Abstract
Single-molecule fluorescence in situ hybridization (smFISH) enables the detection and localization of individual mRNAs in tissue sections with single-molecule resolution while preserving spatial context, and thus, is a useful tool for examining gene expression in biological systems. In particular, the growing reliance on single-cell RNA sequencing (scRNA-seq) to explore cellular heterogeneity has reinvigorated this approach as a validation tool to spatially re-map mRNA expression patterns described in isolated cells to their parent tissue. While use of antibody-based methods, such as indirect immunofluorescence (IIF), remain popular as validation strategies, smFISH often affords superior specificity and maintains congruency with scRNA-seq. Here, we present a detailed protocol that combines multiplexed smFISH using the RNAscope approach with IIF to co-visualize mRNAs and proteins within sections of mouse testes. We provide step-by-step guidelines from testis preparation through visualization that enables mapping of combinations of up to four mRNA/protein targets in frozen sections on the RNAscope platform.
Keywords: smFISH, scRNA-seq, Immunohistochemistry, RNAscope, Spermatogenesis
1. Introduction
In the post-genome era, investigation of multicellular organisms and biological networks underlying organismal function relies on sensitive, high-throughput approaches that can profile the molecular states of cells and tissues. These “omics” approaches can interrogate the DNA, RNA, and protein levels in bulk samples of cells or tissues, generating large-scale data sets which reflect the comprehensive molecular profiles of the starting samples. For instance, RNA-seq using bulk-cell or -tissue preparations detects ensemble average mRNA levels for all genes, but the results may not accurately reflect the profile of any given cell population in the sample [1]. Single-cell RNA-seq (scRNA-seq) solves this problem by resolving gene expression profiles among individual cells [2-7]. Dissociation-based single-cell studies, though, ignore the tissue localization of expression profiles, because source tissues are necessarily dissociated into a single-cell suspension. For instance, we and others have performed exhaustive single-cell RNA-seq to identify comprehensive gene expression phenotypes that correlate with SSC transplantation activity [8,9]. Yet, since this strategy is borne out of dissociated testicular tissue and profiles individual cells, we do not know the location of these SSCs in the testis. Furthermore, it is generally required in the field to validate single-cell transcriptomes using a complementary, tissue-based gene expression method to confirm the reliability of the single-cell transcriptomes and at least partially remap expression profiles in 2D tissue space [8]. Currently, proteins can be readily localized in situ with single-cell resolution using antibody-based approaches (e.g., indirect immunofluorescence) and detected with chromogens or fluorescence, but this approach is wholly dependent upon availability of validated antibody reagents. Moreover, any post-transcriptional gene regulation that takes place during spermatogenesis breaks the correlation between mRNA and protein levels, and, thus, immunostaining for a given protein in testis sections may not accurately represent the pattern of mRNA expression [10]. Direct localization of mRNAs in situ is also possible using hybridization of complementary (antisense) DNA or RNA probes, a process called in situ hybridization (ISH). This has the advantage of direct verification/validation and extension of scRNA-seq results for putative SSCs, any spermatogenic cell, or any cell type in any tissue.
