Abstract
Rift Valley fever virus (RVFV) is an emerging arbovirus found in Africa. While RVFV is pantropic and infects many cells and tissues, viral replication and necrosis within the liver play a critical role in mediating severe disease. The low-density lipoprotein receptor–related protein 1 (Lrp1) is a recently identified host factor for cellular entry and infection by RVFV. The biological significance of Lrp1, including its role in hepatic disease in vivo, however, remains to be determined. Because Lrp1 has a high expression level in hepatocytes, we developed a mouse model in which Lrp1 is specifically deleted in hepatocytes to test how the absence of liver Lrp1 expression affects RVF pathogenesis. Mice lacking Lrp1 expression in hepatocytes showed minimal RVFV replication in the liver, longer time to death, and altered clinical signs toward neurological disease. In contrast, RVFV infection levels in other tissues showed no difference between the two genotypes. Therefore, Lrp1 is essential for RVF hepatic disease in mice.
Lrp1 is essential for Rift Valley fever disease.
INTRODUCTION
The zoonotic arbovirus Rift Valley fever virus (RVFV) causes a serious infectious disease burden on human and animal health in Africa. Humans become infected with RVFV through mosquito bite or by direct contact with infected livestock (1, 2). The associated disease, RVF, is a spectrum of clinical illnesses that often manifests mildly in humans but can progress to more severe outcomes including hemorrhagic fever, panencephalitis, or ocular disease, illustrating the broad tropism for host cell types (3, 4). Severe hepatic manifestations occur during human outbreaks of RVF (3, 5–7). Hepatitis with potential hemorrhaging can appear as early as 2 to 3 days following the onset of symptoms (4, 8). The case fatality rate (CFR) in humans with acute RVF hepatitis approaches 50% compared to the overall CFR of <1% (9). There are currently no approved efficacious drugs for use as prophylactics or therapeutics for RVF in humans. Thus, current treatment is limited to supportive care. Contributing factors leading to severe RVF hepatic disease remains unknown, further limiting the development of safe and efficacious treatment options.
We recently identified low-density lipoprotein receptor (LDLR)–related protein 1 (Lrp1) as an important host protein for RVFV infection (10). We demonstrated an interaction between RVFV glycoprotein Gn and Lrp1, validated the importance of Lrp1 in RVFV infection in vitro, and performed an in vivo proof-of-concept experiment by blocking Lrp1 with a protein ligand, receptor associated protein (RAP). However, the role of Lrp1 in hepatic disease as well as tropism and dissemination of the virus in vivo under relevant exposure routes remain major unanswered questions.
Following subcutaneous exposure to RVFV by mosquito bite, the virus likely infects antigen-presenting cells and travels to lymphoid tissues, where subsequent viremia initiates infection of visceral organs, including the liver and spleen (11–13). Most inbred mouse strains succumb to hepatic disease within 3 to 5 days following footpad infection, and in these cases, infectious virus is found throughout many tissues including the liver, spleen, kidney, heart, and lung, with highest replication in the liver (12, 14–16). A small number of mice may survive the initial hepatic replication and then succumb to neurological disease with high levels of virus in the brain (12, 15).
Germline deletion of Lrp1 is embryonic lethal in mice; therefore, a complete Lrp1 knockout (KO) mouse is not viable, and alternative approaches are warranted (17). Given the vital role of the liver in RVF disease in both preclinical animal models and humans and because Lrp1 is in the LDLR family with known functions mediating cholesterol homeostasis (18–20), we sought to determine the impact of Lrp1 expression in hepatocytes on RVF disease. To this end, we generated a hepatocyte-specific Lrp1 KO model in the C57BL/6 murine background using well-established Cre-Lox technology. Mice with floxed Lrp1 alleles (Lrp1f/f) were crossed with mice expressing Cre recombinase enzyme under the albumin promoter (Alb-cre), resulting in mice that are deleted for Lrp1 only in albumin-expressing hepatocytes. After infection of these mice with RVFV, our data show that Lrp1 KO in hepatocytes led to a near-complete reduction of viral replication in the liver. In addition, we observed a doubling in the survival time of the animals. However, all Lrp1 hepatocyte-specific KO mice succumbed to RVFV infection and displayed neurological signs along with high levels of RVFV in the brain. Together, our data reveal that loss of Lrp1 in hepatocytes prevents mice from developing RVF hepatic disease, which highlights the critical role that Lrp1 protein plays in RVFV infection in liver disease.
RESULTS
Cre recombinase reduces Lrp1 expression, resulting in significant reduction in RVFV infection
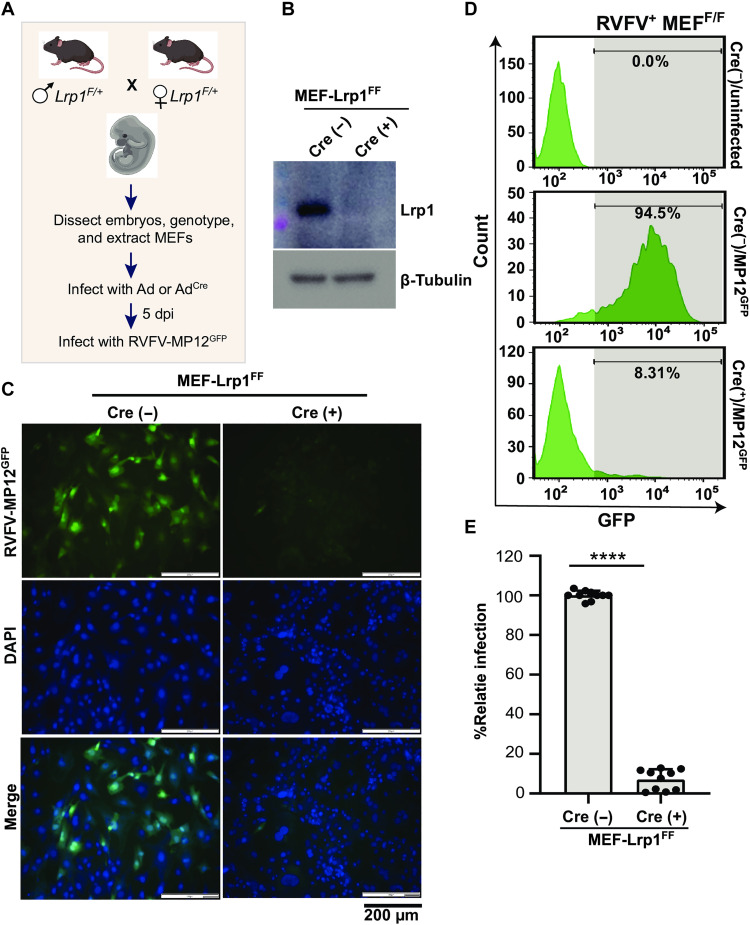
Because germline KO of Lrp1 in mice is embryonic lethal, we used Cre-Lox mouse model technology to generate mice with KO of Lrp1 only in hepatocytes. Initially, we used a CRISPR-cas9 approach to develop Lrp1-floxed mice in a C57BL/6 genetic background (herein Lrp1f/f). Initially, Lrp1f/f mice with germline loxP-flanked Lrp1 alleles were mated to produce embryos from which mouse embryonic fibroblasts (MEFs) were extracted (Fig. 1A). To validate the methodology and confirm the activity of the floxed alleles, Lrp1f/f MEFs were infected with either an adenovirus expressing the cre recombinase enzyme (Adcre) or an empty adenovirus vector (Ad) as control. Treatment of MEFs with Adcre eliminated Lrp1 expression, whereas the control Ad did not (Fig. 1B). Subsequently, cre+ MEFs were infected with the attenuated RVFV MP12 strain [multiplicity of infection (MOI) = 2], which expresses the green fluorescent protein (GFP) reporter protein in place of the nonstructural protein from the S segment (NSs) (MP12GFP). Using both fluorescence microscopy and flow cytometry, cre+ MEFs were mostly negative for GFP by both detection methods, indicating that RVFV MP12GFP infection was reduced to near baseline levels in the absence of Lrp1. The cre− control cells still expressed Lrp1, and >90% were infected by 16 hours postinfection as measured by GFP (Fig. 1, C to E). Thus, cre recombinase effectively excised Lrp1 in MEFs from Lrp1f/f mice, resulting in reduced infection by RVFV.
