Abstract
RNA is a central and universal mediator of genetic information underlying the diversity of cell types and cell states, which together shape tissue organization and organismal function across species and lifespans. Despite numerous advances in RNA sequencing technologies and the massive accumulation of transcriptome datasets across the life sciences1,2, the dearth of technologies that use RNAs to observe and manipulate cell types remains a bottleneck in biology and medicine. Here we describe CellREADR (Cell access through RNA sensing by Endogenous ADAR), a programmable RNA-sensing technology that leverages RNA editing mediated by ADAR to couple the detection of cell-defining RNAs with the translation of effector proteins. Viral delivery of CellREADR conferred specific cell-type access in mouse and rat brains and in ex vivo human brain tissues. Furthermore, CellREADR enabled the recording and control of specific types of neurons in behaving mice. CellREADR thus highlights the potential for RNA-based monitoring and editing of animal cells in ways that are specific, versatile, simple and generalizable across organ systems and species, with wide applications in biology, biotechnology and programmable RNA medicine.
The diversity of cells underlies the diversity of life forms and of physiological systems within individual organisms3. Reading from a singular genome differentially packaged into numerous epigenomes, individual cells of the organism generate distinct transcriptomes as RNA intermediates to implement cell-specific phenotypes and physiology. RNAs are thus the central and universal messengers that convey customized genetic instructions in diverse and individual cell types and cell states that together shape tissue organization and system function across species and lifespans. Indeed, all bodily functions emerge from interactions among cell types, and aberrant cell physiology and function give rise to disease4. Recent advances in single-cell RNA-sequencing (RNA-seq) approaches promise to identify all molecularly-defined cell types in the human body and in many other organisms1. Beyond transcriptome profiling, it is necessary to monitor and manipulate all cell types to identify their specific roles in tissue architecture and system function within organisms. Cell-specific monitoring and intervention is also key to the precision diagnosis and treatment of many human diseases. To achieve these, we need cell type technologies that are specific, facile, scalable, economical and generalizable across species, analogous to gene editing approaches in genetics.
So far, nearly all genetic approaches to accessing cell types rely on DNA-based transcriptional regulatory elements for expressing effector genes (such as markers, sensors and effectors) that mimic cell-specific RNA expression, mostly through germline engineering in only a handful of organisms. However, all germline approaches, including those based on CRISPR5–7, are inherently cumbersome, slow, difficult to scale and generalize, and raise ethical issues, especially in primates and humans8,9. Transcriptional enhancer-based viral vectors have shown promise for targeting cell types in animals10–16, but such enhancers are difficult to identify and validate owing to their complex relationships with target genes and cell types. Ultimately, all DNA- and transcription-based approaches are inherently indirect means of mimicking and leveraging cell-specific RNA expression patterns.
Here we report a new class of RNA-sensing-dependent protein translation technology that bypasses the DNA-based transcription process, and apply it to genetic access of cell types. We developed CellREADR, which harnesses an RNA-sensing and -editing mechanism that is ubiquitous to all animal cells, for detecting specific cellular RNAs and then switching on translation of effector proteins to monitor and manipulate the cell. CellREADR is deployable as a single RNA molecule that uses Watson–Crick base-pairing and is inherently specific, simple, scalable, programmable and generalizable across species.
CellREADR design and implementation
RNA editing is a widespread and robust post-transcriptional mechanism that is essential for the metazoan gene regulatory toolkit implicated in recoding, splicing, microRNA targeting and other RNA-dependent modulatory processes17,18. The most prevalent form of RNA editing is adenosine-to-inosine (A-to-I) conversion, catalysed by ADARs (adenosine deaminases acting on RNA); inosine is subsequently recognized as guanosine (G) by the cellular machinery18. ADARs recognize and are recruited by stretches of base-paired double-stranded RNAs17 (dsRNAs), and can therefore operate as a sequence-guided base-editing machine, which can be harnessed for transcriptome editing19–23. As ADARs are ubiquitous in animal cells24, we designed CellREADR as a single and modular ‘readrRNA’ molecule (Fig. 1a). The 5′ region of readrRNA contains a sensor domain comprising around 300 nucleotides (nt), which is complementary to and thus detects a specific cellular RNA through sequence-specific base-pairing. This sensor domain contains one or more ADAR-editable stop codons that act as a translation switch; we name this region the sense–edit–switch RNA (sesRNA). Downstream of the sesRNA and in-frame with the stop codon is a sequence encoding the self-cleaving peptide T2A, followed in-frame by an effector RNA (efRNA) region that encodes various effector proteins. The entire readrRNA is several kilobases long, depending on which specific sensors and effectors are included, and is thus deliverable to cells through viral vectors or liposome nanoparticles. In cells expressing the target RNA, sesRNA forms dsRNA with the target RNA, which recruits ADARs to assemble an editing complex. At the editable stop codon, ADARs convert A to I, which pairs with the opposing C in the target RNA. This A-to-G substitution converts a UAG stop codon to a UI(G)G tryptophan codon, switching on translation of the efRNA. The in-frame translation generates a fusion protein comprising an N-terminal peptide, T2A and a C-terminal effector, which then self-cleaves by T2A, and releases the functional effector protein (Fig. 1a). readrRNA is expected to remain inert in cells that do not express the target RNA.
Fig. 1 |. CellREADR design and implementation in mammalian cells.
a, Schematic showing the design of CellREADR. CellREADR is a modular readrRNA molecule, consisting of a 5′ sensor domain (sesRNA) and 3′ effector domain (efRNA), separated by a T2A coding region. sesRNA is complementary to a cellular RNA and contains a stop codon that prevents efRNA translation. Base-pairing between sesRNA and target RNA recruits ADARs, which mediate A-to-I editing and convert the UAG stop to a UGG Trp codon, switching on translation of effector protein. b, CAG-tdT encodes tdTomato target RNA and READRtdT-GFP encodes a readrRNA consisting of a BFP sequence followed by sesRNAtdT and efRNAGFP. WPRE and 2a are sequences for a virus post-transcriptional regulatory element and a self-cleaving peptide coding sequence, respectively. pA, polyadenylated tail. c, Cells transfected with both CAG-tdT (tdT) and READRtdT-GFP exhibited robust GFP expression that co-localized with BFP and RFP (bottom). Cells transduced with READRtdT-GFP only (top) or CAG-tdT and READRctrl encoding sesRNActrl (middle) exhibited very low GFP expression. Right, FACS analysis of GFP and RFP expression. The percentage of GFP+ cells is indicated. d, Quantification of CellREADR efficiency by FACS analysis. e–g, The effect of target RNA expression levels on CellREADR. e, rtTA-TRE3g-ChETA (top) is designed so that BFP-ChETA target RNA is transcribed from TRE3g, driven by constitutively expressed rtTA in a tetracycline-concentration-dependent manner. READRChETA-GFP (bottom) is designed to express readrRNA comprising mCherry (mCh) followed by sesRNAChETA and efRNAGFP. f, Increasing tetracycline concentrations resulted in increasing numbers of BFP+ cells, expressed here as a percentage of RFP+ cells (conversion ratio). g, The conversion ratio increased with increasing tetracycline concentration. CAG-ChETA, which results in constitutive expression of BFP-ChETA, served as positive control. READRctrl served as negative control. h,i, ADAR1 is required for CellREADR function. FACS analysis (h) and quantification (i) of wild-type (WT) or ADAR1-KO cells with sesRNAtdT only, sesRNActrl and CAG-tdT, sesRNAtdT and CAG-tdT, or sesRNAtdT and CAG-tdT with overexpression of ADAR2. Arrows indicate GFP+RFP+ populations. j,k, Electropherograms of Sanger sequencing (j) and quantification (k), showing A to G conversion at the intended editing site in different samples. Data in d,f,g,i,k are mean ± s.e.m.; n = 3 independent experiments.
We first made a proof-of-principle version of CellREADR and tested it in the 293T human cell line. We used an expression vector (PGK-tdT) to express the tdTomato gene as an exogenous target RNA and to label transfected cells. We then designed a READR vector (READRtdT-GFP) to express a readrRNA consisting of a sensor region 5′ of the tdTomato sequence embedded with a UAG stop codon (sesRNAtdT), a T2A coding sequence, and a 3′ cassette encoding GFP (Extended Data Fig. 1a,b). Co-transfection of READRtdT-GFP with PGK-tdT resulted in GFP expression in a subset of tdTomato+ cells (Extended Data Fig. 1b,c). To substantiate this result, we replaced the GFP-coding region of the READR vector with that of a tetracycline-inducible transcription activator (tTA2), which drove the expression of a firefly luciferase gene (ffLuc) from the tetracycline response element (TRE3g) promoter. Co-transfection of READRtdT-tTA2 and TRE3g-ffLuc with PGK-tdT resulted in an approximately eightfold induction of luminescence compared with the empty vector control (Extended Data Fig. 1d). We observed sparse and weak GFP or luciferase expression, respectively, from READRtdT-GFP alone or READRtdT-tTA2 and TRE3g-ffLuc when co-transfected with an empty PGK vector (Extended Data Fig. 1b–d), which may have resulted from occasional read-through of the stop codon25 in the sesRNA. Inclusion of an in-frame spacer region of approximately 600 bp before the sesRNA prevented most residual GFP translation; longer spacers decreased the efficacy of readrRNA in the presence of tdTomato target RNA (Extended Data Fig. 2e).
To more quantitatively characterize CellREADR efficiency, we inserted a BFP-encoding cassette upstream of the sesRNAtdT region, which functioned as a spacer as well as a marker of transfected cells (Fig. 1b,c). We co-transfected this READRtdT-GFP construct with PGK-tdT in 293T cells, and used fluorescence-activated cell sorting (FACS) for quantitative analysis. We gated on BFP+ cells to calculate the efficiency of CellREADR, given that every BFP+ cell has the potential to express GFP. In the absence of tdTomato target RNA, READRtdT-GFP transfection resulted in almost no GFP expression (0.31%), indicating the absence of detectable stop read-through. In the presence of tdTomato RNA, the efficiency of READRtdT-GFP (that is, the ratio of GFP+RFP+ cells among RFP+ cells) was 32.6% (Fig. 1c,d), whereas a control READRctrl-GFP vector expressing sesRNA with a scramble coding sequence (sesRNActrl) showed almost no GFP translation (0.46%). The efficiency reached 67.3% when a TRE3g–tTA2 system was used for amplification (Extended Data Fig. 1f). Notably, when Cre or Flp recombinase was used as an effector, CellREADR showed leakiness (Extended Data Fig. 2e,f), which could potentially be mitigated through optimization. Next, we examined whether CellREADR efficiency correlated with target RNA levels. The efficiency of READRtdT-GFP increased with an increasing amount of CAG-tdT vector in 293T cell transfection (Extended Data Fig. 3d). We further designed a tetracycline-inducible expression vector in which a ChETA (a variant of light-gated channelrhodopsin-2) coding sequence was fused to the BFP sequence and driven by the TRE3g promoter; thus the amount of BFP fluorescence indicated the amount of ChETA transcript, which correlated with the tetracycline concentration in the culture medium (Fig. 1e). Increasing the tetracycline concentration and BFP expression levels increased the efficiency of READRChETA-GFP in a ChETA RNA-dependent manner (Fig. 1g and Extended Data Fig. 3e,f). With constitutive promoter-driven BFP-ChETA expression, READRChETA-GFP efficiency reached 41.5% (Fig. 1g and Extended Data Fig. 3g,h). These results demonstrated the dependence of CellREADR efficiency on the amount of target RNA.
To demonstrate the capacity of effector RNAs for mediating physiological function, we constructed READRtdT-Cas9 to express Cas9 and induce efficient genomic DNA editing26 (Extended Data Fig. 2a–c) and READRtdT-taCasp3/TEVp to express caspase-3 and induce apoptosis27 (Extended Data Fig. 2d).
To examine the role of ADAR proteins in CellREADR function, we generated an ADAR1-knockout (KO) 293T cell line using CRISPR–Cas9 (ref. 28) (Extended Data Fig. 4a,b). As ADAR1 is highly expressed, and ADAR2 is barely expressed, in 293T cells22,24,29, ADAR1-KO 293T cells essentially represented an ADAR-null cell line. Removal of ADAR1 largely eliminated the effect of READRtdT-GFP (Fig. 1h,i), which was rescued by exogenous expression of either the p110 or p150 isoform of ADAR1 (Extended Data Fig. 4c,d) or ADAR2 (Fig. 1h,i). Overexpression of ADAR2 in wild-type cells increased READRtdT-GFP efficiency by around 25% (Fig. 1h,i). Sanger sequencing confirmed A-to-I/G editing at the intended site, which converted the stop codon TAG to the tryptophan codon TGG (Fig. 1j,k). Since ADAR expression can be induced by stimulation with interferon29,30, we confirmed this induction in 293T cells and further demonstrated that interferon treatment increased READRtdT-GFP efficiency (Extended Data Fig. 4e–h).
As well as in 293T cells, we demonstrated CellREADR functionality in several other cell lines from different tissues or species, including human HeLa, mouse N2a and mouse KPC1242 cell lines (Extended Data Fig. 3a). Collectively, these results demonstrate that by leveraging cell endogenous ADARs, CellREADR can couple the detection of cellular RNAs to the translation of effector proteins to manipulate the cell in a way that is specific, efficient and robust.
sesRNA properties and programmability
As sesRNA is a key component of CellREADR, we investigated several properties of sesRNAs using the READRtdT-GFP vector (Fig. 2a). sesRNAs shorter than 50 nt were largely ineffective, whereas the length of sesRNAs more than 50 nt in size showed a positive correlation with CellREADR efficiency, with an optimal length of 200–350 nt (Fig. 2a). sesRNAs of around 200 nt tolerated up to 10 mismatches with target RNA (5% of sequence length) or in-frame insertion–deletion mutations (indels) without a major decrease of READRtdT-GFP efficiency (Fig. 2b and Extended Data Fig. 3b); this property confers flexibility for sesRNA design (Methods). However, mismatches near the editing site reduced CellREADR efficiency (Extended Data Fig. 3b). To explore the influence of mutations at different points along the target transcript on sesRNAs design, we used a EF1a-ChETA-tdT expression vector and tested five sesRNAs targeting the promoter and different coding regions. All four sesRNAs targeting transcribed regions exhibited robust efficiencies, whereas the sesRNA targeting the non-transcribed promoter region did not (Fig. 2c). The considerable variation in efficiencies among different sesRNAs for the same RNA target indicates that the specific sequence is an important factor for designing optimal sesRNAs. Finally, we examined whether the inclusion of more stop codons would further increase sesRNA stringency (that is, reduce basal translation in the absence of target RNA) (Fig. 1d). Using luciferase as a sensitive indicator of leaky translation (Fig. 2d), sesRNAs with two stop codons showed reduced leakage with similar efficiency to those with one stop codon (Fig. 2e), although three stop codons substantially reduced the efficiency (Fig. 2f). Together, these results suggest strategies to enhance the stringency, efficiency and flexibility of CellREADR.
Fig. 2 |. Properties of sesRNA.
a, READRBFP-tdT-GFP (top) was used to detect a tdT target RNA (not shown) with sesRNAtdT sequences of different lengths. Expression was quantified by FACS (bottom). b, Effects of mismatches between sesRNA and target RNA base-pairing. Top, black dots indicate mismatches and hollow dots indicate indels in the sesRNA. The number of mismatches and the per cent identity between sesRNAtdT and tdT target RNA are shown for different degrees of mismatch (bottom). c, The effect of sesRNA sensing of different locations and sequences within the target RNA expressed from EF1a-ChETA-tdT; the promoter region was used as negative control. readrRNA efficiencies are shown (bottom). d, To improve stringency, we designed sesRNAtdT with 1, 2 or 3 stop codons (TAG) in the READRtdT-ffLuc vector. e,f, Luminescence of cells transfected with READRtdT-ffLuc only (e) or with READRtdT-ffLuc and CAG-tdT (f). RLU, relative light unit. g, Schematic of dual (middle) and triple (bottom) sensor READRChETA/tdT-GFP vectors with sesRNA arrays targeting a ChETA-tdT fusion transcript (top). h, FACS analysis of cells co-transfected with EF1a-ChETA-tdT and a dual (a–c, c–a and b–a) or triple (c–b–a) READRtdT-GFP vector. Arrows indicate GFP+RFP+ populations. i, Quantification of the efficiency of the dual or triple READRtdT-GFP vectors shown in h. j, Schematic for intersectional targeting of cells expressing two target RNAs (ChETA and tdT) using a dual-sensor READRChETA/tdT-GFP or READRChETA/tdT-ffLuc vector; each sesRNA in the dual-sensor array contains an editable stop codon. k, FACS analysis showed that co-transfection of cells with READRChETA/tdT-GFP and either CAG-ChETA or CAG-tdT resulted in almost complete inhibition of GFP translation, and only triple transfection with all three vectors resulted in GFP expression. Co-transfection of EF1a-ChETA-tdT (resulting in a ChETA-tdT fusion transcript) was used as a positive control. l,m, Quantification by FACS (l) and luciferase assay (m). Data in a–c,e,f,i,l,m are mean ± s.e.m.; n = 3 biological replicates.
As it is based on Watson–Crick base-pairing, CellREADR is inherently programmable, and thus has the potential for joint sensing of two or more different RNAs to access more specific cell types defined by co-expression of more than one RNA. To explore this possibility, we first designed dual or triple sesRNA arrays targeting different regions of the same transcript, with each sesRNA containing an editable stop codon (Fig. 2g,j). All of the tested sesRNA arrays exhibited robust CellREADR efficiency, similar to that of a single sesRNA (Fig. 2h,i). To examine whether CellREADR could detect two target RNAs in a cell, we designed a sesRNA array consisting of sesRNAtdT and sesRNAChETA arranged in tandem (sesRNAtdT/ChETA) (Fig. 2j). Whereas sesRNAtdT and sesRNAChETA individually did not induce induce GFP or luciferase expression with readrRNAtdT/ChETA, we observed GFP expression or increased luminescence when both tdTomato and ChETA vectors were transfected (Fig. 2k–m). This demonstrated the potential for intersectional targeting of more specific cell types based on the programmability of two or more RNA sensors in CellREADR.
Endogenous RNA sensing
Compared with synthetic genes expressing exogenous RNAs, endogenous genes often have more complex genomic structures, including numerous exons, introns and regulatory elements; transcribed endogenous RNAs undergo multiple and elaborate post-transcriptional steps such as splicing and chemical modification before being processed as mature mRNAs. To examine the capacity of CellREADR for detecting cell endogenous RNAs, we selected EEF1A1, a housekeeping gene that is highly expressed in 293T cells, and systematically designed a set of sesRNAs targeting its various exons, introns, 5′ and 3′ untranslated regions (UTRs) and mRNAs (Fig. 3a,b). These sesRNAs were effective in sensing all intended EEF1A1 regions to switch on efRNA translation, with exons being better targets than introns (Fig. 3c). Notably, sesRNAs complementary to an mRNA region joined from two spliced exons achieved similar efficiencies to those contained within an exon, indicating that sesRNA can be designed to target both pre-mRNA and mature mRNA sequences. The efficacy of CellREADR targeting EEF1A1 RNAs increased with the incubation time after cell transfection (Extended Data Fig. 3c).
