Abstract
Sustainable TGF-β1 signaling drives organ fibrogenesis. However, the cellular adaptation to maintain TGF-β1 signaling remains unclear. In this study, we revealed that dietary folate restriction promoted the resolution of liver fibrosis in mice with nonalcoholic steatohepatitis. In activated hepatic stellate cells, folate shifted toward mitochondrial metabolism to sustain TGF-β1 signaling. Mechanistically, nontargeted metabolomics screening identified that α-linolenic acid (ALA) is exhausted by mitochondrial folate metabolism in activated hepatic stellate cells. Knocking down serine hydroxymethyltransferase 2 increases the bioconversion of ALA to docosahexaenoic acid, which inhibits TGF-β1 signaling. Finally, blocking mitochondrial folate metabolism promoted liver fibrosis resolution in nonalcoholic steatohepatitis mice. In conclusion, mitochondrial folate metabolism/ALA exhaustion/TGF-βR1 reproduction is a feedforward signaling to sustain profibrotic TGF-β1 signaling, and targeting mitochondrial folate metabolism is a promising strategy to enforce liver fibrosis resolution.
Keywords: hepatic stellate cells, mitochondrial folate metabolism, TGF-β1 signaling, ALA, DHA, liver fibrosis resolution, SHMT2
Liver fibrosis is the main determinant of mortality in nonalcoholic steatohepatitis (NASH) patients (1, 2, 3) and is characterized by excessive deposition of extracellular matrix, produced by activated hepatic stellate cells (HSCs) and myofibroblasts. TGF-β1 signaling induces HSC activation to promote tissue repair during liver injury, while sustaining TGF-β1 signaling drives the development of liver fibrosis via sustaining HSC activation (4, 5, 6). However, one-half of total activated HSCs may go to an inactive phenotype with downregulation of TGF-β1 signaling during the regression of liver fibrosis (7, 8). Regrettably, to date, the cellular adaptation to modulate TGF-β1 signaling in activated HSCs is still largely unknown.
Folate-mediated one-carbon (1C) metabolism plays an essential role in vital biological processes, including nucleotide, amino acid, and biosynthesis of methyl (9, 10, 11, 12). Dietary folate is essential as deficiency causes neural tube and congenital heart birth defects (13, 14). The effect of folic acid (FA) supplementation on chronic liver diseases is, however, still controversial. On one hand, FA supplementation improved nonalcoholic fatty liver disease primarily by lowering lipid accumulation and oxidative stress (15, 16, 17, 18). On the other hand, FA supplementation did not alleviate liver injury in NASH patients (19). Thus, whether NASH patients should control their folate intake remains to be determined.
Here, we reported that dietary folate restriction promoted the resolution of liver fibrosis in mice with NASH. Depletion of folate aborted TGF-β1 signaling activation and sustainability in HSCs, suggesting that TGF-β1 signaling is necessary but not sufficient for maintaining HSC activation. The underlying mechanism involves a shift in folate consumption to mitochondrial metabolism that causes α-linolenic acid (ALA) exhaustion to sustain TGF-β1 signaling. Our findings highlight, for the first time, a key trigger that tips the balance between fibrosis progression and its homeostasis-based resolution and reveals that targeting mitochondrial folate metabolism is a promising therapeutic strategy to treat liver fibrosis.
Results
Dietary folate restriction promotes the resolution of liver fibrosis in NASH mice
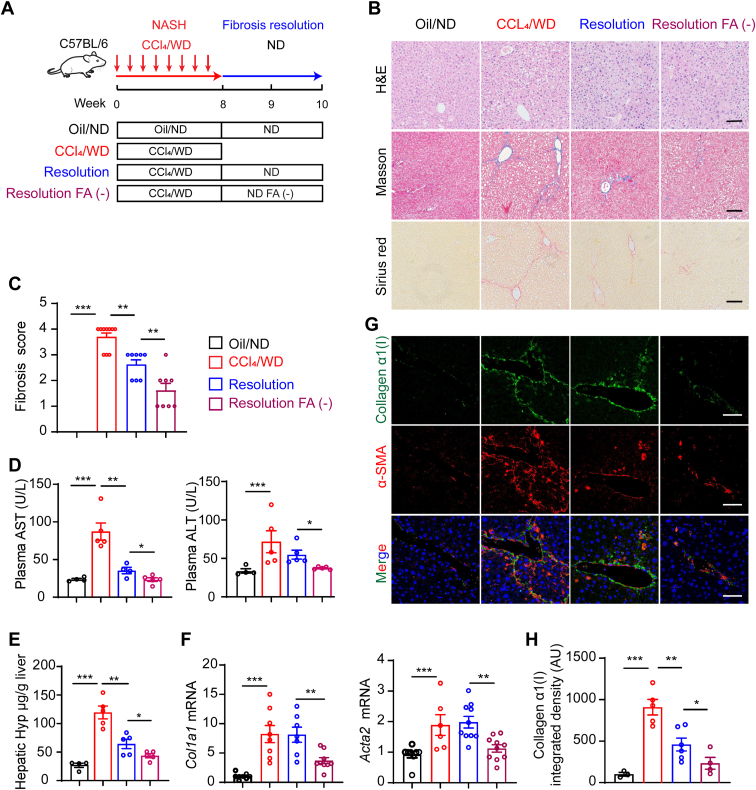
To investigate whether dietary folate restriction affects the resolution of liver fibrosis, a mouse NASH model (20) was built by intraperitoneal injection of CCl4 combined with the consumption of a western diet (WD) containing high-fat, high-fructose, and high-cholesterol for 8 weeks. Then, the mice were fed a folate-depleted diet or a normal diet (ND, 3 mg/kg folate) instead of a WD for two weeks (Fig. 1A). Mice with CCl4/WD-induced NASH were characterized by increases in hepatocyte ballooning, fibrosis, aspartate aminotransferase (AST) and serum alanine aminotransferase (ALT) levels, hydroxyproline content in mouse liver tissue, and profibrotic gene expression (including Acta2 and Col1a1) on week 8 (Fig. 1, B–H). Liver fibrosis was significantly reduced two weeks after CCl4/WD withdrawal, and ALT and AST levels were significantly reduced (Fig. 1, B–H). Surprisingly, Masson and Sirius’s staining of liver sections revealed that the FA-negative (FA (−))-fed mice showed more rapid fibrosis resolution than the mice fed a normal FA diet (Fig. 1, B and C). Furthermore, compared with ND-fed resolution mice, more notable reductions in serum AST and ALT levels, hydroxyproline content, and alpha smooth muscle actin (α-SMA) and collagen α1(I) mRNA were observed in liver tissues of the FA (−) mice (Fig. 1, D–H). These data suggested that after the withdrawal of CCl4/WD, the potential for NASH-related liver fibrosis resolution was found to be not only possible but also promoted by dietary folate restriction.
Figure 1.
Dietary folate restriction promotes hepatic fibrosis resolution in NASH mice.A, experimental design of the fibrosis resolution in mice. B, histological images of H&E, sirius red–stained, and Masson’s trichrome mouse liver tissues (scale bars represent 50 μm). C, Ishak fibrosis stage scores at indicated treatment conditions. D, AST and ALT levels of plasma in mice (n = 5). E, liver tissue hydroxyproline content (n = 5). F, mRNA expression levels of fibrogenic genes (Acta2 and Col1a1) (n = 5). G and H, immunofluorescent staining for collagen α1(I) and α-SMA (scale bars represent 50 μm). The data are shown as mean ± SEM. The statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was tested by student’s unpaired t test. ALT, alanine aminotransferase; AST, aspartate aminotransferase; H&E, hematoxylin-eosin staining; NASH, nonalcoholic steatohepatitis.
FA supplementation sustains TGF-β1 signaling in activated HSCs
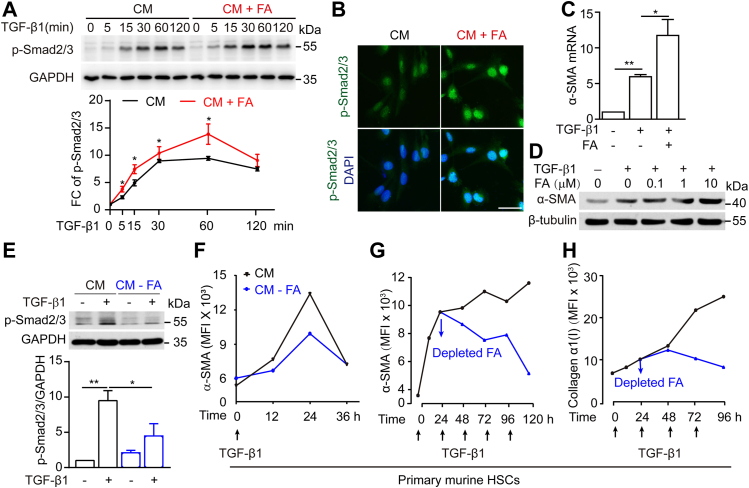
Controlling HSC activation and transdifferentiation into myofibroblasts is considered a promising treatment for liver fibrosis (21, 22, 23, 24). To determine the mechanism by which dietary folate restriction accelerates liver fibrosis resolution in NASH mice, the effect of FA on sustained HSC activation was investigated. Since TGF-β1/Smad2/3 signaling induces HSC activation and drives liver fibrogenesis (25, 26, 27), the time course of TGF-β1 signaling was observed. As shown in Figure 2A, TGF-β1 induced Smad2/3 activation in LX-2 cells cultured in complete medium (CM) with peak activation at 60 min and reduced activation thereafter, suggesting recovery of HSCs via autonomous deactivation of TGF-β1 signaling. In contrast, FA supplementation (10 μM) significantly enhanced Smad2/3 phosphorylation levels and extended its duration time. (Fig. 2A), suggesting that FA increases both the level of HSC activation and its duration. Furthermore, FA supplementation significantly promoted TGF-β1–induced nuclear accumulation of p-Smad2/3 and increased the expression of the downstream gene α-SMA in a dose-dependent manner (Fig. 2, B–D). HSCs cultured in CM without FA showed a decrease of TGF-β1–induced Smad2/3 phosphorylation and α-SMA expression (Fig. 2, E and F). These studies showed that in response to a single TGF-β1 stimulus, primary murine HSCs underwent a quiescence-activation-inactivation process (Fig. 2F). To determine whether the sustainable activation of HSCs in liver fibrosis is due to the repeated stimulation of TGF-β1 activity, the effects of multiple consecutive TGF-β1 stimuli were analyzed, and sustainably increased levels of α-SMA and collagen α1(I) expression were observed. Interestingly, the replacement of CM by CM without FA caused a sharp reduction in the expression of these stimulating proteins, even with continuous TGF-β1 stimulation (Fig. 2, G and H), suggesting that TGF-β1 signaling, considered the key event of HSC activation to date, is determined by folate level.