Fig. 1. Lrp1-depleted MEFs are resistant to RVFV infection.
(A) Schematics showing the generation of mouse embryonic fibroblasts (MEFs) from embryonic day (E) 14.5 embryos obtained by timed mating of Lrp1f/+ mice. dpi, days postinfection. (B) Western blot of Lrp1f/f MEFs infected with adenovirus expressing an empty adenovirus vector (Ad) and the cre recombinase enzyme (Adcre). The blot was probed for lipoprotein receptor–related protein 1 (Lrp1) and β-tubulin. Lrp1-sufficient and Lrp1-deficient MEFs were infected with Rift Valley fever virus (RVFV) MP12GFP at a multiplicity of infection (MOI) of 2 for 16 hours. (C) Fluorescence images show green fluorescent protein (GFP), DAPI (4′,6-diamidino-2-phenylindole), and merged panels. Images were taken at ×20 magnification (scale bars, 200 μm). (D and E) Representative flow cytometry histograms and corresponding analysis of flow cytometry histogram data from MEF infections. Experiment was performed five times in duplicates. Student’s t test was used to determine significance; ****P < 0.0001.
Elimination of Lrp1 in hepatocytes significantly delays death from RVFV infection in vivo
Following validation of reduced RVFV infection in cre+ MEFs from Lrp1f/f mice, we generated hepatocyte-specific Lrp1 KO mice. Albumin is expressed by hepatocytes, and thus using an albumin promoter to express cre recombinase will restrict cre activity to only albumin-expressing cells. Therefore, we bred Lrp1f/f mice with those hemizygous for cre under the albumin promoter (Alb-Cre), thereby generating Lrp1f/fAlb-Cre animals and their littermate Lrp1f/f controls that do not express cre (fig. S1A). Lrp1f/fAlb-Cre mice had nearly undetectable levels of Lrp1 in the liver compared to Lrp1f/f control mice by Western blot and immunofluorescence microscopy (fig. S1, B and C), while Lrp1 expression in the brain, heart, lung, spleen, small intestines, and lymph nodes remained intact (fig. S1, E and F). Thus Lrp1 KO in this model is specific to liver tissue, and the cre recombinase is not leaky. Deletion of Lrp1 in hepatocytes did not affect the expression of other members of the LDLR family such as LDLR and Lrp2 (fig. S1D). In addition, elimination of Lrp1 in hepatocytes had only a small effect on overall liver function as suggested in the literature (20). Of note, cholesterol and alanine aminotransferase are mildly altered compared to Lrp1f/f control mice (fig. S2). Other liver-related blood chemistries including alkaline phosphatase, albumin, bilirubin, and blood urea nitrogen were within the normal range (fig. S2).
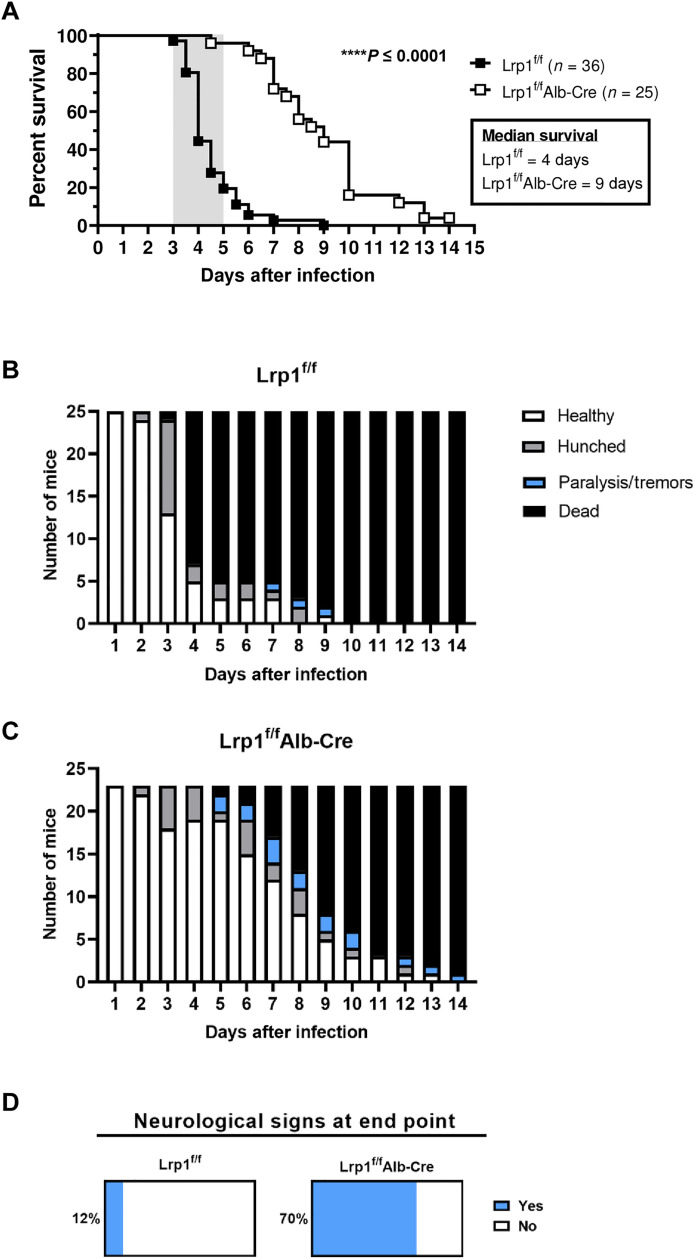
To assess the effect of hepatic Lrp1 deficiency on RVFV infection, we used a well-characterized footpad infection model of RVF, which is commonly used to mimic mosquito bite (21–24). Mixed sex mice of both genotypes (4 to 12 weeks old; n = 25 and 36 for Lrp1f/fAlb-Cre and Lrp1f/f controls, respectively) were inoculated by footpad with 20 plaque-forming units (PFUs) of RVFV ZH501, which corresponds to at least 20 times the 50% lethal dose (LD50)(15). This dose was chosen because it is reproducibly lethal when administered by footpad injection. Most Lrp1f/f control mice succumbed within 3 to 5 days postinfection (dpi) (average survival time: 4 dpi) with <10% surviving beyond 5 dpi (Fig. 2A). The typical clinical window for lethal RVF in C57BL/6 mice is 3 to 5 dpi (shaded area in Fig. 2A) (15). In contrast, 96% of Lrp1f/fAlb-Cre mice survived past the hepatic disease clinical window and succumbed to infection later, resulting in an average survival of 9 dpi (Fig. 2A). This extension in survival was accompanied by differences in clinical signs of disease. Lrp1f/f control mice experienced acute signs of disease such as hunching (Fig. 2B). The Lrp1f/fAlb-Cre mice survived the acute phase and demonstrated signs of neurological disease such as tremors and paralysis (Fig. 2C). Overall, 70% of Lrp1f/fAlb-Cre mice with Lrp1 KO in hepatocytes experienced neurological signs at the time of euthanasia, compared to only 12% of Lrp1f/f control mice (Fig. 2D). Thus, elimination of Lrp1 from hepatocytes extended mouse survival time from a mean of 4 to 9 dpi, and extended survival was associated with increased frequency of neurological signs.
Fig. 2. Elimination of Lrp1 in hepatocytes significantly delays time to death after RVFV infection.
