Abstract
Animal evolution is characterized by frequent turnover of sexually dimorphic traits – new sex-specific characters are gained, and some ancestral sex-specific characters are lost, in many lineages. In insects, sexual differentiation is predominantly cell-autonomous and depends on the expression of the doublesex (dsx) transcription factor. In most cases, cells that transcribe dsx have the potential to undergo sex-specific differentiation, while those that lack dsx expression do not. Consistent with this mode of development, comparative research has shown that the origin of new sex-specific traits can be associated with the origin of new spatial domains of dsx expression. In this report, we examine the opposite situation – a secondary loss of the sex comb, a male-specific grasping structure that develops on the front legs of some drosophilid species. We show that, while the origin of the sex comb is linked to an evolutionary gain of dsx expression in the leg, sex comb loss in a newly identified species of Lordiphosa (Drosophilidae) is associated with a secondary loss of dsx expression. We discuss how the developmental control of sexual dimorphism affects the mechanisms by which sex-specific traits can evolve.
Keywords: Drosophila, Lordiphosa, sexual dimorphism, evolution, doublesex, trait loss, male-specific structures, sex combs
Introduction
Most animals are sexually dimorphic, but the traits that distinguish males from females vary greatly from species to species. This simple observation shows that the evolution of new sex-specific traits is common in animal evolution. The origin and maintenance of sexual dimorphism can be driven by a number of evolutionary forces, most importantly by sexual selection and intragenomic conflict (Andersson, 1994; Bonduriansky and Chenoweth, 2009; Cox and Calsbeek, 2009; Darwin, 1871; van Doorn, 2009; Hill et al., 2019; Lande and Arnold, 1985; Lund-Hansen et al., 2020).
However, the evolution of sexual dimorphism is not a one-way ratchet – many ancestral sex-specific characters are lost, just as new ones are gained (Wiens, 2001). Examples of secondary losses of male-specific, sexually selected traits include the namesake tail “sword” in swordtail fish (Kang et al., 2013), dorsal sailfins in sailfin mollies (Ptacek et al., 2011), conspicuous male pigmentation in phrynosomatid lizards (Wiens, 1999) and manakins (Ribeiro et al., 2015), and vocal sacs and nuptial pads in fanged frogs (Emerson, 1996). The loss of male traits often correlates with the loss of female preferences for these traits (Morris, 1998; Morris et al., 2005; Wiens, 2001). In a swordtail species with absent or reduced male swords, females do not show any preference for longer swords, and may actually prefer swordless males or discriminate against exaggerated swords (Rosenthal et al., 2002; Wong and Rosenthal, 2006). Similarly, the secondary loss of the male sailfin in Poecilia latipunctata correlates with the loss of female visual preference for large-finned males (Ptacek et al., 2011). In other cases, ecological factors may result in selection against sexual dimorphism. For example, pressure from acoustically orienting parasitoid flies has led to the loss or dramatic reduction of male-specific stridulatory and resonator structures in Teleogryllus oceanicus crickets (Bailey et al., 2019; Pascoal et al., 2014; Zuk et al., 2006) – despite sexual selection continuing to favor stridulating males (Tanner et al., 2019).
The molecular mechanisms behind the gain and loss of sex-specific traits may depend on the developmental control of sexual dimorphism. In vertebrate animals, where the gonad non-autonomously controls sex-specific differentiation in the rest of the body by secreting male- or female-specific hormones (but see (Ioannidis et al., 2021) for an important exception), the evolution of somatic sexual traits must be mediated primarily by changes in the tissue-specific responses to hormonal signaling. In swordtails, for example, testosterone analogs induce sword development in females of sword-bearing species, but can only induce small vestigial “swordlets” or do not induce sword development at all in males of swordless species (Gordon et al., 1943; Grobstein, 1942; Sangster, 1948; Schartl et al., 2021; Zander and Dzwillo, 1969). In insects, on the other hand, sexual differentiation is predominantly cell-autonomous – that is, whether a cell will undergo male- or female-specific development depends on the expression of sex-determining genes in that cell itself (Baker and Ridge, 1980; Camara et al., 2008; Kopp, 2012; Robinett et al., 2010). This mode of sexual differentiation has important implications for the evolution of sex-specific traits.
Sex-specific cell differentiation in Drosophila and other insects is mostly directed by the doublesex (dsx) transcription factor, which is spliced into a female-specific isoform (dsxF) in females and a male-specific isoform (dsxM) in males (Baker et al., 1989; Burtis and Baker, 1989; McKeown, 1992; Nagoshi and Baker, 1990; Wexler et al., 2019). The two dsx isoforms have opposite effects on sex differentiation: dsxM promotes the development of male-specific traits and represses female-specific traits, while dsxF promotes female-specific and represses male-specific characters (Baker and Ridge, 1980; Goldman and Arbeitman, 2007; Jursnich and Burtis, 1993; Waterbury et al., 1999). Sex-specific cell differentiation is due to the distinct effects of DsxM and DsxF proteins on target gene expression (Arbeitman et al., 2016; Burtis et al., 1991; Shirangi et al., 2009; Williams et al., 2008). Importantly, dsx is transcribed in controlled spatial patterns (Hempel and Oliver, 2007; Kijimoto et al., 2012; Ledón-Rettig et al., 2017; Loehlin et al., 2010; Palmer and Kronforst, 2020; Rideout et al., 2010; Robinett et al., 2010; Rohner et al., 2021; Sanders and Arbeitman, 2008; Tanaka et al., 2011), and it is this fact that has a profound influence on the evolution of sexual dimorphism. For any sex-specific structure to evolve, dsx must be expressed in the cells that either give rise to that structure or induce it by cell-cell signaling. In tissues that already express dsx, sexual dimorphism can originate if Dsx acquires new downstream targets through cis-regulatory changes that create new, or higher affinity, Dsx binding sites (Shirangi et al., 2009; Williams and Carroll, 2009; Williams et al., 2008). If, on the other hand, the tissue is ancestrally monomorphic and does not express dsx, changes in the spatial regulation of dsx must be a necessary first step. In other words, the origin of new sex-specific traits in insects can be linked to the evolution of new dsx expression domains (Hopkins and Kopp, 2021; Kopp, 2012; Tanaka et al., 2011).
The best-studied example where this mechanism is at work is the origin of male-specific grasping structures on the front (T1) legs in Drosophila and related genera (Kopp, 2011; Rice et al., 2018; Tanaka et al., 2011). One of these, the “sex comb” found in D. melanogaster and its close relatives, develops from mechanosensory bristles that are typically present in both sexes. In females, these precursor bristles retain their default morphology and position. In males, however, the bristle shafts destined to become the “teeth” of the sex comb undergo several modifications, including increased length, thickness, curvature, and melanization. In many species, the entire sex comb also undergoes a coordinated 90-degree rotation, migrating from an originally transverse orientation (perpendicular to the proximo-distal axis of the leg) to a longitudinal one (parallel to the PD axis).
The sex comb is a recent evolutionary innovation that is absent in most Drosophila species (Kopp, 2011). It has been well studied in the Old-World Sophophora clade consisting of the melanogaster and obscura species groups of Drosophila (Fig. 1). In species that primitively lack sex combs, dsx is not expressed in the pupal T1 legs at the corresponding stage of development, consistent with the lack of morphological dimorphism and with the cell-autonomous model of sexual differentiation. In sex comb-bearing species, pupal dsx expression in the T1 leg prefigures the position and size of the sex comb, and dsx acts together with the HOX gene Sex combs reduced (Scr) to determine sex comb morphology (Barmina and Kopp, 2007; Kopp, 2011; Rice et al., 2019; Tanaka et al., 2011).
Figure 1. Origin and secondary loss of sex combs in Lordiphosa.
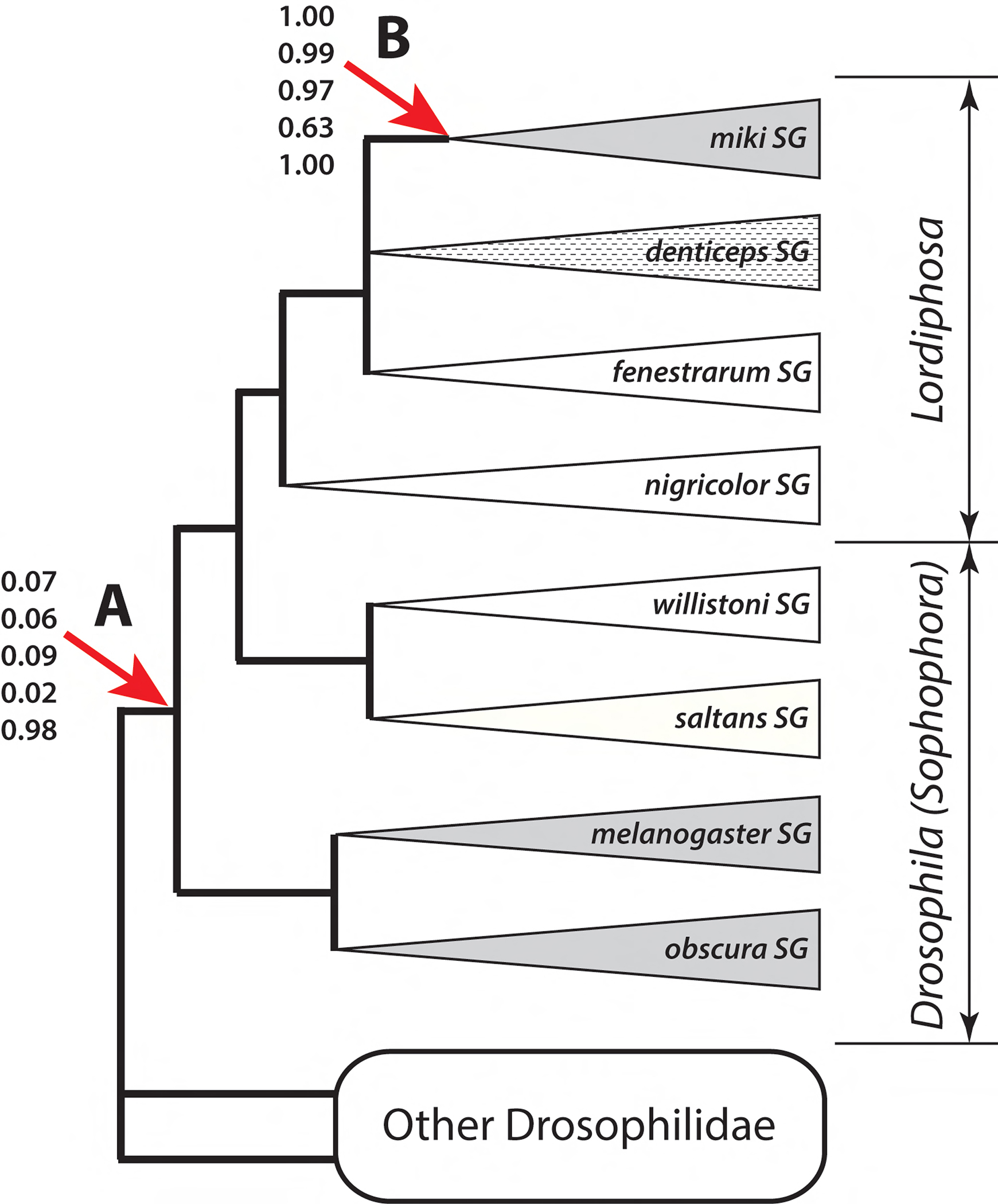
This summary tree shows phylogenetic relationships among Lordiphosa (miki, denticeps, fenestrarum, and nigricolor species groups), New-World Sophophora (willistoni and saltans species groups), and Old-World Sophophora (melanogaster and obscura species groups). See Figure S4 for species-level phylogenetic relationships. The branching order of the miki, denticeps, and fenestrarum groups has not been resolved with confidence. Sex combs are present in all or almost all species of the miki, melanogaster, and obscura species groups (grey triangles), and absent in all species of the willistoni, saltans, nigricolor, and fenestrarum species groups (white triangles). Most species of the denticeps species group have simple sex combs (striped triangle). Numbers at nodes A (last common ancestor of Sophophora + Lordiphosa) and B (last common ancestor of the miki species group) denote the estimated probabilities that the last common ancestor of that clade had sex combs, under five different models of trait evolution in the following order from top to bottom: MK model with unequal rates and strict molecular clock (Fig. S5A), MK model with unequal rates and random local relaxed clock (Fig. S5B), hidden states variable rates model with two latent rate classes (Fig. S5C), modified threshold model with 9 ordered latent states (Fig. S5D), and an approximation of Dollo model (Fig. S5E). See Methods and Table S5 for model descriptions.
