Highlights
-
•
IHNV RtWanju15 isolate is the first virus belonging the JRt Nagano lineage in Jeollabuk-do province.
-
•
There was no difference in pathogenicity between RtWanju15 (Nagano lineage) and RtWanju09 (Shizuoka lineage).
-
•
Parental fish that have never encountered IHNV should be used to rule out the natural adaption of fish to IHNV.
Keywords: Infectious hematopoietic necrosis virus (IHNV), Mass mortality, Rainbow trout, Phylogenetic analysis, Immune-histopathological observation, challenge test
Abstract
In May 2015, a high mortality event in farmed rainbow trout occurred in Jeollabuk-do province in Korea. Histopathological analysis revealed necrosis in the kidney, liver, branchial arch, and gills of moribund fish, and infectious hematopoietic necrosis virus (IHNV) was detected in the lesions by immunohistochemistry. Cytopathic effects were observed in EPC, FHM, and RTG-2 cell lines after inoculation with kidney and spleen tissues and IHNV was detected by reverse transcription polymerase chain reaction (PCR). The amplified PCR product was sequenced, and phylogenetic analysis placed IHNV in the JRt Nagano group. Both in vivo and in vitro trials were performed to compare the virulence properties between RtWanju15 isolate, which causes 100% mortality in imported fry, and a previous isolate RtWanju09 of the JRt Shizuoka group isolated from eggs of healthy broodfish. In vivo challenge with high dose on specific pathogen free (SPF) rainbow trout fry performed in Denmark with isolates RtWanju09, RtWanju15 and DF04/99 isolates showed a survival rates of 60%, 37.5% and 52.5% (average), respectively without statistical difference. The replication efficiency of the two isolates in the in vitro challenge was similar.
1. Introduction
Infectious hematopoietic necrosis (IHN) is a serious viral disease in salmonids (WOAH, 2021; Wolf, 1988) and is listed as a notifiable disease by the World Organization for Animal Health (WOAH, 2021). The causative agent, IHN virus (IHNV) is a member of the genus Novirhabdovirus of the Rhabdoviridae family (WOAH, 2021). The single-stranded virus genome encodes six proteins: nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), non-virion protein (NV), and polymerase (L) (WOAH, 2021). Among them, nucleotide sequences of the G gene are useful for phylogenetic analysis and can be used to classify IHNV isolates for genotyping into L, M, U, E, and J genogroups, which correspond to their geographical location (Kurath et al., 2003; Nishizawa et al., 2006; WOAH, 2021). The JRt Nagano and JRt Shizuoka lineages are both found in Japan. Moreover, RtShiz06s and RtShiz06a isolates of the JRt Shizuoka lineage have been reported to cause higher mortality in rainbow trout (Oncorhynchus mykiss) than RtNag96 and RtNag06a isolates of the JRt Nagano lineage (Mochizuki et al., 2009). It has also been reported that both the JRt Nagano and JRt Shizuoka lineages have been detected in rainbow trout farms in South Korea (Kim et al., 2007).
In 2009, the IHNV RtWanju09 isolate was isolated from eggs contaminated with IHNV in a rainbow trout farm in Jeollabuk-do province, Korea. Broodstock fish were apparently healthy, without any symptoms of IHN. Phylogenetic analysis of the G gene sequence revealed that the IHNV RtWanju09 isolate was grouped into JRt Shizuoka lineage (Kim, 2010). Until 2014, this farm used only its own eggs for the production of rainbow trout. In 2015, the farm acquired rainbow trout fry from a hatchery in the Gangwon-do province in Korea but all fry died within two months of delivery. The Aquatic Animal Disease Judgment Institute did not detect any disease in the list of the Aquatic Animal Disease Control Act in Korea, thus, the farmer sent moribund fish to our laboratory to investigate the cause of the disease.
In this study, we perfomed fish diagnostics on moribund fish using cell culture methods, histopathology, and molecular techniques, we identify the viral disease agent and conducted in vivo and in vitro studies to evaluate a pathogenicity.
2. Materials and methods
2.1. Diseased fish and clinical and laboratory examination
In April 2015, moribund rainbow trout fry (4.8 ± 0.3 g, 32 fish) were collected from a rainbow trout farm in Jeollabuk-do province, Korea (Fig. 1). The fish were farmed in a water flow system at 9–10 °C. Moribund fish were transported to the laboratory for clinical and laboratory analysis. To examine for the presence of infection with parasites, a part of the gills and skin mucus from the fish were sampled and observed under a light microscope. To examine for bacteria, pieces of liver, spleen, and kidney were stamped onto tryptic soy agar (BD Difco, Sparks, MD, USA)
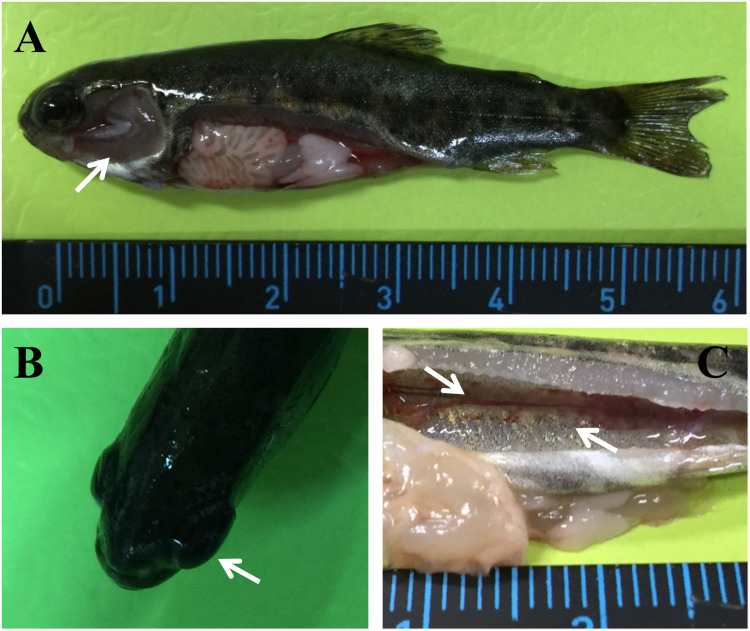
Fig. 1.
