Abstract
Filamin C (FLNC), a large dimeric actin-binding protein in muscle cells, plays a critical role in transmitting force in the cytoskeleton and that between membrane receptors and the cytoskeleton. It performs crucial mechanosensing and downstream mechanotransduction functions via force-dependent interactions with signaling proteins. Mutations in FLNC have been linked to muscle and heart diseases. The mechanical responses of the force-bearing elements in FLNC have not been determined. This study investigated the mechanical responses of FLNC domains and their dimerization interface using magnetic tweezers. Results showed high stability of the N-terminal domains in the rod-1 segment but significant changes in the rod-2 domains in response to forces of a few piconewtons (pN). The dimerization interface, formed by the R24 domain, has a lifetime of seconds to tens of seconds at pN forces, and it dissociates within 1 s at forces greater than 14 pN. The findings suggest the FLNC dimerization interface provides sufficient mechanical stability that enables force-dependent structural changes in rod-2 domains for signaling protein binding and maintains structural integrity of the rod-1 domains.
Introduction
Filamin C (FLNC) is an isoform of the filamin family that is predominantly expressed in striated muscle cells and cardiomyocytes. This protein plays a crucial role in skeletal and cardiac muscle development. It has been extensively studied, and its significance has been established through various research studies.1−7 FLNC is primarily located in the Z-discs, costameres, and intercalated discs of skeletal and cardiac muscle cells, where it acts as a cross-linker between actin filaments and various binding partners, such as integrin, Xin protein, and aciculin.6,8−11 Deficiency, mutations, and truncations of FLNC have been linked to various skeletal and cardiac myopathies, such as myofibrillar skeletal myopathy (MFM), dilated cardiomyopathy (DCM), and arrhythmogenic cardiomyopathy (ACM).12−18 This highlights the critical role that FLNC plays in maintaining the structural integrity and function of skeletal and cardiac muscles.
The three isoforms of filamin, FLNA, FLNB, and FLNC, have similar structural organizations, each forming a V-shaped functional dimer within cells through the C-terminal self-association domain (R24).2,4,7,19,20 The monomeric unit of filamin consists of two N-terminal actin-binding domains (ABDs) followed by 24 immunoglobulin-like (Ig-like) repeats (R1–24), which are separated into rod-1 segment (R1–15), rod-2 segment (R16–23), and the dimerization domain (R24) by two calpain-sensitive hinges located between R15 and R16, and R23 and R24.2,20 The key difference between FLNC and FLNA and -B is the FLNC-specific insertion of 82 amino acids in FLNC R20.6 The rod-1 domains mainly bind to actin filaments and a few other binding partners, such as Cbl-associated protein and ankyrins-G.4,21,22 The rod-2 segment, on the other hand, provides the majority of the binding sites for FLN binding proteins, including signaling proteins, kinases, and membrane receptors.8,23−28 Unlike the linear arrangement of the rod-1 Ig-like repeats, the rod-2 Ig-like repeats are organized in a compact conformation with extensive interactions between neighboring domains that suppress binding by FLN binding factors. It has been suggested that this suppression can be relieved by high enough tensile forces transmitted across FLN dimers, making the rod-2 segment the major player in the mechanotransduction function of FLNs.2,7,29
The FLNC gene plays a crucial role in the development of both skeletal and cardiac muscle. Inadequacies in FLNC, such as mutations and truncations, have been linked to abnormal cell behavior and tissue development. For instance, the deletion of FLNC in cardiomyocytes leads to disordered Z-disk and sarcomere alignments, resulting in decreased cortical tension.30 A particular mutation in the dimerization domain (W2710X) of FLNC, which was found in families affected by MFM, causes the loss of FLNC’s dimerization ability and leads to the formation of aggregates in muscle cells.12 Another mutation in the R20 domain (G2151S) of FLNC was reported in a young patient with severe heart failure, causing abnormal sarcomere organization and misalignment of myofibrils.31
The FLNC dimers are important in transmitting cytoskeletal forces and are involved in both mechanosensing and mechanotransduction. To fully comprehend their functions, it is imperative to comprehend the range of forces transmitted through FLNC dimers as well as the duration of the tension. Moreover, it is important to comprehend the responses of the FLNC Ig-like repeats to these forces, as this information is necessary for a better understanding of the biophysical principles underlying their roles in mechanosensing and mechanotransduction. Despite its significance, this information is lacking in the field.
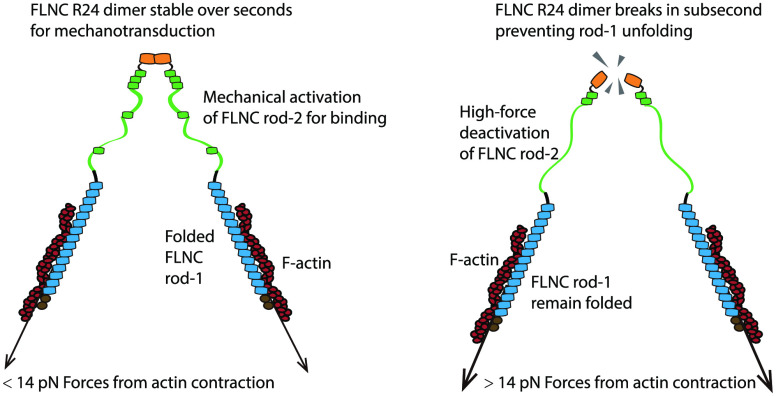
In this research, the behavior of FLNC under tensile forces was analyzed using a magnetic-tweezer setup developed in-house, as described in previous studies.32,33 Our findings demonstrate that the dimerization core of FLNC has a lifespan ranging from a few seconds to tens of seconds when exposed to forces lower than 14 pN. However, it disassembles rapidly when forces surpass 14 pN. Under the 14 pN threshold, the FLNC rod-1 Ig-like domains remain intact, while the FLNC rod-2 Ig-like domains undergo significant unraveling under piconewton forces. These results indicate that the FLNC dimerization interface acts as a mechanical breaker, limiting the transmission of forces within the FLNC dimer to below 14 pN and controlling the structural stability of the Ig-like domains in the rod-1 and rod-2 regions differently (Figure 1).
Figure 1.
Illustration representing the proposed mechanisms for FLNC’s response to tensile forces during mechanical transmission, which results in its mechanosensing and mechanotransduction capabilities. The top panel illustrates FLNC’s connections between membrane receptors and the cytoskeleton, and the bottom panel emphasizes FLNC’s role in cross-linking the cytoskeleton and its binding to signaling proteins.
