ABSTRACT
Highly contagious respiratory illnesses like influenza and COVID-19 pose serious risks to public health. A two-in-one vaccine would be ideal to avoid multiple vaccinations for these diseases. Here, we generated a chimeric receptor binding domain of the spike protein (S-RBD) and hemagglutinin (HA)-stalk-based vaccine for both SARS-CoV-2 and influenza viruses. The S-RBD from SARS-CoV-2 Delta was fused to the headless HA from H1N1 (H1Delta), creating a chimera that forms trimers in solution. The cryo-electron microscopy structure of the chimeric protein complexed with the RBD-targeting CB6 and the HA-stalk-targeting CR9114 antibodies shows that the trimeric protein is stable and accessible for neutralizing antibody binding. Immunization with the vaccine elicited high and long-lasting neutralizing antibodies and effectively protected mice against the challenges of lethal H1N1 or heterosubtypic H5N8, as well as the SARS-CoV-2 Delta or Omicron BA.2 variants. Overall, this study offers a two-in-one universal vaccine design to combat infections caused by both SARS-CoV-2 variants of concern and influenza viruses.
KEYWORDS: Universal vaccine, SARS-CoV-2, Omicron, influenza, subunit vaccine, chimeric antigen, broadly neutralizing antibody
Introduction
Since the twentieth century, humanity has experienced four influenza pandemics (1918 Spanish, 1957 Asian, 1968 Hong Kong, and 2009 swine influenza) and a 1977 Russian epidemic, each resulting in significant economic and human losses [1]. More recently, the world has been being struck by the coronavirus disease (COVID-19) pandemic. These two viral pandemics have had profound impacts on human society, leading to unprecedented economic challenges and tragic loss of life [2,3]. Both diseases, caused by enveloped respiratory virus infections, have similar respiratory illness symptoms and can have fatal consequences. The greatest threat to global public health has been the ongoing COVID-19 pandemic. The effectiveness of current COVID-19 vaccines has been challenged by the ongoing spread of highly transmissible variants of concern (VOCs), including the Alpha, Beta, Gamma, Delta, and Omicron variants, and their capacity for breakthrough infections. Moreover, currently available vaccines may be less effective in protecting vulnerable populations, particularly those who are immunocompromised or elderly, and highlights the need for continued research and development of new vaccines.
The influenza A virus (IAV) is the main cause of the contagious respiratory illness known as influenza [4]. There are 16 subtypes of hemagglutinin (HA) and 9 subtypes of neuraminidase (NA) of IAVs found in mammals and birds, as well as bat-derived H17N10 and H18N11 [5,6]. In addition to the seasonal influenza viruses H1N1 and H3N2, a number of avian IAVs have recently caused sporadic human infections, including H7N9 [7], H7N4 [8], H5N1 [9], H5N6 [10], H5N8 [11], H10N3 [12], and H3N8 [13]. Although vaccination remains the most effective method of preventing influenza-related illness, seasonal influenza vaccine effectiveness ranges from 10% to 60% due to vaccine strains that may not be well matched to circulating strains. Therefore, the development of a vaccine that can protect against both COVID-19 and influenza is urgently needed [14,15].
Ideally, combining two antigens from these two viruses into one chimeric immunogen through rational design can generate a two-in-one vaccine. The receptor binding domain of the SARS-CoV-2 spike protein (S-RBD), which is the main target of neutralizing antibodies, is a component of almost all vaccine candidates [16,17]. Indeed, the RBD can induce broadly neutralizing antibodies against variants [18,19]. For instance, the Delta-based RBD vaccine confers good protection against Omicron subvariants [20]. Therefore, the Delta-RBD is a reasonable choice to combat the current VOCs of SARS-CoV-2.
For influenza, the viral HA protein, which is responsible for receptor binding and membrane fusion, is the major surface antigen that IAV frequently mutates to evade human herd immunity. The stalk region is less prone to mutation and more conserved across diverse influenza subtypes than the globular head domain [21]. By preventing the low pH-induced HA conformational rearrangement, which then prevents membrane fusion, anti-stalk antibodies exhibit much greater cross-subtype neutralizing activity than anti-head antibodies both in vitro and in vivo [22]. These broadly neutralizing antibodies (NAbs) hold much promise for the development of a universal influenza vaccine. Stalk-only or headless HAs have been designed to induce broadly protective anti-stalk antibodies in an effort to reduce the immunodominance of the head domain of the HA [23,24].
Here, we developed a rationally designed vaccine against SARS-CoV-2 and influenza viruses by fusing the S-RBD of delta SARS-CoV-2 to the headless HA of the H1N1 influenza virus. The cryo-electron microscopy (cryo-EM) structure of the chimeric protein in complex with the HA stalk-targeting antibody CR9114 [25] and the RBD-targeting antibody CB6 [26] demonstrates that the trimeric protein is stable and accessible for binding by NAbs. The subunit vaccine induced high levels of NAbs against both SARS-CoV-2 and influenza proteins in mice. In addition, the vaccinated mice had significantly lower respiratory viral loads and were effectively protected against lethal challenges by H1N1 and heterosubtypic H5N8 influenza viruses, as well as SARS-CoV-2 Delta and Omicron BA.2.
Materials and methods
Cells and viruses
Expi293FTM cells (Thermo Fisher Scientific), Vero cells (ATCC), HEK293 T cells (ATCC), and Madin–Darby Canine Kidney (MDCK) cells (ATCC) were cultured at 37°C under 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 10% fetal bovine serum (FBS). All cell lines tested negative for mycoplasma contamination.
For experiments conducted at the Institute of Microbiology, Chinese Academy of Science (IMCAS), a wildtype A/Brisbane/02/2018 (H1N1) virus (GISAID: EPI_ISL_522424) was used as a homologous H1 challenge virus. The heterosubtypic H5N8 challenge virus was a 6:2 re-assortment virus of A/Astrakhan/3212/2020 (H5N8) (GISAID: EPI_ISL_1038924), in which the HA and NA from A/Astrakhan/3212/2020 (H5N8) are encoded in a A/Puerto Rico/8/1934 (GISAID: EPI_ISL_22622) backbone.
For experiments conducted at the Chinese Center for Disease Control and Prevention (China CDC), Delta variant (NPRC 2.192100004) and Omicron variant (BA.2, NPRC 2.192100010) were propagated in Vero cells and titrated by TCID50 assays on Vero cells.
Animals
Specific pathogen-free (SPF) female BALB/c mice were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd. (licensed by Charles River). All mice were allowed free access to water and a standard chow diet and were provided a 12-hour light and dark cycle (temperature: 20–25°C, humidity: 40–70%). All mice used in this study were in good health and were not involved in other experimental procedures. They were housed under SPF conditions in the laboratory animal facilities at IMCAS and the China CDC. Mice were housed with five companions per cage. The challenge studies with SARS-CoV-2 Delta and Omicron (BA.2) variants were conducted in an animal biosafety level 3 (ABSL3) facility at the China CDC. The age of the mice at the time that experiments were performed is indicated in the corresponding figure legends.
Protein expression and purification
In the H1Delta design, the RBD contained S protein residues 319–537 of the SARS-CoV-2 Delta variant (GenBank: OK091006.1) replacing the head region of the A/Victoria/2570/2019 (H1N1) HA protein (GISAID: EPI1801581), and the two pieces were connected by a linker (GGGG). Four mutations (K325C, V338 K, I341 K and R344Q) were introduced in HA1, and eight (I10 T, F63Y, V66I, K68C, F70Y, L73S, R76C and T93C) were introduced in the region of HA2 to maintain the native conformation of the HA stalk region. At positions 76–89, the GCN4 sequence (CMKQIEDKIEEIESK) was used to replace the viral sequence, and a trimer tag (YIPEAPRDGQAYVRKDGEWVLLSTFL) was added at the C-terminus to make the protein more favorable for trimer formation. The signal peptide sequence of the A/Victoria/2570/2019 (H1N1) HA protein (HA protein residues 1–17) was used for protein secretion, and a hexa-His tag was added to the C-terminus to facilitate further purification. The construct was codon-optimized for mammalian cell expression and synthesized by GenScript, China. Then the construct was cloned into the pCAGGS vector and transiently transfected into Expi293FTM cells. After 2 days, the supernatant was collected, and soluble protein was purified by Ni-affinity chromatography using a 5-mL HisTrapTM HP column (GE Healthcare). The samples were further purified via gel filtration chromatography with a SuperdexTM 200 Increase 10/300 GL column (GE Healthcare) in PBS (10 mM Na2HPO4, 1.76 mM KH2PO4, 137 mM NaCl, and 2.7 mM KCl, pH 7.4). The eluted peaks were analyzed by SDS-PAGE for protein size and purity. SDS-PAGE and analytical gel filtration analyses show that the protein had an acceptable yield and purity.