Observation of symptoms from moribund rainbow trout. The arrows show (A) greyish gills, (B) eyes exhibiting exophthalmos, and (C) severe anemia in the kidney.
2.2. Inoculation of organ material from moribund fish to cell cultures
Pieces of spleen, and kidney from the moribund fish were collected in a Petri dish and pooled for inoculation to fish cell lines. Eagle's minimum essential medium (MEM; Gibco, Invitrogen Co.) was added to the tissues, along with 1000 IU/mL penicillin G and 1000 μg/mL streptomycin sulfate. The tissues were homogenized in sterile mortar using pestles and centrifuged at 6000 × g for 10 min at 4 °C. Subsequently, the supernatant was passed through a 0.45 μm syringe filter, and the filtered solution was collected in autoclaved tubes.
Three cell lines (EPC, FHM and RTG-2) were prepared for inoculation of the filtered solution from fish samples. The selected cells were maintained at 20 °C in Eagle's MEM, supplemented with 10% fetal bovine serum, 100 IU/mL penicillin G, and 100 μg/mL streptomycin sulfate. Cells for inoculation of samples were prepared by passing them into new 25 cm2 flasks (Falcon) in 5 mL of MEM and then incubated for 24 h at 20 °C. The 100 μL of the filtered solution were then inoculated onto each cell line and incubated at 15 °C for 7 days. The cells were examined daily for cytopathic effects (CPE) using phase-contrast microscopy (Olympus IX51).
2.3. Observation by transmission electron microscopy (TEM)
The EPC cells exhibiting CPE were pre-fixed in 2.5% glutaraldehyde in PBS at 4 °C overnight. The specimen was post-fixed in 1% OsO4, and dehydrated, embedded in epoxy resin (EMS). The sample was ultrathin-sectioned and stained with uranyl acetate and lead citrate. Finally, microscopic examination was carried out with a JEM 1200-EXII TEM (JEOL LTD., Japan).
2.4. One-step conventional reverse transcription-polymerase chain reaction (RT-PCR) and sequence analysis
Viral RNA was extracted from the culture supernatant using a viral RNA extraction kit (QIAamp® Viral RNA Mini Kit; Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. Briefly, the extracted viral RNA was eluted using 30 μL diethyl pyrocarbonate (DEPC)-treated water. One-step RT-PCR was conducted in 25 μL of the reaction mixture, containing 5 μL of extracted or diluted viral RNA, 10 pM of primers (Table 1), 1 μL of 10 mM dNTPs, 5 μL of 5× RT-PCR reaction buffer (containing 12.5 mM MgCl2), 1 μL of enzyme mix, and DEPC-treated water, using a Qiagen One-step RT-PCR kit (Qiagen, Germany). The PCR conditions for each pathogen were followed as the reference in Table 1. The full open reading frame (ORF) of the IHNV glycoprotein (G) gene was amplified using HG (−31:−12) 5′-AGAACGCAACTCGCAGAGAC-3′ and HG (1602:1622) 5′-GTGGGGAGGAAGTGAAGATTG-3′ (expected size : 1653 bp) (Nishizawa et al., 2006). The PCR products were purified using a Gel Extraction Kit (Qiagen) and subjected to sequencing analysis (Bioneer, South Korea).
Table 1.
Diagnostic primers used in this study.
| Primer name | Sequence (5′ to 3′) | Reference |
|---|---|---|
| IHNV F | AGAGATCCCTACACCAGAGAC | Emmenegger et al. (2000) |
| IHNV R | GGTGGTGTTGTTTCCGTGCAA | |
| VHSV VN F | ATGGAAGGAGGAATTCGTGAAGCG | Snow et al. (2004) |
| VHSV VN R | GCGGTGAAGTGCTGCAGTTCCC | |
| VHSV VN IVa F | ATGGAAGGGGGAATCCGTGCAGCT | Kim (2015) |
| VHSV VN IVa R | GCGGTGAAGTGCTGCAGTTCTC | |
| ISAV F | GACCAGACAAGCTTAGGTAACACAGA | WOAH (2021) |
| ISAV R | GATGGTGGAATTCTACCTCTAGACTTGTA |
The nucleotide sequence of the IHNV G gene from other isolates was obtained from the National Center for Biotechnology Information database (Table 2). Multiple alignments of sequences were performed using ClustalX 2.0, and the results were visualized using the GeneDoc program. Phylogenetic analysis was performed using the neighbor-joining method in the MEGA X program, and bootstrap sampling was repeated 1000 times.
Table 2.
List of IHNV isolates of JRt genogroup for phylogenetic analysis in this study.
