Abstract
The tumor-intrinsic NOD-, LRR- and pyrin domain-containing protein-3 (NLRP3) inflammasome–heat shock protein 70 (HSP70) signaling axis is triggered by CD8+ T cell cytotoxicity and contributes to the development of adaptive resistance to anti–programmed cell death protein 1 (PD-1) immunotherapy by recruiting granulocytic polymorphonuclear myeloid derived suppressor cells (PMN MDSCs) into the tumor microenvironment. Here, we demonstrate that the tumor NLRP3-HSP70 axis also drives the accumulation of PMN-MDSCs into distant lung tissues in a manner that depends on lung epithelial cell Toll-like receptor 4 (TLR4) signaling, establishing a premetastatic niche that supports disease hyperprogression in response to anti–PD-1 immunotherapy. Lung epithelial HSP70-TLR4 signaling induces the downstream Wnt5a-dependent release of granulocyte colony-stimulating factor (G-CSF) and C-X-C motif chemokine ligand 5 (CXCL5), thus promoting myeloid granulopoiesis and recruitment of PMN-MDSCs into pulmonary tissues. Treatment with anti–PD-1 immunotherapy enhanced the activation of this pathway through immunologic pressure and drove disease progression in the setting of Nlrp3 amplification. Genetic and pharmacologic inhibition of NLRP3 and HSP70 blocked PMN-MDSC accumulation in the lung in response to anti–PD-1 therapy and suppressed metastatic progression in preclinical models of melanoma and breast cancer. Elevated baseline concentrations of plasma HSP70 and evidence of NLRP3 signaling activity in tumor tissue specimens correlated with the development of disease hyperprogression and inferior survival in patients with stage IV melanoma undergoing anti–PD-1 immunotherapy. Together, this work describes a pathogenic mechanism underlying the phenomenon of disease hyperprogression in melanoma and offers candidate targets and markers capable of improving the management of patients with melanoma.
INTRODUCTION
The establishment of the premetastatic niche has been implicated as a key step in metastatic cancer progression. Several tumor-derived soluble factors have been described that support various steps involved in creating the premetastatic niche (1, 2). However, conditions in the primary tumor microenvironment that drive the release of these tumor-derived soluble factors and how this process is regulated have remained less clear. Although inflammation has been suggested to be a trigger for both the expression and secretion of tumor-derived soluble factors, the exact underlying molecular mechanism regulating specific tumor-derived soluble factors has not been completely described. In addition, studies addressing how therapeutic interventions may modify the release of tumor-derived soluble factors and ultimately influence the premetastatic niche are lacking. In particular, it is not understood how the currently available checkpoint inhibitor immunotherapies affect tumor-derived soluble factors and the development of the premetastatic niche.
Prior studies have described the phenomenon of hyperprogressive disease (HPD) in select tumors upon treatment with checkpoint inhibitor immunotherapies. Although HPD has been defined on the basis of various parameters, the term generally describes unexpected rapid disease progression upon administration of an anti–programmed cell death protein 1 (PD-1) checkpoint inhibitor (3). Although limited to an estimated 10% of patients with cancer undergoing immunotherapy, HPD represents a devastating complication associated with this treatment modality (4). Prior studies in multiple solid tumor types have shown patients with HPD to have a median overall survival of 4.6 months relative to a median overall survival of 7.6 months in patients without HPD (5). This emphasizes the importance of understanding the underlying molecular pathogenesis of this phenomenon, as it may allow us to identify those tumors more susceptible to this complication and prevent its occurrence. Although some studies have identified immune cell populations or potential genes associated with HPD, several questions regarding the molecular mechanisms underlying these observations remain (6-9). As a result, there is still debate regarding HPD as a complication attributed to checkpoint inhibitor immunotherapies (10). Therefore, we considered the establishment of the distant premetastatic niche as a framework for understanding the mechanistic pathways that drive HPD in response to checkpoint inhibitor immunotherapy.
The Toll-like receptor 4 (TLR4) signaling pathway has been demonstrated to support the development of the premetastatic niche in the lung, leading to the establishment of pulmonary metastases in various malignancies (11). This has included a role for TLR4 in the induction of myeloid cell recruitment to the lung and the generation of hyperpermeable regions within the lung vasculature (11, 12). These roles in the development of the premetastatic niche are consistent with prior genetic studies correlating loss-of-function polymorphisms in TLR4 with improved clinical outcome in patients with metastatic melanoma (13). Despite these observations, the exact mechanistic role of TLR4 in supporting metastatic progression for specific tumor types remains to be fully elucidated.
The NOD-, LRR- and pyrin domain-containing protein 3 (NLRP3) inflammasome has been a focus of investigation associated with several inflammatory disorders and has been primarily studied in myeloid cell populations such as macrophages and dendritic cells (14,15). Upon activation in myeloid cells, NLRP3 oligomerizes with the apoptosis-associated speck-like protein (ASC) containing a caspase recruitment domain adaptor protein to generate a large molecular assembly that catalyzes caspase-1 activation and the downstream release of the proinflammatory cytokines, interleukin-1β (IL-1β) and IL-18. Although prior studies have described an association between the NLRP3 inflammasome with cancer invasion and metastasis, mechanistic insight into the role of tumor-intrinsic NLRP3 in these processes has remained limited (16-18). We recently reported a programmed cell death ligand 1 (PD-L1):NLRP3 signaling axis in tumors that responds to the activation of CD8+ T cell–mediated immunity by stimulating the recruitment of immunosuppressive polymorphonuclear myeloid–derived suppressor cells (PMN-MDSCs) (19, 20). This mechanism was found to drive adaptive resistance to anti–PD-1 immunotherapy in several predinical tumor models, and studies also supported the existence of this mechanism in human melanoma. Consistent with these findings, we also demonstrated that pharmacologic inhibition of the NLRP3 inflammasome augments the efficacy of anti–PD-1 therapy in an anti–PD-1 refractory autochthonous model of melanoma (19).
Although prior work has identified a role for PMN-MDSCs in establishing the premetastatic niche in the lung, the signaling pathways in primary tumors that ultimately regulate PMN-MDSCs accumulation in distant organs have remained less clear (21). Here, we have explored the role of the tumor-intrinsic NLRP3 inflammasome and its primary effector, heat shock protein 70 (HSP70), in cancer metastasis and investigated how this pathway can support HPD in response to anti–PD-1 immunotherapy. We further leveraged this pathway to investigate strategies to identify those patients at risk for developing HPD before initiating checkpoint inhibitor immunotherapy.
RESULTS
Tumor-intrinsic NLRP3 drives PMN-MDSC accumulation in distant tissues
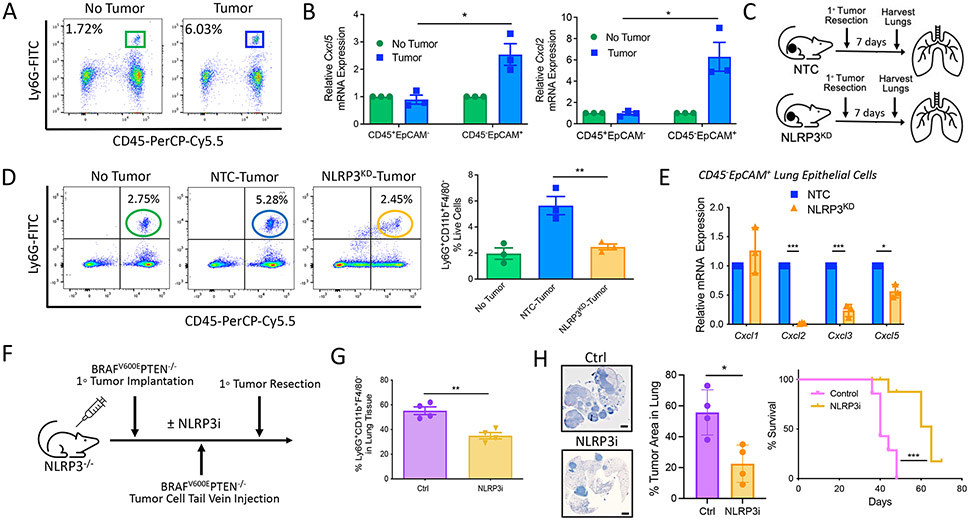
Prior studies have described elevated numbers of circulating neutrophils in tumor-bearing mice relative to healthy controls (22). Consistent with these findings, we also identified an increase in a CD45+CD11b+Ly6G+Ly6CloF4/80− cell population in the lung tissues of transgenic BRAFV600EPTEN−/− mice harboring primary melanomas relative to those BRAFV600EPTEN−/− mice with no active disease (Fig. 1A). Here, we refer to this population as PMN-MDSCs, as we have previously shown that these cells can suppress CD8+ T cell proliferation in vitro and in vivo while also diminishing responses to anti–PD-1 immunotherapy and supporting tumor progression in vivo (fig. S1) (19, 23). Using quantitative real-time polymerase chain reaction (qrt-PCR) analysis of sorted cell populations derived from the harvested lung tissues of BRAFV600EPTEN−/− mice, we further found that the C-X-C motif chemokine receptor 2 (CXCR2)–dependent chemokines, Cxcl5 and Cxcl2, were up-regulated by CD45−EpCAM+ lung epithelial cells only in tumor-bearing hosts (Fig. 1B and fig. S2A). Our previous work showed that the recruitment of PMN-MDSCs into primary tumor tissues was dependent on activation of the tumor-intrinsic NLRP3 inflammasome inducing a HSP70-TLR4-Wnt5a-CXCL5 signaling cascade (fig. S2B) (19). Given these prior findings, we performed syngeneic tumor experiments to evaluate the impact of genetically silencing NLRP3 in a BRAFV600EPTEN−/− melanoma cell line (BRAFV600EPTEN−/−-NLRP3KD) on the accumulation of PMN-MDSCs in lung tissues (Fig. 1C) (19). Primary tumors were resected before metastatic progression, and lung tissues were harvested for flow cytometry analysis 1 week later. This study showed that silencing NLRP3 expression in primary tumors led to a reduction in distant lung PMN-MDSCs to frequencies comparable to non–tumor-bearing mice (Fig. 1D). We were then interested in determining whether the NLRP3 inflammasome expressed by primary melanomas regulated gene expression in distant tissues. Therefore, single-cell suspensions were generated from harvested lung tissues and sorted by fluorescence-activated cell sorting (FACS) to quantitate the expression of CXCR2-dependent chemokines in CD45−EpCAM+ lung epithelial cells by qrt-PCR (fig. S2A). Consistent with our prior findings, genetic silencing of the tumor-expressed NLRP3 inflammasome suppressed Cxcl2, Cxcl3, and Cxcl5 expression in these remote epithelial tissues (Fig. 1E). NLRP3 inflammasome activity in myeloid cells has been implicated in several inflammatory diseases and may contribute to this observed phenomenon (24, 25). To formally exclude a role for myeloid NLRP3 inflammasome activity, additional experiments were performed in NLRP3−/− mice harboring primary BRAFV600E PTEN−/− melanomas that were further subjected to the delivery of BRAFV600EPTEN−/− melanoma cells by tail vein injection. These mice were treated with the pharmacologic NLRP3 inhibitor, MCC950, versus a vehicle control (Fig. 1F). These studies demonstrated that tumor NLRP3 inhibition suppressed the accumulation of PMN-MDSCs in the lungs, enhanced lung CD8+ T cell trafficking, inhibited the establishment of pulmonary metastases, and prolonged the survival of these NLRP3−/− mice (Fig. 1, G and H, and fig. S2C). These findings are further supported by additional experiments demonstrating no differences in primary BRAFV600E PTEN−/− melanoma growth, lung PMN-MDSC accumulation, or distant metastatic progression in NLRP3−/− versus wild-type mice (fig. S3). Together, these data indicate that the tumor-intrinsic NLRP3 inflammasome, rather than the host myeloid NLRP3 inflammasome, promotes the accumulation of PMN-MDSCs in distant lung tissues.
Fig. 1. Tumor-intrinsic NLRP3 promotes PMN-MDSC accumulation in distant lung tissues.
(A) Representative example of frequencies of live+CD45+CD11b+Ly6G+ Ly6CloF4/80− PMN-MDSCs in the lungs of tumor-bearing and non-tumor-bearing autochthonous BRAFV600EPTEN−/− mice. PerCP, peridinin-chlorophyll-protein; FITC, fluorescein isothiocyanate. (B) qrt-PCR analysis of Cxcl5 and Cxcl2 expression by CD45+EpCAM− and CD45−EpCAM+ cell populations derived from the lung tissues of non–tumor-bearing and tumor-bearing BRAFV600EPTEN−/− mice (n = 3). Statistical analysis was performed by two-way ANOVA followed by Sidak’s multiple comparisons test. (C) Experimental schematic to investigate the role of tumor NLRP3 on lung PMN-MDSC accumulation. KD, knockdown; NTC, nontarget control. (D) Flow cytometry analysis of PMN-MDSCs in the lung tissues of non–tumor-bearing, BRAFV600EPTEN−/− tumor–bearing, and BRAFV600EPTEN−/−−NLRP3KD tumor–bearing mice (n = 3). Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test. (E) qrt-PCR analysis of Cxcl1, Cxcl2, Cxcl3, and Cxcl5 expression in FACS-purified CD45−EpCAM+ lung epithelial cells derived from BRAFV600EPTEN−/− tumor–bearing and BRAFV600EPTEN−/−−NLRP3KD tumor–bearing mice (n = 3). (F) Experimental schematic to verify the role of tumor-intrinsic NLRP3 in metastatic progression. NLRP3i, NLRP3 inhibitor. (G) Flow cytometry analysis of PMN-MDSCs in lung tissues of BRAFV600EPTEN−/− tumor–bearing mice after treatment with either NLRP3i or vehicle control (Ctrl; n = 4). (H) Left: Low-magnification imaging and quantification of resected lung tissues after treatment with either NLRP3i or Ctrl. Images are shown at 4×. Scale bars, 2000 μm. Right: Survival curve analysis of BRAFV600EPTEN−/− tumor–bearing mice after treatment with either NLRP3i or Ctrl (n = 5). Statistical analysis for the right panel of (H) was performed by log-rank test. All two-group comparisons were analyzed using unpaired t tests. All data are representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05, **P < 0.005, and ***P < 0.0005.