The fragment of antibody-binding (Fab) of human monoclonal antibodies (mAbs) CR9114 and CB6 were transiently expressed in Expi293FTMcells. The cells were transfected with pCAGGS plasmids containing coding sequences for the immunoglobulin heavy chain and light chain and harvested after 5 days. The supernatant was then collected and purified. Purified H1Delta, CR9114 Fab, and CB6 Fab were ready for immunization and SPR experiments. For structural analyses, H1Delta, CR9114 Fab and CB6 Fab were incubated at the ratio of 1:2:2 and further purified by gel filtration chromatography with a SuperdexTM 200 Increase 10/300 GL column was employed in a buffer composed of 20 mM Tris-HCl (pH8.0) and 150 mM NaCl.
SPR assay
The SPR assays were performed at 25°C using a BIAcore 8 K machine with streptavidin chips (SA chips, GE Healthcare). The buffers for the protein used for kinetic analyses was exchanged to PBST (10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4, 137 mM NaCl, 2.7 mM KCl, and 0.05% Tween 20). Purified H1Delta protein was biotinylated and immobilized on the chip. Serial dilutions of Fabs (CR9114 and CB6) were prepared and flowed over the chip surface. Data were collected over time. The apparent equilibrium dissociation constants (apparent binding affinity, KD) for each antibody were calculated using BIAcore 8000 analysis software (BIAevaluation v3.0). Each set of equilibrium binding responses was fitted to the 1:1 binding model.
Cryo-EM data collection and 3D reconstruction
For the HA-RBD chimeric protein bound to the CB6 Fab and CR9114 Fab, an aliquot of 4 μL solution (0.5 mg/mL) was applied to glow-discharged Quantifiol R 2/1 holey carbon grids and blotted for 2.5 s with a humidity of 90% before being plunged into liquid ethane using a Vitrobot Mark IV (Thermo Fisher). The frozen grid was loaded onto a Titan Krios cryo-transmission electron microscope (Thermo Fisher) that is equipped with a BioQuantum energy filter, operated at 300 kV for data collection. Automatic data collection was performed using EPU software. Movies were recorded with a Gatan K3 direct electron detector in super-resolution counting mode at a pixel size of 1.01 Å. The exposure was performed with a dose rate of 15 e-/pixel/s and an accumulative dose of 50 e-/Å2 for each movie, which was fractionated into 36 sub-frames. The final defocus range of the dataset was approximately – (1.0–2.0) μm. Drift correction for all stacks was performed with MotionCor2 [27]. Initial contrast transfer function (CTF) values for each micrograph were calculated with CTFFIND4.1 [28]. Micrographs with an estimated resolution limit > 6.0 Å were discarded in the initial screening. The subsequent image processing and reconstruction were performed using Relion-3.1 [29] and cryoSPARC [30]. Particles (394,000) were picked from 3,185 micrographs. Then, the picked particles were extracted and subjected to three rounds of reference-free 2D classification in cryoSPARC. A clean dataset with 208,000 particles from good 2D classes were selected, and the initial model was generated by cryoSPARC ab initio. Then, the model was used as a reference in Relion for 3D classification. After the second round of 3D classification without applying symmetry, the predominant class contained a subset of 29,468 good particles. These particles were subjected to 3D refinement, which yielded a reconstruction at ∼13 Å resolution as determined by the Fourier shell correlation (FSC) 0.143 cut-off value. The specific images processing and reconstruction procedures are shown in Figure S2. Due to the fierce flexibility between the global and stalk domain of the chimeric protein, we could only obtain low resolution maps as described above. However, we fitted the crystal structure of the RBD/CB6 complex (PDB: 7C01) or HA/CR9114 (PDB: 5CJQ) into the density maps using CHIMERA [31], which showed a high degree of matching.
Ethics statement
This study was performed in strict accordance with the recommendations described in the Guide for the Care and Use of Laboratory Animals of the Institute of Microbiology, Chinese Academy of Sciences (IMCAS) Ethics Committee. All of the animal experiments were reviewed and approved by the Committee on the Ethics of Animal Experiments of IMCAS and the Ethics Committee of the National Institute for Viral Disease Control and Prevention, China CDC.
Immunization of mice with the H1Delta vaccine
Six-to-eight-week-old female BALB/c mice were immunized intramuscularly (i.m.) at day 0, day 21, and day 42 with H1Delta-Addavax (2 μg/100 μL/mouse or 10 μg/100 μL/mouse) or PBS (control). Serum samples were collected at the indicated times after vaccination and analyzed for antibody responses and neutralizing antibodies as described below.
Challenge of mice with H1N1 and H5N8
Two-to-four weeks after boost immunization, mice were challenged with a lethal dose of influenza A/Brisbane/02/2018 (H1N1) (20 LD50) [32] or A/Astrakhan/3212/2020 (H5N8) (10 LD50) [33]. After challenge, the mice were monitored for 14 days to record body weight changes and survival rates. Weight loss of > 20% was considered as the survival endpoint. For determination of the viral load analysis, three mice were euthanized 3 days following challenge, and the left lobe of the lung was collected. The right lung was used for histopathology analyses.
Determination of lung influenza viral loads
Influenza viral loads in lung homogenates were determined by a TCID50 assay using MDCK cells. Briefly, MDCK cells were seeded in 96-wells flat-bottom plates and incubated overnight (37°C, 5% CO2). The cells were washed with PBS and then incubated for 48 h at 37°C with serial dilutions of the lung homogenates, in quadruplicate, in DMEM supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin, and 2 μg/mL of TPCK-treated trypsin. TCID50 values were calculated using the Reed-Muench method.
Challenge of mice with Delta and Omicron SARS-CoV-2 variants
To evaluate the protection efficacy of vaccine candidates against Delta and Omicron variants, the immunized mice were challenged with 5 × 105 TCID50 of Delta variant (NPRC 2.192100004) or Omicron variant (BA.2, NPRC 2.192100010) via the intranasal route. For Delta variant challenge experiments, the BALB/c mice were transduced intranasally with 8 × 109 vp of Ad5-hACE 5 days before the SARS-CoV-2 infection. Three days post challenge, all mice were euthanized and necropsied, and lung tissues were collected for virus titration and pathological examination. The mice experiments with Delta and Omicron variant challenge were conducted in ABSL3 facilities at the China CDC. SARS-CoV-2-specific qRT-PCR assays were performed using a TaqMan Fast Virus 1-Step Master Mix kit (Thermo Fisher Scientific, USA) on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad, USA) according to the manufacturer’s protocol. Two sets of primers and probes were used to detect a region of the N gene of the viral genome [34] and a region of the E gene of the sgRNA of SARS-CoV-2 [35], respectively, with sequences as follows: gRNA-F, GACCCCAAAATCAGCGAAAT; gRNA-R, TCTGGTTACTGCCAGTTGAATCTG; gRNA-probe, ACCCCGCATTACGTTTGGTGGACC (Omicron variant gRNA-probe: ACTCCGCATTACGTTTGGTGGACC); sgRNA-F, CGATCTCTTGTAGATCTGT TCTC; sgRNA-R, ATATTGCAGCAGTACGCACACA; and sgRNA-probe, ACACTAGCCATCCTTACTGCGCTTCG. Mice lung tissues were stained with H&E for pathological examination. Due to the tight schedule at P3 laboratory, our animal protection experiment for SARS-CoV-2 was postponed for more than two months. However, based on our long-term immunity test results, antibody titers remained at high levels. As a result, we were still able to complete the animal experiment as planned.
