Abstract
Inflammatory bowel diseases (IBDs) are complex chronic disorders of the gastrointestinal tract with the following two subtypes: Crohn's disease and ulcerative colitis. Disease presentation and progression within and across IBDs, especially Crohn's disease, are highly heterogeneous in the location, severity of inflammation, intestinal stenosis and obstruction, and extraintestinal manifestations. Clinical classifications fail to accurately predict the disease course and response to therapies. To date, most IBD genetic associations are derived from individuals of European ancestries, leading to a limitation of the discovery and application of IBD genetics in the rest of the world populations. In this mini-review, we summarize the latest progress of genome-wide association studies of IBD across global ancestries especially the Chinese population, the similarities and differences in genetic architecture between European and East Asian ancestries, as well as, the clinical significances relevant to IBD genetic study.
Keywords: Chinese population, Crohn's disease, East Asian, inflammatory bowel disease, susceptible gene, ulcerative colitis
Introduction
Inflammatory bowel diseases (IBDs) are a group of chronic non-specific inflammatory diseases in the gastrointestinal tract, mainly including ulcerative colitis (UC) and Crohn's disease (CD). The incidence and prevalence of IBD are increasing globally, particularly in China, while the specific pathogenesis of IBD has not been fully elucidated. A series of studies have demonstrated that IBDs are caused by an abnormal immune response to the intestinal microbiota in genetically susceptible individuals.1,2 Genome-wide association studies (GWAS) have identified >240 genetic loci associated with human IBD. However, most IBD genetic associations have been derived using individuals from European populations, with only a few studies of much smaller sample sizes in non-European populations.3,4 In this review, we highlight the latest progress of GWAS of IBD across global ancestries especially in the Chinese population. Moreover, this mini-review also focuses on the similarities and differences in the genetic architecture between European and East Asian ancestries, and the clinical significances relevant to IBD genetic study.
Pathogenesis of IBD
The contemporary view of the pathogenesis of IBD has advanced rapidly, due to recent breakthroughs and emerging applications in human genetics, intestinal mucosal immunology, and microbiome study. A role for genetic risk factors in the development of IBD has been highlighted by the identification of susceptibility loci in GWAS.3, 4 In addition to the genetic contributions to IBD pathophysiology, the mucosal immune system and the commensal microecosystem may also substantially contribute to both the development and progression of IBD. Gut microbes, together with the intestinal epithelial barrier and a variety of immune cells in gut mucosa, form the complex intestinal microecosystem, which does not merely induce immune cell activation and differentiation, but also impacts intestinal homeostasis.5 Previous studies have shown that certain microbiota-derived antigens and metabolites play a vital role in inducing the differentiation of mucosal CD4+ T cells. Polysaccharides produced by Bacteroides fragilis act as a ligand for Toll-like receptor 2 on the surface of CD4+ T cells and participate in type 1 T helper cell (Th1) activation and differentiation.6 However, some intestinal commensal microbes (e.g. Clostridium nucleus, Peptostreptococcus) could induce mucosal CD4+ T cells to differentiate into regulatory T cells (Treg). Evidence also suggests that intestinal C-X3-C motif chemokine receptor 1 (CX3CR1+) phagocytic cells enable the balance between Th1 and Treg cell immune responses by the induction of interleukin 10 (IL-10) production through recognizing symbiotic antigens.7 Further, this process mainly depends on the cellular autophagy related proteins autophagy related 16 like 1 (ATG16L1) and nucleotide binding oligomerization domain containing 2 (NOD2). Consistently, dendritic cells in the intestinal mucosa of patients with ATG16L1 or NOD2 gene deficiency could restrain Treg cell differentiation and IL-10 secretion.8 Moreover, various environmental factors such as smoking, air pollution, hygiene habits, and psychological stress have been identified to be the important factors involved in the development and progression of IBD.2
