Abstract
Positive-strand RNA viruses have been the cause of several recent outbreaks and epidemics, including the Zika virus epidemic in 2015, the SARS outbreak in 2003, and the ongoing SARS-CoV-2 pandemic. On June 18–22, 2022, researchers focusing on positive-strand RNA viruses met for the Keystone Symposium “Positive-Strand RNA Viruses” to share the latest research in molecular and cell biology, virology, immunology, vaccinology, and antiviral drug development.
Keywords: Alphavirus, Enterovirus, Flavivirus, Coronavirus, RNA virus, SARS-CoV-2
Graphical Abstract

Introduction
Positive-strand RNA viruses encompass many well-known and clinically relevant pathogens, including enterovirus 71 (EV-71), hepatitis C virus (HCV), dengue and West Nile virus, and coronaviruses. Given the propensity for these viruses to mutate in addition to their capacity for zoonosis, positive-strand RNA viruses represent constant threats to global health, as evidenced by the recent Zika virus epidemic and the COVID-19 pandemic.
On June 18–22, 2022, researchers focusing on positive-strand RNA viruses met for the Keystone Symposium “Positive-Strand RNA Viruses” to share the latest research in molecular and cell biology, virology, structural biology, immunology, vaccinology, and antiviral drug development. This symposium is the only conference solely dedicated to positive-strand RNA viruses and therefore represented a unique opportunity for researchers to gain insights into this important group of viruses.
Coronavirus RNA exonuclease: implications for antivirals
Mark R. Denison from Vanderbilt University described work on understanding the role of the coronavirus RNA proofreading exonuclease, ExoN, and implications for antivirals. RNA viruses typically have low-fidelity replication compared to DNA-based organisms and therefore usually have limited genome size and complexity. Coronaviruses, however, are different. They have large RNA genomes of approximately 30 kb that encode multiple proteins. They also have higher basal fidelity than other RNA viruses.1 Denison argued that the proofreading activity of the coronavirus protein nsp14, an exoribonuclease, may have been important for the expansion and complexity of the RNA genome in coronaviruses. The activity of nsp14 was first demonstrated in severe acute respiratory syndrome-coronavirus (SARS-CoV) and human CoV. In these studies, nsp14 activity was deemed essential for human CoV viral replication as mutations in the exoribonuclease active motifs decreased viral RNA synthesis and resulted in no viable viruses.2 Later work in Denison’s group showed that nsp14 activity is not essential for all coronaviruses. Mutating the active motifs in nsp14 of murine hepatitis virus (MHV) or SARS-CoV increased mutation frequency, but the viruses were still able to replicate.3,4 In other viruses, including Middle East respiratory syndrome (MERS) -CoV and SARS-CoV-2, mutating the active motif abrogated viral replication. Coronaviruses lacking exonuclease activity (ExoN-) were less fit than wild-type virus and susceptible to agents that the wild-type virus is normally resistant to.5,6 Interestingly, ExoN- viruses were also more susceptible to the nucleotide analog remdesivir, which is approved as an antiviral agent for SARS-CoV-2. These data indicate that nsp14-ExoN is involved in recognizing errors during RNA replication and cooperating with the polymerase during nucleotide selection. Cooperative inhibition of ExoN activity may therefore enhance the activity of nucleoside analogs like remdesivir.7 Denison also discussed the role of nsp14-ExoN in RNA recombination. Coronaviruses normally undergo significant recombination with specific, predictable patterns. In MHV lacking ExoN activity, recombination is decreased, and the recombination patterns are altered. This suggests that not only does ExoN facilitate recombination at distinct genomic regions but also that recombination is a critical, normal component of viral replication.8 Denison’s work may have implications for antivirals that target coronaviruses. For many coronaviruses, targeting ExoN may be sufficient as ExoN- MERS-CoV and SARS-CoV-2 are unable to replicate. For other coronaviruses, targeting ExoN may sensitize the virus to mutagens and/or nucleoside analogs.
Cell Biology of RNA Viruses
Mechanisms of RNA virus exit
Margaret Kielian from the Albert Einstein College of Medicine presented work on understanding viral exit of two small, enveloped positive-sense RNA viruses—rubella virus and chikungunya virus. The rubella virus membrane surrounds the nucleocapsid and contains two transmembrane glycoproteins, E1 and E2. During viral replication, E1 and E2 localize to the endoplasmic reticulum (ER) and, along with the nucleocapsid, are transported and bud into the Golgi complex to form virions.9 Kielian presented unpublished data on a conserved interaction between E2 and the capsid protein that promotes capsid localization to the Golgi. Regarding chikungunya virus, viral assembly and exit typically occur at the plasma membrane, releasing free virus into the extracellular environment. While these viruses can infect new cells, they are also susceptible to neutralization by circulating antibodies. Many viruses have also devised various mechanisms for cell-to-cell transmission, in which a virus infects a target cell via interactions with an infected cell. The virus is thus not subject to neutralizing antibodies and can thus evade the immune system.10 Alphaviruses, like chikungunya virus, are well known for efficient transmission by free virus particles. However, Kielian’s group has observed that alphaviruses can also induce long intercellular extensions (ILE) in infected cells that form stable contacts with neighboring cells.11 She showed unpublished data that support the role of ILEs in cell-to-cell transmission.
Autophagy during EV-D68 infection
William Jackson from the University of Maryland presented work on the role of autophagy in the life cycle of the picornavirus enterovirus D68 (EV-D68) and how interactions with the autophagy machinery may provide a mechanistic rationale for the paralysis some people experience. Autophagy is an important part of cellular homeostasis and organelle turnover. Double membrane autophagosomes engulf their cargo and consecutively fuse with endosomes and lysosomes to create an acidic environment that leads to cargo degradation. Jackson showed that both EV-D68 and poliovirus use the autophagy machinery during replication. Specifically, acidic autophagosomes (amphisomes) promote capsid maturation, the final step in generating infectious virus. If the virus were to remain in an ever-acidifying vesicle, however, it would be degraded. Jackson showed that the viral protease EV-D68 3C prematurely halts autophagy by cleaving the host protein SNAP-29, which mediates autophagosome and lysosome fusion.12 Jackson proposed that the ability of EV-D68 3C to cleave SNAP proteins may also provide a mechanistic explanation for the paralysis the virus causes. In neuronal cells, SNAP-25 regulates neuronal function by mediating acetylcholine release at the synapse. Cleavage of SNAP-25 by botulinum toxin results in flaccid paralysis.13 Jackson’s group has found that EV-D68 3C has similar activity against SNAP25, which could account for muscle weakness prior to neuronal cell death.
Double membrane vesicles in viral infection
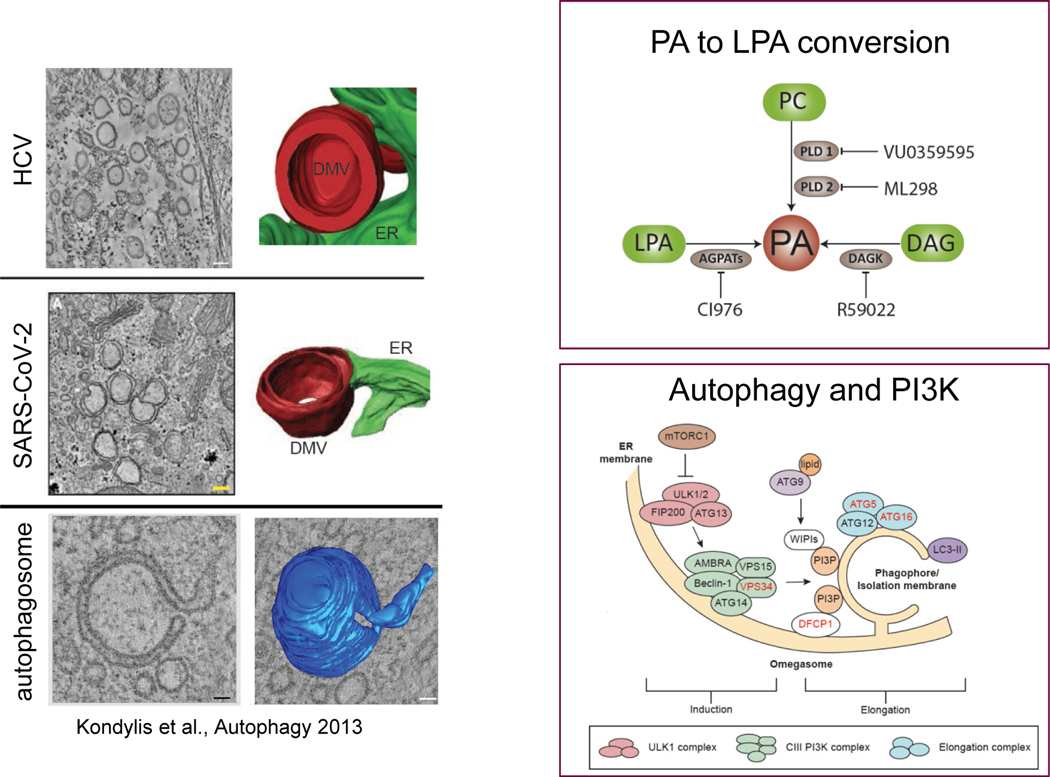
Ralf Bartenschlager from Heidelberg University presented work on understanding the formation of double membrane vesicles (DMV) during SARS-CoV-2 infection (Figure 1). Double membrane vesicles are cytoplasmic structures induced by viral proteins that serve as sites of viral RNA synthesis. Three-dimensional imaging has revealed the presence of ER-derived DMVs scattered throughout SARS-CoV-2-infected cells.14 Since these DMVs resemble structures identified in hepatitis C virus (HCV)-infected cells and autophagosomes, which also form at the ER, Bartenschlager’s group investigated whether the autophagy pathway plays a role in the replication of both viruses. They found that while replication of these two viruses does not require autophagy per se, it does require parts of the autophagy machinery, notably PI3K.15 In a search for cellular lipid pathways involved in DMV formation by HCV and SARS-CoV-2, Bartenschlager’s group isolated DMVs from HCV-infected cells and conducted proteomics. They found that DMVs contain 1-acylglycerol-3-phosphate-O-acyltransferase (AGPAT) enzymes, which catalyze the conversion of the lipid lysophosphatidic acid (LPA) into phosphatidic acid (PA). In both SARS-CoV-2- and HCV-infected cells, AGPAT activity and production of PA were important for viral replication and DMV formation. Possible mechanisms for how PA promotes DMV biogenesis include inducing membrane curvature, recruiting PA-binding cellular or viral proteins, or regulating lipid transport at DMVs.16
Figure 1.