(A) Lrp1f/f and Lrp1f/fAlb-Cre mice were infected with 20 plaque-forming units (PFUs) of RVFV ZH501 by footpad and monitored for survival (n = 61 animals, data were derived from three experiments). Clinical signs of (B) Lrp1f/f mice and (C) Lrp1f/fAlb-Cre mice highlight the frequency of neurological symptoms (note that mice found deceased are not included in this scoring). (D) Percentages of mice across three studies that experienced neurological signs of disease including tremors and paralysis (light blue) on the day of euthanasia in Lrp1f/f and Lrp1f/fAlb-Cre mice. Mice that were found deceased could not be clinically scored and were not included in analysis in (D). Kaplan-Meier survival analysis was completed to assess for significant differences between genotypes. ****P < 0.0001.
Given the stark differences in survival between the genotypes after footpad infection, we infected a cohort of mice intranasally with 20 PFUs of RVFV. Infection through the intranasal route is thought to facilitate direct brain infection (25, 26). Resulting data revealed that there were no differences in survival between Lrpf/f and Lrp1f/fAlb-Cre mice (P = 0.92) following intranasal infection (fig. S4), indicating that brain infection after intranasal inoculation was not affected by lack of Lrp1 in hepatocytes. Both genotypes succumbed to disease with neurological signs with a median survival of 12 days for Lrp1f/f mice and 11 days for Lrp1f/fAlb-Cre mice (fig. S3).
Elimination of Lrp1 in hepatocytes reduces viral burden and inflammation in the liver
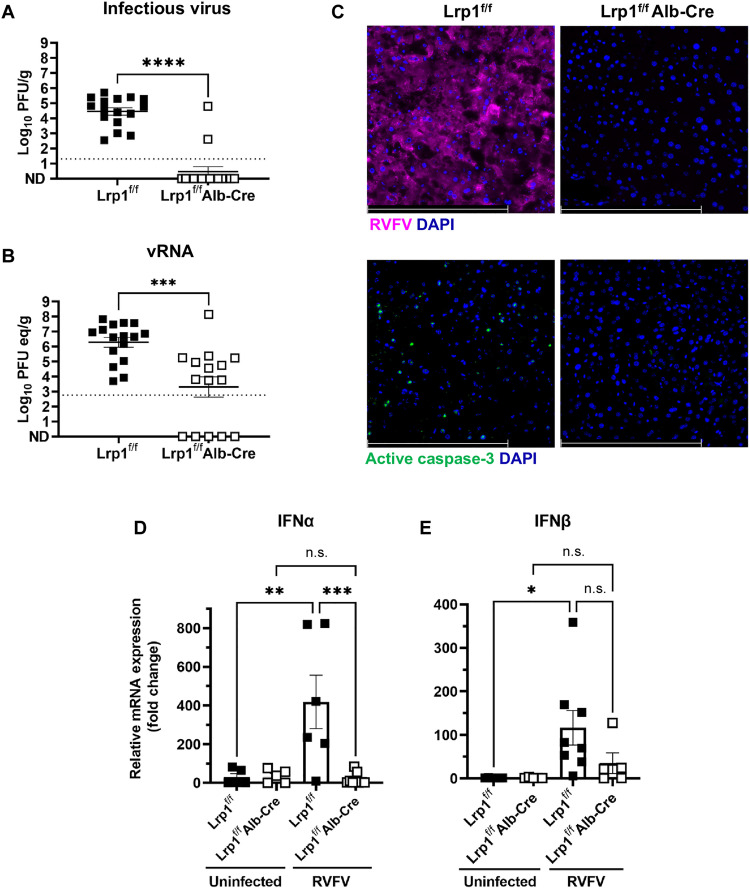
To directly compare tissue viral burden at matched time points, a subset of RVFV-infected mice of both genotypes (n = 16 Lrp1f/fAlb-Cre mice and n = 15 Lrp1f/f controls) were infected via footpad similarly to Fig. 2 and then euthanized at 3 dpi to assess levels of infectious virus and viral RNA (vRNA). The 3-dpi time point was chosen, because most of the control animals would still be alive, and we can directly compare tissue titers between the genotypes. Control animals had average infectious virus levels of 1 × 105 PFU/g liver tissue at 3 dpi, while infectious virus was undetectable in the liver from all but two Lrp1f/fAlb-Cre mice (Fig. 3A). When comparing levels of vRNA by quantitative reverse transcription polymerase chain reaction (qRT-PCR), there was 2.5-log reduction in liver vRNA in Lrp1f/fAlb-Cre mice compared to controls (Fig. 3B). The qRT-PCR assay is more sensitive than a plaque assay, and it can detect noninfectious vRNA. Our data are congruent with the finding that vRNA levels above 104 or 105 PFU/g equivalent are associated with detection of live virus by plaque assay, whereas vRNA levels below that are generally not associated with detectable infectious titers. Fluorescence microscopy with a polyclonal anti-RVFV nucleoprotein (N) antibody revealed extensive staining in Lrp1f/f liver sections and undetectable levels in Lrp1f/fAlb-Cre animals at 3 dpi (Fig. 3C, top panels). Using active caspase-3 as a marker for cellular damage, we found numerous active caspase-3+ cells in the control mice compared to the Lrp1f/fAlb-Cre livers at 3 dpi (Fig. 3C, bottom panels).
Fig. 3. Lrp1 KO in hepatocytes leads to a significant reduction in viral burden in the liver at 3 dpi.
(A) Lrp1f/fAlb-Cre mice (n = 16) have significantly less (A) infectious virus and (B) viral RNA (vRNA) in the liver at 3 dpi than Lrp1f/f controls (n = 15). (C) RVFV nucleoprotein (N) (magenta) and active caspase-3 (green) staining in the liver at 3 dpi in Lrp1f/f and Lrp1f/fAlb-Cre livers (40×). (D) Interferon α (IFNα) and (E) IFNβ relative mRNA expression in the liver of uninfected and RVFV-infected Lrp1f/f and Lrp1f/fAlb-Cre mice at 3 dpi. Bars represent SEM. Statistics were determined by an unpaired t test or a one-way analysis of variance (ANOVA). *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. n.s., not significant. Scale bars, 250 μm. Dotted lines indicate the limit of detection.
Given the reduction in viral replication in the liver of Lrp1f/fAlb-Cre mice, we next assessed levels of vRNA in the serum and other tissues known to support RVFV replication in vivo. In both the serum and brain, vRNA was reduced 10-fold in Lrp1f/fAlb-Cre mice compared to controls (fig. S4A). In contrast, at 3 dpi, there were no significant differences in viral burden between genotypes in the heart, spleen, lung, small intestine, reproductive tissues, cervical lymph nodes, adrenal gland, or leg muscle, suggesting that acute viral infection of these tissues is unaffected by reduced liver infection (fig. S4B).
Expression of type 1 interferons (IFNs) was also assessed in liver tissue at 3 dpi using qRT-PCR, and there were no differences in baseline mRNA expression of either IFN-α or IFN-β in uninfected mice of either genotype (Fig. 3, D and E). However, RVFV-infected Lrp1f/f mice had significantly higher levels of both IFNs compared to uninfected Lrp1f/f mice (Fig. 3, D and E). Expression of IFNs in the liver of infected Lrp1f/fAlb-Cre mice was similar to the baseline levels of uninfected animals.
