities Council Directive (86/609/EEC) and followed Hungarian and local legislation requirements (XXVIII/1998 and 243/1998) and university guidelines regarding the care and use of laboratory animals. This study is also in accordance with ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines/experimental-procedures) and with AVMA Guidelines for the Euthanasia of Animals (https://www.avma.org/resources-tools/avma-policies/avma-guidelines-euthanasia-animals). The experimental protocols were approved by the Institutional Animal Welfare Committee of the University of Szeged (II./1131/2018; date of approval: 30 May 2018).
Supplementary Information
Acknowledgements
This work was supported by a grant from the Ministry of National Resources (GINOP 2.3.2-15-2016-00030 and 2.3.2-15-2016-00034) through the European Union Cohesion Fund. At the time of the experiments, NL and GB were PhD students, partly supported by the Theoretical Medicine Doctoral School, Albert Szent-Györgyi Medical School, University of Szeged. The funders had no role in the study design, data collection and analysis, decision to publish or preparation of the manuscript. Open access was provided by the University of Szeged Open Access Fund (6098).
Abbreviations
- ASP
Aspirin
- BBB
Blood–brain barrier
- CD11b/c
A common epitope shared between CD11b and CD11c (integrin αM and αX chains)
- CNS
Central nervous system
- DAPI
2-(4-Amidinophenyl)-6-indolecarbamidine dihydrochloride
- DMEM
Dulbecco’s Modified Eagle’s Medium
- FBS
Fetal bovine serum
- KYNA
Kynurenic acid
- LPS
Lipopolysaccharide
- PBS
Phosphate-buffered saline
- rpm
Revolutions per minute
- RST
Rosuvastatin
- RT
Room temperature
- SEM
Standard error of the mean
- SZR104
N-(2-(Dimethylamino)ethyl)-3-(morpholinomethyl)-4-hydroxyquinoline-2-carboxamide
Author contributions
M.S. and N.L. contributed equally to this work and should be considered joint first authors. Conceived and designed the experiments: K.G. Performed the experiments: M.S., N.L., K.D. Analyzed the data: M.S., N.L., K.D., G.B. Contributed reagents, materials and/or analysis tools: G.B., A.M., B.L., I.S., L.V., K.G. Wrote the paper: K.G. Edited the paper: K.G. All authors have read and approved the final manuscript.
Funding
Open access funding provided by University of Szeged.
Data availability
Data is contained within the article and Supplementary material.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Melinda Szabo and Noémi Lajkó.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-38107-8.
References
- 1.Kettenmann H, Hanisch U-K, Noda M, Verkhratsky A. Physiology of microglia. Physiol. Rev. 2011;91:461–553. doi: 10.1152/physrev.00011.2010. [DOI] [PubMed] [Google Scholar]
- 2.Prinz M, Priller J. Microglia and brain macrophages in the molecular age: From origin to neuropsychiatric disease. Nat. Rev. Neurosci. 2014;15:300–312. doi: 10.1038/nrn3722. [DOI] [PubMed] [Google Scholar]
- 3.Ginhoux F, Jung S. Monocytes and macrophages: Developmental pathways and tissue homeostasis. Nat. Rev. Immunol. 2014;14:392–404. doi: 10.1038/nri3671. [DOI] [PubMed] [Google Scholar]
- 4.Saijo K, Glass CK. Microglial cell origin and phenotypes in health and disease. Nat. Rev. Immunol. 2011;11:775–787. doi: 10.1038/nri3086. [DOI] [PubMed] [Google Scholar]
- 5.Ling EA, Kaur LC, Yick TY, Wong WC. Immunocytochemical localization of CR3 complement receptors with OX-42 in amoeboid microglia in postnatal rats. Anat. Embryol. (Berl.) 1990;182:481–486. doi: 10.1007/BF00178913. [DOI] [PubMed] [Google Scholar]
- 6.Raivich G, et al. Neuroglial activation repertoire in the injured brain: Graded response, molecular mechanisms and cues to physiological function. Brain Res. Rev. 1999;30:77–105. doi: 10.1016/s0165-0173(99)00007-7. [DOI] [PubMed] [Google Scholar]
- 7.Kata D, et al. Rosuvastatin enhances anti-inflammatory and inhibits pro-inflammatory functions in cultured microglial cells. Neuroscience. 2016;314:47–63. doi: 10.1016/j.neuroscience.2015.11.053. [DOI] [PubMed] [Google Scholar]
- 8.Kata D, et al. A novel pleiotropic effect of aspirin: Beneficial regulation of pro- and anti-inflammatory mechanisms in microglial cells. Brain Res. Bull. 2017;132:61–74. doi: 10.1016/j.brainresbull.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Pérez-Capote K, Serratosa J, Solà C. Excitotoxic and apoptotic neuronal death induce different patterns of glial activation in vitro. J. Neurochem. 2005;94:226–237. doi: 10.1111/j.1471-4159.2005.03183.x. [DOI] [PubMed] [Google Scholar]