Fig. 3 |. Endogenous RNA sensing with CellREADR.
a, Schematic of the human EEF1A1 gene, showing exon (numbered) and intron structures (not to scale), pre-mRNA (middle) and mRNA (bottom). sesRNAEEF1A1 constructs 1 to 7 were designed to sample across the EEF1A1 pre-mRNA and mRNA. b, Schematic of the READREEF1A1-GFP vector for testing the sesRNAEEF1A1 constructs. c, Quantification of the efficiency of sesRNAEEF1A1 1–7 by FACS assay. d, Sensitivity of CellREADR. CellREADR efficiency was quantified for several endogenous cellular RNAs with different expression levels. TPM, transcript per million from RNA-seq data. Between one and three sesRNAs were designed for each target. e, Top, schematic for intersectional targeting of two endogenous RNAs (EEF1A1 and ACTB) using a dual-sensor READREEF1A1/ACTB-GFP vector; each sesRNA in the dual-sensor array contains an editable stop codon. Dual-sensor vectors with scramble sequence (READRCtr1/ctrl-GFP, READREEF1A1/ctrl-GFP or READRCtr1/ACTB-GFP) were used as controls. Bottom, only 293T cells transfected with READRChETA/tdT-GFP resulted in substantial GFP expression. f, RNA-seq analysis shows that CellREADR did not alter the cellular transcriptome. Comparisons of transcriptomes between cells transfected with sesRNAEEF1A1(7) or sesRNActrl (top) and with sesRNAPCNA or sesRNActrl (bottom). Pearson′s correlation coefficient analysis was used to evaluate the differential RNA expression. g, Volcano plot of differential gene expression analysis between sesRNAEEF1A1 and sesRNActrl (top), and sesRNAPCNA and sesRNActrl (bottom). Significantly differentially expressed genes (pink) (adjusted P-value < 0.01 and log2 (fold change) > 2). P-values (Wald test) and adjusted P-values (Benjamini–Hochberg correction) were calculated from DEseq2 for multiple hypothesis testing. h, Transcriptome-wide analysis of the effects of sesRNAPCNA (top) or sesRNAEEF1A1 (bottom) on A-to-I editing by RNA-seq. Pearson′s correlation coefficient analysis was used to evaluate the global RNA editing rate. Data in c–e are mean ± s.e.m. n = 3 biological replicates.
To examine the sensitivity of CellREADR to levels of target RNA, we selected several endogenous RNAs with high (ACTB and PCNA), intermediate (TP53 and XIST) and low (HER2 and ARC) expression levels. CellREADR was able to reliably detect ARC RNA, although at much lower efficiency compared with RNAs expressed at intermediate or high levels (Fig. 3d). For each individual RNA (for example, ACTB or TP53), the sesRNA sequence was a major determinant of RNA sensing (Fig. 3d).
To investigate the detection of cells expressing two specific endogenous RNAs, we designed a sesRNA array consisting of tandemly arranged sesRNAEEF1A1 and sesRNAACTB(sesRNAEEF1A1/ACTB). sesRNAEEF1A1/ACTB triggered robust translation of the effector (GFP) compared with control sesRNA arrays that included one or two scramble sesRNAs (sesRNAEEF1A1/ctrl, sesRNActrl/ACTBand sesRNActrl/ctrl) (Fig. 3e). Thus CellREADR may be able to target more specific cell types using intersectional RNA sensing.
To evaluate the effect of CellREADR on cellular gene expression, we used RNA-seq to compare the transcriptome profiles of 293T cells expressing sesRNAEEF1A or sesRNAPCNA with those expressing a scrambled sesRNActrl. Correlation analysis31 showed that expression of sesRNAEEF1A or sesRNAPCNA had no notable effect on the global transcriptome (Fig. 3f). Differential expression analysis32 revealed that sesRNAPCNA led to a reduction of only one gene (RGPD8) with no clear link to PCNA (Fig. 3g). sesRNAEEF1A1 resulted in an increase of about fourfold in the level of OAS2 mRNA (which encodes 2′-5′-oligoadenylate synthetase 2, an interferon-induced and dsRNA-activated antiviral enzyme) (Fig. 3g) and smaller increases (twofold to fourfold) of several other immune-related mRNAs (Supplementary Table 3). These results suggest that sesRNAs—even those targeting transcripts such as EEF1A1 that are highly expressed—exert no notable activation of cellular innate immune response, unlike transfection with poly(I:C), a synthetic analogue of dsRNA22.
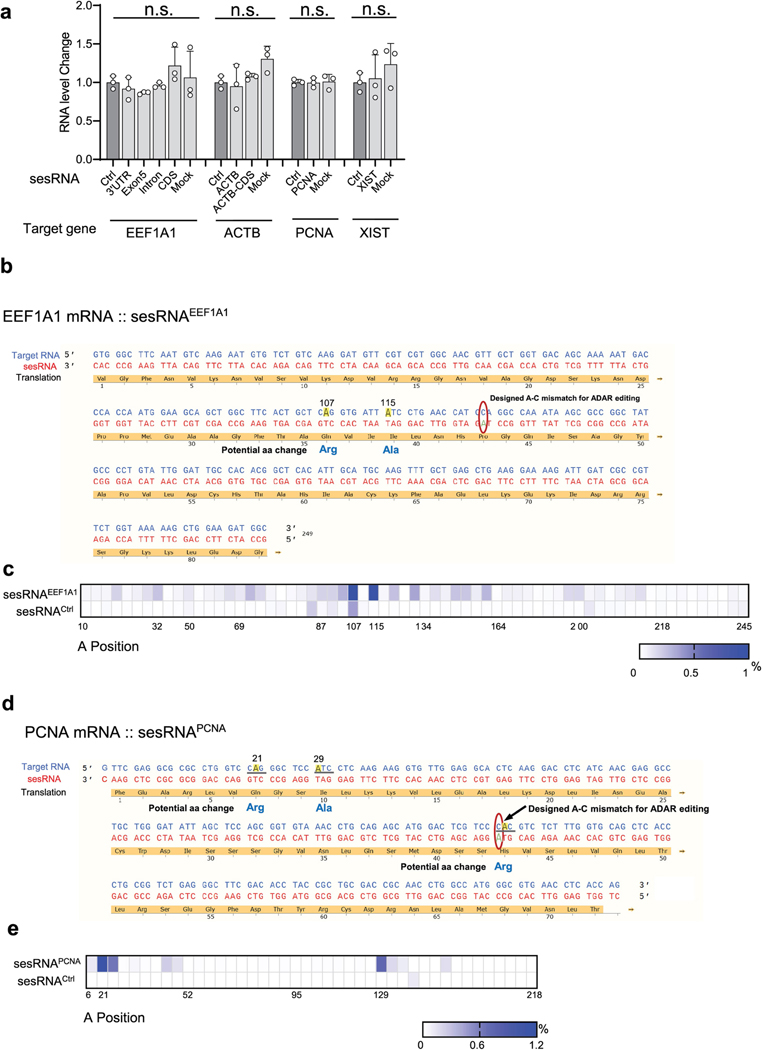
To assess whether CellREADR perturbs RNA editing in cellular transcriptomes, we used REDItools33 to compare the global A-to-I editing sites between sesRNAEEF1A1 or sesRNAPCNA and sesRNActrl groups. Neither the sesRNAPCNA nor sesRNAEEF1A1 group differed from the sesRNActrl group (Fig. 3h), demonstrating that CellREADR had no effects on normal A-to-I editing of cellular transcriptomes. Furthermore, quantitative PCR analysis showed that CellREADR did not alter the levels of target RNA transcripts (Extended Data Fig. 5a); this result also ruled out the possible effect of RNA interference induced by readrRNAs. To evaluate the possibility of unintended editing on target RNA, we examined the adenosine sites within the EEF1A1 RNA targeted by sesRNAEEF1A1 and PCNA RNA targeted by sesRNAPCNA. Very low levels of editing were observed throughout the targeted sequence (Extended Data Fig. 5b–e). Only two or three adenosines in target mRNAs exhibited a higher editing rate, in fewer than 1.2% of the transcripts (Extended Data Fig. 5c,e). Together, these results indicate that endogenous ADARs enable efficient and robust CellREADR functionality with minimum effects on cellular gene expression, RNA editing and target transcripts.
Cell-type targeting in animal tissues
To apply CellREADR to access specific cell types in animal tissues, we designed both singular and binary vector systems (Fig. 4a). In the singular READR vector, a human synapsin (hSYN) promoter drives transcription of mCherry followed by sesRNA and efRNA encoding an smFlag tag and tTA2. This vector directly couples RNA detection with effector gene expression; the efficiency and specificity can be evaluated by colocalization of mCherry, smFlag and the target mRNA or protein. The binary system includes an additional Reporter vector, which contains a TRE3g promoter driving mNeonGreen (mNeon) or other effector proteins in response to tTA2 translated from the READR vector (Fig. 4a). The binary vectors provide amplification of effector gene expression as well as the flexibility to express different effector genes in different cell types by pairing READR and Reporter vectors.
Fig. 4 |. CellREADR targeting, monitoring and manipulation of a neuronal cell type in mice.
a, Schematic of singular and binary adeno-associated virus (AAV) vectors for targeting Ctip2 neurons. W3SL, a sequence combining WPRE and polyA. b, Structure of the mouse Ctip2 gene, showing the location of sesRNACtip2(3). c, S1 cortex injected with READRCtip2(3)-smFlag/tTA2AAVs. Immunofluorescence of Flag (left) and CTIP2 (middle), showing Flag+ READR-labelled cells that were CTIP2+. d, Magnified view of the boxed region in c. Arrowheads show co-labelled cells. e, The specificity of labelling with the READRCtip2(3)singular vector. f, S1 neurons infected with binary READRCtip2vectors expressed mCherry (1); a subset of these in L5b expressed mNeon (2); and their specificity was assessed using CTIP2 immunofluorescence (3–5). The boxed region is expanded in (5). Arrowheads indicate co-labelled cells. g,h, Specificity (g) and efficiency (h) of labelling with READRCtip2(3)binary vectors. i,j, Schematic of paw stimulation (i) and fibre photometry recording (j) in anaesthetized mice. AAV-PTenhancer-tTA2 or AAV-Ctip2-CellREADR-tTA2 was co-injected with AAV-TRE-GCaMP6s into S1 cortex, upper limb area (SSp-ul).k, Heat map of neuronal activity aligned to onset of paw stimulation. l, Left, mean calcium signal aligned to paw stimulation (light grey area). Black traces are from data aligned to shuffled onset times. Shades around the mean trace show s.e.m. Right, peak z-score of responses. m, Schematic of optogenetic stimulation. n, AAV-PTenhancer-tTA2 or AAV-Ctip2-CellREADR-tTA2 was co-injected with AAV-TRE-ChRger2-eYFP into caudal forelimb area (CFA). o, Paw speed before and after light stimulation (blue bar); black traces represent average speed (see Supplementary Videos 1 and 2). p, Paw trajectories during stimulation. Side and front trajectories were normalized to start position. Black traces indicate average trajectories; squares indicate paw positions when stimulation ended. o,p, n = 20 trials for the PT enhancer mouse; n = 16 trials for the Ctip2 CellREADR mouse. Bars in e,g,h,l show mean values. n = 2 mice tested. Mouse drawings in i,m were adapted from scidraw.io under a CC BY 4.0 license (https://scidraw.io/drawing/44).
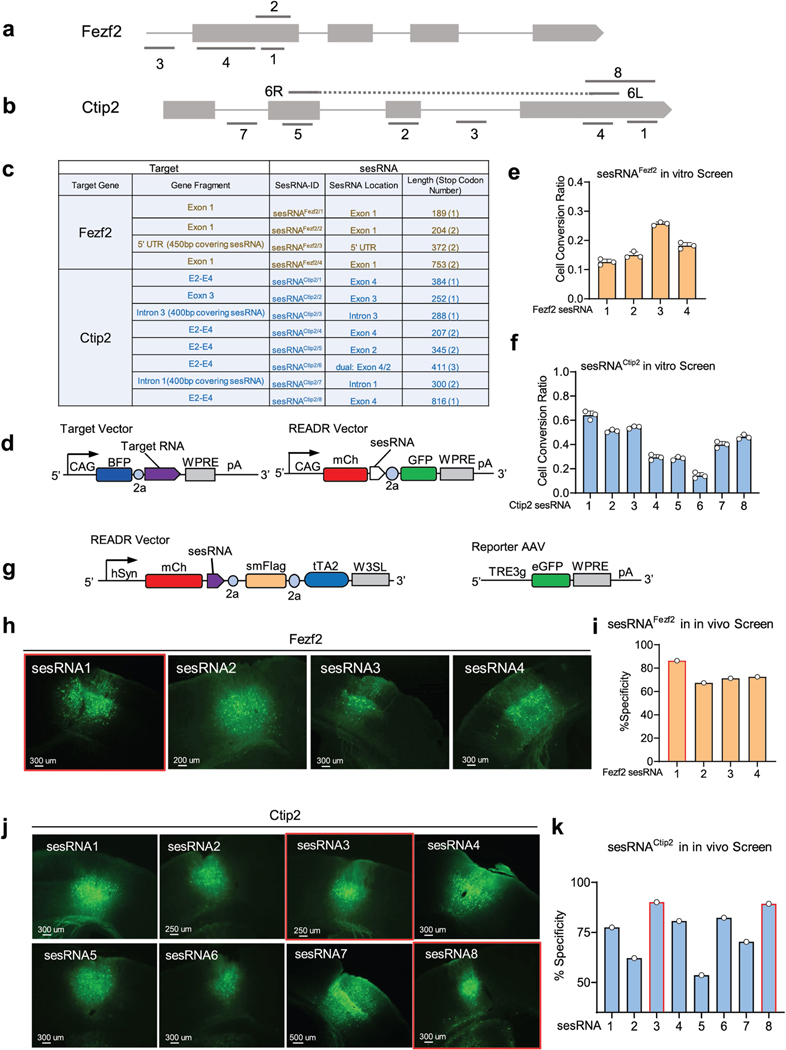
We first designed a set of READR vectors targeting mRNAs that define the major glutamatergic projection neuron types of the mouse cerebral cortex. The zinc-finger transcription factor gene Fezf2 labels the vast majority of layer 5b (L5b) and L6 corticofugal projection neurons (CFPNs) that constitute cortical output channels, and Ctip2 predominantly labels a subset of L5b/L6 CFPNs34,35. To identify appropriate sesRNAs, we first carried out a screen in 293T cells by co-expressing individual sesRNAs with their target sequences (Extended Data Fig. 6a,b,d). We designed four sesRNAs targeting exon 1 or the 5′ UTR of Fezf2 (Extended Data Fig. 6a,c) and eight sesRNAs targeting various exons and introns of Ctip2 (Fig. 4b and Extended Data Fig. 6b,c). These sesRNAs exhibited variable but substantial CellREADR efficiency in 293T cells in the presence of exogenous Fezf2 or Ctip2 target sequences (Extended Data Fig. 6e,f).
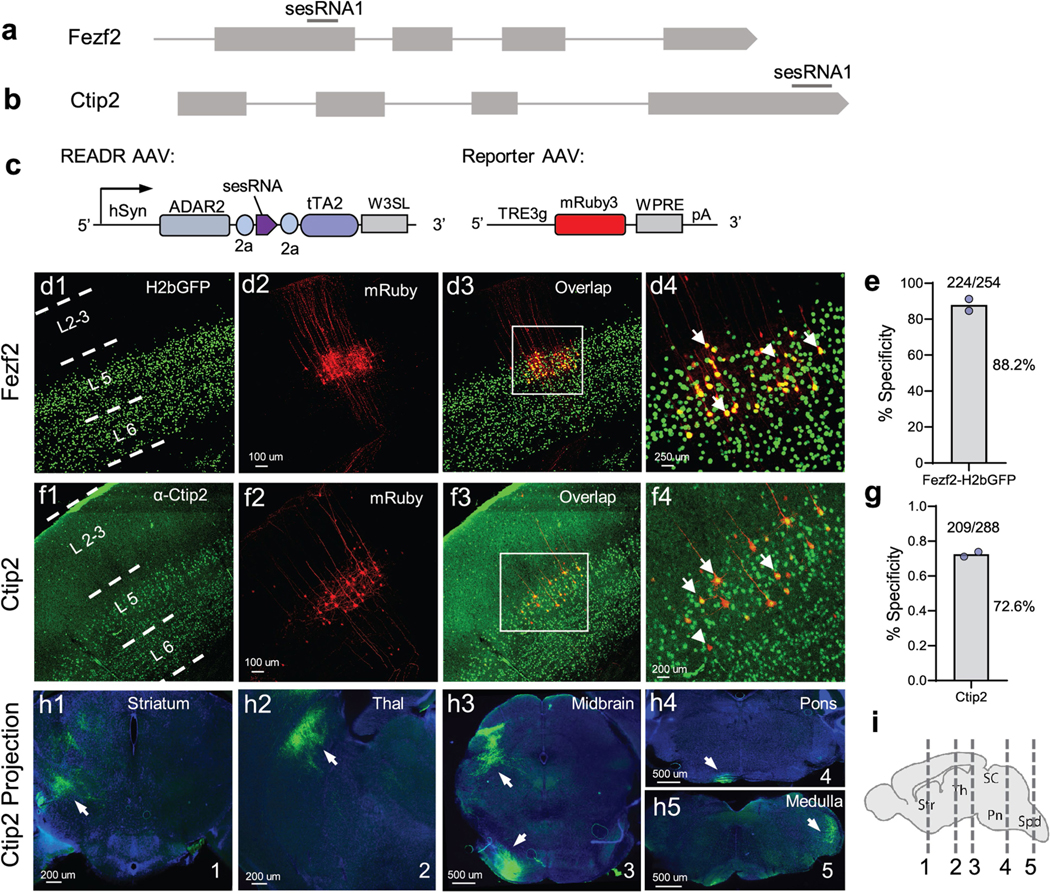
We then used AAV vectors to test these sesRNAs in mouse cortex by focal injection into the whisker barrel somatosensory cortex (S1) or motor cortex (M1). When delivered with binary vectors, Fezf2 sesRNA1 exhibited 86.4% specificity (Extended Data Fig. 6h,i) and Ctip2 sesRNA3 and Ctip2 sesRNA8 exhibited more than 90% specificity, as indicated by staining with CTIP2 antibody (Extended Data Fig. 6j,k). We subsequently focused on further evaluating CTIP2 sesRNA3. In S1 cortex infected with singular READRCtip2/3 AAV, Flag immunofluorescence was concentrated in deep layers (Fig. 4c); CTIP2 and Flag immunolabelling (Fig. 4c,d) demonstrated a specificity of 94.1% (Fig. 4e). In S1 cortex co-infected with binary READRCtip2/3 and ReportermNeon AAVs, mCherry expression from the READR vector labelled cells across cortical layers, whereas mNeon expression from the Reporter vector specifically labelled L5b and some L6 neurons (Fig. 4f); CTIP2 immunofluorescence showed that the labelling had a specificity of 90.4% (Fig. 4g). The efficiency of READRCtip2/3, calculated as the ratio of GFP+ cells among mCherry and CTIP2 double-positive cells, was 84.2% (Fig. 4h).
To test whether ADAR2 overexpression from the READR vector might enhance CellREADR functionality, we replaced mCherry in the READR vector with Adar2 cDNA, and tested Fezf2 and Ctip2 sesRNAs in mouse cortex (Extended Data Fig. 7a–c). Adar2 overexpression did not enhance the specificity or efficiency compared with binary vectors that relied on endogenous ADAR (Extended Data Fig. 7c–i).
In addition to Ctip2 and Fezf2 mRNAs that mark CFPNs, we designed sesRNAs in binary vectors to target several differentially expressed mRNAs36 (Extended Data Fig. 8a,b), including Plxnd1 (expressed in intratelencephalic projection neurons in L2/3 and L5a), Satb2 (expressed in intratelencephalic projection neurons across both upper and lower layers), Rorb (expressed in L4 pyramidal neurons) and Vgat (also known as Slc32a1 and expressed in pan-GABAergic (γ-aminobutyric acid-producing) neurons), respectively. Although only two or three sesRNAs were tested in each case, we were able to identify sesRNAs that achieved 75% or higher specificity (Extended Data Fig. 8c–t). These results indicate that CellREADR is robust and that specific cell-type targeting can be achieved by testing multiple sesRNAs for each target.
To evaluate the long-term effects of CellREADR sesRNAs in vivo, we incubated READRCtip2/3 and control CAG-tdT AAVs in mouse cortex for three months. Quantitative PCR with reverse transcription (RT–qPCR) of nine genes implicated in glia activation and immunogenicity showed no significant change in READRCtip2/3-infected versus control virus-infected tissues (Extended Data Fig. 9).
