Key Points
-
•
Physioxia improves the selectivity of PVA-based mouse HSC cultures.
-
•
Selective HSC cultures deplete GvHD-causing T cells.
Visual Abstract
Abstract
Hematopoietic stem cells (HSCs) are a rare type of hematopoietic cell that can entirely reconstitute the blood and immune system after transplantation. Allogeneic HSC transplantation (HSCT) is used clinically as a curative therapy for a range of hematolymphoid diseases; however, it remains a high-risk therapy because of its potential side effects, including poor graft function and graft-versus-host disease (GVHD). Ex vivo HSC expansion has been suggested as an approach to improve hematopoietic reconstitution in low-cell dose grafts. Here, we demonstrate that the selectivity of polyvinyl alcohol (PVA)-based mouse HSC cultures can be improved using physioxic culture conditions. Single-cell transcriptomic analysis helped confirm the inhibition of lineage-committed progenitor cells in physioxic cultures. Long-term physioxic expansion also afforded culture-based ex vivo HSC selection from whole bone marrow, spleen, and embryonic tissues. Furthermore, we provide evidence that HSC-selective ex vivo cultures deplete GVHD-causing T cells and that this approach can be combined with genotoxic-free antibody-based conditioning HSCT approaches. Our results offer a simple approach to improve PVA-based HSC cultures and the underlying molecular phenotype, and highlight the potential translational implications of selective HSC expansion systems for allogeneic HSCT.
Introduction
Self-renewing multipotent hematopoietic stem cells (HSCs) support the blood and immune systems throughout life.1, 2, 3 HSCs first arise in the mouse embryo at around embryonic day 10.5 (E10.5) and initially localize in the fetal liver before moving to the bone marrow around the time of birth.4 Although the bone marrow microenvironment is the most studied HSC niche, HSCs have also been described in other organs such as the spleen.5,6 HSCs are usually very rare, at an estimated frequency of ∼1:30 000 cells within the adult bone marrow, and only ∼1:300 000 in the adult spleen.7 This has hindered efforts to characterize this biologically interesting stem cell population.
The transplantation of healthy HSCs into a patient (termed HSCT) is a potentially curative therapy for a wide range of blood diseases.8 However, although HSCT has been used for over 60 years, this therapy often remains a last resort treatment because of several safety concerns.9,10 First, allogeneic HSCT currently requires patients to undergo genotoxic preconditioning via radiation therapy and/or chemotherapy to enhance donor engraftment and prevent rejection. Second, the donor and recipient must be sufficiently immune-matched to avoid graft-versus-host disease (GVHD), which is caused by alloreactive T cells that contaminate the donor HSC graft. GVHD represents a serious potential side effect of HSCT, and the donor-recipient immune-matching requirement to avoid it limits the number of potential donors for a transplantation. An ideal HSCT paradigm would involve the availability of large numbers of HSCs so that toxic chemo-irradiative preconditioning would not be required, and cell products lacking GVHD-inducing T cells that would improve donor-recipient immune compatibility.
The biological and clinical importance of HSCs has driven extensive efforts to sustain HSCs ex vivo.11,12 However, for a long time, the stable expansion of HSCs has remained a major challenge in the field. We recently discovered that polymer-based media could support long-term ex vivo expansion of functional mouse HSCs.13,14 Although these cultures were initiated from fluorescence-activated cell sorter–purified CD150+CD34–/loc-Kit+Sca1+Lineage– (CD150+CD34–/loKSL) HSCs, a range of c-Kit+Sca1–Lineage– progenitor cells built up in these cultures over time, reducing the purity of HSCs. Here, we show that by optimizing O2 concentrations for ex vivo HSC cultures, we could limit the buildup of progenitors and mature hematopoietic cells within long-term polyvinyl alcohol (PVA)-based cultures, thereby improving HSC culture selectivity. These optimized culture conditions selectively expanded HSCs from whole bone marrow cells (WBMCs) and whole spleen cells while depleting mature hematopoietic cells, including T cells. Depletion of alloreactive T cells was demonstrated via the transplantation of allogeneic cells, which confirmed that HSC-selective cultures prevented acute GVHD. Finally, we present evidence that these methods can be combined with antibody-mediated inhibition of graft rejection to achieve HSCT, without genotoxic conditioning.
Methods
Mice
All animal experiments were approved by the administrative panel on laboratory animal care at Stanford University or were performed in accordance with UK Home Office regulations. C57BL/6-CD45.2 mice (000664), C57BL/6-CD45.1 mice (PepboyJ; 002014), and Balb/c mice (000651) were purchased from the Jackson Laboratory or bred at the University of Oxford. C57BL/6-CD45.1/CD45.2 mice were bred using C57BL/6-CD45.1 and C57BL/6-CD45.2 mice at Stanford University. Fancd2–/– mice15 were generously provided by Ken Weinberg and bred at Stanford University. All the mice were 8 to 12 weeks old at the starting point of the experiment.
Purified HSC cultures
Immunophenotypic CD150+CD34–/loKSL HSCs were isolated from the pelvic, femur, tibia, and vertebrae of C57BL/6-CD45.1 or C57BL/6-CD45.2 mice using an AriaII cell sorter (BD) and were cultured as described previously.14 Cells were incubated at 5% CO2 and indicated O2 levels using a multigas incubator (Thermo Fisher HERACell 150i or IncuSafe PHC MCO-170M-PE), with complete media changes (performed in regular tissue culture hoods) every 2 to 3 days after the initial 5 days. See supplemental Information for further details.
Unfractionated bone marrow and spleen cultures
Unfractionated WBMCs or whole spleen cells were strained with a 100 μm filter and then plated on CellBIND (Corning) plates in HSC media (see composition in the supplemental Information) and incubated at 5% CO2 and the indicated O2 levels, with complete media changes every 2 or 3 days.
Cytometric analyses
Flow cytometry and cytometry by time of flight (CyTOF) were performed at the indicated time points. Cells were antibody stained (CD201-APC, c-Kit–BV421, Sca1-PE, Gr1-APC/eFluor780, Ter119-APC/eFluor780, CD4-APC/eFluor780, CD8-APC/eFluor780, CD45R-APC/eFluor780, and CD127-APC/eFluor780) for 30 minutes at 4 °C, washed, and analyzed using LSRFortessa (BD) using propidium iodide as a live/dead cell stain. CyTOF was performed as detailed in the supplemental Information.
Transplantation assays
For competitive transplantation assays, 5000 day-28 culture cells derived from C57BL/6-CD45.1 mice were transplanted alongside 1 × 106 C57BL/6-CD45.1/CD45.2 competitor WBMCs into 10 Gy lethally irradiated C57BL/6-CD45.2 recipient mice. Secondary transplantation assays were performed as described earlier, using 2 × 106 WBMCs isolated from primary recipients. For allogeneic transplantation assays into Balb/c mice, fresh or day-28 cultured cells derived from WBMCs and/or whole spleen cells isolated from C57BL/6-CD45.1 mice were transplanted into 7 Gy irradiated Balb/c mice. Hematoxylin and eosin staining was performed on paraformaldehyde-fixed livers by the Stanford Animal Histology Service. For minor allele mismatch transplantation assays in Fancd2–/– mice, day-28 cultured cells derived from WBMCs, CD150+CD48–KSL HSCs, or fresh CD150+CD48-KSL HSCs isolated from C57BL/6-CD45.1 mice were transplanted into Fancd2–/– mice. When indicated, mice were pretreated with 500μg of anti-CD4 (GK1.5; BioXCell) 7 days before transplantation.