- 10.Szabo M, Gulya K. Development of the microglial phenotype in culture. Neuroscience. 2013;241:280–295. doi: 10.1016/j.neuroscience.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 11.Fan Y, et al. Differential regulation of adhesion and phagocytosis of resting and activated microglia by dopamine. Front. Cell. Neurosci. 2018;12:309. doi: 10.3389/fncel.2018.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreutzberg GW. Microglia: A sensor for pathological events in the CNS. Trends Neurosci. 1996;19:312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 13.Pulido-Salgado M, et al. Myeloid C/EBPβ deficiency reshapes microglial gene expression and is protective in experimental autoimmune encephalomyelitis. J. Neuroinflamm. 2017;14:54. doi: 10.1186/s12974-017-0834-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szabo M, Dulka K, Gulya K. Calmodulin inhibition regulates morphological and functional changes related to the actin cytoskeleton in pure microglial cells. Brain Res. Bull. 2016;120:41–57. doi: 10.1016/j.brainresbull.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Vincent C, Siddiqui TA, Schlichter LC. Podosomes in migrating microglia: Components and matrix degradation. J. Neuroinflamm. 2012;9:190. doi: 10.1186/1742-2094-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swartz KJ, During MJ, Freese A, Beal MF. Cerebral synthesis and release of kynurenic acid: An endogenous antagonist of excitatory amino acid receptors. J. Neurosci. 1990;10:2965–2973. doi: 10.1523/JNEUROSCI.10-09-02965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mándi Y, et al. The opposite effect of kynurenic acid and different kynurenic acid analogs on tumor necrosis factor-α (TNF-α) production and tumor necrosis factor-stimulated gene-6 (TSG-6) expression. Front. Immunol. 2019;10:1406. doi: 10.3389/fimmu.2019.01406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Biernacki T, Sandi D, Bencsik K, Vecsei L. Kynurenines in the pathogenesis of multiple sclerosis: Therapeutic perspectives. Cells. 2020;9:1564. doi: 10.3390/cells9061564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zádori D, et al. Kynurenines in chronic neurodegenerative disorders: Future therapeutic strategies. J. Neural Transm. (Vienna) 2009;116:1403–1409. doi: 10.1007/s00702-009-0263-4. [DOI] [PubMed] [Google Scholar]
- 20.Zádori D, et al. Neuroprotective effects of a novel kynurenic acid analogue in a transgenic mouse model of Huntington's disease. J. Neural Transm. (Vienna) 2011;118:865–875. doi: 10.1007/s00702-010-0573-6. [DOI] [PubMed] [Google Scholar]
- 21.Wirthgen E, Hoeflich A, Rebl A, Günther J. Kynurenic acid: The Janus-faced role of an immunomodulatory tryptophan metabolite and its link to pathological conditions. Front. Immunol. 2017;8:1957. doi: 10.3389/fimmu.2017.01957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka M, et al. Mitochondrial impairment: A common motif in neuropsychiatric presentation? The link to the tryptophan-kynurenine metabolic system. Cells. 2022;11:2607. doi: 10.3390/cells11162607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vécsei L, Szalárdy L, Fülöp F, Toldi J. Kynurenines in the CNS: Recent advances and new questions. Nat. Rev. Drug Discov. 2013;12:64–82. doi: 10.1038/nrd3793. [DOI] [PubMed] [Google Scholar]
- 24.Demeter I, et al. A novel kynurenic acid analog (SZR104) inhibits pentylenetetrazole-induced epileptiform seizures. An electrophysiological study. J. Neural Transm. (Vienna) 2012;119:151–154. doi: 10.1007/s00702-011-0755-x. [DOI] [PubMed] [Google Scholar]
- 25.Molnár K, et al. SZR-104, a novel kynurenic acid analogue with high permeability through the blood-brain barrier. Pharmaceutics. 2021;13:61. doi: 10.3390/pharmaceutics13010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lajkó N, et al. Sensitivity of rodent microglia to kynurenines in models of epilepsy and inflammation in vivo and in vitro: Microglia activation is inhibited by kynurenic acid and the synthetic analogue SZR104. Int. J. Mol. Sci. 2020;21:9333. doi: 10.3390/ijms21239333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo M, et al. Kynurenic acid and its analog SZR104 exhibit strong antiinflammatory effects and alter the intracellular distribution and methylation patterns of H3 histones in immunochallenged microglia-enriched cultures of newborn rat brains. Int. J. Mol. Sci. 2022;23:1079. doi: 10.3390/ijms23031079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernández-Arjona M, Grondona JM, Fernández-Llebrez P, López-Ávalos MD. Microglial morphometric parameters correlate with the expression level of IL-1β, and allow identifying different activated morphotypes. Front. Cell. Neurosci. 2019;13:472. doi: 10.3389/fncel.2019.00472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernández-Arjona MDM, Grondona JM, Granados-Durán P, Fernández-Llebrez P, López-Ávalos MD. Microglia morphological categorization in a rat model of neuroinflammation by hierarchical cluster and principal components analysis. Front. Cell. Neurosci. 2017;11:235. doi: 10.3389/fncel.2017.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison H, Young K, Qureshi M, Rowe RK, Lifshitz J. Quantitative microglia analyses reveal diverse morphologic responses in the rat cortex after diffuse brain injury. Sci. Rep. 2017;7:13211. doi: 10.1038/s41598-017-13581-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Priller J, Prinz M. Targeting microglia in brain disorders. Science. 2019;365:32–33. doi: 10.1126/science.aau9100. [DOI] [PubMed] [Google Scholar]
