Abstract
Coxiella burnetii is a Gram-negative intracellular pathogen that causes the debilitating disease Q fever, which affects both animals and humans. The only available human vaccine, Q-Vax, is effective but has a high risk of severe adverse reactions, limiting its use as a countermeasure to contain outbreaks. Therefore, it is essential to identify new drug targets to treat this infection. Macrophage infectivity potentiator (Mip) proteins catalyse the folding of proline-containing proteins through their peptidyl prolyl cis-trans isomerase (PPIase) activity and have been shown to play an important role in the virulence of several pathogenic bacteria. To date the role of the Mip protein in C. burnetii pathogenesis has not been investigated. This study demonstrates that CbMip is likely to be an essential protein in C. burnetii. The pipecolic acid derived compounds, SF235 and AN296, which have shown utility in targeting other Mip proteins from pathogenic bacteria, demonstrate inhibitory activities against CbMip. These compounds were found to significantly inhibit intracellular replication of C. burnetii in both HeLa and THP-1 cells. Furthermore, SF235 and AN296 were also found to exhibit antibiotic properties against both the virulent (Phase I) and avirulent (Phase II) forms of C. burnetii Nine Mile Strain in axenic culture. Comparative proteomics, in the presence of AN296, revealed alterations in stress responses with H2O2 sensitivity assays validating that Mip inhibition increases the sensitivity of C. burnetii to oxidative stress. In addition, SF235 and AN296 were effective in vivo and significantly improved the survival of Galleria mellonella infected with C. burnetii. These results suggest that unlike in other bacteria, Mip in C. burnetii is required for replication and that the development of more potent inhibitors against CbMip is warranted and offer potential as novel therapeutics against this pathogen.
Author summary
The bacterium Coxiella burnetii is a highly infectious organism that is found worldwide and can cause debilitating disease in both animals and humans. Treatment of chronic infections remains very difficult and therefore it is essential to identify new drug targets to treat this infection. The Macrophage infectivity potentiator (Mip) protein has been reported as an attractive anti-virulence target in several pathogenic bacteria but its role has not been investigated in C. burnetii. This study demonstrates that inhibitors of the Mip protein from C. burnetii reduce replication inside host cells, increase the sensitivity of the bacteria to oxidative stress, and exhibit antibiotic properties against both the virulent and avirulent forms of C. burnetii. Furthermore, using a moth larvae infection model, the inhibitors demonstrated protective activity. These results suggest that, unlike other studied bacterial pathogens, C. burnetii requires Mip for replication, and that the further development of potent inhibitors against Mip offer potential as novel therapeutics to treat this infection.
Introduction
Q fever is a worldwide zoonotic disease caused by the bacterial pathogen Coxiella burnetii. The bacterium is highly infectious, with the infectious dose for humans estimated to be as low as 1–10 organisms [1]. Infection with C. burnetii can be asymptomatic and self-limiting. However, approximately 40% of infected individuals present with a severe flu-like illness lasting several weeks. In a minority of cases (~2%) acute infection can develop into persistent focal infections that can lead to serious health complications including endocarditis and vascular infections that will ultimately result in death if left untreated [2]. Treatment of acute infection is usually successful, however the two week recommended doxycycline treatment for acute disease [3,4] is sometimes poorly tolerated [2,5,6]. Complicating this treatment is that diagnosis and clearance of persistent infections is difficult [4,7,8]. Once persistent C. burnetii infection is diagnosed, the prescribed treatment is a combination of doxycycline and hydroxychloroquine lasting 18 or more months. Furthermore, approximately 20% of patients who recover from acute infection suffer for years from chronic fatigue, a condition known as Q fever fatigue syndrome that can severely impact their quality of life [9–12]. An effective human vaccine is available, (Q-VAX, Seqirus Australia), but it is only licenced in Australia and requires pre-vaccination screening as there are severe adverse reactions to the vaccine if the individual has immunity as a result of previous exposure to C. burnetii [13]. This severely limits the use of Q-VAX in the event of an outbreak. Consequently, there is a need to identify new approaches to treat this infection. This can be achieved through gaining a better understanding of both the bacterium’s biology and how it causes disease.
C. burnetii is a Gram-negative obligate intracellular pathogen with a biphasic lifecycle. The organism is shed into the environment by ruminant hosts, such as goats and sheep, in faeces, urine, milk and especially in birth products. The metabolically dormant and highly resilient small cell variant (SCV) of C. burnetii can persist from months to years in the environment [2,14,15]. Upon inhalation of contaminated aerosols, the SCVs are phagocytosed by resident alveolar macrophages and are trafficked through the endocytic pathway. Acidification of the phagocytic vesicle upon lysosomal fusion triggers transition of the SCV into the metabolically active large cell variant (LCV). This activates the Type IV Secretion System (T4SS) [16,17], which is functionally analogous to the Dot/Icm T4SS of Legionella pneumophila [18,19]. Over 130 T4SS effectors have been identified and they act to subvert multiple host pathways. Of particular importance are autophagy and vesicle trafficking, making the phagolysosome compartment, now termed a Coxiella-containing vacuole (CCV), permissive for C. burnetii replication [20–22]. The mature CCV is highly fusogenic, expanding to form one large CCV that can occupy almost the entire cell and support bacterial replication. As the CCV fills with bacteria, the LCVs transition back to the more stable SCVs which are then eventually released by a poorly defined egress [23]. During the replication process, C. burnetii also uses T4SS effector proteins to inhibit programmed cell death, one of several host defence mechanisms used to control pathogen replication [24–26]. In addition to the T4SS, C. burnetii has several other strategies to counteract host defence mechanisms. The bacteria are able to withstand the acidic conditions within the lysosome and resist the action of cationic peptides and lysosomal hydrolases [27]. These organisms are also able to resist macrophage killing mechanisms that involve the release of highly reactive oxidative species. C. burnetii suppress oxidative bursts [28], by expressing radical degrading enzymes and by expressing multiple DNA repair enzymes that serve to maintain genome integrity [29–31].
A major virulence associated protein that has been shown to be important in the pathogenesis of several Gram-negative pathogens is the Macrophage infectivity potentiator (Mip) protein [32,33]. Mip proteins belong to the peptidyl-prolyl cis-trans isomerase (PPIase) superfamily of proteins, also known as immunophilins, which catalyse the slow cis-trans isomerization of prolyl bonds and thereby increase the rate of protein folding. Gene deletion of mip in Legionella spp. significantly reduces the invasion and intracellular replication in human phagocytic cell lines and amoebae [34–39]. Significantly, Mip is essential for full virulence in vivo, with the L. pneumophila mip mutant reported to have reduced survival in a guinea pig infection model [40–42]. A similar report supporting the role of Mip in virulence is demonstrated in Burkholderia pseudomallei, where the loss of Mip results in decreased invasion and survival in J774A.1 macrophages and attenuation in the BALB/c mouse infection model [43].
In recent years immunophilin proteins have received considerable attention as druggable targets [44–47] and consequently there has been increased interest in targeting Mip as a therapeutic option against pathogenic bacteria [32,48–51]. Mip belongs to the FK506 binding proteins (FKBPs) subclass of the immunophilin superfamily, and like other FKBPs, Mip is inhibited by the macrolide antibiotics FK506 and rapamycin. However, the immunosuppressive activity of these compounds makes them unacceptable for use as therapeutic interventions [32]. Several recent publications have reported the synthesis and in vitro testing of Mip inhibitors which contain the important inhibitory component, pipecolic acid, found in FK506 and rapamycin, but lack their immunosuppressive properties. Compounds active against Mips from Chlamydia trachomatis, Neisseria gonorrhoeae, Neisseria meningitidis, and B. pseudomallei have been described, with some showing anti-virulence effects against these organisms [48,50,52–54]. More recently, a further refined subset of pipecolic acid based Mip inhibitors has been developed. These inhibitors have broad spectrum activity against Mip from multiple pathogens including B. pseudomallei, N. meningitidis, Klebsiella pneumoniae and the parasite Leishmania major [55].
The Mip homologue in C. burnetii (CbMip) was first described and characterized by Mo et al. [56]. The CbMip catalytic domain has a high degree of sequence similarity to characterized Mip proteins of other intracellular Gram-negative pathogens including L. pneumophila, B. pseudomallei and C. trachomatis (S1 Fig) [35,43,56,57]. Consequently, CbMip has been considered a virulence factor of C. burnetii [37,58,59]. However, the role of CbMip in C. burnetii pathogenesis has not been directly investigated. In this study, using both the virulent and avirulent forms of C. burnetii Nine Mile RSA439 (C. burnetii NMI and C. burnetii NMII, respectively), the role of the CbMip in C. burnetii pathogenesis, including intracellular replication and growth in axenic media, was investigated. The potential for using inhibitors targeting CbMip to modulate C. burnetii virulence in Galleria mellonella larvae was also explored. The results suggest that unlike in other bacteria, Mip in C. burnetii is required for replication and that inhibitors against CbMip offer potential as novel therapeutics against the bacterium.
Results
Recombinant CbMip PPIase activity is inhibited by SF235 and ANCH37
To first investigate if CbMip has a role in virulence of C. burnetii, validated Mip inhibitors SF235, ANCH37 and AN296 (Fig 1) were utilised, following approaches used to study the role of Mip in another obligate intracellular pathogen, C. trachomatis [54,60]. Although SF235 and ANCH37 are pipecolic acid derivatives that have been optimised against Mip from B. pseudomallei they also display activity against Mips from other bacterial species [50,55,61]. ANCH37 (Fig 1B), like SF235, contains the pipecolic sulphonamide and an amide-based linker to a pyridyl unit, but also contains an additional benzyl moiety which has been shown to increase potency. Initially, the activity of SF235 and ANCH37 against recombinantly expressed CbMip was assessed using a high throughput peptidyl-prolyl cis-trans isomerase assay [62]. Both compounds were found to be inhibitors of CbMip with Ki values of 18±4 μM for SF235 and 4.6±0.9 μM for ANCH37 (S2 Fig). ANCH37 is a racemic mixture consisting of two stereoisomers. The most active compound within the mixture, AN296 (Fig 1C), was prepared [61] and used in the following studies.
Fig 1. Structures of pipecolic acid-based Mip inhibitors, SF235, ANCH37 and AN296.
The synthesis of these inhibitors has been described elsewhere [50,61]).
CbMip inhibitors SF235 and AN296 reduce intracellular replication of C. burnetii
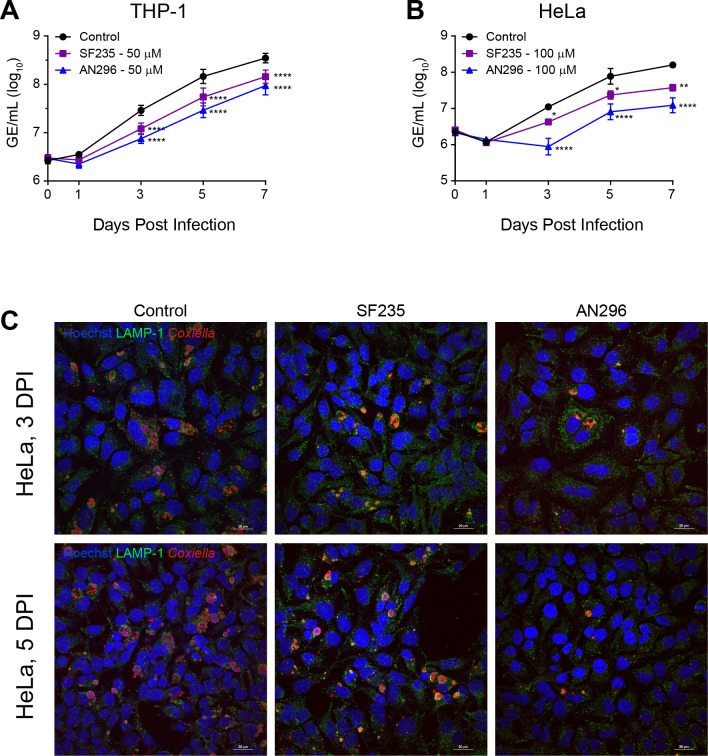
Having confirmed that SF235 and AN296 are potent inhibitors of CbMip, their effect on the intracellular replication of C. burnetii was investigated. C. burnetii NMII was first pretreated with SF235 or AN296 for 1 h, to facilitate complete inhibition of CbMip including any enzyme that may be localized in the cytoplasm [63], and used to infect differentiated THP-1 macrophage cells or HeLa epithelial cervical cancer cells. C. burnetii replication was then monitored by measuring genome equivalents (GE) over 7 days. In the presence of 50 μM AN296, intracellular replication of C. burnetii NMII within THP-1 cells was significantly inhibited (Fig 2A), resulting in a 74%, 80% and 73% (p < 0.0001) reduction in C. burnetii NMII replication compared to the control, 3, 5, and 7 days post infection, respectively. Although slightly less effective, similar results were observed for SF235, where inhibitor treatment (50 μM) resulted in a 58%, 62% and 59% (p < 0.0001) reduction in C. burnetii NMII replication 3, 5, and 7 days post infection, respectively. C. burnetii NMII replication was also significantly reduced in HeLa cells when cultures were treated with higher concentrations of Mip inhibitor. In the presence of 100 μM AN296, intracellular replication of C. burnetii NMII was significantly inhibited in HeLa cells, resulting in a 92%, 90% and 92% (p < 0.0001) reduction in C. burnetii NMII replication 3, 5, and 7 days post infection, respectively (Fig 2B). Furthermore, compared to the untreated control, treatment with 100 μM AN296 appeared to delay the replication kinetics of C. burnetii NMII within HeLa cells by two days. The effect on C. burnetii NMII replication in HeLa cells treated with 100 μM of SF235 was less pronounced but still significant and resulted in a 62%, 70% (p < 0.05) and 76% (p < 0.01) reduction in C. burnetii NMII replication at 3, 5, and 7 days post infection compared to control. HeLa cells were also analysed by immunofluorescence staining at day 3 and 5 post infection and interestingly showed that CCVs were smaller in the presence of SF235 or AN296 compared to the control (Fig 2C). The infection experiments were repeated in THP-1 cells without pre-exposing C. burnetii to the CbMip inhibitors. Addition of 50 μM AN296 after the initial 4 h infection period also significantly inhibited C. burnetii NMII replication within THP-1 cells (S3 Fig), resulting in a 71%, 69% and 63% (p < 0.0001) reduction in C. burnetii NMII replication at 3, 5, and 7 days post infection, respectively. The antibiotic chloramphenicol (31 μM), a potent inhibitor of bacterial protein synthesis and C. burnetii replication [64], was used as a positive control. As expected, addition of chloramphenicol completely inhibited C. burnetii NMII replication within THP-1 cells (S3 Fig). Both SF235 and AN296 were also considered non-toxic to the cells over the course of the infection assay at the concentrations used in each cell-based assay (S4 Fig).
