Abstract
In eukaryotic cells, nonvesicular lipid transport between organelles is mediated by lipid-transfer proteins. Recently, a new class of these lipid transporters has been described to facilitate the bulk of inter-organelle lipid transport at contact sites by forming bridge-like structures with a hydrophobic groove through which lipids travel. Because their predicted structure is composed of repeating β-groove (RBG) domains, they have been named the RBG protein superfamily. Early studies on RBG proteins VPS13 and ATG2 recognized the resemblance of their predicted structures to that of the bacterial Lpt system, which transports newly synthesized lipopolysaccharides (LPS) between the inner and the outer membranes (IMs and OMs) of Gram-negative bacteria. In these didermic bacteria, the IMs and OMs are separated by an aqueous periplasmic compartment that is traversed by a bridge-like structure built with β-jelly roll domains from several Lpt proteins that provides a hydrophobic groove for LPS molecules to travel across the periplasm. Despite structural and functional similarities between RBG proteins and the Lpt system, the bacterial AsmA-like protein family has recently emerged as the likely ancestor of RBG proteins and long sought-after transporters that facilitate the transfer of phospholipids from the IM to the OM. Here, we review our current understanding of the structure and function of bacterial AsmA-like proteins, mainly focusing on recent studies that have led to the proposal that AsmA-like proteins mediate the bulk of phospholipid transfer between the IMs and OMs.
Keywords: LPS, glycerophospholipid, membrane biogenesis, tamB, yhdP, ydbH
Introduction
Gram-negative bacteria such as Escherichia coli are characterized by having an envelope delimited by two concentric membranes (Silhavy et al., 2010). The inner membrane (IM) is a phospholipid bilayer that surrounds the cytoplasm. The outer membrane (OM) is highly asymmetric with an outer leaflet primarily made of lipopolysaccharides (LPS) and an inner leaflet composed of phospholipids (Muhlradt and Golecki, 1975; Kamio and Nikaido, 1976). This asymmetric structure of the OM is responsible for the innate resistance of Gram-negative bacteria to various antibiotics and detergents (Nikaido, 2003). The IM and OM are separated by an aqueous compartment known as the periplasm, which contains many proteins and a thin cell wall composed of peptidoglycan (Silhavy et al., 2010).
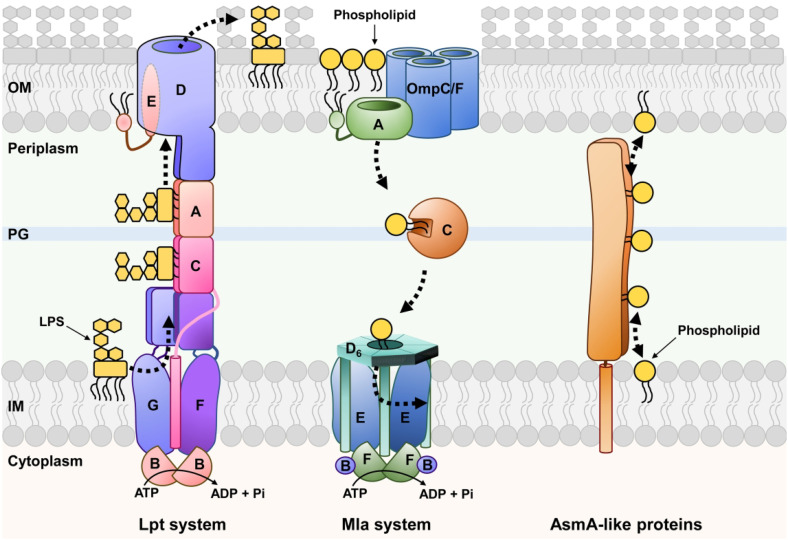
Since the synthesis of lipids and glycolipids occurs in the IM (Osborn et al., 1972, 1974), the growth of the OM (and therefore of Gram-negative bacteria) relies on systems that transport these molecules across the periplasm (Figure 1). These transport systems are thought to generate the lipid asymmetry of the OM by specifically delivering phospholipids and LPS molecules to the inner and outer leaflets of the OM, respectively. In addition, OM phospholipids can be somehow mislocalized to the outer leaflet of the OM, which activates a specific pathway (Mla system) that removes them from the cell surface and transports them to the IM (Malinverni and Silhavy, 2009; Yeow and Chng, 2022). Thus, Gram-negative bacteria transport lipids across the periplasm in an anterograde (IM-to-OM) fashion in order to grow and in a retrograde (OM-to-IM) fashion to maintain lipid asymmetry at the OM.
Figure 1.
Lipid Transport Mechanisms Between the IM and OM of Gram-Negative Bacteria. On the left, the Lpt proteins (labeled A to G) assemble into a bridge-like structure between the IM and OM, The Lpt system transports newly synthesized LPS unidirectionally (represented by a dotted arrow) in an ATP-dependent process. In the middle, the Mla proteins (labeled A to F) and their partners the trimeric β-barrel proteins OmpC or OmpF are shown. The Mla system is responsible for the retrograde transport of phospholipids from the OM to the IM (dotted arrows) in an ATP-dependent manner. On the right, the proposed model for the bidirectional transport (represented by a dotted double arrow) of phospholipids between the IM and OM by AsmA-like proteins. Putative partners are not shown. Refer to the text for more details. PG marks the peptidoglycan cell wall.
Note. IM = inner membrane; OM = outer membrane; LPS = lipopolysaccharide.