RNA-seq analyses
For bulk RNA sequencing (RNA-seq), day-28 cultured cells were stained as indicated in “Cytometric analyses,” CD201+CD150+KSL and Kit+Sca1–Lineage– cells were fluorescence-activated cell sorter–isolated, and RNA was extracted using a Qiagen RNeasy Micro Kit. RNA-seq was performed using Novogene. Single-cell RNA-seq (scRNA-seq) was performed using 10X Genomics v3 reagents, as detailed in the supplemental Information.
Results
Physiological O2 concentrations reduce differentiation in long-term PVA HSC cultures
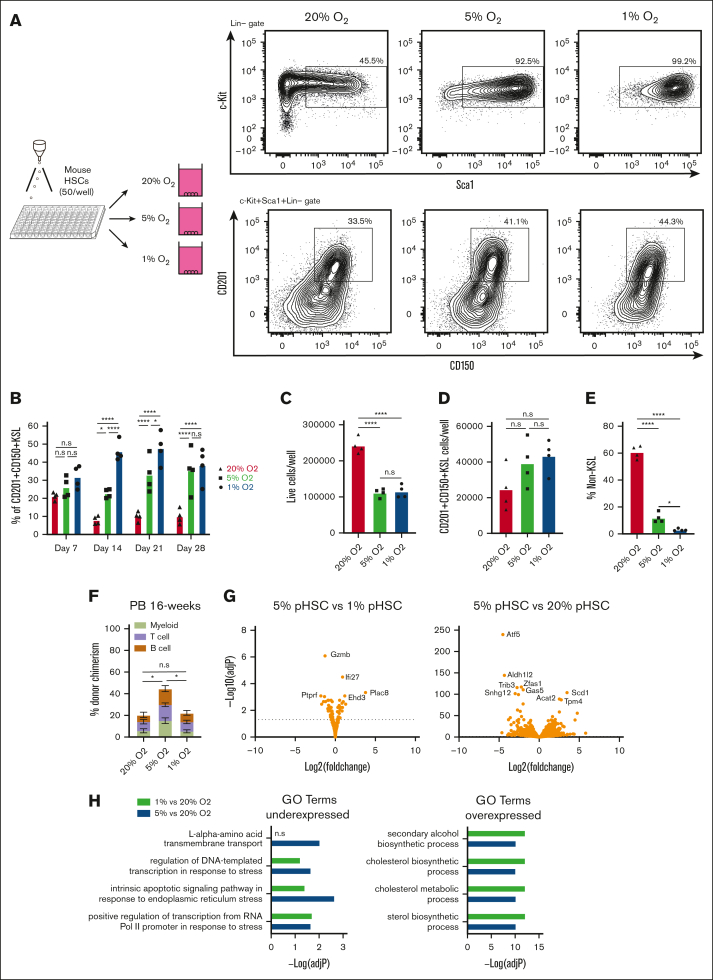
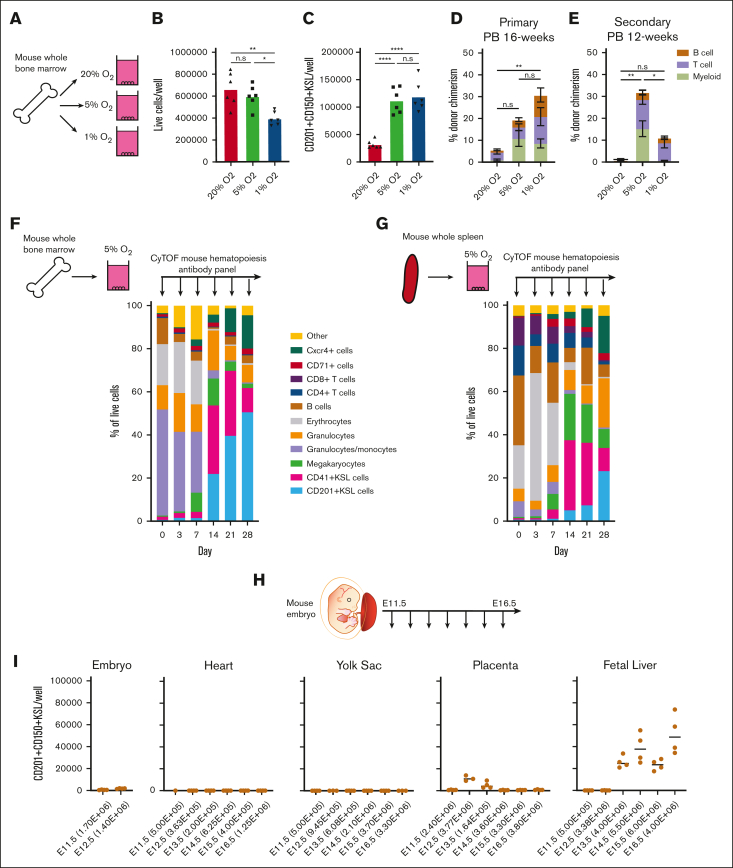
The importance of O2 concentration in cell culture conditions has come to the forefront in discussions about improving the physiological relevance and translational capacity of basic biological findings from ex vivo studies.16, 17, 18 Recapitulating the pericellular O2 found in specific tissue microenvironments has demonstrated a significance in producing data that are both more representative of in vivo physiology and more experimentally robust.19 Direct measurement of pericellular O2 in the bone marrow of mice has helped identify a range from 9.9 to 32 mmHg O2 (1.3%-4.2%);20 long-term HSCs (LT-HSCs) are not found in bone marrow niches with the deepest hypoxia but rather in PO2 conditions from 18 to 19 mmHg (2%-3%).21 We hypothesized that the calibration of O2 concentration to levels that are more representative of those in the bone marrow might further improve our HSC cultures. In our previous studies, HSCs were grown in standard 20% O2 tissue culture incubators, which yield a pericellular O2 concentration of ∼18.4%.21, 22, 23 We compared these HSC culture conditions with cultures grown at 5% O2 and 1% O2 (pericellular O2 concentrations of ∼3.5% and ∼0.7%, respectively; Figure 1A). Flow cytometry of a 4-week culture revealed striking differences in culture composition, with significant increases in the frequency of phenotypic CD201+CD150+KSL populations (Figure 1A-B), which has recently been proposed as the ex vivo HSC fraction.24, 25, 26 This high-frequency of CD201+CD150+KSL cells was quite stable at 30-40% of live cells at low O2 throughout the 4-week culture (Figure 1B).