| Isolate Name | Host | Isolated year | Isolated location | Genbank number | Reference |
|---|---|---|---|---|---|
| Shizu0606 AyTochi86 RtToya80 RtWanju09 RtJe00 RtPy91 RtTochi86 RtShiz06a RtShiz06b G4 RtNag06a RtNag06b RtAichi06 RtAichi06-2 RtNag96 RtGu01 RtUi02 RtWanju15 |
Rainbow trout Ayu Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout Rainbow trout |
2006 1986 1980 2009 2000 1991 1986 2006 2006 1992 2006 2006 2006 2006 1996 2001 2002 2015 |
Shizuoka, Japan Tochigi, Japan Toya, Japan Wanju, Korea Jecheon, Korea Pyeongchang, Korea Tochigi, Korea Shizuoka, Japan Shizuoka, Japan Gifu, Japan Nagano, Japan Nagano, Japan Aichi, Japan Aichi, Japan Nagano, Japan Gumi, Korea Uiseong, Korea Wanju, Korea |
AB510192 AB250933 AB250935 HM021723 AB288205 AB288204 AB250934 AB510193 AB510194 LC372825 AB510195 AB510196 AB510197 AB510198 AB250932 AB288206 AB288207 OM986071 |
Mochizuki et al. (2009) Nishizawa et al. (2006) Nishizawa et al. (2006) Kim. 2010 Kim et al. (2007) Kim et al. (2007) Nishizawa et al. (2006) Mochizuki et al. (2009) Mochizuki et al. (2009) Nishizawa et al. (2006) Mochizuki et al. (2009) Mochizuki et al. (2009) Mochizuki et al. (2009) Mochizuki et al. (2009) Nishizawa et al. (2006) Kim et al. (2007) Kim et al. (2007) The present study |
2.5. Histopathology and immunohistochemistry
Kidney, liver, branchial arch and gills of the affected fish, which were extracted from the moribund fish, fixed in 10% formalin solution. Formalin-fixed tissues were embedded in paraffin wax and sections (4 μm) were stained with haematoxylin and eosin (HE). To detect IHNV in tissue sections, immunohistochemical procedures were performed as described by Kurath et al. (2016). Briefly, the procedures included the use of 5% bovine serum albumin (BSA) in Tris-buffered saline (TBS, pH 7.4) for blocking. The tissue samples were then washed with TBS. An IHNV-nucleoprotein-specific monoclonal antibody, Hyb 136–3 (Fregeneda-Grandes et al., 2009), provided by DTU, Denmark, was used as the primary antibody (1:3000 in TBS with 2.5% BSA). The tissues were incubated overnight at room temperature. The tissues were then washed three times with TBS and incubated with a secondary antibody (biotinylated rabbit anti-mouse Ig (DAKO Cytomation); 1:300). The tissue samples were incubated in streptavidin alkaline phosphatase complex (1:500; code RPN, 1234; Amersham Biosciences, Piscataway, NJ, USA) for 30 min at room temperature. The primary antigen-antibody reaction was visualized by adding fast red salt (F-2768; Sigma-Aldrich, St. Louis, MO, USA) to a fast red substrate solution (Polack and Noorden, 2003).
2.6. Experimental infection trial in rainbow trout
280 Specific pathogen-free (SPF) rainbow trout fry (average weight of 13±0.8 g) were reared at the Technical University of Denmark (Animal experiment permission number: 2013–15–2934–00,976) and divided in 14 10-L tanks at 12 °C. Each viral isolates RtWanju09 (JRt Shizuoka lineage), RtWanju15 (JRt Nagano lineage), and DF04/99 (European group, Fichtner et al., 2000) was tested in duplicate tanks, where rainbow trout were exposed to IHNV by immersion for 5 h in 105 TCID50/mL (high dose) or 103 TCID50/mL (low dose) of the IHNV isolates. The IHNV DF04/99 (which showed high virulence in rainbow trout) was used as positive control. In addition, 2 × 20 fish were included in the study as negative control and exposed to immersion in 10 lt of water with 5 ml of steyle cell culture medium. All experiments were approved of by the Institutional Review Board (animal experiment permission number: DK-2013-15-2934-00976).
Disease occurrence and reduction of survival was monitored daily for 43 days. Fish which succumbed to the infection were tested for IHNV by re-isolation in cell culture and by RT-qPCR. RT-qPCR was performed as a one-step method according to the protocol described by Purcell et al. (2013). Briefly, one-step RT-PCR was conducted in 25 μL of a reaction mixture containing 5 μL of the extracted RNA, 10 pM of primers (F: 5-AGAGCCAAGGCACTGTGCG-3, R: 5-TTCTTTGCGGCTTGGTTGA-3), 12.5 μL of 2 × qPCR master mix, 0.25 μL of the RT/RNase block enzyme mix, and 0.25 μM of a probe (5–6FAM-TGAGACTGAGCGGGACA-NFQ/MGB-3), using a Quantitect Probe RT-PCR kit (Qiagen, Germany), according to the manufacturer's instructions. Forward and reverse primers are indicated by F and R, respectively. RT-PCR was performed using Mx3005P (Stratagene).
To determine whether there is a statistically significant difference of survival probability among experimental groups, R package ``ggsurvplot'' was used for visualizing survival analysis results. Survival comparisons among the groups were performed using Cox proportional hazards regression model. The results were considered statistically significant at p < 0.05. A 95% confidence interval is shown in shadowed area. The dashed line indicates the median survival time (time at which survival probability is 50%).
2.7. Viral replication of IHNV RtWanju09 and IHNV RtWanju15 in fish cell lines
Stock solutions of IHNV RtWanju09 and IHNV RtWanju15 were titrated by 10-fold serial dilutions in MEM and 25 μL/well inoculated on three cell lines (EPC, FHM, and RTG-2) in 96 well plates, with six replicates per experiment. The plates were incubated at 15 °C for 14 days, and the CPE was monitored daily using phase-contrast microscopy. Endpoint titers were calculated as TCID50/mL.
3. Results
3.1. Clinical signs, parasitology, and bacteriology results
Clinically affected fish swam weakly on the farm; they were characterized by skin darkening and pale, greyish gills, and exophthalmia (Fig. 1). Physical examination revealed that the kidney was pale gray and there were signs of severe anemia, but without severe hemorrhage in the organs (Fig. 1). No bacteria were cultured from internal organs using tryptic soy agar or blood heart infusion agar at 15 °C for 2 days and no parasites could be observed by microscopic examination of the gills or mucus from the skin of the fish (data not shown).