ELISA
Influenza virus HA- and SARS-CoV-2 RBD-specific antibodies were determined in sera by ELISAs. Briefly, ELISA plates were coated overnight with 2 μg/mL of HA or RBD recombinant protein in 0.05 M carbonate–bicarbonate buffer, pH 9.6, and blocked with 5% fat-free milk in PBS. Serum samples were twofold serially diluted and added to each well. Plates were incubated with goat anti-mouse IgG-HRP (1:5,000) antibody (Abcam) for 1 h at 37°C and developed with 3,3’,5,5’-tetramethylbenzidine (TMB) substrate. Reactions were stopped with 2 M H2S04, and the absorbance was measured at 450 nm using a microplate reader (PerkinElmer, USA). The endpoint titers were defined as the highest reciprocal dilution of serum to yield an absorbance > 2.1-fold of the background values. Antibody titer below the limit of detection was determined as half the limit of detection.
Pseudotyped virus neutralization assay
The VSV-ΔG-GFP based SARS-CoV-2 pseudotyped virus was constructed as previously described [36]. For neutralization assays, the heat-inactivated (56°C for 30 min) serum samples were serially diluted and incubated with equivalent pseudovirus (1000 transducing units, TU) at 37°C for 1 h. The mixture was transferred to pre-plated Vero cells in 96 well plates in a 100-μL volume. The TU values were read on a CQ1 confocal image cytometer (Yokogawa) after a 15-h incubation.
Histopathology assay
For histopathology, lung tissues from mice were fixed in 4% neutral-buffered formalin for 48 h, embedded in paraffin, and stained with hematoxylin and eosin (H&E). Images were captured using a LEICA Versa 200 and were processed using software K-Viewer 1.5.5.8.
Statistical analysis
Statistical details of experiments can be found in the figure legends. All data plotted with error bars are expressed as means with s.d. unless otherwise indicated. The P values were generated by analysis data with a two-tail unpaired t test using the Prism 7 program (GraphPad Software). For all figures, p values are represented by the following symbols: *p < 0.05, **p < 0.01, and ***p < 0.001.
Result
Rational design and development of the H1Delta protein vaccine
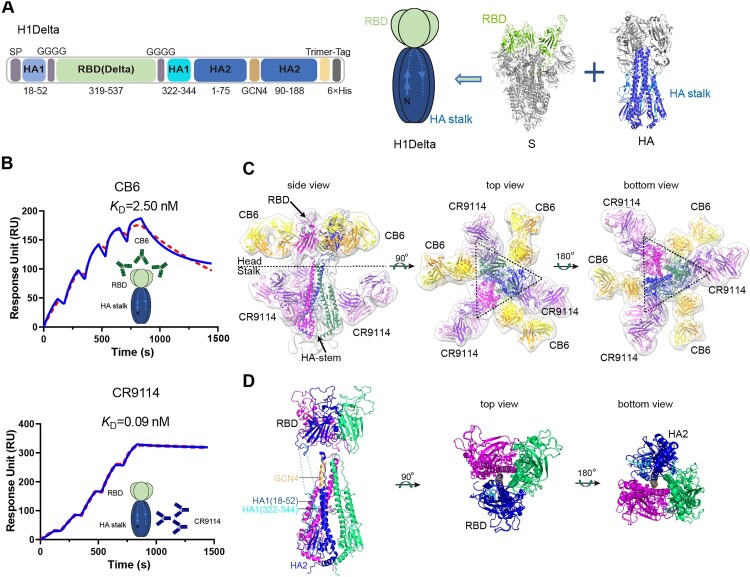
We report a design to combine two antigens together. In the H1Delta design (Figure 1A), the RBD domain contains S protein residues 319–537 of the SARS-CoV-2 Delta variant in place of the head region of the A/Victoria/2570/2019 (H1N1) HA protein (GISAID: EPI1801581), and they were connected by a linker (GGGG). The HA stalk is composed of the N- and C-terminal portions of HA1 and the majority of the HA2 subunit. Four mutations (K325C, V338 K, I341 K and R344Q) were introduced in HA1, and eight (I10 T, F63Y, V66I, K68C, F70Y, L73S, R76C and T93C) were introduced in HA2 to maintain the native conformation of the HA stalk [23]. These included the formation of both an intermolecular (K68C and R76C) disulfide bond and an intramolecular (K325C and T93C) disulfide bond (Supplementary information, Figure S1A). At positions 76–89 of HA2, a GCN4 sequence was used to replace the viral sequence, and a trimer tag was added at the C-terminus to make the protein more favorable for trimer formation [23]. We then expressed the protein in Expi293FTM cells using transient transfection, purified it from the supernatant, and verified it as a trimer (molecular weight ∼ 174 kDa) by analytical gel filtration, reducing and non-reducing SDS-PAGE, and analytical ultracentrifugation (Supplementary information, Figures S1B and S1C). Surface plasmon resonance (SPR) also indicated that the H1Delta protein trimer was correctly folded and could bind both the anti-IAV HA-stalk CR9114 antibody [25] and anti-SARS-CoV-2 S-RBD CB6 antibody [26] (Figure 1B), with affinities similar to those reported previously.
Figure 1.
H1Delta protein vaccine design and structure. (A) Schematic of the H1Delta vaccine design. SP, signal peptide. (B) Representative BIAcore diagrams of H1Delta bound to the SARS-CoV-2 RBD antibody CB6 and influenza HA antibody CR9114. The KD value was calculated using BIAevaluation Version 4.1 (GE Healthcare). (C) Cryo-EM structure of the chimeric protein in complex with the RBD-targeting CB6 antibody and the HA-stalk-targeting CR9114 antibody. (D) Overall structure of the trimeric H1Delta.
Structural analysis of H1Delta by cryo-EM
We next determined the cryo-EM structure of H1Delta in complex with the CR9114 Fab and CB6 Fab (Figure 1C). After image reconstruction by 2D classification and 3D refinement, a 13-Å resolution density map was determined, and the previously published atomic models of the SARS-CoV-2 RBD, RBD/CB6 complex, HA, and HA/CR9114 complex were fitted into the map for rebuilding. The rebuilt models showed their overall conformations. The H1Delta forms a mushroom shaped trimer and can be divided into two parts: a globular head and a stalk region. Three CB6 Fabs bound to three RBD protomers in the globular region, whereas three CR9114 Fabs bound to the headless HA trimer in the stalk region (Figure 1C). This structure demonstrated that the recombinant chimeric immunogen correctly present both the RBD and HA-stalk neutralizing epitopes (Figure 1D). Notably, both the RBD and HA-stalk neutralizing epitopes were well exposed and did not interfere with each other.
Immunogenicity of a H1Delta protein vaccine in mice
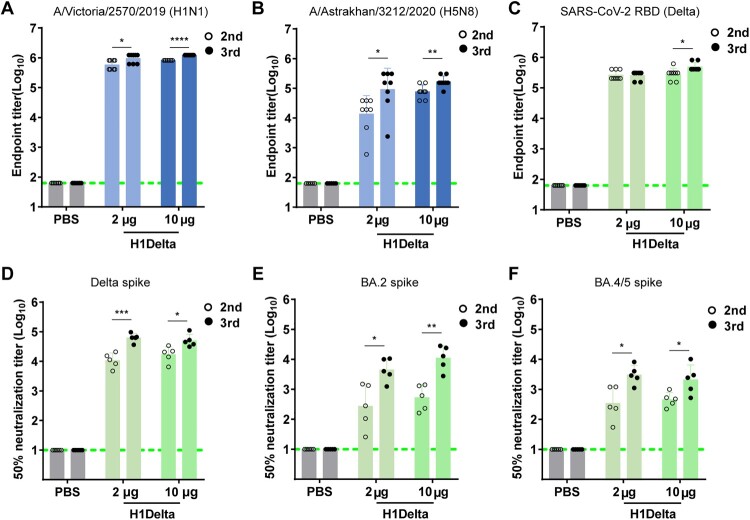
To evaluate the immunogenicity potential of the H1Delta protein against influenza and COVID-19, three immunizations including two groups [high dose (10 μg) and low dose (2 μg)] were given to BALB/c mice, with the combination of AddaVax as adjuvant, 3 weeks apart. PBS plus adjuvant was administered as a control. Serum samples were collected 35 and 56 days after initial vaccination as indicated (Supplementary information, Figure S1D). We first used an ELISA assay to measure H1-specific binding antibodies in the sera of the mice. As shown in Figure 2A, after three immunizations of mice with 2 or 10 μg of H1Delta, the mean endpoint titer of H1-specific IgG rose by nearly 106. As expected, the H1Delta-immunized (10 μg) group elicited cross-reactive H5-specific IgG up to an endpoint titer of ∼105 after the second and third immunizations (Figure 2B).
Figure 2.