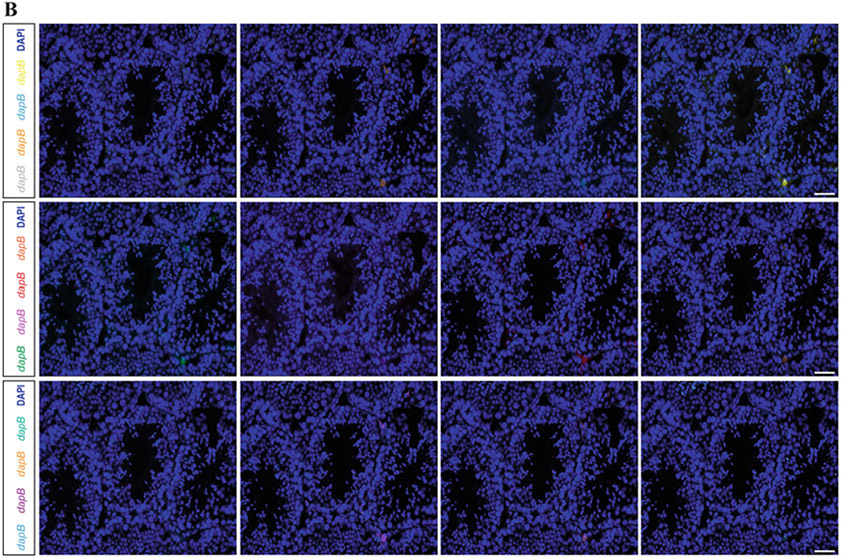
Since the initial reports of the ISH methodology in 1969 by Gall and Pardue [11, 12], numerous advancements have maximized detection speed, sensitivity, and multiplicity of concurrent target detection. RNAscope (Advanced Cell Diagnostics) is a new commercial methodology that provides a single-molecule fluorescent ISH (smFISH) platform to address the challenges of sensitivity and specificity in spatial profiling low-abundance RNA [13-15]. It employs panels of double-Z probes and signal amplification strategy to maximize detection of individual RNA molecules [16]. This protocol describes our optimized strategy to use RNAscope technology to detect transcripts (e.g., Gfra1 and Kit) together with proteins (e.g., DDX4) in testicular cryosections. This strategy read-ily facilitates detection of 1–4 transcripts at a time using the RNA-scope platform and established (off-shelf) or custom probe sets (Fig. 1). Simultaneous detection of 12 RNAs is also possible using the RNAscope HiPlex application, which advances the use of smFISH to dissect the spatial relationships of discrete cell populations in complex tissues like the testis, which we demonstrate using a panel of probes that recognize mRNAs encoding 12 housekeeping gene products (Fig. 2a) versus negative controls (Fig. 2b).
Fig. 1.
Example RNAscope results for spermatogonial markers using mouse testes. smFISH-IHC labeling in the adult mouse testis. Using the described RNAscope method, cryosections from fixed adult testicular tissue was labeled for Gfra1 mRNA [17], Kit mRNA (red), immunostained for DDX4 (white), and counterstained for DAPI (blue). Scale = 50 μm
Fig. 2.
smFISH-12 Plex labeling using RNAscope HiPlex approach in the adult mouse testis. (a) Adult testis tissue sections were hybridized with probes targeting 12 species-specific housekeeping genes and counterstained for DAPI (blue). Scale = 50 μm. (b) Testis sections were also hybridized with a negative control probe recognizing bacterial dapB mRNA (an irrelevant gene) and counterstained for DAPI (blue). Scale = 50 μm
2. Materials
Prepare all solutions using Milli-Q water (prepared by purifying deionized water, to reach a sensitivity of 18 MΩ/cm at 25 °C) unless specified otherwise. Reagents are stored at room temperature, unless specifically noted.
2.1. Testis Preparation and Freezing
10× Dulbecco’s phosphate buffered saline, no calcium chloride, no magnesium chloride (DPBS): 26.6 mM KCl, 14.7 mM KH2PO4, 1379.3 mM NaCl, 80.5 mM Na2 HPO4·7H2O, pH 6.7–7.0.
4% (w/v) Paraformaldehyde in 1× DPBS, pH 7.2. Made fresh and stored at 4 °C for 2–3 days (see Note 1).
Prepare 1× DPBS containing DNase/RNase-Free Distilled Water. Store at 4 °C (see Note 2).
Stock solutions of 10% sucrose (w/v) in 1× DPBS. Store at 4 °C (see Note 3).
Stock solutions of 20% sucrose (w/v) in 1× DPBS. Store at 4 °C (seeNote 3).
Stock solutions of 30% sucrose (w/v) in 1× DPBS. Store at 4 °C (see Note 3).
Optimal Cutting Temperature Compound (O.C.T.).
2.2. RNAscope Reagents and Buffers
RNAscope Multiplex Fluorescent V2 assay kit with AMP 1–3, and TSA Opal dyes for detection of RNA targets [13].
Tyramide Signal Amplification (TSA) Opal dyes 520, 570, 620, and 690 (Akoya Biosciences) (see Notes 4 and 31).