3.2. CPE in fish cell lines
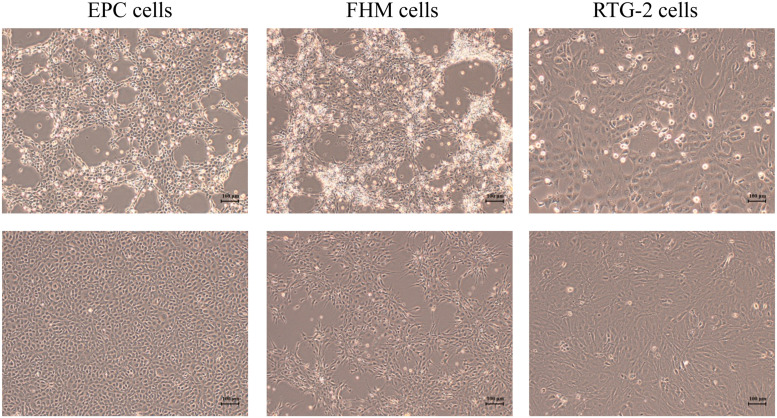
The EPC, FHM, and RTG-2 cell lines were used to isolate infectious viruses from the spleen and kidney of the fish. CPEs began to appear at 2 days post-inoculation in the EPC and FHM cell lines and showed cell rounding, detachment and dead cells at 3 days post-inoculation. In RTG-2 cell line, slight cell rounding was observed at 3 days post-inoculation (Fig. 2). After 5 days post-inoculation, the EPC and FHM cell lines showed full CPE with cells completely detaching from the bottom of the flask. And, full CPE was observed at 7 days post-inoculation in RTG-2 cell line.
Fig. 2.
Fish cell lines were inoculated with spleen and kidney homogenates from moribund fish (upper) and no inoculation was conducted on the control cells (lower). EPC, FHM and RTG-2 cell lines showed cell rounding, detachment and dead cells at 3 days post-inoculation.
3.3. Observation of virus particles by TEM
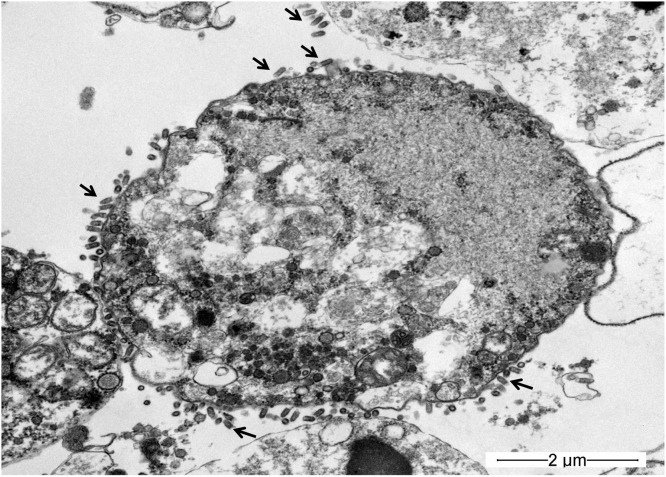
Numerous bullet-shaped virions were observed outside of cell membranes at 5 days post-inoculation. The size of virions was approximately 143–199 nm x 56–73 nm. Virions from cells exhibiting CPE showed morphology typical of rhabdoviruses (Fig. 3).
Fig. 3.
Observation of virus particles by TEM. The arrows show the bullet-shaped rhabdovirus on the EPC cells at 5 days post-inoculation.
3.4. Conventional RT-PCR and genotyping
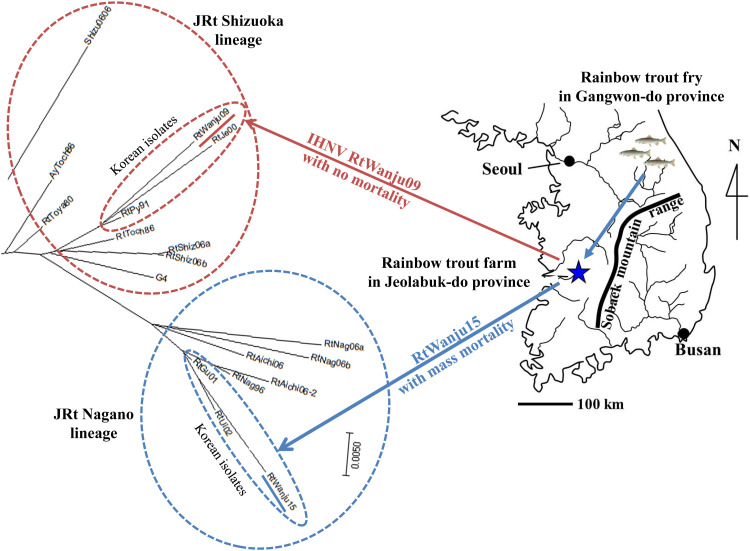
Specific primer sets for viral hemorrhagic septicemia virus (VHSV), infectious salmon anemia virus (ISAV), and IHNV were used to detect the viral genes in total RNA extracted from supernatant of the cells with CPE. Among them, only IHNV was detected (data not shown). The IHNV G gene was detected using the IHNV G ORF primer set and subsequently sequenced (GenBank accession number: OM986071). Sequencing data were analyzed using a phylogenetic tree for IHNV genotyping and the identified sequence was found to belong to the JRt Nagano lineage. The phylogenetic distance of the virus was close to that of the RtGu01 and RtU02 isolates from Gangwon-do province (Fig. 4). This isolate was designated as RtWanju15.
Fig. 4.
Unrooted phylogenetic tree of infectious hematopoietic necrosis virus (IHNV) RtWanju09 and RtWanju15 isolates using full length IHNV G gene sequences.
