Abstract
Sleep disruptions are quite common in psychological disorders, but the underlying mechanism remains obscure. Wolfram syndrome 1 (WS1) is an autosomal recessive disease mainly characterized by diabetes insipidus/mellitus, neurodegeneration and psychological disorders. It is caused by loss-of function mutations of the WOLFRAM SYNDROME 1 (WFS1) gene, which encodes an endoplasmic reticulum (ER)-resident transmembrane protein. Heterozygous mutation carriers do not develop WS1 but exhibit 26-fold higher risk of having psychological disorders. Since WS1 patients display sleep abnormalities, we aimed to explore the role of WFS1 in sleep regulation so as to help elucidate the cause of sleep disruptions in psychological disorders. We found in Drosophila that knocking down wfs1 in all neurons and wfs1 mutation lead to reduced sleep and dampened circadian rhythm. These phenotypes are mainly caused by lack of wfs1 in dopamine 2-like receptor (Dop2R) neurons which act to promote wake. Consistently, the influence of wfs1 on sleep is blocked or partially rescued by inhibiting or knocking down the rate-limiting enzyme of dopamine synthesis, suggesting that wfs1 modulates sleep via dopaminergic signaling. Knocking down wfs1 alters the excitability of Dop2R neurons, while genetic interactions reveal that lack of wfs1 reduces sleep via perturbation of ER-mediated calcium homeostasis. Taken together, we propose a role for wfs1 in modulating the activities of Dop2R neurons by impinging on intracellular calcium homeostasis, and this in turn influences sleep. These findings provide a potential mechanistic insight for pathogenesis of diseases associated with WFS1 mutations.
Author summary
Psychiatric disorders are often accompanied by sleep disruptions, but the causes of theses disruptions are largely unclear. Wolfram syndrome 1 (WS1) is a neurodegenerative disease co-morbid with psychiatric disorders. It is caused by homozygous mutation of the WOLFRAM SYNDROME 1 (WFS1) gene, which encodes a transmembrane protein that localizes to the endoplasmic membrane (ER). Heterozygous mutation carriers do not develop WS1 but display 26-fold higher risk for having psychiatric disorders. WS1 patients also experience sleep abnormalities, therefore here we investigate the role of WFS1 in sleep regulation in hope that this will help us better understand the sleep disruptions associated with psychiatric disorders. Using the fruit fly Drosophila, we found that wfs1 deficiency in wake-promoting dopamine receptor neurons leads to reduced sleep and dampened circadian rhythm. Consistently, pharmacological treatment and genetic interaction analysis reveal that the sleep-modulating role of wfs1 requires dopamine. Furthermore, we found that the sleep reduction caused by wfs1 deficiency is likely due to alteration of ER-mediated calcium homeostasis, which in turn influences neural activity and neurotransmitter release. These findings highlight a role for dopamine and dopamine receptor neurons in WFS1-related sleep disturbances, which may serve as a common mechanism for sleep disruptions associated with psychiatric disorders.
Introduction
Sleep disruptions are common in individuals with psychiatric disorders, and sleep disturbances are risk factors for future onset of depression [1]. However, the mechanism underlying sleep disruptions in psychiatric disorders are largely unclear. Wolfram Syndrome 1 (WS1) is an autosomal recessive neurodegenerative disease characterized by diabetes insipidus, diabetes mellitus, optic atrophy, deafness and psychiatric abnormalities such as severe depression, psychosis and aggression [2–4]. It is caused by homozygous (and compound heterozygous) mutation of the WOLFRAM SYNDROME 1 (WFS1) gene, which encodes wolframin, an endoplasmic reticulum (ER) resident protein highly expressed in the heart, brain, and pancreas. On the other hand, heterozygous mutation of WFS1 does not lead to WS1 but increase the risk of depression by 26 fold [5,6]. A study in mice further confirmed that WFS1 mutation is causative for depression [7]. Consistent with the comorbidity of psychiatric conditions and sleep abnormalities, WS1 patients also experience increased sleep problems compared to individuals with type I diabetes and healthy controls [8]. It has been proposed that sleep symptoms can be used as a biomarker of the disease, especially during relatively early stages, but the mechanisms underlying these sleep disturbances are unclear. Considering that heterozygous WFS1 mutation is present in up to 1% of the population and may be a significant cause of psychiatric disorder in the general population, we decided to investigate the role of wolframin in sleep regulation so as to probe the mechanism underlying sleep disruptions in psychiatric disorders [5,6,9].
Although the wolframin protein does not possess distinct functional domains, a number of ex vivo studies in cultured cells demonstrated a role for it in regulating cellular responses to ER stress and calcium homeostasis, as well as ER-mitochondria cross-talk [10–13]. Mice that lack Wfs1 in pancreatic β cells develop glucose intolerance and insulin deficiency due to enhanced ER stress and apoptosis [14–16]. Knocking out Wfs1 in layer 2/3 pyramidal neurons of the medial prefrontal cortex in mice results in increased depression-like behaviors in response to acute restraint stress. This is accompanied by hyperactivation of the hypothalamic-pituitary-adrenal axis and altered accumulation of growth and neurotrophic factors, possibly due to defective ER function [7]. A more recent study in Drosophila found that knocking down wfs1 in the nervous system does not increase ER stress, but enhances the susceptibility to oxidative stress-, endotoxicity- and tauopathy-induced behavioral deficits and neurodegeneration [17]. Overall, the physiological function of wolframin in vivo, especially in the brain, remains elusive for the most part.
Here we identified a role for wolframin in regulating sleep and circadian rhythm in flies. Wfs1 deficiency in the dopamine 2-like receptor (Dop2R) neurons leads to reduced sleep, while inhibiting dopamine synthesis blocks the effect of wfs1 on sleep, implying that wfs1 influences sleep via dopaminergic signaling. We further found that these Dop2R neurons function to promote wakefulness. Depletion of wfs1 alters neural activity, which leads to increased wakefulness and reduced sleep. Consistent with this, we found that knocking down the ER calcium channel Ryanodine receptor (RyR) or 1,4,5-trisphosphate receptor (Itpr) rescues while knocking down the sarco(endoplasmic)reticulum ATPase SERCA synergistically enhances the short-sleep phenotype caused by wfs1 deficiency, indicating that wolframin regulates sleep by modulating calcium homeostasis [18]. Taken together, our findings provide a potential mechanism for the sleep disruptions associated with WFS1 mutation, and deepen our understanding of the functional role of wolframin in the brain.
Results
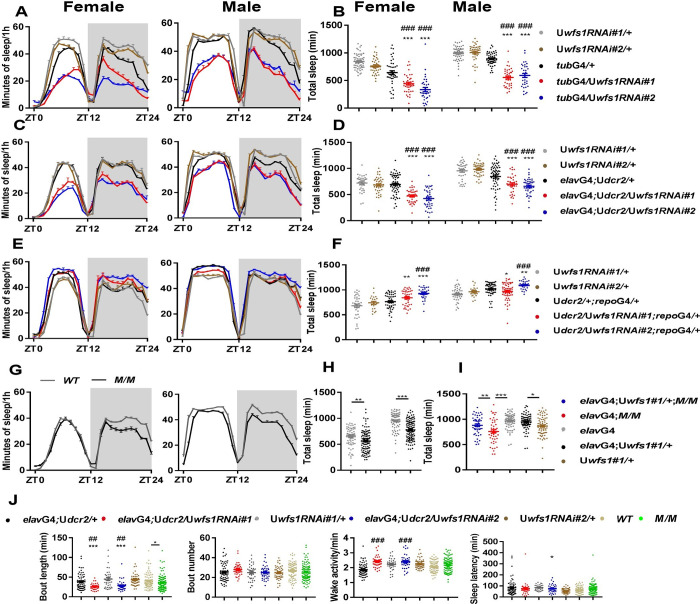
Wfs1 deficiency leads to reduced sleep and dampened locomotor rhythm
A previous study reported that wolframin functions in both neurons and glial cells to regulate climbing ability, life span and neurodegeneration [17]. Therefore, to investigate the consequence of wfs1 deficiency on sleep and circadian rhythm, we knocked down wfs1 in all cells, neurons and glial cells using tubulin(tub)-GAL4, elav-GAL4 and repo-GAL4, respectively [19,20]. Depletion of wfs1 in all cells and neurons result in substantially reduced sleep duration and power of locomotor rhythm which indicates dampened circadian rhythm, while the period of the rhythm is not altered (Fig 1A–1D and Table 1). These phenotypes are consistent between the two independent RNAi lines used, both of which lead to significant reduction of wfs1 mRNA level (S1A Fig). On the other hand, knocking down wfs1 in glial cells did not significantly alter sleep or locomotor rhythm, strongly suggesting that wolframin acts in neurons to regulate sleep and locomotor rhythm (Fig 1E and 1F and Table 1). To ensure that the phenotypes we observed in wfs1 RNAi flies are not merely due to over-expression of RNAi, we adopted a GFP RNAi as a control. Expressing GFP-RNAi does not significantly alter sleep or locomotor rhythm (S1B Fig and Table 1).
Fig 1. Wfs1 deficiency leads to reduced sleep and dampened locomotor rhythm.
The sleep of female and male flies are monitored under 12 h light: 12 h dark (LD) condition. (A, C, E and G) Sleep profile of wfs1 RNAi, mutant and control flies. Gray shade indicates the dark period. (B, D, F and H) Daily sleep duration of wfs1 RNAi, mutant and control flies. (I) Daily sleep duration of wfs1 mutant, pan-neuronal rescue and control flies. (J) Sleep bout length, bout number, wake activity and sleep latency of wfs1 RNAi, mutant and control flies. For comparison between RNAi or over-expression flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, ##P < 0.01, ###P < 0.001; compared to UAS control, *P < 0.05, **P < 0.01, ***P < 0.001. For comparison between mutant vs. control, Mann-Whitney test or Student’s t-test was used: *P < 0.05, **P < 0.01, ***P < 0.001. For comparison between rescue vs. control, one-way ANOVA was used: **P < 0.01. n = 20–80. Error bars represent standard error of the mean (SEM). G4, GAL4; U, UAS; M, wfs1MI14041; ZT, Zeitgeber Time (ZT0 is the time of lights on).