RNAscope probes against mRNAs (see Note 5).
RNAscope Target Retrieval buffer: Prepare 200 mL of 1× Target Retrieval buffer by adding 180 mL of water and 20 mL of 10× Target Retrieval buffer into a staining dish (see Note 6).
1× RNAscope Wash buffer: Prepare 500 mL of wash buffer by transferring 490 mL of water and 10 mL of 50× wash buffer solution into a RNase-free bottle (see Note 7).
Prepare 200 mL of DPBS containing water into a staining dish (see Note 8).
Transfer 200 mL of 100% ethanol (EtOH) into a staining dish.
70% EtOH: Prepare by adding 60 mL of water to 140 mL of 100% EtOH into a staining dish.
50% EtOH: Prepare by adding 100 mL of water to 100 mL of 100% EtOH into a staining dish.
20× Saline Sodium Citrate (SSC): 3 M Sodium Chloride,0.3 M Sodium Citrate in water, pH 7.0.
Prepare 200 mL of 5× SSC buffer containing water into a staining dish (for the optional stopping point in the RNAscope protocol). Store up to 2 months.
2.3. Immunofluorescence (If Necessary)
2.4. General Materials and Equipment
Dumont forceps.
Iridectomy scissors.
RNase & DNase decontaminant spray.
1.5 L RNase-free bottle (see Note 12).
1.5 mL Eppendorf tubes, nuclease free.
15 mL conical tubes, nuclease free.
15 × 15 × 5 mm Cryomolds.
Aluminum foil.
Cryostat (e.g., Leica CM 1860).
Superfrost Plus microscope slides (see Note 13).
24 × 50 mm coverslips.
Tissue-Tek Vertical 24 Slide Rack.
Tissue-Tek Staining Dish.
Microtome/Cryostat blade.
Short razor blade to trim testis blocks.
Slide warmer.
Microscope slide boxes to store pre-cut sections and labeled slides.
Desiccant.
Re-sealable bags to store the microscope slide boxes with pre-cut sections.
Small plastic trays (e.g., autoclaved lids from empty pipette tips boxes) (see Note 14).
Hydrophobic barrier pen.
ProLong Gold antifade mounting medium.
Food steamer (see Note 15).
Hybridization incubator (see Note 16).
Humid chamber (see Note 16).
RNAscope hydrogen peroxide solution.
3. Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1. Testis Freezing and Embedding
Submerge dissected testes in pre-chilled 4% PFA and leave on a rocker for 24 h at 4 °C. Ensure testes are completely covered and avoid air pockets.
Remove fixative and pre-rinse with cold DPBS, then wash twice for 25 min at 4 °C with cold DPBS.
Replace DPBS with cold 10% sucrose, keep testes at 4 °C.
When testes sink to the bottom of the tube, replace 10% sucrose with cold 20% sucrose. Store at 4 °C.
When testes sink to the bottom of the tube, replace 20% sucrose with cold 30% sucrose. Store at 4 °C.
Allow the testes to sink to the bottom of the tube overnight in 30% sucrose.
Label CryoMold 2 with the orientation of the testes and fill with ice-cold O.C.T. without introducing air bubbles (see Note 17).
Fill CryoMold 1 with ice-cold O.C.T. This cryomold will be used to remove excess sucrose.
Using cooled forceps, transfer fixed testes into the CryoMold 1 (see Note 18).
Twirl the testes in the O.C.T., removing as much sucrose solution as possible.
Transfer fixed testes into the O.C.T. of CryoMold 2. Cover the entire testis surface with more ice-cold O.C.T.
Immediately place CryoMold 2 level on dry ice and allow to freeze (see Note 19).
Wrap CryoMold 2 with aluminum foil and store in a labeled resealable plastic bag at −80 °C for long-term storage or wait 2 h before proceeding to cryosectioning (see Note 20).
3.2. Cryosectioning
Before setting the cryostat temperature (−18 to −21 °C), wipe the machine with 70% EtOH.
Rinse a new microtome blade with 100% EtOH, pat dry with paper towels, and set aside.
Transfer O.C.T.-embedded testis blocks from the −80 °C storage to dry ice (see Note 21).