Correlation in the liver-brain titers over time is associated with prolonged survival and neurological disease
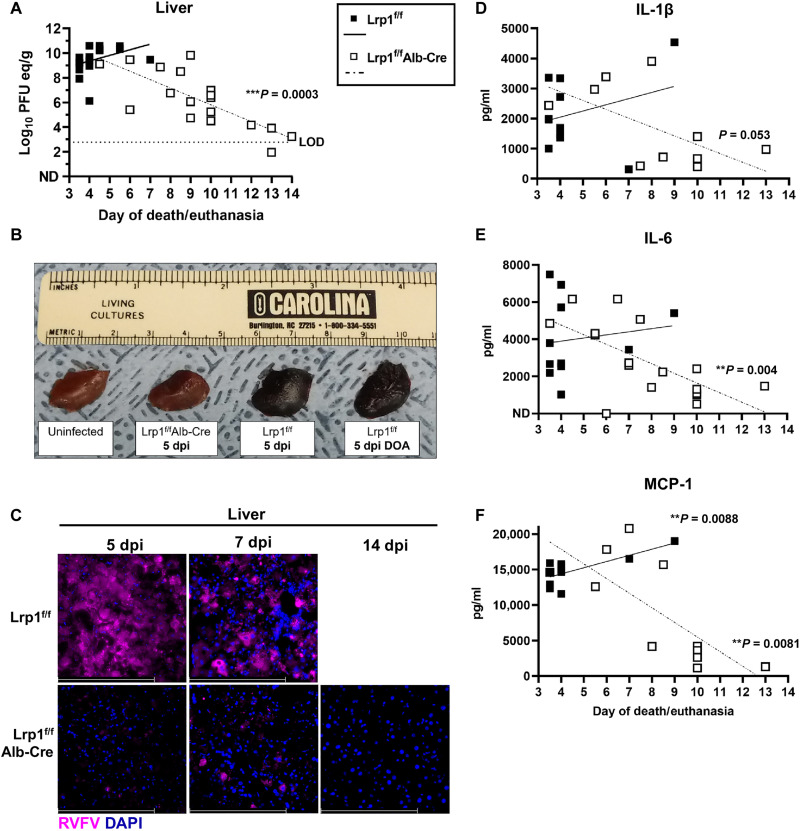
In mice that were infected with 20 PFUs by footpad injection and monitored for survival (Fig. 2A), tissues were collected when the mice met the Institutional Animal Care and Use Committee (IACUC)–approved end point criteria. When measuring vRNA levels by qRT-PCR compared to the day of euthanasia, we found a significant negative correlation (***P = 0.0003) between vRNA titers in the liver over time (Fig. 4A). In the liver, Lrp1f/f control mice succumbed between 3 and 5 dpi and had high liver titers, while Lrp1f/fAlb-Cre mice had lower liver titers the longer they survived the RVFV challenge, with the lowest viral titers at 14 dpi (Fig. 4A). Gross liver pathology showed dark and discolored tissue from Lrp1f/f control mice, while Lrp1f/fAlb-Cre and uninfected control livers displayed normal appearance (Fig. 4B). Fluorescence microscopy revealed abundant RVFV N staining in liver sections from Lrp1f/f control mice compared to limited antigen staining in Lrp1f/fAlb-Cre mice (Fig. 4C).
Fig. 4. Lrp1 KO in hepatocytes reduces RVFV in the liver at time of euthanasia.
(A) vRNA titers in the liver on the day of euthanasia show a significant negative correlation in Lrp1f/fAlb-Cre mice from 3 to 14 dpi. LOD, limit of assay detection. (B) Gross pathology of liver tissue obtained from the indicated genotypes at 5 dpi. DOA, deceased on arrival. (C) RVFV N antigen staining in liver tissue of Lrp1f/f mice compared to Lrp1f/fAlb-Cre mice at 5 and 7 dpi (40×). (D) Interleukin-1β (IL-1β), (E) IL-6, and (F) MCP-1 protein levels in the liver tissue homogenate on the day of euthanasia from 3 to 14 dpi. Scale bars, 250 μm. A linear regression was used to determine whether the slope was significantly nonzero. **P < 0.01; ***P < 0.001.
The inflammatory factors interleukin-1β (IL-1β), IL-6, and monocyte chemoattractang protein (MCP)-1 have been shown in previous studies to be highly expressed in tissues following the RVFV ZH501 infection in mice (16). High levels of these cytokines were found in the liver of mice that died early of hepatic disease, indicating elevated levels of tissue inflammation along with elevated viral replication. All 3 inflammatory factors were negatively correlated with day of death/euthanasia in the livers of Lrp1f/fAlb-Cre mice, indicating that the mice that died later of neurological disease had limited liver inflammation and viral titers (Fig. 4, D to F).
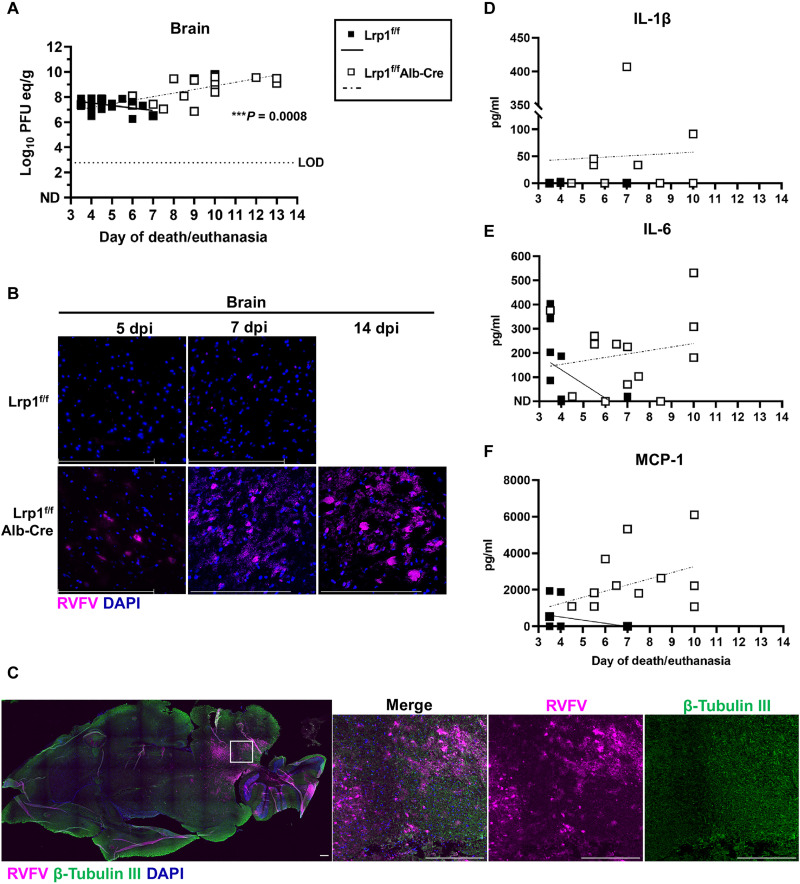
In contrast to the liver, there was a significant positive correlation (***P = 0.0008) over time in brain vRNA titers on the day of euthanasia. The longer the Lrp1f/fAlb-Cre mice survived the RVFV challenge, the higher viral titers were in the brain tissue, with correspondingly lower liver titers (Fig. 5A). The progression of viral replication in the brain was visualized by fluorescence microscopy, as RVFV N staining increased over time in the brain of Lrp1f/fAlb-Cre mice, compared to less viral N protein staining in the brains of Lrp1f/f mice at 7 dpi, as few control mice survived this long (Fig. 5B). We found direct RVFV infection of β-tubulin III+ neurons in the brains of Lrp1f/fAlb-Cre mice at 14 dpi, which likely contributed to the development of the observed neurological disease (Fig. 5C) (27, 28). In contrast to the liver, IL-1β, IL-6, and MCP-1 protein levels were positively correlated in the brains of these same animals, corresponding with the increased viral burden over time (Fig. 5, D to F).
Fig. 5. Mice with Lrp1 KO in hepatocytes succumb to RVF neurological disease.