Susceptible variants are associated with human IBD
Pervious twin studies in IBD have provided evidence of a strong genetic contribution, with heritability of liability estimates of 0.75 for CD and 0.67 for UC.9 More than 240 IBD-related susceptibility loci have been identified by genome-wide sequencing and GWAS, and most of them are co-susceptible to both CD and UC.1 An analysis of 15 IBD GWAS cohorts from European populations has shown that 110 of the 163 IBD loci are common susceptibility genes of CD and UC, which are related to innate and adaptive immunity, including cytokine signal transduction (e.g. IL-23R, IL-10) and immune signal transduction (e.g. interferon gamma (IFNG), Janus kinase 2 (JAK2) , tumor necrosis factor super family 18 (TNFSF18)).10 Specific susceptibility genes for CD are mainly concentrated on the IL-23 pathway (e.g. IL-23R, IL-12B, JAK2, and tyrosine kinase 2 (TYK2)). Several common mutations in IL-23R (e.g. Gly149Arg, Val362Ile, and Arg381Gln) have been found to be significantly associated with CD, but these susceptibility loci are ethnically different, especially the mutant Gly149Arg, which is the most common in the Han population.11 Perianal CD-associated variant rs4151651 in complement factor B (CFB) is a loss-of-function mutation that impairs its cleavage, activation of alternative complement pathway, and pathogen phagocytosis, thus suggesting that targeting the alternative complement pathway may be a novel therapeutic approach in the treatment of perianal CD.12 In addition, a missense variant in the autophagy gene ATG16L1 (rs2241880, Thr300Ala) is observed to be strongly associated with the incidence of CD. Besides, the T300A variant sensitizes ATG16L1 to caspase-3-mediated degradation, thereby suggesting a functional connection between CD, caspase activation, and autophagy.13 Consistently, mice lacking the Atg16l1 gene in intestinal epithelial cells have an exacerbated intestinal injury, characterized by crypt epithelial cell death, heightened inflammation, and decreased survival. However, anti-IFN-γ treatment could abrogate epithelial cell death in Atg16l1ΔIEC mice, suggesting that IFN-γ-targeted therapy may be appropriate for patients with CD with variants in ATG16L1.14
The susceptibility genes of UC are found to be mainly associated with epithelial barrier (e.g. hepatocyte nuclear factor 4 alpha (HNF4A), laminin subunit beta 1 (LAMB1), cadherin 1 (CDH1), and G protein subunit alpha 12 (GNA12)), T cell proliferation and activation (TNFSF14), type I interferon expression (raftlin family member 2 (RFTN2)), and the Toll-like receptor pathway (interferon regulatory factor 5 (IRF5)).11 Further analysis of major histocompatibility complex (MHC) region variation has pointed out that human leukocyte antigen (HLA) shows the strongest correlation with UC.15
Latest progress of genetic study on IBD in Chinese and other East Asian populations
Since most IBD genetic associations have been derived using individuals from European populations, with only a few studies of much smaller sample sizes in non-European populations,10 this strong bias toward European ancestry severely limits our insight into IBD biology and, further, decreases its application amongst most of the world's population. The first trans-ancestry association study of IBD has identified 38 risk loci, including the identified CD gene autophagy related 4B cysteine peptidase (ATG4B), which reinforces the importance of autophagy in CD pathogenesis. Likewise, the importance of epithelial barrier function in IBD pathogenesis is underscored by oncostatin M receptor (OSMR), which modulates a barrier-protective host response in intestinal