In addition to the Old-World Sophophora, sex combs are found in some species of the less well studied genus Lordiphosa, especially in the miki species group (Fartyal et al., 2017; Gao et al., 2011; Katoh et al., 2018; Kopp, 2011; Lee, 1959; Okada, 1956) (Fig. 1). All described members of this group (L. miki, L. stackelbergi (= nipponica), L. magnipectinata, and L. clarofinis) were at some point assigned to the subgenus Sophophora of the genus Drosophila (Bock and Wheeler, 1972; Kikkawa and Peng, 1938; Lee, 1959; Okada, 1956; Wheeler, 1949). They were subsequently transferred to the subgenus Lordiphosa (Lastovka and Maca, 1978; Okada, 1984), which was then elevated to the generic rank by Grimaldi (Grimaldi, 1990). Later phylogenetic analyses identified Lordiphosa as sister to the New-World Sophophora (the willistoni and saltans species groups), making Sophophora paraphyletic (Gao et al., 2011; Hu and Toda, 2000) (Fig. 1). This finding also raised the question whether the sex combs of Old-World Sophophora and Lordiphosa shared a common origin, or evolved independently in the two clades (Kopp, 2011). The cellular mechanism of sex comb development in L. magnipectinata differs from what is seen in the melanogaster and obscura species groups of Sophophora (Atallah et al., 2012). However, this cannot be seen as conclusive evidence of independent evolution, since the cellular mechanisms of sex comb development also differ within the melanogaster species group, despite the unquestionably common origin of sex combs in that lineage (Tanaka et al., 2009).
In this report, we identify a new species of Lordiphosa that has secondarily lost the male sex comb, making its forelegs sexually monomorphic at the morphological level. We show that this species has also lost dsx expression in the T1 leg. This result reinforces the close link between the evolution of sex-specific traits and dsx regulation in insects: just as the origin of new sexually dimorphic traits can be linked to the evolution of new dsx expression domains, the reversion to sexual monomorphism can be caused by tissue-specific loss of dsx expression.
Materials and Methods
1. Lordiphosa collection, identification and culturing
Individuals of the L. clarofinis species complex, including L. clarofinis (Lee, 1959) and three related, undescribed species (provisionally named sp.1, sp.2, and sp.3) were collected in and near forested habitats by net sweeping over herbaceous vegetation dominated by Galinsoga parviflora Gav. (Fig. S1A), Alternanthera philoxeroides (Mart.) Griseb. or Impatiens spp. Collection locations (Fig. S2) are listed in Supplement Table 1. Specimens for phenotypic and phylogenetic analysis were fixed in 70% or 100% alcohol, and then identified and sexed under a stereomicroscope. In addition, isofemale lines were established from wild-caught females following (Atallah et al., 2012), maintained in glass vials (30 mm in diameter, 100 mm in height) plugged with cotton and padded with filter paper and containing a small piece of apple as adult food and shredded leaves and stems of G. parviflora, sterilized by freezing beforehand, as larval food (Fig. S1B–D). The founder female of each line was later subjected to molecular identification using DNA sequences of the internal transcribed spacer-1 (ITS1) region.
To distinguish the four species of the clarofinis complex, we relied on a combination of morphological, molecular, and geographic evidence. Old specimens of L. clarofinis from Korea were obtained from Dr. Nam-Woo Kim (Daegu Haany University) for help with species identification. Except for sp.3, which is known exclusively from Yunnan and is unique in lacking sex combs on male forelegs (see below), the remaining three species of the clarofinis complex are difficult to distinguish by external morphology. There is a slight but distinct differentiation in male genitalia between L. clarofinis and sp.1: ventral postgonite (ventral branch of aedeagal basal process, inner paraphysis) is distally narrow and strongly curved backward in L. clarofinis, but less narrow and only gently curved in sp.1 (Fig. S3). Sp.2, which is geographically restricted to the Sichuan Basin, has smaller sex combs on the first tarsomere than most populations of L. clarofinis (see below). Although the shape of pregonite (paramere, outer paraphysis) is variable in lateral view in L. clarofinis, sp.2 and sp.3, this variation appears to be due to developmental plasticity and was not used in species delimitation. However, all species including sp.2 could be readily distinguished from each other based on DNA sequences of the ITS1 region (Ji et al., 2003), and most of them could also be separated based on mitochondrial COI sequences (see below). L. acongruens (Zhang and Liang, 1992) was collected together with sp.3 in Kunming in May-June of 2018 (Supplement Table 1), cultured in the same manner as the clarofinis complex species, and identified by morphology.
Specimens of L. magnipectinata (Okada, 1956), L. stackelbergi (Duda, 1935), L. mommai (Takada and Okada, 1960) and L. collinella (Okada, 1968) were collected by net sweeping over undergrowth vegetation of spring ephemeral plants in Sapporo (the campus of Hokkaido University and the Jozankei area) and Iwamizawa (Hokkaido) between late May and mid-June of 2019 (Supplement Table 2). Some of the collected specimens were preserved in 100% ethanol and later used for genome sequencing. The remaining individuals, separately for each species, were kept alive in culture vials with standard Drosophila media in an incubator at 18 ± 0.5°C and a photoperiod of 16 h light : 8 h dark (LD 16:8) for several days. To obtain their offspring, the field-collected flies of each species were released into a plastic lidded plate (8.5 cm in diameter, 2.8 cm in height) with decayed leaves and stems of Anemone flaccida F. Schmidt (Ranunculaceae) and soaked filter paper at the bottom. The host plant material had been frozen at −20°C before use to kill all live insects. The female flies were then introduced to lay eggs for approximately 24 h under 18 ± 0.5°C and LD 16:8. After the adult flies were removed from the oviposition plates, the larvae were maintained under the same conditions and fed by adding decayed A. flaccida material.
2. Genome assemblies
The genome assemblies of L. clarofinis, L. stackelbergi, L. magnipectinata, L. collinella, and L. mommai are described in Kim et al. (Kim et al., 2021). We generated similar de novo assemblies for sp.2 and sp.3; we did not sequence the genome of sp.1 due to lack of sufficient material. For L. clarofinis, we sequenced the genome of a strain that was established from a single female collected in Qianling Park, Guiyang City, Guizhou Province, China, in May 2018 and inbred by single-pair, full-sib crosses for 2 generations. For sp.2, we sequenced a non-inbred isofemale line established from a female collected in Longhua Town, Pingshan County, Sichuan Province, China in May 2019. For L. magnipectinata, L. stackelbergi, L. collinella, and L. mommai, genome sequencing was performed using pools of wild-caught individuals collected in May-June 2019 in Sapporo, Hokkaido, Japan (Supplement Table 2). While it was not ideal to sequence pools of non-inbred individuals for genome assembly, we found it necessary to sequence groups of at least 20 flies to obtain sufficient material for Oxford Nanopore sequencing and resorted to computational tools to remove resulting haplotypic duplication in the assembly. For sp.3, we first established a mass culture from a small number of individuals collected in Tanglangchuan, Kunming, in May 2018. An inbred derivative of this strain (b16-1-13) was generated by three generations of single-pair, full-sib crosses, and 25 individuals from this inbred strain were used for Oxford Nanopore long-read sequencing performed by Nextomics Biosciences (Wuhan, China). For sp.2, sequencing was performed following the protocol described by Kim et al. (Kim et al., 2021). Briefly, high molecular weight genomic DNA was extracted from adult flies by phenol-chloroform extraction and prepared for Oxford Nanopore sequencing with a modified version of the SQK-LSK109 ligation kit protocol. Illumina libraries were prepared from a reserved portion of the same gDNA with a modified version of the Nextera XT Library Prep Kit protocol (Baym et al., 2015) and sequenced by Admera Health (South Plainfield, NJ) on a HiSeq 4000 machine. A detailed open-source protocol is provided at Protocols.io (dx.doi.org/10.17504/protocols.io.bdfqi3mw).
We followed the genome assembly workflow described in Kim et al. (Kim et al., 2021) to generate de novo assemblies, with software updated to the latest versions (as of December 2021). Nanopore reads were basecalled with Guppy v5.0.11 using the super-accuracy model. The initial read set was assembled with Flye v2.9 (Kolmogorov et al., 2019). A custom repeat library was built from this initial assembly with RepeatModeler2 (Flynn et al., 2020), then repeat masking was performed with RepeatMasker (Smit et al., 2013) using the custom library only. Duplicated sequences in the assembly were identified and removed with Purge Haplotigs (Roach et al., 2018). Regions annotated as repeats from the previous step were masked while running the Purge Haplotigs pipeline. The purged assembly was then polished with Nanopore reads, with two rounds of Racon (Vaser et al., 2017) and one round of Medaka v1.3.4. A final round of polishing using Illumina reads was performed with Pilon 1.23 (Walker et al., 2014), fixing only base-level (SNP and small indel) errors. Finally, sequences identified by a NCBI Nucleotide BLAST+ 2.12.0+ (Camacho et al., 2009) query as having a subject taxonomy ID matching non-arthropod groups were removed with BlobTools (Laetsch and Blaxter, 2017). No additional scaffolding was performed on the purged contigs.