Cell-type monitoring and control
In addition to specificity, a key feature of any cell-type-discriminating technology is its efficacy for monitoring and manipulating cell function. We tested CellREADR efficacy by applying it to record and control neuronal function in mice. As a benchmark, we compared CellREADR with a transcriptional enhancer-based cell-type-targeting approach. A large scale open chromatin screening and in vivo validation effort identified the mscRE4 enhancer, which labels L5 pyramidal tract (PT) neurons, a subset of CFPNs15. We thus compared the efficacy of mscRE4 enhancer (or PT enhancer) with Ctip2 sesRNA3 using the same tTA-TRE binary vectors for expressing the genetic-encoded calcium indicator GCaMP6s and the light-activated ion channel channelrhodopsin-2 (Fig. 4i–p).
In mice co-infected with PT enhancer-tTA2 and ReporterGCaMP6s AAVs or READRCtip2/3 and ReporterGCaMP6s AAVs in the forelimb somatosensory cortex, mechanical stimulation of the forepaw induced reliable and time-locked increases of GCaMP6s signals in forelimb somatosensory cortex, as measured by fibre photometry, in mice transduced with both PT enhancer and READRCtip2/3 (Fig. 4i–l). This result indicates that effector gene expression levels achieved by CellREADR were sufficient to monitor cell physiology in live animals.
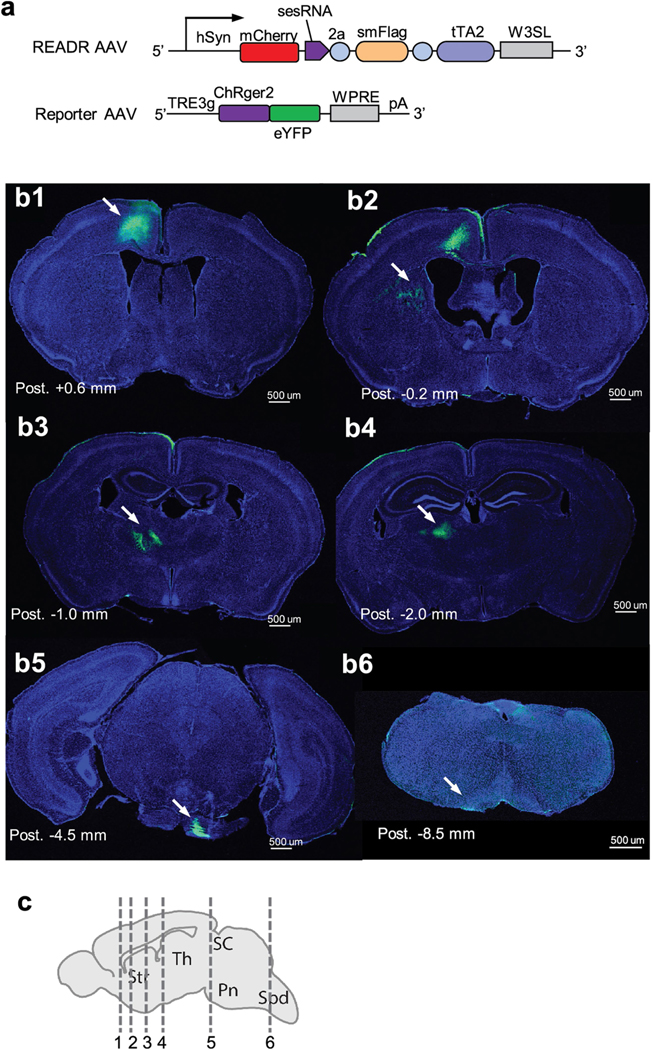
In mice co-infected with PT enhancer-tTA2 and ReporterChRger2-eYFP AAVs or READRCtip2/3 and ReporterChRger2-eYFP AAVs in the CFA, light stimulation of the CFA induced reliable and time-locked forelimb movement, which appeared stronger in mice transduced with READRCtip2/3 compared with those transduced with PT enhancer (Fig. 4m–p and Supplementary Videos 1 and 2). This indicates that the expression of effector genes achieved by CellREADR was sufficient to control cell function in behaving animals. Anatomical analysis of eYFP expression confirmed L5 PT neuron labelling by READRCtip2/3, as revealed by axonal projections in subcortical targets including striatum, thalamus, pons and medulla (Extended Data Fig. 10).
Cell-type targeting across species
To demonstrate the generalizability of CellREADR across mammalian species, we performed a series of experiments in rat and human brains. We first targeted GABAergic neurons in rats using binary AAV vectors targeting Vgat mRNA, a pan-GABAergic transcript (Extended Data Fig. 11b). Co-injection of READRVgat (hSyn-mCherry-sesRNAVgat-smFlag-tTA2) with ReportermNeon (TRE3g-mNeon) AAVs (Extended Data Fig. 11a) into rat S1 barrel cortex and hippocampus specifically labelled GABAergic interneurons in the cortex (Extended Data Fig. 11c) and hippocampus (Extended Data Fig. 11d,e). The specificities, assayed by co-labelling of Vgat mRNA and READRVgat, were 91.7% for cortex and 93.8% for hippocampus, respectively (Extended Data Fig. 11f,g). TLE4 is a conserved transcription factor that marks L6 corticothalamic pyramidal cells in rodents. A sesRNA targeting exon15 of Tle4 mRNA (Extended Data Fig. 11h) delivered by binary READRTle4 AAVs labelled deep layer neurons with 62.5% co-labelling with TLE4 antibody (Extended Data Fig. 11i–k).
We additionally established an organotypic culture platform for testing CellREADR in human neocortical specimens collected during surgery for epilepsy37 (Extended Data Fig. 12a). First, using a GFP construct driven by the hSyn promoter (AAVrg-hSyn-eGFP), we observed dense viral labelling across multiple layers, with a multitude of observed morphologies and a predominance of pyramidal neurons characterized by prominent apical dendrites (Extended Data Fig. 12b,c). We then designed two sesRNAs targeting forkhead box protein P2 mRNA (FOXP2) (Fig. 5a,b); FOXP2 is an evolutionarily conserved gene that is expressed in cortical and striatal projection neurons and implicated in human language skill development38. In situ hybridization and immunostaining indicate that unlike in mouse, where Foxp2 expression is restricted to L6 corticothalamic neurons35, human FOXP2 is expressed across cortical layers (Extended Data Fig. 12d). Five days after applying READRFOXP2 and ReportermNeon AAVs (hSyn-ClipF-sesRNAFOXP2-smV5-tTA2 with TRE3g-mNeon) to human neocortical slices, we observed mNeon-labelled cell bodies in upper and deep layers (Fig. 5c and Extended Data Fig. 12e). FOXP2 immunostaining showed that the binary READRFOXP2 targeted FOXP2-expressing neurons with a specificity of 79.8% (Fig. 5d,e). Labelled neurons also exhibited electrophysiological (Fig. 5f) and morphological (Fig. 5g) properties consistent with glutamatergic pyramidal neurons39. Using two singular READRFOXP2 vectors, each with a different sesRNA, we increased the specificity of targeting FOXP2 neurons to more than 97% (Extended Data Fig. 12f–i).
Fig. 5 |. CellREADR-enabled targeting and recording of human cortical neuron types.
a, Schematic of binary AAV vectors for targeting human neuron types. ClipF, sequence coding for an engineered enzymatic tag here used as a spacer in the READR vector; smV5, sequence coding for spaghetti monster fluorescent protein fused with the epitope tag V5. b, Genomic structure of the human FOXP2 gene and the design of two sesRNAs targeting FOXP2 mRNA. c, READR targeting of human FOXP2 cells. Left, upper layer FOXP2 cells (labelled with mNeon, native fluorescence) targeted with READRFOXP2(1) AAVs in organotypic slices from temporal neocortex. Right, deeper layer FOXP2 cells targeted with READRFOXP2(2) from parietal neocortex. Insets show magnified views of the boxed regions. Wt. matter, white matter. d, Immunostaining of FOXP2 in neurons labelled with READRFOXP2(2). Arrowheads indicate cells labelled with both mNeon (left) and FOXP2 immunofluorescence (middle); the arrow indicates a cell labelled with mNeon only. e, The specificity of READRFOXP2(2) assayed by immunofluorescence. f, Current-clamp recording traces of three mNeon-labelled neurons (top). Input–output curves (bottom) for cells depict similar spiking behaviours. g, Morphology of cell 3 recorded in f. The cell was filled with biocytin and was visualized by silver staining on a bright field. Brown, cyan and purple arrowheads indicate soma, axons and dendrites, respectively. h, SesRNA designed to target exon 2 of human VGAT mRNA. i, An organotypic slice from temporal neocortex infected with AAV-READRVGAT and TRE3g-mNeon and visualized with native fluorescence 8 days after infection. j,k, Morphologies of interneurons labelled by AAV-READRVGAT. l, Colocalization of mNeon native fluorescence (left) with VGAT mRNA detected by in situ hybridization (middle). Arrowheads indicate neurons labelled with both mNeon and in situ hybridization; the arrow indicates a cell labelled with mNeon only. m, The specificity of READRVGAT assayed by VGAT mRNA in situ hybridization. n, Current-clamp recording traces (top) of three labelled neurons, filled with biocytin and visualized with streptavidin. Input–output curves (bottom) depict distinct spiking behaviours: accommodating (blue), fast (green) and delayed onset (pink). o, The morphologies of cells labelled by READRVGAT and recorded in n. Scale bars, 50 μm. Data in e,m are mean values. n = 2 biological replicates.
We also targeted human neocortical GABAergic cells by designing a sesRNA targeting VGAT (Fig. 5h). Using READRVGAT and ReportermNeon, we observed cellular labelling across neocortical layers (Fig. 5i), with labelled cells exhibiting diverse morphologies characteristic of interneurons, including multipolar and smooth dendrites (Fig. 5j), and markedly dense, vertically oriented axons (Fig. 5i,k). In situ hybridization of VGAT mRNA showed a labelling specificity of 76.5% (Fig. 5l,m), which is probably an underestimate owing to the potentially reduced sensitivity of in situ hybridization in cultured ex vivo tissues. In support of this possibility, we observed very few labelled cells with pyramidal morphologies (Fig. 5i–l). Targeted patch clamp recordings of interneurons labelled with both READRVGAT and ReportermNeon seven days after virus application revealed various intrinsic properties, including accommodating, and fast and delayed-onset firing (Fig. 5n). The physiological properties (Fig. 5n) and partially reconstructed morphologies of the patched cells (Fig. 5o) were consistent with those of mammalian neocortical interneurons40. Together, these results demonstrate the cross-species generalizability and utility of CellREADR.
Discussion
The ability to use sequence-programmable RNA sensors as protein translation switches to analyse and manipulate cells define a new generation of molecular and cell engineering tools that will synergize with and expand beyond the ablilities of enhancer-based methods10–16. As it is based on Watson–Crick base-pairing and endogenous ADAR-mediated RNA editing, CellREADR is inherently: (1) specific to cells defined by RNA expression; (2) easy to construct, use and disseminate; (3) scalable for targeting all cells with known RNA expression in any tissue; (4) generalizable to most animal species, including humans; and (5) programmable to achieve intersectional targeting of cells defined by expression of two or more RNAs, as well as multiplexed targeting and manipulation of several cell types in the same tissue. Several aspects of CellREADR can be improved to increase its functionality and versatility. For example, the development of computational algorithms for sesRNA design and their experimental validation should optimize the length and sequence, as well as the position of the stop codon for increased specificity and reliability. Improved readrRNA stability and efRNA translation—for example, by using circular41,42 or chemically modified READR RNAs21,43—should increase its efficacy. Innovations in nucleic acid delivery, such as liposome nanoparticles44, the selective endogenous encapsidation for cellular delivery (SEND) system26 or viral capsid proteins that steer tissue and species tropism45, will enhance the ease and range of CellREADR applications. Finally, we expect CellREADR to apply to other vertebrate and invertebrate species. To facilitate the dissemination and future development of this technology and related resource, we have established a web-based CellREADR portal at https://cellreadr.neuro.duke.edu.
RNA is a universal messenger of gene expression that underlies cell identities and cell states in biological processes across life span and species. CellREADR will enable cell-type-based comparative and evolutionary approaches to discovering general biological principles and accelerate the development of disease models with cell-type resolution, especially using human cells and tissues, for understanding aetiology and mechanisms. With its capacity and flexibility to programme cellular physiology, CellREADR will facilitate next-generation precision diagnostic and therapeutic approaches, such as detecting cancer cells using their RNA markers and eliminating them by inducing programmed cell death or recruiting immune cells46. Whereas gene editing- and RNA editing-based therapeutic approaches38,47 have focused almost exclusively on correcting monogenic disease mutations48,49, many complex disorders such as neuropsychiatric and developmental disorders result from multigenic predispositions that affect the functional wiring of brain circuits and may not be corrected by gene or transcript editing. Modifying and tuning the affected cell types and circuits are rational approaches to treating such conditions50. Synergizing with genome and transcriptome engineering technologies, CellREADR will facilitate research towards elucidating the principles of biological information flow from genotype to phenotype across cell types, and make possible a new generation of programmable cell-specific RNA medicine.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41586-022-05280-1.
Methods
Plasmids
All constructs were generated using standard molecular cloning procedures. Vector backbones were linearized using restriction digestion, and DNA fragment inserts were generated using PCR or gBlock synthesis (Integrated DNA Technologies). Information on all plasmids is included in Supplementary Table 1. All sesRNA inserts were generated by gBlock synthesis (Integrated DNA Technologies), and sesRNA sequences used in this study are included in Supplementary Table 2.
Cell culture and transfection
The HeLa cell line was obtained from the laboratory of A. Krainer. Mouse neuroblastoma Neuro-2a (N2a) cells were purchased from Millipore-Sigma (catalogue (cat. no. 89121404). The HEK293T and KPC1242 cell lines were obtained from the laboratory of D. Fearon. All cell lines have been tested negative for mycoplasma contamination. HEK293T, Hela and N2a cell lines were cultured in Dulbecco′s Modified Eagle Medium (Corning, 10-013-CV) with 10% fetal bovine serum (FBS) (Gibco, 16000036) under 5% CO2 at 37 °C. 1% penicillin-streptomycin was added to the medium. Cells were transfected with Lipofectamine 2000 (Invitrogen, 11668019) DNA transfection reagent according to the manufacturer′s instructions. For interferon treatment, medium containing 1 nM Recombinant Mouse IFNβ (R&D, 8234-MB) was used and changed every 24 h during the transfection process. For tetracycline treatment, medium containing the indicated tetracycline concentration was used 24 h after transfection to replace the tetracycline-free medium and for 48 h before analysis. For apoptosis assay, cells were incubated for 72 h after transfection and the RealTime-Glo Annexin V Apoptosis and Necrosis Assay (Promega JA1011) was used to quantify the apoptosis level.
SURVEYOR assay
PCR of genomic DNA (using primers for DYRK1A: forward, GGAGCTGGTCTGTTGGAGAA; and reverse, TCCCAATCCATAATCCCACGTT) was used to amplify the Cas9 target region from a heterogeneous population of modified and unmodified cells, and the PCR products were reannealed slowly to generate heteroduplexes. The reannealed heteroduplexes were cleaved by SURVEYOR nuclease, whereas homoduplexes were left intact (Alt-R Genome Editing Detection Kit (IDT)). Cas9-mediated cleavage efficiency (percentage of indels) was calculated based on the fraction of cleaved DNA28.
Animals
Six- to twenty-week-old male and female mice were used in this study. The mice were housed under standard laboratory conditions in specific-pathogen-free cages in an animal room at constant temperature (19–23 °C) and regulated humidity under a 12 h:12 h light:dark cycle and received standard laboratory chow and water ad libitum with a maximum of five mice per cage. Wild-type mice were purchased from Jackson Laboratory (C57BL/6J, 000664). Fezf2-creER and Rosa26-LoxpSTOPLoxp-H2bGFP mice were described previously35,51. To label Fezf2+ cortical pyramidal neurons, 200 mg kg−1 tamoxifen (T56648, Sigma) was administered by intraperitoneal injection two days before AAV injection. Long Evans rats (Rattus norvegicus, aged 12 weeks, 50–250 g) were obtained from Charles River. Rats were maintained in a 12 h:12 h light:dark cycle and received standard laboratory chow and water ad libitum with one animal per cage. All animal maintenance and experimental procedures were performed according to Duke University IRB Protocol A241-20-12 for mice, Cold Spring Harbor Laboratory IRB Protocol 18-15-1 for mice and Duke University IRB Protocol A059-19-03 for rats. No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. All experimental procedures were carried out in accordance with NIH guidelines and approved by the Institutional Animal Care and Use Committees of Cold Spring Harbor Laboratory and Duke University.
ADAR1-knockout cell line construction
The ADAR1-knockout cell line was generated via CRISPR–Cas9 genome editing. The guide RNAs (gRNAs) were cloned into pSpCas9 (BB)-2A-puro (pX459) (Addgene, 62988). The gRNA sequences were 5′ GGATACTATTCAAGTCATCTGGG 3′ (gRNA1) and 5′ GTTATTTGAGGCATTTGATG 3′ (gRNA2). In brief, ADAR1-knockout cells were generated with two gRNAs targeting exon 2, which is shared by the p110 and p150 isoforms of ADAR1. HEK293T cells were seeded into 6-well plates and transfected with 1 μg mixture of pX459-gRNA1 and pX459-gRNA2 using Lipofectamine 2000 (Thermo Fisher Scientific, Cat. 11668019). On the next day, all of the medium containing transfection reagents was removed and replaced with fresh medium supplemented with puromycin (final concentration 2 μg ml−1). Fresh medium with puromycin was replaced with old medium every two days, three times. The remaining cells after puromycin selection were collected with TrypLE (Gibco, Cat. 12605036) and distributed in a 96-well plate with 1–2 cells per well. Expanded single clones were screened for ADAR1 deficiency by western blot and disruption of the ADAR1 genomic locus was confirmed by Sanger sequencing.
Flow cytometry analysis
Two days after transfection, cells were collected with TrypLE (Gibco, 12605036), distributed in 96-well round-bottom plate and centrifuged at 500 rpm for 1 min at 4 °C. Supernatant was removed and cells were resuspended with 1% paraformaldehyde (PFA) buffer and incubated at 4 °C overnight or longer time. The cells were then resuspended with 200 μl Flow Cytometry Staining Buffer (Invitrogen, 00-4222-26) before analysis using a LSR Fortessa (BD Biosciences). The Fortessa was operated by FACSDIVA (BD Biosciences) software. Data analysis was performed with FlowJo v10 (FlowJo). For sorting, cells were submitted to the same procedure as for flow cytometry analysis but without 1% PFA treatment and processed using BD FACSAria (BD Biosciences).
CellREADR efficiency for exogenous or endogenous transcripts
To assay CellREADR efficiency with fluorescence reporter genes, HEK293T cells or HEK293T ADAR1-knockout cells were first seeded in 24-well plates. After 24 h, cells were co-transfected with 1.5 μg plasmids in total. Forty-eight hours after transfection, the cells were collected, fixed with 1% PFA and prepared for FACS analysis. To calculate the CellREADR efficiency from the FACS data, the cells with READR vector were gated according to fluorescent protein expression (for example, BFP in Fig. 1b–d) and the efficiency was calculated as the ratio of double-positive cells expressing both efRNA and target RNA to the total number cells expressing the target RNA within the gate. If the spacer was not a fluorescent protein (Figs. 1h and 2), the ratio was calculated directly as the ratio of double-positive cells expressing both efRNA and target RNA among cells expressing the target RNA. For some controls, the CellREADR efficiency was calculated as the percentage of efRNA-expressing cells among all counted cells.
sesRNA design
We used the following procedure for sesRNA design: (1) sesRNA is complementary—that is, antisense, to a specific cellular coding or non-coding RNA sequence; (2) the optimal length of a sesRNA is 200–300 nt; (3) readrRNA consists of a sesRNA and efRNA arranged in a continuous translation reading frame; (4) one or more stop codons (TAG) are placed near the centre of a sesRNA (ranging between 80 and 220 nt from the 5′ end); (5) find 5′-CCA-3′ sequences in the target RNA, which is complementary to a 5′-TGG-3′ sequence in sesRNA, then replace 5′-TGG-3′ with 5′-TAG-3′, so that an A–C mismatch will be introduced when sesRNA base pairs with the target RNA; (6) to ensure that there are no other stop codons in the sesRNA, all other TAG, TAA and TGA sequences in sesRNA are converted to GAG, GAA and GGA, respectively; preferably the converted stop codons should not be near (that is, they should be more than 10 bp from) the TAG defined in step 5; (7) there should be no ATG (initiation codon) after the TAG defined in step 5 to exclude the possibility of unintended translation initiation; (8) sesRNAs can be directed to any region of a cellular transcript, including exons, introns, UTRs, or mature mRNA after splicing. (9) avoid sesRNA with complex secondary structures. sesRNA designs can also be facilitated using the online tool at the CellREADR portal: https://cellreadr.neuro.duke.edu.