Fig 2. Inhibitors of CbMip affect intracellular replication of C. burnetii.
(A and B) Intracellular replication of C. burnetii NMII in the presence of CbMip inhibitor SF235 (purple square), AN296 (blue triangle) or vehicle control (black circle). (A) THP-1 cells with 50 μM of inhibitor (n ═ 6) and (B) HeLa cells with 100 μM of inhibitor (n ═ 3). Error bars represent standard error of the mean. *, p < 0.05: **, p < 0.01; ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test. (C) Representative confocal immunofluorescence (IF) images at 3 and 5 days post-infection (DPI) for HeLa cells. Cells were stained with anti-LAMP1 (green), anti-Coxiella (red), and Hoechst 33258 (blue). Scale bar ═ 20 μm.
The potential essential nature of Mip in C. burnetii
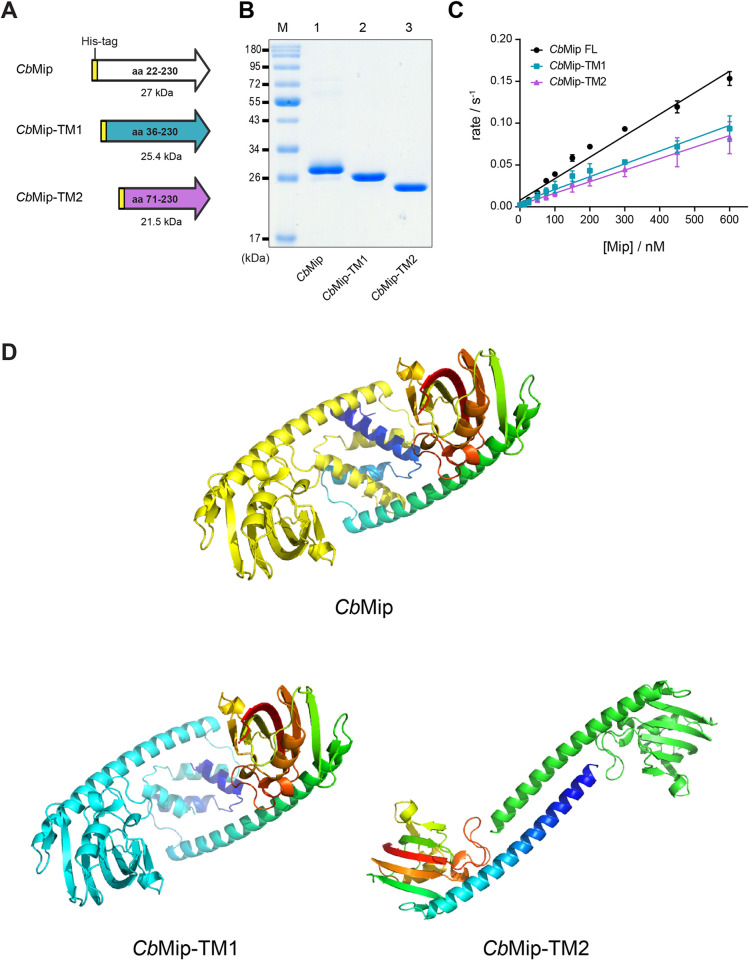
The inhibitory effect of AN296 and SF235 on C. burnetii intracellular replication suggested that Mip may be essential in C. burnetii. To address this directly, attempts were made to delete the mip gene (cbu0630, cbmip) from C. burnetii NMII. However, despite obtaining integration of the pJC-CAT suicide plasmid, all attempts to replace cbmip with a kanamycin resistance cassette were unsuccessful. Furthermore, supporting this hypothesis that cbmip is an essential gene, a transposon insertion library of C. burnetii NMII previously generated by Metters et al. [65] was interrogated for the presence of cbmip insertion mutants. Analysis of the sequencing data revealed two cbmip transposon mutants (S5 Fig), the same number of mutants that would have been expected by chance given the library depth (personal comms; Georgie Metters). Interestingly, both transposon insertion sites were located within the first 200-bp of cbmip and, in both instances, the first ATG sequence following the insertion site was in-frame with the original gene sequence leaving an intact catalytic domain (S5C Fig). The predicted CbMip peptide sequences from these cbmip transposon mutant variants were recombinantly expressed (Fig 3A and 3B) and shown to have retained 50–60% of their PPIase activity (Fig 3C). The kcat/KM values were determined to be, 1.53 ± 0.06 x 105 s-1 M-1, and 1.38 ± 0.07 x 105 s-1 M-1 for the truncated proteins, CbMip-TM1 and CbMip-TM2, respectively as compared to 2.57 ± 0.08 x 105 s-1 M-1 for CbMip. Modelling of CbMip-TM1 and CbMip-TM2 using AlphaFold [66] (Fig 3D) demonstrated that the PPIase domain is likely intact in both truncated proteins, and that CbMip-TM1 may retain dimer formation. However, CbMip-TM2 lacks most of the dimerization domain, and several alternative dimer architectures were modelled. These data suggest that for both cbmip transposon mutants identified in the Metters et al. library, it is very likely that a truncated, but functional, CbMip protein is expressed by these mutants and therefore they do not have a true mip gene disruption, suggesting that CbMip may be essential for C. burnetii growth.
Fig 3. Activity of truncated CbMip proteins.
(A) Schematic diagram of recombinantly expressed truncated CbMip variants. CbMip-TM1 and CbMip-TM2 are truncated CbMip variants that are predicted to be expressed by the C. burnetii cbmip transposon mutants. All truncated CbMip variants contain an N-terminal His-tag like the full length CbMip to facilitate protein purification. (B) Representative Coomassie stained SDS-PAGE gel of purified recombinant truncated CbMip proteins. CbMip proteins were purified by nickel affinity chromatography followed by size exclusion chromatography (SEC) purification. Lane M, Prestained Protein Standard (New England Biolabs) was included for approximation of molecular weight. Lane 1, the expected molecular weight of full length CbMip on a 12% SDS-PAGE gel is 27 kDa; Lane 2, CbMip-TM1 is 25.4 kDa; Lane 3, CbMip-TM2 is 21.5 kDa. (C) kcat/KM of full length and truncated variants of CbMip. The rate of each Mip protein was tested at a range of concentrations and each concentration was tested three times. The kcat/KM values were determined to be 2.57 ± 0.08 x 105 s-1 M-1, 1.53 ± 0.06 x 105 s-1 M-1, and 1.38 ± 0.07 x 105 s-1 M-1 for CbMip, and the truncates CbMip-TM1 and CbMip-TM2, respectively. Error bars show standard errors. (D) The structure of CbMip and the truncated variants CbMip-TM1 and CbMip-TM2 were predicted using AlphaFold2, using the multimer prediction option to model dimers. Structures are presented in cartoon format with one protomer in a single colour (yellow for CbMip, cyan for CbMip-TM1, green for CbMip-TM2) and the other as a rainbow from the N-terminus (blue) to C-terminus (red). Five replicates of the CbMip and CbMip-TM1 models gave very similar structures and dimer architectures. In contrast, five models of CbMip-TM2 gave very similar structures for the FKBP domain, but different architectures for a dimer, indicating that this truncated protein is likely to be monomeric. Images generated using PyMOL v. 2.5.3 (Schrödinger).
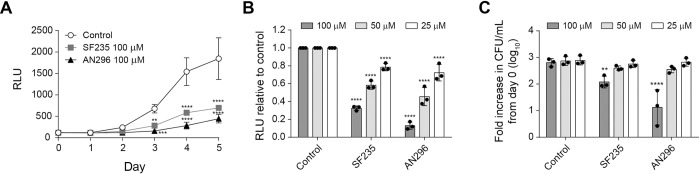
CbMip inhibitors reduce C. burnetii NMII growth in axenic culture in a dose dependant manner
To investigate whether Mip inhibition directly affected C. burnetii growth, a luciferase-expressing derivative of C. burnetii NMII, C. burnetii-lux, was evaluated in axenic media in the presence of different concentrations of SF235 and AN296. In the first instance, due to the pH of the axenic media (4.75) and that the compounds both contain ester linkages, the stability of the compounds was investigated by HPLC chromatography. Using AN296 as an exemplar, it was found to be stable under the acidic conditions of the assay over the entire 7-day period studied (S6 Fig). Proceeding with the axenic experiments, luciferase activity of C. burnetii-lux was inhibited in a dose-dependent manner compared to the control (Figs 4A and S7). Following normalization to DMSO, the presence of SF235 reduced C. burnetii-lux bioluminescence by 68% and 42% (p < 0.0001) at 100 μM and 50 μM, respectively. Treatment with AN296 reduced bioluminescence by 87% and 55% (p < 0.0001) at 100 μM and 50 μM, respectively (Fig 4B). To confirm that the compounds were affecting C. burnetii growth and not simply preventing proper folding and function of the luciferase, the experiment was repeated using wild-type C. burnetii NMII and colony forming units (CFU) were enumerated after 4 days of culture. The fold-increase in C. burnetii NMII CFU/mL (log10) compared to day 0, in the presence of 100 μM or 50 μM of CbMip inhibitor was 19% (p < 0.01) and 54% of the control, respectively, for SF235, and 2.1% (p < 0.0001) and 48% of the control, respectively, for AN296 (Fig 4C). This data demonstrates that SF235 and AN296 directly limit the ability of C. burnetii to replicate in axenic media.
Fig 4. Targeted inhibition of CbMip reduces C. burnetii replication in axenic media in a dose-dependent manner.
(A) Bioluminescent was measured as an indicator of C. burnetii-lux replication. C. burnetii-lux was inoculated at a concentration of 1 × 106 GE/mL into ACCM-2 media with 100 μM of CbMip inhibitors SF235 (grey square), AN296 (closed triangle) or vehicle control (open circle), and growth was monitored over 5 days. Data is presented as RLU (relative light units) with error bars representing the standard deviation (SD) from three independent experiments. (B) C. burnetii-lux replication over 5 days in the presence of 25 μM, 50 μM or 100 μM of SF235 or AN296. Data is presented as RLU relative to the vehicle control with error bars representing SD from at least three independent experiments. (C) Colony forming units per mL was determined for C. burnetii NMII after 4 days of growth in the presence of 25 μM, 50 μM or 100 μM of SF235 or AN296. Data is presented as the fold increase in CFU/mL relative to day 0 with error bars representing the SD from at least three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA followed by Dunnett’s multiple comparison post-test.
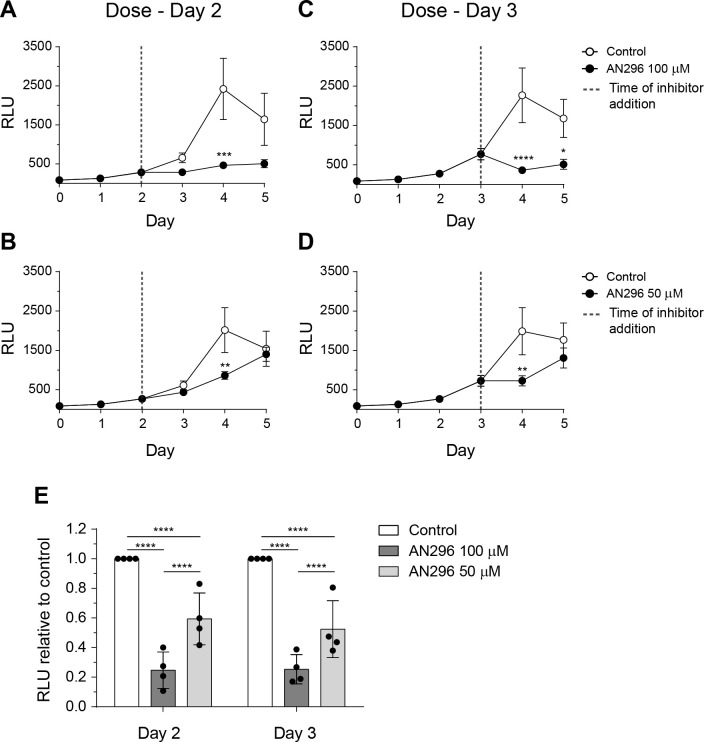
The potential of Mip inhibitors to act on exponentially growing C. burnetii cultures was also investigated using the most effective compound AN296 (Fig 5). Addition of AN296 to the culture on either day 2 (start of logarithmic growth) (Fig 5A and 5B) or day 3 (mid-logarithmic growth) (Fig 5C and 5D) [67] resulted in a significant decrease in bioluminescence. Following normalization to the control, addition of AN296 on day 2 reduced C. burnetii-lux bioluminescence by 75% and 41% (p < 0.0001) at 100 μM and 50 μM (Fig 5E), respectively. Similarly, addition of AN296 on day 3 reduced bioluminescence by 75% and 48% (p < 0.0001) at 100 μM and 50 μM (Fig 5E), respectively. These data demonstrate that AN296 is effective at suppressing the growth of both starter cultures that are predominantly composed of C. burnetii SCVs and actively growing cultures which contain C. burnetii cells that are predominantly LCVs [68]. Together these data suggest that the presence of CbMip inhibitors perturb the ability of C. burnetii to replicate both intracellularly and in axenic media.
Fig 5. Delayed dosing with AN296 impairs C. burnetii replication in axenic media.
Bioluminescence was measured as an indicator of C. burnetii-lux replication. The strain was inoculated at a concentration of 1 × 106 GE/mL into ACCM-2 media and growth was monitored over 5 days. Cultures were dosed with (A+B) 100 μM or (C+D) 50 μM of AN296 (closed circle) or vehicle control (open circle) on (A+C) day 2 or (B+D) day 3 of the growth curve. Data is presented as RLU (relative light units) with error bars represent standard error of the mean from four independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Sidak’s multiple comparison post-test. (E) C. burnetii-lux replication after dosing with 50 μM or 100 μM of AN296 on day 2 or day 3 of the 5 day growth curve. Data is presented as RLU relative to the control with error bars representing SD from at least four independent experiments. ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Tukey’s multiple comparison post-test.