Factors commonly involved in double membrane vesicle formation.
Inferring the evolutionary history of RNA viruses
Tero Ahola from the University of Helsinki discussed the membranous replication structures of RNA viruses and how they may inform the evolutionary history of RNA viruses. The two types of membrane replication structures, DMVs and spherules, are concordant with the new phylogenetic grouping of positive-sense RNA viruses, which is based solely on the conservation of the polymerase. Spherules are always found in members of the phylum Kitrinoviricota (which contains alpha- and flaviviruses) while Pisuviricota (picorna- and nidoviruses) replication sites have more complex morphologies, including DMVs. Ahola proposed that membrane association of the replication complex is a primordial characteristic and may have provided an advantage for virus replication in early eukaryotic cells with internal membranes. Based on structural and functional information on the replication sites of alphaviruses, coronaviruses, and picornaviruses, he asserted that Pisuviricota and Kitrinoviricota evolved membrane association separately and that parallels between these two phyla represent convergent evolution.17
Structural Virology of Positive-Strand RNA Viruses
Alphavirus assembly: interactions within spike
Tuli Mukhopadhyay from Indiana University discussed the viral protein-protein interactions important for alphavirus assembly. Alphaviruses are enveloped viruses with transmembrane trimeric spike proteins that protrude from the virus and mediate viral entry.18 Mukhopadhyay focused on the different conformations of spike and how interactions between the three spike proteins, E1, E2, and E3, change throughout the virus lifecycle. Prior to viral entry, spike exists in a metastable conformation characterized by a large interface between E2 and E1. As the pH decreases, this interface disassociates and induces conformational changes in spike that enable it to adopt its fusogenic form, fuse with the endosomal membrane, and enter the cell.19–21 During viral assembly, interactions between E3 and E2 stabilize the E2-E1 dimer to ensure that the dimer remains intact as the virus travels through the acidifying secretory pathway. As the spikes transit through the Trans-Golgi network, E3 is cleaved off, resulting in the aforementioned metastable spike conformation.22–24 Mukhopadhyay’s group has recently demonstrated the importance of interdimer interactions between E2 and E1 in which a region of E2 is sandwiched between an E1 protein of a neighboring dimer. This interaction is important for generating the trimer of E2-E1 dimers observed on the surface of the virus. Her group generated a chimeric virus that disrupts this interaction in which residues in E2 involved in the interdimer interaction are mutated to the chikungunya sequence while the rest of the virus is wild-type Sindbis virus. This chimeric virus formed defective particles, the phenotype of which was more apparent in mammalian cells than in mosquito cells. Similarly, the chimeric virus displayed growth defects in mammalian but not mosquito cells.25 Second-site revertants identified that glycosylation plays an important role in the assembly of the spike complex.
Structural insights into alphavirus cell entry
Daved Fremont from Washington University discussed attachment factors for different alphaviruses. Fremont’s group, in collaboration with Michael Diamond, has identified Mxra8 as a receptor for several arthritogenic alphaviruses, including chikungunya virus, O’nyong’nyong virus, Mayaro virus, and Ross River virus, among most mammals. 26 The structure of Mxra8 reveals a unique topology consisting of two strand-swapped Ig-like domains. Cryogenic electron microscopy (cryo-EM) structures of Mxra8 in complex with chikungunya viral particles have illustrated how Mxra8 interacts with the viral Spike protein. 27 The structures also reveal why Mxra8 does not support alphavirus infection in cattle. Bovine Mxra8 contains a 15-amino acid insertion, likely a result of DNA replication slippage. These residues form a loop that interferes with the interaction between Mxra8 and the E2-E1 heterodimer. Deleting this insertion from bovine Mxra8 restored binding of chikungunya particles, while adding the insertion into murine Mxra8 blocked virion binding.28 Fremont’s group has also identified LDLRAD3 as the attachment receptor for Venezuelan equine encephalitis virus (VEEV).29 CryoEM structures show that LDLRAD3 binds a cleft formed by two E2-E1 heterodimers. This interaction is reminiscent of that between Mxra8 and Chikungunya virus, even though Mxra8 and LDLRAD3 are not similar from a structural or sequence perspective.27,30
Virus-antibody complex structures
Richard Kuhn from Purdue University presented work on virus–antibody complex structures. These structures can provide insights into antibody binding affinity and potency, mechanisms of neutralization, and antibody cross-reactivity. They may also inform efforts to re-engineer existing antibodies to improve affinity, specificity, and function. Kuhn focused on flaviviruses, specifically dengue virus. Twenty years ago, Kuhn was instrumental in solving the cryo-EM structure of dengue virus, showing that the glycoproteins in mature virions are arranged with icosahedral symmetry with three envelope proteins per asymmetric unit.31 Despite the seeming simplicity of this arrangement, dengue virus structure is quite complex. Kuhn showed that the virions can be quite heterogeneous, which can impact the epitopes available to antibodies. For example, cleavage of the premembrane (prM) structural protein, which typically occurs at the plasma membrane prior to viral release, can be inefficient, resulting in mature, infectious virions that contain prM instead of membrane (M) protein. Kuhn showed cryo-EM structures of two neutralizing antibodies that recognize prM. His group characterized a small panel of prM-reactive antibodies and revealed both cell type- and maturation state-dependent differences in their neutralization potency.32 Other epitopes are found in the loop regions of the E protein.33,34 However, recent evidence suggests that some neutralizing antibodies recognize quaternary epitopes within E proteins, which are only present in the intact virion. These quaternary sites elicit highly potent antibodies, which may prove instrumental in vaccine design.33,34
Structural insights into the mechanism of a SARS-CoV-2 antiviral
Ashleigh Shannon from Bruno Canard’s group at AIX Marseille University presented her work on the structure of the nucleoside analog AT-9010 bound to the SARS-CoV-2 polymerase, nsp12. AT-9010 is the active, triphosphate form of AT-527, also known as bemnifosbuvir, a guanosine nucleoside analog produced by Atea Pharmaceuticals. Cryo-EM structures indicate that AT-9010 binds to two sites within nsp12—the C-terminal RNA-dependent RNA polymerase (RdRp) domain and the N-terminal nucleotidylation (NiRAN) domain. The structure reveals how the incorporation of AT-9010 in the growing RNA chain results in the termination of RNA synthesis by preventing the correct alignment of the incoming NTP. With regard to the NiRAN site, AT-9010 binds in a flipped orientation compared to native NTPs, out-competing native nucleotides and inhibiting NiRAN activity.35
Structural insight into Nsp15 activity
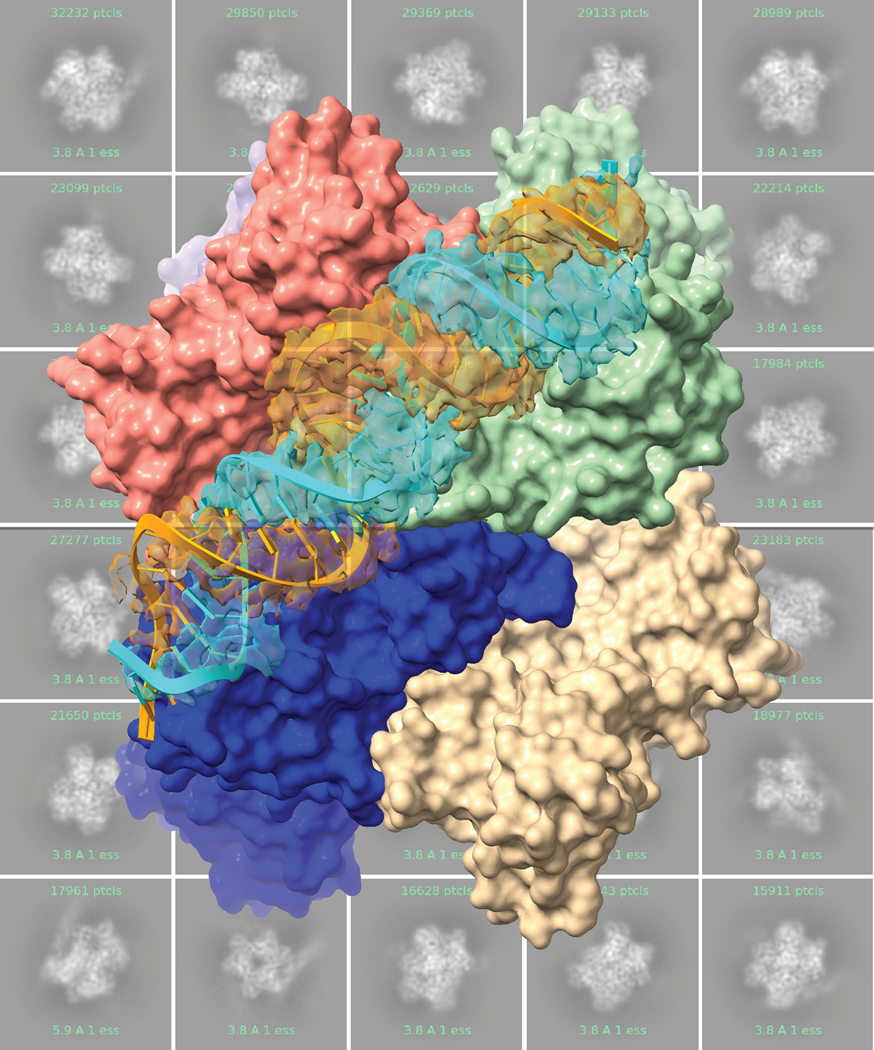
Meredith Frazier, currently at the College of Charleston, presented work from Robin Stanley’s lab at the National Institutes of Health (NIH) on how cryo-EM structures of SARS-CoV-2 Nsp15 reveal insights into nuclease specificity and dynamics (Figure 2). Nsp15 is found in all coronaviruses. It helps to hide replicating viruses from host receptors, thus limiting the host immune response. Hexamerization of Nsp15 is necessary for its endoribonuclease activity. Cryo-EM structures of RNA-free Nsp15 indicate that the protein is fairly flexible. Frazier noted that this flexibility may be important for substrate recognition and/or allosteric communication across the protein.36 Additional structures of the protein in the post-cleavage state bound to single-strand (ss)RNA reveal interactions between neighboring protomers, demonstrating why oligomerization is important for activity.37 More recently, structures of Nsp15 bound to double-strand (ds)RNA show that Nsp15 binds and cleaves dsRNA through the contribution of three protomers, supporting the importance of a hexameric arrangement for binding to dsRNA substrates.38
Figure 2.