HSP70 triggers a TLR4-Wnt5a signaling axis in lung epithelial tissues to promote PMN-MDSC accumulation in the lung
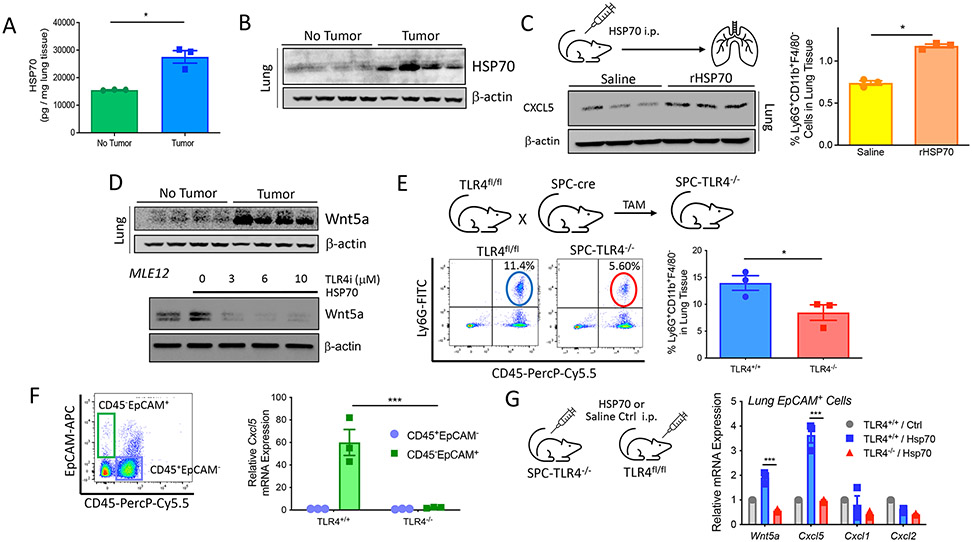
Despite prior reports indicating that the NLRP3 inflammasome can stimulate IL-1β production by melanoma cells, our previous studies have indicated that murine melanoma IL-1β concentrations are negligible relative to IL-1β production by myeloid cell populations in response to NLRP3 inflammasome activation (fig. S4A) (19, 26, 27). Although we were able to detect soluble HSP70 as a secretion product of human melanoma cell lines, we were not able to detect the production of active IL-1β (fig. S4, B and C). This is consistent with additional studies finding that tumor-intrinsic NLRP3 activation elicits the release of higher concentrations of HSP70 relative to IL-1β, the inverse of what is observed in myeloid cell populations in response to various stimuli (fig. S4, D and E). We have previously shown that the tumor-intrinsic NLRP3 inflammasome drives PMN-MDSC recruitment through an autocrine signaling pathway dependent on soluble HSP70 (19). However, we were also able to measure plasma concentrations of HSP70 in both the transgenic BRAFV600EPTEN−/− melanoma model and in patients with melanoma, suggesting that HSP70 could also have systemic effects (19). To address whether primary BRAFV600EPTEN−/− melanomas can affect HSP70 concentrations in distant lung tissues, we performed both enzyme-linked immunosorbent assays (ELISAs) and Western blot analysis, which revealed elevated HSP70 concentrations in the lungs of BRAFV600EPTEN−/− melanoma–bearing mice relative to hosts harboring no primary tumor (Fig. 2, A and B). We were unable to detect differences in HSP70 mRNA concentrations in lung tissues between tumor-bearing and non–tumor-bearing mice based on qrt-PCR analysis, indicating that detectable HSP70 protein was likely derived from the circulation (fig. S5A). To determine whether HSP70 could play the role of a soluble mediator in the induction of CXCR2-dependent chemokines in the distant lung, we delivered recombinant HSP70 (rHSP70) into non–tumor-bearing mice by intraperitoneal injection and measured CXCL5 expression by whole-lung tissue Western blot analysis and PMN-MDSC accumulation in the lung by flow cytometry. These studies showed the delivery of rHSP70 was sufficient to induce CXCL5 expression and promote the accumulation of PMN-MDSCs in lung tissues (Fig. 2C and fig. S5B). Our prior studies have demonstrated that HSP70 signals through TLR4 to induce Wnt5a-mediated CXCR2 chemokine expression in tumor tissues, and other studies have implicated lung TLR4 signaling in the up-regulation of CXCL5 by lung epithelial cells (19, 28). On the basis of these data, whole-lung tissue Western blots were performed to identify an up-regulation in Wnt5a expression by lung tissues in mice harboring a primary melanoma relative to non–tumor-bearing mice (Fig. 2D). Using a murine lung epithelial cell line (MLE12) along with a TLR4 inhibitor, we subsequently confirmed rHSP70 induced Wnt5a expression in a TLR4-dependent manner (Fig. 2D). On the basis of these cumulative findings, we hypothesized that tumor-derived HSP70 signals through TLR4 to promote PMN-MDSC trafficking to these distant tissues. To address the importance of TLR4 signaling in lung epithelial cells in driving this process, we crossed a surfactant protein C (SPC)–Cre–estrogen receptor (ER)T2 transgenic mouse harboring a tamoxifen-inducible Cre recombinase (Cre-ERT2) under the control of the human SFPTC promoter with a mouse harboring a Tlr4 conditional knockout allele (TLR4fl/fl) to generate SPC-Cre-ERT2/TLR4fl/fl offspring (SPC-TLR4−/−; fig. S5C) (29). Using the SPC-TLR4−/− transgenic mouse model and flow cytometry analysis, we found that lung epithelial cell–specific TLR4 deletion suppressed PMN-MDSC accumulation in the lung and increased CD8+ T cell infiltration into these tissues (Fig. 2E and fig. S5D). This observation correlated with a reduction in the expression of both Wnt5a and Cxcl5 by lung epithelial cells upon TLR4 ablation in SPC-TLR4−/− mice (Fig. 2F and fig. S5C). To confirm that these findings in the SPC-TLR4−/− transgenic mice were, in fact, dependent on systemic FISP70, we treated these mice with rHSP70 delivered by intraperitoneal injection and found both Wnt5a and Cxcl5 to be up-regulated only in lung epithelial cells expressing TLR4 (Fig. 2G). These cumulative data indicate that systemic HSP70 triggers a TLR4-Wnt5a signaling axis in lung epithelial tissues to drive PMN-MDSC accumulation in the lungs.
Fig. 2. HSP70 stimulates a TLR4-Wnt5a signaling axis in lung epithelial tissues to drive PMN-MDSC accumulation.
(A) HSP70 ELISA analysis of lung tissues harvested from non–tumor-bearing and tumor-bearing BRAFV600EPTEN−/− mice (n = 3). (B) HSP70 Western blot analysis of lung tissues harvested from non–tumor-bearing and tumor-bearing BRAFV600EPTEN−/− mice (n = 4). (C) CXCL5 Western blot analysis (left) and flow cytometry analysis of PMN-MDSCs (right) in lung tissues after intraperitoneal delivery of normal saline versus recombinant HSP70 (rHSP70; n = 3). i.p., intraperitoneal. (D) Top: Wnt5a Western blot analysis of lung tissues harvested from non–tumor-bearing and tumor-bearing BRAFV600EPTEN−/− mice (n = 4). Bottom: Wnt5a Western blot analysis of MLE12 lung epithelial cell lysates after rHSP70 treatment ± TLR4 inhibitor (TLR4i). (E) Flow cytometry analysis of PMN-MDSCs in the lung tissues of TLR4fl/fl (TLR4+/+) and SPC-TLR4−/− (TLR4−/−) transgenic mice (n = 3) after tamoxifen (TAM) treatment. (F) Cxcl5 qrt-PCR analysis of CD45+EpCAM− and CD45−EpCAM+ cells FACS-sorted from the lung tissues of tumor-bearing TLR4f/f and SPC-TLR4−/− transgenic mice (n = 3). The representative flow cytometry gating strategy is shown on the left. Statistical analysis was performed by two-way ANOVA followed by Sidak’s multiple comparisons test. APC, allophycocyanin. (G) Wnt5a, Cxcl5, Cxcl1, and Cxcl2 qrt-PCR analysis of CD45−EpCAM+ cells FACS-sorted from tumor-bearing TLR4f/f and SPC-TLR4−/− transgenic mice after intraperitoneal delivery of normal saline versus rHSP70 (n = 3). Statistical analysis was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. All two-group comparisons were analyzed using unpaired t tests. All data representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05 and ***P < 0.0005.
ATLR4-Wnt5a signaling axis in lung epithelial tissues promotes pulmonary metastatic progression
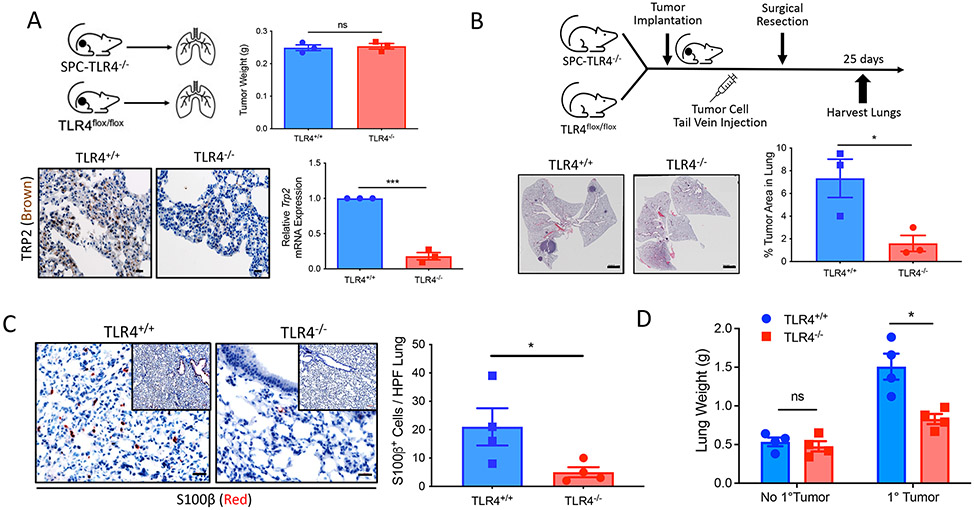
Prior studies have shown gain-of-function polymorphisms of TLR4 to associate with increased metastatic progression and inferior survival in patients with cancer (13, 30, 31). In addition, recent work has further described a role for PMN-MDSCs in establishing a microenvironment in tissues that is more conducive to metastatic progression (1). On the basis of these findings, we investigated the role of lung epithelial TLR4 signaling in metastatic progression to the lung. We therefore implanted the BRAFV600EPTEN−/− melanoma cell line by subcutaneous injection into syngeneic SPC-TLR4−/− and TLR4fl/fl control hosts and resected the primary melanoma tissues before harvesting the lungs 3 weeks later. Whereas the primary tumor tissues exhibited no difference in weight between SPC-TLR4−/− and TLR4fl/fl control mice, lung tissues derived from SPC-TLR4−/− mice were associated with diminished expression of the tyrosine-related peptide 2 (TRP2) melanoma antigen based on both TRP2 immunohistochemistry (IHC) and a previously developed TRP2-targeted qrt-PCR assay for quantifying melanoma metastases (Fig. 3A and fig. S5E) (32). The syngeneic BRAFV600EPTEN−/− melanoma model does not readily metastasize to distant tissues spontaneously. Therefore, to further study the metastasis of this model to the lung, we again implanted the BRAFV600EPTEN−/− melanoma cell line by subcutaneous injection in both SPC-TLR4−/− and TLR4fl/fl control mice to induce PMN-MDSC accumulation at distant tissue sites. These mice were then administered the BRAFV600EPTEN−/− melanoma cell line by tail vein injection, and lung tissues were harvested 25 days later for hematoxylin and eosin (H&E) microscopy; IHC analysis of S100β, a marker associated with melanoma; and lung weight measurements (Fig. 3, B to D). These results further supported the finding that the establishment of lung metastases is dependent on TLR4 signaling in lung epithelial cells and the presence of a primary tumor.
Fig. 3. The TLR4-Wnt5a signaling axis in lung epithelial tissues promotes pulmonary metastatic progression.
(A) Top: Primary BRAFv600EPTEN−/− tumor weight measurements after resection from TLR4f/f (TLR4+/+) and SPC-TLR4−/− (TLR4−/−) transgenic mice (n = 3). Bottom: Representative TRP2 IHC and TRP2 qrt-PCR analysis of lung tissues resected from TLR4f/f and SPC-TLR4−/− transgenic mice bearing BRAFV600EPTEN−/− tumors (n = 3). The IHC images are shown at 40×. Scale bars, 20 μm. ns, not significant. (B) Left: Representative low-magnification imaging of lungs resected from TLR4+/+ and SPC-TLR4−/− transgenic mice after BRAFV600EPTEN−/− tumor tail vein injection. Images are shown at 4×; Scale bars, 2000 μm. Right: Quantification of the area of tumor burden per whole lung (n = 3). (C) Left: Representative S100β IHC of lung tissues resected from TLR4+/+and SPC-TLR4−/− transgenic mice after BRAFV600EPTEN−/− tumor tail vein injection. Images are shown at 40×; Scale bars, 20 μm. Inset images are shown at 10×. Right: S100β-positive cells enumerated per lung based on IHC microscopy. HPF, high-power field. Analysis performed at 40×. (n = 4). (D) Lung weight measurements after resection from TLR4+/+ and SPC-TLR4−/− transgenic mice ± primary BRAFV600EPTEN−/− tumor (n = 4). g, gram. Statistical analysis was performed by two-way ANOVA followed by Sidak’s multiple comparisons test. All two-group comparisons were analyzed using unpaired t tests. All data are representative of two to three independent experiments and expressed as mean values ± SEM. ns, nonsignificant; *P < 0.05 and ***P < 0.0005.
The delivery of tumor cells through tail vein injection circumvents critical steps in the process of metastasis, representing a more artificial system for studying tumor progression. We therefore turned to tumor models capable of spontaneously metastasizing to the lung, including the BRAFV600ECDKN2A−/−PTEN−/− (YUMM1) melanoma model and a tumor model of a different histology, the E0771 breast adenocarcinoma model (33, 34). Our findings in each of these additional models further supported our prior data, demonstrating a critical role for lung epithelial TLR4 signaling in driving PMN-MDSC accumulation and metastatic progression to the lung and demonstrating that this phenomenon is not restricted to BRAFV600EPTEN−/− melanomas (figs. S6 and S7).