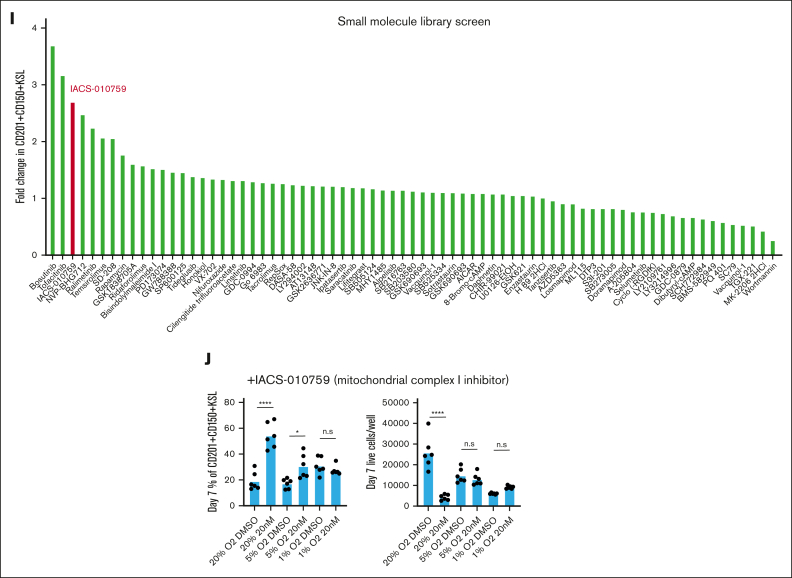
Figure 1.
Improved stability of PVA-based HSC cultures at low O2 concentrations. (A) Schematic diagram of low O2 HSC cultures (left). Mouse CD150+CD34-KSL HSCs were sorted into PVA-based media in 96-well plates (50 cells per well) in 200 μL of media and were cultured for 4 weeks at 20%, 5%, or 1% O2. Representative flow cytometry plots of 4-week HSC-derived cultures (right). c-Kit and Sca1 expression within the lineage– cell fraction (top), and CD201 and CD150 expression within the KSL cell fraction (bottom). (B) Mean frequency of CD201+CD150+KSL cells within the HSC-derived cultures described in panel A (n = 4). (C) Mean number of live cells per well within HSC-derived cultures described in panel A (n = 4). (D) Mean number of CD201+CD150+KSL cells per well within the HSC-derived cultures as described in panel A (n = 4). (E) Mean number of non-KSL cells per well within the HSC-derived cultures described in panel A (n = 4). (F) 16-week donor peripheral blood (PB) chimerism from 4-week-old HSC-derived cultures incubated at 20%, 5%, or 1% O2. Five thousand cells from each culture were transplanted alongside 1 × 106 WBMCs into lethally irradiated recipients (mean ± standard deviation [SD]; n = 8-9). (G) Differential gene expression analysis between 5% O2 pHSC and 1% O2 pHSC samples (left) and between 5% O2 pHSC and 20% O2 pHSC samples (right). Results are displayed as Log2 (fold change) vs –Log (adjusted P value). (H) Gene Ontology (GO) term enrichment analysis for underexpressed and overexpressed genes in the 5% O2 and 20% O2 pHSC samples. (I) Fold change in CD201+CD150+KSL cells relative to control well after a 7-day culture with the indicated compounds at 20% O2. Of the 116 compounds tested, only 74 that supported cell survival/growth are displayed (see supplemental Table 1 for a full list of the compounds). Cell cultures were initiated with 50 CD201+CD150+KSL cells from 3-week HSC cultures. The mean of the 4 wells (from 2 biological replicates) is displayed. (J) Mean frequency of CD201+CD150+KSL cells (left) and mean number of live cells per well (left) within 7-day HSC-derived cultures at 20% O2, 5% O2, or 1% O2, cultured with either dimethyl sulfoxide or the mitochondrial complex I inhibitor IACS-01-759 (20 nM; n = 6). Statistical analysis was performed using analysis of variance (ANOVA). ∗P < .05; ∗∗∗∗P < .0001. n.s., not significant; RNA Pol II, RNA polymerase II.
In terms of absolute cell numbers per well, 1% and 5% O2 reduced the number of live cells by ∼50% (Figure 1C), indicating that similar numbers of CD201+CD150+KSL cells were generated in all cultures (Figure 1D). The increased frequency of CD201+CD150+KSL cells was driven by a decrease in non-KSL cells at low O2 concentrations (Figure 1E). Competitive transplantation assays confirmed the functional capacity of the cells cultured at all O2 concentrations. Compared with 20% O2 cultured cells, 5% O2 cultures displayed an approximately twofold higher donor chimerism at the 16-week end point (Figure 1F; supplemental Figure 1A). In contrast, 1% O2 cultured cells initially displayed chimerism similar to that of 5% O2 cultures at 4-weeks but dropped to levels similar to those of 20% O2 cultures by 16 weeks (Figure 1F; supplemental Figure 1A). The reason for the lower engraftment rate at 1% O2 is unclear.
To characterize the molecular consequences of low O2 concentration culture conditions on HSC expansion, we performed RNA-seq analysis of CD201+CD150+KSL cells from 4-week cultures. The c-Kit+Sca1–Lineage– cells were also analyzed in 20% O2 cultures. Principal component analysis separated CD201+CD150+KSL samples from c-Kit+Sca1–Lineage– samples via principal component 1, while 20% O2 CD201+CD150+KSL samples were separated from 5% and 1% CD201+CD150+KSL samples via principal component 2 (supplemental Figure 1B). When compared with the c-Kit+Sca1–Lineage– samples, gene set enrichment analysis identified enrichment for LT-HSC gene sets in the CD201+CD150+KSL cell samples for all O2 concentrations, whereas the c-Kit+Sca1–Lineage– samples were enriched for the progenitor gene sets (supplemental Figure 1C). These results confirmed that CD201+CD150+KSL cells as phenotypic HSCs (pHSCs) and c-Kit+Sca1–Lineage– cells as phenotypic hematopoietic progenitor cells.
To investigate the transcriptional differences between these samples, we performed a differential gene expression analysis. Between the 5% and 1% O2 pHSC samples, few differentially expressed genes were observed (Figure 1G). By contrast, large differences in gene expression were observed between 20% and 5% O2 pHSCs (Figure 1G). Gene Ontology (GO) term analysis helped identify the upregulation of transcriptional and endoplasmic reticulum stress pathways in 20% O2 pHSCs (Figure 1H), which corresponded to the observed differential expression of the stress response factor Atf5 (Figure 1G). Stress response pathway activity is known to induce HSC differentiation,27,28 which may explain why 20% O2 HSC cultures contained more differentiated cell types than the other cultures. Amino acid transporters were also enriched in 20% pHSCs (relative to 5% O2 pHSCs), whereas sterol and cholesterol metabolism pathways were dominant among the upregulated genes at 5% and 1% O2 (relative to 20% O2 pHSCs; Figure 1H).