A-to-I editing rate by CellREADR
To evaluate the A-to-I editing rate, HEK293T cells were seeded in 12-well plates. At 48 h post-transfection, cells were collected and RNA was extracted. RNAs were purified with RNeasy Mini Kit (Qiagen, 74106) and converted to cDNA using TaqMan Reverse Transcription Reagents (Thermo Fisher, N8080234). PCR products covering the whole sesRNA region were generated with CloneAmp HiFi PCR Premix (Takara, 639298), and purified with NucleoSpin Gel and PCR Clean-up kit (Takara, 74061) for Sanger sequencing. Editing efficacy was calculated as the ratio of Sanger peak heights G/(A + G). Three biological replicates were performed.
Transcriptome-wide RNA-seq analysis
The readrRNActrl or readrRNAPCNA, readrRNAEEF1A1-CDS-expressing plasmids with the red fluorescent protein (tdTomato) expression cassette were transfected into 293T cells. The RFP+ cells (about 5 × 105) were enriched by FACS sorting 48 h after transfection, and RNAs were purified with RNeasy Mini Kit (Qiagen Cat. 74106). To prepare the library, RNA samples were processed with the TrueSeq Stranded mRNA library Prep kit (Illumina, 20020594) and TrueSeq RNA Single Indexes (Illumina, 20020492). Deep sequencing analysis was performed in Illumina NextSeq500 platform at Cold Spring Harbor Laboratory NGS Bioinformatics Center. FastQC (0.11.9) was applied to RNA-seq data to check the sequencing quality. All samples passed quality check were mapped to reference genome (GRCh38-hg38). STAR (v2.7.8a) and RSEM (v1.3.2) were used to align reads and quantify gene expression. TPM values were calculated from RSEM. For all genes with non-zero count in at least one library, the average expression values across biological replicates were compared between samples for detecting differentially expressed genes using DESeq2(v1.34)32. Using normalized counts, we calculated the log2-transformed fold change and obtained the P-value (Wald test) and adjusted P-value (Benjamini–Hochberg correction) for multiple hypothesis testing. Genes with adjusted P-values < 0.01 and log2-transformed fold change > 2 or < 0.5 were identified as significantly differentially expressed. For global A-to-G editing rate analysis, the A-to-G editing rate calculation was performed using RNA editing detection pipeline (REDP), a Python wrapper for REDItools (v2.0)33. Parameters were set to default values. Alignment step was done using STAR (v2.7.8a) with default and coordinate-sorted parameters. Genome reference from Gencode GRCH38.p13 was used and concatenated with known sequences from target genes including EEF1A1 and PCNA.
Western blot
Primary antibodies to ADAR1 (Santa Cruz, sc-271854, 1:500), GFP antibody (Cell Signaling Technology, 2956; 1:10,000), β-actin antibody (Cell Signaling Technology, 3700; 1:10,000) were used in this study for western blots. We used standard western blot protocols. In brief, ~2 × 106 cells were lysed and an equal amount of each lysate was loaded for SDS–PAGE. Then, sample proteins were transferred onto polyvinylidene difluoride membrane (Bio-Rad Laboratories) and immunoblotted with primary antibodies to ADAR1 or GFP, followed by secondary antibody incubation (1:10,000) and exposure.
Luciferase complementation assay
Forty-eight hours after transfection, HEK293T cells expressing firefly luciferase gene were washed in PBS, collected by trituration and transferred to 96-well plates. Promega Luciferase Assay System (Promega, E1500) was used. Firefly luminescence was measured using SpectraMax Multi-Mode Microplate Readers (Molecular Devices).
RT–qPCR
RNA was extracted and purified with RNeasy Mini Kit (Qiagen Cat. 74106). RNA was converted to cDNA using TaqMan Reverse Transcription Reagents (Thermo Fisher, N8080234). RT–qPCR was performed with Taqman probes on the QuantStudio 6 Flex real-time PCR system. The housekeeping gene TBP was used for normalization. The gene probes are purchased from Thermo Fisher Scientific. (Thermo Fisher Scientific, EEF1A1: Hs01591985, PCNA: Hs00427214, XIST: Hs00300535, ACTB: Hs03023943, Mda5 (also known as Ifih1): Mm00459183_m1, Rig-I (also known as Rigi): Mm01216853_m1, Ifnb1: Mm00439552_s1, Il6: Mm00446190_m1, Ccl2: Mm00441242_m1, Fgf2: Mm01285715_m1, Cd40: Mm00441891_m1, Iba1: Mm00479862_g1, Cxcl10: Mm00445235_m1). The housekeeping gene TBP (Thermo Fisher Scientific, human TBP: Hs00427620, mouse Tbp: Mm01277042_m1) was used for normalization.
Virus production
For producing READR and Reporter viruses, HEK293T cells were transiently transfected with READR or Reporter plasmids, AAV serotype plasmids, and pHelper using PEI MAX (Polysciences, 24765). Seventy-two hours after transfection, the cells were collected in cell culture medium, and centrifuged at 4,000 rpm for 15 min. The supernatant was discarded, the pellet was resuspended in cell lysis buffer, frozen and thawed three times using a dry ice/ethanol bath. Cell lysate was centrifuged at 4,000 rpm for 20 min. The contaminating DNA in the supernatant was removed by adding benzonase and was incubated at 37 °C for 30 min. The crude viral preparation was loaded onto an iodixanol density gradient and spun at 60,000 rpm for 90 min using a Beckman Ti70 rotor. After the centrifugation, 3–4 ml crude viral preparation was collected from the 40–60% layer with an 18-gauge needle attached to a 10-ml syringe. The viral crude solution was concentrated to 200–250 μl using the Amicon Ultra-15 centrifugal filter (100 kDa), washed with 8 ml PBS once, and concentrated to an appropriate volume. Aliquots were stored at −80 °C until use. Mouse and rat READR and Reporter viruses were packaged as AAV-DJ or AAV-PHP.eB serotypes. PT enhancer-tTA2 viruses were packaged as PHP.eB using the Addgene vector #163480. Human READR and Reporter viruses were packaged as AAV2-Retro serotype. pAAV(DJ)-hSyn-ADAR2-sesRNAFezf2-tTA2 and pAAV(PHP.eB)-hSyn-ADAR2-sesRNACtip2(1) -tTA2 were from Vigene.
Immunohistochemistry
Mice or rats were anaesthetized (using Avertin) and intracardially perfused with saline followed by 4% PFA in 0.1 M PBS buffer. Following overnight fixation at 4 °C, brains were rinsed three times and sectioned at 50 μm thickness with a Leica 1000s vibratome. Sections were placed in blocking solution containing 10% normal goat serum (NGS) and 0.1% Triton X-100 in 1× PBS for 1 h, then incubated with primary antibodies diluted blocking solution overnight at 4 °C. Anti-GFP (1:1,000; Aves, GFP-1020); anti-CTIP2 (1:250; Abcam 18465); anti-TLE4 (1:500; Scbt: sc-365406); anti-Rorb (1:300; Novus Biologicals NBP1–82532) were used. Sections were rinsed 3 times in PBS and incubated for 1 h at room temperature with corresponding secondary antibodies (1:500; Jackson Immuno Research Labs). Sections were dry-mounted on slides using Fluoromount (Sigma, F4680) mounting medium. For human tissue, after viral expression had reached a peak or following patch clamp recording, cortical slices were fixed overnight in 4% PFA, then rinsed in PBS and stored from 1–7 days in PBS with azide. For biocytin recovery, tissue was permeabilized in PBS with 10% Triton X-100, 5% NGS, 100 μM glycine and 0.5% BSA for 30 min at room temperature on a shaker plate. Primary and secondary antibodies were incubated for 24 h at 4 °C. Biocytin was either developed directly with Alexa 647-conjugated streptavidin, or indirectly using the enzyme metallography method with peroxidase-labelled streptavidin treated with silver ion substrate, which deposits metallic silver at the active site. Recovered cells were imaged on a Keyence BZ-X800 with a 10× or 20× objective. Large overview images of native mNeon fluorescence were made on a Leica SP8 upright confocal microscope. For immunohistochemical labelling of FOXP2 and NeuN, tissue was cryoprotected for at least 24 h in 30% sucrose, then re-sectioned at 40 μm with a sliding microtome. Floating sections were permeabilized in PBS with 1% Triton X-100, 5% NGS, and 100 μM glycine for 30 min at room temperature and labelled overnight at 4 °C with primary antibodies (rabbit anti-FOXP2, 1:500, Abcam 16046; rat anti-Flag, 1:500, Novus NBP1-06712; mouse anti-NeuN, 1:500, Cell Signaling 94403S). For secondary antibodies, tissues were either incubated with biotinylated anti-rabbit antibody (1:1,000, Thermo Fisher 31822) for 2 h at room temperature followed by streptavidin Alexa Fluor 647 for 1 h at room temperature (for FOXP2) or with goat anti-mouse antibody (1:1,000, Thermo Fisher 32723) for 2 h at room temperature (for NeuN). The following secondary antibodies were used: Alexa Fluor 488 AffiniPure Donkey Anti-Chicken IgG (Jackson Immuno Research Labs, 703-005-155), Alexa Fluor 488 AffiniPure Donkey Anti-Rat IgG (Jackson Immuno Research Labs, 712-545-153), Cy3 AffiniPure Donkey Anti-Rabbit IgG (Jackson Immuno Research Labs, 711-165-152), Alexa Fluor 647 AffiniPure Donkey Anti-Mouse IgG (Jackson Immuno Research Labs, 715-605-150), Alexa Fluor 647 AffiniPure Donkey Anti-Rabbit IgG (Jackson Immuno Research Labs, 711-605-152) Alexa Fluor 647 AffiniPure Donkey Anti-Rat IgG (Jackson Immuno Research Labs, 712-605-153).
In situ hybridization
All probes were ordered from Molecular Instruments (mouse Satb2, cat. no. PRM128; mouse Plxnd1, cat. no. PRK885; mouse Fezf2, cat. no. PRA339; mouse Vgat, cat. no. PRE853; rat Vgat, cat. no. PRN024; and human VGAT, cat. no. PRM351). Mouse brain was sliced into 50-μm-thick slices after PFA perfusion fixation and sucrose protection. Hybridization chain reaction in situ was performed via free floating method in a 24-well plate. First, brain slices were exposed to probe hybridization buffer with HCR Probe Set (Molecular Instruments Inc.) at 37 °C for 24 h. Brain slices were washed with probe wash buffer, incubated with amplification buffer and amplified at 25 °C for 24 h. On day 3, brain slices were washed, counter stained if needed.
Stereotaxic viral injection
Adult mice 8 weeks of age or older were anaesthetized by inhalation of 2% isofluorane delivered with a constant air flow (0.4 l min−1). Ketoprofen (5 mg kg−1) and dexamethasone (0.5 mg kg−1) were administered subcutaneously as pre-emptive analgesia and to prevent brain oedema, respectively, prior to surgery, and lidocaine (2–4 mg kg−1) was applied. Mice were mounted in a stereotaxic headframe (Kopf Instruments, 940 series or Leica Biosystems, Angle Two). Stereotactic coordinates were identified. An incision was made over the scalp, a small burr hole drilled in the skull and brain surface exposed. A pulled glass pipette tip of 20–30 μm containing the viral suspension was lowered into the brain; a 500 nl volume of single or mixed viruses was delivered at a rate of 30 nl min−1 using a Picospritzer (General Valve) into 300 μm and 700 μm (1:1 volume) of injection sites; the pipette remained in place for 10 min preventing backflow, prior to retraction, after which the incision was closed with nylon suture thread (Ethilon Nylon Suture) or Tissueglue (3M Vetbond), and animals were kept warm on a heating pad until complete recovery. For rat Tle4 cell-type targeting, a 3:1 mixture of READRTle4 and Reporter AAVs was stereotactically injected into the cortex (0 mm posterior, 3.5 mm lateral, –2 mm ventral for 1,000 nl, and 0 mm posterior, 3.5 mm lateral, –1 mm ventral for 1,000 nl). For rat Vgat targeting, a 3:1 mixture of READRVgat and Reporter AAVs was stereotactically injected into the cortex (−4 mm posterior, 3.5 mm lateral, −2.5 mm ventral for 1,000 nl, and −4 mm posterior, 3.5 mm lateral, –1.5 mm for 1,000 nl). Further immunohistochemistry or in situ hybridization experiments were performed after 3 weeks of virus incubation.
Physiology in mouse
Stereotaxic surgery and viral injection.
All surgeries were performed under aseptic conditions and body temperature was maintained with a heating pad. Standard surgical procedures were used for stereotaxic injection and optical fibre implantation. Mice were anaesthetized with isoflurane (2–5% at the beginning and 0.8–1.2% for the rest of the surgical procedure) and were positioned in a stereotaxic frame and on top of a heating pad maintained at 34–37 °C. Ketoprofen (5 mg kg−1) was administered intraperitonially as analgesic before and after surgery, and lidocaine (2%) was applied subcutaneously under the scalp prior to surgery. Scalp and connective tissue were removed to expose the dorsal surface of the skull. The skin was pushed aside, and the skull surface was cleared using saline. A digital mouse brain atlas was linked to the stereotaxic frame to guide the identification and targeting of different brain areas (Angle Two Stereotaxic System, Leica Biosystems). We used the following coordinates for injections and implantations in the SSp-ul: −0.09 mm posterior from bregma, 2.46 mm lateral from the midline; CFA: 0.37 mm anterior from bregma, 1.13 mm lateral from the midline.
For viral injection, a small burr hole was drilled in the skull and brain surface was exposed. A pulled glass pipette tip of 20–30 μm containing the viral suspension was lowered into the brain; a 500 nl volume was delivered at a rate of 10–30 nl min−1 using a Picospritzer (General Valve); the pipette remained in place for 5 min, preventing backflow prior to retraction. Injection was made at a depth of 700 μm. An optical fibre (diameter 200 μm; NA, 0.39) was then implanted in CFA or SSp-ul for optogenetic activation or fibre photometry respectively. The optical fibre was implanted with its tip touching the brain surface. To fix the optical fibre to the skull, a silicone adhesive (Kwik-Sil, WPI) was applied to cover the hole, followed by a layer of dental cement (C&B Metabond, Parkell), then black instant adhesive (Loctite 426), and dental cement (Ortho-Jet, Lang Dental). A titanium head bar was fixed to the skull around lambda using dental cement. Mice were transferred on a heating pad until complete recovery. Further experiments of optogenetic activation or fibre photometry were performed after 8 weeks of virus incubation.
In vivo optogenetic activation.
We first briefly anaesthetized the mice with isoflurane (2%) to attach a reflective marker on the back of their left paws. Mice were then transferred into a tube, head fixed on a stage, and allowed to fully recover from the anaesthesia before the start of stimulation. A fibre-coupled laser (5-ms pulses, 10–20 mW; λ = 473 nm) was used to stimulate at 20 and 50 Hz and constantly for 0.5 s. The inter-stimulation interval was 9.5 s. Two cameras (FLIR, FL3-U313E4C-C)—one frontal camera and one side camera—were installed on the stage to take high-frame-rate (100 Hz) videos. The two cameras were synchronized by TTL signals controlled by custom-written MATLAB programs (2019a). Videos and TTL-signal states were acquired simultaneously using workflows in Bonsai software. Four LED light lamps were used for illumination (two for each camera).
Behavioural video analysis.
The two cameras were calibrated using the Camera Calibrator app in MATLAB. For left paw tracking using MATLAB, the images from the videos were smoothed with a Gaussian lowpass filter (size 9, sigma 1.8). The centroid of the reflective marker on the paw was first detected by a combination of brightness and hue thresholding, then tracked by feature-based tracking algorithms (PointTracker in Computer Vision Toolbox). The tracking results were validated manually, and errors were corrected accordingly. Trials in which mice made spontaneous movements before stimulation onset (within 0.5 s) were excluded from further analysis.
Paw stimulation.
To stimulate the left paw, mice were lightly anaesthetized with isoflurane (0.75–1%). Body temperature was maintained using a feedback-controlled heating pad. A piezo bender (BA4510, PiezoDrive), driven by a miniature piezo driver (PDu100B, PiezoDrive), was insulated with Kapton tape on which a blunt needle was glued. The tip of the needle was attached to the back of the paw to stimulate it with vibration (100 Hz, sine wave, 1 s). Inter-stimulation interval was 10 s.
In vivo fibre photometry and data analysis.
A commercial fibre photometry system (Neurophotometrics) was used to record calcium activities from SSp-ul. A patch cord (fibre core diameter, 200 μm; Doric Lenses) was used to connect the photometry system with the implanted optical fibre. The intensity of the blue light (λ = 470 nm) for GCaMP excitation was adjusted to ~60 μW at the tip of the patch cord. A violet light (λ = 415 nm, ~60 μW at the tip) was used to acquire the isosbestic control signal to detect calcium-independent artifacts. Emitted signals were band-pass filtered and focused on the sensor of a CMOS camera. Photometry signal and stimulation onset were aligned based on TTL signals generated by a multifunctional I/O device (PCIe-6321, National Instruments). Mean values of signals from the region of interest were calculated and saved by using Bonsai software, and were exported to MATLAB for further analysis.
To process recorded photometry signals, we first performed baseline correction of each signal using the adaptive iteratively reweighted Penalized Least Squares (airPLS) algorithm (https://github.com/zmzhang/airPLS) to remove the slope and low frequency fluctuations in signals. The baseline corrected signals were standardized (z-score) on a trial-by-trial basis using the median value and standard deviation of 5-s baseline period. The standardized 415-nm excited isosbestic signal was fitted to the standardized 470-nm excited GCaMP signal using robust linear regression. Lastly, the standardized isosbestic signal was scaled using parameters of the linear regression and regressed out from the standardized GCaMP signal to obtain calcium dependent signal.
Human organotypic sample preparation
Human neocortical tissues (frontal, parietal, temporal) were obtained from paediatric and adult patients (n = 6; aged 6–60 years) undergoing brain resections for epilepsy. Informed consent was obtained for the donation of tissues under Duke University IRB protocol 00103019, which is reviewed and issued by Duke University Health System (DUHS). According to the IRB protocol, the paediatric patients’ consent are given by their legal guardian or appointed legal representative, and the consent form includes permission to publish de-identified data where the identity of the patients is protected. No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment. Tissue preparation generally followed the methods described in the literature37,52,53. Dissection solution contained (in mM): 75 sucrose, 87 NaCl, 25 glucose, 26 NaHCO3, 2.5 KCl, 1.2 NaH2PO4, 10 MgCl2, 0.5 CaCl2, bubbled with 95/5% O2/CO2 (pH 7.4). Cortex was carefully dissected under ice cold (~4 °C) oxygenated dissection solution to remove the pia. Single gyri were blocked for slicing and cut at 300 μm thickness on a Leica VT1200. Slices were rinsed in Hank’s buffered saline and plated on PTFE membranes (Millipore, PICMORG50) in 6-well plates. Tissue was cultured in a conditioned medium as described40. The conditioned medium also contained anti-biotic/anti-mycotic (1×, Gibco by Thermo Fisher) for the first 7 days in vitro, and cyclosporine (5 μg ml−1) for the entire culture period. HEPES (20 mM) was added to the medium for the first hour after slicing. AAV retrograde (AAVrg) viruses were suspended in conditioned medium and applied directly to the slice surface by pipet on the day of plating. Expression of the mNeon reporter was monitored from day in vitro (DIV) 3 onwards, and tissue was used for immunohistochemistry and patch clamp experiments between DIV 6 and DIV 14.