inflammation. Moreover, many of the identified genes (e.g. lymphocyte antigen 75 (LY75), CD28, C-C motif chemokine ligand 20 (CCL20), NFKB inhibitor zeta (NFKBIZ), aryl hydrocarbon receptor (AHR), and nuclear factor of activated T cells 1 (NFATC1)) modulate the process of T cell activation and immune response. Five more new susceptibility loci (e.g. SHC adaptor protein 1 (SHC1), chromodomain Y like 2 (CDYL2)) for IBD have been identified in GWAS studies in Koreans.3, 16 Non-HLA region susceptibility genes specific to Asian UC patients have also been identified from samples from Japan and North India, including immunoglobulin receptors Fc gamma receptor IIa (FCGR2A), solute carrier family 26 member 3 (SLC26A3), and protein tyrosine phosphatase receptor type C (PTPRC).17, 18
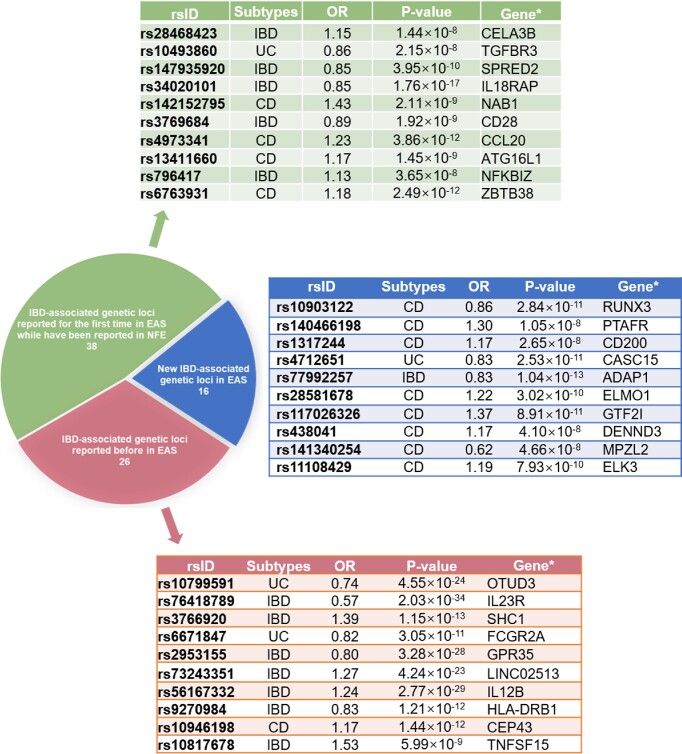
Recently, the largest reported IBD genetic study in China and other East Asian countries has been implemented and includes 5088 Chinese IBD patients and 6279 age- and sex-matched healthy donors, together with Korean and Japanese IBD genetic datasets, thus expanding the number of IBD patients from East Asian countries to 14 393, resulting in the largest IBD genetic study in the East Asian population.19 Most interestingly, this study identified a total of 80 IBD loci in East Asians alone (Fig. 1). Moreover, the study also discovered 81 new genetic loci associated with IBD across both East Asian and European ancestries, thus increasing the number of IBD-associated genetic loci to 320 by comparing 30 713 IBD cases and 338 106 healthy donors from the International IBD Genetics Union and FinnGen.19 Furthermore, Liu and co-authors compared the conditional effect sizes across East Asian and European ancestries for IBD putative causal variants at the locus level, and found that genetic effects for many IBD loci were consistent in both populations, with only a few loci as clear exceptions (e.g. TNFSF15, colony stimulating factor 2 receptor subunit beta (CSF2RB), and IL23R) (Table 1). Although both East Asian and European ancestries had largely consistent genetic effects in the MHC region, the MHC locus showed evidence of significant heterogeneity of the genetic effect across Chinese, Korean, and Japanese ancestries at the locus level. The observed heterogeneity does not necessarily suggest different biology within East Asian ancestries because the MHC locus is a highly complex locus with long-range linkage disequilibrium that spans several megabases. These data have shown that variances supported by IBD-associated loci differ across East Asian and European ancestries, which are, to a greater degree, driven by minor allele frequencies (MAFs) but less by the effect sizes (32% IBD associations have different MAFs, and 22% have different ORs). CD has a greater ancestral dependency than UC, with NOD2 and ATG16L1 as top CD drivers in European ancestries and TNFSF15 in East Asian ancestries, respectively. 19 Interestingly, ATG16L1 as a classical autophagy-regulating protein has been found to be down-regulated in different types of intestinal and accessory tissues in Chinese CD patients.20 Another proteomics study provided a comprehensive characterization of the preclinical inflammatory profile by analyzing plasma samples from individuals who developed UC later in life and identified a prediagnostic protein signature consisting of matrix metallopeptidase 10 (MMP10), C-X-C motif chemokine ligand 9 (CXCL9), CCL11, signaling lymphocytic activation molecule family member 1 (SLAMF1), CXCL11, and monocyte chemotactic protein 1 (MCP-1). 21 The high-throughput profiling of multi-omics analysis has found that fibroblast growth factor 19 (FGF19) and zinc finger and BTB domain containing 16 (ZBTB16) are positively correlated with microbial richness, facilitate a priori determination of optimal therapeutics for patients, and serve as targets for novel therapies. 22 In conclusion, these multi-omics data serve as a blueprint to figure out more variants in different populations and facilitate a priori determination of optimal therapeutics for patients.