Methods
The Design of a Single-Molecule Detector for the Mechanical Stability of FLNC R24 Dimers
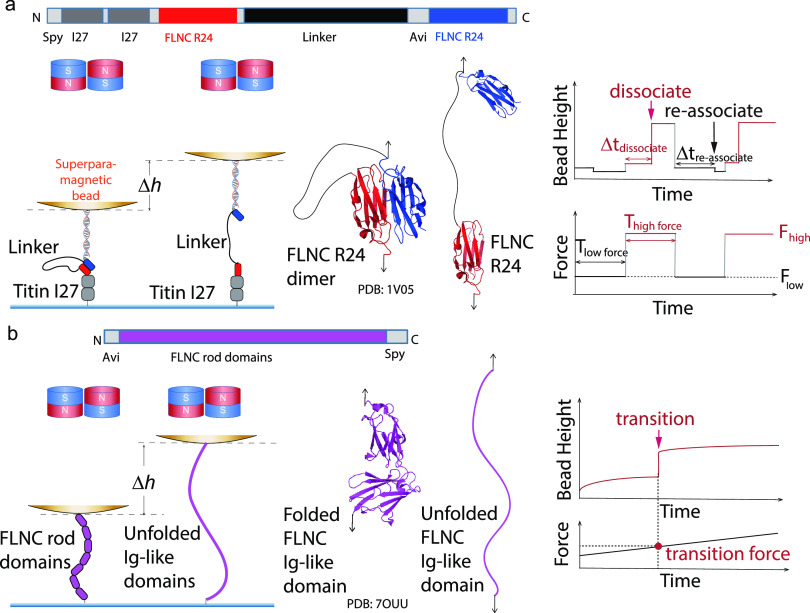
To study the mechanical stability of FLNC R24 dimers, we devised a protein construct that enables us to perform single-molecule assays and directly measure the lifetimes of the dimers under physiologically relevant forces. The construct comprises two R24 monomers linked by a long (155 a.a.) flexible GS-linker, which allows for easy detection of dimer dissociation due to the long extension change that occurs (Figure 2a).34 Additionally, the close proximity of the dissociated R24 monomers facilitates their reassociation under low force, thus improving the experimental efficiency. We also inserted two well-studied titin I27 domains between the N-terminal SPY-tag and the first FLNC R24 domain as a spacer, separating the FLNC R24 dimer from the bottom glass surface. A 1000 bp double-stranded DNA (dsDNA) spacer is placed between the biotin tag at the N-terminus of the second FLNC R24 and a 2.8 μm-diameter superparamagnetic bead (Dynabeads M-270, Invitrogen) (Supporting Information S1, S2, and S3). Using an in-house magnetic-tweezers setup,32 forces are applied to the bead to simulate physiological stretching conditions, causing the dimer to stretch from the N-termini of both domains. The resulting distributions of the dwell lifetimes of FLNC R24 dimers until dissociation are recorded under constant forces, and any dissociation of the dimers or other structural transitions (e.g., FLNC R24 domain unfolding) result in stepwise changes in bead height that equal the extension change.33
Figure 2.
Design of single-molecule detectors for investigating the mechanical stability of FLNC R24 dimers and rod domains using a magnetic-tweezers setup. (a) The single-molecule construct designed to assess the mechanical stability of FLNC R24 dimers. The bead height change (Δh) between the associated R24 dimer and dissociated R24 dimer is measured. These cycles promote the formation of the R24 dimer at low force and induce its dissociation at high force, thereby determining the dwell time (Δtdissociate) of FLNC R24 dimer dissociation under high force conditions. (b) Single-molecule construct designed to evaluate the mechanical stability of FLNC rod domains. The structure of FLNC R14–R15 is shown as an example. The bead height change (Δh) between the folded and unfolded Ig-like domains is measured. The right panel displays the time-varying forces applied to the FLNC rod domain constructs to measure the unfolding forces of FLNC Ig-like domains.
The Design of a Single-Molecule Detector for the Mechanical Stability of FLNC Rod Domains
To investigate the mechanical stability of FLNC rod domains, we designed protein constructs to apply single-molecule assays that directly detect the force at which the FLNC rod domains unfold at a physiologically relevant force loading rate (Figure 2b, Supporting Information S4). The single-molecule constructs consist of several FLNC Ig-like domain repeats whose mechanical stabilities are to be determined (FLNC R1 to R8, R9 to R15, or R16 to R23), a SPY-tag for attaching rod domains to the bottom glass surface, and the Avi-tag for connecting the neutralavidin-coated 2.8 μm-diameter superparamagnetic bead with the single-molecule construct (Supporting Information S1, S2, and S3). Similarly, forces are applied to the beads using an in-house-constructed magnetic-tweezers setup, and the FLNC rod domains undergo structural transitions under the mechanical stretching. The FLNC Ig-like domain repeats unfolding events are detected if the stepwise bead height changes are observed. The distributions of the forces at which the FLNC Ig-like domain unfolding events are recorded under time-varying forces are recorded to examine the mechanical stabilities of FLNC rod domains.
Results
FLNC Dimerization Interface Acts as a Mechanical Breaker during Force Transmission
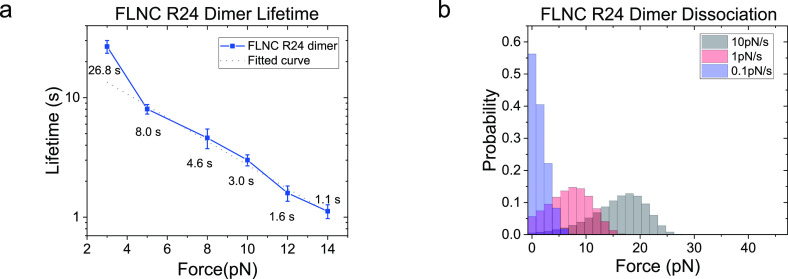
We performed a study on the impact of force on the lifetime of FLNC R24 dimers using magnetic tweezers and a custom-designed single-molecule construct. The design mimics the natural N-to-N terminal stretching geometry of FLNC dimers during force transmission. In our experiment, we applied an initial force of 1 pN, which kept the R24 dimer stable, followed by a jump in the force to measure its lifetime. Our results, displayed in Figure 3, showed that at 3 pN the linked FLNC R24 dimer underwent reversible transitions between the associated and dissociated states, which were indicated by two distinct levels of bead height. The histogram of the dwell times of the dimerized state was fitted with a single-exponential decay function, yielding an average lifetime of τ3 pN = 26.8 ± 3.3 s. As the applied force increased to 8 and 14 pN, the dimer underwent one-way dissociation transitions, with exponential fittings yielding τ8 pN = 4.6 ± 0.9 s and τ14 pN = 1.1 ± 0.2 s, respectively. It is important to note that at 14 pN, the force-bearing R24 domain unfolded concurrently with the dissociation, as evidenced by the step size (inset) in the Supporting Information S5.