- 32.Schwabenland M, et al. Analyzing microglial phenotypes across neuropathologies: A practical guide. Acta Neuropathol. 2021;142:923–936. doi: 10.1007/s00401-021-02370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matcovitch-Natan O, et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- 34.Lee J, Hamanaka G, Lo EH, Arai K. Heterogeneity of microglia and their differential roles in white matter pathology. CNS Neurosci. Ther. 2019;25:1290–1298. doi: 10.1111/cns.13266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francistiová L, Vörös K, Lovász Z, Dinnyés A, Kobolák J. Detection and functional evaluation of the P2X7 receptor in hiPSC derived neurons and microglia-like cells. Front. Mol. Neurosci. 2022;14:793769. doi: 10.3389/fnmol.2021.793769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang IK, et al. Comparative study on high fat diet-induced 4-hydroxy-2E-nonenal adducts in the hippocampal CA1 region of C57BL/6N and C3H/HeN mice. Neurochem. Res. 2009;34:964–972. doi: 10.1007/s11064-008-9846-y. [DOI] [PubMed] [Google Scholar]
- 37.Ayoub AE, Salm AK. Increased morphological diversity of microglia in the activated hypothalamic supraoptic nucleus. J. Neurosci. 2003;23:7759–7766. doi: 10.1523/JNEUROSCI.23-21-07759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shahidehpour RK, et al. Dystrophic microglia are associated with neurodegenerative disease and not healthy aging in the human brain. Neurobiol. Aging. 2021;99:19–27. doi: 10.1016/j.neurobiolaging.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanlua P, Prommahom A, Sricharoenvej S. Increased number of activated microglia in rat spinal cord during early stage of diabetic induction. Folia Morphol. (Warsz) 2020;79:662–671. doi: 10.5603/FM.a2019.0136. [DOI] [PubMed] [Google Scholar]
- 40.Zheng FY, Xiao WH, Bennett GJ. The response of spinal microglia to chemotherapy-evoked painful peripheral neuropathies is distinct from that evoked by traumatic nerve injuries. Neuroscience. 2011;176:447–454. doi: 10.1016/j.neuroscience.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zachary JF, Baszler TV, French RA, Kelley KW. Mouse Moloney leukemia virus infects microglia but not neurons even though it induces motor neuron disease. Mol. Psychiatry. 1997;2:104–106. doi: 10.1038/sj.mp.4000219. [DOI] [PubMed] [Google Scholar]
- 42.Parakalan R, et al. Transcriptome analysis of amoeboid and ramified microglia isolated from the corpus callosum of rat brain. BMC Neurosci. 2012;13:64. doi: 10.1186/1471-2202-13-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett ML, Viaene AN. What are activated and reactive glia and what is their role in neurodegeneration? Neurobiol. Dis. 2021;148:105172. doi: 10.1016/j.nbd.2020.105172. [DOI] [PubMed] [Google Scholar]
- 44.Poles MZ, et al. Kynurenic acid and its synthetic derivatives protect against sepsis-associated neutrophil activation and brain mitochondrial dysfunction in rats. Front. Immunol. 2021;12:717157. doi: 10.3389/fimmu.2021.71715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cox MJ, et al. Tales of 1,008 small molecules: Phenomic profiling through live-cell imaging in a panel of reporter cell lines. Sci. Rep. 2020;10:13262. doi: 10.1038/s41598-020-69354-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin YC, Kofron JL, Traphagen LM. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002;45:4350–4358. doi: 10.1021/jm020155c. [DOI] [PubMed] [Google Scholar]
- 47.Trapotsi MA, et al. Comparison of chemical structure and cell morphology information for multitask bioactivity predictions. J. Chem. Inf. Model. 2021;61:1444–1456. doi: 10.1021/acs.jcim.0c00864. [DOI] [PubMed] [Google Scholar]
- 48.Fülöp F, Szatmári I, Toldi J, Vécsei L. Modifications on the carboxylic function of kynurenic acid. J. Neural Transm. (Vienna) 2012;119:109–114. doi: 10.1007/s00702-011-0721-7. [DOI] [PubMed] [Google Scholar]
- 49.Fülöp, F., Szatmári, I., Toldi, J. & Vécsei, L. Novel types of c-3 substituted kynurenic acid derivatives with improved neuroprotective activity. Patent No: WO2017149333A1 (2017, accessed 31 October 2020). https://patents.google.com/patent/WO2017149333A1/en.