Prior studies have shown that the arrival of hematopoietic cell populations [hematopoietic progenitor cell (HPC)] expressing vascular endothelial growth factor receptor 1 (VEGFR1) in the lung and increased fibronectin expression by local fibroblasts contribute to the formation of a premetastatic niche that subsequently supports metastatic progression (35). Using multiparameter flow cytometry, we indeed observed an increase in a c-kit+CD133+CD34+VEGFR1+ HPC (VEGFR1+ HPC) population in mice harboring BRAFV600E PTEN−/− melanomas relative to non–tumor-bearing controls (fig. S8A). The accumulation of this VEGFR1+ HPC population was also found to be dependent on lung epithelial TLR4 expression and to be enhanced by tumor-intrinsic NLRP3 inflammasome activation (fig. S8). Additional qrt-PCR and IHC studies further demonstrated enhanced fibronectin expression in the lungs of mice bearing BRAFV600EPTEN−/− melanomas and for this up-regulation to be reversed in SPC-TLR4−/− mice, as well as mice treated with an inhibitor to the Wnt signaling regulator, porcupine O-Acyltransferase (PORCN) (ETC-159; fig. S9). Together, these results indicate that the tumor NLRP3 lung epithelial TLR4 axis represents an early step in premetastatic niche development.
Anti–PD-1 immunotherapy drives PMN-MDSC accumulation in the lung through the tumor-intrinsic NLRP3-HSP70 axis
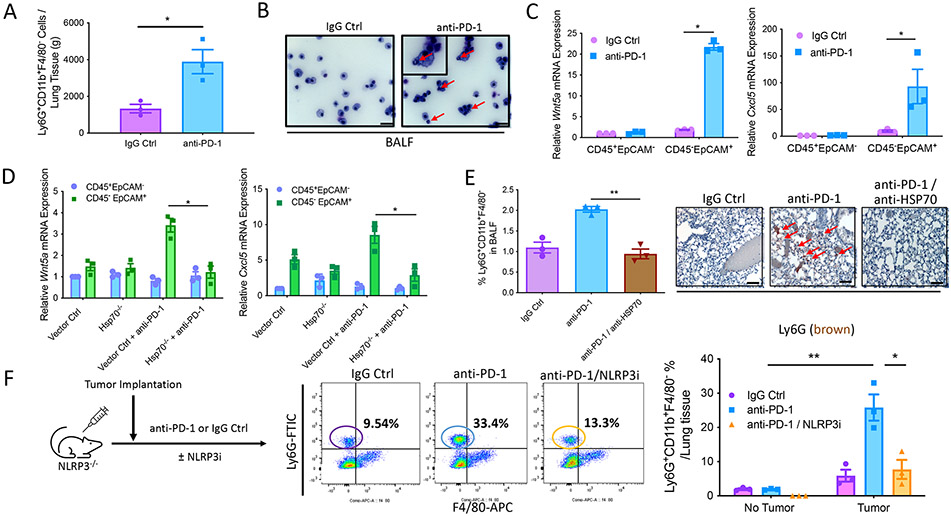
We previously demonstrated that anti–PD-1 immunotherapy promotes the recruitment of PMN-MDSCs into the primary tumor bed by inducing the activation of a tumor-intrinsic NLRP3 inflammasome–HSP70 signaling axis (fig. S2B) (19). Consistent with our previous data, we have further determined that anti–PD-1 immunotherapy drives PMN-MDSC accumulation in the lung based on tissue flow cytometry and bronchoalveolar fluid (BALF) cytology studies, an effect that was only observed in tumor-bearing mice (Fig. 4, A and B, and fig. S10A). These observations were also consistent with studies showing that anti–PD-1 immunotherapy induces the expression of both Wnt5a and Cxcl5 by CD45− EpCAM+ lung epithelial cells only in tumor-bearing mice (Fig. 4C and figs. S2A and S10B). To determine whether tumor-derived HSP70 is responsible for these observed alterations in response to PD-1 blockade, we knocked out HSP70 in the BRAFV600EPTEN−/− melanoma model using CRISPR-Cas9 (BRAFV600EPTEN−/−-HSP70−/−) (19). Although we observed an increase in both Wnt5a and Cxcl5 expression by CD45−EpCAM+ lung epithelial cells in response to anti–PD-1 immunotherapy in BRAFV600E PTEN−/− melanoma–bearing mice, this effect was eliminated in mice harboring BRAFV600EPTEN−/−-HSP70−/− melanomas (Fig. 4D). In line with these data, additional studies further showed an antagonistic antibody to HSP70 suppressed PMN-MDSC numbers in the lung when given in combination with anti–PD-1 immunotherapy (Fig. 4E and fig. S10C). This response to a HSP70 inhibitor was observed to also correlate with a more robust antitumor immune response when administered in combination with anti–PD-1 immunotherapy (fig. S10D). To further determine that anti–PD-1 immunotherapy also required the tumor NLRP3 inflammasome to drive PMN-MDSC accumulation in the lung, we treated BRAFV600EPTEN−/− melanoma–bearing NLRP3−/− mice with anti–PD-1 antibody alone or in combination with the NLRP3 inhibitor, MCC950. Although anti–PD-1 therapy strongly induced the accumulation of PMN-MDSCs into lung tissues in tumor-bearing NLRP3−/− mice based on flow cytometry analysis of both lung tissue and BALF, this effect was largely eliminated by NLRP3 inhibition (Fig. 4F and fig. S10E). Consistent with a role for HSP70-induced Wnt5a in the accumulation of PMN-MDSCs at distant sites, we found anti–PD-1 immunotherapy to enhance Wnt5a expression in lung epithelial cells and for this to be reversed by MCC950 (figs. S11, A and B). In addition, Wnt ligand inhibition phenocopied the observed effect of NLRP3 inhibition on PMN-MDSC recruitment into lung tissues in response to anti–PD-1 immunotherapy (fig. S11C). In line with our prior studies, the genetic deletion of TLR4 specifically in lung epithelial tissues also eliminates PMN-MDSC accumulation in response to anti–PD-1 immunotherapy (fig. S11D). Overall, these findings imply that NLRP3-dependent release of HSP70 from tumors undergoing anti–PD-1 immunotherapy promotes the accumulation of PMN-MDSCs in distant lung tissues through the induction of a local TLR4-Wnt5a signaling pathway.
Fig. 4. Anti–PD-1 immunotherapy promotes PMN-MDSC accumulation in the lung through the tumor-intrinsic NLRP3-HSP70 axis.
(A) Flow cytometry analysis of PMN-MDSCs in the lung tissues of BRAFV600EPTEN−/− tumor-bearing mice after treatment with either IgG isotype control (IgG Ctrl) or anti–PD-1 antibody (n = 3). Statistical analysis was performed by unpaired t test (B) Representative cytological analysis of the bronchoalveolar fluid (BALF) isolated from BRAFV600EPTEN−/− tumor-bearing mice after treatment with either IgG Ctrl or anti–PD-1. Red arrows indicate PMN morphology. Images are shown at 40×. Scale bars, 20 μm. Inset, 100×. (C) Wnt5a and Cxcl5 qrt-PCR analysis of CD45+EpCAM− and CD45−EpCAM+ cells isolated by FACS from lung tissues of BRAFV600EPTEN−/− transgenic mice after treatment with either IgG Ctrl or anti–PD-1 (n = 3). Statistical analysis was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. (D) Wnt5a and Cxcl5 qrt-PCR analysis of CD45+EpCAM− and CD45−EpCAM+ cells isolated by FACS from lung tissues of mice harboring either control BRAFV600EPTEN−/− tumors (Vector Ctrl) or BRAFV600EPTEN−/−HSP70−/− tumors (Hsp70−/−) ± anti–PD-1 treatment (n = 3). Statistical analysis was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. (E) Left: Flow cytometry analysis of PMN-MDSCs in the BALF of BRAFV600EPTEN−/− tumor-bearing mice after treatment with either IgG Ctrl, anti–PD-1, or anti–PD-1 + anti-HSP70 antibody (anti–PD-1/anti-HSP70; n = 3). Right: representative Ly6G IHC of lung tissues harvested from BRAFV600EPTEN−/− tumor–bearing mice after treatment with IgG Ctrl, anti–PD-1, or anti–PD-1/anti-HSP70. Red arrows indicate Ly6G staining. 40×. Scale bars, 20 μm. Statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test. (F) Flow cytometry analysis of PMN-MDSCs in the lung tissues harvested from BRAFV600EPTEN−/− tumor–bearing mice after treatment with IgG Ctrl, anti–PD-1, or anti–PD-1/NLRP3i (n = 3). Statistical analysis was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. All data are representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05 and **P < 0.005.
The tumor-intrinsic NLRP3-HSP70 axis can facilitate HPD in response to anti–PD-1 immunotherapy
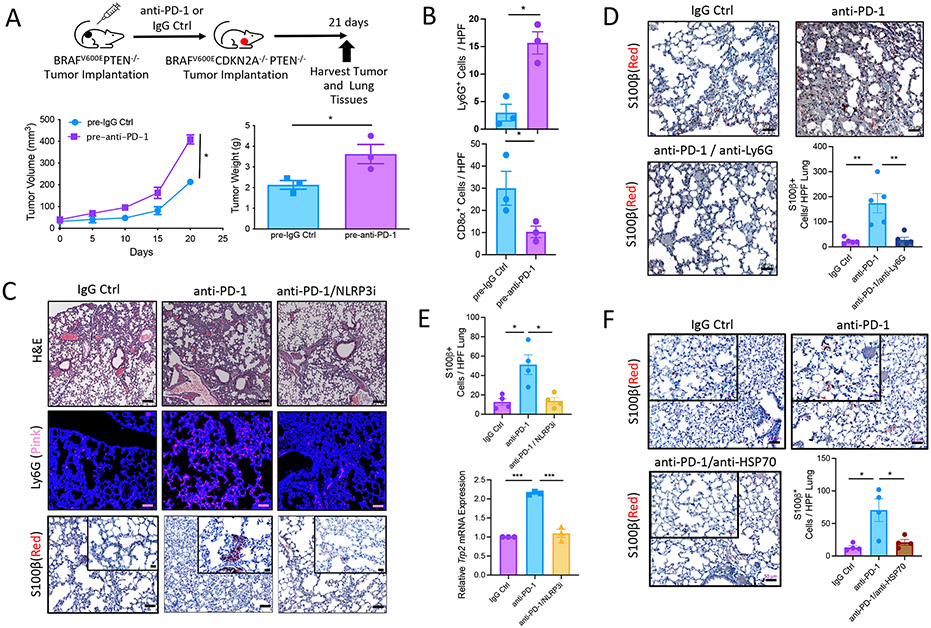
Prior studies have described HPD occurring in response to anti–PD-1 immunotherapy, a phenomenon estimated by some investigators to develop in about 10% of all solid tumors (5). In HPD, tumor burden can markedly increase in response to exposure to anti–PD-1 immunotherapy. Given our prior data describing a role for the tumor-intrinsic NLRP3-HSP70 signaling axis as an important driver of PMN-MDSC accumulation in distant lung tissues in response to anti–PD-1 immunotherapy, we hypothesized that this same tumor-intrinsic signaling pathway can establish an immunologic environment capable of facilitating HPD. To address this question, we first treated transplanted syngeneic BRAFV600E PTEN−/− melanomas with anti–PD-1 therapy. This approach suppressed primary BRAFV600EPTEN−/− melanoma growth as previously observed (36, 37). However, if we subsequently transplanted these same mice harboring anti–PD-1–treated BRAFV600EPTEN−/− melanomas with the YUMM1 melanoma model, we observed evidence of more rapid disease progression relative to those mice previously treated with immunoglobulin G (IgG) control antibody (Fig. 5A). Further IHC studies revealed that those progressive primary YUMM1 melanomas after pretreatment with anti–PD-1 were also found to harbor increased numbers of PMN-MDSCs and decreased frequencies of infiltrating CD8+ T cells (Fig. 5B and fig. S12A). On the basis of these results, we examined the impact of anti–PD-1 immunotherapy on the course of the autochthonous BRAFV600EPTEN−/− melanoma model and sought to determine how PMN-MDSCs influenced the behavior of these tumors. Although a modest therapeutic effect has been generally observed in response to anti–PD-1 immunotherapy in this model, we have uniformly observed disease escape and progression there-after (36, 37). Using the S100β melanoma antigen as a marker, we noted an increase in pulmonary metastasis after anti–PD-1 immunotherapy, a finding that correlated with increased numbers of PMN-MDSCs in the lungs (Fig. 5C). Consistent with our previous data that implicated PMN-MDSCs as playing a substantial role in promoting the development of a distant premetastatic niche, the depletion of circulating PMN-MDSCs nearly eliminated metastatic progression to the lung (Fig. 5D). Further in line with these results, additional studies conducted in the transgenic BRAFV600EPTEN−/− melanoma model demonstrated that the NLRP3 inhibitor, MCC950, suppressed PMN-MDSC populations in the lung while also inhibiting metastatic progression based on both S100β IHC and Trp2 qrt-PCR analysis (Fig. 5, C and E). Suppression in distant lung metastases based on both S100β and TRP2 IHC was also observed with an antagonistic anti-HSP70 antibody, consistent with a role for the NLRP3-HSP70 signaling axis in driving distant metastatic progression (Fig. 5F and fig. S12B). Together, these data suggest that the tumor NLRP3-HSP70 axis can promote metastatic progression in response to anti–PD-1 immunotherapy under select conditions.
Fig. 5. Tumor-intrinsic NLRP3-HPS70 axis promotes HPD in response to anti–PD-1 immunotherapy.
(A) BRAFV600ECDKN2A−/−PTEN−/− tumor volume and weight measurements after treatment of BRAFV600EPTEN−/− tumors with IgG Ctrl (pre-IgG Ctrl) or anti–PD-1 (pre–anti–PD-1; n = 3). (B) Quantification of Ly6G and CD8 IHC of primary BRAFV600ECDKN2A−/−PTEN−/− tumors from the same mice as in (A) (n = 3). (C) Representative hematoxylin and eosin (H&E) microscopy (10×; scale bar, 100 μm), Ly6G IF (20×; scale bar, 50 μm), and 5100β IHC (20×; scale bar, 50 μm. Inset, 40×; scale bar, 20 μm) of lungs derived from transgenic BRAFV600EPTEN−/− mice after IgG Ctrl, anti–PD-1, or anti–PD-1/NLRP3i therapy. (D) Representative images and quantification of S100β IHC of lung tissues derived from transgenic BRAFV600EPTEN−/− mice after IgG Ctrl or anti–PD-1 ± anti-Ly6G antibody depletion (n = 5). Images are shown at 20×. Scale bar, 50 μm. (E) Top: Quantitation of S100β IHC in lung tissue from experiment described in (C) (n = 3 to 4). Bottom: Trp2 qrt-PCR analysis of the lung tissues from experiment described in (C) (n = 3). (F) Representative images and quantification of S100β IHC of lung tissues derived from transgenic BRAFV600EPTEN−/− mice after IgG Ctrl, anti–PD-1, or anti–PD-1/anti-HSP70 treatment (n = 4). 20×; scale bar, 50 μm. For (D) to (F), statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test. All two-group comparisons were analyzed using unpaired t tests. All data are representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05, **P < 0.005, and ***P < 0.0005.