Figure 2.
FA supplementation sustains TGF-β1 signaling activation in HSCs.A, immunoblotting for SMAD2/3 phosphorylation levels of LX-2 cells with or without FA supplementation upon control or TGF-β1(5 ng/ml) treatment for indicated times. B, immunofluorescence detection of p-Smad2/3 levels in cell nuclei after FA supplementation (scale bars represent 40 μm). C and D, mRNA and protein expression analysis of α-SMA in LX-2 cells. LX-2 cells were treated with TGF-β1(5 ng/ml) with or without of FA supplementation. E, Western blots of phosphorylated SMAD2/3 in LX-2 cells with or without FA upon control or TGF-β1(5 ng/ml) treatment for 30 min. F, mouse primary HSCs were cultured with or without FA, then treated with TGF-β1(5 ng/ml) for 36 h. Flow cytometry analysis for α-SMA expression at 0 h, 12 h, 24 h, 36 h. G and H, mouse primary HSCs were stimulated multiple times by TGF-β1(5 ng/ml) at 0 h, 24 h, and 48 h, 72 h, and cells were depleted of FA at 24 h. Flow cytometry analysis for α-SMA and collagen α1(I) protein expression at 0 h, 24 h, 48 h, 72 h, 96 h. The statistical significance (∗p < 0.05, ∗∗p < 0.01) was tested by student’s unpaired t test. CM, complete medium; CM+FA, the addition of folic acid to complete medium; CM-FA, complete medium with folic acid removed; FA, folic acid; HSCs, hepatic stellate cells.
Folate shifts toward mitochondrial metabolism during HSC activation
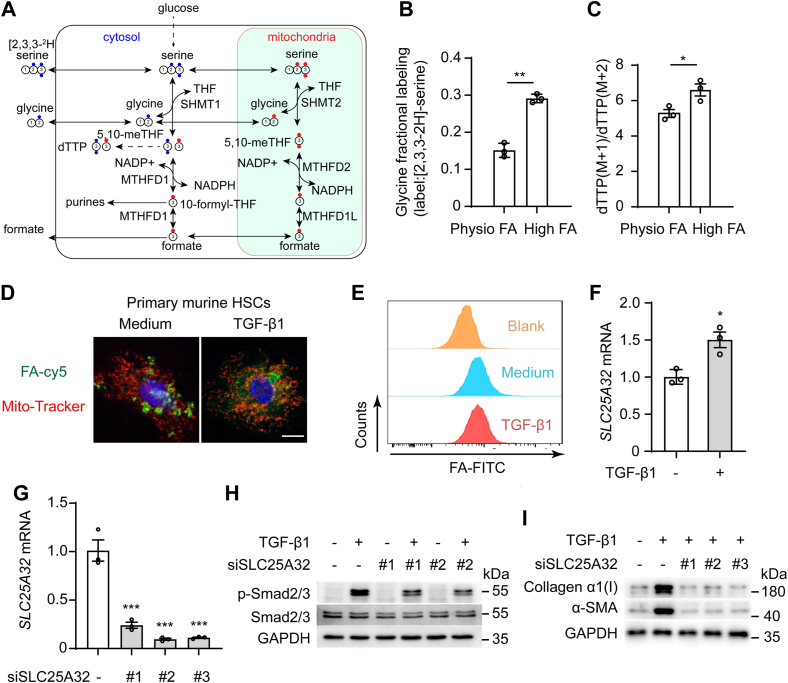
Folate is metabolized in nuclear, cytoplasmic, and mitochondrial metabolic pathways, which can be reprogrammed to adapt to environmental changes (28). To further investigate how folate supports sustainable HSC activation, stable isotope tracing was performed to distinguish cytoplasmic and mitochondrial folate metabolism changes in HSCs (Fig. 3A). The mitochondrial pathway was found to be the predominant contributor of 1C units in LX-2 cells treated with folate administered at a higher level (2 μM) than the physiological level (200 nM) (Fig. 3, B and C), indicating that HSCs’ metabolism switches to rely increasingly on the mitochondrial folate cycle when folate levels are high. Primary mouse HSCs were treated with Cy5-labeled FA and stimulated with TGF-β1, and then, the subcellular distribution of folate was observed. FA accumulated in the mitochondria of the activated HSCs, but it was located mainly in the cytosol of quiescent HSCs (Fig. 3D). The uptake of FA was equivalent to that of LX-2 cells with or without TGF-β1 stimulation (Fig. 3E). These data suggested that the subcellular distribution of folate but not cellular uptake was changed during HSC activation. A mitochondrial folate transporter, encoded by the SLC25A32 gene, has been reported to transfer tetrahydrofolate (THF) into mitochondria (29, 30). TGF-β1 promoted SLC25A32 mRNA expression in LX-2 cells (Fig. 3F). Next, we determined whether mitochondrial translocation of folate affected HSC activation. Knocking down SLC25A32 by siSLC25A32 inhibited TGF-β1–induced Smad2/3 phosphorylation (Fig. 3, G and H). In addition, collagen α1(I) and α-SMA protein expression were suppressed by SLC25A32 knockdown in LX-2 cells (Fig. 3I). These data support this view that mitochondrial folate metabolism is required for sustainable HSC activation.
Figure 3.
Folate shifts toward mitochondrial metabolism for HSC activation.A, when LX-2 cells were fed [2,3,3-2H] serine, thymidine triphosphate (dTTP) with 2H was determined by mass spectrometry. dTTP with one deuterium (M + 1) is produced via the mitochondrial folate metabolism through SHMT2 and with two deuteriums (M + 2) via the cytosolic SHMT1. B, the percentage labeling of intracellular glycine when LX-2 were fed [2,3,3-2H]-serine in high (2 μM) and physiological (200 nM) FA levels. C, the ratio of dTTP M+1 and M+2 when LX-2 cells were fed [2,3,3-2H]]-serine in high and physiological FA levels. D, the subcellular distribution of Cy5-labeled folate in primary murine HSCs with TGF-β1(5 ng/ml) treatment for 36 h (scale bars represent 10 μm). E, flow cytometry analysis of FA-FITC in LX-2 cells after stimulation with control or TGF-β1(5 ng/ml) for 36 h, then treated with FA-Cy5 for 30 min. F, mRNA levels of SLC25A32 in LX-2 cells after treatment with TGF-β1(5 ng/ml) for 24 h. G, knockdown efficiency of three independent SLC25A32 siRNA in LX2 cells. H, immunoblotting for α-SMA and collagen α1(I) proteins expression in knockdown SLC25A32 LX-2 cells treatment with TGF-β1 (5 ng/ml) for 36 h. I, Western blot performed on cell lysates for phosphorylated SMAD2/3 in knockdown SLC25A32 LX-2 cells treated with or without TGF-β1 (5 ng/ml) for 30 min. The statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was tested by student’s unpaired t test. FA, folic acid; HSC, hepatic stellate cell; SHMT, serine hydroxymethyltransferase.