Once the temperature of the Cryostat chamber has reached −18 to −25 °C, transfer testis blocks, microtome blade, and microscope slides into the Cryostat chamber and equilibrate for 1 h (see Note 22).
Position the testis block on a pre-cooled chuck coated with cold O.C.T. (see Note 23).
Cut 7 μm testis sections and collect on cold Superfrost Plus microscope slides (see Note 24).
Allow the slides to air-dry for 1–2 h inside the cryostat.
After drying, immediately store the slides in a slide box with desiccants in a re-sealable bag at −80 °C or proceed immediately to the RNAscope assay (see Note 25).
3.3. Testis Pretreatment
Remove stored slides from the −80 °C freezer or freshly prepared slides from the cryostat and place directly in a staining dish containing DPBS to rinse off the O.C.T. by agitating the slides for 5 min.
Bake slides on a slide warmer for 30 min at 60 °C.
Post-fix testis sections by immersing slides in a plastic tray filled with cold 4% PFA for 15 min at 4 °C (see Note 1).
Tap slides on a paper towel to remove excess fixative and immerse in 50% EtOH for 5 min.
Transfer slides to 70% EtOH for 5 min.
Transfer slides to 100% EtOH for 5 min.
Repeat step 6.
Let slides air dry (flat on the benchtop, face-up) for 5 min.
Add 5–8 drops of RNAscope hydrogen peroxide solution to each slide and incubate for 10 min.
Tap slides on a paper towel to remove excess hydrogen peroxide solution. Submerge slides in a staining dish filled with water and agitate 3–5 times.
3.4. Target Retrieval
Place one staining dish containing 200 mL of Target Retrieval buffer and a second staining dish containing 200 mL of distilled water in the heated steamer (95–99 °C) (see Note 26).
Move slides to the boiling water dish for 10 s, then quickly transfer slides to the boiling Target Retrieval buffer dish for 5 min.
Immediately transfer slides to a staining dish containing RT water for 15 s.
Move slides to a staining dish with fresh 100% EtOH for 3 min. Air-dry the slides.
Using a hydrophobic pen, draw a barrier around the sections (see Note 27). Let it dry for 5 min.
Place the slides in the pre-warm hybridization chamber at 40 °C and add 5 drops of Protease III to cover the sections.
Incubate the slides for 30 min at 40 °C (see Note 28).
Meanwhile, prepare probe mix and only include probes that will be used. Mix 50 volumes of C1 probe with 1 volume of C2, C3, and C4 probes. Mix thoroughly (see Note 29).
Warm probes for 10 min at 40 °C. Cool the probes to RT, then transfer the probes to a centrifuge and perform a quick spin-down.
After the 30 min incubation, wash slides 3–5 times in a dish containing distilled water by moving the slides up and down.
3.5. Hybridization
Place the slides back in the hybridization chamber.
Add 4 drops (120 μL) of probe mix to cover each section and incubate for 2 h at 40 °C.
Wash slides for 2 min in a lid filled with wash buffer. Repeat with fresh buffer.
Tap slides on a paper towel to remove excess wash solution.
***optional stopping point***—store the slides overnight in 5× SSC buffer or proceed with the assay (skip to Subheading 3.6).
3.6. Amplification
Return the slides to the hybridization oven.
Add 4–6 drops of AMP 1 reagent to the slides, and incubate for 30 min at 40 °C (see Note 30).
Wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution and return slides to hybridization oven.
Once in the oven, add 4–6 drops of AMP 2 reagent to the slides. Incubate for 30 min at 40 °C.
Wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution.
Return the slides to the oven.
Add 4–6 drops of AMP 3 reagent and incubate for 15 min at 40 °C.
Remove and wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution.
3.7. Develop C1 Probe (For Example)
Return the slides to the hybridization oven.
Cover sections with 4–6 drops HRP-C1. Leave for 15 min at 40 °C. For example, for the C2 probe use HRP-C2, C3 probe use HRP-C3, and C4 probe use HRP-C4.