Genetic amplification of NLRP3 promotes HPD in response to anti-PD immunotherapy
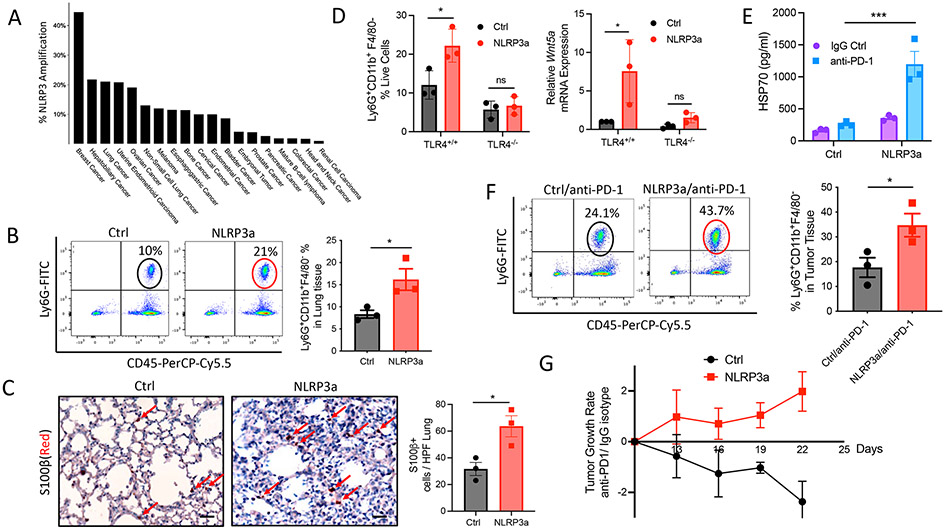
Many tumors exhibit elevated NLRP3 protein expression relative to their normal tissue counterparts (20). Amplification of NLRP3 has been identified in several solid tumor types (Fig. 6A) (38). We therefore hypothesized that genetic alterations that affect the activity of the NLRP3 inflammasome signaling pathway may influence whether a tumor exhibits resistance or HPD in response to anti–PD-1 immunotherapy. To emulate these conditions, we used CRISPR activation (CRISPRa) technology to engineer a BRAFV600E PTEN−/− melanoma cell line to exhibit transcriptional activation of the Nlrp3 gene (BRAFV600EPTEN−/−-NLRP3a; fig. S13A). Consistent with prior studies, Nlrp3 amplification was found to promote tumor growth in vivo as well as tumor cell proliferation and invasion in vitro, suggesting that the NLRP3 inflammasome can contribute to tumor-intrinsic properties of growth and progression (fig. S13, B and C). Additional in vivo studies also demonstrated that BRAFV600EPTEN−/−-NLRP3a melanomas exhibited increased PMN-MDSC accumulation in distant lung tissues (Fig. 6B). In contrast, CD8+ T cell frequencies in local tumor tissues were decreased (fig. S13D). These findings were also found to be associated with enhanced numbers of lung metastases based on S100β IHC (Fig. 6C). Consistent with our previous studies implicating a role for the TLR4-Wnt5a axis in establishing a premetastatic niche in the lung, the impact of tumor NLRP3 amplification on lung PMN-MDSC accumulation, as well as Wnt5a expression, was found to be dependent on lung epithelial TLR4 signaling (Fig. 6D). Additional work further demonstrated that NLRP3 amplification enhanced tumor-dependent HSP70 secretion, although IL-1β concentrations were not affected (Fig. 6E and fig. S13E). These findings are consistent with enhanced tumor PMN-MDSC accumulation and disease progression in response to PD-1 blockade in NLRP3-amplified tumors (Fig. 6, F and G, and fig. S13F). We found a similar response to anti–PD-1 immunotherapy in the E0771 breast cancer model, which we previously determined to express elevated NLRP3 concentrations de novo relative to the NLRP3-amplified BRAFV600E PTEN−/− melanoma model (figs. S7A and S14). These data suggest that tumors harboring genetic alterations associated with enhanced expression or activation of the NLRP3 inflammasome are more likely to exhibit resistance or HPD in response to anti–PD-1 immunotherapy.
Fig. 6. Genetic amplification of NLRP3 promotes HPD in response to anti–PD-1 immunotherapy.
(A) Incidence of NLRP3 amplification in human tumor types based on The Cancer Genome Atlas (TCGA). Data were visualized using cBioPortal. (B) Flow cytometry analysis of PMN-MDSCs in the lungs of Ctrl and NLRP3a tumor–bearing mice (n = 3). (C) Left: S100β IHC of lungs derived from Ctrl and NLRP3a tumor–bearing mice. 20×; scale bars, 50 μm. Red arrows, S100β-positive cells. Right: Quantification of S100β+ cells in the lungs of Ctrl and NLRP3a tumor–bearing mice (n = 3). (D) Left: Flow cytometry analysis of lung tissues resected from TLR4+/+ control and SPC-TLR4−/− mice harboring control BRAFV600EPTEN−/− tumors or BRAFV600EPTEN−/−-NLRP3a tumors (n = 3). Right: Wnt5a qrt-PCR analysis of lung tissues from TLR4+/+ control and SPC-TLR4−/− mice harboring control BRAFV600EPTEN−/− tumors or BRAFV600EPTEN−/−-NLRP3a tumors (n = 3). (E) ELISA analysis of plasma HSP70 concentrations in Ctrl and NLRP3a tumor–bearing mice after IgG Ctrl or anti–PD-1 treatment (n = 3). Statistical analysis was performed by two-way ANOVA followed by Tukey’s multiple comparisons test. (F) Flow cytometry analysis of PMN-MDSCs in the lungs of Ctrl and NLRP3a tumor–bearing mice after anti–PD-1 treatment (n = 3). (G) Ctrl and NLRP3a TGRs after IgG Ctrl or anti–PD-1 treatment. Tumor growth is expressed as a ratio of anti–PD-1 treatment to IgG Ctrl treatment. All two-group comparisons were analyzed using unpaired t tests. All data are representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05 and ***P < 0.0005.
It is noteworthy that we also found that BRAFV600EPTEN−/− melanomas exhibited an elevation in NLRP3 expression in response to anti–PD-1 therapy both in vitro and in vivo, an effect that was enhanced by interferon-γ (IFN-γ) and reversed by the depletion of CD8+ T cells (fig. S15) (19). This finding suggests that increased immunologic pressure may also select for tumors driven for more aggressive behavior through enrichment of tumor cell populations with higher expression of the NLRP3 inflammasome. These findings are reminiscent of those studies describing tolerogenic properties of longstanding exposure to IFN-γ and may reveal a mechanism by which immunoediting may drive tumor escape and metastasis (39).
The HSP70-TLR4 signaling axis in lung epithelial tissues supports primary tumor progression and anti–PD-1 immunotherapy resistance
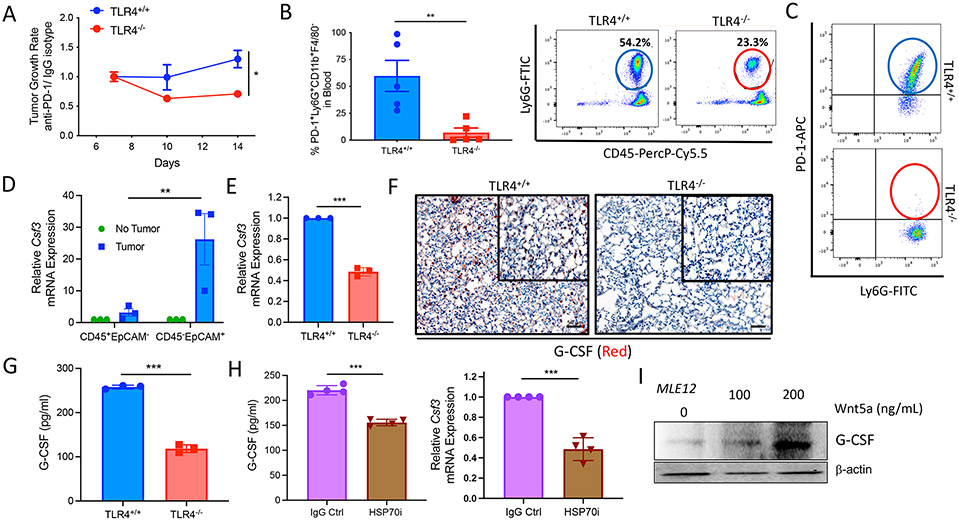
During the course of our studies, we found that primary melanomas responded more favorably to anti–PD-1 immunotherapy in SPC-TLR4−/− hosts relative to TLR4fl/fl control mice (Fig. 7A). This same observation was also made in the E0771 breast cancer model (fig. SI6, A to C). We also found that tumor-bearing TLR4n/n control mice exhibited an increase in the number of circulating Arg1-expressing PD-1+CD45+CD11b+Ly6G+Ly6CloF4/80− PMN-MDSCs in the blood relative to tumor-bearing SPC-TLR4−/− hosts (Fig. 7, B and C, and fig. S16D). Granulocyte colony-stimulating factor (G-CSF) expression by the lung epithelium has been shown to promote neutrophilic inflammation and to induce PD-1 expression by MDSCs (40-42). We found elevated G-CSF expression in the lung epithelium of tumor-bearing versus non–tumor-bearing hosts, and we observed increased frequencies of circulating PMN-MDSCs in TLR4fl/fl control mice versus SPC-TLR4−/− hosts (Fig. 7D and fig. S16B). We therefore hypothesized that the differences observed in the numbers of circulating PD-1+ PMN-MDSCs between SPC-TLR4−/− and TLR4fl/fl control mice are due to alterations in G-CSF expression in the lung epithelium, a finding that we confirmed based on G-CSF–targeted qrt-PCR, IHC, and ELISA results (Fig. 7, E to G). Because we have shown that tumor-derived HSP70 signals through TLR4 in the lung, we performed additional experiments showing that an antagonistic antibody to HSP70 also suppressed lung epithelial G-CSF expression (Fig. 7H). We previously demonstrated that TLR4 signaling can mediate Wnt5a up-regulation in tumor cells and lung epithelial cells (Fig. 2D) (19). Although we found that rHSP70 induced modest G-CSF up-regulation, our in vitro studies indicate that Wnt5a is the more immediate driver of G-CSF in the lung epithelium (Fig. 7I). Together, these results suggest that, in addition to generating a CXCR2 chemokine gradient to attract PMN-MDSCs to lung tissues, the HSP70-TLR4-Wnt5a signaling axis also drives G-CSF–dependent granulopoiesis of PMN-MDSC populations from the bone marrow (fig. S17, A and B). We further speculate that the circulating PD-1+ PMN-MDSC population may also serve as a sink to eliminate anti–PD-1 antibodies from the circulation, thus diminishing the therapeutic efficacy of this agent and contributing to an overall immunotherapy-resistant state.
Fig. 7. The HSP70-TLR4 signaling axis in lung epithelial tissues promotes primary tumor progression and anti–PD-1 immunotherapy resistance.
(A) Control BRAFV600EPTEN−/− TGRs in TLR4+/+ and SPC-TLR4−/− mice during IgG Ctrl or anti–PD-1 treatment. Growth is expressed as a ratio during anti–PD-1 therapy relative to IgG Ctrl (n = 6). (B) Flow cytometry analysis of PMN-MDSCs in the blood of BRAFV600EPTEN−/− tumor-bearing TLR4+/+ and SPC-TLR4−/− mice (n = 5). (C) Representative flow cytometry dot plot of PD-1 expression by circulating PMN-MDSCs in TLR4+/+ and SPC-TLR4−/− transgenic mice (n = 5 mice). (D) Csf3 qrt-PCR analysis of CD45+EpCAM− and CD45−EpCAM+ cells FACS-sorted from the lungs of non–tumor-bearing and BRAFV600EPTEN−/− tumor–bearing mice (n = 3). Csf3 encodes G-CSF. Statistical analysis performed by two-way ANOVA followed by Tukey’s multiple comparisons test. (E) Csf3 qrt-PCR analysis of lung tissues harvested from TLR4+/+ and SPC-TLR4−/− mice (n = 3). (F) Representative G-CSF IHC of lung tissues harvested from TLR4+/+and SPC-TLR4−/− mice (n = 3). 20×; scale bars, 50 μm. (G) G-CSF ELISA analysis of the plasma of TLR4+/+ and SPC-TLR4−/− tumor–bearing transgenic mice (n = 3). (H) Left: G-CSF ELISA analysis of the plasma of transgenic BRAFV600EPTEN−/− mice after anti–PD-1 ± HSP70i treatment (n = 4). Right: Csf3 qrt-PCR analysis of the lung tissues of tumor-bearing transgenic BRAFV600EPTEN−/− mice after anti–PD-1 ± HSP70i treatment (n = 4). (I) Representative G-CSF Western blot analysis of MLE12 lung epithelial cells after treatment with recombinant Wnt5a. All two-group comparisons were analyzed using unpaired t tests. All data are representative of two to three independent experiments and expressed as mean values ± SEM. *P < 0.05, **P < 0.005, and ***P < 0.0005.