Immunogenicity of the H1Delta vaccine in mice. (A) ELISA assays showing the H1-specific IgG titers. (B) ELISA assays showing the H5-specific IgG titers. (C) ELISA assays showing the SARS-CoV-2 Delta-specific IgG titers. (D-F) 50% neutralization titer of pseudotyped virus (Delta, BA.2, and BA.4/5) in serum. P-values were analyzed with two-tail unpaired t test (*p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001).
ELISA-based binding assays demonstrated that vaccinated sera obtained from BALB/c mice after the second and third immunizations exhibited a strong (> 105) binding capacity to the Delta variant RBD (Figure 2C). Meanwhile, we used a panel of pseudotyped viruses (vesicular stomatitis virus backbone) [18,19] displaying SARS-CoV-2 spikes to test the sera neutralizing activities against variants, including Delta, Omicron (BA.2), and Omicron (BA.4/5). H1Delta elicited high levels of NAbs against pseudovirus displaying the Delta spike, with a geometric mean titer (GMT) of 50% neutralization titer (NT50) > 104 after two and three injections (Figure 2D). However, the NT50 GMT was variably reduced against pseudovirus displaying variant spikes. Impressively, 56 days after initial immunization, the NT50 GMT in mice immunized with 10 μg of H1Delta approached ∼10,960 (BA.2) and 3,034 (BA.4/5), respectively (Figures 2E and 2F). These results indicate that the H1Delta protein vaccine is highly effective at eliciting a broad immune response.
Protection efficacy to homologous H1N1 and heterologous H5N8 influenza virus by the H1Delta protein vaccine in mice
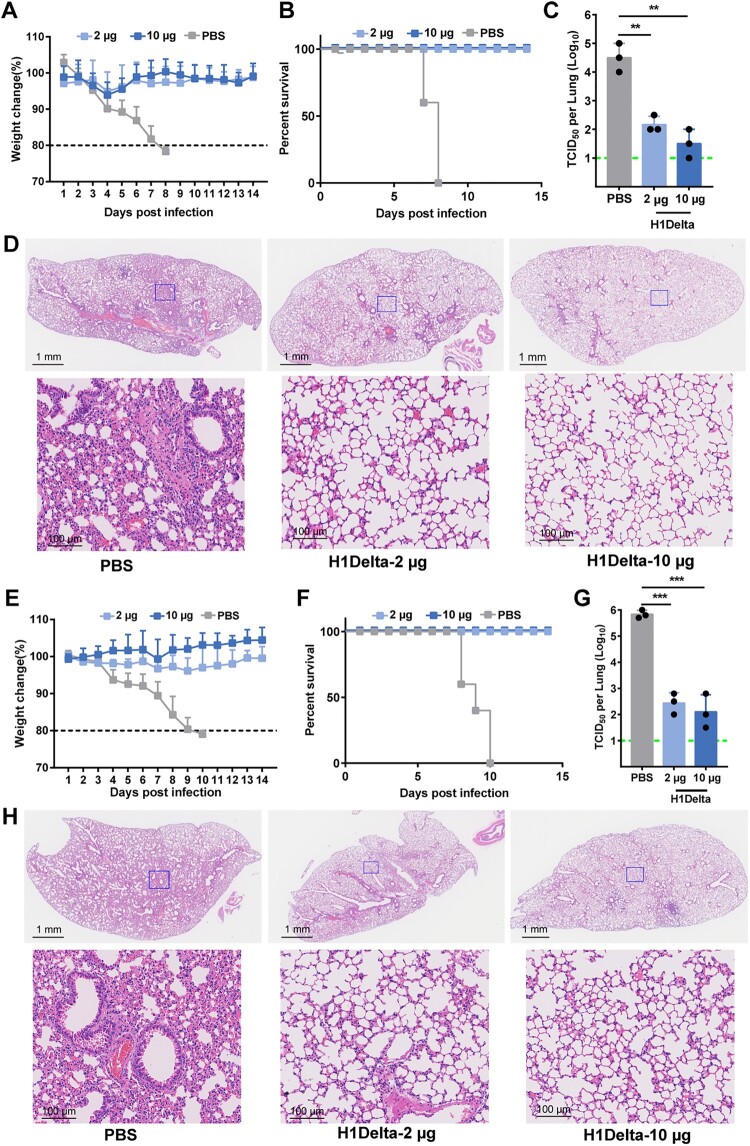
Next, we evaluated the in vivo efficacy of our H1Delta vaccine against H1N1 IAV. BALB/c mice (n = 8) were immunized with a low dose (2 μg) or high dose (10 μg) of H1Delta, with the combination of AddaVax as an adjuvant, three times. PBS plus adjuvant was injected as a control. The immunized mice were then intranasally challenged with 20 LD50 of A/Brisbane/02/2018 (H1N1) virus (Supplementary information, Figure S1E). The H1Delta-vaccinated group exhibited significant high efficacy of protection against lethal H1N1 IAV infection, as evidenced by less weight loss (∼5%) and a 100% survival rate (Figure 3A and B). Compared to the PBS control group, the viral loads in the lungs of the mice in the immunized group decreased significantly (Figure 3C). All lung tissue samples harvested from mice vaccinated with PBS exhibited severe interstitial pneumonia, diffuse inflammatory cell infiltration, pulmonary alveolitis, and necrosis of bronchial epithelial cells (Figure 3D). Milder lesions were observed in mice immunized with H1delta because the pulmonary alveolus was highly visible with lower infiltration of inflammatory cells (Figure 3D). Therefore, H1Delta significantly reduced the lung injury caused by homologous H1N1 IAV.
Figure 3.
Protection efficacy of the H1Delta protein vaccine to homologous H1N1 and heterologous H5N8. (A-D) Groups of 6-to-8-week-old female BALB/c mice (n = 8) were vaccinated with three immunizations of 2 or 10 μg immunogen with an adjuvant of AddaVax in 3-week intervals. PBS with the adjuvant was given as a control. Serum samples were collected 35 and 56 days after initial immunization. Mice were intranasally challenged with 20 LD50 of A/Brisbane/02/2018 (H1N1) virus. Lung tissues were harvested and split for virus titer detection (n = 3) and pathological examination (n = 3), respectively. Vaccine efficacy was assessed by measuring (A) morbidity (weight loss), (B) mortality (survival), (C) lung viral titers on day 3 post challenge, and (D) histological pathology analyses. (E-H) Groups of 6-to-8-week-old female BALB/c mice (n = 8) were vaccinated with three immunizations of 2 or 10 μg immunogen with an adjuvant of AddaVax in 3-week intervals. PBS with the adjuvant was used as a control. Serum samples were collected 35 and 56 days after initial immunization. Mice were intranasally challenged with 10 LD50 of reassortment A/Astrakhan/3212/2020(H5N8) virus. Lung tissues were harvested and split for virus titer detection (n = 3) and pathological examination (n = 3), respectively. Vaccine efficacy was assessed by measuring (E) morbidity (weight loss), (F) mortality (survival), (G) lung viral titers on day 3 post challenge, and (H) histological pathology analyses. Differences were compared using two-tail unpaired t test (**p < 0.01 and ***p < 0.001).
We next assessed whether H1Delta could protect mice against lethal challenge with heterologous H5N8 IAV. BALB/c (n = 8) mice were immunized using the same three-dose vaccination regimen as above (Supplementary information, Figure S1E). The immunized mice were then intranasally challenged with 10 LD50 of reassortant A/Astrakhan/3212/2020(H5N8) virus. Similar to the results obtained during the homologous pH1N1 challenge, weight loss was minimal (< 4%) in all vaccinated groups, whereas humane end points were reached in the PBS control group (Figure 3E and F). Furthermore, vaccinated mice had lower viral loads in the lung compared to PBS controls (Figure 3G). Histopathological analyses also indicated that control mice had extensive lung damage, with consolidated lesions and inflammatory cell infiltration across larger areas. In contrast, the vaccine prevented tissue damage to a large degree, with only minor perivascular and alveolar infiltrates observed in very few areas (Figure 3H). In conclusion, the H1Delta vaccine conferred complete protection from mortality and partial protection from lung damage, even against lethal challenge with a heterologous, potentially pandemic influenza virus.