The SHMT2-/MTHFD2-mediated mitochondrial folate metabolic pathway governs the sustained activation of TGF-β1 signaling
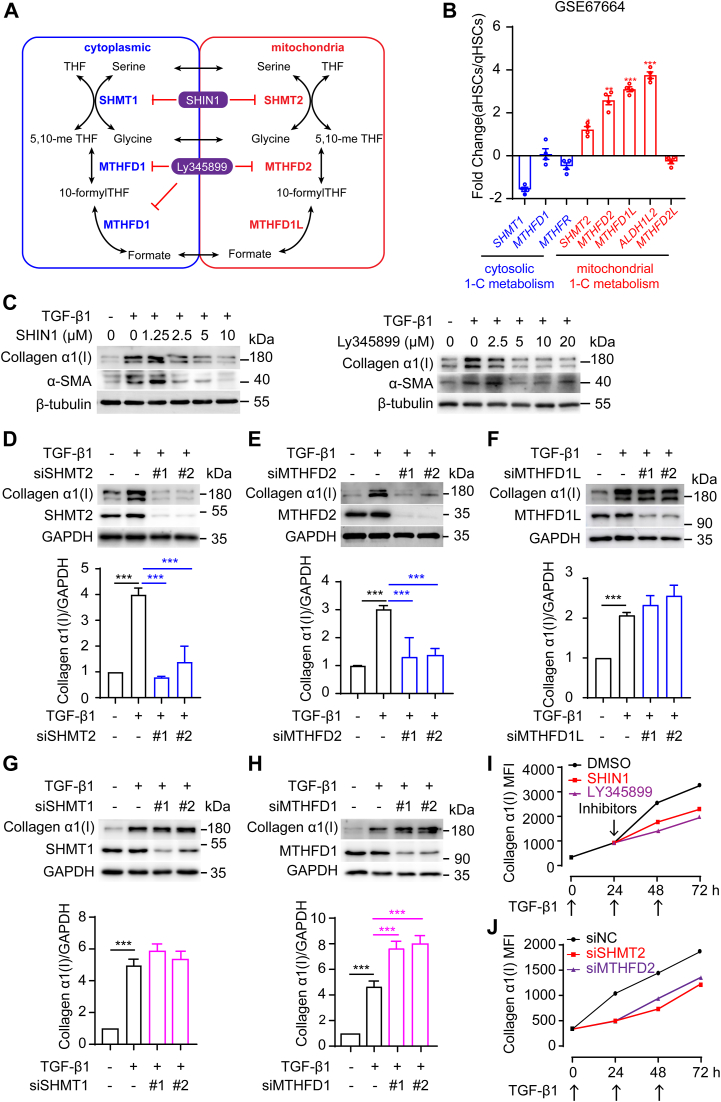
Folate metabolic enzymes in mitochondria are consistently overexpressed in cancer cells. Serine hydroxymethyltransferase 2 (SHMT2) and methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) are two key enzymes involved in mitochondrial 1C metabolism (31, 32, 33). Exploring how activated HSCs switch to relying on mitochondrial 1C flux, we analyzed the expression of key 1C metabolism enzymes in the gene expression dataset (GSE67664) of primary human HSCs. Broad upregulation of mitochondrial 1C metabolism-related genes (SHMT2, MTHFD2, MTHFD1L, ALDH1L2) and downregulated or unchanged cytoplasmic 1C metabolism gene (SHMT1, MTHFD1, and MTHFR) expression was observed in activated HSCs (Fig. 4, A and B). Moreover, the mitochondrial 1C metabolism-related genes SHMT2 and MTHFD2 were markedly upregulated in HSCs from mouse fibrotic liver samples relative to those of the control samples (Fig. S1A). Furthermore, SHMT2 and MTHFD2 expression was significantly upregulated in HSCs but not in hepatocytes from fibrotic liver tissues of NASH mice (Fig. S1B).
Figure 4.
SHMT2-/MTHFD2-mediated mitochondrial folate metabolic pathway sustains TGF-β1 signaling.A, compartmentalization of FA-mediated 1C metabolism. B, qPCR detection of one-carbon metabolism gene expression in primary human HSC datasets from GSE67664. C, Western blot performed on cell lysates for α-SMA and collagen α1(I) after LX-2 cells were pretreated with DMSO or SHIN1 (1.25, 2.5, 5, 10 μM) or LY345899 (2.5, 5, 10, 20 μM) for 6 h, then stimulated with TGF-β1(5 ng/ml) for 24 h. D-H, Western blot for collagen α1(I) proteins expression in knockdown SHMT2, MTHFD2, MTHFD1L, SHMT1, MTHFD1 LX-2 cells treated with TGF-β1(5 ng/ml) for 36 h. I, LX-2 cells were stimulated multiple times by TGF-β1(5 ng/ml) at 0 h, 24 h, 48 h and were pretreated with DMSO or SHIN1 (5 μM) or LY345899 (5 μM) at 24 h. Flow cytometry analysis for collagen α1(I) expression at 0 h, 24 h, 48 h, 72 h. J, knockdown SHMT2 and MTHFD2 LX-2 cells were stimulated multiple times by TGF-β1 (5 ng/ml) at 0 h, 24 h, 48 h. Flow cytometry analysis for collagen α1(I) expression at 0, 24, 48, 72 h. The data are shown as mean ± SEM. The statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was tested by unpaired t test. 1C, one-carbon; FA, folic acid; HSC, hepatic stellate cell; MTHFD, methylenetetrahydrofolate dehydrogenase; SHMT, serine hydroxymethyltransferase.
SHMT2 converts serine into glycine and produces a 1C unit in mitochondria. The role played by the SHMT2-mediated metabolic pathway in liver fibrosis remains largely unknown. Next, two dual inhibitors, SHIN1 (targeting both SHMT1 and SHMT2) and LY345899 (targeting both MTHFD1 and MTHFD2), were used to further investigate how folate metabolism impacts HSC activation and sustainability. Both SHIN1 and LY345899 reduced the TGF-β1–induced protein expression of α-SMA and collagen α1(I) in a dose-dependent manner (Fig. 4C), suggesting that folate-mediated 1C metabolism supports TGF-β1 signaling. To further distinguish the folate metabolism pathways in different subcellular locations with respect to TGF-β1 signaling, we used a genetic approach to knock down the expression of genes encoding different enzymes related to folate-mediated 1C metabolism located in the cytoplasm and mitochondria. Knocking down the SHMT2 or MTHFD2 gene which each encodes a protein located in mitochondria significantly inhibited HSC activation by reducing collagen α1(I) protein level (Fig. 4, D and E). Knocking down MTHFD1L, another gene encoding a core 1C enzyme, and genes encoding other cytoplasmic enzymes, such as SHMT1 and MTHFD1, did not reduce collagen α1(I) expression in activated HSCs (Fig. 4, F–H). Furthermore, inhibition or knockdown of SHMT2 or MTHFD2 decreased collagen α1(I) protein levels in HSCs in which TGF-β1 stimulation was repeated (Fig. 4, I and J). Thus, SHMT2-/MTHFD2-mediated mitochondrial folate metabolism is required for sustainable HSC activation.
Blocking mitochondrial folate metabolism restores HSC homeostasis by preventing ALA exhaustion
SHMT converts glycine (G) to serine (S) using 5,10-methylene-THF in the cytosol and mitochondria, which is a reversible process (34). When serine and glycine were removed from the medium, we found a reduction in the TGF-β1–induced ACTA2 and COL1A1 mRNA and corresponding protein levels (Fig. S2, A and B), and Smad2/3 phosphorylation was attenuated (Fig. S2C), indicating a role for serine-glycine metabolism in TGF-β1 signaling.
We then examined the effect of 1C-derived metabolites on TGF-β1 signaling by adding by-products of folate-medicated 1C metabolism, namely, GSH, SAM, or formate. In the absence of serine glycine, neither the addition of formate nor GSH restored TGF-β1–induced ACTA2 and COL1A1 mRNA or protein expression in LX-2 cells (Fig. S3, A and B). Furthermore, formate addition did not reverse the increases in collagen α1(I) and α-SMA protein expression induced by knocking down SLC25A32 and SHMT2 in LX-2 cells (Fig. S3C). Overall, these findings indicated that formate, GSH, and SAM are not required for the profound SHMT2/MTHFD2 regulation of TGF-β1 signaling.
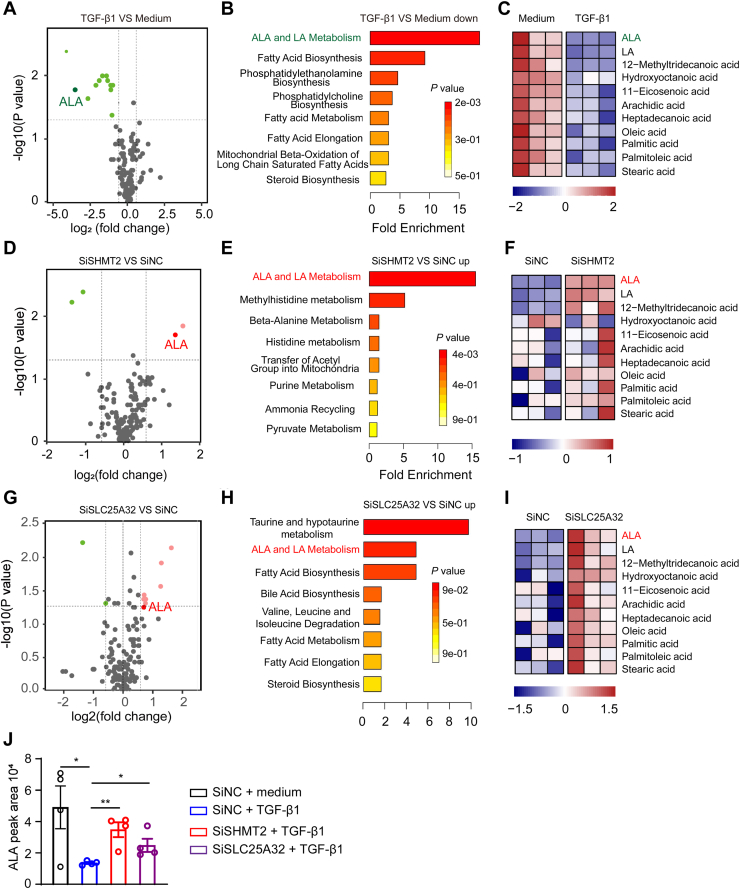
To determine how SHMT2-/MTHFD2-mediated 1C metabolism regulates TGF-β1 signaling, unbiased nontargeted metabolomics analysis was performed to screen key metabolites. LX-2 cells were transfected with siRNA-negative control, siRNA-SHMT2, with or without TGF-β1 stimulation for 24 h to identify differentially abundant metabolites. A total of 156 unique metabolites were identified and quantified by liquid chromatography-tandem mass spectrometry (LC-MS/MS). TGF-β1 reduced the levels of many fatty acids, especially ALA (Fig. 5A). The decrease in metabolites induced by TGF-β1 treatment was most evident in ALA metabolism pathways (Fig. 5, B and C). Interestingly, knocking down SHMT2 increased the ALA level (Fig. 5D) and its metabolism pathways (Fig. 5, E and F). Consistently, knockdown of SLC25A32 to block mitochondrial folate metabolism also increased ALA level and its metabolism pathways. In addition, the major changes in the metabolite ALA level were confirmed by LC-MS. ALA level was increased significantly by knocking down SHMT2 or SLC25A32 in TGF-β1–treated LX-2 cells (Fig. 5J).