To search for pathways that could play a role in the selectivity for CD201+CD150+KSL in low O2 cultures, we screened 116 small molecule inhibitors targeting various intracellular pathways (see supplemental Table 1 for details) using 21-day expanded cells at 20% O2. One of the top hits was IACS-010759, an inhibitor of mitochondrial complex I that plays an important role in aerobic respiration (Figure 1I). Next, we tested this compound in fresh HSCs. At 20% O2, we observed significant increases in CD201+CD150+KSL frequencies and similar reductions in total cell numbers and non-KSL cells as those in our low O2 cultures at 20 nM (Figure 1J). By contrast, little difference was observed when IACS-010759 was added to 5% or 1% O2 HSC cultures. These results suggest that reduced mitochondrial respiratory chain activity may contribute to progenitor cell inhibition at low O2. However, further studies are warranted to assess the metabolic state and dependencies of HSCs and progenitor cells under these culture conditions.
Low O2 concentration alters cellular heterogeneity within HSC cultures
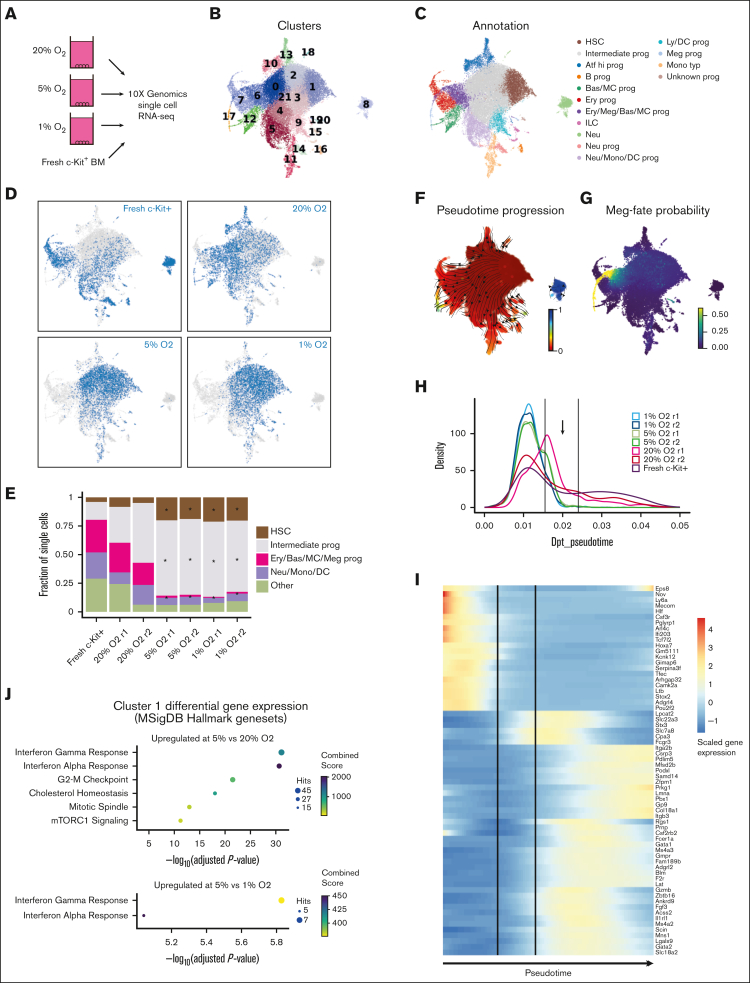
To further investigate the altered cellular heterogeneity within these HSC cultures, we performed scRNA-seq using 4-week 20% O2, 5% O2, and 1% O2 cultures derived from LT-HSCs as well as on a fresh c-Kit–enriched bone marrow sample (Figure 2A). Batch correction using Harmony was performed to integrate fresh and cultured scRNA-seq profiles because the matching populations between samples were highly divergent with clear changes in expression (supplemental Figure 2A). The combined data revealed matching lineage topology and differentiation stages between fresh and cultured cells (supplemental Figure 2B) and identified 21 cell clusters (Figure 2B). We performed manual annotation based on established lineage markers,29 which highlighted the differentiation trajectories toward monocytes, neutrophils, basophils/mast cells, megakaryocytes, erythroids, and lymphoid/B cells (Figure 2C; supplemental Figure 3A). To pinpoint the location of putative HSCs, we analyzed the expressions of known HSC markers (Procr, Mecom, Mllt3, Ly6a, and Hlf; supplemental Figure 3A) and HSC gene signatures (HSCscore30 and RepopSig;24 supplemental Figure 3B-C), which showed the highest values within cluster 1. Comparing the cluster frequency for each sample, we observed striking differences in the quantity/abundance of cells between the high (20%) and low (5% and 1%) O2 cultures, whereas the 5% and 1% patterns were broadly similar (Figure 2D; supplemental Figure 3D). Specifically, we identified an increased abundance of low O2–cultured cells in the HSC and intermediate progenitor clusters accompanied by relative depletion of the more differentiated clusters (Figure 2E). Cells with the highest RepopSig scores were also more abundant at low O2 levels (supplemental Figure 3E).
Figure 2.
Single-cell transcriptomics identifies the molecular consequences of low O2 concentration on HSC cultures. (A) Schematic of the scRNA-seq analysis of hematopoietic stem and progenitor cells (HSPCs) cultured at different O2 concentrations. (B) UMAP projections of all samples with color-coded cluster memberships. (C) Manual annotation of the clusters in panel B based on marker gene expression. (D) Uniform Manifold Approximation and Projection (UMAP) of all cells (gray) and cells from the indicated conditions (blue). In each case, an equal cell number was randomly selected for each sample. (E) Bar plot indicating relative cell abundance in the landscape areas for each sample. Areas were chosen as follows: HSC, cluster 1; Intermediate prog, clusters 0 and 2 to 4; Ery/Bas/MC/Meg prog, clusters 6, 7, and 17; Neu/Mono/DC, clusters 5, 11, and 14; and other, remaining clusters. (F) UMAP projection color-coded with diffusion pseudotime values, overlaid with arrows indicating putative paths of differentiation using random walks estimation with CellRank Pseudotime Kernel. (G) UMAP projection color-coded by the cell fate probability of cells differentiating into the tip of cluster 17 (megakaryocytes). (H) Cell density along the trajectory shown in panel G for each sample and pseudotime values between 0 and 0.05 are shown. Vertical lines indicate the regions of interest where cells at 5% and 1% O2 disappear. (I) Heatmap of genes differentially expressed between the beginning and end of the region of interest shown in panel H. (J) Enrichr gene enrichment analysis of upregulated genes within cluster 1 at 5% O2, compared with 20% (left) and 1% O2 (right). ∗ in panel E indicates a statistically significant change in cell abundance (FDR < 0.05) compared with the 20% O2 condition. Bas, basophil; Ery, erythroid; ILC, innate lymphoid cell; Ly/DC, lymphoid/dendritic cell; meg, megakaryocyte; MC, mast cell; monotyp, typical monocyte; neu, neutrophil; prog, progenitor.