This assembly process resulted in fairly contiguous, highly complete, and accurate assemblies. Both sp.2 and sp.3 sequencing runs had over 50% of the data in reads longer than 25 kbp. These relatively long reads resulted in final assemblies of Lordiphosa sp.2 of size 402.5 Mbp with a contig N50 of 217 Kbp, and L. sp.3 of size 432.7 Mbp with a contig N50 of 1.3 Mbp. While the lower contiguity relative to other drosophilid genome assemblies is expected due to the high genetic diversity contained in the sequenced samples, BUSCO analyses determined the gene content of the assemblies to be highly complete (sp.2: 96.8% complete; sp.3: 98.7% complete) with little duplication (sp.2: 3.8% duplicated; sp.3: 1.2% duplicated). Furthermore, the sizes of each assembly are consistent with previously sequenced Lordiphosa genomes and with estimates based on read depth in single-copy BUSCO genes (Supplementary Table 3). Base-level quality score analysis was performed by aligning Illumina reads back to each genome assembly with BWA (Li and Durbin, 2009), removing PCR duplicates with sambamba (Tarasov et al., 2015), and calling variants with bcftools (Li, 2011). After excluding genomic regions masked by RepeatMasker and filtering sites with base and genotype quality scores less than 30, counts of homozygous non-reference variants (that is, variants contained in the reads that do not appear in the reference) were divided by the total number of callable sites to estimate the error rate, similar to previous work (Kim et al., 2021; Solares et al., 2018). The accuracy of sp. 2 and sp. 3 assemblies was measured to be about 99.97% and 99.99% respectively (or Phred-scaled QV35 and QV39), exceeding the accuracy threshold at which most genes would be expected to have less than 1 error in their coding sequences (Koren et al., 2019). Additional details of all Lordiphosa assemblies are available in Supplementary Table 3.
3. Phylogenetic tree reconstruction
To generate a multi-locus nuclear phylogeny, we selected 250 single-copy genes identified in the genome assemblies of the seven Lordiphosa species and 200 other species of Drosophilidae (Supplementary Table 4). In this phylogeny, each Lordiphosa species was represented by a single genotype. First, we estimated multiple sequence alignments (MSAs) using the L-INS-I strategy in MAFFT v7.453 (Katoh and Standley, 2013), which generates the most accurate MSAs. Sites containing less than 3 non-gap characters were removed. After trimming, the MSA lengths ranged from 325 bp to 16,465 bp with an average length of 2,289 bp. Next, for phylogenetic inference we concatenated 250 MSAs to form a supermatrix containing 572,343 sites in total. We used this supermatrix as a single partition to estimate a maximum-likelihood (ML) tree topology in IQ-TREE v1.6.5 (Nguyen et al., 2015), specifying the GTR+I+G substitution model (Fig. S4A). To estimate the support for each node in the resulting topology, we computed three different reliability measures. We did 1,000 ultrafast bootstrap (UFBoot) replicates (Minh et al., 2013), an approximate likelihood ratio test with the nonparametric Shimodaira–Hasegawa correction (SH-aLRT), and a Bayesian-like transformation of aLRT (Anisimova et al., 2011). Additionally, to account for possible effects of ILS on tree reconstruction accuracy, we estimated a species tree topology in ASTRAL v5.14.7 using individual ML gene trees inferred for each of the 250 loci (Fig. S4B). Here individual gene trees were also estimated with IQ-TREE, again specifying the GTR+I+G substitution model. For the estimated ASTRAL tree we calculated the support of each node using local posterior probabilities (LPP) (Sayyari and Mirarab, 2016).
We implemented the Bayesian algorithm of MCMCTree v4.9h (Yang, 2007) with approximate likelihood computation to estimate divergence times using five fossils for age prior construction, analogous to the calibration scheme A described in (Suvorov et al., 2021). First, we divided our 250 loci into five equal datasets and generated five supermatrices consisting of 50 MSAs each.
We used these datasets to perform the dating analyses. For each of our five datasets, we estimated branch lengths by ML and then the gradient and Hessian matrices around these ML estimates in MCMCTree using the DNA supermatrix and species tree topology estimated by IQ-TREE. Then, we used the gradient and Hessian matrix, which constructs an approximate likelihood function by Taylor expansion (Reis and Yang, 2011), to perform fossil calibration in an MCMC framework. For this step, we specified a GTR+G substitution model with four gamma categories; birth, death and sampling parameters of 1, 1 and 0.1, respectively. To model rate variation, we used an uncorrelated relaxed clock. To ensure convergence, the analysis was run three times independently for each of the 5 datasets for 7 × 106 generations (first 2 ×106 generations were discarded as burn-in), logging every 1,000 generations. Additionally, we performed sampling from the prior distribution only. Convergence was assessed for the combined MCMC runs for each of the 5 datasets using Effective Sample Size criteria (ESS > 100) in Tracer (Rambaut et al., 2018).
Separately from this large taxon sample, we conducted a phylogenetic analysis of the clarofinis species complex where each ingroup species was represented by multiple genotypes (see Supplement Table 1 for collection locations). Two other members of the L. miki species group, L. magnipectinata and L. stackelbergi, were used as outgroups. DNA was extracted from each wild-caught specimen using the TIANamp® Genomic DNA Kit. The mitochondrial COI (cytochrome oxidase subunit I) barcoding region and the nuclear ITS1 (internal transcribed spacer 1) region were amplified and sequenced using the primer pairs LCO1490/HCO2198 (Folmer et al., 1994) and CAS18sF1/CAS5p8sB1d (Ji et al., 2003), respectively. Trace files were edited in the SeqMan module of the DNAStar package 7.1.0 (DNAStar Inc., Madison, WI). The resulting sequences were aligned using the ClustalW algorithm implemented in MEGA7 (Kumar et al., 2016), and a NEXUS haplotype file was generated from the alignment in DnaSP 6 (Rozas et al., 2017). Sequences with 20 or more end gaps on either end were removed from the analysis. Neighbor-joining trees were constructed in MEGA7 from the haplotype files under the Kimura 2-parameter model, and node support was evaluated using 1000 bootstrap replicates.
4. Ancestral character reconstruction
To estimate the probability of sex comb origin and secondary loss, we reconstructed ancestral character states using the time-calibrated trees described above. Sex comb morphology is highly diverse, ranging from massive structures found in the L. miki species group (Fig. 2) and in the ficusphila and montium subgroups of the D. melanogaster species group to a few slightly thickened and weakly pigmented bristles that are barely distinguishable from the homologous female bristles (Kopp, 2011). The latter morphology is widespread in both Sophophora and Lordiphosa. It is found, for example, in most species of the L. denticeps group (Fig. 2) (Fartyal et al., 2017; Katoh et al., 2018), in some species of the fima subgroup (Kopp et al., 2019), and in several other species of the D. melanogaster group including D. setifemur (McEvey, 2009) and D. ironensis. Sex comb is also weakly developed in most species of the ananassae subgroup (Matsuda et al., 2009). Since our focus is on sexual dimorphism rather than any particular aspects of cell differentiation, we considered a species to have sex combs if we could detect any difference in bristle morphology (length, thickness, bluntness, and/or pigmentation) between males and females. By this permissive definition, only three species represented in our phylogenetic analysis lack sex combs entirely: L. sp.3, D. prolongata, and D. majtoi. This distinction is further addressed in the Discussion.
Figure 2. Sex combs and other male-specific leg structures in Lordiphosa.
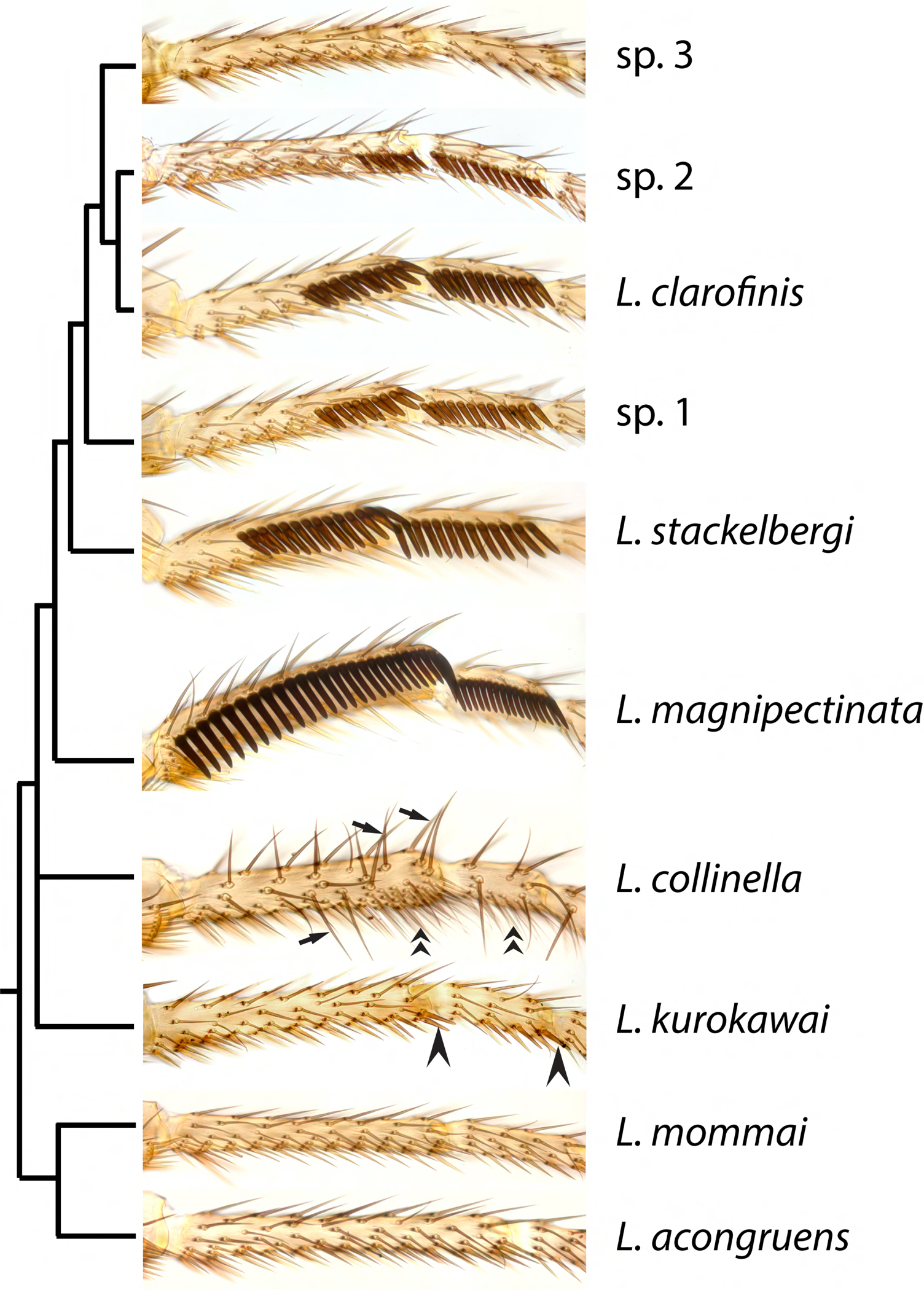
Males of most species of the miki species group, including L. magnipectinata, L. stackelbergi, L. clarofinis, sp.1, and sp.2, have longitudinal sex combs composed of enlarged, blunt, heavily melanized “teeth”, which are modified mechanosensory bristles. The only exception is sp.3, which lacks sex combs. Most species in the denticeps species group, including L. kurokawai, have much smaller and simpler sex combs composed of a few slightly thickened bristles (arrowheads). L. collinella (fenestrarum species group) lacks sex combs, but has other male-specific leg structures: enlarged, pointed dorsal bristles (arrows) and a ventral brush composed of thin, wavy bristles (chevrons). L. mommai and L. acongruens (nigricolor species group) do not show any male-specific leg modifications.