Electrophysiology in human organotypic sample
Cultured human neocortical tissues were transferred to a heated recording bath (32–34 °C) on the platform of an Olympus BX-50 upright microscope. Recording solution contained (in mM): 118 NaCl, 3 KCl, 25 NaHCO3, 1 NaH2PO4, 1 MgCl2, 1.5 CaCl2, 30 glucose fully saturated with 95/5% O2/CO2. Borosilicate patch pipettes were pulled with a resistance of 4.0–5.5 MΩ, and filled with an internal solution containing (in mM):134 potassium gluconate, 10 HEPES, 4 ATP-triphosphate magnesium salt, 3 GTP sodium salt, 14 phosphocreatine, 6 KCl, 4 NaCl, pH adjusted to 7.4 with KOH. Biocytin (0.2%) was added to the internal solution to allow for morphological identification after recording. Patched cells were held in whole-cell mode for a minimum of 12 min to ensure complete filling with biocytin. Cells expressing mNeon were patch-clamped under visual guidance using an Axopatch 700B amplifier (Molecular Devices). Data were digitized with (Digidata 1550B) and recorded with pClamp 10 (Molecular Devices). Membrane voltage was recorded at 100k Hz and lowpass filtered at 10 kHz. Liquid junction potential was not corrected. Pipette capacitance and series resistance were compensated at the start of each recording, and checked periodically for stability of the recording configuration. Intrinsic membrane properties were measured with a −10pA current step. Firing rheobase was measured with a ramp current of 1s duration and 100–300 pA final amplitude. Input–output curves were generated from a series of current step starting at −50 pA and increasing in 10 pA increments until a maximum firing rate was elicited. Data were analysed using NeuroMatic and in-house routines in Igor Pro (Wavemetrics).
Microscopy and image analysis
Cell imaging in tissue culture was performed on ZEISS Axio Observer. Imaging from serially mounted brain sections was performed on a Zeiss LSM 780 or 710 confocal and Nikon Eclipse 90i fluorescence microscope, using ×5, ×63 and ×20 objectives for the tissue, as well as ×5 on a Zeiss Axioimager M2 System equipped with MBF Neurolucida Software (Neurolucida v2020). Quantification and image analysis was performed using Image J/FIJI (v2.0.0) software.
Statistics and reproducibility
Ordinary one-way ANOVA followed by Dunnett’s multiple comparisons with the mean of control was used for analysis in Extended Data Fig. 5. Unpaired two-tailed Student’s t-test was used for analysis in Extended Data Fig. 9. Pearson′s correlation coefficient analysis was used to evaluate the differential RNA expression and editing rate. Data were stored in Microsoft Excel 2016, and further statistics and plotting of graphs were performed with GraphPad Prism 9 (v9.3.1). DESeq2 (v1.34) was used to analyse statistical significance of transcriptome-wide RNA-seq data and calculate P-values (Wald test) and adjusted P-values (Benjamini–Hochberg correction) multiple comparisons. In Extended Data Figs. 1c, 2c and 4b,f, two independent repeats were performed with similar results and representative images were shown. In Fig. 1c and Extended Data Figs. 1b,f, 2b,e,f and 3f,h, at least three independent repeats in HEK293T cells were performed with similar results and representative images were shown. In Figs. 1c,h and 2h,k and Extended Data Fig. 4g, three independent repeats were performed with similar results and representative FACS analysis images were shown. In Fig. 4c,d,f and Extended Data Fig. 7d,f,h, experiments were performed in at least two mice with similar results, in Extended Data Figs. 6h,j, 8d,e,i,j,n,q,r and 10b, the experiment was performed in one mouse and representative images are shown. In Extended Data Fig. 11c–e,i,j, the experiment was performed in one rat and representative images are shown. In Fig. 5c,d,i–l,o and Extended Data Fig. 12c–e, experiments were performed with at least two human samples with similar results, and representative images are shown.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The RNA-seq data are available at NCBI BioProject under accession code PRJNA856413. Genome reference data are available from Gencode GRCH38.p13. Plasmids will be deposited to Addgene. We will provide reagents upon request until they are available from Addgene.
Code availability
The codes for optogenetic activation analyses are available from https://github.com/XuAn-universe/Optogenetic-activation. The codes for fibre photometry analyses are available from https://github.com/XuAn-universe/Fiber-photometry-sensory-stimulation.
Extended Data
Extended Data Fig. 1 |. Design and test of singular and binary CellREADR vectors.
a, Schematic of a singular CellREADR vector. Left, PGK-tdT (top) expresses the tdTomato target RNA from a PGK promoter. READRtdT-GFP (bottom) expresses a READR RNA consisting of sesRNAtdT and efRNAGFP, driven by a CAG promoter. Vertical dashed lines indicate the complementary base-pairing region between tdT mRNA and sesRNAtdT, with sequence surrounding the editable STOP codon shown on the right. At the editing site, the editable adenine in sesRNAtdT (cyan) is mismatched to a cytosine in the tdT mRNA. TdTomato is a tandem repeat of two dTomato genes, thus a tdT RNA contains two copies of target sequence for sesRNAtdT base-pairing. b, Validation of the READRtdT-GFP vector. In 293T cells co-transfected with READRtdT-GFP and PGK-tdT, many cells switched on GFP translation and fluorescence (arrows in top row). In cells co-transfected with control empty vector, few cells showed GFP expression (bottom right). c, GFP expression was further assayed by Western blotting. d, Left, a binary vector design for CellREADR luciferase assay. READRtdT-tTA2 expresses a readrRNA consisting of sesRNAtdT and efRNAtTA2, and TRE3g-ffLuc expresses the luciferase RNA upon tTA2 activation. Right, luciferase activity dramatically increased only in cells transfected with three vectors. Co-transfection of TRE3g-ffLuc with CAG-tTA2, which constitutively expresses tTA2, served as a positive control. e, Schematic of READRtdT-GFP vector in which a spacer sequence is inserted before sesRNAtdT coding region (top). 293T cells were transfected with READRtdT-GFP vector encoding viable length of spacers without (gray) or with (pink) tdT target RNA expression, respectively. Quantification of conversion ratio calculated as percentage of GFP+ cells among RFP+ cells (bottom). f, A binary vector design for CellREADR assay (left). Right, representative images of GFP conversion with binary vectors. In cells co-transfected with sesRNACtrl vector, few GFP+ cells were observed. Conversion percentages are shown on the right. Error bars in d are mean values ± s.e.m. n = 3 independent experiments performed. The bars in e are mean values, n = 2 independent experiments performed. For gel source data in c, see Supplementary Fig. 2.
Extended Data Fig. 2 |. CellREADR enables RNA sensing dependent gene editing and cell ablation, and Cre or Flp as an effort shows leaky activities.
a, left, vector design for CellREADR-mediated and target RNA-dependent gene editing. In READR tdT-Cas9/GFP, a CAG promoter drives expression of BFP followed by sequences coding for sesRNAtdT, Cas9, and eGFP effectors. In another vector, EF1a promoter drives tdT expression and U6b promoter drives the expression of a guide RNA (gRNA) targeting the DYRK1A gene in 293T cells. Right, quantification of READRtdT-Cas9/GFP efficiency as percent of GFP among RFP and BFP expressing cells with or without tdT target RNA. b, Cells transfected with the both U6b-gRNADYRK1A-CAG-tdT and READRtdT-Cas9/GFP showed robust GFP expression co-localized with BFP and RFP (bottom). Cells transfected with READR tdT-Cas9/GFP only (top) showed almost no GFP expression. c, SURVEYOR assay showed Cas9-mediated cleavage in the human DYRK1A locus. DNA cleavage was observed in cell lysates transfected with U6b-gRNADYRK1A-CAG-tdT and READR tdT-Cas9/GFP, but not in U6b-gRNADYRK1A and READRtdT-Cas9/GFP that lacked tdT target RNA. CAG-Cas9 with U6b-gRNADYRK1A cell lysate and 293T cell lysate without plasmid transfection were used as positive control and negative control, respectively. Arrows indicate cleavage products. d, left, vector design for CellREADR-mediated and target RNA-dependent cell death induction. In READR tdT-taCasp3-TEVp, a CAG promoter drives expression of BFP followed by sequences coding for sesRNAtdT and taCasp3-TEVp as effector to induce cell death. Right, cell apoptosis level measured by luminescence was increased in the cells transfected READR tdT-taCasp3-TEVp and EF1a-tdT compared with cells with no tdT RNA. e, Schematic READR vector with Cre coding sequence as effector RNA (left). Representative images of GFP conversion in CellREADR (right). In READRtdT-Cre vector without tdTom RNA, numerous cells showed GFP expression resulting from leaky CRE translation and recombination (right top). Cells transfected with READRtdT-Cre and tdTom RNA showed robust and strong GFP expression (right bottom). Cbh-DIO-eGFP was used as reporter vector for Cre. f, Schematic READR vector with Flp coding sequence as effector RNA (left). Representative images of GFP conversion in CellREADR (right). In READRtdT-Flp vector without tdTom RNA, numerous cells showed GFP expression from leaky FLP translation and recombination (right top). Cells transfected with READRtdT-Flp and tdTom RNA showed robust and strong GFP expression (right bottom). Cbh-fDIO-eGFP was used as reporter vector for Flp. Error bars in a and d are mean values ± s.e.m. n = 3 independent experiments performed.
Extended Data Fig. 3 |. Characterization of sesRNA properties.
a, CellREADR functions in multiple human and mouse cell lines. Schematic of CellREADR vectors. CAG-tdT expresses the tdTomato target RNA from a CAG promoter. READRtdT-GFP expresses a READR RNA consisting of sesRNAtdT and efRNAGFP, driven by a CAG promoter (top). Human (Hela) and mouse (N2a, KPC1242) cell lines showed comparable CellREADR efficiency. Error bars are mean values ± s.e.m. n = 3, n represents the number of independent experiments performed. b, Effects of different nucleotide mismatches between sesRNA and target RNA. Schematic of CellREADR vectors (top, related to Fig. 2b). CellREADR efficiency measured as RFP to GFP conversion ratio with different types of mismatch number, indel or two mismatches within UAG STOP codon. Indel here is one nucleotide insertion or deletion. Error bars are mean values ± s.e.m. n = 3, n represents the number of independent experiments performed. c, CellREADR mediated sensing of endogenous EEF1A1 mRNA and effector translation in 293T cells increased with longer incubation time after transfection. Error bars are mean values ± s.e.m. n = 3, n represents the number of independent experiments performed. d, Vector design of a tri-color CellREADR assay system (top). Quantification of READRtdT-GFP efficiency with increasing amount of CAG-tdT vector used for co-transfection of 293T cells (bottom). e, Schematics of rtTA-TRE3g-ChETA and READRChETA-GFP vectors (also see Fig. 1e). f, Representative images of co-transfected 293Tcells treated with different concentration of tetracycline in culture medium (also see Fig. 1e, f, g). g–h, Co-transfection of READRChETA-GFP with a vector that constitutively expresses ChETA (g) resulted highest conversion to GFP expression cells (h) compared to those in (f).
Extended Data Fig. 4 |. ADAR1 is necessary for CellREADR in 293T cells.
a, Schematic for generating a ADAR1 knockout cell line with CRISPR/Cas9. b, Western blot analysis showing ADAR1 expression in wild-type and no expression in ADAR1 knockout cells. c, Schematics of EF1a-ChETA-tdT and READRtdT-GFP vectors (left), and ADAR1 isoform expression vectors (right). d, Both p110 and p150 ADAR1 isoforms rescued the CellREADR functionality assayed by cell conversion ratio in ADAR1 knockout cells. e–h, INF-b increased CellREADR efficiency and ADAR expression. e, Schematic of EF1a-ChETA-tdT and READRtdT-GFP vectors. f, Western blot analysis showed increased expression of ADAR1-p110 protein and induction of ADAR1-p150 isoform after interferon treatment. g, Representative FACS analysis of GFP and RFP expression with mock (left) or interferon treatment (right). h, Quantification of the READRtdT-GFP efficiency in g, which was increased by interferon treatment. Error bars in d and h are mean values ± s.e.m. n = 3, n represents the number of independent experiments performed. For gel source data, see Supplementary Fig. 2.
Extended Data Fig. 5 |. Effects of CellREADR on targeted mRNA.
a, Quantitative PCR showing that CellREADR-mediated sesRNA expression did not impact the expression levels of targeted RNAs. Error bars are mean values ± s.e.m. n = 3 biological replicates performed. Ordinary one-way ANOVA followed by Dunnett’s multiple comparisons with the mean of control was used for analysis. All the P values are indicated. b, Base-pairing of EEF1A1 mRNA and sesRNAEEF1A1-CDS. EEF1A1 mRNA or sesRNA was represented in blue and red, respectively. Peptide translated from EEF1A1 mRNA was highlighted in brown. Targeted region of EEF1A1 mRNA was analyzed by RNAseq. c, The ratios of A-to-G changes in EEF1A1 mRNA at each adenosine position was quantified and shown in heatmap in both sesRNAEEF1A1-CDS and sesRNACtrl samples. Two adenosines (A107 and A115) showed higher rate of A-to-G editing (c). Off-target editing of two sensitive adenosines can induce potential amino acid change (underlined in b). d, Base-pairing of PCNA mRNA and sesRNAPCNA. PCNA mRNA or sesRNA was represented in blue and red, respectively. Peptide translated from PCNA mRNA was highlighted in brown. Targeted region of PCNA mRNA was analyzed by RNAseq. e, The ratios of A-to-G changes in PCNA mRNA at each adenosine position was quantified and shown in heatmap in both sesRNAPCNA and sesRNACtrl samples. Three adenosines (A21, A29 and A129) showed higher rate of A-to-G editing. Off-target editing of three sensitive adenosines can induce potential amino acid change (underlined in d).
Extended Data Fig. 6 |. Design and screen of sesRNAs targeting Fezf2 and Ctip2 RNAs in vitro and in vivo.
a–b, Genomic structures of mouse Fezf2 (a) and Ctip2 (b) genes with locations of various sesRNAs as indicated. c, List of sesRNAs and Fezf2 and Ctip2 target gene fragments used for sesRNA screen. d, In Target vectors CAG-BFP-Fezf2 or CAG-BFP-Ctip2, a 200–3000 bp genomic region of the Fezf2 or Ctip2 gene containing sequences complementary to a sesRNA in a, b were cloned downstream to the BFP and T2a coding region driven by a CAG promoter. In READR vectors, READRFezf2-GFP or READRCtip2-GFP expresses corresponding sesRNAs shown in a, b. e–f, Quantification of efficiencies READRFezf2-GFP (e) or READRCtip2-GFP (f) as GFP conversion ratio by FACS assay of 293T cells co-transfected with CAG-BFP-Fezf2 or CAG-BFP-Ctip2 target vector, respectively. g, Schematic of binary READR AAV vectors. In READR vector, a hSyn promoter drives expression of mCherry followed by sequences coding for sesRNACtip2, smFlag and tTA2 effectors. In Reporter vector, TRE3g promoter drives mNeon in response to tTA2 from the READR vector. h, Coronal sections of mouse cortex injected with binary READRFezf2 vectors. mNeon indicated READRFezf2 labeled cells. Four Fezf2 sesRNAs were screened. i, Quantification of specificity of 4 Fezf2 sesRNAs in h. For Fezf2 sesRNA in-vivo screen, the specificity of each sesRNA was calculated by co-labeling by READR AAVs and CTIP2 antibody (due to lack of FEZF2 antibody); as Ctip2 represents a subset of Fezf2+ cells (not shown), CTIP2 antibody gives an underestimate of the specificity of Fezf2 sesRNA. SesRNA1 showed highest specificity. j, Coronal sections of mouse cortex injected with binary READRCtip2 vectors. mNeon indicated binary READR labeled cells. Eight Ctip2 sesRNAs were screened. k, Quantification of specificity of 8 sesRNAs in j. The specificity of each sesRNA was calculated by co-labeling by binary READRCtip2 AAVs and CTIP2 antibody (not shown). SesRNA3 and sesRNA8 showed highest specificity. Error bars in e–f are mean values ± s.e.m. n = 3, n represents the number of independent experiments performed. Bars in i and k are values from one mouse performed. Each bar in i and k is the value from one mouse performed (n = 1).
Extended Data Fig. 7 |. CellREADR targeting of PNFezf2 and PNCtip2 types in mouse cortex with ADAR2 overexpression.
a–b, Genomic structures of mouse Fezf2 (a) and Ctip2 (b) genes with locations of sesRNAs as indicated, respectively. c, Schematic of binary AAV vectors for targeting neuron types. In READRFezf2-tTA2, a hSyn promoter drives expression of ADAR2 followed by sequences coding for sesRNAFezf2(1), T2a, and tTA2 effector. In TRE3g-mRuby3, the TRE3g promoter drives mRuby3 in response to tTA2 from the READR virus. d, Image of coronal section from a Fezf2-CreER;LoxpSTOPLoxp-H2bGFP mouse brain, showing the distribution pattern of Fezf2+ PNs in S1 somatosensory cortex (d1). Co-injection of AAVs READRFezf2(1)-tTA2 and TRE3g-mRuby3 specifically labeled PNs in L5b and L6 (d2). Co-labeling by CellREADR AAVs and H2bGFP (d3) with magnified view in (d4). Arrows indicate co-labeled cells; arrowhead shows a neuron labeled by CellREADR AAVs but not by Fezf2-H2bGFP (d4). e, Specificity of READRFezf2(1)-tTA2. f, Coronal section of WT brain immuno-stained with a CTIP2 antibody, showing the distribution pattern of Ctip2+ PNs in S1 cortex (f1). Co-injection of AAVs READRCtip2(1) and TRE3g-mRuby3 specifically labeled PNs in L5b (f2). Co-labeling by CellREADR AAVs and CTIP2 antibody (f3) with magnified view in (f4). Arrows show the co-labeled cells; arrowhead showed a mis-labeled cells by READRCtip2(1) (f4). g, Specificity of READRCtip2(1). h, Axonal projection pattern of AAV READRCtip2(1)-tTA2 and TRE3g-eYFP infected PNs in S1 cortex. Representative images showing projections to striatum (h1), thalamus (h2), midbrain (h3), pons (h4) and medulla (h5) (arrows). i, Schematic locations of coronal sections are shown on the right panel. The bars in e and g are mean values, n = 2 mice performed.