Activation of the tumor-intrinsic NLRP3-HSP70 axis is associated with HPD in patients with melanoma undergoing anti–PD-1 immunotherapy
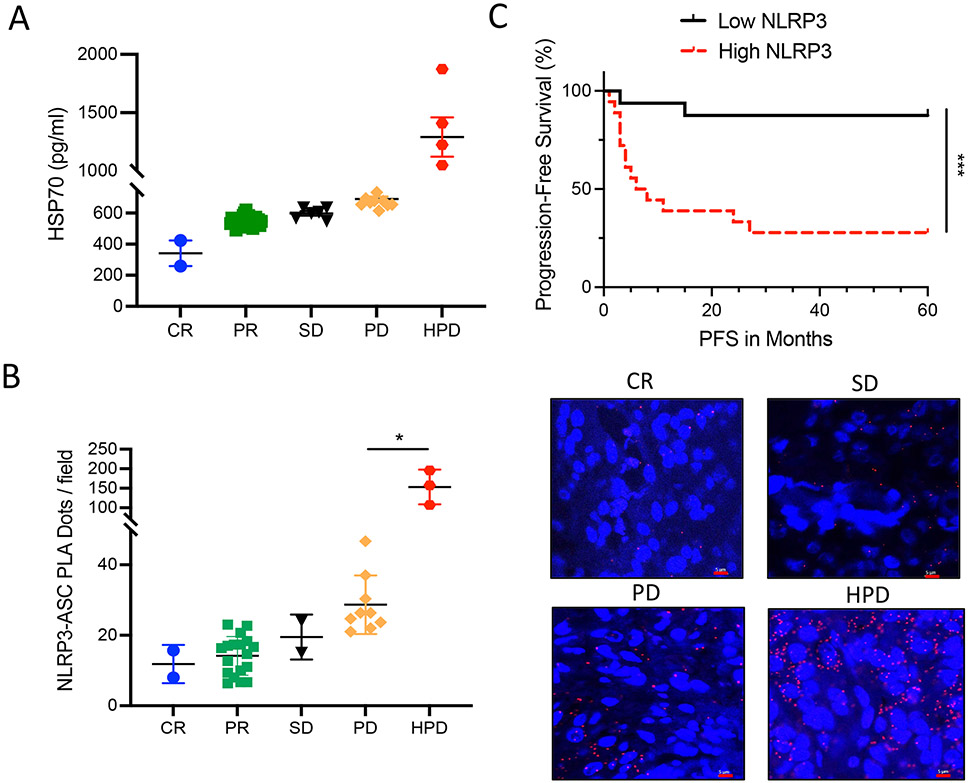
On the basis of our cumulative results suggesting a mechanistic link between the tumor-intrinsic NLRP3-HSP70 axis and metastatic progression in response to anti–PD-1 immunotherapy, we measured baseline plasma HSP70 concentrations in patients with advanced melanoma undergoing checkpoint inhibitor immunotherapy and evaluated these concentrations in terms of treatment response. Whereas previous studies have defined HPD based on differential tumor growth rate (TGR) measurements before and after the initiation of checkpoint inhibitor therapy, prior imaging studies for many of our patients to define TGR before treatment initiation were not available. As a result, we used RECIST1.1 criteria to define HPD based on a twofold increase in tumor burden within 12 weeks of anti–PD-1 initiation. This analysis demonstrated a significant increase in baseline HSP70 concentrations in those patients experiencing HPD relative to all other response groups (P < 0.0001) while undergoing anti–PD-1 immunotherapy (Fig. 8A). Consistent with our prior data, we found no relationship between plasma IL-1β concentration and HPD in this same cohort of patients with melanoma (fig. S18A). On the basis of these results, we sought to quantitate the degree of activation of the tumor-intrinsic NLRP3 inflammasome pathway in clinical tumor specimens to determine whether this may be an indicator of future behavior in response to anti–PD-1 immunotherapy. To accomplish this, we used a PCR-based proximity ligation assay capable of identifying and quantitating NLRP3-ASC molecular interactions in formalin-fixed, paraffin-embedded (FFPE)–based tumor specimens as a surrogate for the activity of the tumor NLRP3 inflammasome pathway (43). Using this approach, we were able to determine that tumors exhibiting evidence of enhanced NLRP3-ASC interactions at baseline were more likely to develop HPD (Fig. 8B). This finding is consistent with the plasma HSP70 data and further indicates that plasma HSP70 concentrations are a reflection of tumor-intrinsic NLRP3 activity. Further work also found that melanomas exhibiting enhanced NLRP3 activity based on the NLRP3-ASC proximity ligation assay, defined as NLRP3-ASC interactions above the median, were associated with a significant reduction in progression-free survival {P = 0.0005, hazard ratio (HR) = 0.12 [95% confidence interval (CI): 0.04 to 0.32]} and overall survival [P = 0.0496, HR = 0.16 (95% CI: 0.04 to 0.70); Fig. 8C and fig. S18B]. These studies in patients with melanoma support the importance of the tumor NLRP3-HSP70 pathway in establishing a distant premetastatic niche through PMN-MDSC recruitment and indicate that this mechanism represents an important underlying driver of HPD in response to checkpoint inhibitor immunotherapy (fig. S19). Together, these findings suggest that quantitative measurements of activation of the NLRP3-HSP70 axis represent a candidate strategy for predicting HPD in patients undergoing anti–PD-1 immunotherapy regimens.
Fig. 8. Activation of the tumor-intrinsic NLRP3-HSP70 axis is associated with hyperprogression in patients with melanoma undergoing anti–PD-1 immunotherapy.
(A) Baseline plasma HSP70 ELISA measurements in patients with advanced melanoma before initiating anti–PD-1 immunotherapy (n = 38). CR, complete responder; PR, partial responder; SD, stable disease; PD, progressive disease; HPD, hyperprogression disease. (B) NLRP3-ASC proximity ligation assay (PLA) analysis of baseline tumor tissues in patients with advanced melanoma before initiating anti–PD-1 immunotherapy (n = 34). Red dots represent NLRP3-ASC interactions. 40×; scale bars, 5 μm. For (A) and (B), statistical analysis was performed by one-way ANOVA followed by Tukey’s multiple comparisons test. (C) Progression-free survival (PFS) analysis of patients with advanced melanoma stratified according to baseline tumor NLRP3-ASC PLA scores. Low NLRP3, below the NLRP3-ASC PLA median. High NLRP3, equal to or above the NLRP3-ASC PLA median. Statistical analysis was performed by log-rank test. All data are representative of two independent experiments and expressed as mean values ± SEM. *P < 0.05 and ***P < 0.0005.
DISCUSSION
We previously described an adaptive resistance mechanism driven by the tumor-intrinsic NLRP3-HSP70 signaling axis that promotes the recruitment of PMN-MDSCs into the tumor microenvironment and suppresses local cytolytic CD8+ T cell activity in response to anti–PD-1 immunotherapy. In this study, we demonstrated that the tumor-intrinsic NLRP3-HSP70 signaling axis can also induce the accumulation of PMN-MDSCs in distant tissues, thereby establishing a niche facilitating metastatic disease progression. Using an inducible lung epithelial cell–specific TLR4 knockout mouse model, we show this effect to be dependent on tumor-derived HSP70 and its ability to trigger a distant TLR4-dependent Wnt5a-CXCL5/G-CSF signaling cascade capable of driving both PMN-MDSC granulopoiesis and recruitment into pulmonary tissues. Given that this mechanism is activated by anti–PD-1 immunotherapy, we conducted additional studies to determine whether this tumor-intrinsic NLRP3-HSP70-TLR4 axis could support the development of HPD. Predinical tumor models of both melanoma and breast adenocarcinoma, as well as clinical studies in patients with advanced melanoma, indicate that this pathway serves as a driver of HPD. This work is consistent with previous findings that have implicated various myeloid cell populations as playing a critical role in promoting metastatic disease progression, and it also highlights HSP70 as a previously undescribed soluble factor released by tumors that systemically drive PMN-MDSC accumulation (21). This relationship between circulating PMN-MDSCs or neutrophils and HPD has been described in previous studies, although the underlying mechanisms linking these phenomena have not been elucidated (44). Just as we have shown HSP70 to signal through TLR4 to up-regulate Wnt5a signaling through an autocrine tumor-intrinsic mechanism in a prior study, this current work shows that circulating HSP70 induces the TLR4-Wnt5a-CXCL5/G-CSF signaling cascade in distant lung epithelial cells (19). These findings are also consistent with prior studies implicating TLR polymorphisms and signaling to support the establishment of pulmonary metastases (13,45). The results presented here are interesting to consider in light of a recent retrospective study that found respiratory diseases and elevated neutrophil:lymphocyte ratios to be associated with disease progression in patients with stage IV melanoma undergoing anti–PD-1 immunotherapy (46). Overall, these findings suggest that there is a continuum between mechanisms of adaptive resistance and HPD in patients undergoing checkpoint inhibitor immunotherapy.
It is noteworthy that our work has identified two pathways by which the tumor NLRP3-HSP70-TLR4 pathway promotes the accumulation of PMN-MDSCs in distant tissues to establish the premetastatic niche necessary for metastatic progression. Tumor-dependent HSP70 drives the up-regulation of both G-CSF and CXCL5 by triggering TLR4 signaling in the lung epithelium. Together, this mechanism drives the release of PMN-MDSCs into the circulation while also establishing a chemokine gradient capable of recruiting these PMN-MDSCs into pulmonary tissues. These studies also revealed that this G-CSF–dependent mechanism induced PD-1 up-regulation by this circulating PMN-MDSC population, a finding suggesting the development of a potential sink for therapeutic anti–PD-1 antibodies and an additional mechanism by which the tumor NLRP3-HSP70-TLR4 axis can contribute to adaptive resistance to anti–PD-1 immunotherapy.
Previous studies have implicated the accumulation of a c-kit+CD133+CD34+VEGFR1+ HPC population, along with enhanced fibronectin expression in the extracellular matrix, as key components necessary for the evolution of the lung premetastatic niche (35). This study further demonstrates that the tumor NLRP3 inflammasome and lung epithelial TLR4 signaling both support the accumulation of this HPC population in pulmonary tissues. Similar to a previous study linking Wnt5a with enhanced fibronectin expression in the lung, this study also shows that lung epithelial TLR4 and Wnt ligand signaling both contribute to fibronectin accumulation within the lung extracellular matrix (47). Together, these data indicate that the tumor NLRP3-HSP70-TLR4 axis represents an early step in establishing the premetastatic niche and, therefore, highlights this pathway as a target for preventing metastatic progression.
Prior studies have concluded that NLRP3 activation in melanoma cells elicits the secretion of the IL-1β proinflammatory cytokine (26,27, 48). However, we found that the activation of the tumor-intrinsic NLRP3 inflammasome promoted the secretion of much higher concentrations of HSP70 relative to IL-1β based on both in vitro and in vivo studies in mice and humans. Our work indicates that IL-1β secretion in response to NLRP3 activation is more prominent relative to HSP70 secretion in myeloid cells such as dendritic cells, suggesting that the HSP70 mediator is more specific to tumor NLRP3 inflammasome activity than IL-1β. These observations indicate that HSP70 represents a promising pharmacologic target for suppressing metastatic progression and enhancing antitumor immunity. This is consistent with our preclinical data demonstrating that an antagonistic antibody specific to HSP70 robustly suppresses PMN-MDSC frequencies in tumor tissues and inhibits disease progression in a treatment-refractory autochthonous model of melanoma.
In addition to characterizing how the tumor-intrinsic NLRP3 inflammasome pathway contributes to metastatic disease progression and demonstrating that this process is driven by PD-1 blockade in various preclinical tumor models, we also present data illustrating that quantitative measures of this pathway correlate with HPD in patients with advanced melanoma undergoing anti–PD-1 immunotherapy. After defining HPD in our patient cohort as a twofold increase in overall tumor burden by week 12 of therapy, we found that baseline plasma concentrations of HSP70 were elevated in patients with HPD relative to all other response groups, including those with progressive disease. We then interrogated melanoma tissue specimens using a proximity ligation assay capable of quantifying NLRP3-ASC binding as a surrogate for activation of the NLRP3 inflammasome. This approach also verified that those tumors exhibiting enhanced NLRP3 activation at baseline developed HPD after the initiation of anti–PD-1 immunotherapy. Although these studies require validation in a larger cohort of patients, these findings (i) further substantiate the NLRP3-HSP70 signaling pathway as a driver of HPD in response to anti–PD-1 immunotherapy and (ii) indicate that assays to quantitate the degree of NLRP3-HSP70 signaling activation may be used to identify those patients at risk for developing HPD as a complication of anti–PD-1 checkpoint inhibitor immunotherapy. Although the definition and incidence of HPD associated with anti–PD-1 immunotherapy continues to be debated, it clearly represents a devastating side effect of our immunotherapy arsenal. By providing an underlying mechanism responsible for driving HPD, these areas of controversy can be resolved by further clinical studies in melanoma and other solid tumors.
It is tempting to speculate that genetic alterations that serve to enhance the activity of the tumor-intrinsic NLRP3 inflammasome will promote the described adaptive resistance mechanism and perhaps drive HPD in response to checkpoint inhibitor immunotherapies (49, 50). In fight of the percentage of tumors exhibiting NLRP3 amplification, this genetic alteration may contribute to the development of this phenotype in various solid tumors treated with anti–PD-1 immunotherapy (38). Our preclinical modeling studies presented here have confirmed that this mechanism could contribute to such a response to checkpoint inhibitor immunotherapy. Whether other somatic or germline mutations involving NLRP3 itself or its regulators may also enhance the activation of this pathway in response to checkpoint blockade remains unknown and is currently under investigation.
The current study is limited in that we only address how the tumor-intrinsic NLRP3-HSP70 pathway influences metastatic progression in the lung. Whether this same mechanism can generate a premetastatic niche in other distant organ tissues remains unclear and requires further experimental investigation. Although we have examined the role of the tumor-intrinsic NLRP3-HSP70 pathway in facilitating adaptive resistance in several preclinical tumor models, our studies investigating the role of this pathway in HPD have been limited to melanoma and breast cancer and require further clinical validation in a larger cohort of patients. How the tumor-intrinsic NLRP3 inflammasome integrates into other biological processes previously described to be associated with immunotherapy resistance, such as mesenchymal transformation, is also unclear and requires additional study.
Together, this work provides insight into the importance of the tumor-intrinsic NLRP3-HSP70 signaling axis in regulating PMN-MDSCs within distant tissues while also highlighting its critical role in modulating responses to immunotherapy. Additional studies are now ongoing to better understand the regulation of this pathway in tumors and how these mechanisms may dictate clinical responses to anti–PD-1 immunotherapy and whether targeting HSP70 may be an effective strategy for overcoming resistance to anti–PD-1 immunotherapy. Overall, these data describe an adaptive resistance mechanism to anti–PD-1 immunotherapy capable of supporting HPD in select settings. Future clinical studies are warranted to test whether markers associated with the NLRP3-HSP70 signaling axis can be used to predict those patients at risk for developing HPD in response to anti–PD-1 immunotherapy while also investigating the role of the tumor NLRP3-HSP70 signaling axis in other cohorts of patients with cancer.