Nsp15 bound to dsRNA.
New models of virus infection
Using organoids to understand inter-species activity
Kwok-Yung Yuen from the University of Hong Kong presented work on developing organoid models to study animal viruses. Over the past 20 years, the potential for animal viruses to jump into human hosts has been illustrated multiple times, from SARS-CoV-1 in 200339–41, MERS-CoV in 201242, and SARS-CoV-2 in 2019.43 Yuen’s group has been instrumental in identifying the animal sources of these viruses and in tracking down ancestral viral strains among natural animal reservoirs.44–48 Over the past year, his team has discovered more than 80 viruses, including more than 30 coronaviruses. Some of these are close relatives to the viruses responsible for the SARS-CoV-1, MERS-CoV, and SARS-CoV-2 outbreaks as well as others.49,50 Often, these viruses are rarely cultured and knowledge about them is limited to their genome. Yuen’s group is developing intestinal and airway organoid models to culture viruses and predict their potential for inter-species jumping. He showed that lung organoids can differentiate between human-adapted avian and swine influenza viruses and non-adapted viruses. Those that had adapted to humans grew better than those that had not.51 Similarly, the SARS-CoV-2 Omicron variant demonstrated higher infectivity and fitness than the wild-type virus in airway organoids.52 In intestinal organoids, SARS-CoV-1 was better than SARS-CoV-2 at infecting cells, consistent with the fact that SARS-CoV-1 is more likely to cause diarrhea among infected individuals.53,54 Yuen’s group has also developed bat intestinal organoids and demonstrated the ability of SARS-CoV-2 to replicate.55 They are now using these organoids to investigate the potential for novel coronaviruses to jump into new species and to test the activity of antivirals.
Organoid models of the maternal-fetal interface
Carolyn Coyne from Duke University discussed work on developing organoid models of the maternal-fetal interface to understand how infection impacts the immunologic crosstalk between the fetal placental tissue and the maternal decidua tissue. Modeling the maternal-fetal interface has been challenging. Easy-to-use, cost-effective models like cell culture do not faithfully recapitulate the interface while more biologically relevant models, like tissue explants, are difficult to establish. In addition, differences between mouse and human placentas mean that results from animal studies cannot always be translated to humans.56 In 2018, organoids derived from early-term human placentas were developed. While these organoids have significant potential to change the field57,58, the regulatory issues surrounding working with fetal tissue are complex and ever-changing. Coyne’s group has developed methods to generate maternal–fetal organoids from full-term human placentas. They have generated matched trophoblast organoids, the fetal cells that comprise the placenta, and decidua organoids, the maternal tissue that interacts with the placenta. She showed that these organoids recapitulate the innate immune signatures of primary cells and tissue explants, including the constitutive secretion of the antiviral interferon, IFN-λ2.59 The group intends to build a large, diverse biobank of these matched organoids. They have also developed chorion organoids and amnion primary epithelial cells and have established various methods to investigate communication between these systems.
Using organoids to model respiratory virus infection
Katja Wolthers from Amsterdam University Medical Center described her work using organoids to model human enteroviruses and parechoviruses. In human airway organoids, infection with various EV-71 strains showed that mutation at position 145 in the VP1 capsid protein was associated with viral infectivity.60 Unpublished work implicates a role for heparan sulfate binding in the increased viral replication seen with VP1–145Q viral strains. This is consistent with human data showing that variation at position 145 is associated with disease severity61 and that heparan sulfate binding variants of EV-71 cause viremia and neuro-infection.62 Wolthers’s group is also using organoids to model EV-D68 infection. Previous work demonstrated that EV-D68, which can cause acute flaccid paralysis, can infect neuronal cells. Similar to EV-71, the infection efficiency of EV-D68 is related to a mutation at site VP1–271, which is involved in heparan sulfate binding.63 Wolthers showed unpublished data modeling EV-D68 infection in brain organoids. Finally, Wolthers’s group is using organoids to model parechovirus infection. Parechovirus is an important cause of sepsis-like illness and central nervous system (CNS) infection in young children, but not much is known about its pathogenesis.64 It is generally assumed that the pathogenesis of parechoviruses is similar to that of enteroviruses, namely that they use the respiratory pathway and gut as viral entry sites and spread to their target organs via the blood and lymph. Wolthers showed that both parechovirus genotypes, PeV-A1 and PeV-A3, can infect the human airway epithelium from the basolateral side65 and the intestinal epithelium.66 The two genotypes showed differences in replication kinetics, cell tropism, and immune response, which may explain the differential symptomology patients experience.66
Modeling neurotropic viral infection in brain organoids
Guo-Li Ming from the University of Pennsylvania presented her work on using brain organoids to model neurotropic viral infections. Ming’s group has developed a protocol to generate region-specific brain organoids, including the forebrain, midbrain, and hindbrain. Forebrain organoids resemble the fetal cortical brain during the second trimester and recapitulate many human-specific features, including human-specific neural stem cells, a rudimentary laminar organization with all six cortical layers, and functional neurons.67,68 Infection of these forebrain organoids with Zika virus reduced the number of neural stem cells and neurons and resulted in morphological changes that resemble features of microcephaly. Ming’s group has used these organoids to screen for neuroprotective and antiviral compounds that may mitigate the effects of Zika infection.67,69–71 To engineer better brain organoids, Ming’s group has developed a slicing method that bypasses the diffusion limit, enabling forebrain organoids to mature further. These organoids resemble the third trimester of human cortical development and contain late-born neural lineages that were not present in other forebrain organoids.72 Finally, Ming described her work on modeling SARS-CoV-2 infection in region-specific brain organoids. She showed that while SARS-CoV-2 showed limited ability to infect cortical, hippocampal, hypothalamic, and midbrain organoids, it robustly infected choroid plexus organoids. These data provide evidence for tropism-specific infection by SARS-CoV-2 and support the use of organoids to study viral infection.73
Camy Guenther from Rushika Perera’s group at Colorado State University presented unpublished work on the impact of Dengue virus infection on lipid metabolites. Jasmine Moshiri from Karla Kirkegaard’s group at Stanford University presented her work on using 3D colonic epithelial organoids to study EV-71 infections in the gut.
Innate immunity and evasion
Innate immunity sits at the crossroads between the body’s intrinsic, initial defenses and acquired adaptive immunity. Viruses have adapted mechanisms to evade many innate immune pathways, resulting in prolonged viral infection and stunting the adaptive immune response. Understanding innate immunity is therefore crucial to understanding the outcome of viral infections. Speakers in this session discussed restriction factors to viral infection and antiviral aspects of innate immunity.
An expanded role for the restriction factor TRIM5ɑ
Sonja Best from the NIAID presented her work on the restriction factor TRIM5ɑ. In nonhuman primates (NHP), TRIM5ɑ robustly blocks HIV infection by recognizing the mature viral capsid. Coating of the capsid by TRIM5ɑ promotes early capsid uncoating and inhibits the early replication events of incoming retroviruses. While TRIM5ɑ had been known for its role in blocking retrovirus infection, little was known about its ability to restrict other types of viruses. Best’s group has shown that TRIM5ɑ can restrict several flaviviruses as well. They tested the ability of TRIM5ɑ to restrict several flaviviruses and found that viruses belonging to the tick-borne encephalitis serogroup were sensitive to TRIM5ɑ while mosquito-borne viruses were not. Additional studies into the mechanism of this effect showed that TRIM5ɑ binds to the viral proteins NS2B/3 during replication and mediates its degradation by the proteasome.74 Best also presented unpublished work illustrating why some flaviviruses are resistant to TRIM5ɑ. Her work shows that TRIM5ɑ can recognize diverse viral molecular patterns and is not limited to recognizing early events and structural proteins, as it does for HIV, but can also recognize nonstructural proteins at later time points in the viral replication cycle.
Anti-coronavirus activity of the restriction factor Ly6e
John Schoggins from the University of Texas Southwestern described his group’s work on understanding how the coronavirus restriction factor, Ly6e, functions in vivo. Schoggins’s group has set up a workflow to discover interferon-stimulated genes and investigate their mechanisms and in vivo functions. Ly6e was discovered in such a screen for inhibitors of 229E coronavirus. It is a small, glycosylphosphatidylinositol-anchored protein found on the cell surface. Schoggins showed that in cell culture, Ly6e inhibits multiple coronavirus strains by impairing spike-mediated fusion, thus blocking viral entry. To further understand the role of Ly6e in vivo, Schoggins’s group developed a mouse model in which Ly6e is knocked out in immune cells. He showed that Ly6e knock-out sensitizes mice to infection with MHV.75 Schoggins showed unpublished data on a role for Ly6e in protecting mice from SARS-CoV-2 infection and pathogenesis.
Probing maternal-fetal immunology via viral infection
Kellie Ann Jurado from the University of Pennsylvania discussed work on understanding innate immunity to congenital Zika virus infection. It is rare for viruses to cause congenital infection as the placenta mounts strong defenses to protect the developing fetus.76 Understanding Zika virus pathogenesis, therefore, offers an opportunity to not only understand the pathogen but also to better understand the host and host immunity. Jurado was involved in the development of a pregnancy mouse model to probe the role of type I interferon during pregnancy. Imaging studies showed that in infected mothers, the maternal-fetal barrier is disrupted in interferon (IFN) heterozygotic fetuses but remains intact in knockout fetuses. These data implicate Type I IFN signaling as a mediator of pregnancy complications and as a key contributor to fetal demise in the setting of Zika infection.77 This work laid the foundation for Jurado’s current research program on understanding maternal-fetal immunology using Zika virus. She presented unpublished data from her independent research lab on the immune regulatory role of IL-27 at the maternal-fetal interface.
Species-specific small regulatory RNA pathways in mosquitoes
Gregory Ebel from Colorado State University presented work on understanding how antiviral innate immune responses in mosquitoes impact virus transmission, with a focus on West Nile virus. Mosquitoes have multiple pathways that mediate viral defense. Ebel focused on two small regulatory RNA pathways (SRRP) in Culex mosquitoes, the exo-siRNA and piwi pathways. Previous work has demonstrated that West Nile virus experiences different evolutionary pressures in birds and mosquitoes. Ebel showed that the induction of RNA interference in mosquitoes creates an environment that favors rare genotypes, thus driving West Nile virus diversity.78,79 Ebel’s group has developed a bioinformatics pipeline to examine SRRP activation in Culex mosquitoes. They found that virus-derived small interfering RNAs induced by West Nile virus infection were primarily exo-siRNAs; no bona fide piRNAs were detected in multiple Culex mosquito tissues.80 This finding was somewhat surprising because several studies have demonstrated a role for piRNAs in antiviral defense in other mosquito species.81–85 However, Ebel’s work demonstrates that in Culex flavivirus systems, the siRNA pathway is more important than the piRNA pathway. Ebel presented unpublished data on the impact of RNA interference on West Nile transmission by Culex mosquitoes.