Table 1. Wfs1 deficiency leads to dampened locomotor rhythm.
| Genotype | Period±SEM (hour) | Power±SEM | Rhythmicity | N |
|---|---|---|---|---|
| Uwfs1RNAi#1/+ | 23.71±0.03 | 63.20±3.82 | 95% | 75 |
| Uwfs1RNAi#2/+ | 23.69±0.03 | 58.70±4.04 | 95% | 65 |
| tubG4/+ | 24.12±0.04 | 74.3±5.99 | 88% | 57 |
| tubG4/Uwfs1RNAi#1 | 24.25±0.13 | 8.00±2.41*** ### | 30% | 27 |
| tubG4/Uwfs1RNAi#2 | 24.28±0.17 | 22.17±5.43** ### | 60% | 15 |
| elavG4 | 23.84±0.04 | 49.42±3.47 | 83% | 77 |
| elavG4;Uwfs1#1/+;MM | 23.71±0.05 | 59.13±5.69$ $ $ | 88% | 48 |
| elavG4;M/M | 23.53±0.11 | 18.00±4.51### | 43% | 42 |
| elavG4;Uwfs1#1/+ | 23.78±0.06 | 74.85±5.41## | 97% | 35 |
| Uwfs1/+ | 23.65±0.04 | 69.75±7.89 | 86% | 35 |
| elavG4;Uwfs1RNAi#1/+ | 23.87±0.06 | 24.35±3.46*** ### | 70% | 43 |
| elavG4;Uwfs1RNAi#2/+ | 23.96±0.11 | 10.24±2.53*** ### | 33% | 36 |
| Udcr2/+;repoG4/+ | 23.92±0.11 | 56.51±7.12 | 91% | 47 |
| Udcr2/Uwfs1RNAi#1;repoG4/+ | 23.69±0.07 | 72.62±7.45 | 100% | 24 |
| Udcr2/Uwfs1RNAi#2;repoG4/+ | 23.79±0.08 | 64.19±6.04 | 100% | 34 |
| Udcr2/+;cryG4-16/+ | 24.90±0.09 | 46.04±7.16 | 86% | 28 |
| Udcr2/Uwfs1RNAi#1;cryG4-16/+ | 24.46±0.08 | 65.14±5.37 | 100% | 26 |
| Udcr2/Uwfs1RNAi#2;cryG4-16/+ | 24.50±0.06 | 63.70±6.61 | 88% | 32 |
| timG4/+ | 24.44±0.04 | 47.15±6.06 | 76% | 33 |
| timG4/wfs1RNAi#1 | 24.33±0.06 | 66.63±7.51 | 84% | 31 |
| timG4/wfs1RNAi#2 | 24.36±0.04 | 76.38±6.72 | 93% | 43 |
| WT | 23.73±0.04 | 50.80±4.85 | 82% | 62 |
| M/M | 23.62±0.04 | 33.31±4.38@@@ | 47% | 95 |
| Dop2RG4 | 23.70±0.04 | 62.19±4.74 | 83% | 84 |
| Dop2RG4;Uwfs1RNAi#1/+ | 23.78±0.05 | 29.69±3.66*** ### | 57% | 90 |
| Dop2RG4;Uwfs1RNAi#2/+ | 24.16±0.32 | 21.35±3.41*** ### | 43% | 82 |
| GαoG4/+ | 23.85±0.09 | 36.60±5.35 | 70% | 37 |
| GαoG4/Uwfs1RNAi#1 | 23.72±0.15 | 7.55±2.86 *** ### | 32% | 28 |
| GαoG4/Uwfs1RNAi#2 | 24.00±0 | 1.41±0.73 *** ### | 5% | 21 |
| UGFPRNAi/+ | 23.61±0.04 | 75.71±10.04 | 95% | 23 |
| tubG4/UGFPRNAi | 24±0.03 | 60.97±7.37 | 82% | 34 |
| elavG4;Udcr2/+ | 23.78±0.07 | 76.02±8.63 | 100% | 20 |
| elavG4;Udcr2/UGFPRNAi | 23.82±0.07 | 77.96±9.19 | 91% | 24 |
| Udcr2/UGFPRNAi;repoG4/+ | 23.78±0.07 | 88.82±8.88## | 100% | 32 |
One-way ANOVA, #/*P < 0.05, ##/**P < 0.01, ###/***P < 0.001. Mann-Whitney test, ###/$ $ $/@@@P < 0.001. # compared with the GAL4 controls; * compared with the UAS controls; $ compared with GAL4 under mutant background; @ compared with WT. G4, GAL4; U, UAS; M, wfs1MI14041.
To further validate that the sleep and circadian disruptions are indeed due to wfs1 deficiency, we assessed the phenotypic effects of wfs1MI14041 mutation which is expected to produce a C-terminal truncated wolframin and may act as a partial loss of function mutation [17]. Homozygous wfs1MI14041 mutants also show significantly reduced sleep and power, consistent with the RNAi results (Fig 1G and 1H and Table 1). Given that the sleep reduction appears to be more prominent in male mutants compared to females, we focused on males for the remainder of this study. To test whether neuronal wolframin expression is sufficient for maintaining sleep and locomotor rhythm, we expressed wfs1 in the neurons of wfs1MI14041 mutants and found that both the reduced sleep and dampened rhythm were rescued in these flies (Figs 1I and S1C and Table 1). Taken together, these series of results indicate that neuronal wolframin is both necessary and sufficient for sleep and locomotor rhythm.
Next, we systematically analyzed the effects of knocking down wfs1 pan-neuronally on sleep structure. wfs1 RNAi lead to decreased sleep both in the light and dark phases, while waking activity is not significantly altered, indicating that the short-sleep phenotype is not caused by hyper-activity (Fig 1J). The sleep reduction is a result of decreased sleep bout length but not bout number, which means that wfs1 depletion perturbs sleep maintenance but not sleep initiation. Consistently, sleep latency is not altered in wfs1 RNAi flies, further implicating that sleep initiation system is intact.
Wfs1 deficiency during development leads to reduced sleep and dampened locomotor rhythm in adults
Neurodegeneration-related symptoms start to occur within the first decade of WS1 patients, with an average age of development ranging from 11–16 years [2–4]. Therefore, we investigated whether the sleep and circadian phenotypes observed in flies that lack wfs1 are caused by deficiency during developmental and/or adult stage. We used a temperature sensitive tubulin (tub)GAL80ts to knock down wfs1 specifically during the adult or developmental stage [21]. tubGAL80ts represses the transcriptional activities of GAL4 at permissive temperature (18°C), thus GAL4-driven transcription can only occur under restrictive temperature (29°C). When RNAi expression is restricted to adults, neither sleep duration nor locomotor rhythm is significantly altered (S2A Fig and S1 Table). On the other hand, when RNAi is expressed exclusively during the developmental stage, sleep is significantly reduced (S2B Fig). We noticed that when flies are raised under 29°C, the adult locomotor rhythm is dampened even for some of the control lines (S1 Table). Therefore, we could not assess the influence of knocking down wfs1 during development on locomotor rhythm.
Previous study has shown that wfs1 deficiency results in significant behavioral deficits and neurodegeneration in flies by 30 days of age [17]. Therefore, we also examined the effects of wfs1 deficiency on sleep and locomotor rhythm in aged flies. We found that only one of the RNAi lines when expressed leads to significantly decreased sleep, and the extent of this reduction appears to be smaller than that of younger flies (S3A–S3D Fig). Aged wfs1 mutants display comparable decrease in sleep compared to that of younger flies (S3E and S3F Fig). Significant reduction of the power of locomotor rhythm can be observed in aged wfs1 RNAi and mutant flies, but the power values are relatively low even for controls which is due to aging-induced dampening of circadian rhythm (S2 Table). Overall, the sleep and locomotor rhythm phenotypes associated with wfs1 deficiency do not appear to be more severe as animals age.
Taken together, these series of findings indicate that wfs1 is required during development for flies to maintain normal sleep and locomotor rhythm when they are adults.
Wfs1 deficiency reduces sleep independent of circadian clock and light/dark condition
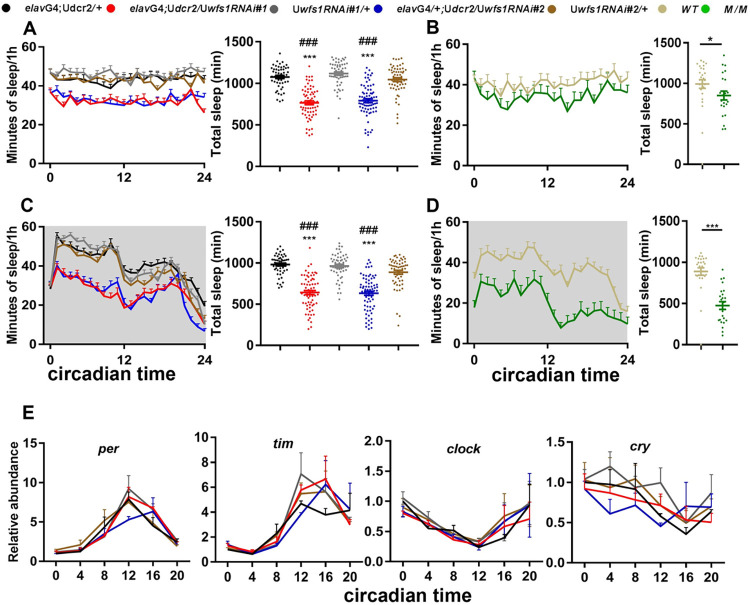
Because of the dampened locomotor rhythm in flies lacking wfs1, we tested whether circadian disruption contributed to the decreased sleep in these flies. We subjected wfs1 RNAi and mutant flies to constant light (LL) condition which rapidly evokes circadian behavioral arrhythmicity and breakdown of the molecular clock [22,23]. We were still able to observe substantial reduction in sleep under LL for both RNAi and mutant flies, indicating that wolframin regulates sleep in a manner that is independent of the circadian clock (Fig 2A and 2B). Since it has been shown that the detrimental effects of LL on the rhythm lasts for a few days into constant darkness (DD), we transferred the flies to DD after LL and again observed decreased sleep in wfs1 RNAi and mutant flies (Fig 2C and 2D) [24]. Taken together, these results demonstrate that the influence of wolframin on sleep does not require the clock or light/darkness. To further validate this, we measured the mRNA levels of core clock genes period (per), timeless (tim) and clock (clk), as well as the circadian photoreceptor cryptochrome (cry) [25]. There is no significant difference between wfs1 RNAi and control flies (Fig 2E).
Fig 2. Wfs1 deficiency reduces sleep independent of circadian clock and light/dark condition.
(A and B) The sleep profile (left panel) and daily sleep duration (right panel) of wfs1 RNAi (A) and mutant flies (B) monitored on the 4th day of constant light, along with relevant controls. (C and D) The sleep profile (left panel) and daily sleep duration (right panel) of wfs1 RNAi (C) and mutant flies (D) monitored on the first day of constant darkness (DD), along with relevant controls. Gray shade indicates the dark period. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, ###P < 0.001; compared to UAS control, ***P < 0.001. For comparison between mutant vs. control, Mann-Whitney test was used: *P < 0.05, ***P < 0.001. n = 23–73. (E) Plots of relative mRNA abundance vs circadian time (CT0, circadian time 0 is the time of subjective lights on) for clock genes determined by qRT–PCR in whole-head extracts. n = 3. For each time series, the value of G4 control at CT0 was set to 1. Error bars represent SEM. G4, GAL4; U, UAS; M, wfs1MI14041.
In addition, we investigated whether wolframin alters sleep homeostasis by assessing recovery sleep after 24h sleep deprivation and observed no significant change between RNAi flies and controls (S4 Fig).
Wolframin maintains sleep and locomotor rhythm via dopaminergic signaling
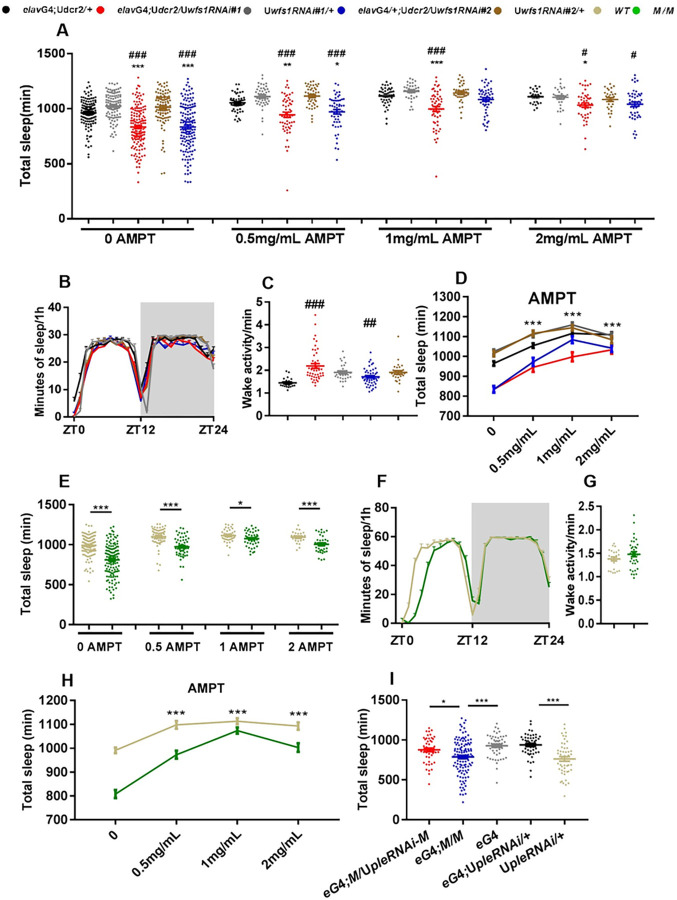
To probe the mechanism by which wfs1 deficiency influences sleep, we adopted a pharmacological approach and treated flies with drugs that target neurotransmitter systems known to regulate sleep, as well as the glutamate release inhibitor riluzole which has been shown to suppress the lifespan phenotype of wfs1 RNAi flies (S5 Fig) [17,26–28]. We found that tyrosine hydroxylase inhibitor α-methyl-para-tyrosine (AMPT) could eliminate the effects of wfs1 deficiency on sleep both in RNAi and mutant flies (Fig 3A–3C and 3E–3G). Further analysis reveals that AMPT increases sleep in a dose-dependent manner, while this increase is more prominent in wfs1 deficient flies (Fig 3D and 3H). These results suggest that the influence of wolframin on sleep requires dopaminergic signaling.