Inhibition of CbMip induces changes on the C. burnetii proteome
To elucidate the phenotypic effects induced by the presence of CbMip inhibitors and the inability to generate a cbmip mutant in C. burnetii, a proteomics approach was used to gain insight into the cellular processes and specific proteins impacted by the inhibition of CbMip. Mid-log phase (day 3) grown C. burnetii NMII were exposed to 100 μM of AN296 for 24 h and compared to control treatments. This revealed only modest changes across the proteome with only 18 proteins significantly altered in response to AN296 treatment (Table 1). Analysis of the KEGG Orthology (KO) categorization associated with these proteins revealed that approximately half of the proteins that underwent significant changes were either unclassified or hypothetical proteins (11/18). Of note however is that two of the proteins that have increased relative abundance, CBU1576 and CBU1686, have been previously identified as putative T4SS effector proteins [69–71]. Interestingly, four other proteins with an increase in relative abundance (RecG, RecQ, PriA and UvrD) were classified as being involved in DNA replication, recombination, and repair. The increased relative abundance of several DNA repair enzymes suggests that C. burnetii NMII may be more sensitive to oxidative stress when grown in the presence of AN296.
Table 1. List of proteins that underwent statistically significant alterations within the proteome of C. burnetii NMII grown in axenic cultures after 24 h of exposure to AN296 (100 μM).
White cells indicate relative increase, grey cells indicate relative decrease compared to the culture treated with vehicle control (DMSO).
| KEGG Orthology (KO)1 | CBU number | Gene name | Gene product | Fold change (Log2) | -Log10 Student’s t-test p-value |
|---|---|---|---|---|---|
| Metabolism | |||||
| Amino acid | CBU1839 | Aminobutyraldehyde dehydrogenase | 1.04042 | 3.22445 | |
| LPS biosynthesis | CBU0142 | lpxC | UDP-3-O-acyl-N-acetylglucosamine deacetylase | 1.24371 | 1.61264 |
| Genetic information processing | |||||
| DNA repair and recombination | CBU0305 | recG | ATP-dependent DNA helicase RecG | 1.38111 | 4.54836 |
| CBU0472 | recQ | ATP-dependent DNA helicase | 1.29853 | 2.68489 | |
| CBU1815 | priA | Primosomal protein N’ (ATP-dependent helicase PriA) | 1.31784 | 4.40603 | |
| CBU2054 | uvrD | DNA helicase II | 1.69821 | 2.45767 | |
| Signalling and cellular processes | |||||
| Other | CBU1490 | higA | Virulence-associated protein I | 1.77358 | 1.92185 |
| Unclassified and Hypothetical proteins | |||||
| CBU0022 | Hypothetical cytosolic protein | 1.10539 | 2.00792 | ||
| CBU0941 | Probable Fe(2+)-trafficking protein | 1.04587 | 1.55144 | ||
| CBU0637 | Coenzyme PQQ synthesis protein C | 1.60759 | 3.47957 | ||
| CBU1576 | Putative T4SS effector protein [71] | 1.11642 | 2.11499 | ||
| CBU1686 | Putative T4SS effector protein [69,70] | 1.17901 | 2.98394 | ||
| CBU1753 | Hypothetical cytosolic protein | 1.23788 | 1.46301 | ||
| CBU2057 | Hypothetical cytosolic protein | 1.1392 | 3.73108 | ||
| CBU0089a | Uncharacterized protein | -2.31933 | 3.5896 | ||
| CBU0110 | Hypothetical exported protein | -1.38947 | 2.67694 | ||
| CBU1095 | Hypothetical exported protein | -1.05242 | 1.663 | ||
| CBU1308 | Phosphohydrolase family protein | -1.25087 | 2.0132 | ||
1 Proteins that were determined to be differentially present by proteomics were manually curated using the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database against the Coxiella burnetii RSA 493 genome (entry number T00124) and assigned a KEGG Orthology (KO). C. burnetii proteins were further clustered according to their predicted functions as reported by the KEGG database.
CbMip inhibition increases C. burnetii sensitivity to oxidative stress
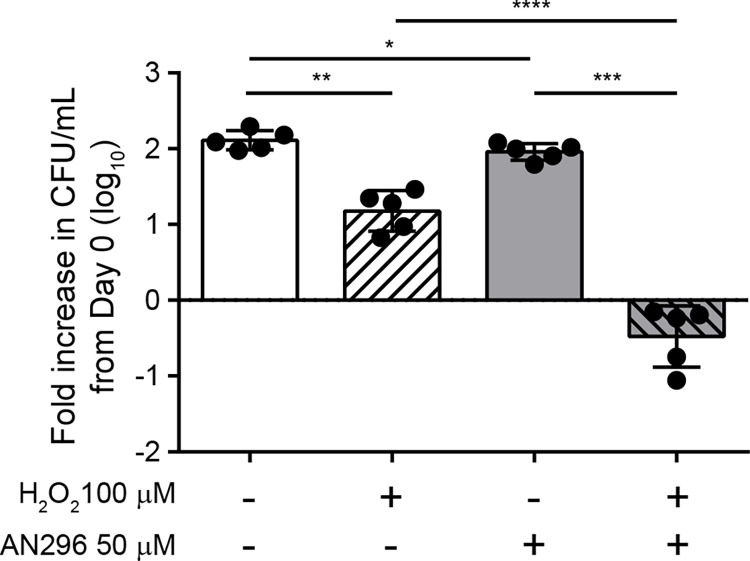
To validate whether Mip inhibition in C. burnetii results in an increased susceptibility to oxidative stress, hydrogen peroxide sensitivity assays were performed. C. burnetii NMII was cultured in axenic media containing no or 100 μM H2O2 and treated with AN296 (50 μM) or DMSO as a control (Fig 6). Enumeration of bacteria after four days of incubation showed that the fold increase in C. burnetii NMII CFU/mL (log10) compared to day 0, was significantly lower for cultures treated with either 100 μM of H2O2 (p < 0.01) or 50 μM AN296 (p < 0.05), which was not unexpected. However, when C. burnetii NMII was exposed to the combined treatment of 100 μM H2O2 and 50 μM AN296 the bacteria failed to replicate in the culture conditions, and lost viability, with an average fold decrease in CFU/mL of 0.48±0.40 (p < 0.0001) (Fig 6). This result demonstrated that the presence of CbMip inhibitor dramatically increases the susceptibility of C. burnetii to oxidative stress.
Fig 6. Inhibition of CbMip increases C. burnetii sensitivity to oxidative stress.
C. burnetii NMII was grown in ACCM-2 media inoculated with either 50 μM AN296 or vehicle control, in the absence or presence of 100 μM H2O2. After 4 days of incubation, cultures were plated out to determine viable colony forming units/mL. Data is presented as the log10 fold increase in CFU/mL from day 0 with error bars representing SD from five independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. p values were determined using one-way ANOVA, followed by Tukey’s multiple comparison post-test.
Inhibition of CbMip stops replication of virulent C. burnetii in axenic culture
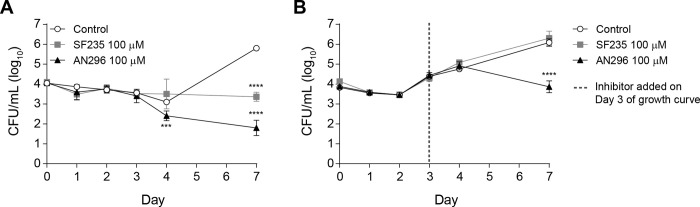
For CbMip inhibitors to be therapeutically useful, they must be active against the virulent, phase I form of C. burnetii. Therefore, the activity of compounds AN296 and SF235 against the phase I parental strain C. burnetii NMI was investigated. The growth of C. burnetii NMI in axenic media while exposed to 100 μM of SF235 or AN296 versus a vehicle control was monitored over 7 days by enumerating CFU/mL for the first 4 days and then on day 7 (Fig 7A). C. burnetii NMI replication was significantly inhibited when grown in the presence of AN296 compared to the control culture at day 4 (p < 0.001) and 7 (p < 0.0001) (Fig 7A). Although slightly less effective, treatment with SF235 also significantly inhibited C. burnetii NMI replication compared to the control on day 7 (p < 0.0001). The inhibitors were also tested against C. burnetii NMI during later stages of growth (Fig 7B). Addition of 100 μM of SF235 to the culture on day 3 had no impact on the growth of C. burnetii NMI compared to the control. However, the addition of 100 μM of AN296 on day 3 significantly inhibited C. burnetii NMI growth compared to the control on day 7 (p < 0.0001). Together these data indicate that SF235 and AN296 also directly impact on the ability of virulent C. burnetii NMI to replicate in axenic media. Given the observed antibacterial effect of AN296 on the growth of C. burnetii NMI, the minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) values were determined for SF235 and AN296 against both the NMI and NMII strains. After 6 days of incubation in ACCM-2 media, the MIC for AN296 was determined to be 100 μM and SF235 exhibited an MIC of 200 μM for both strains (Table 2). The MBCs for both these compounds was determined to be greater than the highest concentration tested (>400 μM). However, significantly fewer bacteria were recovered from cultures treated with 400 μM AN296 compared to cultures treated with SF235, and in both instances the recoverable CFU/mL was lower than those of the starting inoculums.
Fig 7. AN296 is highly potent against virulent C. burnetii NMI in axenic media.
Growth curve of C. burnetii NMI in the presence of CbMip inhibitors. (A) C. burnetii NMI was inoculated at a concentration of 1 × 104 CFU/mL into 5 mL of ACCM-2 media supplemented with 0.50 mM tryptophan and containing 100 μM of CbMip inhibitors, SF235 (grey square), AN296 (closed triangle), or vehicle control (open circle), and growth was monitored over 7 days by enumerating the number of colony forming units per mL in the culture on days 0, 1, 2, 3, 4 and 7. Data is presented as log10 CFU/mL with error bars representing the SD from at least three independent experiments. ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA followed by Dunnett’s multiple comparison post-test. (B) C. burnetii NMI was inoculated at a concentration of 1 × 104 GE/mL into 5 mL of ACCM-2 media supplemented with 0.50 mM tryptophan. After 3 days, cultures were inoculated with 100 μM of CbMip inhibitors, SF235 (grey square), AN296 (closed triangle), or vehicle control (open circle). Growth was monitored over 7 days by enumerating the number of colony forming units per mL in the culture on days 0, 1, 2, 3, 4 and 7. Data is presented as log10 CFU/mL with error bars representing the SD from at least three independent experiments; ****, p < 0.0001. p values were determined using two-way ANOVA followed by Dunnett’s multiple comparison post-test.
Table 2. Antibiotic efficacy of CbMip inhibitors for C. burnetii NMI and C. burnetii NMIIa.
| Compound | C. burnetii NMI | C. burnetii NMII | ||
|---|---|---|---|---|
| MICb | MBCc | MICb | MBCc | |
| SF235 | 100–200 μM | > 200 μM | 100–200 μM | > 400 μM |
| AN296 | 100 μM | > 200 μM | 100 μM | > 400 μM |
aMIC was defined as the lowest concentration of antibiotic required to inhibit bacterial growth in 6 day cultures; MBC was defined as the lowest concentration to give no visible growth when plated onto ACCM-2 agar and incubated for 7 days.
bDetermined by OD in broth culture.
cDetermined by CFU in broth culture.
AN296 reduces intracellular replication of C. burnetii NMII during active infection
Considering the potent effect these CbMip inhibitors had on C. burnetii growth, especially AN296, the therapeutic capacity of this compound was examined further by investigating the effect on C. burnetii NMII replication in THP-1 cells by administering the compound during active infection. Differentiated THP-1 cells were infected with C. burnetii NMII, and AN296 (50 μM) was added to the cell culture medium on day 2 or day 3 of infection. The antibiotic chloramphenicol (31 μM) was used as a positive control. Addition of AN296 even during later stages of C. burnetii NMII infection significantly reduced intracellular replication (Fig 8). Addition of AN296 on day 2 of infection resulted in 60% and 55% (p < 0.0001) reduction in C. burnetii NMII replication compared to the control at 5 and 7 days post infection, respectively (Fig 8A). Addition of AN296 on day 3 of infection resulted in a 47% (p < 0.001) and 53% (p < 0.0001) reduction in C. burnetii NMII replication at 5 and 7 days post infection, respectively (Fig 8B). As expected, addition of chloramphenicol, at day 2 or day 3 of infection stopped further C. burnetii NMII replication (Fig 8). This data shows that inhibitors of CbMip can be used to reduce the replication of C. burnetii NMII during active cell infection.
Fig 8. Delayed dosing with CbMip inhibitors also affects intracellular replication of C. burnetii.
Intracellular replication of C. burnetii NMII in THP-1 cells in the presence of 50 μM AN296 (blue triangle), 31 μM chloramphenicol (Cm) (open circle) or control (closed circle), introduced at (A) day 2 or (B) day 3 of infection. Error bars represent standard error of the mean (n ═ 5). ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test.
AN296 and SF235 improve survival of Galleria mellonella larvae in C. burnetii infection model
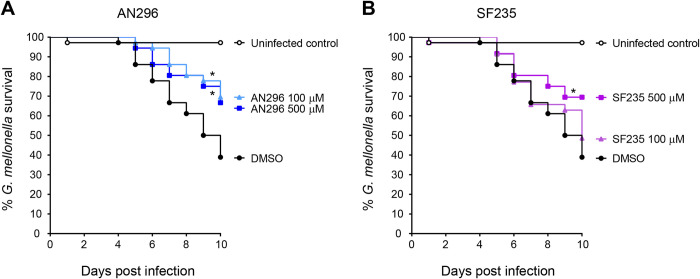
To investigate the efficacy of the Mip compounds in vivo, the Galleria mellonella larva infection model was used. G. mellonella infection with C. burnetii NMII is lethal over a 10-day period and has been used as a model to screen antibiotics [72–74]. Initially AN296 and SF235 were tested for toxicity in G. mellonella larvae. Each larva was inoculated with 10 μL of PBS containing either Mip inhibitor at various concentrations ranging from 10 μM up to 500 μM, or 1% DMSO as a vehicle control. Larvae were monitored daily, for a period of 10 days and demonstrated no significant toxic effects against the compounds at any of the concentrations tested (S8 Fig). From these results, inhibitor concentrations of 100 μM and 500 μM were selected for efficacy studies. Co-administration of AN296 at 100 μM and 500 μM with C. burnetii was found to be protective against C. burnetii induced death as compared to the control (p < 0.05, Fig 9A). Curiously, despite being less effective in assays in vitro, SF235 at 500 μM with C. burnetii was also found to be protective against C. burnetii induced death as compared to the control (p < 0.05, Fig 9B). These studies demonstrate that the use of inhibitors against CbMip is protective against C. burnetii infection in the more complex G. mellonella infection model.