Figure 3.
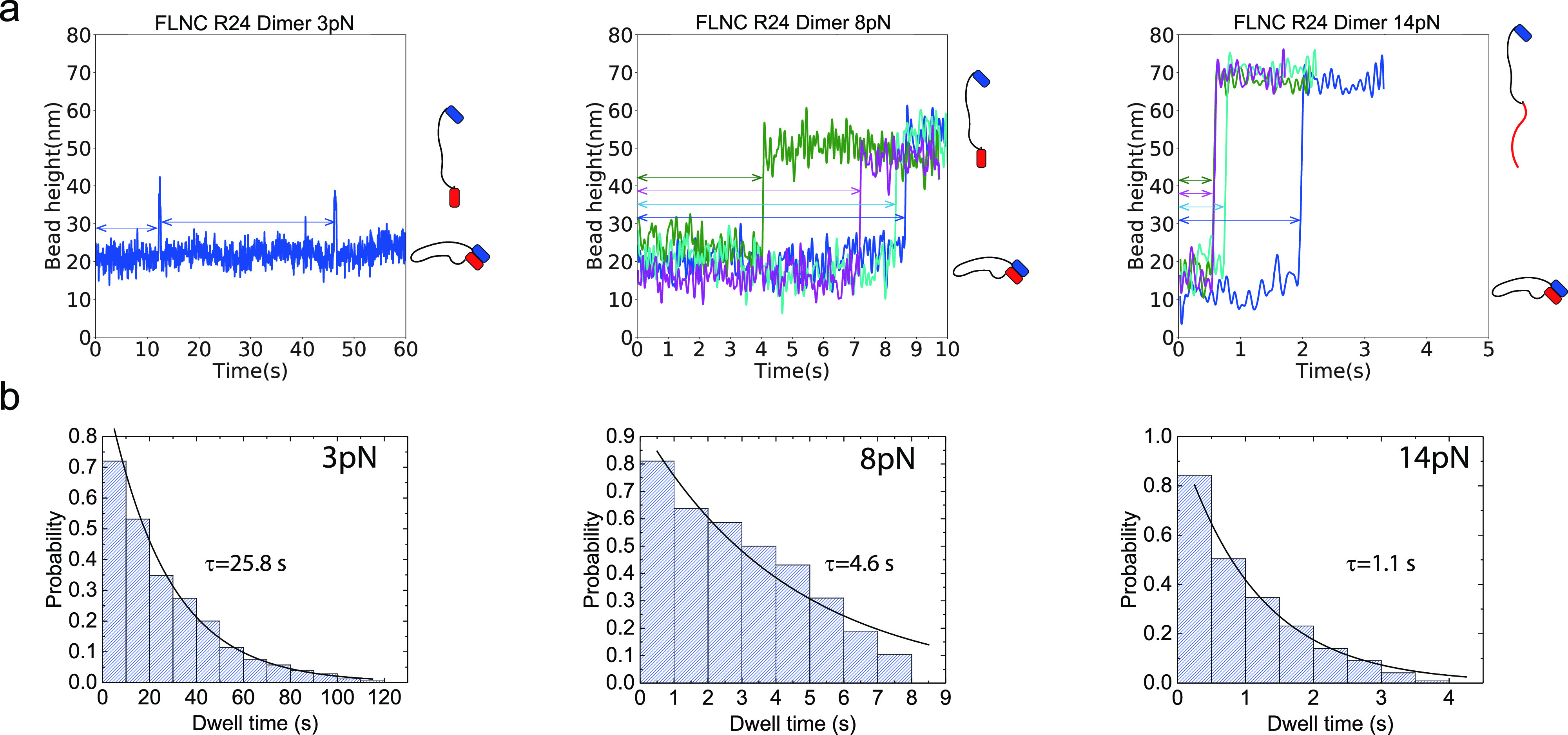
Dwell time measurements of the R24 dimer at different forces. (a) The relationship between bead height and time for the FLNC R24 dimer is shown, with each force–jump cycle depicted in a different color for forces of 8 and 14 pN. The zero time point marks the moment when the applied force transitions from 1 pN to the focused forces of 3, 8, and 14 pN. The right insets in each panel display the red and blue blocks representing the dissociated, associated, and combined dissociated and one unfolded R24 domain states of the FLNC R24 dimer. The double arrows indicate the recorded dwell time in each cycle. (b) Histogram of dwell times is shown, along with the fitted curve that was used to calculate the lifetime of the FLNC R24 dimer at the focused forces (represented as τ) through a single-exponential decay.
The above-mentioned approach was utilized to determine the force-dependent lifetimes over a range of 3 to 14 pN, using six different force values. The results indicated a strong sensitivity to force and showed that the FLNC R24 dimer has a lifetime ranging from seconds to over 10 s at forces lower than 14 pN (Figure 4a and Supporting Information S6), which estimate the FLNC R24 dimer dissociation force distribution (Figure 4b and Supporting Information S7). The error bars, which were obtained through bootstrap sampling of the collected data 200 times, were used to indicate standard deviations (Supporting Information S8). The logarithmic profile of the lifetime, τ(f), shows that the dissociation kinetics overall agrees with Bell’s model excluding the 3 pN data point with short-lived dissociated state that could lead to biased lifetime due to the finite instrument sampling rate (200 Hz).35,36 Fitting with Bell’s model k0eFδ/kBT for forces greater than 3 pN yielded a tempting rate of k0 ≈ 0.04 s–1 and a transition distance of δ ≈ 1.0 nm.
Figure 4.
Stability of the FLNC R24 dimer under mechanical stress. (a) The lifetimes of the dimer at various forces ranging from 3 to 14 pN. The dotted line indicates the best-fitting curve based on Bell’s model. (b) The estimated distribution of dissociation forces for FLNC R24 dimers, considering force loading rates of 10, 1, and 0.1 pN/s, based on the recorded lifetimes.