3.5. Histopathology and immunohistochemistry
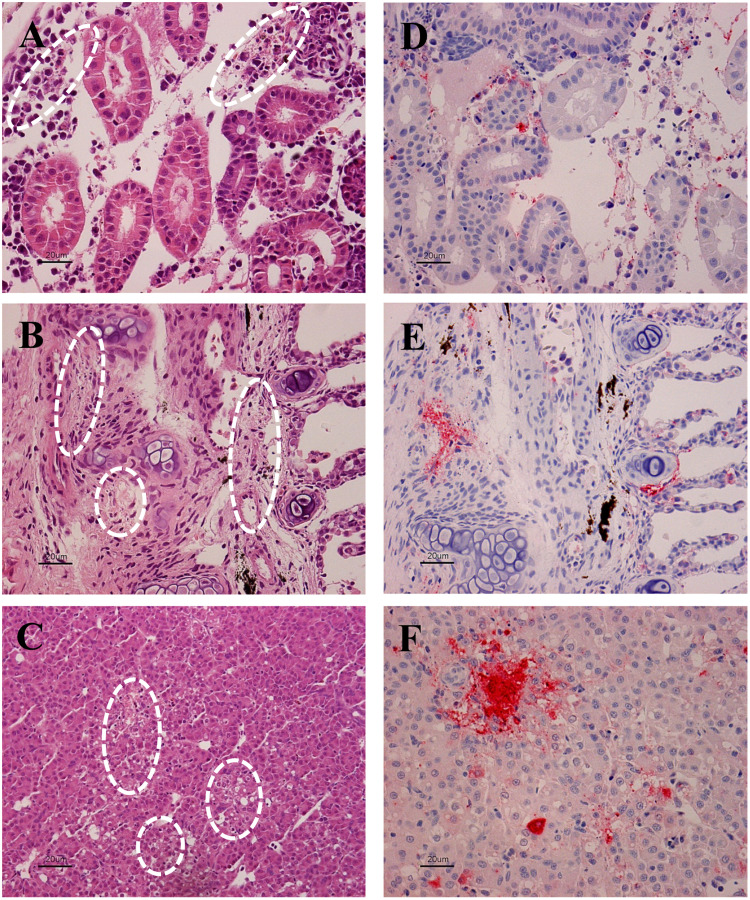
Histopathological examination revealed signs of necrosis of the interstitial hematopoietic tissue in the kidney (arrows in Fig. 5A), branchial arch and gills (Fig. 5B), and liver tissues (arrows in Fig. 5C). Immunohistochemistry using an IHNV-specific antibody revealed viral particles in necrotic lesions of the kidney, liver, branchial arch, and gills of moribund fish (Fig. 5D–F).
Fig. 5.
Histopathological observation of the kidney (A, D), branchial arch and gill (B, E), and liver (C, F) by hematoxylin and eosin staining (left) and immunohistochemistry (right). Necrotic region is indicated by circles. The virus was detected by alkaline phosphatase staining (red color) using an IHNV-specific monoclonal antibody.
3.6. Infection trials
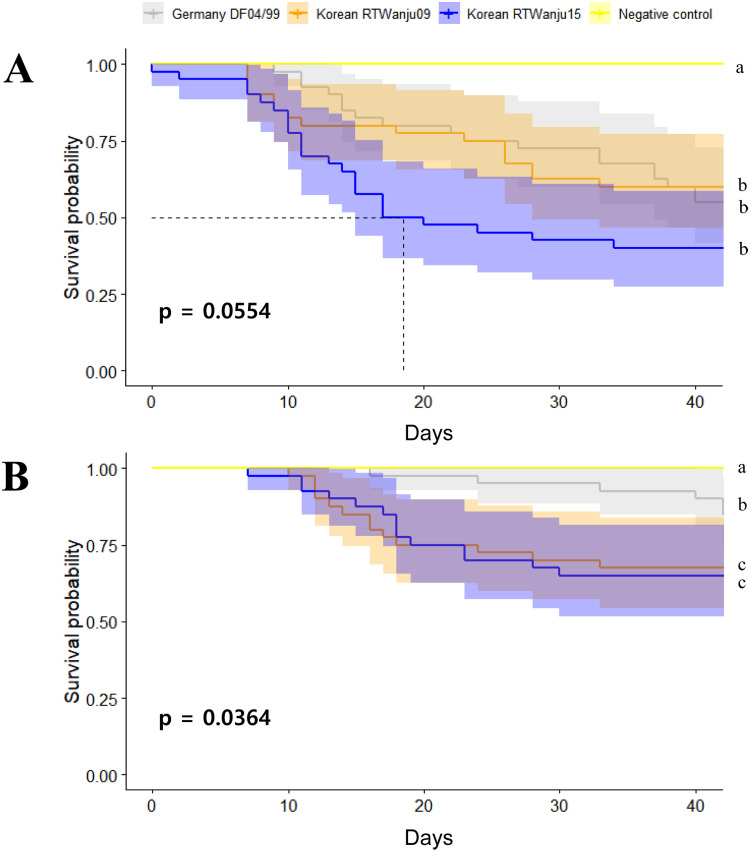
Mortality rates in the duplicate tanks with fish challenged with IHNV RtWanju09, RtWanju15, and DF04/99, respectively were monitored until 43 days post-exposure (Fig. 6). No reduction of survival was observed in the two negative control groups (0 of 20 fish). For the groups challenged with 105 TCID50/mL of IHNV DF04/99, the cumulative survival rates were 45% and 60% (average 52.5%). The cumulative survival in the tanks infected with IHNV RtWanju09 was 65% and 55% (average 60%). The survival in the tanks infected with IHNV RtWanju15 was 35% and 40% (average 37.5%). In high dose groups, there was no significant difference in survival rates between experimental groups (Fig. 6A).
Fig. 6.
Survival rate of rainbow trout infected by immersion with the IHNV isolates RtWanju09, RtWanju15 and DF04/99. Fish were immersed in 105 TCID50/mL (A) or 103 TCID50/mL (B). Mortality was recorded daily until 43 days post-infection and is presented as the survival rate. Different letters indicate statistically significant differences in average of survival rate over the experimental period (p<0.05). Survival comparisons were performed using Cox proportional hazards model.
In the tanks given the low dose of 103 TCID50/mL of virus, the survival rate in the IHNV DF04/99 group was 80% and 90% (average 85%). The survival in the RtWanju09 group was 80% and 55% (average 67.5%). The survival in the RtWanju15 group was 70% and 60% (average 65%). In low dose groups, the survival rates of the RtWanju09 and RtWanju15 showed statistically significant differences from the DF04/99 group, while no significant difference was observed between the RtWanju09 and RtWanju15 groups (Fig. 6B).