Fig 3. Wolframin maintains sleep via dopaminergic signaling.
(A) Daily sleep duration of wfs1 RNAi and control flies fed with different concentrations of AMPT. (B) The sleep profile and (C) wake activity of wfs1 RNAi and control flies fed with 2mg/mL AMPT. (D) The dosage effect of AMPT on daily sleep duration of wfs1 RNAi and control flies. (E) Daily sleep duration of wfs1 mutant and control flies fed with different concentrations of AMPT. (F) The sleep profile and (G) wake activity of wfs1 mutant and control flies fed with 2mg/mL AMPT. (H) The dosage effect of AMPT on daily sleep duration of wfs1 mutant and control flies. (I) Daily sleep duration of wfs1 mutant flies with pan-neuronal knock down of ple. Gray shade indicates the dark period. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, #P < 0.05, ##P < 0.01, ###P < 0.001; compared to UAS control, *P < 0.05, **P < 0.01, ***P < 0.001. For comparison between mutant vs. control, Mann-Whitney test or Student’s t-test was used: *P < 0.05, ***P < 0.001. n = 25–155. For comparison between mutant expressing RNAi vs. control, one-way ANOVA was used: *P < 0.05. Error bars represent SEM. eG4, elavGAL4; G4, GAL4; U, UAS; M, wfs1MI14041; ZT, Zeitgeber Time.
To further validate this, we knocked down the gene encoding tyrosine hydroxylase (ple) using a previously published RNAi line [29]. ple deficiency partially rescues the short-sleep phenotype of wfs1 mutants (Fig 3I). On the other hand, knocking down ple fully rescues the reduced power of locomotor rhythm in wfs1 mutants (S3 Table). We also examined the effects of knocking down wfs1 on the mRNA level of dopamine-related genes and dopamine concentration, but observed no significant difference (S6 Fig).
All in all, these results support the notion that wolframin function to regulate sleep and locomotor rhythm via dopaminergic signaling.
Wolframin acts in Dop2R neurons to maintain sleep via Dop2R signaling
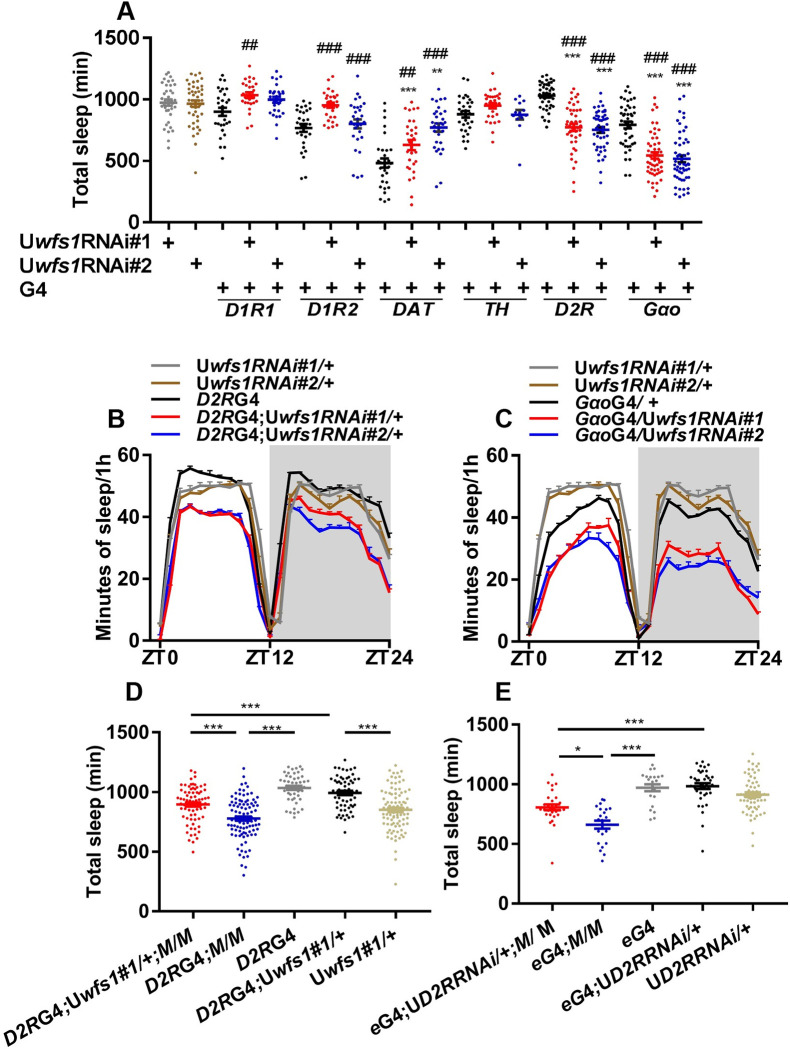
Next, we tested whether wolframin functions in dopaminergic and/or dopamine receptor neurons to regulate sleep. We knocked down wfs1 in dopaminergic neurons using dopamine transporter (DAT)-GAL4 and pale (ple)-GAL4, as well as in various dopamine receptor neurons [30]. We found that lack of wfs1 in Dop2R neurons reduces sleep (Fig 4A and 4B). Because Dop2R has been reported to be a receptor coupled to Goα protein, we knocked down wfs1 in Goα-expressing cells using GAL4 line NP3200 and found this also resulted in reduced sleep (Fig 4A and 4C) [31,32]. We further expressed wfs1 in Dop2R neurons of wfs1MI14041 mutants and found this rescued the decreased sleep, indicating that wolframin expression in Dop2R neurons is both necessary and sufficient for sleep regulation (Fig 4D).
Fig 4. Wolframin acts in Dop2R neurons to maintain sleep via Dop2R signaling.
(A) Daily sleep duration of flies with wfs1 knocked down in dopaminergic neurons, dopamine receptor neurons and Goα+ cells. (B and C) The sleep profile of flies with wfs1 knocked down in Dop2R neurons (B) or Goα+ cells (C). (D) Daily sleep duration of wfs1 mutant, Dop2R neuron rescue and control flies. (E) Daily sleep duration of wfs1 mutant flies with pan-neuronal knock down of Dop2R. Gray shade indicates the dark period. For comparison between RNAi or over-expression flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, ##P < 0.01, ###P < 0.001; compared to UAS control, *P < 0.05, **P < 0.01, ***P < 0.001. For comparison between mutant vs. control, Mann-Whitney test or Student’s t-test was used: *P < 0.05, ***P < 0.001. For comparison between wfs1 rescue vs. controls, one-way ANOVA was used: ***P < 0.001. For comparison between mutant expressing RNAi vs. controls, one-way ANOVA was used: *P < 0.05, ***P < 0.001. n = 13–94. Error bars represent SEM. eGAL4, elavGAL4; D2R, Dop2R; G4, GAL4; U, UAS; M, wfs1MI14041; ZT, Zeitgeber Time.
Consistent with the effects of wolframin on sleep, knocking down wfs1 in Dop2R neurons or Goα+ cells reduces the power of locomotor rhythm, while knocking down wfs1 in clock cells using timGAL4 and cryGAL4-16 do not affect the rhythm (Table 1) [33,34]. Taken together with the result that pan-neuronal knock down of wfs1 does not alter clock gene expression (Fig 2E), we believe that wolframin acts downstream of the clock in the circadian output pathway to regulate locomotor rhythm.
Lastly, we tested whether Dop2R signaling mediates the effects of wolframin on sleep and locomotor rhythm. We found that knocking down Dop2R using a previously published RNAi partially rescues the short-sleep phenotype of wfs1 mutant, but does not significantly alter locomotor rhythm (Fig 4E and S3 Table) [32]. Taken together, these results suggest that wolframin acts in Dop2R neurons and maintains sleep by down-regulating Dop2R signaling.
Dop2R and Goα+ cells act to promote wakefulness
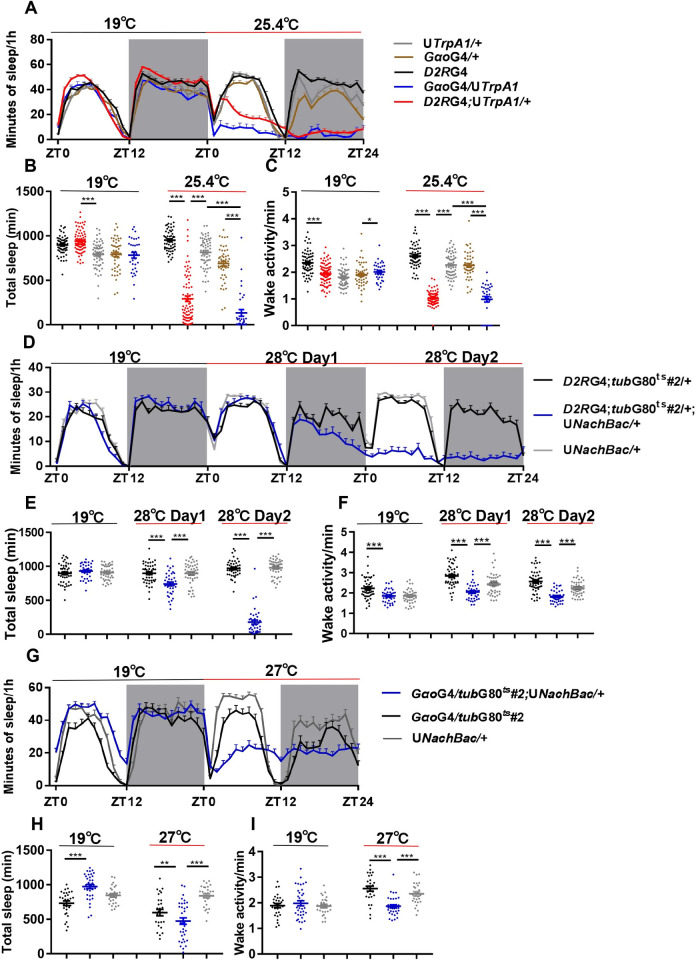
Since the role of Dop2R neurons or Goα-expressing cells in sleep regulation has not been previously reported, we expressed the temperature-gated depolarizing cation channel TrpA1 in these cells to assess the effects of activating them [35]. Activating Dop2R neurons by a higher temperature lead to dramatically reduced sleep, indicating that these cells function to promote wakefulness (Fig 5A and 5B). On the other hand, waking activity is rather decreased when these neurons are activated, which means the enhanced wakefulness is not merely due to hyper activity (Fig 5C). Consistently, activating the Goα+ cells with TrpA1 also resulted in much decreased sleep while waking activity is rather decreased (Fig 5A–5C). In addition, we employed an alternative method to activate these cells by using the bacterial depolarization-activated sodium channel NachBac [36]. Flies expressing NachBac in Dop2R or Goα+ cells did not survive to adulthood. To resolve this issue, we used tubGAL80ts in combination with Dop2RGAL4 or GoαGAL4 to express NachBac specifically during the adult stage. NachBac-expressing flies show significantly reduced sleep while waking activity is decreased (Fig 5D–5I).