FLNC Rod-1 Domains Are Mechanically Stable in the Physiological Force Range
In the following analysis, the mechanical stability of FLNC rod-1 domains, consisting of two sections, R1–R8 and R9–R15, was thoroughly examined. To assess their stability, time-varying forces were applied, increasing from 1 pN to 80 pN. The force–bead height curves were recorded during the process, and stepwise increases in bead height in the curves were used as indicators of domain unfolding events.
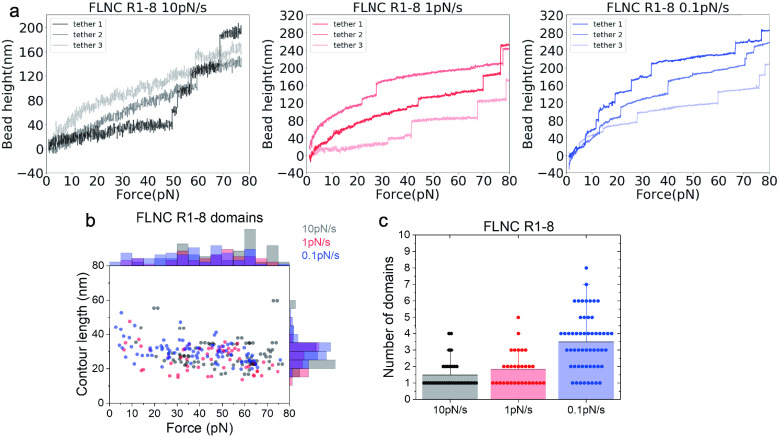
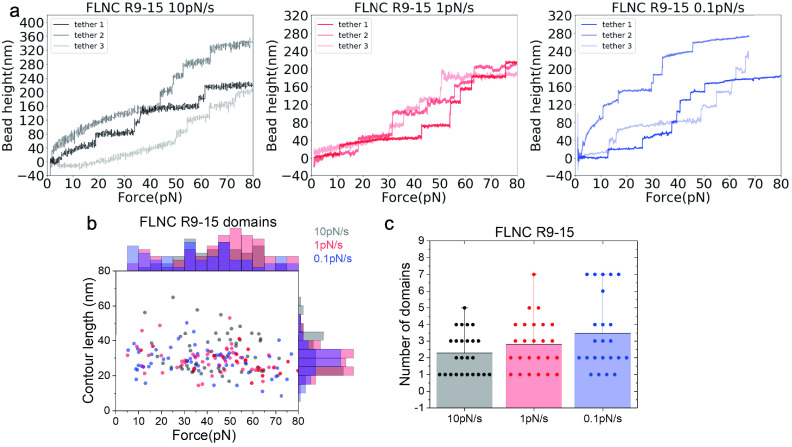
Figures 5 and 6 depict the representative force–height curves of FLNC R1–8 (Figure 5a) and R9–15 (Figure 6a) at different loading rates of 10, 1, and 0.1 pN/s. The results show that the majority of unfolding events occur at forces ranging from 14 to 80 pN across all loading rates (Figures 5b and 6b). In most scans, fewer than eight unfolding events are observed for R1–8 and less than seven for R9–15, suggesting that some domains remain folded up to 80 pN during the force loading process. Consistently, the data also indicate that more domains unfold at lower loading rates, as shown in Figures 5c and 6c. The histogram of contour lengths of the polypeptide released from unfolding events has a center point around 35 nm, matching the expected number of amino acids (around 100) absorbed in individual Ig-like repeats (Supporting Information S9) taking into account the contour length per residue of ≈0.38 nm.37 These results highlight the unfolding events that correspond to the unfolding of individual Ig-like repeats in rod-1 and the varying mechanical stability of the Ig-like repeats. The most significant finding from the unfolding force histograms is that both R1–8 and R9–15 are stable under forces below 14 pN, where the force transmission across FLNC dimers can last seconds to tens of seconds.
Figure 5.
Response of FLNC R1 to R8 domains to increasing time-dependent forces with loading rates of 10, 1, and 0.1 pN/s. (a) The representative curves of the height of the bead versus forces from three independent tethers at each loading rate. Each step signal represents the unfolding of one Ig-like domain. (b) The distribution of unfolding forces and the contour length released with each step signal. (c) The number of domains that unfold during each cycle.
Figure 6.
Response of FLNC R9 to R15 domains to increasing time-dependent forces with loading rates of 10, 1, and 0.1 pN/s. (a) The representative curves of the height of the bead versus forces from three independent tethers at each loading rate. Each step signal represents the unfolding of one Ig-like domain. (b) The distribution of unfolding forces and the contour length released with each step signal. (c) The number of domains that unfold during each cycle.
We have observed in Figures 5b and 6b that the unfolding forces of the rod domains exhibit flat distributions across a broad range of forces, which is due to the combination of unfolding force profiles from different Ig repeats. Additionally, although the majority of the rod-1 domains are mechanically stable, some of them can still unfold at forces as low as a few pN.
FLNC Rod-2 Domains Are Mechanosensitive to pN Forces
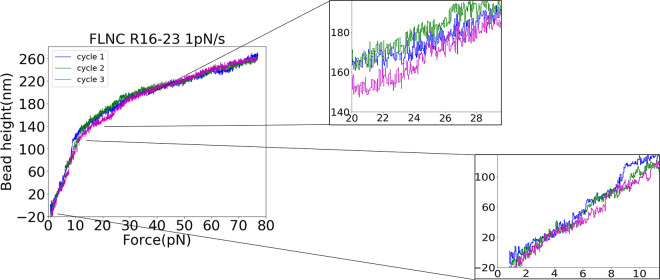
In our investigation of the mechanical stability of FLNC rod-2 domains (R16 to R23), we used force-loading scans to examine the behavior of the domains under a loading of 1 pN/s. As shown in Figure 7, the results of these scans revealed a significant difference in the stability of the rod-2 domains compared to that of the rod-1 domains. Unlike the rod-1 domains, which showed little unfolding at forces below 14 pN (Figures 5b and 6b), the rod-2 domains displayed numerous unfolding events (Figure 7, insets 1 and 2). This highlights the fact that the FLNC rod-2 domains are significantly less mechanically stable than the rod-1 domains, and the unfolding events at forces below 14 pN are particularly noteworthy due to the R24 dimer’s ability to persist for seconds to tens of seconds at forces lower than 14 pN. Here, we would like to highlight that due to the rapid unfolding and refolding rates of the rod-2 domains of FLNC at pN forces and 37 °C, it becomes challenging to capture distinct unfolding and refolding steps with our limited sampling rate. However, clear steps can be observed at 23 °C (Supporting Information S10).