Figure 5.
Mitochondrial folate metabolism induces ALA exhaustion in activated HSCs.A, D, and G, differential metabolites abundance in LX-2 cells transfected with siNC, siSHMT2, or siSLC25A32 24 h after medium or TGF-β1 (5 ng/ml) stimulation (n = 3). B, E, and H, metabolite set enrichment analysis in LX2 cells transfected with siNC, siSHMT2, or siSLC25A32 24 h after medium or TGF-β1 (5 ng/ml) stimulation (n = 3). C, F, and I, heatmap of metabolites in LX-2 cells transfected with siNC, siSHMT2, or siSLC25A32 24 h after medium or TGF-β1 (5 ng/ml) stimulation (n = 3). J, LC-MS analysis of intracellular alpha-linolenic acid in knockdown SHMT2 or SLC25A32 LX-2 cells after treatment with TGF-β1 (5 ng/ml) for 24 h (n = 4). The statistical significance (∗p < 0.05, ∗∗p < 0.01) was tested by student’s unpaired t test. ALA, α-linolenic acid; HSC, hepatic stellate cell; SHMT, serine hydroxymethyltransferase.
Next, lipid droplet accumulation was observed in HSCs treated with siSHMT2 or siSLC25A32 (Fig. S4A). Oxidative respiration increases lipid consumption and reactive oxygen species production (35, 36). Knockdown or inhibition of SHMT2 or MTHFD2 impaired mitochondrial respiration in LX-2 cells (Fig. S4, B and C). Thirteen proteins encoded by mitochondrial DNA (mtDNA) are essential for respiratory chain signaling (37). Defective mtDNA homeostasis can cause overall impairment in mitochondrial complexes I, III, IV, and V (38). mtDNA transcripts were significantly reduced in LX2 cells with SHMT2/MTHFD2 knockdown compared to that in the negative controls (Fig. S4, D–F). These data suggest that SHMT2-mediated mitochondrial folate metabolism promotes mtDNA transcription and oxidative respiration to induce ALA exhaustion.
Bioconversion of ALA into DHA inhibits TGF-β1 signaling by reducing TGFBR1 mRNA expression
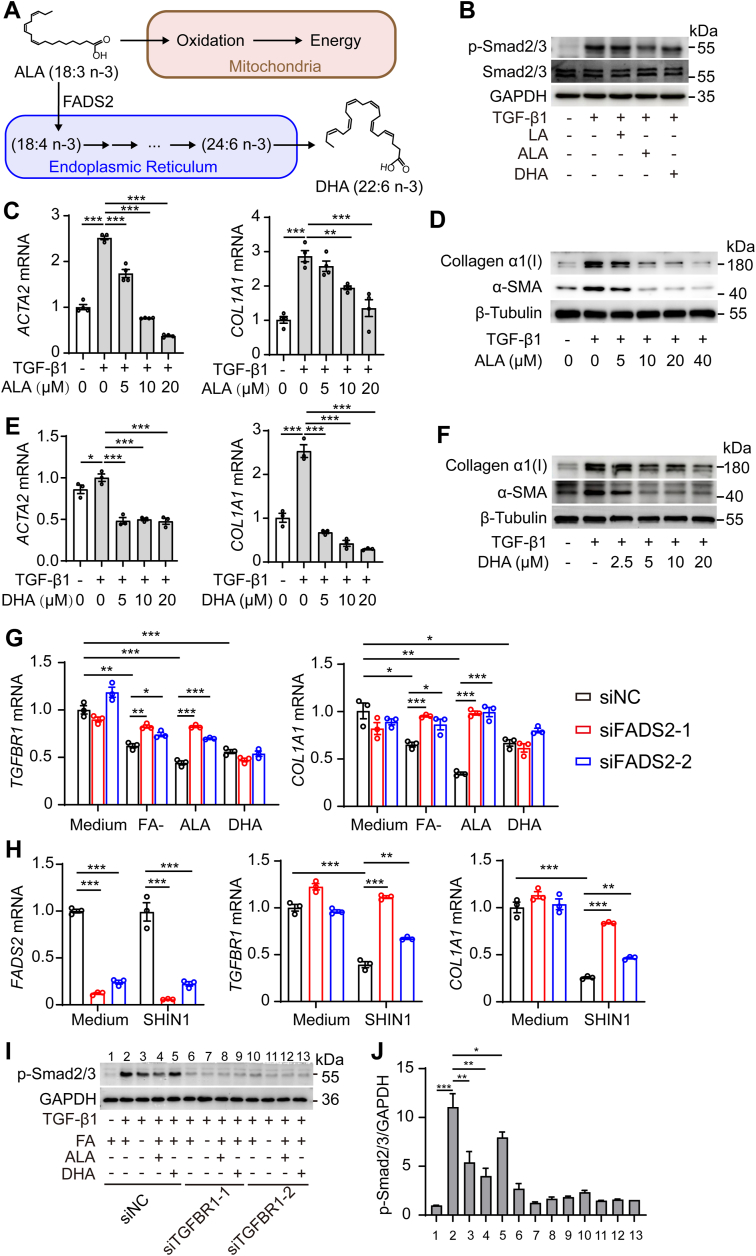
ALA is oxidized to participate in energy metabolism in the mitochondria and also acted as a precursor of n-3 polyunsaturated fatty acids, which were transformed into docosahexaenoic acid (DHA) by elongase and desaturase in the endoplasmic reticulum (Fig. 6A) (39, 40, 41). Next, we sought to determine whether ALA and DHA affect HSC activation. ALA or DHA treatment reduced the SMAD2/3 phosphorylation that had been induced by TGF-β1 (Fig. 6B). This may explain the reduction in TGF-β1–induced Smad2/3 phosphorylation caused by FA deficiency (Fig. 2, A, B, and E). Furthermore, ALA and DHA inhibited TGF-β1–induced α-SMA and collagen α1(I) expression in a dose-dependent manner (Fig. 6, C–F). These data suggest that ALA and DHA inhibit TGF-β1 signaling.
Figure 6.
Bioconversion of ALA into DHA inhibits HSC activation by reducing TGFBR1 mRNA expression.A, ALA is biotransformed to DHA by desaturase in the endoplasmic reticulum. B and C, ACTA2, COL1A1 mRNA, and protein levels in LX2 cells after cells were pretreated with DMSO or ALA (0, 5, 10, 20 μM) for 6 h, then stimulated with TGF-β1 (5 ng/ml) for 24 h. D and E, ACTA2, COL1A1 mRNA, and protein levels in LX2 cells after cells were pretreated with DMSO or DHA (0, 5, 10, 20 μM) for 6 h, then stimulated with TGF-β1 (5 ng/ml) for 24 h. F, immunoblotting for SMAD2/3 phosphorylation(p-Smad2/3) levels in LX-2 cells after pretreatment with DMSO, ALA, LA, and DHA for 6 h, then stimulated with TGF-β1 (5 ng/ml) for 30 min. G, TGFBR1, COL1A1 mRNA levels in LX2 cells transfected with siNC or siFADS2 treated with folic acid–free medium (FA-), ALA (10 μM), DHA (10 μM) for 24 h. H, TGFBR1, COL1A1 mRNA levels in LX2 cells transfected with siNC or siFADS2 treated with SHIN1 (10 μM) for 24 h. Phosphorylation of Smad2/3 levels in LX2 cells transfected with siNC or siFADS2 treated with FA-, ALA (10 μM), DHA (10 μM) for 24 h, then stimulated with TGF-β1 (5 ng/ml) for 30 min. (J) is a quantitative analysis of (I). The statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was tested by student’s unpaired t test. ALA, α-linolenic acid; DHA, docosahexaenoic acid; FADS, fatty acid desaturase; HSC, hepatic stellate cell.