(A) vRNA titers in the brain at euthanasia show a significant positive correlation in Lrp1f/fAlb-Cre mice from 3 to 14 dpi. (B) RVFV staining in the brain tissue of Lrp1f/fAlb-Cre mice compared to Lrp1f/f mice from 5, 7, and 14 dpi (40×). (C) Brain tissue from a Lrp1f/fAlb-cre mice at 12 dpi stained for RVFV N (magenta), β-tubulin III (green), and DAPI (blue; 10×). (D) IL-1β, (E) IL-6, and (F) MCP-1 protein in the brain tissue at euthanasia from 3 to 14 dpi. Scale bars, 250 μm. A linear regression was used to determine whether the slope was significantly nonzero. **P < 0.01.
DISCUSSION
RVFV is a zoonotic RNA virus with a broad tropism for mosquitoes and a range of mammalian species (1, 29–33). Within a host organism, its expansive tropism is evidenced by the presence of infectious virus in most tissues, including the liver, spleen, heart, kidney, brain, and reproductive organs (12, 15, 34). The cellular mechanisms by which RVFV infects many cells and tissue types are unknown. In most laboratory animal species and livestock, the liver is a major target organ, and the high lethality of RVFV is generally caused by widespread tissue damage in the liver due to direct viral cytopathology (12, 35, 36). Following peripheral infection, most mice succumb to hepatic RVF disease between 3 and 5 dpi. While RVFV can be found in the brain and many other tissues at this time, the most pronounced viral burden resides in the liver, which precipitates death of the animal due to organ dysfunction (15).
The discovery of Lrp1 as a host factor for RVFV binding and entry provides a foundation to understand tissue tropism and disease mechanisms in vivo. Lrp1 is a highly conserved and membrane-associated protein that can mediate efficient cellular infection by RVFV across many cell types and species (10). Our previous in vitro work demonstrated a direct interaction between the RVFV glycoprotein Gn and two extracellular domains of Lrp1. Blocking Lrp1 with a high-affinity ligand RAP, monoclonal antibodies, or recombinant Gn all reduced infection of cells from different tissue types and animal species. Biochemical approaches demonstrated a glycosylation-independent interaction between the RVFV glycoprotein Gn and two extracellular cluster domains of Lrp1. A proof-of-concept in vivo experiment demonstrated that mice coadministered intracranially with RVFV and the Lrp1 ligand RAP had significantly enhanced survival than mice that received virus alone. However, we were unable to more extensively study the role of Lrp1 in vivo, because universal Lrp1 deletion is embryonic lethal in mice, and thus germline Lrp1 KO mice are not feasible (17).
To address this and to also provide clarity into the role that Lrp1 plays specifically in hepatic disease in vivo, we generated a mouse model with Lrp1 deleted only in hepatocytes. We used the cre LoxP mouse system to generate hepatocyte-specific KO of Lrp1. We validated the specificity of hepatocyte Lrp1 deletion, along with confirmation that neither expression of other LDLRs nor Lrp1 in other relevant tissues (brain, spleen, heart, lung, intestine, and lymph nodes) were affected. Therefore, this model allows examination of infection, dissemination, and disease progression in mice in the absence of Lrp1 expression in the liver, providing a unique model in which to answer RVFV pathogenesis questions.
Using footpad injection, which remains the standard relevant exposure route, we found that mice with Lrp1 KO in hepatocytes had a remarkably extended survival time and succumbed instead to neurological disease while experiencing the classic signs of RVF encephalitis including tremors and paralysis. We show that the elimination of Lrp1 in this critical cell type shifted the disease course following peripheral RVFV infection, demonstrating its importance for hepatic RVF. Thus, expression of Lrp1 in hepatocytes is necessary for liver infection and early lethal disease after footpad infection with RVFV.
The mechanism underlying extended mouse survival is likely due to nearly complete reduction in viral infection of the liver in the absence of Lrp1. While some low levels of vRNA were found in the liver, our data support a near-complete elimination of infectious virus, with no viral antigen detected in liver samples at 3 dpi. Unexpectedly, elimination of replication in the liver did not prevent overall dissemination of the virus throughout the rest of the animal. Both serum and brain levels of virus were reduced by 10-fold at 3 dpi in liver KO mice, but titers in the heart, lung, spleen, reproductive tissues, cervical lymph node, adrenal gland, small intestine, and leg muscle were unaffected. These data suggest that prevention of liver infection ultimately does not affect viral seeding to the rest of the mouse, which challenges previous assumptions that high virus replication in the liver fuels dissemination to other organs. We show that, in normal mice with intact Lrp1, liver replication proves ultimately fatal for the mouse, but that, in mice deleted for Lrp1 in hepatocytes, the virus is still able to spread and replicate in other tissues in the absence of high liver viral burden. Further studies with the hepatocyte Lrp1 KO mouse model can provide additional specific mechanistic insight into the route of viral dissemination from footpad infection.
Lrp1 is a member of the LDL receptor family, and there is some redundancy in their functions as lipid receptors within the liver (37). Despite this, we demonstrate that Lrp1 is indispensable for RVF hepatic disease, for without it, the virus can no longer efficiently replicate in liver tissue. This also means that other LDLR family members expressed in the liver are unable to compensate for lack of Lrp1, given that LDLR and Lrp2 expression in Lrp1 KO mice is unaffected. Last, our findings further underscore the fact that viral infection of the spleen and other visceral organs is not dependent upon hepatic replication, suggesting a role for additional cell and tissue types in viral dissemination. In summary, using a hepatic Lrp1-deficient mouse model, Lrp1 has a significant biological role as a fundamental cellular factor mediating infection in vivo and influencing the course of RVF disease. Given our previous results showing that human cellular Lrp1 also mediates RVFV infection, insights from the mouse model could provide the basis for further understanding of the pathogenesis of RVFV in humans.
MATERIALS AND METHODS
Biosafety
Work with RVFV ZH501 was completed in biosafety level (BSL)-3 and animal biosafety level (ABSL)-3 conditions in the Division of Select Agents and Toxins–registered space within the Regional Biocontainment Laboratory in the Center for Vaccine Research at the University of Pittsburgh. All RVFV tissue samples were inactivated using approved inactivation protocols before removal from BSL-3/ABSL-3 for further processing. Respiratory protection for laboratory workers was provided by 3M Versaflo Powered Air Purifying Respirators, and all excess infectious material was inactivated by submergence in Vesphene IIse (Steris) diluted to 1:128 for at least 10 min.
Virus
RVFV ZH501 was generated from reverse genetics provided by S. Nichol (Centers for Disease Control and Prevention, Atlanta, GA). For mouse infections, virus was diluted in Dulbecco’s modified Eagle’s medium (DMEM) with 2% FBS (fetal bovine serum) media. Virus was propagated in Vero E6 cells and titered using a standard viral plaque assay (34).
Generation of floxed mice and hepatocyte-specific Lrp1 deletion
The Lrp1 floxed mice were generated at the Genome Engineering & Stem Cell Center and the Transgenic, Knockout and Micro-Injection Core, both at Washington University in St. Louis School of Medicine. The Lrp1-floxed mice were generated by electroporating two CRISPR ribonucleoproteins and two single-stranded oligodeoxynucleotides (ssODNs) as described (38). Briefly, introns 14 and 15 were targeted by guide RNAs (gRNAs), g1: 5′- ggagtccccctagttcttcttgg and g2: 5′-ggagtccccctagttcttcttgg, respectively. The protospacer adjacent motif site for each gRNA is underlined. To insert the loxP sequence, ssODN for g1 had the following sequence: 5′-g*t*aggtaatctgtgggaagagggacctggacacaatcttcaagagaaagagagccaagaGGATCCATAACTTCGTATAGCATACATTATACGAAGTTATagaactagggggactcccaggaagccaaggcttagggagcagaggcctatgatgttct*g*g. The ssODN for g2 had the following sequence: 5′- g*a*gttaagaatagttcctttcttactcgctgtctcgttcctcattcacttgccccatGGATCCATAACTTCGTATAGCATACATTATACGAAGTTATcttcgtgccctcagaactaccccacccccaatcctcctttagccccagagccttctc*t*g. Each asterisk (*) stands for a phosphorothioate bond. Before the transfection of zygotes, single gRNAs (sgRNAs) and ssODNs were first validated in neuro-2a cells for the efficiency of the LoxP integration (>50% efficiency). sgRNAs and ssODNs along with purified Cas9 as ribonucleoprotein complexes were transfected into zygotes, which were then cultured to the blastocyst stage. Of the 548 embryo transfers, 70 pups were born alive, and three pups had LoxP integration at both sites. The LoxP sites were identified in the pups via next-generation sequencing at both integration sites. Mouse genotyping for Lrp1-floxed sites was performed at Transnetyx Inc. (Cordova, TN).