Protection efficacy to SARS-CoV-2 by the H1Delta protein vaccine in mice
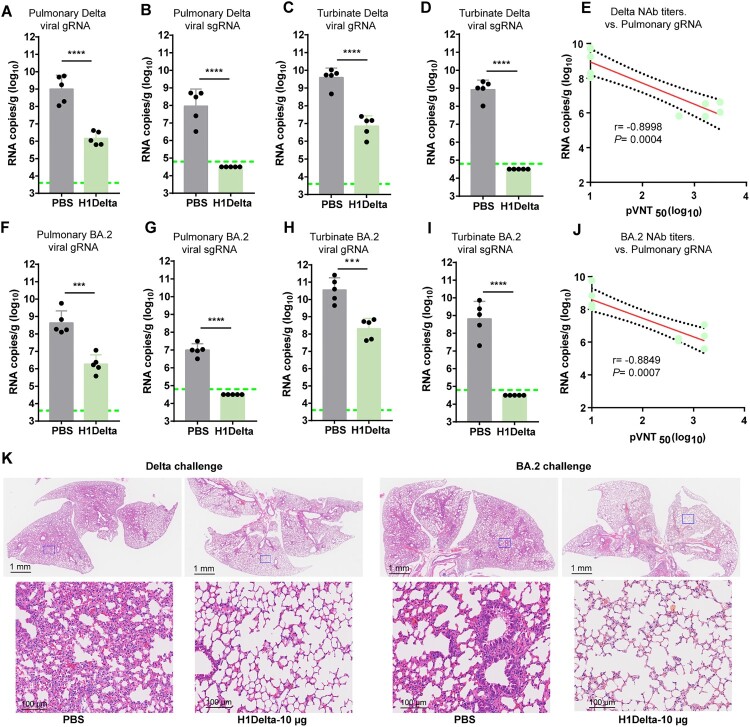
To further explore the protective efficacy of the H1Delta protein vaccine, BALB/c mice (n = 10) immunized with 10 μg vaccine were randomly divided into two groups (n = 5) and challenged with SARS-CoV-2 Delta or Omicron BA.2 variant (Supplementary information, Figures S1F and S1G). Because BALB/c mice are sensitive to Omicron variant but not Delta variant, the mice for the Delta variant challenge were transduced with adenovirus serotype 5 (Ad5) expressing hACE2 five days before virus inoculation [37]. All mice were euthanized and necropsied at 3 days post infection (DPI) to quantify viral genomic (g) RNA and subgenomic (sg) RNA, an indicator of viral replication in the lung and turbinate. For mice challenged with the Delta variant, high levels of viral gRNA were detected in both the lung (average: 1.01 × 109 copies/g) and turbinate (average: 3.91 × 109 copies/g) of PBS-immunized mice (Figure 4A and C). By contrast, significantly reduced viral loads (p < 0.0001) were detected in the lungs (average: 1.45 × 106 copies/g) and turbinates (average: 7.07 × 106 copies/g) of vaccine-immunized mice, with a two-to-three log10 reduction compared to the PBS group (Figure 4A and C). In line with the trends in neutralization, high levels of viral sgRNA were detected in both the lungs (average: 9.13 × 107 copies/g) and the turbinate (average: 8.32 × 108 copies/g) of PBS-treated mice (Figure 4B and D). However, H1Delta-immunized mice displayed undetectable pulmonary and turbinate viral sgRNA, indicating that viral replication was completely controlled (Figure 4B and D). Following vaccination, immune correlates of protection analysis revealed that, according to a linear model, NAb titers strongly correlated with the decline in pulmonary Delta SARS-CoV-2 gRNA (r = −0.8998, p = 0.0004) (Figure 4E). In addition, total anti-RBD IgG titers also show a negative correlation with the viral gRNA load of SARS-CoV-2 (r = −0.9282, p = 0.0001) (Supplementary information, Figure S3A).
Figure 4.
Protection efficacy of the H1Delta protein vaccine to SARS-CoV-2. (A-E) Random selection of five mice in each group that were challenged with 6 × 105 TCID50 of Delta SARS-CoV-2 variant, and (F-G) the other five were challenged with 6 × 105 TCID50 of Omicron (BA.2) variant at 140 days after the primary immunization. Ad5-hACE2 was intravenously administered 5 days before the mice were given the Delta variant challenge. (A) Pulmonary Delta viral gRNA levels were detected by qRT-PCR. (B) Pulmonary Delta viral sgRNA levels were detected by qRT-PCR. (C) Turbinate Delta viral gRNA levels were detected by qRT-PCR. (D) Turbinate Delta viral sgRNA levels were detected by qRT-PCR. (E) Plots showing correlations and corresponding two-sided p values between the pVNT50 of Delta variant (serum samples were collected 140 days after initial immunization) and Delta viral gRNA. (F) Pulmonary Omicron viral gRNA levels were detected by qRT-PCR. (G) Pulmonary Omicron viral sgRNA levels were detected by qRT-PCR. (H) Turbinate Omicron viral gRNA levels were detected by qRT-PCR. (I) Turbinate Omicron viral sgRNA levels were detected by qRT-PCR. (J) Plots showing correlations and corresponding two-sided p values between the pVNT50 of Omicron variant (serum samples were collected 140 days after initial immunization) and Omicron viral gRNA. (K) Histological pathology analyses of lung sections of mice challenged with Delta or Omicron Differences were compared using two-tail unpaired t test (***p < 0.001 and ****p < 0.0001).
For mice challenged with Omicron variant, the averages of pulmonary and turbinate viral gRNA were 4.37 × 108 copies/g and 3.53 × 1010 copies/g in the PBS group but reduced to (236-fold) 1.85 × 106 copies/g and (172-fold) 2.05 × 108 copies/g in the H1Delta vaccine group, respectively (Figure 4F and H). In line with this, the pulmonary and turbinate viral sgRNA loads were detected in all mice in PBS group at high levels (average: 9.98 × 106 copies/g and 6.52 × 108 copies/g) but undetectable in the mice receiving H1Delta vaccine, suggesting the complete control of Omicron viral replication (Figure 4G and I). In the context of Omicron SARS-CoV-2 challenge, NAb titers and pulmonary viral gRNA were also strongly inversely correlated with one another according to a linear model (r = −0.8849, p = 0.0007) (Figure 4J). Similarly, total anti-RBD IgG titers also show a negative correlation with the viral gRNA load of SARS-CoV-2 (r = −0.9044, p = 0.0003) (Supplementary information, Figure S3B).
Histopathological analyses revealed that at 3 DPI with either Delta or Omicron variant, PBS-immunized mice showed moderate-to-severe histopathological changes in lung tissue, including diffuse inflammatory cell infiltration, pulmonary vascular congestion, and the disappearance of alveolar cavities (Figure 4K). As a comparison, mice treated with the H1Delta vaccine exhibited decreased lung injury (Figure 4K). The histopathology results were consistent with the levels of pulmonary and turbinate viral gRNA shown above, demonstrating that H1Delta provided strong protection against both the Delta and Omicron variants.