To probe the apparent block in differentiation caused by low O2 levels, we used estimated pseudotime and the CellRank framework to infer cell fate probabilities for the main differentiation trajectories (Figure 2F-G; supplemental Figure 4A). At low O2 levels, cell density along pseudotime helped confirm a sharp reduction in cell number at ∼0.02 pseudotime mark for both megakaryocyte and neutrophil trajectories (arrows in Figures 2H; supplemental Figure 4B). At this stage, we observed almost no fate separation between the basophil, erythroid, and megakaryocyte trajectories (supplemental Figure 4C-D), whereas the neutrophil trajectory was separated from the other fates (supplemental Figure 4E). Thus, we investigated for the genes that were dynamically expressed during the differentiation stage, with a clear drop in the cell number of neutrophils and megakaryocyte trajectories.
For megakaryocyte trajectory, we identified 75 differentially expressed genes, which showed a stereotypic pattern and order of gene expression changes (Figure 2I). Many of these are known hematopoietic regulators.31 For instance, the expression levels of several HSC-associated genes (eg, Mecom, Hlf, and Ly6a) dropped early on (pseudotime value 0.015). On the other hand, a different set of genes, including several known markers of differentiation (eg, Gata1, Gata2, Ms4a2, and Ms4a3), began to be upregulated during the putative differentiation block stage.
The dynamic genes in the neutrophil trajectory showed a limited overlap with those in the megakaryocyte trajectory, although we observed a similar decrease in the Mecom expression in both (supplemental Figure 4F). Instead, the list of dynamic genes featured genes associated with very early stages of myeloid differentiation, including Mpo, Ctsg, and chemokine Ccl9, appearing just before the region of interest, and more typical neutrophil markers, including Elane and Cebpe, appearing later (supplemental Figure 4F). Together, these results pinpoint critical stages in the transition from the HSC-like state to more differentiated progenitors that are suppressed in low O2 cultures and suggest specific molecular events associated with this transition.
Finally, we performed differential gene expression analysis to investigate O2 concentration-specific transcriptomes within cluster 1 HSCs. We identified upregulation of the interferon response specifically at 5% O2 (Figure 2J; supplemental Figure 4G). This higher inflammatory signaling could be related to the recently described link between inflammatory signaling and ex vivo HSC self-renewal.32 To allow the community to interrogate these data sets. We have generated a website portal for investigating gene expression within these clustersa http://128.232.227.172/Igarashi2022/.
Low O2 concentration can support HSC expansion from unfractionated hematopoietic cell populations
The selectivity of these low O2 cultures prompted us to consider whether culture-based enrichment of HSCs could be achieved from WBMCs (Figure 3A). Although we have previously succeeded in expanding LT-HSCs from c-Kit+ HSPCs, LT-HSC selection from WBMCs was not possible at 20% O2.33 Compared with 20% O2 WBMC cultures, similar number of live cells were generated after 4 weeks at 5% O2, but lower numbers were generated at 1% O2 (Figure 3B). Both 5% O2 and 1% O2 cultures contained approximately threefold more CD201+CD150+KSL cells than the 20% O2 cultures (Figure 3C; supplemental Figure 5A). In the primary competitive transplantation assays, 5% O2 and 1% O2 cultures showed better results than 20% O2 cultures (Figure 3D; supplemental Figure 5B). However, only 5% O2 cultures achieved robust multilineage peripheral blood chimerism in secondary recipients (Figure 3E; supplemental Figure 5C).
Figure 3.
Low O2 cultures expand HSCs from unfractionated WBMCs and embryonic tissues. (A) Schematic diagram of low O2 WBMC cultures. Unfractionated mouse WBMCs were seeded into PVA-based media in 24-well plates (5 × 106 cells per well) in 1 ml of media and cultured for 4 weeks at 20%, 5%, or 1% O2. (B) The mean number of live cells per well within the WBMC-derived cultures described panel in A on day-28 (n = 6). (C) The mean number of CD201+CD150+KSL cells per well within the WBMC-derived cultures described in panel A on day-28 (n = 6). (D) 16-week donor peripheral blood (PB) chimerism from 4-week-old WBMC-derived cultures incubated in 20%, 5%, or 1% O2. Five thousand cells from each culture were transplanted along with 1×106 WBMCs into lethally irradiated recipients (mean ± SD; n = 3-5). (E) Twelve-week donor peripheral blood chimerism after secondary transplantation of WBMCs from primary recipient mice described in panel D (Mean ± SD; n = 3-4). (F) Schematic diagram of the CyTOF time course during 5% O2 WBMC cultures (left) and the frequency of immunophenotypic cell populations at the indicated time points (right). See supplemental Figure 5B for cell immunophenotypes. (G) Schematic diagram of the CyTOF time course during 5% O2 whole spleen cell cultures (left) and the frequency of immunophenotypic cell populations for 28-day cultures (right). supplemental Figure 5B shows cell immunophenotypes. (H) Schematic diagram of selective HSC expansion assay for mouse embryonic tissues. (I) The mean number of CD201+CD150+KSL cells generated from E11.5 to 12.5 embryonic tissue (excluding extraembryonic tissues and fetal liver) and from E11.5 to 16.5 heart, yolk sac, placenta, and fetal liver-derived cultures. The starting cell number are indicated in brackets seeded in 1 mL of media. Statistical analysis was performed using ANOVA.; ∗P < .05; ∗∗P < .01; ∗∗∗∗P < .0001. n.s., not significant.
To track the cellular heterogeneity within these WBMC cultures over time, we performed CyTOF using a hematopoiesis antibody panel (Figure 3F; supplemental Table 2). Given the superior functional results at 5% O2, we focused our analysis on this culture condition. We used unsupervised clustering to resolve cell populations within the cultures and manually annotated these immunophenotypic clusters (supplemental Figure 5D). Clusters in less than 1% of the cell cultures were grouped as “other.” Over the first week, the CD201+KSL fraction was rare (0.5%), but it increased after day 7 and reached more than 50% by day 28 (Figure 1F). The inverse was observed for granulocytes/monocytes and erythroid cell levels, which were initially high and then dropped between days 7 and 14.
To assess a second hematopoietic cell population, we evaluated whole spleen cells in 5% O2 culture using the CyTOF time course assay (Figure 3G; supplemental Figure 5A). Similar to WBMC cultures, CD201+KSL frequencies were initially very low and increased between days 7 and 14, whereas erythroid cells displayed the opposite trend. For fresh spleen cultures, CD4+ and CD8+ T cells were initially present at a frequency of 27% and gradually depleted over time. By day 28, T cells constituted only 2% of the culture. Compared with the WBMC cultures, lower CD201+KSL frequencies were observed in spleen cultures at day 28 (23% vs 50%; Figure 3F-G).
Within both the WBMC and whole spleen cell cultures, we could identify 2 KSL compartments, 1 marked with CD201 (CD201+KSL) and a second marked with CD41 (CD41+KSL; Figure 3F-G; supplemental Figure 5D). Interestingly, although CD201+KSL expanded progressively over 28 days, CD41+KSL was most abundant on days 14 and 21, before dropping at day 28. Further investigation of the Kit+Sca1+Lineage– population, specifically on day 28, suggested that the major population (expressing CD201+) coexpressed CD31 (Pecam) and low levels of CD11b (Mac1) (supplemental Figure 5E). This analysis suggests that additional markers may help resolve the HSC compartment ex vivo.