Lrp1f/f mice were bred with B6-Alb_cre+ hemizygous mice and purchased from the Jackson Laboratory [B6N.Cg-Speer6-ps1Tg(Alb-cre)21Mgn/J, stock: 018961]. For genotyping of cre+ and cre− mice, tissues from the toes, tail, and/or embryos were lysed in DirectPCR Lysis Reagent (Viagen, 101-T) for 30 min at 55°C in presence of proteinase K (0.5 mg/ml). After heat inactivation of the enzyme at 85°C for 5 min, 1 μl of samples were used for PCR for the presence or absence of Alb-Cre using primers; wild-type forward: TGC AAA CAT CAC ATG CAC AC, common reverse: TTG GCC CCT TAC CAT AAC TG, and mutant forward: GAA GCA GAA GCT TAG GAA GAT GG.
Preparation of primary MEFs
Lrp1F/+ mice were intercrossed and time-mated. At gestational day embryonic day (E) 14.5, embryos were harvested and genotyped by PCR using genomic DNA extracted by digesting a limb with proteinase K (0.5 mg/ml) in DirectPCR Lysis Reagent (Viagen, 101-T) for 30 min at 55°C. The genotyped embryos were then minced into small pieces after removing the head and visceral organs and subjected to digestion with 0.25% trypsin/0.02% EDTA (MilliporeSigma, T4049) for 25 min at 37°C. Digested embryos were resuspended and cultured in DMEM supplemented with 10% FBS. After 5 days of culture, adherent cells were harvested as MEFs and cryo-stocked for subsequent use.
Mouse experiments
All experiments complied with the Animal Welfare Act guidelines and were completed with approval from the University of Pittsburgh IACUC (protocol no. 22102000). Mice were housed in individually ventilated cages and had access to food and water ad libitum. Mixed sex mice 4 to 12 weeks old were anesthetized using isoflurane and infected with 20 PFUs of RVFV ZH501 in 20 μl of volume in the right footpad using a 27 ½-gauge needle and a Hamilton syringe. Inoculum was back-titered after infection to confirm the dose. Following the infection, mice fully recovered from anesthesia and were monitored for clinical signs daily up to 14 dpi. A subset of mice was euthanized at 3 dpi to directly compare viral load in the liver, brain, and serum. Blood was collected by cardiac stick to obtain serum. Brain and liver tissue were collected at the timed euthanasia, and a portion of the tissue was saved for virological analysis or Western blotting. A separate portion of each tissue was fixed in 4% paraformaldehyde for 24 hours at 4°C before being processed for histology. Brain and liver tissues were collected and processed similarly from mice that succumbed to infection from 3.5 to 14 dpi. Mice were euthanized using cervical dislocation following isoflurane anesthesia at planned time points or when presenting clinical signs of disease such as weight loss, hunching, ataxia, tremors, or paralysis. Tissues for virological analysis were homogenized and processed by viral plaque assay (VPA) (34) or qRT-PCR (39) as previously described. For blood chemistry, mice were bled retro-orbitally while under anesthesia before RVFV infection or at 3 dpi. A total of 100 μl of blood was run on a VETSCAN VS2 Chemistry Analyzer as per manufacturer’s instructions.
Western blot
Liver or brain tissue collected from mice were homogenized in DMEM with 2% FBS, 2 mM l-glutamine, penicillin (100 U/ml), and 100 μg of streptomycin (D2) media as previously described (39). A total of 100 μl of the homogenate was inactivated in 900 μl of radioimmunoprecipitation assay buffer (Thermo Fisher Scientific, 89901) with 1% Halt Protease Inhibitor (Thermo Fisher Scientific, 78429) for 10 min at room temperature. Samples were removed from biocontainment and centrifuged at 13,500 relative centrifugal force for 20 min. Cellular debris was removed, and a bicinchoninic acid (BCA) assay was completed following the manufacturer’s instructions (Thermo Fisher Scientific, Pierce BCA Protein Assay, 23227). Twenty micrograms of protein from each sample was loaded into a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen, NP0323BOX) and run for 35 min at 165 V. The protein was wet-transferred onto a nitrocellulose membrane (Thermo Fisher Scientific, LC2001) for 1 hour at 10 V. Membranes were blocked for 1 hour at room temperature rocking in 5% nonfat powdered milk diluted in 1× phosphate-buffered saline (PBS) with 0.1% Tween 20 (PBS-T). Following the block, membranes were incubated overnight at 4°C rocking with either rabbit anti-Lrp1 (1:1000; Cell Signaling Technology, cat. no. 64099), rabbit anti-Lrp2 (1:1000; Thermo Fisher Scientific, PA5-67900), mouse anti–β-tubulin (1:1000; MilliporeSigma, T8328), or rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH; 1:1000; Thermo Fisher Scientific, PA1-987) diluted in 5% nonfat milk in PBS-T. The following day, the membranes were washed by rocking in 10 ml of PBS-T three times for 5 min each. Membranes were probed for 1 hour at room temperature rocking with goat anti-rabbit horseradish peroxidase (1:15,000; Thermo Fisher Scientific, 31460) diluted in 5% nonfat milk in PBS-T. The membranes were washed by rocking in 10 ml of PBS-T three times for 5 min each. Each membrane was incubated with the Dura substrate (Thermo Fisher Scientific, 37071) (800 μl of total volume) for 3 min at room temperature. Excess substrate was removed, and membranes were wrapped in Syran wrap before being exposed to film. Film was developed using a Merry X-Ray film processor and scanned using a printer.
Immunofluorescence microscopy
Brain and liver tissues were fixed in 4% paraformaldehyde for at least 24 hours, removed from containment, and washed with 1× PBS. Cassettes were submerged in increasing concentrations of sucrose diluted in dH2O (10, 20, and 40%) for 24 hours each. The tissues were flash-frozen embedded in optimal cutting temperature (OCT) compound and sliced on a cryostat at a thickness of 7 μm. OCT was washed from the slides using 1× PBS with 0.5% bovine serum albumin (PBB) three times. Sections were permeabilized for 15 min at room temperature in 1% Triton X-100. The Triton X-100 was removed from the slides, and they were washed three times with PBB. Slides were blocked using 5% normal goat serum diluted in PBB for 1 hour at room temperature. After three washes with PBB, the slides were incubated with rabbit anti-RVFV N (1:200) (15) and chicken anti–β-tubulin III (1:300; MilliporeSigma, AB9354) diluted in PBB for 1 hour at room temperature. Slides were washed three times with PBB and then incubated with goat anti-rabbit Cy3 (1:1000; Jackson ImmunoResearch, 111-165-144) or goat anti-chicken Alexa Fluor 488 (1:1000; Jackson Immuno, 103-545-155) for 1 hour at room temperature. Slides were washed with 1× PBS three times and incubated with Hoechst stain for 30 s. The Hoechst stain was washed off with 1× PBS, and slides were mounted with coverslips using gelvatol. Slides dried overnight before imaging on a Leica DMi8 wide-field fluorescent microscope at the Center for Vaccine Research. Images were processed using ImageJ. Large images were acquired on a Nikon A1 confocal microscope at the Center for Biologic Imaging at the University of Pittsburgh using a 10× objective with 15% overlap and stitch via blending in NIS-Elements software.