Fig 5. Dop2R and Goα+ cells act to promote wakefulness.
(A-C) The sleep profile (A), daily sleep duration (B), and wake activity (C) of TrpA1 expressing flies and controls under baseline temperature (19°C) and activation temperature (25.4°C). (D-F) The sleep profile (D), daily sleep duration (E), and wake activity (F) of flies expressing NachBac in Dop2R neurons and controls under baseline temperature (19°C) and activation temperature (28°C). (G-I) The sleep profile (G), daily sleep duration (H), and wake activity (I) of flies expressing NachBac in Goα+ cells and controls under baseline temperature (19°C) and activation temperature (27°C). Gray shade indicates the dark period. One-way ANOVA: *P < 0.05, **P < 0.01, ***P < 0.001. n = 23–80. Error bars represent SEM. D2RG4, Dop2RGAL4; G4, GAL4; U, UAS; ZT, Zeitgeber Time.
These results indicate that Dop2R and Goα+ cells act to promote wakefulness, and thus knocking down wfs1 in these cells may enhance their activities, leading to increased wakefulness and decreased sleep.
Wfs1 deficiency decreases sleep by altering the activity of Dop2R neurons
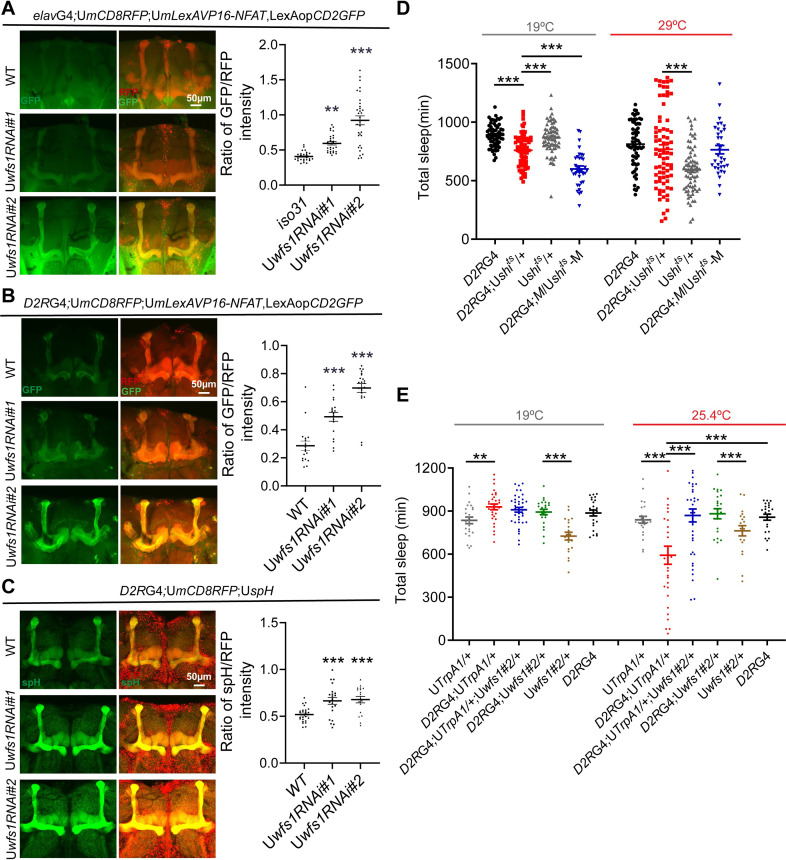
Multiple studies in cultured cells reported roles for wolframin in regulating cellular responses to ER stress and calcium homeostasis, while on the other hand, it has been shown that wfs1 depletion in the fly nervous system does not enhance ER stress [10–13,17]. Based on these work, we speculate that lack of wfs1 results in altered calcium centration in Dop2R/Goα+ cells which in turn leads to altered activity and function. To test this, we used a nuclear factor of activated T cells (NFAT)-based CaLexA reporter [37]. In this system, GFP is expressed in response to sustained neural activity. Since both Dop2RGAL4 and GoαGAL4 are expressed in the mushroom body (MB), an important brain structure for sleep and wake regulation, we focused on the effects of wfs1 deficiency in the MB (S7 Fig) [38–40]. Strikingly, we observed dramatically elevated GFP signals in the MB of wfs1 RNAi flies, implicating increased activity in these neurons (Fig 6A and 6B). To further validate this, we used synapto-pHluorin (spH) which is a fluorescent indicator of neurotransmitter release [41]. spH is localized to synaptic vesicles and fluorescence is increased in a pH-dependent manner when vesicles fuse with the presynaptic membrane. spH fluorescence is significantly increased in MB Dop2R neurons of wfs1 RNAi flies, indicative of enhanced synaptic release likely due to elevated excitability (Fig 6C).
Fig 6. Wfs1 deficiency decreases sleep by altering the activity of Dop2R neurons.
(A) Left panel: representative images of adult fly brain with pan-neuronal expression of CaLexA and mCD8RFP. Right panel: quantification of GFP/RFP signal intensity in MB (two-way ANOVA, n = 28, 28, 32). (B) Left panel: representative images of adult fly brain expressing CaLexA and CD8RFP in Dop2R neurons. Right panel: quantification of GFP/RFP signal intensity in MB (two-way ANOVA, n = 18, 18, 22). (C) Left panel: representative images of adult fly brain with spH expression in Dop2R neurons. Right panel: quantification of spH/RFP signal intensity in MB (two-way ANOVA, n = 26, 22, 18). (D) Daily sleep duration of shits1-expressing wfs1 mutant flies and controls under permissive temperature (19°C) and restrictive temperature (29°C). n = 34–75. (E) Daily sleep duration of flies over-expressing wfs1 and TrpA1, as well as controls under baseline temperature (19°C) and activation temperature (25.4°C). n = 20–39. One-way ANOVA: *P < 0.05, **P < 0.01, ***P < 0.001. Error bars represent SEM. The scale bar represents 50 μm unless indicated otherwise. G4, GAL4; U, UAS; D2RG4, Dop2RGAL4.
We also used the calcium sensor GCaMP6m to monitor intracellular calcium levels [42]. We found that pan-neuronal knock-down of wfs1 significantly reduces GCaMP signal in MB neurons during the earlier half of the day (S8A and S8B Fig). This effect appears to be selective, as GCaMP signal is not altered in the antennal lobe as a control (S8C Fig). Consistent with behavioral data, AMPT treatment blocks the effect of wfs1 deficiency on GCaMP signal (S8D Fig). In addition, we observed reduced GCaMP signal in MB Dop2R neurons of wfs1 RNAi flies (S8E Fig). Moreover, we examined the effect of knocking down wfs1 on dopamine-induced changes of GCaMP signal. We found that dopamine elicits a significantly smaller increase of GCaMP signal in Dop2R neurons of wfs1 RNAi flies (S8F Fig and S1 and S2 Movies).
To determine whether the altered neural activity of Dop2R+ MB neurons is the cause of reduced sleep in wfs1 deficient flies, we first adopted a MB247GAL80 which inhibits GAL4 activity in MB (S9A Fig) [43]. This attenuates the short-sleep phenotype of wfs1 RNAi flies, which indicates that lack of wfs1 in MB contributes to the decreased sleep in these flies (S9B Fig). We next tested whether silencing Dop2R neurons could rescue the short-sleep phenotype of wfs1 mutants by expressing a temperature-sensitive mutant dynamin (shits1) which blocks synaptic vesicle recycling at restrictive temperature (29°C) [44]. While wfs1 mutants expressing shits1 display reduced sleep under permissive temperature (19°C), this sleep phenotype is rescued under restrictive temperature (Fig 6D). Consistently, over-expressing wfs1 can rescue the decreased sleep caused by activating Dop2R neurons (Fig 6E). In addition, we tested for genetic interaction between activation of Dop2R neurons and wfs1 heterozygous mutation but did not observe significant synergistic effect (S10 Fig).
Taken together, these results indicate that the sleep loss associated with wfs1 deficiency is caused by altered excitability of Dop2R neuron.
Wfs1 interacts with genes that control ER calcium store to regulate sleep
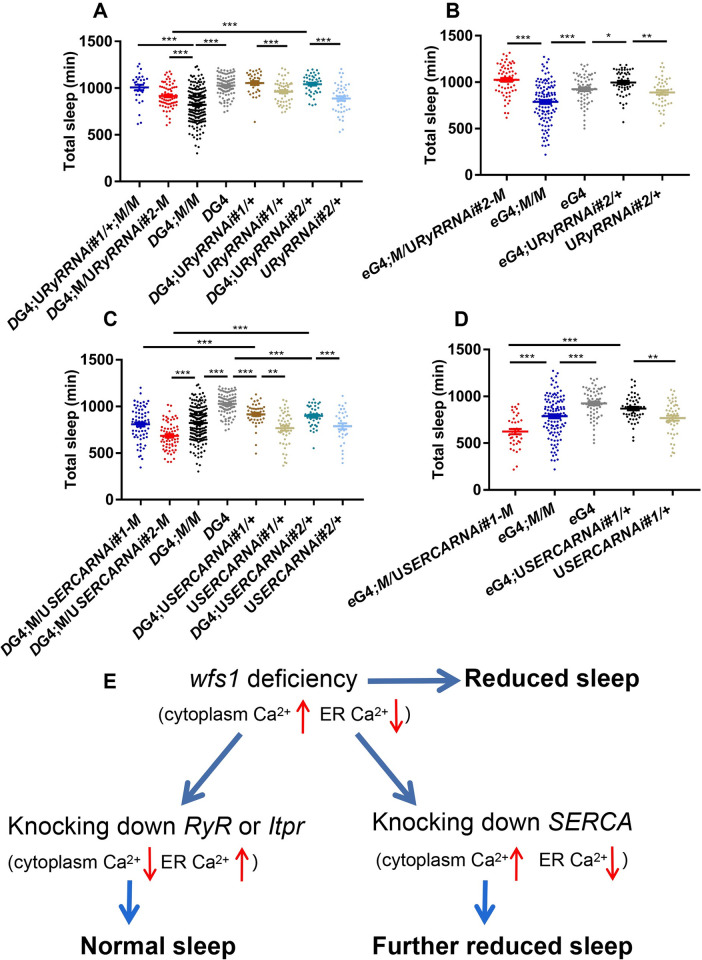
Given the effects of wfs1 deficiency on intracellular calcium level which should largely reflect changes of cytoplasmic calcium level, we next tested whether this altered cytoplasmic calcium homeostasis is the cause of sleep reduction [45]. Pharmacological experiments suggested that wolframin may modulate cytoplasmic calcium level via Ryanodine receptor (RyR), a gated calcium channel that controls the release of calcium from ER [46]. Therefore, we tested for genetic interaction between wfs1 and RyR in sleep regulation. We used two independent RNAi lines to knock down RyR in Dop2R neurons of wfs1 mutant flies, and found that one line (RNAi#1) fully rescued the short sleep phenotype while the other line (RNAi#2) partially rescued this phenotype (Figs 7A and S11A). On the other hand, knocking down RyR alone does not result in altered sleep duration. We also conducted pan-neuronal knock-down in wfs1 mutants using these two RNAi lines. While RNAi#1 does not produce progenies when expressed pan-neuronally, RNAi#2 leads to a substantial lengthening of sleep duration when expressed pan-neuronally (Fig 7B).
