Supplemental Digital Content is Available in the Text.
Quantitative assessment of intracellular protein kinase A pathway activity in differentiating human induced pluripotent stem cell–derived nociceptive sensory neurons reveals a protracted onset of opioid receptor–mediated inhibitory signaling.
Keywords: Human iPSC-derived nociceptive sensory neurons, PKA signaling, High-content screening, Nociceptors, iPSC, Opioids
Abstract
Induced pluripotent stem cells (iPSCs) have enabled the generation of various difficult-to-access cell types such as human nociceptors. A key challenge associated with human iPSC-derived nociceptors (hiPSCdNs) is their prolonged functional maturation. While numerous studies have addressed the expression of classic neuronal markers and ion channels in hiPSCdNs, the temporal development of key signaling cascades regulating nociceptor activity has remained largely unexplored. In this study, we used an immunocytochemical high-content imaging approach alongside electrophysiological staging to assess metabotropic and ionotropic signaling of large scale–generated hiPSCdNs across 70 days of in vitro differentiation. During this period, the resting membrane potential became more hyperpolarized, while rheobase, action potential peak amplitude, and membrane capacitance increased. After 70 days, hiPSCdNs exhibited robust physiological responses induced by GABA, pH shift, ATP, and capsaicin. Direct activation of protein kinase A type II (PKA-II) through adenylyl cyclase stimulation with forskolin resulted in PKA-II activation at all time points. Depolarization-induced activation of PKA-II emerged after 35 days of differentiation. However, effective inhibition of forskolin-induced PKA-II activation by opioid receptor agonists required 70 days of in vitro differentiation. Our results identify a pronounced time difference between early expression of functionally important ion channels and emergence of regulatory metabotropic sensitizing and desensitizing signaling only at advanced stages of in vitro cultivation, suggesting an independent regulation of ionotropic and metabotropic signaling. These data are relevant for devising future studies into the development and regulation of human nociceptor function and for defining time windows suitable for hiPSCdN-based drug discovery.
1. Introduction
Pain is the most prominent cause of morbidity and disability across the world.40,60 While prevalence for chronic pain is high,5,37,38,40,52,60,73,76,80 treatment satisfaction is low.73 Most research on sensory neurons in general has so far been conducted in rodent systems.50 Accordingly, the lack of novel therapeutics is partly attributed to a lack of understanding of the human nociceptive system. Indeed, the expression and functionality of, eg, voltage-gated sodium channels differ considerably between rodent and human nociceptive neurons.13,48,62,83 Furthermore, human dorsal root ganglia (DRG) neurons show a broader coexpression of sensory neuron subtype markers and less subtype specificity, indicating a more polymodal function of primate nociceptors.62,67,68 Last but not least, human-specific transcriptional profiles not described in other species have been reported.54
Opioids are currently the most effective and thus among the most frequently prescribed analgesic drugs. The problem of deleterious side effects such as obstipation, fatigue, addiction, and often life-threatening respiratory depression44,71,72 remains and is accompanied by a lack of evidence for analgesic efficacy for long-term therapy.14,26 There is a huge need for alternative analgesics. Mechanisms to reduce side effects and to increase long-term efficacy require disease models using human nociceptive neurons to understand the underlying regulatory mechanisms.
Opioids modulate the activity of the central and peripheral nervous system on various levels and can also affect other organ systems. In the peripheral nervous system, they mainly reduce nociceptive neuron activity. They exert their action through the classical µ-, δ-, and κ-opioid peptide receptors (MOP, DOP, and KOP, respectively).39,46 Signaling through these G protein–coupled receptors (GPCRs) increases hyperpolarizing potassium channel activity, reduces voltage-gated calcium channels, and reduces cyclic adenosin monophosphate (cAMP)-mediated protein kinase A (PKA) activity.33,44,58,70 Nociceptive neuron activity can be increased by a plethora of factors such as inflammatory interleukins, prostaglandins, and growth factors. Many of those factors, including depolarization7,27,29 as recently shown by us, sensitize through intracellular cascades such as the PKA pathway.3,7,20,22,23,27,29,36,59,65,77,82 Accordingly, detecting PKA-II activity with conformation-specific antibodies in rodent or iPSC-derived nociceptors allows to monitor PKA-mediated sensitization stimuli as well as desensitizing opioid activity.6,7,29–35,78
Protocols to generate human nociceptors from human pluripotent stem cells (PSCs) have been developed based on small-molecule treatment, transcription factor overexpression, or combinations of both.4,12,28,41,42,51,55,63,66,75 Such iPSC-derived nociceptors reflect a large range of nociceptive properties and have been used successfully to model neuropathic pain phenotypes in vitro.8,47,49,53,64 Many of these studies focused on ion channels involved in nociceptor electrical activity. By contrast, metabotropic signaling as well as sensitizing or desensitizing pathways have not received much attention. This issue is of special importance because human iPSC-derived sensory neurons show a very protracted in vitro differentiation. In this study, we set out to identify the dynamics of protein marker expression, electrophysiological functionality, and metabotropic receptor activity and its impact on PKA signaling in hiPSC-derived nociceptors across 70 days of in vitro differentiation.
2. Methods
2.1. Induced pluripotent stem cell culture and quality control
Main experiments were performed on the male iPSC line UKBi013-A (https://hpscreg.eu/cell-line/UKBi013-A), and additional experiments were performed on the female iPSC line UKBi018-A (https://hpscreg.eu/cell-line/UKBi018-A). The use of iPSC lines was approved by the Ethics Committee of the Medical Faculty of the University of Bonn (approval number 275/08), and informed consent was obtained from the patients. iPSCs were cultured in StemMACS iPS-Brew (Miltenyi Biotec, Bergisch Gladbach, Germany, 130-104-368) and split with 0.5 mM EDTA (Sigma Aldrich, St. Louis, MO, E6511) during maintenance. Quality control of iPSCs for pluripotency through Tra1-60 flow cytometric analysis and genomic integrity through single-nucleotide polymorphism (SNP) analysis was performed on a routine basis before differentiation, as previously described.19 All detected aberrations were reviewed according to the following criteria: no detected copy number variation (CNV) should be bigger than 500,000 bp. All genes affected by a detected CNV were filtered against a gene list, composed of human genes from the gene ontology terms “neural crest development,” “sensory perception of pain,” as well as additional relevant marker genes. Only iPSCs that did not show any overlap with the quality control list were used for further experiments.
2.2. Differentiation of induced pluripotent stem cell–derived nociceptors
Differentiation of iPSCs into nociceptors was performed as previously described12 with slight modifications. In detail, single-cell iPSCs were seeded at suitable densities (UKBi013-A: 3 × 104 cells/cm2; UKBi018-A: 7.5 × 104 cells/cm2) in StemMAC iPS-Brew in the presence of 10 µM ROCK inhibitor Y-27632 (Cell Guidance Systems, Cambridge, United Kingdom, SM02) on Geltrex-coated T175 flasks (Thermo Fisher Scientific, Waltham, MA, A1413202) at day -1. Twenty-four hours after plating, media were changed to differentiation media. Neuronal differentiation was induced by dual SMAD inhibition, 100 nM LDN 193189 (Axon Medchem, Reston, VA, 1509) and 10 µM SB 431542 (Biozol, Eching, Germany, TRC-S1546600), from day 0 to day 6. To specify the differentiating cells into sensory neurons, 3 µM CHIR 99021 (Miltenyi Biotec, 130104172), 10 µM SU5402 (Sigma Aldrich, SML0443), and 10 µM DAPT (Axon Medchem, 1484) were added to the culture from day 3 to day 14. Two basal media were used during differentiation: medium 1 consists of Knock-Out DMEM (Thermo Fisher Scientific, 10829-018) with 20% Knock-Out Serum Replacement (Thermo Fisher Scientific, 10828-028), 2 mM (1x) GlutaMAX (Thermo Fisher Scientific, 35050038), 100 µM (1x) NEAA (Thermo Fisher Scientific, 11140035), and 0.02 mM 2-Mercaptoethanol (Thermo Fisher Scientific, 31350010). Medium 2 consists of Neurobasal Medium (Thermo Fisher Scientific, 21103-049) supplemented with 1% N2 supplement (Thermo Fisher Scientific, 17502-048), 2% B27 supplement (Thermo Fisher Scientific, 17504044), 2 mM (1x) GlutaMAX (Thermo Fisher Scientific, 35050038), and 0.02 mM 2-Mercaptoethanol (Thermo Fisher Scientific, 31350010). Between day 0 and day 3, cells were kept in medium 1, from day 4 to day 5, cells were kept in 75% medium 1 and 25% medium 2, for day 6, cells were kept in 50% medium 1 and 50% medium 2, from day 7 to 9, cells were kept in 25% medium 1 and 75% medium 2, and from day 10 to 14, differentiating cells were kept in 100% medium 2. At day 14 of differentiation, cells were dissociated with Accutase (Thermo Fisher Scientific, A11105-01) and frozen in cold CryoStor CS10 freezing medium (Sigma Aldrich, C2874-100 ML) at −80°C. After 24 hours, frozen cells were transferred to a liquid nitrogen tank for long-term storage.
2.3. Flow cytometric analysis of pluripotency and neural crest cell identity
Mouse anti-TRA1-60 (Millipore, Burlington, MA, MAB4360) in combination with a goat anti-mouse IgM 488 (Life Tech, Carlsbad, CA, A21042) secondary antibody was used according to the manufacturer's protocol to assess the percentage of pluripotent cells in the culture before differentiation.
Allophycocyanin (APC)-conjugated recombinant anti-CD57 antibody (Miltenyi Biotec, 130-111-811) costained with phycoerythrin (PE)-conjugated recombinant anti-CD271 antibody (Miltenyi Biotec, 130-091-885) was used according to the manufacturer's protocol to assess the percentage of HNK1+/p75+ neural crest cells at day 6 of sensory neuron differentiation. Flow cytometric analysis was performed using a BD Accuri (Becton, Dickinson and Company, Franklin Lakes, NJ).
2.4. Thawing, seeding, and maturing human induced pluripotent stem cell–derived nociceptors
Frozen iPSC-derived nociceptors were thawed, counted, and plated at an appropriate density (Table 1) in maturation medium consisting of Neurobasal A Medium (Thermo Fisher Scientific, 10888022) supplemented with 1% N2 supplement (Thermo Fisher Scientific, 17502-048), 2% B27 supplement (Thermo Fisher Scientific, 17504044), 2 mM (1x) GlutaMAX (Thermo Fisher Scientific, 35050038) and 0.02 mM 2-Mercaptoethanol (Thermo Fisher Scientific, 31350010), 12 µg/mL Gentamycin (Thermo Fisher Scientific, 15710049), 200 µM Ascorbic acid (Sigma Aldrich, A4544), 0.1 µg/mL human recombinant Laminin (BioLamina, LN521), 10 ng/mL GDNF (Cell Guidance Systems, GFH2), 10 ng/mL BDNF (Cell Guidance Systems, GFH1), 10 ng/mL NGF (Peprotech, Cranbury, NJ, 450-01), and 10 ng/mL NT3 (Peprotech, 450-03) supplemented with 10 µM ROCK inhibitor Y-27632 (Cell Guidance Systems, SM02). Media were changed after 24 hours to remove ROCK inhibitor. After 3 to 4 days, cells were treated with 1 µg/mL Mitomycin C (Sigma Aldrich, M4287) for 2 hours at 37°C to inactivate proliferative cells. Medium was changed twice per week. Cells were cultivated for up to 70 days. Specific maturation time points are indicated in all figures.
Table 1.
Human induced pluripotent stem cell–derived nociceptor seeding densities.
| Application | Seeding density and format |
|---|---|
| HCS microscopy | 40.000 cells per well in a 96-W MTP (Greiner) |
| High-resolution microscopy | 40.000 cells per coverslip (Neuvitro, GG-12-PLO-Laminin) |
| Patch clamp electrophysiology | 50.000 cells per coverslip (Neuvitro, GG-12-PLO-Laminin) |
| FLIPR calcium assay | 40.000 cells per well in a 96-W MTP (Greiner) |
FLIPR, fluorescent imaging plate reader; HCS, high-content screening.
2.5. Mouse dorsal root ganglia neuron cultures
All animal experiments were performed with male C57BL/6N mice at the University Cologne. Mice were kept on a 12-hour light/dark cycle and provided with food and water ad libitum. A total of 3 newborn and 14 six-week-old mice were sacrificed. Effort was made to minimize the number of animals used and their suffering. All animal experiments were performed under strict consideration of the 3R framework and in accordance with the German animal welfare law and approved by the Landesamt für Natur, Umwelt, und Verbraucherschutz Nordrhein-Westfalen (LANUV 4.18.003). Dorsal root ganglia were prepared from 2 different age groups (newborn and 6-week-old mice). The mice were sacrificed by CO2 intoxication, and DRGs were removed, pooled, and incubated in Neurobasal A (Invitrogen, Waltham, MA, 12349-015)/B27 medium (Invitrogen, 17504) containing collagenase P (Roche, Penzberg, Germany 11213873001) (10 U/mL, 1 hour, 37°C, 5% CO2). The DRGs were dissociated by trituration with fire-polished Pasteur pipettes. Axon stumps and disrupted cells were removed by bovine serum albumin (BSA) gradient centrifugation (15% BSA, 120 g, 8 minutes). Viable cells were resuspended in NeurobasalA/B27 medium, plated on poly-L-ornithin (0.1 mg/mL, Sigma Aldrich, P2533)/laminin (5 µg/mL, Invitrogen, 23017015)–precoated 96-well imaging plates (Greiner, Kremsmünster, Austria, 655090), and incubated overnight (37°C, 5% CO2).