Sex comb presence/absence matrix was analyzed under five different models of trait evolution: (1) a simple MK model (Lewis, 2001) with unequal rates and a strict molecular clock; (2) an MK model with unequal rates and a random local relaxed clock (RLC; (Drummond and Suchard, 2010)); (3) a hidden states variable rates model with two latent rate classes (Beaulieu et al., 2013). The latter model assumes that each of the two binary character states (trait present vs trait absent) can exist in two discrete, not directly observable rate classes (“fast evolving / likely to change” vs ”slow evolving / not likely to change”). We also used (4) an approximation of a Dollo model made by assuming that the rate of trait loss is more than 300 times greater than the rate of gain; and (5) a modified threshold model similar to Felsenstein’s (Felsenstein, 2005). This modified threshold model assumes that the gain of a trait proceeds sequentially through 9 ordered latent states, such that each state can only transition to its nearest neighbor state towards or away from the trait. This implies that a species that lacks the trait can exist in any of the 9 “trait-absent” states that are not directly observable. For species that lack sex combs, we assume a uniform distribution over these latent states.
All models were implemented and analyzed in RevBayes (Höhna et al., 2016), and RevBayes MCMC outputs were analyzed in R using phytools (Revell, 2012). See Supplement Table 5 for prior distribution assumptions for all model parameters. Under each of the above models, except RLC, and for each of the four trees inferred from different sets of genes, MCMC was run for 50,000 cycles with a sampling rate of 1 in 50 to produce at least 200 ESS for all parameters. Because the RLC model takes significantly longer to run, we ran the MCMC for a set amount of time instead of set number of cycles. This also achieved > 200 ESS for all RLC analyses. Two independent chains were run to confirm convergence to the same posterior. Sensitivity to prior specification was assessed by comparing the marginal posterior to marginal prior for each parameter. 100 stochastic character evolution histories were simulated during one of the MCMC chains for each of the models by sampling every tenth sample from the posterior distribution. The resulting simmap files of 100 character histories mapped to a fixed tree were then analyzed to infer the ancestral states at nodes and along branches as well as the number and type of transitions. For the RLC model, we checked sensitivity to the prior on the frequency of rate shifts by comparing inferences under a prior model with expected number of rate shifts of 2 and that of 10 and obtained very similar inferences.
5. Adult leg imaging and phenotypic analysis
T1 legs of adult flies were dissected from the distal tibia down, mounted in Hoyers media between two coverslips, and photographed on a Leica DM5000 microscope with a Leica DC500 camera under brightfield illumination. Images were processed in Photoshop to remove the background and match exposure and color balance across species. Multiple males were examined for each species, and their sex comb phenotypes were found to be consistent within each species. We never observed any sex comb teeth in sp.3 despite examining several hundred wild-caught and lab-raised males. To examine intraspecific variation and interspecific differences in sex comb size in the remaining species of the L. clarofinis complex species, T1 legs of field-collected males were mounted and the numbers of sex comb teeth on the first and second tarsomeres were counted under the microscope.
6. Pupal leg staining
Temporary laboratory cultures of each species were established from wild-caught individuals as described above. Species identification was confirmed using a combination of morphological and molecular data. For L. clarofinis, we used strains collected in Zhanggou, Gaoqiao, Emeishan, Sichuan Province, China in May 2018, and in Longhua Town, Pingshan County, Sichuan in May 2019. For sp.2, the laboratory strain was also collected in Longhua in May 2019. For sp.3, we used strains collected in Kunming, Yunnan Province, China in May 2018 and in May 2019. For L. acongruens, we used cultured F1 progeny of flies collected in western Kunming in 2018 together with sp.3. For L. magnipectinata, L. stackelbergi, and L. collinella, the cultured strains were collected in Sapporo, Hokkaido, Japan in May 2019.
Specimens for staining were collected from lab cultures as white prepupae (0–1 hours After Puparium Formation, APF) and sexed based on the presence/absence of testes, which could be seen through the body wall. Sexed pupae were placed on a wet Kimwipe in a closed Petri dish and aged at room temperature until dissection. We observed unusually large individual variation in the rate of development within each species. Thus, instead of targeting narrow time windows, we dissected many pupae that appeared to be at the desired developmental stage based on external morphology and determined the timing of sex comb development after the fact based on the morphology of transverse bristle rows (TBRs) in the same leg. This approach allowed us to align corresponding developmental stages between species and sexes.
The dorsal half and posterior third of each pupa were cut away and remaining tissues were fixed for 30 minutes in 4% paraformaldehyde (Electron Microscopy Sciences) in 0.1 M Tris-HCl/0.3 M NaCl (pH 7.4) (TN). Fixation was followed by three washes in TN buffer. Further dissections were carried out in 0.1M Tris-HCl/0.3M NaCl (pH 7.4) containing 0.5% Triton X-100 (TNT). Legs were removed from the pupal cuticle by first tearing the cuticle around the dorsal femur/tibia boundary and then pulling out the tibial and tarsal segments. Tissues were then treated with Image-iT FX Signal Enhancer (Invitrogen) for 30min and washed three times for 15 min each in TNT. Samples were incubated with the primary antibodies overnight at 4°C. The primary antibodies used were monoclonal mouse anti-Dsx (Mellert et al., 2012) (1:5 dilution in TNT) and rat-Elav-7E8A10 (O’Neill et al., 1994) (1:10 dilution in TNT), both obtained from the Developmental Studies Hybridoma Bank. Legs were then washed in TNT buffer four times and incubated in the secondary antibodies overnight at 4°C. The secondary antibodies were Invitrogen/Thermo Fisher 488 mouse # A32723 preabsorbed against rat and Invitrogen/Thermo Fisher 555 rat #A21434 preabsorbed against mouse (1:200 dilution in TNT). Stained samples were washed four times in TNT and mounted in Fluoromount 50 (Southern Biotech). Fluorescent images were collected on an Olympus FV1000 confocal microscope at UC Davis or an Olympus FV1000 confocal microscope at the Research Institute of Resource Insects, Chinese Academy of Forestry, Kunming. The 40X lens was used, with the gain adjusted for the dynamic range of each sample. For L. clarofinis, sp.3, and L. acongruens, we imaged at least 10 males per species; fewer individuals were examined for sp.1 and sp.2. To assemble leg images, signals from non-epidermal tissues were removed from each confocal section, and Z-series projection was produced using ImageJ.
Results
1. Sex comb evolution in Lordiphosa
Sex combs are present in almost all Old-World Sophophora (the D. melanogaster and D. obscura species groups) but are absent in all New-World Sophophora (the willistoni and saltans groups) (Fig. 1). The Lordiphosa genus is the sister lineage to the New-World Sophophora (Gao et al., 2011; Kim et al., 2021; Suvorov et al., 2022). Within Lordiphosa, sex combs are found in all described members of the miki species group and all members of the denticeps species group except L. medogensis Katoh and Gao, 2018 (Fartyal et al., 2017; Gao et al., 2011; Katoh et al., 2018) (Fig. 1). Although the phylogeny of Lordiphosa is not fully resolved, neither the miki nor the denticeps species groups are basal within that genus. Thus, it is possible that sex combs evolved independently two or three times: once in the Old-World Sophophora, and either once or twice in Lordiphosa. Independent origin of sex combs in Sophophora and Lordiphosa would be consistent with the differences in cellular mechanisms that underlie sex comb development in the L. miki species group compared to Sophophora (Atallah et al., 2012; Tanaka et al., 2009). However, it is also possible that sex combs evolved once at the base of the (Sophophora + Lordiphosa) clade, and subsequently underwent many independent losses: in the New-World Sophophora, in the L. nigricolor species group, in the L. fenestrarum species group, and in a few isolated species of the D. melanogaster and L. denticeps species groups (Fartyal et al., 2017; Kopp, 2011). To distinguish between these scenarios, we used a whole-genome dataset consisting of 250 single-copy BUSCO genes (572,343 total sites) to reconstruct the phylogeny of >200 drosophilid species and estimate the probability of the single-gain and multiple-gains models of sex comb evolution.
Under most models of character evolution, our analysis suggests that sex combs originated independently in Lordiphosa and in Old-World Sophophora (Fig. S5 A–D). The probability that the last common ancestor of both clades had sex combs is 0.09 under the hidden states variable rates model (Beaulieu et al., 2013), and 0.02–0.07 under the MK models (Drummond and Suchard, 2010; Lewis, 2001) and the threshold model with latent ordered states (Fig. 1). As expected, enforcing an approximate Dollo model by assuming a highly informative prior where the rate of trait loss is more than 300 times higher than the rate of trait gain leads to a different conclusion, namely that the sex comb originated only once at the base of all Drosophilidae and was subsequently lost many times (Fig. S5 E). This result may be partly driven by biased taxon sampling, since the Old-World Sophophora are heavily overrepresented in the available set of genomes and therefore in the BUSCO phylogeny (Kim et al., 2021; Suvorov et al., 2022). However, unless we assume this extreme model of trait evolution, our analysis supports independent origin of sex combs in Lordiphosa, separately from the Old-World Sophophora (Fig. 1, Fig. S5 A–D).