Fig 7. Wfs1 interacts with genes that control ER calcium store to regulate sleep.
(A and B) Daily sleep duration of wfs1 mutant flies with RyR knocked down in Dop2R neurons (A) or all neurons (B). (C and D) Daily sleep duration of wfs1 mutant flies with SERCA knocked down in Dop2R neurons (C) or all neurons (D). For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used. For comparison between mutants vs. controls, Student’s t-test was used. *P < 0.05, **P < 0.01, ***P < 0.001. For comparison between mutant expressing RNAi vs. control, one-way ANOVA was used: ***P < 0.001. n = 30–195. Error bars represent SEM. DG4, Dop2RGAL4; eG4, elavGAL4; U, UAS; M, wfs1MI14041. (E) Model illustrating the genetic interactions between wfs1 and RyR/Itpr/SERCA.
Previous study has also implicated that inositol 1,4,5-trisphosphate (IP3) receptor (IP3R), another channel that releases calcium from ER, mediates the influences of WFS1 deficiency on ER calcium homeostasis [47,48]. Therefore, we tested for genetic interaction between wfs1 and Itpr which encodes IP3R. Similar to RyR, knocking down Itpr pan-neuronally using a previously published RNAi line leads to a substantial lengthening of sleep duration in wfs1 mutants (S12 Fig) [49].
Since the genetic interactions between RyR/Itpr and wfs1 suggest that RyR and Itpr function in opposite direction of wfs1 to regulate sleep, we also tested for genetic interaction between wfs1 and SERCA, which pumps calcium into ER and has been previously shown to directly bind with wolframin [50]. In contrary to RyR and Itpr, we found that knocking down SERCA (with one of the two independent RNAi lines, RNAi #2) in Dop2R neurons enhances the short-sleep phenotype of wfs1MI14041, while knocking down SERCA alone does not alter sleep duration (Figs 7C and S11B). For panneuronal knock-down we were not able to obtain flies expressing RNAi#2, while expression of RNAi#1 results in reduction of sleep on wfs1 mutant but not wild-type background (Fig 7D).
Taken together, these results demonstrate that RyR and Itpr deficiency rescues or reverses the sleep reduction in wfs1 mutants, while SERCA deficiency synergistically enhances the sleep reduction. This suggests that wolframin and SERCA function together in the same direction to regulate sleep, while RyR and IP3R act in the opposite direction.
Discussion
Sleep problems have been reported in WS1 patients [8]. Their scores on Pediatric Sleep Questionnaire are more than 3 times higher than healthy controls and doubled compared to individuals with type I diabetes, indicating that the sleep issues are not merely due to metabolic disruptions. Indeed, our work here suggests that the sleep problems in human patients are of neural origin, specifically in the wake-promoting Dop2R neurons. Given that the rebound sleep is not significantly altered in wfs1 depleted flies, we believe that lack of wfs1 does not shorten sleep duration by impairing the sleep homeostasis system. Instead, wfs1 deficiency leads to excessive wakefulness which in turn results in decreased sleep. Considering that heterozygous WFS1 mutation is present in up to 1% of the population, it would be interesting to examine whether these heterozygous mutations contribute to sleep disruptions in the general population [9].
In mouse, chick, quail and turtle, Wfs1 has been shown to be expressed in brain regions where dopamine receptor Drd1 is expressed [51]. D1-like dopamine receptor binding is increased while striatal dopamine release is decreased in Wfs1-/- mice [51–53]. Our results here also implicate a role for wolframin in dopamine receptor neurons and that lack of wfs1 impacts dopaminergic signaling, as the effects of wfs1 deficiency on both sleep and MB calcium concentration is blocked by the tyrosine hydroxylase inhibitor AMPT. Both Dop2RGAL4 and GoαGAL4 exhibit prominent expression in the MB, and to be more specific, in the α and β lobes of MB (S7C and S7D Fig). Previous studies have shown that dopaminergic neurons innervate wake promoting MB neurons, and here we found Dop2R and Goα+ cells to be wake-promoting as well [54]. Therefore, we suspect that wolframin functions in MB Dop2R/Goα+ neurons to influence sleep. Taken together, these findings suggest an evolutionarily conserved role of wolframin within the dopaminergic system. As this system is also important for sleep/wake regulation in mammals, it is reasonable to suspect that wolframin functions in mammals to modulate sleep by influencing the dopaminergic tone as well [55].
MB neural activity appears to be enhanced in wfs1 deficient flies based on the results obtained using CaLexA and spH reporters. This elevated activity is consistent with our behavioral data, as activation of Dop2R/Goα+ cells reduces sleep, similar to the effects of wfs1 deficiency. Moreover, silencing Dop2R neurons rescues the short-sleep phenotype of wfs1 mutants, while over-expressing wfs1 restores the decreased sleep induced by activation of Dop2R neurons. These findings suggest that wolframin functions to suppress the excitability of MB Dop2R neurons, which in turn reduces wakefulness and promotes sleep. Comparable cellular changes have been observed in SERCA mutant flies. Electric stimulation leads to an initial increase followed by prolonged decrease of calcium concentration in mutant motor nerve terminal compared to the control, while action potential firing is increased in the mutants [56]. This series of results underpin the importance of ER calcium homeostasis in determining membrane excitability and thus neural function.
GCaMP6 monitoring reveals that wfs1 deficiency selectively reduces fluorescence signal in the MB both under baseline condition and after dopamine treatment, which should reflect a reduction of cytosolic calcium level that is usually associated with decreased excitability [45]. Previous studies have shown that lack of wolframin leads to increased basal calcium level in neural progenitor cells derived from induced pluripotent stem (iPS) cells of WS1 patients and primary rat cortical neurons, but after stimulation the rise of calcium concentration is smaller in Wfs1 deficient neurons, resulting in reduced calcium level compared to controls [12,46]. Similarly, evoked calcium increase is also diminished in fibroblasts of WS1 patients and MIN6 insulinoma cells with WFS1 knocked down [13,50]. Notably, wolframin has been shown to bind to calmodulin (CaM) in rat brain, and is capable of binding with calcium/CaM complex in vitro and in transfected cells [57]. This may undermine the validity of using GCaMP to monitor calcium level in wfs1 deficient animals and cells, and could potentially account for the contradictory data acquired using CaLexA vs GCaMP.
It is intriguing that in our study here wfs1 deficiency appears to selectively impair the function of Dop2R/Goα+ neurons. It has been shown that in the rodent brain Wfs1 is enriched in layer II/III of the cerebral cortex, CA1 field of the hippocampus, central extended amygdala, striatum, and various sensory and motor nuclei in the brainstem [58–61]. Wfs1 expression starts to appear during late embryonic development in dorsal striatum and amygdala, and the expression quickly expands to other regions of the brain at birth [61]. We suspect that in flies wfs1 may be enriched in Dop2R/Goα+ cells during a critical developmental period, and that sufficient level of wolframin is required for their maturation and normal functioning in adults. Another possibility is that these cells are particularly susceptible to calcium dyshomeostasis induced by loss of wfs1. Indeed, this is believed to be an important cause of selective dopaminergic neuron loss in Parkinson’s Disease, as dopaminergic neurons are unique in their autonomic excitability and selective dependence on calcium channel rather than sodium channel for action potential generation [62]. We reason that Dop2R/Goα+ neurons may also be more sensitive to abnormal intracellular calcium concentration, making them particularly vulnerable to wfs1 deficiency. The pathogenic mechanism underlying the neurodegeneration of WS1 is quite complex, possibly involving brain-wide neurodegenerative processes and neurodevelopmental dis-regulations [4]. Our findings here provide some evidence supporting a role for altered dopaminergic system during development. Obviously, much more needs to be done to test these hypotheses.
The precise role of wolframin in ER calcium handling is not yet clear. It has been shown in human embryonic kidney (HEK) 293 cells that knocking down WFS1 reduces while over-expressing WFS1 increases ER calcium level [63]. The authors concluded that wolframin upregulates ER calcium concentration by increasing the rate of calcium uptake. Consistently, we found by genetic interaction that knocking down RyR or Itpr (which act to reduce ER calcium level and thus knocking down either one will increase ER calcium level) rescues the short-sleep phenotype caused by wfs1 mutation, while knocking down SERCA (which acts to increase ER calcium level and thus knocking down this gene will reduce ER calcium level) synergistically enhances the short-sleep phenotype. Based on the results of these genetic interactions, we propose that lack of wfs1 increases cytosolic calcium while decreasing ER calcium, leading to hyperexcitability of Dop2R neurons and thus reduced sleep (Fig 7F). Knocking down RyR or Itpr decreases cytosolic calcium and increases ER calcium, counteracting the influences of wfs1 deficiency and thus rescuing the short-sleep phenotype. On the other hand, knocking down SERCA further increases cytosolic calcium and decreases ER calcium, rendering an enhancement of the short-sleep phenotype. In line with this, study conducted in neural progenitor cells derived from iPS cells of WS1 patients demonstrated that pharmacological inhibition of RyR can prevent cell death caused by WFS1 mutation [46]. In addition, inhibiting the function of IP3R may mitigate ER stress in wolframin deficient cells [47,48]. One caveat is that SERCA protein level is increased in primary islets isolated from Wfs1 conditional knock-out mice, as well as in MIN6 cells and neuroblastoma cell line with WFS1 knocked down [50]. We reason this may be a compensatory increase to make up for the reduced ER calcium level due to wolframin deficiency. We do acknowledge that the hypothesis proposed in Fig 7F is not supported by our GCaMP data, which indicates decreased cytosolic calcium level in Dop2R neurons of wfs1 deficient flies. We suspect that since the sleep phenotype associated with lack of wfs1 is of developmental origin, it is possible there is an initial increase of cytosolic calcium during critical developmental period in wfs1 deficient flies and this influences the function of Dop2R neurons in adults. Clearly, further characterizations need to be done to fully elucidate this issue, and preferably another calcium indicator independent of the GCaMP system should be employed.
In conclusion, here we identify a role for wolframin in the wake-promoting Dop2R neurons. wfs1 depletion in these cells lead to impaired calcium homeostasis and altered neural activity, which in turn leads to enhanced wakefulness and reduced sleep. Our study may provide some insights for the mechanisms underlying the sleep disruptions in individuals with WFS1 mutation, as well as for the pathogenesis of WS1.
Materials and methods
Fly strains
Flies were raised on standard cornmeal food at 25°C and ~50% humidity under 12h light/12h dark (LD) cycles. All strains were obtained from Bloomington Stock Center TsingHua Fly Center, and Vienna Drosophila Resource Center, or as gifts from colleagues. All fly crosses were carried out at 25°C unless noted otherwise. The following fly strains were used in this study: Dop2RGAL4, Dop1R1GAL4, Dop1R2GAL4, DATGAL4 and THGAL4 which were generated in Dr. Yi Rao’s laboratory [30], UAS-wfs1RNAi#1 (TH02454.N), UAS-wfs1RNAi#2 (TH201500967.S), wfs1MI14041 (BDSC:59250), UASwfs1#1 (BDSC: 8357), UASwfs1#2 (BDSC: 8356), UAS-GFPRNAi (BDSC: 9331), UAS-pleRNAi (BDSC: 25796), UAS-Dop2RRNAi (VDRC: V11471), UASNachBac (BDSC:9467), UAS-shits1(BDSC: 44222), UASTrpA1 (BDSC: 26263), UAS-SERCARNAi#1 (THU2107), UAS-SERCARNAi#2 (THU5676), UAS-RyRRNAi#1 (TH02531.N), UAS-RyRRNAi#2 (THU5738), UAS-ItprRNAi (BDSC: 25937), isogenic w1118 (BDSC: 5905), UASmLexA-VP16-NFAT (BDSC: 66542), UASspH [41], UASGCaMP6m (BDSC: 42750), UASmCD8GFP (BDSC: 32186), UASmCD8RFP (BDSC: 32219), repoGAL4 (BDSC: 7415), elavGAL4 (BDSC: 458), tubGAL4 [64], timGAL4 (BDSC: 7126), cryGAL4-16 (BDSC: 24514), GoαGAL4 (BDSC: 104410), 247GAL80 (BDSC: 64306), UASdcr2 (BDSC: 24650), tubGAL80ts#1 (BDSC: 7017) and tubGAL80ts#2 (BDSC: 7019). All flies used for sleep monitoring were backcrossed with the isogenic w1118 strain for at least 5 times. All experiments were conducted in male flies unless otherwise specified.