Extended Data Fig. 8 |. CellREADR targeting of additional cortical neuron types in the mouse.
a–b, Expression level and laminar distribution of cortical cell type markers in mice. a, Group plot of selected genes in transcriptomic cell type clusters, based on dataset from the Allen Institute for Brain Science. Gene expression level and cortical distribution were shown. b, Gene expression level of several major cell type marker genes. Each dot indicates the marker gene RNA expression level in cell type cluster. Error bars are mean values ± s.e.m. (n = 8 for Fezf2, n = 8 for Ctip2, n = 28 for SatB2, n = 15 for PlxnD1, n = 7 for Tle4, n = 7 for Foxp2, n = 13 for Rorb, n = 60 for vGAT). PT (pyramidal tract), IT(intratelencephalic) and CT (Corticothalamic) indicate three main excitatory cortical neuronal types. The plots were generated from mouse cortical scRNAseq online tool (https://celltypes.brain-map.org/rnaseq/mouse/v1-alm) with published scRNAseq data. c, Genomic structures of the mouse Satb2 gene with location of a sesRNA as indicated (top). Bottom, Cell labeling pattern in S1 by co-injection of binary vectors described in Fig. 4i. AAVs READRSatb2 and TRE3g-mNeon labeled cells in both upper and deep layers (d1). Satb2 mRNA in-situ hybridization (d2). Co-labeling by READRSatb2 and Satb2 mRNA (d3). e, Magnified view of boxed region in d3. Arrowheads indicate co-labeled cells. f, Satb2 mRNA expression pattern in S1 cortex at P56 from the Allen Mouse Brain Atlas. g, Specificity of READRSatb2 measured as the percent of Satb2+ cells among mNeon cells. h, Genomic structures of the mouse PlxnD1 gene with location of a sesRNA as indicated. i–j, AAVs READRPlxnD1 and TRE3g-mNeon labeled cells in upper layers and L5a in S1 (i1). PlxnD1 mRNA in-situ hybridization (i2). Co-labeling by READRPlxnD1 AAVs and PlxnD1 mRNA (i3). j, Magnified view of boxed region in i3. Arrowheads show the co-labeled cells. k, PlxnD1 mRNA expression in P56 S1 cortex from the Allen Mouse Brain Atlas. l, Specificity of READRPlxnD1 measured as the percent of PlxnD1+ cells among mNeon cells. m, Genomic structures of the mouse Rorb gene with location of a sesRNA. n, AAVs READRRorb and TRE3g-mNeon labeled cells in layer 4 (n1, n3). DAPI staining indicated laminar structure (n2). mNeon labeling pattern is consistent with Rorb mRNA expression in P56 S1 cortex from the Allen Mouse Brain Atlas (o). p, Genomic structures of mouse the vGAT gene with location of a sesRNA as indicated. q-r, binary READRvGAT and TRE3g-mNeon labeled cells (q1). vGAT mRNA in-situ hybridization. (q2). Co-labeling by READRvGAT AAVs and vGAT mRNA (q3). r, Magnified view of rectangle in (q3). Arrowheads show the co-labeled cells. s, vGAT mRNA expression in P56 S1 cortex from the Allen Mouse Brain Atlas. t, Specificity of READRvGAT measured as the percent of vGAT + cells among mNeon cells. Each bar in g, l and t is the value from one mouse performed (n = 1).
Extended Data Fig. 9 |. Assessment of cortical cellular immune responses following long-term expression of CellREADR vectors.
a, Schematic of evaluation of the long-term effects of CellREADR in vivo. For each mouse, READRCtip2(3) or CAG-tdT control AAVs were injected into S1 cortex and incubate for three months. Fresh brains were dissected and small pieces of cortical tissue at the injection site were collected. Quantitative PCR was performed immediately. S1 tissues of mice without viral injection were used as control. b, RNA expression level changes of nine genes implicated in glia activation and immunogenicity. Error bars are mean values ± s.e.m. n = 3 biological replicates in different mice performed. Unpaired two-tailed Student’s t-test was used for analysis and P values were given.
Extended Data Fig. 10 |. Axonal projection pattern of L5/6 CFPNs in caudal forelimb motor area targeted by AAVs READRCtip2 and TRE3g-ChRger2-eYFP.
a, Schematic of binary AAV vectors of READRCtip2(3)-smFlag/tTA2 and TRE3g-ChRger2-eYFP for optogenetic activation and axonal projection tracing. b, Axonal projection pattern of CFPNs infected in CFA. Representative images showing projections to striatum (b2), thalamus (b3, b4), pons (b5) and medulla (b6, arrows). c, Schematic locations of coronal sections in b are shown.
Extended Data Fig. 11 |. CellREADR targeting of neuron types in rat.
a, Schematic of binary AAV vectors for cell type targeting in rat. In READR vector, a hSyn promoter drives expression of mCherry followed by sequences coding for sesRNA, smFlag and tTA2 effectors. Along with READR, a Reporter vector drives mNeonG expression from a TRE3g promoter in response to tTA2 from the READR vector. b, Genomic structures of the rat vGAT gene with location of a sesRNA as indicated. c, AAVs READRvGAT and TRE3g-mNeon were injected into cortical deep layer and hippocampus. Binary vectors labeled cells shown in cortex (c1). c2, Magnified view of boxed region in c1. vGAT mRNAs were labeled by in-situ hybridization (c3). Co-labeling by mNeon and vGAT mRNA (c4). Arrows showed the co-labeled cells. d, Cell labeling pattern in the hippocampus CA1 region by co-injection of AAVs READRvGAT and TRE3g-mNeon. e, Magnified view of boxed region in d. Arrows indicate co-labeled cells. f–g, Specificity of rat READRvGAT in rat cortex (f) and hippocampus (g) measured as the percent of vGAT+ cells among mNeon cells. h, Genomic structures of the rat Tle4 gene with the location of a sesRNA as indicated. i–j, AAVs READRTle4 and TRE3g-mNeon co-injected into the rat motor cortex labeled cells concentrated in deep layers (i1). Tle4+ PNs were labeled by TLE4 antibody staining (i2, 3). j, Magnified view of the boxed region in i. Arrows indicate co-labeled cells by CellREADR and TLE4 antibody. k, Specificity of rat READRTle4 in rat cortex, measured as the percent of TLE4 + cells among mNeon cells. Each bar in f, g and k is the value from one rat performed (n = 1).
Extended Data Fig. 12 |. CellREADR vector targeting of neuron types in human cortical ex vivo tissues.
a, Schematic of organotypic platform for of human cortical ex vivo tissues. Left panel was adapted from “Brain (lateral view)”, by BioRender.com (2022). Retrieved from https://app.biorender.com/biorender-templates. b, Schematic of a hSyn-eGFP viral construct used to drive widespread neuronal cell labeling. c. AAVrg-hSyn-eGFP labeled cells were distributed across all layers, and exhibited diverse morphologies (c1). Insets from c1 (c2) and c2 (c3, 4) depict numerous cells with pyramidal morphologies, including prominent vertically oriented apical dendrites. d, FOXP2 expression in human neocortex. FOXP2 mRNA expression pattern taken from the Allen Institute human brain-map (specimen# 4312), showing upper and deep layer expression (arrows) (d1). FOXP2 immunostaining in the current study (magenta) also demonstrated both upper and deep labeling (d2, 4). NeuN immunostaining (red) depicting cortical neurons (d3, 4). Dashed lines delineate pia and white matter. e, READRFOXP2(1) labeling in an organotypic slice derived from the same tissue used in d. Overview of bright field and mNeon native fluorescence in the organotypic slice demonstrating highly restricted labeling, as compared to that observed in d. Inset from e2 (e3) depicting morphologies of upper layer pyramidal neurons. f, Schematics of two singular vectors of READRFOXP2. In READRFOXP2(1), the hSyn promoter drives an expression cassette encoding ClipF, sesRNA1, smV5, and tTA2. In READRFOXP2(2), the hSyn promoter drives an expression cassette encoding ClipF, sesRNA2, smFlag, and FlpO. g–h, Seven days after application of READRFOXP2(2) AAV on DIV 1, tissue was fixed and stained with antibodies against FOXP2 and FLAG (g). h, Boxed region from g. FLAG-labeled cells from READRFOXP2(2) exhibited relatively small somata with short apical dendrites (h1–h3, arrowheads). Non-specific background fluorescence signals (e.g. a blood vessel-like profile) are indicated by thin arrows (h2). i, Quantification of CellREADR specificity measured as the percentage of V5+ cells (for READRFOXP2(1)) and FLAG+ cells (for READRFOXP2(2)) labeled by FOXP2 immunostaining, respectively. Each bar in i is the value from one human brain sample performed (n = 1).
Extended Data Table. 1.
Plasmids used in this study.
| plasmids | Figure index | Constructs (backbone) | Note on construct |
|---|---|---|---|
| 1 | Fig 1b–d | pCAG-tdTom | Addgene# 83029 |
| 2 | pCAG-BFP-sesRNA-2a-eGFP | Backbone from Addgene# 100844, Gcamp6s was replaced with BFP-sesRNA-2a-eGFP | |
| 3 | Fig 1e–g | pTRE-BFP-2a-Cheta-pPGK-puro-2a-rtTA | Backbone from Addgene# 71782, BFP-2a-Cheta was inserted after TRE elements |
| 4 | pCAG-BFP-2a-Cheta | Backbone from Addgene# 100844, Gcamp6s was replaced with BFP-2a Cheta | |
| 5 | pCAG-mCherry-sesRNA-2a-eGFP | Backbone from Addgene# 100844, Gcamp6s was replaced with mCherry-sesRNA-2a-eGFP | |
| 6 | Fig 1h–i | pCAG-tdTom | |
| 7 | pCAG-spacer(2.1kb)-2a-sesRNA-2a-eGFP | from plasmids#5 spacer sequence was from CasRx coding regions of Addgene# 109049, 2.1kb from ATG start | |
| 8 | pCAG-ADAR2-sesRNA-2a-eGFP | from plasmids#5, ADAR2 sequence was from Addgene# 117929 | |
| 9 | Fig 1j–k | pCAG-tdTom | |
| 10 | pCAG-ClipF-sesRNA-2a-eGFP | from plasmids#5, ClipF sequence was from Addgene# 174554 | |
| 11 | pCAG-ADAR2-sesRNA-2a-eGFP | ||
| 12 | Fig 2a–d | pCAG-BFP-sesRNA-2a-eGFP | |
| 13 | pEF1a-Cheta-tdTom | Addgene# 37756 | |
| 14 | Fig 2e | pCAG-BFP-sesRNA-2a-ffLuc | from plasmids#8, ffLuc sequence was from Addgene# 71394 |
| 15 | Fig 2f | pEF1a-Cheta-tdTom | |
| 16 | pCAG-BFP-sesRNA-2a-ffLuc | ||
| 17 | Fig 2g–i | pEF1a-Cheta-tdTom | |
| 18 | pCAG-BFP-sesRNA-2a-eGFP | ||
| 19 | Fig 2j–m | pEF1a-Cheta-tdTom | |
| 20 | pEF1a-Cheta | From Addgene# 37756, Cheta-tdTom was replaced with Cheta | |
| 21 | pCAG-tdTom | ||
| 22 | pCAG-BFP-sesRNA-2a-eGFP | ||
| 23 | pCAG-BFP-sesRNA-2a-ffLuc | ||
| 24 | Fig 3b–e | pCAG-BFP-sesRNA-2a-eGFP | |
| 25 | Fig 3f–h | pCAG-ClipF-sesRNA-tTA | Backbone from Addgene# 112668,rtTA was replaced with ClipF-sesRNA-tTA |
| 26 | Fig 4a–c | pAAV-hSyn-mCherry-sesRNA-2a-smFlag-2a-tTA2 | smFlag from Addgene #59756 |
| 27 | pAAV-TRE3g-mNeonGreen | ||
| 28 | pAAV-TRE3g-GCaMP6s | GCaMP6s from Addgene #100844 | |
| 29 | pAAV-TRE3g-ChRger2-eYFP | ChRger2-eYFP from Addgene #127239 | |
| 30 | pAAV-TRE3g-eGFP | TRE3g backbone was kindly gifted from Nelson lab at Brandeis University | |
| 31 | Fig 5 | pAAV-hSyn-ClipF-sesRNA-2a-smFlag-tTA2 | ClipF sequence was from Addgene# 174554 |
| 32 | pAAV-TRE3g-mNeonGreen | TRE3g backbone was kindly gifted from Nelson lab at Brandeis University | |
| 33 | Extended Data Fig 1b, c | PKG-tdTom | |
| 34 | CAG-sesRNA-2a-eGFP | ||
| 35 | PKG-Empty | ||
| 36 | Extended Data Fig 1e, f | PKG-tdTom | |
| 37 | CAG-sesRNA-2a-tTA2 | ||
| 38 | TRE3g-ffLuc | TRE3g backbone was kindly gifted from Nelson lab at Brandeis University | |
| 39 | Extended Data Fig 1 g, h | pCAG-spacer-sesRNA-2a-eGFP | Spacer sequence was from CasRx coding regions of Addgene# 109049, different length was used from ATG start as indicated |
| 40 | pCAG-tdTom | ||
| Extended Data Fig 1 i, j | pCAG-BFP-sesRNA-2a-tTA2 | ||
| TRE3g-eGFP | |||
| 41 | Extended Data Fig 2 | pCAG-BFP-sesRNA-2a-Cas9–2a-eGFP | Cas9 from addgene# 62988 |
| 42 | pCAG-BFP-sesRNA-2a-tcaCasp3-TEVp | tcaCasp3-TEVp from addgene# 45580 | |
| 43 | Extended Data Fig 3c–f | pTRE-BFP-2a-Cheta-pPGK-puro-2a-rtTA | |
| 44 | pCAG-BFP-2a-Cheta | ||
| 45 | pCAG-mCherry-sesRNA-2a-eGFP | ||
| 46 | Extended Data Fig 4 | CMV-ADAR1-p110 | From addgene# 117928, GFP coding region was deleted |
| 47 | CMV-ADAR1-p150 | From addgene# 117927, GFP coding region was deleted | |
| 48 | Extended Data Fig 9 e, f | pCAG-BFP-2a-TargetGene | Target Gene was amplified from mice genomic DNA, Cheta was used as a control. Details in Extended Data Fig.10 |
| 49 | pCAG-mCherry-sesRNA-2a-eGFP | ||
| 50 | Extended Data Fig 9 h–k | pAAV-hSyn-mCherry-sesRNA-2a-smFlag-2a-tTA2 | |
| 51 | pAAV-TRE3g-mNeonGreen | ||
| 52 | Extended Data Fig 10 | pAAV-hSyn-ADAR2–2a-sesRNA-tTA2 | |
| 53 | pAAV-TRE3g-mRuby3 | ||
| 54 | pAAV-TRE3g-eGFP | ||
| 55 | Extended Data Fig 12, 15 | pAAV-hSyn-mCherry-sesRNA-2a-smFlag-2a-tTA2 | |
| 56 | Extended Data Fig 16 | pAAV-hSyn-ClipF-sesRNA-2a-smFlag-FlpO | FlpO from previous study (Matho et al Nature 2011) |
| 57 | Extended Data Fig 16 | pAAV-hSyn-ClipF-sesRNA-2a-smV5-tTA2 | smV5 from addgene # 71817 |
Extended Data Table. 2.
SesRNA used in this study.
| Figure Index | sesRNA name | Target gene | sesRNA position | Leng th | Stop Codon No. | sesRNA sequence |
|---|---|---|---|---|---|---|
| Fig. 1–2, Extended Data Fig. 1–6 | tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 1, Extended Data Fig. 1–6 | Ctrl | Not Target | N/ | 195 | 1 | aggcaagccctacgagggcacccagaccatgagaatcaaggtggtcgagggcggccctctccccttcgccttcgacatcctggctactagcttcctctacggcagcaagaccttca tcaaccacacccagggcatccccgacttcttcaagcagtccttccctgagggcttcacatgggagagagtcaccacata |
| Fig. 1, Extended Data Fig. 1–6 | Cheta | Cheta | CDS | 228 | 1 | agccctcggggaaggacagcttcttgtaatcggggatgtcggcggggtgcttcacgtacgccttggagccgtacaggaactggggggacaggatgtcccaggcgaagggcagg gggccgcccttggtcaccttcagcttagcggtctgggtgccctcgtaggggcggccctcgccctcgccctcgatctcgaactcgtggccgttcatggagccctccatgcgcacct |
| Fig. 2a | 25nt-tdT | tdTomato | CDS | 25 | 1 | cctcccagcccatagtcttcttcta |
| Fig. 2a | 50nt-tdT | tdTomato | CDS | 50 | 1 | gctcggtggaggcctcccagcccatagtcttcttctgcattacggggccg |
| Fig. 2a | 100nt-tdT | tdTomato | CDS | 100 | 1 | tcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttcttctgcattacggggccgtcgggggggaagttggtgccgca |
| Fig. 2a | 150nt-tdT | tdTomato | CDS | 150 | 1 | gggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttcttctgcattacggggccgtcgggggggaagttggtg ccgcgcatcttcaccttgtagatcagcgtgccgt |
| Fig. 2a | 300nt-tdT | tdTomato | CDS | 300 | 1 | Gcagttgcacgggcttcttggccatgtagatggtctggaactccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggt acaggcgctcggtggaggcctcccagcccatagtcttcttctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg aggagtcctgggtcacggtcaccagaccgccgtcctcgaagttcatcacgcgctcccactggaagc |
| Fig. 2a | 400nt-tdT | tdTomato | CDS | 400 | 1 | tgtccagcttggtgtccacgtagtagtagccgggcagttgcacgggcttcttggccatgtagatggtctggaactccaccaggtagtggccgccgtccttcagcttcagggcctggt ggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttcttctgcattacggggccgtcgggggggaagttggtgccgcgcatc ttcaccttgtagatcagcgtgccgtcctgcagggaggagtcctgggtcacggtcaccagaccgccgtcctcgaagttcatcacgcgctcccactggaagccctcggggaaggaca gcttcttgtaatcggggatgtcggcggggtgcttcacgtacgccttggagcg |
| Fig. 2b, Extended Data Fig. 6 | 1mm-A-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccgtcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tcggcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 1mm-B-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccttctgcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgctttacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 2mm-A-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccatcccatagtcttct tctgcgttacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 2mm-B-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccttctgcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgctttacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 4mm-A-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagcggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgtgggggtacaggcgctcggtggtggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttctagatcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 4mm-B-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgccgtccttcggcttcagggcctggtggatctcgcccttcagaacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtagggggggaagttggtgccgcgcatcttcaccttgtagagcagcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 10mm-A-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtggtggccgccgtccttcatcttcagggactggtggatctcgcccttcagcacgccgtcgagggggtacaggcgcgcggtggaggcctcccagcccatagtcttct tcggcattacggggacgtcgggggggaagttggtgctgcgcatcttcacattgtagatcatcgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 10mm-B-tdT | tdTomato | CDS | 195 | 1 | actccaccaggtagtggccgacgtccttcagcttcaggtcctggtggatctcgcccttccgcacgccgtcgcggtggtacaggcgcccggtggaggtctcccagcccatagtcttctt cttcattacggggccgtcggggggtaagttggggccgcgcatcttcaccttgtagatcagtgtgccgtcctgcaggg |