Meanwhile, only prepare the fluorophores that will be used. For example, we will assign Opal 520 to the C1 channel since the lowest expressed gene is targeted in this channel. If Opal 690 is used instead, you may need to use a higher concentration. Prepare a 1:750 stock of Opal 520 (see Note 31).
Wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution.
Add 200 μL of Opal 520 solution per slide and incubate for 30 min at 40 °C in oven.
Wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution.
Place slides in the oven and add 4–6 drops HRP blocker covering the slides.
Incubate for 15 min at 40 °C.
Wash slides for 2 min in a plastic tray containing wash buffer.
Tap slides on a paper towel to remove excess wash solution.
If labeling only the C1-probe, skip to Subheading 3.9 Counterstain or if combined with immunofluorescence (skip to Subheading 3.8). However, if labeling another channel, repeat steps 1–13 until the desired channels have been developed.
3.8. Immunofluorescence (If Necessary)
Add 4 °C cold permeabilization buffer solution to the slides for 2 h at 4 °C inside a humid chamber.
Remove the permeabilization buffer solution.
Add the primary antibody in permeabilization buffer solution to the slide (see Note 32).
Place slides in a humid chamber and incubate overnight at 4 °C.
Wash the slides with DPBST twice for 5 min.
Add the secondary antibody in blocking buffer to the slide.
Allow it to sit for 45 min at RT in a humid chamber.
Tap slides on a paper towel to remove secondary antibody mix (see Note 33).
Wash the slides with DPBST twice for 5 min.
Proceed to Subheading 3.9.
3.9. DAPI Counterstain
Stain with DAPI by adding 4 drops of DAPI solution to the slide and incubate for 30 s.
Quickly tap the slide on a paper towel to remove DAPI and add 1–2 drops of ProLong Gold antifade mounting medium.
Coverslip each slide and allow slides to sit in dark for 30 min to overnight.
Store slides in a slide box at 4 °C (see Note 34).
4. Notes
Using 4% PFA older than 2 days will decrease staining quality.
The DPBS made with nuclease-free water will be used to wash the fixative off the testes and to create different concentrations of sucrose.
Vortexing the sucrose solution helps to dissolve the sucrose crystals; follow this with 0.2 μm filtering. Make fresh sucrose solution in DPBS with nuclease-free water for optimal results.
To visualize genes of interest, we suggest conjugating probes to the following TSA-linked fluorophore dyes: Opal 520 (FITC range), Opal 570 (Cy3 range), Opal 620 (Texas Red range), and Opal 690 (Cy 5.5 range). In our experience, it is best to assign lowly expressed genes to the FITC channel and abundantly expressed genes in the Cy5.5 channel (Akoya Biosciences).
Target RNA species are detected by Advanced Cell Diagnostics propriety double Z probe design and signal amplification strategy, contributing to its sensitivity and specificity of target-specific signals. For example, we labeled for Gfra1 and Kit transcripts in adult mouse testicular sections (see Fig. 1).
Target retrieval buffer is used to expose RNA epitopes masked by the fixation process.
Warm 50× wash buffer for 15 min at 40 °C before diluting to 1×.
To rinse the O.C.T. off the testis section before starting the assay, dunk the slide several times for 5 min in 1× DPBS.
For detection of the target protein antigen, permeabilization of the testes is required for the antibody to gain entry into the cell and recognize the antigen.
For experiments involving smFISH and immunostaining, it is important to take into consideration the protease treatment and subsequent wash steps, which may reduce the robustness of antibody labeling. For example, we have found that the antibody dilution required for detecting DDX4 protein (see Fig. 1) was lower (higher antibody concentration) than in our standard immunostaining protocols. Optimization of mRNA-protein co-labeling includes changing the antibody dilution and altering protease incubation times to preserve RNAscope detection reagents and probes. Additionally, for quantitative analysis, DDX4 is a useful marker for image segmentation to delimit individual cells. For more information on quantitative or semi-quantitative assessment of RNAscope staining, please refer to ACD’s website (https://acdbio.com/services/quantitative-analysis).
Indirect immunofluorescence uses a secondary antibody to detect the primary antibody. The secondary antibody is specific for immunoglobulins derived from the host species for the primary antibody.