Sleep and locomotor rhythm analysis
Drosophila Activity Monitor system (Trikinetics) was used to analyze fly sleep and locomotor rhythm. Flies 3~5 days old were used for experiments unless otherwise specified. Flies were entrained under LD at 25°C for 3 days, and then their activities in the next 4 days under LD condition were analyzed, followed by 7 days of DD. Sleep is defined as 5 min consecutive inactivity. Sleep was analyzed with Counting Macro written in Excel (Microsoft) following previously published protocol [65]. Waking activity is calculated by dividing daily total activity during waking period by the daily duration of waking period. Chi-squared periodogram analyses of period, power, and significance values during DD was carried out using ClockLab (Actimetrics) software. For DD rhythmicity, rhythmic flies were defined as those with chi-squared power-significance ≥10. %Rhythmic is calculated as the percentage of flies of a given genotype that exhibit power-significance ≥10. Period calculations considered all flies with power-significance ≥10.
To knock down wfs1 only in adult stage, flies were raised at 19°C and sleep and locomotor rhythm were monitored at 27–29°C. To knock down wfs1 only in developmental stage, flies were raised at 29°C and sleep and locomotor rhythm were monitored at 19°C. TrpA1, NachBac and shits1 flies were raised at 19°C and baseline sleep was monitored at 19°C. Temperature was then raised at lights on to 25.4°C (for TrpA1), 27°C (for NachBac) and 29°C (for shits) for further sleep monitoring.
Sleep Deprivation
Mechanical sleep deprivation was performed at 25°C using a multi-tube vortexer (VWR) modified by TriKinetics to house DAM2 activity monitors. After 3 days of LD entrainment and 1 day of baseline sleep recording, the multi-tube vortexer delivered 10s-long vibrations at random intervals centered around 60 s (± 30 s). The intensity of the vortexer was set to 4. Flies that displayed sleep loss ≥ 90% during the day of sleep deprivation were used to analyze the recovery sleep. The amount of sleep lost was calculated by subtracting the sleep duration during sleep deprivation from that of baseline sleep. The sleep loss percentage was calculated by amount of sleep lost dividing baseline sleep duration. Recovery sleep was calculated by the sleep duration in the first two hours of the day after deprivation subtracting that of the day before deprivation.
Drug treatment
For pharmacological experiments, drugs were mixed in the fly food at indicated concentrations and the flies were fed with these food for the entire period during sleep monitoring. Concentrations of different drugs are as follows: 0.5–2 mg/ml AMPT (Sigma), 5 mg/ml L-DOPA (BBI Life Sciences), or 10 Mm ethanolamine-O-sulphate (Tokyo Chemical Industry), 10 mg/ml nipecotic acid (Sigma), 5 mM riluzole (Tokyo Chemical Industry), and 1% alcohol as a vehicle control for riluzole (Sinopharm Chemical Reagent Co., Ltd).
ELISA
30 fly heads per sample were homogenized for 1 min with an automated tissue homogenizer (Shanghai Jinxin) at 4°C in 300 μl lysis buffer (0.01N HCl, 1mM EDTA, 4mM sodium metabisulfite). The samples were then centrifuged at 13,680 g for 30 min at 4°C and supernatants were collected. 100 μl supernatant was used per reaction to measure dopamine level using Dopamine Research ELISA following the manufacturer’s instructions (Labor Diagnostika Nord).
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Approximately 20–30 7-day-old flies were collected and frozen immediately on dry ice. For analysis of clock gene expression, flies were entrained for at least 3 days in LD and then collected at the indicated time points. Fly heads were isolated and homogenized in Trizol reagent (Life Technologies). Total RNA was extracted and qRT-PCR conducted following our previously published procedures [66]. The sequences of primers used are in S4 Table.
Confocal imaging
For live imaging experiments (GCaMP and spH), flies 2–3 days old were collected into tubes with standard food and maintained under LD for 3 days. For AMPT treatment, flies 2–3 days old were treated with 2mM AMPT for 3 days in LD. On day 4, flies were anesthetized with CO2 and brains were dissected at the indicated time points in PBS. The dissected brain samples were transferred into Drosophila adult hemolymph-like saline solution on glass slide and sealed with cover slide. The duration for dissection and microscopy should be completed within 2 hours for each time point. Images were captured with Olympus FV3000 confocal microscopy with a 20X objective lens. Time-series images were collected within a period of 250 seconds at 4 Hz, and the resolution was set as 256 X 256 pixels. After capturing baseline images, dopamine hydrochloride (Sigma, 2 mM) was applied using a pipette. Maximal GCaMP intensity was normalized to that of the baseline.
For immunostaining and GFP imaging, flies were anesthetized with CO2 and dissected and fixed with 4% paraformaldehyde diluted in PBS. The brains were then fixed with 4% paraformaldehyde for 30 minutes. Samples were washed with 1×PBT (1×PBS + 0.3% TritonX-100) for 3 times and dissected further to remove additional debris in 1×PBS solution. For immunostaining, brain samples were then blocked in 1×PBT solution with 5% fetal bovine serum (Hyclone) for 30 minutes and subsequently stained with mouse anti-nc82 (1:100, DSHB). After 3×10 min PBT rinses, the brains were incubated with goat anti-mouse-Cy5 (1:500, Abcam) at room temperature for 2 h. Then the brains were rinsed 3×10 min in PBS and mounted and imaged using Olympus FV3000 confocal microscope with a 20X objective lens.
Images were acquired using the same settings (power, gain, offset) for each experiment. GCaMP, GFP, spH and RFP fluorescence intensity were measured and quantified by ImageJ software. The mushroom body and antennal lobe border was traced manually.
Statistical analysis
One-way ANOVA (Prism Graphpad) was used to compare the differences between multiple genotypes. For data that fit normal distribution, Student’s t-test (Prism Graphpad) was used to compare the difference between two genotypes. For data that do not fit normal distribution, Mann-Whitney test (Prism Graphpad) was used to compare the difference between two genotypes.
Supporting information
(A) and (C) Relative mRNA abundance of wfs1 determined by qRT–PCR in whole-head extracts. (B) Daily sleep duration of GFP RNAi flies. n = 20–35. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, #P < 0.05; compared to UAS control, ***P < 0.001. For (A) and (C), the value of GAL4 control at was set to 1. n = 4–6. Error bars represent SEM. Mann-Whitney test: compared to GAL4 control, #P < 0.05, ##P < 0.01; compared to UAS control, *P < 0.05, **P < 0.01. G4, GAL4; U, UAS.
(TIF)
Daily sleep duration of flies with wfs1 knocked down in adult stage (A) or developmental (B) stage. n = 31–126. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, #P < 0.05, ###P < 0.001; compared to UAS control, **P < 0.01, ***P < 0.001. G4, GAL4; U, UAS; G80, GAL80.
(TIF)
(A, C and E) The sleep profile of wfs1 RNAi flies, wfs1 mutants and controls collected 30 days post-eclosion. (B, D and F) The sleep profile of flies, wfs1 mutants and controls collected 30 days post-eclosion. Gray shade indicates the dark period. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to GAL4 control, #P < 0.05, ###P < 0.001; compared to UAS control, **P < 0.01, ***P < 0.001. For comparison between mutant vs. control, Mann-Whitney test or Student’s t-test was used: ***P < 0.001. n = 26–73. Error bars represent SEM. G4, GAL4; U, UAS; M, wfs1MI14041; ZT, Zeitgeber Time.
(TIF)
(A) Sleep profile of wfs1 RNAi and control flies the day before, during and after sleep mechanical deprivation. White and black bars indicate light and dark period, respectively. (B) Left panel: sleep duration the day before sleep deprivation. Middle panel: percentage of sleep lost on the day of sleep deprivation. Right panel: The recovery sleep during the first 2 h on the day immediately after sleep deprivation. One-way ANOVA: compared to GAL4 control, ###P < 0.001; compared to UAS control, *P < 0.05, **P < 0.01, ***P < 0.001. n = 18–49. Error bars represent SEM. G4, GAL4; U, UAS; ZT, Zeitgeber Time.
(TIF)
Daily sleep duration of wfs1 RNAi, mutant and control flies treated with the indicated drugs. Alcohol is used as a vehicle control for riluzole, while all other drugs were directly dissolved in the food. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used. For comparison between mutants expressing RNAi vs. mutant control, Student’s t-test was used. ***P < 0.001. n = 21–67. Error bars represent SEM. G4, GAL4; U, UAS; M, wfs1MI14041; NipA, nipecotic acid; EOS, ethanolamine-O-sulphate.
(TIF)
(A-G) Relative mRNA abundance of genes involved in dopamine signaling in whole-head extracts, determined by qRT-PCR. For each experiment, the value of G4 control was set to 1. (H) Dopamine concentration in whole head extracts determined by ELISA. Error bars represent SEM. n = 3–6. Mann-Whitney test: compared to GAL4 control, #P < 0.05. G4, GAL4; U, UAS; M, wfs1MI14041.
(TIF)
(A and B) Representative image of adult fly brain with mCD8GFP expression in Dop2R (A) or Goα+ (B) cells. (C) and (D) are enlarged image of MB shown in (A) and (C), respectively. The brains are stained with antibody against BRUCHPILOT (NC82) to label axons. Red arrowhead indicates the MB. The scale bar represents 50 μm and 17 μm, respectively. D2RG4, Dop2RGAL4; G4, GAL4; U, UAS.
(TIF)
Representative live image of adult fly brain with pan-neuronal expression of GCaMP6m and mCD8RFP. Brain samples were collected and dissected at the indicated time points. (B) Quantification of GCaMP6m/RFP signal intensity in MB (two-way ANOVA, n = 16–20). (C) Left panel: representative live image of adult fly brain with pan-neuronal expression of GCaMP6m and mCD8RFP with the antennal lobe indicated by the white rectangle. Brain samples were collected and dissected at ZT6. Right panel: quantification of antennal lobe GCaMP6m/RFP intensity (one-way ANOVA, n = 22, 20, 22). (D) Left panel: representative live image of adult fly brain with pan-neuronal expression of GCaMP6m and mCD8RFP. The flies were fed with 2 mM AMPT for 3 days. Brain samples were collected and dissected at ZT6. Right panel: quantification of GCaMP6m/RFP signal intensity in MB (two-way ANOVA, n = 20). (E) Left panel: representative live image of adult fly brain with GCaMP6m expressed in Dop2R neurons. Brain samples were collected and dissected at ZT6. Right panel: quantification of MB GCaMP6m signal intensity normalized to the control (Student’s t-test, n = 26, 28). (F) Left panel: time-series GCaMP6m intensity of adult fly brain treated with dopamine. GCaMP6m intensity was normalized to the baseline level. Right panel: quantification of maximum GCaMP6m intensity after dopamine treatment (Student’s t-test, n = 17, 10). Error bars represent SEM. *P < 0.05, **P < 0.01, ***P < 0.001. The scale bar represents 50 μm unless indicated otherwise. G4, GAL4; U, UAS; D2RGAL4, Dop2RGAL4. ZT, Zeitgeber Time.