MATERIALS AND METHODS
Study design
The primary objective of this work was to investigate the role of the tumor-intrinsic NLRP3 inflammasome and its downstream effector, HSP70, in PMN-MDSC–mediated premetastatic niche development in distant tissues and to determine whether this pathway may also contribute to the development of HPD in response to checkpoint inhibitor immunotherapy. The overall goal was to identify pharmacologic targets and biomarkers capable of improving our ability to detect those patients at risk for developing HPD and to improve our management of this complication associated with checkpoint inhibitor immunotherapy. The study included laboratory-controlled in vitro cell culture experiments, in vivo animal experiments, studies using clinical specimens derived from patients with stage IV melanoma undergoing anti–PD-1 immunotherapy, and in silico analysis of an existing tumor tissue database. The impact of the tumor-intrinsic NLRP3 inflammasome on PMN-MDSCs was measured using flow cytometry, cytology, IHC, immunofluorescence (IF), and qrt-PCR analysis; the impact on tumor progression and metastasis was measured on the basis of primary tumor size measurements, primary tumor weight, lung weight, H&E microscopy, qrt-PCR, IHC, cell proliferation, and cell invasion assays. Activation of the NLRP3 pathway was measured using Western blot, qrt-PCR, ELISA, and a NLRP3-ASC proximity ligation assay. Mice were treated with pharmacologic inhibitors of the NLRP3 inflammasome and HSP70, and PMN-MDSCs were depleted by antibody-dependent cellular cytotoxicity. The NLRP3-HSP70 signaling axis was manipulated using both CRISPR-Cas9 and CRISPRa. Tlr4 was genetically deleted specifically in lung epithelial cells, and Nlrp3 was deleted systemically using transgenic mouse systems. For animal experiments, an even distribution of male and female mice was randomly assigned into treatment groups. Sample size was determined on the basis of an a probability of 0.05, a power of 0.8, and an effect of at least 1.2× SD. Preliminary in vivo studies have shown that sample sizes need to be at least six or greater per group in syngeneic tumor model systems, whereas eight or greater are necessary in autochthonous tumor model systems. Fewer sample numbers per group were found to be sufficient depending on analysis end points. All experiments were performed independently at least three times except where noted. All experiments were conducted in a blinded fashion where analysis was independent of any intervention when feasible. All outliers have been included in the data presented.
Clinical samples
All patients provided written informed consent for use of biological specimens on an ongoing Institutional Review Board–approved clinical specimen acquisition protocol at Duke Cancer Institute designed to investigate immunotherapy resistance (NCT02694965). Baseline (week 0) FFPE tissues were collected from 34 patients with untreated stage IV melanoma, and baseline plasma samples were collected from 38 patients with untreated stage IV melanoma before initiating anti–PD-1 monotherapy at Duke Cancer Institute (table SI). Treatment response at week 12 and every 12 weeks there-after was determined on the basis of independent radiologic review of computed tomography (CT) imaging using RECIST1.1 criteria. HPD was defined as a ≥2-fold increase in total tumor burden by week 12 CT imaging.
Animal studies
C57BL/6J (C57, H-2b; stock number: 000664), B6.CgBraftm1Mmcm Ptentm1HwuTg(Tyr-cre/ERT2)13Bos/BosJ (BrafV600EPten−/−, H-2b; stock number 012328), B6(Cg)-Tlr4tm1.2Karp/J(TLR4fl/fl; stock number: 24872), B6.129S-Sftpctm1(cre/ERT2)B1h/J(SPC-CreERT2; stock number: 28054), and C57BL/6Tg(TcraTcrb)1100Mjb/J (OT-1, H-2b; stock number: 003831) mice were obtained from the Jackson Laboratories. M. Shinohara (Duke University) provided B6.129S6-Nlrp3tm1Bhk/J(NLRP3KO) mice. SPC-Cre-ERT2 mice were crossed with TLR4fl/fl mice to generate SPC-Cre-ERT2/TLR4fl/fl offspring (SPC-TLR4−/−). Conditional knockout of Tlr4 was confirmed by PCR per protocol provided by the Jackson Laboratory and by flow cytometry. Mice were treated with 100 μl of tamoxifen (20 mg/ml; Sigma-Aldrich, CAS no. 10540-29-1) by intraperitoneal delivery daily × 5 consecutive days for a total dose of 75 mg/kg (51). All experimental groups included randomly chosen littermates of both sexes, ages 6 to 10 weeks, and of the same strain. All animal experiments were performed on the basis of a protocol approved by the Institutional Animal Care and Use Committee at Duke University Medical Center.
Autochthonous tumor studies
B6.Cg-Braftm1Mmcm ptentm1Hwu Tg(Tyr-cre/ERT2 H-2b)13Bos/BosJ transgenic mice were subdermally injected with 4-hydroxytomoxfen (38.75 μg per mouse; Sigma-Aldrich, H6278–50MG CCF) to induce primary melanoma development at the base of the tail. Mice were randomly assigned to treatment cohorts until tumor volumes reached 64 mm3 (36, 37, 52). For select experiments, mice were treated with the following agents: NLRP3 inhibitor, MCC950 (Invivogen, inh-mcc) at 10 mg/kg i.p. injection every other day; anti–PD-1 antibody (Bio X Cell, BE0146) or rat IgG2a isotype control (Bio X Cell, BE0089) at 200 μg i.p. every 3 days; anti-Ly6G antibody (Bio X Cell, BE0075-1), initial dose at 200 μg per mouse followed by 100 μg per mouse per day × 2; and HSP70 monoclonal antibody (3A3, MA3-006) at 5 μg i.p. every 3 days. Primary tumor volumes were monitored by orthogonal caliper measurements every 3 days. Tumor volume was calculated according to the formula: cm3 = [(length, cm) × (width, cm)2]/2.
Syngeneic tumor studies
BRAFV600EPTEN−/− BRAFV600EPTEN−/−NTC, BRAFV600E PTEN−/−-NLRP3KD, BRAFV600EPTEN−/−-NLRP3a, BRAFV600E PTEN−/−-Ctrl, and BRAFV600ECDKN2A−/− PTEN−/− cell lines (0.5 × 105 to 1 × 106 cells) were implanted by subcutaneous injection into the base of the tail or chest of syngeneic C57BL/6 mice. The E0771 cell line (0.25 × 106 to 0.5 × 106) was injected into the mammary fat pad of syngeneic C57BL/6 mice in 0.1 ml of sterile saline using a 27-gauge needle. Tumor growth was monitored by caliper measurement every 3 days, and treatment was initiated when tumor volumes reached 64 or 90 mm3, depending on the study. Once tumor volume reached 500 to 600 mm3 in select experiments, mice were anesthetized using 2% isoflurane using an anesthesia mask, and primary tumor tissues were resected as previously described (53). Wounds were managed with surgical clips, and an antiseptic iodine solution was applied to the site. After surgery, animals were monitored under a heat lamp until fully recovered.
Cell lines and culture conditions
BRAFV600EPTEN−/−-NLRP3a and BRAFV600EPTEN−/−-Ctrl cell lines were generated using the established CRISPR amplification technique (54). The NLRP3 and control CRISPRa plasmids (Santa Cruz Biotechnology, sc-432122-ACT) were packaged into a lentiviral vector in human embryonic kidney 293T cells as previously prescribed (55). Clones were selected using a combination of antibiotics including puromycin (Santa Cruz Biotechnology, sc-108071), hygromycin B (Santa Cruz Biotechnology, sc-29067), and blasticidin S HCL (Santa Cruz Biotechnology, sc-495389). BRAFV600EPTEN−/−-NLRP3KD, BRAFV600EPTEN−/−-OVA, and BRAFV600EPTEN−/−-NTC cell lines were generated as described previously (19). The BRAFV600EPTEN−/− (male, BPD6) cell line was also generated previously (36). Stable cell lines were selected by puromycin resistance (Sigma-Aldrich, P8833). MLE12(CRL-2110) and BRAFV600ECDKN2A−/−PTEN−/−(YUMM1, CRL-3363) cell fines were purchased from the American Type Culture Collection. The E0771 mammary tumor cell fine was provided by D. McDonnell (Duke University). All BRAFV600EPTEN−/− cell fines and E0771 were maintained at 37°C in Dulbecco’s modified Eagle’s medium (Invitrogen) with 2 mM l-glutamine supplemented with 10% fetal bovine serum (FBS) and penicillin (100 U/ml). MLE12 cell fine was maintained in Dulbecco's medium : Ham's F12, 50:50 mix, insulin 0.005 mg/mL, transferrin 0.01 mg/mL, sodium selenite 30 nM, hydrocortisone 10 nM, beta-estradiol 10 nM, HEPES 10 mM, L-glutamine 2 mM (HITES) medium supplemented with 2% FBS. For select in vitro experiments, cell fines were treated with Wnt5a (100 to 200 ng/ml; R&D Systems/Bio-Techne, 645-WN-010), IFN-γ (100 ng/ml; BioAbChem, 42-IFNg), anti–PD-L1 antibody (1 to 2 μg/ml), TLR4 inhibitor (3 to 10 μM; Invivogen, tlrl-cli95), or MCC950 NLRP3 inhibitor (2.5 to 10 μM; Invivogen, inh-MCC). All cell fines were tested mycoplasma-free by Duke University Cell Culture Facility shared services.
Tumor cell CD8+ T cell coculture assays and CD8+ T cell proliferation assays
Naïve CD8+ T cells were isolated from the spleens of OT-1 transgenic mice by magnetic bead CD8 purification according to the manufacturer’s instructions (Miltenyi Biotec, 130-104-075) and activated with IL-2 (100 U/ml; PeproTech, 212-12) and SIINFEKL peptide (1 μg/ml; New England Peptide, BP10-915) for 3 days. Activated OT-1 CD8+T cells were incubated with BRAFV600EPTEN−/−-OVA cells in the presence and the absence of anti–PD-1 antibody (1 μg/ml) with or without IFN-γ antibody (MAB4851; 4 to 20 ng/ml) for 72 hours at a tumor cell:CD8+ T cell ratio of 1:5. For the ex vivo CD8+ T cell proliferation assay, splenocytes were isolated from IgG isotype control- and anti-Ly6G antibody–treated mice. Then, single cells were labeled with carboxyfluorescein succinimidyl ester (CFSE; Thermo Fisher Scientific, C34554) and cultured in 96-well flat-bottom plates for either 3 or 6 days at 37°C in RPMI 1640 medium supplemented with 10% FBS, penicillin, and streptomycin. Cells were harvested, and activated CD8+ T cells were quantitated on the basis of flow cytometry. For in vitro PMN-MDSC suppression T cell proliferation assays, naïve CD8+ T cells were purified from lymph nodes, labeled with CFSE, and stimulated with anti-CD3/anti-CD28 beads (Thermo Fisher Scientific, 1116D) at a 1:1 bead to cell ratio, in the presence or the absence of PMN-MDSCs (1:3 T cell/PMN-MDSC ratio) for 4 days. Cells were harvested on day 4, and activated CD8+ T cells were quantitated on the basis of flow cytometry.
Cell invasion and proliferation assays
A Matrigel invasion chamber with an 8-μm transwell membrane (Corning, 354480) was used to examine the relative invasion properties of cell fines. BRAFV600EPTEN−/−-NLRP3a and BRAFV600E PTEN−/−-Ctrl cell fines (5 × 104 cells) were seeded in the upper chamber in serum-free conditions, and FBS was added to the lower chamber. After 48 hours, transwell inserts were washed and cells were fixed with methanol and stained with crystal violet [1% (w/v)] for 10 min before microscopic quantification. For cellular proliferation assays, cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (Promega, G4000) as per the manufacturer’s instructions. Absorbance was measured at 590 nm using a Tecan plate.
Tumor and lung tissue cell isolation
Tumors were resected at a similar size and mechanically disaggregated by a gentleMACS dissociator (Miltenyi Biotec) and then digested with serum-free RPMI 1640 containing collagenase IV (1 mg/ml; Sigma-Aldrich), hyaluronidase (0.1 mg/ml; Sigma-Aldrich), and deoxyribonuclease (20 U/ml; Sigma-Aldrich) on a shaker at 250 rpm at 37°C for 30 min (36). Resected lung tissues were mechanically separated by gentleMACS (lung mode) and enzymatically digested using the same enzyme digestion solution and the same conditions for 20 min. Cell suspensions were filtered using a 70-μm filter, washed with flow buffer (phosphate-buffered saline, 2 mM EDTA, and 2% FBS), and red blood cells were lysed using lysis buffer (Sigma-Aldrich) for 10 min at room temperature.
RNA isolation and qrt-PCR analysis
Small representative tissue specimens were harvested from tumors and lungs and stored in RNAlater at −80°C. Tissues were lysed in RLT buffer by using a gentleMACS dissociator (Miltenyi Biotec), and cell fines were lysed in RLT buffer and stored at −80°C. Lung tissue–derived cells were sorted by FACS based on their surface marker expression and lysed in RLT buffer before storing at −80°C. Total RNA was isolated by the RNeasy Plus Mini Kit (QIAGEN, 74134), and RNA was quantified by NanoDrop. RNA (500 to 1000 ng) was used in cDNA synthesis (iScript Reverse Transcription Supermix, Bio-Rad, 1708841). qrt-PCR was performed using an ABI7500 Real-Time PCR system (Life Technologies). All qrt-PCR reactions were performed using validated primers and SsoAdvanced Universal SYBR Green Super Mix (Bio-Rad, 1725271) or SsoAdvance Universal Probes Supermix (Bio-Rad, 1725281). For developing a Trp2 qrt-PCR assay to measure melanoma metastases in lung tissues, 1 × 102 to 1 × 106 BRAFV600E PTEN−/− melanoma cells were admixed with a single-cell suspension generated from a single lung lobe and processed for qrt-PCR analysis using Trp2-specific TaqMan probes. Relative Trp2 mRNA expression was correlated with the number of admixed melanoma cells, and both high sensitivity and specificity were achieved. All data were normalized to Actb expression and relative gene expression was quantitated on the basis of the 2-ΔΔCt method. Relative mRNA expression of the following genes were measured: Arg1, Csf3, Cxcl1, Cxcl2, Cxcl3, Cxcl5, Hsp70, Il1b, Fn1, Spp1, Tgfb, Tgfbr2, Trp2, and Wnt5a. The primer sequences for these genes are located in table S2.