2. dsx expression associated with sex comb development in Lordiphosa
All species of the nigricolor species group, which represents the most basal lineage within Lordiphosa (Gao et al., 2011; Kim et al., 2021), lack sex combs. We examined leg development and dsx expression in L. acongruens, a representative of this species group (Fig. 1, 2). We found that, at the stage when sex combs develop in other species of Lordiphosa and in Old-World Sophophora, Dsx expression is absent in both males and females of L. acongruens (Fig. 3A). This is consistent with the lack of Dsx expression at this stage in the New-World Sophophora, which also lack sex combs (Rice et al., 2019; Tanaka et al., 2011), as well as with the absence of sex-specific chemosensory bristles in Lordiphosa (Kopp and Barmina 2022). Dsx expression is also absent at earlier, prepupal stages in L. acongruens (Fig. 3H).
Figure 3. Dsx expression in Lordiphosa T1 legs.
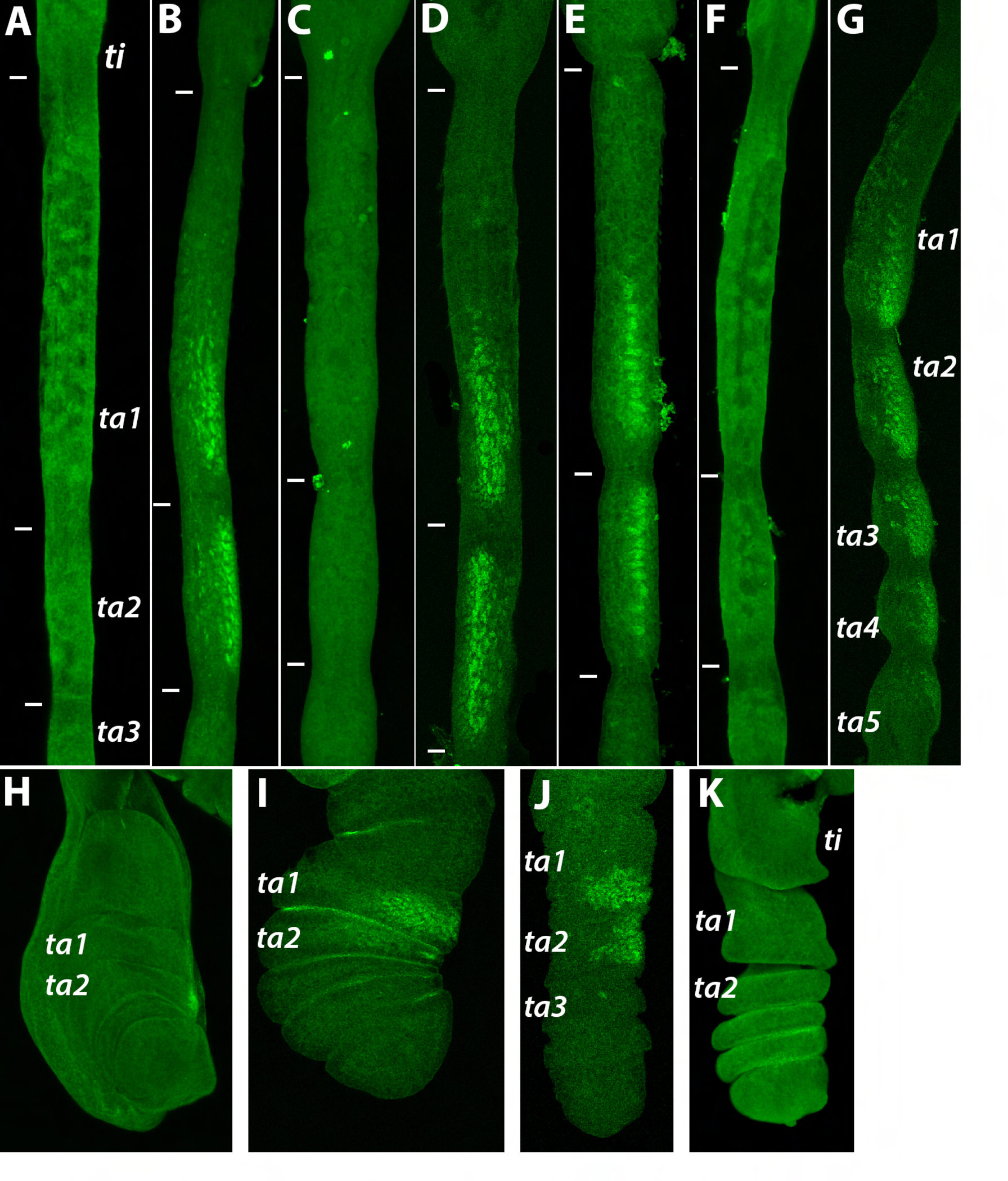
Proximal is up, distal is down. Tibia (ti) and tarsal segments 1–5 (ta1–ta5) are indicated in some panels; in other panels, the boundaries between these segments are shown with white tick marks. Panels A–G show pupal legs at stages equivalent to 24–30 hours After Puparium Formation (APF) in D. melanogaster. A. L. acongruens male; Dsx protein is absent. B. L. clarofinis male; Dsx is expressed on the anterior-ventral leg surface in the distal part of ta1 and all of ta2. C. L. clarofinis female; no Dsx expression is seen at this stage. D. L. sp.1 male; Dsx expression is similar to L. clarofinis. E. L. sp.2 male; Dsx expression is similar to L. clarofinis. F. L. sp.3 male; no Dsx expression is observed. G. L. collinella male; Dsx staining is present in all tarsal segments. H. The T1 leg imaginal disc of the white prepupa (0 hrs APF) of L. acongruens male; no Dsx expression is observed. I. Prepupal leg of L. sp.2 male at the stage equivalent to ~2–3 hrs APF in D. melanogaster; Dsx is expressed on the anterior-ventral surface of ta1 and ta2. J. Prepupal leg of L. sp.2 male at the stage equivalent to ~6–7 hrs APF in D. melanogaster. K. Prepupal leg of L. sp.3 male at the stage equivalent to ~5–6 hrs APF in D. melanogaster; no Dsx expression is seen.
All species of the L. miki species group, except sp.3, have well-developed sex combs reminiscent of the largest sex combs seen in the melanogaster species group of Sophophora (Atallah et al., 2012; Kopp, 2011) (Fig. 2). We observed Dsx expression in the presumptive sex comb region of the prepupal and pupal T1 legs of L. clarofinis, sp.1, and sp.2 (Fig. 3 B, D, E, I, J). As in the Old-World Sophophora (Tanaka et al., 2011), the spatial domain of Dsx expression corresponds to sex comb position. Also as in Sophophora, Dsx expression at the pupal stage is strong in males but absent in females (Fig. 3 B, C). Thus, despite the apparently independent evolution of sex combs and the differences in the cellular mechanisms of their development in Lordiphosa vs Sophophora (Atallah et al., 2012), in both cases their origin is associated with a novel spatial domain of dsx expression.
Sex combs are absent in the L. fenestrarum species group. However, L. collinella, a member of this group, has different sexually dimorphic traits on the T1 leg. In this species, a sparse ventral brush and a set of enlarged dorsal spines, both derived from mechanosensory bristles, are present in males but not in females (Fig. 2). As with the sex comb, we find that the presence of these structures in adult males is prefigured by male-specific expression of Dsx at the pupal stage (Fig. 3G). This consistent association of novel sex-specific traits with newly evolved domains of dsx expression is predicted by the cell-autonomous model of sexual differentiation in insects (Hopkins and Kopp, 2021; Kopp, 2011; Ledón-Rettig et al., 2017).
3. Secondary loss of the sex comb in the clarofinis species complex
Lordiphosa clarofinis was originally described from type specimens collected in Kongju, South Korea (Lee, 1959). Our field surveys in China and Japan during the last two decades identified a much larger geographical range for L. clarofinis than previously known, covering the central, southern, and southwestern China except for Yunnan (Table S1 and Fig. S2). These surveys also revealed the presence of three new forms closely resembling L. clarofinis. L. sp.1 aff. clarofinis is found in Japan and characterized by ventral postgonite that is distally broader and has a gentler backward curvature than in L. clarofinis (Fig. S3). L. sp.2 aff. clarofinis is restricted to the Sichuan Basin, and L. sp.3 aff. clarofinis occurs exclusively in Yunnan. The combination of morphological characters and molecular evidence (Fig. S6) suggests that L. clarofinis, sp.2, and sp.3 are distinct, but we did not test whether they are reproductively isolated from each other.
L. sp.3 aff. clarofinis is unique in the miki species group in completely lacking sex combs on both the first and the second tarsomeres (Fig. 2). The other three species of the clarofinis complex have sex combs on both tarsomeres but differ in the number of sex comb teeth, especially on the first tarsomere. The sex combs of L. clarofinis and sp.1 are similar in size, and both are significantly larger than in sp.2 (Fig. S7). An exception is found in the Sichuan population of L. clarofinis, which shows sex combs that are similar in size to sp.2, which is sympatric with this population, rather than to 10 other populations of L. clarofinis, which are allopatric with sp.2 (Fig. S7). The reason for the sympatric convergence between L. clarofinis and sp.2 is unknown.
We used single-copy BUSCO genes to reconstruct the phylogeny of the miki species group (the clade represented in our analysis by the clarofinis species complex, L. stackelbergi, and L. magnipectinata). First, a neighbor-joining tree was constructed separately from each of 10 different loci, using L. mommai (nigricolor species group) as outgroup. All 10 genes supported the ((L. clarofinis, sp.2) sp.3) tree topology (genome data are not available for sp.1). Nine loci supported (magnipectinata (stackelbergi (clarofinis complex))) topology; one supported ((magnipectinata, stackelbergi) (clarofinis complex)) topology. The combined 250-locus BUSCO dataset (see above) produced a credible set of only one tree: root-(magnipectinata (stackelbergi (sp.3 (clarofinis, sp.2)))) with 100% bootstrap and aLRT support at each node (Fig. S4A); the same topology is also observed in the ASTRAL tree (Fig. S4B).