Fig 9. Administration of CbMip inhibitors improves survival of G. mellonella larvae in C. burnetii infection model.
C. burnetii NMII were incubated in PBS containing 1% DMSO and 0, 100 μM or 500 μM of (A) AN296 or (B) SF235 for 1 h at 37°C prior to challenge of G. mellonella larva (n ═ 12 per group) with the mixture. Larvae were challenged with a single dose of 106 GE of C. burnetii NMII with or without CbMip inhibitors and survival was assessed every 24 h for 10 days. Uninfected controls received PBS containing 1% DMSO. Kaplan-Meier survival curves were determined from three independent experiments. Data was analysed using the log-rank (Mantel-Cox) test with Bonferroni correction for multiple comparisons with a significance level of 0.05.
Discussion
The Mip homologue in C. burnetii (CbMip) was first described and characterised by Mo et al. more than 25 years ago [56]. Mature CbMip is approximately 24 kDa in size and has a predicted pI of 10.7. Similar to many other Mip proteins, CbMip consists of two distinct domains, an N-terminal dimerization domain linked by an alpha helix to the C-terminal domain containing the FKBP fold exhibiting PPIase activity which is inhibited by rapamycin [56]. This protein likely forms a homodimer on the outer membrane of the bacterial cell and may also be secreted, at least in axenic media [32,75]. CbMip is reported to be expressed at comparable levels in both LCVs and SCVs [76] and studies focused on developing putative vaccine candidates or serodiagnostic markers of C. burnetii infection have identified CbMip as a highly immunogenic protein in mice and, interestingly, it is seroreactive in Q-fever patients, showing that CbMip is also expressed in vivo [77–80]. However, despite these numerous studies on the CbMip protein, very little is known about the role of the protein in C. burnetii pathogenesis.
Previous studies investigating the functional role of Mip, either through mutagenesis or inhibition, in facultative intracellular pathogens such as L. pneumophila and B. pseudomallei [35,43,54,81], reported that loss of Mip activity had no impact on growth in the cultivation medium. Therefore, it was initially unexpected that C. burnetii growth in axenic media was impacted by the presence of inhibitors for CbMip. However, since C. burnetii is an obligate intracellular pathogen, to facilitate growth outside of cells, ACCM-2 media has been formulated to mimic the intracellular environment and is unlike the highly permissive growth medium used to culture facultative intracellular pathogens. Using inhibitors against CbMip we observed a dose dependent effect on C. burnetii replication in axenic media. These compounds do not merely stop the bacteria from replicating but lead to a loss in bacterial counts suggesting that an outcome of blocking CbMip activity is a bactericidal effect on C. burnetii. This observation may also explain why attempts to mutate cbmip have not been successful. Attempted targeted mutagenesis was unsuccessful after achieving the merodiploid intermediate; normally the most challenging point to reach using the loop in/loop out protocol for targeted gene inactivation in C. burnetii [82]. Resolution of the integrated plasmid through sucrose selection only ever yielded wild-type revertants. It cannot be ruled out that this region of the genome is less amenable to homologous recombination, thereby making the generation of a cbmip mutant an extremely rare event. However, comparison with an alternative mutagenesis approach which relies on a different molecular mechanism for mutagenesis was done. The Himar1-based transposon system developed for C. burnetii mutagenesis [83] involves the use of a transposable element and accompanying transposase to direct random, nonspecific integration of the transposon (Tn) at T/A base pairs, and has proven to be a successful mutagenesis method for generating mutant libraries in C. burnetii [21,65,84]. A Tn mutant of cbmip was not present in the libraries reported by Newton et al. and Martinez et al. however it is noted that although the Martinez library generated over 3,000 mutants, only 26.6% of the CDS were mutated. The more recent 10,000 transposon mutant library generated by Metters et al. [65] identified two transposon hits in the cbmip open reading frame. However, both mutants would permit transcription of a truncated protein with an intact PPIase domain. In this current study, these truncated CbMip variants were shown to be catalytically active, through recombinant expression. This data is consistent with a study by Mo et al. who reported that the cbmip transcript contains two internal translation re-initiation sites to facilitate expression of truncated CbMip analogues with a catalytically active FKBP domain in C. burnetii [63]. These two internal translation re-initiation sites are also retained in the surviving transposon mutants. These data suggest that the mutants in the Metters et al. library retain some CbMip activity and therefore cannot be considered true cbmip mutants. As the truncated CbMips reported here and earlier all lack a signal sequence, this suggests that the essential role of CbMip is intracellular and requires FKBP activity. The absence of a catalytically inactive mutant in published C. burnetii Tn mutant libraries and the inability to generate a cbmip mutant using available genetic tools, combined with the findings that high concentrations of small molecule inhibitors against CbMip are bactericidal against C. burnetii, strongly indicate that cbmip may be an essential gene in C. burnetii.
Proteomic analysis of C. burnetii cultures exposed to the inhibitor AN296, at concentrations that inhibited replication but did not reduce bacterial viability counts, revealed that 18 proteins were differentially abundant. One significant change was the increase in four DNA repair proteins, which suggested that the cells were under stress. ACCM-2 media has been reported to represent a high oxidative stress environment [85] and DNA repair is considered part of the oxidative defence mechanisms of C. burnetii [30]. The increase in DNA repair enzymes led to the hypothesis that the bacterial cells were subject to oxidative stress when incubated with CbMip inhibitors. This theory was validated using a hydrogen peroxide sensitivity assay where C. burnetii cells were exposed to a low concentration of H2O2 [86] in the absence or presence of CbMip inhibitor at concentrations that still permitted replication. This experiment showed that C. burnetii, in the presence of CbMip inhibitors, becomes very sensitive to H2O2, suggesting that inhibition of CbMip leads to an increased susceptibility to oxidative stress, resulting in the bacterium responding by upregulating the expression of several DNA repair systems. Professional phagocytes are more efficient at using oxidative burst to kill pathogens [87,88], and as such the proteomics and H2O2 sensitivity results explain why lower concentrations of CbMip inhibitors (50 μM), were more effective at inhibiting C. burnetii replication in THP-1 cells than HeLa cells. This may also be the reason why the inhibitors are also effective at inhibiting C. burnetii replication at later stages of the infection.
Similar effects of Mip inhibition have been observed in another obligate intracellular pathogen, C. trachomatis, where exposure to the macrolides, FK506 and rapamycin, or pipecolic acid-based inhibitors, PipN3 and PipN4, resulted in inhibition of intracellular replication and reduced infectivity of C. trachomatis progeny [54,60]. Even though the Mip protein from C. trachomatis, CtMip, has been well studied [54,57,60,89–91], to date no ctmip mutant has been described and characterized for C. trachomatis. Although an axenic culture medium that supports the propagation of C. trachomatis has not yet been reported, a medium that supports the metabolic activity and survival of C. trachomatis outside of the host has been developed [92]. It would be interesting to test if inhibitors of CtMip would have bactericidal activity against C. trachomatis similar to that observed for C. burnetii. Such investigation would shed light on whether Mip plays an indispensable role in the lifecycle of obligate intracellular pathogens.
This study demonstrated that AN296 was effective in vitro at inhibiting the replication of C. burnetii post-exposure in THP-1 cells. Addition of AN296 to cells, 2 or 3 days post exposure to C. burnetii, resulted in a significant decrease in intracellular replication. To further validate AN296 for therapeutic potential, it was screened in the more complex G. mellonella infection model. This model serves as an excellent starting point for therapeutic studies as the larvae can be maintained at human body temperature (37°C), they have functional homologues to several components of the mammalian innate immune system, and are equally susceptible to both phase I and phase II C. burnetii strains [74]. In addition, the toxicity and efficacy of several antibiotics tested in the G. mellonella model have been shown to correlate with the mouse model [93,94] and G. mellonella larvae also have the capacity to metabolise compounds, another factor that must be taken into consideration when taking new compounds forward as potential therapeutics [95–97]. Importantly the inhibitors were non-toxic to G. mellonella larvae at the highest dose tested. AN296 was found to protect larvae from infection with C. burnetii NMII and was effective for at least ten days. Of note is that despite being less effective in assays in vitro, SF235 was also found to be protect G. mellonella against C. burnetii induced death.
Mip proteins, and FKBPs in general, have been reported to act on several protein targets [47,98,99]. Therefore, it is unsurprising that inhibition of CbMip activity results in significant changes in the abundance of multiple proteins, which cumulatively can act to reduce pathogen replication. One also cannot at this stage rule out the effect these inhibitors may have on host FKBPs. From the results obtained, SF235 and ANCH37 (AN296) have reduced potency against CbMip (18±4 μM and 4.6±0.9 μM, respectively) as compared to the values reported against BpMip to which they were originally developed (0.29+0.06 μM and 0.12 μM, respectively) [55,61]. Despite this, these compounds present excellent starting points for development into more potent and stable molecules for use in further studies including those in vivo.
The data presented warrants further investigation into the exact role that CbMip plays in C. burnetii biology. In addition, since CbMip inhibitors SF235 and AN296 were found to also be effective against the virulent phase I strain, these data support the further development of CbMip inhibitors for increased potency and specificity for CbMip have potential as novel therapeutics against Q fever.
Materials and methods
Reagents
Cell culture media, reagents and fetal bovine serum (FBS) were obtained from Gibco (ThermoFisher Scientific). General chemicals were obtained from Sigma-Aldrich/Merck Millipore unless otherwise stated.
Culture for bacterial strains and mammalian cell lines
The bacterial strains and plasmids used in this study are shown in S1 Table. E. coli strains were grown in Luria-Bertani medium and antibiotics were added as necessary to the following final concentrations: ampicillin, 100 μg/mL; kanamycin, 50 μg/mL. Both C. burnetii NMI (the infectious phase I variant, RSA493), and C. burnetii NMII (the avirulent phase II variant, RSA439, clone 4) were used in the growth experiments in axenic culture, and C. burnetii NMII was used for mutagenesis, cell infection assays and the G. mellonella infection model.
C. burnetii was routinely grown in acidified citrate cysteine medium-2 (ACCM-2) media from Sunrise Science Products (San Diego, CA) at 37°C in 5% CO2 and 2.5% O2 [100] to stationary phase (6–7 days) before harvesting for infection or growth assays. When required chloramphenicol and kanamycin were used in C. burnetii ACCM-2 cultures at 3 μg/mL and 350 μg/mL, respectively. ACCM-2 agarose plates were prepared using UltraPure Agarose (Invitrogen) and additionally supplemented with 0.50 mM L-tryptophan [101].
All manipulations of C. burnetii NMI were carried out in a class III microbiological safety cabinet complying with British Standard EN12469:2000 and all studies were risk assessed and approved by Dstl’s Biosafety Committee.
THP-1 human monocytic cells and HeLa human carcinoma cells were maintained in RPMI 1640 supplemented with GlutaMAX and 10% (v/v) heat-inactivated FBS at 37°C in 5% CO2 unless otherwise described.
Quantification of C. burnetii genome equivalents
For studies using C. burnetii NMII, genome equivalents (GE) of C. burnetii were quantified using the Quant-iT PicoGreen dsDNA assay kit (ThermoFisher Scientific) or by quantitative PCR (qPCR), using ompA specific primers, as previously described [102,103]. PicoGreen assays were performed following the manufacturers protocol and results were read using the POLARStar plate reader (BMG Labtech), data were processed using the MARS analysis software (BMG Labtech) and analyzed using Microsoft Excel. Quantification by qPCR was performed using a QuantStudio 3 real-time PCR system (Applied Biosystems, Thermo Fisher Scientific). QuantStudio Design and Analysis Software was used to generate standard curves and perform initial analysis. Data was exported to Microsoft Excel for further analysis. For studies using C. burnetii NMI, the CFU/mL of single use frozen C. burnetii stocks was quantified by plating out in serial dilutions on ACCM-2 agar plates which were then left for 7–10 days before viability counts were performed.
Growth assays in axenic media
Studies involving Phase I strain, C. burnetii NMI
Growth curves. Vented flasks containing 5 mL of ACCM-2 media supplemented with 0.50 mM L-tryptophan and containing either 100 μM of SF235 or AN296 or DMSO vehicle control were inoculated at 1 × 104 CFU/mL using freshly thawed stocks of C. burnetii NMI. Cultures were incubated statically at 37°C in a Galaxy 170 R incubator (New Brunswick Scientific) adjusted to 5 % CO2 and 2.5 % O2. Samples of 100 μL were removed at days 0, 1, 2, 3, 4 and 7 and plated in serial dilutions on ACCM-2 agar plates which were then left for 7–10 days before viability counts were performed. For delayed dosing experiments, the assay was prepared without inhibitors as described above and on day 3 of the growth curve, 25 μL of inhibitor or DMSO diluted in media was added to each culture.
Studies involving Phase II strains, C. burnetii NMII
Luminescence assays. Stationary phase (6–7 day) ACCM-2 cultures of the luciferase-expressing C. burnetii NMII strain, C. burnetii-lux, were quantified using the PicoGreen assay and appropriately diluted to 1 × 106 genome equivalents (GE)/mL in white 96-well plates (Corning) in 0.1 mL of fresh ACCM-2 medium per well, in the presence of varying concentrations of Mip inhibitor. Bioluminescence was measured every 24 h over 5 days by a POLARStar plate reader (BMG Labtech). For delayed dosing experiments, the assay was prepared without inhibitors as described above and 25 μL of inhibitor or DMSO diluted in ACCM-2 was added to each well on day 2 or day 3 of the assay.
CFU assay. Using C. burnetii NMII and clear 96-well plates (Corning), the growth assay was prepared as described above for the Luminescence assay. Following 4 days of incubation, samples from test wells were taken and plated in serial dilutions on ACCM-2 agar plates which were then left for 7 days before viability counts were performed.
Sensitivity to hydrogen peroxide assays
Stationary phase (6–7 day) ACCM-2 cultures of C. burnetii NMII were quantified using the PicoGreen assay and appropriately diluted to 1.0 × 106 genome equivalents (GE)/mL in clear 96-well plates (Corning) in 0.1 mL of fresh ACCM-2 medium containing either 50 μM AN296 or DMSO vehicle control, in the absence or presence of 100 μM H2O2. The plate was incubated for 4 days before the cultures were plated out to determine viable CFU/mL.