(TIF)
(A) Representative image of adult fly brain with mCD8GFP expression in non-MB Dop2R neurons. The brains are stained with antibody against BRUCHPILOT (NC82) to label axons. The scale bar represents 50 μm. (B) Daily sleep duration of wfs1RNAi flies and controls. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: compared to UAS control, **P < 0.01, ***P < 0.001. n = 18–39. Error bars represent SEM. D2RG4, Dop2RGAL4; U, UAS; G80, GAL80.
(TIF)
(A and B) Daily sleep duration (A) and wake activity (B) of wfs1 heterozygous mutant flies with Dop2R neurons activated by TrpA1. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used: **P < 0.01, ***P < 0.001. For comparison between mutants expressing TrpA1 vs. mutant control, Student’s t-test was used: ***P < 0.001. n = 24–45. Error bars represent SEM. D2RG4, Dop2RGAL4; U, UAS; M, wfs1MI1404.
(TIF)
(A and B) Relative mRNA abundance of RyR (A) and SERCA (B) determined by qRT-PCR in whole-head extracts. Female fly heads were used for this entire experiment as we were not able to obtain male elavGAL4;URyRRNAi#1/+ flies. We were not able to obtain either male or female elavGAL4;USERCARNAi#2/+ flies. For each experiment, the value of G4 control at was set to 1. Error bars represent SEM. n = 3–5. Mann-Whitney test: *P < 0.05, **P < 0.01. G4, GAL4; U, UAS.
(TIF)
Daily sleep duration of wfs1 mutant flies with Itpr knocked down in all neurons. For comparison between RNAi flies vs. UAS/GAL4 controls, one-way ANOVA was used. For comparison between mutants expressing RNAi vs. mutant control, Student’s t-test was used: **P < 0.01, ***P < 0.001. n = 38–116. Error bars represent SEM. eG4, elavGAL4; U, UAS; M, wfs1MI14041.
(TIF)
(AVI)
(AVI)
(XLSX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
We would like to thank Drs. Magaret Ho, Yi Rao, Liming Wang, Junhai Han and Yi Zhong for kindly providing flies used in this study.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was supported by grants from the Ministry of Science and Technology of China (STI 2030-Major Projects 2021ZD0203200-02), as well as Natural Science Foundation of China (31930021 and 32022035) to LZ. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Rumble ME, White KH, Benca RM. Sleep Disturbances in Mood Disorders. Psychiatr Clin North Am. 2015;38(4):743–59. doi: 10.1016/j.psc.2015.07.006 . [DOI] [PubMed] [Google Scholar]
- 2.Inoue H, Tanizawa Y, Wasson J, Behn P, Kalidas K, Bernal-Mizrachi E, et al. A gene encoding a transmembrane protein is mutated in patients with diabetes mellitus and optic atrophy (Wolfram syndrome). Nat Genet. 1998;20(2):143–8. doi: 10.1038/2441 . [DOI] [PubMed] [Google Scholar]
- 3.Strom TM, Hortnagel K, Hofmann S, Gekeler F, Scharfe C, Rabl W, et al. Diabetes insipidus, diabetes mellitus, optic atrophy and deafness (DIDMOAD) caused by mutations in a novel gene (wolframin) coding for a predicted transmembrane protein. Hum Mol Genet. 1998;7(13):2021–8. doi: 10.1093/hmg/7.13.2021 . [DOI] [PubMed] [Google Scholar]
- 4.Rigoli L, Bramanti P, Di Bella C, De Luca F. Genetic and clinical aspects of Wolfram syndrome 1, a severe neurodegenerative disease. Pediatr Res. 2018;83(5):921–9. doi: 10.1038/pr.2018.17 . [DOI] [PubMed] [Google Scholar]
- 5.Swift RG, Polymeropoulos MH, Torres R, Swift M. Predisposition of Wolfram syndrome heterozygotes to psychiatric illness. Mol Psychiatry. 1998;3(1):86–91. doi: 10.1038/sj.mp.4000344 . [DOI] [PubMed] [Google Scholar]
- 6.Swift M, Swift RG. Psychiatric disorders and mutations at the Wolfram syndrome locus. Biol Psychiatry. 2000;47(9):787–93. doi: 10.1016/s0006-3223(00)00244-4 . [DOI] [PubMed] [Google Scholar]
- 7.Shrestha P, Mousa A, Heintz N. Layer 2/3 pyramidal cells in the medial prefrontal cortex moderate stress induced depressive behaviors. eLife. 2015;4. doi: 10.7554/eLife.08752 ; PubMed Central PMCID: PMC4566133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bischoff AN, Reiersen AM, Buttlaire A, Al-Lozi A, Doty T, Marshall BA, et al. Selective cognitive and psychiatric manifestations in Wolfram Syndrome. Orphanet J Rare Dis. 2015;10:66. doi: 10.1186/s13023-015-0282-1 ; PubMed Central PMCID: PMC4450481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swift RG, Perkins DO, Chase CL, Sadler DB, Swift M. Psychiatric disorders in 36 families with Wolfram syndrome. Am J Psychiatry. 1991;148(6):775–9. doi: 10.1176/ajp.148.6.775 . [DOI] [PubMed] [Google Scholar]
- 10.Osman AA, Saito M, Makepeace C, Permutt MA, Schlesinger P, Mueckler M. Wolframin expression induces novel ion channel activity in endoplasmic reticulum membranes and increases intracellular calcium. J Biol Chem. 2003;278(52):52755–62. doi: 10.1074/jbc.M310331200 . [DOI] [PubMed] [Google Scholar]
- 11.Fonseca SG, Fukuma M, Lipson KL, Nguyen LX, Allen JR, Oka Y, et al. WFS1 is a novel component of the unfolded protein response and maintains homeostasis of the endoplasmic reticulum in pancreatic beta-cells. J Biol Chem. 2005;280(47):39609–15. doi: 10.1074/jbc.M507426200 . [DOI] [PubMed] [Google Scholar]
- 12.Cagalinec M, Liiv M, Hodurova Z, Hickey MA, Vaarmann A, Mandel M, et al. Role of Mitochondrial Dynamics in Neuronal Development: Mechanism for Wolfram Syndrome. PLoS Biol. 2016;14(7):e1002511. doi: 10.1371/journal.pbio.1002511 ; PubMed Central PMCID: PMC4951053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Angebault C, Fauconnier J, Patergnani S, Rieusset J, Danese A, Affortit CA, et al. ER-mitochondria cross-talk is regulated by the Ca(2+) sensor NCS1 and is impaired in Wolfram syndrome. Sci Signal. 2018;11(553). doi: 10.1126/scisignal.aaq1380 . [DOI] [PubMed] [Google Scholar]
- 14.Ishihara H, Takeda S, Tamura A, Takahashi R, Yamaguchi S, Takei D, et al. Disruption of the WFS1 gene in mice causes progressive beta-cell loss and impaired stimulus-secretion coupling in insulin secretion. Hum Mol Genet. 2004;13(11):1159–70. doi: 10.1093/hmg/ddh125 . [DOI] [PubMed] [Google Scholar]
- 15.Riggs AC, Bernal-Mizrachi E, Ohsugi M, Wasson J, Fatrai S, Welling C, et al. Mice conditionally lacking the Wolfram gene in pancreatic islet beta cells exhibit diabetes as a result of enhanced endoplasmic reticulum stress and apoptosis. Diabetologia. 2005;48(11):2313–21. doi: 10.1007/s00125-005-1947-4 . [DOI] [PubMed] [Google Scholar]
- 16.Yamada T, Ishihara H, Tamura A, Takahashi R, Yamaguchi S, Takei D, et al. WFS1-deficiency increases endoplasmic reticulum stress, impairs cell cycle progression and triggers the apoptotic pathway specifically in pancreatic beta-cells. Hum Mol Genet. 2006;15(10):1600–9. doi: 10.1093/hmg/ddl081 . [DOI] [PubMed] [Google Scholar]
- 17.Sakakibara Y, Sekiya M, Fujisaki N, Quan X, Iijima KM. Knockdown of wfs1, a fly homolog of Wolfram syndrome 1, in the nervous system increases susceptibility to age- and stress-induced neuronal dysfunction and degeneration in Drosophila. PLoS Genet. 2018;14(1):e1007196. doi: 10.1371/journal.pgen.1007196 ; PubMed Central PMCID: PMC5794194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pozzan T, Rizzuto R, Volpe P, Meldolesi J. Molecular and cellular physiology of intracellular calcium stores. Physiol Rev. 1994;74(3):595–636. doi: 10.1152/physrev.1994.74.3.595 . [DOI] [PubMed] [Google Scholar]
- 19.Lin DM, Goodman CS. Ectopic and increased expression of Fasciclin II alters motoneuron growth cone guidance. Neuron. 1994;13(3):507–23. doi: 10.1016/0896-6273(94)90022-1 . [DOI] [PubMed] [Google Scholar]
- 20.Sepp KJ, Schulte J, Auld VJ. Peripheral glia direct axon guidance across the CNS/PNS transition zone. Dev Biol. 2001;238(1):47–63. doi: 10.1006/dbio.2001.0411 . [DOI] [PubMed] [Google Scholar]
- 21.McGuire SE, Mao Z, Davis RL. Spatiotemporal gene expression targeting with the TARGET and gene-switch systems in Drosophila. Sci STKE. 2004;2004(220):pl6. doi: 10.1126/stke.2202004pl6 . [DOI] [PubMed] [Google Scholar]
- 22.Konopka RJ, Pittendrigh C, Orr D. Reciprocal behaviour associated with altered homeostasis and photosensitivity of Drosophila clock mutants. J Neurogenet. 1989;6(1):1–10. doi: 10.3109/01677068909107096 . [DOI] [PubMed] [Google Scholar]
- 23.Price JL, Dembinska M.E., Young M.W., and Rosbash M. Suppression of PERIOD protein abundance and circadian cycling by the Drosophila clock mutation timeless. EMBO J. 1995;14:4044–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nave C, Roberts L, Hwu P, Estrella JD, Vo TC, Nguyen TH, et al. Weekend Light Shifts Evoke Persistent Drosophila Circadian Neural Network Desynchrony. J Neurosci. 2021;41(24):5173–89. doi: 10.1523/JNEUROSCI.3074-19.2021 ; PubMed Central PMCID: PMC8211545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patke A, Young MW, Axelrod S. Molecular mechanisms and physiological importance of circadian rhythms. Nature reviews Molecular cell biology. 2020;21(2):67–84. doi: 10.1038/s41580-019-0179-2 . [DOI] [PubMed] [Google Scholar]
- 26.Pendleton RG, Rasheed A, Paluru P, Joyner J, Jerome N, Meyers RD, et al. A developmental role for catecholamines in Drosophila behavior. Pharmacol Biochem Behav. 2005;81(4):849–53. doi: 10.1016/j.pbb.2005.06.008 . [DOI] [PubMed] [Google Scholar]
- 27.Van Swinderen B, Andretic R. Dopamine in Drosophila: setting arousal thresholds in a miniature brain. Proc Biol Sci. 2011;278(1707):906–13. doi: 10.1098/rspb.2010.2564 ; PubMed Central PMCID: PMC3049062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ki Y, Lim C. Sleep-promoting effects of threonine link amino acid metabolism in Drosophila neuron to GABAergic control of sleep drive. eLife. 2019;8. doi: 10.7554/eLife.40593 ; PubMed Central PMCID: PMC6636906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayersdorfer F, Voigt A, Schneuwly S, Botella JA. Dopamine-dependent neurodegeneration in Drosophila models of familial and sporadic Parkinson’s disease. Neurobiol Dis. 2010;40(1):113–9. doi: 10.1016/j.nbd.2010.02.012 . [DOI] [PubMed] [Google Scholar]