In essence, having an OM makes Gram-negative bacteria face the same challenges that eukaryotic cells do with lipid transfer between two organelles. Nevertheless, while in eukaryotic cells lipid transport between organelles can be mediated by vesicular trafficking and lipid-transfer proteins, the bacterial periplasm is too small to accommodate vesicular transport since it has an average width of 25 nm (Asmar et al., 2017). Therefore, IM–OM lipid transfer is mediated by lipid-transfer proteins that facilitate two general types of lipid transport: (1) transport mediated by soluble chaperones that shuttle one lipid molecule at a time such as in the Mla system for retrograde transport of phospholipids and (2) transport mediated by long bridge-like structures that connect the IM and OM and can accommodate multiple lipid molecules at a time such as in the Lpt system that transports LPS (Lundstedt et al., 2021; Yeow and Chng, 2022; Figure 1). Interestingly, the Mla and Lpt systems are energized by ATP-binding cassette (ABC) transporters to mediate unidirectional transport against the concentration gradient (Sherman et al., 2018; Owens et al., 2019; Low et al., 2021; Tang et al., 2021), but the bulk of IM–OM phospholipid transport occurs bidirectionally through diffusive flow (Osborn et al., 1972; Jones and Osborn, 1977). Furthermore, while the Lpt and Mla systems were identified almost 2 decades ago, the transporters of phospholipids to the OM had remained elusive until recently. In this review, we highlight the work that has led to the proposal that some, but not all, members of the AsmA-like protein family transport phospholipids in Gram-negative bacteria between the IM and OM. We also discuss how redundancy between AsmA-like paralogs made their discovery difficult, and why they have been proposed to be the ancestor of the repeating β-groove (RBG) proteins that mediate organelle-to-organelle lipid transport in eukaryotic cells.
Overview of Lipid Transport Mechanisms Between the IM and OM of Gram-Negative Bacteria
As mentioned above, the Lpt system transports newly synthesized LPS from the IM to the outer leaflet of the OM (for a recent and extensive review on Lpt, see Lundstedt et al., 2021). This system has components (LptA-LptG proteins) in every cellular compartment that assemble into a single transport machine that bridges the IM and OM (Figure 1). LPS transport starts at the IM with the LptB2CFG ABC transporter, which uses ATP binding and hydrolysis to extract LPS molecules from the outer leaflet of the IM and place them onto the Lpt periplasmic bridge (Owens et al., 2019). This bridge is composed of β-jelly roll domains from the LptFCAD proteins, which assemble into a long structure that provides a continuous hydrophobic groove that accommodates the acyl chains of LPS molecules through the aqueous periplasm (Figure 2; Okuda et al., 2012). When LPS arrives at the β-jelly roll domain of LptD, it likely induces the lateral opening of the β-barrel domain of LptD. This opening would allow the translocation of LPS across the OM by having the acyl chains of the glycolipid in the hydrophobic core of the OM while its polysaccharide component travels through the hydrophilic lumen of the LptD β-barrel (Botos et al., 2016). LPS transport is unidirectional and powered against its concentration gradient by the continuous extraction of LPS from the IM by the LptB2CFG ABC transporter at the IM, which presumably creates a stream of LPS molecules along the periplasmic Lpt bridge (Okuda et al., 2012; Sherman et al., 2018).
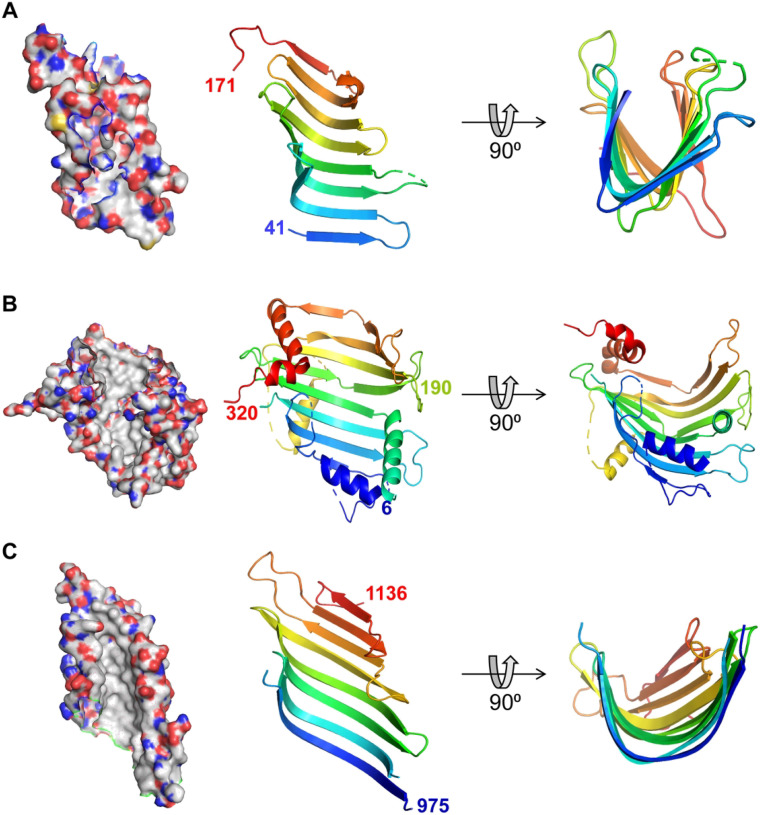
Figure 2.