To determine how ALA and DHA regulate TGF-β1 signaling in HSCs, we investigated gene expression changes in HSCs induced by SHMT2 knockdown. Kyoto Encyclopedia of Genes and Genomes analysis of differential gene expression revealed that the profibrotic signaling pathway was downregulated and that activation of many biosynthesis pathways was upregulated in HSCs treated with siSHMT2 (Fig. S5A). Furthermore, gene set enrichment analyses showed a correlation of gene expression in knockdown SHMT2 cells with TGF-β1 signaling (Fig. S5B). The expression of genes related to TGF-β1 signaling in SHMT2-knockdown LX-2 cells were downregulated, and TGFBR1 was the most downregulated gene related to TGF-β1 signaling (Fig. S5C). Moreover, the knockdown of SHMT2 reduced TGFBR1 mRNA and protein expression (Fig. S5, D and E).
To further validate that the suppression effect of SHMT2 knockdown on TGF-β1 signaling is mediated by increased ALA and DHA levels, we examined the effect of ALA and DHA on TGFBR1 mRNA expression. FA deficiency, ALA, and DHA reduced TGFBR1 mRNA expression in HSCs (Fig. 6G). Does ALA inhibit the expression of TGFBR1 directly or does it act by converting it into DHA? Fatty acid desaturase 2 (FADS2) is the key enzyme responsible for converting ALA into DHA. We explored this question by knocking down FADS2 in HSCs. Knockdown FADS2 significantly blocked the reductions of TGFBR1 and COL1A1 mRNA induced by FA deficiency and ALA, but not DHA relative to the negative control (Fig. 6G). Furthermore, knockdown of FADS2 also rescued the decrease in TGFBR1 and COL1A1 mRNA due to SHMT2 inhibition to block mitochondrial folate metabolism (Fig. 6H). These results indicate that ALA inhibits the expression of TGFBR1 mRNA through bioconversion to DHA rather than acting on its own. Furthermore, we observed that FA deficiency, ALA, and DHA did not reduce the Smad2/3 signals after knocking down TGFBR1 in HSCs (Fig. 6, I and J). Taken together, the increase in ALA to DHA bioconversion induced by blocking SHMT2-mediated mitochondrial folate metabolism inhibited HSC activation and sustainability by controlling TGF-β1 signaling.
Targeting SHMT2/MTHFD2 promotes the resolution of liver fibrosis in NASH mice
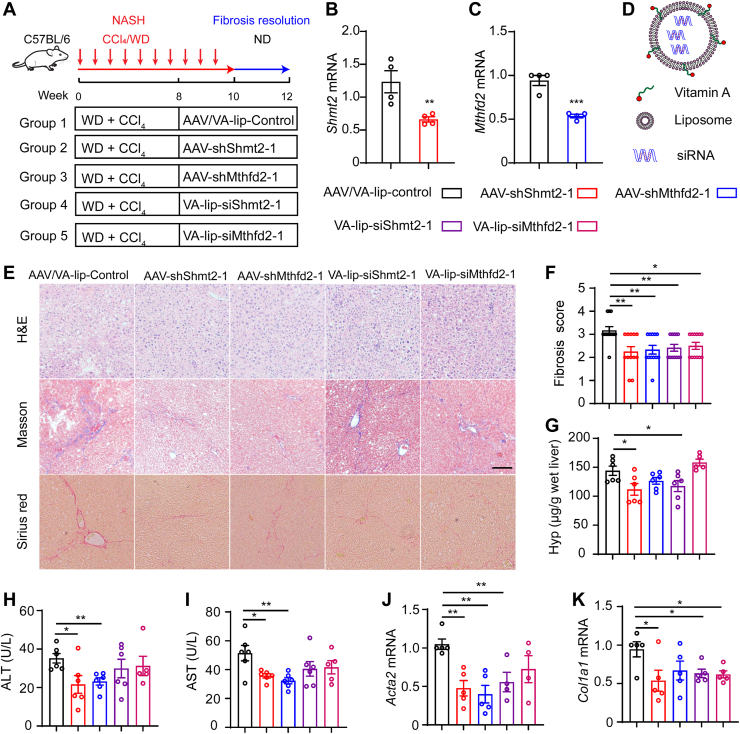
To uncover the effect of SHMT2-/MTHFD2-mediated mitochondrial folate metabolism on fibrosis resolution in vivo, we examined fibrosis resolution in NASH mice with knockdown of Shmt2 and Mthfd2 (Fig. 7A). We generated an adeno-associated virus carrying shRNA-Shmt2-GFP and shRNA-Mthfd2-GFP, which was used to knock down Shmt2 and Mthfd2 in liver cells (Fig. 7, B and C), and we established vitamin A (VA)-coupled liposome nanoparticles to deliver Shmt2 and Mthfd2 siRNA to knockdown SHMT2 and MTHFD2 expression in HSCs (Fig. 7D). Sirius red staining and Masson staining of liver tissues revealed that liver-specific knockdown of Shmt2 or Mthfd2 in mice either by adeno-associated virus–carrying siRNA or VA-liposome delivery caused a significant decrease in fibrosis after two weeks of treatment compared to control mice (Figs. 7, E and F and S6, A and B). Knocking down Shmt2 or Mthfd2 markedly decreased serum ALT and AST levels and the hydroxyproline content in liver tissue (Figs. 7, G–I and S6, C–E). Consistently, the mRNA levels of profibrotic genes were reduced in Shmt2- or Mthfd2-knockdown mice (Figs. 7, J and K and S6, F and G). Furthermore, knockdown of Shmt2 decreased Tgfbr1 mRNA levels (Fig. S6H). These data suggested that targeting mitochondrial folate metabolism in HSCs is a promising therapeutic strategy to promote liver fibrosis resolution.
Figure 7.
Knockdown of SHMT2/MTHFD2 promotes liver fibrosis resolution in NASH mice.A, schematic illustrating animal experimental design. B and C, liver Shmt2 and Mthfd2 mRNA levels. Ad-shRNA–medicated Shmt2 and Mthfd2 knockdown in mouse liver tissue (n = 4). D, schematic representation of vitamin A–coupled liposomes nanoparticle for the delivery of siRNA. E, representative image of H&E, Masson, and sirius red–stained paraffin-embedded sections of mouse liver tissues (scale bar represents 50 μm). F, Ishak fibrosis stage scores at indicated treatment conditions (n = 6). G, hepatic hydroxyproline content (n = 6). H and I, ALT and AST levels of plasma (n = 6). J and K, hepatic mRNA expression of fibrogenic genes Acta2 and Col1a1 (n = 5). The statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001) was tested by student’s unpaired t test. AST, aspartate aminotransferase; ALT, alanine aminotransferase; H&E, hematoxylin-eosin staining; MTHFD, methylenetetrahydrofolate dehydrogenase; NASH, nonalcoholic steatohepatitis; SHMT, serine hydroxymethyltransferase.
Discussion
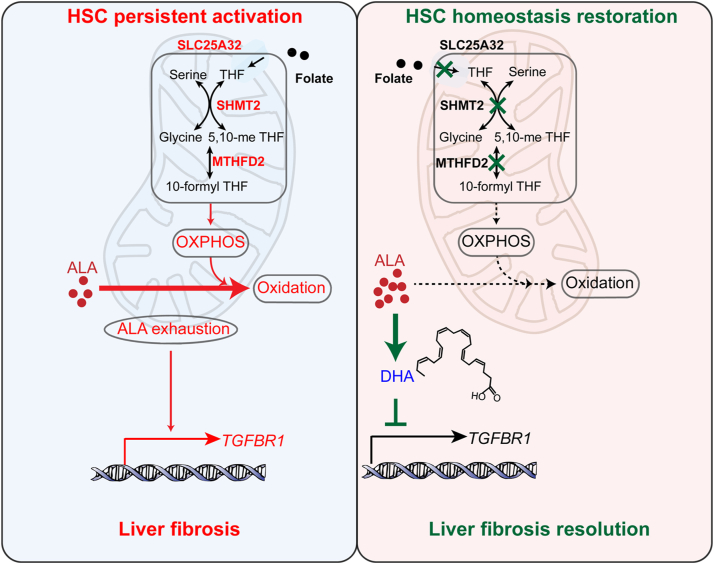
Controlling the sustainable TGF-β1 signaling for resolving liver fibrosis remains a challenge, as the mechanism underlying the maintenance of TGF-β1 signaling is still unclear. In this study, we revealed that mitochondrial folate metabolism/ALA exhaustion/TGF-βR1 reproduction is a feedforward signaling to sustain profibrotic TGF-β1 signaling. Notably, SHMT2-/MTHFD2-mediated mitochondrial folate metabolism controls n-3 polyunsaturated fatty acid metabolism for HSC sustainable activation (Fig. 8). Importantly, targeting SHMT2/MTHFD2 is a promising therapeutic strategy for the treatment of liver fibrosis. Moreover, our study suggests that NASH patients should pay more attention to folate intake.
Figure 8.
Mechanism of mitochondrial folate metabolism in hepatic stellate cells to maintain TGF-β1 signaling. Mitochondrial folate metabolism/α-linolenic acid (ALA) exhaustion/TGF-βR1 reproduction is a feedforward signaling to sustain profibrotic TGF-β1 signaling. Targeting mitochondrial folate metabolism blocks TGF-β1 signaling sustainability and enforces HSC homeostasis restoration. HSC, hepatic stellate cell.