Figure 1.
IBD-associated genetic loci in Chinese and other East Asian (EAS) populations. There are 80 genetic loci significantly associated with CD, UC, or both in the EAS population (P < 5 × 10−8). Among these 80 genetic loci, 16 genes are new IBD-associated loci, and 26 genes have been identified in the EAS population previously. A total of 54 genes were first reported in the EAS population, while 38 of these 54 loci have been reported in the non-Finnish European (NFE) population previously. The three tables show the most important IBD-associated loci based on the odd ratios (ORs) and P values. ORs and P values are from the inverse-variance-weighted fixed-effect meta-analysis (two-tailed) including all East Asian samples. *Nearest gene to the index variant.
Table 1.
Comparative genetic effect of putative causal variants across East Asian and European populations.
| RS_ID | Gene | POP | Phenotype | EUR_AF | EAS_AF | PIP | Effect_cond_EUR | SE_cond_EUR | Effect_cond_EAS | SE_cond_EAS | P_het | Sig_het |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs7556897 | CCL20 | EAS | CD | 0.3429 | 0.2024 | 0.409 | 0.0347 | 0.0172 | 1.93E-01 | 0.0315 | 1.08E-05 | adj.P < 0.05 |
| rs6856616 | LINC02513 | EAS | CD | 0.0596 | 0.2321 | 0.874 | 0.1109 | 0.0315 | 2.84E-01 | 0.0254 | 1.83E-05 | adj.P < 0.05 |
| rs7043505 | TNFSF15 | EAS | CD | 0.4503 | 0.6895 | 0.488 | 0.0310144 | 0.0271579 | 4.28E-01 | 0.0392266 | 9.28E-17 | adj.P < 0.05 |
| rs3812565 | CARD9 | EUR | CD | 0.3549 | 0.1885 | 0.81471455 | 0.178248 | 0.0163733 | 4.00E-03 | 0.0284994 | 1.15E-07 | adj.P < 0.05 |
| rs7915475 | AC067752.1 | EUR | CD | 0.3012 | 0.1468 | 0.52844728 | -0.105578 | 0.0182737 | 0.0274657 | 0.0374278 | 0.00140178 | adj.P < 0.05 |
| rs11195128 | DUSP5 | EAS | CD | 0.3201 | 0.1478 | 0.949 | 0.113 | 0.0172 | 2.82E-01 | 0.0348 | 1.39E-05 | adj.P < 0.05 |
| rs7307562 | LRRK2 | EUR | CD | 0.393 | 0.494 | 0.9999 | -0.0813516 | 0.0165517 | 0.00591389 | 0.0227686 | 0.0019344 | adj.P < 0.05 |
| rs9889296 | CCL2 | EUR | CD | 0.3231 | 0.5496 | 0.51160938 | -0.1523 | 0.0182 | -3.33E-02 | 0.023 | 4.96E-05 | adj.P < 0.05 |
| rs2427537 | ZBTB46 | EAS | CD | 0.4702 | 0.0437 | 0.967 | 0.0127391 | 0.0169923 | -2.21E-01 | 0.0552614 | 5.38E-05 | adj.P < 0.05 |
| rs12628495 | CSF2RB | EAS | CD | 0.0895 | 0.3393 | 0.519 | -0.0476638 | 0.0263475 | 2.51E-01 | 0.026149 | 9.03E-16 | adj.P < 0.05 |
| rs4655215 | RNF186 | EUR | UC | 0.291 | 0.47 | 1 | -0.133951 | 0.0186741 | -0.0273661 | 0.0291071 | 0.00205579 | adj.P < 0.05 |
The sample sizes used to derive ORs in East Asian and European populations were 22 828 and 40 266 for CD, and 22 318 and 45 975 for UC, respectively. Only non-Finnish European samples were used in the European analysis. Cochran's Q test (two-sided) was used for testing heterogeneity. We chose the number of putative causal variants tested for the correction such as an adjusted (adj.) P < 0.05 for CD or UC. EAS, East Asian population; EUR, European population.