2.6. Stimulation of human induced pluripotent stem cell–derived nociceptors and dorsal root ganglia neurons for high-content screening microscopy
For high-content screening (HCS) microscopy, hiPSCdN and rodent sensory neuron cultures were stimulated in 96-well imaging plates with 100 µL of maturation medium per well. Compounds used in this study were as follows: serotonin (5-HT, 10 mM in dH2O) and fentanyl (10 mM in dH2O) were purchased from Sigma-Aldrich; cicaprost (PGI2 analog, 10 mM stock in DMSO) was from Cayman Chemicals (Ann Arbor, MI); and forskolin (10 mM stock in DMSO), [D-Ala2]-deltorphin II (1 mM in dH2O/0.1% BSA), dynorphin A (1 mM in dH2O/0.1% BSA), [Met5]-enkephalin acetate salt (1 mM in dH2O/0.1% BSA), and nociceptin (1 mM in dH2O/0.1% BSA) were purchased from Tocris (Bristol, United Kingdom). All drugs were prepared as stocks and stored at −70°C or −20°C until use.
For stimulation, compounds were dissolved in 12.5 μL of PBS in 96-well V-bottom plates (NerbePlus, Winsen, Germany, 10-111-0000), prewarmed to 37°C, mixed with 50 μL of medium from the culture wells, and added back to the same wells. Stimulations were performed with automated 8-channel pipettes (Eppendorf, Hamburg, Germany) at low dispense speed on heated (37°C) blocks. The cells were fixed for 10 minutes at room temperature (RT) by adding 100 μL of 8% paraformaldehyde resulting in a final concentration of 4%.
2.7. Immunofluorescence staining
Fixed cells were permeabilized with 0.3% T-X100 (Sigma Aldrich, T8787) in PBS for 15 minutes at RT. After washing, unspecific binding was blocked for 1 hour at RT using PBS supplemented with 5% normal donkey serum (NDS, Sigma Aldrich, D9663) and 0.1% T-X100. Primary antibodies were incubated overnight at 4°C in PBS supplemented with 1% NDS and 0.1% T-X100 (BRN3A: 1:200, Merck, Darmstadt, Germany, AB5945; ISLET1: 1:100, abcam, Cambridge, United Kingdom, ab86501; RIIβ: 1:100, BD Biosciences, Franklin Lakes, NJ, 610625; Peripherin: 1:200, Santa Cruz Biotechnology, Dallas, TX, sc-7604; Nav1.8: 1:100, Alomone labs, Jerusalem, Israel, ASC-016, TrpV1: 1:100, Alomone labs, ACC-030; S100β: 1:200, abcam, ab52642). Cells were washed twice with PBS and incubated with secondary antibodies for 1 hour at RT. Secondary antibodies were diluted 1:500 in PBS supplemented with 1% NDS and 0.1% T-X100 (donkey antirabbit IgG-Alexa Fluor 488: Thermo Fisher, A-21206, donkey antimouse IgG-Alexa Fluor 594: Thermo Fisher, A-21203, donkey antigoat IgG-Alexa Fluor 647: Thermo Fisher, A-21447). After washing, coverslips were mounted on high-precision cover glasses using Fluoromount-G with DAPI (Invitrogen, 00-4959-52). The cover glasses with the mounted coverslips were fixed on microscope slides and imaged through the cover glass. Extracellular markers were stained accordingly, but without the permeabilization step and without T-X100 in any of the incubation and blocking solutions (Nav1.7: 1:100, abcam, ab85015; TrkA: 1:50, Alomone labs, ANT-018; hRet: 1:100, R&D Systems, Minneapolis, MN, AF1485). Images for quantification purposes were taken with the InCell Analyzer 2200 plate microscope (GE Healthcare, Chicago, IL) in a randomized and automated manner. High-resolution images were taken with ZEISS Axio Imager Z1 equipped with an Apotom 1.0 (Zeiss, Oberkochen, Germany).
For immunofluorescence staining for HCS imaging, the fixed cells were washed twice with PBS. After blocking and permeabilization (2% NDS [Dianova, Hamburg, Germany, 017-000-121), 1% BSA (Sigma Aldrich, A9418), 0.1% Triton X-100 (VWR, Radnor, PA, 112298.0101), 0.05% Tween 20 (Sigma, P9416]) for 1 hour at RT, cultures were incubated with the respective primary antibodies: chicken polyclonal antibodies against UCHL1 (Novus, Centennial, CO, NB110-58872, only for DRG neurons), rabbit monoclonal anti-RIIα (phospho-Ser96) (1:1000, clone 151, Abcam, ab32390), and mouse monoclonal anti-PKA RIIβ (1:1000, BD Transduction Laboratories, Franklin Lakes, NJ, 610625). The primary antibodies were diluted in 1% BSA in PBS and stored at 4°C overnight. Subsequent to 3 washes with PBS (10 minutes, RT), cells were incubated with secondary Alexa dye–coupled antibodies (1:1000) and DAPI (50 ng/mL) for 1 hour at RT, protected from light. As secondary antibodies, highly cross-adsorbed Alexa-Fluor −647 (Life Technologies, A31571), −555 (Invitrogen, A32732), and −488 (Invitrogen, A32931) were used in 1:1000 dilutions. After 3 washes with PBS (30 minutes, RT), wells were rinsed with 0.1% Triton X-100 in PBS (5 minutes), followed by 20 minutes of incubation with NeuroTrace 500/525 Green Fluorescent Nissl Stain solution diluted in PBS (1:500, Invitrogen, N21480) at RT. After Nissl staining, wells were rinsed with PBS (5 minutes), 0.1% Triton X-100 in PBS (5 minutes), and again 3 times with PBS (30 minutes). Finally, 96-well plates were filled with PBS, sealed, and stored at 4°C. Before scanning, plates were allowed to equilibrate to RT, protected from light.
2.8. High-content screening microscopy
Stained cultures in 96-well plates were scanned using a Cellomics ArrayScan XTI (Thermo Fisher Scientific) with a light-emitting diode light source. Images were acquired with a 20× (hiPSCdN) or 10× (DRG neurons) objective (Zeiss) and analyzed using the Cellomics software package. In brief, images of Nissl (hiPSCdN)/UCHL1 (DRG neurons) staining were background corrected (low-pass filtration), converted to binary image masks (fixed threshold), segmented (geometric method), and cells were identified by the object selection parameters: size: 10 to 300 µm2 (hiPSCdN 48 hours), 10 to 600 µm2 (hiPSCdN 3 weeks), 10 to 800 µm2 (hiPSCdN 5 weeks), 10 to 1000 µm2 (hiPSCdN 8 weeks), and 80 to 7500 µm2 (DRG neurons); circularity (perimeter 2/4π area): 1 to 3; length-to-width ratio: 1 to 3 (hiPSCdN) or 1 to 2 (DRG neurons); average intensity: 300 to 5000 (48 hours-5 weeks), 300 to 10,000 (8 weeks), and 800 to 12,000 (DRG); total intensity: 500 − 1 × 109 (hiPSCdN) and 2 × 105 − 5 × 107 (DRG). These image masks were then overlaid on images obtained at other fluorescence wavelengths to quantify signal intensities. To calculate spillover between fluorescence channels, 3 respective controls were prepared for each triple staining: (1) Nissl/UCHL1 alone, (2) Nissl/UCHL1 + antibody 1, and (3) Nissl/UCHL1 + antibody 2. Processing of raw fluorescence data was automated using R.74 Raw single-cell data of the controls were used to calculate the slope of best-fit straight lines by linear regression, which was then used to compensate spillover, as previously described.61 For analysis, raw mean values of replicates were normalized to a mean baseline value from all untreated wells. One-dimensional and 2-dimensional probability density plots were generated using R packages.74 Gating of subpopulations was performed by setting thresholds at local minima of probability density plots.
2.9. Electrophysiology
2.9.1. Current-clamp recordings
Whole-cell current-clamp recordings were performed with Axopatch-200B amplifier (Molecular Devices, San Jose, CA), which was interfaced by an A/D converter (Digidata 1440, Molecular Devices) to a PC running PClamp software (version 10, Molecular Devices). Pipette electrodes (GB150TF-8P, Science Products, Hofheim, Germany) were fabricated using a vertical puller (Narishige PC-10, Narishige, Tokyo, Japan). Recordings were performed in a bath solution containing the following: 140 mM NaCl (ROTH, Karlsruhe, Germany, 9265.2), 3 mM KCl (ROTH, 6781.3), 2 mM CaCl2 (Sigma Aldrich, C5080), 1 mM MgCl2 (Sigma Aldrich, M8266), 25 mM D-Glucose (Sigma Aldrich, 47829), and 10 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethane sulphonic acid) (ROTH, 9105.3) (pH 7.4 adjusted with NaOH; osmolality 310-320 mOsm). For recordings of membrane potential, the patch pipette contained the following solution: 140 mM potassium gluconate (Fluka, 60245), 5 mM HEPES (ROTH, 9105.3), 0.16 mM EGTA (ethylene glycol bis(β-aminoethylether) tetraaceticacid) (ROTH, 3054.3), 5 mM MgCl2 (Sigma Aldrich, M8266), and 5 mM phosphocreatine disodium salt (Sigma Aldrich, P7936) (pH 7.3 adjusted with KOH, osmolality 290 mOsm). Action potentials were induced by several 500 milliseconds square current injections of increasing amplitude. Recordings were sampled at a rate of 100 kHz and low-pass filtered at 10 kHz (4-pole lowpass Bessel filter). The first action potential evoked by the square pulse protocol was used to analyze the action potential properties. Finally, spontaneous neuronal activity was monitored for 2 minutes in a gap-free recording. Data were analyzed using ClampFit software (Molecular Devices).
2.9.2. Voltage-clamp recordings
Whole-cell patch-clamp experiments were performed with a HEKA EPC 10 patch-clamp amplifier and using Patchmaster software v2x73 (HEKA Elektronik Dr. Schulze GmbH, Ludwigshafen, Germany). The sampling rate was 50 kHz, and a Bessel filter with frequency borders of 3 to 1000 Hz was used. Borosilicate patch electrodes (Harvard Apparatus, Holliston, MA) manufactured with a DMZ-universal puller (Zeitz-Instrumente GmbH, Planegg, Germany) and resistances of 3 to 5 MΩ were used. The electrodes were filled with an internal solution containing the following (in mM): 130 KCI, 2 MgCI2, 0.5 CaCl2, 5 BAPTA, 10 HEPES, 3 Na2ATP, and pH 7.2 adjusted with KOH. The extracellular solution contained the following (in mM): 150 NaCl, 3 KCl, 2 CaCl2, 2 MgCI2, 10 HEPES, and 10 glucose. The pH was adjusted to 7.4 with NaOH. After achieving whole-cell configuration, the cell was voltage-clamped at a holding membrane potential of −60 mV. Afterwards, the neurons were recorded for ligand-gated ion channels in voltage-clamp. After a stable potential was observed, the various ligands (ATP 10 µM—2 seconds; γ-aminobutyric acid (GABA) 100 µM—10 seconds; pH5.3—10 seconds) were directly applied to the cell by a multivalve perfusion system (ALA Scientific Instruments, New York, NY), and the hyperpolarization effect on each neuron was monitored. Patch-clamp data were analyzed using the Patchmaster online measuring tool.
2.10. Capsaicin fluorescent imaging plate reader assay
The fluorescent imaging plate reader (FLIPR) assay was conducted as described in the study conducted by Damann et al.,16 with minor adaptations. Cells were plated in clear bottom black 96-well plates (Corning Cellbind Surface Assay Plates No. 3340, Corning, NY) at a density of 40,000 cells/well, and 49 up to 58 days after differentiation, the resulting peripheral sensory neurons were tested in the Fluorescent Imaging Plate Reader (FLIPR). On the day of the experiment and before dye loading, the neurons were inspected under the microscope to identify wells with a detached neuronal layer, which were excluded from further analysis. In the following, medium was replaced, and the cells were incubated with Fluo-4 (Molecular Probes, Eugene, OR) and 0.01 vol % Pluronic F127 (Molecular Probes) in Hanks buffered saline solution (HBSS, Gibco Invitrogen) for 30 minutes at 37°C. The plates were then washed with HBSS buffer and, after a further incubation for 15 minutes at room temperature, used in the FLIPR assay for Ca2+ measurement (wavelength λex = 488 nm, λem = 540 nm).
Two FLIPR protocols were designed to test for agonistic activity by capsaicin and the inhibition of capsaicin-stimulated TRPV1 activation by preincubation with a fixed concentration of capsazepine (10 µM), respectively. KCl was used to test for nonspecific activation of the fluorescence assay. The final KCl concentration selected was 100 mM. Capsaicin (3× concentrated for a 50-µL addition to reach a total volume of 150 µL; stock concentration 10 mM in DMSO, Sigma, M2028) as well as KCl (3× concentrated for a 50-µL addition to reach a total volume of 150 µL; 300 mM in HBSS/0.3% DMSO, Merck, 1.04936.1000) were added to the cells, and the fluorescence was measured. The highest test concentration of capsaicin was 10 µM; all other concentrations were generated by 1 to 10 dilution steps in HBSS that a final DMSO concentration of 0.3% was not exceeded. In separate wells, cells were treated outside the instrument with 10 µM of the specific TRPV1 antagonist capsazepine 6 minutes before capsaicin stimulation to test for the specificity of the reaction. The overall detection time for the run was 6 minutes. For data analysis, FLIPR raw data (fluorescence units) were normalized to the response elicited by 10 μM capsaicin. The quantification for approximative half maximal effective concentration (EC50) values was performed by measuring the highest fluorescence intensity (FC, fluorescence counts) over time. EC50 values were calculated using GraphPad Prism (GraphPad Software, San Diego, CA).
3. Statistics
Data sets are presented using appropriate summary statistics as indicated in the figure legends. Statistical analyses were performed using GraphPad Prism (GraphPad Software). Statistical analyses with respective post hoc tests and appropriate corrections for multiple comparisons were performed as described in detail in the figure legends. P < 0.05 was considered statistically significant.