- 50.Lőrinczi B, Csámpai A, Fülöp F, Szatmári I. Synthesis of new C-3 substituted kynurenic acid derivatives. Molecules. 2020;25:937. doi: 10.3390/molecules25040937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dulka K, Nacsa K, Lajkó N, Gulya K. Quantitative morphometric and cell-type-specific population analysis of microglia-enriched cultures subcloned to high purity from newborn rat brains. IBRO Neurosci. Rep. 2021;10:119–129. doi: 10.1016/j.ibneur.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saura J, Tusell JM, Serratosa J. High-yield isolation of murine microglia by mild trypsinization. Glia. 2003;44:183–189. doi: 10.1002/glia.10274. [DOI] [PubMed] [Google Scholar]
- 53.Robinson AP, White TM, Mason DW. Macrophage heterogeneity in the rat as delineated by two monoclonal antibodies MRC OX-41 and MRC OX-42, the latter recognizing complement receptor type 3. Immunology. 1986;57:239–247. [PMC free article] [PubMed] [Google Scholar]
- 54.Wang CC, Wu CH, Shieh JY, Wen CY, Ling EA. Immunohistochemical study of amoeboid microglial cells in fetal rat brain. J. Anat. 1996;189:567–574. [PMC free article] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Karperien, A. FracLac User’s Guide (2001). http://rsbweb.nih.gov/ij/plugins/fraclac/FLHelp/Introduction.htm.
- 57.Karperien A, Ahammer H, Jelinek HF. Quantitating the subtleties of microglial morphology with fractal analysis. Front. Cell. Neurosci. 2013;7:3. doi: 10.3389/fncel.2013.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abràmoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- 59.Karperien AL, Jelinek HF, Buchan AM. Box-counting analysis of microglia form in schizophrenia. Alzheimer’s disease and affective disorder. Fractals. 2008;16:103–107. doi: 10.1142/s0218348x08003880. [DOI] [Google Scholar]
- 60.Karperien, A., Jelinek, H.F. & Milosevic, N.T. Lacunarity analysis and classification of microglia in neuroscience. In 8th European Conference on Mathematical and Theoretical Biology MS#88 (2011). 10.13140/2.1.3576.6082.
- 61.Fujita H, et al. Effects of GM-CSF and ordinary supplements on the ramification of microglia in culture: A morphometrical study. Glia. 1996;18:269–281. doi: 10.1002/(sici)1098-1136(199612)18:4<269::aid-glia2>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 62.Karperien AL, Jelinek HF. Fractal, multifractal, and lacunarity analysis of microglia in tissue engineering. Front. Bioeng. Biotechnol. 2015;3:51. doi: 10.3389/fbioe.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schindelin J, et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bisong, E. Google colaboratory. In Building Machine Learning and Deep Learning Models on Google Cloud Platform. Apress, Berkeley, CA, USA (2019). 10.1007/978-1-4842-4470-8_7.
- 65.van Rossum, G. Python reference manual. In Department of Computer Science [CS]. Centrum Wiskunde & Informatica, Amsterdam, the Netherlands, ISSN 0169-118X (1995).
- 66.Virtanen P, et al. SciPy 1.0: Fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 68.McKinney, W. Data structures for statistical computing in Python. In Proceedings of the 9th Python in Science Conference, vol. 445 (van der Walt, S. & Millman, J. eds.) 56–61 (2010). 10.25080/Majora-92bf1922-00a.
- 69.Harris CR, et al. Array programming with NumPy. Nature. 2020;585:357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data is contained within the article and Supplementary material.