A multitrait locus associated with sarbecovirus pathogenesis
Alexandra Schäfer from Ralph Baric’s lab at the University of North Carolina presented work on identifying a multitrait locus that regulates sarbecovirus pathogenesis. Schäfer used the genetically diverse collaborative cross mice to identify strains that demonstrate a range of phenotypes to SARS-CoV infection in terms of weight loss, viral titers, and mortality. These phenotypes were associated with a quantitative trait locus that clustered in a region on chromosome 9.86 Several of these genes display synteny with genes identified in a human GWAS study that correlate with COVID-19 disease severity.87,88 Schäfer showed that knocking out two of the six ortholog genes in this region, CCR9 and CXCR6, prevented recovery from SARS-CoV-2 infection, increased inflammatory response and mortality, and was associated with prolonged lung dysfunction. This study validates the collaborative cross as a platform to identify and validate novel susceptibility genes that regulate emerging virus pathogenesis across species.86
Immune Correlates of Protection
Characterizing the SARS-CoV-2 immune response: examples from Singapore
Lisa F.P. Ng from A*STAR gave an overview of how several research groups worked together to characterize the SARS-CoV-2-mediated immune response, including the mechanisms underlying severe disease progression, biomarkers related to diagnosis, mild, and severe disease, and humoral responses. Early in the pandemic, A*STAR infectious diseases labs worked to understand the cytokine response to SARS-CoV-2 infection to understand the mechanism of severe disease. They compared the cytokine profile of uninfected controls and infected patients with varying disease severity and found systemic upregulation of several inflammatory cytokines in those with severe disease.89 They also followed the systemic cytokine response in convalescent patients after recovery and showed that patients exhibit a disrupted cytokine profile up to six months after infection.90 Additional research identified a cellular profile associated with acute COVID-19 disease characterized by a population of inflammatory monocytes and neutrophils. The study suggested that the ratio of immature neutrophils to a specific T-cell population could serve as an early marker of severe COVID-19.91 Finally, Ng described work on the immune response to the COVID-19 vaccines. Analyses of the memory B-cell response among vaccinated individuals showed that while those with breakthrough infections (primarily of the Delta variant) did not have an inferior plasma antibody response they did have a lower memory B-cell response. They also showed that those who experienced breakthrough infection with the Delta variant had a muted inflammatory response that was similar to that of uninfected individuals.92 Similar data for the Omicron variant helped to inform the country’s public health response to the pandemic as Omicron swept the world. Given that the majority of the Singapore population was vaccinated, and that Omicron infection produces mild systemic inflammation, policymakers were comfortable relaxing many COVID-19 protocols despite rising cases. Ng stressed that this is one example of how seemingly basic research can impact public policy.
Understanding the deubiquitinating activity of coronavirus proteases
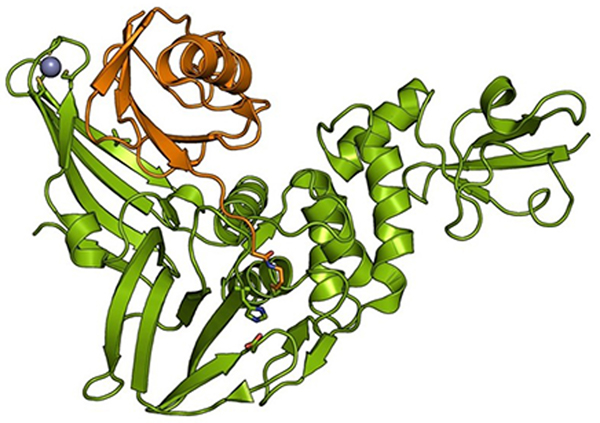
Marjolein Kikkert from Leiden University Medical Center presented work on how the coronavirus papain-like protease (PLpro) modulates the innate immune response (Figure 3). PLpro cleaves the viral polypeptide to generate mature proteins. It also has deubiquitinating (DUB) activity. As many innate immune pathways are regulated by ubiquitination, Kikkert’s group hypothesized that the DUB activity of PLpro may inhibit innate immune signaling. Kikkert showed unpublished data on the impact of blocking PLpro DUB activity on MERS-CoV replication and disease outcome in a mouse model. The potential of DUB-less MERS-CoV as a vaccine was also explored. She additionally presented unpublished work on developing ubiquitin-based inhibitors of PLpro as potential antivirals.
Figure 3.
Structure of MERS-CoV PLpro (in green) in complex with ubiquitin (orange), adapted from Bailey-Elkin et al. J Biol Chem. 2014.
Immune responses to SARS-CoV-2 infection and COVID-19 vaccines
Alessandro Sette from the La Jolla Institute for Immunology presented work on understanding immune protection mediated by SARS-CoV-2 infection and vaccination. Early in the pandemic, there was fear that coronaviruses in general were nonimmunogenic and that a vaccine would not be feasible. Fortunately, initial studies confirmed strong humoral and T-cell responses against the spike protein, which has been the focus of vaccine efforts, as well as other viral antigens.93 In follow-up studies, SARS-CoV-2 immune memory following natural infection was shown to persist for at least six months, with most individuals maintaining substantial immune memory eight months after infection. Immune memory was found to be complex and heterogeneous, with different kinetics for T-cell, memory B-cell, and antibody responses.94 As the virus mutates and variants emerge, Sette stressed that we must be specific when we discuss protection against SARS-CoV-2. Protection against infection is largely mediated by neutralizing antibodies, which are largely ineffective against new variants. In contrast, T cells play a key role in protecting against severe disease. Thousands of T-cell epitopes have been identified across the SARS-CoV-2 genome, enabling the T-cell response to remain robust against emerging variants.95–100 Sette’s group has compared the immune memory profile to four COVID-19 vaccines. They found that mRNA vaccines are consistently the most immunogenic; however, antibody levels wane over time. For all the vaccines tested, T-cell responses were fairly stable over the 6-month period. These results are consistent with the high degree of protection against hospitalization and death that we continue to see for the COVID-19 vaccines even as protection against infection wanes.101
Antibody correlates of protection from dengue virus infection
Eva Harris from the University of California, Berkeley presented work on defining Fc-associated antibody correlates of protection from dengue virus (DENV) infection. She described key findings from the Pediatric Dengue Cohort Study, a 19-year study from 2004 to the present of 4000 children aged 2 to 17 years in Managua, Nicaragua. The study contains samples collected from individuals before and after infections, enabling researchers to look for markers of both asymptomatic and symptomatic infections. One of the greatest risk factors for severe dengue disease is subsequent infection with a different DENV serotype. Within the cohort study, researchers compared samples from individuals prior to a secondary symptomatic versus asymptomatic infection with DENV serotype 3. They found that while both groups had similar levels of neutralizing antibodies, asymptomatic individuals had higher pre-existing total anti-DENV antibody levels. Profiling the antibodies’ Fc effector functions revealed that higher antibody-dependent complement deposition and ability to mediate virolysis via anti-envelope (E) and anti-nonstructural protein 1 (NS1) antibodies were associated with subsequent asymptomatic infection.102 These data highlight the importance of antibody Fc functions in protection from symptomatic DENV infection.102 Harris also presented data from an ongoing study within the cohort to tease out Fc antibody functions that correlate with subsequent classic dengue and severe dengue disease, finding that antibodies that mediated complement deposition and virolysis were protective against subsequent severe disease.
Targeting hypoxia inducible factor in SARS-CoV-2 infection
Peter Wing from the University of Oxford presented work on the impact of hypoxia inducible factors (HIFs) on SARS-CoV-2 infection. HIFs impact a wide range of viruses in a context-specific manner.103 Wing showed that activating HIFs inhibits SARS-CoV-2 entry via a reduction in ACE2 expression. Hypoxia also inhibits later events in the viral life cycle. Inducing HIF expression after viral entry inhibited the establishment of replication complexes.104 The previous work demonstrated the impact of HIF on SARS-CoV-2 in lung epithelial cells. Wing presented unpublished data translating these results into a hamster model of SARS-CoV-2 infection.
Innate immune signaling at the maternal-fetal interface
Julie Eggenberger from Michael Gale’s lab at the University of Washington presented work on understanding the maternal decidual response to Zika infection. Earlier work has shown that Zika virus can infect multiple cell types in the maternal decidual tissues, which make direct contact with the fetal-derived placenta.105 Eggenberger has established several ex vivo and in vitro model systems to study the decidua and innate immune signaling within the tissue. She presented unpublished data on Zika virus and replication within these decidual tissue and cell models.
Dietary and environmental effects of coronavirus infection
Siddharth Krishnamurthy from Yasmine Belkaid’s lab at the NIH presented unpublished work on the dietary and environmental factors that impact coronavirus infection of intestinal cells. Many zoonotic viruses infect the intestinal cells of their hosts and must therefore adapt to the mucosal environment. Krishnamurthy developed a novel model of intestinal murine coronavirus infection and diet and found that virus could only infect specific intestinal compartments when mice were fed specific diets. Krishnamurthy showed that this expanded tissue tropism was likely mediated by the binding of non-spike envelope proteins binding additional attachment sites on the target cells. By understanding how viruses from zoonotic families adapt to intestinal niches, Krishnamurthy hopes to better understand why certain viruses are better poised for spillover.
Viral Evolution and Epidemiology
Coronavirus spike-independent infection of bat cells
Marcus Mah from Gavin Smith’s group at Duke-NUS Medical School presented research on understanding coronavirus evolution by infecting bat cells. Of the seven coronaviruses known to infect humans, five have bat ancestral origins. Mah focused on human coronavirus 229E (HCoV-229E). Mah serially passaged six viruses isolated from human clinical samples in Rhinolophus lepidus bat kidney cells. Next generation sequencing revealed deletions in the spike region as the viruses were passaged through the bat cells (Figure 4). Additional work confirmed that the passaged viruses can infect and replicate within bat cells without the spike protein, which is required for human infection.106 This work is consistent with previous findings that some natural bat coronaviruses do not bind to orthologs of their cognate receptor.107 It also supports a theory proposed several years ago that ancestral coronaviruses only contain the machinery for nonspecific fusion and evolved the domains of the spike protein that provide the specificity to target cognate receptors.108
Figure 4.
