Sugarcane is a major food and bioenergy crop globally. It produces ~80% of sugar consumed worldwide, with Brazil and India together accounting for 61% of world sugarcane production in 2021 [1]. Globally, sugarcane is the 5th largest crop by production value and acreage, and it is also the second largest bioenergy crop [1,2]. Modern sugarcane is an interspecific hybrid (Saccharum species hybrid) of wild progenitor species Saccharum officinarum (2n = 80; x = 10) and Saccharum spontaneum (2n = 40 to 130; x = 8) [3]. This genetically complex polyploid crop with varied chromosome numbers (100 to 130) has one of the largest genomes (~10 kb) among plants, making sugarcane breeding considerably slow and challenging.
Sugarcane breeding involves visual clonal selection combined with manual screening for cane stalk weight and cane sugar content through a 10- to 12-year-long multistage selection scheme with disease screening incorporated toward the end of the selection program. Globally, the rate of sugarcane yield improvement realized at commercial crop production level through breeding in recent decades remains considerably lower than that of other major crops, and in some breeding programs, genetic gain appears to have plateaued [1].
Long breeding cycle, practical difficulties for extensive phenotyping of breeding populations, low narrow-sense heritability of economically important traits, large complex polyploid genome with high heterozygosity, and genotype–environment–management interaction effects have been attributed to low rate of genetic gain. More specifically, the high biomass of sugarcane plants makes accurate detailed phenotyping logistically very challenging, which compromises selection accuracy. This is particularly so in the early stages of selection confounded by large interplot competition effects caused by small single- or 2-row plots [4]. Thus, accurate, cost-effective, and high-throughput phenotyping offers an excellent opportunity for more precise estimation of true yield potential of sugarcane clones in breeding trials, a major bottleneck for fast-tracking sugarcane improvement [5].
Recognizing the persisting slow yield improvement from sugarcane breeding and the accelerated genetic gains realized through molecular marker-assisted selection (MAS) in various other crops [6,7], some of the leading sugarcane industries invested substantial resources for sugarcane genome sequencing and MAS in the past 3 decades [8]. Over this period, sugarcane DNA marker systems have gradually evolved from the initial hybridization-based [9] to the current DNA-sequence-derived single-nucleotide polymorphism (SNP) markers, facilitated by high-throughput next-generation sequencing technologies [8]. The rapid advancements in DNA sequencing and marker technologies led to the creation of genotyping systems for whole-genome profiling, such as genomic selection (GS), which further strengthened marker discovery and marker-trait association studies. GS is a robust genotyping method capable of using large number of trait-linked DNA makers (e.g., SNP markers) spread across the whole genome and provides a more robust estimation of the genetic merit of a clone (for economically important traits) than previously achieved. Multiple independent studies found that it can be used as an effective high-throughput genetic screen for selecting elite sugarcane clones in breeding programs [8]. Using SNP markers, Deomano et al. [10], Yadav et al. [11], and, more recently, O’Connell et al. [12] proved the relative advantages of GS over conventional selection for cane yield, sugar content, and disease resistance. However, despite the long history of molecular marker discovery, application of MAS, including GS, in sugarcane breeding is yet to be realized.
Reliable, accurate phenotyping of large genetically diverse genotyped populations on a regular basis is required to implement GS in sugarcane breeding. While experimental data and modeling have shown the predictive power (estimation of clone genetic merit) and potential variety development application of GS in sugarcane, its success in breeding for complex quantitative traits such as cane yield and sugar content, which are predominantly controlled by nonadditive genetic effects (i.e., interactions among genes or alleles that are not readily passed on from parents to progeny) [11], depends on, at least in part, accurate, high-throughput, and cost-effective phenotyping. High-throughput phenomics thus will greatly reduce the cost and increase the efficiency of creating reliable GS training sets and phenotypic data required for building statistically robust clone performance prediction models needed for selection [8,10]. Further, on an ongoing basis, these GS prediction models need to be refined with the inevitable change in genetic background of breeding populations and the target production environments with diverse agroecological conditions.