4. Results
4.1. Early expression of maturation markers in cryopreservable human induced pluripotent stem cell–derived nociceptive sensory neurons
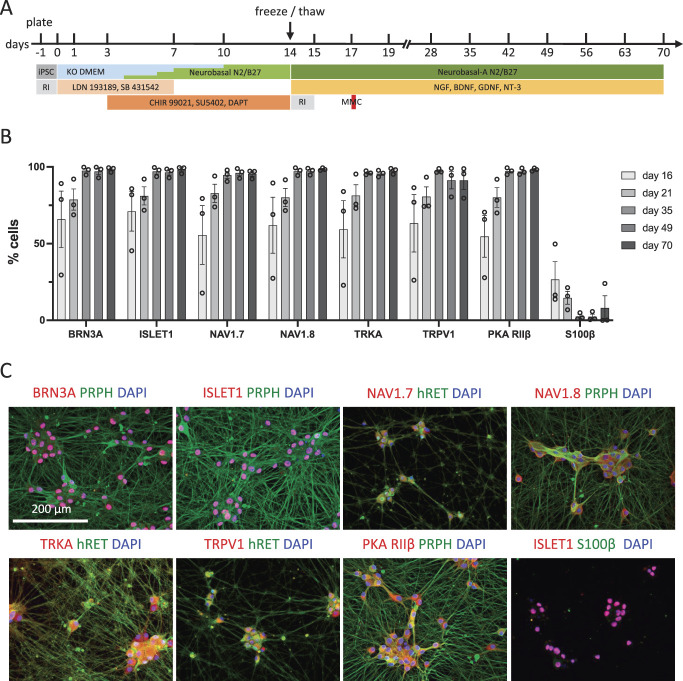
Nociceptive sensory neurons were generated from human iPSC following the protocol by Chambers et al.12 Conditions were optimized to set up a robust, scalable, and efficient differentiation and maturation process of the used iPSC lines. To that end, cell preparation was split into a first phase encompassing the generation of cryopreservable immature nociceptors after 14 days of differentiation, followed by a standardized maturation phase for up to 70 days (Fig. 1A). The starting cell density was optimized for each used iPSC line. To that end, multiple iPSC seeding densities ranging from 2 × 104 to 12 × 104 cells/cm2 were tested. The optimal seeding density was determined based on successful generation of peripheral neurons and the purity of the obtained cultures. Following the regimen of dual SMAD inhibition,11,12 treatment with LDN193189 and SB431542 was extended to day 6 of differentiation, and the application of differentiation-relevant small molecules (CHIR99021, DAPT, SU5402) was started at day 3 instead of day 2 and prolonged until day 14 instead of day 10 (Fig. 1A). Before entering differentiation, iPSCs were validated for pluripotency based on TRA1-60 expression (Fig. S1A, available at http://links.lww.com/PAIN/B776) and genomic integrity through high-resolution SNP analysis (data not shown). Formation of neuroectoderm was induced by dual SMAD inhibition, using BMP (bone morphogenetic protein) and TGF-β (transforming growth factor beta) signaling inhibitors LDN 193189 and SB 431542. Differentiating cells were further specified towards the sensory neural lineage by inhibition of GSK3, FGF, and Notch signaling using CHIR 99021, SU5402, and DAPT, respectively (Fig. 1A). An intermediate neural crest stage was confirmed by detection of coexpression of HNK1 and p75 after 6 days of in vitro differentiation (Fig. S1B, available at http://links.lww.com/PAIN/B776). To enable large-scale provision of up to 1 billion cells, immature sensory neurons were harvested after 14 days of differentiation and frozen at 1 to 10 × 106 cells/mL in CryoStor CS10 freezing medium at −80°C. For long-term storage, cryopreserved cells were transferred to liquid nitrogen after 24 hours.
Figure 1.
Generation and validation of nociceptive sensory neurons. (A) Timeline of the differentiation and maturation protocol. (B) Quantification of peripheral neuronal, nociceptor, and glial marker expression of cryopreserved hiPSCdNs over the time course of in vitro differentiation (mean ± SEM, N = 3). (C) Immunostaining of the sensory neuron markers BRN3A, ISLET1, PRPH, Nav1.7, and Nav1.8, the nociceptor markers TRKA, TRPV1, and PKAR, as well as the glial marker S100β in hiPSCdN neuron cultures after 70 days of differentiation. hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; iPSCs, induced pluripotent stem cells; MMC, mitomycin C; RI, ROCK inhibitor. BRN3A, brain-specific homeobox/POU domain protein 3A; ISLET1, ISL LIM homeobox 1; PRPH, Peripherin, Nav1.7, Voltage-Gated Sodium Channel Alpha Subunit Nav1.7; Nav1.8, Voltage-Gated Sodium Channel Alpha Subunit Nav1.8; TRKA, Tropomyosin receptor kinase A; TRPV1, transient receptor potential cation channel subfamily V member 1 ; PKA RIIβ, Protein Kinase A regulatory subunit RIIβ; KO DMEM, Knock Out Dulbecco's Modified Eagle Medium; NGF, Nerve growth factor; BDNF, Brain-derived neurotrophic factor; GDNF, Glial cell line-derived neurotrophic factor; NT-3; Neurotrophin-3.
For subsequent applications, cells were thawed, seeded in suitable culture formats, and matured in the presence of neurotrophic factors for up to day 70 (Table 1; Fig. 1A). Cells were treated with mitomycin C 3 to 4 days after thawing to maintain neuronal cultures and inhibit proliferation of non-neuronal cells. Already 48 hours after thawing at day 17 of in vitro culture, 65.9% ± 18.9% and 71.0% ± 12.9% of the cells showed expression of the transcription factors BRN3A and ISLET1, respectively. At this time point, 55.6% ± 19.1% and 62.0% ± 18.3% of the cells stained positive for the peripheral voltage-gated sodium ion channels Nav1.7 and Nav1.8, respectively; the nociceptor marker proteins TRKA, TRPV1, and PKA-RIIβ were detected in 59.4% ± 18.6%, 63.3% ± 18.7%, and 54.7% ± 13.5% of the cells, respectively (Fig. 1C, S2, S3, available at http://links.lww.com/PAIN/B776). Expression of these markers increased with further maturation, plateauing at day 35 of in vitro maturation. At the same time, variability between different cultures decreased with time. After day 70 of in vitro maturation, expression of all these markers approached 100% (BRN3A: 98.4 ± 0.9%; ISLET1: 98.2 ± 1.3%, Nav1.7: 95.5 ± 1.6%; Nav1.8 98.5 ± 0.5%; TRKA: 97.5 ± 1.2%; TRPV1: 91.3 ± 5.9%; PKA-RIIβ: 98.3 ± 0.6%; Figs. 1B and C, S2, S3, available at http://links.lww.com/PAIN/B776). HiPSCdNs typically grew in clusters, which generated a dense hub and spoke architecture of strongly peripherin-positive processes. The number of S100β-positive presumptive glial cells sharply dropped after mitomycin C treatment on day 17 from 26.7 ± 11.6% after day 16 to levels well below 10% at ≥day 35 of maturation (Figs. 1B and C, S2, S3, available at http://links.lww.com/PAIN/B776). Thus, the in vitro differentiation protocol used enables robust generation of hiPSCdNs with a largely homogeneous marker expression profile.
4.2. Time-dependent electrophysiological maturation and ligand-gated ion channel functionality
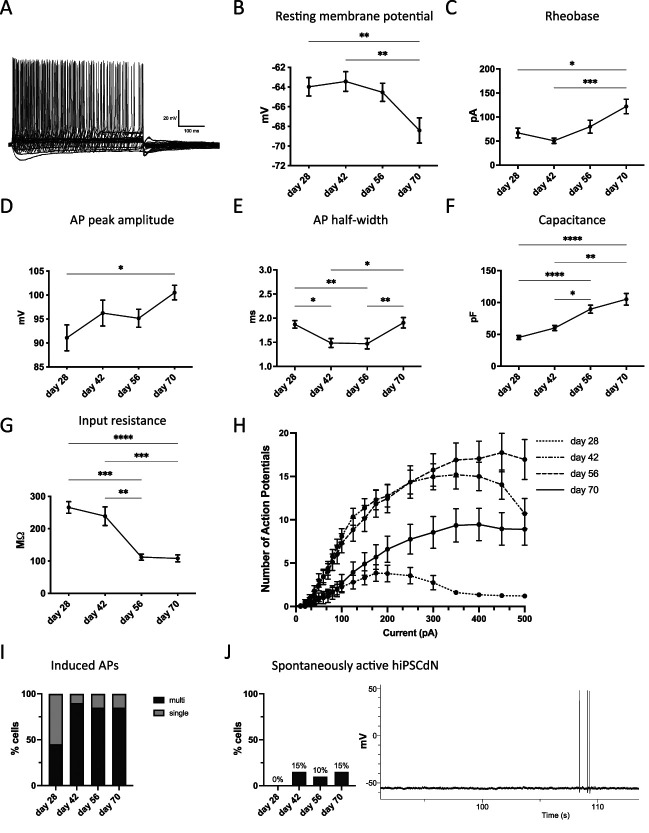
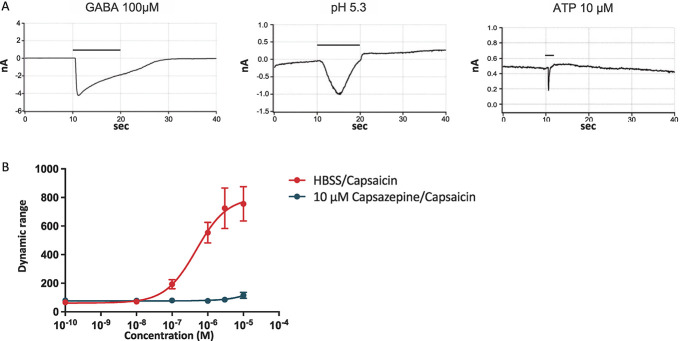
Human iPSC-derived nociceptors were characterized by whole-cell current-clamp recordings after 28, 42, 56, and 70 days of differentiation (Fig. 2, S4, available at http://links.lww.com/PAIN/B776). Furthermore, functionality of ligand-gated ion channels was confirmed by voltage-clamp recordings or FLIPR calcium assay at day 70 of in vitro culture (Fig. 3). For each tested time point, 20 cells were analyzed by current-clamp recordings. The resting membrane potential gradually decreased from −63.98 ± 0.93 mV at day 28 in culture to −68.41 ± 1.28 mV at day 70 (mean ± SEM, Fig. 2B). Rheobase increased from 67.00 ± 10.13 pA at day 28 to 122.00 ± 15.02 pA at day 70 in culture (Fig. 2C). Simultaneously, the action potential peak amplitude increased from 91.08 ± 2.7 mV at day 28 to 96.24 ± 2.70 mV at day 42 and 100.50 ± 1.52 mV at day 70 (Fig. 2D). The action potential half-width decreased from 1.87 ± 0.08 milliseconds at day 28 to 1.49 ± 0.09 milliseconds and 1.47 ± 0.11 milliseconds at day 42 and day 56, respectively, followed by an increase back to 1.90 ± 0.11 milliseconds at day 70 (Fig. 2E). The capacitance of the iPSC-derived nociceptors doubled from 45.13 ± 3.12 pF at day 28 to 89.67 ± 6.3 pF at day 56 and further increased to 105.20 ± 9.22 pF at day 70 of in vitro culture (Fig. 2F). The input resistance of hiPSCdNs decreased over time from 265.90 ± 18.08 at day 28 to 108.20 ± 10.83 MΩ at day 70 (Fig. 2G). Action potential firing frequency on current injection was found to strongly increase up until day 56, followed by a slight decrease at day 70 of maturation (Fig. 2H). The number of neurons capable of firing multiple action potentials on current injections increased from 50% at day 28 to 85% to 90% between day 42 and day 70 of maturation (Fig. 2I). Spontaneous activity was assessed by a 2-minute gap-free recording and only detected in a minority of the analyzed neurons (10%-15%) after at least 42 days of maturation with a very low frequency below 0.05 Hz (Fig. 2J). The presence and functionality of ligand-gated channels was verified in voltage-clamp or by FLIPR calcium assays (Fig. 3). After 70 days in culture, hiPSCdNs showed evoked currents after application of 100 µM GABA, pH shift (to pH 5.3), and 10 µM ATP (Fig. 3A). Furthermore, using a fluorescent imaging plate reader (FLIPR) assay, functionality of the heat-sensitive receptor TRPV1 was tested after 70 days of in vitro maturation. We examined the response of hiPSCdNs to increasing concentrations of capsaicin (0.0001 µM-10 µM). Concentrations greater than 0.1 µM capsaicin induced an increase in fluorescence. The activation of hiPSCdNs by capsaicin was fully blocked by preincubation with 10 µM of the TRPV1-specific antagonist capsazepine (Fig. 3B), indicating functional expression of TRPV1. Taken together, these data indicate that the iPSCdNs undergo protracted functional maturation that continues to at least 70 days of in vitro differentiation.
Figure 2.
Electrophysiological properties develop over time in hiPSCdNs. Electrophysiological properties of hiPSCdNs derived from iPSC line UKBi013-A. (A) Exemplary trace of a multiple action potential firing hiPSCdN after 70 days of in vitro differentiation. (B–G) Electrophysiological parameters change and develop over the time of maturation: resting membrane potential (B), rheobase (C), action potential (AP) peak amplitude (D), AP half-width (E), capacitance (F), and input resistance (G). Values are represented as mean ± SEM. N = 20 patched cells per time point. The Friedman test with the Dunn multiple comparison test; *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. (H) Action potential firing behavior in response to a series of 500 milliseconds current steps from 0 to 500 pA. Data are represented as mean values ± SEM. (I) Quantification of induced action potential firing behavior over the time course of in vitro maturation. (J) Quantification of spontaneous firing behavior over the time course of in vitro differentiation and representative trace of a spontaneously active hiPSCdN after 70 days of in vitro cultivation (not corrected for liquid junction potential). hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; iPSCs, induced pluripotent stem cells.
Figure 3.