The Ct value of IHNV gene detection from dead fish was calculated by real-time RT-PCR. The challenge tests of dead fishes for high- and low-dose IHNV Germany DF04/99 showed Ct values of 26.15 and 26.56, respectively. The Ct values of two fish exposed to high doses of IHNV RTWanju15 were 27.34 and 26.86, respectively. The Ct value from dead fish treated with a low dose of IHNV RtWanju15 was 26.49, and that from a dead fish treated with a low dose of IHNV RtWanju09 was 27.16.
3.7. Replication of IHNV in fish cell lines
No statistically significant differences in viral titers were observed between the two Korean IHNV isolates, RtWanju09 and RtWanju15 (Table 3). The onset of viral replication was slower in RTG-2 cells than in EPC and FHM cells, but the final titers observed 14 days after inoculation only varied by 1 log. Six days post-inoculation, viral titers stabilized and did not increase significantly in the following 8 days.
Table 3.
The development of viral titers of RtWanju09 and RtWanju15 in three cell lines. Titers given as log10 TCID50/mL.
| Observation Days | IHNV RtWanju09 | IHNV RtWanju15 | ||||||
|---|---|---|---|---|---|---|---|---|
| EPC | FHM | RTG-2 | EPC | FHM | RTG-2 | |||
| 3 6 10 14 |
6.05 6.55 6.8 7.05 |
5.3 6.05 6.05 6.05 |
3.8 5.55 6.05 6.05 |
5.8 6.8 6.8 7.05 |
5.8 6.3 6.8 6.8 |
3.55 5.05 5.3 6.05 |
||
4. Discussion
IHNV RtWanju09 belonging to JRt Shizuoka lineage was isolated in 2009 from rainbow trout eggs in a farm where fish did not show any clinical signs of IHN (Kim, 2010). No fish had been introduced to the farm from other areas until 2014 and no IHN outbreaks were observed during the intervening period. However, in 2015 the farm chose to import rainbow trout fry from Gangwon-do province, which had no records of any specific diseases. In the following two months (March to May), all 20,000 imported rainbow trout fry died at the farm. In this study, we detected IHNV from moribund fish by cell culture, observation by TEM and RT-PCR, and we observed that the virus was pathogenic by immunohistochemistry, which showed virus-specific destruction of hematopoietic tissues.
Phylogenetic analysis of the IHNV G gene revealed that the IHNV isolated in the present study, IHNV RtWanju15, belonged to the JRt Nagano lineage, thus the fish were not infected by the IHNV RtWanju09 of the JRt Shizuoka lineage. Our study is the first to detect and isolate the IHNV JRt Nagano lineage from rainbow trout in the Jeollabuk-do province in Korea. Furthermore, phylogenetic analysis and tracing of fish transport demonstrated that the virus was introduced to the farm by purchase of fish from the Gangwon-do province. Therefore, the pathogenicity of two isolates, RtWanju09 and RtWanju15, which were isolated in the same region but belong to different lineages, was examined.
There was no case of IHN in Korea before 1990, but the first outbreak (RtPy91) was reported in a hatchery in Gangwon-do in 1991 (Park et al., 1993). It was found that RtJe00 isolated from Jecheon, Gangwon-do in 2000, including RtPy91 isolate, belonged to the JRt Shizuoka lineage (Kim et al., 2007). RtGu01 and RtUi02 isolated from the Gyeongsangbuk-do in 2001 and 2002, respectively revealed that they belong to the JRt Nagano lineage supporting that two JRt lineages have regional distribution (Kim et al., 2007). Kim (2010) determined that IHNV first reported in Jeollabuk-do belonged to JRt Shizuoka lineage and originated from the Gangwon-do. Phylogenetic studies and pathogenicity comparison on IHNV isolates from Gangwon-do, it was found that not only JRt Shizuoka lineage but also JRt Nagano lineage existed in Gangwon-do (Cha et al., 2012; Kim et al., 2021; Lee et al., 2021). The IHNV JRt Shizuoka lineage was reported in Gangwon-do in 1991, it has spread to Jeollabuk-do. Similarly, IHNV JRt Nagano lineage, which was detected in the Gyeongsangbuk-do, has moved to the Gangwon-do region.
The virulence of some IHNV isolates belonging to JRt Shizuoka lineage in Asia was reported to be higher than that of the JRt Nagano lineage in rainbow trout (Mochizuki et al., 2009). Mochizuki et al. (2009) also suggested that the IHNV isolate ChAb76 caused no mortality in rainbow trout. However, in the 1980s, ChAb76 and RtNag76 isolates belonging to genogroup U showed 70% mortality in Oncorhynchus masou and rainbow trout. It was speculated that the significant difference in virulence to rainbow trout of the IHNV ChAb76 isolate was due to natural selection of survivors under aquaculture conditions (Mochizuki et al., 2009). In this study, IHNV RTWanju09 was isolated from eggs of asymptomatic fish in Jeollabuk-do province in 2009, and until 2014, no IHN outbreaks occurred in Jeollabuk-do province. However, this study revealed its virulence in SPF fish from Denmark. Similarly, IHN (RtWanju15) only occurred in the rainbow trout farm in Jeollabuk-do province that purchased 20,000 fish fry from a hatchery in the Gangwon-do province. IHN did not occur in any of the rainbow trout farms in the Gangwon-do province, although the same batch of fish was distributed to other rainbow trout farms in the same province. Therefore, it is thought that IHN adapted to the host and environment, which is why RTWanju09 and RTWanju15 were pathogenic but did not cause disease in Jeollabuk-do and Ganwon-do province where they originated from. Mortality caused by IHNV RTWanju15 depends on environmental stress by physical handling, acute environmental changes, which could decrease host immunity (LaPatra, 1998). IHNV targets not only the hematopoietic, but also the immunological tissues of the kidney and spleen, thereby weakening host immunity.