Schematic of nucleotide sequences within the spike and ORF4 gene regions. Colored filled rectangles represent parts of the genome in C2R1, C2R5, and C2R10 viruses that are present in the respective passage. Virus isolates names are in bold. Horizontal black lines indicate deleted parts of the genome with the respective size denoted by Δ, starts and ends of the deletions are indicated by the respective nucleotide position.
Understanding SARS-CoV-2 variant advantages
Rita Meganck described unpublished work from her time in Ralph Baric’s lab at the University of North Carolina on understanding why certain SARS-CoV-2 variants won out over prior variants. She showed replication data in several human airway models for SARS-CoV-2 variants isolated from patient samples. Her data illustrate the different growth kinetics and viral tropism between variants.
Genetic heterogeneity in alphaviruses
Donghoon Chung from the University of Louisville presented unpublished data on the role of genetic heterogeneity in alphavirus populations and how it varies within the infectious population and the population as a whole.
Viral Evolution and Epidemiology
Real-time genomic surveillance of SARS-CoV-2
Sebastian Maurer-Stroh from A*STAR described the efforts to provide real-time virus genomic surveillance for SARS-CoV-2. The COVID-19 pandemic marks the first time that real-time surveillance of viral sequences has been available to track an ongoing pandemic. As of mid-2022, the Global Initiative on Sharing Avian Influenza Data (GISAID) has received over 11 million SARS-CoV-2 genomes, contributed by 100 countries within one month of collection. Maurer-Stroh stressed that the success of GISAID stems from the foundation of trust it had established with the scientific community. All the data in the database are free and publicly available, but submitters do not forfeit their rights to the data. Submitted sequences undergo a quality control step and are annotated with quality metrics. The database also contains rich meta-data that includes fields for sequencing technology, sample history, and clinical trial information. Maurer-Stroh emphasized that GISAID is not only a repository for sequences but also provides various tools that enable researchers to see where in the world similar sequences are found and to identify potential emerging variants. The tools can provide insight on whether a mutation is a founder or marker mutation, which can help to identify where a virus comes from, or a phenotypic mutation, which can impact viral fitness. The tool CoVsurver provides researchers with resources and data on the reported impact of different mutations and ranks and predicts which variants are most likely to spread. Similar platforms have been developed for other priority platforms, including influenza (EpiFlu™), RSV (EpiRSV), and monkeypox (EpiPox).
Linking SARS-CoV-2 variant epidemiology to virus biology
Raul Andino from the University of California, San Francisco presented work on understanding entry of coronaviruses and linking epidemiological data from the pandemic to cell biology insights. Andino’s and Kelly’s groups conducted a study of vaccinated and unvaccinated individuals with Delta or Omicron infection. They found that vaccination has little impact on viral load or viral shedding in those who experienced a breakthrough Delta infection and that Delta had a longer duration of viral shedding compared to other lineages. Omicron infection was associated with reduced viral shedding compared to Delta but had higher shedding early in infection, around days 2 and 3.109 To understand how Omicron replaced Delta if it sheds less, Andino’s and Jackson’s groups turned to cell biology. Using human nasal epithelium organoids–air–liquid interface cultures, they found that the virus infects few cells early in infection and observed a spike in infection around 48 hours. They also documented a key role for cilia in early infection and microvilli in later infection. Andino put forth a model in which the mucus layer initially protects cells from infection. At this time point, viruses associate with the cilia, and some can enter the cell. As the virus replicates, newly synthesized viruses associate with actin-rich microvilli. Through mechanisms not yet defined, the virus promotes extension and branching of the microvilli, which enables it to travel through the periciliary layer and reach new cells. They identified two kinases, PAK1/2 and PAK4, that are activated during infection and modulate microvilli extension. Inhibiting either of these enzymes impacted infection at later time points. Regarding Omicron, Andino showed that Omicron establishes infection earlier and spreads faster than previous variants. By 24 hours, when other variants are still establishing infection, Omicron has already spread.110 Andino’s work demonstrates that cilia and microvilli are important pro-virus organelles in the nasal tract and provides insight into the factors that have enabled Omicron to supplant prior variants.
The impact of prior exposure on SARS-CoV-2 evolution
Bette Korber from Los Alamos National Laboratory discussed how pre-existing immunity has shaped the evolution of SARS-CoV-2. SARS-CoV-2 has quickly gone through different variants. However, the transition between variants has not been uniform around the world. For example, in the UK and Europe, there was a clear simple pattern of variant transitions from Alpha to Beta, Delta, and Omicron. However, in other regions of the world, the transition to Alpha had not occurred before the Delta variant came in and dominated. At the time of the conference, the Omicron variant had supplanted all previous variants around the world, and Omicron subvariants, namely BA.5, had begun taking hold. GISAID data show that Omicron BA.5 is the most quickly expanding variant on every continent. Korber’s group has been following variants of interest and found that almost all variants that were expanding had mutations in the N-terminal domain (NTD) and receptor-binding domain (RBD) as well as positive charge additions near the furin cleavage site. Korber showed that selection for transition in variants can result from either enhanced infectivity and/or neutralizing antibody resistance. The recurrence of both NTD and RBD mutations in key epitope regions indicates that immune escape has impacted which variants prevail. This immune evasion was a major selective force that enabled Omicron to predominate. Korber also presented unpublished work on how her group is using the recurrence of mutations between variants to model serum neutralization profiles to predict neutralization of future variants.
Coronaviruses and Respiratory Viruses
Potential mechanisms for long COVID
SARS-CoV-2 emerged as a respiratory virus in late 2019. As we have learned more about COVID-19, it has become apparent the virus impacts nearly every organ system and that a portion of patients can experience persistent symptoms long after the virus is cleared, a phenomenon known as long COVID.
Stanley Perlman from the University of Iowa presented work on understanding the mechanisms behind long COVID and the neurological sequelae of COVID-19. Perlman focused on anosmia, or loss of smell, which was recognized early in the pandemic as an identifiable feature of COVID-19. Perlman proposed that chronic anosmia in people who have recovered from COVID-19 may serve as a marker for an increased risk of neurological sequelae or neurodegenerative disorders.111 Perlman’s group is using mouse models to better understand the mechanism of SARS-CoV-2-mediated olfactory system dysfunction. After confirming that the virus does induce anosmia in mice, they looked at which cells in the olfactory system the virus is infecting. Perlman showed that SARS-CoV-2 does not infect the olfactory neurons, which transmit olfactory signals to the CNS, but rather infect sustentacular cells, which are important for neuron homeostasis. They also noted that infection damaged the cilia in the olfactory epithelium, downregulated the expression of olfactory receptors, and increased the expression of pro-inflammatory cytokines and chemokines. In addition, persistent changes in the olfactory bulb in the brain were observed even though no virus was detected in the brain. Based on these results, Perlman’s group has put forth a model in which infection of the sustentacular cells in the nasal cavity disrupts olfactory neuron function potentially via inflammatory mediators, which impacts the perception of smell and, potentially, sites connected to the olfactory bulb within the brain.112
Reverse genetics systems to probe SARS-CoV-2 variant characteristics
Pei-Yong Shi from the University of Texas Medical Branch discussed the three SARS-CoV-2 reverse genetics systems his group has developed. Typically, reverse genetics systems employ infectious complementary DNA (cDNA) viral clones. Due to the size of SARS-CoV-2, Shi’s group has created an infectious cDNA clone of the virus using seven cDNA fragments. These fragments are ligated for in vitro transcription of viral genomic RNA. The viral RNA transcript is transfected into cells to create recombinant virus with desired mutations.113 Shi’s group has engineered reporter genes into the viral genome that enabled them to develop high-throughput neutralization assay114, which has been used in dose-finding studies of COVID-19 vaccines.115 They also developed a second, trans-complementation system that makes the virus more accessible to researchers. The system results in a single-round infectious SARS-CoV-2 virus that can be safely used in biosafety level 2 labs.116 The third system they developed deletes viral accessory genes not required for replication, resulting in an attenuated virus.117 Shi showed how they have used their reverse genetic systems to understand the viral fitness and transmission of variants. He showed that D614G, an early variant that emerged in 2020, increased viral replication through improved infectivity in cells and primary airway culture. In hamsters, the mutant preferentially enhanced fitness in the upper respiratory tract, leading to enhanced transmission but did not impact disease severity.118 By making the single point mutations of the nine mutations found in the Alpha variant, Shi’s group pinpointed N501Y as a key mutation that increases ACE2 binding, thus enhancing infection and transmission.119 Similar studies on the Delta variant highlighted the importance of the P681R mutation in enhancing viral replication by improving S1/S2 cleavage.120
Cell membrane remodeling during viral infection
Montserrat Bárcena from Leiden University Medical Center presented work on cell membrane remodeling during viral infection. These modified intracellular membranes, known as viral replication organelles, play a key role in viral replication. In particular, they can support viral RNA synthesis by concentrating relevant factors, increasing synthesis efficiency, and/or helping viral elements evade innate immune sensing.121 Bárcena focused on the complicated vesiculotubular networks formed by viruses of the Nidovirales order. Her group is studying coronaviruses and arteriviruses, two distantly related families within Nidovirales, in the hopes of uncovering common features across the order. Electron tomography studies have allowed Bárcena’s group to visualize the membrane networks induced by these viruses, revealing hundreds of interconnected DMVs derived from the ER.117–119 Later work confirmed that viral RNA synthesis occurs at DMVs, likely in their interior.122–124 Using in situ cryo-EM125, Bárcena’s group discovered double-membrane spanning pore complexes that connect the lumen of the DMVs with the cytosol. The complexes contain a hexameric core made of the coronaviral protein nsp3. These DMV-spanning pores appear to be a common feature across nidoviruses. Bárcena proposed that these pores may couple viral RNA synthesis, export, and encapsidation.126
Responses to SARS-CoV-2 in bat cells
Sophie-Marie Aicher from Nolwenn Jouvenet’s lab at the Institute Pasteur Paris presented work on understanding bat species-specific molecular interactions and innate immune responses to SARS-CoV-2. Bats are reservoirs for numerous RNA viruses. While SARS-CoV-2 has not directly been found in bats, the closest known ancestor is a bat beta coronavirus with a sequence identity of 96.8% to SARS-CoV-2. Aicher has established a panel of bat cell models, including primary cells and continuous cell lines of various bat tissues from several bat species. She induced exogenous expression of the human ACE2 receptor to render the cells permissive to SARS-CoV-2 infection and assessed the ability of the virus to replicate and produce infectious virions. Aicher showed that infection with SARS-CoV-2 resulted in distinct species-specific phenotypes in terms of viral replication and release of infectious virions. Further analyses of the molecular mechanisms involved showed that bat cells initiate a potent IFN response upon infection that the virus is unable to evade.127
A QTQTN motif in spike affects replication and disease
Michelle Vu from Vineet Menachery’s lab at the University of Texas Medical Branch presented work on a unique motif found upstream of the SARS-CoV-2 furin cleavage site in the spike protein. The stalk domain of spike contains two cleavage sites that are processed by host proteases and initiate viral entry. Previous work in Menachery’s lab showed that the furin cleavage site, which is unique to SARS-CoV-2, is important for pathogenesis. Structural modeling predictions indicate that deleting this site shortens a loop in spike. This attenuated viral replication in respiratory epithelial cells, reduced spike processing, and attenuated disease in a hamster model.128 Vu’s work has focused on a motif upstream of the furin cleavage site that is only found in SARS-CoV-2 and a closely related bat coronavirus. Structural modeling predictions indicate that deleting that QTQTN motif not only shortens a loop in spike but also makes it more rigid. Deleting this motif attenuated viral replication in vitro in human airway epithelial cells and attenuated disease in vivo in a hamster model. Vu showed that the QTQTN motif is involved in spike processing; deleting it impacts the ability of host proteases to access the furin cleavage site. The motif also contains a glycosylation site that is important for protease interaction and TMPRSS2-mediated entry.129
Probing SARS-CoV-2-mediated inflammation in the lung
Devin James Kenney from Florian Douam’s lab at Boston University presented work on understanding the innate immune cells that drive inflammation and viral clearance in the lung during SARS-CoV-2 infection. To achieve this, Douam’s group has developed a panel of humanized mouse models. Kenney showed that, in a mouse model with engrafted human lung tissue, the tissue is permissive to SARS-CoV-2 infection. In contrast, a co-engrafted model that contains both human lung and a human immune system is protected from infection and severe histopathology. Proteomics and single-cell RNAseq analyses revealed a gene signature of 11 genes whose expression correlated with lung tissue protection. Kenney identified that these genes were upregulated in inflammatory and regulatory macrophages found in the lung grafts in the co-engrafted model during infection.130 He also showed unpublished work that utilized single-cell RNA-seq approaches and in vivo depletion experiments to identify the macrophage populations likely driving viral clearance in infected human lung tissues.