Structural Similarity Between Periplasmic Lpt Proteins and Chorein-N Domain Containing Proteins. Cartoon and surface representations of (A) the crystal structure of Escherichia coli LptA (PDB ID: 2R1A (Suits et al., 2008)) showing residues from 41 to 171, and (B) the crystal structure of Chaetomium thermophilum VPS13 protein (PDB ID: 6CBC (Kumar et al., 2018)) from residue 6 to 320, containing a Chorein-N domain. According to HHpred, AsmA-like proteins are similar up to around residue 190 (labeled). (C) Part of TamB (residues 963–1138) crystal structure from E. coli (PDB ID: 5VTG (Josts et al., 2017)), demonstrating a hydrophobic β-taco fold that resembles the Lpt proteins. Surface representations (panels on the left) are colored according to atoms (carbons are white, oxygen is red, nitrogen is blue, and sulfur is yellow) to highlight the hydrophobic interior of the pocket. Cartoon representations (middle and right panels) are colored from blue at the N-terminus to red at the C-terminus. PyMOL Molecular Graphics System Version 2.5.1 (Schrödinger, LCC) was used to generate structure images.
The Mla system, which facilitates the retrograde transport of phospholipids that mislocalize to the cell surface, is also powered by an IM ABC transporter (Figure 1; Malinverni and Silhavy, 2009; Powers and Trent, 2019; Low et al., 2021; Lundstedt et al., 2021; Tang et al., 2021; Yeow and Chng, 2022). However, unlike the Lpt system, Mla utilizes a periplasmic chaperone (MlaC) that shuttles a single phospholipid molecule from the OM lipoprotein MlaA to the MlaD6E2F2B2 IM complex (Ercan et al., 2019). Structurally, the Mla and Lpt systems are also very different. The MlaA protein has a donut-like shape with a hydrophobic central hole that is thought to allow the transit of phospholipids from the outer leaflet of the OM to the periplasmic MlaC protein (Abellon-Ruiz et al., 2017). MlaC has a hydrophobic pocket that can accommodate the acyl chains of a phospholipid molecule (Ekiert et al., 2017; Ercan et al., 2019). After transiting through the periplasm, MlaC transfers its cargo to MlaD, which then transfers it to a hydrophobic pocket in the MlaE dimer of the ABC transporter (Chi et al., 2020; Coudray et al., 2020; Tang et al., 2021). Notably, the bacterial Mla system is orthologous to the TGD system that transports lipids between the chloroplast outer and inner envelope membranes (Hurlock et al., 2014).
Decades ago, it was shown that intermembrane trafficking of the bulk of phospholipids occurs bidirectionally in Gram-negative bacteria. Liposome fusion experiments in Salmonella typhimurium showed that, unlike LPS, phospholipids could flow bidirectionally between the IM and OM (Osborn et al., 1972; Jones and Osborn, 1977). This diffusional flow of phospholipids was proposed to occur through zones of adhesion (known as Bayer's junctions) between the IM and OM because Manfred Bayer had previously identified such structures in electron micrographs (Bayer, 1968; Osborn et al., 1972; Jones and Osborn, 1977). Pulse-labeling experiments later demonstrated that phospholipids rapidly equilibrate between the IM and OM in E. coli (Donohue-Rolfe and Schaechter, 1980; Langley et al., 1982). The bidirectional lipid transfer was dependent on the proton motive force but not on ATP or lipid synthesis. It was then postulated that translocation might occur at IM–OM adhesion zones and that the proton motive force might be required for the development of these zones, or flipping of newly synthesized phospholipids across the IM (Donohue-Rolfe and Schaechter, 1980; Langley et al., 1982). However, in 1990, Bayer's junctions were proven to be artifacts of microscopy fixation (Kellenberger, 1990), marking the start of a long search for the phospholipid transporters needed to build the OM.
Linking AsmA-Like Proteins to Intermembrane Phospholipid Transport
After decades of searching, YhdP was the first protein to be associated with the diffusive flow of phospholipids from the IM to the OM (Grimm et al., 2020). Using live-cell microscopy, anterograde phospholipid trafficking was monitored in an E. coli mlaA* mutant strain that has elevated LPS synthesis because of a gain of function in MlaA (Grimm et al., 2020). The mlaA* allele is dominant negative and encodes a two-codon deletion. The resulting MlaA* protein does not perform its normal function of removing mislocalized phospholipids from the outer leaflet of the OM; instead, it is predicted that the altered structure of the mutant protein allows the transfer of phospholipids from the inner to the outer leaflet of the OM (Sutterlin et al., 2016). This mislocalization of phospholipids triggers a response that increases LPS synthesis. Since LPS only makes the outer leaflet of the OM, phospholipid transfer to the OM is also increased in this mutant. Because LPS and phospholipid synthesis share a common precursor, this mutant lyses at a high rate upon entering a stationary phase of growth because lipid synthesis decreases as nutrients become scarce. Specifically, when mlaA* cells enter the stationary phase, the high rate of LPS transport demands a level of phospholipids transport to the OM that is higher than their synthesis at the IM, which leads to the lethal shrinking of the IM. A Tn-Seq screen for delayed death of mlaA* mutants during the stationary phase identified YhdP. In yhdP mlaA* mutants, the IM-to-OM flow of phospholipids was shown to be decreased, implicating YhdP in anterograde phospholipid transport across the Gram-negative envelope (Grimm et al., 2020).