Human induced pluripotent stem cell–derived nociceptors express functional ligand-gated channels and receptors. (A) hiPSCdNs were found to be responsive in voltage-clamp to the application of 100 µM GABA, a shift to pH 5.4, and the application of 10 µM ATP after 70 days of in vitro culture. Black bars indicate compound application, followed by a washout. (B) Fluorescent Imaging Plate Reader (FLIPR) calcium assay revealed reactivity to the TRPV1 stimulating compound capsaicin in a dose-dependent manner that was fully blocked by application of the TRPV1 antagonist capsazepine after 70 days of differentiation. hiPSCdNs, human induced pluripotent stem cell–derived nociceptors. GABA, γ-aminobutyric acid; ATP, adenosine triphosphate; TRPV1, transient receptor potential cation channel subfamily V member 1; HBSS, Hanks' balanced salt solution.
4.3. Receptor-independent intracellular protein kinase A type II sensitization signaling matures early in human induced pluripotent stem cell–derived nociceptors
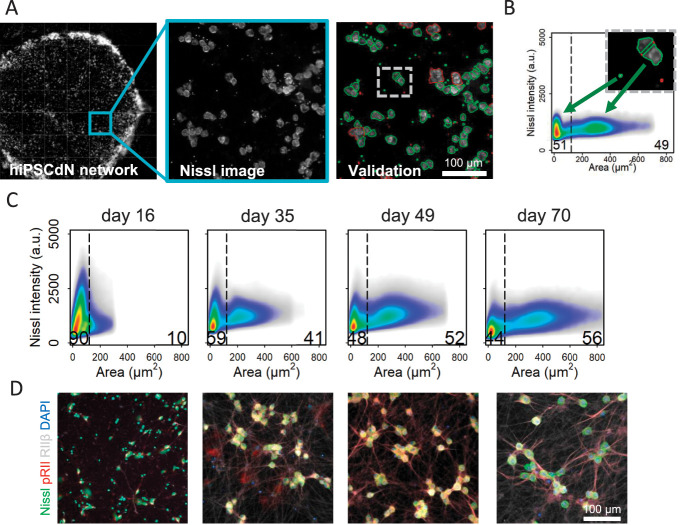
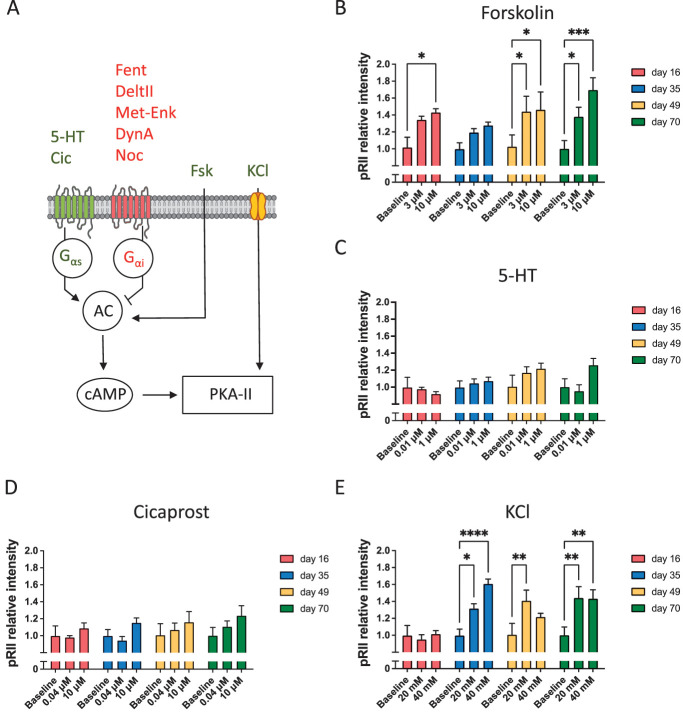
A large number of diverse stimuli such as prostaglandins, serotonin, epinephrine, opioids, and neuronal depolarization modulate the activity of protein kinase A type II (PKA-II) in nociceptive neurons. When PKA-II signaling emerges in hiPSCdNs is not known. We adapted an HCS microscopy–based method for monitoring the activation process of endogenous PKA-II signaling activity6,7,29–35,78 in hiPSCdN cultures after 16 days, 35 days, 49 days, and 70 days of differentiation. Software-based identification of objects in neuronal cultures with dense neurite networks was achieved by establishing fluorescence detection of Nissl staining of neuronal cell bodies for automated image analysis (Fig. 4A).
Figure 4.
High-Content Screening (HCS) microscopy quantifies single neurons on a full population basis. (A) Pipeline of automated HCS object identification. Green or red encircled cells indicate automatically selected or rejected objects, respectively. (B) Size-dependent identification of neuronal cell bodies. Black line delimitates 2 subpopulations according to cell size. On the left side of the threshold, the smaller cells as well as cellular debris and on the right side, the larger mature neuronal cells were identified (threshold cell body area is always = 120 µm2). (C) Size of identified neuronal cell bodies increases over the maturation time. N = 3 independent cell preparations, data were pooled from all cell culture conditions for the respective time point, >482,000 neurons/time point were included in the 2D density plots. (D) Representative HCS images of hiPSCdN cultures after 16 days, 35 days, 49 days, and 70 days of in vitro culture for cell body marker (Nissl, green), PKA-II activity (pRII, red), nociceptor marker (PKA-RIIβ, grey), and cell nuclei (DAPI, blue). hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; PKA-II, protein kinase A type II. PKA-RIIβ, Protein Kinase A regulatory subunit RIIβ, DAPI, 4',6-diamidino-2-phenylindole.
Using the automated imaging analysis objects of distinct sizes were observed (Figs. 4B–D, S5, available at http://links.lww.com/PAIN/B776). A lower cutoff of 10 µm2 was set to discriminate cells from debris. Following this protocol, we identified objects within a size range between 10 and approximately 800 µm2. The proportion of cells with a cross-sectional area above a threshold of 120 µm2 increased gradually over time from 10.2% ± 2.63% (after 16 days) to 56% ± 3.41% (after 70 days; Figs. 4C and D, S5, available at http://links.lww.com/PAIN/B776). Similar to the recorded increase in cell membrane capacity (Fig. 2F), objects above the threshold increased in size with time of maturation (from 172.80 ± 0.17 µm2 at day 16 to 402.10 ± 0.48 µm2 at day 70) (Fig. S6; Suppl. Table 2, available at http://links.lww.com/PAIN/B776). Functionally, no differences were observed between responses to stimulations of large and small-sized objects (data not shown). For better comparability with electrophysiological experiments described earlier, which are generally performed on larger neurons but not on very small objects, only quantification of objects above the threshold is shown in subsequent functional analyses.
Immunofluorescent detection of the phosphorylated regulatory RII subunit of PKA (pRII) is a surrogate measurement for the activity of PKA-II (Fig. 5A).32 High-content screening microscopy–based measurement of pRII intensity changes allows quantification of endogenous PKA-II responses in hiPSC-derived nociceptors over the time course of in vitro maturation (Figs. 5B–E). First, mediators known for their nociceptor sensitizing activity were analyzed27 (forskolin7,9,29,30,32–34,78, 5HT9,32,34, KCl29,78, PGI2/cicaprost9,34, opioids33). Cells were stimulated for 5 minutes, fixed, and immunocytochemically analyzed for pRII response on a single-cell level. Forskolin is membrane permeable and directly activates cytoplasmic adenylyl cyclases independent of membrane receptors and their downstream signaling pathways (Fig. 5A). Forskolin induced pRII signals of similar magnitude at all time points, which reached statistical significance in all but the day 35 group (Fig. 5B). At 70 days, we noticed a concentration-dependent increase of pRII intensity when comparing the responses with 3 µM and 10 µM forskolin, respectively, and confirmed this in hiPSCdNs derived from a genetically distinct iPSC line (UKBi018-A; Fig. 5B, Fig. S7A, available at http://links.lww.com/PAIN/B776).
Figure 5.
PKA-II intensity in human iPSC-derived nociceptors exposed to pronociceptive mediators. (A) Schematic diagram of proalgesic mediators (green) and analgesic ligands (red) used in this study. Components of the figure were adapted from Servier Medical Art (https ://smart .servi er.com/#). (B) Forskolin (Fsk, 3 and 10 μM) induces a constant pRII response across the tested time points of maturation in hiPSCdNs derived from iPSC line UKBi013-A. (C and D) Stimulation with GPCRs ligands, serotonin (5-HT, 0.01 and 1 μM), and cicaprost (Cic, 0.04 and 10 μM) resulted in a sensitization regulation that showed a trend to increase with maturation time. (E) KCl (20 and 40 mM) induced a significant pRII response only after 35 days, followed by a constant sensitization for longer time points. Values are represented as mean values ± SEM; N = 3 independent cell culture preparations with 9 to 15 individual data points per conditions, >9000 neurons/condition. All values are shown as “pRII relative intensities.” All data were normalized to the baseline control. Two-way ANOVA with Bonferroni multiple comparison test; *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001 indicate significance levels between conditions compared with the baseline control of the respective time point. ANOVA, analysis of variance; hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; iPSC, induced pluripotent stem cells; PKA-II, protein kinase A type II. GPCR, G protein-coupled receptors; 5-HT, 5-hydroxytryptamine / serotonin; pRII, phosphorylated regulatory RII subunit of PKA; cAMP, cyclic adenosine monophosphate; KCI, potassium chloride.
By contrast, stimulation of hiPSCdNs with the GPCR ligands, serotonin (5-HT), and cicaprost (Cic, more stable prostacyclin PGI2 analogue) resulted in no significant pRII responses, although a slight trend towards increased responses was detectable after longer maturation times (Figs. 5C and D). Previously, we identified depolarization to be able to induce pRII sensitization signaling.7,29 Indeed, depolarization by KCl induced a significant pRII response also in hiPSCdNs at all maturation stages beyond day 16 (Fig. 5E).
These data indicate that robust PKA-II signaling can be initiated as early as day 16 by membrane-permeating pharmacological treatment as well as by depolarization in hiPSCdN. By contrast, receptor ligands did not show significant induction of PKA-II activation during the observed maturation time of 70 days.
4.4. Inhibitory effects of opioids on protein kinase A type II signaling emerge only after prolonged in vitro maturation in human induced pluripotent stem cell–derived nociceptors
While opioids are standard drugs for pain therapy, the expression and signaling capability of MOP, KOP, DOP, and the nociceptin/orphanin FQ peptide receptor (NOP) in hiPSCdNs has not been analyzed yet. Immunocytochemical staining indicates expression of all 3 classical opioid receptors (MOP, KOP, and DOP) as well as NOP in hiPSCdN cultures throughout the maturation time (Fig. 6A, S8, available at http://links.lww.com/PAIN/B776). We then stimulated the cells with receptor agonists and measured their inhibitory impact on forskolin (3 µM)–induced pRII-immunofluorescence signals (Figs. 6B–G), as previously described for rodent DRG neurons.6 Activation of opioid receptors inhibits adenylyl cyclase–driven cAMP generation and thus decreases PKA-II activity (Fig. 5A). Stimulation of opioid receptors did not result in reduced PKA-II activity after maturation of 16, 35, or 49 days. Only after maturation for 70 days, inhibitory effects were detected. Application of fentanyl (MOP agonist, Fig. 6C) resulted in a significant reduction of the forskolin-induced PKA-II activity for both concentrations tested (5 and 10 µM). Likewise, application of nociceptin (NOP agonist, Fig. 6D) induced a significant reduction of the forskolin-induced PKA-II activity for low (5 µM) and high (20 µM) concentrations tested. The DOP agonist Deltorphin II, on the contrary, showed an almost significant reduction of the PKA-II activity at 20 µM (P = 0.0506), but not at 10 µM (Fig. 6E). Dynorphin A (KOP agonist, Fig. 6F) and Met-Enkephalin (DOP/MOP agonist, Fig. 6G) did not show significant inhibitory effects on PKA-II activation at any tested time point. To corroborate opioid signaling in hiPSC-derived nociceptors, we confirmed the fentanyl response in hiPSdNs generated from a second female donor (Fig. S7B, available at http://links.lww.com/PAIN/B776). Of interest, the inhibitory effect of fentanyl in hiPSCdNs at day 70 was comparable in effect size with its inhibitory activity in DRG neurons derived from 6-week-old mice (Fig. 7). Collectively, these data show that functional modulation of PKA-II signaling through the classical opioid receptor MOP as well as NOP receptors in hiPSC-derived nociceptors generated by small molecule–based differentiation requires prolonged in vitro maturation of approximately 70 days.
Figure 6.