| Fig. 2b, Extended Data Fig. 6 | 20mm-tdT | tdTomato | CDS | 195 | 1 | actccaccaggcagtggccgacgtccttcggcttcaggtcctggcggatctcgcccttccgcacgccgccgcggtggtacaggcgctcagtggaggtctcccagcccatagtcttca tcttcattacggggccgtcgcggggtaagttggggccgcccatcttcacgttgtagatcagtgtgccgacctgcaggg |
| Extended Data Fig. 6 | 1nt-insert in frame | tdTomato | CDS | 196 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtgcgaggcctcccagcccatagtcttc ttctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Extended Data Fig. 6 | 1nt-insert out frame | tdTomato | CDS | 196 | 1 | Actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcagtacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagaatcagcgtgccgtcctgcaggg |
| Extended Data Fig. 6 | 1nt-del in frame | tdTomato | CDS | 194 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacgggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Extended Data Fig. 6 | mm (TGC-TAG) | tdTomato | CDS | 196 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggc |
| Extended Data Fig. 6 | mm (TGT-TAG) | tdTomato | CDS | 255 | 1 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatggtcttct tctgcattacggggccgtcgggggggaagttggtagcgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggaggagtcctgggtcacggtcaccagaccgccgtcctcg aagttcatcacgcgctcccact |
| Fig. 2c | Promotor | Cheta-tdTom | Promotor | 238 | 1 | agttttaaacagagaggaatctttgcagctaatggaccttctaggtctggaaaggagtgggaattggctccggtgcccgtcagtgggcagagcgcacatcgcccacagtccccga gaagttagggggaggggtcggcaattgaaccggtgcctagagaaggtggcgcggggaaaactgggaaagtgaagtcgtgtactggctccgcctttttcccgagggtgggggag aaccgtatat |
| Fig. 2c | Cheta1 | Cheta-tdTom | CDS | 1 | ggtggttttgcggatatctccgtgaatcaatatgtgctcgtggatcaggacgcgcaggtagtgtcccaacaacccccaacaatttttactcatcagatcaataatcgtgtgacctacg gtggagccatagacgctcaggacgccaaaaccttcgggccccaaaatgaagagaattaggaacataccccagctcacgaaaaacagccatgccatgccggtcacgacctggcg gcaccgaccctttggcacagt | |
| Fig. 2c | Cheta2 | Cheta-tdTom | CDS | 1 | ggttctcctgctgtagtcgttgctcaggccggtgaggttgctcaggcggataaggatgacaggacaagtgatgcagccaggttgcatagcgcagccactgcacccggtgtcctgtA gcaaggtagagcaagagggattcttaaactcaaaaaagaactcgagaatcaccttaaccatttcaatggcgcacacatagatctcctcccagccgcatgtagatttccaggtttgg taggcatagaacatcagcagcaaaatgctgaatcctgctgcaagccactgcaggacatt | |
| Fig. 2c | tdT1 | Cheta-tdTom | CDS | 195 | 1 | Actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2c | tdT2 | Cheta-tdTom | CDS | 228 | 1 | agccctcggggaaggacagcttcttgtaatcggggatgtcggcggggtgcttcacgtacgccttggagccgtacaggaactggggggacaggatgtcccaggcgaagggcagg gggccgcccttggtcaccttcagcttAgcggtctgggtgccctcgtaggggcggccctcgccctcgccctcgatctcgaactcgtggccgttcatggagccctccatgcgcacct |
| Fig. 2d | 2TAG | Cheta-tdTom | CDS | 255 | 2 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtagcgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggaggagtcctgggtcacggtcaccagaccgccgtcctcg aagttcatcacgcgctcccact |
| Fig. 2d | 3TAG | Cheta-tdTom | CDS | 255 | 3 | actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtagaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtagcgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggaggagtcctgggtcacggtcaccagaccgccgtcctcg aagttcatcacgcgctcccact |
| Fig. 2g–i | Cheta1-tdT1 | Cheta-tdTom | CDS | 447 | 2 | Ggtggttttgcggatatctccgtgaatcaatatgtgctcgtggatcaggacgcgcaggtagtgtcccaacaacccccaacaatttttactcatcagatcaataatcgtgtgacctac ggtggagccatagacgctcaggacgccaaaaccttcgggccccaaaatgaagagaattaggaacataccccagctcacgaaaaacagccatgccatgccggtcacgacctggc ggcaccgaccctttggcacagtActccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtgga ggcctcccagcccatagtcttcttctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcaggg |
| Fig. 2g–i | tdT1-Cheta1 | Cheta-tdTom | CDS-UTR | 447 | 2 | Actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggGgtggttttgcggatatctccgtgaatcaatatgtgctc gtggatcaggacgcgcaggtagtgtcccaacaacccccaacaatttttactcatcagatcaataatcgtgtgacctacggtggagccatagacgctcaggacgccaaaaccttcg ggccccaaaatgaagagaattaggaacataccccagctcacgaaaaacagccatgccatgccggtcacgacctggcggcaccgaccctttggcacagt |
| Fig. 2g–i | Cheta1-Cheta2 | Cheta-tdTom | CDS | 543 | 2 | ggtggttttgcggatatctccgtgaatcaatatgtgctcgtggatcaggacgcgcaggtagtgtcccaacaacccccaacaatttttactcatcagatcaataatcgtgtgacctacg gtggagccatagacgctcaggacgccaaaaccttcgggccccaaaatgaagagaattaggaacataccccagctcacgaaaaacagccatgccatgccggtcacgacctggcg gcaccgaccctttggcacagtggttctcctgctgtagtcgttgctcaggccggtgaggttgctcaggcggataaggatgacaggacaagtgatgcagccaggttgcatagcgcag ccactgcacccggtgtcctgtAgcaaggtagagcaagagggattcttaaactcaaaaaagaactcgagaatcaccttaaccatttcaatggcgcacacatagatctcctcccagc cgcatgtagatttccaggtttggtaggcatagaacatcagcagcaaaatgctgaatcctgctgcaagccactgcaggacatt |
| Fig. 2g–i | tdT-Cheta1-cheta2 | Cheta-tdTom | CDS | 738 | 3 | Actccaccaggtagtggccgccgtccttcagcttcagggcctggtggatctcgcccttcagcacgccgtcgcgggggtacaggcgctcggtggaggcctcccagcccatagtcttct tctgcattacggggccgtcgggggggaagttggtgccgcgcatcttcaccttgtagatcagcgtgccgtcctgcagggggtggttttgcggatatctccgtgaatcaatatgtgctc gtggatcaggacgcgcaggtagtgtcccaacaacccccaacaatttttactcatcagatcaataatcgtgtgacctacggtggagccatagacgctcaggacgccaaaaccttcg ggccccaaaatgaagagaattaggaacataccccagctcacgaaaaacagccatgccatgccggtcacgacctggcggcaccgaccctttggcacagtggttctcctgctgtagt cgttgctcaggccggtgaggttgctcaggcggataaggatgacaggacaagtgatgcagccaggttgcatagcgcagccactgcacccggtgtcctgtAgcaaggtagagcaa gagggattcttaaactcaaaaaagaactcgagaatcaccttaaccatttcaatggcgcacacatagatctcctcccagccgcatgtagatttccaggtttggtaggcatagaacat cagcagcaaaatgctgaatcctgctgcaagccactgcaggacatt |
| Fig.3 | EEF1A1-3’ UTR | Human-EEF1A1 | 3’ UTR | 225 | 1 | cttcagtctccacccactcagtgtggggaaactccatcgcataaaacccctccccccaacctaaagacgacgtactccaaaagctcgagaactaatcgaggtgcctggacggc gcccggtactccgtagagtcacatgaagcgacggctgaggacggaaaggcccttttcctttgtgtgggtgactcacccgcccgctctcccgagcgccgcgtcctccattttg |
| Fig.3 | EEF1A1-5’ UTR | Human-EEF1A1 | 5’ UTR | 249 | 1 | ctcatagggtgtacactagccactacacttatttcttatgtcatggcaaatagtcaactttcactgcccagtcattttaacccacgtttcaacatgcacatcccagtaatttagaaa cattttgtttccaaagattcacttaacattggtttagcaacaggaagctttctatgcaacacaaggactcagtttttggcctgttttagtgacaggcaatcagcaacatgctgcat ttctctccagtgttgt |
| Fig.3 | EEF1A1-intron | Human-EEF1A1 | Intron | 228 | 1 | gtaaggattaagagtcgttacttggttactaaaacacaaactccagcttcaatttccttgtccccagcccttaattagcagtttccactttacaactccaagtccaaagtgatttta gtcactttgggttacagaagcaaccaaaaatcaaacttttataagtcggatcttaactatgaacatccaaatctactcactagcaatacgattacagaagtcaccaaaagc |
| Fig.3 | EEF1A1-Exon3 | Human-EEF1A1 | Exon 3 | 249 | 1 | gttagcacttggctccagcatgttgtcaccattccaaccagaaattggcacaaatgctactgtgtcggggttgtagccaattttcttaatgtaagtgctgacttccttaacaatttc ctcatatctcttctggctgtagggtagctcagtggaatccattttgttaacaccgacaatgagttgtttcacacccagtgtgtaagccagaagggcatgctctcgggtctgcccat tcttggagataccagc |
| Fig.3 | EEF1A1-Exon5 | Human-EEF1A1 | Exon 5 | 249 | 1 | agtgaagccagctgcttccattggtgggtcatttttgctgtcaccagcaacgttgccacgacgaacatccttgacagacacattcttgacattgaagcccacattgtccccagga agagcttcactcaaagcttcatggtgcatttcgacagattttacttccgttgtaacgttgactagagcaaaggtgaccaccataccgggtttgagaacaccagtctccactcggc caacaggaacagtaccaat |
| Fig.3 | EEF1A1-eie | Human-EEF1A1 | Exon 2 | 249 | 1 | gtctgtctcatatcacgaacagcaaagcgacctattaaaaaaaaagttaattattacccaaagtactgttcagttgtatttttcatctttaacacaacttttttacatttaagtagt catccttacccaaaggtggatagtctgagaagctctcaacacacataggcttgccaggaaccatatcaacaatggcagcatcaccagacttcaagaattgagggccatcttcc agctttttaccagaacggc |
| Fig.3 | EEF1A1-CDS | Human-EEF1A1 | CDS | 249 | 1 | gccatcttccagctttttaccagaacggcgatcaatcttttccttcagctcagcaaacttgcatgcaatgtgagccgtgtggcaatccaatacaggggcatagccggcgcttattt ggcctagatggttcaggataatcacctgagcagtgaagccagctgcttccattggtgggtcatttttgctgtcaccagcaacgttgccacgacgaacatccttgacagacacat tcttgacattgaagcccac |
| Fig.3 | Xist-1 | Human-XIST | lncRNA | 234 | 1 | aaaatatggaggacgtgtcaagaagacactaggagaaagtatagaattgaaaaaaatattttatggaatttaggtgatttttttaaagaaatacgccataaagggtgttagg ggactagaaaatgttctagaaagaaccccaagtgcagagagatcttcagtcaggaagcttccagccccgagagagtaagaaatatggctgcagcagcgaattgcagcgctt taagaactgaa |
| Fig.3 | Xist-2 | Human- XIST |
lncRNA | 210 | 1 | caattgggactgagcattttaactgtccaacaaaagacgggttgtctgcgaccccctgctgaatgcaaatggggttcagagtgctgtctaatccaataggtaggattattggca attagaattttactcaatgcaaagagttttaggtggccaacacagtacacaagaaaagatcactactcaaaattgggactgggactacagaagcaa |
| Fig.3 | PCNA-E1 | Human-PCNA | Exon 1 | 219 | 1 | ctggtgaggttcacgcccatggccaggttgcggtcgcagcgggaggtgtcgaagccctcagaccgcagggtgagctgcaccaaagagacgtaggacgagtccatgctctgc aggtttacaccgctggagctaatatcccagcaggcctcgttgatgaggtccttgagtgcctccaacaccttcttgaggatggagccctggaccaggcgcgcctcgaac |
| Fig.3 | ActB-CDS | Human-ActB | CDS | 249 | 1 | CTGGGTGCCAGGGCAGTGATCTCCTTCTGCATCCTGTCGGCAATGCCAGGGTACATGGTGGTGCCGCCAGACAGCACTGTGTTGGCG TACAGGTCTTTGCGGATGTCCACGTCACACTTCATGATGGAGTTGAAGGTAGTTTCGTAGATGCCACAGGACTCCATGCCCAGGAAG GAAGGCTGGAAGAGTGCCTCAGGGCAGCGGAACCGCTCATTGCCAATGGTGATGACCTGGCCGTCAGGCAGCTCG |
| Fig.3 | ActB-2 Exon2 | Human-ActB | Exon 2 | 237 | 1 | gggtcatcttctcgcggttggccttggggttcaggggggcctcggtcagcagcacggggtgctcctcgggagccacacgcagctcattgtagaaggtgtggtgccagattttct ccatgtcgtcccagttggggacgatgccgtgctcgatagggtacttcagggggaggatgcctctcttgctctgggcctcgtcgcccacataggaatccttctgacccatgcccac catcacgc |
| Fig.3 | TP53-1-E3 | Human-TP53 | Exon 3 | 279 | 1 | cgtgcaagtcacagacttggctgtcccagaatgcaagaagcccagacggaaaccgtagctgccctggtaggttttctgggaagggacagaagatggcaggggccaggagg gggctggtgcaggggccgccggtgtaggagctgctAgtgcaggggccacggggggagcagcctctggcattctgggagcttcatctggacctgggtcttcagtgaaccattg ttcaatatcgtccggggacagcatcaaatcatccattgcttgggacggcaaggggga |
| Fig.3 | TP53-2-5UTR | Human-TP53 | 5’ UTR | 240 | 1 | gagatggagtctctctctgtcacctgggctggagcacagtggcatgatctcagctcactgcaacctctaccttccgggttcaagccattctcctgcctcagtctcccgagtagctA ggattacaggcgagtaccaccacacccagctaatttttgtatttttagtagagacagggctttgcatgttggccaggctggtctcgaactccttacttcaggtgatcggcccgcct cagcctcc |
| Fig.3 | HER2-1-E27-1 | Human-HER2 | Exon 27 | 354 | 1 | tcacactggcacgtccagacccaggtactctgggttctctgccgtaggtgtccctttgaaggtgctgggtggagccccccgctctggtgggtcctggtcccagtaatagaggttg tcgaaggctAggctgaaggcaggaggagggtggggctgaggggcagctcctccctggggtgtcaagtactcggggttctccacggcacccccaaaggcaaaaacgtctttg acgaccccattcttccctggggagagagtcttgggcctttccagagtggcaccagcaggtcgggcagcaggcagagggccctctcggggcgaagggggctggggccgaaca tctggctggttcacata |
| Fig.3 | HER2-2-E27-2 | Human-HER2 | Exon 27 | 354 | 2 | tcacactggcacgtccagacccaggtactctgggttctctgccgtaggtgtccctttgaaggtgctgggtggagccccccgctctggtgggtcctggtcccagtaatagaggttg tcgaaggctAggctgaaggcaggaggagggtggggctgaggggcagctcctccctggggtgtcaagtactcggggttctccacggcacccccaaaggcaaaaacgtctttg acgaccccattcttccctAgggagagagtcttgggcctttccagagtggcaccagcaggtcgggcagcaggcagagggccctctcggggcgaagggggctggggccgaac atctggctggttcacata |
| Fig.3 | HER2-3-E3-1 | Human-HER2 | Exon 3 | 273 | 1 | cagtggccctggtcagctctgaataaccaagagaaggtttcaatgacggtgaaggccacctgtgaggcttcgaagctgcagctcccgcaggcctcctggggaggcccctgtg acaggggtggtattgttcagcgggtctccattgtctggcacggccagggcatagttgtcctcaaagagctgggtgcctcgcacaatccgcagcctctgcagtAggacctgcctc acttggttgtgagcgatgagcacgtagccctgcacctcctggatatc |
| Fig.3 | ARC-1-CDS | Human-ARC | CDS | 219 | 1 | agaacacctcgcgccacacgtgcatctcgcgcttggcccagcgctccaggttggcgatggtctcctggcagcggcacaggcaggccttggtggacttcttccagcgctgcgag tcgctcgtAggcacgtagccgtccaggttgctctccagcttcccgaccgaccggtgcagccccttcagctcgcgctccacctgcttggacacctcggccagcaggt |
| Fig.3 | ARC-2-CDS | Human-ARC | CDS | 265 | 1 | cgcaggaaacgcttgagcttgggctgcagggtgcccaccacgtactggatgatctcctcctcgtccgcgtccacgtggagcgtctggtacaggtcccgcttgcgccacaggaa ctAgtccagcggctcgccctgcttctgcggcaggtccagctcgcgctgggtggcctctcgggacagcgtgccctcgctgtactgcaggaactccttcttgaactccacccagttc ttcacggagccctgcttgaactcccaccacttcttgg |
| Fig.3 | ARC-3-CDS | Human-ARC | CDS | 276 | 1 | cttctgctctgggtgtcccatgagcttggtggccgctggcccctgccatccggacacttggttttctggatgtccagagggcctgggtgttgtagcagaatgaggaagctggggt gttctgtgatggcctgggagtgtggagggtggggtccaggccggaaagacagacggagggagggtgatgggtAgagccctgcaggggggagggctctttgccgctgggtg cggtgctcacatctggatgacacgagtgtgtgcgagtccgcccggagctcg |
| Extended Data Fig. 9 | Ctip2-ses1 | Mouse-Ctip2 | Exon 4 | 384 | 1 | gaagcgcaggctgccgttctcagaggaatgttccgacgatgtggcgaaaggcgactggcgcgcatccgtgaagcccaggaatgggtccttcatgaagtggcgcgatgctgc gtagcccacgagccactgcgagtacacgttctcagatgggatgagggcggcaggtagcagctccaggtctttctccaccttgatgcgcttggccgcgtgcagcgcgggaccac cgagcccagggctgggcagcggtgctggcttgcgtgggaagagagctggaaagggctctgcgcctggcgcgaaggccccgccgcgcccgttcactgcacccggtgcacccg cgtccccacagccaacagctccggcatcacccgtgtcgcctgcacgctt |
| Extended Data Fig. 9 | Ctip2-ses2 | Mouse-Ctip2 | Exon 3 | 252 | 1 | caccgcagcggctctgttaccggacaactcacactggcatccaaagggagcctccatctgaccctcaccctgagtcccgtcgcccgagatggggcgtgcaccacagcaaggc agcgggaaacagggtaggagaacgcccagggcacgcagaggggaagtaatcacggaggtgggagggtgggaagaggaggcagctatgggggccatcgatggcagctg ggcaggcctgcacggccctggagaaaaaacaat |
| Fig. 4, Extended Data Fig. 9 | Ctip2-ses3 | Mouse-Ctip2 | Intron 3 | 288 | 1 | acattaagaatcggtctgttcccaggagttctgtctccaaggtcacaccatgtctgggaaggaaaactgttccttcacgcaaatgctcgcagcaacttcccagtaccctttatcct tgtgtagccacctgcctcgctctctacactgcagagactgatgaagcactgtgagccagggccatggagcaggagaagggctgctttcacggaaccacagtcagcaagacat cggggattacgtgatttccagcgagggttcaagtatcgagtccataagaaaggtgagtcc |
| Extended Data Fig. 9 | Ctip2-ses4 | Mouse-Ctip2 | Exon 4 | 207 | 2 | ttccgacgatgtggcgaaaggcgactggcgcgcatccgtgaagcccaggaatgggtccttcatgaagtggcgcgatgctgcgtagcccacgagccactgcgagtacacgttc tcagataggatgagggcggcaggtagcagctccaggtctttctccaccttgatgcgcttggccgcgtgcagcgcgggaccaccgagcccagggct |
| Extended Data Fig. 9 | Ctip2-ses5 | Mouse-Ctip2 | Exon 2 | 345 | 2 | tgcaatgttctcctgcttgggacagatgcctttcgtgggggacaggaggtggtcatcttcatcaggggtgacctggatcccgatctccactagctcagatactctcctgagctca gagcgagaggagggaggtagactgctcttgtccaggaccttgtcgtagcaggggcccaggcctccacactgtttcttcttgtgctctataaaaaccaggatgtcccccagcgg gaagttcatctgacactggccacaggtgaggagatcagggtcggggcctcccaccatcagccccaggctgctaggctcctctatctccagaccctcgtcttcctcgaggatggt agc |
| Extended Data Fig. 9 | Ctip2-ses6 | Mouse-Ctip2 | Mix: Exon 4/2 | 411 | 3 | ttccgacgatgtggcgaaaggcgactggcgcgcatccgtgaagcccaggaatgggtccttcatgaagtggcgcgatgctgcgtagcccacgagccactgcgagtacacgttc tcagataggatgagggcggcaggtagcagctccaggtctttctccaccttgatgcgcttggccgcgtgcagcgcgggaccaccgagcccagggcttttgtggtcatcttcatca ggggtgacctggatcccgatctccactggctcagatactctcctgagctcagagcgagaggagggaggtagactgctcttgtccaggaccttgtcgtagcaggggcccaggcc tccacactgtttcttcttgtgctctataaaaaccaggatgtcccccagcgggaagttcatctgacactggcc |
| Extended Data Fig. 9 | Ctip2-ses7 | Mouse-Ctip2 | Intron 1 | 300 | 2 | agtcctccagaaatatacagatgcattgtgcccgcccggcacacaggaaaggaagaacaccgactgagacataggagaagggtgtcaaaatattgtttggtcagaaaaga aacaaaaagggacaaaggaaataaactgagaagcaaaacaaaatagggataggcggggggcaaactgaaattgaaattgagaattcaaactaagcagggaggggtgc tatccctgggagggatccactgcccgcctcgagcaggcctggcaaggcctctgctccttacagcctttgataaaaataataaa |
| Fig. 4, Extended Data Fig. 9 | Ctip2-ses8 | Mouse-Ctip2 | Exon 4 | 765 | 2 | gagcttgctcgcctgcgagcacgcatggtcgcacagctggcacttgtagggcttctcgcccgtgtggctgcgccggtgcacgatgagattgctctggaacttgaaggtcttgcc gcagaactcacaggacttgctcttggcaggcggctgcggtggcggtgtgcccgcaggcatgggtggcagcggtggcgtgctgaggaacggggacttgggactgggctggaa agggttcagcagccggtgcatagggttgccacggcctggggacacgggcggcggcgtggagctgttgccggccagttctcgcagccgccgggagaagtccatggcaggag agtctatggccatggggttcaggcgcatgactcggtcgaaggcactggggtgctgggccacgagccccatctcctctgcactgaggcggtgtgggtccaagtgatggcgtagc ggtgggctgaagagcggtggcgtacctagcaagcggccctcaccgaagccagggtggtcccgcaggatggggcccgtcatgcgcagcaggttgaaaggattgctgtccccc aggaaattcatgagtggggactgcgccacggtctccggcccgagcggtggcgggatggtgagcctgggcgtgagcgaggtgctggccggcccaggctccaggtagattcgg aagccatgtgtgttctgtgcgtgctgcagcaggaaccaggcgctgttgaagggctgcttgcatgttgtgcaaatgtagctggaaggctcatcttt |
| Extended Data Fig. 9 | Fezf2-ses1 | Mouse-Fezf2 | Exon1 | 189 | 1 | cgcagccaggctggccagtttggcgttctccagcagaaaaagcttggggtgagcagccagggaagtgggggcctgtgcgttgaggaggccggataggaaaaggtggcctc cgaggagctccgaaggtgggtaagcggtggagtccaggtagttgaagtagtagagagagccgctggcaggcagccccac |
| Extended Data Fig. 9 | Fezf2-ses2 | Mouse-Fezf2 | Exon1 | 204 | 2 | gaacttgtccgcagccaggctggccagtttggcgttctccagcagaaaaagcttggggtgagcagccagggaagtgggggcctgtgcgttgaggaggccggataggaaaa ggtggcctccgaggagctccgaaggtaggtaagcggtggagtccaggtagttgaagtagtagagagagccgctggcaggcagccccacagcctg |