RNase-free bottles will be used to store wash buffer, 5× SSC buffer, and Milli-Q water.
Using Superfrost Plus charged microscope slides is essential to prevent detachment of testis sections during the various processing steps. In our experience, slides older than 3 years are unsuitable.
We used lids from empty pipette tip boxes for the post-fixation and wash steps in the RNAscope protocol. Two slides can fit in a single lid. Sterilize lids with 70% EtOH before and after use.
For the target retrieval step, we recommend using the Oster Food Steamer (model 5712). After each use, empty the water and wipe down the steamer with 100% EtOH and RNase decontaminant spray to prevent microbial growth.
We created a hybridization chamber utilizing a large glass desiccator fitted with a metal plate and an incubator. The metal place serves as a support for the slides inside the hybridization chamber. We place soaking wet filter paper beneath the plate to create a humid environment. It is important to note that there are many potential options for the humid chamber, including commercial options, plastic food storage containers, and pipette tip boxes; all will work, provided the environment maintains high humidity. It is important to wipe down the chamber with 100% EtOH and RNase decontaminant spray before each use to prevent bacterial growth. For immunostaining, we have found that a humid chamber constructed from a pipette tip box is sufficient.
Before embedding testes, place O.C.T. on ice for >30 min or store at 4 °C for 2 h.
Place sterilized forceps on dry ice for >30 min to minimize temperature changes in the testes that could negatively impact RNA quality within the tissue and reduce the likelihood of successful RNAscope detection.
Create a flat surface on the dry ice to prevent freezing testes in an undesired orientation.
Storing testis blocks in an air-tight bag and wrapping them with foil beforehand will prevent testes from drying out.
Transporting testis blocks on dry ice will prevent temperature fluctuations and ensure RNA quality.
Typically, a Cryostat chamber temperature of −18 to −25 °C is sufficiently cold to section mouse testes. However, depending on the model of the cryostat used, set Cryostat chamber temperature according to the manufacturer’s recommendations for testicular tissue. Testis blocks, microtome blades, and microscope slides must be equilibrated to Cryostat chamber temperature before sectioning to avoid RNA degradation.
Trim testis blocks to a desirable size using a standard razor blade.
Before collecting testis cryosections, warm the back (or non-charged side) of the slide with the palm of your hand for a few seconds. Several testis cryosections can be positioned within a central square (0.75″ × 0.75″) on a slide and be sufficiently covered by RNAscope reagents. Be sure to avoid the overlap of O.C.T. from adjacent cryosections with the testis tissue.
Store slide boxes inside large resealable bags. From our experience, slides should be used within 1 week of sectioning. Longer storage times may increase autofluorescence and reduce specific labeling.
With a clean thermometer, check the temperature. The steamer approximately takes 30 min to reach a temperature of 95–99 °C. Do not allow the reagents to sit longer than 30 min in the warm steamer.
Draw a hydrophobic barrier (0.75″ × 0.75″) on a clean slide and use it as a guide for drawing barriers on the experimental slides. During the assay, before you draw the barrier, place the guide slide beneath the experimental slide and draw the barrier.
The length of Protease III treatment varies with the developmental age of the animal. In our experience, adult mouse testes are optimally pre-treated with a 30 min Protease incubation at 40 °C, while postnatal day (P)6 mouse testes will be sufficiently pre-treated with only a 15–20 min incubation at RT using 3–4 drops of Protease III. This step should be optimized for each sample type. The kit comes with Protease Plus, Protease III, and Protease IV, each of which vary in digestion strength from weak, moderate, to strong.
Gene-specific probes are designed to recognize specific RNA targets and can be visualized by their HRP linkage to one of four fluorescent channels, C1, C2, C3, and C4. These channels are subsequently developed by assigning different TSA-linked fluorophore dyes to the probes and therefore genes of interest can be mixed and matched to Opal 520, Opal 570, Opal 620, and Opal 690. Probes of the same channel should not be combined. Probe mixes can be stored at 4 °C for 6 months.
Equilibrate all the amplification reagents and HRP blockers to RT.