Folate deficiency leads to anemia, carcinogenesis, and neural tube defects in newborns (42, 43, 44). Whether FA supplementation is beneficial in patients with chronic liver disease is unclear. FA supplementation has been shown to improve nonalcoholic fatty liver disease by reducing lipid accumulation in hepatocytes (17), whereas its role in liver fibrosis has received less attention. Mahamid et al reported that low serum levels of folate and B12 correlate with the severity of NASH patients (45). Our study suggested for the first time that FA impedes fibrosis resolution in NASH mice. In addition, folate may participate in the pathogenesis of idiopathic pulmonary fibrosis (46). Thus, folate metabolism may play opposite roles in different stages of the same disease. FA supplementation in patients with fibrosis may hinder fibrosis resolution, and therefore FA supplementation may need to be tightly controlled according to the context.
Multiple types of metabolic pathways are active in the liver, including 1C and lipid metabolism pathways. These pathways are not isolated and may engage in crosstalk. For example, the FA-mediated three-step 1C transfer process is involved in phosphatidylcholine synthesis (47). Polyunsaturated fatty acid metabolism serves an essential role in both normal physiology and pathological states (48). Crosstalk between components of folate metabolism and polyunsaturated fatty acid metabolism has rarely been investigated. For the first time, in this study, an increase in ALA level was observed in HSCs after mitochondrial folate metabolism was inhibited. SHMT2 is considered a central link between mitochondrial folate metabolism and polyunsaturated fatty acid metabolism. Therefore, the potential mechanism of the effect of folate may involve the knockdown of SHMT2, which reduces mitochondrial function and inhibits polyunsaturated fatty acid catabolism. We also found that the blockade of mitochondrial folate metabolism induced mitochondrial dysregulation in HSCs. However, the underlying mechanisms still need further exploration.
TGF-β1 signaling is a master pathway that drives organ fibrosis (27). In this study, we revealed that TGF-β1 signaling is necessary but not sufficient for maintaining HSC activation. Namely, a single TGF-β1 stimulation usually led to only a transient increase in HSC activity. In contrast, chronic TGF-β1 stimulation sustains HSC activation, which may mask the resolution of liver fibrosis. Metabolic adaptation during HSC activation may sustain TGF-β1 signaling as that is regulated by several metabolites. For example, 5-methoxy tryptophan downregulates TGF-β1 signaling (49). Creatine promotes TGF-β1 signaling in cancer (50). In HSCs, polyunsaturated fatty acids suppress TGF-β1 signaling by inhibiting the phosphorylation of SMAD2/3, but the mechanism remains unknown (51, 52). Our study revealed that n-3 polyunsaturated fatty acids inhibit TGF-β1 signaling by downregulating TGFBR1 mRNA expression. How do n-3 polyunsaturated fatty acids regulate TGFBR1 mRNA expression? We speculate that DHA may regulate TGF-β1 signaling by modifying certain protein levels after protein fatty acylation, which regulates various cell signaling pathways (53).
Taken together, our findings suggest that mitochondrial folate metabolism is a key metabolic pathway for maintaining HSC activation and impeding liver fibrosis resolution. Targeting mitochondrial folate metabolism to restore HSC homeostasis is a promising therapeutic strategy for the treatment of liver fibrosis. SHMT2/MTHFD2 regulate polyunsaturated fatty acid metabolism and are promising antifibrotic targets. Our study also calls attention to FA fortification in people with organ fibrosis.
Experimental procedures
Reagents
Antibodies were purchased from the following resources: antibodies to phospho-SMAD2/3 (8828S), SMAD2/3 (8685S), SHMT1 (80715S), SHMT2 (33443S) from Cell Signaling Technology; antibodies to α-Actin (1A4) (ab150301) and TGFBR1(ab31013) from Abcam; antibodies to MTHFD1 (67670-1-1g), MTHFD1L (16113-1-AP) from Proteintech; antibodies to collagen 1α(I) from Southern Biotech; antibodies to GAPDH (AM1021B), Tubulin (M20005M) from Abmart; antibodies to α-Actin (1A4) (SC-32251) from Santa Cruz Biotechnology; antibody to MTHFD2 (GTX104990-S) from Gene Tex.
FA-free RPMI 1640 (27016021), high glucose Dulbecco's modified Eagle medium (DMEM, 06-1055-57-1ACS) was purchased from Gibico. Linoleic acid (L105575), DHA (HY-B2167), SHIN1 (HY-101943), LY345899 (HY-112066) was purchased from MCE. FA (F8758), SAM (A7007), Oligomycin A (75351), FCCP (C2920), rotenone (R8875), antimycin (A8674) was purchased from Sigma-Aldrich. L-serine (M101131), L-methionine (S137887), linolenic acid (L105575) were purchased from Aladdin. Mitotracker Red (M7512), LipidTOX (H34476) was purchased from Molecular Probes. Cy5-labeled FA (PG2-FAS5-2K) was purchased from Nanocs. TRizol Reagent (9109) was purchased from Takara. L-serine (2,3,3-D3) (DLM-582) was purchased from Cambridge Isotope Laboratories.
Cell culture
LX-2 cell line was obtained from Xiang Ya Central Experiment Laboratory. Cells were cultured in DMEM containing 2% fetal bovine serum. DMEM containing 20% fetal bovine serum was used to culture mouse primary HSCs. Cells were cultured in an incubator at 37 °C and 5% (v/v) CO2.
Mouse experiments
The eight-week-old male C57BL/6N mice were obtained from Charles River. Experiments used male mice between 8 weeks of age. All mouse Experimental procedures in this study were approved by the Animal Care Committee of Nanjing University. For fibrosis resolution studies, mice were stimulated with CCl4 and fed with WDs containing high-fat, high-fructose, and high-cholesterol for 8 weeks (20), as soon as mice were fed a folate-depletion diet or ND were allowed to recover from fibrosis after CCl4 and WD discontinuation. To knock down SHMT2 or MTHFD2 in a liver-specific manner, an adeno-associated virus targeting SHMT2 or MTHFD2 was administered by intravenous injection two weeks before stimulus withdrawal. The knockdown of SHMT2 or MTHFD2 in HSC, VA-coupled liposomes carrying siRNAs were injected intravenously into mice three times a week before stimulus withdrawal. VA-lip–siRNA complex was produced according to the previous method. Briefly, siRNA of mouse SHMT2 and MTHFD2 were synthesized by Genscript.
Primary liver cell isolation
The primary mouse liver cells were isolated from the mouse liver as described (54). Mouse liver was perfused by the hepatic portal vein with HBSS and EGTA buffer for approximately 10 min at 37 °C. After that, transfer the tubing to the pronase/collagenase buffer. After digestion of the liver takes approximately 5 to 10 min, collect the liver and transfer it to cold HBSS. The collected cell was filtered by 100 μm cell filter and centrifuge for 5 min at a speed of 50g at 4 °C. The supernatant and pellet were collected in separate tubes. The hepatocytes are in the pellet. The supernatant is nonparenchymal liver cells. For the supernatant, a density gradient–based separation was performed to obtain HSCs. The other nonparenchymal cells are obtained separately by different centrifugation according to a previous protocol. Collection of the cells is obtained for subsequent experiments.
Biochemical analyses
Serum AST and alanine aminotransferase (ALT) were tested by commercial assay kit according to standard procedures. The assay kit was obtained from the Jiancheng Company.
RNA extraction and qPCR analysis
Cells or frozen liver were lysed in 1 ml triazole reagent (Invitrogen). The total mRNA was extracted and reverse transcribed into complementary DNA (cDNA) using the Reverse Transcription Kits (Vazyme, R223-01). The real-time PCR was performed. Amplification reactions were set up in 10 μl volumes containing primers, cDNA, and SYBR Green. All qPCR primer sequences in this study are listed in Table S1.
Western blotting
Cells were lysed using lysis buffer. Protein was separated by 10% SDS-PAGE and transferred to polyvinylidene fluoride membranes. After blocking with 5% skimmed milk for 1 h, the membranes were incubated with the primary antibodies at 4 °C overnight. Next day, the membranes were incubated with the horseradish peroxidase–conjugated secondary antibodies for 1 h at room temperature, then scanned using AMERSHAM ImageQuant 800 (Cytiva).
Immunofluorescence microscopy
All paraffin-embedded slides of liver samples were deparaffinized, rehydrated, and antigen retrieved with sodium citrate antigen retrieval solution. The liver tissue side was permeated using 0.2% Triton X-100 using 0.2% Triton X-100. Then, were incubated with 5% goat serum for 1 h at RT. Subsequently, the tissue slides were incubated with the primary antibody overnight at 4 °C. Primary antibodies were used as follows: α-SMA (1:100, Santa Cruz, SC-32251), α-SMA (1:100, Abcam, ab150301), collagen α1(I) (1:100, Southern Biotech, 1310-30), TGFBR1 (1:100, Abcam, ab31013). After that, the slide was washed and incubated with the indicated fluorescent secondary antibodies for 2 h. The liver tissue sections were stained with diamidino-phenyl-indole (DAPI). Immunofluorescence images were captured using a confocal laser microscope (Leica). Quantifications were performed using ImageJ.