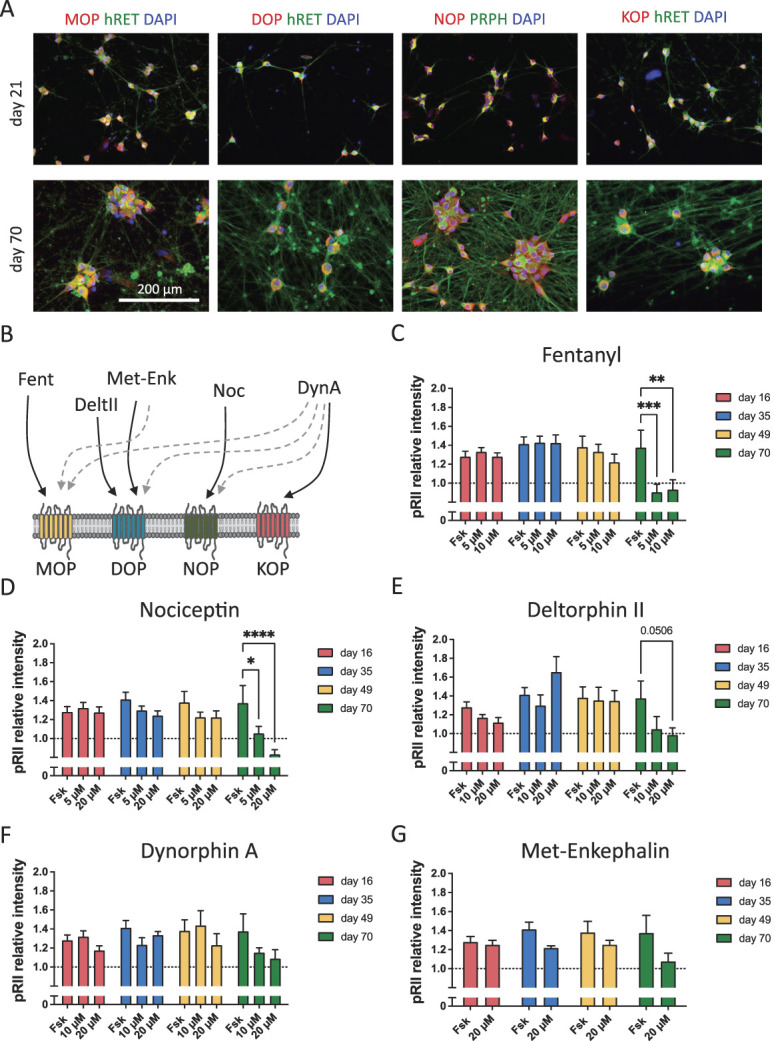
Opioid receptor signaling develops nonlinearly over maturation time in human iPSC-derived nociceptors. (A) Representative images of hiPSCdN expressing the opioid receptors MOP, KOP, DOP, and NOP after 21 and 70 days of differentiation. (B) Selected opioid receptor ligands used in this study and their respective target receptor (Fent 5 Fentanyl, DeltII 5 Deltorphin II, Met-Enk5Met-Enkephalin, Noc5Nociceptin, and DynA5Dynorphin A).Main affinity is indicated by solid arrows, while lower affinity is indicated by dashed arrows [adapted from Refs. 20, 46]. Components of the figure were adapted from Servier Medical Art (https ://smart .servi er.com/#). (C–G) pRII intensities after forskolin (Fsk) stimulation and application of analgesic compounds in hiPSCdN derived from iPSC line UKBi013-A. Fentanyl (C, MOP agonist) nociceptin (D, NOP agonist) showed significant reduction of forskolin for both concentrations tested. Deltorphin II (E, DOP agonist) only for the higher concentration, while dynorphin A (F, KOP agonist) and Met-Enkephalin (G, DOP/MOP agonist) did not show inhibitory effects on PKA-II activation at any tested time point. All values are shown as “pRII relative intensities.” All data were normalized to the baseline control, baseline levels are indicated as a dashed line. Two-way ANOVA with Bonferroni multiple comparison test; *P < 0.05; **P < 0.01; and ***P < 0.001 indicate significance levels between conditions compared with the forskolin stimulation only at the respective time point. Data are represented as mean values ± SEM; N = 3 to 4 independent cell culture preparations with 5 to 15 individual data points per conditions, >5000 neurons/condition. ANOVA, analysis of variance; hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; iPSC, induced pluripotent stem cells; PKA-II, protein kinase A type II.
DAPI, 4',6-diamidino-2-phenylindole; MOP, µ-opioid peptide receptor; KOP, κ-opioid peptide receptor ; DOP, δ-opioid peptide receptor; pRII, phosphorylated regulatory RII subunit of PKA; PRPH, peripherin; hRET, human receptor tyrosine kinase.
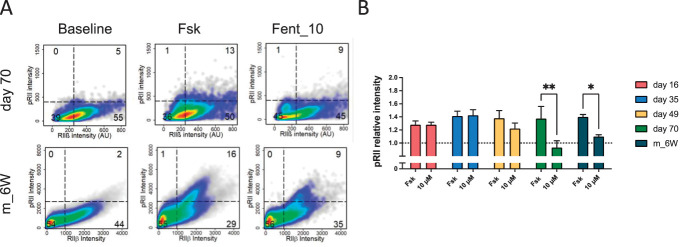
Figure 7.
Opioid receptor signaling in hiPSCdN and mouse DRG neurons. (A) Cell density plots showing pRII/RIIβ–labeled 70 days differentiated hiPSCdN derived from iPSC line UKBi013-A and DRG neurons from 6-week-old mice (m_6W) stimulated with 3 µM forskolin (Fsk) or 3 µM forskolin in combination with 10 µM fentanyl (Fent_10). Dashed lines indicate gating thresholds to discriminate between RIIβ (−) and RIIβ (+) neurons. The opioid influences only the RIIβ (+) cells of 6-week-old mouse DRGs, while in the case of 70 days differentiated hiPSCdN, we can observe that both RIIβ (−) and RIIβ (+) are influenced by fentanyl. (B) Fold changes of pRII intensities for all time points in hiPSCdN cultures and DRG neurons of 6-week-old mice (m_6W). Fentanyl (10 μM) shows similar efficiency between the mouse data and 70-day-old hiPSCdN cells only, suggesting that functional MOP signaling is fully established at this respective time point and not earlier. All values are shown as “pRII relative intensities.” All data were normalized to the baseline control, and baseline levels are indicated as a dashed line. Two-way ANOVA with Bonferroni multiple comparison test; *P < 0.05; **P < 0.01; and ***P < 0.001 indicate significance levels between conditions compared with the forskolin stimulation of the respective time point. Data are represented as mean values ± SEM; N = 14 animals for m_6W, >7000 neurons/condition; N = 3 independent cell culture preparations with 5 to 15 individual data points per conditions (hiPSCdN), >5000 neurons/condition. ANOVA, analysis of variance; DRG, dorsal root ganglia; hiPSCdNs, human induced pluripotent stem cell–derived nociceptors; iPSC, induced pluripotent stem cells. MOP, μ-opioid peptide receptor; pRII, phosphorylated regulatory RII subunit of PKA; RIIβ, total regulatory RII subunit of PKA.
5. Discussion
Human iPSC-derived nociceptors offer the potential to investigate molecular and cellular mechanisms in this otherwise hardly accessible type of sensory neuron. In contrast to, eg, exogenous expression of human target proteins in immortalized cell lines, this in vitro system provides a more authentic representation of cell intrinsic regulatory mechanisms involving, among others, intracellular signaling networks. Because the regulatory interplay between ionotropic and metabotropic signaling is key to the understanding of activity of nociceptive neurons,21,27,29 we focused our analysis of the hiPSC-derived nociceptors on these 2 properties.
5.1. Authenticiy of human induced pluripotent stem cell–derived nociceptors as a model system for human nociceptive neurons
Multiple protocols have been published for the differentiation of human embryonic, as well as induced pluripotent stem cells into mixed sensory neuron cultures and defined sensory neuron subtypes.41 Strategies used include extrinsic factor–driven differentiation,1,12,18,51 “forward programming” by transcription factor overexpression,55 and combined approaches.4,17,28,66 The most frequently used paradigm was introduced by Chambers et al.12 This protocol uses a dual SMAD inhibition (inhibition of BMP and TGF-β signaling) in a cell density–dependent manner to induce early neuroectoderm, followed by further patterning into peripheral nociceptive neurons.11 Stem cell–derived nociceptors generated in this manner have been successfully used for disease modeling8,15,45,47,49,81 and personalized therapy studies.53 We established a large-scale differentiation scheme based on this protocol. We were able to generate reproducible batches of up to 1 billion nociceptive sensory neurons that express key markers for sensory neurons (BRN3A, ISLET1, PRPH, hRET, NAV1.7, and NAV1.8) as well as nociceptors (TRKA, PKARIIβ, and TRPV1). Contaminating glial cells were gradually removed over cultivation time after a treatment with the antiproliferative drug mitomycin C. High-content screening microscopy revealed that plated hiPSCdNs have a mean cross-sectional area of 402.1 ± 0.48 µm2, which, in case of an ideal circle, corresponds to a mean diameter of 22.6 ± 0.78 µm after 70 days of differentiation. The diameter of human DRG neurons was previously reported to range from 20 to 100 µm, and the size of human TrkA-positive nociceptors was found to range from 300 to 700 µm2 in cryosections of DRG tissue.24,62 This suggests that hiPSCdNs recapitulate important morphological features of small-diameter human nociceptive sensory neurons. The generated human nociceptors are electrophysiologically active, and our data obtained from 70-day-old hiPSCdNs largely recapitulate findings in primary human fetal and adult DRG neurons for resting membrane potential, action potential peak amplitude, and membrane capacitance (Suppl. Table 1, available at http://links.lww.com/PAIN/B776).10,25,56 The action potential half-width of primary fetal and adult human DRG neurons was reported to range from 3 to 7 milliseconds,10,25,56 whereas in our hiPSCdNs, it was found to be 1.9 milliseconds for the iPSC-derived nociceptors (Suppl. Table 1, available at http://links.lww.com/PAIN/B776). The expression of functional GABA, P2X3, ASIC, and TRPV1 receptors observed in our hiPSCdNs is in line with previously published expression patterns and functional traits of human stem cell–derived nociceptive sensory neuron cultures, as well as with data generated from primary human adult and embryonic DRG neurons.4,10,12,25,41,56,63,79 While we cannot assure that our iPSC-derived nociceptive sensory neurons reflect authentic DRG neurons, these data indicate that they exhibit protein expression profiles and functional traits compatible with primary nociceptors and are thus suitable for assessing nociceptive metabotropic signaling.
5.2. High-content screening microscopy reveals differential emergence of pain-related signaling pathways across hiPSCdN differentiation
We introduced HCS microscopy for the analysis of nociceptive neurons including endogenous opioid signaling.9,29,33,34,43 A complete digitalization of cell cultures as performed in the process of HCS microscopy analysis allows quantification of very large numbers of cells at the single-cell level, including assessment of heterogeneity. This approach enabled us to monitor sensitization signaling activity of an entire population of cultured hiPSCdNs for up to 70 days. This vastly extends previous studies where HCS microscopic analysis of iPSC-derived sensory neurons has only been performed up to 39 days of differentiation, with a restricted focus on calcium imaging in response to voltage-gated sodium channel modulators.69 By contrast, we applied an HCS microscopy approach to study how metabotropic intracellular sensitization signaling changes during long-term maturation of hiPSCdN. We quantified the expression of PKA-RIIβ, a subgroup marker predominantly expressed in nociceptive neurons, as well as a kinase required for pain sensitization (PKA-II)30 and found it to be expressed in virtually all neurons after day 35 of in vitro culture. In line with the increasing membrane capacitance of hiPSCdN, an increasing soma size was observed by HCS measurements. While activation of GPCRs by 5-HT or cicaprost showed only a tendency of PKA-II response after 35 days of differentiation, forskolin induced a robust pRII response already after 16 days by direct activation of adenylyl cyclase. These data indicate that functionality of the intracellular signaling machinery develops early, while the receptor input level develops gradually with the hiPSCdN maturation. KCl stimulation showed a significant increase of the pRII response from 16 to 35 days, with subsequent plateauing. This suggests that not only GPCRs but also voltage-gated channels are dynamically regulated throughout maturation.
Sensory neurons are differentiated in vivo from neural crest cells at the transition from the first to the second trimester of pregnancy around gestational weeks 10 to 15.57 The opioid receptor signaling system in humans is already established before birth, and maternal opioid use can lead to severe developmental defects, affecting among others somatosensory cortex, hippocampus, or heart.2 Of interest, the opioid receptor agonists fentanyl and nociceptin showed inhibitory activity only after 70 days of neuronal differentiation, ie, the longest time point of in vitro differentiation examined. The effect at this stage is comparable with the inhibitory effect of fentanyl observed in mouse DRG neurons. Application of the KOP agonist dynorphin A did not counteract forskolin-induced PKA-II signaling. Thus, studies addressing the regulatory network of sensitizing and inhibitory GPCRs may require maturation times of up to 70 days. Expression of all 4 opioid peptide receptors was already detected as early as 21 days of differentiation. However, receptor functionality was detected only after 70 days. This contrast between expression and function suggests that the receptors themselves or key proteins of the downstream signaling pathway might be differentially regulated across hiPSCdN maturation or that intracellular transport and integration of receptors into the cell membrane changes over time. Further studies are required to decipher this phenomenon. Such studies might further gain by including single-cell RNA sequencing to extend the single-cell imaging data to the transcriptomic level and to assess heterogeneity and potential subpopulations among the hiPSCdN.
6. Conclusion
In summary, we present a thorough time course analysis of differentiating hiPSCdNs including sensory and nociceptor marker expression, electrophysiological properties as well as proalgesic and analgesic intracellular PKA signaling. While intracellular PKA signaling emerges early, inhibitory opioid receptor signaling was not observed before 70 days of in vitro maturation. A fully functional cell model is a key prerequisite for disease modeling and drug discovery. The onset of inhibitory opioid receptor signaling in hiPSCdNs at advanced stages of in vitro differentiation highlights the importance of careful time course analyses to identify windows suitable for the analysis of respective pathways and their exploitation for drug screening applications and personalized medicine.
Conflict of interest statement
O. Brüstle is a cofounder and shareholder of LIFE & BRAIN GmbH. All other authors have no conflict of interest to declare.
Appendix A. Supplemental digital content
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B776.
Supplemental video content
An audio abstract associated with this article can be found at http://links.lww.com/PAIN/B777.
Acknowledgments
The authors thank Vanessa Frickel for technical support. This research was supported by the European Regional Development Fund (NeuRoWeg) grants EFRE-0800407 and EFRE-0800408 to O. Brüstle and EFRE-0800384 to T. Hucho.
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Web site (www.painjournalonline.com).
P. Röderer and A. Belu contributed equally.
O. Brüstle and T. Hucho contributed equally.
Contributor Information
Pascal Röderer, Email: pascal.roederer@uni-bonn.de.
Andreea Belu, Email: andreea.a.belu@gmail.com.
Luzia Heidrich, Email: lheidrich@lifeandbrain.com.
Maike Siobal, Email: maike.siobal@uk-koeln.de.
Jörg Isensee, Email: joerg.isensee@uk-koeln.de.
Jonathan Prolingheuer, Email: jproling@smail.uni-koeln.de.