Clinical significance of IBD genetic study
Genetic studies allow us to better understand the clinical significance of IBD, including the pathogenesis, disease risk, and classification. Based on a broad array of studies, available evidence has demonstrated that host genetics is associated with IBD development, particularly with disease behaviors, locations, and medical response. A previous study has reported that CD patients could be classified into three phenotypes (ileal, colonic, and ileocolonic lesions) according to three variants (NOD2, MHC, and macrophage stimulating 1 (MST1)).23 NOD2 has been found to act as a vital risk factor in the pathogenesis of CD, whose mutations are also associated with specific disease phenotypes (e.g. ileocoecal disease, ileocoecal resection, structuring, and perianal disease) in subsequent studies. Therapeutic drug monitoring in patients with a mutation in the NOD2 gene has also shown more frequent tumor necrosis factor (TNF) trough levels in the subtherapeutic range and lower anti-TNF trough levels compared to patients with no mutation in NOD2. In addition, these patients might require higher doses of anti-TNF agents in order to achieve sufficient anti-TNF trough levels.24 Anti-TNF therapies are the most widely used biologic drugs for treating IBD, but repeated or long-term administration could induce the formation of anti-drug antibodies, thus increasing the risk of treatment failure. In a GWAS of a population of European descent, the variant HLA-DQA1*05 has been observed to present a 2-fold increased risk of development of antibodies against infliximab and adalimumab in CD patients, regardless of concomitant immunomodulator use. Therefore, testing patients for HLA-DQA1*05 might help physicians to decide whether patients should receive anti-TNF and combination immunomodulator therapy.25 Recent studies identified four nudix hydrolase 15 (NUDT15) coding variants (p.Arg139Cys, p.Arg139His, p.Val18Ile, and p.Val18_Val19insGlyVal) that appear as a loss of nucleotide diphosphatase activity. NUDT15 inactivates thiopurine metabolites and decreases thiopurine cytotoxicity in vitro, and patients with defective NUDT15 alleles show excessive levels of thiopurine active metabolites and toxicity. These results indicate that NUDT15 variation plays a determinant part in thiopurine intolerance.26 Further studies in the Chinese population also confirmed that NUDT15 variants (p.Arg139Cys, p.Val18Ile, and p.Val18_Val19insGlyVal) are risk factors for thiopurine-induced leukopenia. Combined detection of the three variants could increase the predictive sensitivity of thiopurine-induced leukopenia and help to distinguish early leukopenia in heterozygotes.27 Therefore, NUDT15 variants are recommend to be detected before initiating thiopurine drugs in Chinese patients with IBD. Dose monitoring by NUDT15 variant detection is promising for future individualized therapy. A long duration of IBD may increase the risk in colorectal cancer. Whole-exome sequencing analyses of IBD and colorectal cancer have revealed that IBD-associated tumors have a different mutation spectrum. Several genes are mutated more frequently or uniquely in IBD-associated tumors, such as SRY-box transcription factor 9 (SOX9) and E1A binding protein p300 (EP300), which encode proteins in the wingless-type MMTV integration site (WNT) pathway, indicating an important role for WNT signaling in the development of colorectal tumors in IBD patients.28 Primary sclerosing cholangitis (PSC) is a highly comorbid phenotype with IBD. Of note, 6 of the 14 loci associated with PSC and IBD display strong evidence of a shared causal variant with UC, CD, or both (MST1, IL21, histone deacetylase 7 (HDAC7), SH2B adaptor protein 3 (SH2B3), CD226, and proteasome assembly chaperone 1 (PSMG1)). Although comorbid gastrointestinal inflammation seen in the majority of PSC patients cannot be fully explained by shared genetic risk, the biliary and intestinal inflammation seen specifically in PSC should be studied to advance our understanding of the disease and improve clinical outcome for patients with this devastating disorder.29
Conclusions and perspectives
This mini-review provides a breakthrough in the genetic characteristics of IBD that clarifies genetic variants associated with IBD using the largest samples to date from the East Asian population including Chinese IBD patients. Many new IBD-associated loci discovered from this study are driven by variants with elevated MAF in the East Asian population, which highlights the potential of recruiting global populations in genetics studies to identify new disease associations. This will help us to better understand the pathogenesis and provide potential precision medicine in these emerging regions with high incidence of IBD. Although the data have shed some light on improving the statistical power for the East Asian population compared with European studies, fine-mapping resolution and the ability to compare genetic effects across different ancestries are fairly limited, especially at loci hosting multiple independent associations. Additionally, a comparison of genetic effects can only proceed in variants shared across East Asian and European populations. It is unreliable to evaluate extremely rare putative causal variants for their causal roles in East Asian or European populations. Rapidly expanding omics analytical platforms and techniques represent a paradigm shift in molecular epidemiological research. Advances in multi-omics analysis pave the path for advances in molecular epidemiology of IBD, which may have important implications for the prediction of IBD onset and preventive strategies. Multi-omics analysis with genes is also needed to screen rare variants in non-European populations, particularly in Chinese IBD patients.