For FA distribution in mouse HSCs, primary HSCs were treated with TGF-β1 (5 ng/ml) for 48 h. Cells were cultured for 12 h in FA-free medium and then incubated with 2 μM FA-labeled cy5 and Mitotracker Red probe (100 nM, Invitrogen). Cells were fixed and stained with DAPI for 5 min using confocal laser microscopy to capture images of cells and to observe the distribution of FA in mouse HSCs. For Bodipy and lipidTOX assays, fixed LX-2 cells were incubated in Bodipy (1:5000, Invitrogen) and LipidTOX (1:10,000, Invitrogen) for 30 min at RT. Next, the fixed HSCs were gently washed and incubated with DAPI (1:10,000). Images were taken on the Olympus microscope.
Flow cytometry
For uptake of HSCs in FA assays, cells were treated with TGF-β1 (5 ng/ml) for 48 h. Then, cells were starved of FA for 12 h and then were incubated with FA-labeled FITC for 30 min. The fluorescence density of LX-2 cells was measured by the FACSCanto flow cytometer. For intracellular staining of type I collagen, LX-2 cells were fixed with eBioscience Fixation/Permeabilization buffer for 30 min. Then, HSCs were incubated with Type I Collagen-AF488 for 1 h. Finally, cells were gently washed and resuspended with FACS buffer. The fluorescence intensity of proteins was measured by FACSCanto flow cytometer and analyzed using Flow Jo software 10 (https://www.flowjo.com/solutions/flowjo/downloads/).
Isotope tracing
LX-2 cells were cultured in a high (2 μM) or physiological (200 nM) folate medium for at least two weeks. Isotope tracing assay was carried out by feeding LX2 cells with [2,3,3-2H]-serine. LX-2 cells were stably cultured for 36 h in a [2,3,3-2H] serine medium, and the medium was changed 2 h before sample collection. Metabolism was quenched by adding 5:3:2 (volume ratio) methanol:acetonitrile:water. The collected samples were stored at −80 °C until further analysis. LC-MS was performed by LipidALL Technologies.
Seahorse assays
Using an XF96 Analyzer (Seahorse Bioscience) to determine the oxygen consumption rate (OCR). LX-2 cells were seeded in XF 96-well microplates (Seahorse Bioscience) and transfected with SHMT2 or MTHFD2 siRNA for 48 h or treated with SHIN1 (0, 2, 4 μM) or LY345899 (0, 2, 4 μM) for 24 h. The culture medium was removed, and LX-2 cells were washed with a 200 μl assay medium containing XF base medium with 25 mM glucose, 1 mM sodium pyruvate, and 2 mM L-glutamine. Then, cells were cultured for 1 h without CO2 before OCR analysis. The Seahorse 96 optical cartridge was equilibrated using an equilibration solution. The mitochondrial stress test was measured by injecting 1 μM oligomycin, 1 μM FCCP, 0.5 μM rotenone/antimycin into each cartridge port. The OCR was normalized to the amount based on the protein content measured by Bradford assays.
siRNA transfection
For Western blotting and qPCR experiments, LX-2 cells were seeded in plates and then transfected with 20 nM siRNA targeting SLC25A32, SHMT1, SHMT2, MTHFD1, MTHFD1L, and MTHFD2 using RNAiMAX Lipofectamine (Invitrogen) according to standard protocol. All siRNAs targeting sequence in the study are listed in Table S2.
RNA-seq and analysis
Total RNA from the LX-2 cells treated with SHMT2 or scrambled control siRNA were extracted using the Trizol method using the RNA Nano 6000 Assay Kit (Agilent Technologies) to determine RNA integrity. And then the mRNA library was prepared for transcriptome sequencing. cDNA library was sequenced using an Illumina Novaseq platform. Reads were aligned to the human reference genome using Hisat2 v2.0.5. The read numbers mapped to each gene were counted using featureCounts v1.5.0-p3. The DESeq2 R package was used as a differential expression analysis. cDNA library preparation and transcriptome sequencing in the Novogene Laboratories (Novogene, China). The RNA-seq data were deposited in NCBI’s Gene Expression Omnibus. This GEO Series accession number is GSE210718.
Statistics analysis
The results are indicated as the mean ± SEM and plotted using GraphPad Prism soft. Data statistical analysis was performed using the unpaired Student’s t-test. Significance was considered to be ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, respectively. Genome enrichment analysis and gene ontology analysis is done in R using the limma package. The heat maps in this study were generated in RStudio with a heatmap R package. Other graphs were generated in GraphPad Prism.
Data availability
The RNA sequencing data were deposited in NCBI’s Gene Expression Omnibus. This GEO Series accession number is GSE210718. Data are available in the manuscript.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank all other members in the Wu lab for helpful discussion. This study was supported by the National Natural Science Foundation of China (2193005, 81722047, 81874317); Fundamental Research Funds for the Central Universities (020814380161, China).
Author contributions
Y. G., B. Z., S. X., Z. Z., W. L., T. W., M. Y., X. S., and Y. T. investigation; Q. X. and X. W. conceptualization; Y. G., B. Z., and S. X. methodology; Y. G. and B. Z., data curation; Y. G. writing–original draft; Y. G., B. Z., and X. W. visualization; Q. X. and X. W. supervision; Q. X., and X. W. writing–review and editing.
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Contributor Information
Qiang Xu, Email: molpharm@163.com.
Xingxin Wu, Email: xingxin.wu@nju.edu.cn.
Supporting information
References
- 1.Angulo P., Kleiner D.E., Dam-Larsen S., Adams L.A., Bjornsson E.S., Charatcharoenwitthaya P., et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology. 2015;149:389–397.e10. doi: 10.1053/j.gastro.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diehl A.M., Day C. Cause, pathogenesis, and treatment of nonalcoholic Steatohepatitis. N. Engl. J. Med. 2017;377:2063–2072. doi: 10.1056/NEJMra1503519. [DOI] [PubMed] [Google Scholar]
- 3.Vilar-Gomez E., Calzadilla-Bertot L., Wai-Sun Wong V., Castellanos M., Aller-de la Fuente R., Metwally M., et al. Fibrosis severity as a determinant of cause-specific mortality in patients with advanced nonalcoholic fatty liver disease: a multi-national cohort study. Gastroenterology. 2018;155:443–457.e17. doi: 10.1053/j.gastro.2018.04.034. [DOI] [PubMed] [Google Scholar]
- 4.Tsuchida T., Friedman S.L. Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:397–411. doi: 10.1038/nrgastro.2017.38. [DOI] [PubMed] [Google Scholar]
- 5.Higashi T., Friedman S.L., Hoshida Y. Hepatic stellate cells as key target in liver fibrosis. Adv. Drug Deliv. Rev. 2017;121:27–42. doi: 10.1016/j.addr.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim K.K., Sheppard D., Chapman H.A. TGF-β1 signaling and tissue fibrosis. Cold Spring Harb. Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a022293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Troeger J.S., Mederacke I., Gwak G.-Y., Dapito D.H., Mu X., Hsu C.C., et al. Deactivation of hepatic stellate cells during liver fibrosis resolution in mice. Gastroenterology. 2012;143:1073–1083.e22. doi: 10.1053/j.gastro.2012.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kisseleva T., Cong M., Paik Y., Scholten D., Jiang C., Benner C., et al. Myofibroblasts revert to an inactive phenotype during regression of liver fibrosis. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9448–9453. doi: 10.1073/pnas.1201840109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang M., Vousden K.H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer. 2016;16:650–662. doi: 10.1038/nrc.2016.81. [DOI] [PubMed] [Google Scholar]
- 10.Locasale J.W. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 2013;13:572–583. doi: 10.1038/nrc3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stover P.J., Field M.S. Trafficking of intracellular folates. Adv. Nutr. 2011;2:325–331. doi: 10.3945/an.111.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tibbetts A.S., Appling D.R. Compartmentalization of Mammalian folate-mediated one-carbon metabolism. Annu. Rev. Nutr. 2010;30:57–81. doi: 10.1146/annurev.nutr.012809.104810. [DOI] [PubMed] [Google Scholar]
- 13.Beaudin A.E., Stover P.J. Insights into metabolic mechanisms underlying folate-responsive neural tube defects: a minireview. Birth Defects Res. A. Clin. Mol. Teratol. 2009;85:274–284. doi: 10.1002/bdra.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey L.B., Berry R.J. Folic acid supplementation and the occurrence of congenital heart defects, orofacial clefts, multiple births, and miscarriage. Am. J. Clin. Nutr. 2005;81:1213S–1217S. doi: 10.1093/ajcn/81.5.1213. [DOI] [PubMed] [Google Scholar]
- 15.da Silva R.P., Kelly K.B., Al Rajabi A., Jacobs R.L. Novel insights on interactions between folate and lipid metabolism. Biofactors. 2014;40:277–283. doi: 10.1002/biof.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen K.E., Wu Q., Wang X., Deng L., Caudill M.A., Rozen R. Steatosis in mice is associated with gender, folate intake, and expression of genes of one-carbon metabolism. J. Nutr. 2010;140:1736–1741. doi: 10.3945/jn.110.124917. [DOI] [PubMed] [Google Scholar]