Improving the reverse genetics workflow
Brett Lindenbach from Yale University discussed his collaboration with Yale colleague Farren Isaacs on improving the reverse genetics workflow for RNA viruses. Reverse genetics explores the impact of genotype on phenotypes. Typically, this involved creating libraries of viral mutants and characterizing their infectivity and other phenotypes. The standard workflow for creating these libraries consists of introducing cDNA viral clones into a bacterial plasmid. Creating a robust library of viral mutants can be inefficient and time-consuming via this workflow. In addition, some coronavirus and flavivirus cDNAs are unstable in bacterial plasmids. Methods to overcome this instability, such as divide-and-conquer-based approaches and low copy number vector cells have their pros and cons.131 Lindenbach described how his lab has used the eukaryotic multiplex automated genome editing (eMAGE) system, which enables precise DNA editing in yeast cells132,133, to create complex viral libraries in SARS-CoV-2.
Vaccines and Therapeutics
Novel antibody therapies for COVID-19 and dengue virus
Yukiko Nishida from Chugai Pharmaceutical discussed joint research between the company and A*STAR on developing antibody therapies for COVID-19 and dengue virus. The Chugai Pharmabody Research center, established in Singapore in 2012, has developed several proprietary antibody engineering technologies to optimize drug candidates for clinical purposes. Starting with a neutralizing COVID-19 antibody discovered by A*STAR’s Singapore Immunology Network (SIgN) that blocks RBD/ACE2 binding134, the Chugai Pharmabody Research center employed antibody engineering techniques to increase antibody neutralization activity and improve drug-like characteristics, such as pharmacokinetics and manufacturability. The COVID-19 clinical candidate antibody, 5A6CCS1-SG1095ACT3, has mutations that reduce binding to human FcyRs to reduce the risk of antibody-dependent enhancement of infection as well as mutations that increase binding to FcRn to increase the plasma half-life. Nishida showed that the antibody neutralized all SARS-CoV-2 variants tested and significantly reduced lung viral load in a hamster infection model.135 Chugai Pharmabody Research and SIgN are also working on developing an anti-dengue antibody therapy. In 2017, SIgN researchers identified a human anti-dengue antibody, SIgN-3C, that neutralizes all four dengue virus serotypes.136 Researchers at the Chugai Pharmabody Research center introduced mutations into the antibody to reduce antibody-dependent enhancement, which is known to increase the risk of severe dengue disease. The antibody is currently being developed for clinical purposes.
Effect of age on vaccine-induced immunity
Laurent Rénia from the Lee Kong Chian School of Medicine presented the efforts of PROTECT, a core group of infectious disease clinicians and scientists in Singapore established in early 2020 to address the pandemic. Rénia showed how the group was instrumental in developing new serologic assays for SARS-COV-2.137 In particular, research identifying SARS-CoV-2-specific epitopes on the spike protein led to a serologic test to detect COVID-19 antibodies.138 Additional work showed that high levels of COVID-19 antibodies were associated with more severe disease.139 A separate, flow-cytometry-based assay, S-flow, can detect cell-surface expressed spike and has been used to detect antibody production in infected patients as well as to identify borderline or unknown cases.140,141 The focus of Rénia’s talk, however, was on a recently published study, the SCOPE project, which investigated the effect of age and viral variants on mRNA vaccine-induced immunity. The study consisted of approximately 300 healthcare workers and elderly participants who received the 2-dose regimen of the Pfizer mRNA vaccine. While the vaccine induced high immunogenicity, it waned over time. In addition, approximately one-quarter of participants, most of whom were over 60 years of age, were low antibody responders. Fortunately, the antibody response was boosted with a third dose. In addition to being less likely to respond, elderly participants were also slower to respond; however, their response was more stable over time compared to younger participants. Similar to other reports, there was poor cross-reactivity against the Delta and Omicron variants. Encouragingly, there was no age-based difference in T-cell response. The data demonstrate the importance of booster doses to maintain antibody levels and highlight the fact that the elderly continues to be a high-risk population, even with effective vaccines.142
Targeting neurotrophic alphaviruses
Evan Williams from Colleen Jonsson’s lab at the University of Tennessee presented unpublished work on a new antiviral chemotype, BDGR-164. The small molecule conferred 100%, 95%, and 70% prophylactic protection against lethal intranasal challenge of Venezuelan, Eastern, or Western equine encephalitis virus (V/E/WEEV) in BALB/c, respectively. Infectious viral titer in the brain was reduced five (W/EEEV) and eight (VEEV) logs following prophylactic treatment at 4 days post infection. Transcriptome analyses of the brains of mice at 4 days post-infection revealed that treatment resulted in a significant reduction in viral transcript level and host responses.
Acknowledgments
Work conducted in the Douam Lab is supported in part by a start-up fund and Peter Paul Career Development Professorship from Boston University (to F.D.), grants from the National Institutes of Health (K22 AI144050; to F.D.) and by a Clinical and Translational Science Award from the National Center for Advancing Translational Sciences of the National Institutes of Health (UL1 TR001430; to F.D.).
M. Kielian is supported by Grants AIR01075647 and GMR01057454.
L.F.P. Ng and L. Renia were supported by the Biomedical Research Council (BMRC), A*CRUSE (Vaccine monitoring project), the A*ccelerate GAP-funded project (ACCL/19-GAP064-R20H-H) from Agency of Science, Technology and Research (A*STAR), Singapore National Medical Research Council COVID-19 Research Fund (COVID19RF-001; COVID19RF-007; COVID19RF-0008; COVID19RF-060), US Food and Drug Administration (#75F40120C00085) and A*STAR COVID-19 Research funding (H/20/04/g1/006).
Work by Sonja Best is supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Yukiko Nishida was supported by the Global Health Innovative Technology Fund (GHIT Grant G2016–217.
Footnotes
Competing Interests
A. Sette is a consultant for Gritstone Bio, Flow Pharma, Moderna, AstraZeneca, Qiagen, Avalia, Fortress, Gilead, Sanofi, Merck, RiverVest, MedaCorp, Turnstone, NA Vaccine Institute, Gerson Lehrman Group, and Guggenheim; La Jolla Institute for Immunology has filed for patent protection for various aspects of T-cell epitope and vaccine design work.
L. Renia: a patent application for the S-flow assay has been filed (Singapore patent #10202009679P: A Method of Detecting Antibodies And Related Products).