The key question for our analysis is whether the sex comb was lost secondarily in sp.3 aff. clarofinis. To address this question, we estimated the probability that the last common ancestor of the Lordiphosa miki species group had sex combs. We found this probability to be 0.99–1.00 under the Dollo and MK models (Drummond and Suchard, 2010; Lewis, 2001) and 0.97 under the hidden states variable rates model (Beaulieu et al., 2013) (Fig. 1). An outlier result in this analysis was produced by the threshold model with latent ordered states, which accommodates more gradual transitions between the “present” and “absent” states of discrete characters. However, even this model puts the probability of secondary loss at 0.63 (Fig. 1). Thus, we conclude that the sex comb was present in the last common ancestor of the clarofinis species complex, but was lost secondarily in sp.3 after its divergence from the common ancestor of L. clarofinis and sp.2.
Separately, we constructed neighbor-joining trees of 140 mitochondrial COI haplotypes isolated from 650 individuals of six Lordiphosa species, and 36 nuclear ITS1 haplotypes sampled from 653 individuals (Table S1). Both analyses lend strong support to the monophyly of the L. clarofinis species complex with respect to the outgroups (L. magnipectinata and L. stackelbergi) (Fig. S6). L. sp.1 aff. clarofinis is placed as most basal within the complex, although with weak support. The relationship among the remaining three species is not fully resolved. In the COI tree, L. clarofinis is well supported as monophyletic, but sequences from L. sp.2 aff. clarofinis and L. sp.3 aff. clarofinis form an intermingled clade, with neither species appearing as monophyletic and some haplotypes shared between sp.2 and sp.3 (Fig. S6 A). In the ITS1 tree, each of the four focal species is monophyletic. However, sp.2 and sp.3 are placed as sister taxa to the exclusion of L. clarofinis (Fig. S6 B), in contrast to the multilocus BUSCO phylogeny that places L. clarofinis as sister to sp.2 (Fig. S4 A, B). The incongruence between ITS1 and BUSCO tree topologies could be due to ancestral lineage sorting, post-speciation gene flow, or simply to the small amount of data in the ITS1 dataset. The COI gene tree is most consistent with hybridization between sp.2 and sp.3 resulting in interspecific introgression of mitochondria, which is known to occur in Drosophila (Bachtrog et al., 2006; Llopart et al., 2014; Nunes et al., 2010; Turelli et al., 2018). If such hybridization has indeed occurred, it could also have affected some nuclear loci, including genes that control sex comb development. However, the tree reconstructed from the ~20 kb dsx non-coding region supported the same relationships as the BUSCO tree: root-(magnipectinata (stackelbergi (sp.3 (clarofinis, sp.2)))). Most importantly for this study, sp.3 occupies a derived position within the miki species group and the clarofinis complex in all analyses, confirming a secondary loss of the sex comb in this species.
4. Secondary loss of dsx expression in sp.3 correlates with loss of the sex comb.
In both males and females of sp.3, no Dsx expression is seen in the T1 legs at the pupal stages when sex combs are developing in L. clarofinis and sp.2 (Fig. 3F). In most Drosophila species, including some that lack sex combs, dsx is also expressed in the T1 legs earlier, in late third instar larvae and prepupae (Tanaka et al., 2011). This early expression is necessary for the development of sex-specific chemosensory bristles as well as the sex comb (Rice et al., 2019). In sp.3., however, no Dsx expression is seen at the prepupal stage when chemosensory bristles are specified (Fig. 3K). This is consistent with the absence of sex-specific chemosensory bristles in all examined species of Lordiphosa, which stands in contrast with the majority of Sophophora and Drosophila species (Kopp and Barmina, 2022). We conclude that the secondary loss of the sex comb in sp.3 coincides with the loss of dsx expression in the presumptive sex comb region. In addition, the secondary loss of sex-specific chemosensory bristles observed in Lordiphosa may also correlate with the loss of the early phase of dsx expression in prepupal legs.
Discussion
Our results show that the relationship between dsx expression and the evolution of sexual dimorphism goes both ways: while the origin of novel sex-specific traits is sometimes linked to new domains of dsx expression, the loss of sexually dimorphic structures can be associated with the loss of dsx expression. This linkage is a logical consequence of the mainly cell-autonomous control of sexual differentiation in insects: with rare exceptions, cells that transcribe dsx have the potential to differentiate in sex-specific ways, while those that lack dsx expression do not (Camara et al., 2008; Hopkins and Kopp, 2021; Kopp, 2012; Ledón-Rettig et al., 2017). It will be interesting to see whether the pattern of gains and losses of dsx expression extends to other models where sex-specific traits have been both gained and lost – such as beetle horns (Emlen et al., 2005; Moczek et al., 2006), Lepidopteran androconia (Prakash and Monteiro, 2020; Simmons et al., 2012; Valencia-Montoya et al., 2021), or Batesian mimicry in swallowtail butterflies (Kunte, 2009; Palmer and Kronforst, 2020).
However, the gain and loss of dsx expression is not the only mechanism by which sex-specific traits can be gained and lost. In many cases, dsx expression in the tissue can be conserved, and the tissue can retain the potential for sexually dimorphic development, even as particular sex-specific characters are gained and lost. In such scenarios, the gain and loss of sex-specific traits is caused by evolutionary changes in the response of downstream target genes to dsx, rather than to changes at the dsx locus itself. This can happen through changes in Dsx binding sites in the regulatory regions of those downstream targets – as, for example, in the evolution of sexually dimorphic abdominal pigmentation (Gompel and Carroll, 2003; Kopp et al., 2000; Rogers et al., 2013; Williams et al., 2008), or in the synthesis of sex-specific pheromones in oenocyte cells (Shirangi et al., 2009). In fact, this mechanism may operate in the evolution of sex combs, as well. In D. prolongata (melanogaster species group), the sex comb was lost, but dsx continues to be expressed and controls the development of sex-specific chemosensory bristles in the T1 leg (Luecke et al., 2022). Although complete secondary loss of sex combs (defined in this study as the absence of any detectable sexual dimorphism) is relatively rare, extreme reductions of the sex comb are more common. In the D. melanogaster species group alone, various degrees of reduction are seen in D. setifemur, D. ironensis, D. flavohirta, and some species of the ananassae, fima, and takahashii subgroups (Grimaldi et al., 2015; Kopp, 2011; Kopp et al., 2019; Matsuda et al., 2009; McEvey, 2009). Since these species still retain some level of sexual dimorphism, we expect that their attenuated sex comb morphology results from changes in dsx target genes rather than a loss of dsx expression.
Finally, the development of sexually dimorphic structures is not always cell-autonomous. For example, the development of the male-specific Muscle of Lawrence (MOL) in Drosophila depends not on the sex of the myoblasts that give rise to that muscle, but rather on the sex of the motoneuron that innervates it, and specifically on the expression of the fruitless (fru) gene in that neuron (Lawrence and Johnston, 1986; Nojima et al., 2010; Usui-Aoki et al., 2000). MOL has also undergone many gains and losses in Drosophila evolution (Gailey et al., 1997; Liang et al., 2021), which could be caused by changes either in the differentiation of the inducing motoneuron or in the response of the developing muscle to its synaptic output. Unsurprisingly, the genetic basis of trait evolution will be biased by the mode of its development.
Supplementary Material
References
- Andersson MB (1994). Sexual selection (Princeton, N.J.: Princeton University Press; ). [Google Scholar]
- Anisimova M, Gil M, Dufayard J-F, Dessimoz C, and Gascuel O (2011). Survey of Branch Support Methods Demonstrates Accuracy, Power, and Robustness of Fast Likelihood-based Approximation Schemes. Systematic Biology 60, 685–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman MN, New FN, Fear JM, Howard TS, Dalton JE, and Graze RM (2016). Sex Differences in Drosophila Somatic Gene Expression: Variation and Regulation by doublesex. G3 (Bethesda) 6, 1799–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah J, Watabe H, and Kopp A (2012). Many ways to make a novel structure: a new mode of sex comb development in Drosophilidae. Evolution & Development 14, 476–483. [DOI] [PubMed] [Google Scholar]
- Bachtrog D, Thornton K, Clark A, and Andolfatto P (2006). Extensive introgression of mitochondrial DNA relative to nuclear genes in the Drosophila yakuba species group. Evolution Int J Org Evolution 60, 292–302. [PubMed] [Google Scholar]
- Bailey NW, Pascoal S, and Montealegre-Z F (2019). Testing the role of trait reversal in evolutionary diversification using song loss in wild crickets. Proc Natl Acad Sci U S A 116, 8941–8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, and Ridge KA (1980). Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics 94, 383–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Burtis K, Goralski T, Mattox W, and Nagoshi R (1989). Molecular genetic aspects of sex determination in Drosophila melanogaster. Genome 31, 638–645. [DOI] [PubMed] [Google Scholar]
- Barmina O, and Kopp A (2007). Sex-specific expression of a HOX gene associated with rapid morphological evolution. Dev Biol 311, 277–286. [DOI] [PubMed] [Google Scholar]
- Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, and Kishony R (2015). Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10, e0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu JM, O’Meara BC, and Donoghue MJ (2013). Identifying hidden rate changes in the evolution of a binary morphological character: the evolution of plant habit in campanulid angiosperms. Syst Biol 62, 725–737. [DOI] [PubMed] [Google Scholar]
- Bock IR, and Wheeler MR (1972). The Drosophila melanogaster species group. Univ. Texas Publs 7, 1–102. [Google Scholar]
- Bonduriansky R, and Chenoweth SF (2009). Intralocus sexual conflict. Trends Ecol Evol 24, 280–288. [DOI] [PubMed] [Google Scholar]
- Burtis KC, and Baker BS (1989). Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56, 997–1010. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Coschigano KT, Baker BS, and Wensink PC (1991). The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. Embo J 10, 2577–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camara N, Whitworth C, and Van Doren M (2008). The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol 83, 65–107. [DOI] [PubMed] [Google Scholar]
- Cox RM, and Calsbeek R (2009). Sexually antagonistic selection, sexual dimorphism, and the resolution of intralocus sexual conflict. Am Nat 173, 176–187. [DOI] [PubMed] [Google Scholar]
- Darwin C (1871). The descent of man, and selection in relation to sex (London: J. Murray; ). [Google Scholar]