The Opal dyes have been reconstituted according to the manufacturer’s instructions and stored at 2–8 °C. Prepare diluted fluorophore stocks using the provided TSA buffer and store in the dark at 2–8 °C. Diluted stocks may be reused for up to a month. Effective fluorophore dilutions range between 1:750 and 1:1000. If a smaller probe volume is used (e.g., 50 μL), we suggest using a fluorophore concentration of 1:750.
For antibody optimization, we recommend carrying out the RNAscope assay until the protease step, and then proceeding with Subheading 3.8. In doing so, one can minimize the use of expensive RNAscope detection reagents and probes. We advise users to include multiple cryosections for optimizing antibody concentrations. Additionally, positive and negative controls should be used in all experiments.
We prepared a secondary antibody dilution of 1:200 in blocking buffer.
After the assay, slides can be imaged after 8 h and within 2 weeks.
Acknowledgments
This work was supported by NIH grants R01 HD90007 and U01 DA054179.
References
- 1.Di Carlo D, Tse HTK, Gossett DR (2012) Introduction: Why analyze single cells? Methods Mol Biol 853:1–10. 10.1007/978-1-61779-567-1_1 [DOI] [PubMed] [Google Scholar]
- 2.Velte EK, Niedenberger BA, Serra ND, Singh A, Roa-De La Cruz L, Hermann BP et al. (2019) Differential RA responsiveness directs formation of functionally distinct spermatogonial populations at the initiation of spermatogenesis in the mouse. 146(12):dev.173088 [pii]. 10.1242/dev.173088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N et al. (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5):377–382. 10.1038/nmeth.1315 [DOI] [PubMed] [Google Scholar]
- 4.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J et al. (2011) Tumour evolution inferred by single-cell sequencing. Nature 472(7341):90–94. 10.1038/nature09807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P et al. (2011) Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21(7):1160–1167. 10.1101/gr.110882.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Picelli S, Bjorklund AK, Faridani OR, Sagasser S, Winberg G, Sandberg R (2013) Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat Methods 10(11):1096–1098. 10.1038/nmeth.2639 [DOI] [PubMed] [Google Scholar]
- 7.Hashimshony T, Wagner F, Sher N, Yanai I (2012) CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2(3):666–673. 10.1016/j.celrep.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 8.Suzuki S, Diaz VD, Hermann BP (2019) What has single-cell RNA-seq taught us about mammalian spermatogenesis? Biol Reprod 101(3):617–634. 10.1093/biolre/ioz088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC et al. (2018) The mammalian spermatogenesis single-cell transcriptome, from spermatogonial stem cells to spermatids. Cell Rep 25(6):1650–1667. e8. 10.1016/j.celrep.2018.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB et al. (2015) Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol Reprod 92(2):54. 10.1095/biolreprod.114.125757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gall JG, Pardue ML (1969) Formation and detection of RNA-DNA hybrid molecules in cytological preparations. Proc Natl Acad Sci U S A 63(2):378–383. 10.1073/pnas.63.2.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pardue ML, Gall JG (1969) Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A 64(2):600–604. 10.1073/pnas.64.2.600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newkirk SJ, Lee S, Grandi FC, Gaysinskaya V, Rosser JM, Vanden Berg N et al. (2017) Intact piRNA pathway prevents L1 mobilization in male meiosis. Proc Natl Acad Sci U S A 114(28):E5635–E5E44. 10.1073/pnas.1701069114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takei N, Nakamura T, Kawamura S, Takada Y, Satoh Y, Kimura AP et al. (2018) High-sensitivity and high-resolution in situ hybridization of coding and long non-coding RNAs in vertebrate ovaries and testes. Biol Proced Online 20:6. 10.1186/s12575-018-0071-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li K, Xu J, Luo Y, Zou D, Han R, Zhong S et al. (2021) Panoramic transcriptome analysis and functional screening of long noncoding RNAs in mouse spermatogenesis. Genome Res 31(1):13–26. 10.1101/gr.264333.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A et al. (2012) RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn 14(1):22–29. 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S et al. (2018) A comprehensive roadmap of murine spermatogenesis defined by single-cell RNA-Seq. Dev Cell 46(5):651–667. e10. 10.1016/j.devcel.2018.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]