Given the selectivity of these culture conditions, we wondered whether this system could be used to assay the HSC expansion capacity. Therefore, we turned to embryonic development to determine when expandable potential could be first observed. As definitive HSCs are reported to be first seen from E10.5,4 we evaluated the expansion potential of various embryonic organs at time points from E11.5 to E16.5 (Figures 3H). Our results largely mirrored the reported localization of HSCs in the developing embryo, with pHSCs expanding from the fetal liver from E13.5 but not from the yolk sac34 (Figure 3I). Consistent with the reported transient HSC activity in the placenta,35,36 pHSCs expanded from E12.5 and E13.5 placentas but not from those at later time points. The total live cell numbers (supplemental Figure 5F) also correlated with the presence of pHSCs in these cultures (Figure 3I), demonstrating culture selectivity. However, we did not detect an expansion potential in the embryo from E11.5 to E12.5, suggesting that our assay is selective for a subset of mature HSCs. These results suggest that this culture system may become a useful assay for HSC expansion potential, although further work is needed to functionally validate these findings.
Ex vivo selective culture depletes GVHD-causing T cells
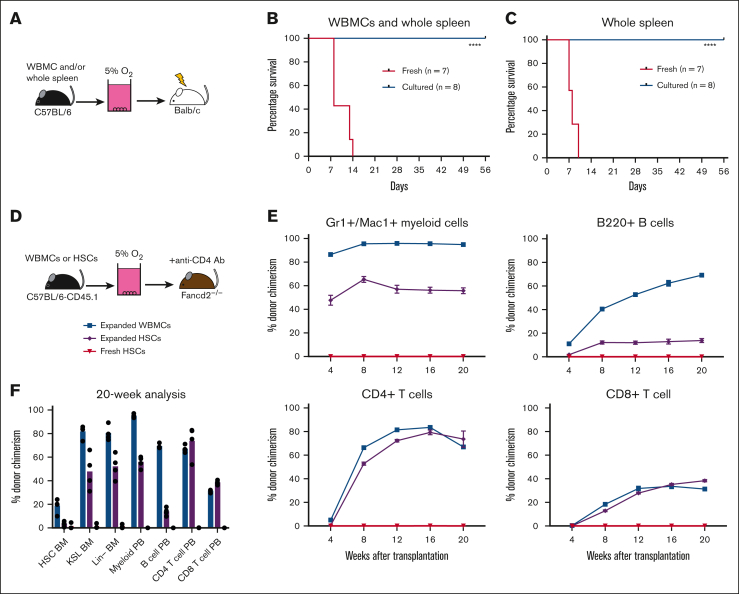
In allogeneic HSCT, alloreactive T cells are a major cause of GVHD. Based on the robust concomitant depletion of T cells and expansion of HSCs in our whole spleen cell cultures, we hypothesized that our low O2 ex vivo HSC expansion culture conditions would allow for engraftment after allogeneic transplantation while avoiding GVHD. Therefore, we tested this in an acute GVHD model, in which C57BL/6 donor cells were transplanted into irradiated allogeneic Balb/c recipients (Figure 4A). To induce acute GVHD in this model, a mixture of WBMCs and whole spleen cells was cotransplanted into irradiated recipients. Consistent findings from with other reports,37 transplantation of fresh spleen/WBMC led to acute GVHD in 2 weeks (Figure 4B; supplemental Figure 6A). By contrast, recipients of cultured spleen/WBMCs, or even expanded spleen only, survived long-term (Figure 4B-C) and displayed multilineage donor peripheral blood chimerism (supplemental Figure 6B). We further confirmed that the loss of T cells was responsible for the survival of these recipients by transplantation of fresh T cells with cultured spleen/WBMCs. The addition of fresh T cells led to rapid GVHD (supplemental Figure 6C). Together, these results confirm that low O2 HSC-selective media conditions lead to the depletion of mature immune cells and enrichment of transplantable HSPCs.
Figure 4.
Low O2 selective HSC cultures help avoid GVHD. (A) Schematic diagram of allogeneic transplantation assay. Unfractionated WBMCs and/or spleen cells from C57BL/6 mice were transplanted into irradiated Balb/c mice before or after 4-week PVA-based culture. (B) Survival of Balb/c recipients in the assay described in panel B, after transplantation of 5 × 106 WBMCs and 5 × 106 whole spleen cells (fresh or cultured) from C57BL/6 mice (n = 7-8). Statistical analysis was performed using the Mantel-Cox test. (C) Survival of Balb/c recipients in the assay described in panel B, after the transplantation of 5 × 106 whole spleen cells (fresh or cultured) from C57BL/6 mice (n = 7-8). Statistical analysis was performed using Mantel-Cox test. (D) Schematic diagram of antibody conditioning transplantation assay. Four-week cultured WBMCs (derived from 20 × 106 WBMCs), 4-week cultured HSCs (derived from 500 HSCs), or 5000 fresh HSCs from C57BL/6-CD45.1 mice were transplanted into Fancd2–/–-CD45.2 mice 7 days after treatment with the anti-CD4 (GK1.5) antibody. (E) Donor chimerism in the Fancd2–/– recipient mice at 4 or 20 weeks with peripheral blood Mac1+Gr1+ myeloid cells (top left), B220+ B cells (top right), CD4+CD3+ T cells (bottom left), and CD8+CD3+ T cells (bottom right) (n = 4). (F) Donor chimerism in the Fancd2–/– bone marrow and peripheral blood compartments of recipients described in panel F (n = 4). ∗∗∗∗P < .0001.
Finally, we evaluated whether we could combine this GVHD-free HSC expansion method with antibody-based inhibition of immune rejection in a relevant allogeneic HSCT mouse model (Figure 4D). Given the toxicities of bone marrow conditioning regimens and GVHD in patients with Fanconi anemia receiving allogeneic HSC transplantation, we selected a minor allele mismatch Fancd2–/– HSCT mouse model for this proof-of-concept study. Minor mismatches between wild-type C57BL/6 and Fancd2–/– mice lead to rejection of C57BL/6 donor cells when transplanted without conditioning.38 However, immune rejection can be inhibited by anti-CD4 antibody-based conditioning. We initially confirmed that anti-CD4 antibody treatment was necessary for engraftment of C57BL/6 cells (expanded from 5 × 106 WBMCs) in Fancd2–/– mice (supplemental Figure 6D). This experiment helped confirm that donor chimerism was dependent on anti-CD4 conditioning but only achieved ∼50% donor chimerism.
We repeated the transplantation experiments using cells expanded from 20 × 106 WBMCs for anti-CD4–conditioned Fancd2–/– mice. We achieved robust levels (>80%) of myeloid donor chimerism even within 4 weeks, which remained stably high over 20 weeks (Figure 4E-F). Notably, we observed lower levels of donor chimerism in the bone marrow HSC compartment than those in the downstream progenitor and peripheral blood compartments. These results suggest a selective advantage in the ability of wild-type HSCs to support hematopoiesis, as compared with Fancd2–/– HSCs, and correspond with the reported reduced fitness of Fancd2–/– HSCs.38 By contrast, 5000 freshly purified HSCs failed to engraft in Fancd2–/–mice, whereas 4-week expanded cultures derived from 500 HSCs also displayed robust donor chimerism (Figure 4E-F). It is currently unclear why freshly purified HSCs failed to engraft, but it is likely due to the low efficiency of engraftment in the nonconditioned setting and the need for supraphysiological HSC numbers for engraftment without bone marrow conditioning.39 Together, these results highlight the potential translational implications for selective HSC expansion cultures.