- van Doorn GS (2009). Intralocus sexual conflict. Ann N Y Acad Sci 1168, 52–71. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, and Suchard MA (2010). Bayesian random local clocks, or one rate to rule them all. BMC Biology 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson SB (1996). Phylogenies and Physiological Processes—The Evolution of Sexual Dimorphism in Southeast Asian Frogs. Systematic Biology 45, 278–289. [Google Scholar]
- Emlen DJ, Marangelo J, Ball B, and Cunningham CW (2005). Diversity in the weapons of sexual selection: horn evolution in the beetle genus Onthophagus (Coleoptera: Scarabaeidae). Evolution Int J Org Evolution 59, 1060–1084. [PubMed] [Google Scholar]
- Fartyal RS, Sati PC, Pradhan S, Kandpal MC, Toda MJ, Chatterjee RN, Singh BK, and Bhardwai A (2017). A review of the genus Lordiphosa Basden in India, with descriptions of four new species from the Himalayan region (Diptera, Drosophilidae). Zookeys 49–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J (2005). Using the quantitative genetic threshold model for inferences between and within species. Philos Trans R Soc Lond B Biol Sci 360, 1427–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, and Smit AF (2020). RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A 117, 9451–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, and Vrijenhoek R (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol 3, 294–299. [PubMed] [Google Scholar]
- Gailey DA, Ohshima S, Santiago SJ, Montez JM, Arellano AR, Robillo J, Villarimo CA, Roberts L, Fine E, Villella A, et al. (1997). The muscle of lawrence in Drosophila: a case of repeated evolutionary loss. Proc Natl Acad Sci U S A 94, 4543–4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao JJ, Hu YG, Toda MJ, Katoh T, and Tamura K (2011). Phylogenetic relationships between Sophophora and Lordiphosa, with proposition of a hypothesis on the vicariant divergences of tropical lineages between the Old and New Worlds in the family Drosophilidae. Mol Phylogenet Evol 60, 98–107. [DOI] [PubMed] [Google Scholar]
- Goldman TD, and Arbeitman MN (2007). Genomic and functional studies of Drosophila sex hierarchy regulated gene expression in adult head and nervous system tissues. PLoS Genet 3, e216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gompel N, and Carroll SB (2003). Genetic mechanisms and constraints governing the evolution of correlated traits in drosophilid flies. Nature 424, 931–935. [DOI] [PubMed] [Google Scholar]
- Gordon M, Cohen H, and Nigrelli R (1943). A Hormone-Produced Taxonomic Character in Platypoecilus maculatus Diagnostic of Wild P. xiphidium. American Naturalist 77, 569–572. [Google Scholar]
- Grimaldi DA (1990). A phylogenetic, revised classification of genera in the Drosophilidae (Diptera). Bulletin of the AMNH ; no. 197. Drosophilidae (Diptera). [Google Scholar]
- Grimaldi D, Ginsberg PS, Thayer L, McEvey S, Hauser M, Turelli M, and Brown B (2015). Strange Little Flies in the Big City: Exotic Flower-Breeding Drosophilidae (Diptera) in Urban Los Angeles. PLOS ONE 10, e0122575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grobstein C (1942). Endocrine and developmental studies of gonopod differentiation in certain poeciliid fishes. J Exp Zool 89, 305–328. [Google Scholar]
- Hempel LU, and Oliver B (2007). Sex-specific DoublesexM expression in subsets of Drosophila somatic gonad cells. BMC Developmental Biology 7, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MS, Reuter M, and Stewart AJ (2019). Sexual antagonism drives the displacement of polymorphism across gene regulatory cascades. Proceedings of the Royal Society B: Biological Sciences 286, 20190660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhna S, Landis MJ, Heath TA, Boussau B, Lartillot N, Moore BR, Huelsenbeck JP, and Ronquist F (2016). RevBayes: Bayesian Phylogenetic Inference Using Graphical Models and an Interactive Model-Specification Language. Syst Biol 65, 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins BR, and Kopp A (2021). Evolution of sexual development and sexual dimorphism in insects. Current Opinion in Genetics & Development 69, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y-G, and Toda MJ (2000). Polyphyly of Lordiphosa and its relationships in Drosophilinae (Diptera: Drosophilidae). Systematic Entomology 25, 1–17. [Google Scholar]
- Ioannidis J, Taylor G, Zhao D, Liu L, Idoko-Akoh A, Gong D, Lovell-Badge R, Guioli S, McGrew MJ, and Clinton M (2021). Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine adult secondary sex characteristics. PNAS 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Zhang D-X, and He L (2003). Evolutionary conservation and versatility of a new set of primers for amplifying the ribosomal internal transcribed spacer regions in insects and other invertebrates. Molecular Ecology Notes 3, 581–585. [Google Scholar]
- Jursnich VA, and Burtis KC (1993). A positive role in differentiation for the male doublesex protein of Drosophila. Dev Biol 155, 235–249. [DOI] [PubMed] [Google Scholar]
- Kang JH, Schartl M, Walter RB, and Meyer A (2013). Comprehensive phylogenetic analysis of all species of swordtails and platies (Pisces: Genus Xiphophorus) uncovers a hybrid origin of a swordtail fish, Xiphophorus monticolus, and demonstrates that the sexually selected sword originated in the ancestral lineage of the genus, but was lost again secondarily. BMC Evolutionary Biology 13, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, and Standley DM (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol Biol Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh TK, Zhang G, Toda MJ, Zhang WX, and Gao JJ (2018). The Lordiphosa denticeps species group (Diptera: Drosophilidae) in China, with redescriptions of four known species and descriptions of nine new species. Zootaxa 4471, 37–75. [DOI] [PubMed] [Google Scholar]
- Kijimoto T, Moczek AP, and Andrews J (2012). Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proc Natl Acad Sci U S A 109, 20526–20531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa H, and Peng FT (1938). Drosophila species of Japan and adjacent localities. Japanese Journal of Zoology 7, 507–552. [Google Scholar]
- Kim BY, Wang JR, Miller DE, Barmina O, Delaney E, Thompson A, Comeault AA, Peede D, D’Agostino ER, Pelaez J, et al. (2021). Highly contiguous assemblies of 101 drosophilid genomes. Elife 10, e66405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M, Yuan J, Lin Y, and Pevzner PA (2019). Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37, 540–546. [DOI] [PubMed] [Google Scholar]
- Kopp A (2011). Drosophila Sex Combs as a Model of Evolutionary Innovations. Evol Dev 13, 504–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A (2012). Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet 28, 175–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopp A, Duncan I, Godt D, and Carroll SB (2000). Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 408, 553–559. [DOI] [PubMed] [Google Scholar]
- Kopp A, Barmina O, and Prigent SR (2019). Phylogenetic position of the Drosophila fima and dentissima lineages, and the status of the D. melanogaster species group. Mol Phylogenet Evol 139, 106543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Phillippy AM, Simpson JT, Loman NJ, and Loose M (2019). Reply to ‘Errors in long-read assemblies can critically affect protein prediction.’ Nat Biotechnol 37, 127–128. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, and Tamura K (2016). MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunte K (2009). The Diversity and Evolution of Batesian Mimicry in Papilio Swallowtail Butterflies. Evolution 63, 2707–2716. [DOI] [PubMed] [Google Scholar]
- Laetsch DR, and Blaxter ML (2017). BlobTools: Interrogation of genome assemblies. F1000Research 6, 1287. [Google Scholar]
- Lande R, and Arnold SJ (1985). Evolution of mating preference and sexual dimorphism. J Theor Biol 117, 651–664. [DOI] [PubMed] [Google Scholar]
- Lastovka P, and Maca J (1978). European species of the Drosophila subgenus Lordiphosa (Diptera, Drosophilidae). Acta Ent. Bohemoslovaca 75, 404–420. [Google Scholar]
- Lawrence PA, and Johnston P (1986). The muscle pattern of a segment of Drosophila may be determined by neurons and not by contributing myoblasts. Cell 45, 505–513. [DOI] [PubMed] [Google Scholar]
- Ledón-Rettig CC, Zattara EE, and Moczek AP (2017). Asymmetric interactions between doublesex and tissue- and sex-specific target genes mediate sexual dimorphism in beetles. Nat Commun 8, 14593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TJ (1959). On a new species, “Drosophila clarofinis” sp. nov. Korean Journal of Zoology 2, 7–9. [Google Scholar]
- Lewis PO (2001). A likelihood approach to estimating phylogeny from discrete morphological character data. Syst Biol 50, 913–925. [DOI] [PubMed] [Google Scholar]
- Li H (2011). A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Katoh T, Sato K, Yamamoto D, and Wen S (2021). Evolution of a neuromuscular sexual dimorphism in the Drosophila montium species group. Sci Rep 11, 15272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llopart A, Herrig D, Brud E, and Stecklein Z (2014). Sequential adaptive introgression of the mitochondrial genome in Drosophila yakuba and Drosophila santomea. Molecular Ecology 23, 1124–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loehlin DW, Oliveira DC, Edwards R, Giebel JD, Clark ME, Cattani MV, van de Zande L, Verhulst EC, Beukeboom LW, Munoz-Torres M, et al. (2010). Non-coding changes cause sex-specific wing size differences between closely related species of Nasonia. PLoS Genet 6, e1000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke D, Rice G, and Kopp A (2022). Sex-specific evolution of a Drosophila sensory system via interacting cis- and trans-regulatory changes. 2022.01.12.475924. [DOI] [PMC free article] [PubMed]
- Lund-Hansen KK, Abbott JK, and Morrow EH (2020). Feminization of complex traits in Drosophila melanogaster via female-limited X chromosome evolution*. Evolution 74, 2703–2713. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Ng C-S, Doi M, Kopp A, and Tobari YN (2009). Evolution in the Drosophila ananassae species subgroup. Fly 3, 1–13. [DOI] [PubMed] [Google Scholar]
- McEvey SF (2009). Taxonomic Review of the Australian Drosophila setifemur Species Group, a New Name for the D. dispar Species Group (Diptera: Drosophilidae). Records of the Australian Museum 61, 31–38. [Google Scholar]
- McKeown M (1992). Sex differentiation: the role of alternative splicing. Curr Opin Genet Dev 2, 299–303. [DOI] [PubMed] [Google Scholar]
- Mellert DJ, Robinett CC, and Baker BS (2012). doublesex functions early and late in gustatory sense organ development. PLoS One 7, e51489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minh BQ, Nguyen MAT, and von Haeseler A (2013). Ultrafast Approximation for Phylogenetic Bootstrap. Mol Biol Evol 30, 1188–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moczek AP, Cruickshank TE, and Shelby A (2006). When ontogeny reveals what phylogeny hides: gain and loss of horns during development and evolution of horned beetles. Evolution Int J Org Evolution 60, 2329–2341. [PubMed] [Google Scholar]
- Morris M (1998). Further examination of female preference for vertical bars in swordtails: preference for “no bars” in a species without bars. Journal of Fish Biology. [Google Scholar]