- 30.Deng B, Li Q, Liu X, Cao Y, Li B, Qian Y, et al. Chemoconnectomics: Mapping Chemical Transmission in Drosophila. Neuron. 2019;101(5):876–93 e4. doi: 10.1016/j.neuron.2019.01.045 . [DOI] [PubMed] [Google Scholar]
- 31.Bredendiek N, Hutte J, Steingraber A, Hatt H, Gisselmann G, Neuhaus EM. Go alpha is involved in sugar perception in Drosophila. Chem Senses. 2011;36(1):69–81. doi: 10.1093/chemse/bjq100 . [DOI] [PubMed] [Google Scholar]
- 32.Zhou M, Chen N, Tian J, Zeng J, Zhang Y, Zhang X, et al. Suppression of GABAergic neurons through D2-like receptor secures efficient conditioning in Drosophila aversive olfactory learning. Proc Natl Acad Sci U S A. 2019;116(11):5118–25. doi: 10.1073/pnas.1812342116 ; PubMed Central PMCID: PMC6421402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95(5):669–79. Epub 1998/12/09. doi: 10.1016/s0092-8674(00)81637-2 [pii]. . [DOI] [PubMed] [Google Scholar]
- 34.Emery P, Stanewsky R, Hall JC, Rosbash M. A unique circadian-rhythm photoreceptor. Nature. 2000;404(6777):456–7. Epub 2000/04/13. doi: 10.1038/35006558 . [DOI] [PubMed] [Google Scholar]
- 35.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454(7201):217–20. doi: 10.1038/nature07001 ; PubMed Central PMCID: PMC2730888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nitabach MN, Wu Y, Sheeba V, Lemon WC, Strumbos J, Zelensky PK, et al. Electrical hyperexcitation of lateral ventral pacemaker neurons desynchronizes downstream circadian oscillators in the fly circadian circuit and induces multiple behavioral periods. J Neurosci. 2006;26(2):479–89. Epub 2006/01/13. doi: 10.1523/JNEUROSCI.3915-05.2006 [pii] . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Masuyama K, Zhang Y, Rao Y, Wang JW. Mapping neural circuits with activity-dependent nuclear import of a transcription factor. J Neurogenet. 2012;26(1):89–102. doi: 10.3109/01677063.2011.642910 ; PubMed Central PMCID: PMC3357894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joiner WJ, Crocker A, White BH, Sehgal A. Sleep in Drosophila is regulated by adult mushroom bodies. Nature. 2006;441(7094):757–60. doi: 10.1038/nature04811 . [DOI] [PubMed] [Google Scholar]
- 39.Pitman JL, McGill JJ, Keegan KP, Allada R. A dynamic role for the mushroom bodies in promoting sleep in Drosophila. Nature. 2006;441(7094):753–6. doi: 10.1038/nature04739 . [DOI] [PubMed] [Google Scholar]
- 40.Sitaraman D, Aso Y, Jin X, Chen N, Felix M, Rubin GM, et al. Propagation of Homeostatic Sleep Signals by Segregated Synaptic Microcircuits of the Drosophila Mushroom Body. Curr Biol. 2015;25(22):2915–27. doi: 10.1016/j.cub.2015.09.017 ; PubMed Central PMCID: PMC4654684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miesenbock G. Synapto-pHluorins: genetically encoded reporters of synaptic transmission. Cold Spring Harb Protoc. 2012;2012(2):213–7. doi: 10.1101/pdb.ip067827 . [DOI] [PubMed] [Google Scholar]
- 42.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499(7458):295–300. doi: 10.1038/nature12354 ; PubMed Central PMCID: PMC3777791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee T, Luo L. Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron. 1999;22(3):451–61. doi: 10.1016/s0896-6273(00)80701-1 [DOI] [PubMed] [Google Scholar]
- 44.Waddell S, Armstrong JD, Kitamoto T, Kaiser K, Quinn WG. The amnesiac gene product is expressed in two neurons in the Drosophila brain that are critical for memory. Cell. 2000;103(5):805–13. doi: 10.1016/s0092-8674(00)00183-5 . [DOI] [PubMed] [Google Scholar]
- 45.Zhong C, Schleifenbaum J. Genetically Encoded Calcium Indicators: A New Tool in Renal Hypertension Research. Front Med (Lausanne). 2019;6:128. doi: 10.3389/fmed.2019.00128 ; PubMed Central PMCID: PMC6585435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lu S, Kanekura K, Hara T, Mahadevan J, Spears LD, Oslowski CM, et al. A calcium-dependent protease as a potential therapeutic target for Wolfram syndrome. Proc Natl Acad Sci U S A. 2014;111(49):E5292–301. doi: 10.1073/pnas.1421055111 ; PubMed Central PMCID: PMC4267371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.MacMillan D, McCarron JG. Regulation by FK506 and rapamycin of Ca2+ release from the sarcoplasmic reticulum in vascular smooth muscle: the role of FK506 binding proteins and mTOR. Br J Pharmacol. 2009;158(4):1112–20. doi: 10.1111/j.1476-5381.2009.00369.x ; PubMed Central PMCID: PMC2785532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hara T, Mahadevan J, Kanekura K, Hara M, Lu S, Urano F. Calcium efflux from the endoplasmic reticulum leads to beta-cell death. Endocrinology. 2014;155(3):758–68. doi: 10.1210/en.2013-1519 ; PubMed Central PMCID: PMC3929724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong CO, Chen K, Lin YQ, Chao Y, Duraine L, Lu Z, et al. A TRPV channel in Drosophila motor neurons regulates presynaptic resting Ca2+ levels, synapse growth, and synaptic transmission. Neuron. 2014;84(4):764–77. doi: 10.1016/j.neuron.2014.09.030 ; PubMed Central PMCID: PMC4254599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zatyka M, Da Silva Xavier G, Bellomo EA, Leadbeater W, Astuti D, Smith J, et al. Sarco(endo)plasmic reticulum ATPase is a molecular partner of Wolfram syndrome 1 protein, which negatively regulates its expression. Hum Mol Genet. 2015;24(3):814–27. doi: 10.1093/hmg/ddu499 ; PubMed Central PMCID: PMC4291252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tekko T, Lakspere T, Allikalt A, End J, Kolvart KR, Jagomae T, et al. Wfs1 is expressed in dopaminoceptive regions of the amniote brain and modulates levels of D1-like receptors. PLoS One. 2017;12(3):e0172825. doi: 10.1371/journal.pone.0172825 ; PubMed Central PMCID: PMC5436468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matto V, Terasmaa A, Vasar E, Koks S. Impaired striatal dopamine output of homozygous Wfs1 mutant mice in response to [K+] challenge. J Physiol Biochem. 2011;67(1):53–60. doi: 10.1007/s13105-010-0048-0 . [DOI] [PubMed] [Google Scholar]
- 53.Visnapuu T, Plaas M, Reimets R, Raud S, Terasmaa A, Koks S, et al. Evidence for impaired function of dopaminergic system in Wfs1-deficient mice. Behav Brain Res. 2013;244:90–9. doi: 10.1016/j.bbr.2013.01.046 . [DOI] [PubMed] [Google Scholar]
- 54.Sitaraman D, Aso Y, Rubin GM, Nitabach MN. Control of Sleep by Dopaminergic Inputs to the Drosophila Mushroom Body. Front Neural Circuits. 2015;9:73. doi: 10.3389/fncir.2015.00073 ; PubMed Central PMCID: PMC4637407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lazarus M, Chen JF, Urade Y, Huang ZL. Role of the basal ganglia in the control of sleep and wakefulness. Curr Opin Neurobiol. 2013;23(5):780–5. doi: 10.1016/j.conb.2013.02.001 ; PubMed Central PMCID: PMC3683373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanyal S, Consoulas C, Kuromi H, Basole A, Mukai L, Kidokoro Y, et al. Analysis of conditional paralytic mutants in Drosophila sarco-endoplasmic reticulum calcium ATPase reveals novel mechanisms for regulating membrane excitability. Genetics. 2005;169(2):737–50. doi: 10.1534/genetics.104.031930 ; PubMed Central PMCID: PMC1449089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yurimoto S, Hatano N, Tsuchiya M, Kato K, Fujimoto T, Masaki T, et al. Identification and characterization of wolframin, the product of the wolfram syndrome gene (WFS1), as a novel calmodulin-binding protein. Biochemistry. 2009;48(18):3946–55. doi: 10.1021/bi900260y . [DOI] [PubMed] [Google Scholar]
- 58.Takeda K, Inoue H, Tanizawa Y, Matsuzaki Y, Oba J, Watanabe Y, et al. WFS1 (Wolfram syndrome 1) gene product: predominant subcellular localization to endoplasmic reticulum in cultured cells and neuronal expression in rat brain. Hum Mol Genet. 2001;10(5):477–84. doi: 10.1093/hmg/10.5.477 . [DOI] [PubMed] [Google Scholar]
- 59.Luuk H, Koks S, Plaas M, Hannibal J, Rehfeld JF, Vasar E. Distribution of Wfs1 protein in the central nervous system of the mouse and its relation to clinical symptoms of the Wolfram syndrome. J Comp Neurol. 2008;509(6):642–60. doi: 10.1002/cne.21777 . [DOI] [PubMed] [Google Scholar]
- 60.Kawano J, Fujinaga R, Yamamoto-Hanada K, Oka Y, Tanizawa Y, Shinoda K. Wolfram syndrome 1 (Wfs1) mRNA expression in the normal mouse brain during postnatal development. Neurosci Res. 2009;64(2):213–30. doi: 10.1016/j.neures.2009.03.005 . [DOI] [PubMed] [Google Scholar]
- 61.Tekko T, Lillevali K, Luuk H, Sutt S, Truu L, Ord T, et al. Initiation and developmental dynamics of Wfs1 expression in the context of neural differentiation and ER stress in mouse forebrain. Int J Dev Neurosci. 2014;35:80–8. doi: 10.1016/j.ijdevneu.2014.03.009 . [DOI] [PubMed] [Google Scholar]
- 62.Huang L, Xue Y, Feng D, Yang R, Nie T, Zhu G, et al. Blockade of RyRs in the ER Attenuates 6-OHDA-Induced Calcium Overload, Cellular Hypo-Excitability and Apoptosis in Dopaminergic Neurons. Front Cell Neurosci. 2017;11:52. doi: 10.3389/fncel.2017.00052 ; PubMed Central PMCID: PMC5334509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takei D, Ishihara H, Yamaguchi S, Yamada T, Tamura A, Katagiri H, et al. WFS1 protein modulates the free Ca(2+) concentration in the endoplasmic reticulum. FEBS Lett. 2006;580(24):5635–40. doi: 10.1016/j.febslet.2006.09.007 . [DOI] [PubMed] [Google Scholar]
- 64.Nabel-Rosen H, Volohonsky G, Reuveny A, Zaidel-Bar R, Volk T. Two isoforms of the Drosophila RNA binding protein, how, act in opposing directions to regulate tendon cell differentiation. Developmental cell. 2002;2(2):183–93. doi: 10.1016/s1534-5807(01)00118-6 . [DOI] [PubMed] [Google Scholar]
- 65.Pfeiffenberger C, Lear BC, Keegan KP, Allada R. Processing sleep data created with the Drosophila Activity Monitoring (DAM) System. Cold Spring Harb Protoc. 2010;2010(11):pdb prot5520. Epub 2010/11/03. doi: 10.1101/pdb.prot5520 . [DOI] [PubMed] [Google Scholar]
- 66.Bu B, He W, Song L, Zhang L. Nuclear Envelope Protein MAN1 Regulates the Drosophila Circadian Clock via Period. Neurosci Bull. 2019;35(6):969–78. doi: 10.1007/s12264-019-00404-6 ; PubMed Central PMCID: PMC6864020. [DOI] [PMC free article] [PubMed] [Google Scholar]