Acknowledgments
This work was partially supported by the Shanghai Hospital Development Center Foundation (Grant No. SHDC12022118) and the National Natural Science Foundation of China (Grant No. 91942312).
Contributor Information
Han Gao, Center for Inflammatory Bowel Disease Research and Department of Gastroenterology, The Shanghai Tenth People's Hospital, Tongji University, Shanghai 200072, China.
Zhanju Liu, Center for Inflammatory Bowel Disease Research and Department of Gastroenterology, The Shanghai Tenth People's Hospital, Tongji University, Shanghai 200072, China.
Conflict of interest
The authors have no conflicts of interest to disclose. In addition, as an Editorial Board Member of Precision Clinical Medicine, the corresponding author Zhanju Liu was blinded from reviewing and making decision on this manuscript.
References
- 1. de Lange KM, Moutsianas L, Lee JCet al. . Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–61. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Collaborators GBDIBD . The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990-2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol Hepatol. 2020;5:17–30. doi: 10.1016/S2468-1253(19)30333-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jung S, Ye BD, Lee HSet al. . Identification of three novel susceptibility loci for inflammatory bowel disease in Koreans in an extended genome-wide association study. J Crohn's Colitis. 2021;15:1898–907. doi: 10.1093/ecco-jcc/jjab060. [DOI] [PubMed] [Google Scholar]
- 4. Brant SR, Okou DT, Simpson CLet al. . Genome-wide association study identifies African-specific susceptibility loci in African Americans with inflammatory bowel disease. Gastroenterology. 2017;152:206–217.e2. e2. doi: 10.1053/j.gastro.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen H, Li H, Liu Z.. Interplay of intestinal microbiota and mucosal immunity in inflammatory bowel disease: a relationship of frenemies. Therap Adv Gastroenterol. 2020;13:175628482093518. doi: 10.1177/1756284820935188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Scott NA, Andrusaite A, Andersen Pet al. . Antibiotics induce sustained dysregulation of intestinal T cell immunity by perturbing macrophage homeostasis. Sci Transl Med. 2018;10, doi: 10.1126/scitranslmed.aao4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim M, Galan C, Hill AAet al. . Critical role for the microbiota in CX(3)CR1(+) intestinal mononuclear phagocyte regulation of intestinal T cell responses. Immunity. 2018;49:151–163.e5. e5. doi: 10.1016/j.immuni.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chu H, Khosravi A, Kusumawardhani IPet al. . Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–20. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gordon H, Trier Moller F, Andersen Vet al. . Heritability in inflammatory bowel disease: from the first twin study to genome-wide association studies. Inflamm Bowel Dis. 2015;21:1428–34. doi: 10.1097/MIB.0000000000000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jostins L, Ripke S, Weersma RKet al. . Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellinghaus D, Jostins L, Spain SLet al. . Analysis of five chronic inflammatory diseases identifies 27 new associations and highlights disease-specific patterns at shared loci. Nat Genet. 2016;48:510–8. doi: 10.1038/ng.3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Akhlaghpour M, Haritunians T, More Set al. . Genetic coding variant in complement factor B (CFB) is associated with increased risk for perianal Crohn's disease and leads to impaired CFB cleavage and phagocytosis. Gut. 2023;0:1–13. doi: 10.1136/gutjnl-2023-329689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Murthy A, Li Y, Peng Iet al. . A Crohn's disease variant in Atg16l1 enhances its degradation by caspase 3. Nature. 2014;506:456–62. doi: 10.1038/nature13044. [DOI] [PubMed] [Google Scholar]