YhdP belongs to the Pfam AsmA-like protein clan CL0401 and, for unknown reasons, is necessary for maintaining the barrier function of the OM (Sonnhammer et al., 1997; Mitchell et al., 2017; Mitchell et al., 2018). Interestingly, E. coli encodes six AsmA-like paralogs (YhdP, TamB, YdbH, AsmA, YicH, and YhjG) that vary in size but share a common predicted structure (Figure 3). The founding member of this family, AsmA, was identified genetically because its loss suppresses defects in the folding of OM β-barrel proteins (Misra and Miao, 1995). Although its function is yet to be determined, loss of AsmA changes OM lipid homeostasis by decreasing LPS levels (Deng and Misra, 1996). Another paralog, TamB, was also initially linked to OM β-barrel protein assembly. TamB forms complexes with Omp85-family OM proteins BamA, TamA, and TOC75 in various bacteria and chloroplasts (Iqbal et al., 2016; Chen et al., 2018; Stubenrauch and Lithgow, 2019). Omp85-family proteins fold and insert β-barrel proteins into the OM of Gram-negative bacteria, mitochondria, and chloroplasts (Gentle et al., 2004; Gross et al., 2021). Accordingly, earlier studies proposed a role for the translocation and assembly module (composed of TamA and TamB) complex in the assembly of a subset of OM β-barrel proteins (Stubenrauch et al., 2016; Josts et al., 2017; Bialer et al., 2019). However, the role of TamB in OM protein biogenesis has been challenged by multiple studies (Gallant et al., 2008; Azari et al., 2013; Yu et al., 2017; Bernstein, 2019).
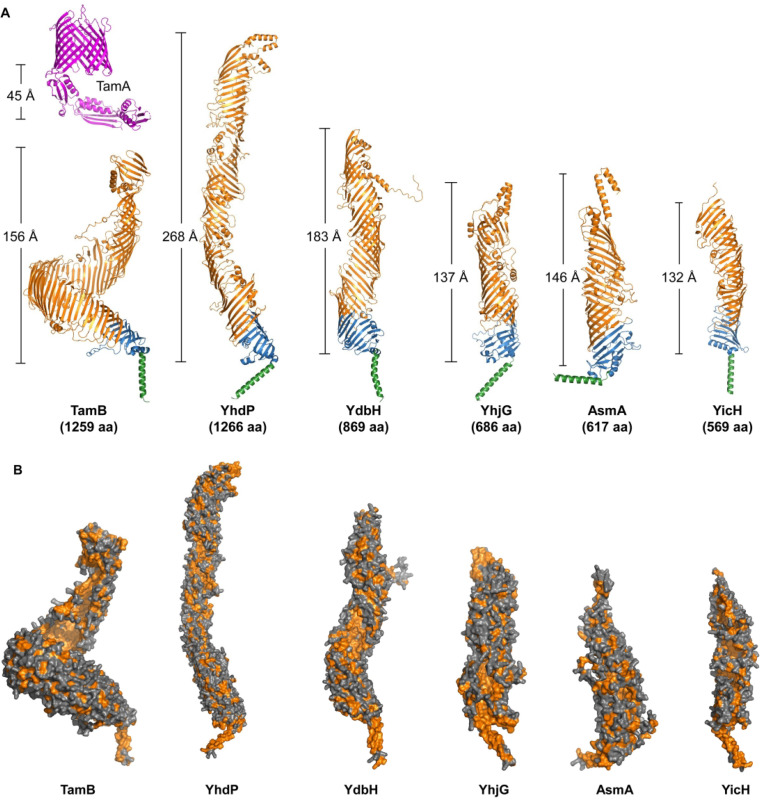
Figure 3.
Predicted Structures of Six AsmA-Like Proteins in Escherichia Coli. (A) Cartoon representations of AlphaFold's model structures for TamB, YhdP, YdbH, YhjG, AsmA, and YicH (Jumper et al., 2021). Each AsmA-like protein contains a transmembrane helix at the N-terminus (in green) predicted to serve as their IM anchor, a periplasmic Chorein-N domain (in blue), and a periplasmic AsmA-like domain (in orange). The approximate length of the periplasmic region for each AsmA-like protein was estimated using PyMOL and shown. At the top of TamB's predicted structure, the crystal structure of its partner TamA from E. coli (PDB ID: 4C00 (Gruss et al., 2013)) is shown (in magenta) along with the estimated length of its periplasmic domain. (B) Surface representations of AsmA-like family proteins. Their periplasmic domains form a groove-like structure lined with hydrophobic amino acids (shown in orange). PyMOL Molecular Graphics System Version 2.5.1 (Schrödinger, LCC) was used to generate structure images.
Architecture of AsmA-Family Proteins
AsmA-like proteins are conserved in didermic bacteria, mitochondria, and chloroplasts (Levine, 2019; Zhang et al., 2019). Bacterial AsmA-like proteins are anchored to the IM by an N-terminal α-helix and have a large periplasmic domain (Figure 3). Initial bioinformatic analyses on the periplasmic region identified a 100–150-residue Chorein-N domain followed by an AsmA-like domain of variable length among paralogs (Levine, 2019). Since (1) Chorein-N domains exist in eukaryotic lipid-transfer proteins VPS13 and ATG2 (Figures 2B and 3; McEwan and Ryan, 2022), (2) AsmA-like domains were predicted to be composed of repeats of β-taco domains with a structure similar to the β-jellyroll domains of the periplasmic Lpt bridge (Figures 2 and 3; Josts et al., 2017), and (3) YhdP affected anterograde phospholipid transport in cells (Grimm et al., 2020), AsmA-like proteins became candidates for IM–OM lipid transporters.