Elke Janocha, Email: elke@wjanocha.de.
Markus Valdor, Email: markus.valdor@googlemail.com.
Silke Hagendorf, Email: Silke.Hagendorf@grunenthal.com.
Gregor Bahrenberg, Email: Gregor.Bahrenberg@grunenthal.com.
Thoralf Opitz, Email: topitz@uni-bonn.de.
Michaela Segschneider, Email: michaela.segschneider@uni-bonn.de.
Simone Haupt, Email: simonehaupt@outlook.de.
Anja Nitzsche, Email: anja_nitzsche@yahoo.de.
Oliver Brüstle, Email: oliver.bruestle@uni-bonn.de.
References
- [1].Alshawaf AJ, Viventi S, Qiu W, D'Abaco G, Nayagam B, Erlichster M, Chana G, Everall I, Ivanusic J, Skafidas E, Dottori M. Phenotypic and functional characterization of peripheral sensory neurons derived from human embryonic stem cells. Sci Rep 2018;8:603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Asdjodi S, Rubarth RB, Hardy J, Lee H. The effects of opioids during pregnancy: a literature review. Georg Med Rev 2020;4:16759. [Google Scholar]
- [3].Bhave G, Zhu W, Wang H, Brasier DJ, Oxford GS, Gereau RW. cAMP-dependent protein kinase regulates desensitization of the capsaicin receptor (VR1) by direct phosphorylation. Neuron 2002;35:721–31. [DOI] [PubMed] [Google Scholar]
- [4].Boisvert EM, Engle SJ, Hallowell SE, Liu P, Wang Z-W, Li X-J. The specification and maturation of nociceptive neurons from human embryonic stem cells. Sci Rep 2015;5:16821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cabrera-León A, Cantero-Braojos MÁ, Garcia-Fernandez L, Guerra De Hoyos JA. Living with disabling chronic pain: results from a face-to-face cross-sectional population-based study. BMJ Open 2018;8:e020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cai S, Moutal A, Yu J, Chew LA, Isensee J, Chawla R, Gomez K, Luo S, Zhou Y, Chefdeville A, Madura C, Perez-Miller S, Bellampalli SS, Dorame A, Scott DD, François-Moutal L, Shan Z, Woodward T, Gokhale V, Hohmann AG, Vanderah TW, Patek M, Khanna M, Hucho T, Khanna R. Selective targeting of NaV1.7 via inhibition of the CRMP2-Ubc9 interaction reduces pain in rodents. Sci Transl Med 2021;13:eabh1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cann M, Kuzmenkov A, Isensee J, Andreev‐Andrievskiy A, Peigneur S, Khusainov G, Berkut A, Tytgat J, Vassilevski A, Hucho T. Scorpion toxin MeuNaTxα‐1 sensitizes primary nociceptors by selective modulation of voltage‐gated sodium channels. FEBS J 2021;288:2418–35. [DOI] [PubMed] [Google Scholar]
- [8].Cao L, McDonnell A, Nitzsche A, Alexandrou A, Saintot PP, Loucif AJC, Brown AR, Young G, Mis M, Randall A, Waxman SG, Stanley P, Kirby S, Tarabar S, Gutteridge A, Butt R, McKernan RM, Whiting P, Ali Z, Bilsland J, Stevens EB. Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci Transl Med 2016;8:335ra56. [DOI] [PubMed] [Google Scholar]
- [9].Carbajal AG, Ebersberger A, Thiel A, Ferrari L, Acuna J, Brosig S, Isensee J, Moeller K, Siobal M, Rose-John S, Levine J, Schaible H-G, Hucho T. Oncostatin M induces hyperalgesic priming and amplifies signaling of cAMP to ERK by RapGEF2 and PKA. J Neurochem 2021;157:1821–37. [DOI] [PubMed] [Google Scholar]
- [10].Caviedes P, Ault B, Rapoport SI. Replating improves whole cell voltage clamp recording of human fetal dorsal root ganglion neurons. J Neurosci Methods 1990;35:57–61. [DOI] [PubMed] [Google Scholar]
- [11].Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 2009;27:275–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chambers SM, Qi Y, Mica Y, Lee G, Zhang XJ, Niu L, Bilsland J, Cao L, Stevens E, Whiting P, Shi SH, Studer L. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 2012;30:715–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chang W, Berta T, Kim YH, Lee S, Lee SY, Ji RR. Expression and role of voltage-gated sodium channels in human dorsal root ganglion neurons with special focus on Nav1.7, species differences, and regulation by paclitaxel. Neurosci Bull 2018;34:4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chou R, Turner JA, Devine EB, Hansen RN, Sullivan SD, Blazina I, Dana T, Bougatsos C, Deyo RA. The effectiveness and risks of long-term opioid therapy for chronic pain: a systematic review for a National Institutes of health pathways to prevention workshop. Ann Intern Med 2015;162:276–86. [DOI] [PubMed] [Google Scholar]
- [15].Clark AJ, Kugathasan U, Baskozos G, Priestman DA, Fugger N, Lone MA, Othman A, Chu KH, Blesneac I, Wilson ER, Laurà M, Kalmar B, Greensmith L, Hornemann T, Platt FM, Reilly MM, Bennett DL. An iPSC model of hereditary sensory neuropathy-1 reveals L-serine-responsive deficits in neuronal ganglioside composition and axoglial interactions. Cell Rep Med 2021;2:100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Damann N, Bahrenberg G, Stockhausen H, Habermann CJ, Lesch B, Frank-Foltyn R, Lee J, Ann J, Christoph T. In vitro characterization of the thermoneutral transient receptor potential vanilloid-1 (TRPV1) inhibitor GRTE16523. Eur J Pharmacol 2020;871:172934. [DOI] [PubMed] [Google Scholar]
- [17].Desiderio S, Vermeiren S, Van Campenhout C, Kricha S, Malki E, Richts S, Fletcher EV, Vanwelden T, Schmidt BZ, Henningfeld KA, Pieler T, Woods CG, Nagy V, Verfaillie C, Bellefroid EJ. Prdm12 directs nociceptive sensory neuron development by regulating the expression of the NGF Receptor TrkA. Cell Rep 2019;26:3522–36.e5. [DOI] [PubMed] [Google Scholar]
- [18].Dionisi C, Rai M, Chazalon M, Schiffmann SN, Pandolfo M. Primary proprioceptive neurons from human induced pluripotent stem cells: a cell model for afferent ataxias. Sci Rep 2020;10:7752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Elanzew A, Nießing B, Langendoerfer D, Rippel O, Piotrowski T, Schenk F, Kulik M, Peitz M, Breitkreuz Y, Jung S, Wanek P, Stappert L, Schmitt RH, Haupt S, Zenke M, König N, Brüstle O. The StemCellFactory: a modular system integration for automated generation and expansion of human induced pluripotent stem cells. Front Bioeng Biotechnol 2020;8:580352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].England S, Bevan S, Docherty RJ. PGE2 modulates the tetrodotoxin-resistant sodium current in neonatal rat dorsal root ganglion neurones via the cyclic AMP-protein kinase A cascade. J Physiol 1996;495:429–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ferreira SH, Nakamura M, de Abreu Castro MS. The hyperalgesic effects of prostacyclin and prostaglandin E2. Prostaglandins 1978;16:31–7. [DOI] [PubMed] [Google Scholar]
- [22].Fitzgerald EM, Okuse K, Wood JN, Dolphin AC, Moss SJ. cAMP-dependent phosphorylation of the tetrodotoxin-resistant voltage-dependent sodium channel SNS. J Physiol 1999;516:433–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gangadharan V, Kuner R. Pain hypersensitivity mechanisms at a glance. Dis Model Mech 2013;6:889–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Haberberger RV, Barry C, Dominguez N, Matusica D. Human dorsal root ganglia. Front Cell Neurosci 2019;13:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Han C, Estacion M, Huang J, Vasylyev D, Zhao P, Dib-Hajj SD, Waxman SG. Human Na v 1.8: enhanced persistent and ramp currents contribute to distinct firing properties of human DRG neurons. J Neurophysiol 2015;113:3172–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Häuser W, Bock F, Hüppe M, Nothacker M, Norda H, Radbruch L, Schiltenwolf M, Schuler M, Tölle T, Viniol A, Petzke F, Bär KJ, Baerwald C, Beintker M, Büntzel J, Elling-Audersch C, Freys S, Gnass I, Havemann-Reinecke U, Hupfer K, Kellner U, Lahmann C, Marziniak M, Müller G, Petri H, Rody A, Schäfer M, Schöffel D, Thieme V, Tronnier V, Wolter D, Ziegler D. Recommendations of the second update of the LONTS guidelines: long-term opioid therapy for chronic noncancer pain. Schmerz 2020;34:204–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hucho T, Levine JD. Signaling pathways in sensitization: toward a nociceptor cell biology. Neuron 2007;55:365–76. [DOI] [PubMed] [Google Scholar]
- [28].Hulme AJ, McArthur JR, Maksour S, Miellet S, Ooi L, Adams DJ, Finol-Urdaneta RK, Dottori M. Molecular and functional characterization of Neurogenin-2 induced human sensory neurons. Front Cell Neurosci 2020;14:600895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Isensee J, van Cann M, Despang P, Araldi D, Moeller K, Petersen J, Schmidtko A, Matthes J, Levine JD, Hucho T. Depolarization induces nociceptor sensitization by CaV1.2-mediated PKA-II activation. J Cell Biol 2021;220:e202002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Isensee J, Diskar M, Waldherr S, Buschow R, Hasenauer J, Prinz A, Allgöwer F, Herberg FW, Hucho T. Pain modulators regulate the dynamics of PKA-RII phosphorylation in subgroups of sensory neurons. J Cell Sci 2014;127:216–9. [DOI] [PubMed] [Google Scholar]
- [31].Isensee J, Hucho T. High-content imaging of immunofluorescently labeled TRPV1-positive sensory neurons. Methods Mol Biol 2019;1987:111–24. [DOI] [PubMed] [Google Scholar]
- [32].Isensee J, Kaufholz M, Knape MJ, Hasenauer J, Hammerich H, Gonczarowska-Jorge H, Zahedi RP, Schwede F, Herberg FW, Hucho T. PKA-RII subunit phosphorylation precedes activation by cAMP and regulates activity termination. J Cell Biol 2018;217:2167–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Isensee J, Krahé L, Moeller K, Pereira V, Sexton JE, Sun X, Emery E, Wood JN, Hucho T. Synergistic regulation of serotonin and opioid signaling contribute to pain insensitivity in Nav1.7 knockout mice. Sci Signal 2017;10:eaah4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Isensee J, Schild C, Schwede F, Hucho T. Crosstalk from cAMP to ERK1/2 emerges during postnatal maturation of nociceptive neurons and is maintained during aging. J Cell Sci 2017;130:2134–46. [DOI] [PubMed] [Google Scholar]
- [35].Isensee J, Wenzel C, Buschow R, Weissmann R, Kuss AW, Hucho T. Subgroup-elimination transcriptomics identifies signaling proteins that define subclasses of TRPV1-positive neurons and a novel paracrine circuit. PLoS One 2014;9:e115731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jeske NA, Por ED, Belugin S, Chaudhury S, Berg KA, Akopian AN, Henry MA, Gomez R. A-kinase anchoring protein 150 mediates transient receptor potential family V type 1 sensitivity to phosphatidylinositol-4,5-bisphosphate. J Neurosci 2011;31:8681–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Johannes CB, Le TK, Zhou X, Johnston JA, Dworkin RH. The prevalence of chronic pain in United States adults: results of an internet-based survey. J Pain 2010;11:1230–9. [DOI] [PubMed] [Google Scholar]
- [38].Kennedy J, Roll JM, Schraudner T, Murphy S, McPherson S. Prevalence of persistent pain in the U.S. adult population: new data from the 2010 national health interview survey. J Pain 2014;15:979–84. [DOI] [PubMed] [Google Scholar]