In the present study, the virulence of IHNV RtWanju09 (Kim, 2010) and RtWanju15 was compared in a completely different batch of rainbow trout (SPF fish in Denmark) to confirm or rule out the possibility of natural adaption of fish to IHNV. Survival rate of rainbow trout was lower at 105 TCID50/mL than when challenged with 103 TCID50/mL, regardless of the virus genotype, showing a dose dependent manner. DF04/99 isolate was used as a positive control group because it had high pathogenicity in rainbow trout (Fichtner et al., 2000). However, the results of in vivo challenge showed that DF04/99 was similar to RtWanju09 in high dose group, and induced the lowest mortality among the three isolates in low dose group. LaPatra et al. (1993) reported that the mortality caused by IHNV is different according to virus concentration and a decreasing susceptibility of salmonids with increasing size and age. It is speculated that the reason DF04/99 was less virulent in this study was because they used 10 g fish for infection trial in Fichtner et al. (2000) and we used bigger fish (13 g) for infection trial in this study.
In high dose groups, the newly isolated IHNV RtWanju15 showed 50% of survival rate on the 19 days post exposure, showing the highest virulence among the experimental groups. Even after 6 weeks of exposure, RtWanju15 still caused a low survival rate of 37.5%, compared to RtWanju09 and DF04/99. However, the p-value between the three groups was 0.0554, indicating no statistical significance in survival rate. In low dose groups, RtWanju15 and RtWanju09 showed similar survival rates (65% and 67.5%, respectively), and showed significant results compared to the 85% survival rate of DF04/99 (p = 0.0364). These results indicate that there is no significant difference in pathogenicity between RtWanju15 (JRt Nagano lineage) and RtWanju09 (JRt Shizuoka lineage) on rainbow trout. We recommend that parental fish that have never encountered IHNV should be used when testing for virulence-naïve SPF fish.
The Ct values yielded by samples collected from moribund fish exposed to different IHNV isolates and at different doses were very similar, supporting the detection of a similar copy number of the IHNV RNA target. There was no difference in viral replication in isolates of fish exposed to various IHNV infections or cultured cells after challenge with 105 TCID50/mL or 103 TCID50/mL of IHNV. In addition, there was no difference in the ability of the IHNV RtWanju15 and RtWanju09 to induce CPE. Thus, it appears that the viral titers in cell cultures were almost equal to the titers in dead fish, regardless of genogroups as European and Asian isolates.
At present, IHN is one of the most serious viral diseases in rainbow trout and Atlantic salmon farms (Kurath et al., 2016), and is listed as a notifiable viral disease by the World organisation for Animal Health (WOAH, 2021). The virulence changes of IHNV have been demonstrated by several researchers. Mochizuki et al. (2009) demonstrated that IHNV in Japan has changed pathogenicity over 30 years and is still changing virulence in rainbow trout farms environment by G gene diversity. Troyer et al. (2000) also observed high diversity in rainbow trout aquaculture in North America. Recently, it has been reported that the pathogenicity of Italian isolates of IHNV has increased rapidly (Abbadi et al., 2021). Although IHN causes high mortality in rainbow trout farms (Saksida, 2006), it has not been listed in the Aquatic Animal Disease Control Act in Korea. High vigilance for IHN is of utmost importance regardless of genotype. Therefore, the inclusion of IHN in disease control act is a fundamental requirement to gain control over this disease for the benefit of fish health, along with the prosperity of fish farmers in Korea as well as all other salmonid fish farming countries.
CRediT authorship contribution statement
Hyoung Jun Kim: Conceptualization, Methodology, Software, Data curation, Visualization, Writing – original draft. Niels Jørgen Olesen: Writing – review & editing. Ole Bendik Dale: Resources, Validation. Young Chul Kim: Validation, Formal analysis. Tae Sung Jung: Writing – review & editing. Niccolò Vendramin: Resources, Methodology. Se Ryun Kwon: Validation, Formal analysis, Project administration, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
We would like to acknowledge Torunn Taksdal, Argelia Cuenca, Christina Flink Desler, and Tine Moesgaard Iburg for their excellent technical and scientific support.
Funding
This work was supported by Korea Institute of Marine Science & Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries, Korea [grant number 201903922] and the National Institute of Fisheries Science [grant number R2023059].
Data availability
The data that has been used is confidential.