Relative quantitation of type I IFNs
cDNA was made from RNA isolated from RVFV-infected or uninfected liver homogenate of Lrp1f/f and Lrp1f/fAlb-Cre tissue at 3 dpi as previously described (40). Using TaqMan Multiplex Master Mix (Applied Biosystems), TaqMan Gene Expression Assay kits were used for IFNα and IFNβ (IFNα: Thermo Fisher Scientific, 4331182 and IFNβ: Thermo Fisher Scientific, 4331182), and data were normalized to GAPDH (Thermo Fisher Scientific, 4448489) and from uninfected liver tissue of both genotypes. A QuantStudio 6 (Thermo Fisher Scientific) was used for amplification using the following parameters: 20-s hold step at 95°C, followed by a 1-s 95°C amplification step, and a 20-s 60° hold step repeated 40×.
ELISAs
IL-6, IL-1β, and MCP-1 enzyme-linked immunosorbent assays (ELISAs) were performed on homogenized liver or brain samples diluted 1:10 in D2 media. The assay was run according to the manufacturer’s instructions (Ancillary Kit: R&D Systems, DY008B and cytokines: R&D Systems, D6050, MLB00C, and MJE00B, respectively).
Statistics
Statistical analyses were performed using GraphPad Prism v9.
Acknowledgments
We acknowledge S. Barrick for animal study coordination. We thank members of the Hartman, Amarasinghe, and Leung groups for their support at various stages of this study. We also thank the Genome Engineering and iPSC Center GEiC at the Washington University in St. Louis for cell line engineering services as well as the members of the Transgenic, Knockout and Micro-Injection Core in the Department of Pathology & Immunology at the Washington University in St. Louis.
Funding: National Institutes of Health grants R01 AI161765 (G.K.A. and A.L.H.), R01 AI169850 (G.K.A. and A.L.H.), R56 AI171920 (G.K.A. and A.L.H.), R21 AI163603 (G.K.A. and A.L.H.), and R01 AI130152 (T.E.).
Author contributions: Conceptualization, methodology, investigation, and visualization: All authors. Supervision: D.W.L., T.E., G.K.A., and A.L.H. Funding acquisition: G.K.A. and A.L.H. Writing (original draft): M.M.S. and A.L.H. Writing (review and editing): M.M.S., S.S.G., D.W.L., T.E., G.K.A., and A.L.H.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
REFERENCES AND NOTES
- 1.M. Kwaśnik, W. Rożek, J. Rola, Rift valley fever–A growing threat to humans and animals. J. Vet. Res. 65, 7–14 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.R. Métras, W. J. Edmunds, C. Youssouffi, L. Dommergues, G. Fournié, A. Camacho, S. Funk, E. Cardinale, G. le Godais, S. Combo, L. Filleul, H. Youssouf, M. Subiros, Estimation of rift valley fever virus spillover to humans during the Mayotte 2018-2019 epidemic. Proc. Natl. Acad. Sci. U.S.A. 117, 24567–24574 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.L. W. Laughlin, J. M. Meegan, L. J. Strausbaugh, D. M. Morens, R. H. Watten, Epidemic rift valley fever in Egypt: Observations of the spectrum of human illness. Trans. R. Soc. Trop. Med. Hyg. 73, 630–633 (1979). [DOI] [PubMed] [Google Scholar]
- 4.T. Ikegami, S. Makino, The pathogenesis of rift valley fever. Viruses 3, 493–519 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Al-Hazmi, E. A. Ayoola, M. Abdurahman, S. Banzal, J. Ashraf, A. El-Bushra, A. Hazmi, M. Abdullah, H. Abbo, A. Elamin, E.-T. Al-Sammani, M. Gadour, C. Menon, M. Hamza, I. Rahim, M. Hafez, M. Jambavalikar, H. Arishi, A. Aqeel, Epidemic rift valley fever in Saudi Arabia: A clinical study of severe illness in humans. Clin. Infect. Dis. 36, 245–252 (2003). [DOI] [PubMed] [Google Scholar]
- 6.T. A. Madani, Y. Y. Al-Mazrou, M. H. Al-Jeffri, A. A. Mishkhas, A. M. Al-Rabeah, A. M. Turkistani, M. O. Al-Sayed, A. A. Abodahish, A. S. Khan, T. G. Ksiazek, O. Shobokshi, Rift Valley fever epidemic in Saudi Arabia: Epidemiological, clinical, and laboratory characteristics. Clin. Infect. Dis. 37, 1084–1092 (2003). [DOI] [PubMed] [Google Scholar]
- 7.D. J. van Velden, J. D. Meyer, J. Olivier, J. H. Gear, B. McIntosh, Rift Valley fever affecting humans in South Africa: A clinicopathological study. S. Afr. Med. J. 51, 867–871 (1977). [PubMed] [Google Scholar]
- 8.E. Javelle, A. Lesueur, V. Pommier de Santi, F. de Laval, T. Lefebvre, G. Holweck, G. A. Durand, I. Leparc-Goffart, G. Texier, F. Simon, The challenging management of Rift Valley fever in humans: Literature review of the clinical disease and algorithm proposal. Ann. Clin. Microbiol. Antimicrob. 19, 4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.D. Wright, J. Kortekaas, T. A. Bowden, G. M. Warimwe, Rift Valley fever: Biology and epidemiology. J. Gen. Virol. 100, 1187–1199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. S. Ganaie, M. M. Schwarz, C. M. McMillen, D. A. Price, A. X. Feng, J. R. Albe, W. Wang, S. Miersch, A. Orvedahl, A. R. Cole, M. F. Sentmanat, N. Mishra, D. A. Boyles, Z. T. Koenig, M. R. Kujawa, M. A. Demers, R. M. Hoehl, A. B. Moyle, N. D. Wagner, S. H. Stubbs, L. Cardarelli, J. Teyra, A. McElroy, M. L. Gross, S. P. J. Whelan, J. Doench, X. Cui, T. J. Brett, S. S. Sidhu, H. W. Virgin, T. Egawa, D. W. Leung, G. K. Amarasinghe, A. L. Hartman, Lrp1 is a host entry factor for Rift Valley fever virus. Cell 184, 5163–5178.e24 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.P.-Y. Lozach, A. Kühbacher, R. Meier, R. Mancini, D. Bitto, M. Bouloy, A. Helenius, DC-SIGN as a receptor for phleboviruses. Cell Host Microbe 10, 75–88 (2011). [DOI] [PubMed] [Google Scholar]
- 12.D. R. Smith, K. E. Steele, J. Shamblin, A. Honko, J. Johnson, C. Reed, M. Kennedy, J. L. Chapman, L. E. Hensley, The pathogenesis of Rift Valley fever virus in the mouse model. Virology 407, 256–267 (2010). [DOI] [PubMed] [Google Scholar]
- 13.K. Terasaki, S. Makino, Interplay between the virus and host in Rift Valley fever pathogenesis. J. Innate Immun. 7, 450–458 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. M. Bales, D. S. Powell, L. M. Bethel, D. S. Reed, A. L. Hartman, Choice of inbred rat strain impacts lethality and disease course after respiratory infection with Rift Valley fever virus. Front. Cell. Infect. Microbiol. 2, 105 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H. N. Cartwright, D. J. Barbeau, A. K. McElroy, Rift Valley fever virus is lethal in different inbred mouse strains independent of sex. Front. Microbiol. 11, 1962 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. K. Gray, M. N. Worthy, T. L. Juelich, S. L. Agar, A. Poussard, D. Ragland, A. N. Freiberg, M. R. Holbrook, Chemotactic and inflammatory responses in the liver and brain are associated with pathogenesis of Rift Valley fever virus infection in the mouse. PLOS Negl. Trop. Dis. 6, e1529 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.J. Herz, D. E. Clouthier, R. E. Hammer, LDL receptor-related protein internalizes and degrades uPA-PAI-1 complexes and is essential for embryo implantation. Cell 71, 411–421 (1992). [DOI] [PubMed] [Google Scholar]