Duration and long-lasting protection of the humoral response induced by the H1Delta protein vaccine
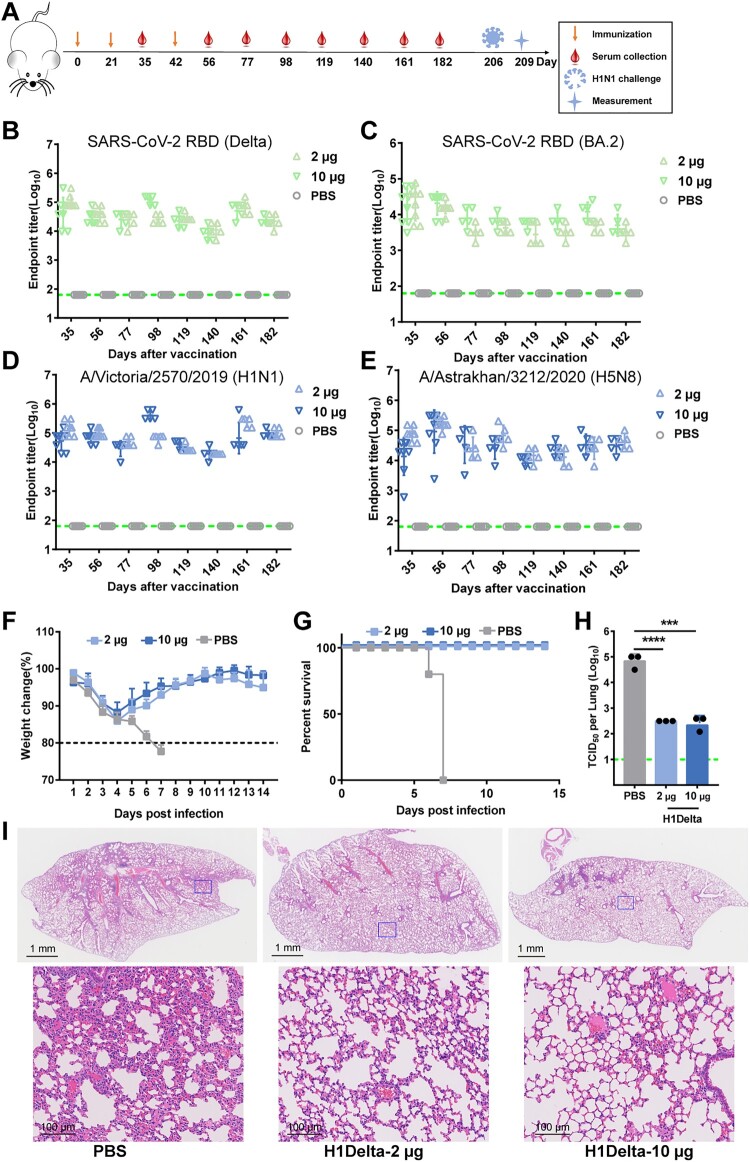
To evaluate the durability and length of protection of H1Delta, BALB/c mice were bled at designated times to explore the kinetics of the induced humoral response (Figure 5A). Notably, at 182 days after the initial immunization, the Delta RBD-specific antibody titer remained > 104 (Figure 5B). Although the antibody titer against the BA.2 RBD decreased slightly, it was near 104, and the influenza H1- and H5-specific antibody titers were both > 104 (Figure 5C, D and E). The mice were ultimately challenged with 20 LD50 of A/Brisbane/02/2018 (H1N1) virus 7 months post vaccination. We found that the body weights of both the 2 μg and 10 μg vaccination group mice showed a moderate decrease (<15%) at 1 DPI but a faster increase after 4 DPI compared to the placebo group (Figure 5F and G). The lungs of H1Delta-immunized mice displayed significantly reduced infectious viral burden (Figure 5H). Histopathological examination revealed severe bronchopneumonia and interstitial pneumonia in the placebo mice. In contrast, only very mild bronchopneumonia was observed in the H1Delta-immunized mice (Figure 5I). These data demonstrated that our H1Delta protein is an effective candidate vaccine and can induce a persistent antibody response and protection.
Figure 5.
Long-lasting protection of the H1Delta protein vaccine. (A) Time course of H1Delta vaccine immune antibody monitoring, viral challenge, and measurement. (B-E) ELISA assay showing the SARS-CoV-2 Delta (B), BA.2 (C), IAV H1N1 (D), and H5N8 (E) specific IgG titers. (F) Body weight changes at 14 days after virus infection. (G) Mouse survival rates were monitored for 14 days. (H) Virus titers in lungs were detected. (I) Histological pathology analyses of lung sections of mice challenged. Differences were compared using two-tail unpaired t test (***p < 0.001 and ****p < 0.0001).
Discussion
It is clear that the SARS-CoV-2 and influenza viruses pose serious risks to public health. It is highly likely that the two viruses will co-circulate globally and regular vaccination against both viruses will be required to minimize the impact of these infections. While the seasonality of SARS-CoV-2 is not yet known, it is reasonable to consider combining vaccination efforts for both viruses to make interventions more cost-effective. Several randomized controlled trials have been started to investigate the potential of providing simultaneous protection against SARS-CoV-2 and influenza viruses by co-administering licensed and/or experimental vaccines against both viruses. While the immune responses generated by these combination vaccines against both viruses are generally comparable to those produced by each vaccine individually, it is anticipated that the public may be less accepting of receiving two different vaccines together compared to being willing to receive either vaccine separately [38]. Nevertheless, the bivalent vaccine has significant potential to offer improved protection, ensure quality control during production, and be easy to use, making it an attractive option for future vaccine development. Although strategies have been proposed to develop universal vaccines against individual viruses [23,24,37,39,40], a combined “universal” vaccine to prevent these two respiratory virus diseases is urgently needed. During the preparation of this manuscript, Cao K. et al. described a chimpanzee adenovirus 68-based vaccine (AdC68-CoV/Flu) that targets SARS-CoV-2 and H7N9 IAV and uses a chimeric immunogen comprised of the S-RBD, the HA2 of H7N9 IAV, and ferritin [41]. However, the N- and C-terminal portions of HA1 are missing from their design. The portions of HA1 involved in the conserved conformational epitopes targeted by broadly targeting NAbs against the HA-stalk, including CR9114, CR6261, FI6, and C179 [25,32,42–46], could help the immunogen remain in the prefusion conformation [47]. Lack of HA1 components could be one of the reasons why the immunogen was not secreted, and we suspect that the recombinant protein might not exist as a stable trimer as ours does [41]. In addition, live-attenuated influenza virus was used for virus vector-based SARS-CoV-2 vaccine development, and intracellular RBD [48,49] or transmembrane RBD [50] were encoded to induce an immune response. However, a unique subunit vaccine is urgently needed. We therefore seek to develop a stable, straightforward-to-produce protein that exposes native-like epitopes for broad-spectrum NAb recognition.
The high viral diversity of IAV poses a challenge to influenza vaccines. Enhancing the cross-reactivity of the immune response is the key to develop a universal influenza vaccine. Targeting the more conserved stalk region is one strategy for stimulating broadly reactive antibodies against the large diversity of HAs [51]. Stalk-directed antibodies display cross-reactivity between subtypes, and even between IAV and type B viruses [52–54]. To induce these stalk-directed antibodies, researchers have investigated a number of vaccination strategies. In spite of this, the immunodominant head domain makes it difficult to induce antibodies against the immuno-subdominant stalk domain [55]. Efforts to overcome this challenge include the development of “headless” HA constructs [23,24,56], hyperglycosylation of the head domain [57], and the development of chimeric [58–61] or mosaic HAs [62] via sequential immunization with “exotic” heads. No one, however, has ever attempted to replace the head with proteins from other viruses. In contrast, the S-RBD is a viable immunogen for COVID-19 vaccine development with few side effects. Numerous studies demonstrate that modified RBD proteins, such as an RBD Fc dimer [63], tandem-repeat dimer [37,64], HR (heptad-repeat sequence)-induced trimer [65], and multivalent nanoparticles can produce more neutralizing antibodies, suggesting that polymeric protein vaccines may stimulate immune responses more potently than monomeric antigens.
In this study, we developed a rationally designed bivalent vaccine against SARS-CoV-2 and influenza viruses using the S-RBD and HA stalk, which we named H1Delta. To preserve a broad spectrum of HA-stalk epitopes and maintain a stable trimer conformation, stabilizing H1 stalk mutations were based on a previous report [23]. The H1Delta protein was correctly folded as a trimer and could be recognized by both the anti-influenza HA-stalk antibody CR9114 and the anti-SARS-CoV-2 S protein antibody CB6, according to the results of analytical ultracentrifugation and SPR assays. Seeing is believing, and the cryo-EM structure of H1Delta in complex with the CR9114 and the CB6 Fabs demonstrates that the trimeric protein is stable and accessible to binding by NAbs. It was encouraging to observe that each RBD monomer is in a relatively stable state. We speculate that this RBD trimer design further enhances its immunogenicity like the SARS-CoV-2 S trimer. The RBD antibody levels produced by this vaccine and our previous dimeric RBD vaccine are comparable [37,64]. Antigenicity data suggest the presentation of native-like conserved epitopes for recognition by broadly neutralizing NAbs. Furthermore, mice immunized with the vaccine had significantly lower respiratory viral loads and were effectively protected against both lethal H1N1 and heterosubtypic H5N8 influenza viruses, as well as the SARS-CoV-2 Delta and Omicron BA.2 VOCs. Notably, long-term durability of high-level antibodies and protection were observed for at least 180 days with only a slight decrease. The longevity of the antibodies against RBD was as good as, if not superior to, those produced by the multivalent nanoparticle vaccine [66]. However, we did not establish a strict control to systematically compare the immunological effect induced by the RBD monomer, HA headless construct, and RBD trimer, which might be done in the future. Whether the two immunogens from two viruses have synergistic effects for inducing immune responses also requires further study. In addition, further studies are needed to evaluate the performance of our chimeric antigen in human populations with pre-existing immunity to SARS-CoV-2 and IAV. Nonetheless, we believe that our chimeric antigen has the potential to overcome the challenges posed by pre-existing immunity through its targeting of the highly conserved regions of both viruses, and could be an important tool in the fight against the COVID-19 pandemic and future influenza or coronavirus outbreaks. More importantly, antibodies of both COVID-19 and influenza declined in a very short period and repeated vaccinations are recommended. Two recent publications of the IAV HA-stalk alone vaccines support our HA-stalk-targeting vaccine for pre-existing immunity population [67,68].