- 17.Sid V., Wu N., Sarna L.K., Siow Y.L., House J.D., O K. Folic acid supplementation during high-fat diet feeding restores AMPK activation via an AMP-LKB1-dependent mechanism. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015;309:R1215–R1225. doi: 10.1152/ajpregu.00260.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sid V., Siow Y.L., O K. Role of folate in nonalcoholic fatty liver disease. Can J. Physiol. Pharmacol. 2017;95:1141–1148. doi: 10.1139/cjpp-2016-0681. [DOI] [PubMed] [Google Scholar]
- 19.Charatcharoenwitthaya P., Levy C., Angulo P., Keach J., Jorgensen R., Lindor K.D. Open-label pilot study of folic acid in patients with nonalcoholic steatohepatitis. Liver Int. 2007;27:220–226. doi: 10.1111/j.1478-3231.2006.01404.x. [DOI] [PubMed] [Google Scholar]
- 20.Tsuchida T., Lee Y.A., Fujiwara N., Ybanez M., Allen B., Martins S., et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J. Hepatol. 2018;69:385–395. doi: 10.1016/j.jhep.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang S.-S., Tang X.T., Lin M., Yuan J., Peng Y.J., Yin X., et al. Perivenous stellate cells are the main source of myofibroblasts and cancer-associated fibroblasts formed after chronic liver injuries. Hepatology. 2021;74:1578–1594. doi: 10.1002/hep.31848. [DOI] [PubMed] [Google Scholar]
- 22.Schwabe R.F., Tabas I., Pajvani U.B. Mechanisms of fibrosis development in nonalcoholic Steatohepatitis. Gastroenterology. 2020;158:1913–1928. doi: 10.1053/j.gastro.2019.11.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X., Xu J., Rosenthal S., Zhang L.-J., McCubbin R., Meshgin N., et al. Identification of lineage-specific transcription factors that prevent activation of hepatic stellate cells and promote fibrosis resolution. Gastroenterology. 2020;158:1728–1744.e14. doi: 10.1053/j.gastro.2020.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tacke F., Trautwein C. Mechanisms of liver fibrosis resolution. J. Hepatol. 2015;63:1038–1039. doi: 10.1016/j.jhep.2015.03.039. [DOI] [PubMed] [Google Scholar]
- 25.Derynck R., Zhang Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 26.Shi Y., Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 27.Meng X.-M., Nikolic-Paterson D.J., Lan H.Y. TGF-β: the master regulator of fibrosis. Nat. Rev. Nephrol. 2016;12:325–338. doi: 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 28.Lee W.D., Pirona A.C., Sarvin B., Stern A., Nevo-Dinur K., Besser E., et al. Tumor reliance on cytosolic versus mitochondrial one-carbon flux depends on folate availability. Cell Metab. 2021;33:190–198.e196. doi: 10.1016/j.cmet.2020.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Lawrence S.A., Hackett J.C., Moran R.G. Tetrahydrofolate recognition by the mitochondrial folate transporter. J. Biol. Chem. 2011;286:31480–31489. doi: 10.1074/jbc.M111.272187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Titus S.A., Moran R.G. Retrovirally mediated complementation of the glyB phenotype. Cloning of a human gene encoding the carrier for entry of folates into mitochondria. J. Biol. Chem. 2000;275:36811–36817. doi: 10.1074/jbc.M005163200. [DOI] [PubMed] [Google Scholar]
- 31.Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., et al. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee G.Y., Haverty P.M., Li L., Kljavin N.M., Bourgon R., Lee J., et al. Comparative oncogenomics identifies PSMB4 and SHMT2 as potential cancer driver genes. Cancer Res. 2014;74:3114–3126. doi: 10.1158/0008-5472.CAN-13-2683. [DOI] [PubMed] [Google Scholar]
- 33.Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ducker G.S., Chen L., Morscher R.J., Ghergurovich J.M., Esposito M., Teng X., et al. Reversal of cytosolic one-carbon flux compensates for loss of the mitochondrial folate pathway. Cell Metab. 2016;23:1140–1153. doi: 10.1016/j.cmet.2016.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu L., Zhang K., Sandoval H., Yamamoto S., Jaiswal M., Sanz E., et al. Glial lipid droplets and ROS induced by mitochondrial defects promote neurodegeneration. Cell. 2015;160:177–190. doi: 10.1016/j.cell.2014.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z., Zhao X., Shang W., Liu Y., Ji J.F., Liu J.P., et al. Pyrroline-5-carboxylate synthase senses cellular stress and modulates metabolism by regulating mitochondrial respiration. Cell Death Differ. 2021;28:303–319. doi: 10.1038/s41418-020-0601-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvo S.E., Mootha V.K. The mitochondrial proteome and human disease. Annu. Rev. Genomics Hum. Genet. 2010;11:25–44. doi: 10.1146/annurev-genom-082509-141720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X., Du M., Li D., Wen S., Xie J., Li Y., et al. Mutations in FASTKD2 are associated with mitochondrial disease with multi-OXPHOS deficiency. Hum. Mutat. 2020;41:961–972. doi: 10.1002/humu.23985. [DOI] [PubMed] [Google Scholar]
- 39.Brenna J.T. Efficiency of conversion of alpha-linolenic acid to long chain n-3 fatty acids in man. Curr. Opin. Clin. Nutr. Metab. Care. 2002;5:127–132. doi: 10.1097/00075197-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 40.Whelan J. Dietary stearidonic acid is a long chain (n-3) polyunsaturated fatty acid with potential health benefits. J. Nutr. 2009;139:5–10. doi: 10.3945/jn.108.094268. [DOI] [PubMed] [Google Scholar]
- 41.Fekete K., Decsi T. Long-chain polyunsaturated fatty acids in inborn errors of metabolism. Nutrients. 2010;2:965–974. doi: 10.3390/nu2090965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stover P.J. Physiology of folate and vitamin B12 in health and disease. Nutr. Rev. 2004;62 doi: 10.1111/j.1753-4887.2004.tb00070.x. [DOI] [PubMed] [Google Scholar]
- 43.De Wals P., Tairou F., Van Allen M.I., Uh S.-H., Lowry R.B., Sibbald B., et al. Reduction in neural-tube defects after folic acid fortification in Canada. N. Engl. J. Med. 2007;357:135–142. doi: 10.1056/NEJMoa067103. [DOI] [PubMed] [Google Scholar]
- 44.Cario H., Smith D.E.C., Blom H., Blau N., Bode H., Holzmann K., et al. Dihydrofolate reductase deficiency due to a homozygous DHFR mutation causes megaloblastic anemia and cerebral folate deficiency leading to severe neurologic disease. Am. J. Hum. Genet. 2011;88:226–231. doi: 10.1016/j.ajhg.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahamid M., Mahroum N., Bragazzi N.L., Shalaata K., Yavne Y., Adawi M., et al. Folate and B12 levels correlate with histological severity in NASH patients. Nutrients. 2018;10:440. doi: 10.3390/nu10040440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu Y., Hao C., Zhai R., Yao W. Folate and macrophage folate receptor-β in idiopathic pulmonary fibrosis disease: the potential therapeutic target? Biomed. Pharmacother. 2020;131 doi: 10.1016/j.biopha.2020.110711. [DOI] [PubMed] [Google Scholar]
- 47.Da Silva R.P., Eudy B.J., Deminice R. One-carbon metabolism in fatty liver disease and fibrosis: one-carbon to rule them all. J. Nutr. 2020;150:994–1003. doi: 10.1093/jn/nxaa032. [DOI] [PubMed] [Google Scholar]
- 48.Wang M., Ma L.-J., Yang Y., Xiao Z., Wan J.-B. n-3 Polyunsaturated fatty acids for the management of alcoholic liver disease: a critical review. Crit. Rev. Food Sci. Nutr. 2019;59:S116–S129. doi: 10.1080/10408398.2018.1544542. [DOI] [PubMed] [Google Scholar]
- 49.Fang L., Chen H., Kong R., Que J. Endogenous tryptophan metabolite 5-Methoxytryptophan inhibits pulmonary fibrosis by downregulating the TGF-β/SMAD3 and PI3K/AKT signaling pathway. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118399. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L., Zhu Z., Yan H., Wang W., Wu Z., Zhang F., et al. Creatine promotes cancer metastasis through activation of Smad2/3. Cell Metab. 2021;33:1111–1123.e4. doi: 10.1016/j.cmet.2021.03.009. [DOI] [PubMed] [Google Scholar]
- 51.Hu S., Bae M., Park Y.K., Lee J.Y. n-3 PUFAs inhibit TGFbeta1-induced profibrogenic gene expression by ameliorating the repression of PPARgamma in hepatic stellate cells. J. Nutr. Biochem. 2020;85 doi: 10.1016/j.jnutbio.2020.108452. [DOI] [PubMed] [Google Scholar]
- 52.Enguita M., Razquin N., Pamplona R., Quiroga J., Prieto J., Fortes P. The cirrhotic liver is depleted of docosahexaenoic acid (DHA), a key modulator of NF-κB and TGFβ pathways in hepatic stellate cells. Cell Death Dis. 2019;10:14. doi: 10.1038/s41419-018-1243-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu S.-H., He X.-D., Nie J., Hou J.-L., Wu J., Liu X.-Y., et al. Methylene-bridge tryptophan fatty acylation regulates PI3K-AKT signaling and glucose uptake. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu X., Wu X., Ma Y., Shao F., Tan Y., Tan T., et al. CUG-binding protein 1 regulates HSC activation and liver fibrogenesis. Nat. Commun. 2016;7 doi: 10.1038/ncomms13498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA sequencing data were deposited in NCBI’s Gene Expression Omnibus. This GEO Series accession number is GSE210718. Data are available in the manuscript.