Figure 7.
Mechanical responses of FLNC rod-2 domains, as represented by the height of the bead against forces with a loading rate of 1 pN/s. The use of different colors indicates varying force cycles. The stepwise signals in the zoomed-in region demonstrate the structural transitions of FLNC rod-2 domains.
For both rod-1 and rod-2 domains subjected to repeated pulling, we held the molecule at 1 pN for 60 s to allow the unfolded domains to refold. In the subsequent force-loading cycle, we observed similar contour lengths being released at similar force ranges, indicating that the domains were likely refolded into their native state. It is worth noting that over time, some of the domains gradually lose their ability to refold during the repeated cycles of unfolding. This leads to a longer extension at lower forces, as observed in our experiments. To ensure that the majority of the domains refolded, we restricted the number of cycles per tether to 5 cycles. The sample sizes of recorded unfolding events of rod-1 and rod-2 domains are provided in Supporting Information S11.
Comparison of the Mechanical Stability between FLNC Dimerization Interface and Rod Domains
To gain insight into the mechanisms
of force transmission in the FLNC dimer and its effect on structural
transitions in the force-bearing rod domains, we compare the mechanical
stability of the FLNC dimerization interface and the rod domains under
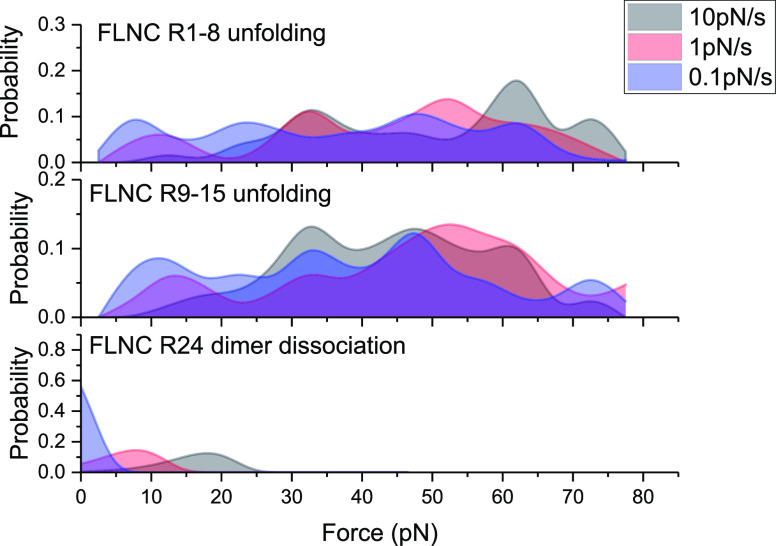
similar force loading conditions. Figure 8 illustrates the probability of domain unfolding
for the R1–R8 (top panel) and R9–R15 (middle panel)
rod-1 domains, which were obtained from single-molecule experiments
performed at three different loading rates. Figure 8 bottom panel depicts the probability of
dissociation of the R24 dimerization interface, which was calculated
from the force-dependent lifetime data shown in Figure 4, using the formula 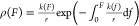 . The results demonstrate that the majority
of rod-1 domains unfold at forces significantly higher than those
required to dissociate the R24 dimerization interface.
. The results demonstrate that the majority
of rod-1 domains unfold at forces significantly higher than those
required to dissociate the R24 dimerization interface.
Figure 8.
A comparison of the unfolding force distributions of FLNC rod-1 domains and the dissociation force distributions of FLNC R24 dimers demonstrates the superior mechanical stability of FLNC rod-1 domains over that of FLNC R24 dimers.
In stark contrast to the rod-1 domains, as depicted in Figures 5 and 6, the rod-2 domains (R16–23) undergo unfolding at forces that fall within a similar range where the R24 dimer dissociates. This observation is made under the same loading rate of 1 pN/s. This contrast highlights a significant difference in the mechanical stability of the rod-2 domains compared to that at the dimerization interface. It suggests that the rod-2 domains exhibit weaker stability compared to the stability at the dimerization interface.
The results, when combined, paint a picture of how the rod-1 domains can maintain their structural integrity due to their greater mechanical stability compared to the dimerization interface. On the other hand, the rod-2 domains experience significant structural alterations in response to changes in force within the pN range, as they have a comparable or weaker mechanical stability compared to the dimerization interface.
Discussion
The objective of this study is to examine the mechanical responses of the FLNC rod-1 domains, rod-2 domains, and the R24 dimerization interface. The findings indicate that the FLNC R24 dimerization interface can retain its dimer complex structure for several to tens of seconds when subjected to forces below 14 pN. However, it dissociates rapidly within 1 s when exposed to forces above 14 pN. The majority of FLNC rod-1 domains demonstrate mechanical stability under forces below 14 pN at physiological loading rates. Conversely, FLNC rod-2 domains exhibit significantly weaker mechanical stability, as indicated by frequent domain unfolding at forces below 14 pN.
The lifetime of the R24 dimer is determined by the magnitude of the force applied, which determines the range of force transmitted across the FLNC dimer in the process of mechanotransduction. Mechanotransduction is based on the binding and unbinding of factors that are influenced by force and occur within a time frame ranging from seconds to minutes depending on various factors such as the concentration of binding factors and the strength of interactions (Supporting Information S12).38−42 The significance of a few seconds lifetime over pN forces lies in two reasons: First, pN forces are necessary to activate mechanosensing domains in various mechanosensing proteins, such as talin rod domains,43 α-catenins,44,45 and FLNC rod-2 domains shown in this study, which requires seconds to reach physiologically relevant loading rates on the order of pN per second. Second, the binding of signaling proteins to activated mechanosensing proteins takes time. In aqueous solutions, typical diffusion-limited binding rates range from 107 to 109 M–1 s–1. However, in cells, the binding rate is typically 1–2 orders of magnitude lower due to the crowded environment. Therefore, at μM concentrations, the time scale associated with binding takes seconds or longer. The discovery that the FLNC dimer has lifetimes of over a second only when forces are below 14 pN suggests that the R24 dimer interface functions as a force-dependent breaker, limiting the range of physiologically relevant forces to within 14 pN.