| Extended Data Fig. 9 | Fezf2-ses3 | Mouse-Fezf2 | 5’ UTR | 372 | 2 | gcacctagtcggcccggtcacccatttgcattcaaaggaacagaagcagaacaaagtgcacccaagggtaccaaattaccgcccattaacccggcgcgcaaaggaaccca ccagctattgccctaggactatgaaagggggaagaagggggaggctgtacaaaggaaaggagaaggggtagaagaaaggagtctcaaattaacactcccagcccttccc tagacccaggccttcgccacgcgtgctgggaactgggcactgaaagagcaaagtgaaagagggccggaagaaccacctcgcttttacctctttcccccaccgccaaggaga tgcgttccgagccatgcagcgtgtctcttctgtcacattttg |
| Extended Data Fig. 9 | Fezf2-ses4 | Mouse-Fezf2 | Exon1 | 753 | 2 | cttgccgcacacttcgcaggtgaagtttttgggtttgctgtcagtagagccccccgggagtttgctgtggctcttgactccccctcgttcagctgtcaaggccgagttctccttcag cacctgctccagtggcgcatgcaagcgctccttatggggataggaagctgggtgggggaacttgtccgcagccaggctggccagtttggcgttctccagcagaaaaagcttg gggtgagcagccagggaagtgggggcctgtgcgttgaggaggccggataggaaaaggtggcctccgaggagctccgaaggtaggtaagcggtggagtccaggtagttga agtagtagagagagccgctggcaggcagccccacagcctggttgatgacctgcggtttgatgaccctgccggcgggcagcgcagaaggcgcgaggcccagttcggccttgc agcacacgccacagttggttttgcacaagccgctggcgccgcacactggggccccaccgccgctgcctcctcctccaccgccgcccgcccggaggctgctcttccagaactcc gagtaactgagcagcgtcttggacggcacctcgtagccgagaggctggagagggatcatacagggcagcggcgagcagaggttgagcagtttcttgctctggctgctgtctg cctcgaacgcagcaggccggggctcgaaaggcgctcggggctcggacgtcttggccatgatgcgctcgatagagaaagc |
| Extended Data Fig. 12 | Satb2-sesRNA | Mouse-SatB2 | CDS | 708 | 2 | ttatctctggtcggtttcggccggtgcagccttgctcttgtcggcattctcttcctcggcctccaccttgtacatctcctccgagccttcctcactgtcattctcctcggactcggtca gcagctcctcgtccttatactctgccacgtccacggcggagcccaggtgctccttcagcttgccatggtgcttcacgtggtacctctggttctggaagaacttgatgatggtgtgt ttggggagatccagctgggcggagagtgtgtggatggcttcctggtcgggatagaggcctacatcatggatgaagctttgaaggatgcccagggcttccaaagagatctttg tgcgagaccgaggctttttggcacagctgtcttctgttagaggaggcggtaggggtgcttcttctctgggaggggagctctccttagttggctgggactgctgtcgatgaagga cctgcacgggctcaggtgggagctggaccacaggctgcatgcgttcactgtggtgaggtcgagattcttcctcatagatcacatcccgctcaggttggggaagattcaggaaa cggcggatggtgcagagattctcccaaagggtgcggttttctgggctggggttctccttccaacgaagcagttcgcaaagccagccctgacttttgtttgcagccactttggca aacagggcttgagacacctt |
| Extended Data Fig. 12 | plxnD1-sesRNA | Mouse-PlxnD1 | exon1 | 294 | 1 | gcggggcaggcagatgcgcgccagcaggctgcgcgcctggctgtccttgtcgcccgcacgcgcctcgctgttgagcgccaggtacgcgtagggctgcgcgctgtgcggtagc accgccgggtgcaagaaggcacgcacgaagcccagcttgtgctgctccttggcgccctgcttgatcttcaggatgttatcgtccgacgggttaaggtcgaaggtgaagagctt ggccaagtctccacgcgcgtccagggagcggatagcgatctcgggcgtgttctcgaagcggtggtcttc |
| Extended Data Fig. 12 | Rorb-sesRNA | Mouse-Rorb | exon4 | 393 | 1 | tacgatctcagacattgttatcccgggagcgaactgcccgttgttgaaagagctataggtaaacaagttgggtacagatgtgaggtcatagataggttcttgctttatctgtttg atcccagtcatgtccagctcctgactgatcgggagacggctgaccggaatctatgctgtaataaccttcggacttaggcaggtcaatgcgtgcccgttggcgtatgtgccgccg gtctcggtgttcaggttgctgaggccattgctaatgctgctgctgtacaccctggcgagggcctccgcctccccactctgctgctgccgctgctcctgcagcctttgctgaggctt ctgcacctcagcatacaggctgtcccgctgcttcttggacatcctccc |
| Extended Data Fig. 12 | vGAT-sesRNA | Mouse-vGAT | exon1 | 300 | 2 | cctggtccttggagccagagggtggcagaggagcgccgccgcgctgataatgaatgtctccctcgacgggagcttctgcgccctcgtctccgcagggctcgccttccgatttca ggatgtccatctgcaggccctggcgatgctcaaagtcgagatcgtcgcagtgcgcgaagcccaccgcttcctcatcggtagccgcctgaaaccccatcctagcaaacatgccg ctcaccttggcctgggacttgttggacacggaggtggccacattggtcagcttgctgcggagcagggtggcca |
| Extended Data Fig. 15 | Tle4-sesRNA | Rat-Tle4 | exon15 | 194 | 2 | atgagaagcagaccttggagtcggggctgttggccagagcatagcaggcaggggctgaggacgtcagctctgccttgttgcgtggggttggagctgccaggtcccagatag acagtgtactagcttcccctccaacaattaaggtgcggccatcagggagcaatctgcaggagcggatgtagttatccctgt |
| Extended Data Fig. 15 | vGAT-sesRNA | Rat-vGAT | exon2 | 255 | 1 | ggccacatcgaagaagacctggtgccacagcagcttgcgccagagaagacgcaagtggaagaggctgggcagcaggaagcagaggccggctcccgtgaggctgcccgtg aggcccatgagcagcgcgaagtgtagcacgtagatggccatgagcagcgtgaagaccaccagcgcgcagcgcagcgtcagcccccaggacttaaggcgaccgtcgccacc gtagcaggcggggaagaaggcacgactgccttcctg |
| Fig. 5, Extended Data Fig. 16 | FOXP2-1-sesRNA | Human-FOXP2-1 | CDS | 264 | 1 | ggtaagctctgtggggatgtctccaacatattcttcgacatggtgacactagacaccagatttagaggtttgggagatggtttgggctctgagggtcgcatgtgcaagtaggtc atcattgctggaagacgttcgcgttctttagaaagctgtatttctaactgttgcaccacctgcatttgcactcgacacggagcagtgcttcggtcatccaatgcgtgttcattgtta aggtgctttaaaaactgtccaaaatcttcacaa |
| Fig. 5, Extended Data Fig. 16 | FOXP2-2-sesRNA | Human-FOXP2-2 | CDS | 279 | 2 | ctggccatttgcaaactccatggccatagagagtgtgagaggccccagtctcctcatgtgacgagctgtctcgtcttgcacttagaactgaagactgtccattcactatagaat gatgagttattggtggtggtgctttagaagtgttggaggaggaagtcgaggaggaattgtgagtggtgcggtctagccctccatgttgaatgccattgtcttccatactgtgaa ctccagtcacttctttccataactgctgaatctcagcaggacttaagccag |
| Fig. 5, Extended Data Fig. 16 | VGAT-sesRNA | Human-VGAT | Exon2 | 202 | 1 | aggacttcaggcgcccgtcgccgctgtagcaggccgggaaaaaggcgcggctgccttcctggaagagcgacttctccagcacctcgacagcggcaaagaatagcagagga taggacaacagcgccttggccaccagaaagatgttgaccacggcgcggatggagccgggcaggttatccgtgatgacctccttggtctcgtc |
Extended Data Table. 3.
| Gene# | Gene Name | Immune/Interferon Response Related? | Fold Change (Log2) |
|---|---|---|---|
| OAS2 | 2’-5’-Oligoadenylate Synthetase 2 | Yes | 2.36 |
| OAS1 | 2’-5’-Oligoadenylate Synthetase 1 | Yes | 1.93 |
| APOL6 | Apolipoprotein L6 | No | 1.83 |
| IFI27 | Interferon Alpha Inducible Protein 27 | Yes | 1.82 |
| OAS3 | 2’-5’-Oligoadenylate Synthetase 3 | Yes | 1.81 |
| MX2 | MX Dynamin Like GTPase 2 | Yes | 1.8 |
| XAF1 | XIAP Associated Factor 1 | Yes | 1.75 |
| IFIT3 | Interferon Induced Protein With Tetratricopeptide Repeats 3 | Yes | 1.53 |
| DDX60L | DExD/H-Box 60 Like | Yes | 1.5 |
| IFI44 | Interferon Induced Protein 44 | Yes | 1.49 |
| RSAD2 | Radical S-Adenosyl Methionine Domain Containing 2 |
Yes | 1.43 |
| PARP10 | Poly(ADP-Ribose) Polymerase Family Member 10 | No | 1.4 |
| IFIT1 | Interferon Induced Protein With Tetratricopeptide Repeats 1 | Yes | 1.4 |
| TRIM22 | Tripartite Motif Containing 22 | Yes | 1.33 |
| IFI44L | Interferon Induced Protein 44 Like | Yes | 1.29 |
| LRRC37A | Leucine Rich Repeat Containing 37A | No | 1.27 |
| IFIH1 | Interferon Induced With Helicase C Domain 1 |
Yes | 1.21 |
| BISPR | BST2 Interferon Stimulated Positive Regulator |
Yes | 1.21 |
| IFIT2 | Interferon Induced Protein With Tetratricopeptide Repeats 2 | Yes | 1.21 |
| UBA7 | Ubiquitin Like Modifier Activating Enzyme 7 | No | 1.19 |
| CMPK2 | Cytidine/Uridine Monophosphate Kinase 2 | No | 1.19 |
| ISG15 | ISG15 Ubiquitin Like Modifier | Yes | 1.19 |
| DDX58 | DExD/H-Box Helicase 58 | Yes | 1.19 |
| SAMD9 | Sterile Alpha Motif Domain Containing 9 | No | 1.18 |
| OASL | 2’-5’-Oligoadenylate Synthetase Like | Yes | 1.15 |
| LOC100996724 | PDE4DIP Pseudogene 2 | No | 1.15 |
| DDX60 | DExD/H-Box Helicase 60 | Yes | 1.14 |
| PARP9 | Poly(ADP-Ribose) Polymerase Family Member 9 | Yes | 1.12 |
| MX1 | MX Dynamin Like GTPase 1 | Yes | 1.06 |
| PTBP2 | Polypyrimidine Tract Binding Protein 2 | No | 1.04 |
| IFI6 | Interferon Alpha Inducible Protein 6 | Yes | 1.04 |
| EPSTI1 | Epithelial Stromal Interaction 1 | No | 1.03 |
Supplementary Material
Acknowledgements
We thank L. Wan for providing the HeLa cell line; M. Tadross, A. West, K. Meyer, A. Zador and S. Soderling for comments on the manuscript; J. Hatfield and B.-x. Han for animal preparation; and R. Utama for bioinformatic analysis. This work was supported in part by NIMH grants 1DP1MH129954-01 and 5U19MH114821-03 to Z.J.H. D.G.S. was supported by the NINDS K12 Neurosurgery Research Career Development Program K12 Award and the Klingenstein-Simons Foundation. Extended Data Fig. 12a was generated using BioRender.
Footnotes
Competing interests Z.J.H. and Y.Q. have filed a provisional patent application on CellREADR technology through Duke University. The other authors declare no competing interests.
Additional information
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41586-022-05280-1.
Peer review information Nature thanks Botond Roska and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Reprints and permissions information is available at http://www.nature.com/reprints.
References
- 1.Hu BC The human body at cellular resolution: the NIH Human Biomolecular Atlas Program. Nature 574, 187–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.BRAIN Initiative Cell Census Network (BICCN). A multimodal cell census and atlas of the mammalian primary motor cortex. Nature 598, 86–102 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arendt D. et al. The origin and evolution of cell types. Nat. Rev. Genet 17, 744–757 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Regev A. et al. The Human Cell Atlas. eLife 6, e27041 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jinek M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baylis F, Darnovsky M, Hasson K. & Krahn TM Human germ line and heritable genome editing: the global policy landscape. CRISPR J. 3, 365–377 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Feng G. et al. Opportunities and limitations of genetically modified nonhuman primate models for neuroscience research. Proc. Natl Acad. Sci. USA 117, 24022–24031 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mich JK et al. Functional enhancer elements drive subclass-selective expression from mouse to primate neocortex. Cell Rep. 34, 108754 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartl D, Krebs AR, Juttner J, Roska B. & Schubeler D. Cis-regulatory landscapes of four cell types of the retina. Nucleic Acids Res. 45, 11607–11621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juttner J. et al. Targeting neuronal and glial cell types with synthetic promoter AAVs in mice, non-human primates and humans. Nat. Neurosci 22, 1345–1356 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Nair RR, Blankvoort S, Lagartos MJ & Kentros C. Enhancer-driven gene expression (EDGE) enables the generation of viral vectors specific to neuronal subtypes. iScience 23, 100888 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vormstein-Schneider D. et al. Viral manipulation of functionally distinct interneurons in mice, non-human primates and humans. Nat. Neurosci 23, 1629–1636 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graybuck LT et al. Enhancer viruses for combinatorial cell-subclass-specific labeling. Neuron 109, 1449–1464.e13 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimidschstein J. et al. A viral strategy for targeting and manipulating interneurons across vertebrate species. Nat. Neurosci 19, 1743–1749 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eisenberg E. & Levanon EY A-to-I RNA editing—immune protector and transcriptome diversifier. Nat. Rev. Genet 19, 473–490 (2018). [DOI] [PubMed] [Google Scholar]
- 18.Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat. Rev. Mol. Cell Biol. 17, 83–96 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abudayyeh OO et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katrekar D. et al. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 16, 239–242 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merkle T. et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol 37, 133–138 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Qu L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol 37, 1059–1069 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Rauch S. et al. Programmable RNA-guided RNA effector proteins built from human parts. Cell 178, 122–134.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan MH et al. Dynamic landscape and regulation of RNA editing in mammals. Nature 550, 249–254 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beier H. & Grimm M. Misreading of termination codons in eukaryotes by natural nonsense suppressor tRNAs. Nucleic Acids Res. 29, 4767–4782 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Segel M. et al. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 373, 882–889 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang CF et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell 153, 896–909 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran FA et al. Genome engineering using the CRISPR–Cas9 system. Nat. Protoc 8, 2281–2308 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung H. et al. Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamers MM, van den Hoogen BG & Haagmans BL ADAR1: “editor-in-chief” of cytoplasmic innate immunity. Front. Immunol 10, 1763 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pertea M, Kim D, Pertea GM, Leek JT & Salzberg SL Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc 11, 1650–1667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Love MI, Huber W. & Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lo Giudice C, Tangaro MA, Pesole G. & Picardi E. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat. Protoc 15, 1098–1131 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Lodato S. et al. Gene co-regulation by Fezf2 selects neurotransmitter identity and connectivity of corticospinal neurons. Nat. Neurosci 17, 1046–1054 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matho KS et al. Genetic dissection of the glutamatergic neuron system in cerebral cortex. Nature 598, 182–187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hodge RD et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 573, 61–68 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eugene E. et al. An organotypic brain slice preparation from adult patients with temporal lobe epilepsy. J. Neurosci. Methods 235, 234–244 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anzalone AV, Koblan LW & Liu DR Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol 38, 824–844 (2020). [DOI] [PubMed] [Google Scholar]
- 39.Mitric M. et al. Layer- and subregion-specific electrophysiological and morphological changes of the medial prefrontal cortex in a mouse model of neuropathic pain. Sci Rep. 9, 9479 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang ZJ & Paul A. The diversity of GABAergic neurons and neural communication elements. Nat. Rev. Neurosci 20, 563–572 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yi Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol 40, 946–955 (2022). [DOI] [PubMed] [Google Scholar]
- 42.Katrekar D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol 40, 938–945 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Monian P. et al. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol 40, 1093–1102 (2022). [DOI] [PubMed] [Google Scholar]
- 44.Hou X, Zaks T, Langer R. & Dong Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bedbrook CN, Deverman BE & Gradinaru V. Viral strategies for targeting the central and peripheral nervous systems. Annu. Rev. Neurosci 41, 323–348 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Joyce JA & Fearon DT T cell exclusion, immune privilege, and the tumor microenvironment. Science 348, 74–80 (2015). [DOI] [PubMed] [Google Scholar]
- 47.Aquino-Jarquin G. Novel engineered programmable systems for ADAR-mediated RNA editing. Mol. Ther. Nucleic Acids 19, 1065–1072 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cox DB, Platt RJ & Zhang F. Therapeutic genome editing: prospects and challenges. Nat. Med 21, 121–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doudna JA The promise and challenge of therapeutic genome editing. Nature 578, 229–236 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser T, Zhou Y. & Feng G. Animal models for neuropsychiatric disorders: prospects for circuit intervention. Curr. Opin. Neurobiol 45, 59–65 (2017). [DOI] [PubMed] [Google Scholar]
- 51.He M. et al. Strategies and tools for combinatorial targeting of GABAergic neurons in mouse cerebral cortex. Neuron 92, 555 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Schwarz N. et al. Human cerebrospinal fluid promotes long-term neuronal viability and network function in human neocortical organotypic brain slice cultures. Sci. Rep 7, 12249 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz N. et al. Long-term adult human brain slice cultures as a model system to study human CNS circuitry and disease. eLife 8, e48417 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data are available at NCBI BioProject under accession code PRJNA856413. Genome reference data are available from Gencode GRCH38.p13. Plasmids will be deposited to Addgene. We will provide reagents upon request until they are available from Addgene.