- 14. Foerster EG, Tsang DKL, Goyal Set al. . ATG16L1 protects from interferon-gamma-induced cell death in the small intestinal crypt. Mucosal Immunology. 2023;16:135–52. doi: 10.1016/j.mucimm.2023.02.001. [DOI] [PubMed] [Google Scholar]
- 15. Naito T, Okada Y.. HLA imputation and its application to genetic and molecular fine-mapping of the MHC region in autoimmune diseases. Semin Immunopathol. 2022;44:15–28. doi: 10.1007/s00281-021-00901-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang SK, Hong M, Oh Het al. . Identification of loci at 1q21 and 16q23 that affect susceptibility to inflammatory bowel disease in Koreans. Gastroenterology. 2016;151:1096–9. doi: 10.1053/j.gastro.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 17. Charbit-Henrion F, Parlato M, Malamut Get al. . Intestinal immunoregulation: lessons from human mendelian diseases. Mucosal Immunol. 2021;14:1017–37. doi: 10.1038/s41385-021-00398-3. [DOI] [PubMed] [Google Scholar]
- 18. Snell A, Segal J, Limdi Jet al. . Inflammatory bowel disease in India: challenges and opportunities. Frontline Gastroenterol. 2021;12:390–6. doi: 10.1136/flgastro-2020-101500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu Z, Liu R, Gao Het al. . Genetic architecture of the inflammatory bowel diseases across East Asian and European ancestries. Nat Genet. 2023;55:796–806. doi: 10.1038/s41588-023-01384-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gao X, Sun R, Jiao Net al. . Integrative multi-omics deciphers the spatial characteristics of host-gut microbiota interactions in Crohn's disease. Cell Reports Medicin, 2023:4;101050., doi: 10.1016/j.xcrm.2023.101050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bergemalm D, Andersson E, Hultdin Jet al. . Systemic inflammation in preclinical ulcerative colitis. Gastroenterology. 2021;161:1526–39.doi: 10.1053/j.gastro.2021.07.026. [DOI] [PubMed] [Google Scholar]
- 22. Lee JWJ, Plichta D, Hogstrom Let al. . Multi-omics reveal microbial determinants impacting responses to biologic therapies in inflammatory bowel disease. Cell Host Microbe. 2021;29:1294–304.doi: 10.1016/j.chom.2021.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cleynen I, Boucher G, Jostins Let al. . Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet North Am Ed. 2016;387:156–67. doi: 10.1016/S0140-6736(15)00465-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schäffler H, Geiss D, Gittel Net al. . Mutations in the NOD2 gene are associated with a specific phenotype and lower anti-tumor necrosis factor trough levels in Crohn's disease. J Dig Dis. 2018;19:678–84. doi: 10.1111/1751-2980.12677. [DOI] [PubMed] [Google Scholar]
- 25. Sazonovs A, Kennedy NA, Moutsianas Let al. . HLA-DQA1*05 carriage associated with development of anti-drug antibodies to infliximab and adalimumab in patients with Crohn's disease. Gastroenterology. 2020;158:189–99. doi: 10.1053/j.gastro.2019.09.041. [DOI] [PubMed] [Google Scholar]
- 26. Moriyama T, Nishii R, Perez-Andreu Vet al. . NUDT15 polymorphisms alter thiopurine metabolism and hematopoietic toxicity. Nat Genet. 2016;48:367–73. doi: 10.1038/ng.3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chao K, Wang X, Cao Qet al. . Combined detection of NUDT15 variants could highly predict thiopurine-induced leukopenia in Chinese patients with inflammatory bowel disease: A multicenter analysis. Inflamm Bowel Dis. 2017;23:1592–9. doi: 10.1097/MIB.0000000000001148. [DOI] [PubMed] [Google Scholar]
- 28. Robles AI, Traverso G, Zhang Met al. . Whole-exome sequencing analyses of inflammatory bowel disease-associated colorectal cancers. Gastroenterology. 2016;150:931–43. doi: 10.1053/j.gastro.2015.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ji SG, Juran BD, Mucha Set al. . Genome-wide association study of primary sclerosing cholangitis identifies new risk loci and quantifies the genetic relationship with inflammatory bowel disease. Nat Genet. 2017;49:269–73. doi: 10.1038/ng.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]