Here, we transduced replication-defective adenoviruses encoding human ACE2 into BALB/c mice through intranasal injection and established hACE2-transduced mice as an infection model. Despite being a well-established mouse model, transduced mice may show mouse-to-mouse variance in hACE2 expression and the possibility for pulmonary tissue and bronchial inflammation associated with AdV delivery. hACE2 knock-in mice or a hamster model may be more suitable for SARS-CoV-2 infection. In addition, the immunogenicity and protection efficacy of the H1Delta chimeric vaccine were not evaluated in a ferret or non-human primate model due to the limited animal supply at this moment.
Our findings indicate that this design could elicit broadly neutralizing NAbs against SARS-CoV-2 Delta and Omicron VOCs, as well as cross-protection against group 1 IAVs such as H1N1 and H5N8. This rational design would thus be a feasible approach for rapidly adapting group 2 IAVs, influenza B viruses, and new SARS-CoV-2 variants in the future. In conclusion, this study proposes a proof-of-concept two-in-one vaccine strategy to fight infections brought on by the new SARS-CoV-2 variants and influenza viruses.
Supplementary Material
Acknowledgments
We thank all of the staff at the Cryo-EM Center of the Southern University of Science and Technology for their assistance. We thank Tianyi Zheng (Zhejiang University), Linjie Li, and Yuxuan Han (University of Chinese Academy of Sciences) for help in protein and qPCR sample preparation. We are grateful to Zheng Fan at the Institute of Microbiology, Chinese Academy of Sciences (IMCAS) for technical assistance with SPR experiments.
Funding Statement
This work was supported by National Key Research and Development Projects of the Ministry of Science and Technology of China: [Grant Number 2020YFA0907102, 2021YFC2301400, 2021YFC2301300); Strategic Priority Research Program of CAS: [Grant Number XDB29010202, XDB29040203]; CAS Project for Young Scientists in Basic Research: [Grant Number YSBR-010]; the National Natural Science Foundation of China: [Grant Number 82122040].
Disclosure statement
Y.L.L., T.H., Y.B., B.Z., H.S., and G.F.G. are listed in the patent as the inventors of the influenza-COVID-19 chimeric protective vaccine. All other authors declare no competing interests.
Author’s contributions
G.F.G., H.S., and B.Z. conceived the project and designed the experiments. S.L., Y.X., and P.H. solved the cryo-EM structures. T.H. expressed and purified the proteins with the help of X.J. Y.B., C.Z., Y.H., and Y.F.L. prepared the influenza viruses. Y.L.L., T.H., and X.W. performed the immunization, evaluation, and influenza virus challenge experiments. Y.L.L., T.H., X.W., Y.L., R.Z., and X.Z. performed the neutralization assays using the VSV-based pseudoviruses. G.W., P.L., K.X., W.L., X.Y.L., and X.C.L. coordinated the challenge experiment at the China CDC ABSL-3. H.S., Y.L.L., T.H., and P.L. analyzed the data. H.S. and Y.L.L. wrote the manuscript. G.F.G. revised the manuscript.
References
- 1.Shi Y, Wu Y, Zhang W, et al. Enabling the ‘host jump’: structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol. 2014 Dec;12(12):822–31. doi: 10.1038/nrmicro3362 [DOI] [PubMed] [Google Scholar]
- 2.Smith GJ, Vijaykrishna D, Bahl J, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459(7250):1122–1125. doi: 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- 3.Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao GF. From “A”IV to “Z”IKV: attacks from emerging and Re-emerging pathogens. Cell. 2018;172(6):1157–1159. doi: 10.1016/j.cell.2018.02.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Y, Wu Y, Tefsen B, et al. Bat-derived influenza-like viruses H17N10 and H18N11. Trends Microbiol. 2014 Apr;22(4):183–191. doi: 10.1016/j.tim.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu WJ, Wu Y, Bi YH, et al. Emerging HxNy influenza A viruses. Cold Spring Harb Perspect Med. 2022 Feb;12(2):a038406. doi: 10.1101/cshperspect [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Bi Y, Wang J, et al. Human infections with recently-emerging highly pathogenic H7N9 avian influenza virus in China. J Infect. 2017;75(1):71–75. doi: 10.1016/j.jinf.2017.04.001 [DOI] [PubMed] [Google Scholar]
- 8.Tong XC, Weng SS, Xue F, et al. First human infection by a novel avian influenza A(H7N4) virus. J Infect. 2018 Sep;77(3):249–257. doi: 10.1016/j.jinf.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 9.de Jong MD, Simmons CP, Thanh TT, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12(10):1203–1207. doi: 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Y, Tan S, Yang Y, et al. Clinical and immunological characteristics of human infections with H5N6 avian influenza virus. Clin Infect Dis. 2019;68(7):1100–1109. doi: 10.1093/cid/ciy681 [DOI] [PubMed] [Google Scholar]
- 11.Pyankova OG, Susloparov IM, Moiseeva AA, et al. Isolation of clade 2.3.4.4b A(H5N8), a highly pathogenic avian influenza virus, from a worker during an outbreak on a poultry farm, Russia, December 2020. Euro Surveill. 2021 Jun;26(24):1560–7917. doi: 10.2807/1560-7917.ES.2021.26.24.2100439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Niu S, Zhang B, et al. The whole genome analysis for the first human infection with H10N3 influenza virus in China. J Infect 2021 Jun 27;S0163-4453(21)00318-2. doi: 10.1016/j.jinf.2021.06.021 [DOI] [PubMed] [Google Scholar]
- 13.Yang R, Sun H, Gao F, et al. Human infection of avian influenza A H3N8 virus and the viral origins: a descriptive study. Lancet Microbe. 2022;3(11):e824–e834. doi: 10.1016/S2666-5247(22)00192-6 [DOI] [PubMed] [Google Scholar]
- 14.Uyeki TM, Hui DS, Zambon M, et al. Influenza. Lancet. 2022;400(10353):693–706. doi: 10.1016/S0140-6736(22)00982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sayers DR. Severe acute respiratory syndrome coronavirus 2 vaccine boosters: An influenza vaccine perspective. Mil Med. 2022 Aug 9;usac243. doi: 10.1093/milmed/usac243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dai L, Gao GF.. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao GF. Science-based COVID-19 vaccine development. Natl Sci Rev. 2021;8(10):nwab193, doi: 10.1093/nsr/nwab193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X, Li D, Ruan W, et al. Effects of a prolonged booster interval on neutralization of Omicron variant. N Engl J Med. 2022;386(9):894–896. doi: 10.1056/NEJMc2119426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao X, Zhang R, Qiao S, et al. Omicron SARS-CoV-2 neutralization from inactivated and ZF2001 vaccines. N Engl J Med. 2022;387(3):277–280. doi: 10.1056/NEJMc2206900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qu L, Yi Z, Shen Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185(10):1728–1744.e16. doi: 10.1016/j.cell.2022.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu R, Ekiert DC, Krause JC, et al. Structural basis of preexisting immunity to the 2009 H1N1 pandemic influenza virus. Science. 2010 Apr 16;328(5976):357–360. doi: 10.1126/science.1186430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skehel JJ, Wiley DC.. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–569. doi: 10.1146/annurev.biochem.69.1.531 [DOI] [PubMed] [Google Scholar]
- 23.Impagliazzo A, Milder F, Kuipers H, et al. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science. 2015;349(6254):1301–1306. doi: 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- 24.Yassine HM, Boyington JC, McTamney PM, et al. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med. 2015;21(9):1065–1070. doi: 10.1038/nm.3927 [DOI] [PubMed] [Google Scholar]
- 25.Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science (New York, NY). 2012 Sep 14;337(6100):1343–1348. doi: 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi R, Shan C, Duan X, et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584(7819):120–124. doi: 10.1038/s41586-020-2381-y [DOI] [PubMed] [Google Scholar]