Parallel to genomics technologies, sensor-based phenotyping advanced relatively rapidly in the past 2 decades. High-throughput phenotyping has been used extensively for improving crop agronomy and yield forecasting in many crops including sugarcane [13,14]. However, application of phenomics in commercial crop breeding is in its infancy, despite the fact that phenotyping forms the core of breeding and that it is very resource-intense. Interestingly, at least in grain crops, much of the research driving phenotyping innovations that are likely to transform breeding comes from the application of phenomics to unravel the hitherto intractable aspects of crop physiology and plant development [15]. For instance, phenomics-assisted yield prediction using indirect traits has been established in cereals [16], and its potential breeding applications have been emphasized in recent reviews [15]. Practically, a large number of plant growth, developmental, physiological, and leaf spectral attributes as well as certain tissue chemical parameters associated with biomass accumulation and crop yield can be quantified using high-throughput phenotyping platforms, some of which are now commercially available. However, not all biomass- or yield-linked indirect traits are suitable for clone selection in breeding. Ideally, indirect traits appropriate for breeding application must have at least some of the attributes shown in Figure, including high genetic correlation with and contribution to yield, directly linked to key physiological or developmental processes controlling growth and/or yield, high heritability, large genetic variation, and amenable to high-throughput phenotyping. Thus, selection systems incorporating appropriately chosen indirect yield traits may facilitate greater genetic gains, and economic return as heritability of such traits can be higher than yield itself [5].
Figure.
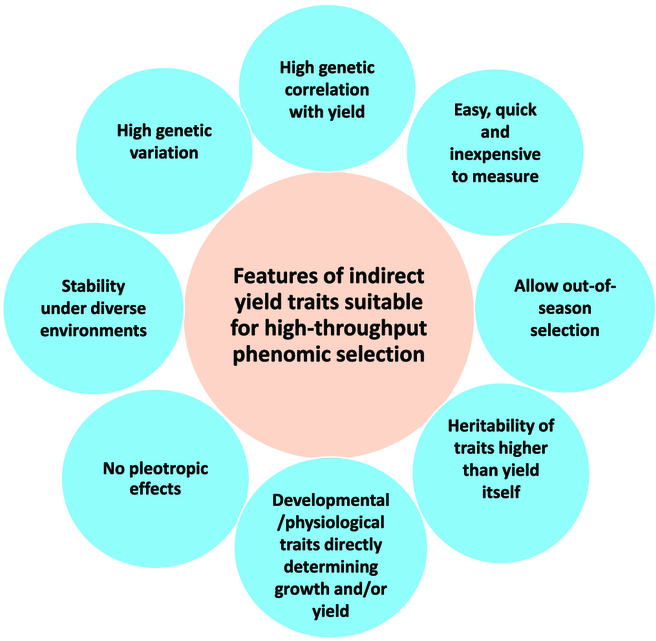
Desirable attributes of yield-associated indirect selection traits with potential for accelerated genetic improvement of sugarcane through breeding. A potential trait used for selection may have one or multiple attributes shown in this figure.
Practically many of the previously intractable indirect yield-related traits are now readily amenable to high-throughput phenotyping. For instance, high-throughput phenotyping of various features of canopy development and canopy architecture, leaf light interception, canopy reflectance, plant height, leaf area, crop growth rate, canopy conductance, canopy temperature, leaf senescence, and photosynthesis has been reported [15]. In addition, many of these surrogate traits have been successfully used to develop prediction models for cereal crop biomass, water use, radiation use efficiency, crop photosynthesis, nitrogen accumulation, and yield [15,17]. Although not advanced to the extent as seen in grain crops, substantial efforts on application of phenomics in sugarcane agriculture have been reported (Table) [5,14,18]. Sugarcane is an ideal crop for field phenomics. Its large, high biomass stem is the harvested plant part, takes 12 to 22 months to mature, grown mainly in rain-fed marginal lands, and, thus, experiences recurrent climatic extremes, especially water deficit, widespread incidence of pests and diseases, and, above all, there is a real need for accurate, high-resolution, high-throughput phenotyping to accelerate genetic gain from sugarcane breeding.
Table.
Traits amenable to high-throughput phenotyping (HTP) and their potential for coapplication with GS for sugarcane breeding. The applicability of HTP and /or GS for these traits has been demonstrated in sugarcane experimentallya.