Discussion
In this report, we demonstrate that the selectivity of PVA-based mouse HSC expansion cultures can be improved by optimizing the O2 concentrations. Furthermore, we discovered that HSCs expand at the expense of GVHD-causing T cells under optimized low O2 conditions. In addition, we present proof-of-concept data for the combined use of expanded HSCs and antibody-mediated inhibition of immunologic rejection in allogeneic HSCT. However, further work will be needed to determine the translational potential of this study. For example, we will need to investigate whether antibody conditioning for HLA-mismatched alloHSCT could be combined with these approaches.40 In addition, T-cell–containing splenocytes were used in our acute GVHD assays, whereas peripheral blood products are used in the clinical setting.
The risk of GVHD associated with alloHSCT has led to a major research effort in autologous HSCT gene therapies, in which a patient’s HSCs are collected, gene-corrected, and returned to reconstitute a healthy hematopoietic system.41,42 However, this therapeutic strategy is still challenging and expensive and patient access is usually limited to a few specialized centers. Our results suggest that if HSC-selective expansion methods can be translated into humans and the risk of GVHD becomes avoidable, GvHD-free allogeneic HSCT could provide a competitive approach to autologous HSCT gene therapy. These results also highlight the potential caveats of allogeneic HSCT. In hematologic malignancies, allo–T-cell–mediated graft-versus-tumor effects are essential for long-term remission after HSCT.43 However, expanded HSC products could be combined with T-cell add-back strategies, and there may be advantages of being able to separate the dose of HSPCs from that of T cells.
In addition to the translational implications of this study, we hope that our findings will support further investigations of HSC biology and hematopoiesis. The long-term ex vivo HSC culture system provides a tractable ex vivo model system for studying HSC self-renewal and lineage commitment. In summary, we identified physioxia as optimal for PVA-based HSC expansion cultures and demonstrated that these methods are highly selective for HSCs compared with more mature hematopoietic cell types, with implications for both basic and translational stem cell biology.
Conflict-of-interest disclosure: H.N. is a cofounder and shareholder in Celaid, Megakaryon and Century Therapeutics. A.C.W. is a consultant for Graphite Bio and ImmuneBridge. A.C. discloses financial interests in the following entities working in the rare genetic disease space: Beam Therapeutics, Decibel Therapeutics, Editas Medicine, Global Blood Therapeutics, GV, Lyrik Pharma, Magenta Therapeutics, and Spotlight Therapeutics. IK is currently employed by Xap Therapeutics. The remaining authors declare no competing financial interests.
Acknowledgments
The authors thank the Stanford Stem Cell Institute FACS Core and WIMM Flow Cytometry Core for flow cytometry access, the Stanford Animal Histology Service for histology, and CZ-Biohub for performing next-generation sequencing. A.C.W. acknowledges support from the Kay Kendall Leukemia Fund, Medical Research Council, National Institutes of Health (K99HL150218), Leukemia and Lymphoma Society (3385-19), and Edward P. Evans Foundation. H.N. was supported by the National Institutes of Health (R01DK116944 and R01HL147124), the Ludwig Foundation, the Stinehart-Reed Foundation, and the Japan Society for the Promotion of Science. K.J.I. acknowledges the support from the National Science Foundation. T.-K.T. acknowledges the support from The Agency for Science, Technology and Research, Singapore. J.B. acknowledges support from the Swedish Research Council and Assar Gabrielsson Foundation (2017-0034). I.K. and B.G. were supported by the Wellcome Trust, Cancer Research UK, and the Medical Research Council. J.W.H. was supported by an National Institutes of Health T32 fellowship (T32CA196585) and an American Cancer Society Roaring Fork Valley postdoctoral fellowship (PF-20-032-01-CSM). The sponsors of this study are organizations that support science in general. They had no role in gathering, analyzing, or interpreting the data.
Authorship
Contribution: K.J.I., I.K., Y.Y.C., D.K., T.-K.T., J.B., I.H., P.Y.H., H.M.K., J.W.H., and K.N. designed and performed the experiments, analyzed the data, and reviewed and edited the manuscript; G.P.N., K.S.B., A.C., B.G., and H.N. designed the experiments, analyzed the data, and reviewed and edited the manuscript; and A.C.W. designed and performed the experiments, analyzed the data, and wrote the manuscript.
Footnotes
∗K.J.I. and I.K. are joint first authors.
Raw and processed data reported in this article have been deposited in the Gene Expression Omnibus database (accession numbers GSE207743, GSE175400, and GSE207740).
Data are available on request from the corresponding author, Adam C. Wilkinson (adam.wilkinson@imm.ox.ac.uk).
The full-text version of this article contains a data supplement.