- 27.Zheng SQ, Palovcak E, Armache JP, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017 Apr;14(4):331–332. doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rohou A, Grigorieff N.. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J Struct Biol. 2015;192(2):216–221. doi: 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zivanov J, Nakane T, Forsberg BO, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018 Nov 9;7:e42166. doi: 10.7554/eLife.42166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Punjani A, Rubinstein JL, Fleet DJ, et al. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14(3):290–296. doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 31.Pettersen EF, Goddard TD, Huang CC, et al. UCSF Chimera? A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 32.Sui J, Hwang WC, Perez S, et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol. 2009 Mar;16(3):265–273. doi: 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boyoglu-Barnum S, Hutchinson GB, Boyington JC, et al. Glycan repositioning of influenza hemagglutinin stem facilitates the elicitation of protective cross-group antibody responses. Nat Commun. 2020;11(1):791. doi: 10.1038/s41467-020-14579-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chandrashekar A, Liu J, Martinot AJ, et al. SARS-CoV-2 infection protects against rechallenge in rhesusmacaques. Science. 2020;369(6505):812–817. doi: 10.1126/science.abc4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 36.Domachowske J, Madhi SA, Simoes EAF, et al. Safety of nirsevimab for RSV in infants with heart or lung disease or prematurity. N Engl J Med. 2022;386(9):892–894. doi: 10.1056/NEJMc2112186 [DOI] [PubMed] [Google Scholar]
- 37.Xu K, Gao P, Liu S, et al. Protective prototype-Beta and Delta-Omicron chimeric RBD-dimer vaccines against SARS-CoV-2. Cell. 2022;185(13):2265–2278.e14. doi: 10.1016/j.cell.2022.04.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Domnich A, Orsi A, Trombetta CS, et al. COVID-19 and seasonal influenza vaccination: cross-protection, Co-administration, combination vaccines, and hesitancy. Pharmaceuticals (Basel, Switzerland). 2022 Mar 8;15(3):322. doi: 10.3390/ph15030322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koff WC, Berkley SF.. A universal coronavirus vaccine. Science. 2021;371(6531):759. doi: 10.1126/science.abh0447 [DOI] [PubMed] [Google Scholar]
- 40.Martinez DR, Schafer A, Leist SR, et al. Chimeric spike mRNA vaccines protect against Sarbecovirus challenge in mice. Science. 2021;373(6558):991–998. doi: 10.1126/science.abi4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao K, Wang X, Peng H, et al. A single vaccine protects against SARS-CoV-2 and influenza virus in mice. J Virol 2022 Feb 23;96(4):e0157821. doi: 10.1128/jvi.01578-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Throsby M, van den Brink E, Jongeneelen M, et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PloS one. 2008;3(12):e3942. doi: 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ekiert DC, Bhabha G, Elsliger MA, et al. Antibody recognition of a highly conserved influenza virus epitope. Science. 2009 Apr 10;324(5924):246–251. doi: 10.1126/science.1171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- 45.Hai R, Krammer F, Tan GS, et al. Influenza viruses expressing chimeric hemagglutinins: globular head and stalk domains derived from different subtypes. J Virol. 2012;86(10):5774–5781. doi: 10.1128/JVI.00137-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dreyfus C, Ekiert DC, Wilson IA.. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol. 2013;87(12):7149–7154. doi: 10.1128/JVI.02975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steel J, Lowen AC, Wang TT, et al. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio. 2010 May 18;1(1):e00018–10. doi: 10.1128/mBio.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu F, Zhuang C, Chu K, et al. Safety and immunogenicity of a live-attenuated influenza virus vector-based intranasal SARS-CoV-2 vaccine in adults: randomised, double-blind, placebo-controlled, phase 1 and 2 trials. The Lancet Respiratory Medicine. 2022;10(8):749–760. doi: 10.1016/S2213-2600(22)00131-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Wang P, Yuan L, et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Science Bulletin. 2022;67(13):1372–1387. doi: 10.1016/j.scib.2022.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaparian RR, Harding AT, Hamele CE, et al. A virion-based combination vaccine protects against influenza and SARS-CoV-2 disease in mice. J Virol 2022 Aug 10;96(15):e0068922. doi: 10.1128/jvi.00689-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu NC, Wilson IA.. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol. 2018 Feb;25(2):115–121. doi: 10.1038/s41594-018-0025-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Joyce MG, Wheatley AK, Thomas PV, et al. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell. 2016;166(3):609–623. doi: 10.1016/j.cell.2016.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corti D, Voss J, Gamblin SJ, et al. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science. 2011;333(6044):850–856. doi: 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- 54.Dreyfus C, Laursen NS, Kwaks T, et al. Highly conserved protective epitopes on influenza B viruses. Science. 2012;337(6100):1343–1348. doi: 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Biswas A, Chakrabarti AK, Dutta S.. Current challenges: from the path of “original antigenic sin” towards the development of universal flu vaccines. Int Rev Immunol. 2020;39(1):21–36. doi: 10.1080/08830185.2019.1685990 [DOI] [PubMed] [Google Scholar]
- 56.van der Lubbe JEM, Verspuij JWA, Huizingh J, et al. Mini-HA is superior to full length hemagglutinin immunization in inducing stem-specific antibodies and protection against group 1 influenza virus challenges in mice. Front Immunol. 2018;9:2350. doi: 10.3389/fimmu.2018.02350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eggink D, Goff PH, Palese P.. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J Virol 2014;88(1):699–704. doi: 10.1128/JVI.02608-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krammer F, Pica N, Hai R, et al. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J Virol 2013;87(12):6542–6550. doi: 10.1128/JVI.00641-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nachbagauer R, Feser J, Naficy A, et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat Med. 2021;27(1):106–114. doi: 10.1038/s41591-020-1118-7 [DOI] [PubMed] [Google Scholar]
- 60.Liu WC, Nachbagauer R, Stadlbauer D, et al. Sequential immunization with live-attenuated chimeric hemagglutinin-based vaccines confers heterosubtypic immunity against influenza A viruses in a preclinical ferret model. Front Immunol. 2019;10:756. doi: 10.3389/fimmu.2019.00756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu WC, Nachbagauer R, Stadlbauer D, et al. Chimeric hemagglutinin-based live-attenuated vaccines confer durable protective immunity against Influenza A viruses in a preclinical ferret model. Vaccines (Basel). 2021 Jan 11;9(1):40. doi: 10.3390/vaccines9010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Broecker F, Liu STH, Suntronwong N, et al. A mosaic hemagglutinin-based influenza virus vaccine candidate protects mice from challenge with divergent H3N2 strains. NPJ Vaccines. 2019;4:31. doi: 10.1038/s41541-019-0126-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun S, Cai Y, Song TZ, et al. Interferon-armed RBD dimer enhances the immunogenicity of RBD for sterilizing immunity against SARS-CoV-2. Cell Res. 2021;31(9):1011–1023. doi: 10.1038/s41422-021-00531-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dai L, Zheng T, Xu K, et al. A Universal Design of Betacoronavirus Vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3):722–733.e11. doi: 10.1016/j.cell.2020.06.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He C, Yang J, Hong W, et al. A self-assembled trimeric protein vaccine induces protective immunity against Omicron variant. Nat Commun. 2022;13(1):5459. doi: 10.1038/s41467-022-33209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W, Huang B, Zhu Y, et al. Ferritin nanoparticle-based SARS-CoV-2 RBD vaccine induces a persistent antibody response and long-term memory in mice. Cell Mol Immunol. 2021;18(3):749–751. doi: 10.1038/s41423-021-00643-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Andrews SF, Cominsky LY, Shimberg GD, et al. An influenza H1 hemagglutinin stem-only immunogen elicits a broadly cross-reactive B cell response in humans. Sci Transl Med. 2023;15(692):eade4976. doi: 10.1126/scitranslmed.ade4976 [DOI] [PubMed] [Google Scholar]
- 68.Widge AT, Hofstetter AR, Houser KV, et al. An influenza hemagglutinin stem nanoparticle vaccine induces cross-group 1 neutralizing antibodies in healthy adults. Sci Transl Med. 2023;15(692):eade4790. doi: 10.1126/scitranslmed.ade4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.