| Traits | Genotyping technology | Marker systems b | Sensor systems | Application | Genomics-phenomics integration potential | Current status | |
|---|---|---|---|---|---|---|---|
| Genomics | Phenomics | ||||||
| Cane yield | Cane yield | AAS array, DArT, and GBS | SNP markers and DArT markers | Multidimensional RGB, LiDAR, and multispectral imaging | Crop yield improvement, crop growth assessment | High | Limited institutional capacity and marker cost still limit wider application; targeted implementation efforts underway |
| Sugar content | Sugar content | AAS array, DArT, and GBS | SNP markers and DArT markers | NIR | Crop yield improvement | Medium to high | As above, hyperspectral imaging research for HTP |
| Fiber content | Fiber content | AAS array, DArT, and GBS | SNP markers and DArT markers | NIR | Stalk quality and energy cane | Medium to high | Moderate high-throughput currently |
| Stalk height, stalk number, and biomass | Stalk height, stalk number, and biomass | AAS array, DArT, and GBS | SNP markers and DArT markers | LiDAR and multidimensional RGB | Crop yield, stalk weight, rationability, and crop establishment | Medium to high | Used for crop growth monitoring and crop management |
| Canopy cover and canopy architecture | LiDAR and multidimensional RGB | Canopy filling, crop gap, light interception, and water use | Limited genomics research | Extensively used for crop growth modeling and crop monitoring | |||
| Brown rust | Brown rust | AAS array and DArT | Bru gene | RGB and multispectral imaging | Brown rust resistance | High | Currently limited implementation in breeding programs |
| Orange rust, smut, and red rot | AAS array | SNP markers | Diseases resistance | Limited phenomics research | Application of smut markers for GS underway | ||
| Flowering | Flowering | AAS array | SNP markers | RGB and multispectral imaging | Flowering time and flowering propensity | High | Breeding application not reported |
| Canopy conductance and canopy temperature | IR temperature sensors and thermal camera | Crop water use and thermal and water stress tolerance | Little genomics research on this trait | A valuable trait for breeding sugarcane for water-limited environments | |||
| Early-stage crop vigor | RGB and multispectral imaging | Early-stage clone selection in breeding | Little genomics research on this trait | Value of this trait for improving early-stage clone selection demonstrated | |||
These are the most frequently tested sugarcane DNA marker systems, but other polymerase-chain-reaction-based and single-gene marker systems with high-throughput screening potential have been reported for sugarcane by different groups.
AAS array, Affimetrix Axiom SNP array; DArT, Diversity Array Technology; GBS, genotyping by sequencing; RGB, red, green and blue; LiDAR, light detection and ranging; NIR, near-infrared.
Increasing stem biomass and cane sugar content are the ultimate objectives of sugarcane breeding for sugar production, while boosting total above-ground biomass with increased fiber content forms the target for energy cane breeding. Sugarcane growth is highly responsive to N and water supply, and one of the initial and common applications of sugarcane aerial field phenomics was tracking leaf N for N fertilization management [14]. While this agronomic application becomes very popular, its value in breeding comes from the positive relationship of leaf N content, leaf chlorophyll content, and sugarcane radiation use efficiency, which, in turn, determines biomass production. Similarly, research conducted by Basnayake et al. [19] using genetically broad-based sugarcane populations showed that canopy conductance and its surrogate trait canopy temperature have strong genetic correlation with cane yield across a range of growing conditions and that canopy temperature could be used as a predictor of sugarcane yield. Building on these studies, Natarajan et al. [5] constructed an indirect selection index for yield prediction using a multitude of secondary yield traits and showed its superiority over conventional clone selection under single-row and multirow (pure stand) conditions in large commercial sugarcane breeding trials in Australia. This pioneering study clearly established the potential of high-throughput field phenotyping of yield-associated indirect traits for improving clonal selections in sugarcane breeding.
The ongoing progress and investments in next-generation sequencing technologies, genomics, marker discovery, marker systems development, sequencing the genome of sugarcane and related species, bioinformatics and big data analytics, and investments in implementing GS and phenomics in some commercial sugarcane breeding programs are suggesting that MAS and high-throughput field phenomics will become an integral part of sugarcane breeding in at least in large sugarcane industries. Accelerated yield gain from GS-assisted sugarcane selection is very evident [10–12]. Integrating field phenomics into GS-assisted sugarcane breeding will further boost the realized rate of genetic gain and yield improvement. Drought tolerant maize developed for US maize industry forms the first example of discovery to product delivery (drought tolerant maize) from a breeding program integrating genomics, phenomics, and trait and crop modeling [20]. The remarkable progress made in developing sugarcane GS system [8,10–12], high-throughput phenomics-assisted screening of sugarcane for various crop performance traits [5,18], and sugarcane trait/crop modeling [2] now presents a unique opportunity to bring together and exploit these powerful tools and technologies to transform sugarcane breeding for the future.
Acknowledgments
Funding: This study was supported by the grants from National Natural Science Foundation of China (project nos. 32001484, 32201910, and 32260715) by the Ministry of Science and Technology of the People’s Republic of China; Major Research Program Fund (project nos. AB22035028, AA22117002, and 2023GXNSFDA026051) by the Department of Science and Technology, Guangxi Provincial Government, Guangxi, and Guangxi Academy of Agricultural Sciences; and China Agriculture Research System Projects (project nos. CARS-170107 and CARS-170711). Author contributions: T.L., X.L., and P.L. conceived the study. T.L. and X.L. prepared the first draft. P.L. revised and finalized the manuscript. Competing interests: The authors declare that they have no competing interests.
References
- 1.FAO, FAOSTAT database. 2023. http://www.fao.org/faostat/en/#data/QC.