- Morris MR, Moretz JA, Farley K, and Nicoletto P (2005). The role of sexual selection in the loss of sexually selected traits in the swordtail fish Xiphophorus continens. Animal Behaviour 69, 1415–1424. [Google Scholar]
- Nagoshi RN, and Baker BS (1990). Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev 4, 89–97. [DOI] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, and Minh BQ (2015). IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol Biol Evol 32, 268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Kimura K, Koganezawa M, and Yamamoto D (2010). Neuronal synaptic outputs determine the sexual fate of postsynaptic targets. Curr Biol 20, 836–840. [DOI] [PubMed] [Google Scholar]
- Nunes MDS, Wengel PO-T, Kreissl M, and Schlötterer C (2010). Multiple hybridization events between Drosophila simulans and Drosophila mauritiana are supported by mtDNA introgression. Mol Ecol 19, 4695–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T (1956). Systematic study of Drosophilidae and allied families of Japan (Tokyo: Gihodo Co.). [Google Scholar]
- Okada T (1984). New or little known species of Drosophila (Lordiphosa) with taximetrical analyses (Diptera, Drosophilidae). Kontyu 52, 565–575. [Google Scholar]
- O’Neill EM, Rebay I, Tjian R, and Rubin GM (1994). The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell 78, 137–147. [DOI] [PubMed] [Google Scholar]
- Palmer DH, and Kronforst MR (2020). A shared genetic basis of mimicry across swallowtail butterflies points to ancestral co-option of doublesex. Nat Commun 11, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascoal S, Cezard T, Eik-Nes A, Gharbi K, Majewska J, Payne E, Ritchie MG, Zuk M, and Bailey NW (2014). Rapid convergent evolution in wild crickets. Curr Biol 24, 1369–1374. [DOI] [PubMed] [Google Scholar]
- Prakash A, and Monteiro A (2020). Doublesex Mediates the Development of Sex-Specific Pheromone Organs in Bicyclus Butterflies via Multiple Mechanisms. Mol Biol Evol 37, 1694–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek MB, Childress MJ, Petersen JA, and Tomasso AO (2011). Phylogenetic Evidence for the Gain and Loss of a Sexually Selected Trait in Sailfin Mollies. ISRN Zoology 2011, e251925. [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, and Suchard MA (2018). Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Systematic Biology 67, 901–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis M. dos, and Yang Z (2011). Approximate Likelihood Calculation on a Phylogeny for Bayesian Estimation of Divergence Times. Molecular Biology and Evolution 28, 2161–2172. [DOI] [PubMed] [Google Scholar]
- Revell LJ (2012). phytools: an R package for phylogenetic comparative biology (and other things). Methods in Ecology and Evolution 3, 217–223. [Google Scholar]
- Ribeiro RD, McCormack JE, Álvarez HG, Carrasco L, Grether GF, Mena-Olmedo P, Sedano R, Smith TB, and Karubian J (2015). Loss of sexual dimorphism is associated with loss of lekking behavior in the green manakin Xenopipo holochora. Journal of Avian Biology 46, 307–314. [Google Scholar]
- Rice G, Barmina O, Hu K, and Kopp A (2018). Evolving doublesex expression correlates with the origin and diversification of male sexual ornaments in the Drosophila immigrans species group. Evol Dev 20, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice GR, Barmina O, Luecke D, Hu K, Arbeitman M, and Kopp A (2019). Modular tissue-specific regulation of doublesex underpins sexually dimorphic development in Drosophila. Development 146, dev178285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Dornan AJ, Neville MC, Eadie S, and Goodwin SF (2010). Control of sexual differentiation and behavior by the doublesex gene in Drosophila melanogaster. Nat Neurosci 13, 458–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach MJ, Schmidt SA, and Borneman AR (2018). Purge Haplotigs: allelic contig reassignment for third-gen diploid genome assemblies. BMC Bioinformatics 19, 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett CC, Vaughan AG, Knapp JM, and Baker BS (2010). Sex and the Single Cell. II. There Is a Time and Place for Sex. PLoS Biol 8, e1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers WA, Salomone JR, Tacy DJ, Camino EM, Davis KA, Rebeiz M, and Williams TM (2013). Recurrent modification of a conserved cis-regulatory element underlies fruit fly pigmentation diversity. PLoS Genet 9, e1003740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohner PT, Linz DM, and Moczek AP (2021). Doublesex mediates species-, sex-, environment- and trait-specific exaggeration of size and shape. Proceedings of the Royal Society B: Biological Sciences 288, 20210241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal GG, Ryan MJ, and Wagner WEJ (2002). Secondary loss of preference for swords in the pygmy swordtail Xiphophorus nigrensis (Pisces: Poeciliidae). Anim Behav 63, 37–45. [Google Scholar]
- Rozas J, Ferrer-Mata A, Sánchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, and Sánchez-Gracia A (2017). DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol Biol Evol 34, 3299–3302. [DOI] [PubMed] [Google Scholar]
- Sanders LE, and Arbeitman MN (2008). Doublesex establishes sexual dimorphism in the Drosophila central nervous system in an isoform-dependent manner by directing cell number. Dev Biol 320, 378–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangster W (1948). A Study of the Quantitative Effects of Ethynyl Testosterone upon the Sword and Gonopodium of Xiphophorus Hellerii. Physiological Zoology 21, 134–147. [DOI] [PubMed] [Google Scholar]
- Sayyari E, and Mirarab S (2016). Fast Coalescent-Based Computation of Local Branch Support from Quartet Frequencies. Molecular Biology and Evolution 33, 1654–1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schartl M, Kneitz S, Ormanns J, Schmidt C, Anderson JL, Amores A, Catchen J, Wilson C, Geiger D, and Du K (2021). The developmental and genetic architecture of the sexually selected male ornament of swordtails. Current Biology 31, 911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirangi TR, Dufour HD, Williams TM, and Carroll SB (2009). Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol 7, e1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons RB, Weller SJ, and Johnson SJ (2012). The Evolution of Androconia in Mimetic Tiger Moths (Noctuoidea: Erebidae: Arctiinae: Ctenuchina and Euchromiina). Annals of the Entomological Society of America 105, 804–816. [Google Scholar]
- Smit AFA, Hubley R, and Green P (2013). RepeatMasker Open-4.0.
- Solares EA, Chakraborty M, Miller DE, Kalsow S, Hall K, Perera AG, Emerson JJ, and Hawley RS (2018). Rapid Low-Cost Assembly of the Drosophila melanogaster Reference Genome Using Low-Coverage, Long-Read Sequencing. G3 Genes|Genomes|Genetics 8, 3143–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’Agostino ERR, Price DK, Waddell P, Lang M, Courtier-Orgogozo V, et al. (2021). Widespread introgression across a phylogeny of 155 Drosophila genomes. Current Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvorov A, Kim BY, Wang J, Armstrong EE, Peede D, D’Agostino ERR, Price DK, Waddell P, Lang M, Courtier-Orgogozo V, et al. (2022). Widespread introgression across a phylogeny of 155 Drosophila genomes. Curr Biol 32, 111–123.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, and Kopp A (2009). Distinct developmental mechanisms underlie the evolutionary diversification of Drosophila sex combs. Proceedings of the National Academy of Sciences 106, 4764–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Barmina O, Sanders LE, Arbeitman MN, and Kopp A (2011). Evolution of Sex-Specific Traits through Changes in HOX-Dependent doublesex Expression. PLoS Biol 9, e1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner JC, Swanger E, and Zuk M (2019). Sexual signal loss in field crickets maintained despite strong sexual selection favoring singing males. Evolution 73, 1482–1489. [DOI] [PubMed] [Google Scholar]
- Tarasov A, Vilella AJ, Cuppen E, Nijman IJ, and Prins P (2015). Sambamba: fast processing of NGS alignment formats. Bioinformatics 31, 2032–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turelli M, Cooper BS, Richardson KM, Ginsberg PS, Peckenpaugh B, Antelope CX, Kim KJ, May MR, Abrieux A, Wilson DA, et al. (2018). Rapid Global Spread of wRi-like Wolbachia across Multiple Drosophila. Curr Biol 28, 963–971.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui-Aoki K, Ito H, Ui-Tei K, Takahashi K, Lukacsovich T, Awano W, Nakata H, Piao ZF, Nilsson EE, Tomida J, et al. (2000). Formation of the male-specific muscle in female Drosophila by ectopic fruitless expression. Nat Cell Biol 2, 500–506. [DOI] [PubMed] [Google Scholar]
- Valencia-Montoya WA, Quental TB, Tonini JFR, Talavera G, Crall JD, Lamas G, Busby RC, Carvalho APS, Morais AB, Oliveira Mega N, et al. (2021). Evolutionary trade-offs between male secondary sexual traits revealed by a phylogeny of the hyperdiverse tribe Eumaeini (Lepidoptera: Lycaenidae). Proceedings of the Royal Society B: Biological Sciences 288, 20202512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaser R, Sović I, Nagarajan N, and Šikić M (2017). Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 27, 737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury JA, Jackson LL, and Schedl P (1999). Analysis of the doublesex female protein in Drosophila melanogaster: role on sexual differentiation and behavior and dependence on intersex. Genetics 152, 1653–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler J, Delaney EK, Belles X, Schal C, Wada-Katsumata A, Amicucci MJ, and Kopp A (2019). Hemimetabolous insects elucidate the origin of sexual development via alternative splicing. Elife 8, e47490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler MR (1949). Taxonomic studies on the Drosophilidae. Univ. Texas Publs 4920, 157–195. [Google Scholar]
- Wiens JJ (1999). Phylogenetic evidence for multiple losses of a sexually selected character in phrynosomatid lizards. Proceedings of the Royal Society of London. Series B: Biological Sciences 266, 1529–1535. [Google Scholar]
- Wiens JJ (2001). Widespread loss of sexually selected traits: how the peacock lost its spots/.Trends Ecol Evol 16, 517–523. [Google Scholar]
- Williams TM, and Carroll SB (2009). Genetic and molecular insights into the development and evolution of sexual dimorphism. Nature Reviews 10, 797–804. [DOI] [PubMed] [Google Scholar]
- Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, and Carroll SB (2008). The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 134, 610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong BBM, and Rosenthal GG (2006). Female disdain for swords in a swordtail fish. Am Nat 167, 136–140. [DOI] [PubMed] [Google Scholar]
- Yang Z (2007). PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol 24, 1586–1591. [DOI] [PubMed] [Google Scholar]
- Zander CD, and Dzwillo M (1969). Untersuchungen zur Entwicklung und Vererbung des Caudalfortsatzes der Xiphophorus-Arten (Pisces). Zeitschr. Wissensch. Zool. 178, 275–315. [Google Scholar]
- Zhang WX, and Liang XC (1992). Seven new species of the subgenus Lordiphosa of Drosophila (Diptera: Drosophilidae). Acta Zoologica Sinica 17, 473–482. [Google Scholar]
- Zuk M, Rotenberry JT, and Tinghitella RM (2006). Silent night: adaptive disappearance of a sexual signal in a parasitized population of field crickets. Biol Lett 2, 521–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