Importantly, our findings demonstrate that the FLNC rod-2 domains are highly responsive to tensile forces within a range of 14 pN. This sensitivity results in significant structural reorganization as the forces change. The rod-2 segments of the FLN family proteins are considered to be the primary region responsible for mechanosensing and mechanotransduction. Studies of FLNA have shown that this region serves as a binding site for a majority of its binding partners. Hence, the sensitivity to forces in the pN range suggests that the binding to the rod-2 region is sensitively regulated by mechanical forces, including the activation and deactivation of binding (Supporting Information S13).46,47 The presence of extensive domain–domain interactions in the rod-2 region leads to autoinhibition of binding to various signaling proteins and membrane receptors. However, force may facilitate the binding by breaking these domain–domain interactions, exposing the binding sites. This was shown in previous studies of factors’ binding to FLNA rod-2 domains.46,48−50 On the other hand, force-dependent deactivation is also possible as the domain unfolds when force further increases, potentially leading to a loss of binding sites. Therefore, the mechanical stability of the rod-2 domains implies complex, biphasic regulation of FLNC-mediated mechanotransduction functions.
In comparison to the FLNC rod-2 domains, the majority of the FLNC rod-1 domains maintain stability when subjected to force changes within 14 pN at normal physiological force loading rates. These rod-1 domains are believed not only to participate in actin binding but also to offer binding sites for several signaling proteins, as shown by previous studies.4,21,22 The robust mechanical stability of the FLNC rod-1 domains indicates that they provide secure binding sites for their binding partners, which are not sensitive to changes in the tensile forces. This suggests that the binding sites within the FLNC rod domains are divided into a mechanically sensitive rod-2 region and a nonsensitive rod-1 region.
The FLNC R24 dimer exhibits characteristic behavior when subjected to tensile forces, acting as a mechanical breaker rather than a highly stable lock. Unlike FLNC rod-1 domains, the R24 dimers are less robust, which allows them to break in order to protect the rod-1 domains from unraveling under high stress. Despite this, the R24 dimers have a lifetime of several seconds to tens of seconds when subjected to forces below 14 pN, making them stable enough for FLNC rod-2 domains to perform their role in mechanotransduction. These mechanical properties of the FLNC R24 dimer enable it to dynamically reorganize the cytoskeleton by breaking apart and also allow FLNC to function robustly in mechanotransduction.
The results of this study raise several questions for further investigation. The mechanical stability of the R24 dimerization interface is key to maintaining the distinct structural integrity of the rod-2 and rod-1 domains and their binding to factors. It is not clear if this stability of the R24 dimer is preserved in the FLNA and FLNB isoforms. The varying mechanical stability of the rod-2 and rod-1 domains in FLNC is in line with previous findings in FLNA,51 but it remains to be determined if this is also a common property of FLNB. Moreover, many FLN mutations and truncations have been linked to various diseases. Future studies can explore the impact of disease-related mutations, such as how these mutations affect the mechanical stability of FLN domains and their binding interactions.
Acknowledgments
We would like to thank Dr. Fumihiko Nakamura (Tianjin University, China) for stimulating discussions. We would also like to thank MBI Protein Expression Core facility for protein expression.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.3c02303.
Additional experimental details and protein construct sequences; physiological force-loading rates; illustration of transitions in the FLNC R24 dimer; dwell time measurements of the FLNC R24 dimer under three additional forces; Bell’s model fitting of force-dependent dissociation rates; bootstrapping of recorded dwell times of FLNC R24 dimers under constant forces; calculation of released contour length based on the unfolding step size of FLNC Ig-like repeats; FLNC rod-2 domain unfolding events at 23 °C; sample size of recorded unfolding events of rod-1 and rod-2 domains; the physiological importance of tension duration over seconds time scale; force-dependent activation and the high-force deactivation of FLNC rod-2 domains; related references (PDF)
This research is supported by the Singapore Ministry of Education Academic Research Fund Tier 3 (MOE Grant No. MOET32021-0003), the Singapore Ministry of Education Academic Research Funds Tier 2 (MOE Grant Nos. MOE-T2EP50220-0015, MOE2019-T2-02-014), and National Research Foundation, Prime Ministers Office, Singapore, and the Ministry of Education under the Research Centres of Excellence program.
The authors declare no competing financial interest.
Supplementary Material
References
- Stossel T. P.; Condeelis J.; Cooley L.; Hartwig J. H.; Noegel A.; Schleicher M.; Shapiro S. S. Filamins as integrators of cell mechanics and signalling. Nat. Rev. Mol. Cell Biol. 2001, 2, 138–145. 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- Nakamura F.; Stossel T. P.; Hartwig J. H. The filamins: organizers of cell structure and function. Cell adhesion & migration 2011, 5, 160–169. 10.4161/cam.5.2.14401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z.-w.; Xu W.-f.; Davie E. W.; Chung D. W. Molecular cloning of human ABPL, an actin-binding protein homologue. Biochemical and biophysical research communications 1998, 251, 914–919. 10.1006/bbrc.1998.9506. [DOI] [PubMed] [Google Scholar]
- Thompson T. G.; Chan Y.-M.; Hack A. A.; Brosius M.; Rajala M.; Lidov H. G.; McNally E. M.; Watkins S.; Kunkel L. M. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J. Cell Biol. 2000, 148, 115–126. 10.1083/jcb.148.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Ven P. F.; Obermann W. M.; Lemke B.; Gautel M.; Weber K.; Fürst D. O. Characterization of muscle filamin isoforms suggests a possible role of γ-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil. Cytoskeleton 2000, 45, 149–162. . [DOI] [PubMed] [Google Scholar]