- 2.Singels A, Jones MR, Lumsden TG. Potential for sugarcane production under current and future climates in South Africa: Sugar and ethanol yields, and crop water use. Sugar Tech. 2023;25(2):473–481. [Google Scholar]
- 3.Ram B, Hemaprabha G, Singh BD, Appunu C. History and current status of sugarcane breeding, germplasm development and molecular biology in India. Sugar Tech. 2022;24(1):4–29. [Google Scholar]
- 4.Jackson P, McRae TA. Selection of sugarcane clones in small plots: Effects of plot size and selection criteria. Crop Sci. 2001;41(2):315–322. [Google Scholar]
- 5.Natarajan S, Basnayake J, Wei X, Lakshmanan P. High-throughput phenotyping of indirect traits for early-stage selection in sugarcane breeding. Remote Sensing. 2019;11(24): Article 2952. [Google Scholar]
- 6.Collard BC, Mackill DJ. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos Trans R Soc Lond Ser B Biol Sci. 2008;363(1491):557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Varshney RK, Bohra A, Yu J, Graner A, Zhang Q,Sorrells ME. Designing future crops: Genomics-assisted breeding comes of age. Trends Plant Sci. 2021;26(6):631–649. [DOI] [PubMed] [Google Scholar]
- 8.Aitken KS. History and development of molecular markers for sugarcane breeding. Sugar Tech. 2022;24(1):341–353. [Google Scholar]
- 9.Da Silva JA, Sorrells ME, Burnquist WL, Tanksley SD. RFLP linkage map and genome analysis of Saccharum spontaneum. Genome. 1993;36(4):782–791. [DOI] [PubMed] [Google Scholar]
- 10.Deomano E, Jackson P, Wei X, Aitken K, Kota R, Pérez-Rodríguez P. Genomic prediction of sugar content and cane yield in sugar cane clones in different stages of selection in a breeding program, with and without pedigree information. Mol Breed. 2020;40(4): Article 38. [Google Scholar]
- 11.Yadav S, Jackson P, Wei X, Ross EM, Aitken K, Deomano E, Atkin F, Hayes BJ, Voss-Fels KP. Accelerating genetic gain in sugarcane breeding using genomic selection. Agronomy. 2020;10(4): Article 585. [Google Scholar]
- 12.O'Connell A, Deo J, Deomano E, Wei X, Jackson P, Aitken KS, Manimekalai R, Mohanraj K, Hemaprabha G, Ram B, et al. Combining genomic selection with genome-wide association analysis identified a large-effect QTL and improved selection for red rot resistance in sugarcane. Front Plant Sci. 2022;13: Article 1021182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roitsch T, Cabrera-Bosquet L, Fournier A, Ghamkhar K, Jiménez-Berni J, Pinto F, Ober ES. Review: New sensors and data-driven approaches—A path to next generation phenomics. Plant Sci. 2019;282:2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia AP, Umezu CK, Moriones Polania EC, Dias Neto AF, Rossetto R, Albiero D. Sensor-based technologies in sugarcane agriculture. Sugar Tech. 2022;24(3):679–698. [Google Scholar]
- 15.Furbank RT, Jimenez-Berni JA, George-Jaeggli B, Potgieter AB,Deery DM. Field crop phenomics: Enabling breeding for radiation use efficiency and biomass in cereal crops. New Phytol. 2019;223(4):1714–1727. [DOI] [PubMed] [Google Scholar]
- 16.Messina CD, Podlich D, Dong Z, Samples M, Cooper M. Yield-trait performance landscapes: From theory to application in breeding maize for drought tolerance. J Exp Bot. 2011;62(3):855–868. [DOI] [PubMed] [Google Scholar]
- 17.Deery DM, Jones HG. Field phenomics: Will it enable crop improvement? Plant Phenomics. 2021;2021: Article 9871989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Phuphaphud A, Saengprachatanarug K, Posom J, Taira E, Panduangnate L. Prediction and classification of energy content in growing cane stalks for breeding programmes using visible and shortwave near infrared. Sugar Tech. 2022;24(5):1497–1509. [Google Scholar]
- 19.Basnayake J, Jackson PA, Inman-Bamber NG, Lakshmanan P. Sugarcane for water-limited environments. Variation in stomatal conductance and its genetic correlation with crop productivity. J Exp Bot. 2015;66(13):3945–3958. [DOI] [PubMed] [Google Scholar]
- 20.Cooper M, Gho C, Leafgren R, Tang T, Messina C. Breeding drought-tolerant maize hybrids for the US corn-belt: Discovery to product. J Exp Bot. 2014;65(21):6191–6204. [DOI] [PubMed] [Google Scholar]