MIC and MBC assays
Studies involving Phase I strain, C. burnetii NMI
Vented flasks containing 5 mL of ACCM-2 media supplemented with 0.50 mM L-tryptophan and containing increasing concentrations of Mip inhibitor were inoculated at 1 × 105 CFU/mL using freshly thawed stocks of C. burnetii NMI. Cultures were incubated statically at 37°C for 6 days. The MIC was determined both by measuring the OD and then by plating out each broth to obtain a viable bacterial count. The MBC was determined by plating out neat broth from flasks with no visible growth onto ACCM-2 plates.
Studies involving Phase II strains, C. burnetii NMII
Stationary phase (6–7 day) ACCM-2 cultures of C. burnetii NMII were quantified using the PicoGreen assay and appropriately diluted to 2.5 × 105 genome equivalents (GE)/mL in clear 24-well plates (Nunc) in 1 mL of fresh ACCM-2 medium supplemented with 0.50 mM L-tryptophan per well, in the presence of increasing concentrations of Mip inhibitor, and then incubated for 6 days. The MIC was determined both by measuring the OD and then by plating out each broth to obtain a viable bacterial count. The MBC was determined by plating out neat broth of wells with no visible growth onto ACCM-2 plates.
Mutagenesis of cbmip (CBU0630)
Mutagenesis of cbmip (cbu0630) was attempted in C. burnetii NMII following the loop in/loop out method for targeted gene inactivation described by Beare et al. [82]. All oligonucleotides used in this study are shown in S2 Table. The 2-kb fragments of the 5′- and 3′-end-flanking regions of cbmip were first amplified from genomic DNA by PCR using the upstream and downstream oligonucleotide pairs cbu0630-up F and cbu0630-up R, and cbu0630-down F and cbu0630-down R, respectively. These two PCR fragments were purified and joined together by strand overlapping extension (SOE) PCR [104], using primers cbu0630-up F and cbu0630-down R, to generate a PCR product containing a unique internal NotI site between the 5′- and 3′- flanking regions and flanked by BamHI and SalI restriction sites. This 4-kb fragment was cloned into BamHI/SalI-digested pJC-CAT, generating plasmid pJC-CAT::cbu0630-prep. The 1169P-Kan cassette was then amplified from pJB-Kan by PCR using oligonucleotides P1169-Kan-NotI F and P1169-Kan-NotI R, treated with NotI and cloned into NotI-digested pJC-CAT::cbu0630-prep to create the deletion plasmid pJC-CAT::cbu0630-Kan. This construct was electroporated into C. burnetii NMII and chloramphenicol and kanamycin resistant colonies were successfully isolated and confirmed as cbu0630 integrants through PCR analysis of isolated genomic DNA. Following sucrose selection, a limited number of very small colonies were isolated however they either failed to grow or were confirmed as wild-type revertants.
Cloning and expression of CbMip
The nucleotide sequence encoding C. burnetii mip without the predicted N-terminal signal peptide (nucleotides 64 to 690, corresponding to amino acids 22–230) was synthesized with codon optimization for expression in E. coli by GenScript (Piscataway, NJ) and was received as plasmid pMK-cbMipOpt. The cbmip coding sequence was amplified by PCR from pMK-cbMipOpt as a template using primers CbMip_NcoI F and CbMip_BamHI R to introduce flanking NcoI and BamHI sites. The amplified DNA fragment was first subcloned into pCR-Blunt II-TOPO and then excised by NcoI and BamHI restriction digest and cloned into the same restriction sites of the expression vector pETM-11 (European Molecular Biology Laboratory, [105]), which encodes an N-terminal hexahistidine tag followed by a TEV site, to generate pETM-11-cbMip.
Truncated cbmip gene constructs corresponding to transposon mutants, cbmip-TM1 (nucleotides 102 to 690, corresponding to amino acids 36–230) and cbmip-TM2 (nucleotides 213 to 690, corresponding to amino acids 71–230), were generated following the same methodology described above for the full length cbmip using the appropriate primer pairs for each truncated construct. After transformation into chemically competent E. coli TOP10, positive clones were verified by DNA sequencing (AGRF, Australia, or Macrogen, South Korea). The codon optimized nucleotide sequence of cbmip and the protein sequences for each construct are listed in S3 Table.
Protein production and purification
Recombinant plasmids were transformed into the E. coli strain BL21(DE3)pLysS and grown with shaking in 1 L LB medium supplemented with 50 μg/mL kanamycin at 37°C to an optical density at 600 nm of 0.4, whereupon protein production was induced for 2 h by the addition of 1.0 mM IPTG. Cells were cooled on ice for 15 min and harvested by centrifugation at 3,000 x g for 15 min at 4°C, and cell pellets were stored at -20°C until further processing. The cell pellets were resuspended in 10 mL/g Buffer A (50 mM HEPES, 150 mM NaCl, 10% (v/v) glycerol, 25 mM imidazole, pH 7.5) with the addition of lysozyme (0.2 mg/mL, Sigma-Aldrich), complete EDTA-free protease inhibitor cocktail (Roche) and deoxyribonuclease (DNAse I, 1 μg/mL, Roche), lysed using an EmulsiFlex C5 homogeniser (Avestin) and clarified at 24,000 x g for 30 min at 4°C. The soluble supernatant was passed through a 0.22 μm filter and then applied onto a 5 mL HisTrap HP column (Cytiva) that had been pre-equilibrated with Buffer A using an ÄKTA Start (Cytiva). After washing with Buffer A, the 6xHis tagged protein was eluted using a gradient of 1–100% Buffer B (50 mM HEPES, 150 mM NaCl, 10% (v/v) glycerol, 500 mM imidazole, pH 7.5). Fractions containing the protein of interest were pooled, concentrated using centrifugal filter (Amicon Ultra, MWCO 10 kDa). This was followed by size-exclusion chromatography on a HiLoad 16/600 Superdex 75pg (GE Healthcare Life Sciences) column in 50 mM HEPES, 150 mM NaCl, 10% (v/v) glycerol, pH 7.5. Again, fractions containing the protein of interest were pooled and concentrated using centrifugal filter. Proteins were assessed as having >95% purity by SDS–PAGE. The concentration was calculated from the A280 and the theoretical extinction coefficient calculated using the ProtParam tool from ExPASy. Protein aliquots were snap frozen in liquid nitrogen and stored at -80°C.
Enzymatic assays
Purified recombinant CbMip proteins were tested for PPIase activity in an enzyme assay by measuring the cis-trans isomerization of the tetrapeptide Suc-Ala-Phe-Pro-Phe-p-nitroanilide (Bachem #4016001). The peptidyl-prolyl cis-trans isomerase assays were performed using a protease-coupled assay as previously described [55,62]. Data were determined for enzyme at 0, 6.25, 12.5, 25, 50, 75, 100, 150, 200, 300, 450, 600 nM, with three replicates. Data were analysed as previously described [55]. Briefly, the kobs was calculated using one-phase association, upon which the kenz was determined using the equation kenz = kobs—kuncatalysed. The specificity constant, kcat/KM, was fit to the equation; kcat/KM = kenz/[PPIase] using linear regression in GraphPad Prism v. 9.0.1. The inhibition constant (Ki) of SF235 and ANCH37 against CbMip were determined using a revised Morrison equation as previously described [55]. All enzymatic data were fitted using Graphpad Prism v. 9.0.1.
Protein modeling with AlphaFold2
Protein models were made with AlphaFold v.2.2.0 [66], running on a local server using the 2022-03-03 database. Models were made in multimeric mode with an input of two copies of the full length CbMip with the signal sequence removed, CbMip-TM1, or CbMip-TM2. Five models were made for each input sequence, superimposed using PyMOL v.2.5.3 (Schrödinger). Models were manually inspected, and figure prepared, using PyMOL.
C. burnetii intracellular growth assays
Growth assays in THP-1 and HeLa cells were performed similarly to those described previously [103]. Briefly, THP-1 cells were seeded at 5 × 105 into 24-well flat-bottom tissue culture plates (Nunc) and differentiated with 10 nM phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich) for 3 days. HeLa cells were seeded at 5 × 104 cells/well and incubated for 24 h prior to infection. Samples for immunofluorescence (IF) were seeded onto 13-mm sterile glass coverslips. C. burnetii NMII cultures were quantified as described above and diluted appropriately into RPMI 1640 supplemented with 5% (v/v) FBS to infect THP-1 cells at a multiplicity of infection (MOI) of 5 and HeLa cells at a MOI of 50. Diluted C. burnetii NMII were pre-treated for 1 h at 37°C with defined concentrations of Mip inhibitor or DMSO vehicle control (final concentration of DMSO was 0.1%) and subsequently 0.5 mL aliquots per well were used to infect cells. Upon infection, cells were centrifuged at 500 x g for 5 min and then incubated for 4 h at 37°C and 5% CO2 before being washed once in PBS to remove extracellular bacteria and supplemented with fresh medium containing Mip inhibitor or DMSO as control. At indicted time points, cells were either fixed for microscopy or lysed with H2O and collected for quantification of C. burnetii intracellular replication. For day 1, 3, 5 and 7 day post infection samples, media from each duplicate well was pooled and collected alongside the lysed cells. Following lysis, samples were centrifuged at 17,000 x g for 20 min at 4°C before gDNA was extracted from the resulting pellet using the Quick-DNA Miniprep kit/Zymo gDNA extraction kit (Zymo Research) and quantified by qPCR.
Immunofluorescence microscopy
At indicated time points, infected cells were fixed for 20 min at room temperature with 4% (w/v) paraformaldehyde (in PBS). After washing in PBS, samples were first blocked in PBS containing 5% (v/v) FBS and 0.05% (w/v) saponin (Sigma-Aldrich) (blocking buffer). Samples were then stained using primary antibodies diluted in blocking buffer: 1:50 anti-LAMP-1 clone H4A3, supernatant (H4A3 was deposited to the DSHB by August, J.T. / Hildreth, J.E.K. (DSHB Hybridoma Product H4A3)) and 1:10,000 rabbit anti-C. burnetii antibodies (Roy Laboratory, Yale University). Secondary antibodies, anti-mouse Alexa Fluor 488 and anti-rabbit Alexa Fluor 568 (Thermo Fisher Scientific) were used at a dilution of 1:3,000 in blocking buffer. DNA was stained using Hoechst 33258 (1:10,000; Thermo Fisher Scientific) and coverslips were mounted onto glass slides using ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Fluorescence microscopy was performed using a Nikon A1Si confocal microscope, and images were acquired using NIS-Elements software (Nikon).
Inhibitor cytotoxicity assays
Cytotoxicity induced by SF235 and AN296 over the time period of a C. burnetii intracellular growth assay was assessed in both HeLa and THP-1 cells.
IC50 determination
The cytotoxicity of the inhibitors themselves, in the absence of C. burnetii infection, was investigated by determining their IC50 values at several time points, for up to 6 days of incubation. Cell viability was determined colorimetrically using the Cell proliferation reagent WST-1 (Roche, Basel, Switzerland). Cells were seeded in triplicate in 96-well plates. HeLa cells were prepared as a stock solution of 1 × 104 cells/mL, wells were then seeded with 100 μL of diluted stock solution as appropriate for the incubation time point (t1 = 100%; t2 = 50%; t3 = 30%; t4 = 25% and t5 = 20%). Similarly, THP-1 cells were prepared as a stock solution of 5 × 105 cells/mL. The two inhibitors were diluted according to their individual water solubility in cell media to give a maximum concentration of 400 μM for SF235 and 200 μM for AN296 and 2% DMSO. This working concentration was serially diluted 1:1 in media and a 100 μL aliquot of each inhibitor concentration was added to the cells, resulting in the final inhibitor concentrations ranging from 0.391 μM to 200 μM for SF235 and 0.391 μM to 100 μM for AN296. The final concentration of DMSO was 1% at the highest concentration of inhibitor tested and less at each of the dilutions. A dilution series of DMSO (starting at 1%) served as a control for each cell line at each assessed time point. Untreated cells were used as controls. The cells were incubated at 37°C and 5% CO2 for the indicated period of time. To determine cell viability, 10 μL of WST-1 was added to each well according to the manufacturer’s instructions. After 1 h (HeLa cells) or 4 h (THP-1 cells) of incubation, the absorbance of the soluble formazan product at 450 nm and the background at 630 nm were determined using a Tecan infinite 200Pro microplate reader (Tecan Trading AG, Männedorf, Switzerland). Data were analysed and IC50 values were determined using GraphPad Prism v9.01.
LDH release assay
The inhibitor concentrations tested were the same as those used in the C. burnetii intracellular growth assays in THP-1 cells (50 μM) and the control was 0.1% DMSO. LDH release was measured after incubating cells with inhibitors for 4 h (representing the day 0 infection time point), 28 h (day 1), 76 h (day 3) and 124 h (day 5) using the Roche LDH Cell Cytotoxicity kit (#11644793001), following the manufacturer’s instructions. At each indicated time point, a positive control (Triton X-100) and negative control (media only) were included in in triplicate. All results are presented as the mean of three independent experiments containing two technical repeats. Cytotoxicity was calculated as the percentage of lactate dehydrogenase (LDH) measured in Triton X-100 for that assay and are represented relative to the control (0.1% DMSO).
Infection of Galleria mellonella larvae
G. mellonella were maintained and infected as previously reported [74,103]. Larvae were grown in-house and kept at 30°C in the dark until use. To test the toxicity of the inhibitors, 50 mM stock solutions of compound dissolved in DMSO were diluted in PBS to the desired concentrations and the final concentration of DMSO was kept at 1% (v/v). A 10 μL aliquot was injected into the right proleg of G. mellonella larvae, which were then kept isolated at 37°C in the dark. Survival was assessed every 24 h for 10 days. To assess the effect of Mip inhibitors on C. burnetii pathogenicity, 108 GE/mL of C. burnetii NMII was incubated in the presence of 0, 100 μM or 500 μM of compound in PBS with a final concentration of 1% DMSO, at 37°C for 1 h. A 10 μL aliquot of this mixture was then injected into larvae and survival tracked for 10 days as above. All treatment groups started with 12 larvae. Kaplan-Meier survival curves were determined from three independent experiments. Data was analysed using the log-rank (Mantel-Cox) test with Bonferroni correction for multiple comparisons with a significance level of 0.05.