- [39].Kieffer BL, Evans CJ. Opioid receptors: from binding sites to visible molecules in vivo. Neuropharmacology 2009;56:205–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kyu HH, Abate D, Abate KH, Abay SM, Abbafati C, Abbasi N, Abbastabar H, Abd-Allah F, Abdela J, Abdelalim A, Abdollahpour I, Abdulkader RS, Abebe M, Abebe Z, Abil OZ, Aboyans V, Abrham AR, Abu-Raddad LJ, Abu-Rmeileh NME, Accrombessi MMK, Acharya D, Acharya P, Ackerman IN, Adamu AA, Adebayo OM, Adekanmbi V, Ademi Z, Adetokunboh OO, Adib MG, Adsuar JC, Afanvi KA, Afarideh M, Afshin A, Agarwal G, Agesa KM, Aggarwal R, Aghayan SA, Agrawal A, Ahmadi A, Ahmadi M, Ahmadieh H, Ahmed MB, Ahmed S, Aichour AN, Aichour I, Aichour MTE, Akinyemiju T, Akseer N, Al-Aly Z, Al-Eyadhy A, Al-Mekhlafi HM, Al-Raddadi RM, Alahdab F, Alam K, Alam T, Alashi A, Alavian SM, Alene KA, Alijanzadeh M, Alizadeh-Navaei R, Aljunid SM, Alkerwi A, Alla F, Allebeck P, Alonso J, Alsharif U, Altirkawi K, Alvis-Guzman N, Aminde LN, Amini E, Amiresmaili M, Ammar W, Amoako YA, Anber NH, Andrei CL, Androudi S, Animut MD, Anjomshoa M, Ansha MG, Antonio CAT, Anwari P, Arabloo J, Aremu O, Ärnlöv J, Arora A, Arora M, Artaman A, Aryal KK, Asayesh H, Ataro Z, Ausloos M, Avila-Burgos L, Avokpaho EFGA, Awasthi A, Ayala Quintanilla BP, Ayer R, Azzopardi PS, Babazadeh A, Badali H, Balakrishnan K, Bali AG, Banach M, Banoub JAM, Barac A, Barboza MA, Barker-Collo SL, Bärnighausen TW, Barquera S, Barrero LH, Bazargan-Hejazi S, Bedi N, Beghi E, Behzadifar M, Behzadifar M, Bekele BB, Bekru ET, Belachew AB, Belay YA, Bell ML, Bello AK, Bennett DA, Bensenor IM, Berhane A, Bernabe E, Bernstein RS, Beuran M, Beyranvand T, Bhala N, Bhatt S, Bhaumik S, Bhutta ZA, Biadgo B, Biehl MH, Bijani A, Bikbov B, Bilano V, Bililign N, Bin Sayeed MS, Bisanzio D, Bjorge T, Bleyer A, Bobasa EM, Bou-Orm IR, Boufous S, Bourne R, Brady OJ, Brant LC, Brayne C, Brazinova A, Breitborde NJK, Brenner H, Briant PS, Briko AN, Britton G, Brugha T, Buchbinder R, Busse R, Butt ZA, Cahuana-Hurtado L, Campuzano Rincon JC, Cano J, Cárdenas R, Carrero JJ, Carter A, Carvalho F, Castaneda-Orjuela CA, Rivas JC, Castro F, Catalá-López F, Cercy KM, Cerin E, Chaiah Y, Chang JC, Charlson FJ, Chattu VK, Chiang PPC, Chitheer A, Choi JYJ, Christensen H, Christopher DJ, Chung SC, Cicuttini FM, Cirillo M, Collado-Mateo D, Cooper C, Cortesi PA, Cortinovis M, Cousin E, Criqui MH, Cromwell EA, Cross M, Crump JA, Daba AK, Dachew BA, Dadi AF, Dandona L, Dandona R, Dargan PI, Daryani A, Das Gupta R, Das Neves J, Dasa TT, Davitoiu DV, De La Hoz FP, De Leo D, De Neve JW, De Steur H, Degefa MG, Degenhardt L, Deiparine S, Demoz GT, Denova-Gutiérrez E, Deribe K, Dervenis N, Des Jarlais DC, Dey S, Dharmaratne SD, Dhimal M, Dinberu MT, Dirac MA, Djalalinia S, Doan L, Dokova K, Doku DT, Dorsey ER, Doyle KE, Driscoll TR, Dubey M, Dubljanin E, Duken EE, Duncan BB, Duraes AR, Ebrahimi H, Ebrahimpour S, Echko MM, Edessa D, Edvardsson D, Effiong A, Eggen AE, Ehrlich JR, El Bcheraoui C, El-Khatib Z, Elyazar IRF, Enayati A, Endalifer ML, Endries AY, Er B, Erskine HE, Eskandarieh S, Esteghamati A, Esteghamati S, Fakhim H, Faramarzi M, Fareed M, Farhadi F, Farid TA, Sá Farinha CSE, Farioli A, Faro A, Farzadfar F, Fazaeli AA, Feigin VL, Fentahun N, Fereshtehnejad SM, Fernandes E, Fernandes JC, Ferrari AJ, Ferreira ML, Filip I, Fischer F, Fitzmaurice C, Foigt NA, Foreman KJ, Frank TD, Fukumoto T, Fullman N, Fürst T, Furtado JM, Gakidou E, Gall S, Gallus S, Ganji M, Garcia-Basteiro AL, Gardner WM, Gebre AK, Gebremedhin AT, Gebremichael TG, Gelano TF, Geleijnse JM, Genova-Maleras R, Geramo YCD, Gething PW, Gezae KE, Ghadami MR, Ghadiri K, Ghasemi-Kasman M, Ghimire M, Ghoshal AG, Gill PS, Gill TK, Ginawi IA, Giussani G, Gnedovskaya E V., Goldberg EM, Goli S, Gómez-Dantés H, Gona PN, Gopalani SV, Gorman TM, Goulart AC, Goulart BNG, Grada A, Grosso G, Gugnani HC, Guillemin F, Guo Y, Gupta PC, Gupta R, Gupta R, Gupta T, Gutiérrez RA, Gyawali B, Haagsma JA, Hachinski V, Hafezi-Nejad N, Bidgoli HH, Hagos TB, Hailegiyorgis TT, Haj-Mirzaian A, Haj-Mirzaian A, Hamadeh RR, Hamidi S, Handal AJ, Hankey GJ, Hao Y, Harb HL, Harikrishnan S, Haririan H, Haro JM, Hassankhani H, Hassen HY, Havmoeller R, Hay RJ, Hay SI, Hedayatizadeh-Omran A, Heibati B, Hendrie D, Henok A, Heredia-Pi I, Herteliu C, Heydarpour F, Heydarpour P, Hibstu DT, Hoek HW, Hoffman HJ, Hole MK, Rad EH, Hoogar P, Hosgood HD, Hosseini SM, Hosseinzadeh M, Hostiuc M, Hostiuc S, Hotez PJ, Hoy DG, Hsairi M, Htet AS, Huang JJ, Iburg KM, Ikeda CT, Ilesanmi OS, Irvani SSN, Irvine CMS, Islam SMS, Islami F, Jacobsen KH, Jahangiry L, Jahanmehr N, Jain SK, Jakovljevic M, James SL, Jayatilleke AU, Jeemon P, Jha RP, Jha V, Ji JS, Johnson CO, Jonas JB, Jonnagaddala J, Shushtari ZJ, Joshi A, Jozwiak JJ, Jungari SB, Jürisson M, Kabir Z, Kadel R, Kahsay A, Kalani R, Kanchan T, Kar C, Karami M, Karami Matin B, Karch A, Karema C, Karimi N, Karimi SM, Kasaeian A, Kassa DH, Kassa GM, Kassa TD, Kassebaum NJ, Katikireddi SV, Kaul A, Kawakami N, Kazemi Z, Kazemi Karyani A, Keighobadi MM, Keiyoro PN, Kemmer L, Kemp GR, Kengne AP, Keren A, Khader YS, Khafaei B, Khafaie MA, Khajavi A, Khalid N, Khalil IA, Khan EA, Khan MS, Khan MA, Khang YH, Khater MM, Khazaei M, Khoja AT, Khosravi A, Khosravi MH, Kiadaliri AA, Kidanemariam ZT, Kiirithio DN, Kim C-I, Kim D, Kim YE, Kim YJ, Kimokoti RW, Kinfu Y, Kisa A, Kissimova-Skarbek K, Knudsen AKS, Kocarnik JM, Kochhar S, Kokubo Y, Kolola T, Kopec JA, Kosen S, Kotsakis GA, Koul PA, Koyanagi A, Krishan K, Krishnaswami S, Krohn KJ, Defo BK, Bicer BK, Kumar GA, Kumar M, Kuzin I, Lad DP, Lad SD, Lafranconi A, Lalloo R, Lallukka T, Lami FH, Lang JJ, Langan SM, Lansingh VC, Latifi A, Lau KMM, Lazarus J V., Leasher JL, Ledesma JR, Lee PH, Leigh J, Leili M, Leshargie CT, Leung J, Levi M, Lewycka S, Li S, Li Y, Liang X, Liao Y, Liben ML, Lim LL, Lim SS, Limenih MA, Linn S, Liu S, Looker KJ, Lopez AD, Lorkowski S, Lotufo PA, Lozano R, Lucas TCD, Lunevicius R, Lyons RA, Ma S, Macarayan ERK, Mackay MT, Maddison ER, Madotto F, Maghavani DP, Mai HT, Majdan M, Majdzadeh R, Majeed A, Malekzadeh R, Malta DC, Mamun AA, Manda AL, Manguerra H, Mansournia MA, Mantilla Herrera AM, Mantovani LG, Maravilla JC, Marcenes W, Marks A, Martins-Melo FR, Martopullo I, März W, Marzan MB, Massano J, Massenburg BB, Mathur MR, Maulik PK, Mazidi M, McAlinden C, McGrath JJ, McKee M, McMahon BJ, Mehata S, Mehrotra R, Mehta KM, Mehta V, Mejia-Rodriguez F, Mekonen T, Melese A, Melku M, Memiah PTN, Memish ZA, Mendoza W, Mengistu G, Mensah GA, Mereta ST, Meretoja A, Meretoja TJ, Mestrovic T, Miazgowski B, Miazgowski T, Millear AI, Miller TR, Mini GK, Mirarefin M, Mirica A, Mirrakhimov EM, Misganaw AT, Mitchell PB, Mitiku H, Moazen B, Mohajer B, Mohammad KA, Mohammadi M, Mohammadifard N, Mohammadnia-Afrouzi M, Mohammed MA, Mohammed S, Mohebi F, Mokdad AH, Molokhia M, Monasta L, Montanez JC, Moosazadeh M, Moradi G, Moradi M, Moradi-Lakeh M, Moradinazar M, Moraga P, Morawska L, Velásquez IM, Morgado-Da-Costa J, Morrison SD, Moschos MM, Mousavi SM, Mruts KB, Muche AA, Muchie KF, Mueller UO, Muhammed OS, Mukhopadhyay S, Muller K, Mumford JE, Murthy GVS, Musa KI, Mustafa G, Nabhan AF, Nagata C, Nagel G, Naghavi M, Naheed A, Nahvijou A, Naik G, Najafi F, Nam HS, Nangia V, Nansseu JR, Neamati N, Negoi I, Negoi RI, Neupane S, Newton CRJ, Ngunjiri JW, Nguyen AQ, Nguyen G, Nguyen HT, Nguyen HLT, Nguyen HT, Nguyen LH, Nguyen M, Nguyen NB, Nguyen SH, Nichols E, Ningrum DNA, Nixon MR, Nomura S, Noroozi M, Norrving B, Noubiap JJ, Nouri HR, Shiadeh MN, Nowroozi MR, Nsoesie EO, Nyasulu PS, Odell CM, Ofori-Asenso R, Ogbo FA, Oh IH, Oladimeji O, Olagunju AT, Olagunju TO, Olivares PR, Olsen HE, Olusanya BO, Olusanya JO, Ong KL, Ong SK, Oren E, Ortiz A, Ota E, Otstavnov SS, Overland S, Owolabi MO, Mahesh PA, Pacella R, Pakhare AP, Pakpour AH, Pana A, Panda-Jonas S, Park EK, Park J, Parry CDH, Parsian H, Pasdar Y, Patel S, Patil ST, Patle A, Patton GC, Paturi VR, Paudel D, Paulson KR, Pearce N, Pereira A, Pereira DM, Perico N, Pesudovs K, Petzold M, Pham HQ, Phillips MR, Pigott DM, Pillay JD, Piradov MA, Pirsaheb M, Pishgar F, Plana-Ripoll O, Polinder S, Popova S, Postma MJ, Pourshams A, Poustchi H, Prabhakaran D, Prakash S, Prakash V, Prasad N, Purcell CA, Qorbani M, Quistberg DA, Radfar A, Rafay A, Rafiei A, Rahim F, Rahimi K, Rahimi Z, Rahimi-Movaghar A, Rahimi-Movaghar V, Rahman M, Ur Rahman MH, Rahman MA, Rahman SU, Rai RK, Rajati F, Ranjan P, Rao PC, Rasella D, Rawaf DL, Rawaf S, Reddy KS, Reiner RC, Reitsma MB, Remuzzi G, Renzaho AMN, Resnikoff S, Rezaei S, Rezai MS, Ribeiro ALP, Roberts NLS, Robinson SR, Roever L, Ronfani L, Roshandel G, Rostami A, Roth GA, Rothenbacher D, Rubagotti E, Sachdev PS, Sadat N, Sadeghi E, Saeedi Moghaddam S, Safari H, Safari Y, Safari-Faramani R, Safdarian M, Safi S, Safiri S, Sagar R, Sahebkar A, Sahraian MA, Sajadi HS, Salam N, Salama JS, Salamati P, Saleem Z, Salimi Y, Salimzadeh H, Salomon JA, Salvi SS, Salz I, Samy AM, Sanabria J, Sanchez-Nino MD, Santomauro DF, Santos IS, Santos JV, Santric Milicevic MM, Sao Jose BP, Sardana M, Sarker AR, Sarmiento-Suárez R, Sarrafzadegan N, Sartorius B, Sarvi S, Sathian B, Satpathy M, Sawant AR, Sawhney M, Saxena S, Schaeffner E, Schmidt MI, Schneider IJC, Schutte AE, Schwebel DC, Schwendicke F, Scott JG, Sekerija M, Sepanlou SG, Serván-Mori E, Seyedmousavi S, Shabaninejad H, Shafieesabet A, Shahbazi M, Shaheen AA, Shaikh MA, Shams-Beyranvand M, Shamsi M, Sharafi H, Sharafi K, Sharif M, Sharif-Alhoseini M, Sharma J, Sharma R, She J, Sheikh A, Shi P, Shibuya K, Shiferaw MS, Shigematsu M, Shiri R, Shirkoohi R, Shiue I, Shokoohinia Y, Shokraneh F, Shoman H, Shrime MG, Si S, Siabani S, Sibai AM, Siddiqi TJ, Sigfusdottir ID, Sigurvinsdottir R, Silva DAS, Silva JP, Silveira DGA, Singam NSV, Singh JA, Singh NP, Singh V, Sinha DN, Skiadaresi E, Skirbekk V, Sliwa K, Smith DL, Smith M, Filho AMS, Sobaih BH, Sobhani S, Soofi M, Sorensen RJD, Soyiri IN, Sposato LA, Sreeramareddy CT, Srinivasan V, Stanaway JD, Starodubov VI, Stein DJ, Steiner C, Steiner TJ, Stokes MA, Stovner LJ, Subart ML, Sudaryanto A, Sufiyan MB, Sulo G, Sunguya BF, Sur PJ, Sykes BL, Sylaja PN, Sylte DO, Szoeke CEI, Tabarés-Seisdedos R, Tabuchi T, Tadakamadla SK, Tandon N, Tassew SG, Tavakkoli M, Taveira N, Taylor HR, Tehrani-Banihashemi A, Tekalign TG, Tekelemedhin SW, Tekle MG, Temsah MH, Temsah O, Terkawi AS, Tessema B, Teweldemedhin M, Thankappan KR, Theis A, Thirunavukkarasu S, Thomas N, Tilahun B, To QG, Tonelli M, Topor-Madry R, Torre AE, Tortajada-Girbés M, Touvier M, Tovani-Palone MR, Towbin JA, Tran BX, Tran KB, Troeger CE, Tsadik AG, Tsoi D, Tudor Car L, Tyrovolas S, Ukwaja KN, Ullah I, Undurraga EA, Updike RL, Usman MS, Uthman OA, Vaduganathan M, Vaezi A, Valdez PR, Varavikova E, Varughese S, Vasankari TJ, Venketasubramanian N, Villafaina S, Violante FS, Vladimirov SK, Vlassov V, Vollset SE, Vos T, Vosoughi K, Vujcic IS, Wagnew FS, Waheed Y, Wang Y, Wang YP, Weiderpass E, Weintraub RG, Weiss DJ, Weldegebreal F, Weldegwergs KG, Werdecker A, West TE, Westerman R, Whiteford HA, Widecka J, Wijeratne T, Williams HC, Wilner LB, Wilson S, Winkler AS, Wiyeh AB, Wiysonge CS, Wolfe CDA, Woolf AD, Wyper GMA, Xavier D, Xu G, Yadgir S, Yahyazadeh Jabbari SH, Yamada T, Yan LL, Yano Y, Yaseri M, Yasin YJ, Yeshaneh A, Yimer EM, Yip P, Yisma E, Yonemoto N, Yoon SJ, Yotebieng M, Younis MZ, Yousefifard M, Yu C, Zadnik V, Zaidi Z, Zaman S Bin, Zamani M, Zandian H, Zar HJ, Zenebe ZM, Zipkin B, Zhou M, Zodpey S, Zucker I, Zuhlke LJ, Murray CJL. Global, regional, and national disability-adjusted life-years (DALYs) for 359 diseases and injuries and healthy life expectancy (HALE) for 195 countries and territories, 1990-2017: a systematic analysis for the global burden of disease study 2017. Lancet 2018;392:1859–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lampert A, Bennett DL, McDermott LA, Neureiter A, Eberhardt E, Winner B, Zenke M. Human sensory neurons derived from pluripotent stem cells for disease modelling and personalized medicine. Neurobiol Pain 2020;8:100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lee G, Chambers SM, Tomishima MJ, Studer L. Derivation of neural crest cells from human pluripotent stem cells. Nat Protoc 2010;5:688–701. [DOI] [PubMed] [Google Scholar]