Supplementary Material
References
- 1.Wilkinson AC, Igarashi KJ, Nakauchi H. Haematopoietic stem cell self-renewal in vivo and ex vivo. Nat Rev Genet. 2020;21(9):541–554. doi: 10.1038/s41576-020-0241-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eaves CJ. Hematopoietic stem cells: concepts, definitions, and the new reality. Blood. 2015;125(17):2605–2613. doi: 10.1182/blood-2014-12-570200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132(4):631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dzierzak E, Speck NA. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol. 2008;9(2):129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505(7483):327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303–320. doi: 10.1038/s41580-019-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morita Y, Iseki A, Okamura S, Suzuki S, Nakauchi H, Ema H. Functional characterization of hematopoietic stem cells in the spleen. Exp Hematol. 2011;39(3):351–359.e3. doi: 10.1016/j.exphem.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Copelan EA. Hematopoietic stem-cell transplantation. N Engl J Med. 2006;354(17):1813–1826. doi: 10.1056/NEJMra052638. [DOI] [PubMed] [Google Scholar]
- 9.Chabannon C, Kuball J, Bondanza A, et al. Hematopoietic stem cell transplantation in its 60s: a platform for cellular therapies. Sci Transl Med. 2018;10(436) doi: 10.1126/scitranslmed.aap9630. [DOI] [PubMed] [Google Scholar]
- 10.Hill GR, Betts BC, Tkachev V, Kean LS, Blazar BR. Current concepts and advances in graft-versus-host disease immunology. Annu Rev Immunol. 2021;39:19–49. doi: 10.1146/annurev-immunol-102119-073227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilkinson AC, Nakauchi H. Stabilizing hematopoietic stem cells in vitro. Curr Opin Genet Dev. 2020;64:1–5. doi: 10.1016/j.gde.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar S, Geiger H. HSC niche biology and HSC expansion ex vivo. Trends Mol Med. 2017;23(9):799–819. doi: 10.1016/j.molmed.2017.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilkinson AC, Ishida R, Kikuchi M, et al. Long-term ex vivo haematopoietic-stem-cell expansion allows nonconditioned transplantation. Nature. 2019;571(7763):117–121. doi: 10.1038/s41586-019-1244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson AC, Ishida R, Nakauchi H, Yamazaki S. Long-term ex vivo expansion of mouse hematopoietic stem cells. Nat Protoc. 2020;15(2):628–648. doi: 10.1038/s41596-019-0263-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houghtaling S, Timmers C, Noll M, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17(16):2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mantel CR, O'Leary HA, Chitteti BR, et al. Enhancing hematopoietic stem cell transplantation efficacy by mitigating oxygen shock. Cell. 2015;161(7):1553–1565. doi: 10.1016/j.cell.2015.04.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobayashi H, Morikawa T, Okinaga A, et al. Environmental optimization enables maintenance of quiescent hematopoietic stem cells ex vivo. Cell Rep. 2019;28(1):145–158.e9. doi: 10.1016/j.celrep.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 18.Wilkinson AC, Yamazaki S. The hematopoietic stem cell diet. Int J Hematol. 2018;107(6):634–641. doi: 10.1007/s12185-018-2451-1. [DOI] [PubMed] [Google Scholar]
- 19.Al-Ani A, Toms D, Kondro D, Thundathil J, Yu Y, Ungrin M. Oxygenation in cell culture: critical parameters for reproducibility are routinely not reported. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0204269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spencer JA, Ferraro F, Roussakis E, et al. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508(7495):269–273. doi: 10.1038/nature13034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christodoulou C, Spencer JA, Yeh SCA, et al. Live-animal imaging of native haematopoietic stem and progenitor cells. Nature. 2020;578(7794):278–283. doi: 10.1038/s41586-020-1971-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pavlacky J, Polak J. Technical feasibility and physiological relevance of hypoxic cell culture models. Front Endocrinol (Lausanne) 2020;11:57. doi: 10.3389/fendo.2020.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhdanov AV, Ogurtsov VI, Taylor CT, Papkovsky DB. Monitoring of cell oxygenation and responses to metabolic stimulation by intracellular oxygen sensing technique. Integr Biol (Camb) 2010;2(9):443–451. doi: 10.1039/c0ib00021c. [DOI] [PubMed] [Google Scholar]
- 24.Che JLC, Bode D, Kucinski I, et al. Identification and characterization of in vitro expanded hematopoietic stem cells. EMBO Rep. 2022;23(10) doi: 10.15252/embr.202255502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becker HJ, Ishida R, Wilkinson AC, et al. A single cell cloning platform for gene edited functional murine hematopoietic stem cells. bioRxiv. 2022 [Google Scholar]
- 26.Zhang Q, Konturek-Ciesla A, Yuan O, Bryder D. Ex vivo expansion potential of murine hematopoietic stem cells: a rare property only partially predicted by phenotype. bioRxiv. 2022 doi: 10.7554/eLife.91826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Galen P, Kreso A, Mbong N, et al. The unfolded protein response governs integrity of the haematopoietic stem-cell pool during stress. Nature. 2014;510(7504):268–272. doi: 10.1038/nature13228. [DOI] [PubMed] [Google Scholar]
- 28.Kruta M, Sunshine MJ, Chua BA, et al. Hsf1 promotes hematopoietic stem cell fitness and proteostasis in response to ex vivo culture stress and aging. Cell Stem Cell. 2021;28(11):1950–1965.e6. doi: 10.1016/j.stem.2021.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dahlin JS, Hamey FK, Pijuan-Sala B, et al. A single-cell hematopoietic landscape resolves 8 lineage trajectories and defects in Kit mutant mice. Blood. 2018;131(21):e1–e11. doi: 10.1182/blood-2017-12-821413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamey FK, Gottgens B. Machine learning predicts putative hematopoietic stem cells within large single-cell transcriptomics data sets. Exp Hematol. 2019;78:11–20. doi: 10.1016/j.exphem.2019.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkinson AC, Gottgens B. Transcriptional regulation of haematopoietic stem cells. Adv Exp Med Biol. 2013;786:187–212. doi: 10.1007/978-94-007-6621-1_11. [DOI] [PubMed] [Google Scholar]
- 32.Chagraoui J, Lehnertz B, Girard S, et al. UM171 induces a homeostatic inflammatory-detoxification response supporting human HSC self-renewal. PLoS One. 2019;14(11) doi: 10.1371/journal.pone.0224900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ochi K, Morita M, Wilkinson AC, Iwama A, Yamazaki S. Non-conditioned bone marrow chimeric mouse generation using culture-based enrichment of hematopoietic stem and progenitor cells. Nat Commun. 2021;12(1):3568. doi: 10.1038/s41467-021-23763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganuza M, Chabot A, Tang X, et al. Murine hematopoietic stem cell activity is derived from pre-circulation embryos but not yolk sacs. Nat Commun. 2018;9(1):5405. doi: 10.1038/s41467-018-07769-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gekas C, Dieterlen-Lièvre F, Orkin SH, Mikkola HKA. The placenta is a niche for hematopoietic stem cells. Dev Cell. 2005;8(3):365–375. doi: 10.1016/j.devcel.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 36.Ottersbach K, Dzierzak E. The murine placenta contains hematopoietic stem cells within the vascular labyrinth region. Dev Cell. 2005;8(3):377–387. doi: 10.1016/j.devcel.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Schroeder MA, DiPersio JF. Mouse models of graft-versus-host disease: advances and limitations. Dis Model Mech. 2011;4(3):318–333. doi: 10.1242/dmm.006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrakasan S, Jayavaradhan R, Ernst J, et al. KIT blockade is sufficient for donor hematopoietic stem cell engraftment in Fanconi anemia mice. Blood. 2017;129(8):1048–1052. doi: 10.1182/blood-2015-12-689083. [DOI] [PubMed] [Google Scholar]
- 39.Shimoto M, Sugiyama T, Nagasawa T. Numerous niches for hematopoietic stem cells remain empty during homeostasis. Blood. 2017;129(15):2124–2131. doi: 10.1182/blood-2016-09-740563. [DOI] [PubMed] [Google Scholar]
- 40.George BM, Kao KS, Kwon HS, et al. Antibody conditioning enables MHC-mismatched hematopoietic stem cell transplants and organ graft tolerance. Cell Stem Cell. 2019;25(2):185–192.e3. doi: 10.1016/j.stem.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morgan RA, Gray D, Lomova A, Kohn DB. Hematopoietic stem cell gene therapy: progress and lessons learned. Cell Stem Cell. 2017;21(5):574–590. doi: 10.1016/j.stem.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dever DP, Porteus MH. The changing landscape of gene editing in hematopoietic stem cells: a step towards Cas9 clinical translation. Curr Opin Hematol. 2017;24(6):481–488. doi: 10.1097/MOH.0000000000000385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negrin RS. Graft-versus-host disease versus graft-versus-leukemia. Hematology Am Soc Hematol Educ Program. 2015;2015:225–230. doi: 10.1182/asheducation-2015.1.225. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.