Proteomic analysis
Stationary phase ACCM-2 cultures of C. burnetii NMII grown up directly from glycerol stocks were quantified using the PicoGreen assay and appropriately diluted to 1 × 106 genome equivalents (GE)/mL into 20 mL of fresh ACCM-2. Cultures were incubated for 72 h prior to the addition of 100 μM AN296 or vehicle control. Final DMSO concentration was kept at 0.2% (v/v) for all cultures. After 24 h, cultures were harvested by centrifugation and washed twice with PBS. Total protein was extracted from C. burnetii cell pellets using chloroform and methanol, following previously published procedures [106]. Total protein content was assessed using a bicinchoninic acid protein assay (Pierce, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Protein clean-up and in-solution digestion
Quantified precipitated samples were resuspended in 100 μL of 5% SDS by boiling for 10 min at 95°C. Samples were then reduced with 10 mM DTT for 10 min at 95°C and then alkylated with 40 mM chloroacetamide for 1 h in the dark. Reduced/alkylated samples were then cleaned up using Micro S-traps (https://protifi.com/pages/s-trap) according to the manufacturer’s instructions. Samples were then digested for 4 h with trypsin/lys-c (~1:25 protease/protein ratio), collected and dried. Dried samples were then further cleaned up with home-made high-capacity StageTips composed of 1 mg Empore C18 material (3 M) and 5 mg of OLIGO R3 reverse phase resin (Thermo Fisher Scientific) as described [107,108]. Columns were wet with Buffer B (0.1% formic acid, 80% acetonitrile) and conditioned with Buffer A* (0.1% TFA, 2% acetonitrile) prior to use. Acidified samples were loaded onto conditioned columns, washed with 10 bed volumes of Buffer A* and bound peptides were eluted with Buffer B before being dried then stored at -20°C.
LFQ-based quantitative proteome liquid chromatography-mass spectrometry
Dried proteome digests were re-suspended in Buffer A* and separated using a two-column chromatography set up composed of a PepMap100 C18 20 mm x 75 μm trap and a PepMap C18 500 mm x 75 μm analytical column (Thermo Fisher Scientific). Samples were concentrated onto the trap column at 5 μL/min for 5 min with Buffer A (0.1% formic acid, 2% DMSO) and then infused into an Orbitrap Q-Exactive plus Mass Spectrometer (Thermo Fisher Scientific) at 300 nL/min via the analytical column using a Dionex Ultimate 3000 UPLC (Thermo Fisher Scientific). 135-min analytical runs were undertaken by altering the buffer composition from 2% Buffer B (0.1% formic acid, 77.9% acetonitrile, 2% DMSO) to 22% B over 105 min, then from 22% B to 40% B over 10 min, then from 40% B to 80% B over 5 min. The composition was held at 80% B for 5 min, and then dropped to 2% B over 2 min before being held at 2% B for another 8 min. The Q-Exactive Mass Spectrometer was operated in a data-dependent mode automatically switching between the acquisition of a single Orbitrap MS scan (375–1400 m/z, maximal injection time of 50 ms, an Automated Gain Control (AGC) set to a maximum of 3 × 106 ions and a resolution of 70k) and 15 Orbitrap MS/MS HCD scans (stepped NCE of 28;30;34, a maximal injection time of 65 ms, an AGC set to a maximum of 1 × 105 ions and a resolution of 17.5k).
Mass spectrometry data analysis
Proteome datasets were processed using MaxQuant (v1.6.17.0.) [109] and searched against the C. burnetii strain RSA 493 / Nine Mile phase I and Dugway 5J108-111 proteomes (Uniprot accession: UP000002671 and UP000008555, respectively). Both the Dugway 5J108-111 and RSA 493 / Nine Mile phase I proteomes were included to enable the mapping / incorporation of Uniprot assignment from both strains. Searches were undertaken using “Trypsin” enzyme specificity with carbamidomethylation of cysteine as a fixed modification. Oxidation of methionine and acetylation of protein N-termini were included as variable modifications and a maximum of 2 missed cleavages allowed. To enhance the identification of peptides between samples, the Match between Runs option was enabled with a precursor match window set to 2 min and an alignment window of 20 min with the label free quantitation (LFQ) option enabled [110]. The resulting outputs were processed within the Perseus (v1.6.0.7) analysis environment [111] to remove reverse matches and common protein contaminates prior to further analysis. For LFQ comparisons biological replicates were grouped and missing values were then imputed based on the observed total peptide intensities with a range of 0.3σ and a downshift of 2.5σ using Perseus. Student t-tests were undertaken to compare the proteomes between groups. The resulting MS data and search results have been deposited into the PRIDE ProteomeXchange Consortium repository [112,113] with the dataset identifier PXD036679.
Synthesis of pipecolic acid based Mip inhibitors
Inhibitors SF235, ANCH37 and AN296 were prepared as previously described [50,61] (Australian Patent Application No. PCT/AU2023/050201).
Stability assessment of AN296 at pH = 4.75
For this purpose, Buffer C (S4 Table) was prepared which mimicked the inorganic components of ACCM-2 media and was adjusted to pH 4.75 using citrate buffer. A 100 μM stock solution of AN296 was then prepared in Buffer C and incubated at 37°C for a period of 7 days, corresponding to cell assay conditions with sampling performed daily. Analysis of 50 μL aliquots using HPLC involved an isocratic method with a mobile phase composition of MilliQ/ACN (45%/55% (v/v)) with a flow rate of 1 mL/min and UV-metric detection at 250 nm and 260 nm. A Knauer Eurospher II 100–5 C18 H (150x4.6 mm) (KNAUER Wissenschaftliche Geräte GmbH, Berlin, Deutschland) column was used as the stationary phase. Daily measurements were performed on 3 samples with 2 injections each, as well as AN296 in methanol for retention time control. The first analysis was performed immediately after dilution and before incubation and thus served as a reference point for calculating the content.
Chemicals and reference substances
All reagents were of analytical grade. HPLC grade acetonitrile from VWR International GmbH (Darmstadt, Germany). Water for HPLC was purified using a Milli-Q purification system by Merck Millipore (Schwalbach, Germany).
Apparatus
HPLC experiments were performed on an Agilent 1100 modular chromatographic system (Agilent technologies, Waldbronn, Germany) consisting of a vacuum degasser (G1322A), a binary pump (G1312A), an autosampler (G1313A), a thermostated column compartment (G1316A) and a diode array detector (G1315B). Agilent ChemStation Rev C 01.10 software was used for data processing. For incubation a Grant Boekel HIS25 Incubator (Grant Instruments, Cambridge, England) was used.
Statistical analysis
All numerical results were analysed using Microsoft Excel 2010. Statistical analyses were performed using GraphPad Prism, v. 9.0.1.
Supporting information
(A) Multiple sequence alignment of Mip proteins from C. trachomatis (CtMip), B. pseudomallei (BpMip) C. burnetii (CbMip) and L. pneumophila (LpMip) using CLUSTAL O (1.2.4). The start of the highly conserved catalytic/PPIase domain is indicated by an open triangle. The essential amino acid (Asp) in LpMip for PPIase activity is conserved in all proteins and is indicated by a closed triangle. (B) Percentage identity matrix of the PPIase domain of Mip from C. trachomatis (CtMip-cat), B. pseudomallei (BpMip-cat) C. burnetii (CbMip-cat) and L. pneumophila (LpMip-cat) using CLUSTAL 2.1.
(TIF)
Data were collected in a single experiment for CbMip as described in the experimental section. Each inhibitor concentration of SF235 (closed circle) and ANCH37 (blue square) was tested three times. Data were fitted to equation as described previously [55]. Results are representative of at least two experiments conducted on separate days with different preparations of inhibitor. Error bars show standard error of the mean.
(TIF)
Intracellular replication of C. burnetii NMII in THP-1 cells in the presence of 50 μM AN296 (blue triangle), 31 μM chloramphenicol (Cm) (open circle) or control (closed circle), introduced after the 4 h infection period. Error bars represent standard error of the mean (n ═ 5). ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test.
(TIF)
(A) IC50 values for SF235 and AN296 and DMSO after incubation for the indicated periods of time were determined in THP-1 and HeLa cells using the Cell Proliferation Reagent WST-1. (B) Cytotoxicity was measured after THP-1 cells were incubated with SF235 (50 μM), AN296 (50 μM), chloramphenicol (cm, 31 μM) or control (0.1% DMSO) for the indicated period of time using Roche LDH Cell Cytotoxicity kit. No significant difference in cytotoxicity was found between the inhibitors and the control (0.1% DMSO). Data is presented as percent cytotoxicity relative to the control with error bars representing the SD from three independent experiments; p values were determined using two-way ANOVA followed by Dunnett’s multiple comparison post-test.
(TIF)
Overview of transposon mutants generated in the Coxiella burnetii NMII Metters et al. library. Confirmed transposon insertion sites are indicated by red vertical lines. All TA insertion sites, possible locations where the transposable element can intergrate, are indicated by black vertical lines. (A) Transposon insertion sites around the cbmip (cbu0630) genetic locus. (B) Enhanced view of transposon insertion sites within the cbmip gene. Two out of a possible 37 mutants were identified in the transposon mutant library. (C) Detailed analysis of transposon insertion sites in cbmip. In lower case text is the DNA coding sequence for the C. burnetii mip gene (cbu0630) of strain C. burnetii NMII. Below, in upper case text, is the encoded amino acid sequence, green box indicates the PPIase catalytic domain. The two transposon insertion sites (TA) identified in the transposon mutant library are indicated by blue triangles. The first ATG following the transposon site is indicated by red boxes. In both instances of transposon insertion in the cbmip sequence, the downstream ATG from which protein translation would initiate is in-frame with the original full-length gene and therefore would result in the production of a truncated CbMip protein in the C. burnetii cbu0630 transposon mutants.
(TIF)
The stability of compound AN296 under acidic conditions (pH of 4.75), was investigated by HPLC chromatography. A stock solution of AN296 (100 μM) was prepared in Buffer C (composition in S4 Table) which mimicked the inorganic components of ACCM-2 media and was incubated at 37°C for a period of 7 days. Data is presented as the amount of AN296 remaining compared to day 0 (at 250 nm and 260 nm), with error bars representing the standard deviation.
(TIF)
Bioluminescence was measured as an indicator of C. burnetii-lux replication. The strain was inoculated at a concentration of 1 × 106 GE/mL into ACCM-2 media with (A) 50 μM or (B) 25 μM of CbMip inhibitors SF235 (grey square), AN296 (closed triangle) or vehicle control (open circle) and grown over 5 days. Data is presented as RLU (relative light units) with error bars representing the standard deviation from three independent experiments. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test.
(TIF)
Each larva received a single 10 μL injection of in PBS containing either (A) AN296 or (B) SF235 or vehicle control into the right proleg. Inhibitors were tested at increasing concentrations starting at 10 μM and increasing up to 50 μM, 250 μM and 500 μM (groups of n ═ 10). Larvae were monitored everyday via twitching response over 10 days. All but two larvae, which received 10 μM of AN296, survived the entire duration of the experiment.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(XLSX)
Acknowledgments
The H4A3 monoclonal antibody developed by August, J.T. / Hildreth, J.E.K was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. The authors acknowledge the facilities, and the scientific and technical assistance of Microscopy Australia at the Centre for Microscopy, Characterisation & Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments. We would like to thank Nathan Unsworth and Chun-Qiang Liu from Defence Science and Technology Group for their generous gift of plasmid pMK-cbMipOpt. The bacterial expression vector pETM11 was a gift from Dr Gunter Stier (EMBL, Heidelberg, Germany). We would like to thank William Stuart (University of Exeter) for setting up and maintaining the AlphaFold server. We would like to thank Professor Richard Titball for critical reading of the manuscript.
Data Availability
All relevant data are within the manuscript, supporting files and the MS dataset has been deposited into the PRIDE ProteomeXchange Consortium repository, the dataset identifier is PXD036679.