To date, only a fragment of E. coli TamB containing C-terminal amino acids 963–1138 has been crystallized, revealing a structure with a β-groove with a hydrophobic interior (Figure 2C; Josts et al., 2017). However, AlphaFold predicts that all six E. coli paralogs form a long bridge-like structure of various lengths and superhelicity that contains a hydrophobic groove resembling that of eukaryotic RBG lipid-transfer proteins (Figure 3; Jumper et al., 2021; Neuman et al., 2022). Notably, while the structure of RBG domains, five antiparallel β-strands and a loop, is generally conserved in eukaryotic RBG proteins, the number of β-strands is more variable in the repeats found in AsmA-like proteins. Based on the similarities of their predicted structures and function (discussed below), AsmA-like proteins have been proposed to be the ancestors of eukaryotic RBG proteins (Neuman et al., 2022).
Functional Analyses of AsmA-Like Proteins
Since YhdP was a candidate phospholipid transporter but not essential for OM biogenesis (Mitchell et al., 2017, 2018; Grimm et al., 2020), it was important to test if YhdP was functionally redundant with any of the other five AsmA-like paralogs that E. coli encodes. An in-depth functional characterization of AsmA-like proteins indeed revealed that the three largest paralogs (YhdP, TamB, and YdbH) are redundant in performing a function that is essential for growth and OM lipid homeostasis in E. coli (Ruiz et al., 2021; Douglass et al., 2022). In contrast, the additive effects in OM permeability observed when introducing a deletion allele of asmA into either a ΔyhdP or ΔtamB mutant demonstrated that AsmA functions independently of YhdP and TamB (Ruiz et al., 2021). To date, there are no clues as to the function of YicH and YhjG.
Functional redundancy between YhdP and TamB was revealed by strong synthetic defects observed when combining the ΔtamB and ΔyhdP alleles (Ruiz et al., 2021; Douglass et al., 2022). Unlike the single mutants, a ΔtamB ΔyhdP double mutant exhibits severe defects in envelope-related phenotypes such as OM permeability, cell shape, and increased tendency to lyse. Interestingly, lysis of ΔtamB ΔyhdP cells could be fully suppressed when two systems that normally remove mislocalized phospholipids from the cell surface (the Mla system and the PldA lipase) were eliminated (Ruiz et al., 2021). Thus, in the absence of YhdP and TamB, E. coli grows significantly better if it also lacks systems that remove phospholipids from the OM. These results linked YhdP and TamB to OM lipid homeostasis and agreed with the idea that YhdP and TamB transport phospholipids to the OM.
If YhdP and TamB transport phospholipids to the OM, E. coli must have at least one additional redundant system since the OM is essential for growth. Genetic suppressor analyses identified the redundant system, the AsmA-like paralog YdbH (Ruiz et al., 2021). Specifically, gain-of-function alleles of ydbH suppressed OM defects in the ΔtamB ΔyhdP double mutant. Redundancy between TamB, YhdP, and YdbH was further supported by the synthetic lethality caused by their combined loss (Ruiz et al., 2021; Douglass et al., 2022). In fact, TamB, YhdP, or YdbH, are the only AsmA-like paralogs to be individually sufficient for supporting the growth of E. coli but are not equivalent since their loss confers different phenotypes (Ruiz et al., 2021). Although we lack a direct biochemical demonstration of function, 32P-labeled phospholipids accumulated in the IM when YhdP was depleted in a ΔydbH ΔtamB double mutant (Douglass et al., 2022). Together, the aforementioned in vivo studies, predicted structures of AsmA-like proteins, and their similarities with eukaryotic RBG proteins support that TamB, YhdP, and YdbH are redundant and directly facilitate anterograde transport of phospholipids between the IM and OM.
Functional Partners of AsmA-Like Proteins
The eukaryotic lipid-transfer proteins VPS13 and ATG2 facilitate the bulk transfer of lipids between organelles at membrane contact sites (Valverde et al., 2019; Leonzino et al., 2021; Noda, 2021; Adlakha et al., 2022). These proteins interact with peripheral and/or integral membrane proteins at both membranes and function in collaboration with lipid scramblases (Ghanbarpour et al., 2021; Adlakha et al., 2022). As noted above, TamB physically interacts with OM β-barrel partners of the Omp85 family in bacteria (BamA or TamA) and chloroplasts (TOC75; Iqbal et al., 2016; Chen et al., 2018; Stubenrauch and Lithgow, 2019). This interaction is functionally relevant since the loss of either TamB or TamA results in the same phenotypes in E. coli (Ruiz et al., 2021). In agreement, TamB's predicted structure suggests that it is too short to bridge the periplasm on its own but could do so in a complex with TamA (Figure 3A). However, whether TamA is solely anchoring TamB to the OM or providing additional functional support remains untested. In contrast, even though each TamB and YhdP has approximately 1,260 residues, the superhelicity of YhdP is predicted to be significantly lower than that of TamB, making YhdP's predicted structure long enough to span the periplasm (Figure 3A). YdbH, on the other hand, only has 869 residues and is predicted to need a partner to reach the OM. Future work should therefore focus on identifying putative physical partners of AsmA-like proteins.
Model for Phospholipid Transport by AsmA-Like Proteins
Our current model suggests that TamA-TamB, YhdP, and YdbH form bridge-like structures that mediate the diffusional flow of phospholipids between the IM and OM (Ruiz et al., 2021; Douglass et al., 2022). The hydrophobic groove that runs along their predicted structures likely shields fatty acids of multiple phospholipids as they pass through the periplasm, akin to how the Lpt bridge functions in LPS transport between the IM and OM. However, TamB, YhdP, and YdbH are predicted to facilitate the bidirectional diffusive flow of phospholipids instead of the unidirectional LPS transport by Lpt (Figure 1). Although the nature of the functional redundancy between TamB, YhdP, and YdbH remains unknown, it explains why the mechanism for, and factors involved in the bulk of phospholipid transfer between the IM and OM transport were difficult to identify for so long despite significant advances made in the identification and characterization of factors involved in the biogenesis of other envelope components.