- van der Ven P. F.; Wiesner S.; Salmikangas P.; Auerbach D.; Himmel M.; Kempa S.; Hayeß K.; Pacholsky D.; Taivainen A.; Schröder R.; et al. Indications for a novel muscular dystrophy pathway: γ-filamin, the muscle-specific filamin isoform, interacts with myotilin. J. Cell Biol. 2000, 151, 235–248. 10.1083/jcb.151.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z.; Nakamura F. Structure and function of filamin C in the muscle Z-disc. International Journal of Molecular Sciences 2020, 21, 2696. 10.3390/ijms21082696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gontier Y.; Taivainen A.; Fontao L.; Sonnenberg A.; van der Flier A.; Carpen O.; Faulkner G.; Borradori L. The Z-disc proteins myotilin and FATZ-1 interact with each other and are connected to the sarcolemma via muscle-specific filamins. Journal of cell science 2005, 118, 3739–3749. 10.1242/jcs.02484. [DOI] [PubMed] [Google Scholar]
- van der Ven P. F.; Ehler E.; Vakeel P.; Eulitz S.; Schenk J. A.; Milting H.; Micheel B.; Fürst D. O. Unusual splicing events result in distinct Xin isoforms that associate differentially with filamin c and Mena/VASP. Experimental cell research 2006, 312, 2154–2167. 10.1016/j.yexcr.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Linnemann A.; van der Ven P. F.; Vakeel P.; Albinus B.; Simonis D.; Bendas G.; Schenk J. A.; Micheel B.; Kley R. A.; Fürst D. O. The sarcomeric Z-disc component myopodin is a multiadapter protein that interacts with filamin and α-actinin. European journal of cell biology 2010, 89, 681–692. 10.1016/j.ejcb.2010.04.004. [DOI] [PubMed] [Google Scholar]
- Molt S.; Bührdel J. B.; Yakovlev S.; Schein P.; Orfanos Z.; Kirfel G.; Winter L.; Wiche G.; van der Ven P. F.; Rottbauer W.; et al. Aciculin interacts with filamin C and Xin and is essential for myofibril assembly, remodeling and maintenance. Journal of cell science 2014, 127, 3578–3592. 10.1242/jcs.152157. [DOI] [PubMed] [Google Scholar]
- Vorgerd M.; Van der Ven P. F.; Bruchertseifer V.; Löwe T.; Kley R. A.; Schröder R.; Lochmüller H.; Himmel M.; Koehler K.; Fürst D. O.; et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. American Journal of Human Genetics 2005, 77, 297–304. 10.1086/431959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürst D. O.; Goldfarb L. G.; Kley R. A.; Vorgerd M.; Olivé M.; van der Ven P. F. Filamin C-related myopathies: pathology and mechanisms. Acta neuropathologica 2013, 125, 33–46. 10.1007/s00401-012-1054-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruparelia A. A.; Oorschot V.; Ramm G.; Bryson-Richardson R. J. FLNC myofibrillar myopathy results from impaired autophagy and protein insufficiency. Human molecular genetics 2016, 25, 2131–2142. 10.1093/hmg/ddw080. [DOI] [PubMed] [Google Scholar]
- Brodehl A.; Gaertner-Rommel A.; Milting H. FLNC (Filamin-C) A new (er) player in the field of genetic cardiomyopathies. Circulation: Cardiovascular Genetics 2017, 10, e001959. 10.1161/CIRCGENETICS.117.001959. [DOI] [PubMed] [Google Scholar]
- Hall C. L.; Akhtar M. M.; Sabater-Molina M.; Futema M.; Asimaki A.; Protonotarios A.; Dalageorgou C.; Pittman A. M.; Suarez M. P.; Aguilera B.; et al. Filamin C variants are associated with a distinctive clinical and immunohistochemical arrhythmogenic cardiomyopathy phenotype. International journal of cardiology 2020, 307, 101–108. 10.1016/j.ijcard.2019.09.048. [DOI] [PubMed] [Google Scholar]
- Song S.; Shi A.; Lian H.; Hu S.; Nie Y. Filamin C in cardiomyopathy: From physiological roles to DNA variants. Heart Failure Reviews 2022, 27, 1–13. 10.1007/s10741-021-10172-z. [DOI] [PubMed] [Google Scholar]
- Powers J. D.; Kirkland N. J.; Liu C.; Razu S. S.; Fang X.; Engler A. J.; Chen J.; McCulloch A. D. Subcellular Remodeling in Filamin C Deficient Mouse Hearts Impairs Myocyte Tension Development during Progression of Dilated Cardiomyopathy. International journal of molecular sciences 2022, 23, 871. 10.3390/ijms23020871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel M.; Van Der Ven P. F.; Stöcklein W.; Fürst D. O. The limits of promiscuity: isoform-specific dimerization of filamins. Biochemistry 2003, 42, 430–439. 10.1021/bi026501+. [DOI] [PubMed] [Google Scholar]
- Kesner B. A.; Milgram S. L.; Temple B. R.; Dokholyan N. V. Isoform divergence of the filamin family of proteins. Molecular biology and evolution 2010, 27, 283–295. 10.1093/molbev/msp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M.; Liu J.; Cheng A.; DeYoung S. M.; Saltiel A. R. Identification of CAP as a costameric protein that interacts with filamin C. Molecular biology of the cell 2007, 18, 4731–4740. 10.1091/mbc.e07-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiweilidan Y.; Klauza I.; Kordeli E. Novel interactions of ankyrins-G at the costameres: the muscle-specific Obscurin/Titin-Binding-related Domain (OTBD) binds plectin and filamin C. Exp. Cell Res. 2011, 317, 724–736. 10.1016/j.yexcr.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Tu Y.; Wu S.; Shi X.; Chen K.; Wu C. Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell 2003, 113, 37–47. 10.1016/S0092-8674(03)00163-6. [DOI] [PubMed] [Google Scholar]
- Murray J. T.; Campbell D. G.; Peggie M.; Alfonso M.; Cohen P. Identification of filamin C as a new physiological substrate of PKBα using KESTREL. Biochem. J. 2004, 384, 489–494. 10.1042/BJ20041058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lypowy J.; Chen I.-Y.; Abdellatif M. An Alliance between Ras GTPase-activating Protein, Filamin C, and RasGTPase-activating Protein SH3 Domain-binding Protein Regulates MyocyteGrowth. J. Biol. Chem. 2005, 280, 25717–25728. 10.1074/jbc.M414266200. [DOI] [PubMed] [Google Scholar]
- Nakagawa K.; Sugahara M.; Yamasaki T.; Kajiho H.; Takahashi S.; Hirayama J.; Minami Y.; Ohta Y.; Watanabe T.; Hata Y.; et al. Filamin associates with stress signalling kinases MKK7 and MKK4 and regulates JNK activation. Biochem. J. 2010, 427, 237–245. 10.1042/BJ20091011. [DOI] [PubMed] [Google Scholar]