Funding Statement
Funding for HJN’s research team is supported by NHMRC Idea’s grant 2010841. This work was supported by Dstl contract DSTLX-1000051512 to NJH. IHN, CHJ and GM were funded by the UK Ministry of Defence. This work was supported by the North Atlantic Treaty Organization (NATO), Brussels, Belgium grant SPS 984835, and the German Research Foundation (DFG, Bonn, Germany; grant SFB 630) for the development of Mip inhibitors against L. pneumophila and B. pseudomallei, respectively, and The Federal Ministry of Education and Research for the development of Mip inhibitors against T. cruzi and B. pseudomallei, given to UH. This paper includes research that was supported by DMTC Limited (Australia) to MST, AWD, NMB, JI and EAK. The authors have prepared this paper in accordance with the intellectual property rights granted to partners from the original DMTC project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brooke RJ, Kretzschmar ME, Mutters NT, Teunis PF. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect. Dis. 2013;13:488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eldin C, Melenotte C, Mediannikov O, Ghigo E, Million M, Edouard S, et al. From Q fever to Coxiella burnetii infection: a paradigm change. Clin. Microbiol Rev. 2017;30(1):115–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson A, Bijlmer H, Fournier PE, Graves S, Hartzell J, Kersh GJ, et al. Diagnosis and management of Q fever—United States, 2013: recommendations from CDC and the Q Fever Working Group. MMWR Recomm. Rep. 2013;62(RR-03):1–30. [PubMed] [Google Scholar]
- 4.Melenotte C, Million M, Raoult D. New insights in Coxiella burnetii infection: diagnosis and therapeutic update. Expert Rev. Anti Infect. Ther. 2020;18(1):75–86. [DOI] [PubMed] [Google Scholar]
- 5.Maurin M, Raoult D. Q fever. Clin. Microbiol. Rev. 1999;12(4):518–53. doi: 10.1128/CMR.12.4.518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saunders DL, Garges E, Manning JE, Bennett K, Schaffer S, Kosmowski AJ, et al. Safety, tolerability, and compliance with long-term antimalarial chemoprophylaxis in American soldiers in Afghanistan. The American journal of tropical medicine and hygiene. 2015;93(3):584–90. doi: 10.4269/ajtmh.15-0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kampschreur LM, Wegdam-Blans MC, Wever PC, Renders NH, Delsing CE, Sprong T, et al. Chronic Q fever diagnosis- consensus guideline versus expert opinion. Emerg. Infect. Dis. 2015;21(7):1183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kersh GJ. Antimicrobial therapies for Q fever. Expert Rev. Anti Infect. Ther. 2013;11(11):1207–14. doi: 10.1586/14787210.2013.840534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keijmel SP, Delsing CE, Bleijenberg G, van der Meer JWM, Donders RT, Leclercq M, et al. Effectiveness of long-term doxycycline treatment and cognitive-behavioral therapy on fatigue severity in patients with Q fever fatigue syndrome (Qure Study): A randomized controlled trial. Clin. Infect. Dis. 2017;64(8):998–1005. doi: 10.1093/cid/cix013 [DOI] [PubMed] [Google Scholar]
- 10.Marmion BP, Shannon M, Maddocks I, Storm P, Penttila I. Protracted debility and fatigue after acute Q fever. Lancet. 1996;347(9006):977–8. doi: 10.1016/s0140-6736(96)91469-5 [DOI] [PubMed] [Google Scholar]
- 11.Morroy G, Keijmel SP, Delsing CE, Bleijenberg G, Langendam M, Timen A, et al. Fatigue following acute Q-fever: A systematic literature review. PLoS One. 2016;11(5):e0155884. doi: 10.1371/journal.pone.0155884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reukers DFM, van Jaarsveld CHM, Akkermans RP, Keijmel SP, Morroy G, van Dam ASG, et al. Impact of Q-fever on physical and psychosocial functioning until 8 years after Coxiella burnetii infection: An integrative data analysis. PLoS One. 2022;17(2):e0263239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woldeyohannes SM, Perkins NR, Baker P, Gilks CF, Knibbs LD, Reid SA. Q fever vaccine efficacy and occupational exposure risk in Queensland, Australia: A retrospective cohort study. Vaccine. 2020;38(42):6578–84. doi: 10.1016/j.vaccine.2020.08.006 [DOI] [PubMed] [Google Scholar]
- 14.Angelakis E, Raoult D. Q Fever. Vet. Microbiol. 2010;140(3–4):297–309. doi: 10.1016/j.vetmic.2009.07.016 [DOI] [PubMed] [Google Scholar]
- 15.Cross AR, Baldwin VM, Roy S, Essex-Lopresti AE, Prior JL, Harmer NJ. Zoonoses under our noses. Microbes Infect. 2019;21(1):10–9. doi: 10.1016/j.micinf.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Newton HJ, McDonough JA, Roy CR. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One. 2013;8(1):e54566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton P, Thomas DR, Reed SCO, Lau N, Xu B, Ong SY, et al. Lysosomal degradation products induce Coxiella burnetii virulence. Proc. Natl. Acad. Sci. U. S. A. 2020;117(12):6801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seshadri R, Paulsen IT, Eisen JA, Read TD, Nelson KE, Nelson WC, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 2003;100(9):5455–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol. Microbiol. 2003;49(4):965–76. [DOI] [PubMed] [Google Scholar]
- 20.Crabill E, Schofield WB, Newton HJ, Goodman AL, Roy CR. Dot/Icm-translocated proteins important for biogenesis of the Coxiella burnetii-containing vacuole identified by screening of an effector mutant sublibrary. Infect. Immun. 2018;86(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, et al. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog. 2014;10(7):e1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burette M, Bonazzi M. From neglected to dissected: How technological advances are leading the way to the study of Coxiella burnetii pathogenesis. Cell. Microbiol. 2020;22(4):e13180. [DOI] [PubMed] [Google Scholar]
- 23.Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J. Bacteriol. 2004;186(21):7344–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunha LD, Ribeiro JM, Fernandes TD, Massis LM, Khoo CA, Moffatt JH, et al. Inhibition of inflammasome activation by Coxiella burnetii type IV secretion system effector IcaA. Nat. Commun. 2015;6:10205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luhrmann A, Roy CR. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect. Immun. 2007;75(11):5282–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Voth DE, Howe D, Heinzen RA. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect. Immun. 2007;75(9):4263–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghigo E, Colombo MI, Heinzen RA. The Coxiella burnetii parasitophorous vacuole. Adv. Exp. Med. Biol. 2012;984:141–69. [DOI] [PubMed] [Google Scholar]
- 28.Hill J, Samuel JE. Coxiella burnetii acid phosphatase inhibits the release of reactive oxygen intermediates in polymorphonuclear leukocytes. Infect. Immun. 2011;79(1):414–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dragan AL, Voth DE. Coxiella burnetii: international pathogen of mystery. Microbes Infect. 2020;22(3):100–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mertens K, Lantsheer L, Ennis DG, Samuel JE. Constitutive SOS expression and damage-inducible AddAB-mediated recombinational repair systems for Coxiella burnetii as potential adaptations for survival within macrophages. Mol. Microbiol. 2008;69(6):1411–26. [DOI] [PubMed] [Google Scholar]
- 31.Mertens K, Samuel JE. Defense mechanisms against oxidative stress in Coxiella burnetii: adaptation to a unique intracellular niche. Adv. Exp. Med. Biol. 2012;984:39–63. [DOI] [PubMed] [Google Scholar]
- 32.Scheuplein NJ, Bzdyl NM, Kibble EA, Lohr T, Holzgrabe U, Sarkar-Tyson M. Targeting protein folding: a novel approach for the treatment of pathogenic bacteria. J. Med. Chem. 2020;63(22):13355–88. doi: 10.1021/acs.jmedchem.0c00911 [DOI] [PubMed] [Google Scholar]
- 33.Unal CM, Steinert M. FKBPs in bacterial infections. Biochim. Biophys. Acta. 2015;1850(10):2096–102. doi: 10.1016/j.bbagen.2014.12.018 [DOI] [PubMed] [Google Scholar]
- 34.Arakaki N, Higa F, Koide M, Tateyama M, Saito A. Induction of apoptosis of human macrophages in vitro by Legionella longbeachae through activation of the caspase pathway. J. Med. Microbiol. 2002;51(2):159–68. [DOI] [PubMed] [Google Scholar]
- 35.Cianciotto NP, Eisenstein BI, Mody CH, Toews GB, Engleberg NC. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 1989;57(4):1255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cianciotto NP, Fields BS. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. U. S. A. 1992;89(11):5188–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doyle RM, Steele TW, McLennan AM, Parkinson IH, Manning PA, Heuzenroeder MW. Sequence analysis of the mip gene of the soilborne pathogen Legionella longbeachae. Infect. Immun. 1998;66(4):1492–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Connell WA, Bangsborg JM, Cianciotto NP. Characterization of a Legionella micdadei mip mutant. Infect. Immun. 1995;63(8):2840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasch J, Unal CM, Klages A, Karsli U, Heinsohn N, Brouwer R, et al. Peptidyl-Prolyl-cis/trans-isomerases Mip and PpiB of Legionella pneumophila contribute to surface translocation, growth at suboptimal temperature, and infection. Infect. Immun. 2019;87(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cianciotto NP, Eisenstein BI, Mody CH, Engleberg NC. A mutation in the mip gene results in an attenuation of Legionella pneumophila virulence. J. Infect. Dis. 1990;162(1):121–6. [DOI] [PubMed] [Google Scholar]
- 41.Kohler R, Fanghanel J, Konig B, Luneberg E, Frosch M, Rahfeld JU, et al. Biochemical and functional analyses of the Mip protein: influence of the N-terminal half and of peptidylprolyl isomerase activity on the virulence of Legionella pneumophila. Infect. Immun. 2003;71(8):4389–97. doi: 10.1128/IAI.71.8.4389-4397.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wagner C, Khan AS, Kamphausen T, Schmausser B, Unal C, Lorenz U, et al. Collagen binding protein Mip enables Legionella pneumophila to transmigrate through a barrier of NCI-H292 lung epithelial cells and extracellular matrix. Cell. Microbiol. 2007;9(2):450–62. [DOI] [PubMed] [Google Scholar]
- 43.Norville IH, Harmer NJ, Harding SV, Fischer G, Keith KE, Brown KA, et al. A Burkholderia pseudomallei macrophage infectivity potentiator-like protein has rapamycin-inhibitable peptidylprolyl isomerase activity and pleiotropic effects on virulence. Infect. Immun. 2011;79(11):4299–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dunyak BM, Gestwicki JE. Peptidyl-proline isomerases (PPIases): Targets for natural products and natural product-inspired compounds. J. Med. Chem. 2016;59(21):9622–44. doi: 10.1021/acs.jmedchem.6b00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feng X, Pomplun S, Hausch F. Recent progress in FKBP ligand development. Curr. Mol. Pharmacol. 2015;9(1):27–36. doi: 10.2174/1874467208666150519113313 [DOI] [PubMed] [Google Scholar]
- 46.Hopkins S, Gallay P. Cyclophilin inhibitors: an emerging class of therapeutics for the treatment of chronic hepatitis C infection. Viruses. 2012;4(11):2558–77. doi: 10.3390/v4112558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolos JM, Voll AM, Bauder M, Hausch F. FKBP ligands-where we are and where to go? Front. Pharmacol. 2018;9:1425. doi: 10.3389/fphar.2018.01425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pomplun S, Sippel C, Hahle A, Tay D, Shima K, Klages A, et al. Chemogenomic profiling of human and microbial FK506-binding proteins. J. Med. Chem. 2018;61(8):3660–73. doi: 10.1021/acs.jmedchem.8b00137 [DOI] [PubMed] [Google Scholar]
- 49.Rasch J, Theuerkorn M, Unal C, Heinsohn N, Tran S, Fischer G, et al. Novel cycloheximide derivatives targeting the moonlighting protein Mip exhibit specific antimicrobial activity against Legionella pneumophila. Front. Bioeng. Biotechnol. 2015;3:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seufert F, Kuhn M, Hein M, Weiwad M, Vivoli M, Norville IH, et al. Development, synthesis and structure-activity-relationships of inhibitors of the macrophage infectivity potentiator (Mip) proteins of Legionella pneumophila and Burkholderia pseudomallei. Bioorg. Med. Chem. 2016;24(21):5134–47. [DOI] [PubMed] [Google Scholar]
- 51.Unal CM, Steinert M. Microbial peptidyl-prolyl cis/trans isomerases (PPIases): virulence factors and potential alternative drug targets. Microbiology and molecular biology reviews: MMBR. 2014;78(3):544–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Begley DW, Fox D 3rd, Jenner D, Juli C, Pierce PG, Abendroth J, et al. A structural biology approach enables the development of antimicrobials targeting bacterial immunophilins. Antimicrob. Agents Chemother. 2014;58(3):1458–67. doi: 10.1128/AAC.01875-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Juli C, Sippel M, Jager J, Thiele A, Weiwad M, Schweimer K, et al. Pipecolic acid derivatives as small-molecule inhibitors of the Legionella MIP protein. J. Med. Chem. 2011;54(1):277–83. [DOI] [PubMed] [Google Scholar]
- 54.Reimer A, Seufert F, Weiwad M, Ebert J, Bzdyl NM, Kahler CM, et al. Inhibitors of macrophage infectivity potentiator-like PPIases affect neisserial and chlamydial pathogenicity. International journal of antimicrobial agents. 2016;48(4):401–8. doi: 10.1016/j.ijantimicag.2016.06.020 [DOI] [PubMed] [Google Scholar]
- 55.Iwasaki J, Lorimer DD, Vivoli-Vega M, Kibble EA, Peacock CS, Abendroth J, et al. Broad-spectrum in vitro activity of macrophage infectivity potentiator inhibitors against Gram-negative bacteria and Leishmania major. J. Antimicrob. Chemother. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mo YY, Cianciotto NP, Mallavia LP. Molecular cloning of a Coxiella burnetii gene encoding a macrophage infectivity potentiator (Mip) analogue. Microbiology (Reading). 1995;141 (Pt 11):2861–71. [DOI] [PubMed] [Google Scholar]
- 57.Lundemose AG, Rouch DA, Birkelund S, Christiansen G, Pearce JH. Chlamydia trachomatis Mip-like protein. Mol Microbiol. 1992;6(17):2539–48. doi: 10.1111/j.1365-2958.1992.tb01430.x [DOI] [PubMed] [Google Scholar]
- 58.Porter SR, Czaplicki G, Mainil J, Guatteo R, Saegerman C. Q Fever: current state of knowledge and perspectives of research of a neglected zoonosis. Int J Microbiol. 2011;2011:248418. doi: 10.1155/2011/248418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Santiago AE, Mann BJ, Qin A, Cunningham AL, Cole LE, Grassel C, et al. Characterization of Francisella tularensis Schu S4 defined mutants as live-attenuated vaccine candidates. Pathogens and disease. 2015;73(6):ftv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lundemose AG, Kay JE, Pearce JH. Chlamydia trachomatis Mip-like protein has peptidyl-prolyl cis/trans isomerase activity that is inhibited by FK506 and rapamycin and is implicated in initiation of chlamydial infection. Mol. Microbiol. 1993;7(5):777–83. [DOI] [PubMed] [Google Scholar]
- 61.Scheuplein N, Bzdyl NM, Lohr T, Kibble E, Hasenkopf A, Herbst C, et al. Analysis of structure–activity-relationships of novel inhibitors of the macrophage infectivity potentiator (Mip) proteins of Neisseria meningitidis, Neisseria gonorrhoeae, and Burkholderia pseudomallei. J. Med. Chem. 2023: accepted 13 June 2023. doi: 10.1021/acs.jmedchem.3c00458 [DOI] [PubMed] [Google Scholar]
- 62.Vivoli M, Renou J, Chevalier A, Norville IH, Diaz S, Juli C, et al. A miniaturized peptidyl-prolyl isomerase enzyme assay. Anal. Biochem. 2017;536:59–68. doi: 10.1016/j.ab.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 63.Mo YY, Seshu J, Wang D, Mallavia LP. Synthesis in Escherichia coli of two smaller enzymically active analogues of Coxiella burnetii macrophage infectivity potentiator (CbMip) protein utilizing a single open reading frame from the cbmip gene. Biochem. J. 1998;335 (Pt 1):67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Howe D, Melnicakova J, Barak I, Heinzen RA. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell. Microbiol. 2003;5(7):469–80. [DOI] [PubMed] [Google Scholar]