- [43].Loos C, Moeller K, Fröhlich F, Hucho T, Hasenauer J. A hierarchical, data-driven approach to modeling single-cell populations predicts latent causes of cell-to-cell variability. Cell Syst 2018;6:593–603.e13. [DOI] [PubMed] [Google Scholar]
- [44].Machelska H, Celik MÖ. Advances in achieving opioid analgesia without side effects. Front Pharmacol 2018;9:1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].McDermott LA, Weir GA, Themistocleous AC, Segerdahl AR, Blesneac I, Baskozos G, Clark AJ, Millar V, Peck LJ, Ebner D, Tracey I, Serra J, Bennett DL. Defining the functional role of NaV1.7 in human nociception. Neuron 2019;101:905–19.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].McDonald J, Lambert DG. Opioid receptors. BJA Educ 2015;15:219–24. [Google Scholar]
- [47].Meents JE, Bressan E, Sontag S, Foerster A, Hautvast P, Rösseler C, Hampl M, Schüler H, Goetzke R, Le TKC, Kleggetveit IP, Le Cann K, Kerth C, Rush AM, Rogers M, Kohl Z, Schmelz M, Wagner W, Jørum E, Namer B, Winner B, Zenke M, Lampert A. The role of Nav1.7 in human nociceptors: insights from human induced pluripotent stem cell–derived sensory neurons of erythromelalgia patients. PAIN 2019;160:1327–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Middleton SJ, Barry AM, Comini M, Li Y, Ray PR, Shiers S, Themistocleous AC, Uhelski ML, Yang X, Dougherty PM, Price TJ, Bennett DL. Studying human nociceptors: from fundamentals to clinic. Brain 2021;144:1312–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mis MA, Yang Y, Tanaka BS, Gomis-Perez C, Liu S, Dib-Hajj F, Adi T, Garcia-Milian R, Schulman BR, Dib-Hajj SD, Waxman SG. Resilience to pain: a peripheral component identified using induced pluripotent stem cells and dynamic clamp. J Neurosci 2019;39:382–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Mogil JS. Animal models of pain: progress and challenges. Nat Rev Neurosci 2009;10:283–94. [DOI] [PubMed] [Google Scholar]
- [51].Münst S, Koch P, Kesavan J, Alexander-Mays M, Münst B, Blaess S, Brüstle O. In vitro segregation and isolation of human pluripotent stem cell-derived neural crest cells. Methods 2018;133:65–80. [DOI] [PubMed] [Google Scholar]
- [52].Nahin RL. Estimates of pain prevalence and severity in adults: United States, 2012. J Pain 2015;16:769–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Namer B, Schmidt D, Eberhardt E, Maroni M, Dorfmeister E, Kleggetveit IP, Kaluza L, Meents J, Gerlach A, Lin Z, Winterpacht A, Dragicevic E, Kohl Z, Schüttler J, Kurth I, Warncke T, Jorum E, Winner B, Lampert A. Pain relief in a neuropathy patient by lacosamide: proof of principle of clinical translation from patient-specific iPS cell-derived nociceptors. EBioMedicine 2019;39:401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nguyen MQ, von Buchholtz LJ, Reker AN, Ryba NJ, Davidson S. Single-nucleus transcriptomic analysis of human dorsal root ganglion neurons. Elife 2021;10:e71752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Nickolls AR, Lee MM, Espinoza DF, Szczot M, Lam RM, Wang Q, Beers J, Zou J, Nguyen MQ, Solinski HJ, AlJanahi AA, Johnson KR, Ward ME, Chesler AT, Bönnemann CG. Transcriptional programming of human mechanosensory neuron subtypes from pluripotent stem cells. Cell Rep 2020;30:932–46.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Nieminen K, Suarez-Isla BA, Rapoport SI. Electrical properties of cultured dorsal root ganglion neurons from normal and trisomy 21 human fetal tissue. Brain Res 1988;474:246–54. [DOI] [PubMed] [Google Scholar]
- [57].Quinn RK, Drury HR, Lim R, Callister RJ, Tadros MA. Differentiation of sensory neuron lineage during the late first and early second trimesters of human foetal development. Neuroscience 2021;467:28–38. [DOI] [PubMed] [Google Scholar]
- [58].Raehal KM, Schmid CL, Groer CE, Bohn LM. Functional selectivity at the μ-opioid receptor: implications for understanding opioid analgesia and tolerance. Pharmacol Rev 2011;63:1001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Rathee PK, Distler C, Obreja O, Neuhuber W, Wang GK, Wang SY, Nau C, Kress M. PKA/AKAP/VR-1 module: a common link of Gs-mediated signaling to thermal hyperalgesia. J Neurosci 2002;22:4740–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Rice ASC, Smith BH, Blyth FM. Pain and the global burden of disease. PAIN 2016;157:791–6. [DOI] [PubMed] [Google Scholar]
- [61].Roederer M. Compensation in flow cytometry. Curr Protoc Cytometry 2002;24:14.1-1.14.20. [DOI] [PubMed] [Google Scholar]
- [62].Rostock C, Schrenk-Siemens K, Pohle J, Siemens J. Human vs. mouse nociceptors – similarities and differences. Neuroscience 2018;387:13–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saito-Diaz K, Street JR, Ulrichs H, Zeltner N. Derivation of peripheral nociceptive, mechanoreceptive, and proprioceptive sensory neurons from the same culture of human pluripotent stem cells. Stem Cell Rep 2021;16:446–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Schinke C, Fernandez Vallone V, Ivanov A, Peng Y, Körtvelyessy P, Nolte L, Huehnchen P, Beule D, Stachelscheid H, Boehmerle W, Endres M. Modeling chemotherapy induced neurotoxicity with human induced pluripotent stem cell (iPSC)-derived sensory neurons. Neurobiol Dis 2021;155:105391. [DOI] [PubMed] [Google Scholar]
- [65].Schnizler K, Shutov LP, Van Kanegan MJ, Merrill MA, Nichols B, McKnight GS, Strack S, Hell JW, Usachev YM. Protein kinase A anchoring via AKAP150 is essential for TRPV1 modulation by forskolin and prostaglandin E2 in mouse sensory neurons. J Neurosci 2008;28:4904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Schrenk-Siemens K, Wende H, Prato V, Song K, Rostock C, Loewer A, Utikal J, Lewin GR, Lechner SG, Siemens J. PIEZO2 is required for mechanotransduction in human stem cell–derived touch receptors. Nat Neurosci 2015;18:10–6. [DOI] [PubMed] [Google Scholar]
- [67].Shiers S, Klein RM, Price TJ. Quantitative differences in neuronal subpopulations between mouse and human dorsal root ganglia demonstrated with RNAscope in situ hybridization. PAIN 2020;161:2410–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Shiers SI, Sankaranarayanan I, Jeevakumar V, Cervantes A, Reese JC, Price TJ. Convergence of peptidergic and non-peptidergic protein markers in the human dorsal root ganglion and spinal dorsal horn. J Comp Neurol 2021;529:2771–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Stacey P, Wassermann AM, Kammonen L, Impey E, Wilbrey A, Cawkill D. Plate-based phenotypic screening for pain using human iPSC-derived sensory neurons. SLAS Discov 2018;23:585–96. [DOI] [PubMed] [Google Scholar]
- [70].Stein C. Opioid receptors. Annu Rev Med 2016;67:433–51. [DOI] [PubMed] [Google Scholar]
- [71].Stein C, Lang LJ. Peripheral mechanisms of opioid analgesia. Curr Opin Pharmacol 2009;9:3–8. [DOI] [PubMed] [Google Scholar]
- [72].Strang J, Volkow ND, Degenhardt L, Hickman M, Johnson K, Koob GF, Marshall BDL, Tyndall M, Walsh SL. Opioid use disorder. Nat Rev Dis Primers 2020;6:3–28. [DOI] [PubMed] [Google Scholar]
- [73].Survey 2017 pain alliance Europe: Pain Alliance Europe, 2017. Available at: https://pae-eu.eu/survey-2017-pain-alliance-europe/. Accessed December 17, 2020. [Google Scholar]
- [74].Team RDC. R: a language and environment for statistical computing. Vienna, Austria, 2011. [Google Scholar]
- [75].Thier MC, Hommerding O, Panten J, Pinna R, García-González D, Berger T, Wörsdörfer P, Assenov Y, Scognamiglio R, Przybylla A, Kaschutnig P, Becker L, Milsom MD, Jauch A, Utikal J, Herrmann C, Monyer H, Edenhofer F, Trumpp A. Identification of embryonic neural plate border stem cells and their generation by direct reprogramming from adult human blood cells. Cell Stem Cell 2019;24:166–82.e13. [DOI] [PubMed] [Google Scholar]
- [76].Tsang A, Von Korff M, Lee S, Alonso J, Karam E, Angermeyer MC, Borges GLG, Bromet EJ, de Girolamo G, de Graaf R, Gureje O, Lepine JP, Haro JM, Levinson D, Oakley Browne MA, Posada-Villa J, Seedat S, Watanabe M. Common chronic pain conditions in developed and developing countries: gender and age differences and comorbidity with depression-anxiety disorders. J Pain 2008;9:883–91. [DOI] [PubMed] [Google Scholar]
- [77].Woolf CJ, Ma Q. Nociceptors-noxious stimulus detectors. Neuron 2007;55:353–64. [DOI] [PubMed] [Google Scholar]
- [78].Yang NJ, Isensee J, Neel DV, Quadros AU, Zhang H-XB, Lauzadis J, Liu SM, Shiers S, Belu A, Palan S, Marlin S, Maignel J, Kennedy-Curran A, Tong VS, Moayeri M, Röderer P, Nitzsche A, Lu M, Pentelute BL, Brüstle O, Tripathi V, Foster KA, Price TJ, Collier RJ, Leppla SH, Puopolo M, Bean BP, Cunha TM, Hucho T, Chiu IM. Anthrax toxins regulate pain signaling and can deliver molecular cargoes into ANTXR2+ DRG sensory neurons. Nat Neurosci 2022;25:168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Young GT, Gutteridge A, Fox HD, Stevens EB, Wilbrey AL, Cao L, Cho LT, Brown AR, Benn CL, Friedman JH, Kammonen LR, Bictash M, Whiting P, Bilsland JG. Characterizing human stem cell-derived sensory neurons at the single-cell level reveals their ion channel expression and utility in pain research. Mol Ther 2014;22:1530–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zajacova A, Grol-Prokopczyk H, Zimmer Z. Pain trends among american adults, 2002–2018: patterns, disparities, and correlates. Demography 2021;58:711–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zeltner N, Fattahi F, Dubois NC, Saurat N, Lafaille F, Shang L, Zimmer B, Tchieu J, Soliman MA, Lee G, Casanova JL, Studer L. Capturing the biology of disease severity in a PSC-based model of familial dysautonomia. Nat Med 2016;22:1421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang X, Li L, McNaughton PA. Proinflammatory mediators modulate the heat-activated ion channel TRPV1 via the Scaffolding protein AKAP79/150. Neuron 2008;59:450–61. [DOI] [PubMed] [Google Scholar]
- [83].Zhang X, Priest BT, Belfer I, Gold MS. Voltage-gated Na + currents in human dorsal root ganglion neurons. Elife 2017;6:e23235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental digital content associated with this article can be found online at http://links.lww.com/PAIN/B776.
An audio abstract associated with this article can be found at http://links.lww.com/PAIN/B777.