Conclusions and Future Directions
Recent studies have made significant advancements in our understanding of intermembrane phospholipid transport between the IM and OM in Gram-negative bacteria. An important knowledge gap has been bridged and many fundamental questions have emerged. Why E. coli and many other bacteria encode functionally redundant AsmA-like proteins is unknown. Do variations in expression, cargo preference, and/or cellular localization explain this redundancy? What drives transport? Do they function bidirectionally? What proteins, if any, are their functional partners? Do they interact at the IM with lipid scramblases and floppases or with the machinery involved in lipid synthesis? How can YdbH substitute for the larger TamB and YhdP proteins? How is their function coordinated with other envelope biogenesis factors to ensure the balanced growth of different envelope layers? These questions together with biochemical research aiming to test their interaction with lipids and reconstituting their function need to be addressed so that we can understand their essential function in bacterial physiology.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the National Institute of General Medical Sciences (grant number GM100951).
ORCID iD: Natividad Ruiz https://orcid.org/0000-0002-6369-2206
References
- Abellon-Ruiz J, Kaptan SS, Basle A, Claudi B, Bumann D, Kleinekathofer U, van den Berg B. (2017). Structural basis for maintenance of bacterial outer membrane lipid asymmetry. Nat Microbiol 2, 1616–1623. 10.1038/s41564-017-0046-x. [DOI] [PubMed] [Google Scholar]
- Adlakha J, Hong Z, Li P, Reinisch KM. (2022). Structural and biochemical insights into lipid transport by VPS13 proteins. Journal of Cell Biology 221, 10.1083/jcb.202202030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmar AT, Ferreira JL, Cohen EJ, Cho SH, Beeby M, Hughes KT, Collet JF. (2017). Communication across the bacterial cell envelope depends on the size of the periplasm. PLoS Biology 15, e2004303. 10.1371/journal.pbio.2004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azari F, Nyland L, Yu C, Radermacher M, Mintz KP, Ruiz T. (2013). Ultrastructural analysis of the rugose cell envelope of a member of the Pasteurellaceae family. Journal of Bacteriology 195, 1680–1688. 10.1128/JB.02149-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer ME. (1968). Areas of adhesion between wall and membrane of Escherichia coli. Journal of General Microbiology 53, 395–404. 10.1099/00221287-53-3-395. [DOI] [PubMed] [Google Scholar]
- Bernstein HD. (2019). Type V secretion in Gram-negative bacteria. EcoSal Plus 8, 10.1128/ecosalplus.ESP-0031-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialer MG, Ruiz-Ranwez V, Sycz G, Estein SM, Russo DM, Altabe S, Sieira R, Zorreguieta A. (2019). Mapb, the Brucella suis TamB homologue, is involved in cell envelope biogenesis, cell division and virulence. Scientific Reports 9, 2158. 10.1038/s41598-018-37668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botos I, Majdalani N, Mayclin SJ, McCarthy JG, Lundquist K, Wojtowicz D, Barnard TJ, Gumbart JC, Buchanan SK. (2016). Structural and functional characterization of the LPS transporter LptDE from gram-negative pathogens. Structure 24, 965–976. 10.1016/j.str.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YL, Chen LJ, Chu CC, Huang PK, Wen JR, Li HM. (2018). TIC236 links the outer and inner membrane translocons of the chloroplast. Nature 564, 125–129. 10.1038/s41586-018-0713-y. [DOI] [PubMed] [Google Scholar]
- Chi X, Fan Q, Zhang Y, Liang K, Wan L, Zhou Q, Li Y. (2020). Structural mechanism of phospholipids translocation by MlaFEDB complex. Cell Research 30, 1127–1135. 10.1038/s41422-020-00404-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudray N, Isom GL, MacRae MR, Saiduddin MN, Bhabha G, Ekiert DC. (2020). Structure of bacterial phospholipid transporter MlaFEDB with substrate bound. Elife 9, 10.7554/eLife.62518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Misra R. (1996). Examination of AsmA and its effect on the assembly of Escherichia coli outer membrane proteins. Molecular Microbiology 21, 605–612. 10.1111/j.1365-2958.1996.tb02568.x. [DOI] [PubMed] [Google Scholar]
- Donohue-Rolfe AM, Schaechter M. (1980). Translocation of phospholipids from the inner to the outer membrane of Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 77, 1867–1871. 10.1073/pnas.77.4.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglass MV, McLean AB, Trent MS. (2022). Absence of YhdP, TamB, and YdbH leads to defects in glycerophospholipid transport and cell morphology in Gram-negative bacteria. PLoS Genetics 18, e1010096. 10.1371/journal.pgen.1010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Isom GL, Greenan G, Ovchinnikov S, Henderson IR, Cox JS, Vale RD. (2017). Architectures of lipid transport systems for the bacterial outer membrane. Cell 169, 273–285 e217. 10.1016/j.cell.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercan B, Low WY, Liu X, Chng SS. (2019). Characterization of interactions and phospholipid transfer between substrate binding proteins of the OmpC-Mla system. Biochemistry 58, 114–119. 10.1021/acs.biochem.8b00897. [DOI] [PubMed] [Google Scholar]