- 65.Metters G, Hemsley C, Norville I, Titball R. Identification of essential genes in Coxiella burnetii. Microb Genom. 2023;9(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–9. doi: 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vallejo Esquerra E, Yang H, Sanchez SE, Omsland A. Physicochemical and nutritional requirements for axenic replication suggest physiological basis for Coxiella burnetii niche restriction. Frontiers in cellular and infection microbiology. 2017;7:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Omsland A, Cockrell DC, Howe D, Fischer ER, Virtaneva K, Sturdevant DE, et al. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. U. S. A. 2009;106(11):4430–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beare PA, Sandoz KM, Larson CL, Howe D, Kronmiller B, Heinzen RA. Essential role for the response regulator PmrA in Coxiella burnetii type 4B secretion and colonization of mammalian host cells. J. Bacteriol. 2014;196(11):1925–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lifshitz Z, Burstein D, Peeri M, Zusman T, Schwartz K, Shuman HA, et al. Computational modeling and experimental validation of the Legionella and Coxiella virulence-related type-IVB secretion signal. Proc. Natl. Acad. Sci. U. S. A. 2013;110(8):E707–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, et al. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J. Bacteriol. 2013;195(17):3914–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kuba M, Neha N, De Souza DP, Dayalan S, Newson JPM, Tull D, et al. Coxiella burnetii utilizes both glutamate and glucose during infection with glucose uptake mediated by multiple transporters. Biochem. J. 2019;476(19):2851–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martinez E, Allombert J, Cantet F, Lakhani A, Yandrapalli N, Neyret A, et al. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc. Natl. Acad. Sci. U. S. A. 2016;113(23):E3260–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norville IH, Hartley MG, Martinez E, Cantet F, Bonazzi M, Atkins TP. Galleria mellonella as an alternative model of Coxiella burnetii infection. Microbiology (Reading). 2014;160(Pt 6):1175–81. [DOI] [PubMed] [Google Scholar]
- 75.Stead CM, Omsland A, Beare PA, Sandoz KM, Heinzen RA. Sec-mediated secretion by Coxiella burnetii. BMC Microbiol. 2013;13:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seshadri R, Hendrix LR, Samuel JE. Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect. Immun. 1999;67(11):6026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Seshu J, McIvor KL, Mallavia LP. Antibodies are generated during infection to Coxiella burnetii macrophage infectivity potentiator protein (Cb-Mip). Microbiology and immunology. 1997;41(4):371–6. [DOI] [PubMed] [Google Scholar]
- 78.Vigil A, Ortega R, Nakajima-Sasaki R, Pablo J, Molina DM, Chao CC, et al. Genome-wide profiling of humoral immune response to Coxiella burnetii infection by protein microarray. Proteomics. 2010;10(12):2259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiong X, Wang X, Wen B, Graves S, Stenos J. Potential serodiagnostic markers for Q fever identified in Coxiella burnetii by immunoproteomic and protein microarray approaches. BMC Microbiol. 2012;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang GQ, Samuel JE. Identification and cloning potentially protective antigens of Coxiella burnetii using sera from mice experimentally infected with Nine Mile phase I. Ann. N. Y. Acad. Sci. 2003;990:510–20. [DOI] [PubMed] [Google Scholar]
- 81.Leuzzi R, Serino L, Scarselli M, Savino S, Fontana MR, Monaci E, et al. Ng-MIP, a surface-exposed lipoprotein of Neisseria gonorrhoeae, has a peptidyl-prolyl cis/trans isomerase (PPIase) activity and is involved in persistence in macrophages. Mol. Microbiol. 2005;58(3):669–81. [DOI] [PubMed] [Google Scholar]
- 82.Beare PA, Heinzen RA. Gene inactivation in Coxiella burnetii. Methods Mol. Biol. 2014;1197:329–45. [DOI] [PubMed] [Google Scholar]
- 83.Beare PA, Howe D, Cockrell DC, Omsland A, Hansen B, Heinzen RA. Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J. Bacteriol. 2009;191(5):1369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez E, Cantet F, Bonazzi M. Generation and multi-phenotypic high-content screening of Coxiella burnetii transposon mutants. J Vis Exp. 2015(99):e52851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dresler J, Klimentova J, Pajer P, Salovska B, Fucikova AM, Chmel M, et al. Quantitative proteome profiling of Coxiella burnetii reveals major metabolic and stress differences under axenic and cell culture cultivation. Front. Microbiol. 2019;10:2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bitew MA, Hofmann J, De Souza DP, Wawegama NK, Newton HJ, Sansom FM. SdrA, an NADP(H)-regenerating enzyme, is crucial for Coxiella burnetii to resist oxidative stress and replicate intracellularly. Cell. Microbiol. 2020;22(5):e13154. [DOI] [PubMed] [Google Scholar]
- 87.Flannagan RS, Cosio G, Grinstein S. Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 2009;7(5):355–66. doi: 10.1038/nrmicro2128 [DOI] [PubMed] [Google Scholar]
- 88.Lam GY, Huang J, Brumell JH. The many roles of NOX2 NADPH oxidase-derived ROS in immunity. Semin. Immunopathol. 2010;32(4):415–30. doi: 10.1007/s00281-010-0221-0 [DOI] [PubMed] [Google Scholar]
- 89.Neff L, Daher S, Muzzin P, Spenato U, Gulacar F, Gabay C, et al. Molecular characterization and subcellular localization of macrophage infectivity potentiator, a Chlamydia trachomatis lipoprotein. J. Bacteriol. 2007;189(13):4739–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lundemose AG, Rouch DA, Penn CW, Pearce JH. The Chlamydia trachomatis Mip-like protein is a lipoprotein. J. Bacteriol. 1993;175(11):3669–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lundemose AG, Birkelund S, Fey SJ, Larsen PM, Christiansen G. Chlamydia trachomatis contains a protein similar to the Legionella pneumophila mip gene product. Mol. Microbiol. 1991;5(1):109–15. [DOI] [PubMed] [Google Scholar]
- 92.Omsland A, Sager J, Nair V, Sturdevant DE, Hackstadt T. Developmental stage-specific metabolic and transcriptional activity of Chlamydia trachomatis in an axenic medium. Proc. Natl. Acad. Sci. U. S. A. 2012;109(48):19781–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Clay KA, Hartley MG, Armstrong S, Bewley KR, Godwin K, Rayner E, et al. Evaluation of the efficacy of doxycycline, ciprofloxacin, levofloxacin, and co-trimoxazole using in vitro and in vivo models of Q Fever. Antimicrob. Agents Chemother. 2021;65(11):e0067321. doi: 10.1128/AAC.00673-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ignasiak K, Maxwell A. Galleria mellonella (greater wax moth) larvae as a model for antibiotic susceptibility testing and acute toxicity trials. BMC Res. Notes. 2017;10(1):428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piatek M, Sheehan G, Kavanagh K. Galleria mellonella: The versatile host for drug discovery, in vivo toxicity testing and characterising host-pathogen interactions. Antibiotics (Basel). 2021;10(12). doi: 10.3390/antibiotics10121545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thomas RJ, Hamblin KA, Armstrong SJ, Muller CM, Bokori-Brown M, Goldman S, et al. Galleria mellonella as a model system to test the pharmacokinetics and efficacy of antibiotics against Burkholderia pseudomallei. International journal of antimicrobial agents. 2013;41(4):330–6. doi: 10.1016/j.ijantimicag.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 97.Venthur H, Lizana P, Manosalva L, Rojas V, Godoy R, Rocha A, et al. Analysis of glutathione-S-transferases from larvae of Galleria mellonella (Lepidoptera, Pyralidae) with potential alkaloid detoxification function. Front. Physiol. 2022;13:989006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ghartey-Kwansah G, Li Z, Feng R, Wang L, Zhou X, Chen FZ, et al. Comparative analysis of FKBP family protein: evaluation, structure, and function in mammals and Drosophila melanogaster. BMC Dev Biol. 2018;18(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Perrucci GL, Gowran A, Zanobini M, Capogrossi MC, Pompilio G, Nigro P. Peptidyl-prolyl isomerases: a full cast of critical actors in cardiovascular diseases. Cardiovasc. Res. 2015;106(3):353–64. doi: 10.1093/cvr/cvv096 [DOI] [PubMed] [Google Scholar]
- 100.Omsland A, Beare PA, Hill J, Cockrell DC, Howe D, Hansen B, et al. Isolation from animal tissue and genetic transformation of Coxiella burnetii are facilitated by an improved axenic growth medium. Appl. Environ. Microbiol. 2011;77(11):3720–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sanchez SE, Vallejo-Esquerra E, Omsland A. Use of axenic culture tools to study Coxiella burnetii. Curr. Protoc. Microbiol. 2018;50(1):e52. [DOI] [PubMed] [Google Scholar]
- 102.Jaton K, Peter O, Raoult D, Tissot JD, Greub G. Development of a high throughput PCR to detect Coxiella burnetii and its application in a diagnostic laboratory over a 7-year period. New Microbes New Infect. 2013;1(1):6–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kuba M, Neha N, Newton P, Lee YW, Bennett-Wood V, Hachani A, et al. EirA is a novel protein essential for intracellular replication of Coxiella burnetii. Infect. Immun. 2020;88(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32(2):e19. doi: 10.1093/nar/gnh014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dummler A, Lawrence AM, de Marco A. Simplified screening for the detection of soluble fusion constructs expressed in E. coli using a modular set of vectors. Microb. Cell Fact. 2005;4:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen Y, Guenther JM, Gin JW, Chan LJG, Costello Z, Ogorzalek TL, et al. Automated "Cells-To-Peptides" sample preparation workflow for high-throughput, quantitative proteomic assays of microbes. J. Proteome Res. 2019;18(10):3752–61. doi: 10.1021/acs.jproteome.9b00455 [DOI] [PubMed] [Google Scholar]
- 107.Ishihama Y, Rappsilber J, Mann M. Modular stop and go extraction tips with stacked disks for parallel and multidimensional Peptide fractionation in proteomics. J. Proteome Res. 2006;5(4):988–94. doi: 10.1021/pr050385q [DOI] [PubMed] [Google Scholar]
- 108.Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat. Protoc. 2007;2(8):1896–906. doi: 10.1038/nprot.2007.261 [DOI] [PubMed] [Google Scholar]
- 109.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature biotechnology. 2008;26(12):1367–72. doi: 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- 110.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Molecular & cellular proteomics: MCP. 2014;13(9):2513–26. doi: 10.1074/mcp.M113.031591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nature methods. 2016;13(9):731–40. doi: 10.1038/nmeth.3901 [DOI] [PubMed] [Google Scholar]
- 112.Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47(D1):D442–D50. doi: 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(22):11033. doi: 10.1093/nar/gkw880 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Multiple sequence alignment of Mip proteins from C. trachomatis (CtMip), B. pseudomallei (BpMip) C. burnetii (CbMip) and L. pneumophila (LpMip) using CLUSTAL O (1.2.4). The start of the highly conserved catalytic/PPIase domain is indicated by an open triangle. The essential amino acid (Asp) in LpMip for PPIase activity is conserved in all proteins and is indicated by a closed triangle. (B) Percentage identity matrix of the PPIase domain of Mip from C. trachomatis (CtMip-cat), B. pseudomallei (BpMip-cat) C. burnetii (CbMip-cat) and L. pneumophila (LpMip-cat) using CLUSTAL 2.1.
(TIF)
Data were collected in a single experiment for CbMip as described in the experimental section. Each inhibitor concentration of SF235 (closed circle) and ANCH37 (blue square) was tested three times. Data were fitted to equation as described previously [55]. Results are representative of at least two experiments conducted on separate days with different preparations of inhibitor. Error bars show standard error of the mean.
(TIF)
Intracellular replication of C. burnetii NMII in THP-1 cells in the presence of 50 μM AN296 (blue triangle), 31 μM chloramphenicol (Cm) (open circle) or control (closed circle), introduced after the 4 h infection period. Error bars represent standard error of the mean (n ═ 5). ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test.
(TIF)
(A) IC50 values for SF235 and AN296 and DMSO after incubation for the indicated periods of time were determined in THP-1 and HeLa cells using the Cell Proliferation Reagent WST-1. (B) Cytotoxicity was measured after THP-1 cells were incubated with SF235 (50 μM), AN296 (50 μM), chloramphenicol (cm, 31 μM) or control (0.1% DMSO) for the indicated period of time using Roche LDH Cell Cytotoxicity kit. No significant difference in cytotoxicity was found between the inhibitors and the control (0.1% DMSO). Data is presented as percent cytotoxicity relative to the control with error bars representing the SD from three independent experiments; p values were determined using two-way ANOVA followed by Dunnett’s multiple comparison post-test.
(TIF)
Overview of transposon mutants generated in the Coxiella burnetii NMII Metters et al. library. Confirmed transposon insertion sites are indicated by red vertical lines. All TA insertion sites, possible locations where the transposable element can intergrate, are indicated by black vertical lines. (A) Transposon insertion sites around the cbmip (cbu0630) genetic locus. (B) Enhanced view of transposon insertion sites within the cbmip gene. Two out of a possible 37 mutants were identified in the transposon mutant library. (C) Detailed analysis of transposon insertion sites in cbmip. In lower case text is the DNA coding sequence for the C. burnetii mip gene (cbu0630) of strain C. burnetii NMII. Below, in upper case text, is the encoded amino acid sequence, green box indicates the PPIase catalytic domain. The two transposon insertion sites (TA) identified in the transposon mutant library are indicated by blue triangles. The first ATG following the transposon site is indicated by red boxes. In both instances of transposon insertion in the cbmip sequence, the downstream ATG from which protein translation would initiate is in-frame with the original full-length gene and therefore would result in the production of a truncated CbMip protein in the C. burnetii cbu0630 transposon mutants.
(TIF)
The stability of compound AN296 under acidic conditions (pH of 4.75), was investigated by HPLC chromatography. A stock solution of AN296 (100 μM) was prepared in Buffer C (composition in S4 Table) which mimicked the inorganic components of ACCM-2 media and was incubated at 37°C for a period of 7 days. Data is presented as the amount of AN296 remaining compared to day 0 (at 250 nm and 260 nm), with error bars representing the standard deviation.
(TIF)
Bioluminescence was measured as an indicator of C. burnetii-lux replication. The strain was inoculated at a concentration of 1 × 106 GE/mL into ACCM-2 media with (A) 50 μM or (B) 25 μM of CbMip inhibitors SF235 (grey square), AN296 (closed triangle) or vehicle control (open circle) and grown over 5 days. Data is presented as RLU (relative light units) with error bars representing the standard deviation from three independent experiments. **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. p values were determined using two-way ANOVA, followed by Dunnett’s multiple comparison post-test.
(TIF)
Each larva received a single 10 μL injection of in PBS containing either (A) AN296 or (B) SF235 or vehicle control into the right proleg. Inhibitors were tested at increasing concentrations starting at 10 μM and increasing up to 50 μM, 250 μM and 500 μM (groups of n ═ 10). Larvae were monitored everyday via twitching response over 10 days. All but two larvae, which received 10 μM of AN296, survived the entire duration of the experiment.
(TIF)
(DOC)
(DOC)
(DOC)
(DOC)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript, supporting files and the MS dataset has been deposited into the PRIDE ProteomeXchange Consortium repository, the dataset identifier is PXD036679.