Western blot analysis
Tissues or cells were lysed in NP-40 lysis buffer (Sigma-Aldrich) supplemented with a complete protease inhibitor and phosphatase inhibitor (Roche). After lysing in Laemmli sample buffer, equal volumes of lysates were separated using 10 or 15% SDS–polyacrylamide gel electrophoresis and then transferred to a polyvinylidene difluoride membrane (Bio-Rad Laboratories Inc., 162017). After blocking for 30 min in tris-buffered saline (TBS) containing 0.1% Tween 20 and 5% milk, the membranes were probed with various primary antibodies followed by horseradish peroxidase (HRP)–conjugated secondary antibodies. Primary antibodies included anti–β-actin mouse monoclonal (1:3000, clone c4; Santa Cruz Biotechnology, sc-47778), anti-NLRP3 rabbit monoclonal (1:1000, clone D4D8T; Cell Signaling Technology, 15101S; 1:1000, clone EPR23073-96; Abcam, 270499), anti–Caspase-1–p20 mouse monoclonal (1:500, clone Casper-1; Adipogen, AG-20B-0042-C100), anti-HSP70 mouse monoclonal (1:1000, clone C92F3A-5; Santa Cruz Biotechnology, sc-66048), anti-CXCL5 goat polyclonal (1:1000, clone P50228; R&D Systems, AF433), anti-Wnt5a mouse monoclonal (1:1000, clone A5; Santa Cruz Biotechnology, sc-365370), anti–G-CSF rabbit monoclonal (1:1000, clone ABAC-3; Bosterbio, M02280-1), and anti–IL-1β mouse monoclonal (1:1000, clone 3A6; Cell Signaling Technology, 12242). Immunoreactivity was visualized using chemiluminescence substrate (Thermo Fisher Scientific, 34095/34075) and imaged by a ChemiDoc XRSplus System (Bio-Rad). All uncut Western blot images are presented in data file S1.
Enzyme-linked immunosorbent assay
CXCL5, HSP70, G-CSF, and IL-1β concentrations in culture supernatants and mouse plasma samples were determined by appropriate mouse ELISA kits (CXCL5: R&D Systems, MΧ000; HSP70: R&D Systems, DYC1663-2; G-CSF: R&D Systems, DY414-05; IL-1β: R&D Systems, DY401-05). Human plasma HSP70 and IL-1β concentrations were measured in a blinded fashion using the human DuoSet assay (R&D Systems, HSP70: DY1663, IL-1β: DY201-05). All ELISAs were performed according to the manufacturer’s protocol.
Flow cytometry analysis
Cells (1 to 2 × 106) were diluted in 150 μl of flow buffer per well of V bottom culture plates, incubated with the LIVE/DEAD Fixable Violet Dead Cell Stain Kit (0.5 μl/liter × 106 cells; Thermo Fisher Scientific, L34964A) on ice for 20 min, washed twice, and incubated with Fc block (2 μg/ml; anti-CD16/CD32) before staining with appropriate conjugated antibodies. Cells were washed, resuspended in 2% paraformaldehyde for 15 min at 4°C, and analyzed using a BD FACSCanto flow cytometry system. Compensation was performed using fluorescence minus one controls. Cells were characterized using the following combinations of cell surface markers after gating on viable single-cell populations. All antibodies were obtained from commercial vendors and used at 1 μg/1 × 106 cells: anti-Mouse CD11b, phycoerythrin (PE) conjugated (clone: MIH5; BD Pharmingen, 558091); anti-Mouse Ly6G-GRl, fluorescein iso-thiocyanate (FITC) conjugated (clone: RB6-8C5; BD Pharmingen, 5532127); anti-Mouse Ly6G, FITC conjugated (clone: 1A8; BioLegend,127605); anti-Mouse Ly-6G, FITC conjugated (clone: 1A8-Ly6g; Thermo Fisher Scientific, 11-9668-82), anti-Mouse F4/80, allophycocyanin (APC) conjugated (clone: BM8; BD Pharmingen, 560408); anti-Mouse CD45, PerCP-Cy5.5 conjugated (clone: 145-201; BD Pharmingen, 551163); anti-Mouse CD326/Ep-CAM, FITC conjugated (clone: G8.8; BioLegend.118207); anti-Mouse CD326/Ep-CAM, APC conjugated (clone: G8.8; BD Biosciences, 563478); anti-Mouse CD90.2, FITC conjugated (clone: 53-2.1; BD Biosciences, 553003); anti-Mouse CD8α, brilliant violet (BV) 510 conjugated (clone: 53-6.7; BD Pharmingen, 563068); anti-Mouse CD3e, PerCP-Cy5.5 conjugated (clone: 145-201; BD Pharmingen, 551163); anti-Mouse CD8α, APC conjugated (clone: 53-6.7; BD Biosciences, 553035); anti-Mouse PD-1/CD279, PE conjugated (clone: 29F.1A12; BD Biosciences, 568250); anti-Mouse Ly6C, PE-Cy7 conjugated (clone: AL-21; BD Pharmingen, 560593); anti-Mouse VEGFR1, PE conjugated (clone: 141522; Novus Biologicals, FAB4711P); anti-Mouse CD133, PE-Cy7 conjugated (clone: 315-201; BioLegend, 141209); anti-Mouse CD34, FITC conjugated (clone: RAM34; BD Biosciences, 560238); and anti-Mouse CD117/c-kit, PerCP-Cy5.5 conjugated (clone: 2B8; BD Biosciences, 560557). Cell populations were characterized on the basis of the following marker profiles: Ly6G+ PMN-MDSCS: CD45+CD11b+Ly6GhiLy6CloF4/80−; tumor-associated macrophages: CD45+ CD11b+Ly6G−F4/80+; type II lung epithelial cells: CD45–CD90.2−EpCAM+; CD8+ T cells: CD45+CD3ε+CD8α+CD44+/−; and bone marrow–derived hematopoietic cells: c-kit+VEGR1+CD133+CD34+. FACS was performed using a Beckman Astrios Cell Sorter at cell densities of 1 × 106 cells/ml in 1% RPMI supplemented with penicillin/streptomycin. Flow cytometry data were analyzed using FlowJo software vl0.3.
IHC and IF analysis
Paraffin sections (5 μm) from primary melanomas and lung tissues were processed using standard protocols for IHC and IF staining. Tissues were permeabilized by incubation in 0.4% Triton X-100 in TBS for 20 min. The following primary antibodies were incubated for 18 hours at 4°C: anti-Wnt5a, mouse monoclonal (1:200, clone: A-5; Santa Cruz Biotechnology, 365370), anti-Ly6G, rabbit monoclonal (1:100, clone: EPR22909-135; Abcam, ab238132), S100β, rabbit polyclonal (1:500 or 1:250, clone: 6285; Novus Biologicals, NBP1-87102), anti-NLRP3, rabbit polyclonal (1:500, clone: 114548; Novus Biologicals, NBP2-12446), anti–G-CSF, rabbit monoclonal (1:250, clone ABAC-3; Bosterbio, M02280-1), anti-fibronectin, mouse monoclonal (1:200, clone: TV-1; Novus Biologicals, NBP2-32849), and rabbit-specific HRP/AEC IHC Detection Kit–Micro-polymer (Abcam, ab236468). Anti-rat polymers (1:2000) were used as secondary antibodies and the chromogen detection system for IHC. For IF, goat anti-rabbit Alexa Fluor 564 (1:3000) and goat anti-mouse Alexa Fluor 488 (1:3000) were incubated at room temperature for 1 hour. Sections were imaged with an Axio Imager upright microscope. PMN-MDSC quantitation by Ly6G staining and CD8+ T cell quantitation by CD8α staining were performed at ×20 magnification. Six to eight random fields were averaged per section over three to four sections per specimen. Area calculations for lung tumor burden based on S100β staining was performed at ×20 magnification of two to three sections per specimen and quantified (tumor burden area/total lung area) using ImageJ software.
Proximity ligation assay
Human tumor tissues (5 μm) were deparaffinized, rehydrated, and subjected to antigen retrieval using standard procedures. Tissues were permeabilized by incubation in 0.4% Triton X in TBS for 20 min and incubated with a blocking solution containing 0.1% bovine serum albumin in TBS and 0.05% Tween solution for 30 min at room temperature. Slides were then incubated overnight with a cocktail of human NLRP3/NALP3 (amino acids 540 to 689) antibody (1:200, 0.2 mg/ml; R&D Systems, AF7010) and anti-human ASC and mouse monoclonal antibody (1:200, 0.2 mg/ml; Santa Cruz Biotechnology, sc-514414) at 4°C. Then, a mixture of 1× DuoLink in situ proximity ligation assay probe anti-mouse PLUS and 1× Duolink proximity ligation assay probe anti-goat MINUS (Sigma-Aldrich, DU092001-30RXN) was added to the section and incubated for 1 hour in a humidity chamber preheated to 37°C. Ligase solution was added to each sample and incubated for an additional 30 min in a humidity chamber at 37°C. Amplification solution (35 μl) was added to each slide and incubated for 100 min in the humidity chamber at 37°C. Last, mounting medium with 4’,6-diamidino-2-phenylindole was added, and a coverslip was placed. Images were taken by a SP5 Leica confocal microscope. ImageJ software was used to quantify fluorescent spots in a blinded fashion based on three fields per tissue section at ×40 magnification and averaged over two tissue sections per tissue sample.
The Cancer Genome Atlas data analysis
NLRP3 amplification was quantitated in different cancer types using The Cancer Genome Atlas (TCGA) database. Data were visualized using cBioPortal for Cancer Genomics. Analysis based on 2922 samples in the Pan-Cancer Analysis of Whole Genomes (International Cancer Genome Consortium/TCGA) (56).
Statistics
All raw, individual-level data are presented in data file S2. Specific statistical tests are reported in the figure legends. GraphPad Prism 9 Windows version was used for all statistical analyses. Normal data distribution was verified using the D’Agostino-Pearson normality test. Unpaired t tests were used to compare mean differences between control and treatment groups. One- or two-way analyses of variance (ANOVAs) followed by Tukey’s multiple comparisons test or Sidak’s multiple comparisons test, respectively, were performed to analyze data containing three or more groups. Progression-free and overall survival in patients with stage IV melanoma undergoing anti–PD-1 immunotherapy was analyzed using a log-rank test. All tests were two sided with P values reported. A P value of less than 0.05 was considered significant. All quantitative data are presented as the means ± SEM.
Supplementary Material
Acknowledgments:
We would like to thank M. Shinohara (Duke University) for providing the NLRP3−/− mouse strain and D. McDonnell (Duke University) for providing the E0771 breast cancer cell line for our studies. We would also like to thank K. Johnson, C. A. Wiggs, and E. Bolch on the Duke Melanoma Clinical Research Team for assistance in obtaining outside melanoma tissues specimens.
Funding:
This work was supported, in part, by a NIH R37CA249085 (to BAH.), a 2021 Conquer Cancer-Bristol-Myers Squibb Advanced Clinical Research Award in Immune Checkpoint Inhibitor Therapy (to B.A.H.), the Ross Bierkan Melanoma Research Fund (to B.A.H., A.K.S.S., and G.M.B.), a Duke University Health Scholar Award (to B.A.H.), a Duke Strong Start Award (to B.A.H.), a Merck & Co. Pre-Clinical Award (to B.A.H.), a NIH R37CA249085-02S1 (to B.T.), a Damon Runyon Physician Scientist Award (to N.C.D.), and a NIH F32CA247067 (to M.P.P.).
Footnotes
Competing interests: B.A.H. receives research funding from Merck & Co., Tempest Therapeutics, Leap Therapeutics, Sanofi, and Exicure Therapeutics; is a consultant/advisory board member for G1 Therapeutics; and has received honoraria from Novartis, Merck & Co., and HMP Education. B.A.H. and B.T. are inventors on patent application (DU7650PROV) submitted by Duke University that covers the tumor-intrinsic NLRP3 inflammasome signaling pathway as a genetic and functional biomarker for immunotherapy response. A.K.S.S. receives research funding from Bristol-Myers Squibb, Ideaya, ImmunoCore, Merck & Co, Nektar Therapeutics, and Replimmune and serves as a consultant/advisory board member for lovance, Novartis, Pfizer, and Regeneron. G.M.B. receives research funding from Istari Oncology, Delcath, Oncosec Medical, Replimmune, and Checkmate Pharmaceuticals. The other authors declare that they have no competing interests.
Data and materials availability: All data associated with this study are in the paper or the Supplementary Materials. All material transfer agreement requests should be sent to C. Bradney, Ph.D., Director, Office of Research Contracts, Duke University (curtis.bradney@duke.edu).