- Gallant CV, Sedic M, Chicoine EA, Ruiz T, Mintz KP. (2008). Membrane morphology and leukotoxin secretion are associated with a novel membrane protein of Aggregatibacter actinomycetemcomitans. Journal of Bacteriology 190, 5972–5980. 10.1128/JB.00548-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentle I, Gabriel K, Beech P, Waller R, Lithgow T. (2004). The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. Journal of Cell Biology 164, 19–24. 10.1083/jcb.200310092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbarpour A, Valverde DP, Melia TJ, Reinisch KM. (2021). A model for a partnership of lipid transfer proteins and scramblases in membrane expansion and organelle biogenesis. Proceedings of the National Academy of Sciences of the United States of America 118. 10.1073/pnas.2101562118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm J, Shi H, Wang W, Mitchell AM, Wingreen NS, Huang KC, Silhavy TJ. (2020). The inner membrane protein YhdP modulates the rate of anterograde phospholipid flow in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 117, 26907–26914. 10.1073/pnas.2015556117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross LE, Klinger A, Spies N, Ernst T, Flinner N, Simm S, Ladig R, Bodensohn U, Schleiff E. (2021). Insertion of plastidic beta-barrel proteins into the outer envelopes of plastids involves an intermembrane space intermediate formed with Toc75-V/OEP80. Plant Cell 33, 1657–1681. 10.1093/plcell/koab052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss F, Zahringer F, Jakob RP, Burmann BM, Hiller S, Maier T. (2013). The structural basis of autotransporter translocation by TamA. Nature Structural & Molecular Biology 20, 1318–1320. 10.1038/nsmb.2689. [DOI] [PubMed] [Google Scholar]
- Hurlock AK, Roston RL, Wang K, Benning C. (2014). Lipid trafficking in plant cells. Traffic 15, 915–932. 10.1111/tra.12187. [DOI] [PubMed] [Google Scholar]
- Iqbal H, Kenedy MR, Lybecker M, Akins DR. (2016). The TamB ortholog of Borrelia burgdorferi interacts with the beta-barrel assembly machine (BAM) complex protein BamA. Molecular Microbiology 102, 757–774. 10.1111/mmi.13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones NC, Osborn MJ. (1977). Translocation of phospholipids between the outer and inner membranes of Salmonella typhimurium. Journal of Biological Chemistry 252, 7405–7412. [PubMed] [Google Scholar]
- Josts I, Stubenrauch CJ, Vadlamani G, Mosbahi K, Walker D, Lithgow T, Grinter R. (2017). The structure of a conserved domain of TamB reveals a hydrophobic beta taco fold. Structure 25, 1898–1906 e1895. 10.1016/j.str.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamio Y, Nikaido H. (1976). Outer membrane of Salmonella typhimurium: Accessibility of phospholipid head groups to phospholipase c and cyanogen bromide activated dextran in the external medium. Biochemistry 15, 2561–2570. 10.1021/bi00657a012. [DOI] [PubMed] [Google Scholar]
- Kellenberger E. (1990). The ‘Bayer bridges’ confronted with results from improved electron microscopy methods. Molecular Microbiology 4, 697–705. 10.1111/j.1365-2958.1990.tb00640.x. [DOI] [PubMed] [Google Scholar]
- Kumar N, Leonzino M, Hancock-Cerutti W, Horenkamp FA, Li P, Lees JA, Wheeler H, Reinisch KM, De Camilli P. (2018). VPS13A And VPS13C are lipid transport proteins differentially localized at ER contact sites. Journal of Cell Biology 217, 3625–3639. 10.1083/jcb.201807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley KE, Hawrot E, Kennedy EP. (1982). Membrane assembly: Movement of phosphatidylserine between the cytoplasmic and outer membranes of Escherichia coli. Journal of Bacteriology 152, 1033–1041. 10.1128/jb.152.3.1033-1041.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonzino M, Reinisch KM, De Camilli P. (2021). Insights into VPS13 properties and function reveal a new mechanism of eukaryotic lipid transport. Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids 1866, 159003. doi: 10.1016/j.bbalip.2021.159003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP. (2019). Remote homology searches identify bacterial homologues of eukaryotic lipid transfer proteins, including Chorein-N domains in TamB and AsmA and Mdm31p. BMC Mol Cell Biol 20, 43. 10.1186/s12860-019-0226-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low WY, Thong S, Chng SS. (2021). ATP disrupts lipid-binding equilibrium to drive retrograde transport critical for bacterial outer membrane asymmetry. Proceedings of the National Academy of Sciences of the United States of America 118. 10.1073/pnas.2110055118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundstedt E, Kahne D, Ruiz N. (2021). Assembly and maintenance of lipids at the bacterial outer membrane. Chemical Reviews 121, 5098–5123. 10.1021/acs.chemrev.0c00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinverni JC, Silhavy TJ. (2009). An ABC transport system that maintains lipid asymmetry in the Gram-negative outer membrane. Proceedings of the National Academy of Sciences of the United States of America 106, 8009–8014. 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Ryan KM. (2022). ATG2 And VPS13 proteins: Molecular highways transporting lipids to drive membrane expansion and organelle communication. FEBS Journal 289, 7113–7127. 10.1111/febs.16280. [DOI] [PubMed] [Google Scholar]
- Misra R, Miao Y. (1995). Molecular analysis of asmA, a locus identified as the suppressor of OmpF assembly mutants of Escherichia coli K-12. Molecular Microbiology 16, 779–788. 10.1111/j.1365-2958.1995.tb02439.x. [DOI] [PubMed] [Google Scholar]