- Juo L.-Y.; Liao W.-C.; Shih Y.-L.; Yang B.-Y.; Liu A.-B.; Yan Y.-T. HSPB7 interacts with dimerized FLNC and its absence results in progressive myopathy in skeletal muscles. Journal of cell science 2016, 129, 1661–1670. 10.1242/jcs.179887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier M. P.; Alderson T. R.; de Villiers C. P.; Nicholls D.; Gastall H. Y.; Allison T. M.; Degiacomi M. T.; Jiang H.; Mlynek G.; Fürst D. O.; et al. HspB1 phosphorylation regulates its intramolecular dynamics and mechanosensitive molecular chaperone interaction with filamin C. Science Advances 2019, 5, eaav8421. 10.1126/sciadv.aav8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razinia Z.; Mäkelä T.; Ylänne J.; Calderwood D. A. Filamins in mechanosensing and signaling. Annual review of biophysics 2012, 41, 227–246. 10.1146/annurev-biophys-050511-102252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guergueltcheva V.; Peeters K.; Baets J.; Ceuterick-de Groote C.; Martin J.; Suls A.; De Vriendt E.; Mihaylova V.; Chamova T.; Almeida-Souza L.; et al. Distal myopathy with upper limb predominance caused by filamin C haploinsufficiency. Neurology 2011, 77, 2105–2114. 10.1212/WNL.0b013e31823dc51e. [DOI] [PubMed] [Google Scholar]
- Fung E.; Guo D.; Zhu W.; Ahmadabadi B.; Lee C.; Teekakirikul P.; Zhao H. Functional Validation of a Pathogenic Missense Variant in Human Filamin C Cardiomyopathy through Disruption of a Zebrafish Homologue Recapitulates Cardiac Disease. Journal of Heart and Lung Transplantation 2021, 40, S237. 10.1016/j.healun.2021.01.681. [DOI] [Google Scholar]
- Chen H.; Fu H.; Zhu X.; Cong P.; Nakamura F.; Yan J. Improved high-force magnetic tweezers for stretching and refolding of proteins and short DNA. Biophysical journal 2011, 100, 517–523. 10.1016/j.bpj.2010.12.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X.; Zeng X.; Lu C.; Yan J. Studying the mechanical responses of proteins using magnetic tweezers. Nanotechnology 2017, 28, 414002. 10.1088/1361-6528/aa837e. [DOI] [PubMed] [Google Scholar]
- Pudas R.; Kiema T.-R.; Butler P. J. G.; Stewart M.; Ylänne J. Structural basis for vertebrate filamin dimerization. Structure 2005, 13, 111–119. 10.1016/j.str.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Stigler J.; Rief M. Hidden Markov Analysis of Trajectories in Single-Molecule Experiments and the Effects of Missed Events. ChemPhysChem 2012, 13, 1079–1086. 10.1002/cphc.201100814. [DOI] [PubMed] [Google Scholar]
- Jacobson D. R.; Perkins T. T. Correcting molecular transition rates measured by single-molecule force spectroscopy for limited temporal resolution. Phys. Rev. E 2020, 102, 022402. 10.1103/PhysRevE.102.022402. [DOI] [PubMed] [Google Scholar]
- Erickson H. P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl. Acad. Sci. U. S. A. 1994, 91, 10114–10118. 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.; Yu M.; Yan J. Direct single-molecule quantification reveals unexpectedly high mechanical stability of vinculintalin/α-catenin linkages. Science Advances 2019, 5, eaav2720. 10.1126/sciadv.aav2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le S.; Yu M.; Yan J. Phosphorylation reduces the mechanical stability of the α-catenin/β-catenin complex. Angew. Chem. 2019, 131, 18836–18842. 10.1002/ange.201911383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Yao M.; Baker K. B.; Gough R. E.; Le S.; Goult B. T.; Yan J. Force-dependent interactions between talin and full-length vinculin. J. Am. Chem. Soc. 2021, 143, 14726–14737. 10.1021/jacs.1c06223. [DOI] [PubMed] [Google Scholar]
- Liu J.; Le S.; Yao M.; Huang W.; Tio Z.; Zhou Y.; Yan J. Tension Gauge Tethers as Tension Threshold and Duration Sensors. ACS sensors 2023, 8, 704–711. 10.1021/acssensors.2c02218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J.; Ge C.; Jothikumar P.; Yuan Z.; Liu B.; Bai K.; Li K.; Rittase W.; Shinzawa M.; Zhang Y.; et al. A TCR mechanotransduction signaling loop induces negative selection in the thymus. Nature immunology 2018, 19, 1379–1390. 10.1038/s41590-018-0259-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.; Goult B. T.; Klapholz B.; Hu X.; Toseland C. P.; Guo Y.; Cong P.; Sheetz M. P.; Yan J. The mechanical response of talin. Nat. Commun. 2016, 7, 11966. 10.1038/ncomms11966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M.; Qiu W.; Liu R.; Efremov A. K.; Cong P.; Seddiki R.; Payre M.; Lim C. T.; Ladoux B.; Mege R.-M.; et al. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 2014, 5, 4525. 10.1038/ncomms5525. [DOI] [PubMed] [Google Scholar]
- Pang S. M.; Le S.; Kwiatkowski A. V.; Yan J. Mechanical stability of αT-catenin and its activation by force for vinculin binding. Molecular biology of the cell 2019, 30, 1930–1937. 10.1091/mbc.E19-02-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni L.; Stigler J.; Pelz B.; Ylänne J.; Rief M. Dynamic force sensing of filamin revealed in single-molecule experiments. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 19679–19684. 10.1073/pnas.1211274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M.; Schütze K.; Moriwaki Y.; Heo L.; Ovchinnikov S.; Steinegger M. ColabFold: making protein folding accessible to all. Nat. Methods 2022, 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lad Y.; Kiema T.; Jiang P.; Pentikäinen O. T.; Coles C. H.; Campbell I. D.; Calderwood D. A.; Ylänne J. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. EMBO journal 2007, 26, 3993–4004. 10.1038/sj.emboj.7601827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pentikäinen U.; Ylänne J. The regulation mechanism for the auto-inhibition of binding of human filamin A to integrin. Journal of molecular biology 2009, 393, 644–657. 10.1016/j.jmb.2009.08.035. [DOI] [PubMed] [Google Scholar]
- Chen H. S.; Kolahi K. S.; Mofrad M. R. Phosphorylation facilitates the integrin binding of filamin under force. Biophysical journal 2009, 97, 3095–3104. 10.1016/j.bpj.2009.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Zhu X.; Cong P.; Sheetz M. P.; Nakamura F.; Yan J. Differential mechanical stability of filamin A rod segments. Biophysical journal 2011, 101, 1231–1237. 10.1016/j.bpj.2011.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.