REFERENCES AND NOTES
- 1.Liu Y, Cao X, Characteristics and significance of the pre-metastatic niche. Cancer Cell 30, 668–681 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Peinado H, Zhang H, Matei IR, Costa-Silva B, Hoshino A, Rodrigues G, Psaila B, Kaplan RN, Bromberg JF, Kang Y, Bissell MJ, Cox TR, Giaccia AJ, Erler JT, Hiratsuka S, Ghajar CM, Lyden D, Pre-metastatic niches: Organ-specific homes for metastases. Nat. Rev. Cancer 17, 302–317 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Champiat S, Ferrara R, Massard C, Besse B, Marabelle A, Soria J-C, Ferté C, Hyperprogressive disease: Recognizing a novel pattern to improve patient management. Nat. Rev. Clin. Oncol 15, 748–762 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Ferrara R, Mezquita L, Texier M, Lahmar J, Audigier-Valette C, Tessonnier L, Mazieres J, Zalcman G, Brosseau S, Le Moulec S, Leroy L, Duchemann B, Lefebvre C, Veillon R, Westeel V, Koscielny S, Champiat S, Ferté C, Planchard D, Remon J, Boucher M-E, Gazzah A, Adam J, Bria E, Tortora G, Soria J-C, Besse B, Caramella C, Hyperprogressive disease in patients with advanced non-small cell lung cancer treated with PD-1/PD-L1 inhibitors or with single-agent chemotherapy. JAMA Oncol. 4, 1543–1552 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champiat S, Dercle L, Ammari S, Massard C, Hollebecque A, Postel-Vinay S, Chaput N, Eggermont A, Marabelle A, Soria J-C, Ferté C, Hyperprogressive disease is a new pattern of progression in cancer patients treated by anti-PD-1/PD-L1. Clin. Cancer Res 23, 1920–1928 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Kato S, Goodman A, Walavalkar V, Barkauskas DA, Sharabi A, Kurzrock R, Hyperprogressors after immunotherapy: Analysis of genomic alterations associated with accelerated growth rate. Clin. Cancer Res 23, 4242–4250 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamada T, Togashi Y, Tay C, Ha D, Sasaki A, Nakamura Y, Sato E, Fukuoka S, Tada Y, Tanaka A, Morikawa H, Kawazoe A, Kinoshita T, Shitara K, Sakaguchi S, Nishikawa H, PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. U.S.A 116, 9999–10008 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang DH, Chung C, Sun P, Lee DH, Lee S-I, Park D, Koh JS, Kim Y, Yi HS, Lee JE, Circulating regulatory T cells predict efficacy and atypical responses in lung cancer patients treated with PD-1/PD-L1 inhibitors. Cancer Immunol. Immunother 71, 579–588 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Russo GL, Moro M, Sommariva M, Cancila V, Boeri M, Centonze G, Ferro S, Ganzinelli M, Gasparini P, Huber V, Milione M, Porcu L, Proto C, Pruneri G, Signorelli D, Sangaletti S, Sfondrini L, Storti C, Tassi E, Bardelli A, Marsoni S, Torri V, Tripodo C, Colombo MP, Anichini A, Rivoltini L, Balsari A, Sozzi G, Garassino MC, Antibody-Fc/FcR interaction on macrophages as a mechanism for hyperprogressive disease in non-small cell lung cancer subsequent to PD-1/PD-L1 blockade. Clin. Cancer Res 25, 989–999 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Matos I, Martin-Liberal J, García-Ruiz A, Hierro C, Ochoa de Olza M, Viaplana C, Azaro A, Vieito M, Braña I, Mur G, Ros J, Mateos J, Villacampa G, Berche R, Oliveira M, Alsina M, Elez E, Oaknin A, Munoz-Couselo E, Carles J, Felip E, Rodón J, Tabernero J, Dienstmann R, Perez-Lopez R, Garralda E, Capturing hyperprogressive disease with immune-checkpoint inhibitors using RECIST 1.1 criteria. Clin. Cancer Res 26, 1846–1855 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Hiratsuka S, Watanabe A, Sakurai Y, Akashi-Takamura S, Ishibashi S, Miyake K, Shibuya M, Akira S, Aburatani H, Maru Y, The S100A8–serum amyloid A3–TLR4 paracrine cascade establishes a pre-metastatic phase. Nat. Cell Biol 10, 1349–1355 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Hiratsuka S, Ishibashi S, Tomita T, Watanabe A, Akashi-Takamura S, Murakami M, Kijima H, Miyake K, Aburatani H, Maru Y, Primary tumours modulate innate immune signalling to create pre-metastatic vascular hyperpermeability foci. Nat. Commun 4, 1853 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gast A, Bermejo JL, Claus R, Brandt A, Weires M, Weber A, Plass C, Sucker A, Hemminki K, Schadendorf D, Kumar R, Association of inherited variation in Toll-like receptor genes with malignant melanoma susceptibility and survival. PLOS ONE 6, e24370 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Booshehri LM, Hoffman HM. CAPS and NLRP3. J. Clin. Immunol 39, 277–286 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanson KV, Deng M, Ting JP-Y, The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol 19, 477–489 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moossavi M, Parsamanesh N, Bahrami A, Atkin SL, Sahebkar A, Role of the NLRP3 inflammasome in cancer. Mol. Cancer 17, 158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Kong H, Zeng X, Liu W, Wang Z, Yan X, Wang H, Xie W, Activation of NLRP3 inflammasome enhances the proliferation and migration of A549 lung cancer cells. Oncol. Rep 35, 2053–2064 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Wang H, Luo Q, Feng X, Zhang R, Li J, Chen F, NLRP3 promotes tumor growth and metastasis in human oral squamous cell carcinoma. BMC Cancer 18, 500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Theivanthiran B, Evans KS, DeVito NC, Plebanek M, Sturdivant M, Wachsmuth LP, Salama AK, Kang Y, Hsu D, Balko JM, Johnson DB, Starr M, Nixon A, Holtzhausen A, Hanks BA, A tumor-intrinsic PD-L1/NLRP3 inflammasome signaling pathway drives resistance to anti-PD-1 immunotherapy. J. Clin. Invest 130, 2570–2586 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theivanthiran B, Haykal T, Cao L, Holtzhausen A, Plebanek M, DeVito NC, Hanks BA, Overcoming immunotherapy resistance by targeting the tumor-intrinsic NLRP3-HSP70 signaling axis. Cancers (Basel) 13, 4753 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y, Ding Y, Guo N, Wang S, MDSCs: Key criminals of tumor pre-metastatic niche formation. Front. Immunol 10, 172 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaul ME, Fridlender ZG, Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol 16, 601–620 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Bronte V, Brandau S, Chen SH, Colombo MP, Frey AB, Greten TF, Mandruzzato S, Murray PJ, Ochoa A, Ostrand-Rosenberg S, Rodriguez PC, Sica A, Umansky V, Vonderheide RH, Gabrilovich DI, Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun 7,12150 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stackowicz J, Gaudenzio N, Serhan N, Conde E, Godon O, Marichal T, Starkl P, Balbino B, Roers A, Bruhns P, Jönsson F, Moguelet P, Georgin-Lavialle S, Broderick L, Hoffman HM, Galli SJ, Reber LL, Neutrophil-specific gain-of-function mutations in Nlrp3 promote development of cryopyrin-associated periodic syndrome. J. Exp. Med 218, e20201466 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brydges SD, Mueller JL, McGeough MD, Pena CA, Misaghi A, Gandhi C, Putnam CD, Boyle DL, Firestein GS, Horner AA, Soroosh P, Watford WT, O’Shea JJ, Kastner DL, Hoffman HM, Inflammasome-mediated disease animal models reveal roles for innate but not adaptive immunity. Immunity 30, 875–887 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tengesdal IW, Menon DR, Osborne DG, Neff CP, Powers NE, Gamboni F, Mauro AG, D’Alessandro A, Stefanoni D, Henen MA, Mills TS, De Graaf DM, Azam T, Vogeli B, Palmer BE, Pietras EM, DeGregori J, Tan A-C, Joosten LAB, Fujita M, Dinarello CA, Marchetti C, Targeting tumor-derived NLRP3 reduces melanoma progression by limiting MDSCs expansion. Proc. Natl. Acad. Sci. U.S.A 118, e2000915118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto M, Liu W, Luo Y, Tanaka A, Cai X, Norris DA, Dinarello CA, Fujita M, Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1β. J. Biol. Chem 285, 6477–6488 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jia H, Sodhi CP, Yamaguchi Y, Lu P, Martin LY, Good M, Zhou Q, Sung J, Fulton WB, Nino DF, Prindle T Jr., Ozolek JA, Hackam DJ, Pulmonary epithelial TLR4 activation leads to lung injury in neonatal necrotizing Enterocolitis. J. Immunol 197, 859–871 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gui Y-S, Wang L, Tian X, Feng R, Ma A, Cai B, Zhang H, Xu K-F, SPC-Cre-ERT2 transgenic mouse for temporal gene deletion in alveolar epithelial cells. PLOS ONE 7, e46076 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omrane I, Baroudi O, Kourda N, Bignon YJ, Uhrhammer N, Desrichard A, Medimegh I, Ayari H, Stambouli N, Mezlini A, Bouzayenne H, Marrakchi R, Benammar-Elgaaid A, Bougatef K, Positive link between variant Toll-like receptor 4 (Asp299Gly and Thr399Ile) and colorectal cancer patients with advanced stage and lymph node metastasis. Tumour Biol. 35, 545–551 (2014). [DOI] [PubMed] [Google Scholar]
- 31.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, Amigorena S, Ryffel B, Barrat FJ, Saftig P, Levi F, Lidereau R, Nogues C, Mira J-P, Chompret A, Joulin V, Clavel-Chapelon F, Bourhis J, André F, Delaloge S, Tursz T, Kroemer G, Zitvogel L, Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat. Med 13, 1050–1059 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Kyjacova L, Saup R, Rothley M, Schmaus A, Wagner T, Bosserhoff A, Garvalov BK, Thiele W, Sleeman JP, Quantitative detection of disseminated melanoma cells by Trp-1 transcript analysis reveals stochastic distribution of pulmonary metastases. J. Clin. Med 10, 5459 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ewens A, Mihich E, Ehrke MJ, Distant metastasis from subcutaneously grown E0771 medullary breast adenocarcinoma. Anticancer Res. 25, 3905–3915 (2005). [PubMed] [Google Scholar]
- 34.Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, Davies MA, Grossmann AH, VanBrocklin MW, McMahon M, Holmen SL, AKT1 activation promotes development of melanoma metastases. Cell Rep. 13, 898–905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaplan RN, Riba RD, Zacharoulis S, Bramley AH, Vincent L, Costa C, MacDonald DD, Jin DK, Shido K, Kerns SA, Zhu Z, Hicklin D, Wu Y, Port JL, Altorki N, Port ER, Ruggero D, Shmelkov SV, Jensen KK, Rafii S, Lyden D, VEGFR1-positive haematopoietic bone marrow progenitors initiate the pre-metastatic niche. Nature 438, 820–827 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtzhausen A, Zhao F, Evans K, Tsutsui M, Orabona C, Tyler DS, Hanks BA, Melanoma-derived Wnt5a promotes local dendritic-cell expression of IDO and immunotolerance: Opportunities for pharmacologic enhancement of immunotherapy. Cancer Immunol. Res 3, 1082–1095 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao F, Evans K, Xiao C, DeVito N, Theivanthiran B, Holtzhausen A, Siska PJ, Blobe G, Hanks BA, Stromal fibroblasts mediate Anti-PD-1 resistance via MMP-9 and dictate TGFβ inhibitor sequencing in melanoma. Cancer Immunol. Res 6, 1459–1471 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, Moorhead M, Chaudhuri S, Tomsho LP, Peters BA, Pujara K, Cordes S, Davis DP, Carlton VE, Yuan W, Li L, Wang W, Eigenbrot C, Kaminker JS, Eberhard DA, Waring P, Schuster SC, Modrusan Z, Zhang Z, Stokoe D, de Sauvage FJ, Faham M, Seshagiri S, Diverse somatic mutation patterns and pathway alterations in human cancers. Nature 466, 869–873 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Benci JL, Xu B, Qiu Y, Wu TJ, Dada H, Twyman-Saint Victor C, Cucolo L, Lee DSM, Pauken KE, Huang AC, Gangadhar TC, Amaravadi RK, Schuchter LM, Feldman MD, Ishwaran H, Vonderheide RH, Maity A, Wherry EJ, Minn AJ, Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 167, 1540–1554.e12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koyama S, Sato E, Masubuchi T, Takamizawa A, Kubo K, Nagai S, Izumi T, Alveolar type II-like cells release G-CSF as neutrophil chemotactic activity. Am. J. Physiol 275, L687–L693 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Kim Y-M, Kim H, Lee S, Kim S, Lee J-U, Choi Y, Park HW, You G, Kang H, Lee S, Park JS, Park Y, Park H-S, Park C-S, Lee S-W, Airway G-CSF identifies neutrophilic inflammation and contributes to asthma progression. Eur. Respir. J 55, 1900827 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Strauss L, Mahmoud MAA, Weaver JD, Tijaro-Ovalle NM, Christofides A, Wang Q, Pal R, Yuan M, Asara J, Patsoukis N, Boussiotis VA, Targeted deletion of PD-1 in myeloid cells induces antitumor immunity. Sci Immunol 5, eaay1863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alam MS, Proximity ligation assay (PLA). Curr. Protoc. Immunol 123, e58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Varnier R, Garrivier T, Hafliger E, Favre A, Coutzac C Spire C, Rochefort P, Sarabi M, Desseigne F, Guibert P, Cattey-Javouhey A, Funk-Debleds P, Mastier C, Buisson A, Pérol D, Trédan O, Blay JY, Phelip J-M, de la Fouchardiere C, Hyperprogressive disease after combined anti-PD-L1 and anti-CTLA-4 immunotherapy for MSI-H/dMMR gastric cancer: A case report. Front. Oncol 11, 756365 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Gu Y, Han Y, Zhang Q, Jiang Z, Zhang X, Huang B, Xu X, Zheng J, Cao X, Tumor exosomal RNAs promote lung pre-metastatic niche formation by activating alveolar epithelial TLR3 to recruit neutrophils. Cancer Cell 30, 243–256 (2016). [DOI] [PubMed] [Google Scholar]
- 46.Di Pietro FR, Verkhovskaia S, Mastroeni S, Carbone ML, Abeni D, Di Rocco CZ, Samà N, Zappalà AR, Marchetti P, De Galitiis F, Failla CM, Fortes C, Clinical predictors of response to anti-PD-1 first-line treatment in a single-centre patient cohort: A real-world study. Clin. Oncol. (R. Coll. Radiol.) 34, e18–e24 (2022). [DOI] [PubMed] [Google Scholar]
- 47.Kumawat K, Menzen MH, Bos IS, Baarsma HA, Borger P, Roth M, Tamm M, Halayko AJ, Simoons M, Prins A, Postma DS, Schmidt M, Gosens R. Noncanonical WNT-5A signaling regulates TGF-β-induced extracellular matrix production by airway smooth muscle cells. FASEB J. 27, 1631–1643 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Tengesdal IW, Dinarello A, Powers NE, Burchill MA, Joosten LAB, Marchetti C, Dinarello CA, Tumor NLRP3-derived IL-1β drives the IL-6/STAT3 axis resulting in sustained MDSC-mediated immunosuppression. Front. Immunol 12, 661323 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang Q, Wu J, Zhao X, Shen S, Zhu C, Liu T, Cui X, Chen L, Wei C, Cheng P, Cheng W, Wu A, Establishment of tumor inflammasome clusters with distinct immunogenomic landscape aids immunotherapy. Theranostics 11, 9884–9903 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Angelicola S, Ruzzi F, Landuzzi L, Scalambra L, Gelsomino F, Ardizzoni A, Nanni P, Lollini P-L, Palladini A, IFN-γ and CD38 in hyperprogressive cancer development. Cancers (Basel) 13, 309 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sohal DS, Nghiem M, Crackower MA, Witt SA, Kimball TR, Tymitz KM, Penninger JM, Molkentin JD, Temporally regulated and tissue-specific gene manipulations in the adult and embryonic heart using a tamoxifen inducible Cre protein. Circ. Res 89, 20–25 (2001). [DOI] [PubMed] [Google Scholar]
- 52.Zhao F, Xiao C, Evans KS, Theivanthiran T, DeVito N, Holtzhausen A, Liu J, Liu X, Boczkowski D, Nair S, Locasale JW, Hanks BA, Paracrine Wnt5a-β-Catenin signaling triggers a metabolic program that drives dendritic cell tolerization. Immunity 48, 147–160.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pulaski BA, Ostrand-Rosenberg S, Mouse 4T1 breast tumor model. Curr. Protoc. Immunol Chapter 20, Unit 20 22 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, Repurposing CRISPR as an RNA guided platform for sequence specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joung J, Kirchgatterer PC, Singh A, Cho JH, Nety SP, Larson RC, Macrae RK, Deasy R, Tseng Y-Y, Maus MV, Zhang F, CRISPR activation screen identifies BCL-2 proteins and B3GNT2 as drivers of cancer resistance to T cell-mediated cytotoxicity. Nat. Commun 13, 1606 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.