- 18.J. Terrand, V. Bruban, L. Zhou, W. Gong, Z. el Asmar, P. May, K. Zurhove, P. Haffner, C. Philippe, E. Woldt, R. L. Matz, C. Gracia, D. Metzger, J. Auwerx, J. Herz, P. Boucher, LRP1 controls intracellular cholesterol storage and fatty acid synthesis through modulation of Wnt signaling. J. Biol. Chem. 284, 381–388 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.A. Laatsch, M. Merkel, P. J. Talmud, T. Grewal, U. Beisiegel, J. Heeren, Insulin stimulates hepatic low density lipoprotein receptor-related protein 1 (LRP1) to increase postprandial lipoprotein clearance. Atherosclerosis 204, 105–111 (2009). [DOI] [PubMed] [Google Scholar]
- 20.A. N. Hamlin, S. Chinnarasu, Y. Ding, X. Xian, J. Herz, A. Jaeschke, D. Y. Hui, Low-density lipoprotein receptor-related protein-1 dysfunction synergizes with dietary cholesterol to accelerate steatohepatitis progression. J. Biol. Chem. 293, 9674–9684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.H. M. Lazear, J. Govero, A. M. Smith, D. J. Platt, E. Fernandez, J. J. Miner, M. S. Diamond, A mouse model of zika virus pathogenesis. Cell Host Microbe 19, 720–730 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. L. Rossi, R. B. Tesh, S. R. Azar, A. E. Muruato, K. A. Hanley, A. J. Auguste, R. M. Langsjoen, S. Paessler, N. Vasilakis, S. C. Weaver, Characterization of a novel murine model to study zika virus. Am. J. Trop. Med. Hyg. 94, 1362–1369 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.I. Martin-Martin, P. C. Valenzuela Leon, L. Amo, G. Shrivastava, E. Iniguez, A. Aryan, S. Brooks, B. B. Kojin, A. E. Williams, S. Bolland, H. Ackerman, Z. N. Adelman, E. Calvo, Aedes aegypti sialokinin facilitates mosquito blood feeding and modulates host immunity and vascular biology. Cell Rep. 39, 110648 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.H. N. Cartwright, D. J. Barbeau, J. D. Doyle, E. Klein, M. T. Heise, M. T. Ferris, A. K. McElroy, Genetic diversity of collaborative cross mice enables identification of novel rift valley fever virus encephalitis model. PLOS Pathog. 18, e1010649 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Lacote, C. Tamietti, M. Chabert, M. P. Confort, L. Conquet, C. Pulido, N. Aurine, C. Baquerre, A. Thiesson, B. Pain, M. de Las Heras, M. Flamand, X. Montagutelli, P. Marianneau, M. Ratinier, F. Arnaud, Intranasal exposure to Rift Valley fever virus live-attenuated strains leads to high mortality rate in immunocompetent mice. Viruses 14, 2470 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.N. R. Hum, F. A. Bourguet, A. Sebastian, D. Lam, A. M. Phillips, K. R. Sanchez, A. Rasley, G. G. Loots, D. R. Weilhammer, MAVS mediates a protective immune response in the brain to Rift Valley fever virus. PLOS Pathog. 18, e1010231 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.M. Ludlow, J. Kortekaas, C. Herden, B. Hoffmann, D. Tappe, C. Trebst, D. E. Griffin, H. E. Brindle, T. Solomon, A. S. Brown, D. van Riel, K. C. Wolthers, D. Pajkrt, P. Wohlsein, B. E. E. Martina, W. Baumgärtner, G. M. Verjans, A. D. M. E. Osterhaus, Neurotropic virus infections as the cause of immediate and delayed neuropathology. Acta Neuropathol. 131, 159–184 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.J. R. Albe, D. A. Boyles, A. W. Walters, M. R. Kujawa, C. M. McMillen, D. S. Reed, A. L. Hartman, Neutrophil and macrophage influx into the central nervous system are inflammatory components of lethal Rift Valley fever encephalitis in rats. PLOS Pathog. 15, e1007833 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Rissmann, N. Kley, R. Ulrich, F. Stoek, A. Balkema-Buschmann, M. Eiden, M. H. Groschup, Competency of amphibians and reptiles and their potential role as reservoir hosts for Rift Valley fever virus. Viruses 12, 1206 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.W. C. Wilson, I. J. Kim, J. D. Trujillo, S. Y. Sunwoo, L. E. Noronha, K. Urbaniak, D. S. McVey, B. S. Drolet, I. Morozov, B. Faburay, E. E. Schirtzinger, T. Koopman, S. V. Indran, V. Balaraman, J. A. Richt, Susceptibility of white-tailed deer to Rift Valley fever virus. Emerg. Infect. Dis. 24, 1717–1719 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D. Gora, T. Yaya, T. Jocelyn, F. Didier, D. Maoulouth, S. Amadou, T. D. Ruel, J. P. Gonzalez, The potential role of rodents in the enzootic cycle of Rift Valley fever virus in Senegal. Microbes Infect. 2, 343–346 (2000). [DOI] [PubMed] [Google Scholar]
- 32.F. G. Davies, L. Karstad, Experimental infection of the African buffalo with the virus of Rift Valley fever. Tropl. Anim. Health Prod. 13, 185–188 (1981). [DOI] [PubMed] [Google Scholar]
- 33.A. Evans, F. Gakuya, J. T. Paweska, M. Rostal, L. Akoolo, P. J. Van Vuren, T. Manyibe, J. M. Macharia, T. G. Ksiazek, D. R. Feikin, R. F. Breiman, M. K. Njenga, Prevalence of antibodies against Rift Valley fever virus in Kenyan wildlife. Epidemiol. Infect. 136, 1261–1269 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.A. L. Caroline, M. R. Kujawa, T. D. Oury, D. S. Reed, A. L. Hartman, Inflammatory biomarkers associated with lethal Rift Valley fever encephalitis in the lewis rat model. Front. Microbiol. 6, 1509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.L. Odendaal, S. J. Clift, G. T. Fosgate, A. S. Davis, Lesions and cellular tropism of natural Rift Valley fever virus infection in adult sheep. Vet. Pathol. 56, 61–77 (2019). [DOI] [PubMed] [Google Scholar]
- 36.L. Odendaal, A. S. Davis, G. T. Fosgate, S. J. Clift, Lesions and cellular tropism of natural Rift Valley fever virus infection in young lambs. Vet. Pathol. 57, 66–81 (2020). [DOI] [PubMed] [Google Scholar]
- 37.G. W. Go, A. Mani, Low-density lipoprotein receptor (LDLR) family orchestrates cholesterol homeostasis. Yale J. Biol. Med. 85, 19–28 (2012). [PMC free article] [PubMed] [Google Scholar]
- 38.M. F. Sentmanat, J. M. White, E. Kouranova, X. Cui, Highly reliable creation of floxed alleles by electroporating single-cell embryos. BMC Biol. 20, 31 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.C. M. McMillen, N. Arora, D. A. Boyles, J. R. Albe, M. R. Kujawa, J. F. Bonadio, C. B. Coyne, A. L. Hartman, Rift Valley fever virus induces fetal demise in Sprague-Dawley rats through direct placental infection. Sci. Adv. 4, eaau9812 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. M. McMillen, D. A. Boyles, S. G. Kostadinov, R. M. Hoehl, M. M. Schwarz, J. R. Albe, M. J. Demers, A. L. Hartman, Congenital Rift Valley fever in Sprague Dawley rats is associated with diffuse infection and pathology of the placenta. PLoS Negl. Trop. Dis. 16, e0010898 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4