References
- Abbadi M., Gastaldelli M., Pascoli F., Zamperin G., Buratin A., Bedendo G., Toffan A., Panzarin V. Increased virulence of Italian infectious hematopoietic necrosis virus (IHNV) associated with the emergence of new strains. Virus Evol. 2021;7:veab056. doi: 10.1093/ve/veab056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha S.J., Jung Y.H., Lee H.Y., Jung J.Y., Cho H.J., Park M.S. Genotype distribution of infectious haematopoietic necrosis virus (IHNV) in Korea. J. Fish Pathol. 2012;25:143–150. doi: 10.7847/jfp.2012.25.3.143. [DOI] [Google Scholar]
- Emmenegger E.J., Meyers T.R., Burton T.O., Kurath G. Genetic diversity and epidemiology of infectious hematopoietic necrosis virus in Alaska. Dis. Aquat. Org. 2000;40:163–176. doi: 10.3354/dao040163. [DOI] [PubMed] [Google Scholar]
- Fichtner D., Bergmann S., Enzmann P.J., Granzow H., Schütze H., Mock D., Schäfer J.W. Isolation and characterisation of a variant strain of infectious haematopoietic necrosis (IHN) virus. Bull. Eur. Assoc. Fish Pathol. 2000;20:135–142. [Google Scholar]
- Fregeneda-Grandes J.M., Skall H.F., Olesen N.J. Antibody response of rainbow trout with single or double infections involving viral haemorrhagic septicaemia virus and infectious haematopoietic necrosis virus. Dis. Aquat. Organ. 2009;83:23–29. doi: 10.3354/dao01993. [DOI] [PubMed] [Google Scholar]
- Kim H.J. Phylogenetic analysis of infectious hematopoietic necrosis virus (IHNV) isolated from cultured rainbow trout Oncorhynchus mykiss in Korea. Fish Pathol. 2010;23:1–8. [Google Scholar]
- Kim H.J. Validation of the sensitivities of one-step and two-step reverse-transcription PCR methods for detection of viral hemorrhagic septicemia virus (VHSV) IVa isolates from cultured olive flounder in Korea. Aquaculture. 2015;448:359–364. doi: 10.1016/j.aquaculture.2015.06.034. [DOI] [Google Scholar]
- Kim S.S., Kim K.I., Yoo H.K., Han Y.S., Jegal M.E., Byun S.G., Lim H.J., Park J.S., Kim Y.J. Differential virulence of infectious hematopoietic necrosis virus (IHNV) isolated from salmonid fish in Gangwon Province, Korea. Fish Shellfish Immunol. 2021;119:490–498. doi: 10.1016/j.fsi.2021.10.038. [DOI] [PubMed] [Google Scholar]
- Kim W.S., Oh M.J., Nishizawa T., Park J.W., Kurath G., Yoshimizu M. Genotyping of Korean isolates of infectious hematopoietic necrosis virus (IHNV) based on the glycoprotein gene. Arch. Virol. 2007;152:2119–2124. doi: 10.1007/s00705-007-1027-9. [DOI] [PubMed] [Google Scholar]
- Kurath G., Garver K.A., Troyer R.M., Emmenegger E.J., Einer-Jensen K., Anderson E.D. Phylogeography of infectious haematopoietic necrosis virus in North America. J. Gen. Virol. 2003;84:803–814. doi: 10.1099/vir.0.18771-0. [DOI] [PubMed] [Google Scholar]
- Kurath G., Winton J.R., Dale O.B., Purcell M.K., Falk K., Busch R.A. Atlantic salmon, Salmo salar L. are broadly susceptible to isolates representing the North American genogroups of infectious hematopoietic necrosis virus. J. Fish Dis. 2016;39:55–67. doi: 10.1111/jfd.12323. [DOI] [PubMed] [Google Scholar]
- LaPatra S.E., Fryer J.L., Rohovec J.S. Virulence comparison of different electropherotypes of infectious hematopoietic necrosis virus. Dis. Aquat. Org. 1993;16:115–120. doi: 10.3354/dao016115. [DOI] [Google Scholar]
- Lee C.J., Kim K.I., Han Y.S., Jegal M.E., Kim Y.J. Comparison of the pathogenicity of infectious hematopoietic necrosis virus genotypes isolated from rainbow trout in Gangwon Province. J. Life Sci. 2021;31:574–580. doi: 10.5352/JLS.2021.31.6.574. [DOI] [Google Scholar]
- Mochizuki M., Kim H.J., Kasai H., Nishizawa T., Yoshimizu M. Virulence change of infectious hematopoietic necrosis virus against rainbow trout Oncorhynchus mykiss with viral molecular evolution. Fish Pathol. 2009;44:159–165. doi: 10.3147/JSFP.44.159. [DOI] [Google Scholar]
- Nishizawa T., Kinoshita S., Kim W.S., Higashi S., Yoshimizu M. Nucleotide diversity of Japanese isolates of infectious hematopoietic necrosis virus (IHNV) based on the glycoprotein gene. Dis. Aquat. Organ. 2006;71:267–272. doi: 10.3354/dao071267. [DOI] [PubMed] [Google Scholar]
- Park M.A., Sohn S.G., Lee S.D., Chun S.K., Park J.W., Fryer J.L., Hah Y.C. Infectious haematopoietic necrosis virus from salmonids cultured in Korea. J. Fish Dis. 1993;16:471–478. doi: 10.1111/j.1365-2761.1993.tb00880.x. [DOI] [Google Scholar]
- Polack J.M., Van Noorden S. BIOS Scientific Publishers Limited; Oxford, UK: 2003. Introduction to Immunocytochemistry. [Google Scholar]
- Purcell M.K., Thompson R.L., Garver K.A., Hawley L.M., Batts W.N., Sprague L., Sampson C., Winton J.R. Universal reverse-transcriptase real-time PCR for infectious hematopoietic necrosis virus (IHNV) Dis. Aquat. Organ. 2013;106:103–115. doi: 10.3354/dao02644. [DOI] [PubMed] [Google Scholar]
- Saksida S.M. Infectious haematopoietic necrosis epidemic (2001 to 2003) in farmed Atlantic Salmon Salmo salar in British Columbia. Dis. Aquat. Organ. 2006;72:213–223. doi: 10.3354/dao072213. [DOI] [PubMed] [Google Scholar]
- Snow M., Bain N., Black J., Taupin V., Cunningham C.O., King J.A., Skall H.F., Raynard R.S. Genetic population structure of marine viral haemorrhagic septicaemia virus (VHSV) Dis. Aquat. Organ. 2004;61:11–21. doi: 10.3354/dao061011. [DOI] [PubMed] [Google Scholar]
- Troyer R.M., LaPatra S.E., Kurath G. Genetic analyses reveal unusually high diversity of infectious haematopoietic necrosis virus in rainbow trout aquaculture. J. Gen. Virol. 2000;81:2823–2832. doi: 10.1099/0022-1317-81-12-2823. [DOI] [PubMed] [Google Scholar]
- WOAH (World Organisation for Animal Health) OIE; Paris: 2021. Manual of Diagnostic Tests For Aquatic Animal Diseases. [Google Scholar]
- Wolf K. In: Fish Viruses and Fish Viral Diseases. Wolf K., editor. Cornell University Press; Ithaca: 1988. Infectious hematopoietic necrosis; pp. 83–114. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that has been used is confidential.