References
- 1.Smith EC, Sexton NR & Denison MR 2014. Thinking Outside the Triangle: Replication Fidelity of the Largest RNA Viruses. Annu. Rev. Virol 1: 111–132. [DOI] [PubMed] [Google Scholar]
- 2.Minskaia E, Hertzig T, Gorbalenya AE, et al. 2006. Discovery of an RNA virus 3’->5’ exoribonuclease that is critically involved in coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U. S. A 103: 5108–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckerle LD, Lu X, Sperry SM, et al. 2007. High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol 81: 12135–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eckerle LD, Becker MM, Halpin RA, et al. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6: e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith EC, Blanc H, Surdel MC, et al. 2013. Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9: e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graham RL, Becker MM, Eckerle LD, et al. 2012. A live, impaired-fidelity coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med 18: 1820–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Agostini ML, Andres EL, Sims AC, et al. 2018. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio 9: e00221–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gribble J, Stevens LJ, Agostini ML, et al. 2021. The coronavirus proofreading exoribonuclease mediates extensive viral recombination. PLoS Pathog. 17: e1009226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das PK & Kielian M. 2021. Molecular and Structural Insights into the Life Cycle of Rubella Virus. J. Virol JVI.02349–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong P, Agosto LM, Munro JB, et al. 2013. Cell-to-cell transmission of viruses. Curr. Opin. Virol 3: 44–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martinez MG & Kielian M. 2016. Intercellular Extensions Are Induced by the Alphavirus Structural Proteins and Mediate Virus Transmission. PLoS Pathog. 12: e1006061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corona AK, Saulsbery HM, Corona Velazquez AF, et al. 2018. Enteroviruses Remodel Autophagic Trafficking through Regulation of Host SNARE Proteins to Promote Virus Replication and Cell Exit. Cell Rep. 22: 3304–3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardner AP & Barbieri JT 2018. Light Chain Diversity among the Botulinum Neurotoxins. Toxins 10: E268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cortese M, Lee J-Y, Cerikan B, et al. 2020. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe 28: 853–866.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twu W-I, Lee J-Y, Kim H, et al. 2021. Contribution of autophagy machinery factors to HCV and SARS-CoV-2 replication organelle formation. Cell Rep. 37: 110049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabata K, Prasad V, Paul D, et al. 2021. Convergent use of phosphatidic acid for hepatitis C virus and SARS-CoV-2 replication organelle formation. Nat. Commun 12: 7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahola T. 2019. New Phylogenetic Grouping of Positive-Sense RNA Viruses Is Concordant with Replication Complex Morphology. mBio 10: e01402–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Button JM, Qazi SA, Wang JC-Y, et al. 2020. Revisiting an old friend: new findings in alphavirus structure and assembly. Curr. Opin. Virol 45: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voss JE, Vaney M-C, Duquerroy S, et al. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468: 709–712. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Jose J, Xiang Y, et al. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468: 705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons DL, Vaney M-C, Roussel A, et al. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427: 320–325. [DOI] [PubMed] [Google Scholar]
- 22.Sjöberg M, Lindqvist B. & Garoff H. 2011. Activation of the alphavirus spike protein is suppressed by bound E3. J. Virol 85: 5644–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder AJ & Mukhopadhyay S. 2012. The alphavirus E3 glycoprotein functions in a clade-specific manner. J. Virol 86: 13609–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uchime O, Fields W. & Kielian M. 2013. The role of E3 in pH protection during alphavirus assembly and exit. J. Virol 87: 10255–10262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren SC, Qazi SA, Towell B, et al. 2022. Mutations at the Alphavirus E1’-E2 Interdimer Interface Have Host-Specific Phenotypes. J. Virol 96: e0214921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang R, Kim AS, Fox JM, et al. 2018. Mxra8 is a receptor for multiple arthritogenic alphaviruses. Nature 557: 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basore K, Kim AS, Nelson CA, et al. 2019. Cryo-EM Structure of Chikungunya Virus in Complex with the Mxra8 Receptor. Cell 177: 1725–1737.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim AS, Zimmerman O, Fox JM, et al. 2020. An Evolutionary Insertion in the Mxra8 Receptor-Binding Site Confers Resistance to Alphavirus Infection and Pathogenesis. Cell Host Microbe 27: 428–440.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma H, Kim AS, Kafai NM, et al. 2020. LDLRAD3 is a receptor for Venezuelan equine encephalitis virus. Nature 588: 308–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basore K, Ma H, Kafai NM, et al. 2021. Structure of Venezuelan equine encephalitis virus in complex with the LDLRAD3 receptor. Nature 598: 672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn RJ, Zhang W, Rossmann MG, et al. 2002. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell 108: 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pierson TC, Fremont DH, Kuhn RJ, et al. 2008. Structural insights into the mechanisms of antibody-mediated neutralization of flavivirus infection: implications for vaccine development. Cell Host Microbe 4: 229–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sevvana M. & Kuhn RJ 2020. Mapping the diverse structural landscape of the flavivirus antibody repertoire. Curr. Opin. Virol 45: 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sevvana M, Rogers TF, Miller AS, et al. 2020. Structural Basis of Zika Virus Specific Neutralization in Subsequent Flavivirus Infections. Viruses 12: E1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon A, Fattorini V, Sama B, et al. 2022. A dual mechanism of action of AT-527 against SARS-CoV-2 polymerase. Nat. Commun 13: 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pillon MC, Frazier MN, Dillard LB, et al. 2021. Cryo-EM structures of the SARS-CoV-2 endoribonuclease Nsp15 reveal insight into nuclease specificity and dynamics. Nat. Commun 12: 636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frazier MN, Dillard LB, Krahn JM, et al. 2021. Characterization of SARS2 Nsp15 nuclease activity reveals it’s mad about U. Nucleic Acids Res. 49: 10136–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frazier MN, Wilson IM, Krahn JM, et al. 2022. Flipped Over U: Structural Basis for dsRNA Cleavage by the SARS-CoV-2 Endoribonuclease. 2022.03.02.480688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peiris JSM, Lai ST, Poon LLM, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet Lond. Engl 361: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ksiazek TG, Erdman D, Goldsmith CS, et al. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med 348: 1953–1966. [DOI] [PubMed] [Google Scholar]
- 41.Drosten C, Günther S, Preiser W, et al. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med 348: 1967–1976. [DOI] [PubMed] [Google Scholar]
- 42.Zaki AM, van Boheemen S, Bestebroer TM, et al. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med 367: 1814–1820. [DOI] [PubMed] [Google Scholar]
- 43.Bchetnia M, Girard C, Duchaine C, et al. 2020. The outbreak of the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): A review of the current global status. J. Infect. Public Health 13: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guan Y, Zheng BJ, He YQ, et al. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302: 276–278. [DOI] [PubMed] [Google Scholar]
- 45.Woo PC, Lau SK & Yuen K. 2006. Infectious diseases emerging from Chinese wet-markets: zoonotic origins of severe respiratory viral infections. Curr. Opin. Infect. Dis 19: 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tu C, Crameri G, Kong X, et al. 2004. Antibodies to SARS coronavirus in civets. Emerg. Infect. Dis 10: 2244–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lau SKP, Woo PCY, Li KSM, et al. 2005. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. U. S. A 102: 14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Shi Z, Yu M, et al. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310: 676–679. [DOI] [PubMed] [Google Scholar]
- 49.Zhou P, Fan H, Lan T, et al. 2018. Fatal swine acute diarrhoea syndrome caused by an HKU2-related coronavirus of bat origin. Nature 556: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lednicky JA, Tagliamonte MS, White SK, et al. 2021. Independent infections of porcine deltacoronavirus among Haitian children. Nature 600: 133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou J, Li C, Sachs N, et al. 2018. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl. Acad. Sci. U. S. A 115: 6822–6827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chiu MC, Li C, Liu X, et al. 2022. A bipotential organoid model of respiratory epithelium recapitulates high infectivity of SARS-CoV-2 Omicron variant. Cell Discov. 8: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao X, Li C, Liu X, et al. 2021. Human Intestinal Organoids Recapitulate Enteric Infections of Enterovirus and Coronavirus. Stem Cell Rep. 16: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Parasa S, Desai M, Thoguluva Chandrasekar V, et al. 2020. Prevalence of Gastrointestinal Symptoms and Fecal Viral Shedding in Patients With Coronavirus Disease 2019: A Systematic Review and Meta-analysis. JAMA Netw. Open 3: e2011335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhou J, Li C, Liu X, et al. 2020. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat. Med 26: 1077–1083. [DOI] [PubMed] [Google Scholar]
- 56.Ander SE, Diamond MS & Coyne CB 2019. Immune responses at the maternal-fetal interface. Sci. Immunol 4: eaat6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Turco MY, Gardner L, Kay RG, et al. 2018. Trophoblast organoids as a model for maternal-fetal interactions during human placentation. Nature 564: 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Turco MY, Gardner L, Hughes J, et al. 2017. Long-term, hormone-responsive organoid cultures of human endometrium in a chemically defined medium. Nat. Cell Biol 19: 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang L, Semmes EC, Ovies C, et al. 2022. Innate immune signaling in trophoblast and decidua organoids defines differential antiviral defenses at the maternal-fetal interface. 2021.03.29.437467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Sanden SMG, Sachs N, Koekkoek SM, et al. 2018. Enterovirus 71 infection of human airway organoids reveals VP1–145 as a viral infectivity determinant. Emerg. Microbes Infect 7: 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chang S-C, Li W-C, Chen G-W, et al. 2012. Genetic characterization of enterovirus 71 isolated from patients with severe disease by comparative analysis of complete genomes. J. Med. Virol 84: 931–939. [DOI] [PubMed] [Google Scholar]
- 62.Tseligka ED, Sobo K, Stoppini L, et al. 2018. A VP1 mutation acquired during an enterovirus 71 disseminated infection confers heparan sulfate binding ability and modulates ex vivo tropism. PLoS Pathog. 14: e1007190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sooksawasdi Na Ayudhya S, Meijer A, Bauer L, et al. 2020. Enhanced Enterovirus D68 Replication in Neuroblastoma Cells Is Associated with a Cell Culture-Adaptive Amino Acid Substitution in VP1. mSphere 5: e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sridhar A, Karelehto E, Brouwer L, et al. 2019. Parechovirus A Pathogenesis and the Enigma of Genotype A-3. Viruses 11: E1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karelehto E, Cristella C, Yu X, et al. 2018. Polarized Entry of Human Parechoviruses in the Airway Epithelium. Front. Cell. Infect. Microbiol 8: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.García-Rodríguez I, van Eijk H, Koen G, et al. 2021. Parechovirus A Infection of the Intestinal Epithelium: Differences Between Genotypes A1 and A3. Front. Cell. Infect. Microbiol 11: 740662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qian X, Nguyen HN, Song MM, et al. 2016. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 165: 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qian X, Jacob F, Song MM, et al. 2018. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc 13: 565–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tang H, Hammack C, Ogden SC, et al. 2016. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 18: 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu M, Lee EM, Wen Z, et al. 2016. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med 22: 1101–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yoon K-J, Song G, Qian X, et al. 2017. Zika-Virus-Encoded NS2A Disrupts Mammalian Cortical Neurogenesis by Degrading Adherens Junction Proteins. Cell Stem Cell 21: 349–358.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qian X, Su Y, Adam CD, et al. 2020. Sliced Human Cortical Organoids for Modeling Distinct Cortical Layer Formation. Cell Stem Cell 26: 766–781.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jacob F, Pather SR, Huang W-K, et al. 2020. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 27: 937–950.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiramel AI, Meyerson NR, McNally KL, et al. 2019. TRIM5α Restricts Flavivirus Replication by Targeting the Viral Protease for Proteasomal Degradation. Cell Rep. 27: 3269–3283.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pfaender S, Mar KB, Michailidis E, et al. 2020. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat. Microbiol 5: 1330–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Megli CJ & Coyne CB 2022. Infections at the maternal-fetal interface: an overview of pathogenesis and defence. Nat. Rev. Microbiol 20: 67–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yockey LJ, Jurado KA, Arora N, et al. 2018. Type I interferons instigate fetal demise after Zika virus infection. Sci. Immunol 3: eaao1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brackney DE, Beane JE & Ebel GD 2009. RNAi targeting of West Nile virus in mosquito midguts promotes virus diversification. PLoS Pathog. 5: e1000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Grubaugh ND, Rückert C, Armstrong PM, et al. 2016. Transmission bottlenecks and RNAi collectively influence tick-borne flavivirus evolution. Virus Evol. 2: vew033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rückert C, Prasad AN, Garcia-Luna SM, et al. 2019. Small RNA responses of Culex mosquitoes and cell lines during acute and persistent virus infection. Insect Biochem. Mol. Biol 109: 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Morazzani EM, Wiley MR, Murreddu MG, et al. 2012. Production of virus-derived ping-pong-dependent piRNA-like small RNAs in the mosquito soma. PLoS Pathog. 8: e1002470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schnettler E, Donald CL, Human S, et al. 2013. Knockdown of piRNA pathway proteins results in enhanced Semliki Forest virus production in mosquito cells. J. Gen. Virol 94: 1680–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Varjak M, Maringer K, Watson M, et al. 2017. Aedes aegypti Piwi4 Is a Noncanonical PIWI Protein Involved in Antiviral Responses. mSphere 2: e00144–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tassetto M, Kunitomi M, Whitfield ZJ, et al. 2019. Control of RNA viruses in mosquito cells through the acquisition of vDNA and endogenous viral elements. eLife 8: e41244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goic B, Stapleford KA, Frangeul L, et al. 2016. Virus-derived DNA drives mosquito vector tolerance to arboviral infection. Nat. Commun 7: 12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schäfer A, Leist SR, Gralinski LE, et al. 2022. A Multitrait Locus Regulates Sarbecovirus Pathogenesis. mBio e0145422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Severe Covid-19 GWAS Group, Ellinghaus D, Degenhardt F, et al. 2020. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med 383: 1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pairo-Castineira E, Clohisey S, Klaric L, et al. 2021. Genetic mechanisms of critical illness in COVID-19. Nature 591: 92–98. [DOI] [PubMed] [Google Scholar]
- 89.Young BE, Ong SWX, Ng LFP, et al. 2021. Viral Dynamics and Immune Correlates of Coronavirus Disease 2019 (COVID-19) Severity. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am 73: e2932–e2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ong SWX, Fong S-W, Young BE, et al. 2021. Persistent Symptoms and Association With Inflammatory Cytokine Signatures in Recovered Coronavirus Disease 2019 Patients. Open Forum Infect. Dis 8: ofab156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carissimo G, Xu W, Kwok I, et al. 2020. Whole blood immunophenotyping uncovers immature neutrophil-to-VD2 T-cell ratio as an early marker for severe COVID-19. Nat. Commun 11: 5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tay MZ, Rouers A, Fong S-W, et al. 2022. Decreased memory B cell frequencies in COVID-19 delta variant vaccine breakthrough infection. EMBO Mol. Med 14: e15227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Grifoni A, Weiskopf D, Ramirez SI, et al. 2020. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell 181: 1489–1501.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dan JM, Mateus J, Kato Y, et al. 2021. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science 371: eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tarke A, Sidney J, Kidd CK, et al. 2021. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med 2: 100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grifoni A, Sidney J, Vita R, et al. 2021. SARS-CoV-2 human T cell epitopes: Adaptive immune response against COVID-19. Cell Host Microbe 29: 1076–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tarke A, Coelho CH, Zhang Z, et al. 2022. SARS-CoV-2 vaccination induces immunological T cell memory able to cross-recognize variants from Alpha to Omicron. Cell 185: 847–859.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Keeton R, Tincho MB, Ngomti A, et al. 2022. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature 603: 488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gao Y, Cai C, Grifoni A, et al. 2022. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med 28: 472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Madelon N, Heikkilä N, Sabater Royo I, et al. 2022. Omicron-Specific Cytotoxic T-Cell Responses After a Third Dose of mRNA COVID-19 Vaccine Among Patients With Multiple Sclerosis Treated With Ocrelizumab. JAMA Neurol. 79: 399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Z, Mateus J, Coelho CH, et al. 2022. Humoral and cellular immune memory to four COVID-19 vaccines. Cell 185: 2434–2451.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Dias AG, Atyeo C, Loos C, et al. 2022. Antibody Fc characteristics and effector functions correlate with protection from symptomatic dengue virus type 3 infection. Sci. Transl. Med 14: eabm3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Liu PJ, Balfe P, McKeating JA, et al. 2020. Oxygen Sensing and Viral Replication: Implications for Tropism and Pathogenesis. Viruses 12: E1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wing PAC, Keeley TP, Zhuang X, et al. 2021. Hypoxic and pharmacological activation of HIF inhibits SARS-CoV-2 infection of lung epithelial cells. Cell Rep. 35: 109020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.El Costa H, Gouilly J, Mansuy J-M, et al. 2016. ZIKA virus reveals broad tissue and cell tropism during the first trimester of pregnancy. Sci. Rep 6: 35296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mah MG, Linster M, Low DH, et al. 2022. Spike-independent infection of human coronavirus 229E in bat cells. 2021.09.18.460924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Starr TN, Zepeda SK, Walls AC, et al. 2022. ACE2 binding is an ancestral and evolvable trait of sarbecoviruses. Nature 603: 913–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li F. 2016. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol 3: 237–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Garcia-Knight M, Anglin K, Tassetto M, et al. 2022. Infectious viral shedding of SARS-CoV-2 Delta following vaccination: a longitudinal cohort study. 2022.05.15.22275051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu C-T, Lidsky PV, Xiao Y, et al. 2022. SARS-CoV-2 Replication in Airway Epithelia Requires Motile Cilia and Microvillar Reprogramming. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xydakis MS, Albers MW, Holbrook EH, et al. 2021. Post-viral effects of COVID-19 in the olfactory system and their implications. Lancet Neurol. 20: 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zheng J, Wong L-YR, Li K, et al. 2021. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature 589: 603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xie X, Muruato A, Lokugamage KG, et al. 2020. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 27: 841–848.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Muruato AE, Fontes-Garfias CR, Ren P, et al. 2020. A high-throughput neutralizing antibody assay for COVID-19 diagnosis and vaccine evaluation. Nat. Commun 11: 4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mulligan MJ, Lyke KE, Kitchin N, et al. 2020. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature 586: 589–593. [DOI] [PubMed] [Google Scholar]
- 116.Zhang X, Liu Y, Liu J, et al. 2021. A trans-complementation system for SARS-CoV-2 recapitulates authentic viral replication without virulence. Cell 184: 2229–2238.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Liu Y, Zhang X, Liu J, et al. 2022. A live-attenuated SARS-CoV-2 vaccine candidate with accessory protein deletions. Nat. Commun 13: 4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Plante JA, Liu Y, Liu J, et al. 2021. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 592: 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Liu Y, Liu J, Plante KS, et al. 2022. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 602: 294–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu Y, Liu J, Johnson BA, et al. 2022. Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. Cell Rep. 39: 110829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Romero-Brey I. & Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6: 2826–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Snijder EJ, Limpens RWAL, de Wilde AH, et al. 2020. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 18: e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Knoops K, Kikkert M, van den Worm SHE, et al. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6: e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Knoops K, Bárcena M, Limpens RWAL, et al. 2012. Ultrastructural characterization of arterivirus replication structures: reshaping the endoplasmic reticulum to accommodate viral RNA synthesis. J. Virol 86: 2474–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Narayan K. & Subramaniam S. 2015. Focused ion beams in biology. Nat. Methods 12: 1021–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wolff G, Limpens RWAL, Zevenhoven-Dobbe JC, et al. 2020. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369: 1395–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Aicher S-M, Streicher F, Chazal M, et al. 2022. Species-Specific Molecular Barriers to SARS-CoV-2 Replication in Bat Cells. J. Virol 96: e0060822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Johnson BA, Xie X, Bailey AL, et al. 2021. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591: 293–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Vu MN, Lokugamage KG, Plante JA, et al. 2022. QTQTN motif upstream of the furin-cleavage site plays a key role in SARS-CoV-2 infection and pathogenesis. Proc. Natl. Acad. Sci. U. S. A 119: e2205690119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kenney DJ, O’Connell AK, Turcinovic J, et al. 2022. Humanized mice reveal a macrophage-enriched gene signature defining human lung tissue protection during SARS-CoV-2 infection. Cell Rep. 39: 110714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lindenbach BD 2022. Reinventing positive-strand RNA virus reverse genetics. Adv. Virus Res 112: 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Barbieri EM, Muir P, Akhuetie-Oni BO, et al. 2017. Precise Editing at DNA Replication Forks Enables Multiplex Genome Engineering in Eukaryotes. Cell 171: 1453–1467.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Liang Z, Metzner E. & Isaacs FJ 2020. Advanced eMAGE for highly efficient combinatorial editing of a stable genome. 2020.08.30.256743. [Google Scholar]
- 134.Asarnow D, Wang B, Lee W-H, et al. 2021. Structural insight into SARS-CoV-2 neutralizing antibodies and modulation of syncytia. Cell 184: 3192–3204.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kuramochi T, Gan SW, Ho AWS, et al. 2022. Comprehensive engineering of a therapeutic neutralizing antibody targeting SARS-CoV-2 spike protein to neutralize escape variants. mAbs 14: 2040350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Xu M, Zuest R, Velumani S, et al. 2017. A potent neutralizing antibody with therapeutic potential against all four serotypes of dengue virus. NPJ Vaccines 2: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee CY-P, Lin RTP, Renia L, et al. 2020. Serological Approaches for COVID-19: Epidemiologic Perspective on Surveillance and Control. Front. Immunol 11: 879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Poh CM, Carissimo G, Wang B, et al. 2020. Two linear epitopes on the SARS-CoV-2 spike protein that elicit neutralising antibodies in COVID-19 patients. Nat. Commun 11: 2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Amrun SN, Lee CY-P, Lee B, et al. 2020. Linear B-cell epitopes in the spike and nucleocapsid proteins as markers of SARS-CoV-2 exposure and disease severity. EBioMedicine 58: 102911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Goh YS, Chavatte J-M, Lim Jieling A, et al. 2021. Sensitive detection of total anti-Spike antibodies and isotype switching in asymptomatic and symptomatic individuals with COVID-19. Cell Rep. Med 2: 100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Goh YS, Ng LFP & Renia L. 2021. A flow cytometry-based assay for serological detection of anti-spike antibodies in COVID-19 patients. STAR Protoc. 2: 100671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Renia L, Goh YS, Rouers A, et al. 2022. Lower vaccine-acquired immunity in the elderly population following two-dose BNT162b2 vaccination is alleviated by a third vaccine dose. Nat. Commun 13: 4615. [DOI] [PMC free article] [PubMed] [Google Scholar]