- Mitchell AM, Srikumar T, Silhavy TJ. (2018). Cyclic enterobacterial common antigen maintains the outer membrane permeability barrier of Escherichia coli in a manner controlled by YhdP. mBio 9, 10.1128/mBio.01321-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell AM, Wang W, Silhavy TJ. (2017). Novel RpoS-dependent mechanisms strengthen the envelope permeability barrier during stationary phase. Journal of Bacteriology 199. 10.1128/JB.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlradt PF, Golecki JR. (1975). Asymmetrical distribution and artifactual reorientation of lipopolysaccharide in the outer membrane bilayer of Salmonella typhimurium. European Journal of Biochemistry 51, 343–352. 10.1111/j.1432-1033.1975.tb03934.x. [DOI] [PubMed] [Google Scholar]
- Neuman SD, Levine TP, Bashirullah A. (2022). A novel superfamily of bridge-like lipid transfer proteins. Trends in Cell Biology 32, 962–974. 10.1016/j.tcb.2022.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. (2003). Molecular basis of bacterial outer membrane permeability revisited. Microbiology and Molecular Biology Reviews 67, 593–656. 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda NN. (2021). Atg2 and Atg9: Intermembrane and interleaflet lipid transporters driving autophagy. Biochimica et Biophysica Acta, Molecular and Cell Biology of Lipids 1866, 158956. 10.1016/j.bbalip.2021.158956. [DOI] [PubMed] [Google Scholar]
- Okuda S, Freinkman E, Kahne D. (2012). Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338, 1214–1217. 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn MJ, Gander JE, Parisi E. (1972). Mechanism of assembly of the outer membrane of Salmonella typhimurium. Site of synthesis of lipopolysaccharide. Journal of Biological Chemistry 247, 3973–3986. [PubMed] [Google Scholar]
- Osborn MJ, Rick PD, Lehmann V, Rupprecht E, Singh M. (1974). Structure and biogenesis of the cell envelope of Gram-negative bacteria. Annals of the New York Academy of Sciences 235, 52–65. 10.1111/j.1749-6632.1974.tb43256.x. [DOI] [PubMed] [Google Scholar]
- Owens TW, Taylor RJ, Pahil KS, Bertani BR, Ruiz N, Kruse AC, Kahne D. (2019). Structural basis of unidirectional export of lipopolysaccharide to the cell surface. Nature 567, 550–553. 10.1038/s41586-019-1039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MJ, Trent MS. (2019). Intermembrane transport: Glycerophospholipid homeostasis of the Gram-negative cell envelope. Proceedings of the National Academy of Sciences of the United States of America 116, 17147–17155. 10.1073/pnas.1902026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Davis RM, Kumar S. (2021). Yhdp, TamB, and YdbH are redundant but essential for growth and lipid homeostasis of the Gram-negative outer membrane. mBio 12, e0271421. 10.1128/mBio.02714-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman DJ, Xie R, Taylor RJ, George AH, Okuda S, Foster PJ, Needleman DJ, Kahne D. (2018). Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 359, 798–801. 10.1126/science.aar1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. (2010). The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology 2, a000414. 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R. (1997). Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins 28, 405–420. [DOI] [PubMed] [Google Scholar]
- Stubenrauch C, Belousoff MJ, Hay ID, Shen HH, Lillington J, Tuck KL, Peters KM, Phan MD, Lo AW, Schembri MA, et al. (2016). Effective assembly of fimbriae in Escherichia coli depends on the translocation assembly module nanomachine. Nat Microbiol 1, 16064. 10.1038/nmicrobiol.2016.64. [DOI] [PubMed] [Google Scholar]
- Stubenrauch CJ, Lithgow T. (2019). The TAM: A translocation and assembly module of the beta-barrel assembly machinery in bacterial outer membranes. EcoSal Plus 8. 10.1128/ecosalplus.ESP-0036-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suits MD, Sperandeo P, Deho G, Polissi A, Jia Z. (2008). Novel structure of the conserved Gram-negative lipopolysaccharide transport protein A and mutagenesis analysis. Journal of Molecular Biology 380, 476–488. 10.1016/j.jmb.2008.04.045. [DOI] [PubMed] [Google Scholar]
- Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, Silhavy TJ. (2016). Disruption of lipid homeostasis in the gram-negative cell envelope activates a novel cell death pathway. Proceedings of the National Academy of Sciences of the United States of America 113, E1565–E1574. 10.1073/pnas.1601375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Chang S, Qiao W, Luo Q, Chen Y, Jia Z, Coleman J, Zhang K, Wang T, Zhang Z, et al. (2021). Structural insights into outer membrane asymmetry maintenance in Gram-negative bacteria by MlaFEDB. Nature Structural & Molecular Biology 28, 81–91. 10.1038/s41594-020-00532-y. [DOI] [PubMed] [Google Scholar]
- Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, Reinisch KM, Melia TJ. (2019). ATG2 Transports lipids to promote autophagosome biogenesis. Journal of Cell Biology 218, 1787–1798. 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeow J, Chng SS. (2022). Of zones, bridges and chaperones - phospholipid transport in bacterial outer membrane assembly and homeostasis. Microbiology (Reading) 168, 10.1099/mic.0.001177. [DOI] [PubMed] [Google Scholar]
- Yu J, Li T, Dai S, Weng Y, Li J, Li Q, Xu H, Hua Y, Tian B. (2017). A tamB homolog is involved in maintenance of cell envelope integrity and stress resistance of Deinococcus radiodurans. Scientific Reports 7, 45929. 10.1038/srep45929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Wu S, Boehlein SK, McCarty DR, Song G, Walley JW, Myers A, Settles AM. (2019). Maize defective kernel5 is a bacterial TamB homologue required for chloroplast envelope biogenesis. Journal of Cell Biology 218, 2638–2658. 10.1083/jcb.201807166. [DOI] [PMC free article] [PubMed] [Google Scholar]